Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer. Patients with TNBC have poor overall survival because of limited molecular therapeutic targets. Recently, exosomes have been recognized as key mediators in cancer progression, but the molecular components and function of TNBC-derived exosomes remain unknown. The main goal of this study was to reveal the proteomic landscape of serum exosomes derived from ten patients with TNBC and 17 healthy donors to identify potential therapeutic targets. Using a tandem mass tag–based quantitative proteomics approach, we characterized the proteomes of individual patient-derived serum exosomes, identified exosomal protein signatures specific to patients with TNBC, and filtered out differentially expressed proteins. Most importantly, we found that the tetraspanin CD151 expression levels in TNBC-derived serum exosomes were significantly higher than those exosomes from healthy subjects, and we validated our findings with samples from 16 additional donors. Furthermore, utilizing quantitative proteomics approach to reveal the proteomes of CD151-deleted exosomes and cells, we found that exosomal CD151 facilitated secretion of ribosomal proteins via exosomes while inhibiting exosome secretion of complement proteins. Moreover, we proved that CD151-deleted exosomes significantly decreased the migration and invasion of TNBC cells. This is the first comparative study of the proteomes of TNBC patient–derived and CD151-deleted exosomes. Our findings indicate that profiling of TNBC-derived exosomal proteins is a useful tool to extend our understanding of TNBC, and exosomal CD151 may be a potential therapeutic target for TNBC.
Keywords: TNBC, exosomes, proteomics, exosomal CD151
Abbreviations: DEP, differentially expressed protein; FBS, fetal bovine serum; FC, fold change; FDR, false discovery rate; HER2, human epidermal growth factor receptor 2; NTA, nanoparticle tracking analysis; PD, Proteome Discoverer; TBST, Tris-buffered saline containing 0.1% Tween-20; TEM, transmission electron microscopy; TMT, tandem mass tag
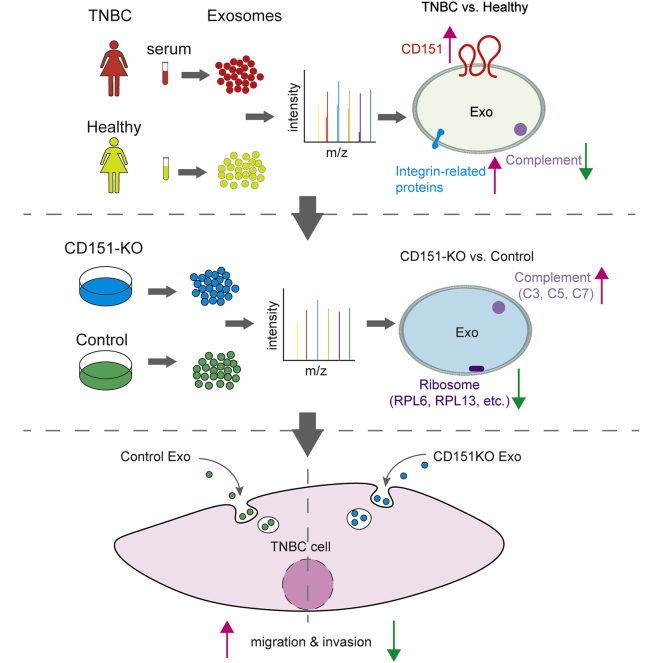
Graphical Abstract
Highlights
-
•
Quantitative proteomics of TNBC patient serum-derived exosomes.
-
•
CD151 is significantly enriched in the TNBC patient serum-derived exosomes.
-
•
CD151 regulates the secretion of ribosomal and complement proteins via exosomes.
-
•
Exosomal CD151 promotes TNBC cell migration and invasion.
In Brief
Using a quantitative proteomics approach, Li et al. characterized the proteomes of triple-negative breast cancer (TNBC) patient–derived serum exosomes and found the tetraspanin CD151 to be significantly enriched. Proteomic analysis of CD151-deleted exosomes and cells showed regulation of ribosomal and complement protein secretion. CD151-deleted exosomes were shown to significantly decrease the migration and invasion of TNBC cells, indicating that exosomal CD151 may be a potential therapeutic target for TNBC.
Triple-negative breast cancer (TNBC), characterized by lack of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 (HER2) expression, accounts for 10 to 20% of all breast cancers (1) and is an aggressive and invasive subtype of breast cancer with a high proliferation rate (2). Clinically, because of the lack of molecular targets, treatment of TNBC is limited and patients have a poorer prognosis and overall survival in comparison with patients with other types of breast cancer (3). Therefore, there is a clinical imperative to develop targeted TNBC therapies.
Exosomes are released from multiple cell types, are present in biological fluids, and have been identified as important mediators of tumor progression (4, 5, 6). Exosomes carry specific repertories of proteins, nucleic acids, and lipids, and the cargo of an exosome is an indicator of its cell of origin and can be exchanged between different cell types, mediating distant cellular communication and thus affecting physiological and pathological processes (7).
TNBC-derived exosomes have been found to play a key role in cancer progression. Specifically, HCC1806 TNBC cell–derived exosomes induced nontumorigenic MCF10A breast cell proliferation (8), and exosomes derived from TNBC cell line Hs578Ts(i)8 significantly increased the proliferation, migration, and invasion capacities of three recipient cell lines (SKBR3, MDA-MB-231, and HCC1954) (9). Meanwhile, specific microRNA expression patterns were identified in plasma exosomes of TNBC and HER2-positive breast cancer patient, showing that the levels of exosomal miR-376c and miR-382 were significantly higher in patients with TNBC compared with HER2-positive breast cancer patients, and the levels of exosomal miR-374 were associated with a higher tumor size in patients with TNBC (10). In addition, some exosomal proteins were found to play a role in mediating tumor progression. For example, Rab27a was found to facilitate secretion of exosomes to support tumor growth (11), and integrins α6β4 and α6β1 were found to be expressed on the exosomes of TNBC cell lines and to facilitate lung tropism (4). Hence, profiling the protein signatures of exosomes could be a promising strategy in TNBC therapy. However, few reports describe the components of TNBC-derived exosomes (12, 13, 14), and the potential for the utilization of proteomes of TNBC-derived exosomes in TNBC therapy remains to be determined.
In the present study, we compared the full protein expression signatures of serum exosomes derived from patients with TNBC and healthy donors using quantitative proteomics. Utilizing this expression profile, we found that CD151 was highly enriched in TNBC-derived exosomes. We also demonstrated that the expression levels of CD151 were high in TNBC cell line MDA-MB-231. In order to identify the contribution of exosomal CD151, we knocked out the protein in MDA-MB-231 cells, and we revealed that CD151 regulated protein secretion via exosomes by using quantitative proteomics. In particular, CD151 favored sorting of ribosomal proteins and integrins, rather than complement proteins, into exosomes. Moreover, exosomal CD151 derived from MDA-MB-231 cells promoted TNBC cell migration and invasion. We believe that our proteomic profile of exosomes derived from patients with TNBC and healthy donors will have profound effects on knowledge of TNBC, and exosomal CD151 will be a valuable target for TNBC therapy.
Experimental Procedures
Experimental Design and Statistical Rationale
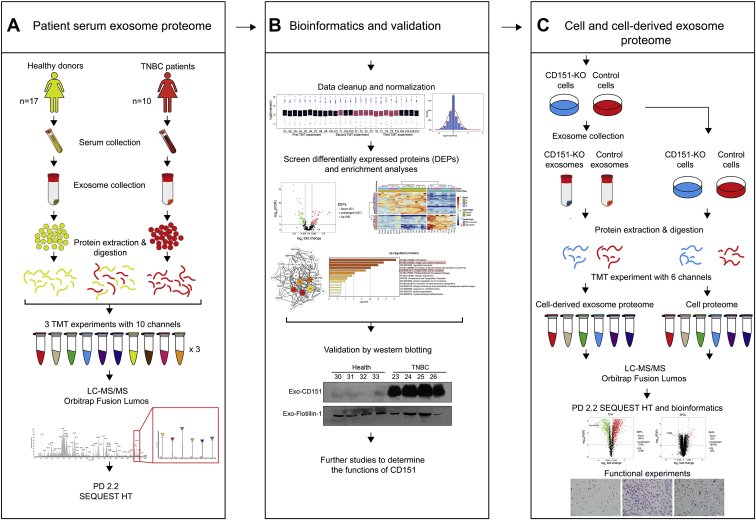
The main goal of this study was designed to reveal the proteomic landscape of exosomes derived from patients with TNBC and healthy subjects to identify potential therapeutic targets. The donated serum of about ten patients with TNBC and 17 healthy subjects was used in proteomics analysis. We analyzed every donor sample–derived exosomes individually as independent biological replicates. Additional 24 participants (12 patients with TNBC and 12 healthy donors) donated serum, and 16 participants (eight patients with TNBC and eight health donors) donated serum exosomes; these were used for data validation. CD151 expression levels were found significantly high in TNBC-derived exosomes. Quantitative proteomics approach was further performed to reveal the proteomes of CD151-deleted exosomes and cells (Fig. 1).
Fig. 1.
Workflow for characterization of TNBC-related changes in serum and cell line–derived exosome proteome.A, five hundred microliter of serum was collected from ten patients with TNBC and 17 healthy donors, and exosomes were purified by ultracentrifugation. Exosomes were lysed, and 100 μg of exosomal protein was digested. The peptides in each individual sample and the pooled samples were randomly distributed and labeled across three 10-plex TMT experiments. After labeling, samples were mixed within each TMT experiment and separated into seven fractions. Tandem MS data were obtained using an Orbitrap Fusion Lumos instrument, and proteins were identified using Proteome Discoverer 2.2. B, the serum exosome proteome data were normalized and analyzed. DEPs were determined by t test and further characterized through gene enrichment and network analysis. Representative protein CD151 was validated by Western blotting. C, CD151 was knocked out in MDA-MB-231 cells by using CRISPR–Cas9. Quantitative proteomics approach was performed to reveal the proteomes of CD151-deleted exosomes and cells. DEP, differentially expressed protein; TMT, tandem mass tag; TNBC, triple-negative breast cancer.
