Abstract
Therapeutic oligonucleotides (TOs) represent one of the most promising drug candidates in the targeted cancer treatment due to their high specificity and capability of modulating cellular pathways that are not readily druggable. However, efficiently delivering of TOs to cancer cellular targets is still the biggest challenge in promoting their clinical translations. Emerging as a significant drug delivery vector, nanoparticles (NPs) can not only protect TOs from nuclease degradation and enhance their tumor accumulation, but also can improve the cell uptake efficiency of TOs as well as the following endosomal escape to increase the therapeutic index. Furthermore, targeted and on-demand drug release of TOs can also be approached to minimize the risk of toxicity towards normal tissues using stimuli-responsive NPs. In the past decades, remarkable progresses have been made on the TOs delivery based on various NPs with specific purposes. In this review, we will first give a brief introduction on the basis of TOs as well as the action mechanisms of several typical TOs, and then describe the obstacles that prevent the clinical translation of TOs, followed by a comprehensive overview of the recent progresses on TOs delivery based on several various types of nanocarriers containing lipid-based nanoparticles, polymeric nanoparticles, gold nanoparticles, porous nanoparticles, DNA/RNA nanoassembly, extracellular vesicles, and imaging-guided drug delivery nanoparticles.
Keywords: Therapeutic oligonucleotides, Nanoparticles, Anti-cancer, Targeted delivery, Clinical translation
Graphical abstract
Highlights
-
•
Therapeutic TOs are most promising drug candidates for cancer therapy.
-
•
Successful delivery of TOs is the biggest challenge for clinic translation.
-
•
Nanocarriers have revolutionized and greatly promote the delivery of TOs in vivo.
-
•
LNPs and extracellular vesicles are most commonly developed for TOs delivery.
-
•
New generation of delivery systems for TOs still need to be further developed.
1. Introduction
Therapeutic oligonucleotides (TOs), short DNA or RNA oligomers, are an emerging category of drugs that can either interact with disease-associated genes via complementary Watson-Crick base pairing or recognize target proteins through the formation of three-dimensional secondary structures in a sequence-specific manner. With the rapid development of molecular biology tools, an extensive target gene profile responsible for the occurrence and development of specific diseases are being revealed, offering attractive opportunities for the development of oligonucleotide-mediated gene regulations for direct treatments of diseases or drug sensitizations. Comparing with the traditional small molecular drugs, TOs exhibit unique advantages such as the high selectivity to the target, possibility to target traditionally undruggable targets, capabilities to achieve personalized medicine, and negligible toxicity towards normal tissues [1]. TOs can be engineered without difficulty to prevent the flow of genetic information at all levels (replication, transcription, transformation, and recombination). Owing to their extraordinary controllability and adaptive structure, TOs is also commonly used for specific recognition and binding to cognate target molecules with high affinities to inhibit their functions, for example, through receptor-ligand interactions, or stimulating immune responses [2]. Since the first approval of antisense oligonucleotide drug, Fomivirsen, in the 1990s for the treatment of cytomegalovirus retinitis in individuals with AIDS [3], numerous TOs have now been approved from the FDA during the last two decades [[4], [5], [6]].
Gene based cancer therapy has been regarded as one of the most promising cancer treatments in the past several decades [7,8]. Some specific oligonucleotides, which have been regarded as the most efficient and natural gene regulation tools, such as antisense oligonucleotides (ASO), small interfering RNA (siRNA), microRNA (miRNA), messenger RNA (mRNA), aptamers, DNAzymes and immunoregulatory oligonucleotides have shown powerful theranostic activity against various cancers [[9], [10], [11], [12], [13], [14], [15]]. It should be noted however, deficient stability towards nuclease-mediated degradation and poor delivery efficiency to target organs of TOs seriously limited their wide-spread usage in clinic. As for cancer therapy, it is even more difficult for TOs to enter the tumor tissue and perform their anti-tumor activity because of the tumor microenvironment and physical barriers, which are totally different from the normal tissue [16]. To improve the stability of TOs, much efforts have been put into the chemical modifications of TOs on the internucleotide linkages [[17], [18], [19]], where a nonbridging oxygen is replaced by a sulphur atom. An in-depth discussion of these issues is outside the scope of this review and more information can be found elsewhere [20,21]. To overcome delivery obstacles, numerous strategies have been developed by designing versatile nanoparticles (NPs) that can load and transport TOs to the specific target in a programmable way [[22], [23], [24]]. Some of these NPs have positively-charged moieties that can form NPs-TOs assembly through electrostatic interactions, while others have the hydrophilic porous and hollow structures that can directly load TOs. By introducing specific functional groups to NPs that can sense the tumor microenvironment, programmable delivery and targeted release of TOs could be achieved. In the past decades, remarkable progresses have been made in the research of NPs-based delivery of TOs for cancer treatment [[25], [26], [27], [28], [29]].
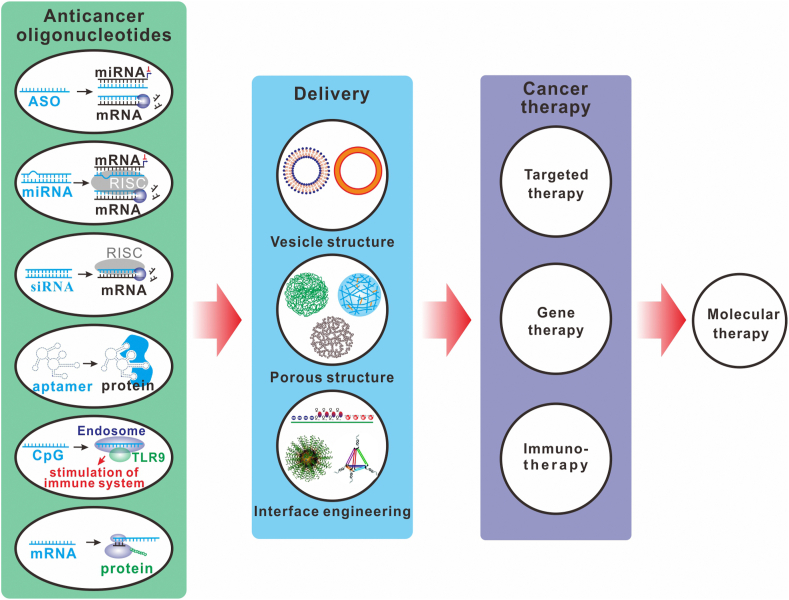
This review provides a comprehensive overview on the recent progress of nanocarrier-based systems for targeted delivery of TOs. Particularly we will focus on those TOs that have the therapeutic or modulatory activities specifically for cancer treatment (Scheme 1). Following a brief introduction of TO categorizations and the obstacles that prevent the clinical transformation of TOs, we will give a systematic overview on the TOs delivery based on the most widely used types of nanocarriers, including lipid-based nanoparticles, polymeric nanoparticles, gold nanoparticles, porous nanoparticles, DNA/RNA nanoassembly, extracellular vesicles, and imaging-guided drug delivery nanoparticles.
Scheme 1.
Nanocarrier-based systems for delivery of oligonucleotides for molecular therapy of cancers. Abbreviations: ASO, Antisense oligonucleotides; miRNA, microRNA; mRNA, messenger RNA; siRNA, small interfering RNA; RISC, RNA-induced silencing complex; TLR9, Toll-like receptors 9.
2. Classifications of the therapeutic oligonucleotides
Generally, therapeutic oligonucleotides are DNA or RNA strands that can be categorized into seven main groups: Antisense oligonucleotides, small interference RNA, microRNA, messenger RNA, DNAzymes, aptamers, and immunoregulatory oligonucleotides according to their different mechanisms in disease treatment.
Antisense oligonucleotides (ASOs) ASOs are defined as a series of short single-stranded oligonucleotides that contain 16 to 20 nucleotides and are complementary to different classes of RNA [30]. Synthetic ASOs bind to RNAs via Watson-Crick base pairing and realize their capabilities in two main ways: RNA degradation or occupancy-only mechanisms [31]. RNA targeted degradation is one of the most classic ways of ASO to achieve gene inhibition with the help of ribonuclease H1 (RNase H1) and argonaute 2 endonuclease (Ago2) in the nucleus [32]. ASO usually contains a central 8 to 10 bases DNA gap called ‘gapmers’, which is the substrate of RNase H [33]. Occupancy-only mechanisms, also called space blocking mechanism, which will not lead to the targeted RNA degradation, but through change the RNA processing, such as cause exon skipping, or promote exon inclusion and also the subcellular localization of RNA to realize the gene silencing [30,34]. Antisense approach inhibits the expression of the certain disease-related gene is a promising therapeutic method because of the increased specificity and reduced side-effect toxicity [35]. ASOs are interesting in cancer therapy because their possibilities of decreasing the expression of oncogenic genes and targeting non-coding RNAs [36]. There are several ASOs based drugs currently in phase I clinical trials, such as STAT3 transcriptional inhibitor Danvatirsen (AZD9150) [37], TGF-β signal pathway inhibitor Trabedersen (OT-101) [38], the clusterin mRNA targeting second-generation ASO Custirsen (OGX-011) [39] and etc.
Small interference RNA (siRNA) and microRNA (miRNA) can be regarded as two types of RNA interference (RNAi) that follow different pathways, which are forms of double stranded RNA (dsRNA)-mediated gene silencing at the transcriptional, posttranscriptional, and/or translational levels [40,41]. Both endogenous microRNAs and chemically synthesized small interference RNAs regulate gene expression with the help of Argonaute-containing RISC complexes [42,43]. miRNAs are noncoding RNAs with approximately 22 nucleotides, which can regulate protein-coding genes by partially binding to the 3′-untranslated region (3′UTR) of their mRNA targets. Generally, miRNAs inhibit target gene by translational repression, but they also increasing mRNA decay via deadenylation and decapping, resulting in degradation of these mRNAs or translational inhibition [44]. Since the recognition of miRNA and target mRNA can be realized by only a few consecutive complementary base paring, thus each miRNA can regulate the expression of multiple mRNAs and one of the target gene is regulated by multiple miRNAs at the same time. This may be a problem due to the lack of selectivity, while on the other hands, it provides a coordinate regulation of a gene clusters [21]. siRNA is a 22 to 25 basepair long RNA with a dinucleotide overhang at the 3’. siRNA consists of an antisense strand and a complementary sense strand, the antisense strand functionalize the activity of gene silencing, while the complementary sense strand are key factor that can help the intracellular transport of the antisense strand to RNA endonuclease Ago2 [31]. Therefore, RNA degradation is the most predominant manner that siRNAs realize their function. However, it's not like miRNAs, siRNAs cause cleavage but not translational suppression of target mRNA once bind to the targeted sequences [45,46]. The dysregulation of miRNAs expression at cellular level is always considered as one of the most important causes of tumorigenesis and cancer development [47]. This makes the supplementary of downregulated miRNA with miRNA mimics and inactivate the upregulated miRNA with antimiRNAs to be a common strategy for cancer therapy. The regulation of tumor related transcription factors and signaling pathway regulators with RNAi are also emerged as good way for cancer therapy. In 2018, the first RNAi based drug Onpattro (patisiran) [48] was approved by FDA in the application of Hereditary Transthyretin Amyloidosis treatment, and there are more than 20 siRNA, miRNA mimics and antimiRNAs in clinical trials [49,50].
Messenger RNA (mRNA) mRNA, especially in vitro transcribed messenger RNA (IVT-mRNA) has also been considered as oligonucleotides in past several years. As a bridge between DNA and protein in eukaryotic cells holds great potential in vaccine development, protein replacement therapies and genetic diseases treatment, including cancers [51,52]. Compared with plasmid DNA and protein drugs, mRNA is much safer and more efficient. Firstly, mRNA functions outside the nucleus, which can avoid the potential insertion into the genome; secondly, the temporary bioactivity of mRNA makes the protein expression more controllable; thirdly, one mRNA molecule can generate multiple copies of a protein [53,54]. The immunogenicity, stability, translatability and also intracellular delivery of mRNA are the key factors for mRNA-based drug therapy [55,56]. With the modification of untranslated regions (UTRs), rare codons in protein-coding sequences and RNA bases, both enhanced mRNA stability, translatability and decreased immunostimulatory activity can be achieved [[57], [58], [59], [60], [61]]. And the in vitro transcribed messenger RNA (IVT-mRNA) has been developed as a new drug class for diverse therapeutic applications in the past several decades [51,62,63]. Whereas, methods and intracellular delivery system remain the major challenge for the broad application of mRNA therapeutics [55].
DNAzymes DNAzymes are catalytically DNA molecules, mimicking a diverse range of enzymes. Some DNAzymes can bind to and cleave the mRNA of targeted genes, and some others can effectively catalyze RNA ligation or phosphorylation [64]. RNA-cleaving DNAzymes, which can cleave the sequence between any unpaired purine and pyrimidine of mRNA transcripts under physiological conditions [65], have gained more and more attention in the application of diagnosis and treatment owing to their perfect stability, modifiability and multiple selection of combined application [66]. Compared to ribozymes, DNAzymes are easier to synthesis, more stable and much cheaper in the application of disease treatment. In recent years, DNAzymes has been regarded as a good candidate for cancer therapy [66,67].
Aptamers Aptamers, which are origin from Latin aptus (fit) and Greek meros (part), represent an emerging class of therapeutics. Aptamers, the folded three-dimensional structures of which can recognize various targets including small molecules, peptides, proteins and cells with high affinity are short single-stranded DNA or RNA molecules with 20–100 nucleotides [68,69]. Aptamers are referred to as “synthetic antibodies”, they bind to biological molecules via a very specific manner but not act through sequence-specific complementary paring. Comparing with antibodies however, aptamers have displayed more advantages in the design of moieties for the target recognition, e.g. they are much smaller and more stable than antibodies and can be chemically-modified in a defined and precise way, and most importantly they are non-immunogenic [70,71]. In contrast to ASOs or siRNAs, which must be delivered inside the cell, aptamers can target either extracellular or intracellular proteins [72]. Nowadays, aptamers have become an effective tool in the development of targeted delivery systems for both diagnostic and therapeutic purposes, and some specific aptamers, like AS1411, have attracted much attention in clinic for exploring their potential against a variety of cancer targets [72].
Immunoregulatory oligonucleotides Except the above mentioned TOs, there is a special type of oligonucleotides, which act by modulating immunity for treating diseases in a targeted and specific manner. Immunoregulatory oligonucleotides either stimulate the immunity via binding to toll-like receptors (TLRs, e.g. TLR3, TLR4, TLR7/8, and TLR9) for innate immune recognition of pathogens, or antagonize TLRs to regulate aberrant immunity for treating autoimmune disorders [73,74]. Among these oligonucleotides, unmethylated cytosine-phosphate-guanine (CpG)-containing oligodeoxynucleotide, as TLR9 agonists, has become an effective and widely-used adjuvant in developing vaccines for cancer treatment [[75], [76], [77]].
3. Obstacles that prevent the clinical transformation of TOs for cancer therapy
Generally, the instability, reticuloendothelial systems (RES) clearance and renal excretion of nucleic acids in blood circulation are main challenges that limit the clinical transition of TOs. The main reason for TOs’ instability is the presence of abundant nuclease in the plasma. Obviously, considerable TOs will be degraded rapidly once being injected into the blood because the DNases and RNases widely exist in the plasma and tissues. Meanwhile, the mononuclear phagocytes of the RES can also eliminate TOs since the prominent role of RES in host is defense [85]. Because TOs are artificially synthesized in vitro, when they are administrated in vivo, they may face the risk of being labeled and recognized as ‘foreign invaders’ followed by uptake and clearance by RES, especially the Kupffer cells lined in the liver sinusoids and macrophages in the marginal zone of the spleen. In addition, molecules in the sizes of 3–6 nm or smaller are easily filtered by kidney [86]. A significantly large number of TOs, e.g. siRNA and uncharged morpholino antisense oligonucleotides are mainly in this size range and thus will be rapidly excreted out by the renal route [87]. Therefore, rational TOs delivery systems must be able to evade or at least to minimize the RES clearance and renal excretion. However, the tumor tissue appears to be smarter than the normal tissue in escaping from drug treatment, which brings another challenge for TOs in the application of cancer treatment.
Limited drug penetration within tumor stroma In tumor tissues, the dense and aberrant extracellular matrix (ECM), a non-cellular meshwork consisting of structural proteins, glycoproteins, and proteoglycans [88], may significantly limit the diffusion of drugs to the deep site of tumors. This matter is particularly important for TO delivery because of their macromolecular nature. Compared to healthy tissues, where blood flow and lymphatic drainage are well-balanced [89], in tumor tissues, the lymphatic drainage is blocked while vasculature is extremely leaky, resulting in an elevated interstitial fluid pressure (IFP) [90]. The IFP gradually increased with the distance from the vessel, which hinders the homogeneous distribution of macromolecular drugs or nanomedicines throughout the whole tumor [[91], [92], [93], [94]]. Taken the above together, both the dense ECM and IFP limited the drug penetration within tumor stroma. Strategies to overcome this limitation include degradation of the tumor ECM proteins or downregulating their expression [95,96], opening epithelial junctions [97], vessel normalization [98], and remodeling the tumor stroma through targeting of tumor-associated macrophage [99,100]. Moreover, the tumor microenvironment conditions, such as lower pH that are considered as the consequence of the accumulation of acid metabolites and oxygen deficit at the tumor site might also the reason of failed TOs delivery [101].
Cell/organelle membrane barriers To realize the anti-tumor activity, almost all of the different types of TOs must be internalized into the tumor cells. Among these TOs, only aptamers can bind to the target cell membrane proteins via their loop structures, followed by cell uptake through receptor-mediated endocytosis (RME). While for most of other unmodified TOs, the electrostatic repulsion between the negatively-charged TOs and as well as the negatively-charged cell membrane prevent TOs passing through the cell membrane [102]. By means of transport vectors or using nanocarriers, TOs can be internalized by cells through specific uptake mechanism [103,104]. However, the poor permeability of tumor cell plasma membrane caused by changes of membrane lipid organization and increased sphingolipids and cholesterol membrane content make it even more difficult for TOs to enter cells [105]. For example, ABC efflux pumps transporters such as the P-glycoprotein (P-gp), MRP1 and BCRP correlated to drug resistant have always been found to be upregulated in cancer cells [106]. The decreased influx transporters, such as solute carriers (SLC) on the membrane of tumor cells also make a great challenge for achieving considerable drug levels in cells in the similar way [107]. And it should also be noted that the internalized TOs are mostly trapped in the endo/lysosomal compartment, which means that additional strategies related to endo/lysosomal escape must be considered especially for TOs with the target located in the cytoplasm or even nucleus, e.g. antisense oligonucleotides, siRNA/miRNA, and mRNA. Over the past decades, plenty of strategies have been developed to overcome the problem of endo/lysosomal confinement. Proton absorbing materials, such as peptides with arginine, lysine and histidine [108], have a high buffering capacity between pH 7.2 and 5.0 and thus have been extensively utilized to design systems for lyso/endosomal escape. These materials act through the proposed mechanism of ‘proton sponge effect’ [109], which involves an extensive inflow of ions and water pouring into the microenvironment of endosomal thus consequently leads to the release of the entrapped components form the disruptive endosomal membrane. As for delivering TOs to the nucleus, the nucleus membrane may serve as another barrier to prevent TO entering the nucleus. Nuclear transport frequently occurs through nuclear pore complexes, nonetheless, TOs condensates are hard to pass through nuclear pore complexes (NPC) due to their large size [110]. For dividing cells, TOs can get in the nucleus during mitosis when the permeability barrier eliminates. Whereas, in non-dividing cells, TOs must pass through the nuclear membrane via the NPC, and the NPC only allows entry of molecules with the size up to 9 nm and less than 40 kDa through free diffusion [111]. Regarding to larger macromolecules, varieties of nuclear localization signal (NLS) peptides have been developed in an attempt to deliver TOs into the nucleus through NPC, which is an energy-dependent process [112]. NLSs are short clusters of amino acids which can bind to TOs either through covalent attachment or by noncovalent electrostatic interaction and mediate transnuclear transport through the NPC.
Blood-brain barrier Delivering drugs to brain tumors is much more difficult because of the existence of the blood-brain barrier (BBB) [113], which consists of tightly linked endothelial cells supported by a network of pericytes and astrocytes and is impermeable to most of molecules as small as sucrose. Theoretically, macromolecular TOs cannot directly pass through the BBB to enter the brain parenchyma. Many attempts have been applied to deliver drugs across the BBB, such as using a hypertonic solution to temporarily open the tight junctions of the endothelial cells [114], using ultrasound to open BBB [115,116], and applying a vasodilator to enhance the vascular permeability [117]. However, the invasive disruption of BBB structures may bring up serious toxicity to brain parenchyma or undergo unrecoverable side effects. Therefore, designing new delivery systems with the capacity to across the BBB is still a big challenge, especially for noninvasively administrations of TOs for brain cancer therapy.
The challenge of lyso/endosomal trap Because both of the naked TOs drugs and the TOs drugs carried with liposome delivery system or conjugate delivery system need to enter into the cell in the form of endocytosis for them to function [118]. Thus, the final destination of all delivered TOs are decided by lyso/endosomal. However, most of the TOs in the endosomes that are produced by cell uptaking will be trapped inside of the lysosomes after the fusing of endosomes and lysosomes following the acidification and maturation of the endosomes [119,120]. This leads to the final degradation of these trapped TOs because of the certain enzymes in the lysosomes. Currently, the adding of pH sensitive polymers, liposomes and CPP (Cell Caring Peptide) to the delivery system are widely used to increase the lyso/endosome escaping [121]. Whereas, the instability, high-sensitivity to the environment (especially the acidic microenvironment of tumor tissues) and also the lower targeting of pH-responsive delivery carriers remain the main problems that need to be solved. In addition, the discovery of GalNAC (N-Acetyl-d-galactosamine) delivery systems also gives a better solution for the delivery of TOs drugs targeting the liver [122]. But it cannot be used for the delivering to other target organs, because there are no similar expression level and recycling rate receptors as ASGPR has been found in other tissues. Therefore, other more efficient delivery systems, which can improve the delivery and utilization efficiency of TOs drugs by increasing endosomal escape are still needed to promote the clinical application of TOs drugs for cancer therapy [123].
Off-target effects The off-target effects mainly consists of two reasons: one of the off-target effects is caused by RES clearance and renal excretion in blood circulation; the other reason is the off-target effect of some TOs. For example, the antisense strand of siRNA can not only mediate the silencing of homologous genes, but also cause the inhibition of some non-homologous genes through miRNA pathway. Meanwhile, the sense strand mediates the silencing of its homologous genes, causing the off-target effect mediated by the sense strand [124]. Otherwise, unmodified double-stranded RNA can also lead to the activation of innate immune response [125]. All of these off target effects will lead to toxic and side effects of TOs drugs and still need to be solved in the clinical application for cancer therapy.
In summary, the continuous improvement of non-viral nanoparticle carriers has provided a great opportunity for the application of TOs in cancer therapy, but the inefficiency of delivery carriers and lack of targeting are still the main problems restricting their clinical application.
4. Nanoparticles-based delivery of TOs
Generally, the loading of TOs to nanocarriers can be approached via physical or chemical methods. Negatively-charged TOs can either form complex with cationic nanocarriers via electrostatic interactions [126] or be loaded within the hollow structured nanocarrier with a hydrophilic inner cavity [127]. Besides, TOs can also directly bind to surface of metal nanoparticle through the facile Au–S chemistry [128,129]. Compared to the free TOs, TOs in nano formulations are much more advantageous: (1) NPs improve the stability of TOs. Except the chemical modifications of TO framework, using specific nanocarriers to form TO-NPs complex is an alternative way to improve the stability of TOs due to the shielding effect of nanoparticles. In addition, densely packing oligonucleotides on the NP surface has been reported as an effective way to prevent TOs from degradation by nuclease [130]. (2) NPs enhance the tumor accumulation of TOs. In the past 30 years, the enhanced permeability and retention (EPR) effect has become a central principle of passive targeting that drives the development of various nanomedicines for tumor-targeted drug delivery [131], though the importance and extent of EPR effect in human patients have been heavily debated in recent years [132,133]. Through relational design of NP size and shape, enhanced tumor accumulation of nanomedicines could be achieved [[134], [135], [136]]. (3) NPs improve cell uptake efficiency of TOs. Numerous studies have shown that NPs can enter cells as well as to translocate across the cell membranes through different cell uptake mechanisms depending on the structure of NPs [119,120]. Furthermore, through the surface functionalization of NPs with cancer-cell-targeting ligands [137], significant enhancement of cell uptake with high specificity can be achieved. (4) NPs can achieve targeted and on-demand drug release. To minimize the toxicity of nanomedicines to normal tissues, NPs can be designed with the capability to release their payloads only in the tumor tissues via responding to the tumor-specific microenvironments, such as hypoxia [138], acidic pH [139], irregular redox status [140], and so on. In addition, some smart NPs are capable of responding to external stimuli, like light [141,142], magnetic field [143], and ultrasound [144], during which process, controlled drug location and timing of administration, termed on-demand drug release could be realized.
Over the past few decades, striking progresses have been made in the advance of nanocarrier-based systems for targeted delivery of all types of DNA/RNA oligonucleotides. TOs can be loaded by nanocarriers through a variety of strategies, including physical encapsulations, electrostatic condensation, covalent binding, and Watson-Crick base-pairing. In the following sections, we will give a comprehensive overview on these progresses in terms of different formulations of nanocarriers: lipid (Section 4.1) and polymeric nanoparticles (Section 4.2), gold nanoparticles (Secion 4.3), porous nanomaterials (Section 4.4), DNA nanoassembly (Secion 4.5), extracellular vesicles (Secion 4.6), and imaging-guided drug delivery systems (Section 4.7).
4.1. Lipid-based nanoparticles
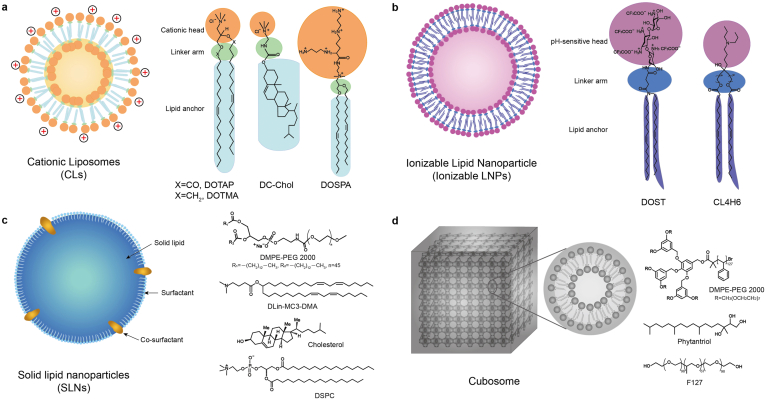
In the past decades, lipid-based nanoparticles (LNPs), composed of various lipid compositions and ratios with different structures, have shown great potential as delivery vehicles for TOs, containing antisense oligonucleotides, siRNA, and microRNA analogs [145]. To improve the stability and increase the delivery efficiency of LNPs, some helper lipids with specific geometry are usually added to LNPs [24], including the cone-shaped dioleoylphosphatidylethanolamine (DOPE, can promote endosomal release of TOs), cylindrical-shaped lipid phosphatidylcholine (can improve the stability of bilayer) and cholesterol (can improve the intracellular delivery of TOs and in vivo stability of LNPs). According to the structure and components of lipids, LNPs can be classified into four types: cationic liposomes, ionizable LNPs, solid LNPs, and cubosomes (Fig. 1).
Fig. 1.
Structural features of lipid-based nanoparticles, including cationic liposomes (a), ionizable lipid nanoparticles (b), solid lipid nanoparticles (c), and cubosomes (d).
4.1.1. Cationic liposomes
Cationic liposomes (CLs) are vesicular structures mainly consisted of the positively charged lipids, which can interact with negatively charged DNA, forming cationic lipid/DNA complexes called lipoplexes [146,147]. Due to their high nucleic acids loading capacity, favorable interactions with cell membranes and versatile design of cationic lipids, which provides an efficient tool in screening liposomal formulations to achieve optimal gene therapy, CLs have gained much attention as gene delivery vectors for decades [145]. CLs are composed of a cationic hydrophilic head group, a hydrophobic lipid anchor group, and a linker arm bridged in between. Based on the structure of cationic head group, the cationic lipids can be classified into three major categories, (1) monovalent lipids, such as N (1-(2,3-dioleyloxy) propyl)-N,N,N-trimethylammonium chloride (DOTMA) [148] and 1,2-dioleyl-3-trimethylammonium-propane (DOTAP) [149], (2) multivalent lipids such as 2,3-dioleyloxy-N-(2(sperminecarboxaminino)ethyl)-N,N-dime-thyl-1-propanaminium trifluroacetate (DOSPA) and dioctadecylamidoglycylspermine (DOGS) [150], and (3) cationic lipid derivatives, such as 3β-(N-(N′,N'-dimethylaminoethane)-carbamoyl) cholesterol (DC-Chol) [151]. As for the linker arm, numerous studies have shown that the structure and orientation of linker bondshave a significant influence on the chemical stability and biodegradability of cationic head lipid, thus affecting their transfection efficiency and cytotoxicity [152,153]. Detail information based on how the structure of linker in cationic lipids influence the gene delivery efficacy of CLs can be found in a recent review [154].
CLs have demonstrated great potential in earlier studies in the targeted transportations of TOs [155,156]. To increase the DNA loading efficiency, massive cationic lipid are usually required in the formation of cationic liposomes [157]. However, high dose of cationic lipid administered in vivo might induce serious systematic toxicity. In addition, the strong interactions with serum proteins may lead to the rapid clearance of cationic liposomes from the circulation, decreasing the bioavailability [158].
Surface functionalization of nanoparticles (NPs) with PEG, or PEGylation, is one of the most extensively used strategies to enhance the stability and then prolong the fluid circulation of nanodrug delivery systems in vivo through reducing the opsonin adsorption on NPs [145]. However, it should be noted that PEGylation can prevent the NPs from interacting with the plasma membrane thus decrease their uptake efficiency by target cells. As for LNPs, inclusion of a PEGylating lipid can additionally hinder the fusion of LNPs with endosomal membrane upon internalization into cells, which seriously limits the delivery efficacy of cytosol-targeting therapeutics, e.g. siRNA [159].
