Graphical abstract
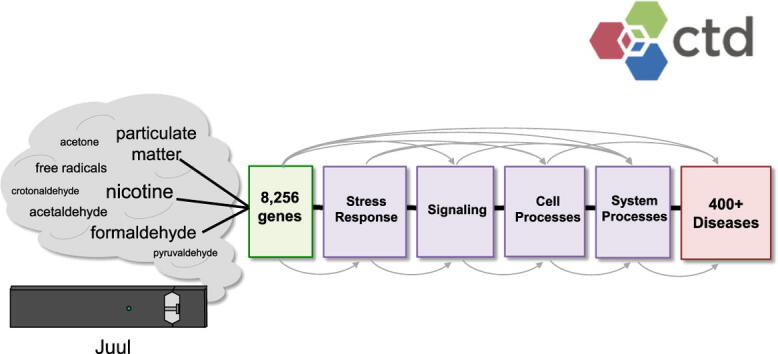
Abbreviations: A, acetaldehyde; AC, acetone; C, crotonaldehyde; CGPD, chemical-gene-phenotype-disease; COPD, chronic obstructive pulmonary disease; Cr, chromium; CTD, Comparative Toxicogenomics Database; F, formaldehyde; FR, free radicals; M, marker/mechanism relationship; MIE, molecular initiating event; MOA, mode-of-action; Mn, manganese; N, nicotine; NAFFCAPP, nicotine, acetaldehyde, formaldehyde, free radicals, crotonaldehyde, acetone, pyruvaldehyde, and particulate matter chemical mixture; NAFP, nicotine, acetaldehyde, formaldehyde, particulate matter chemical mixture; nAChR, nicotinic acetylcholine receptor; Ni, nickel; P, pyruvaldehyde; Pb, lead; PM, particulate matter; ROS, reactive oxygen species; Zn, zinc
Keywords: E-cigarettes, Vaping, Juul, Respiratory disease, Database, Environmental exposure
Highlights
-
•
Vaping chemicals of Juul aerosols were analyzed using CTD.
-
•
CTD can be used to predict disease associations of Juul aerosol chemicals.
-
•
Predictive pathways that relate vaping aerosols to diseases are described.
Abstract
There is a critical need to understand the health risks associated with vaping e-cigarettes, which has reached epidemic levels among teens. Juul is currently the most popular type of e-cigarette on the market. Using the Comparative Toxicogenomics Database (CTD; http://ctdbase.org), a public resource that integrates chemical, gene, phenotype and disease data, we aimed to analyze the potential molecular mechanisms of eight chemicals detected in the aerosols generated by heating Juul e-cigarette pods: nicotine, acetaldehyde, formaldehyde, free radicals, crotonaldehyde, acetone, pyruvaldehyde, and particulate matter. Curated content in CTD, including chemical-gene, chemical-phenotype, and chemical-disease interactions, as well as associated phenotypes and pathway enrichment, were analyzed to help identify potential molecular mechanisms and diseases associated with vaping. Nicotine shows the most direct disease associations of these chemicals, followed by particulate matter and formaldehyde. Together, these chemicals show a direct marker or mechanistic relationship with 400 unique diseases in CTD, particularly in the categories of cardiovascular diseases, nervous system diseases, respiratory tract diseases, cancers, and mental disorders. We chose three respiratory tract diseases to investigate further, and found that in addition to cellular processes of apoptosis and cell proliferation, prioritized phenotypes underlying Juul-associated respiratory tract disease outcomes include response to oxidative stress, inflammatory response, and several cell signaling pathways (p38MAPK, NIK/NFkappaB, calcium-mediated).
1. Introduction
Though electronic cigarettes were introduced as aids for smoking cessation more than a decade ago (Dinardo and Rome, 2019), youth-targeted flavors, packaging and marketing have contributed to epidemic levels of teen vaping in recent years (Farzal et al., 2019). Health consequences of exposure to vaping chemicals and molecular mechanisms underlying vaping-related illnesses are largely unknown.
Juul e-cigarettes are a compact, closed system nicotine-delivery device that were introduced in 2015 and quickly became the most popular vaping device with year-over-year growth of nearly 700% and a 50% market share (Kavuluru et al., 2019), prompting concern that their popularity among youth is a public health crisis (Walley et al., 2019). Because of their dominant market position and popularity among youth, we chose Juul e-cigarettes as the primary subject of this analysis. We set out to investigate the gene interactions, phenotype, pathway, and disease associations of chemicals detected in aerosols generated by heating Juul e-cigarettes connected to a digital puffing machine (Talih et al., 2019) or a Human Puff Profile Cigarette Smoking Machine (Reilly et al., 2019). Chemicals detected by these two methods include nicotine, acetaldehyde, formaldehyde, free radicals, crotonaldehyde, acetone, pyruvaldehyde, and particulate matter. Additional evidence for the presence of nicotine, acetaldehyde, formaldehyde, reactive oxygen species, acetone, pyruvaldehyde and particulate matter have also recently been described in Juul aerosols (Muthumalage et al., 2019, Mallock et al., 2020, Azimi et al., 2021). Concentrations of these chemicals in Juul emissions are listed in Table 1.
Table 1.
