Abstract
Haptophytes are biogeochemically and industrially important protists with underexplored genomic diversity. We present a nuclear genome assembly for the class Pavlovales, which was assembled with PacBio long-read data into highly contiguous sequences. We sequenced strain Diacronema lutheri NIVA-4/92, formerly known as Pavlova lutheri, because it has established roles in aquaculture and has been a key organism for studying microalgal lipid biosynthesis. Our data show that D. lutheri has the smallest and most streamlined haptophycean genome assembled to date, with an assembly size of 43.503 Mb and 14,446 protein-coding genes. Together with its high nuclear GC content, Diacronema is an important genus for investigating selective pressures on haptophyte genome evolution, contrasting with the much larger and more repetitive genome of the coccolithophore Emiliania huxleyi. The D. lutheri genome will be a valuable resource for resolving the genetic basis of algal lipid biosynthesis and metabolic remodeling that takes place during adaptation and stress response in natural and engineered environments.
Keywords: protist, haptophyte, lipid metabolism, biotechnology, PacBio sequencing
Significance
Haptophytes are evolutionarily significant protists, yet they are underrepresented in genomic studies and several clades have no available genomes. We used third generation long-read sequencing to assemble a high-contiguity genome for Diacronema lutheri and provide an initial investigation of the nuclear genome architecture of the class Pavlovales. Our results are of value for understanding the evolution of haptophytes, algal lipid metabolism, and for strain improvement in biotechnology.
Introduction
Haptophytes comprise a major proportion of the phytoplankton community that globally have large-scale impacts on carbon cycling and ocean biogeochemistry (Liu et al. 2009; Gutowska et al. 2017; Heureux et al. 2017). They include over 300 characterized species, with hundreds more strains detected in ocean metabarcoding studies (Kim et al. 2011; Gran‐Stadniczeñko et al. 2017). The genus Tisochrysis and Pavlova (Diacronema) are especially valuable for the aquaculture and biotechnology industries, where they supply food and essential lipids for farmed fish and shellfish (Shah et al. 2018). Further developments in large-scale microalgae cultivation could expand the production of sustainable foods, oils, and plastic replacement materials in the future (Cottrell et al. 2020; Naduthodi et al. 2021).
Despite their impact and their intriguing evolutionary history, high-quality haptophyte genome sequences remain scarce, and the available data do not reflect their diversity or new discoveries (Burki et al. 2012; Sibbald and Archibald 2017; Kawachi et al. 2021). To bridge this knowledge gap, we assembled and annotated a high-quality genome for Diacronema lutheri (Pavlova lutheri), for the purpose of understanding its architecture, sequence evolution, and capacity to synthesize diverse natural products, particularly lipids.
The Pavlovales, which include the genus Pavlova, Diacronema, Rebecca, and Exanthemachrysis, invariably occupy the outer branches of haptophyte phylogenies, some considerable evolutionary distance from the coccolithophores, Chrysochromulina and Phaeocystis clades (Egge et al. 2015; Edvardsen et al. 2016; Song et al. 2021). Pavlova sp. cells are about five microns in size with a short haptonema, their swimming motion driven by two flagella of slightly unequal length. They are unusual among microalgae as a combined source of eicosapentaenoic acid (20:5n−3) and docosahexaenoic acid (22:6n−3) that are integrated into different types of structural and storage lipids (Meireles et al. 2003). Pavlova sp. also synthesize additional betaine lipids including 1,2-diacylglyceryl-3-O-carboxyhydroxymethylcholine (DGCC) and 1,2-diaclyglyceryl-3-O-2′-(hydroxmethyl)-(N, N, N-trimethyl)-β-alanine (DGTA), plus some unusual dihydroxylated sterols (pavlovols) that are unique to the class (Volkman 2016; Li-Beisson et al. 2019; Marcellin-Gros et al. 2020).