On the one hand, the quaternary amine cationic lipid can supply high charge density to improve the loading of negatively charged oligonucleotides. On the other hand, the tertiary amine cationic lipid can provide a pH responsiveness for optimal delivery of oligonucleotides, therefore, Yung et al. developed a pH-sensitive carrier, QTsome, based on a combination of quaternary amine and tertiary amine cationic lipids [160]. Quaternary amine based cationic lipids are permanently charged, while tertiary amine based cationic lipids are mostly free of charge at neutral pH conditions and get fully ionized only at acidic pH, enhancing its ability to escape from the endosome compartment. QTsome loaded with TOs (AM-21) targeting to miR-21, a gene perticipated in multiple pathways regulating tumor progression and chemotherapy resistance, displayed high colloidal stability and good fusogenic activity in the endosome. Additionally, in vivo evaluations with tumor-bearing mice revealed that QTsome/AM-21 upregulated miR-21 target genes thus induced tumor regression, improved anti-tumor efficacy with chemo-gene therapy and prolonged survival.
4.1.2. Ionizable LNPs
LNPs containing ionizable cationic lipids were developed to address the dilemma of CLs regarding their off-target effect and systemic toxicity induced by the highly charged cationic lipids despite their high drug loading efficiency. The functions of ionizable part are required to meet a number of tough challenges during the LNP-based TO delivery. Firstly, the lipoplexes is positively charged and adsorb negatively charged oligonucleotides forming the nanoparticle under the acidic conditions. Secondly, the ionizable part endows the LNPs with a distinct acid-dissociation constant (pKa) so that the surface of LNPs is close to neutral when they are at physiological condition. Thirdly, the lipid must assume a positive charge in the acidified endosome for purpose of interacting with naturally anionic phospholipids in the endosomal membrane and destabilize it [161]. Therefore two important factors should be taken into account when designing a proper ionizable lipoplexes: (1) the pKa value of cationic head group, which is generally used to determine the pH condition for lipid gets protonation or deprotonation; (2) the capabilities of these ionizable lipids to induce a nonbilayer phase structure (hexagonal HII) upon protonation when they interact with endosomal membrane [145]. In recent years, ionizable LNPs have gained much notice owing to their negligible toxicity compared to CLs and versatility of structural variations of ionizable lipid, which provides an effective tool in screening optimal formulations for TO delivery [162]. Habrant et al. reported a series of novel ionizable carriers derivative from naturally occurring aminoglycoside tobramycin, which could form complex with various types of TOs, including mRNA, DNA, and siRNA [163]. After transfection potency evaluation of these carriers with structural changes at the level of linker and hydrophobic domain, the authors found that the lead molecules carrying biodegradable diester linkers exhibited the best transfection efficiency across all tested nucleic acids and cell types. Except the delivery of single formulation of TOs, ionizable LNPs can also be used to co-deliver multiple types of TOs. For example, Ball et al. co-formulated siRNA and mRNA that targeted to diseases-associated genes in a single lipidoid nanoparticle formulation, which consists of an ionizable amine-containing lipidoid, cholesterol, DSPC, DOPE, and PEG-lipid. They found that simultaneous delivery of siRNA and mRNA enhanced the efficacy of both drugs compared to their single counterparts, and with the addition of a negatively charged “helper polymer”, polystyrenesulfonate, the efficiency of the LNP drug delivery platforms have been dramatically improved [164].
4.1.3. Solid LNP
Solid lipid nanoparticles (SLNs) typically contain a hydrophobic solid matrix core, which is usually comprised of biodegradable lipid components, and a surfactant shell designed to stabilize the SLNs. The lipids used to prepare SLN mainly comprise fatty acids (stearic acid), steroids (cholesterol), waxes (cetyl palmitate), monoglycerides, diglycerides and triglycerides. Different lipids and surfactants can affect the particle size, surface charge, long-term stability during storage, encapsulation efficiency and release profile [165]. Since 1990, SLNs have been developed as an alternative to nanoparticles, liposomes, and microparticles to deliver therapeutics because of their excellent biocompatibility and combinatorial advantages of polymeric nanoparticles, fat emulsions, and liposomes [166,167]. Furthermore, the preparation process without organic solvents makes the SLNs be of great potential for future clinical context.
A significant feature of SLNs is their ability to deliver hydrophilic and hydrophobic drugs depending on the preparation methods. Therefore, SLNs are promising carriers to co-deliver TOs and other types of therapeutics, such as small molecular drugs and biomacromolecules (e.g. polysaccharides, vaccine antigens) for combined therapy [[168], [169], [170]]. For example, Shi et al. developed a dual-drug-containing nano-vehicles for simultaneous delivering an endogenous microRNA (miR-34a) and paclitaxel (PTX) for synergistic cancer treatment. Results showed that the co-delivery system (miSLNs-34a/PTX) with an average size of approximately 220 nm could protect both miR-34a and PTX from degradation during the circulation. Furthermore, miSLNs-34a/PTX showed synergistic anticancer efficacy, where they displayed a much higher efficiency in inhibiting B16F10-bearing tumor growth compared to their single drug-loaded SLNs counterparts [171]. In another study, Kucukturkmen et al. utilized the high-pressure homogenization method to construct a cationic SLNs for co-delivering anti-miR-21 oligonucleotide, a TO targeting glioma-proliferation- and drug-resistance-associated gene (miR-21), and pemetrexed, a multi-targeted antifolate agent for treatment of brain tumors. A loading efficiency over 90% and the controlled release of pemetrexed were achieved, and cellular internalization of anti-miR-21 oligonucleotide/pemetrexed-coloaded SLNs by U87MG human glioblastoma cells was significantly higher and much more effective than that of free pemetrexed [172]. Although SLNs provides a feasibility to incorporate various oligonucleotides and lipophilic drugs with high payload, the preparation process of SLNs should be very carefully designed, considering that the stress and strain associated with the homogenization process may cause the fragile DNA/RNA, or other bioactive molecules degradation.
4.1.4. Cubosomes
Cubosomes are colloidally stable cubic LNPs, which typically contain amphiphilic lipids with a unique internal bicontinuous cubic two-phase structure [173,174]. Since the first report by K. Larsson in 1989 [175], cubosomes have been regarded as promising nanovehicles for different routes of drug administration [176]. Cubosomes have a high internal and external surface area for loading both small molecular drugs and bioactive DNA/RNAs, owing to their continuously compartmentalized self-assemblies with hydrophilic and hydrophobic domains. Compared to conventional spherical LNPs, the unique curved structure of cubosomes provides an additional advantage for cytosolic delivery of siRNA, mRNA or any other TOs that needs to be escaped from endosomes.
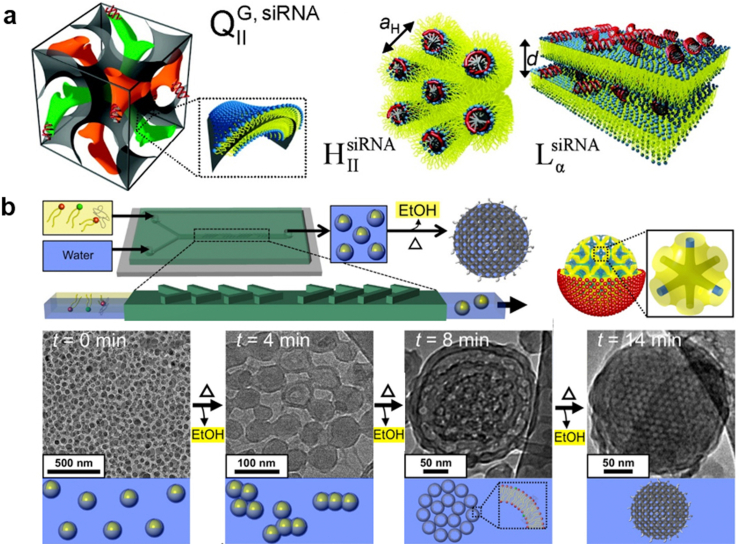
The cubosome-based TO-delivery system was first reported by Leal et al. where they developed an inverse gyroid bicontinuous cubic nanostructure composed of nonionic lipid glycerol monooleates (GMO) and small amounts of univalent cationic lipid DOTAP, which facilitated the incorporation of siRNA within its water channels [177]. Remarkable gene silencing was achieved due to the improved endosomal escape, which could be attributed to the formation of dynamical transient pores driven by the positive Gaussian modulus of the cubic phase membrane. In addition, the cubic lipid nanostructures did not show negative implications on cell viability and plasma membrane integrity because of their low charge densities. Interestingly, they found that the cubic (QIIG, siRNA) and the inverted hexagonal (HIIsiRNA) phase containing GMO exhibited higher total silencing and lower nonspecific silencing than the lamellar (LαsiRNA) phase (Fig. 2a).
Fig. 2.
Cubosomes-based NPs for siRNA delivery. (a) Schematic depiction of the cubic (QIIG, siRNA) phase, the inverted hexagonal (HIIsiRNA) phase, and the lamellar (LαsiRNA) phase of cationic liposomes/siRNA complexes, Reproduced with permission [177]. Copyright 2010, American Chemical Society; (b) Microfluidic synthesis of cubosomes and their formation mechanisms, Reproduced with permission [178]. Copyright 2018, American Chemical Society.
Ultrachilled sonication method was exclusively successful in the formation of cubosomes in earlier studies, while the obtained particle size was usually in micro scale and their size distributions were too broad, which were adverse for their clinical transitions. To overcome this obstacle, Kim et al. developed a microfluidic nano-manufacturing device to synthesize cubosomes based on GMO/DOTAP/GMO-PEG lipid mixture (Fig. 2b). The microfluidic device allows rapid mixing (0.6 s) of lipid/ethanol and water solutions, leading to the formation of 50 nm ethanol-in-water emulsion droplets that stabilized by the lipid layer. After the evaporation of ethanol, membranes self-organized into periodic bicontinuous cubic arrays to form cubosomes of approximately 200 nm with extremely narrow size distribution (PDI = 0.04). Moreover, when the amount of a steric stabilizer GMO-PEG was increased from 1 to 2 mol %, cubosomes with sizes as small as 75 nm were obtained without loss of internal structure. Finally, the small size cubosomes demonstrated a significant role in terms of delivering and eliciting specific gene knockdown in targeted cells [178].
4.2. Polymeric nanoparticles
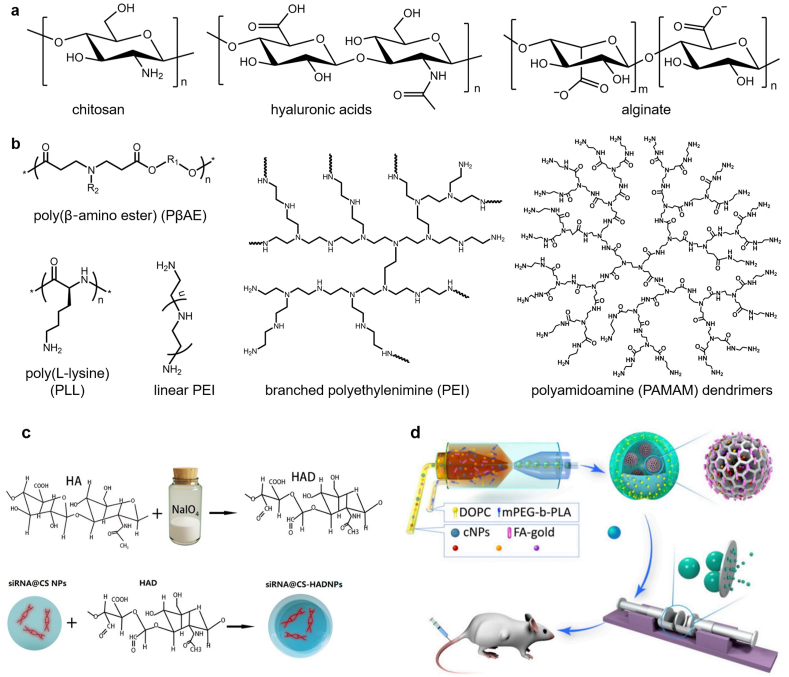
Nanoparticles constructed from polymers, termed polymeric nanoparticles (PNPs), have been extensively explored in the anti-tumor delivery of various drug formulations due to their flexibility in tuning the physical-chemical properties for efficient drug loading and controllable release. The structural versatility endows PNPs with great opportunities in carrying TOs through multiple strategies, e.g. electro-adsorption by cationic polymer micelles, physical encapsulation within the cavity of polymer vesicles, or hybridization to PNPs constructed from amphiphilic DNA block copolymers via Watson-Crick base pairing [179], in pursuit of the optimal anti-tumor efficacy. Based on the polymer source of origin, PNPs can be divided into two categories: natural-origin polymers (Fig. 3a) based NPs and synthetic polymers (Fig. 3B) based NPs.
Fig. 3.
Typical polymers used for TOs delivery. (a) Natural–origin polymers-based NPs; (b) Synthetic polymers-based NPs; (c) Chitosan-hyaluronic acid dialdehyde nanoparticles (CS-HAD-NPs) for Bcl-2 siRNA delivery in the application of bladder cancer therapy [180]. Copyright 2021, Elsevier B.V.; (d) Schematic of core-shell structure based nanoparticles for drug delivery [181]. Copyright 2019, United States National Academy of Sciences.
4.2.1. Natural-origin polymers-based NPs
Natural-origin polymers, like chitosan, hyaluronic acids, and alginate (Fig. 3a) have received considerable interest for TO delivery due to their degradability, biocompatibility, and low cost [182]. Chitosan (CS) is a linear polysaccharide with free amino groups on the lateral chain, which can bind to TOs through electro-static interactions in a simple but efficient way. Chitosan NPs are biodegradable and have low immunogenicity, and their solubility as well as the biocompatibility can be largely improved through modifications of CSs with small molecules or polymers. For example, Sun et al. found that PEGylated CS had superior structural stability in the physical environment compared to unmodified CS nanoparticles [183]. PEG-CS NPs could effectively deliver siRNA to the targeting breast cancer cells and reduced the growth of xenograft tumors of 4T1 cells in vivo. In another study, Corbet et al. modified CS NPs with PEG through a simple non-covalent method, the surface of which was functionalized with the prototypical RGD peptidomimetic (RGDp) for tumor targeting [184]. The optimized RGDp/PEG-CS nanoformulations, with naphthyridine-containing RGDp randomly coupled to the PEG chain by clip photochemistry and the use of a lipophilic linker, have shown great capability in delivering siRNAs targeting to metabolic biomarkers of cancer cells: lactate and the glutamine transporters, leading to significant antitumor effects. Hyaluronic acid (HA) is an anionic, nonsulfated glycosaminoglycan that is the main component of the extracellular matrix (ECM). Due to their capability to target CD44-overexpressing tumor cells, HA has gained significant interest in the targeted anticancer therapy. In our recent work, targeting CD44 for bladder cancer treatment was achieved by delivering the Bcl-2 siRNA with self-crosslinkable chitosan-hyaluronic acid dialdehyde nanoparticles (Fig. 3c) [180]. It should be noted that HA cannot directly interact with oligonucleotides because of their electrostatic repulsion. However, this obstacle can be overcome by chemically modifying HA with specific groups for oligonucleotides conjugation or adding a third component of cationic polymers (polycations) to condense oligonucleotides [185]. In the latter situation, HA molecules usually locate on the surface of oligonucleotides-polycation complex, which endows the complex with the tumor targeting ability. Similar to HA, alginate, a natural polysaccharide consisted of α-d-mannuronic acid and β-l-guluronic acid, is another non-condensation oligonucleotides delivery system. In the presence of divalent cations (such as Ca2+, Ba2+), alginate nanogels with oligonucleotides-encapsulation capability can be obtained by precisely manipulating the diffusion of cations from a large outer reservoir into alginate solution [186]. Alternatively, alginate nanogel can be designed as a biocompatible shell decorated on the surface of polycation-DNAs complex, which can not only protect DNAs from nuclease degradation, but also enhance the transfection efficacy compared to non-decorated counterparts.
4.2.2. Synthetic polymers-based NPs
Although natural polymers have shown excellent biocompatibility, their difficulties in controlling the differences between batches, their poor mechanical properties and limited processing capabilities mainly restrict their clinical transition. Comparing with natural polymers, synthetic polymers are recently being more attractive as carriers for TOs delivery owing to their reproducible properties on the basis of molecular weight, degradation, mechanical properties, and structural tunability to achieve efficient TOs loading and targeted release. The most frequently used synthetic polymers for TO delivery contain but are not limited to polyethylenimine (PEI), poly(l-lysine) (PLL), poly(β‐amino ester) (PβAE), polyamidoamine (PAMAM) dendrimers (Fig. 3b) [[187], [188], [189], [190], [191], [192], [193], [194], [195]]. One common feature of these polymers is that they all contain primary or tertiary amino groups that are protonable. After protonation, these polycations can form much small-sized polyion complexes (PICs) or polyplexes in aqueous media with anionic oligonucleotides through electrostatic interactions. The structural variability of polycations (e.g. the molecular weight, the charge density, the degree of branching) can be precisely tuned via controlling the polymerization conditions, which provides a feasibility for researchers to screen PNP formulations to achieve the best transfection efficiency. To efficiently condense TOs into nanoparticles, dense positive charged polycations with high molecular weight are generally required, which usually cause serious toxicity however. In contrast, low molecular weight polycations with low density of positive charges usually show poor efficacy in delivering TOs into cytosols. To overcome this dilemma, people used natural polyphenols, which have strong binding affinity with DNAs or RNAs via non-covalent ionic interactions, to achieve the condensation of nucleic acids by polycations with low molecular weight [196]. Effective TOs delivery with minimal toxicity was thus approached.
Using diblock copolymer PEG-b-PLL, Hayashi et al. investigated the influence of oligonucleotide rigidity on PIC formation by complexing the polymer with single-stranded RNA (ssRNA) or dsRNA. The PICs prepared from the flexible ssRNA showed a two-step assembly behavior: (1) formation of minimal charge-neutralized units (unit PICs); (2) formation of PIC micelles from unit PICs. In contrast, The PICs prepared from siRNA remained in the unit PICs stage throughout the measured range of concentrations, indicating that the rigidity of ionomers could be used as a pivotal structural parameter to stabilize the structure of the primary ion‐complex [187]. To overcome the instability of rigid siRNA-loaded micellar PICs, Naito et al. directly conjugated the siRNA to PEG-b-PLL through a reversible linker, tetravalent 3-fluorophenylboronic acid (FPBA), which can be replaced by the adenosine triphosphate (ATP) in a concentration‐dependent manner. At a low ATP concentration (<0.3 mM), the FPBA-crosslinked PIC micelles were quite stable, while at a higher ATP concentration (~3 mM, cytoplasmic condition), PIC micelles rapidly dissociated and released the siRNA payloads [192]. Except the strategies of loading TOs in the core of NPs, TOs can also be covalently immobilized to the surface of NPs without losing their potency. For instance, Chan et al. proposed a micellar NP based on the dual functionalized poly(d,l-lactide-co-2-methyl-2-carboxytrimethylene carbonate)-graft-poly(ethylene glycol), which was sequentially decorated with the targeting trastuzumab antibodies and siRNAs or antisense oligonucleotides (AOs) on the exterior PEG corona via the orthogonal Click reactions [197]. The targeted delivery systems are as effective as Lipofectamine in gene silencing without the associated potential toxicity of the latter. Moreover, the diblock copolymers can also be used as the shell structure of composite nanoparticles to achieve multiple drug delivery. In our previous work, a photothermal-responsive nanosized hybrid polymersome drug delivery system had been established for hydrophobic anticancer drugs delivery, and it can also be used for magnetic nanoparticles, DNA, or antibodies delivery to realized more specialized applications (Fig. 3d) [181].
As another widely developed biodegradable polymers with broadly adjustable structural diversity, poly(β-amino ester)s (PβAEs), mainly synthesized from a variety of diacrylates, amino-alcohols, and end-capping monomers by Michael addition reactions, have shown excellent efficiency in gene delivery [198]. A large number of PβAES libraries can be synthesized and screened with high throughput, generally with a simple synthesis procedure, to determine the optimal structures, resulting in efficient gene delivery to a wide range of cell types range of cell types [199]. To improve the stability of RNAi in serum, Dosta et al. synthesized hydrophobized versions of PβAEs using hexylamine, hexadecylamine, and cholesterol. The obtained polyplexes were stable against plasma proteins for more than 48 h and much higher transfection efficiency was achieved compared to non-hydrophobized PβAEs [194]. Besides the application in single gene delivery, PβAEs can also act as a potential carrier for combined delivery of TOs with proteins for synergistic cancer treatment. For example, Rui et al. prepared a novel class of hyperbranched PβAEs including both cationic and anionic charges through polymer end-capping with carboxylate ligands, and realized the gene editing function by co-delivery of gene-targeting short guide RNA (sgRNA) and Cas9 ribonucleoproteins (RNPs) [195]. The authors speculated that the introduction of carboxylate ligands can enhance the interactions between polymers and proteins, thus was beneficial for RNPs encapsulation. Furthermore, they found that the hydrophobicity of polymer end-group affected protein complexation, endocytosis of nanoparticles and escape from endosomes. Using optimized formulation of polyplexes, successful delivery of sgRNA/RNPs in vitro and in vivo and high levels of gene editing at relatively low RNP doses were achieved, highlighting the robustness and therapeutic potential of these nanocarriers.
Except the linear or hyper-branched polymers, tree-like dendritic polymers, termed dendrimers, are also promising carriers for TO delivery. In general, dendrimers mainly contain three distinct parts [200]: (1) a central core, where a dendrimer growth begins; (2) an inner shell, consisting of repetitive branching units (named as generations); and (3) an outer shell, comprised of numerous terminal functionalities attached to the surface. A representative example of dendrimer is polyamidoamine (PAMAM) dendrimer, which is consisted of interconnected ethylenediamine molecules through electrostatic interactions with oligonucleotides to form complexes [201]. It was found that the interactions of electrostatic oligodeoxynucleotide-dendrimer complexes were sensitive to pH as well as the ionic strength, and the maximal interaction occurred at low pH and ionic strength [202]. PAMAM dendrimers have inner cavities and peripheral functional groups, which that can be modified into physically embedding or covalently conjugate drugs with high curative effect [203], so as to become a promising platform for the co-delivery of genes and other drugs [188,204,205]. In addition, PAMAM dendrimers can be combined with other polymers to approach specific functionalities, such as sustained delivery of TO. For instance, Segovia et al. proposed a PAMAM dendrimer-based hydrogel system, embedded with siRNA loaded PβAE-NPs [206]. Local and continuous delivery of siRNA was achieved and the composite hydrogel displayed nearly twice the transfection efficiency in vitro compared to the most effective commercially transfection reagents. It should be noted however, PAMAM dendrimers with lower generations (G0-G4) usually have size smaller than 10 nm, which may face the risk of rapid renal clearance, while higher-generation dendrimers may produce higher cytotoxicity [207]. To decrease the cytotoxicity and improve the transfection efficiency, strategies focusing on the surface engineering of PAMAM dendrimers with specific moieties, e.g. lipids, peptides, amino acids, polymers, have been extensively explored [208,209]. Alternatively, interests have been put into the composite systems, where small sized PAMAM dendrimers were used as one of the building components in couple with other NP formulations. The fabricated composite delivery systems could sense the tumor microenvironment and programmable drug delivery and deep tumor penetration could be achieved [190,193,210,211].
4.3. Gold nanoparticles
Gold nanoparticles (AuNPs) is another broadly developed nanocarrier for targeted delivery of TOs in a variety of cancer cell lines due to their (1) precise control over sizes in the range of 2–200 nm; (2) facile surface functionalization with any kinds of TOs; (3) tunable localized surface plasmon resonance (LSPR) in a broad wavelength spectrum from visible to near IR region; (4) ease of qualitative and quantitative analysis in the complicated biological environment.
4.3.1. Spherical gold nanoparticles
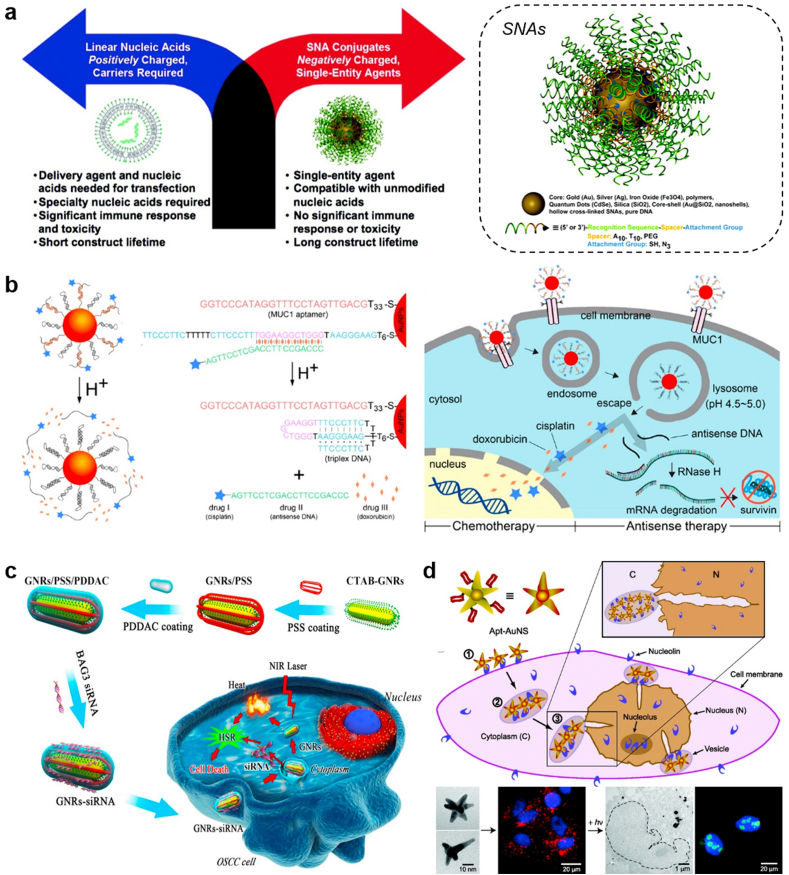
In 1996, the Mirkin group constructed a spherical form of nucleic acids using the AuNPs template, termed as spherical nucleic acids (SNAs) [212]. The densely and highly oriented packing of DNAs on the surface of AuNPs sterically prevent DNA degradation by nucleases [130], while the hybridization capability of ssDNA on SNAs to its complimentary sequence did not decline. Moreover, SNAs can enter cells in high numbers via scavenger receptor mediated endocytosis without the assistance of transfection reagents [213,214], offering a new paradigm for gene regulation (Fig. 4a) that is different with traditional strategies [215], where negatively charged nucleic acids are always need to be pre-complexed with positively-charged carriers to enter cells. Up to now, dozens of SNAs have been developed and evaluated by Mirkin group for their potential in rapid diagnostics of cancers, as well as for various diseases that are difficult to address with traditional therapeutic strategies [129,216]. For example, in one earlier study, they fabricated the RNAi-based SNA targeting oncogenes (Bcl2L12) which is traditionally untargetable by small molecules or antibodies in glioblastoma multiforme (GBM) pathogenesis [217]. Upon intracranial or intravenous administration, Bcl2L12-SNAs could cross the blood-brain barrier (BBB), penetrate the glioma, and promote apoptosis of glioma cells by enhancing caspase and p53 activities. In another study, they fabricated immunomolulatory oligonucleotides-based SNAs to stimulate (IS–SNAs) or regulate (IR-SNAs) immunity by engaging TLRs [218]. Compared with the free oligonucleotides, IS-SNAs exhibited an 80-fold increase in responses to model antigens, which IR-SNAs have reduced fibrosis score in mice with nonalcoholic steatohepatitis by 30%. Recently, they expanded the type of SNAs by choosing different core materials, including liposome [[219], [220], [221]], polymeric micelles [222,223], proteins [224], and metal organic framework [225,226]. Such well-defined three-dimensional SNAs exhibit distinctive properties that apart from those of both the nanoparticles and nucleic acids form they derive, such as minimal immunogenicity [227,228], negligible cytotoxicity and undetectable off-target effects [229].
Fig. 4.
Gold nanoparticle-based platforms for TOs delivery. (a) Spherical nucleic acids offer a different paradigm for gene regulation. Reproduced with permission [215], Copyright 2012, American Chemical Society; (b) Multidrug-loaded nanoswitch and its intracellular pH-responsive multidrug delivery and release. Reproduced with permission [233], Copyright 2019, American Chemical Society; (c) BAG3 siRNA loaded gold nanorods to improve the PTT efficiency via silencing the heat-shock response. Reproduced with permission [238], Copyright 2016, Elsevier Ltd.; (d) Nucleolin-mediated, active trafficking of AS1411-conjugated gold nanostars to the cancer cell nucleus. Reproduced with permission [241], Copyright 2012, American Chemical Society.
In addition to the use as the single formation for cancer treatment, TOs-loaded Au nanospheres can also be combined with chemotherapeutics to achieve enhanced anticancer therapy through synergistic effect between TOs and chemotherapeutics [[230], [231], [232]]. In an earlier study, Kim and his coworkers constructed a pH-responsive gold nanocluster formed from 13 nm of spherical Au NPs grafted with two types of oligodeoxynucleotides (ODNs), bcl-2 antisense ODN and i-motif binding ODN (iBO, a four-stranded DNA secondary structure that partially complementary to the i-motif, which usually forms in acidic conditions with cytosine-rich sequences) [231]. At neutral pH, addition of the i-motifs as the linker strands clumped Au NPs together through the partial hybridization of i-motif and iBO, and this duplex formation facilitate the loading of anticancer drug Dox by intercalation. After being internalized to the endosomes, however, the cluster could rapidly disassemble because of the formation of i-motif structure of the linker strands within acidic environment, which resulted in exposure of antisense ODNs to mRNA and enhanced Dox release for cancer cell killing. Compared with the single AuNPs as the drug carrier, size-tunable gold nanoclusters designed in this study provided more opportunities for both TOs and chemotherapeutics loading and stimuli-responsive drug release capacities [232].
Recently, Chen et al. developed a DNA-based nanodevice capable of releasing antisense DNA and chemotherapeutics responding to a pH range of approximately 5.0–7.0 [233]. The nanodevice was fabricated from AuNPs functionalized with the sequence-specific DNA complex, consisting of an anti-MUC1 aptamer (function as the targeting ligand), a conformation switchable DNA sequence in responsive to pH change, and antisense DNA (asDNA) targeting the cancer-associated survivin mRNA (Fig. 4b). The model drug Dox could be effectively loaded in the nanodevice via intercalation in the double strand region of the DNA complex. In lysosomes (pH ~4.5–5.0), the switchable DNA strand rapidly changed the conformation from linear (pH 7.4) to triplex, leading to the release of Dox and asDNA for synergistic cancer cell killing. Finally, the nanoswitch displayed an efficient gene silencing and a significant tumor-growth inhibition in the tumor-bearing mouse model.