Patient Serum Samples
Serum samples from patients with TNBC (n = 30; aged 36–88 years; median age, 52) were obtained from College of Basic Medical Sciences, Navy Medical University (supplemental Table S1). Serum samples from healthy individuals (n = 37; aged 31–71 years; median age, 53) were also obtained from College of Basic Medical Sciences, Navy Medical University. Healthy individuals did not have any female-related disorders or malignant diseases. All samples were obtained from fasting individuals. None of the patients were treated with neoadjuvant therapy or other therapy prior to biopsy. Serum separator tubes were used to collect blood samples and then centrifuged at 3000g for 10 min to extract the serum, and the serum was stored at −80 °C until analyzed. A list of samples used for different experiments is included in supplemental Table S2. The Shanghai Changhai Hospital Ethics Committee approved the studies, and we obtained written informed consent from all participants. All the procedures followed the Declaration of Helsinki principles.
Isolation of Exosomes Derived from Human Serum Samples
Exosomes were isolated from serum samples by differential centrifugation combined with filtering, as described in a previous study, with minor modifications (15). Briefly, 500 μl of serum samples were thawed on ice, and then these samples were diluted in 15 ml PBS followed by filtering through a 0.22-μm pore filter (Millipore) and subsequent ultracentrifugation at 150,000g for 3 h at 4 °C using a 70 Ti rotor (Beckman Coulter). Afterward, the pellet was washed in 15 ml PBS, and a second ultracentrifugation step was carried out at 150,000g for 3 h at 4 °C. Finally, the pellet was resuspended in 100 μl PBS and stored at −80 °C.
Transmission Electron Microscopy Assay
Samples (0.1 μg/μl) were tested in a Tecnai G2 20 Twin transmission electron microscope (FEI; 200 kV). Exosomes were fixed to 200 mesh carbon-layered copper grids (Beijing Zhongjingkeyi Technology) for up to 90 s. Surplus material was drained by blotting, and the samples were negatively stained with 10 μl of uranyl acetate solution.
Western Blotting
Samples containing 15 μg of exosomal proteins derived from the serum or cell lysates of a study subject were denatured with SDS (Sigma) loading buffer, boiled for up to 5 min, separated by SDS-PAGE, and transferred onto a polyvinylidene fluoride membrane (Bio-Rad) at 30 V for 60 min. Next, the membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 1 h and subsequently incubated overnight at 4 °C with primary antibodies against TSG101 (sc-7964; Santa Cruz Biotechnology), HSP70 (4873S; CST), Alix (2171S; CST), CD63 (ab59479; Abcam), Calreticulin (12238T; CST), CD151 (96282S; CST), ITGB1 (4706S; CST), Flotillin-1 (610820; BD), β-actin (ab3280; Abcam), RPL6 (15387; Proteintech), RPL13 (11271; Proteintech), RPL24 (17082; Proteintech), RPS3A (14123; Proteintech), RPS8 (18228; Proteintech), RPS10 (14894; Proteintech), C3 (21337; Proteintech), C5 (66634; Proteintech), and C7 (17642; Proteintech). The membranes were washed four times with TBST and incubated with horse radish peroxide–conjugated secondary antibodies at 37 °C for 1 h. Finally, the membranes were washed with TBST four additional times, after which immunoreactive bands were detected with an ECL Kit (Millipore).
Nanoparticle Tracking Analysis
Exosomes were diluted 1:1000 in PBS and analyzed with the ZetaView PMX 110 (Particle Metrix) equipped with a 405-nm laser to determine the size and concentration of exosomes. Nanoparticles were illuminated by the laser, and their movement was captured for 1 min. Using the nanoparticle tracking analysis (NTA) software (ZetaView, version 8.02.28), videos were analyzed to provide size distribution profiles and particle concentrations.
Tryptic Digestion
First, the protein concentration of each sample was determined using the Pierce Bicinchoninic Acid Protein Kit (Thermo Scientific). Next, 100 μg protein from each sample was diluted in 8 M urea, 100 mM dithiothreitol was used to reduce disulfide bonds for 30 min at 37 °C, and 200 mM iodoacetamide was used to alkylate cysteines for 30 min. Protein was digested with LysC for 3 h at 37 °C and trypsin overnight at 37 °C. Digestion was terminated by adding trifluoroacetic acid to 0.5%, and digested proteins were desalted, dried, and dissolved in 100 mM TEAB buffer.
Tandem Mass Tag Labeling
For serum exosomes, tandem mass tag (TMT) 10-plex reagent (90110; Thermo Scientific) was used as previously described with minor modifications (16). Three 10-plex sets were used to label 27 individual samples, and each set included one pooled sample. A pooled sample was prepared for normalization between runs by combining 4 μg of peptides from each individual sample. For MDA-MB-231 cells and cell-derived exosomes, TMT 6-plex reagent (90061; Thermo Scientific) was used to label CD151-KO cells and exosomes. About 41 μl anhydrous acetonitrile was used to dissolve the TMT reagents, after which 25 μg of peptides in 50 μl volume of TEAB buffer was labeled with 10 μl of TMT reagent and incubated for 3 h. The reaction was terminated by the addition of 4 μl of 5% hydroxylamine followed by incubation for 15 min. Each set mixture was mixed, and the mixture was dried and stored at −80 °C.
High-pH Reversed-phase Tip Fractionation
The TMT-labeled peptide mixture was separated by high-pH reversed-phase tip fractionation (17). The peptides were resuspended with 100 μl of 25 mM NH4COOH, pH 10, and loaded onto the C18 material. Bound peptides were eluted by 200 μl acetonitrile gradient buffer (10, 12.5, 15, 17.5, 20, 22.5, 25, and 50% acetonitrile) in 25 mM NH4COOH at pH 10. For serum exosome proteome, the 10 and 50% acetonitrile fractions were pooled, resulting in seven fractions in one TMT experiment and a total of 21 fractions in all three TMT experiments. For cell-derived exosome proteome, the 10 and 50% acetonitrile fractions were pooled, and the 12.5 and 25% acetonitrile fractions were pooled, resulting in a total of six fractions in TMT experiment with six channels. These fractions were dried and stored at −20 °C until LC–MS/MS measurement.
LC–MS/MS Analysis
TMT-labeled peptides were reconstituted in 0.1% formic acid and delivered to an Acclaim PepMap 75 μm × 2 cm NanoViper C18, 3 μm column (Thermo Scientific) with mobile phase containing 0.1% formic acid at 8 μl/min flow rate by EASY-nLC 1200 system. Then, the trap column was switched to a PepMap RSLC C18, 2 μm, 75 μm × 25 cm column (Thermo Scientific). Peptides were separated in the second dimension using a 194 min acetonitrile gradient 6 to 90% buffer (0.1% formic acid in 80% acetonitrile) at 300 nl/min flow rate. Tandem MS data were collected using an Orbitrap Fusion Lumos Tribrid instrument (Thermo Scientific). The general mass spectrometric settings included MS1 orbitrap resolution = 120,000; MS2 orbitrap resolution = 50,000; high-energy collision dissociation fragmentation (for MS/MS), and 37% collision energy.
Database Search Parameters and Acceptance Criteria
Protein identifications, protein quantifications, and database searches were performed by Proteome Discoverer (PD), version 2.2.0.388 (Thermo Scientific). For each of the TMT experiments, raw files from the fractions were merged and searched with the SEQUEST HT search engine with a Homo sapiens Swiss-Prot UniProt protein database downloaded on February 2019 (95,556 entries). Searches were configured with static modifications on lysines and N terminus (+229.163 Da) for the TMT reagents, carbamidomethyl on cysteines (+57.021 Da), dynamic modifications for oxidation of methionine residues (+15.995 Da), precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.02 Da, the maximum of missed trypsin cleavage sites of 2, the minimum peptide length of 6, and the maximum peptide length of 144. The false discovery rate (FDR) was calculated using Percolator algorithm provided by PD. FDR of 1% was applied at the peptide and protein levels, and at least one unique peptide was used for protein identification. For TMT experiment with ten channels, two normalization methods were employed for the 30-plex experiment (three TMT experiments with ten channels each) to obtain accurate data for protein quantification. The total reporter ion intensity was applied to normalize the reporter ion intensities within each 10-plex experiment, and the pooled internal channel was utilized to normalize reporter ion intensities among different TMT experiments. For TMT experiment with six channels, the total reporter ion intensity was used to normalize the reporter ion intensities within 6-plex experiment.
Bioinformatics Analysis
The screening of differentially expressed proteins (DEPs) was carried out by using the “statistical analysis” tool of Metaboanalyst (https://www.metaboanalyst.ca/). Significant p value was obtained by Student's t test (patients with TNBC versus health donors). Proteins with FDR-adjusted p value <0.01 and fold change (FC) >1.5 were identified as DEPs. For Gene Ontology analysis, Metascape was used to convert UniProtKB accessions into Entrez Gene IDs (18), and Entrez Gene IDs were mapped to Gene Ontology terms for cellular components by FunRich software (version 3.1.3) (19, 20). Volcano plots and Venn diagrams were drawn using R package ggplot2 (version 3.2.1) (21) and R package VennDiagram (version 1.6.20) (22). Heat maps were generated using hierarchical clustering analysis based on Euclidean distance and R package pheatmap (version 1.0.12) (23). Enrichment of functions and signaling pathways of the target proteins was determined using Metascape (http://metasape.org) (18) and the enrichKEGG function in the R package clusterProfiler (version 3.12.0) (24). The STRING database (http://string-db.org) was applied to identify protein networks, which were visualized by the plugin cytoHubba (version 0.1) (25) in Cytoscape software (version 3.8.2 [Cytoscape Consortium]) (26).