Concentrations of Chemicals in Juul Emissions.
| mean per 15 puffs (Talih et al., 2019) | range per puff (Reilly et al., 2019) | |
|---|---|---|
| Nicotine, mg | 2.07 | 0.154 – 0.188 |
| Acetaldehyde, μg | 6.05 | not detected |
| Formaldehyde, μg | 0.56 | 0.12 – 0.26 |
| Free Radicals, pmol | not assessed | 5.47 – 6.13 |
| Crotonaldehyde, μg | 0.85 | not assessed |
| Acetone, μg | 24.9 | 0.17 – 0.22 |
| Pyruvaldehyde, μg | 0.95 | not assessed |
| Particulate Matter, mg | 38.9 | not assessed |
CTD is a public scientific resource, wherein PhD-level scientists manually curate the scientific literature for data on chemicals, genes and diseases and integrate this data with select public data sets to help determine the molecular mechanisms underlying chemically-influenced diseases (Davis et al., 2019). The external data sets integrated with CTD-curated data include OMIM, MeSH, NCBI Gene, GO, KEGG and Reactome Pathway databases (Davis et al., 2009, Davis et al., 2019). Thus, when CTD biocurators curate data providing direct evidence that a chemical interacts with a gene, that data is linked to other associated gene attributes, such as annotated molecular functions, cellular location, phenotypes, pathways, and diseases. We distinguish between the concepts of phenotype and disease, designating a phenotype as a biological outcome that is not inherently a disease, such as an alteration in blood pressure, whereas hypertension is a disease. This operational distinction facilitates integration of chemical-induced phenotypic and disease outcomes from the literature, and provides insight into the pre-disease state (Davis et al., 2018). CTD also generates transitive chemical-disease inferences by integrating independently curated chemical-gene, gene-disease, and chemical-disease interactions. Therefore, a previously unrecognized relationship may become evident when a direct chemical-gene statement is combined with a direct gene-disease statement to generate a chemical-disease inference (inferred via the shared gene). CTD statistically ranks these inferences to facilitate hypothesis development (King et al., 2012).
To facilitate identification of intermediate steps in the pathway from a chemical exposure to a disease outcome, CTD can be used to computationally link Chemical-Gene interactions with Phenotypes and Disease outcomes (“CGPD-tetramers”). These tetramers represent building blocks that can be assembled into larger chemical-induced pathways to design potential mechanism/mode-of-action (MOA), progressing from subcellular to system-wide processes (Davis et al., 2020). As several recent publications describe multiple impacts of e-cigarettes on pulmonary endpoints (Thirión-Romero et al., 2019), we used CTD to analyze relationships among Juul aerosol chemicals, interacting genes, phenotypes and respiratory illnesses. Such interactions provide intermediate points that may represent key events in the building of chemical-directed pathways linking Juul e-cigarettes to respiratory diseases.
2. Materials and methods
2.1. CTD data version and web tools
Analysis was performed using CTD public data available in October 2020 (revision 16329). CTD is updated with new content on a monthly basis; consequently, counts described herein may change over time. CTD’s public analytical and visualization tools were used (http://ctdbase.org/tools/) in subsequent analysis, including Batch Query, Set Analyzer, MyVenn, and Chemical-Phenotype Interaction Query. Default values were used for corrected p-values (threshold 0.01). For all data downloads, a filter was used to return data for exact input query terms only.
2.2. Juul chemicals
We analyzed a set of primary compounds previously reported in Juul aerosols (Reilly et al., 2019, Talih et al., 2019), including (CTD accession identifier): Nicotine (D009538), Acetaldehyde (D000079), Formaldehyde (D005557), Free Radicals (D005609), Crotonaldehyde (also known as 2-butenal, C012796), Acetone (D000096), Pyruvaldehyde (also known as methylglyoxal, D011765), and Particulate Matter (D052638). Though there are multiple types of free radicals produced by Juul e-cigarettes, we limited our study to Reactive Oxygen Species (D017382), since Juul aerosols have been shown to generate significant amounts of acellular reactive oxygen species (ROS) (Muthumalage et al., 2019). We refer to this entire chemical set by the abbreviation NAFFCAPP.
2.3. Chemical relationships in CTD
CTD’s ‘Batch Query’ tool (http://ctdbase.org/tools/batchQuery.go) was used to retrieve chemical-disease relationships by inputting the list of NAFFCAPP chemical terms, selecting ‘Disease Associations’ as output, and chemical-disease relationships were sorted by direct evidence for marker/mechanistic relationships among the respective chemicals and diseases. To retrieve chemical-gene relationships, the NAFFCAPP chemical terms were used as input in the ‘Batch Query’ tool, and curated gene interactions were downloaded. Output was sorted and duplicate genes were eliminated to yield 8,256 unique genes that were subsequently used as input in CTD’s ‘Set Analyzer’ tool (http://ctdbase.org/tools/analyzer.go) and queried for enriched pathways, as previously described (Davis et al., 2013). Statistical enrichment of a pathway indicates that the fraction of genes annotated to it in a test set is significantly larger than the fraction of genes annotated to it in the genome.
2.4. Determination of key event relationships
CTD tools ‘Batch Query’ and ‘MyVenn’ (http://ctdbase.org/tools/myVenn.go) were used to identify relationships that link NAFFCAPP chemicals to respiratory outcomes for three respiratory tract diseases: Pulmonary Fibrosis (D011658), Asthma (D001249), and Lung Neoplasms (D008175). Specifically, direct disease associations obtained from CTD’s ‘Batch Query’ of the NAFFCAPP chemical set were sorted by disease name, and genes in the inference network that infer the respective chemicals to these diseases were combined into a single data set of 295 genes. These genes support the direct curated ‘M’ (marker/mechanism) relationship between Juul aerosols and these specific diseases, and identify potential molecular initiating events. Prioritized phenotypes were determined as those that are independently associated with the NAFFCAPP chemicals and the respiratory disease examples by combining output from CTD’s ‘Batch Query’ and ‘MyVenn’ analysis tools. A batch query was performed using the four chemicals (nicotine, acetaldehyde, formaldehyde, and particulate matter) that show direct relationships to the disease examples as input and selecting curated phenotype associations as output, resulting in 552 unique phenotypes. Secondly, GO/phenotype terms that are annotated to each of the 295 genes were downloaded (940 phenotypes). ‘MyVenn’ was used to compare phenotypes annotated to the chemicals with phenotypes annotated to the genes, selecting ‘Other’ as input type, to identify 248 prioritized phenotypes common to both sets.