4.3.2. Non-spherical gold nanoparticles
Compared to sphereical AuNPs, non-spherical AuNPs (e.g. nanorods, nanostars, nanocubes), also called anisotropic AuNPs, have gained more interest in recent years in targeted delivery of TOs for anticancer therapy. Firstly, the anisotropic character of non-spherical AuNPs allows the LSPR peak of AuNPs shift from the visible region to the near infrared region (NIR), which enables researchers design NIR-responsive TO delivery systems to achieve deep tumor penetration and therefore enhance therapeutic index of TOs; Secondly, because of their optical response, anisotropic AuNPs show excellent optical signal enhancement, which can be used in the design of sensors, in addition to acting as the drug carrier, to probe the delivery as well as therapeutic efficiency of TOs in real time; Thirdly, due to the electron-phonon interactions, the NIR radiation absorbed by the anisotropic AuNPs surface can be rapidly converted into heat, which have been widely used in NIR-based photothermal therapy or photothermal/TOs combination therapy; Fourthly, numerous studies have shown that some specific anisotropic AuNPs have higher cellular uptake efficiency compared to spherical AuNPs, and the intracellular transportation of TO payloads with higher anticancer therapeutic index can also be realized by the anisotropic AuNPs.
The precise tunability in terms of sizes and aspect ratios of gold nanorods (AuNRs) provides many attractive optical properties for sensitive bioimaging and remote-controlled TOs delivery [234]. AuNRs are usually synthesized by seed-mediated reduction of tetrachloroauric acid in the presence of cetyltrimethylammonium bromide (CTAB) [235]. It is not easy to attach thiolated DNA on AuNRs is not easy because CTAB can be tightly adsorbed on AuNRs surface, which prevents the formation of an Au–S bond between thiolated DNA and AuNRs. Therefore, to further expand the potential of AuNRs in TOs delivery, the efficient and facile DNA-functionalization strategies must be developed. In pursuit of this purpose, Li et al. used mPEG-SH to replace CTAB on the surface of AuNRs and Tween 20 to assist the replacement process and stabilize AuNRs, and this complete functionalization could be finished within 1 h [236]. In another study, Wang et al. presented a potent strategy for loading siRNA duplexes onto AuNRs based on the dithiocarbamate chemistry [237]. They systematically evaluated the bioactivity of AuNRs–siRNA complexes against eGFP-producing ovarian cancer cells (SKOV-3). Efficient knockdown was achieved by on-demand release of DTC-anchored siRNA upon femtosecond-pulsed laser irradiation. Interestingly, using the same AuNRs, the knockdown activity of DTC-anchored siRNA was much higher than that of thiol-anchored siRNA. Non-invasive NIR photothermal therapy (PTT) based on AuNRs has become a promising tool in cancer treatment. To minimize the hyperthermia-induced collateral damage to normal tissues, a mild laser power or agent to produce moderate heat is suggested. However, mild PTT can trigger the heat shock response (HSR) in cancer cells, upregulating the expression of heat shock proteins and inhibiting the pathways of cell apoptosis. To overcome this dilemma, Wang et al. loaded siRNA targeting BAG3 (an HSR-associated gene) on AuNRs (Fig. 4c), which can effectively deliver the siRNA into cancer cells in silencing the heat-shock response [238]. The in vivo studies demonstrated that AuNRs-siRNA could effectively reduce the HSR in cancer cells and were sensitive to PTT following enhanced cell apoptosis under moderate laser irradiation.
The gold nanostars (AuNS) with a multi-branched morphology is another type of anisotropic AuNPs that have been widely developed in the targeted delivery of anticancer TOs. Comparing with gold nanorods, the synthesis of AuNS is much easier and greener: they can be synthesized by the ‘one-port, seedless’ reduction of Au3+ in biocompatible Good's buffer (e.g. HEPES, MOPS, EPPS) and do not need the toxic surfactant for stabilization, which make the post-synthesis loading of TOs more efficient [239]. From this aspect, AuNS seems more suitable than AuNRs when developed as the TO vehicle. In the past ten years, the Odom group developed a series of AuNS-based system for targeted delivery of TOs [[240], [241], [242], [243], [244], [245], [246]]. In one earlier study, they fabricated the AS1411 (a nucleolin specific aptamer)-conjugated AuNS and directly observed that the nanoconstructs could be locatedin the nucleus and cause major changes in nuclear phenotype through nuclear envelope invaginations (Fig. 4d), leading to the apoptosis of cancer cells [241]. Later on, they found that AS1411@AuNS exhibited notable anticancer effects in a group of 12 cancer cell lines [240]. In another work, they fabricated lysosomal targeting nanoconstructs composed of anti-HER2 aptamer (human epidermal growth factor receptor 2, HApt) grafted onto AuNS surface [243]. Within lysosomes, HER2 could be degraded by enzymes under acidic pH condition, leading to apoptosis of cancer cells. This work demonstrates that by targeting lysosomes and utilizing biomarkers for lysosomal degradation, the known shortcomings of therapeutics in nanoscale-the inactivation of TOs due to the entrapment of nanoconstructs in endo/lysosomes-can be overcome. Recently, the same group investigated how the surface curvature of AuNPs decorated with immunostimulatory oligonucleotides CpG influence the immune activation effects of CpG-AuNPs [244]. Interestingly, they found that the mixed-curvature constructs (nanostars) produce a relatively high percentage of hollow endosomes and a higher immune cell response than constant-curvature constructs (nanospheres) constructs. This work highlights that the local organizations of CpG inside endosomes, for pursuing optimal immune responses and intracellular delivery, can be controlled by using the anisotropic AuNS. Recently, our group found that the HApt-conjugated AuNS could also act as the promising carrier for incorporation of small molecular drugs (e.g. anticancer doxorubicin, Dox) to achieve on-demand drug delivery and enhanced toxicity against cancer cells [247].
Ultrasmall (~2 nm) AuNPs can directly deliver oligonucleotides to the nucleus without any additional nuclear-targeting functional ligands, which can interfere with the transcription process of related genes, thereby affecting the life activities of cells, and thus showing great anti-tumor potential. However, their low degree of net cellular uptake because of exocytosis and rapid clearance from the body limited the in vivo therapeutic efficacy [248,249]. To address these problems, Huo et al. designed a sunflower-like gold-DNA nanostructure (~200 nm), which was assembled from 2 nm AuNPs modified by the complementary silent sequence of the c-myc oncogene [250]. The nanosunflowers had a strong NIR absorption ability, and could decompose and release ultrasmall Au NPs under NIR irradiation. Increased cellular uptake, tunable gene silencing, and controlled tumor inhibition were successfully achieved by synergistically regulating the pre-incubation time and the NIR irradiation time point, the cell uptake was obviously increased while the adjustable gene silencing and a significant inhibitory effect on tumors were achieved.
Except the covalent immobilization strategies, TOs can also physically absorb to the surface of AuNPs pre-modified with the cationic polymers through electrostatic interactions. For example, Lei et al. developed a siRNA-loaded gold nanocluster system (GNC-siRNA complex) to target the interactions between tumor cells andneuron against cancer via depleting the nerve growth factor (NGF) [251], a kind of neurotrophic factor that actively promote the growth of neurites and stimulate neurogenesis, contributing to the survival, proliferation, invasion, and metastasis of tumors [[252], [253], [254], [255]]. GNCs that constituted by a straight but effective method endowed the complex with a high loading capacity of siRNA which significantly reduced NGF expression as well as the neurite sprouting, and eventually inhibited the tumor growth by activating a variety of downstream protein kinases. This study showed an attractive therapeutic direction to cancer treatment. In another study, Yi and coworkers reported a sub-50nm nanoassembly for target delivery of siRNA to cancer stem-like cells (CSCs). The nanostructure was prepared via a two-step assembling process, where lipoic acid-modified, glucose-installed poly(ethylene glycol)-block-poly(l-lysine) was first associated with siRNA via electrostatic interaction to form an unimer polyion complex (uPIC), which was further decorated on 20-nm Au NPs through Au–S bonding. As the core, Au NPs make it easier and more precise to obtain sub-50nm nanostructures compare to other formulations of nanocarriers for further efficient permeation of tumor tissues. The nanoassembly elicited significantly gene silencing effect and successfully suppressed the growth of the CSC-rich orthotopic breast tumor [256].
4.4. Porous nanomaterials
Porous nanomaterials are composed of a solid framework with porous structures and large surface area, which allow the high-efficiency encapsulation of drug molecules within the pores and facile modification of interest functional groups on the surface. The rapid development of synthetic methodologies of porous nanomaterials in recent years has largely expended the delivery applicability of drug moieties, either hydrophobic or hydrophilic, from traditional small molecules to more potent macromolecular drugs, like proteins and nucleic acids. Moreover, the pores inside SiNPs allow high-efficiently loading both oligonucleotides and compounds for combined cancer treatments. Numerous studies have indicated that through rational design of the pore size and morphology, porous nanomaterials can a promising vehicle in carrying TOs for the targeted cancer therapy. Generally, the widely developed porous nanomaterials for TOs delivery can be classified into three types: porous silicon nanoparticles, mesoporous silica nanoparticles and mental-organic framework. Compared to other types of nanoparticles, porous nanomaterials have uniquely mesoporous structures, high drug-loading capacity, high active surface area and potential for the development of gated supports for on‐demand delivery applications, and other unique characteristics that make them great potential alternatives for cancer treatment applications [[257], [258], [259]].
4.4.1. Porous silicon nanoparticles (pSiNPs)
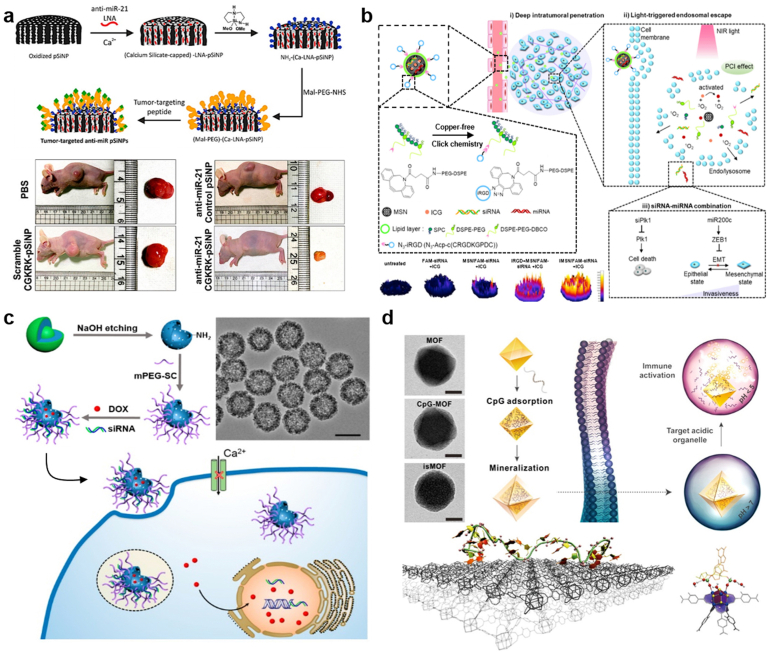
Porous silicon nanoparticles (pSiNPs) can be synthesized either through physical methods, including the pulsed laser ablation of silicon wafer and the heat decomposition of silanes, or through chemical strategies which usually involve in the reduction of silicon halides by specific reducing agents (e.g. sodium naphthalenide, lithium naphthalenide, lithium–aluminum hydride and zintl salts). In a recent study, Bertucci et al. constructed a microRNA therapeutics-encapsulated pSiNPs with the tumor-homing peptide displaying on the surface (Fig. 5a) to achieve efficient cancer treatment targeting to miR-21, an oncogenic miRNA overexpressed in many tumors [260]. By means of the calcium silicate-trapping chemistry, a significantly high encapsulation efficiency (97 ± 2%) of anti-miR-21 oligonucleotides within pSiNPs (17 wt%) was obtained, and the tumor-targeting pSiNPs displayed a significant growth inhibitory effect on ovarian tumor via down-regulating the expression level of miR-21.
Fig. 5.
Targeted delivery of TOs based on porous nanoparticles. (a) Preparation of tumor-targeted anti-miR porous silicon nanoparticles (pSiNPs). Reproduced with permission [260], Copyright 2019, American Chemical Society; (b) NIR-Triggered RNA co-delivery by tumor-penetrating mesoporous silica nanoparticles (iMSNs) for siPlk1/miR-200c combination therapy. Reproduced with permission [268], Copyright 2020, American Chemical Society; (c) Fabrication of Dox/siRNA-loaded mesoporous silica nanoparticles to regulate Ca2+ signaling for drug-resistant breast cancer therapy. Reproduced with permission [271], Copyright 2019, American Chemical Society; (d) Coordination-based method for preparing immunostimulatory DNA–MOFs. Reproduced with permission [290], Copyright 2017, American Chemical Society.
pSiNPs are degradable in organisms, and the produced free silicon atoms can be converted into non-toxic silicic acid, which can be effectively metabolized and eliminated by the human body. However, the freshly prepared pSiNPs are prone to rapid degradation in aqueous medium, and thus post-modifications (e.g. surface functionalization with polymers) are needed to improve their stability. For instance, Kafshgari et al. constructed a chitosan-decorated pSiNP system for TO delivery [261], where the chitosan coating could significantly improve sustained oligonucleotide release and enhance the internalization of oligonucleotide-loaded pSiNPs across the cell membrane. Furthermore, the biocompatibility and non-inflammatory properties of pSiNPs were well maintained through in vivo evaluations. In another study [262], Tong et al. established a polyethyleneimine (PEI)-capped pSiNPs system to achieve high-capacity loading for delivery of siRNA, targeting to the gene of multidrug resistance-associated protein 1 (MRP1), an overexpressed chemoresistance-associated biomarker in glioblastoma multiforme (GBM). Optimized siRNA release (70% released over 48 h) and effective knockdown of MRP1 expression in GBM by 30% were observed. The authors demonstrated that MRP1-siRNA loaded pSiNPs could successfully silence MRP1 in GBM tumors 48 h post-injection and a significant reduction of GBM proliferation was observed, which may be mediated by the decrease of MRP1 transmembrane transport and subsequent leading to cell cycle arrest.
Except directly using as the TOs delivery vehicle, pSiNPs can also be combined with other NP formulations to overcome the shortages of each single formulation. For example, it has been reported that the fusogenic liposomes, which can directly fuse with cell membrane to avoid insufficient endocytotic sequestration and subsequent lysosomal degradation, generally suffer from low loading capability of TOs and non-negligible leakage during circulation in vivo. In order to overcome this obstacle, Kim et al. designed a novel delivery system, termed fusogenic nanoparticles (FNPs) [263], which harnessed together a fusogenic lipid and a pSiNP core, the latter of which could efficiently encapsulate siRNAs via the Ca2+ precipitation strategy with minimal leakage [264]. FNPs demonstrated a receptor‐independent endocytosis cell uptake mechanism, and showed great potential for sensitizing cancer cells to chemotherapy in gene therapy and immunotherapy in polarizing macrophages towards the tumorigenic M1 phenotype.
4.4.2. Mesoporous silica nanoparticles (MSNs)
Silica is an oxide form of silicon with the chemical formula SiO2. SiO2-NPs are usually synthesized by the hydrolysis of the tetraethyl orthosilicate (TEOS) in an alkaline medium. Using the commercially available alkoxysilanes or halosilanes, the surface of SiO2-NPs is facile to functionalize with amino, mercapto, epoxy or acryl groups to facilitate the immobilizations of fluorophores, targeting moieties or drugs. By adding surfactants (such as cetyl trimethylammonium bromide), micelle forming type materials, polymers, or other dopants to the synthesis solutions, mesoporous silica nanoparticles (MSNs) can be obtained [265]. It has been demonstrated that the increased delivery efficiency of MSNs can be achieved by tailoring their size, shape and external surface functionalization [266]. A significant advantage of MSNs in TO delivery is their controllable porous structures and surface functionalization, endowing them with extremely high TO-loading capacity. Taking siRNA for example, Möller, et al. once reported a core-shell mesoporous silica nanoparticle system with multifunctional polymer caps, where they demonstrated a loading capacity of siRNA could reach as high as 380 μg mg−1 [267].
Recently, Wang et al. developed a novel MSNs platform, termed iMSNs (Fig. 5b) [268], in which the large internal surface area and pore volume make it easy to carry various therapeutics. On the one hand, stabilized structures endowed iMSNs a high loading efficacy for co-delivery of mRNA and siRNA via modification with a surface lipid layer, which conjugated to the iRGD peptide. On the other hand, a photosensitizer, ICG, was loaded into the iMSNs for in situ generations of ROS to realize NIR triggered cytoplasmic RNA release. Thereby, this co-delivery nanoplatform improved therapeutic effect by silencing multiple target genes and cellular pathways of tumor cells simultaneously acted on eliminating the primary tumor and inhibiting the metastatic process. In addition to cellular generation of ROS, cytosolic Ca2+ is also a vital regulator of signal transduction in various cancer processes containing proliferation, tumor genesis, and migration [269,270]. The controlled increase of cytosolic Ca2+ concentration through the plugging of the T-type Ca2+ channel with antagonists or siRNA can inhibit cancerous proliferation, while the specific delivery of antagonists or delicate siRNA to tumor site remains an inevitable challenge. Wang et al. fabricated the mesoporous silica nanocapsules (MSNCs) for cocktail delivery of both doxorubicin (DOX) and siRNA targeting T-type Ca2+ channels concurrently (Fig. 5c) [271]. They demonstrated that the accumulation of intracellular drug concentration led to the decrease of cytosolic Ca2+ concentration, downregulated expression of Ca2+ channel and specifically induced of G0/G1 arrest of the cell cycle, finally exhibited an extraordinary synergistic therapeutic effect for multidrug resistant breast cancer. Therefore, the delivery of TOs acted on T-type Ca2+ channels could be a strategy with great potential for multidrug resistant cancer treatment with MSNCs.
In another report, silica nanoparticles demonstrated a well performance in overcoming the shortages of nucleic acid nanoparticles (NANPs) when considering their targeted delivery in vivo. NANPs have been identified as a novel but promising scaffold in incoprorating TOs for anticancer purpose due to their excellent capability of tailoring DNA/RNA sequences in a programmable manner. However, the susceptibility of NANPs to enzymatic degradation as well as their inability to cross biological membranes seriously limited their clinical transition. To overcome these problems of NANPs, Juneja et al. combined NANPs with MSNPs via the electrostatic association, and the formed nanoconstructs could not only effectively deliver NANPs to their targe sites but also synergize with other therapeutic agents to achieve enhanced anticancer effect [272].
4.4.3. Metal-organic framework
Mental-organic framework (MOF), a type of porous hybrid materials establish from metal irons or clusters bridged by organic ligands, has been widely developed and exploited in diverse fields, including gas storage and separation [273,274], catalysis [275], biosensing [276], and biomedicine [277,278]. When exploited as the drug delivery platform, MOFs show promising prospects and advantages for the treatment of cancer, which include but not limited in the following aspects: i) the large surface area and precisely controllable porous structure endow the MOFs with a high drug loading capacity; ii) the structure of MOFs can be well adjusted to achieve stimuli-responsive drug release with a high specificity in tumor tissues; iii) MOFs are biodegradable and have shown good biocompatibility in a numerous studies [279,280]. When used as the drug delivery vehicle of TOs, MOFs show extra advantages, e.g. the metal elements that form the MOF structure may also be an active component synergistically taking part in the therapeutic action of TOs. Therefore, MOFs have received a much broader attention in recent years, as an alternative delivery system to the existing nanocarriers in the targeted delivery of TOs [[281], [282], [283], [284]].
It should be noted that the traditional MOFs used for loading small molecular drugs were no longer effective in encapsulating TOs within their internal pores, due to the macromolecular nature of TOs [285]. To overcome this obstacle, Michelle et al. [286]. recently established a highly porous zirconium-based MOF structure, named NU-100, an optimal structure selected by molecular simulations, offering a porosity compatible with siRNA. The authors demonstrated the successful loading of the siRNAs within the internal pore of MOF, not on the surface of the framework, via the fluorescence-lifetime imaging microscopy. In addition, the combined encapsulation of species that were able to open up endosomes with siRNA@MOF complex was proved as an effective way to deliver siRNA to cytoplasm and achieved sufficient gene knockdown. Different from the method of synthesizing large TO molecules and infiltrating them into the MOFs with large pores, Liu and coworkers developed a facile and versatile strategy to prepare MOF‐based biomimetic co-delivery system that can greatly increase drug loading efficiency [287]. The antisense oligonucleotides that target the antiapoptotic protein Bcl-2 (G3139) and doxorubicin were co-assembled into a uniform NP formulation (DOX/Fe‐G) with the aid of FeII ions. Subsequent biomineralization of the DOX/Fe-G surface with a thin shell of ZIF-8 MOFs was used to produce the final core-shell form of the nanoplatform (DOX/Fe-G@Z), which exhibited effective both in vitro and in vivo. Subsequently, the thin shell of ZIF-8 MOFs were mineralized on the surface of DOX/Fe‐G to produce the final core-shell nanostructures (DOX/Fe‐G@Z), which exhibited effective tumor growth inhibition both in vitro and in vivo. The authors emphasized that the current straightforward biomimetic approach could greatly engender broad opportunities of MOF for practical uses.
Apart from the physically encapsulation method, DNA molecules can also be loaded in MOFs via covalent conjugation [288] or coordination chemistry-based strategies [289,290]. In 2014, Mirkin group reported the first MOF-DNA conjugate [288], which was realized by the Cu-free strain promoted click reaction between the azide-functionalized zirconium based framework, UiO-66-N3 and the dibenzylcyclooctyne-modified DNA. Compared with bear MOF particles of comparable size, DNA conjugation increased MOF stability in aqueous NaCl and enhanced cellular uptake efficiency. Later on, they developed a more general strategy to functionalize MOF nanoparticles with oligonucleotides at high density through modifying the terminal of oligonucleotide with a phosphate [289], which could coordinate with a series of unsaturated metal nodes on their respective MOF structure. Note that the modification of oligonucleotides produced additional cost and may interfere the function of oligonucleotides, particularly when they are active drug molecules. To overcome this limitation, Wang et al. constructed a method based on intrinsic coordination for preparation of MOF-DNA complex from naïve oligonucleotides, which could be adsorbed on MOFs through multivalent binding between the phosphate backbone and zirconium centers (Fig. 5d) [290]. Moreover, biomineralizing MOF-DNA with a calcium phosphate (CaP) exoskeleton could endow the nanoconstruct with physiologically-stimuli-responsive DNA-release properties: at acidic endo/lysosomes, CaP dissolved under acidic pH conditions, generating phosphate ions which could efficiently display DNA from MOFs. Using immunostimulatory CpG as the model payload, MOF-CpG@CaP displayed high cellular uptake and organelle specificity, as well as spatiotemporal control of the immune response triggered by Toll-like receptors (TLR).
DNAzymes, a type of synthetic single-stranded DNA molecules that can catalyze some biological reactions similar to RNAzymes and proteins [291], have recievedgreat attention in the treatment of cancers owing to their capabilities of inhibiting multiple tumorigenic processes via the catalytic cleavage of oncogene targets when supplied with enough specific cofactors [292]. However, the low efficacy of cellular uptake and inadequate cofactor supply prevents the in-depth development of DNAzyme as an effective drug candidate to clinic. To address these shortcomings, Wang et al. [293]. designed a self-sufficient nanosystem for DNAzyme delivery based on the zeolitic imidazolate framework-8 (ZIF-8) nanoparticle, a zeolitic imidazolate framework, capable of disassembling at acidic endo/lysosomes accompanied by the continuous release of the DNAzyme payloads and the cofactor Zn2+, and finally promoting the DNAzyme-mediated gene silencing. This study showed a good example of designing smart gene delivery systems based on the specific intracellular biocatalytic reactions that have important therapeutic relevance, representing a new direction in the drug design. In another study [294], the same group designed a DNAzyme-based delivery system, called DNAzyme nanosponges, consisting of a DNAzyme–substrate scaffold fabricated by the rolling circle polymerizations and several ZnO nanoparticles. Synergistically enhanced therapeutic performance was achieved by the pH-responsive dissolution of ZnO into Zn2+, which further mediated nonviolent DNAzyme-catalyzed cleavage of DNA scaffolds and thereby realizing precise and effictive drug delivery.
Similar to other types of nanocarriers, endosomal escape should also be carefully considered in the rational design of TO-loaded MOFs. To realize the endo/lysosomal escape and enhance the TOs delivery to the intracellular target, Lin et al. designed a photo-controllable MOF nanoswitch fabricated from ZIF-8 nanoparticles with the photothermal agent indocyanine green (ICG) encapsulated within the MOF pores and the siRNAs immobilized on the surface through electrostatic adsorption [295]. Upon mild laser irradiation, large amount of siRNA was released intracellularly accompanied by the dissociation of MOF complex into protonatable 2-methylimidazalo and osmotic rupturing Zn2+ ions, which cooperatively led to endo/lysosomal rupture. The nanoswitch displayed a high degree of spatiotemporal control of gene transfer for cancer site-specific RNA interference. Interestingly, the dissociation of MOF complex caused the recovery of ICG fluorescence which facilitated the real-time monitoring of siRNA delivery.
In summary, the physicochemical characteristics of MOFs as well as their variability of coordination metal ions and organic ligands make them promising nanocarriers for TOs delivery. High drug load efficacy and stimuli-responsive drug release can be achieved through MOF structural optimization and rational design. Meanwhile, TOs delivery based on MOFs has attracted more and more interest in the combination of gene therapy with other therapeutic strategies, e.g. chemotherapy, photodynamic therapy, photothermal therapy, as well as immunotherapy owing to the well-controlled porous structure of MOF.
4.5. DNA/RNA nanoassembly
Due to their programmable and predictable capability of hybridization with their complimentary sequences, DNA/RNA molecules have attracted more and more interest in the construction of nanoassembly for the smart delivery of TOs. Strategies developed for fabricating DNA/RNA nanoassembly mainly include DNA origami [296], single-stranded tile (SST) DNA nanostructures [297], and single-stranded (ss) DNA/RNA origami [298]. Importantly, TOs themselves can not only exhibit the therapeutic effect but also serve as the part of building block in the fabrication of DNA/RNA nanoassembly, increasing their targeted efficacy. In brief, DNA/RNA nanoassembly as an ideal TOs delivery nannocarriers has the following characteristics. Firstly, the excellent biocompatibility of the DNA/RNA entities; Secondly, the vigorous self-assembly capability through Watson and Crick base-pairing interactions; Thirdly, designable and controllable precise structures with addressable modification sites; Fourthly, the facile functionalization methods for targeting ligands and TOs. Up to now, numerous DNA/RNA nanostructures have been applied in the delivery of TOs like siRNAs, mRNA, and immune modulators for gene therapy, target therapy, and immunotherapy.
4.5.1. siRNA-hybridized DNA nanoassembly
siRNAs as a class of potent therapeutics for various cancer treatment from aberrant gene expression, remain to be precluded from cell permeability and resistance to degradations [299]. Despite an appropriate manner to overcome the inevitable leakage of synthetic siRNA and off-target toxicity, the current intracellular generation (i.e. in situ production in target cells) of siRNA by a spontaneous collision between complementary strands confined the assembly efficiencies owing to the lack of controllability and the slow reaction rate [300]. Lee et al. designed a DNA tetrahedron nanoformulations for site-specific hybridization of siRNA [301]. Due to the addressability of the DNA tetrahedron nanostructures which have a nick site in the middle of each edge thus could be further used to tether siRNA strands (Fig. 6a). Using such a self-assembled DNA nanostructure, the sizes, the spatial orientations as well as densities of targeting ligands on the surface of NPs could be precisely controlled, providing an efficient tool for screening the best formulation for optimal cancer therapy. Ren et al. designed a cationic folic acid modified polyethyleneimine capped DNA nanoplatform that constructed by two pairs of DNA/RNA hybrids alternately hybridized to adjacent sites on a DNA scaffold [302]. miRNA in the cytoplasm of cancer cells initiated the generation process of siRNA which called DNA templated synthetic reaction, further lead to the hybridization of two anchored DNA strands and continuous siRNA generation without the involvement of exotic enzymes and catalyst, while the precise arrangement sequences guaranteed the high reaction efficiency. This mRNA triggered DNA nanoplatform showed highly efficient siRNA in situ generation and enhanced inhibition of miRNA-specific tumor cells, thus can be easily generalized for precise gene therapy.
Fig. 6.
TOs delivery based on the DNA/RNA nanoassembly. (a) Self-assembled DNA tetrahedral nanostructure for targeted siRNA delivery (arrow head represents 5 end of the nucleic acid strand). Reproduced according to Ref. [301].; (b) Stepwise self-assembly of D-PGM for three-receptor-mediated cell identification and targeted cancer therapy. Reproduced with permission [305], Copyright 2020, American Chemical Society; (c) Self-assembly of CpG-bearing DNA tetrahedrons and their multivalent immunostimulatory effect evaluations. Reproduced with permission [306], Copyright 2011, American Chemical Society. (d) Nucleic acid nanoparticles (NANPs) differing in size, shape, composition, connectivity, and sequence complementarity for immunological recognition investigations. Reproduced with permission [312], Copyright 2018, American Chemical Society.
4.5.2. Aptamer-hybridized DNA nanoassembly
Due to the high-specificity of aptamers in recognizing targeted cells, incorporation of aptamers to the DNA nanoassembly has become a common strategy to increase the targeting efficiency of these nanocarriers. In one earlier study, Wu et al. constructed a poto-crosslinked DNA nanogel (AptNAs), the framework of which consisted of sgc8-aptamers targeting leukemia cells, ASOs targeting multidrug resistance genes, and DNA building blocks [303]. Due to their programmable design in molecular level and excellent selectivity in targeted recognition and transportation, AptNAs displayed a highly-specific toxicity against cancer cells and a significantly down-regulated expression of drug efflux pump protein P-gp, which synergistically enhanced the anti-cancer effect. Later on, the same group reported another DNA nanogel for targeted gene regulation. Different with previous one, the DNA nanogel was crosslinked via a disulfide linking unit with two different Y-shaped building units with sticky sequence distributed at the terminal of each arm [304]. Aptamers targeting A549 lung cancer cells, ASOs targeting c-raf-1 mRNA (a cancer-associated biomarker) and DNAzymes targeting lung-cancer-proliferation-associated matrix metalloproteinase-9 could be hybridized to the DNA framework in a site-specific manner. The formed DNA nanogels could sense the intracellular GSH, resulting in the release of ASOs and DNAzymes to exert their roles in cancer cell inhibition.