Immunogold Labeling and Electron Microscopy
The grids were placed into blocking buffer for 1 h and then into a solution of the primary antibody at the appropriate dilution overnight at 4 °C (anti-CD151, 96283S, CST; 1:500 dilution). Subsequently, PBS was used to rinse the grids, which were floated on drops of the secondary antibody attached to 10-nm gold particles (AURION) at room temperature for 1.5 h. The grids were rinsed with PBS and placed into 2.5% glutaraldehyde in 0.1 M phosphate buffer for 15 min. Finally, the grids were stained with uranyl acetate and viewed under a Tecnai G2 20 Twin transmission electron microscope (FEI; 200 kV).
Cell Lines and Cell Culture
Triple-negative human metastatic breast carcinoma cells (BT549, MDA-MB-468, and MDA-MB-231) and estrogen receptor expression breast carcinoma cells (T47D and MCF7) were purchased from American Type Culture Collection. MCF-7, MDA-MB-468, and MDA-MB-231 cells were maintained in Dulbecco's modified Eagle's medium with the addition of 10% (v/v) fetal bovine serum (FBS), 100 μg/ml streptomycin, and 100 U/ml penicillin. T47D cells were grown in RPMI1640 supplemented with 10% FBS, 10 μg/ml insulin, 100 U/ml penicillin, and 100 μg/ml streptomycin. BT549 cells were grown in RPMI1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured in a humidified incubator with 5% CO2 at 37 °C and confirmed to be free of mycoplasma contamination.
KO of CD151 by CRISPR–Cas9 in Cells
CD151-KO cells were generated using the CRISPR–Cas9 method in MDA-MB-231 cells. Guide RNAs (5′-TGAGTGGATCCGCTCACAGG or 5′-GCTGGTAGTAGGCGTAGGCG) targeting a sequence within the exon of the CD151 gene were cloned into lentiCRISPRv2 (plasmid no. 52961). Stable CD151-KO cells were selected in the presence of 2 μg/ml puromycin (Sigma–Aldrich) for 5 days and assayed for expression of CD151 protein by Western blotting.
Isolation of Exosomes from Cells
Exosomes were isolated from cell supernatants by differential centrifugation as described in a previous study, with minor modifications (15). Cells were cultured in T225 (225 cm2) flasks in Dulbecco's modified Eagle's medium supplemented with 10% FBS until they reached a confluency of 80%, after which cells were grown in serum-free medium for 2 days. Next, supernatants were collected and centrifuged for 10 min at 1000g to remove cell contamination, followed by centrifugation for 30 min at 10,000g to remove apoptotic bodies and cellular debris, after which the media were filtered using a 0.22-μm pore filter (Millipore). Finally, exosomes were collected by ultracentrifugation for 2 h at 100,000g (45 Ti rotor; Beckman), followed by washing in 70 ml PBS and ultracentrifugation for 2 h at 100,000g to produce pellets.
Exosome Labeling and Transfer Assay
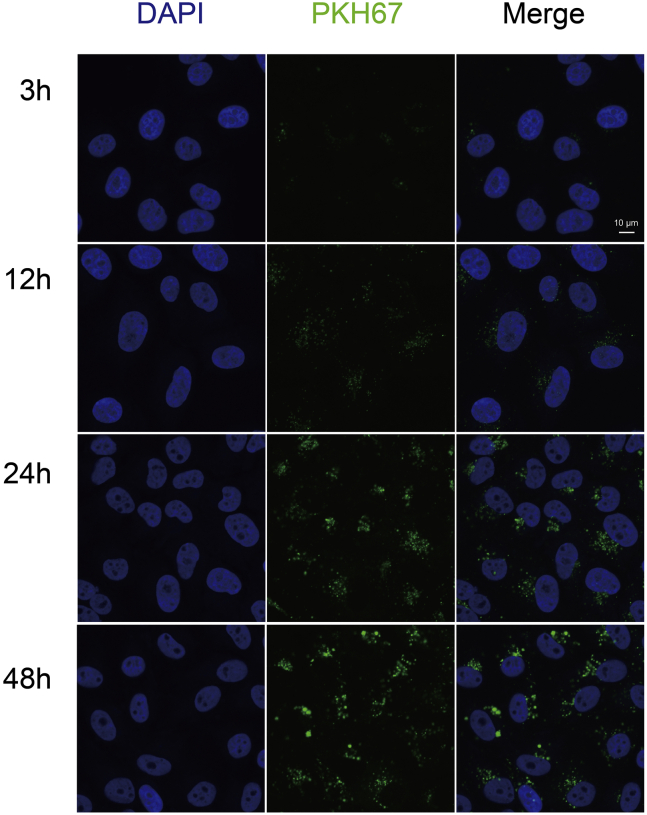
Exosomes were labeled with a green fluorescent dye (PKH67; Sigma–Aldrich) according to the manufacturer's instructions. After labeling, exosomes were washed with PBS and ultracentrifuged for 2 h at 100,000g to remove excess dye. Cells were incubated with 5 μg of PKH67-labeled exosomes at 37 °C for 3, 12, 24, or 48 h. Uptake of PKH67-labeled exosomes was analyzed using a Nikon confocal microscope (A1RSi; Nikon).
Migration and Invasion Assays
For migration assay, 2 × 105 MDA-MB-468 and 1 × 105 MDA-MB-231 cells were plated in transwell chambers (6-well transwell chambers, 8-μm pore size; 3428; Corning). For invasion assay, 4 × 104 MDA-MB-468 and 2 × 104 MDA-MB-231 cells were plated in matrigel precoated transwell chambers (24-well transwell chambers, 8-μm pore size; 354480; Corning). The medium in inserts was free of FBS, whereas the medium out was supplemented with 10% FBS. Exosomes were added to the bottom chambers at a concentration of 20 μg/ml, and PBS was used as a control treatment. After 24 h of incubation, cells on the upper side of the membrane were wiped off with a cotton swab and the lower side of the membrane was fixed with methanol and stained with crystal violet. Representative fields were photographed, and the number of cells per field was counted by ImageJ software (U.S. National Institutes of Health).
Statistical Analysis
All functional experiments were repeated independently at least three times. Data were analyzed using GraphPad Prism 8 software (GraphPad Software, Inc) and shown as mean ± SEM. Multiple sets of data were analyzed by one-way or two-way ANOVA, and unpaired Student's t test was used for analyses of two sets of data. Significant differences are marked with asterisks (∗p < 0.05; ns, not significant).
Results
Identification and Characterization of Serum Exosomes
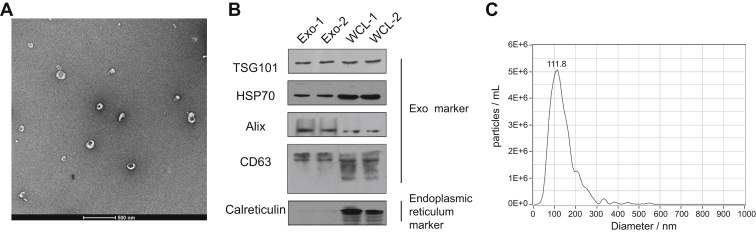
Exosomes were isolated from the serum samples of healthy donors by ultracentrifugation. To determine the relative purity and morphology of isolated exosomes, we performed transmission electron microscopy (TEM), immunoblotting, and NTA to characterize serum exosomes. TEM analysis showed that the isolated nanovesicles had typical cup-shaped structures and a diameter within the range of 30 to 150 nm (Fig. 2A). Western blotting analysis showed that exosomal protein markers TSG101, HSP70, Alix, and CD63 were found in the serum samples (Fig. 2B), and calreticulin, an endoplasmic reticulum resident protein, was only detected in the MDA-MB-231 whole-cell lysates. NTA revealed that the concentration of particles was approximately 5.7 × 107 following a 2000-fold dilution, and the mode size was approximately 111.8 nm (Fig. 2C), which was consistent with the size of exosomes. Taken together, these results indicate that the exosomes were well characterized by various assays and highly purified.
Fig. 2.
Verification of the identity of serum exosomes isolated from healthy donors.A, transmission electron micrographs of the isolated exosomes revealed typical cup-shaped structures with a diameter of approximately 30 to 150 nm. The scale bar represent 500 nm. B, Western blotting analysis of the exosomes showed that TNBC serum-derived exosomes expressed traditional exosome markers TSG101, HSP70, Alix, and CD63, but not endoplasmic reticulum marker calreticulin, compared with whole-cell lysates (WCLs). MDA-MB-231 WCL was used as a control. C, NTA of serum exosomes revealed a similar exosome mode (111.8 nm). NTA, nanoparticle tracking analysis; TNBC, triple-negative breast cancer.
Global Proteome Profiling of Exosomes Derived From Patients With TNBC and Healthy Donors
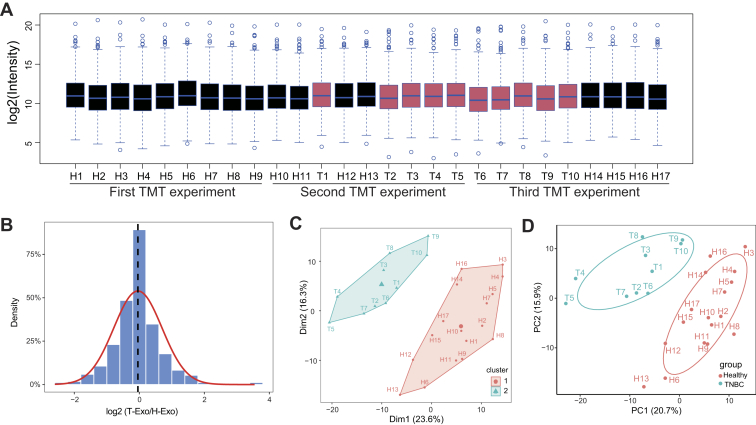
In order to examine the exosomal proteome changes in patients with TNBC and healthy donors, we randomized our samples (10 patients with TNBC and 17 healthy subjects) across three 10-plex TMT experiments and included one identical pooled sample in each TMT 10-plex analysis (Fig. 1A and supplemental Table S3). The total reporter ion intensity was applied to normalize the reporter ion intensities within each 10-plex experiment, and the pooled internal channel was utilized to normalize reporter ion intensities among different TMT experiments. To validate the effect of normalization, the distributions for normalized intensity of quantified proteins in each sample (Fig. 3A) and the log2 expression ratios between the groups after normalization (Fig. 3B) were presented. The median of normalized data were almost at the same level, and the mean of log2 ratio neared 0, indicating good standardization. We compared the reproducibility of the samples using Pearson correlation and obtained a mean correlation of 0.907 by calculating the correlation between the three separate technical replicates (Mix1, Mix2, and Mix3) in different TMT groups (supplemental Fig. S1). Subsequently, we asked whether exosomal proteome was different between healthy donors and patients with TNBC. The hierarchical clustering of proteomic data showed that TNBC and healthy samples were clustered separately (Fig. 3C), and the principle component analysis showed that TNBC and healthy samples were also separated by the biplot of principal components 1 and 2 (Fig. 3D), supporting their differential expression patterns. Thus, these data suggest that TNBC patient- and healthy donor–derived exosomes undergo global alterations in protein expression.