CGPD-tetramers are novel information blocks that link Chemical-Gene interactions with Phenotype and Disease outcomes. They are computationally generated by integrating five independently curated data sets in CTD: chemical-gene interactions, chemical-phenotype interactions, gene-GO/phenotype associations, chemical-disease associations, and gene-disease associations (Davis et al., 2020). We constructed CGPD-tetramers for the NAFFCAPP set of chemicals with respect to the three respiratory tract diseases (pulmonary fibrosis, asthma, and lung neoplasms). Shared chemicals, genes and phenotypes among the CGPD tetramers were compared to help elucidate potential mechanistic pathways.
There were 81 phenotypes common to the 248 prioritized phenotypes and the 112 phenotypes shared among CGPD tetramers for pulmonary fibrosis, asthma, and lung neoplasms. CTD’s “Chemical-Phenotype Interaction Query” tool was used to select phenotypes that have been annotated to Respiratory System.
3. Results
3.1. Disease associations of Juul aerosol chemicals
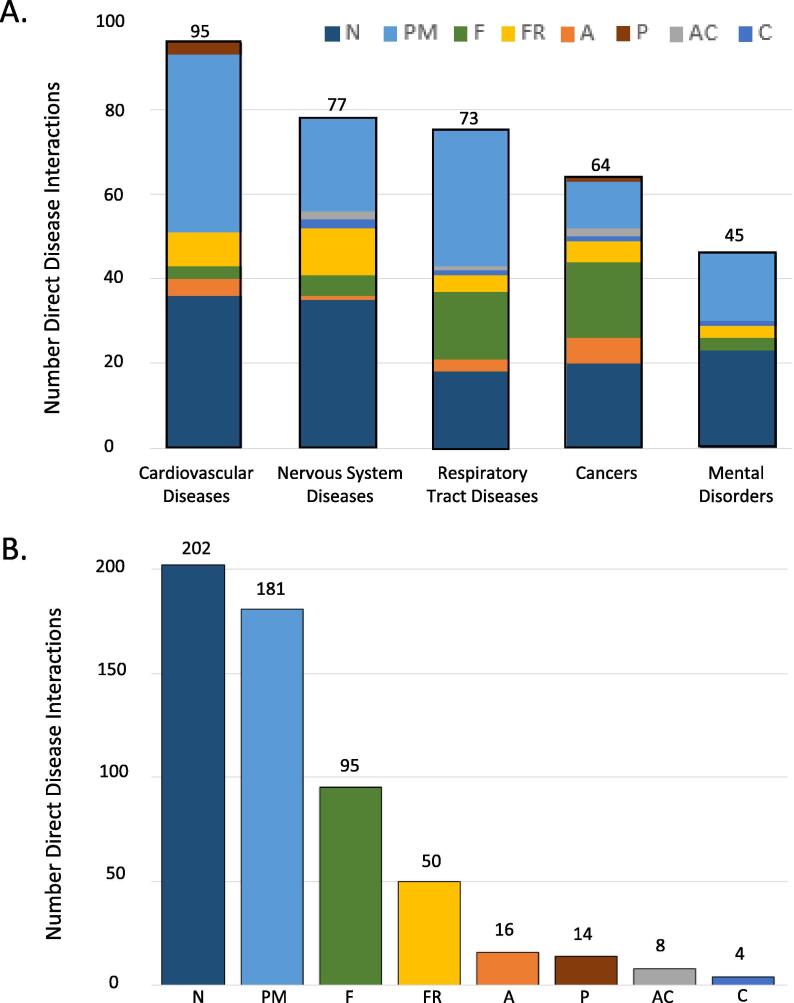
The NAFFCAPP set of chemicals detected in Juul aerosols have direct relationships with 400 diseases in CTD, which can be grouped to 35 disease categories, with some diseases mapping to more than one category (e.g., lung neoplasms maps to both respiratory tract diseases and cancers). The top five disease groups are cardiovascular, nervous system, respiratory tract, cancers, and mental disorders (Fig. 1A). Cardiovascular disease, the disease group with the highest number of relationships, includes 95 marker or mechanistic relationships between six chemicals and 69 cardiovascular outcomes such as hypertension, stroke, myocardial infarction and atherosclerosis. The highest number of chemical-disease relationships in this category is attributed to particulate matter, followed by nicotine. Examples of direct disease relationships in the category of nervous system diseases are Alzheimer Disease, Parkinson Disease, and seizures, with the greatest number of relationships for nicotine. Respiratory tract diseases such as lung neoplasms, pulmonary fibrosis, asthma, and pneumonia show more than 70 direct relationships with NAFFCAPP chemicals, with the highest number attributed to particulate matter. All eight of the NAFFCAPP chemicals contribute to direct relationships with one or more cancers, such as lung, breast and stomach neoplasms, with the greatest number of direct relationships for nicotine and formaldehyde. In the category of mental disorders, there are numerous direct associations between NAFFCAPP chemicals and autistic disorder, cognition disorders and depressive disorders, with the highest number of direct associations for nicotine. All individual marker or mechanistic chemical-disease relationships for these chemicals are provided (Supplemental Table S1).
Fig. 1.
Curated Disease Associations to Chemicals in Juul Aerosols: nicotine (N), particulate matter (PM), formaldehyde (F), free radicals (FR), acetaldehyde (A), pyruvaldehyde (P), acetone (AC), crotonaldehyde (C). (A) The top five categories of diseases with direct relationships to chemicals in Juul aerosols are shown, detailing the number of curated relationships in these categories and distribution by chemical. (B) Numbers of direct marker/mechanistic relationships in all disease categories attributed to each chemical constituent in Juul aerosols.
Besides the top disease groups related to NAFFCAPP chemicals, individual chemicals were examined for disease associations and the total number of direct relationships (Fig. 1B). Nicotine is associated with the most curated disease interactions (2 0 2), followed by particulate matter (1 8 1). Formaldehyde and free radicals are associated with 50–100 diseases, while the remaining chemicals each have less than 20 curated disease relationships.