Recently, a combination of different signaling circuits consisting of diverse aptamers has also shown great potential in tumor cell-specific therapeutic applications. Nevertheless, due to the ingenious molecular structure of combinational logic circuits and the highly complex 3D nanostructure, such combinatorial molecular circuits based on multiaptamer have not been applied to DNA nanostructures for drug delivery of cancer treatment, especially for high drug loading nanocarriers with tumor-specific targeting. Ouyang and coworkers developed a triple-receptor targeting precision-guided missile DNA nanovehicle (D-PGM) consisting of specific recognition (GC) and drug carrier (WH) compartment (Fig. 6b) [305]. The GC is a DNA logic nanocircuit composed of three self-assembled ssDNA partially complementary to various aptamers in a highly organized manner, while the WH is a rod-shaped 3D DNA origami assembled from four oligonucleotides as drug carrier. More accurate and effective drug delivery achieved under similar cells conditions when the GC binds to three aptamers and dissociates in an organized manner compared to traditional single or dual targeting nanovehicles. Thus, such a precise triple-receptors based DNA nanostructure provided enhanced diagnostic and therapeutic accuracy and represented a potential strategy for personalized treatment of cancer.
4.5.3. Immunostimulatory oligonucleotides-incorporated DNA nanoassembly
Fan's group designed a DNA tetrahedron structure as a nanocarrier for delivery of immunoregulatory oligonucleotides. Due to the advantage of the programmability of DNA tetrahedron structure, they can selectively connect different numbers of CpG motifs to one DNA tetrahedron structure (Fig. 6c), which significantly enhanced the immunostimulatory effect of CpG because of the multivalent interactions between CpG and the toll-like receptor 9 (TLR9) [306]. Recently, Christopher et al. proposed a replicable ssRNA origami (RNA-OG) technology, through which a long RNA molecule can be programmed to self-assemble into various custom-shaped nanostructures. For instance, polyinosinic/polycytidylic acid (PolyIC), a well-known dsRNA analog, have shown efficacy in cancer immunotherapy through Toll-like receptors 3 (TLR3) triggered innate immune and entered clinical trials [307]. Nevertheless, these analogs-induced high concentrations of type-I interferons (IFNs) lead to systemic toxicity cannot be ignored. Recently, Qi et al. reported RNA origami nanostructures produced from a single strand RNA with high accuracy and yield while do not require as many short synthetic DNA strands as traditional DNA origamis [308]. They demonstrated that achieving a continued antitumor immunity that depends on the presence and levels of CD8+ T and NK cells. Compared with PolyIC administration, the levels of IFNs produced systemically is much lower as well as the systemic toxicity, and the nature of immunosuppression in the peritoneal cavity can be reduced. The RNA origami could be developed as a powerful and safe nanomedicine for cancer immunotherapy.
Linear short hairpin RNAs (shRNAs) have exhibited enhanced stability and induced immune response in vivo compared to siRNA, which is generally used for multidrug delivery [309]. The multiple mechanisms of drug resistance like drug efflux and inhibition of apoptosis, severely limit the effective intracellular release of nanodrugs in cancer cells, resulting in insufficient therapeutic effect. At the same time, owing to the excellent abilities of multidrug co-loading and editable size and morphology of DNA nanostructures, Liu and coworkers fabricated a precisely self-assembled DNA origami containing chemotherapy drug loading nanostructure and two kinds of linear shRNAs outside [310]. The DNA nanoplatform realized the co-delivery of various drugs and eventually achieved a synergistic anticancer effect among diverse cargoes: shRNA silenced the multidrug-resistant associated genes which reduced the drug efflux thus led to the accumulation of chemodrugs and induced apoptosis of tumor cells, while chemodrugs played intrinsic cytotoxicity. By integrating TOs with DNA nanoplatform and collaborating with other drugs, it is possible to make the treatment effective.
Despite tremendous advances in the development of DNA/RNA nanoassembly, its immunotoxicity, one of the major obstacles to traditional TOs clinical transformation, has not been fully characterized. Guo and coworkers demonstrated that the immunogenicity of RNA nanoassembly is depending on the size, morphology, and specific sequence. Importantly, they found RNA polygons that do not extend at the vertices are immune inert. Thus, the immunogenicity of RNA nanoassembly can be adjusted to produce minimum immune response as a safe therapeutic nanovehicle, or a strong immune response as a vaccine adjuvant for cancer immunotherapy [311]. These conclusions were further confirmed by Hong and his coworkers [312]. In their study, they systematically studied the immune recognition capabilities of representative DNA/RNA NANPs with different parameters (Fig. 6d), and found that type III interferons (IFN-III) is additional biomarkers of nucleic acid nanoparticles (NANPs) after their internalization by phagocytic cells, compared to traditional DNA/RNA nanoassembly. Furthermore, the immunomodulatory effect of NANPs relies on multiple physicochemical properties, including size, structure (planar or three-dimensional construction), composition, and connectivity. Predictably, NANPs can be programmed as a prototype of specific molecular language for communication with immune system and regulation of immune response.
4.6. Extracellular vesicles
Extracellular vesicles (EVs) are cell-derived membranous particles that can be naturally released from a cell, but unlike a cell, EVs cannot replicate. EVs are present in various body fluids [313], and are crucial carriers that participate in intercellular communication during tissue homeostasis and repair under physiological conditions [314,315]. According to the size and origin, EVs are classified in to four main subtypes (Scheme 2), including endosome-derived exosomes, plasma-membrane-derived microvesicles, large oncosomes (tumor-derived large vesicles), and apoptotic bodies [316,317]. The contents of the cargos loaded in EVs can distinguish from those in the originating cells, which unveil a selective loading process [318]. Prolonged EVs in circulation can, therefore, act as robust, minimally invasive, specific biomarkers for diagnosing a variety of diseases, including cancers [319,320]. Owing to their low immunogenicity and capacity to cross biological barriers [321], EVs has been identified as a promising vehicle in transporting a variety of cargos, such as lipids, proteins, DNA/RNA species, for cancer treatments [[322], [323], [324], [325]]. Since endogenous RNA species have been reported to be definitively present in EVs, EVs showed superior biocompatibility and delivery efficiency for TOs compared to synthetic nanoparticles [326,327]. Generally, EVs that are utilized for the delivery of TOs in the application of cancer therapy can be categorized in to natural sources and engineered sources.
Scheme 2.
Schematic illustration of subtypes of extracellular vesicles.
4.6.1. Natural EVs
EVs can be secreted by almost all cell subtypes, and there are some natural secreted EVs that contain therapeutic RNA/DNA can be used for cancer therapy directly. For example, the mesenchymal stem cells (MSCs) derived miR-143, miR-133b, miR-9-3p rich exosomes can be used for the treatment of prostate cancer, glioma and bladder cancer via their targeting proteins [[328], [329], [330]]. And some nature killer (NK) cell derived EVs that contain miRNAs, such as miR-186 and miR-3607–3p can also be used for neuroblastoma and pancreatic cancer treatment [331,332]. Moreover, the natural derived EVs are also considered as ideal nanocarriers for synthesized TOs owing to their nucleic acid loading capacity and membrane permeability. Based on this, several micRNA mimics and siRNAs have been reported to be loaded into HEK293 and dendritic cells derived exosomes for cancer therapy [320,333,334]. And the most commonly used methods for TOs loading are transfection reagent, ultrasound, freeze-thaw cycle and electroporation [[335], [336], [337], [338]].
4.6.2. Engineered EVs
Compared to natural EVs, the delivery of TOs with engineered EVs are more potential because of their enhanced targeting and customized diversity functions [339]. Recipient cells can internalize EVs through a variety of pathways, including membrane fusion, receptor-mediated endocytosis, lipid-raft-mediated endocytosis or micropinocytosis. It has been reported that the natural targeting ability of EVs to recipient cells is determined by the specific biomarkers (e.g. tetraspanin, integrin) derived from the producer cells [340]. For example, EVs displaying Tspan8 (tetraspanin molecular family)/integrin α4 complex was able to selectively target EVs to cells in the pancreas [341]. Furthermore, unlike negative EVs that interact only with the dendritic cells, CD63-positive EVs mainly target to neuronal and glial cells [342]. While for EVs derived from the human embryonic kidney cells (HEK293T), the isolated EVs are absence of integrin markers [343,344], and thus have limited targeting effect towards the recipient cells [345]. To increase the specificity of HEK293T-EVs, Pi et al. decorated EVs with an arrow-shaped three-way junction RNA nanoparticles (pRNA-3WJ), containing a cholesterol for membrane anchoring, a fluorophore for imaging, and a ligand for tumor cell targeting [346]. The resulting ligand-displaying EVs were able to specifically deliver siRNA to cells, and showed effective inhibition of tumor growth in the three cancer models studied in this research.
As for drug delivery vehicles, the potential interaction between the natural bioactive payloads and therapeutics may interfere with the desired therapeutic effects. To address these hurdles, several strategies, such as genetic manipulation and functionalization after secretion, have been developed recently [[347], [348], [349], [350]]. Nevertheless, these additional modifications, in some ways, compromise the very crucial components in exosome-cell interactions and drug delivery [351]. On this account, Yerneni et al. proposed a scalable, practical approach for rapidly and efficiently manipulating engineer exosomes with oligonucleotide conjugates [352]. With a conjugated cholesterol moiety, the single-stranded DNA (ssDNA) can be tethered to an external lipid bilayer of exosomes, while the complementary DNA can easily attach to various functions such as reactive functional groups and small molecules. The confocal micrographs showed that the aptamer AS1411-tethered exosomes preferred to bind to nucleolin on MIAPaCa2 (human pancreatic cancer cells) cell surface rather than HEK293 (normal cell), and was thereby able to circumvent the inhibitory effects of heparin and methyl-β-cyclodextrin resulting in altering the specific targeting capability. Meanwhile, ssDNA tethered exosomes achieved spatially restricted apoptosis in PCI-13 cells in virtue of bioprinting through live/dead staining. The oligonucleotide tethers have shown their facile, robust, and general versatility in the augmentation of the extracellular vesicle that is independent of its cell origin, and was conducive to the clinic transformation of gene-drug delivery based on exosome.
4.7. Imaging-guided drug delivery systems (IGDD)
Imaging-guided drug delivery (IGDD) systems, referring to the combination of drug targeting and imaging, have emerged as the essential component of personalized cancer treatment. IGDD can be employed to visualize and quantify the biodistributions of drugs, and to non-invasively assess their therapeutic efficacy. In the field of TOs delivery, IGDD systems can significantly improve the targeting delivery efficiency of TOs and make the cancer treatment more precise. In the following section, we will introduce three typical IGDD systems based on the nanoparticles or nanoassembly that can not only behave as the drug vehicle, but also display imaging properties.
4.7.1. Iron oxide nanoparticles
Iron oxide nanoparticles (IONPs) are a class of inorganic nanoparticles with interesting properties like small volume, large surface area, magnetic and superparamagnetic [353]. IONPs can be formed by placed in a specific magnetic field, which allows heat to be absorbed by electromagnetic current waves in an alternating magnetic field. The unique properties of IONPs have been exploited for cancer diagnostics, as well as drug delivery [354]. As for delivery of TOs, the targeting and efficacy remains a significant obstacle to explore their clinical contex. Jiang et al. developed a magnetic siRNA delivery nanosystem composed of a lipid-like shell (termed lipidoid) and an ION core [355]. They found that the size of lipidoid-coated IONs affected the delivery efficacy of DNA, while there is no obvious difference in the delivery of siRNA. In addition, enhanced targeting delivery of TOs was achieved with the external magnetic field compared to commercial Lipofectamine 2000. The lipidoid IONs delivery platform proposed here provides an optimal strategy for magnetic guided targeted delivery, and the opportunity to combine the advantages of gene therapy with MRI and magnetic hyperthermia.
In recent years, membrane-derived vesicles have been used as natural analogs of liposomes for drug delivery. The capsule has a component of the source cell membrane, which makes it inherently biocompatible and slightly immunogenic in vivo. Several studies have proposed some biomimetic delivery platforms from various cell membranes, including stem cells [356], platelets [357,358], leukocytes [359], and cancer cells [360,361], to apply in personalized biomedicine for cancer treatment. Mu and coworkers employed the first mesenchymal stem cells membrane-coated IONPs for siRNA delivery. IONPs@PDA exhibited favorable photothermal capability and high siRNA loading efficiency, which maintained the magnetic resonance imaging (MRI) inherited and photothermal effects after membrane coated (Fig. 7a) [362]. The membrane-coated nanovehicles were endowed with excellent blood circulation and remarkably tumor targeting property. siRNA induced Plk1 gene knockdown significantly inhibited the bioactivity of endogenous Plk1 gene and cause obvious ablation of tumor, therefore achieved MRI-guided gene therapy and photothermal therapy.
Fig. 7.
TOs delivery based on imaging-guided drug delivery systems. (a) MSC membrane-coated Fe3O4@PDA–siRNA NPs for the tumor-targeting siRNA delivery. Reproduced with permission [362], Copyright 2018, American Chemical Society; (b) si-GPX4/Pt-codelivery IONPs platform for the synergistic treatment of glioblastoma. Reproduced with permission [366], Copyright 2020, American Chemical Society. (c) Targeting Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded miRNAs by CDs-mediated delivery of locked nucleic acid (LNA) for treating KSHV-induced cancers. Reproduced with permission [375], Copyright 2020, American Chemical Society.
Compare to the non-invasive trigger, the endogenous stimulatory, hypoxia, plays a key role in solid tumor development and resistance to therapy, providing compelling target sites for arresting tumor progression [363]. For instance, the monocarboxylate transporters (MCTs) upregulate in primary and metastatic tumors owing to the excessive intracellular concentration of lactic acid from active glycolysis. Besides, the elevated reactive oxygen species (ROS) in tumor cells make it more vulnerable to oxidative assault. These hallmarks present a novel direction for combinatorial cancer therapy. Liu and coworkers constructed amorphous iron oxide (AIO) nanoparticle-based siRNA delivery nanovehicles (siRNA@NPs) for co-targeting two cancer hallmarks [364]. siRNA loaded AIO NPs via electrostatic interactions are coated by biocompatible lipid and responsive to acidic pH after cellular uptake, further, the released iron ions induced endosomal membrane oxidation through the osmotic pressure in which leads to the effective endosomal escape of siRNA. Hence, the intracellular lactate/H+ accumulated via siRNA-mediated MCT4 silencing while facilitated more H2O2 production triggered by hypoxia, to amplify the oxidative damage procession: reacted with iron ions, H2O2 produced highly reactive and toxic •OH which completely aggravated oxidative stress of tumor cells and subsequent cancerous cell death. This innovative nanosystem achieved effective gene silencing and also exhibited MRI-mediated tracking of siRNA, which prompts the applications of TOs in theranostic nanomedicine.
In addition to the capability of directly transporting drug payloads to the desired targets, IONPs can also act as an effective and easily available carrier for the intracellular delivery of iron (Fe2+/Fe3+), which can take part in the iron-dependent programmed cell death through a process called Fenton reaction, an innovative strategy that has been identified recently in the treatment of malignant cancers [365]. Zhang et al. designed a targeting system based on the porous IONP that could deliver irons, chemotherapeutic cisplatin (Pt), and the siRNA targeting glutathione peroxidase 4 (si-GPX4) for the elimination of glioblastoma multiforme (GBM) [366]. The co-delivery system displayed a highly efficient synergistic effect (Fig. 7b): on the one hand, Pt induced apoptosis and simultaneously produced H2O2, together with the irons (Fe2+/Fe3+) released from IONP significantly increased the ROS generation (remarkably •OH) via the Fenton reaction within GBM cells; while on the other hand, si-GPX4 can effectively know down the negative regulator (GPX4) acted on the ferroptotic process. Finally, the co-delivery system showed an outstanding therapeutic effect against GBM model in vivo.
4.7.2. Carbon dots (CDs)
Carbon dots (CDs), also called carbon quantum dots or carbon nanodots, are organic compounds-derived carbon-based materials with the size below 10 nm. As emerging luminescent materials, CDs have shown great potential in the construction of multimodal imaging agents as well as in the delivery of oligonucleotides because of their remarkable photo-stability, high quantum yield, tunable emission, broad excitation spectra, cost-effective synthetic method, and low toxicity [[367], [368], [369]]. When used as the delivery vehicle for TOs, the surface of CDs must be modified with specific moieties for TOs’ binding. By modifying the CDs surface with cationic polymer, like polyethylenimine (PEI), poly-l-lysine, and poly(amidoamine) (PAMAM) [370], CDs exhibit water solubility and accomplish surface passivation, as well as stable binding efficacy of TOs which further released after internalization by the targeted cells [371,372]. For instance, Kim et al. synthesized fluorescent PEI-passivated CDs (CDs-PEI) for delivery of siRNA targeting the breast cancer cells [373]. The CDs-based RNAi displayed a high efficacy in gene knockdown both in vitro and in vivo and a clear intracellular tracking of the gene delivery process could be approached without additional fluorescent conjugates.
It has been reported that 15% of human cancer are associated with the virus-infections [374]. Targeting oncogenic viruses-encoded miRNAs represent a very promising therapeutic approach. Recently, Ju et al. used CDs as the carrier to load the locked nucleic acids (LNA)-based oligonucleotides, through electrostatic interaction, for specific knockdown of virus miRNAs (Fig. 7c) [375]. LNA oligonucleotides with an increased binding affinity exhibited the ability to re-induce the degradation of target miRNAs, leading to enhanced apoptosis and slowing down the process of proliferation. Moreover, since cancer viruses reside in viral cancer cells rather than in normal cells, targeting miRNAs of oncogenic viruses is an ideal approach for combating virus-induced cancers, and provided the specific delivery of TOs without additional modification of targeting ligands.
Apart from luminescence-based imaging, CDs-based gene carriers can also be designed to achieve multi-modal imaging. For example, He et al. fabricated novel Gd-doped CDs grafted with cationic polymers to achieve the combined gene delivery and magnetic resonance (MR)/fluorescence dual modal imaging [376]. The multifunctional CDs showed much higher transfection efficiency than PEI 25 kDa and could realize spatiotemporal monitoring of the gene delivery process via fluorescence imaging. In addition, the Gd-doped CDs displayed an improved MR imaging performance, where the longitudinal relaxation rates were at least 3- times higher than that of the clinically used Gd reagent (Gd-DTPA).
4.7.3. Aggregation-induced emission (AIE) nanodots
Discovered by Tang's group, aggregation-induced emission luminogens (AIEgens) have shown great potential in the real-time monitoring of biological dynamics [377]. AIEgens are non-fluorescence when they are dispersed in the solution in a single molecular state, while they will display strong fluorescent emission upon forming aggregates because of the restriction of intramolecular rotation. Nowadays, various AIEgens-based nanoscale fluorescent probes, namely AIE nanodots, have been widely developed as the novel IGDD system due to their high sensitivity and spatio-temporal resolution [378,379]. As for the TOs delivery, AIE nanodots can either be fabricated by incorporating AIE dyes to the preformed nanoparticles or through self-assembling AIE-moiety-containing amphiphilic conjugates [[380], [381], [382], [383]].
In one earlier study, Hu et al. reported the first AIE nanodots system for siRNA delivery [381]. The AIE nanodots were prepared by physically encapsulating AIE dyes to mixed polymeric micelles from Pluronics F127 and mPEG-DSPE, which were further decorated with a positively-charged polymer PAH for siRNA loading. The authors monitored the intracellular delivery of siRNA by means of AIE-nanodots and demonstrated that the AIE-nanodots@PHA/siRNA complex could significantly suppressed the expression of the pancreatic cancer-associated K-ras gene. To overcome the endo/lysosomal entrapment of gene vectors, Yuan et al. designed a novel AIE nanodot assembled from a ROS-responsive co-polymer [380], which consists of a photosensitizer TPECM with AIE characteristics and DNA-binding oligoethylenimine co-conjugated in the polymer backbone via the photocleavable aminoacrylate linker. When incubated with cells, the nanodots/DNA complex were mostly found in the endo/lysosomes. While upon visible-light irradiation, TPECM produced ROS, leading to the disruption of the endo/lysosomal membranes and cleavage of the polymer which thus caused the DNA release for transcription. In a recent study, the same group reported another type of AIE-nanodots which also demonstrated a ROS-responsive endo/lysosomal escape properties [382]. Different with previous study, here they covalently immobilized the anti-apoptotic Bcl-2 antisense oligonucleotides (OSA) to the AIE nanodots via the Click reaction. AIE-based SNAs enabled the real-time monitoring of the biodistributions of OSA in vivo. Upon light irradiation, the intracellular SNAs dissociated and the released OSA caused the degradation of the Bcl-2 mRNA. In vivo evaluations indicated that combined treatment of AIE-based SNAs with light significantly inhibited the tumor growth with no effect to the healthy tissues. Recently, Cheng et al. synthesized an amphiphilic peptide-AIEgens conjugate for simultaneously visualizing and delivering antisense oligonucleotides (ASO) to nucleus [383]. The peptide sequence contains three parts: a peptide segment targeting integrin-overexpressed cancer cells, a nuclear localization signaling peptide for nuclear-targeting delivery of ASO, and a positively-charged membrane-penetrating peptide for both ASO-loading and endosomal escape purposes. The multifunctional peptide-AIEgens assembly not only enabled the visualization of the ASO delivery process in real-time, but also exhibited superior Bcl-2 suppression in targeted cell lines and obvious anti-tumor efficacy in vivo.
4.8. Tumor microenvironment responsive TOs delivery
The microenvironment differences between tumor and normal tissues provide a new perspective for controllable release of oligonucleotides within tumor sites. In accordance with several characters of tumor microenvironment (TME), such as the weak acidic tumor matrix, reductive environments, elevated intracellular reactive oxygen species (ROS) and up-regulated expression of enzymes, a variety of smart drug delivery systems that can sense these TME characters have been developed in the past decades [[384], [385], [386]]. In this section, we will give a brief introduction on the recent advances of TME-responsive nanocarriers for TOs delivery.
To modulate the immunosuppressive TME and boost the immune response, Jia et al. prepared an acid-/reduction-dual responsive polyplexes (DRPs), which were composed of the block copolymer mPEG-PLA-PHis-ss-PEI, siRNA against the programmed death ligand-1 (siPD-L1) and the metabolic regulator resveratrol [387]. After being transported to the acidic lysosomes, polyhistine (PHis) was protonated and the structure of DRPs was disrupted. The exposure of the disulfide bonds to the reductive environment led to the simultaneous release of siPD-L1 and resveratrol. Results showed that the DRPs treatments not only upregulated mitochondrial oxidative phosphorylation but also increased the infiltration of CD4+/CD8+ T cells to tumor tissues by re-shaping the immunosuppressive TME. In combination with siPD-L1, significant anti-tumor effect was observed.
As for the polycations-based TOs delivery systems, when the complex entered cells, effective separation of TOs from the complex is beneficial for hybridization to their target sequences. To achieve this purpose, Liu et al. designed a ROS-responsive charge-reversal DNA delivery system. They synthesized a boronic acid-pendanted quaternary ammonium-containing polymer, namely B-PDEAEA, to complex with DNAs [388]. Upon exposure to ROS, the oxidization of boronic acid groups occurred, followed by a cascade reaction, resulting in the DNA release due to the charge-reversal of polymers from positive to negative. The transfection efficiency of ROS responsive B-PDEAEA polymer is more than 15,000 times higher than that of non-responsive PDEAEA counterparts. In another study, Xin et al. synthesized PEG modified copolymers which also contains quaternary ammonium and boronic acid, named PEG-B-PAEBEA, for co-delivery of miR-34a, a well-known noncoding RNA oncogene targets, and volasertib, an inhibitor of polo-like kinase 1 that is overexpressed in many cancer types [140]. Based on similar ROS-liable dissociation of polyplexes accompanied by the TOs release, PEG-B-PAEBEA significantly upregulated the expression of miR-34a and dysregulated related oncogenes, resulting in efficient tumor growth inhibition.
Matrix metalloproteinases (MMPs), usually high-expressed in the extracellular matrix (ECM) of tumor tissues [389,390], are also excellent targets for the design of TME-responsive drug delivery systems. MMP-2, a protease involved in the degradation of tumor ECM, is relatively overexpressed in almost all tumors. In one earlier study, Hatakeyama et al. developed a PEG-peptide-lipid conjugate for TOs delivery. They used the MMP-2 sensitive peptide GGGVPLSLYSGGGG as the linker between PEG and lipid DOPE to fabricate the envelope-type liposomes (PPD-MEND) [391]. PPD-MEND exhibited a long blood circulation property, while once accumulated in tumor tissues, the peptides were cleaved by MMP-2, resulting in the deshielding of PEG which facilitated the internalization of TOs by tumor cells. The transfection activity of TOs by PPD-MEND was approximately 3 times higher than that of PEGylated MEND without the peptide linker. In another study, Huang et al. fabricated a pH-/MMP-2-dual responsive lipid nanoparticles, the surface of which was modified with the positively-charged cell-penetrating peptide pre-masked by a negatively-charged peptide at neutral pH. These two peptides were linked by the third MMP-2-cleavable peptides. At acidic tumor site, the masking peptides became charge-neutral and separated from the nanoparticle surface by the cleavage of MMP-2. Subsequently, efficient cellular uptake, endosomal escape and gene transfection were successfully achieved [392]. Except MMP-2, other ECM enzymes, such as MMP-9 and hyaluronidase, responsive systems have also been designed to address the challenges of TOs delivery [393,394]. These studies suggest that enzyme-sensitive short peptides are delicate designs in fabrication of TOs delivery for cancer gene therapy.
5. Nanocarrier based anticancer oligonucleotide in clinic
Both the supplementary of deficient anti-tumor gene and the silencing of tumor driver gene are pursued as approaches for cancer therapy. While the basic requirement for anticancer oligonucleotide drugs is that the oligonucleotides must be delivered to the target position safely and efficiently. The ideal TOs nanocarriers for clinical use are expected to have the ability of immune evasion, target cell or tissue delivering and high efficiency of cell entry [395]. Despite years of development, the high cytotoxicity and immunogenicity as well as problems such as high cost, limited size and poor loading capability are still the main limitations for viral vectors in the application of cancer therapy. Among the non-viral nanocarries, such as lipids and lipid derived materials, polymers, porous nanomaterials, MOF and many other biomaterials that have been developed for TOs delivery in the past several years, the lipid-based nanoparticles (LNPs) were the most widely studied and have the greatest potential for clinical application [51,63]. In 2018, the first lipid based nucleic acid drug Patisiran has been approved by the US FDA [48]. In addition, there are several other lipids based nanocarriers have been tested in preclinical studies (Table 1, Table 2) [[396], [397], [398], [399]].
Table 1.
Cancer therapy-associated TOs in clinical trials.
| TOs types | Name | Targets | Target disease | Clinical Phase |
|---|---|---|---|---|
| ASO | AZD9150 [78] | STAT3 | Lymphoma | Phase Ⅱ |
| siRNA | ALN-VSP02 [79] | Kinesin spindle protein and VEGF | Cancer | Phase Ⅰ |
| TKM-080301 [80] | Polo-like kinase 1 | Hepatocellular Carcinoma | Phase Ⅰ/Ⅱ | |
| siG12D-LODER [81] | KRAS | Pancreatic cancer | Phase Ⅱ | |
| DOPC-encapsulated siRNA [82] | EphA2 | Cancer | Phase Ⅰ | |
| Atu027 [83] | protein kinae N3 | Pancreatic cancer | Phase Ⅰb/Ⅱa | |
| mRNA-4157 [84] | Cancer cells | Cancer | Phase Ⅰ |
Abbreviations: TOs, therapeutic oligonucleotides; ASO, antisense oligonucleotides; siRNA, small interference RNA; DOPC, 1,2-Dioleoyl-sn-glycero-3-phosphocholine; STAT3, signal transducer and activator of transcription 3; VEGF, vascular endothelial growth factor; KRAS, Kirsten rat sarcoma viral oncogene homolog; EphA2, ephrin type-A receptor 2.
Table 2.
Nanocarrier-based TO-formulations for cancer therapy in clinical trails.
| Delivery system | Name | TOs subtypes | Target | Disease | Clinical phase |
|---|---|---|---|---|---|
| LNP | siRNA-EphA2-DOPC [404] | siRNA | EphA2 | Advanced solid tumors | Ⅰ |
| TKM-080301 [80] | siRNA | PLK1 | Advanced hepatocellular carcinoma | Ⅰ/Ⅱ | |
| DCR-MYC [405] | siRNA | MYC | Hepatocellular carcinoma | Ib/2 | |
| ALN-VSP02 [19] | siRNA | VEGF | Solid tumors | Ⅰ | |
| lipo-MERIT [406] | mRNA | TAA | Melanoma | Ⅰ | |
| TNBC-MERIT [407] | mRNA | TAA | Triple negative breast cancer | Ⅰ | |
| mRNA-2416 [400] | mRNA | OX40L | Advanced solid tumor | Ⅰ/Ⅱ | |
| Pbi-shRNA-STMN1 [408] | shRNA | STMN1 | Advanced liver cancer, solid tumor | Ⅰ | |
| MTL-CEBPA [409] | saRNA | CEBPA | Liver cancer | Ⅰ | |
| EV | iExosomes [410] | siRNA | KRASG12D mutation | Pancreatic cancer | Ⅰ |
| AuNP | NU-0129 [411] | RNA | BCL2L12 | Glioblastoma multiforme | Ⅰ |
Abbreviations: LNP, lipid-based nanoparticles; EV, extracellular vesicle; AuNP, gold nanoparticles; EphA2, Ephrin type-A receptor 2; PLK1, serine/threonine-protein kinase PLK1; MYC, MYC Proto-Oncogene; VEGF, vascular endothelial growth factor; TAA, Tumor-associated antigen; OX40L, tumor necrosis factor ligand superfamily member 4; STMN1, human stathmin 1; CEBPA, CCAAT/enhancer binding protein alpha; KRASG12D, GTPase KRas G12D; BCL2L12, Bcl-2-like protein 12.
However, the preferred accumulation in liver of lipid based materials caused by the interaction with apolipoprotein E is the main defect that limits its further application [400,401]. Polymeric materials, such as polyamines, polypeptides, and block polymers are not as clinically advanced for nucleic acid delivery as lipid based materials because some additional challenges related to polydispersity, toxicity and biodegradation [51,402]. Other materials, including cell derived vehicles, gold nanoparticles and DNA/RNA nanotechnology were also used for TOs delivery, but the applications in TOs-based therapies still need to be further explored [401].