Fig. 3.
TMT-based quantitative proteomics analysis of exosomes derived from patients with TNBC and healthy donors.A, boxplot analysis of the intensity (log2) of all proteins across 27 samples. The intensities of all quantified proteins in three TMT experiments were normalized and plotted by the R boxplot function (median protein intensity in each sample was shown as a blue line, TNBC sample as red body, and healthy sample as black body). B, density plot of TNBC/healthy donor intensity ratio (log2) for all proteins. For all quantified protein, the intensity of protein in all samples was normalized and the log2 intensity ratio between TNBC and healthy samples was calculated. These data were plotted as a distribution frequency and Gaussian curve fitted by using R package ggplot2. The red curve shows the normal distribution (μ = −0.04; δ = 0.74), and the dashed line shows the mean value obtained from Gaussian distribution. C, hierarchical clustering of all samples. The intensities of quantified proteins from all samples were log2 transformed, and hierarchical clustering analysis was performed by using hclust algorithm of eclust function in the R package factoextra (1.0.7). D, principle component (PC) analysis of normalized signals from proteomic data. A scatter plot for PC 1 and 2 is shown by using the prcomp function in R. Each dot represents the sample derived from an individual human (“H” and “T” represent the individual healthy donor and patient with TNBC). Different sample types are indicated by color. TMT, tandem mass tag; TNBC, triple-negative breast cancer.
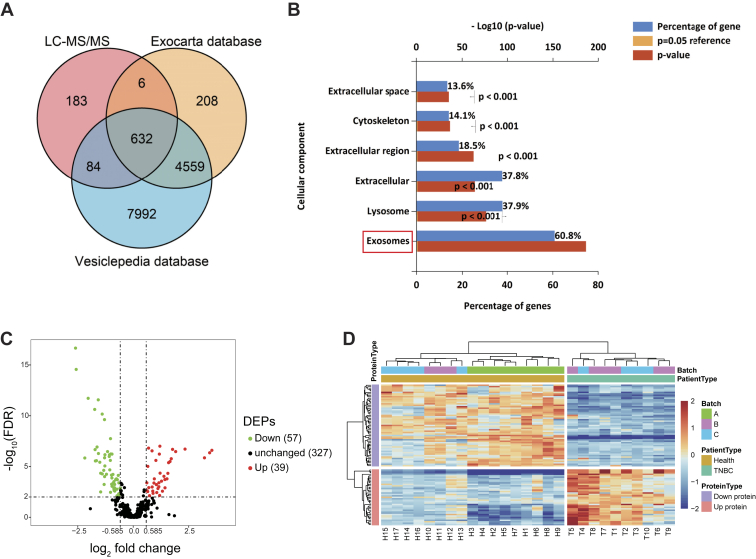
A total of 1050 proteins with an average of 712 per TMT experiment were identified in TNBC-derived exosomes (supplemental Table S4). For these proteins, there was an overlap of 632 Entrez Gene IDs in the Vesiclepedia (http://www.microvesicles.org) and Exocarta (http://www.exocarta.org) databases (Fig. 4A). Common exosomal markers, such as CD9, CD63, CD81, HSPA8, and Alix, were shown to be present in our purified exosomes by MS data analysis (supplemental Fig. S2). Furthermore, by using the FunRich tool, the identified proteins were found to be localized predominantly in exosomes (60.8%) (Fig. 4B), consistent with the typical location of exosomal proteins. Among the remaining proteins, 37.9% were lysosome-associated proteins and 37.8% were extracellular proteins. A further 18.5% were located in the extracellular region, whereas 14.1% were cytoskeleton-associated proteins. Taken together, these results confirmed that these vesicles were exosomes.
Fig. 4.
Global profiling of proteins encapsulated in TNBC-derived exosomes using MS.A, Venn diagrams of proteins identified in TNBC-derived exosomes by MS. The total number of proteins identified was compared with published results for exosomal and extracellular vesicle proteins from the Exocarta and Vesiclepedia databases. B, intracellular locations of all identified proteins assigned by the FunRich 3.1.3 tool. C, volcano plot of quantified proteins constructed from log2 fold change (x-axis) and −log10 q value (y-axis) data. The threshold for determining DEPs is indicated by the dashed lines (adjusted p value < 0.01, fold change > 1.5). Red dots indicate significantly upregulated proteins, and green dots indicate significantly downregulated proteins in TNBC serum exosomes. D, hierarchical clustering analysis of a heat map of the 96 DEPs was performed based on Euclidean distance by the R package “pheatmap,” revealing that the molecular profile of each group is unique, with the individuals in each group being closest together. The red color indicates increased protein abundance, and the blue color indicates decreased protein abundance. DEP, differentially expressed protein; TNBC, triple-negative breast cancer.
In order to identify DEPs, we used group t test and identified 96 DEPs for TNBC-derived exosomal protein–related changes, respectively (FC > 1.5; FDR < 0.01) (supplemental Table S5). The distributions of statistical significance (−log10 FDR) and change (log2 FC) for all quantified proteins were represented by volcano plots (Fig. 4C). Further analysis of hierarchical clustering revealed that individual samples were classified into two clusters (Figs. 3C and 4D). Although there was a slight batch effect observed for batch A, the biological parameters (patient group and health group) clustered closely (Fig. 4D), which indicated that the data were suitable for extraction of meaningful biological information. At the same time, these results also showed that the expression levels of proteins varied within each group, indicating that there were differences between individuals.
Enrichment Analyses and Protein–Protein Interaction Networks of DEPs
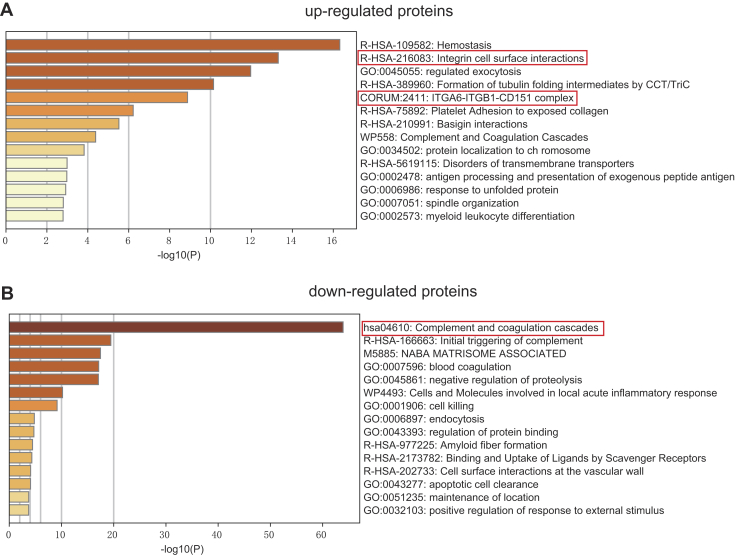
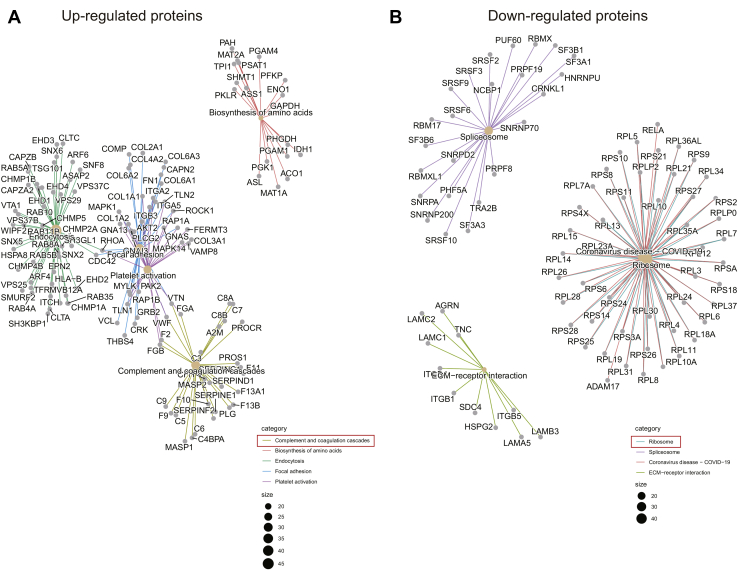
To explore the functions of the identified DEPs, we analyzed the proteomes of exosomes derived from patients with TNBC and healthy donors using Metascape. Upregulated DEPs with high expression levels in TNBC-derived exosomes were highly enriched in genes related to hemostasis, integrin cell surface interactions, regulated exocytosis, and formation of tubulin folding intermediates by chaperonin containing TCP-1/TCP-1 ring complex and ITGA6–ITGB1–CD151 complex (Fig. 5A and supplemental Table S6). Among them, integrin cell surface interactions and ITGA6–ITGB1–CD151 complex are closely related to tumor cell invasion and metastasis (4). These findings indicate that TNBC-derived exosomes may play a significant role in integrin-regulated tumor metastasis by transporting cellular information. In contrast, downregulated DEPs with low expression levels in exosomes derived from TNBC subjects were significantly enriched in proteins related to the complement and coagulation cascades (Fig. 5B and supplemental Table S7). In addition, downregulated DEPs were found to be involved in the initial triggering of complement, blood coagulation, negative regulation of proteolysis, cell killing, and endocytosis. To obtain a better idea of the biological pathways engaged in TNBC-derived exosomes, Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed using the R package clusterProfiler (supplemental Fig. S3). The DEPs were highly enriched in pathways associated with focal adhesion and the complement and coagulation cascades.