A few studies have amplified and sequenced individual Pavlova sp. genes (Tonon et al. 2003; Robert et al. 2009), but the majority of the nuclear genome remains unexplored. Nosenko et al. (2007) used pulsed-field electrophoresis to estimate a modest genome size of 20.7 and 28.7 Mb for Diacronema sp. and Pavlova gyrans, respectively, so we expected a comparable result. Here, we assembled the genome of strain “Pavlova sp. NIVA-4/92,” which we identify as D. lutheri (synonymous with P. lutheri) based on its mitochondrial, plastid, and 18S sequences (Hulatt et al. 2020). We primarily used long PacBio reads at high coverage with the aim to comprehensively unravel key biosynthetic pathways, resolve evolutionary relationships among genes, and determine mechanisms controlling triacylglycerol biosynthesis and adaptive lipid remodeling (Cañavate et al. 2017; Wei et al. 2017). Our data will also be valuable for applied studies of genome-informed strain improvement and models of cell metabolic flux.
Results and Discussion
Genome Size and Quality
The D. lutheri nuclear genome assembly is 43,502,671 bp in size and contains 14,446 annotated protein-coding genes, making it the most compact among sequenced haptophytes (table 1). The assembly consists of 103 contigs, with approximately half of the total sequence length contained in 16 contigs. These high-contiguity sequences reflect the advances in long-read sequencing technology compared with earlier Illumina and 454-based methods. The theoretical coverage of the PacBio reads is ×368, and in practice the nuclear genome coverage is most commonly about ×315. The genome size is 1.5–2 times larger than that previously predicted for other related Pavlovales, but the contig lengths are within the expected chromosome size range of 0.18–4 Mb (Nosenko et al. 2007). Analysis of the genome sequence with BUSCO v.4 identified 80.8% of core eukaryotic genes were complete and only 0.4% of these were duplicated (table 1). Compared with the other haptophyte assemblies, these scores support a rather complete genome with minimal sequence duplication.
Table 1.
Comparison of Four Published Haptophyte Genomes with Diacronema lutheri NIVA-4/92.
| Diacronema lutheri | Tisochrysis lutea | Chrysochromulina tobin | Chrysochromulina parva | Emiliania huxleyi | |
|---|---|---|---|---|---|
| JAGTXO010000000 | TisoV1 | GCA_001275005.1 | GCA_002887195.1 | GCF_000372725.1 | |
| Assembly length (contigs) (Mb) | 43.503 | 57.719 | 59.073 | 65.765 | 155.931 |
| Total number contigs | 103 | 9,930 | 3,412 | 8,362 | 16,921 |
| Scaffolds | — | 7,695 | — | — | 7,795 |
| Contig N50 | 16 | 1,970 | 798 | 1,243 | 1,314 |
| Contig L50 (kb) | 852.26 | 8.07 | 24.11 | 16.05 | 29.72 |
| Longest contig | 3.042 Mb | 726.925 kb | 121.428 kb | 101.752 kb | 299.609 kb |
| GC content (± contig) | 73.25% | 58.67% | 63.37% | 63.58% | 65.67% |
| ±1.34% | ±2.95% | ±2.74% | ±3.99% | ±4.13% | |
| Method | PacBio + Illumina | Illumina | Illumina + 454 | Illumina | Sanger |
| Complete (%) | 80.80 | 68.30 | 62.00 | 72.90 | 51.80 |
| Complete, single copy (%) | 80.40 | 65.90 | 61.60 | 72.50 | 37.30 |
| Complete, duplicated (%) | 0.40 | 2.40 | 0.40 | 0.40 | 14.50 |
| Fragmented (%) | 6.30 | 11.40 | 7.50 | 7.10 | 16.10 |
| Missing | 12.90% | 20.30% | 30.50% | 20.00% | 32.10% |
| Genesa | 14,446 | 20,582 | 16,777 | 28,138 | 30,569 |
| Annotation method | BRAKER2 | MAKER2 | MAKER2 | MAKER2 | JGI Annotation Pipeline |
| Reference | This study | Carrier et al. (2018) | Hovde et al. (2015) | Hovde et al. (2019) | Read et al. (2013) |
Note.—Assembly statistics are based on contigs for comparability. BUSCO v.4 was run on the genome sequences with the “eukaryote_odb10” data set.