6. Conclusions and Future Perspectives
In the past decades, as researchers gradually realized that the nanoscale non-viral carriers can be exploited as fascinating delivery systems for oligonucleotides-based therapeutics such as short ssDNA/RNA, as well as siRNA, numerous research projects have been carried out to find the best material and formula to achieve higher delivery and therapeutic effects against cancers. Strategies to enhance TOs delivery and therapeutic effects from molecular modification (chemical, terminal, and ribose sugar modification, etc.) to conjugates (lipid, GalNAc, antibody and aptamer conjugates, etc.) have significantly promoted the clinical development of TOs. However, there is still a long way to the clinical transformation of synthetic TOs. For each type of nanocarrier, there are both advantages and disadvantages (Table 3) in targeted TOs delivery, which should be carefully considered before choosing optimal formulation to achieve the best efficacy. Up to now, only ten TOs are in regulatory approval list of FDA and most of the them are targeting to the liver, one of the central metabolic organs [403]. Versatile nanoscale carriers, as summarized in this review, can improve the stability of TOs, enhance their cell uptake efficiency, increase the cytosolic delivery and achieve sustained drug release via smart design of nanocarriers with programmable linkage. The utility of highly optimized combinations of TOs, targeting ligands and nanocarriers enables therapeutic molecules to reach previously inaccessible target tissues/cellular compartments. From the current challenges and emerging trends in delivery of TOs in nanoscale, two appealing directions may hold great potential in promoting the clinical transitions of TOs.
Table 3.
Advantages and disadvantages of different types of nanocarriers in the TOs delivery.
| Nanocarriers | Advantages | Disadvantages |
|---|---|---|
| Lipid-based nanoparticles |
|
|
| Polymeric nanoparticles |
|
|
| Gold nanoparticles |
|
|
| Porous nanomaterials |
|
|
| DNA/RNA nanoassembly |
|
|
| Extracellular Vesicles |
|
|
Smart Delivery of TOs by Stimuli-responsive Nanocarriers. The unique properties of tumor microenvironment (for instance, acid environment, redox state, hypoxia, and specific enzymatic activity) offer a variety of stimulus to realize more complex ‘smart’ delivery. Therefore, what the important aspect of the stimuli-responsive solutions is that the TOs functions in particular action site. To further improve the efficiency of treatment, enzymes/acid-activate ligands and endogenous or exogenous stimulus-responsive nanostructure can be used to release captured nanoparticles to the tumor site. Tumor targeting ligands appear on unshielded nanoparticles, and redirecting particles into the tumor through secondary targeting could be the solution to this challenge. Several stimuli-responsive nanomedicines are yet remaining at the clinical phase, such as Opaxia [412], ThermoDox [413], and AuroShell [414]. Until now, the only responsive nanomedicine, Visudyne, was approved by the FDA utilized in photodynamic therapy. Obviously, many approved drug formulations have been fully evaluated and adequately improved while an important feature of these clinical nanocarriers is their simple formulation. Therefore, as for the majority of stimuli-responsive nanocarriers for TOs delivery that were of sophisticated assemblies and compositional deliberations, the balance between these smart TOs delivery systems and scaling up to the manufacturing scale is the key to prompt the clinical application of responsive nanomedicines.
Combination of Therapies of TOs with Other Emerging Treatments. Combination therapies involve two or more treatments targeting cancer-associated signaling or pathways, and is generally better than the mono-therapy approach in combating cancer [415], considering the multiple mechanisms of cancer occurrence, development, and drug resistance. It has been proved that the combination therapy with in a synergistic manner can decrease the therapeutic dosage of each individual drug (or treatment) and reverse multi-drug resistance [416]. Recent advances in a variety of multifunctional nanocarriers is expected to extend the success of traditional combination therapy to a novel class of therapeutic categories, like chemo-gene [417], photo-gene [418], and immune-gene therapy [419,420]. These strategies have greatly enhanced the effectiveness of single gene therapy and are beneficial to accelerate the clinical transformation of anticancer TOs to a higher extent.
CRediT authorship contribution statement
Lei Wu: Writings and revisions of “Introduction”, “4.2 Polymeric nanoparticles”, “4.3 Gold nanoparticles”, “4.4 Porous nanomaterials”, “4.5 DNA/RNA nanoassembly”, “4.7 Imaging-guided drug delivery systems (IGDD)”, and “4.8 Tumor microenvironment responsive TOs delivery”; Organization of Table 3. Wenhui Zhou: Writings of “2 Classifications of the therapeutic oligonucleotides”, “4.6 Extracellular Vesicles”, and “5 Nanocarrier based anticancer oligonucleotide in clinic” sections; Table 1 and Table 2 organizations; Copyright applications for all cited Figures. Lihua Lin: Writings and revisions of “4.1 Lipid-based nanoparticles” section. Anhong Chen: Writing of “3 Obstacles that prevent the clinical transformation of TOs for cancer therapy” section. Jing Feng: Writings polishing. Xiangmeng Qu: Supervision, Writing – review & editing, of whole manuscript, focusing on the manuscript organizations for different sections. Hongbo Zhang: Supervision, Writing – review & editing, of whole manuscript, focusing on manuscript organizations and aspects related to the clinical translations. Jun Yue: Supervision, Writing – review & editing, of whole manuscript, focusing on key issues related to different nanoparticle formulations; Writings of “Abstract” and “6 Conclusions and Future Perspectives” sections.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the Natural Science Foundation of China (81871472), Natural Science Foundation of Guangdong Province (Project No. 2019A1515010696 and 2021A1515012333), Shenzhen Municipal Science, Technology and Innovation Commission (Project No. JCYJ20190807163003704), “100 Talents Program” of the start-up foundation from Sun Yat-sen University, Academy of Finland (328933) and Sigrid Jusélius Foundation.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xiangmeng Qu, Email: quxm5@mail.sysu.edu.cn.
Hongbo Zhang, Email: hongbo.zhang@abo.fi.
Jun Yue, Email: yuejun3@mail.sysu.edu.cn.
References
- 1.Smith C.I.E., Zain R. Therapeutic oligonucleotides: state of the art. In: Insel P.A., editor. vol. 59. 2019. pp. 605–630. (Annual Review of Pharmacology and Toxicology). [DOI] [PubMed] [Google Scholar]
- 2.Amreddy N., Babu A., Muralidharan R., Panneerselvam J., Srivastava A., Ahmed R., Mehta M., Munshi A., Ramesh R. Chapter five - recent advances in nanoparticle-based cancer drug and gene delivery. In: Tew K.D., Fisher P.B., editors. Advances in Cancer Research. Academic Press; 2018. pp. 115–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitravene Study Group A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with aids. Am. J. Ophthalmol. 2002;133:467–474. doi: 10.1016/s0002-9394(02)01327-2. [DOI] [PubMed] [Google Scholar]
- 4.Chi K.N., Yu E.Y., Jacobs C., Bazov J., Kollmannsberger C., Higano C.S., Mukherjee S.D., Gleave M.E., Stewart P.S., Hotte S.J. A phase I dose-escalation study of apatorsen (OGX-427), an antisense inhibitor targeting heat shock protein 27 (Hsp27), in patients with castration-resistant prostate cancer and other advanced cancers. Ann. Oncol. 2016;27:1116–1122. doi: 10.1093/annonc/mdw068. [DOI] [PubMed] [Google Scholar]
- 5.Schultheis B., Strumberg D., Santel A., Vank C., Gebhardt F., Keil O., Lange C., Giese K., Kaufmann J., Khan M., Drevs J. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J. Clin. Oncol. 2014;32:4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- 6.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H.R., Jung E., Cho S., Jeon Y.J., Kim Y. Targeting non-oncogene addiction for cancer therapy. Biomolecules. 2021;11 doi: 10.3390/biom11020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libutti S.K. Recording 25 years of progress in cancer gene therapy. Canc. Gene Ther. 2019;26:345–346. doi: 10.1038/s41417-019-0121-y. [DOI] [PubMed] [Google Scholar]
- 9.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 10.Meng H.M., Liu H., Kuai H., Peng R., Mo L., Zhang X.B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016;45:2583–2602. doi: 10.1039/c5cs00645g. [DOI] [PubMed] [Google Scholar]
- 11.Kalimuthu S., Gangadaran P., Rajendran R.L., Zhu L., Oh J.M., Lee H.W., Gopal A., Baek S.H., Jeong S.Y., Lee S.W., Lee J., Ahn B.C. A new approach for loading anticancer drugs into mesenchymal stem cell-derived exosome mimetics for cancer therapy. Front. Pharmacol. 2018;9:1116. doi: 10.3389/fphar.2018.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y., Guo S., Wu L., Chen P., Wang L., Liu Y., Ju H. Near-infrared boosted ROS responsive siRNA delivery and cancer therapy with sequentially peeled upconversion nano-onions. Biomaterials. 2019;225:119501. doi: 10.1016/j.biomaterials.2019.119501. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y.X., Wang Y., Blake S., Yu M., Mei L., Wang H., Shi J. RNA nanotechnology-mediated cancer immunotherapy. Theranostics. 2020;10:281–299. doi: 10.7150/thno.35568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muniyan S., Rachagani S., Parte S., Halder S., Seshacharyulu P., Kshirsagar P., Siddiqui J.A., Vengoji R., Rauth S., Islam R., Mallya K., Datta K., Xi L., Das A., Teply B.A., Kukreja R.C., Batra S.K. Sildenafil potentiates the therapeutic efficacy of docetaxel in advanced prostate cancer by stimulating NO-cGMP signaling. Clin. Canc. Res. : an official journal of the American Association for Cancer Research. 2020;26:5720–5734. doi: 10.1158/1078-0432.CCR-20-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh J.M., Venters C.C., Di C., Pinto A.M., Wan L., Younis I., Cai Z., Arai C., So B.R., Duan J., Dreyfuss G. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 2020;11:1. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasan N., Baselga J., Hyman D.M. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duschmale J., Hansen H.F., Duschmale M., Koller E., Albaek N., Moller M.R., Jensen K., Koch T., Wengel J., Bleicher K. In vitro and in vivo properties of therapeutic oligonucleotides containing non-chiral 3' and 5' thiophosphate linkages. Nucleic Acids Res. 2020;48:63–74. doi: 10.1093/nar/gkz1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W., De Hoyos C.L., Migawa M.T., Vickers T.A., Sun H., Low A., Bell T.A., 3rd, Rahdar M., Mukhopadhyay S., Hart C.E., Bell M., Riney S., Murray S.F., Greenlee S., Crooke R.M., Liang X.H., Seth P.P., Crooke S.T. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019;37:640–650. doi: 10.1038/s41587-019-0106-2. [DOI] [PubMed] [Google Scholar]
- 19.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 20.Welch J.J., Dean D.A., Nilsson B.L. Synthesis and application of peptide-siRNA nanoparticles from disulfide-constrained cyclic amphipathic peptides for the functional delivery of therapeutic oligonucleotides to the lung. Methods Mol. Biol. 2021;2208:49–67. doi: 10.1007/978-1-0716-0928-6_4. [DOI] [PubMed] [Google Scholar]
- 21.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boisguerin P., Deshayes S., Gait M.J., O'Donovan L., Godfrey C., Betts C.A., Wood M.J., Lebleu B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv. Drug Deliv. Rev. 2015;87:52–67. doi: 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ming X., Laing B. Bioconjugates for targeted delivery of therapeutic oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:81–89. doi: 10.1016/j.addr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Antimisiaris S., Mourtas S., Papadia K. Targeted si-RNA with liposomes and exosomes (extracellular vesicles): how to unlock the potential. Int. J. Pharm. 2017;525:293–312. doi: 10.1016/j.ijpharm.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 26.Li Q., Zhao D., Shao X., Lin S., Xie X., Liu M., Ma W., Shi S., Lin Y. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery. ACS Appl. Mater. Interfaces. 2017;9:36695–36701. doi: 10.1021/acsami.7b13328. [DOI] [PubMed] [Google Scholar]
- 27.Ding Y., Jiang Z., Saha K., Kim C.S., Kim S.T., Landis R.F., Rotello V.M. Gold nanoparticles for nucleic acid delivery. Mol. Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu B., Tang C., Yin C. Enhanced antitumor efficacy of folate modified amphiphilic nanoparticles through co-delivery of chemotherapeutic drugs and genes. Biomaterials. 2014;35:6369–6378. doi: 10.1016/j.biomaterials.2014.04.095. [DOI] [PubMed] [Google Scholar]
- 29.Tai W.Y., Gao X.H. Noncovalent tagging of siRNA with steroids for transmembrane delivery. Biomaterials. 2018;178:720–727. doi: 10.1016/j.biomaterials.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Bennett C.F. Therapeutic antisense oligonucleotides are coming of age. Annu. Rev. Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 31.Goyon A., Yehl P., Zhang K. Characterization of therapeutic oligonucleotides by liquid chromatography. J. Pharmaceut. Biomed. Anal. 2020;182 doi: 10.1016/j.jpba.2020.113105. [DOI] [PubMed] [Google Scholar]
- 32.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metabol. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2017;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aartsma-Rus A., van Ommen G.-J.B. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonetti C., Zupi G. Targeting different signaling pathways with antisense oligonucleotides combination for cancer therapy. Curr. Pharmaceut. Des. 2007;13:463–470. doi: 10.2174/138161207780162917. [DOI] [PubMed] [Google Scholar]
- 36.Quemener A.M., Bachelot L., Forestier A., Donnou-Fournet E., Gilot D., Galibert M.D. The powerful world of antisense oligonucleotides: from bench to bedside. Wiley Interdiscip Rev RNA. 2020;11 doi: 10.1002/wrna.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiah J.V., Grandis J.R., Johnson D.E. Targeting STAT3 with proteolysis targeting chimeras and next-generation antisense oligonucleotides. Mol. Canc. Therapeut. 2021;20:219–228. doi: 10.1158/1535-7163.MCT-20-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Cruz O.J., Qazi S., Hwang L., Ng K., Trieu V. Impact of targeting transforming growth factor beta-2 with antisense OT-101 on the cytokine and chemokine profile in patients with advanced pancreatic cancer. OncoTargets Ther. 2018;11:2779–2796. doi: 10.2147/OTT.S161905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beer T.M., Hotte S.J., Saad F., Alekseev B., Matveev V., Flechon A., Gravis G., Joly F., Chi K.N., Malik Z., Blumenstein B., Stewart P.S., Jacobs C.A., Fizazi K. Custirsen (OGX-011) combined with cabazitaxel and prednisone versus cabazitaxel and prednisone alone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel (AFFINITY): a randomised, open-label, international, phase 3 trial. Lancet Oncol. 2017;18:1532–1542. doi: 10.1016/S1470-2045(17)30605-8. [DOI] [PubMed] [Google Scholar]
- 40.Mello C.C., Conte D. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 41.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 43.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 44.Bayraktar R., Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Canc. Metastasis Rev. 2018;37:33–44. doi: 10.1007/s10555-017-9724-7. [DOI] [PubMed] [Google Scholar]
- 45.de Fougerolles A., Vornlocher H.-P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 47.Goodall G.J., Wickramasinghe V.O. RNA in cancer. Nat. Rev. Canc. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 48.Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.P., Vita G., Attarian S., Plante-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 49.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 50.Wang F., Zuroske T., Watts J.K. RNA therapeutics on the rise. Nat. Rev. Drug Discov. 2020;19:441–442. doi: 10.1038/d41573-020-00078-0. [DOI] [PubMed] [Google Scholar]
- 51.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berraondo P., Martini P.G.V., Avila M.A., Fontanellas A. Messenger RNA therapy for rare genetic metabolic diseases. Gut. 2019;68:1323–1330. doi: 10.1136/gutjnl-2019-318269. [DOI] [PubMed] [Google Scholar]
- 53.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 54.Xiong Q., Lee G.Y., Ding J., Li W., Shi J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018;11:5281–5309. doi: 10.1007/s12274-018-2146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu A.M., Choi Y.H., Tu M.J. RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol. Rev. 2020;72:862–898. doi: 10.1124/pr.120.019554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granot Y., Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics-An innate immune system standpoint. Semin. Immunol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 57.Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wojtczak B.A., Sikorski P.J., Fac-Dabrowska K., Nowicka A., Warminski M., Kubacka D., Nowak E., Nowotny M., Kowalska J., Jemielity J. 5'-Phosphorothiolate dinucleotide cap analogues: reagents for messenger RNA modification and potent small-molecular inhibitors of decapping enzymes. J. Am. Chem. Soc. 2018;140:5987–5999. doi: 10.1021/jacs.8b02597. [DOI] [PubMed] [Google Scholar]
- 60.Li B., Luo X., Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjugate Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 61.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11 doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhdanov V.P. Intracellular RNA delivery by lipid nanoparticles: diffusion, degradation, and release. Biosystems. 2019;185:104032. doi: 10.1016/j.biosystems.2019.104032. [DOI] [PubMed] [Google Scholar]
- 64.Xu Z.J., Yang L.F., Sun L.Q., Cao Y. Use of DNAzymes for cancer research and therapy. Chin. Sci. Bull. 2012;57:3404–3408. doi: 10.1007/s11434-012-5380-z. [DOI] [Google Scholar]
- 65.Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou W., Ding J., Liu J. Theranostic DNAzymes, Theranostics. 2017;7:1010–1025. doi: 10.7150/thno.17736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi L., Wu W., Duan Y., Xu L., Li S., Gao X., Liu B. Carrier-free hybrid DNA nanoparticles for light-induced self-delivery of functional nucleic acid enzymes. ACS Nano. 2021;15:1841–1849. doi: 10.1021/acsnano.0c10045. [DOI] [PubMed] [Google Scholar]
- 68.Zhu G., Chen X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018;134:65–78. doi: 10.1016/j.addr.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao L., Qi X., Yan X., Huang Y., Liang X., Zhang L., Wang S., Tan W. Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J. Am. Chem. Soc. 2019;141:17493–17497. doi: 10.1021/jacs.9b09292. [DOI] [PubMed] [Google Scholar]
- 70.Li X., Yang Y., Zhao H., Zhu T., Yang Z., Xu H., Fu Y., Lin F., Pan X., Li L., Cui C., Hong M., Yang L., Wang K.K., Tan W. Enhanced in vivo blood-brain barrier penetration by circular tau-transferrin receptor bifunctional aptamer for tauopathy therapy. J. Am. Chem. Soc. 2020;142:3862–3872. doi: 10.1021/jacs.9b11490. [DOI] [PubMed] [Google Scholar]
- 71.Nimjee S.M., White R.R., Becker R.C., Sullenger B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ireson C.R., Kelland L.R. Discovery and development of anticancer aptamers. Mol. Canc. Therapeut. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 73.Cooper C., Mackie D. Hepatitis B surface antigen-1018 ISS adjuvant-containing vaccine: a review of HEPLISAV safety and efficacy. Expert Rev. Vaccines. 2011;10:417–427. doi: 10.1586/erv.10.162. [DOI] [PubMed] [Google Scholar]
- 74.Zhu X., Nishimura F., Sasaki K., Fujita M., Dusak J.E., Eguchi J., Fellows-Mayle W., Storkus W.J., Walker P.R., Salazar A.M., Okada H. Toll like receptor-3 ligand poly-ICLC promotes the efficacy of peripheral vaccinations with tumor antigen-derived peptide epitopes in murine CNS tumor models. J. Transl. Med. 2007;5:10. doi: 10.1186/1479-5876-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kandimalla E.R., Bhagat L., Li Y., Yu D., Wang D., Cong Y.P., Song S.S., Tang J.X., Sullivan T., Agrawal S. Immunomodulatory oligonucleotides containing a cytosine-phosphate-2'-deoxy-7-deazaguanosine motif as potent toll-like receptor 9 agonists. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6925–6930. doi: 10.1073/pnas.0501729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krieg A.M. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 77.Shirota H., Tross D., Klinman D.M. CpG oligonucleotides as cancer vaccine adjuvants. Vaccines (Basel) 2015;3:390–407. doi: 10.3390/vaccines3020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reilley M.J., McCoon P., Cook C., Lyne P., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Curran M., Liu Q., Zhou T., Schmidt J., Jo M., Lee S.J., Yamashita M., Hughes S.G., Fayad L., Piha-Paul S., Nadella M.V.P., Xiao X., Hsu J., Revenko A., Monia B.P., MacLeod A.R., Hong D.S. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. J Immunother Cancer. 2018;6:119. doi: 10.1186/s40425-018-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tabernero J., Shapiro G.I., LoRusso P.M., Cervantes A., Schwartz G.K., Weiss G.J., Paz-Ares L., Cho D.C., Infante J.R., Alsina M., Gounder M.M., Falzone R., Harrop J., White A.C., Toudjarska I., Bumcrot D., Meyers R.E., Hinkle G., Svrzikapa N., Hutabarat R.M., Clausen V.A., Cehelsky J., Nochur S.V., Gamba-Vitalo C., Vaishnaw A.K., Sah D.W., Gollob J.A., Burris H.A., 3rd First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Canc. Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 80.El Dika I., Lim H.Y., Yong W.P., Lin C.C., Yoon J.H., Modiano M., Freilich B., Choi H.J., Chao T.Y., Kelley R.K., Brown J., Knox J., Ryoo B.Y., Yau T., Abou-Alfa G.K. An open-label, multicenter, phase I, dose escalation study with phase II expansion cohort to determine the safety, pharmacokinetics, and preliminary antitumor activity of intravenous TKM-080301 in subjects with advanced hepatocellular carcinoma. Oncol. 2019;24 doi: 10.1634/theoncologist.2018-0838. 747-e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Titze-de-Almeida R., David C., Titze-de-Almeida S.S. The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm. Res. (N. Y.) 2017;34:1339–1363. doi: 10.1007/s11095-017-2134-2. [DOI] [PubMed] [Google Scholar]
- 82.Landen C.N., Jr., Chavez-Reyes A., Bucana C., Schmandt R., Deavers M.T., Lopez-Berestein G., Sood A.K. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Canc. Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 83.Schultheis B., Strumberg D., Kuhlmann J., Wolf M., Link K., Seufferlein T., Kaufmann J., Feist M., Gebhardt F., Khan M., Stintzing S., Pelzer U. vol. 12. A Randomized Phase Ib/IIa Study; Cancers (Basel): 2020. (Safety, Efficacy and Pharcacokinetics of Targeted Therapy with the Liposomal RNA Interference Therapeutic Atu027 Combined with Gemcitabine in Patients with Pancreatic Adenocarcinoma). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burris H.A., Iii, Patel M.R., Cho D.C., Clarke J.M., Gutierrez M., Zaks T.Z., Frederick J., Hopson K., Mody K., Binanti-Berube A., Robert-Tissot C., Cowens K., Breton B., Zhong S., Zhou H., Cohen P.S., Keating K., Meehan R.S., Gainor J.F. A phase 1, open-label, multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in subjects with resected solid tumors and in combination with pembrolizumab in subjects with unresectable solid tumors (Keynote-603) Journal of Global Oncology. 2019;5 doi: 10.1200/JGO.2019.5.suppl.93. 93-93. [DOI] [Google Scholar]
- 85.Gordon S., Taylor P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 86.Haraldsson B., Nystrom J., Deen W.M. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol. Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 87.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eble J.A., Niland S. The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis. 2019;36:171–198. doi: 10.1007/s10585-019-09966-1. [DOI] [PubMed] [Google Scholar]
- 89.Jain R.K., Martin J.D., Stylianopoulos T. The role of mechanical forces in tumor growth and therapy. Annu. Rev. Biomed. Eng. 2014;16:321–346. doi: 10.1146/annurev-bioeng-071813-105259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stylianopoulos T., Munn L.L., Jain R.K. Reengineering the physical microenvironment of tumors to improve drug delivery and efficacy: from mathematical modeling to bench to bedside. Trends Cancer. 2018;4:292–319. doi: 10.1016/j.trecan.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stapleton S., Milosevic M., Tannock I.F., Allen C., Jaffray D.A. The intra-tumoral relationship between microcirculation, interstitial fluid pressure and liposome accumulation. J. Contr. Release. 2015;211:163–170. doi: 10.1016/j.jconrel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 92.Jain R.K. Delivery of molecular and cellular medicine to solid tumors. Adv. Drug Deliv. Rev. 1997;26:71–90. doi: 10.1016/s0169-409x(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 93.Baxter L.T., Jain R.K. Transport of fluid and macromolecules in tumors. I. Role of interstitial pressure and convection. Microvasc. Res. 1989;37:77–104. doi: 10.1016/0026-2862(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 94.Eetezadi S., Ekdawi S.N., Allen C. The challenges facing block copolymer micelles for cancer therapy: in vivo barriers and clinical translation. Adv. Drug Deliv. Rev. 2015;91:7–22. doi: 10.1016/j.addr.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 95.Beyer I., Li Z., Persson J., Liu Y., van Rensburg R., Yumul R., Zhang X.B., Hung M.C., Lieber A. Controlled extracellular matrix degradation in breast cancer tumors improves therapy by trastuzumab. Mol. Ther. 2011;19:479–489. doi: 10.1038/mt.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ganesh S., Gonzalez Edick M., Idamakanti N., Abramova M., Vanroey M., Robinson M., Yun C.O., Jooss K. Relaxin-expressing, fiber chimeric oncolytic adenovirus prolongs survival of tumor-bearing mice. Canc. Res. 2007;67:4399–4407. doi: 10.1158/0008-5472.CAN-06-4260. [DOI] [PubMed] [Google Scholar]
- 97.Bocsik A., Walter F.R., Gyebrovszki A., Fulop L., Blasig I., Dabrowski S., Otvos F., Toth A., Rakhely G., Veszelka S., Vastag M., Szabo-Revesz P., Deli M.A. Reversible opening of intercellular junctions of intestinal epithelial and brain endothelial cells with tight junction modulator peptides. J Pharm Sci. 2016;105:754–765. doi: 10.1016/j.xphs.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 98.Martin J.D., Seano G., Jain R.K. Normalizing function of tumor vessels: progress, opportunities, and challenges. Annu. Rev. Physiol. 2019;81:505–534. doi: 10.1146/annurev-physiol-020518-114700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi I.K., Strauss R., Richter M., Yun C.O., Lieber A. Strategies to increase drug penetration in solid tumors. Front Oncol. 2013;3:193. doi: 10.3389/fonc.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abyaneh H.S., Regenold M., McKee T.D., Allen C., Gauthier M.A. Towards extracellular matrix normalization for improved treatment of solid tumors. Theranostics. 2020;10:1960–1980. doi: 10.7150/thno.39995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiao Y., Wan J., Zhou L., Ma W., Yang Y., Luo W., Yu Z., Wang H. Stimuli-responsive nanotherapeutics for precision drug delivery and cancer therapy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11 doi: 10.1002/wnan.1527. [DOI] [PubMed] [Google Scholar]
- 102.Jones C.H., Chen C.K., Ravikrishnan A., Rane S., Pfeifer B.A. Overcoming nonviral gene delivery barriers: perspective and future. Mol. Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Juliano R.L., Ming X., Carver K., Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Therapeut. 2014;24:101–113. doi: 10.1089/nat.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linnane E., Davey P., Zhang P., Puri S., Edbrooke M., Chiarparin E., Revenko A.S., Macleod A.R., Norman J.C., Ross S.J. Differential uptake, kinetics and mechanisms of intracellular trafficking of next-generation antisense oligonucleotides across human cancer cell lines. Nucleic Acids Res. 2019;47:4375–4392. doi: 10.1093/nar/gkz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peetla C., Vijayaraghavalu S., Labhasetwar V. Biophysics of cell membrane lipids in cancer drug resistance: implications for drug transport and drug delivery with nanoparticles. Adv. Drug Deliv. Rev. 2013;65:1686–1698. doi: 10.1016/j.addr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoffmann E.K., Lambert I.H. Ion channels and transporters in the development of drug resistance in cancer cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130109. doi: 10.1098/rstb.2013.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gonen N., Assaraf Y.G. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist. Updates. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 108.Asayama S., Sudo M., Nagaoka S., Kawakami H. Carboxymethyl poly(L-histidine) as a new pH-sensitive polypeptide to enhance polyplex gene delivery. Mol. Pharm. 2008;5:898–901. doi: 10.1021/mp800094b. [DOI] [PubMed] [Google Scholar]