Fig. 5.
Pathway enrichment analyses of DEPs. The 20 most significantly enriched pathways and biological processes in the proteomic data of upregulated proteins (A) and downregulated proteins (B) were identified by Metascape. DEP, differentially expressed protein.
Proteins that play a crucial role in biological processes are defined as hub proteins, and the regulation of other proteins is often affected by these proteins. To find hub proteins in the aforementioned enriched pathways, the proteins associated with these pathways were inputted into the STRING database to construct protein–protein interaction networks, and we performed maximal clique centrality algorithm in cytoHubba, and the proteins with the top five maximal clique centrality values were filtered as hub proteins (supplemental Fig. S4). The top proteins ranked in the complement and coagulation cascades pathway consist of C8, VTN, C5, and C9. Interestingly, in the integrin cell surface interactions and focal adhesion pathway, ITGB1 was the top one protein ranked by the algorithm. In addition, in the ITGA6–ITGB1–CD151 complex, exosomal integrins (ITGA6 and ITGB1) have been shown to promote lung metastasis (4), so the hub protein CD151 may play an important role in TNBC by regulating integrin secretion.
Validation of MS Results by Immunoblots
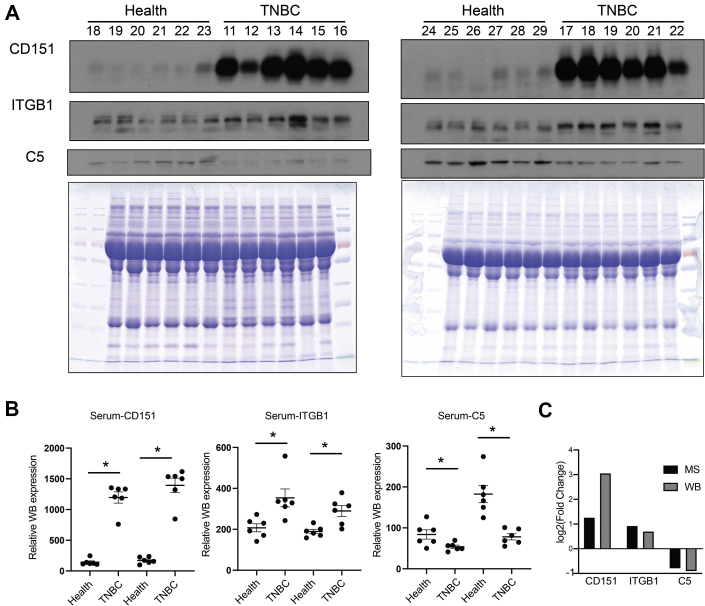
To validate the MS results, the hub protein C5 in the complement and coagulation cascades pathway, ITGB1 in the integrin cell surface interactions pathway, and CD151 in the ITGA6–ITGB1–CD151 complex were selected for validation. Serum samples from an additional 12 patients with TNBC and 12 healthy donors were independently evaluated by Western blotting. Compared with the healthy donor samples, the relative expression levels of CD151 and ITGB1 were increased in the TNBC serum samples, whereas C5 levels were decreased, consistent with the proteomics results (Fig. 6, A–C).
Fig. 6.
Western blotting was performed to validate the MS results.A, serum from additional 12 patients with TNBC and 12 healthy donors was independently assessed by Western blotting. A Coomassie blue–stained gel containing total proteins was used as a loading control. B, the relative gene expression levels were determined separately for each treatment as the mean ± SEM (n = 6), ∗p < 0.05. C, validation of selected proteins indicated that the results from MS generally agreed well with the Western blotting results. ns, not significant; TNBC, triple-negative breast cancer.
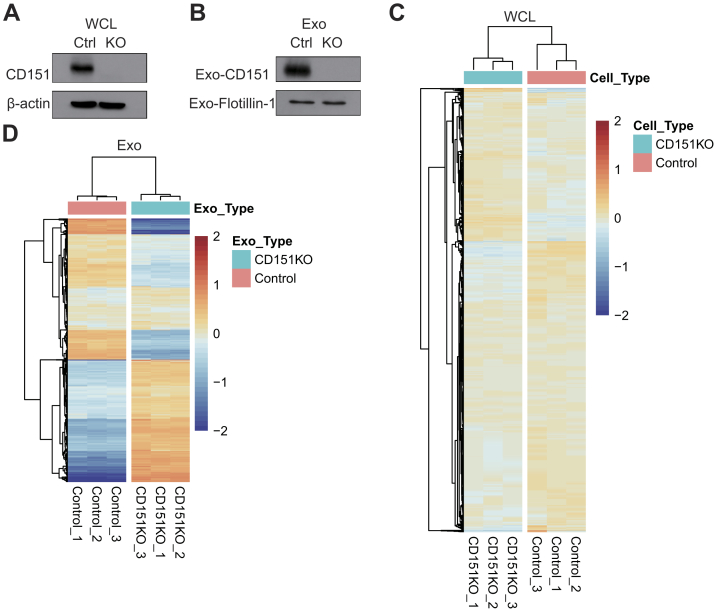
The Exosomal CD151 Is Significantly Enriched in the TNBC Patient-derived and MDA-MB-231 Cell–Derived Exosomes
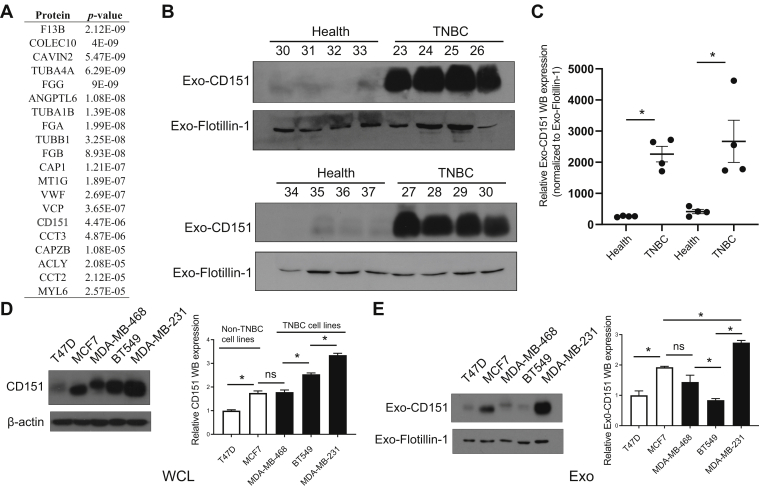
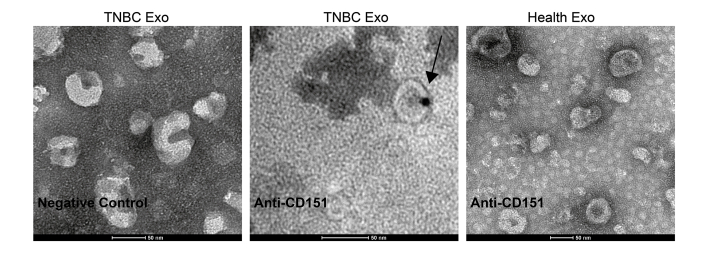
In total, 96 significantly modulated proteins were identified in TNBC-derived exosomes, respectively. Top 20 significantly upregulated exosomal proteins of TNBC were shown in Figure 7A. Among which, CD151, which has accumulating evidence about its role in cancer, was chosen for further investigation. CD151, a member of the tetraspanin superfamily, is upregulated in various cancers (27). Moreover, CD151 is highly expressed in the TNBC subtype and can promote the proliferation, migration, and invasion of breast cancer cells by modulating the activities of laminin-binding integrins α3β1, α6β4, and α6β1 (28, 29, 30). In addition, CD151 was found to be highly enriched in exosomes derived from pancreatic ductal adenocarcinoma cell lines and patients with lung cancer (31, 32). However, the role of CD151 in exosomes of TNBC has never been reported. CD151 expression in exosomes derived from TNBC serum samples, breast cancer cell lysate, and exosomes derived from breast cancer cells was validated (Fig. 7, B–E). As shown in Figure 7, B and C, exosomal CD151 expression was significantly increased in the TNBC patient–derived exosomes. The results of immunogold TEM further confirmed that CD151 was expressed in TNBC-derived exosomes compared with normal volunteer-derived exosomes (supplemental Fig. S5). These results indicate that exosomal CD151 could be used as a biomarker to separate patients with TNBC from healthy subjects. In addition, we tested CD151 expression levels in a panel of five breast cancer cell lines. As shown in Figure 7D, cellular CD151 levels were especially upregulated in TNBC cell line MDA-MB-231. In addition, exosomal CD151 levels were evaluated in exosomes derived from those breast cancer cell lines. Among those breast cancer cell–derived exosomes, exosomal CD151 was significantly enriched in exosomes secreted from TNBC cell line MDA-MB-231 (Fig. 7E).
Fig. 7.
Expression of CD151 in exosomes derived from patients with TNBC and TNBC cell lines.A, top 20 upregulated proteins identified in TNBC patient-derived exosomes ranked by p value. Significance of their upregulation in TNBC patient–derived exosomes is listed accordingly. B, immunoblot was performed to access the expression levels of exosomal CD151 expression in additional patients with TNBC and healthy donors. “30 to 37” and “23 to 30” represent the individual healthy donors and patients with TNBC. Flotillin-1 was used as a loading control. C, the relative quantified results of the immunoblots are represented as mean ± SEM (n = 4). ∗p < 0.05. D, immunoblot analysis of CD151 expression in WCLs of breast cancer cell lines (non-TNBC cell lines: T47D and MCF7; TNBC cell lines: MDA-MB-468, BT549, and MDA-MB-231). Total protein (15 μg) was loaded onto the gels. β-actin was used as a control for equal loading. E, immunoblot analysis of CD151 expression in exosomes derived from breast cancer cell lines. Total protein (15 μg) was loaded onto the gels. Flotillin-1 was used as a control for equal loading. ns, not significant; TNBC, triple-negative breast cancer; WCLs, whole-cell lysates.