Structural annotation of genes are as reported in the corresponding manuscripts, which were annotated with different methods.
The D. lutheri nuclear genome assembly has a high 73.25% GC content that is reflected in the raw PacBio subreads and in the Illumina reads (supplementary figs. 1 and 2, Supplementary Material online). It surpasses that of the coccolithophore Emiliania huxleyi (65.67%) and is among the highest observed in eukaryotic cells. Understanding the selective mechanisms driving this elevated GC skew might help explain patterns in haptophyte evolution, and could also support detection of cryptic picoplanktonic haptophytes in metagenomes (Liu et al. 2009; Edvardsen et al. 2016).
Repetitive Elements
Approximately 22.9% of the D. lutheri genome is repetitive, substantially less than the E. huxleyi genome, of which 64% was classified as repeats (Read et al. 2013). Long-terminal repeats (LTRs) and secondarily long-interspersed terminal repeats (LINEs) comprised the majority of the annotated repeat elements in D. lutheri, representing 32.7% and 3.8% of masked bases, respectively (supplementary table 1, Supplementary Material online).
Gene Annotations
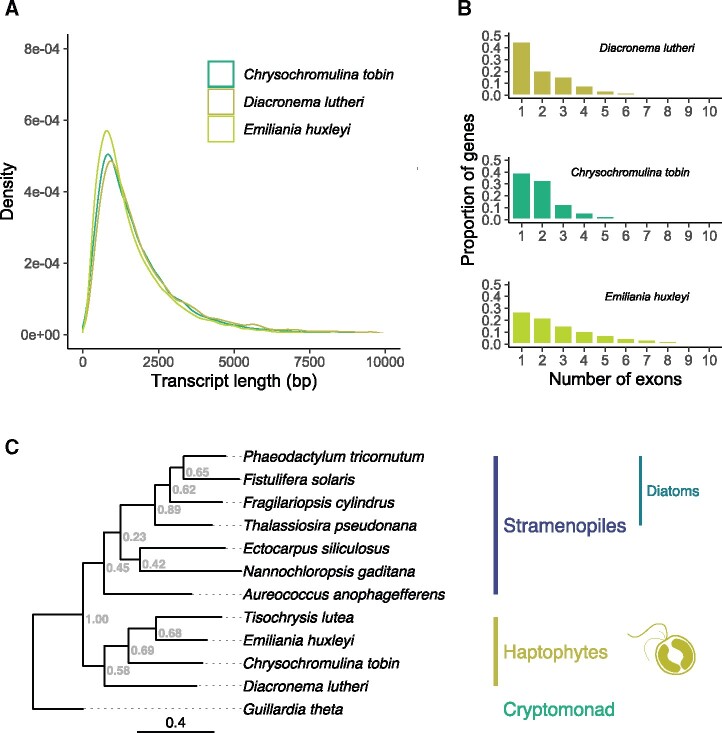
The total length of protein-coding nucleotides is 26.62 Mb which represents 61.2% of the genome. The gene length and exon counts of D. lutheri were compared with the structural annotations of E. huxleyi and Chrysochromulina tobin (fig. 1A and B). Single-exon genes account for 45% of the D. lutheri coding sequences, with fewer genes containing a single intron (21%) or multiple introns (34%). The C. tobin genome encodes fewer single-exon genes whereas E. huxleyi, with the largest sequenced genome, encodes only 27% single-exon genes, with 50% of genes containing two or more introns. Such variation raises questions on patterns of genome-wide intron gain and loss in haptophytes, and the extent to which posttranscriptional regulation by alternative splicing is prevalent across different clades.
Fig. 1.
(A) Gene transcript length distributions for Diacronema lutheri annotated in this study compared with two other haptophytes, Emiliania huxleyi and Chrysochromulina tobin. (B) The number of exons per gene for the same three genomes, expressed as a proportion of the total number of annotated genes. (C) The species tree of 12 protists bearing red alga derived plastids, including four sequenced haptophytes, seven stramenopiles, and one cryptomonad. The tree is derived from 1,152 orthogroups with at least one gene copy from each species. Branch lengths represent substitutions per site and support values are derived from the STAG algorithm implemented in OrthoFinder2.