- 109.Zhang Y., Satterlee A., Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol. Ther. 2012;20:1298–1304. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Collas P., Alestrom P. Nuclear localization signal of SV40 T antigen directs import of plasmid DNA into sea urchin male pronuclei in vitro. Mol. Reprod. Dev. 1996;45:431–438. doi: 10.1002/(SICI)1098-2795(199612)45:4<431::AID-MRD4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 111.Der Aa M.A.E.M.V., Mastrobattista E., Oosting R.S., Hennink W.E., Koning G.A., Crommelin D.J.A. The nuclear pore complex: the gateway to successful nonviral gene delivery. Pharmaceut. Res. 2006;23:447–459. doi: 10.1007/s11095-005-9445-4. [DOI] [PubMed] [Google Scholar]
- 112.Wente S.R. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 113.Arvanitis C.D., Ferraro G.B., Jain R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Canc. 2020;20:26–41. doi: 10.1038/s41568-019-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rapoport S.I. Osmotic opening of the blood-brain barrier: principles, mechanism, and therapeutic applications. Cell. Mol. Neurobiol. 2000;20:217–230. doi: 10.1023/a:1007049806660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen C.C., Sheeran P.S., Wu S.Y., Olumolade O.O., Dayton P.A., Konofagou E.E. Targeted drug delivery with focused ultrasound-induced blood-brain barrier opening using acoustically-activated nanodroplets. J. Contr. Release : official journal of the Controlled Release Society. 2013;172:795–804. doi: 10.1016/j.jconrel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carpentier A., Canney M., Vignot A., Reina V., Beccaria K., Horodyckid C., Karachi C., Leclercq D., Lafon C., Chapelon J.Y., Capelle L., Cornu P., Sanson M., Hoang-Xuan K., Delattre J.Y., Idbaih A. Clinical trial of blood-brain barrier disruption by pulsed ultrasound. Sci. Transl. Med. 2016;8:343re2. doi: 10.1126/scitranslmed.aaf6086. [DOI] [PubMed] [Google Scholar]
- 117.Davis B., Tang J., Zhang L., Mu D., Jiang X., Biran V., Vexler Z., Ferriero D.M. Role of vasodilator stimulated phosphoprotein in VEGF induced blood-brain barrier permeability in endothelial cell monolayers. Int. J. Dev. Neurosci. 2010;28:423–428. doi: 10.1016/j.ijdevneu.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blanco E., Shen H., Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Behzadi S., Serpooshan V., Tao W., Hamaly M.A., Alkawareek M.Y., Dreaden E.C., Brown D., Alkilany A.M., Farokhzad O.C., Mahmoudi M. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 2017;46:4218–4244. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Foroozandeh P., Aziz A.A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale research letters. 2018;13:339. doi: 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ovais M., Mukherjee S., Pramanik A., Das D., Mukherjee A., Raza A., Chen C. Designing stimuli-responsive upconversion nanoparticles that exploit the tumor microenvironment. Adv. Mater. 2020;32 doi: 10.1002/adma.202000055. [DOI] [PubMed] [Google Scholar]
- 122.Debacker A.J., Voutila J., Catley M., Blakey D., Habib N. Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol. Ther. 2020;28:1759–1771. doi: 10.1016/j.ymthe.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang T., Shigdar S., Shamaileh H.A., Gantier M.P., Yin W., Xiang D., Wang L., Zhou S.F., Hou Y., Wang P., Zhang W., Pu C., Duan W. Challenges and opportunities for siRNA-based cancer treatment. Canc. Lett. 2017;387:77–83. doi: 10.1016/j.canlet.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 125.Hur S. Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Loh X.J., Lee T.C., Dou Q., Deen G.R. Utilising inorganic nanocarriers for gene delivery. Biomaterials science. 2016;4:70–86. doi: 10.1039/c5bm00277j. [DOI] [PubMed] [Google Scholar]
- 127.Pakunlu R.I., Wang Y., Saad M., Khandare J.J., Starovoytov V., Minko T. In vitro and in vivo intracellular liposomal delivery of antisense oligonucleotides and anticancer drug. J. Contr. Release : official journal of the Controlled Release Society. 2006;114:153–162. doi: 10.1016/j.jconrel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 128.Love J.C., Estroff L.A., Kriebel J.K., Nuzzo R.G., Whitesides G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 129.Rosi N.L., Giljohann D.A., Thaxton C.S., Lytton-Jean A.K., Han M.S., Mirkin C.A. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 130.Seferos D.S., Prigodich A.E., Giljohann D.A., Patel P.C., Mirkin C.A. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maeda H., Wu J., Sawa T., Matsumura Y., Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J. Contr. Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 132.Park K. The beginning of the end of the nanomedicine hype. J. Contr. Release : official journal of the Controlled Release Society. 2019;305:221–222. doi: 10.1016/j.jconrel.2019.05.044. [DOI] [PubMed] [Google Scholar]
- 133.Wilhelm S., Tavares A.J., Dai Q., Ohta S., Audet J., Dvorak H.F., Chan W.C.W. Analysis of nanoparticle delivery to tumours. Nature Reviews Materials. 2016;1 doi: 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- 134.Perry J.L., Reuter K.G., Luft J.C., Pecot C.V., Zamboni W., DeSimone J.M. Mediating passive tumor accumulation through particle size, tumor type, and location. Nano Lett. 2017;17:2879–2886. doi: 10.1021/acs.nanolett.7b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Toy R., Peiris P.M., Ghaghada K.B., Karathanasis E. Shaping cancer nanomedicine: the effect of particle shape on the in vivo journey of nanoparticles. Nanomedicine. 2014;9:121–134. doi: 10.2217/nnm.13.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y.R., Lin R., Li H.J., He W.L., Du J.Z., Wang J. Strategies to improve tumor penetration of nanomedicines through nanoparticle design. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11 doi: 10.1002/wnan.1519. [DOI] [PubMed] [Google Scholar]
- 137.Srinivasarao M., Low P.S. Ligand-targeted drug delivery. Chem. Rev. 2017;117:12133–12164. doi: 10.1021/acs.chemrev.7b00013. [DOI] [PubMed] [Google Scholar]
- 138.Alimoradi H., Matikonda S.S., Gamble A.B., Giles G.I., Greish K. Hypoxia responsive drug delivery systems in tumor therapy. Curr. Pharmaceut. Des. 2016;22:2808–2820. doi: 10.2174/1381612822666160217130049. [DOI] [PubMed] [Google Scholar]
- 139.Juang V., Chang C.H., Wang C.S., Wang H.E., Lo Y.L. pH-responsive PEG-shedding and targeting peptide-modified nanoparticles for dual-delivery of irinotecan and microRNA to enhance tumor-specific therapy. Small. 2019;15 doi: 10.1002/smll.201903296. [DOI] [PubMed] [Google Scholar]
- 140.Xin X., Lin F., Wang Q., Yin L., Mahato R.I. ROS-responsive polymeric micelles for triggered simultaneous delivery of PLK1 inhibitor/miR-34a and effective synergistic therapy in pancreatic cancer. ACS Appl. Mater. Interfaces. 2019;11:14647–14659. doi: 10.1021/acsami.9b02756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qin L., Yan P., Xie C., Huang J., Ren Z., Li X., Best S., Cai X., Han G. Gold nanorod-assembled ZnGa2O4:Cr nanofibers for LED-amplified gene silencing in cancer cells. Nanoscale. 2018;10:13432–13442. doi: 10.1039/c8nr03802c. [DOI] [PubMed] [Google Scholar]
- 142.Huschka R., Zuloaga J., Knight M.W., Brown L.V., Nordlander P., Halas N.J. Light-induced release of DNA from gold nanoparticles: nanoshells and nanorods. J. Am. Chem. Soc. 2011;133:12247–12255. doi: 10.1021/ja204578e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Global Burden of Disease Cancer C., Fitzmaurice C., Abate D., Abbasi N., Abbastabar H., Abd-Allah F., Abdel-Rahman O., Abdelalim A., Abdoli A., Abdollahpour I., Abdulle A.S.M., Abebe N.D., Abraha H.N., Abu-Raddad L.J., Abualhasan A., Adedeji I.A., Advani S.M., Afarideh M., Afshari M., Aghaali M., Agius D., Agrawal S., Ahmadi A., Ahmadian E., Ahmadpour E., Ahmed M.B., Akbari M.E., Akinyemiju T., Al-Aly Z., AlAbdulKader A.M., Alahdab F., Alam T., Alamene G.M., Alemnew B.T.T., Alene K.A., Alinia C., Alipour V., Aljunid S.M., Bakeshei F.A., Almadi M.A.H., Almasi-Hashiani A., Alsharif U., Alsowaidi S., Alvis-Guzman N., Amini E., Amini S., Amoako Y.A., Anbari Z., Anber N.H., Andrei C.L., Anjomshoa M., Ansari F., Ansariadi A., Appiah S.C.Y., Arab-Zozani M., Arabloo J., Arefi Z., Aremu O., Areri H.A., Artaman A., Asayesh H., Asfaw E.T., Ashagre A.F., Assadi R., Ataeinia B., Atalay H.T., Ataro Z., Atique S., Ausloos M., Avila-Burgos L., Avokpaho E., Awasthi A., Awoke N., Ayala Quintanilla B.P., Ayanore M.A., Ayele H.T., Babaee E., Bacha U., Badawi A., Bagherzadeh M., Bagli E., Balakrishnan S., Balouchi A., Barnighausen T.W., Battista R.J., Behzadifar M., Behzadifar M., Bekele B.B., Belay Y.B., Belayneh Y.M., Berfield K.K.S., Berhane A., Bernabe E., Beuran M., Bhakta N., Bhattacharyya K., Biadgo B., Bijani A., Bin Sayeed M.S., Birungi C., Bisignano C., Bitew H., Bjorge T., Bleyer A., Bogale K.A., Bojia H.A., Borzi A.M., Bosetti C., Bou-Orm I.R., Brenner H., Brewer J.D., Briko A.N., Briko N.I., Bustamante-Teixeira M.T., Butt Z.A., Carreras G., Carrero J.J., Carvalho F., Castro C., Castro F., Catala-Lopez F., Cerin E., Chaiah Y., Chanie W.F., Chattu V.K., Chaturvedi P., Chauhan N.S., Chehrazi M., Chiang P.P., Chichiabellu T.Y., Chido-Amajuoyi O.G., Chimed-Ochir O., Choi J.J., Christopher D.J., Chu D.T., Constantin M.M., Costa V.M., Crocetti E., Crowe C.S., Curado M.P., Dahlawi S.M.A., Damiani G., Darwish A.H., Daryani A., das Neves J., Demeke F.M., Demis A.B., Demissie B.W., Demoz G.T., Denova-Gutierrez E., Derakhshani A., Deribe K.S., Desai R., Desalegn B.B., Desta M., Dey S., Dharmaratne S.D., Dhimal M., Diaz D., Dinberu M.T.T., Djalalinia S., Doku D.T., Drake T.M., Dubey M., Dubljanin E., Duken E.E., Ebrahimi H., Effiong A., Eftekhari A., El Sayed I., Zaki M.E.S., El-Jaafary S.I., El-Khatib Z., Elemineh D.A., Elkout H., Ellenbogen R.G., Elsharkawy A., Emamian M.H., Endalew D.A., Endries A.Y., Eshrati B., Fadhil I., Fallah Omrani V., Faramarzi M., Farhangi M.A., Farioli A., Farzadfar F., Fentahun N., Fernandes E., Feyissa G.T., Filip I., Fischer F., Fisher J.L., Force L.M., Foroutan M., Freitas M., Fukumoto T., Futran N.D., Gallus S., Gankpe F.G., Gayesa R.T., Gebrehiwot T.T., Gebremeskel G.G., Gedefaw G.A., Gelaw B.K., Geta B., Getachew S., Gezae K.E., Ghafourifard M., Ghajar A., Ghashghaee A., Gholamian A., Gill P.S., Ginindza T.T.G., Girmay A., Gizaw M., Gomez R.S., Gopalani S.V., Gorini G., Goulart B.N.G., Grada A., Ribeiro Guerra M., Guimaraes A.L.S., Gupta P.C., Gupta R., Hadkhale K., Haj-Mirzaian A., Haj-Mirzaian A., Hamadeh R.R., Hamidi S., Hanfore L.K., Haro J.M., Hasankhani M., Hasanzadeh A., Hassen H.Y., Hay R.J., Hay S.I., Henok A., Henry N.J., Herteliu C., Hidru H.D., Hoang C.L., Hole M.K., Hoogar P., Horita N., Hosgood H.D., Hosseini M., Hosseinzadeh M., Hostiuc M., Hostiuc S., Househ M., Hussen M.M., Ileanu B., Ilic M.D., Innos K., Irvani S.S.N., Iseh K.R., Islam S.M.S., Islami F., Jafari Balalami N., Jafarinia M., Jahangiry L., Jahani M.A., Jahanmehr N., Jakovljevic M., James S.L., Javanbakht M., Jayaraman S., Jee S.H., Jenabi E., Jha R.P., Jonas J.B., Jonnagaddala J., Joo T., Jungari S.B., Jurisson M., Kabir A., Kamangar F., Karch A., Karimi N., Karimian A., Kasaeian A., Kasahun G.G., Kassa B., Kassa T.D., Kassaw M.W., Kaul A., Keiyoro P.N., Kelbore A.G., Kerbo A.A., Khader Y.S., Khalilarjmandi M., Khan E.A., Khan G., Khang Y.H., Khatab K., Khater A., Khayamzadeh M., Khazaee-Pool M., Khazaei S., Khoja A.T., Khosravi M.H., Khubchandani J., Kianipour N., Kim D., Kim Y.J., Kisa A., Kisa S., Kissimova-Skarbek K., Komaki H., Koyanagi A., Krohn K.J., Bicer B.K., Kugbey N., Kumar V., Kuupiel D., La Vecchia C., Lad D.P., Lake E.A., Lakew A.M., Lal D.K., Lami F.H., Lan Q., Lasrado S., Lauriola P., Lazarus J.V., Leigh J., Leshargie C.T., Liao Y., Limenih M.A., Listl S., Lopez A.D., Lopukhov P.D., Lunevicius R., Madadin M., Magdeldin S., El Razek H.M.A., Majeed A., Maleki A., Malekzadeh R., Manafi A., Manafi N., Manamo W.A., Mansourian M., Mansournia M.A., Mantovani L.G., Maroufizadeh S., Martini S.M.S., Mashamba-Thompson T.P., Massenburg B.B., Maswabi M.T., Mathur M.R., McAlinden C., McKee M., Meheretu H.A.A., Mehrotra R., Mehta V., Meier T., Melaku Y.A., Meles G.G., Meles H.G., Melese A., Melku M., Memiah P.T.N., Mendoza W., Menezes R.G., Merat S., Meretoja T.J., Mestrovic T., Miazgowski B., Miazgowski T., Mihretie K.M.M., Miller T.R., Mills E.J., Mir S.M., Mirzaei H., Mirzaei H.R., Mishra R., Moazen B., Mohammad D.K., Mohammad K.A., Mohammad Y., Darwesh A.M., Mohammadbeigi A., Mohammadi H., Mohammadi M., Mohammadian M., Mohammadian-Hafshejani A., Mohammadoo-Khorasani M., Mohammadpourhodki R., Mohammed A.S., Mohammed J.A., Mohammed S., Mohebi F., Mokdad A.H., Monasta L., Moodley Y., Moosazadeh M., Moossavi M., Moradi G., Moradi-Joo M., Moradi-Lakeh M., Moradpour F., Morawska L., Morgado-da-Costa J., Morisaki N., Morrison S.D., Mosapour A., Mousavi S.M., Muche A.A., Muhammed O.S.S., Musa J., Nabhan A.F., Naderi M., Nagarajan A.J., Nagel G., Nahvijou A., Naik G., Najafi F., Naldi L., Nam H.S., Nasiri N., Nazari J., Negoi I., Neupane S., Newcomb P.A., Nggada H.A., Ngunjiri J.W., Nguyen C.T., Nikniaz L., Ningrum D.N.A., Nirayo Y.L., Nixon M.R., Nnaji C.A., Nojomi M., Nosratnejad S., Shiadeh M.N., Obsa M.S., Ofori-Asenso R., Ogbo F.A., Oh I.H., Olagunju A.T., Olagunju T.O., Oluwasanu M.M., Omonisi A.E., Onwujekwe O.E., Oommen A.M., Oren E., Ortega-Altamirano D.D.V., Ota E., Otstavnov S.S., Owolabi M.O., P A.M., Padubidri J.R., Pakhale S., Pakpour A.H., Pana A., Park E.K., Parsian H., Pashaei T., Patel S., Patil S.T., Pennini A., Pereira D.M., Piccinelli C., Pillay J.D., Pirestani M., Pishgar F., Postma M.J., Pourjafar H., Pourmalek F., Pourshams A., Prakash S., Prasad N., Qorbani M., Rabiee M., Rabiee N., Radfar A., Rafiei A., Rahim F., Rahimi M., Rahman M.A., Rajati F., Rana S.M., Raoofi S., Rath G.K., Rawaf D.L., Rawaf S., Reiner R.C., Renzaho A.M.N., Rezaei N., Rezapour A., Ribeiro A.I., Ribeiro D., Ronfani L., Roro E.M., Roshandel G., Rostami A., Saad R.S., Sabbagh P., Sabour S., Saddik B., Safiri S., Sahebkar A., Salahshoor M.R., Salehi F., Salem H., Salem M.R., Salimzadeh H., Salomon J.A., Samy A.M., Sanabria J., Santric Milicevic M.M., Sartorius B., Sarveazad A., Sathian B., Satpathy M., Savic M., Sawhney M., Sayyah M., Schneider I.J.C., Schottker B., Sekerija M., Sepanlou S.G., Sepehrimanesh M., Seyedmousavi S., Shaahmadi F., Shabaninejad H., Shahbaz M., Shaikh M.A., Shamshirian A., Shamsizadeh M., Sharafi H., Sharafi Z., Sharif M., Sharifi A., Sharifi H., Sharma R., Sheikh A., Shirkoohi R., Shukla S.R., Si S., Siabani S., Silva D.A.S., Silveira D.G.A., Singh A., Singh J.A., Sisay S., Sitas F., Sobngwi E., Soofi M., Soriano J.B., Stathopoulou V., Sufiyan M.B., Tabares-Seisdedos R., Tabuchi T., Takahashi K., Tamtaji O.R., Tarawneh M.R., Tassew S.G., Taymoori P., Tehrani-Banihashemi A., Temsah M.H., Temsah O., Tesfay B.E., Tesfay F.H., Teshale M.Y., Tessema G.A., Thapa S., Tlaye K.G., Topor-Madry R., Tovani-Palone M.R., Traini E., Tran B.X., Tran K.B., Tsadik A.G., Ullah I., Uthman O.A., Vacante M., Vaezi M., Varona Perez P., Veisani Y., Vidale S., Violante F.S., Vlassov V., Vollset S.E., Vos T., Vosoughi K., Vu G.T., Vujcic I.S., Wabinga H., Wachamo T.M., Wagnew F.S., Waheed Y., Weldegebreal F., Weldesamuel G.T., Wijeratne T., Wondafrash D.Z., Wonde T.E., Wondmieneh A.B., Workie H.M., Yadav R., Yadegar A., Yadollahpour A., Yaseri M., Yazdi-Feyzabadi V., Yeshaneh A., Yimam M.A., Yimer E.M., Yisma E., Yonemoto N., Younis M.Z., Yousefi B., Yousefifard M., Yu C., Zabeh E., Zadnik V., Moghadam T.Z., Zaidi Z., Zamani M., Zandian H., Zangeneh A., Zaki L., Zendehdel K., Zenebe Z.M., Zewale T.A., Ziapour A., Zodpey S., Murray C.J.L. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749–1768. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He H., Du L., Guo H., An Y., Lu L., Chen Y., Wang Y., Zhong H., Shen J., Wu J., Shuai X. Redox responsive metal organic framework nanoparticles induces ferroptosis for cancer therapy. Small. 2020 doi: 10.1002/smll.202001251. [DOI] [PubMed] [Google Scholar]
- 145.Wang Y., Miao L., Satterlee A., Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv. Drug Deliv. Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wasungu L., Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Contr. Release : official journal of the Controlled Release Society. 2006;116:255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 147.Liu X., Huang G. Formation strategies, mechanism of intracellular delivery and potential clinical applications of pH-sensitive liposomes. Asian J. Pharm. Sci. 2013;8:319–328. doi: 10.1016/j.ajps.2013.11.002. [DOI] [Google Scholar]
- 148.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alexander M.Y., Akhurst R.J. Liposome-medicated gene transfer and expression via the skin. Hum. Mol. Genet. 1995;4:2279–2285. doi: 10.1093/hmg/4.12.2279. [DOI] [PubMed] [Google Scholar]
- 150.Remy J.S., Sirlin C., Vierling P., Behr J.P. Gene transfer with a series of lipophilic DNA-binding molecules. Bioconjugate Chem. 1994;5:647–654. doi: 10.1021/bc00030a021. [DOI] [PubMed] [Google Scholar]
- 151.Gao X., Huang L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 152.de Lima M., Neves S., Filipe A., Duzgunes N., Simoes S. Cationic liposomes for gene delivery: from biophysics to biological applications. Curr. Med. Chem. 2003;10:1221–1231. doi: 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- 153.Bhattacharya S., Dileep P.V. Cationic oxyethylene lipids. Synthesis, aggregation, and transfection properties. Bioconjugate Chem. 2004;15:508–519. doi: 10.1021/bc0340215. [DOI] [PubMed] [Google Scholar]
- 154.Zhi D., Bai Y., Yang J., Cui S., Zhao Y., Chen H., Zhang S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 2018;253:117–140. doi: 10.1016/j.cis.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 155.Sun X., Chen Y., Zhao H., Qiao G., Liu M., Zhang C., Cui D., Ma L. Dual-modified cationic liposomes loaded with paclitaxel and survivin siRNA for targeted imaging and therapy of cancer stem cells in brain glioma. Drug Deliv. 2018;25:1718–1727. doi: 10.1080/10717544.2018.1494225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Diao L., Tao J., Wang Y., Hu Y., He W. Co-delivery of dihydroartemisinin and HMGB1 siRNA by TAT-modified cationic liposomes through the TLR4 signaling pathway for treatment of lupus nephritis. Int. J. Nanomed. 2019;14:8627–8645. doi: 10.2147/IJN.S220754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zelphati O., Szoka F.C., Jr. Intracellular distribution and mechanism of delivery of oligonucleotides mediated by cationic lipids. Pharm. Res. (N. Y.) 1996;13:1367–1372. doi: 10.1023/a:1016026101195. [DOI] [PubMed] [Google Scholar]
- 158.Xia Y., Sun J., Liang D. Aggregation, fusion, and leakage of liposomes induced by peptides. Langmuir. 2014;30:7334–7342. doi: 10.1021/la501618f. [DOI] [PubMed] [Google Scholar]
- 159.Hatakeyama H., Akita H., Harashima H. The polyethyleneglycol dilemma: advantage and disadvantage of PEGylation of liposomes for systemic genes and nucleic acids delivery to tumors. Biol. Pharm. Bull. 2013;36:892–899. doi: 10.1248/bpb.b13-00059. [DOI] [PubMed] [Google Scholar]
- 160.Yung B.C., Li J., Zhang M., Cheng X., Li H., Yung E.M., Kang C., Cosby L.E., Liu Y., Teng L., Lee R.J. Lipid nanoparticles composed of quaternary amine-tertiary amine cationic lipid combination (QTsome) for therapeutic delivery of AntimiR-21 for lung cancer. Mol. Pharm. 2016;13:653–662. doi: 10.1021/acs.molpharmaceut.5b00878. [DOI] [PubMed] [Google Scholar]
- 161.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 162.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Habrant D., Peuziat P., Colombani T., Dallet L., Gehin J., Goudeau E., Evrard B., Lambert O., Haudebourg T., Pitard B. Design of ionizable lipids to overcome the limiting step of endosomal escape: application in the intracellular delivery of mRNA, DNA, and siRNA. J. Med. Chem. 2016;59:3046–3062. doi: 10.1021/acs.jmedchem.5b01679. [DOI] [PubMed] [Google Scholar]
- 164.Ball R.L., Hajj K.A., Vizelman J., Bajaj P., Whitehead K.A. Lipid nanoparticle formulations for enhanced Co-delivery of siRNA and mRNA. Nano Lett. 2018;18:3814–3822. doi: 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- 165.Blasi P., Giovagnoli S., Schoubben A., Ricci M., Rossi C. Solid lipid nanoparticles for targeted brain drug delivery. Adv. Drug Deliv. Rev. 2007;59:454–477. doi: 10.1016/j.addr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 166.Pardeike J., Hommoss A., Muller R.H. Lipid nanoparticles (SLN, NLC) in cosmetic and pharmaceutical dermal products. Int. J. Pharm. 2009;366:170–184. doi: 10.1016/j.ijpharm.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 167.Paliwal R., Paliwal S.R., Kenwat R., Kurmi B.D., Sahu M.K. Solid lipid nanoparticles: a review on recent perspectives and patents. Expert Opin. Ther. Pat. 2020;30:179–194. doi: 10.1080/13543776.2020.1720649. [DOI] [PubMed] [Google Scholar]
- 168.Das S., Chaudhury A. Recent advances in lipid nanoparticle formulations with solid matrix for oral drug delivery. AAPS PharmSciTech. 2011;12:62–76. doi: 10.1208/s12249-010-9563-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim B.D., Na K., Choi H.K. Preparation and characterization of solid lipid nanoparticles (SLN) made of cacao butter and curdlan. Eur. J. Pharmaceut. Sci. 2005;24:199–205. doi: 10.1016/j.ejps.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 170.Erel-Akbaba G., Carvalho L.A., Tian T., Zinter M., Akbaba H., Obeid P.J., Chiocca E.A., Weissleder R., Kantarci A.G., Tannous B.A. Radiation-induced targeted nanoparticle-based gene delivery for brain tumor therapy. ACS Nano. 2019;13:4028–4040. doi: 10.1021/acsnano.8b08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shi S., Han L., Deng L., Zhang Y., Shen H., Gong T., Zhang Z., Sun X. Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J. Contr. Release. 2014;194:228–237. doi: 10.1016/j.jconrel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 172.Kucukturkmen B., Bozkir A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and miR-21 antisense oligonucleotide to glioblastoma cells. Drug Dev. Ind. Pharm. 2018;44:306–315. doi: 10.1080/03639045.2017.1391835. [DOI] [PubMed] [Google Scholar]
- 173.Barriga H.M.G., Holme M.N., Stevens M.M. Cubosomes: the next generation of smart lipid nanoparticles? Angew. Chem. 2019;58:2958–2978. doi: 10.1002/anie.201804067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.La Y., Park C., Shin T.J., Joo S.H., Kang S., Kim K.T. Colloidal inverse bicontinuous cubic membranes of block copolymers with tunable surface functional groups. Nat. Chem. 2014;6:534–541. doi: 10.1038/nchem.1946. [DOI] [PubMed] [Google Scholar]
- 175.Larsson K. Cubic lipid-water phases: structures and biomembrane aspects. J. Phys. Chem. 1989;93:7304–7314. [Google Scholar]
- 176.Karami Z., Hamidi M. Cubosomes: remarkable drug delivery potential. Drug Discov. Today. 2016;21:789–801. doi: 10.1016/j.drudis.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 177.Leal C., Bouxsein N.F., Ewert K.K., Safinya C.R. Highly efficient gene silencing activity of siRNA embedded in a nanostructured gyroid cubic lipid matrix. J. Am. Chem. Soc. 2010;132:16841–16847. doi: 10.1021/ja1059763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim H., Sung J., Chang Y., Alfeche A., Leal C. Microfluidics synthesis of gene silencing cubosomes. ACS Nano. 2018;12:9196–9205. doi: 10.1021/acsnano.8b03770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schnitzler T., Herrmann A. DNA block copolymers: functional materials for nanoscience and biomedicine. Acc. Chem. Res. 2012;45:1419–1430. doi: 10.1021/ar200211a. [DOI] [PubMed] [Google Scholar]
- 180.Liang Y., Wang Y., Wang L., Liang Z., Li D., Xu X., Chen Y., Yang X., Zhang H., Niu H. Self-crosslinkable chitosan-hyaluronic acid dialdehyde nanoparticles for CD44-targeted siRNA delivery to treat bladder cancer. Bioact Mater. 2021;6:433–446. doi: 10.1016/j.bioactmat.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang H., Cui W., Qu X., Wu H., Qu L., Zhang X., Makila E., Salonen J., Zhu Y., Yang Z., Chen D., Santos H.A., Hai M., Weitz D.A. Photothermal-responsive nanosized hybrid polymersome as versatile therapeutics codelivery nanovehicle for effective tumor suppression. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7744–7749. doi: 10.1073/pnas.1817251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Malafaya P.B., Silva G.A., Reis R.L. Natural-origin polymers as carriers and scaffolds for biomolecules and cell delivery in tissue engineering applications. Adv. Drug Deliv. Rev. 2007;59:207–233. doi: 10.1016/j.addr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 183.Sun P., Huang W., Jin M., Wang Q., Fan B., Kang L., Gao Z. Chitosan-based nanoparticles for survivin targeted siRNA delivery in breast tumor therapy and preventing its metastasis. Int. J. Nanomed. 2016;11:4931–4945. doi: 10.2147/IJN.S105427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Corbet C., Ragelle H., Pourcelle V., Vanvarenberg K., Marchand-Brynaert J., Preat V., Feron O. Delivery of siRNA targeting tumor metabolism using non-covalent PEGylated chitosan nanoparticles: identification of an optimal combination of ligand structure, linker and grafting method. J. Contr. Release. 2016;223:53–63. doi: 10.1016/j.jconrel.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 185.Serrano-Sevilla I., Artiga A., Mitchell S.G., De Matteis L., de la Fuente J.M. Natural polysaccharides for siRNA delivery: nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules. 2019:24. doi: 10.3390/molecules24142570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jain M.A.S. Calcium alginate microparticles as a non-condensing DNA delivery and transfection system for macrophages. Pharmaceut. Eng. 2012:42–49. [Google Scholar]
- 187.Hayashi K., Chaya H., Fukushima S., Watanabe S., Takemoto H., Osada K., Nishiyama N., Miyata K., Kataoka K. Influence of RNA strand rigidity on polyion complex formation with block catiomers. Macromol. Rapid Commun. 2016;37:486–493. doi: 10.1002/marc.201500661. [DOI] [PubMed] [Google Scholar]
- 188.Zhang X., Pan J., Yao M., Palmerston Mendes L., Sarisozen C., Mao S., Torchilin V.P. Charge reversible hyaluronic acid-modified dendrimer-based nanoparticles for siMDR-1 and doxorubicin co-delivery. Eur. J. Pharm. Biopharm. : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V. 2020;154:43–49. doi: 10.1016/j.ejpb.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang M., Wu B., Tucker J.D., Lu P., Lu Q. A combinatorial library of triazine-cored polymeric vectors for pDNA delivery in vitro and in vivo. J. Mater. Chem. B. 2017;5:3907–3918. doi: 10.1039/c6tb03311c. [DOI] [PubMed] [Google Scholar]
- 190.Wang J., Cooper R.C., Yang H. Polyamidoamine dendrimer grafted with an acid-responsive charge-reversal layer for improved gene delivery. Biomacromolecules. 2020;21:4008–4016. doi: 10.1021/acs.biomac.0c00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pan J., Attia S.A., Subhan M.A., Filipczak N., Mendes L.P., Li X., Kishan Yalamarty S.S., Torchilin V.P. Monoclonal antibody 2C5-modified mixed dendrimer micelles for tumor-targeted codelivery of chemotherapeutics and siRNA. Mol. Pharm. 2020;17:1638–1647. doi: 10.1021/acs.molpharmaceut.0c00075. [DOI] [PubMed] [Google Scholar]
- 192.Naito M., Ishii T., Matsumoto A., Miyata K., Miyahara Y., Kataoka K. A phenylboronate-functionalized polyion complex micelle for ATP-triggered release of siRNA. Angew. Chem. 2012;51:10751–10755. doi: 10.1002/anie.201203360. [DOI] [PubMed] [Google Scholar]