In addition to directly curated relationships between Juul aerosol chemicals and diseases, all eight NAFFCAPP chemicals have inferred relationships to diseases in CTD generated by integration of chemical-gene and gene-disease statements. These transitive inferred associations between NAFFCAPP chemicals and 3,262 additional diseases provide indirect evidence of chemical-disease relationships, and are statistically ranked with an inference score. While the greatest number of inferred associations overall are attributed to nicotine, formaldehyde and particulate matter, all NAFFCAPP chemicals contribute to these inferred relationships, providing rationale for further investigation.
3.2. Enriched data sets
Molecular pathways that may contribute to Juul aerosol-induced diseases were identified using CTD’s ‘Set Analyzer’ tool. Collectively, the NAFFCAPP chemical set interacts with 8,256 genes. These genes were analyzed to survey the pathways annotated to them. Of 916 significantly enriched pathways, five of the top ten pathways are related to the immune system and/or signaling, while additional pathways include those related to metabolism and gene expression. Many similarities exist among the gene sets contributing to these pathways. For example, all genes annotated to Signaling by Interleukins (REACT:R-HSA-449147) belong to a subset of the genes annotated to Cytokine Signaling in Immune System (REACT:R-HSA-1280215), which in turn is a subset of genes contributing to the Immune System (REACT:R-HSA-168256) pathway. Genes that interact with nicotine, formaldehyde, and particulate matter account for 434/442 (98%) genes annotated to Cytokine Signaling in Immune System, yet genes that interact with all eight NAFFCAPP chemicals contribute to this pathway, and some genes, such as TNF, interact with all eight NAFFCAPP chemicals. To provide insight into the pathways that may be affected by each of the NAFFCAPP chemicals, this analysis was repeated with genes that interact with each of the Juul aerosol chemicals individually. Genes annotated to each of the top 10 significantly enriched pathways were summed for each chemical (Table 2).
Table 2.
Significantly Enriched Pathways of NAFFCAPP-interacting Genes.
| Enriched Pathway | Pathway ID | Total number unique NAFFCAPP genes annotated to pathway | Particulate Matter | Formaldehyde | Nicotine | Free Radicals-Reactive Oxygen Species | Acetaldehyde | Pyruvaldehyde | Crotonaldehyde | Acetone |
|---|---|---|---|---|---|---|---|---|---|---|
| Immune System | REACT:R-HSA-168256 | 1,084 | 722 | 541 | 276 | 144 | 62 | 41 | 43 | 7 |
| Metabolism | REACT:R-HSA-1430728 | 1,071 | 680 | 464 | 209 | 89 | 48 | 22 | 0 | 0 |
| Signal Transduction | REACT:R-HSA-162582 | 1,101 | 697 | 511 | 300 | 126 | 76 | 31 | 45 | 0 |
| Innate Immune System | REACT:R-HSA-168249 | 681 | 468 | 304 | 176 | 105 | 40 | 29 | 23 | 0 |
| Gene Expression | REACT:R-HSA-74160 | 837 | 429 | 467 | 144 | 53 | 29 | 0 | 0 | 0 |
| Metabolism of proteins | REACT:R-HSA-392499 | 735 | 456 | 334 | 143 | 56 | 24 | 0 | 0 | 0 |
| Cytokine Signaling in Immune system | REACT:R-HSA-1280215 | 442 | 308 | 235 | 149 | 80 | 40 | 26 | 30 | 6 |
| Metabolic pathways | KEGG:hsa01100 | 605 | 362 | 281 | 111 | 47 | 32 | 0 | 0 | 0 |
| Disease | REACT:R-HSA-1643685 | 458 | 323 | 198 | 123 | 58 | 32 | 14 | 0 | 0 |
| Signaling by Interleukins | REACT:R-HSA-449147 | 325 | 245 | 168 | 124 | 68 | 35 | 24 | 26 | 6 |
3.3. Developing mechanistic pathways for Juul aerosol-induced adverse outcomes
We leveraged CTD curated content with two methods to help prioritize relationships that link Juul chemical stressors with adverse respiratory outcomes, using pulmonary fibrosis, asthma, and lung neoplasms as specific examples of respiratory tract diseases.
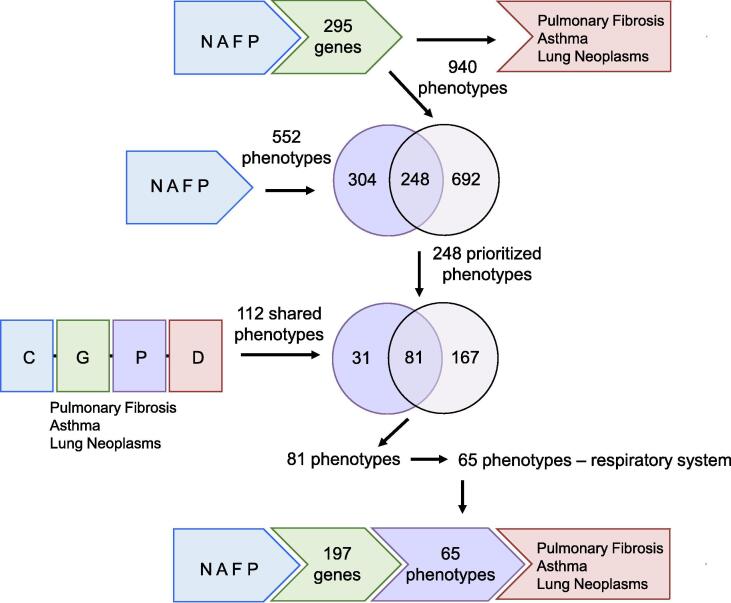
First, in CTD, there are direct relationships between NAFFCAPP chemicals and pulmonary fibrosis, asthma, and lung neoplasms via four chemicals: nicotine, formaldehyde, acetaldehyde, and particulate matter. In addition, these four chemicals collectively interact with 295 genes that also have direct relationships with these same diseases, enabling the identification of potential molecular initiating events in pathways that connect Juul aerosol chemicals with these respiratory tract diseases. To identify underlying biological processes that contribute to these gene-disease relationships, we compared the 552 phenotypes curated to these four chemicals and the 940 phenotypes annotated to the 295 genes. There were 248 phenotypes common to both sets that may contribute to underlying disease pathways; they are directly influenced by the chemicals in Juul aerosols and independently annotated to genes in the inference network that link vaping aerosols to these respiratory tract diseases (Fig. 2). These 248 phenotypes are prioritized for biological processes that may participate in the MOA of Juul aerosols; representative phenotypes include oxidative demethylation, T cell migration, and mucus secretion.