In total 9,498 of the 14,446 protein-coding genes received at least one gene ontology (GO) identification. An initial survey of genes related to lipid metabolism identified 25 proteins with annotated desaturase activity, six with elongase activity, and 54 with acyltransferase functions. Thirty-seven tRNAs decoding the standard 20 amino acids were annotated by tRNAscan-SE, and an additional 42 ncRNAs were annotated by Infernal/Rfam.
Gene Orthology Comparative Genomics
The amino acid sequences from the whole genome of D. lutheri were compared with those of three available haptophyte data sets, and with eight further data sets from more distantly related red-plastid bearing species, using OrthoFinder2 (Emms & Kelly 2019). From the four haptophyte sequence sets and a total of 89,703 genes, 749 orthogroups contained single-copy genes, whereas 3,438 orthogroups contained at least one gene ortholog from each species. For the expanded 12 species set of amino acid sequences, a total of 213,864 genes were distributed among 25,757 orthogroups, of which 1,152 orthogroups were common to all 12 genomes, but only five were single-copy orthologs found in all organisms. Figure 1C displays the species-tree using the amino acid sequences from the 12 genomes.
Conclusions
We assembled a new haptophyte genome for the class Pavlovales with long PacBio reads to build high contiguity sequences. The genome size, gene, and GC content of D. lutheri places the Pavlovales as an important clade for understanding selective processes and genome streamlining among ecologically and biogeochemically important haptophytes. Our results will be more fully exploited through investigation of lipid metabolism, metabolic modeling, and strain improvement for industrial bioprocesses.
Materials and Methods
Cell Culture Preparation
Strain “Pavlova sp. NIVA-4/92” was obtained from the Norwegian Culture Collection of Algae (NORCCA). This species reportedly originates from Oslofjord, Norway, and has been held in culture since 1989. Cells were cultivated in f/2 medium (Guillard and Ryther 1962) using 0.2 µm filtered and autoclaved seawater containing the antibiotics ampicillin, kanamycin, and streptomycin. Clonal cultures were obtained by cell-sorting with an Astrios EQ flow cytometer (University of Colorado Cancer Center, Denver, CO). Cell cultures were prepared in 500 ml bioreactors bubbled with filtered air containing 1% CO2. In the late exponential phase, the cells were collected and pelleted by centrifugation, then flash-frozen in liquid N2 and stored at −80 °C.
DNA Sequencing
High molecular weight DNA was extracted from cells and the fragments were size selected at over 30 kb by Arizona Genomics Institute (Tucson, AZ). After SMRTbell library preparation, sequencing was performed on a PacBio Sequel system using three 1M SMRT cells with v2.1 chemistry and 10 h movies. The raw data were processed with the command line tools from SMRTLink v.5.1 and the total yield was 993,273 subreads (16.6 Gb) with N50 length 23.458 kb. The longest subread was 92.675 kb. The GC content of subreads initially indicated a nuclear genome with a high GC content of ∼70% (supplementary fig. 1, Supplementary Material online). Short-read sequencing was performed with an Illumina MiSeq producing 250 bp paired-end reads on one v.2 flow cell, yielding 6.55 Gb of reads. The Illumina sequences were trimmed with Trimmomatic (Bolger et al. 2014) and quality checked by FastQC (Babraham Bioinformatics).
RNA Samples and Sequencing
Three independent RNA sequence libraries were obtained from two different experiments. Experiment 1 comprised a set of pooled Erlenmeyer flask cultures exposed to six different stress conditions (control, low-nutrient, low-temperature, low salinity, darkness, high light) to express the maximum number of genes. Experiment 2 was a bioreactor study from which two representative RNA samples (one control and one phosphorus-limited treatment) were selected. In each case, RNA was extracted from cell pellets using Trizol reagent and chloroform, followed by an RNA Clean & Concentrate mini-column preparation (Zymo Research, Irvine, CA). Illumina sequencing was performed by Novogene (Beijing, China) Ltd, yielding 21.4, 28.4 and 20.0 million cleaned and trimmed 150 bp paired-end strand-specific reads (fragments) from each of the three libraries, respectively.