- 193.Liu J., Li H.J., Luo Y.L., Chen Y.F., Fan Y.N., Du J.Z., Wang J. Programmable delivery of immune adjuvant to tumor-infiltrating dendritic cells for cancer immunotherapy. Nano Lett. 2020;20:4882–4889. doi: 10.1021/acs.nanolett.0c00893. [DOI] [PubMed] [Google Scholar]
- 194.Dosta P., Ramos V., Borrós S. Stable and efficient generation of poly(β-amino ester)s for RNAi delivery. Molecular Systems Design & Engineering. 2018;3:677–689. doi: 10.1039/c8me00006a. [DOI] [Google Scholar]
- 195.Rui Y., Wilson D.R., Choi J., Varanasi M., Sanders K., Karlsson J., Lim M., Green J.J. Carboxylated branched poly(beta-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci Adv. 2019;5 doi: 10.1126/sciadv.aay3255. eaay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang H., Wang C., Zou Y., Hu J., Li Y., Cheng Y. Natural polyphenols in drug delivery systems: current status and future challenges. Giant. 2020;3 doi: 10.1016/j.giant.2020.100022. [DOI] [Google Scholar]
- 197.Chan D.P., Deleavey G.F., Owen S.C., Damha M.J., Shoichet M.S. Click conjugated polymeric immuno-nanoparticles for targeted siRNA and antisense oligonucleotide delivery. Biomaterials. 2013;34:8408–8415. doi: 10.1016/j.biomaterials.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 198.Iqbal S., Qu Y., Dong Z., Zhao J., Rauf Khan A., Rehman S., Zhao Z. Poly (β‐amino esters) based potential drug delivery and targeting polymer; an overview and perspectives (review) Eur. Polym. J. 2020;141 doi: 10.1016/j.eurpolymj.2020.110097. [DOI] [Google Scholar]
- 199.Keeney M., Ong S.G., Padilla A., Yao Z., Goodman S., Wu J.C., Yang F. Development of poly(beta-amino ester)-based biodegradable nanoparticles for nonviral delivery of minicircle DNA. ACS Nano. 2013;7:7241–7250. doi: 10.1021/nn402657d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhu J., Shi X. Dendrimer-based nanodevices for targeted drug delivery applications. J. Mater. Chem. B. 2013;1:4199–4211. doi: 10.1039/c3tb20724b. [DOI] [PubMed] [Google Scholar]
- 201.Lyu Z., Ding L., Huang A.Y.T., Kao C.L., Peng L. Poly(amidoamine) dendrimers: covalent and supramolecular synthesis. Materials Today Chemistry. 2019;13:34–48. doi: 10.1016/j.mtchem.2019.04.004. [DOI] [Google Scholar]
- 202.Mikiciuk-Olasik M.M.-P.E. Dendrimers in drug delivery. 2016. [DOI]
- 203.Abedi-Gaballu F., Dehghan G., Ghaffari M., Yekta R., Abbaspour-Ravasjani S., Baradaran B., Dolatabadi J.E.N., Hamblin M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl Mater Today. 2018;12:177–190. doi: 10.1016/j.apmt.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pan J., Attia S.A., Subhan M.A., Filipczak N., Mendes L.P., Li X., Kishan Yalamarty S.S., Torchilin V.P. Monoclonal antibody 2C5-modified mixed dendrimer micelles for tumor-targeted codelivery of chemotherapeutics and siRNA. Mol. Pharm. 2020;17:1638–1647. doi: 10.1021/acs.molpharmaceut.0c00075. [DOI] [PubMed] [Google Scholar]
- 205.Pan J., Mendes L.P., Yao M., Filipczak N., Garai S., Thakur G.A., Sarisozen C., Torchilin V.P. Polyamidoamine dendrimers-based nanomedicine for combination therapy with siRNA and chemotherapeutics to overcome multidrug resistance. Eur. J. Pharm. Biopharm. : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V. 2019;136:18–28. doi: 10.1016/j.ejpb.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Segovia N., Pont M., Oliva N., Ramos V., Borros S., Artzi N. Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Advanced healthcare materials. 2015;4:271–280. doi: 10.1002/adhm.201400235. [DOI] [PubMed] [Google Scholar]
- 207.Janaszewska A., Lazniewska J., Trzepinski P., Marcinkowska M., Klajnert-Maculewicz B. Cytotoxicity of dendrimers. Biomolecules. 2019;9 doi: 10.3390/biom9080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yang J., Zhang Q., Chang H., Cheng Y. Surface-engineered dendrimers in gene delivery. Chem. Rev. 2015;115:5274–5300. doi: 10.1021/cr500542t. [DOI] [PubMed] [Google Scholar]
- 209.Song Z., Liang X., Wang Y., Han H., Yang J., Fang X., Li Q. Phenylboronic acid-functionalized polyamidoamine-mediated miR-34a delivery for the treatment of gastric cancer. Biomaterials science. 2019;7:1632–1642. doi: 10.1039/c8bm01385c. [DOI] [PubMed] [Google Scholar]
- 210.Wang J., Cooper R.C., He H., Li B., Yang H. Polyamidoamine dendrimer microgels: hierarchical arrangement of dendrimers into micrometer domains with expanded structural features for programmable drug delivery and release. Macromolecules. 2018;51:6111–6118. doi: 10.1021/acs.macromol.8b01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Li J., Han Y., Chen Q., Shi H., Ur Rehman S., Siddiq M., Ge Z., Liu S. Dual endogenous stimuli-responsive polyplex micelles as smart two-step delivery nanocarriers for deep tumor tissue penetration and combating drug resistance of cisplatin. J. Mater. Chem. B. 2014;2:1813–1824. doi: 10.1039/c3tb21383h. [DOI] [PubMed] [Google Scholar]
- 212.Mirkin C.A., Letsinger R.L., Mucic R.C., Storhoff J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 213.Patel P.C., Giljohann D.A., Daniel W.L., Zheng D., Prigodich A.E., Mirkin C.A. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Choi C.H., Hao L., Narayan S.P., Auyeung E., Mirkin C.A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cutler J.I., Auyeung E., Mirkin C.A. Spherical nucleic acids. J. Am. Chem. Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 216.Callmann C.E., Cole L.E., Kusmierz C.D., Huang Z., Horiuchi D., Mirkin C.A. Tumor cell lysate-loaded immunostimulatory spherical nucleic acids as therapeutics for triple-negative breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2020;117:17543–17550. doi: 10.1073/pnas.2005794117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jensen S.A., Day E.S., Ko C.H., Hurley L.A., Luciano J.P., Kouri F.M., Merkel T.J., Luthi A.J., Patel P.C., Cutler J.I., Daniel W.L., Scott A.W., Rotz M.W., Meade T.J., Giljohann D.A., Mirkin C.A., Stegh A.H. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006839. 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Radovic-Moreno A.F., Chernyak N., Mader C.C., Nallagatla S., Kang R.S., Hao L., Walker D.A., Halo T.L., Merkel T.J., Rische C.H., Anantatmula S., Burkhart M., Mirkin C.A., Gryaznov S.M. Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Banga R.J., Chernyak N., Narayan S.P., Nguyen S.T., Mirkin C.A. Liposomal spherical nucleic acids. J. Am. Chem. Soc. 2014;136:9866–9869. doi: 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Guan C., Chernyak N., Dominguez D., Cole L., Zhang B., Mirkin C.A. RNA-based immunostimulatory liposomal spherical nucleic acids as potent TLR7/8 modulators. Small. 2018:14. doi: 10.1002/smll.201803284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ferrer J.R., Sinegra A.J., Ivancic D., Yeap X.Y., Qiu L., Wang J.J., Zhang Z.J., Wertheim J.A., Mirkin C.A. Structure-dependent biodistribution of liposomal spherical nucleic acids. ACS Nano. 2020;14:1682–1693. doi: 10.1021/acsnano.9b07254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Banga R.J., Meckes B., Narayan S.P., Sprangers A.J., Nguyen S.T., Mirkin C.A. Cross-linked micellar spherical nucleic acids from thermoresponsive templates. J. Am. Chem. Soc. 2017;139:4278–4281. doi: 10.1021/jacs.6b13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang C., Hao L., Calabrese C.M., Zhou Y., Choi C.H., Xing H., Mirkin C.A. Biodegradable DNA-brush block copolymer spherical nucleic acids enable transfection agent-free intracellular gene regulation. Small. 2015;11:5360–5368. doi: 10.1002/smll.201501573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Samanta D., Ebrahimi S.B., Kusmierz C.D., Cheng H.F., Mirkin C.A. Protein spherical nucleic acids for live-cell chemical analysis. J. Am. Chem. Soc. 2020;142:13350–13355. doi: 10.1021/jacs.0c06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang S., Chen Y., Wang S., Li P., Mirkin C.A., Farha O.K. DNA-functionalized metal-organic framework nanoparticles for intracellular delivery of proteins. J. Am. Chem. Soc. 2019;141:2215–2219. doi: 10.1021/jacs.8b12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang S., McGuirk C.M., d'Aquino A., Mason J.A., Mirkin C.A. Metal-organic framework nanoparticles. Adv. Mater. 2018;30 doi: 10.1002/adma.201800202. [DOI] [PubMed] [Google Scholar]
- 227.Massich M.D., Giljohann D.A., Seferos D.S., Ludlow L.E., Horvath C.M., Mirkin C.A. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol. Pharm. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zheng D., Giljohann D.A., Chen D.L., Massich M.D., Wang X.Q., Iordanov H., Mirkin C.A., Paller A.S. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Cutler J.I., Zhang K., Zheng D., Auyeung E., Prigodich A.E., Mirkin C.A. Polyvalent nucleic acid nanostructures. J. Am. Chem. Soc. 2011;133:9254–9257. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yue R., Chen M., Ma N. Dual MicroRNA-triggered drug release system for combined chemotherapy and gene therapy with logic operation. ACS Appl. Mater. Interfaces. 2020;12:32493–32502. doi: 10.1021/acsami.0c09494. [DOI] [PubMed] [Google Scholar]
- 231.Day H.A., Pavlou P., Waller Z.A. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014;22:4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 232.Kim J., Lee Y.M., Kang Y., Kim W.J. Tumor-homing, size-tunable clustered nanoparticles for anticancer therapeutics. ACS Nano. 2014;8:9358–9367. doi: 10.1021/nn503349g. [DOI] [PubMed] [Google Scholar]
- 233.Chen X., Chen T., Ren L., Chen G., Gao X., Li G., Zhu X. Triplex DNA nanoswitch for pH-sensitive release of multiple cancer drugs. ACS Nano. 2019;13:7333–7344. doi: 10.1021/acsnano.9b03846. [DOI] [PubMed] [Google Scholar]
- 234.Chen C.C., Lin Y.P., Wang C.W., Tzeng H.C., Wu C.H., Chen Y.C., Chen C.P., Chen L.C., Wu Y.C. DNA-gold nanorod conjugates for remote control of localized gene expression by near infrared irradiation. J. Am. Chem. Soc. 2006;128:3709–3715. doi: 10.1021/ja0570180. [DOI] [PubMed] [Google Scholar]
- 235.Li P., Wu Y., Li D., Su X., Luo C., Wang Y., Hu J., Li G., Jiang H., Zhang W. Seed-mediated synthesis of tunable-aspect-ratio gold nanorods for near-infrared photoacoustic imaging. Nanoscale research letters. 2018;13:313. doi: 10.1186/s11671-018-2734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Li J.X., Zhu B.Q., Zhu Z., Zhang Y.C., Yao X.J., Tu S., Liu R.D., Jia S.S., Yang C.Y.J. Simple and rapid functionalization of gold nanorods with oligonucleotides using an mPEG-SH/tween 20-assisted approach. Langmuir : the ACS journal of surfaces and colloids. 2015;31:7869–7876. doi: 10.1021/acs.langmuir.5b01680. [DOI] [PubMed] [Google Scholar]
- 237.Wang J.X., Thomas M., Lin P., Cheng J.X., Matei D.E., Wei A. siRNA delivery using dithiocarbamate-anchored oligonucleotides on gold nanorods. Bioconjugate Chem. 2019;30:443–453. doi: 10.1021/acs.bioconjchem.8b00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang B.K., Yu X.F., Wang J.H., Li Z.B., Li P.H., Wang H., Song L., Chu P.K., Li C. Gold-nanorods-siRNA nanoplex for improved photothermal therapy by gene silencing. Biomaterials. 2016;78:27–39. doi: 10.1016/j.biomaterials.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 239.Xie J.P., Lee J.Y., Wang D.I.C. Seedless, surfactantless, high-yield synthesis of branched gold nanocrystals in HEPES buffer solution. Chem. Mater. 2007;19:2823–2830. doi: 10.1021/cm0700100. [DOI] [Google Scholar]
- 240.Dam D.H.M., Culver K.S.B., Odom T.W. Grafting aptamers onto gold nanostars increases in vitro efficacy in a wide range of cancer cell types. Mol. Pharm. 2014;11:580–587. doi: 10.1021/mp4005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dam D.H.M., Lee J.H., Sisco P.N., Co D.T., Zhang M., Wasielewski M.R., Odom T.W. Direct observation of nanoparticle-cancer cell nucleus interactions. ACS Nano. 2012;6:3318–3326. doi: 10.1021/nn300296p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dam D.H.M., Lee R.C., Odom T.W. Improved in vitro efficacy of gold nanoconstructs by increased loading of G-quadruplex aptamer. Nano Lett. 2014;14:2843–2848. doi: 10.1021/nl500844m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lee H., Dam D.H.M., Ha J.W., Yue J., Odom T.W. Enhanced human epidermal growth factor receptor 2 degradation in breast cancer cells by lysosome-targeting gold nanoconstructs. ACS Nano. 2015;9:9859–9867. doi: 10.1021/acsnano.5b05138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lee K., Huang Z.N., Mirkin C.A., Odom T.W. Endosomal organization of CpG constructs correlates with enhanced immune activation. Nano Lett. 2020;20:6170–6175. doi: 10.1021/acs.nanolett.0c02536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yue J., Feliciano T.J., Li W.L., Lee A., Odom T.W. Gold nanoparticle size and shape effects on cellular uptake and intracellular distribution of siRNA nanoconstructs. Bioconjugate Chem. 2017;28:1791–1800. doi: 10.1021/acs.bioconjchem.7b00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yue J., Pallares R.M., Cole L.E., Coughlin E.E., Mirkin C.A., Lee A., Odom T.W. Smaller CpG-conjugated gold nanoconstructs achieve higher targeting specificity of immune activation. ACS Appl. Mater. Interfaces. 2018;10:21920–21926. doi: 10.1021/acsami.8b06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wu D.L., Zhao P., Wu L., Lin L.H., Yu G.Y., Xu L.L., Yue J. Aptamer-functionalized gold nanostars for on-demand delivery of anticancer therapeutics. Acs Appl Bio Mater. 2020;3:4590–4599. doi: 10.1021/acsabm.0c00499. [DOI] [PubMed] [Google Scholar]
- 248.Huo S., Jin S., Ma X., Xue X., Yang K., Kumar A., Wang P.C., Zhang J., Hu Z., Liang X.J. Ultrasmall gold nanoparticles as carriers for nucleus-based gene therapy due to size-dependent nuclear entry. ACS Nano. 2014;8:5852–5862. doi: 10.1021/nn5008572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jiang Y., Huo S., Mizuhara T., Das R., Lee Y.W., Hou S., Moyano D.F., Duncan B., Liang X.J., Rotello V.M. The interplay of size and surface functionality on the cellular uptake of sub-10 nm gold nanoparticles. ACS Nano. 2015;9:9986–9993. doi: 10.1021/acsnano.5b03521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Huo S., Gong N., Jiang Y., Chen F., Guo H., Gan Y., Wang Z., Herrmann A., Liang X.J. Gold-DNA nanosunflowers for efficient gene silencing with controllable transformation. Sci Adv. 2019;5:eaaw6264. doi: 10.1126/sciadv.aaw6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lei Y., Tang L., Xie Y., Xianyu Y., Zhang L., Wang P., Hamada Y., Jiang K., Zheng W., Jiang X. Gold nanoclusters-assisted delivery of NGF siRNA for effective treatment of pancreatic cancer. Nat. Commun. 2017;8:15130. doi: 10.1038/ncomms15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Aloe L., Rocco M.L., Balzamino B.O., Micera A. Nerve growth factor: role in growth, differentiation and controlling cancer cell development. J. Exp. Clin. Canc. Res. 2016;35:116. doi: 10.1186/s13046-016-0395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhu Z., Kleeff J., Kayed H., Wang L., Korc M., Buchler M.W., Friess H. Nerve growth factor and enhancement of proliferation, invasion, and tumorigenicity of pancreatic cancer cells. Mol. Carcinog. 2002;35:138–147. doi: 10.1002/mc.10083. [DOI] [PubMed] [Google Scholar]
- 254.Miknyoczki S.J., Lang D., Huang L., Klein-Szanto A.J.P., Dionne C.A., Ruggeri B.A. Neurotrophins and Trk receptors in human pancreatic ductal adenocarcinoma: expression patterns and effects onIn vitro invasive behavior. Int. J. Canc. 1999;81:417–427. doi: 10.1002/(sici)1097-0215(19990505)81:3<417::Aid-ijc16>3.0.Co;2-6. [DOI] [PubMed] [Google Scholar]
- 255.Schneider M.B., Standop J., Ulrich A., Wittel U., Friess H., Andren-Sandberg A., Pour P.M. Expression of nerve growth factors in pancreatic neural tissue and pancreatic cancer. J. Histochem. Cytochem. 2001;49:1205–1210. doi: 10.1177/002215540104901002. [DOI] [PubMed] [Google Scholar]
- 256.Yi Y., Kim H.J., Zheng M., Mi P., Naito M., Kim B.S., Min H.S., Hayashi K., Perche F., Toh K., Liu X., Mochida Y., Kinoh H., Cabral H., Miyata K., Kataoka K. Glucose-linked sub-50-nm unimer polyion complex-assembled gold nanoparticles for targeted siRNA delivery to glucose transporter 1-overexpressing breast cancer stem-like cells. J. Contr. Release : official journal of the Controlled Release Society. 2019;295:268–277. doi: 10.1016/j.jconrel.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 257.Vivero-Escoto J.L., Huxford-Phillips R.C., Lin W. Silica-based nanoprobes for biomedical imaging and theranostic applications. Chem. Soc. Rev. 2012;41:2673–2685. doi: 10.1039/c2cs15229k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yu Y., Duan J., Yu Y., Li Y., Liu X., Zhou X., Ho K.F., Tian L., Sun Z. Silica nanoparticles induce autophagy and autophagic cell death in HepG2 cells triggered by reactive oxygen species. J. Hazard Mater. 2014;270:176–186. doi: 10.1016/j.jhazmat.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 259.Kneuer C., Sameti M., Bakowsky U., Schiestel T., Schirra H., Schmidt H., Lehr C.M. A nonviral DNA delivery system based on surface modified silica-nanoparticles can efficiently transfect cells in vitro. Bioconjugate Chem. 2000;11:926–932. doi: 10.1021/bc0000637. [DOI] [PubMed] [Google Scholar]
- 260.Bertucci A., Kim K.H., Kang J., Zuidema J.M., Lee S.H., Kwon E.J., Kim D., Howell S.B., Ricci F., Ruoslahti E., Jang H.J., Sailor M.J. Tumor-targeting, MicroRNA-silencing porous silicon nanoparticles for ovarian cancer therapy. ACS Appl. Mater. Interfaces. 2019;11:23926–23937. doi: 10.1021/acsami.9b07980. [DOI] [PubMed] [Google Scholar]
- 261.Kafshgari M.H., Delalat B., Tong W.Y., Harding F.J., Kaasalainen M., Salonen J., Voelcker N.H. Oligonucleotide delivery by chitosan-functionalized porous silicon nanoparticles. Nano Research. 2015;8:2033–2046. doi: 10.1007/s12274-015-0715-0. [DOI] [Google Scholar]
- 262.Tong W.Y., Alnakhli M., Bhardwaj R., Apostolou S., Sinha S., Fraser C., Kuchel T., Kuss B., Voelcker N.H. Delivery of siRNA in vitro and in vivo using PEI-capped porous silicon nanoparticles to silence MRP1 and inhibit proliferation in glioblastoma. J. Nanobiotechnol. 2018;16:38. doi: 10.1186/s12951-018-0365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kim B., Sun S., Varner J.A., Howell S.B., Ruoslahti E., Sailor M.J. Securing the payload, finding the cell, and avoiding the endosome: peptide-targeted, fusogenic porous silicon nanoparticles for delivery of siRNA. Adv. Mater. 2019;31 doi: 10.1002/adma.201902952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kang J., Joo J., Kwon E.J., Skalak M., Hussain S., She Z.G., Ruoslahti E., Bhatia S.N., Sailor M.J. Self-sealing porous silicon-calcium silicate core-shell nanoparticles for targeted siRNA delivery to the injured brain. Adv. Mater. 2016;28:7962–7969. doi: 10.1002/adma.201600634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liberman A., Mendez N., Trogler W.C., Kummel A.C. Synthesis and surface functionalization of silica nanoparticles for nanomedicine. Surf. Sci. Rep. 2014;69:132–158. doi: 10.1016/j.surfrep.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Malvindi M.A., Brunetti V., Vecchio G., Galeone A., Cingolani R., Pompa P.P. SiO2 nanoparticles biocompatibility and their potential for gene delivery and silencing. Nanoscale. 2012;4:486–495. doi: 10.1039/c1nr11269d. [DOI] [PubMed] [Google Scholar]
- 267.Moller K., Muller K., Engelke H., Brauchle C., Wagner E., Bein T. Highly efficient siRNA delivery from core-shell mesoporous silica nanoparticles with multifunctional polymer caps. Nanoscale. 2016;8:4007–4019. doi: 10.1039/c5nr06246b. [DOI] [PubMed] [Google Scholar]
- 268.Wang Y., Xie Y., Kilchrist K.V., Li J., Duvall C.L., Oupicky D. Endosomolytic and tumor-penetrating mesoporous silica nanoparticles for siRNA/miRNA combination cancer therapy. ACS Appl. Mater. Interfaces. 2020;12:4308–4322. doi: 10.1021/acsami.9b21214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.An J., Zhang K., Wang B., Wu S., Wang Y., Zhang H., Zhang Z., Liu J., Shi J. Nanoenabled disruption of multiple barriers in antigen cross-presentation of dendritic cells via calcium interference for enhanced chemo-immunotherapy. ACS Nano. 2020;14:7639–7650. doi: 10.1021/acsnano.0c03881. [DOI] [PubMed] [Google Scholar]
- 270.Marchi S., Giorgi C., Galluzzi L., Pinton P. Ca(2+) fluxes and cancer. Mol Cell. 2020;78:1055–1069. doi: 10.1016/j.molcel.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 271.Wang S., Liu X., Chen S., Liu Z., Zhang X., Liang X.J., Li L. Regulation of Ca(2+) signaling for drug-resistant breast cancer therapy with mesoporous silica nanocapsule encapsulated doxorubicin/siRNA cocktail. ACS Nano. 2019;13:274–283. doi: 10.1021/acsnano.8b05639. [DOI] [PubMed] [Google Scholar]
- 272.Juneja R., Vadarevu H., Halman J., Tarannum M., Rackley L., Dobbs J., Marquez J., Chandler M., Afonin K., Vivero-Escoto J.L. Combination of nucleic acid and mesoporous silica nanoparticles: optimization and therapeutic performance in vitro. ACS Appl. Mater. Interfaces. 2020;12:38873–38886. doi: 10.1021/acsami.0c07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Li J.R., Kuppler R.J., Zhou H.C. Selective gas adsorption and separation in metal-organic frameworks. Chem. Soc. Rev. 2009;38:1477–1504. doi: 10.1039/b802426j. [DOI] [PubMed] [Google Scholar]
- 274.Millward A.R., Yaghi O.M. Metal-organic frameworks with exceptionally high capacity for storage of carbon dioxide at room temperature. J. Am. Chem. Soc. 2005;127:17998–17999. doi: 10.1021/ja0570032. [DOI] [PubMed] [Google Scholar]
- 275.Lee J., Farha O.K., Roberts J., Scheidt K.A., Nguyen S.T., Hupp J.T. Metal-organic framework materials as catalysts. Chem. Soc. Rev. 2009;38:1450–1459. doi: 10.1039/b807080f. [DOI] [PubMed] [Google Scholar]
- 276.Zhu H., Yin R., Han F., Guan J., Zhang X., Mao X., Zhao L., Li Q., Hou X., Bi K. Characterization of chemical constituents in Zhi-Zi-Da-Huang decoction by ultra high performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Separ. Sci. 2014;37:3489–3496. doi: 10.1002/jssc.201400728. [DOI] [PubMed] [Google Scholar]
- 277.Simon-Yarza T., Mielcarek A., Couvreur P., Serre C. Nanoparticles of metal-organic frameworks: on the road to in vivo efficacy in biomedicine. Adv. Mater. 2018;30 doi: 10.1002/adma.201870281. [DOI] [PubMed] [Google Scholar]
- 278.Zhang Y., Wang F.M., Ju E.G., Liu Z., Chen Z.W., Ren J.S., Qu X.G. Metal-organic-framework-based vaccine platforms for enhanced systemic immune and memory response. Adv. Funct. Mater. 2016;26:6454–6461. doi: 10.1002/adfm.201600650. [DOI] [Google Scholar]
- 279.Wu M.X., Yang Y.W. Metal-organic framework (MOF)-Based drug/cargo delivery and cancer therapy. Adv. Mater. 2017;29 doi: 10.1002/adma.201606134. [DOI] [PubMed] [Google Scholar]
- 280.Meng J., Liu X., Niu C., Pang Q., Li J., Liu F., Liu Z., Mai L. Advances in metal-organic framework coatings: versatile synthesis and broad applications. Chem. Soc. Rev. 2020;49:3142–3186. doi: 10.1039/c9cs00806c. [DOI] [PubMed] [Google Scholar]
- 281.Zhang J., He M., Nie C., He M., Pan Q., Liu C., Hu Y., Chen T., Chu X. Biomineralized metal-organic framework nanoparticles enable a primer exchange reaction-based DNA machine to work in living cells for imaging and gene therapy. Chem. Sci. 2020;11:7092–7101. doi: 10.1039/d0sc00339e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhuang J., Gong H., Zhou J., Zhang Q., Gao W., Fang R.H., Zhang L. Targeted gene silencing in vivo by platelet membrane-coated metal-organic framework nanoparticles. Sci Adv. 2020;6 doi: 10.1126/sciadv.aaz6108. eaaz6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wang Y., Yan J., Wen N., Xiong H., Cai S., He Q., Hu Y., Peng D., Liu Z., Liu Y. Metal-organic frameworks for stimuli-responsive drug delivery. Biomaterials. 2020;230:119619. doi: 10.1016/j.biomaterials.2019.119619. [DOI] [PubMed] [Google Scholar]
- 284.He C., Lu K., Liu D., Lin W. Nanoscale metal-organic frameworks for the co-delivery of cisplatin and pooled siRNAs to enhance therapeutic efficacy in drug-resistant ovarian cancer cells. J. Am. Chem. Soc. 2014;136:5181–5184. doi: 10.1021/ja4098862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Chen Q., Xu M., Zheng W., Xu T., Deng H., Liu J. Se/Ru-Decorated porous metal-organic framework nanoparticles for the delivery of pooled siRNAs to reversing multidrug resistance in taxol-resistant breast cancer cells. ACS Appl. Mater. Interfaces. 2017;9:6712–6724. doi: 10.1021/acsami.6b12792. [DOI] [PubMed] [Google Scholar]
- 286.Teplensky M.H., Fantham M., Poudel C., Hockings C., Lu M., Guna A., Aragones-Anglada M., Moghadam P.Z., Li P., Farha O.K., Bernaldo de Quirós Fernández S., Richards F.M., Jodrell D.I., Kaminski Schierle G., Kaminski C.F., Fairen-Jimenez D. A highly porous metal-organic framework system to deliver payloads for gene knockdown. Inside Chem. 2019;5:2926–2941. doi: 10.1016/j.chempr.2019.08.015. [DOI] [Google Scholar]
- 287.Liu B., Hu F., Zhang J., Wang C., Li L. A biomimetic coordination nanoplatform for controlled encapsulation and delivery of drug-gene combinations. Angew. Chem. 2019;58:8804–8808. doi: 10.1002/anie.201903417. [DOI] [PubMed] [Google Scholar]
- 288.Morris W., Briley W.E., Auyeung E., Cabezas M.D., Mirkin C.A. Nucleic acid-metal organic framework (MOF) nanoparticle conjugates. J. Am. Chem. Soc. 2014;136:7261–7264. doi: 10.1021/ja503215w. [DOI] [PubMed] [Google Scholar]
- 289.Wang S.Z., McGuirk C.M., Ross M.B., Wang S.Y., Chen P.C., Xing H., Liu Y., Mirkin C.A. General and direct method for preparing oligonucleotide-functionalized metal-organic framework nanoparticles. J. Am. Chem. Soc. 2017;139:9827–9830. doi: 10.1021/jacs.7b05633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wang Z., Fu Y., Kang Z., Liu X., Chen N., Wang Q., Tu Y., Wang L., Song S., Ling D., Song H., Kong X., Fan C. Organelle-specific triggered release of immunostimulatory oligonucleotides from intrinsically coordinated DNA-metal-organic frameworks with soluble exoskeleton. J. Am. Chem. Soc. 2017;139:15784–15791. doi: 10.1021/jacs.7b07895. [DOI] [PubMed] [Google Scholar]
- 291.Kumar S., Jain S., Dilbaghi N., Ahluwalia A.S., Hassan A.A., Kim K.H. Advanced selection methodologies for DNAzymes in sensing and healthcare applications. Trends Biochem. Sci. 2019;44:190–213. doi: 10.1016/j.tibs.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 292.Dass C.R., Choong P.F.M., Khachigian L.M. DNAzyme technology and cancer therapy: cleave and let die. Mol. Canc. Therapeut. 2008;7:243–251. doi: 10.1158/1535-7163.Mct-07-0510. [DOI] [PubMed] [Google Scholar]
- 293.Wang H., Chen Y., Wang H., Liu X., Zhou X., Wang F. DNAzyme-loaded metal-organic frameworks (MOFs) for self-sufficient gene therapy. Angew. Chem. 2019;58:7380–7384. doi: 10.1002/anie.201902714. [DOI] [PubMed] [Google Scholar]
- 294.Wang J., Wang H., Wang H., He S., Li R., Deng Z., Liu X., Wang F. Nonviolent self-catabolic DNAzyme nanosponges for smart anticancer drug delivery. ACS Nano. 2019;13:5852–5863. doi: 10.1021/acsnano.9b01589. [DOI] [PubMed] [Google Scholar]
- 295.Lin G., Zhang Y., Zhang L., Wang J., Tian Y., Cai W., Tang S., Chu C., Zhou J., Mi P., Chen X., Liu G. Metal-organic frameworks nanoswitch: toward photo-controllable endo/lysosomal rupture and release for enhanced cancer RNA interference. Nano Research. 2020;13:238–245. doi: 10.1007/s12274-019-2606-2. [DOI] [Google Scholar]
- 296.Hong F., Zhang F., Liu Y., Yan H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017;117:12584–12640. doi: 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- 297.Wei B., Dai M., Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Han D., Qi X., Myhrvold C., Wang B., Dai M., Jiang S., Bates M., Liu Y., An B., Zhang F., Yan H., Yin P. Single-stranded DNA and RNA origami. Science. 2017;358 doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Groves B., Chen Y.J., Zurla C., Pochekailov S., Kirschman J.L., Santangelo P.J., Seelig G. Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol. 2016;11:287–294. doi: 10.1038/nnano.2015.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lee H., Lytton-Jean A.K., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M., Peng C.G., Charisse K., Borodovsky A., Manoharan M., Donahoe J.S., Truelove J., Nahrendorf M., Langer R., Anderson D.G. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ren K., Zhang Y., Zhang X., Liu Y., Yang M., Ju H. In situ SiRNA assembly in living cells for gene therapy with MicroRNA triggered cascade reactions templated by nucleic acids. ACS Nano. 2018;12:10797–10806. doi: 10.1021/acsnano.8b02403. [DOI] [PubMed] [Google Scholar]