Fig. 2.
Methodology to determine Juul-Affected Phenotypes in Respiratory Tract Disease Outcomes. The four Juul aerosol chemicals: nicotine, acetaldehyde, formaldehyde, and particulate matter (NAFP) show marker or mechanistic associations with pulmonary fibrosis, asthma, and lung neoplasms, which are independently supported by inferred relationships via 295 genes that interact with one or more Juul aerosol chemicals, and are independently curated to one or more of the three diseases. As well, the four chemicals directly modulate 552 phenotypes, while the 295 genes are independently annotated to 940 phenotypes. There were 248 phenotypes common to both sets. Furthermore, chemical-gene-phenotype-disease (CGPD) tetramers were computationally generated among the four NAFP chemicals and pulmonary fibrosis, asthma, and lung neoplasms, resulting in 112 shared phenotypes among the three sets of CGPDs. Comparison between the two sets of phenotypes reveal an intersection of 81 phenotypes, of which 65 are annotated to respiratory system, and are associated with 197 genes. These genes represent potential molecular initiating events in the mode-of-action of Juul chemicals on respiratory outcomes.
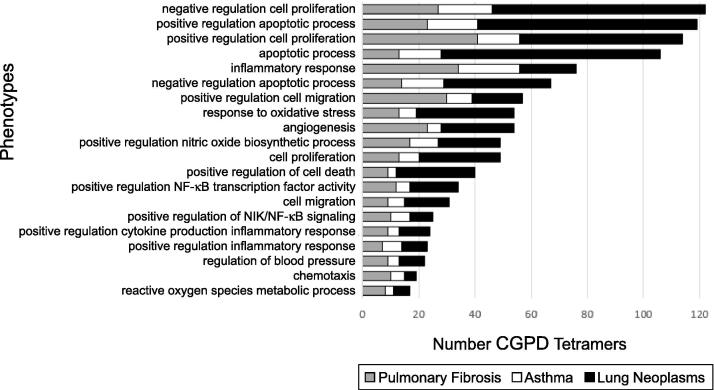
Second, we used CTD to predict molecular initiating events and phenotypes that link Juul aerosols to respiratory tract disease outcomes by generating computational associations between Chemical-Gene interactions and associated Phenotypes and Disease (CGPD-tetramers), using NAFFCAPP chemicals and the same respiratory tract disease examples, pulmonary fibrosis, asthma, and lung neoplasms. This bioinformatics approach yielded 830, 505, and 1,401 CGPD-tetramers for the three diseases, respectively (Supplemental Table S2). CGPD-tetramers for the three respiratory diseases share 112 phenotypes, of which 81 (72%) were identical to those found by filtering priority phenotypes for respiratory tract diseases using the first method. These 81 phenotypes were further restricted to those that have been previously annotated to ‘Respiratory System’ using CTD’s Anatomy module, resulting in 65 highly prioritized phenotypes (Fig. 2). These 65 phenotypes are highlighted by several key relationships: first, they are shared among pulmonary fibrosis, asthma, and lung neoplasm CGPD-tetramers; second, they contain curated chemical-phenotype annotations with nicotine, acetaldehyde, formaldehyde or particulate matter; third, they contain curated gene-phenotype annotations in CTD with NAFP-interacting genes; and fourth, they are supported by imported GO annotations for NAFP-interacting genes that align with phenotypes in CTD. These phenotypes were ranked by frequency in which they appear in pulmonary fibrosis, asthma and lung neoplasm CGPD-tetramers, and the top 20 most commonly predicted phenotypes are shown (Fig. 3). In addition to important roles for cell proliferation and apoptosis, this analysis highlights potentially important roles for inflammatory response, response to oxidative stress, cell migration, cytokine production involved in inflammatory response and chemotaxis, linking Juul aerosols and respiratory tract diseases; these highly prioritized phenotypes represent potential candidate events in the MOA of Juul aerosol chemicals.
Fig. 3.
The top 20 most common prioritized phenotypes for Juul aerosol chemicals associated with pulmonary fibrosis, asthma and lung neoplasms. CGPD-tetramers were computationally generated for four chemicals in Juul aerosols (nicotine, formaldehyde, acetaldehyde, and particulate matter), interacting genes, intermediate phenotypes, and three respiratory tract diseases (pulmonary fibrosis, asthma and lung neoplasms). A total of 65 phenotypes were prioritized as shared among the CGPD-tetramers for the three target respiratory diseases, and annotated to the specific chemicals, genes in the inference network, and respiratory system, and the 20 most frequent phenotypes are presented as number of CGPD-tetramers per phenotype for each disease.