CD151 Regulates the Secretion of Many Proteins via Exosomes
Given that CD151 affects the subcellular distribution and biochemical organization of α6 integrins (28), we performed experiments to determine whether CD151 regulates intracellular trafficking of other proteins and controls secretion of these proteins via exosomes. To test the functionality of exosomal CD151, we knocked out CD151 using CRISPR–Cas9 in MDA-MB-231 cells. Western blotting was performed on CD151KO (231-CD151KO) and control cells (231-Control). CD151 protein expression was completely abolished in 231-CD151KO, indicating that CD151 was successfully knocked out (Fig. 8A), and the same result was observed in exosomes (231-CD151KO-Exo versus 231-Control-Exo) (Fig. 8B). Next, stable isotope 6-plex TMT labeling of exosomal and cellular proteins was performed. We quantified 2032 exosomal proteins (supplemental Table S8) and 6849 cellular proteins (supplemental Table S9) by quantitative proteomics. For cellular proteome and cell-derived exosome proteome, we obtained a mean correlation of 0.999 and 0.998 by Pearson correlation (supplemental Fig. S6). We found that the profiles of exosomal proteins in 231-CD151KO-Exo and 231-Control-Exo differed markedly, but those proteins in 231-CD151KO and 231-Control cells differed slightly (Fig. 8, C and D). These results suggest that CD151 likely regulates the secretion of many proteins via exosomes rather than controlling their expression in cells.
Fig. 8.
CD151 regulates the secretion of many proteins via exosomes. Western blot analysis of WCLs (A) and exosomes (B) showing CD151 was only present in the WT WCLs and exosomes compared with CD151KO WCLs and exosomes. β-actin and flotillin-1 were used as loading controls for (A) and (B), respectively. Heat map of all quantified protein in WCLs (C) and exosomes (D) performed based on Euclidean distance by the package “pheatmap” in R, revealing that the profiles of total cellular protein were alike, but the profiles of total exosomal proteins were obviously different. The heat map input was the relative intensities of whole quantified proteins. The intensity of each protein in each sample was divided by the average intensity of the same protein in all six samples, and log2 transformation was done to scale the heat map input. The red color indicates increased protein abundance, and the blue color indicates decreased protein abundance. WCLs, whole-cell lysates.
Exosomal CD151 Promotes Ribosomal Protein Secretion and Inhibits Complement Protein Secretion via Exosomes
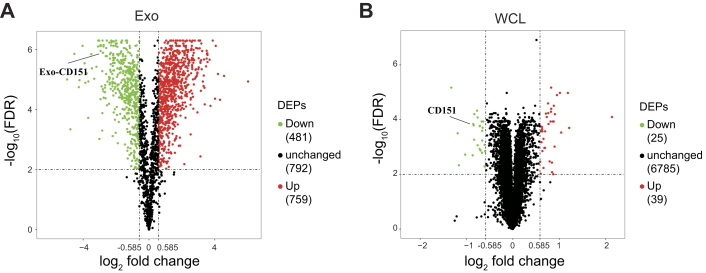
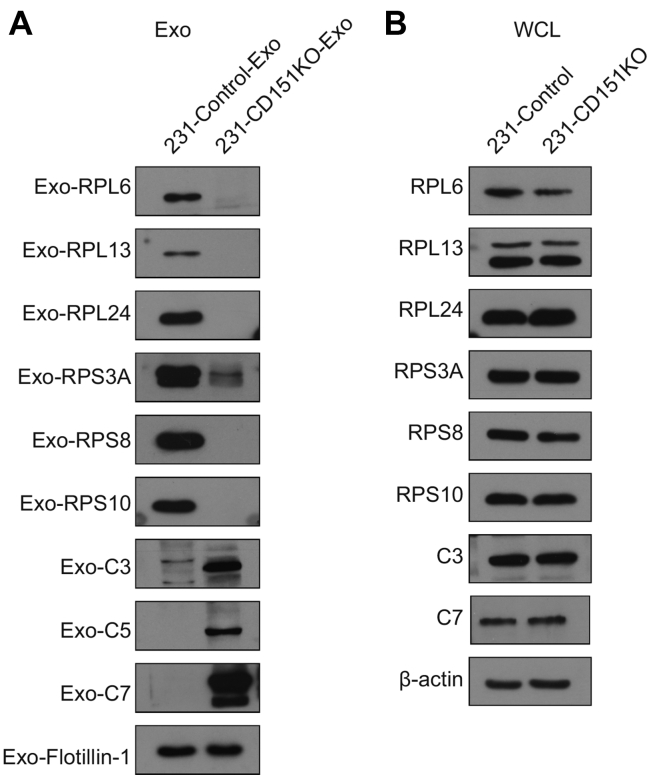
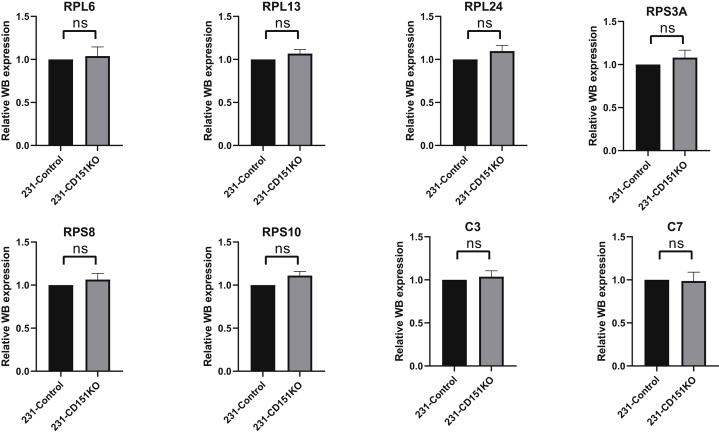
Among the quantified exosomal proteins, 759 proteins showed high (>1.5) KO/WT ratios (Fig. 9A and supplemental Table S10), suggesting that CD151 inhibited secretion of these exosomal proteins. In contrast, 481 proteins showed low (<0.667) KO/WT ratios (Fig. 9A and supplemental Table S10), suggesting that CD151 enhanced secretion of these exosomal proteins. However, among the quantified intracellular proteins (6849), only 64 DEPs were identified (Fig. 9B and supplemental Table S11). This finding provides additional information suggesting that CD151 regulates the secretion of a large number of proteins via exosomes rather than regulating their expression. The Kyoto Encyclopedia of Genes and Genomes analysis of exo-DEPs suggested that DEPs were involved in the complement and coagulation cascades, coronavirus disease 2019, extracellular matrix–receptor interaction, endocytosis, and ribosomes (supplemental Fig. S7). Specifically, highly expressed proteins in 231-CD151KO-Exo were enriched in the complement and coagulation cascades, biosynthesis of amino acids, endocytosis, focal adhesion, and platelet activation (Fig. 10A). Here, the pathways of the complement and coagulation cascades captured our attention, because complement proteins have been found to be highly enriched in exosomes from healthy donors compared with exosomes from patients with TNBC. These results indicate that CD151 likely inhibits secretion of complement-related proteins via exosomes in patients with TNBC. Among the downregulated Exo-DEPs, 46 are ribosomal proteins (Fig. 10B). To investigate whether secretion of these proteins was regulated by CD151, six ribosomal proteins (RPL6, RPL13, RPL24, RPS3A, RPS8, and RPS10) and three complement proteins (C3, C5, and C7) were investigated. The six selected ribosomal proteins displayed lower expression levels in 231-CD151KO-Exo compared with 231-Control-Exo, but the three complement proteins showed higher expression levels in 231-CD151KO-Exo compared with 231-Control-Exo (Fig. 11A), consistent with the proteomics data. In addition, there was no obvious difference between 231-CD151KO cells and 231-Control cells (Fig. 11B and supplemental Fig. S8). These findings suggest that exosomal CD151 promotes ribosomal protein secretion and inhibits complement protein secretion via exosomes.
Fig. 9.
Volcano plot of quantified proteins of WCLs and exosomes after knocking out CD151.A, volcano plot of quantified proteins of exosomes. B, volcano plot of quantified proteins of WCLs. The threshold for determining DEPs is indicated by the dashed lines (adjusted p value < 0.01, fold change > 1.5). Red dots indicate significantly upregulated proteins, and green dots indicate significantly downregulated proteins. DEPs, differentially expressed proteins; WCLs, whole-cell lysates.
Fig. 10.
KEGG enrichment analysis of DEPs in exosomes derived from CD151KO cells and control cells. KEGG analysis of upregulated proteins (A) and downregulated proteins (B) in exosomes by the R package clusterProfiler. DEPs, differentially expressed proteins; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Fig. 11.
Western blot analysis of ribosomal proteins and complement proteins. Ribosomal and complement proteins of exosomes (A) and WCLs (B). Flotillin-1and β-actin were used as loading controls for (A) and (B), respectively. WCLs, whole-cell lysates.
Exosomal CD151 Promotes TNBC Cell Migration and Invasion In vitro
Previous studies have shown that exosomes can modulate the function of recipient cells by transferring their cargoes to recipient cells (33). Therefore, before investigating whether exosomal CD151 promotes TNBC cell migration and invasion, uptake of these exosomes by recipient cells was validated (Fig. 12). Nearly all recipient cells showed a green signal 12 h after the addition of labeled exosomes. These results demonstrate that the uptake of these exosomes by recipient cells is very efficient.
Fig. 12.