Genome Assembly and Polishing
The PacBio data were used for genome assembly, whereas the Illumina sequences were used only for polishing the assembled contigs. PacBio subreads were assembled with CANU v.1.7 and the options “minReadLength = 3,000” “corOutCoverage = 100” “correctedErrorRate = 0.04” (Koren et al. 2017). The organelle sequences were identified and extracted from the whole genome assembly and finished separately (Hulatt et al. 2020). To minimize errors in the assembled sequences the contigs were polished three times with the PacBio command line tools in SMRTLink v.5.1 (Pacific Biosciences, Menlo Park, CA), where the raw subreads were aligned to the contigs with BLASR (Chaisson and Tesler 2012) and polished with the ARROW hidden Markov model to high consensus accuracy. Next, the 250-bp PE Illumina reads were used for final polishing to eliminate potential remaining small indels and single base errors. To do this the Illumina reads were aligned to the contigs using BWA-MEM (Li 2013), polished using Pilon for three rounds (Walker et al. 2014), and subsequently polished using FreeBayes (Garrison and Marth 2012) for three rounds. Genome coverage by the Illumina data was approximately 150-fold on an average.
Genome Curation
The initial assembly contained 136 contigs with a total length of 45.09 MB. To objectively identify and remove potentially erroneous, short or duplicated sequences derived from low-abundance or contaminant reads, the PurgeHaplotigs pipeline was applied (Roach et al. 2018). Raw PacBio subreads were mapped to the genome using Minimap2 with the options “-ax map-pb” (Li 2018) and spurious contigs were removed by defining lower, mid, and upper coverage limits. This process eliminated 33 relatively short sequences of total length 1.6 Mb and average length 48 kb, or about twice the N50 read length. The curated assembly was finally assessed for possible remaining contamination using BLAST against the “nt” database followed by manual inspection of top hits, but no further contigs were removed.
Genome Quality Assessment
Genome quality was monitored through the assembly and curation process using BUSCO (Seppey et al. 2019) and results presented in this manuscript are from BUSCO v.4.0.2 and the “eukaryote_odb10” collection of 255 conserved core eukaryotic genes. For comparative purposes, BUSCO was run on the genome sequences of D. lutheri and four other haptophyte assemblies with the optimized “–long” two-pass option and otherwise default BLAST settings.
Structural Annotation of Genes
To characterize repetitive regions a custom repeat library was constructed de novo using REPEATMODELER with all contigs over 100 kb (Smit et al. 2015a). The genome sequence was soft-masked using REPEATMASKER with the option “-xsmall” (Smit et al. 2015b). Gene structural annotation was subsequently performed with the BRAKER2 pipeline using RNA-seq evidence combined with AUGUSTUS and GENEMARK-ES for gene prediction (Bruna et al. 2021). The three RNA-seq libraries were aligned individually to the genome using STAR v.2.7.3a (Dobin et al. 2013) (supplementary table 2, Supplementary Material online) and the braker.pl pipeline was provided the RNA-seq read alignments and run with the “–softmasking” option.
Functional Annotation of Genes
To assign functions to the protein-coding sequences three different methods were used in parallel and the consensus results were collected: 1) INTERPROSCAN-5 was used to search for conserved protein signatures (Jones et al. 2014), 2) Protein sequences were searched with BlastP against the curated SwissProt database (Boeckmann et al. 2003), and 3) EGGNOG-MAPPER was used for orthology assignment, running emapper.py with DIAMOND alignment (Huerta-Cepas et al. 2017). Transfer RNAs were annotated with tRNAscan-SE v.2.0.7 with recommended settings for eukaryote genome annotation (Chan and Lowe 2019). Noncoding RNAs were annotated with INFERNAL (Nawrocki and Eddy 2013) and the Rfam library of covariance models (Kalvari et al. 2018).