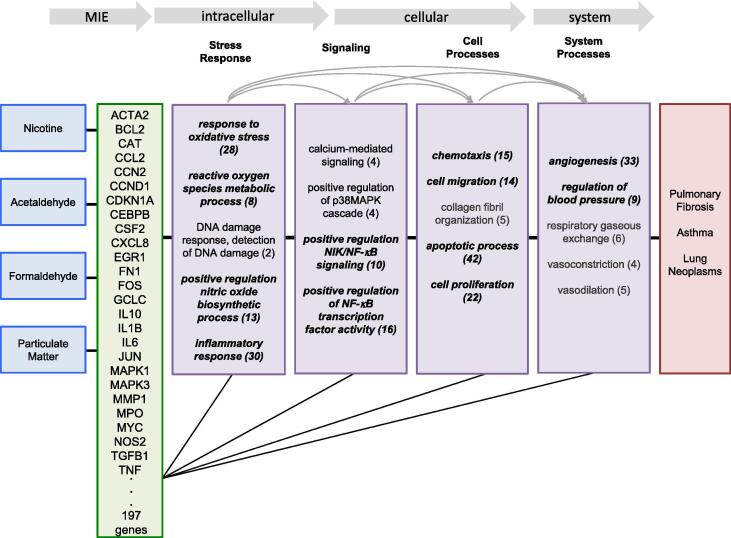
To further address molecular initiation events contributing to the prioritized phenotypes, genes affected by Juul aerosol chemicals were analyzed. A total of 20,073 chemical-gene interactions were downloaded, corresponding to 8,256 unique genes and 459 different types of chemical-gene interactions, such as chemical-induced changes in mRNA expression, protein phosphorylation, protein activity, and secretion. Curated interactions between the four chemicals NAFP and 197 genes prioritized as potential molecular initiating events were aligned with computationally-generated CGPDs (Supplemental Table S3). By integrating CTD chemical-gene interactions with these prioritized phenotypes, predictive mechanistic pathways can be constructed that associate nicotine, acetaldehyde, formaldehyde and particulate matter with these three respiratory outcomes (Fig. 4). For example, these four chemicals interact with 197 genes, in potential molecular initiating events represented by 26 genes that interact with all four chemicals and are annotated to one or more of the priority phenotypes. These phenotypes were mapped to subcellular, cellular or system processes that align with locations of potential key events in the sequential molecular pathways. Genes that are annotated to multiple phenotypes interrelate and connect phenotypes along intermediate pathways to the disease outcomes, helping to identify candidate events in predictive Juul MOAs.
Fig. 4.
Predictive mechanistic pathways that relate Juul aerosol chemicals to representative respiratory outcomes, generated by integrating CTD content. Chemical-gene interactions between nicotine, acetaldehyde, formaldehyde and particulate matter and 197 genes represent potential molecular initiating events (MIE) that link the chemical toxicants to pulmonary fibrosis, asthma and lung neoplasms, and are represented by 26 genes that interact with all four of the chemicals. Nineteen phenotypes that are directly modulated by these chemicals and are annotated to genes they interact with represent potential intermediate steps along predictive mechanistic pathways, and align with intracellular, cellular, and system processes. All of the phenotypes were prioritized as key contributors to the pathways via four types of supporting evidence: 1) curated chemical-phenotype interaction 2) curated gene-phenotype annotation 3) imported gene-GO annotation 4) computational generation of chemical-gene-phenotype-disease tetramers. Phenotypes shown in bold italic were among the 20 most frequent phenotypes in computationally generated CGPD tetramers. Numbers in parentheses represent the total number of genes of the 197 potential MIEs associated with each phenotype, with associations designated by solid black lines. Curved gray arrows indicate phenotypes that are interrelated via shared genes.
4. Discussion
4.1. Disease associations of Juul aerosol chemicals
This study investigates predictive disease associations of chemicals in Juul aerosols and underlying pathways. Using CTD, eight chemicals detected in Juul aerosols (nicotine, acetaldehyde, formaldehyde, free radicals/reactive oxygen species, crotonaldehyde, acetone, pyruvaldehyde, and particulate matter) were analyzed for interacting genes, intermediary phenotypes, pathways and disease associations. Top disease categories associated with these chemicals in CTD are cardiovascular diseases, nervous system diseases, respiratory tract diseases, neoplasms, and mental disorders, and several recent studies support associations between e-cigarettes and disease risks in these same categories, including thrombosis (Ramirez et al., 2020), ischemic stroke (Sifat et al., 2018), asthma (Clapp and Jaspers, 2017), cancer risk (Canistro et al., 2017), and depression (Leventhal et al., 2016).
Specific examples of Juul aerosol-induced disease parameters have been reported in humans, rats and mice. In a randomized crossover design, young, healthy, nonsmokers showed increased mean arterial pressure and heart rate, and decreased muscle sympathetic nerve activity following inhalation of Juul e-cigarettes, but not non-nicotine placebo e-cigarettes (Gonzalez and Cooke, 2021). Acute exposure to Juul aerosols led to impaired endothelial function in rats comparable to cigarette smoke (Rao et al., 2020). Three months of Juul aerosol exposure to mice induced dysregulation of glutamatergic system activity in mesolimbic brain regions, as evidenced by differential effects on several targets of the glutamatergic system in the nucleus accumbens and hippocampus (Alhaddad et al., 2020).
4.2. Chemicals contributing to disease associations.
Based on curated chemical-disease interactions in CTD, three chemicals emerge as key contributors to potential Juul-induced disease outcomes: nicotine, formaldehyde, and particulate matter (Supplemental Tables S1–S3). These three chemicals show mechanistic relationships with 364 diseases in CTD.
Recent evidence has shown that cytotoxicity of Juul aerosols strongly correlates with nicotine concentration (Omaiye et al., 2019). In CTD, nicotine is associated with 1,170 genes and 202 diseases. Nicotine has long been known to be addictive (National Academy of Sciences, 2018), as well as play a key role in the induction and progression of cardiovascular disorders (Balakumar and Kaur, 2009). Nicotinic acetylcholine receptors (nAChR) have been shown to regulate cell proliferation and inhibit apoptosis, key biological processes that are related to cancer (Gotts et al., 2019). Altered expression of the ACE2 protein (the putative receptor for the COVID-19 virus) in TH2 cells exposed to nicotine, in addition to changes in nicotinic receptor signaling and activation of inflammatory cytokines has led to recent speculation that nicotine exposure may increase cardiopulmonary risk to COVID-19 (Olds and Kabbani, 2020).