Intercellular trafficking of exosomes among cell lines as determined by isolated exosomes labeled with PKH67 dye. Shown here are isolated exosomes derived from MDA-MB-231 cells and tested for uptake in MDA-MB-468 cells. Images were taken 3, 12, 24, and 48 h after exosome addition using a confocal microscope.
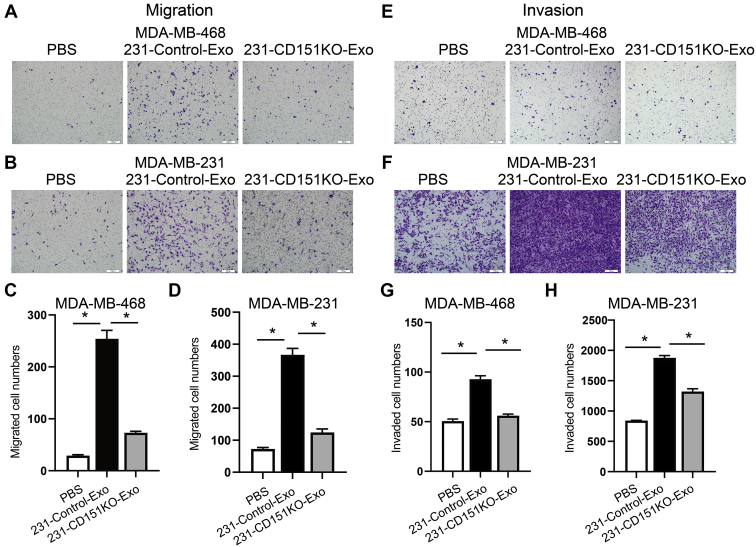
To verify whether exosomal CD151 promotes TNBC cell mobility and invasiveness in vitro, TNBC cell lines (MDA-MB-468 and MDA-MB-231) were cocultured with PBS, control exosomes, and CD151KO exosomes for 24 h. Transwell assays showed that MDA-MB-468 and MDA-MB-231 cells incubated with 231-Control-Exo exhibited enhanced cell migration and invasion ability compared with PBS control group (Fig. 13). Depletion of exosomal CD151 in exosomes decreased MDA-MB-468 and MDA-MB-231 cell mobility (Fig. 13, A–D), and its depletion also downregulated the invasive potential of MDA-MB-468 and MDA-MB-231 cells compared with 231-Control-Exo group (Fig. 13, E–H). Therefore, these results suggest that TNBC-derived exosomes promote TNBC cell migration and invasion in vitro, in part by exosomal CD151.
Fig. 13.
Exosomal CD151 promotes TNBC cell migration and invasion.A–D, 231-CD151KO-Exo decreases migration of MDA-MB-468 (A and C) and MDA-MB-231 (B and D) cells compared with 231-Control-Exo group. E–H, 231-CD151KO-Exo decreases invasion of MDA-MB-468 (E and G) and MDA-MB-231 (F and H) cells compared with 231-Control-Exo group. Migrated cell number and invaded cell number was determined by ImageJ software. Error bars represent mean ± SEM. Significance was determined using one-way ANOVA (∗p < 0.05). ns, not significant; TNBC, triple-negative breast cancer.
Discussion
Despite the established roles of exosomes in various types of cancer, few studies have focused attention on system-wide proteomics analysis of exosomes, particularly for serum exosomes in patients with TNBC, as compared with other kinds of cancer (12, 13, 34). However, the establishment of a more comprehensive view of the proteomic profiles of the exosomes of patients with TNBC and exploration of the functions of DEPs in exosomes would be of great value to clinicians and researchers. Furthermore, targeting the DEPs of exosomes may lead to new treatments for TNBC.
In this study, 1050 exosomal proteins were identified from patients with TNBC by carrying out a 10-plex TMT-based LC–MS/MS quantitative analysis. As far as we know, this is the first study of the serum exosomes of patients with TNBC based on this technique. Thus, our current finding provides the advantages of individualized precise quantification and maximized protein coverage for TNBC-derived exosomes. Furthermore, a total of 96 DEPs were filtered out and analyzed using the Metascape software tool (18). The identified DEPs were found to be involved in the integrin cell surface interactions, regulated exocytosis, ITGA6–ITGB1–CD151 complex, and the complement and coagulation cascades.
Exosomes contain a variety of different bioactive molecules representing their originated cells, thus providing useful biomarkers (35). Exosomal glypican-1 has been identified as a potential noninvasive diagnostic to detect pancreatic cancer (15). Exosomal PD-L1 was also identified as a predictor for anti-PD-1 therapy (36, 37). In this study, we identified CD151 as a protein highly expressed in exosomes derived from the serum of patients with TNBC and the culture supernatants of TNBC cells, suggesting that the increased expression of CD151 in serum exosomes could be a diagnostic biomarker for TNBC.
CD151 is an important transmembrane scaffolding protein and interacts with a wide number of integrin receptors such as α6β1, α6β4, α3β1, and α7β1 (38). Recruitment of these integrins and other partner proteins leads to form tetraspanin-enriched microdomains (39). CD151 has been demonstrated to activate extracellular signal–regulated kinases 1 and 2 and protein kinase B (Akt) signaling to promote nontumorigenic mammary epithelial cell proliferation (40). The function of CD151 in promoting cancer metastasis has been revealed in breast cancer (41), osteosarcoma (42), and hepatocellular carcinoma (43). In contrast, CD151 represses prostate cancer by antagonizing cell proliferation, epithelial–mesenchymal transition, and integrins (44). Despite the evidence implicating the complex roles of CD151 in different cancers, its involvement in TNBC remains unclear. Here, we showed for the first time the high CD151 expression in exosomes of patients with TNBC, suggesting its unexplored functions in the crosstalk with neighboring and distant cells during TNBC metastasis. Although CD151 has been found in exosomes derived from pancreatic ductal adenocarcinoma cell lines and patients with lung cancer (31, 32), functional roles in tumor progression are not yet known. In the current study, we used quantitative proteomics to reveal the proteomes of CD151KO and control exosomes and found that exosomal CD151 regulated the secretion of numerous proteins via exosomes. In particular, exosomal CD151 facilitated secretion of ribosomal proteins via exosomes while inhibiting exosome secretion of complement proteins. Previously, as a regulator of protein trafficking, the only known function of CD151 was to function in the internalization of its associated integrins via its endocytosis sorting motif (36). Thus, our findings have significantly expanded the known role of CD151. In addition, combining with proteomics results of TNBC patient–derived exosomes that complement proteins in TNBC patient–derived exosomes were expressed at relatively low levels, these results further confirmed that exosomal CD151 inhibits exosome secretion of complement proteins.
Emerging evidence suggests that exosomes mainly exert their function by transferring DNA, RNA, and proteins to recipient cells, thereby modulating gene expression and the function of recipient cells (33). The proteins transferred by exosomes can ultimately change the gene expression and functions of recipient cells, thus participating in tumor formation, proliferation, and metastasis (4, 5). However, the communication of TNBC cells via exosomes is unclear. In the present study, TNBC cell lines MDA-MB-468 (low invasive potential) and MDA-MB-231 (high invasive potential) were used. We proved that MDA-MB-231–derived exosomal CD151 promoted not only TNBC cell with low invasive potential but also TNBC cell with high invasive potential migration and invasion.
Our study has identified that patients with TNBC have elevated levels of exosomal CD151 in the serum and proved that CD151 appears to be the major mechanism by which exosomes promote TNBC cell migration and invasion. Therefore, developing exosomal CD151 as a serum-based biomarker could be attractive, and targeting exosome carrying CD151 should be considered in TNBC therapeutic strategy. In the future, we will try to develop effective small molecules to block CD151 into exosomes. Such molecules could have the potential to act alone or in combination with current molecule inhibitors GW4869 and Nexinhib-20 to inhibit exosome packaging and secretion (45, 46), to yield benefits for patients with TNBC.
Conclusions
In conclusion, we have performed proteomic profiling of serum exosomes in a prospective study including patients with TNBC and healthy donors. We found that complement proteins were lowly expressed, whereas CD151 was highly expressed in serum exosomes from patients with TNBC compared with those of healthy patients, and we demonstrated the functions of exosomal CD151 in regulating protein secretion and promoting TNBC cell migration and invasion. We believe that our profile of TNBC patient–derived serum exosomes is a useful tool that provides important insight into the role of exosomes and establishes exosomal CD151 as a potential therapeutic target for TNBC.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (47) with the dataset identifier PXD027696.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank the other members of Ji laboratory for helpful discussions. We thank our colleagues Drs Yingchun Hu and Chunyan Shan at the National Center for Protein Science at Peking University for assistance with transmission electron microscopic imaging and fluorescence microscopic imaging. We thank our colleagues Drs Qi Zhang and Dong Liu at the National Center for Protein Science at Peking University for assistance with MS. This work was supported by the National Natural Science Foundation of China (grant number: 31670832); National Key Research and Development Program of China (grant number: 2016YFA0500301); and funds from the Qidong-SLS Innovation and the State Key Laboratory of Protein and Plant Gene Research, College of Life Science, Peking University.
Author contributions
S. L., P. S., X. L., and J. J. methodology; S. L., X. L., S. Y., H. P., Z. L., P. Y., Q. Z., and Q. W. formal analysis; S. L. and X. L. investigation; S. L. writing–original draft; X. L. writing–review and editing; S. L., X. L., S. Y., H. P., Z. L., P. Y., Q. Z., and Q. W. visualization.
Contributor Information
Pingping Shen, Email: ppshen@nju.edu.cn.
Xizhou Li, Email: lixizhou721@126.com.
Jianguo Ji, Email: jijg@pku.edu.cn.
Supplemental Data
Supplemental Figure S5.
Supplemental Figure S8.