- 303.Wu C., Han D., Chen T., Peng L., Zhu G., You M., Qiu L., Sefah K., Zhang X., Tan W. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J. Am. Chem. Soc. 2013;135:18644–18650. doi: 10.1021/ja4094617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Li J., Zheng C., Cansiz S., Wu C., Xu J., Cui C., Liu Y., Hou W., Wang Y., Zhang L., Teng I.T., Yang H.H., Tan W. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 2015;137:1412–1415. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ouyang C., Zhang S., Xue C., Yu X., Xu H., Wang Z., Lu Y., Wu Z.S. Precision-guided missile-like DNA nanostructure containing warhead and guidance control for aptamer-based targeted drug delivery into cancer cells in vitro and in vivo. J. Am. Chem. Soc. 2020;142:1265–1277. doi: 10.1021/jacs.9b09782. [DOI] [PubMed] [Google Scholar]
- 306.Li J., Pei H., Zhu B., Liang L., Wei M., He Y., Chen N., Li D., Huang Q., Fan C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 307.Christopher M.E., Wong J.P. Use of toll-like receptor 3 agonists against respiratory viral infections. Anti-Inflammatory Anti-Allergy Agents Med. Chem. 2012;10:327–338. doi: 10.2174/1871523011109050327. [DOI] [Google Scholar]
- 308.Qi X., Liu X., Matiski L., Rodriguez Del Villar R., Yip T., Zhang F., Sokalingam S., Jiang S., Liu L., Yan H., Chang Y. RNA origami nanostructures for potent and safe anticancer immunotherapy. ACS Nano. 2020;14:4727–4740. doi: 10.1021/acsnano.0c00602. [DOI] [PubMed] [Google Scholar]
- 309.Li J., Wang Y., Zhu Y., Oupicky D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Contr. Release : official journal of the Controlled Release Society. 2013;172:589–600. doi: 10.1016/j.jconrel.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Liu J., Song L., Liu S., Zhao S., Jiang Q., Ding B. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. 2018;57:15486–15490. doi: 10.1002/anie.201809452. [DOI] [PubMed] [Google Scholar]
- 311.Guo S., Li H., Ma M., Fu J., Dong Y., Guo P. Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Mol. Ther. Nucleic Acids. 2017;9:399–408. doi: 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Hong E., Halman J.R., Shah A.B., Khisamutdinov E.F., Dobrovolskaia M.A., Afonin K.A. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018;18:4309–4321. doi: 10.1021/acs.nanolett.8b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kalra H., Drummen G.P., Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int. J. Mol. Sci. 2016;17:170. doi: 10.3390/ijms17020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.van Dommelen S.M., Vader P., Lakhal S., Kooijmans S.A., van Solinge W.W., Wood M.J., Schiffelers R.M. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J. Contr. Release : official journal of the Controlled Release Society. 2012;161:635–644. doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 315.Colombo M., Raposo G., Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 316.Zaborowski M.P., Balaj L., Breakefield X.O., Lai C.P. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 318.Villarroya-Beltri C., Baixauli F., Gutierrez-Vazquez C., Sanchez-Madrid F., Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin. Canc. Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 320.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 321.Elsharkasy O.M., Nordin J.Z., Hagey D.W., de Jong O.G., Schiffelers R.M., Andaloussi S.E., Vader P. Extracellular vesicles as drug delivery systems: why and how? Adv. Drug Deliv. Rev. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 322.Zhou X., Xie F., Wang L., Zhang L., Zhang S., Fang M., Zhou F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020;17:323–334. doi: 10.1038/s41423-020-0391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schubert A., Boutros M. Extracellular vesicles and oncogenic signaling. Mol Oncol. 2021;15:3–26. doi: 10.1002/1878-0261.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ono K., Sogawa C., Kawai H., Tran M.T., Taha E.A., Lu Y., Oo M.W., Okusha Y., Okamura H., Ibaragi S., Takigawa M., Kozaki K.I., Nagatsuka H., Sasaki A., Okamoto K., Calderwood S.K., Eguchi T. Triple knockdown of CDC37, HSP90-alpha and HSP90-beta diminishes extracellular vesicles-driven malignancy events and macrophage M2 polarization in oral cancer. J. Extracell. Vesicles. 2020;9:1769373. doi: 10.1080/20013078.2020.1769373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Angioni R., Liboni C., Herkenne S., Sanchez-Rodriguez R., Borile G., Marcuzzi E., Cali B., Muraca M., Viola A. CD73(+) extracellular vesicles inhibit angiogenesis through adenosine A2B receptor signalling. J. Extracell. Vesicles. 2020;9:1757900. doi: 10.1080/20013078.2020.1757900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Chaput N., Thery C. Exosomes: immune properties and potential clinical implementations. Semin. Immunopathol. 2011;33:419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 327.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9 doi: 10.1038/ncb1596. 654-U72. [DOI] [PubMed] [Google Scholar]
- 328.Che Y., Shi X., Shi Y., Jiang X., Ai Q., Shi Y., Gong F., Jiang W. Exosomes derived from miR-143-overexpressing MSCs inhibit cell migration and invasion in human prostate cancer by downregulating TFF3. Mol. Ther. Nucleic Acids. 2019;18:232–244. doi: 10.1016/j.omtn.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 329.Xu H., Zhao G., Zhang Y., Jiang H., Wang W., Zhao D., Hong J., Yu H., Qi L. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/beta-catenin signaling pathway by targeting EZH2. Stem Cell Res. Ther. 2019;10:381. doi: 10.1186/s13287-019-1446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Cai H., Yang X., Gao Y., Xu Z., Yu B., Xu T., Li X., Xu W., Wang X., Hua L. Exosomal MicroRNA-9-3p secreted from BMSCs downregulates ESM1 to suppress the development of bladder cancer. Mol. Ther. Nucleic Acids. 2019;18:787–800. doi: 10.1016/j.omtn.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Neviani P., Wise P.M., Murtadha M., Liu C.W., Wu C.H., Jong A.Y., Seeger R.C., Fabbri M. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Canc. Res. 2019;79:1151–1164. doi: 10.1158/0008-5472.CAN-18-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sun H., Shi K., Qi K., Kong H., Zhang J., Dai S., Ye W., Deng T., He Q., Zhou M. Natural killer cell-derived exosomal miR-3607-3p inhibits pancreatic cancer progression by targeting IL-26. Front. Immunol. 2019;10:2819. doi: 10.3389/fimmu.2019.02819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 333.Limoni S.K., Moghadam M.F., Moazzeni S.M., Gomari H., Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl. Biochem. Biotechnol. 2019;187:352–364. doi: 10.1007/s12010-018-2813-4. [DOI] [PubMed] [Google Scholar]
- 334.Liang G., Zhu Y., Ali D.J., Tian T., Xu H., Si K., Sun B., Chen B., Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020;18:10. doi: 10.1186/s12951-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shtam T.A., Kovalev R.A., Varfolomeeva E.Y., Makarov E.M., Kil Y.V., Filatov M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Fuhrmann G., Serio A., Mazo M., Nair R., Stevens M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Contr. Release : official journal of the Controlled Release Society. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 337.Kim M.S., Haney M.J., Zhao Y., Mahajan V., Deygen I., Klyachko N.L., Inskoe E., Piroyan A., Sokolsky M., Okolie O., Hingtgen S.D., Kabanov A.V., Batrakova E.V. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655–664. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Sato Y.T., Umezaki K., Sawada S., Mukai S.A., Sasaki Y., Harada N., Shiku H., Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6:21933. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhang X., Zhang H., Gu J., Zhang J., Shi H., Qian H., Wang D., Xu W., Pan J., Santos H.A. Engineered extracellular vesicles for cancer therapy. Adv. Mater. 2021 doi: 10.1002/adma.202005709. [DOI] [PubMed] [Google Scholar]
- 340.Murphy D.E., de Jong O.G., Brouwer M., Wood M.J., Lavieu G., Schiffelers R.M., Vader P. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019;51:1–12. doi: 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Rana S., Yue S., Stadel D., Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012;44:1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 342.Laulagnier K., Javalet C., Hemming F.J., Chivet M., Lachenal G., Blot B., Chatellard C., Sadoul R. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 2018;75:757–773. doi: 10.1007/s00018-017-2664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.van Dongen H.M., Masoumi N., Witwer K.W., Pegtel D.M. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol. Mol. Biol. Rev. 2016;80:369–386. doi: 10.1128/MMBR.00063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M., Guo B., Evers B.M., Guo P. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Didiot M.C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R., Echeverria D., Raj L., Morrissey D.V., DiFiglia M., Aronin N., Khvorova A. Exosome-mediated delivery of hydrophobically modified siRNA for huntingtin mRNA silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Gao X., Ran N., Dong X., Zuo B., Yang R., Zhou Q., Moulton H.M., Seow Y., Yin H. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat0195. [DOI] [PubMed] [Google Scholar]
- 349.Armstrong J.P.K., Stevens M.M. Strategic design of extracellular vesicle drug delivery systems. Adv. Drug Deliv. Rev. 2018;130:12–16. doi: 10.1016/j.addr.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kooijmans S.A.A., Fliervoet L.A.L., van der Meel R., Fens M., Heijnen H.F.G., van Bergen En Henegouwen P.M.P., Vader P., Schiffelers R.M. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J. Contr. Release : official journal of the Controlled Release Society. 2016;224:77–85. doi: 10.1016/j.jconrel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 351.Christianson H.C., Svensson K.J., van Kuppevelt T.H., Li J.P., Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yerneni S.S., Lathwal S., Shrestha P., Shirwan H., Matyjaszewski K., Weiss L., Yolcu E.S., Campbell P.G., Das S.R. Rapid on-demand extracellular vesicle augmentation with versatile oligonucleotide tethers. ACS Nano. 2019;13:10555–10565. doi: 10.1021/acsnano.9b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Anderson S.D., Gwenin V.V., Gwenin C.D. Magnetic functionalized nanoparticles for biomedical, drug delivery and imaging applications. Nanoscale research letters. 2019;14:188. doi: 10.1186/s11671-019-3019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Colombo M., Carregal-Romero S., Casula M.F., Gutierrez L., Morales M.P., Bohm I.B., Heverhagen J.T., Prosperi D., Parak W.J. Biological applications of magnetic nanoparticles. Chem. Soc. Rev. 2012;41:4306–4334. doi: 10.1039/c2cs15337h. [DOI] [PubMed] [Google Scholar]
- 355.Jiang S., Eltoukhy A.A., Love K.T., Langer R., Anderson D.G. Lipidoid-coated iron oxide nanoparticles for efficient DNA and siRNA delivery. Nano Lett. 2013;13:1059–1064. doi: 10.1021/nl304287a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Toledano Furman N.E., Lupu-Haber Y., Bronshtein T., Kaneti L., Letko N., Weinstein E., Baruch L., Machluf M. Reconstructed stem cell nanoghosts: a natural tumor targeting platform. Nano Lett. 2013;13:3248–3255. doi: 10.1021/nl401376w. [DOI] [PubMed] [Google Scholar]
- 357.Dong H., Du S.R., Zheng X.Y., Lyu G.M., Sun L.D., Li L.D., Zhang P.Z., Zhang C., Yan C.H. Lanthanide nanoparticles: from design toward bioimaging and therapy. Chem. Rev. 2015;115:10725–10815. doi: 10.1021/acs.chemrev.5b00091. [DOI] [PubMed] [Google Scholar]
- 358.Idris N.M., Jayakumar M.K., Bansal A., Zhang Y. Upconversion nanoparticles as versatile light nanotransducers for photoactivation applications. Chem. Soc. Rev. 2015;44:1449–1478. doi: 10.1039/c4cs00158c. [DOI] [PubMed] [Google Scholar]
- 359.Xuan M., Shao J., Dai L., He Q., Li J. Macrophage cell membrane camouflaged mesoporous silica nanocapsules for in vivo cancer therapy. Advanced healthcare materials. 2015;4:1645–1652. doi: 10.1002/adhm.201500129. [DOI] [PubMed] [Google Scholar]
- 360.Yu Z., Zhou P., Pan W., Li N., Tang B. A biomimetic nanoreactor for synergistic chemiexcited photodynamic therapy and starvation therapy against tumor metastasis. Nat. Commun. 2018;9:5044. doi: 10.1038/s41467-018-07197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kroll A.V., Fang R.H., Jiang Y., Zhou J., Wei X., Yu C.L., Gao J., Luk B.T., Dehaini D., Gao W., Zhang L. Nanoparticulate delivery of cancer cell membrane elicits multiantigenic antitumor immunity. Adv. Mater. 2017;29 doi: 10.1002/adma.201703969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Mu X., Li J., Yan S., Zhang H., Zhang W., Zhang F., Jiang J. siRNA delivery with stem cell membrane-coated magnetic nanoparticles for imaging-guided photothermal therapy and gene therapy. ACS Biomater. Sci. Eng. 2018;4:3895–3905. doi: 10.1021/acsbiomaterials.8b00858. [DOI] [PubMed] [Google Scholar]
- 363.Wilson W.R., Hay M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Canc. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 364.Liu Y., Ji X., Tong W.W.L., Askhatova D., Yang T., Cheng H., Wang Y., Shi J. Engineering multifunctional RNAi nanomedicine to concurrently target cancer hallmarks for combinatorial therapy. Angew. Chem. 2018;57:1510–1513. doi: 10.1002/anie.201710144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Su Y., Zhao B., Zhou L., Zhang Z., Shen Y., Lv H., AlQudsy L.H.H., Shang P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Canc. Lett. 2020;483:127–136. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 366.Zhang Y., Fu X., Jia J., Wikerholmen T., Xi K., Kong Y., Wang J., Chen H., Ma Y., Li Z., Wang C., Qi Q., Thorsen F., Wang J., Cui J., Li X., Ni S. Glioblastoma therapy using codelivery of cisplatin and glutathione peroxidase targeting siRNA from iron oxide nanoparticles. ACS Appl. Mater. Interfaces. 2020;12:43408–43421. doi: 10.1021/acsami.0c12042. [DOI] [PubMed] [Google Scholar]
- 367.Sun Y.P., Zhou B., Lin Y., Wang W., Fernando K.A., Pathak P., Meziani M.J., Harruff B.A., Wang X., Wang H., Luo P.G., Yang H., Kose M.E., Chen B., Veca L.M., Xie S.Y. Quantum-sized carbon dots for bright and colorful photoluminescence. J. Am. Chem. Soc. 2006;128:7756–7757. doi: 10.1021/ja062677d. [DOI] [PubMed] [Google Scholar]
- 368.Mohammadinejad R., Dehshahri A., Sagar Madamsetty V., Zahmatkeshan M., Tavakol S., Makvandi P., Khorsandi D., Pardakhty A., Ashrafizadeh M., Ghasemipour Afshar E., Zarrabi A. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Contr. Release : official journal of the Controlled Release Society. 2020;325:249–275. doi: 10.1016/j.jconrel.2020.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Mohammadinejad R., Dadashzadeh A., Moghassemi S., Ashrafizadeh M., Dehshahri A., Pardakhty A., Sassan H., Sohrevardi S.M., Mandegary A. Shedding light on gene therapy: carbon dots for the minimally invasive image-guided delivery of plasmids and noncoding RNAs - a review. J. Adv. Res. 2019;18:81–93. doi: 10.1016/j.jare.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Kesharwani P., Iyer A.K. Recent advances in dendrimer-based nanovectors for tumor-targeted drug and gene delivery. Drug Discov. Today. 2015;20:536–547. doi: 10.1016/j.drudis.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wang L., Wang X., Bhirde A., Cao J., Zeng Y., Huang X., Sun Y., Liu G., Chen X. Carbon-dot-based two-photon visible nanocarriers for safe and highly efficient delivery of siRNA and DNA. Advanced healthcare materials. 2014;3:1203–1209. doi: 10.1002/adhm.201300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Liu C., Zhang P., Zhai X., Tian F., Li W., Yang J., Liu Y., Wang H., Wang W., Liu W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials. 2012;33:3604–3613. doi: 10.1016/j.biomaterials.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 373.Kim S., Choi Y., Park G., Won C., Park Y.-J., Lee Y., Kim B.-S., Min D.-H. Highly efficient gene silencing and bioimaging based on fluorescent carbon dots in vitro and in vivo. Nano Research. 2016;10:503–519. doi: 10.1007/s12274-016-1309-1. [DOI] [Google Scholar]
- 374.Sarid R., Gao S.J. Viruses and human cancer: from detection to causality. Canc. Lett. 2011;305:218–227. doi: 10.1016/j.canlet.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Ju E., Li T., Liu Z., da Silva S.R., Wei S., Zhang X., Wang X., Gao S.J. Specific inhibition of viral MicroRNAs by carbon dots-mediated delivery of locked nucleic acids for therapy of virus-induced cancer. ACS Nano. 2020;14:476–487. doi: 10.1021/acsnano.9b06333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.He X., Luo Q., Zhang J., Chen P., Wang H.J., Luo K., Yu X.Q. Gadolinium-doped carbon dots as nano-theranostic agents for MR/FL diagnosis and gene delivery. Nanoscale. 2019;11:12973–12982. doi: 10.1039/c9nr03988k. [DOI] [PubMed] [Google Scholar]
- 377.Qian J., Tang B.Z. AIE luminogens for bioimaging and theranostics: from organelles to animals. Inside Chem. 2017;3:56–91. doi: 10.1016/j.chempr.2017.05.010. [DOI] [Google Scholar]
- 378.Gao Y., Zheng Q.C., Xu S., Yuan Y., Cheng X., Jiang S., Kenry Q. Yu, Song Z., Liu B., Li M. Theranostic nanodots with aggregation-induced emission characteristic for targeted and image-guided photodynamic therapy of hepatocellular carcinoma. Theranostics. 2019;9:1264–1279. doi: 10.7150/thno.29101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Chen X., Gao H., Deng Y., Jin Q., Ji J., Ding D. Supramolecular aggregation-induced emission nanodots with programmed tumor microenvironment responsiveness for image-guided orthotopic pancreatic cancer therapy. ACS Nano. 2020;14:5121–5134. doi: 10.1021/acsnano.0c02197. [DOI] [PubMed] [Google Scholar]
- 380.Yuan Y., Zhang C.J., Liu B. A photoactivatable AIE polymer for light-controlled gene delivery: concurrent endo/lysosomal escape and DNA unpacking. Angew. Chem. 2015;54:11419–11423. doi: 10.1002/anie.201503640. [DOI] [PubMed] [Google Scholar]
- 381.Hu R., Yang C., Wang Y., Lin G., Qin W., Ouyan Q., Law W.-C., Nguyen Q.T., Yoon H.S., Wang X., Yong K.-T., Tang B.Z. Aggregation-induced emission (AIE) dye loaded polymer nanoparticles for gene silencing in pancreatic cancer and their in vitro and in vivo biocompatibility evaluation. Nano Research. 2014;8:1563–1576. doi: 10.1007/s12274-014-0642-5. [DOI] [Google Scholar]
- 382.Shi L., Wu W., Duan Y., Xu L., Xu Y., Hou L., Meng X., Zhu X., Liu B. Light-induced self-escape of spherical nucleic acid from endo/lysosome for efficient non-cationic gene delivery. Angew. Chem. 2020;59:19168–19174. doi: 10.1002/anie.202006890. [DOI] [PubMed] [Google Scholar]
- 383.Cheng Y., Sun C., Liu R., Yang J., Dai J., Zhai T., Lou X., Xia F. A multifunctional peptide-conjugated AIEgen for efficient and sequential targeted gene delivery into the nucleus. Angew. Chem. 2019;58:5049–5053. doi: 10.1002/anie.201901527. [DOI] [PubMed] [Google Scholar]
- 384.Shim M.S., Xia Y. A reactive oxygen species (ROS)-responsive polymer for safe, efficient, and targeted gene delivery in cancer cells. Angew. Chem. 2013;52:6926–6929. doi: 10.1002/anie.201209633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Wu J., Chen J., Feng Y., Tian H., Chen X. Tumor microenvironment as the "regulator" and "target" for gene therapy. J. Gene Med. 2019;21 doi: 10.1002/jgm.3088. [DOI] [PubMed] [Google Scholar]
- 386.Li Y., Gao J., Zhang C., Cao Z., Cheng D., Liu J., Shuai X. Stimuli-responsive polymeric nanocarriers for efficient gene delivery. Top. Curr. Chem. 2017;375:27. doi: 10.1007/s41061-017-0119-6. [DOI] [PubMed] [Google Scholar]
- 387.Jia L., Gao Y., Zhou T., Zhao X.L., Hu H.Y., Chen D.W., Qiao M.X. Enhanced response to PD-L1 silencing by modulation of TME via balancing glucose metabolism and robust co-delivery of siRNA/Resveratrol with dual-responsive polyplexes. Biomaterials. 2021;271:120711. doi: 10.1016/j.biomaterials.2021.120711. [DOI] [PubMed] [Google Scholar]
- 388.Liu X., Xiang J., Zhu D., Jiang L., Zhou Z., Tang J., Liu X., Huang Y., Shen Y. Fusogenic reactive oxygen species triggered charge-reversal vector for effective gene delivery. Adv. Mater. 2016;28:1743–1752. doi: 10.1002/adma.201504288. [DOI] [PubMed] [Google Scholar]
- 389.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Qiu N., Gao J., Liu Q., Wang J., Shen Y. Enzyme-responsive charge-reversal polymer-mediated effective gene therapy for intraperitoneal tumors. Biomacromolecules. 2018;19:2308–2319. doi: 10.1021/acs.biomac.8b00440. [DOI] [PubMed] [Google Scholar]
- 391.Hatakeyama H., Akita H., Ito E., Hayashi Y., Oishi M., Nagasaki Y., Danev R., Nagayama K., Kaji N., Kikuchi H., Baba Y., Harashima H. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 392.Huang S., Shao K., Kuang Y., Liu Y., Li J., An S., Guo Y., Ma H., He X., Jiang C. Tumor targeting and microenvironment-responsive nanoparticles for gene delivery. Biomaterials. 2013;34:5294–5302. doi: 10.1016/j.biomaterials.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 393.Li J., Ge Z., Liu S. PEG-sheddable polyplex micelles as smart gene carriers based on MMP-cleavable peptide-linked block copolymers. Chem Commun (Camb) 2013;49:6974–6976. doi: 10.1039/c3cc43576h. [DOI] [PubMed] [Google Scholar]
- 394.Liang K., Bae K.H., Lee F., Xu K., Chung J.E., Gao S.J., Kurisawa M. Self-assembled ternary complexes stabilized with hyaluronic acid-green tea catechin conjugates for targeted gene delivery. J. Contr. Release : official journal of the Controlled Release Society. 2016;226:205–216. doi: 10.1016/j.jconrel.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 395.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 396.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S., Theisen M., Hong S.J., Zhou J., Rajendran R., Levy B., Howell R., Besin G., Presnyak V., Sabnis S., Murphy-Benenato K.E., Kumarasinghe E.S., Salerno T., Mihai C., Lukacs C.M., Chandler R.J., Guey L.T., Venditti C.P., Martini P.G.V. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 2018;24:2520. doi: 10.1016/j.celrep.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 397.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., Shah R.R., Shah A., Ling D., Growe J., Pink M., Rohde E., Wood K.M., Salomon W.E., Harrington W.F., Dombrowski C., Strapps W.R., Chang Y., Morrissey D.V. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 398.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson O., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Kulkarni J.A., Cullis P.R., van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Therapeut. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 401.Zhao W., Hou X., Vick O.G., Dong Y. RNA delivery biomaterials for the treatment of genetic and rare diseases. Biomaterials. 2019;217:119291. doi: 10.1016/j.biomaterials.2019.119291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nature Reviews Materials. 2017;2 doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 403.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Wagner M.J., Mitra R., McArthur M.J., Baze W., Barnhart K., Wu S.Y., Rodriguez-Aguayo C., Zhang X., Coleman R.L., Lopez-Berestein G., Sood A.K. Preclinical mammalian safety studies of EPHARNA (DOPC nanoliposomal EphA2-targeted siRNA) Mol. Canc. Therapeut. 2017;16:1114–1123. doi: 10.1158/1535-7163.MCT-16-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Miller A.J., Chang A., Cunningham P.N. Chronic microangiopathy due to DCR-MYC, a myc-targeted short interfering RNA. Am. J. Kidney Dis. 2020;75:513–516. doi: 10.1053/j.ajkd.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 406.Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A.N., Omokoko T., Kranz L.M., Diken M., Kreiter S., Haas H., Attig S., Rae R., Cuk K., Kemmer-Bruck A., Breitkreuz A., Tolliver C., Caspar J., Quinkhardt J., Hebich L., Stein M., Hohberger A., Vogler I., Liebig I., Renken S., Sikorski J., Leierer M., Muller V., Mitzel-Rink H., Miederer M., Huber C., Grabbe S., Utikal J., Pinter A., Kaufmann R., Hassel J.C., Loquai C., Tureci O. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 407.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Phadke A.P., Jay C.M., Wang Z., Chen S., Liu S., Haddock C., Kumar P., Pappen B.O., Rao D.D., Templeton N.S., Daniels E.Q., Webb C., Monsma D., Scott S., Dylewski D., Frieboes H.B., Brunicardi F.C., Senzer N., Maples P.B., Nemunaitis J., Tong A.W. In vivo safety and antitumor efficacy of bifunctional small hairpin RNAs specific for the human Stathmin 1 oncoprotein. DNA Cell Biol. 2011;30:715–726. doi: 10.1089/dna.2011.1240. [DOI] [PubMed] [Google Scholar]
- 409.Sarker D., Plummer R., Meyer T., Sodergren M.H., Basu B., Chee C.E., Huang K.W., Palmer D.H., Ma Y.T., Evans T.R.J., Spalding D.R.C., Pai M., Sharma R., Pinato D.J., Spicer J., Hunter S., Kwatra V., Nicholls J.P., Collin D., Nutbrown R., Glenny H., Fairbairn S., Reebye V., Voutila J., Dorman S., Andrikakou P., Lloyd P., Felstead S., Vasara J., Habib R., Wood C., Saetrom P., Huber H.E., Blakey D.C., Rossi J.J., Habib N. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-alpha, in patients with advanced liver cancer: a first-in-human, multicenter, open-label, phase I trial. Clin. Canc. Res. : an official journal of the American Association for Cancer Research. 2020;26:3936–3946. doi: 10.1158/1078-0432.CCR-20-0414. [DOI] [PubMed] [Google Scholar]
- 410.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Klebowski B., Depciuch J., Parlinska-Wojtan M., Baran J. Applications of noble metal-based nanoparticles in medicine. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19124031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Li C., Wallace S. Polymer-drug conjugates: recent development in clinical oncology. Adv. Drug Deliv. Rev. 2008;60:886–898. doi: 10.1016/j.addr.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Yang W., Lee J.C., Chen M.H., Zhang Z.Y., Bai X.M., Yin S.S., Cao K., Wang S., Wu W., Yan K. Thermosensitive liposomal doxorubicin plus radiofrequency ablation increased tumor destruction and improved survival in patients with medium and large hepatocellular carcinoma: a randomized, double-blinded, dummy-controlled clinical trial in a single center. J. Canc. Res. Therapeut. 2019;15:773–783. doi: 10.4103/jcrt.JCRT_801_18. [DOI] [PubMed] [Google Scholar]
- 414.Stern J.M., Kibanov Solomonov V.V., Sazykina E., Schwartz J.A., Gad S.C., Goodrich G.P. Initial evaluation of the safety of nanoshell-directed photothermal therapy in the treatment of prostate disease. Int. J. Toxicol. 2016;35:38–46. doi: 10.1177/1091581815600170. [DOI] [PubMed] [Google Scholar]
- 415.Bayat Mokhtari R., Homayouni T.S., Baluch N., Morgatskaya E., Kumar S., Das B., Yeger H. Combination therapy in combating cancer. Oncotarget. 2017;8:38022–38043. doi: 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Wei X., Wang Y., Xiong X., Guo X., Zhang L., Zhang X., Zhou S. Codelivery of a π-π stacked dual anticancer drug combination with nanocarriers for overcoming multidrug resistance and tumor metastasis. Adv. Funct. Mater. 2016;26:8266–8280. doi: 10.1002/adfm.201603336. [DOI] [Google Scholar]
- 417.Zhu L., Guo Y., Qian Q., Yan D., Li Y., Zhu X., Zhang C. Carrier-free delivery of precise drug-chemogene conjugates for synergistic treatment of drug-resistant cancer. Angew. Chem. 2020 doi: 10.1002/anie.202006895. [DOI] [PubMed] [Google Scholar]
- 418.Gong X., Li R., Wang J., Wei J., Ma K., Liu X., Wang F. A smart theranostic nanocapsule for spatiotemporally programmable photo-gene therapy. Angew. Chem. 2020 doi: 10.1002/anie.202008413. [DOI] [PubMed] [Google Scholar]
- 419.Correale P., Saladino R.E., Giannarelli D., Giannicola R., Agostino R., Staropoli N., Strangio A., Del Giudice T., Nardone V., Altomonte M., Pastina P., Tini P., Falzea A.C., Imbesi N., Arcati V., Romeo G., Caracciolo D., Luce A., Caraglia M., Giordano A., Pirtoli L., Necas A., Amler E., Barbieri V., Tassone P., Tagliaferri P. Distinctive germline expression of class I human leukocyte antigen (HLA) alleles and DRB1 heterozygosis predict the outcome of patients with non-small cell lung cancer receiving PD-1/PD-L1 immune checkpoint blockade. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Kashyap A.S., Thelemann T., Klar R., Kallert S.M., Festag J., Buchi M., Hinterwimmer L., Schell M., Michel S., Jaschinski F., Zippelius A. Antisense oligonucleotide targeting CD39 improves anti-tumor T cell immunity. J Immunother Cancer. 2019;7:67. doi: 10.1186/s40425-019-0545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]