The second key contributor to potential Juul aerosol-induced outcomes, formaldehyde, interacts with 3,927 genes in CTD, detailed in 4,502 gene interactions, and shows marker/mechanistic relationships with 95 unique diseases. Formaldehyde is an established carcinogen, according to the International Agency for Research on Cancer (National Academy of Sciences, 2018). In long-term studies, formaldehyde has shown carcinogenic effects on various organs and tissues, and produced an increase in the total number of malignant tumors in experimental animals (Soffritti et al., 2002). In addition, formaldehyde has been shown to cause an increased risk of myeloid leukemia (Schwilk et al., 2010), and there is substantial evidence that it is capable of causing DNA damage and mutagenesis (National Academy of Sciences, 2018).
Particulate matter has long been recognized as a leading contributor to global disease burden (Cohen et al., 2017, Costa, 2018), with impacts on cardiovascular and respiratory health (Fiordelisi et al., 2017, Losacco and Perillo, 2018, Rajagopalan et al., 2018). In CTD, particulate matter has been shown to interact with 4,739 genes, participate in 10,187 gene interactions, and contribute to 181 direct disease relationships with marker/mechanism evidence (child terms of particulate matter such as smoke, soot and dust were excluded from this analysis).
The remaining chemicals detected in Juul aerosols, acetaldehyde, free radicals, crotonaldehyde, acetone, and pyruvaldehyde are associated with some of the same diseases as nicotine, formaldehyde and particulate matter, but also show direct relationships with 39 unique diseases including diabetes mellitus type 1 and amyotrophic lateral sclerosis.
4.3. Individual chemical-gene interactions.
Analysis of genes that interact with NAFFCAPP chemicals identifies potential molecular initiating events along pathways of vaping-induced disease outcomes and the generation of testable hypotheses. For example, particulate matter has been shown to induce EGR1 expression leading to inflammatory cytokine production and mucus hyperproduction in airway epithelium via the NF-κB and activator protein pathways (Xu et al., 2018). Analysis of individual chemical-gene interactions of the NAFP chemical set in CTD (Supplemental Table S3) reveals that nicotine is also capable of inducing EGR1 mRNA and protein expression, while formaldehyde and acetaldehyde can increase EGR1 mRNA expression. This suggests that Juul pods with reduced nicotine (3% vs 5%) may continue to induce EGR1 and consequent downstream effects due to the formation of carbonyls acetaldehyde and formaldehyde.
Inspection of individual chemical-gene interactions can also lead to testable hypotheses of alternate e-liquids in vaping products. Aerosols generated by the Juul device with a modified e-liquid containing 60:40 propylene glycol:glycerol with and without citral (to compare with other commercially available e-liquids) showed significantly increased levels of formaldehyde and free radical production (Reilly et al., 2019). Data in CTD show that particulate matter, formaldehyde, and reactive oxygen species collectively interact with nine mucin genes (MUC1, MUC16, MUC19, MUC2, MUC3A, MUC4, MUC5AC, MUC5B, and MUCL3), altering both mRNA and protein expression. Thus, increases in formaldehyde and free radical production may also alter mucin production and downstream effects. These chemical-mediated changes in mucin expression provide mechanistic steps that may contribute to alterations in mucin secretion that are observed in cigarette and e-cigarette users (Reidel et al., 2018).
4.4. Potential mechanistic pathways
Contributing factors to underlying pathways between Juul aerosols and representative adverse respiratory outcomes were analyzed in three ways: selection of significantly enriched pathways of the genes that interact with NAFFCAPP chemicals, determination of priority phenotypes that are annotated to NAFFCAPP chemicals as well as the genes they interact with, and computational generation of CGPD-tetramers. These three methods were used to strengthen evidence for contributing events in the pathways, and to avoid missing terms that may not yet have all five lines of supporting evidence that are required to generate CGPD-tetramers. Significantly enriched pathways include several related to the immune system and cytokine signaling in the immune system.
Beyond the ubiquitous roles of cell proliferation and apoptosis to disease pathways, several phenotypes emerged as playing potential key roles in the link between Juul aerosols and respiratory outcomes: oxidative stress, inflammatory responses, and cell signaling. Biological processes related to oxidative stress emerged as having the most annotations to NAFFCAPP chemicals. These were supported by NAFFCAPP-interacting gene annotations and CGPD-tetramers. While numerous reviews detail the contributions of the oxidative stress pathway to lung diseases such as asthma and chronic obstructive pulmonary disease (COPD) (Barnes, 2017, de Groot et al., 2019), this work integrates these oxidative stress phenotypes in the broader context of potential upstream and downstream events in the pathway. Phenotypes that contributed the highest number of terms to CGPD-tetramers were related to inflammation and immune responses. Immune system (REACT:R-HAS-168256) was also the most significantly enriched pathway of all the NAFFCAPP-interacting genes.
Several lines of evidence support a potential role for aberrant signaling underlying Juul-induced adverse outcomes, including significant enrichment of signaling pathways of NAFFCAPP-interacting genes, changes in gene expression, protein activity and secretion of cytokines by NAFFCAPP chemicals, and computational generation of CGPD-tetramers that include numerous signaling phenotypes. Numerous cytokines and chemokines including CXCL8, IFNG, IL1B, IL2, IL4, IL6, IL10 and TNF interact with five or more of the chemicals in the NAFFCAPP set, suggesting that multiple chemicals contribute to the pathway and disease endpoints. Cytokine-cytokine receptor interaction (KEGG:hsa04060) is also a significantly enriched pathway with genes annotated to all eight NAFFCAPP chemicals, with nearly half (47%) of the genes annotated to this pathway overlapping with genes annotated to Signaling by Interleukins (REACT:R-HSA-449147). Importance of the cytokine-cytokine receptor interaction pathway is supported by clinical studies showing that it is one of four pathways that overlapped between comparisons of differentially expressed genes in nasal biopsies of e-cigarette users vs. non-smokers and cigarette users vs. non-smokers (Martin et al., 2016).