References
- 1.O'Brien K.M., Cole S.R., Tse C.K., Perou C.M., Carey L.A., Foulkes W.D., Dressler L.G., Geradts J., Millikan R.C. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin. Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Goh C.Y., Wyse C., Ho M., O'Beirne E., Howard J., Lindsay S., Kelly P., Higgins M., McCann A. Exosomes in triple negative breast cancer: Garbage disposals or Trojan horses? Cancer Lett. 2020;473:90–97. doi: 10.1016/j.canlet.2019.12.046. [DOI] [PubMed] [Google Scholar]
- 4.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 6.Boelens M.C., Wu T.J., Nabet B.Y., Xu B., Qiu Y., Yoon T., Azzam D.J., Twyman-Saint Victor C., Wiemann B.Z., Ishwaran H., Ter Brugge P.J., Jonkers J., Slingerland J., Minn A.J. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 8.Ozawa P.M.M., Alkhilaiwi F., Cavalli I.J., Malheiros D., de Souza Fonseca Ribeiro E.M., Cavalli L.R. Extracellular vesicles from triple-negative breast cancer cells promote proliferation and drug resistance in non-tumorigenic breast cells. Breast Cancer Res. Treat. 2018;172:713–723. doi: 10.1007/s10549-018-4925-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien K., Rani S., Corcoran C., Wallace R., Hughes L., Friel A.M., McDonnell S., Crown J., Radomski M.W., O'Driscoll L. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur. J. Cancer. 2013;49:1845–1859. doi: 10.1016/j.ejca.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Stevic I., Muller V., Weber K., Fasching P.A., Karn T., Marme F., Schem C., Stickeler E., Denkert C., van Mackelenbergh M., Salat C., Schneeweiss A., Pantel K., Loibl S., Untch M. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018;16:179. doi: 10.1186/s12916-018-1163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobrie A., Krumeich S., Reyal F., Recchi C., Moita L.F., Seabra M.C., Ostrowski M., Théry C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 12.Kavanagh E.L., Lindsay S., Halasz M., Gubbins L.C., Weiner-Gorzel K., Guang M.H.Z., McGoldrick A., Collins E., Henry M., Blanco-Fernández A., O Gorman P., Fitzpatrick P., Higgins M.J., Dowling P., McCann A. Protein and chemotherapy profiling of extracellular vesicles harvested from therapeutic induced senescent triple negative breast cancer cells. Oncogenesis. 2017;6 doi: 10.1038/oncsis.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palazzolo G., Albanese N.N., Di Cara G., Gygax D., Vittorelli M.L., Pucci-Minafra I. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012;32:847–860. [PubMed] [Google Scholar]
- 14.Gangoda L., Liem M., Ang C.S., Keerthikumar S., Adda C.G., Parker B.S., Mathivanan S. Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics. 2017;17 doi: 10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- 15.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plubell D.L., Wilmarth P.A., Zhao Y., Fenton A.M., Minnier J., Reddy A.P., Klimek J., Yang X., David L.L., Pamir N. Extended multiplexing of tandem mass tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol. Cell. Proteomics. 2017;16:873–890. doi: 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruprecht B., Zecha J., Zolg D.P., Kuster B. High pH reversed-phase micro-columns for simple, sensitive, and efficient fractionation of proteome and (TMT labeled) phosphoproteome digests. Methods Mol. Biol. 2017;1550:83–98. doi: 10.1007/978-1-4939-6747-6_8. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., Bacic A., Hill A.F., Stroud D.A., Ryan M.T., Agbinya J.I. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 20.Pathan M., Keerthikumar S., Chisanga D., Alessandro R., Ang C.S., Askenase P., Batagov A.O., Benito-Martin A., Camussi G., Clayton A., Collino F., Di Vizio D., Falcon-Perez J.M., Fonseca P., Fonseka P. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles. 2017;6:1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villanueva R.A.M., Chen Z.J. ggplot2: Elegant graphics for data analysis, 2nd edition. Meas. Interdiscip. Res. Perspect. 2019;17:160–167. [Google Scholar]
- 22.Chen H., Boutros P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szekely G.J., Rizzo M.L. Hierarchical clustering via joint between-within distances: Extending Ward's minimum variance method. J. Classif. 2005;22:151–183. [Google Scholar]
- 24.Yu G.C., Wang L.G., Han Y.Y., He Q.Y. clusterProfiler: An R Package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8 Suppl 4(Suppl 4) doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su G., Morris J.H., Demchak B., Bader G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 2014;47:1–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanska H.M., Berditchevski F. Tetraspanins in human epithelial malignancies. J. Pathol. 2011;223:4–14. doi: 10.1002/path.2779. [DOI] [PubMed] [Google Scholar]
- 28.Yang X.H., Richardson A.L., Torres-Arzayus M.I., Zhou P., Sharma C., Kazarov A.R., Andzelm M.M., Strominger J.L., Brown M., Hemler M.E. CD151 accelerates breast cancer by regulating alpha 6 integrin function, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon M.J., Park S., Choi J.Y., Oh E., Kim Y.J., Park Y.H., Cho E.Y., Nam S.J., Im Y.H., Shin Y.K., Choi Y.L. Clinical significance of CD151 overexpression in subtypes of invasive breast cancer. Br. J. Cancer. 2012;106:923–930. doi: 10.1038/bjc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng X., Li Q., Hoff J., Novak M., Yang H., Jin H., Erfani S.F., Sharma C., Zhou P., Rabinovitz I., Sonnenberg A., Yi Y., Stipp C.S., Kaetzel D.M., Hemler M.E. Integrin-associated CD151 drives ERBB2-evoked mammary tumor onset and metastasis. Neoplasia. 2012;14:678–689. doi: 10.1593/neo.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castillo J., Bernard V., San Lucas F.A., Allenson K., Capello M., Kim D.U., Gascoyne P., Mulu F.C., Stephens B.M., Huang J., Wang H., Momin A.A., Jacamo R.O., Katz M., Wolff R. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018;29:223–229. doi: 10.1093/annonc/mdx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandfeld-Paulsen B., Jakobsen K.R., Bæk R., Folkersen B.H., Rasmussen T.R., Meldgaard P., Varming K., Jørgensen M.M., Sorensen B.S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016;11:1701–1710. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 33.Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: Communication from a distance. Dev. Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Moon P.G., Lee J.E., Cho Y.E., Lee S.J., Jung J.H., Chae Y.S., Bae H.I., Kim Y.B., Kim I.S., Park H.Y., Baek M.C. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin. Cancer Res. 2016;22:1757–1766. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 35.Demory Beckler M., Higginbotham J.N., Franklin J.L., Ham A.J., Halvey P.J., Imasuen I.E., Whitwell C., Li M., Liebler D.C., Coffey R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., Xia H., Man Q., Zhong W., Antelo L.F., Wu B. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poggio M., Hu T.Y., Pai C.C., Chu B., Belair C.D., Chang A., Montabana E., Lang U.E., Fu Q., Fong L., Blelloch R. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177:414–427. doi: 10.1016/j.cell.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterk L.M.T., Geuijen C.A.W., van den Berg J.G., Classen N., Weening J.J., Sonnenberg A.J. Association of the tetraspanin CD151 with the laminin-binding integrins alpha3beta1, alpha6beta 1, alpha6beta4 and alpha7beta1 in cells in culture and in vivo. J. Cell Sci. 2002;115:1161–1173. doi: 10.1242/jcs.115.6.1161. [DOI] [PubMed] [Google Scholar]
- 39.Zoller M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 40.Novitskaya V., Romanska H., Dawoud M., Jones J.L., Berditchevski F. Tetraspanin CD151 regulates growth of mammary epithelial cells in three-dimensional extracellular matrix: Implication for mammary ductal carcinoma in situ. Cancer Res. 2010;70:4698–4708. doi: 10.1158/0008-5472.CAN-09-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadej R., Romanska H., Kavanagh D., Baldwin G., Takahashi T., Kalia N., Berditchevski F. Tetraspanin CD151 regulates transforming growth factor beta signaling: Implication in tumor metastasis. Cancer Res. 2010;70:6059–6070. doi: 10.1158/0008-5472.CAN-09-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z.Y., Wang F., Li Q., Zhang H.F., Cui Y., Ma C.B., Zhu J.J., Gu X.Y., Sun Z.G. CD151 knockdown inhibits osteosarcoma metastasis through the GSK-3 beta/beta-catenin/MMP9 pathway. Oncol. Rep. 2016;35:1764–1770. doi: 10.3892/or.2015.4517. [DOI] [PubMed] [Google Scholar]
- 43.Ke A.W., Shi G.M., Zhou J., Huang X.Y., Shi Y.H., Ding Z.B., Wang X.Y., Devbhandari R.P., Fan J. CD151 amplifies signaling by integrin alpha 6 beta 1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140:1629–1641.e15. doi: 10.1053/j.gastro.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Han R.B., Hensley P.J., Li J.M., Zhang Y., Stark T.W., Heller A., Qian H., Shi J.F., Liu Z.Y., Huang J.A., Jin T.C., Wei X.W., Zhou B.P., Wu Y.D., Kyprianou N. Integrin-associated CD151 is a suppressor of prostate cancer progression. Am. J. Transl. Res. 2020;12:1428–1442. [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson J.L., Ramadass M., He J., Brown S.J., Zhang J., Abgaryan L., Biris N., Gavathiotis E., Rosen H., Catz S.D. Identification of neutrophil exocytosis inhibitors (nexinhibs), small molecule inhibitors of neutrophil exocytosis and inflammation: DRUGGABILITY OF THE SMALL GTPase Rab27a. J. Biol. Chem. 2016;291:25965–25982. doi: 10.1074/jbc.M116.741884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luberto C., Hassler D.F., Signorelli P., Okamoto Y., Sawai H., Boros E., Hazen-Martin D.J., Obeid L.M., Hannun Y.A., Smith G.K. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem. 2002;277:41128–41139. doi: 10.1074/jbc.M206747200. [DOI] [PubMed] [Google Scholar]
- 47.Ma J., Chen T., Wu S.F., Yang C.Y., Bai M.Z., Shu K.X., Li K.L., Zhang G.Q., Jin Z., He F.C., Hermjakob H., Zhu Y.P. iProX: An integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (47) with the dataset identifier PXD027696.