Genome Sequences and Gene Orthology
Three haptophyte genome data sets were obtained from NCBI GenBank (E. huxleyi assembly GCA_000372725.1; C. tobin assembly GCA_001275005.1; Chrysochromulina parva assembly GCA_002887195.1) and one data set for Tisochrysis lutea was obtained from SEANOE (assembly v1; https://www.seanoe.org/data/00361/47171/; last accessed April 6, 2021; doi:10.17882/47171). Amino acid coding sequences for a further eight species were obtained from NCBI GenBank (Thalassiosira pseudonana CCMP1335 GCA_000149405.2; Phaeodactylum tricornutumGCA_000150955.2; Aureococcus anophagefferensGCA_000186865.1; Ectocarpus siliculosusGCA_000310025.1; Guillardia theta CCMP2712 GCA_000315625.1; Nannochloropsis gaditana B-31 GCA_000569095.1; Fragilariopsis cylindrusGCA_001750085.1; Fistulifera solarisGCA_002217885.1).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by a Marie Skłodowska-Curie Individual Fellowship to C.J.H. (Grant No. 749910) under the European Union’s Horizon 2020 research and innovation program. The project was hosted by the Posewitz lab at Colorado School of Mines, Golden, CO and the authors are thankful for support by the National Center for Genome Resources (NCGR) New Mexico and UNINETT Sigma-2 Compute Infrastructure (Norway) (Project No. NN9634K).
Data Availability
The Diacronema lutheri genome assembly is available in NCBI GenBank via BioProject number PRJNA725470. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAGTXO000000000. The version described in this article is version JAGTXO010000000. DNA sequence reads are deposited in the SRA under BioSample accession SAMN18879650. The genome assembly is also hosted at Dryad: https://doi.org/10.5061/dryad.5qfttdz55.
Literature Cited
- Boeckmann B, et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31(1):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M.. 2021. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3(1):lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert J-F, Keeling PJ.. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 279(1736):2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañavate JP, Armada I, Hachero‐Cruzado I.. 2017. Interspecific variability in phosphorus‐induced lipid remodelling among marine eukaryotic phytoplankton. New Phytol. 213(2):700–713. [DOI] [PubMed] [Google Scholar]
- Carrier G, et al. 2018. Draft genomes and phenotypic characterization of Tisochrysis lutea strains. Toward the production of domesticated strains with high added value. Algal Res. 29:1–11. [Google Scholar]
- Chaisson MJ, Tesler G.. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Kollmar M, editor. Gene Prediction New York: Humana. p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell RS, Blanchard JL, Halpern BS, Metian M, Froehlich HE.. 2020. Global adoption of novel aquaculture feeds could substantially reduce forage fish demand by 2030. Nat Food. 1(5):301–308. [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardsen B, Egge ES, Vaulot D.. 2016. Diversity and distribution of haptophytes revealed by environmental sequencing and metabarcoding – a review. Perspect Phycol. 3(2):77–91. [Google Scholar]
- Egge ES, et al. 2015. Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high‐throughput sequencing. Mol Ecol. 24(12):3026–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelley S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907.
- Gran-Stadniczeñko S, Šupraha L, Egge ED, Edvardsen B.. 2017. Haptophyte diversity and vertical distribution explored by 18S and 28S ribosomal RNA gene metabarcoding and scanning electron microscopy. J Eukaryot Microbiol. 64(4):514–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RR, Ryther JH.. 1962. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol. 8:229–239. [DOI] [PubMed] [Google Scholar]
- Gutowska MA, et al. 2017. Globally important haptophyte algae use exogenous pyrimidine compounds more efficiently than thiamin. MBio. 8(5):e01459–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heureux AM, et al. 2017. The role of Rubisco kinetics and pyrenoid morphology in shaping the CCM of haptophyte microalgae. J Exp Bot. 68(14):3959–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovde BT, et al. 2015. Genome sequence and transcriptome analyses of Chrysochromulina tobin: metabolic tools for enhanced algal fitness in the prominent order Prymnesiales (Haptophyceae). PLoS Genet. 11(9):e1005469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovde BT, et al. 2019. Chrysochromulina: genomic assessment and taxonomic diagnosis of the type species for an oleaginous algal clade. Algal Res. 37:307–319. [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 34(8):2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulatt CJ, Wijffels RH, Viswanath K, Posewitz MC.. 2020. The complete mitogenome and plastome of the haptophyte Pavlova lutheri NIVA-4/92. Mitochondrial DNA B Resour. 5(3):2748–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, et al. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46(D1):D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M, et al. 2021. Rappemonads are haptophyte phytoplankton. Curr Biol. 31(11):2395–2403. [DOI] [PubMed] [Google Scholar]
- Kim E, et al. 2011. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci U S A. 108(4):1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997.
- Li H.2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Thelen J, Fedosejevs E, Harwood JL.. 2019. The lipid biochemistry of eukaryotic algae. Prog Lipid Res. 74:31–68. [DOI] [PubMed] [Google Scholar]
- Liu H, et al. 2009. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci U S A. 106(31):12803–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellin-Gros R, Piganeau G, Stien D.. 2020. Metabolomic insights into marine phytoplankton diversity. Marine Drugs. 18(2):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles LA, Guedes AC, Malcata FX.. 2003. Lipid class composition of the microalga Pavlova lutheri: eicosapentaenoic and docosahexaenoic acids. J Agric Food Chem. 51(8):2237–2241. [DOI] [PubMed] [Google Scholar]
- Naduthodi MIS, Claassens NJ, D’Adamo S, van der Oost J, Barbosa MJ.. 2021. Synthetic biology approaches to enhance microalgal productivity. Trends Biotechnol. doi: 10.1016/j.tibtech.2020.12.010. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T, Boese B, Bhattacharya D.. 2007. Pulsed-field gel electrophoresis analysis of genome size and structure in Pavlova gyrans and Diacronema sp. (Haptophyta). J Phycol. 43(4):763–767. [Google Scholar]
- Read BA, et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499(7457):209–213. [DOI] [PubMed] [Google Scholar]
- Roach MJ, Schmit SA, Borneman AR.. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert SS, et al. 2009. Isolation and characterisation of a Δ5-fatty acid elongase from the marine microalga Pavlova salina. Mar Biotechnol (NY). 11(3):410–418. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Shah MR, et al. 2018. Microalgae in aquafeeds for a sustainable aquaculture industry. J Appl Phycol. 30(1):197–213. [Google Scholar]
- Sibbald SJ, Archibald JM.. 2017. More protist genomes needed. Nat Ecol Evol. 1(5):145–143. [DOI] [PubMed] [Google Scholar]
- Smit AFAHubley R.. RepeatModeler Open-1.0. 2008–2015. Available from: https://www.repeatmasker.org/faq.html. Accessed July 2021. [Google Scholar]
- Smit AFAHubley R, Green P.. RepeatMasker Open-4.0. 2013–2015. Available from: http://www.repeatmasker.org. Accessed July 2021. [Google Scholar]
- Song H, Chen Y, Liu F, Chen N.. 2021. Large differences in the haptophyte Phaeocystis globosa mitochondrial genomes driven by repeat amplifications. Front Microbiol. 12:676447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon T, Harvey D, Larson TR, Graham IA.. 2003. Identification of a very long chain polyunsaturated fatty acid Δ4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 553(3):440–444. [DOI] [PubMed] [Google Scholar]
- Volkman JK.2016. Sterols in microalgae. In: Borowitzka MA, Beardall J, Raven JA, editors. The Physiology of Microalgae. Cham, Switzerland: Springer International Publishing. p. 485–505. [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, et al. 2017. A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol Biofuels. 10:174–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Diacronema lutheri genome assembly is available in NCBI GenBank via BioProject number PRJNA725470. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAGTXO000000000. The version described in this article is version JAGTXO010000000. DNA sequence reads are deposited in the SRA under BioSample accession SAMN18879650. The genome assembly is also hosted at Dryad: https://doi.org/10.5061/dryad.5qfttdz55.