In addition to supporting evidence for priority phenotypes generated from pathway enrichment and chemical- and gene-phenotype annotations, CGPD-tetramers linking Juul aerosol chemicals to respiratory disease outcomes also revealed new priority phenotypes that can be tested. For example, ‘memory’ and ‘learning’ emerged as phenotypes associated with NAFFCAPP chemicals and adverse respiratory outcomes (Supplemental Table S2). Cigarette smoking has also been shown to negatively impact executive function in older adults, an effect that is synergized by lung diseases (Amini et al., 2020). Thus, CGPD-tetramers linking Juul aerosol chemicals to interacting genes and cognitive phenotypes can identify specific genes to further study and explore for smoking-induced and vaping-induced cognitive issues.
Integration of curated chemical-gene interactions in CTD with prioritized phenotypes can help construct predictive MOAs that link molecular initiating events with key events towards disease outcomes (Davis et al., 2018). Here, we show representative interactions between nicotine, acetaldehyde, formaldehyde and particulate matter with 26 genes that affect phenotypes along predictive disease pathways linking these chemicals to pulmonary fibrosis, asthma and lung neoplasm endpoints. These biological processes are directly influenced by one or more of the four chemicals and are independently associated with the same genes by GO annotations. Alignment of phenotypes from subcellular events to system-wide processes can highlight relationships between potential key events that can be tested as intermediate end points for risk assessment, and help to fill in mechanistic gaps between these chemicals and disease outcomes.
4.5. Strengths and limitations
Environmental exposures to toxic insults such as e-cigarette aerosols affect human health, but the mechanisms are largely unknown. CTD provides a unique resource that integrates primary literature on chemical-gene, chemical-disease, and gene-disease interactions, with phenotype information, GO annotations, pathway information, and exposure studies, culminating in over 45 million toxicogenomic relationships (Davis et al., 2021). Analysis of eight chemicals detected in Juul aerosols yielded interactions with 8,256 unique genes, described in over 20,000 chemical-gene interactions. CTD tools promote the analysis of these interactions in ways that can build mechanistic pathways and help to fill in molecular knowledge gaps between the chemical toxicants and disease outcomes.
While this study begins to look at the gene, phenotype, pathway and disease relationships associated with chemical constituents of e-cigarette emissions, several limitations remain. Chemical composition of the inhaled aerosol and levels is still under investigation, and may depend on several factors. Several studies have shown that potentially toxic metals (nickel, chromium, lead, manganese and zinc) are detected in e-cigarette emissions, and may originate from the coils that heat the e-liquids as well as joints and wires (Aherrera et al., 2017, Olmedo et al., 2018), yet toxic metals were not assessed by Talih (Talih et al., 2019). Though acrolein has been detected in vaping aerosols at a concentration of 0.07–4.19 micrograms per 15 puffs (National Academies of Sciences et al., 2018), and in aerosols from initial and modified Juul devices in Europe (Mallock et al., 2020), acrolein in Juul emissions was not assessed by Reilly or detected by Talih, and may depend on the puffing regimen analyzed (Reilly et al., 2019, Talih et al., 2019). Further, this analysis does not take into account possible interactions among the chemicals studied.
Besides chemical composition of the aerosol, levels of these chemicals and metabolites in e-cigarette users are also under investigation. Numerous studies have measured the concentration of nicotine in humans after vaping Juuls, ranging from 9.8 mg/ml plasma per 10 puffs (Maloney et al., 2021) to 31 ng/ml serum after 10 min puffing (Yingst et al., 2019). Biomarkers of exposure and cardiopulmonary injury were measured for acetaldehyde and formaldehyde in mice after propylene glycol:vegetable glycerin-derived (PG-VG) aerosol exposure (ingredients of e-cigarette liquids including Juul). PG-VG exposure significantly increased post-exposure urinary acetate (a metabolite of acetaldehyde), and exposure to formaldehyde or PG-VG-derived aerosol stimulated significant pulmonary irritation and endothelial dysfunction (Jin et al., 2021). These findings support the presence of these chemicals in vivo following exposure to Juul aerosols.
Effects of vaping are dependent on abundant factors in addition to the chemical constituents and type of device, including nicotine concentration (Juul is available with 3% and 5% nicotine strength: https://www.juul.com/resources/all-about-tobacco-menthol-juulpods), vaping patterns (such as length of inhale/exhale, puff volume, frequency of vaping and time to first puff), coil resistance, product age, composition, battery output (ohms), user age, weight, metabolism, health, and genetics (National Academies of Sciences et al., 2018). In addition to variations in device and user, analyses of chemical-gene-phenotype-disease associations are limited by CTD-curated content. CTD is updated monthly, and with continued curation of publications related to vaping chemicals, associations among these chemicals with phenotypes, pathways and diseases will continue to be updated.
5. Conclusions
We describe analysis of Juul aerosol chemicals in CTD, including disease associations, gene interactions, enriched phenotype and pathway relationships, and prioritized events along predictive pathways to representative respiratory adverse outcomes. Cardiovascular diseases, nervous system diseases, respiratory tract diseases, cancers, and mental disorders were the most abundant categories of disease associations, with the highest number of relationships attributed to nicotine, particulate matter and formaldehyde. Several predictive mechanistic pathways were generated, based on chemical- and gene-annotated phenotypes in conjunction with CGPD-tetramers. Integration of CTD data and computational generation of CGPD-tetramers can help to fill molecular knowledge gaps and generate testable hypotheses to better understand the effects of Juul aerosol chemicals and the effects of vaping.
CRediT authorship contribution statement
Cynthia J. Grondin: Conceptualization, Formal analysis, Data curation, Writing – original draft, Writing - review & editing. Allan Peter Davis: Data curation, Writing - review & editing. Jolene A. Wiegers: Software, Writing - review & editing. Thomas C. Wiegers: Software, Writing - review & editing. Daniela Sciaky: Data curation, Writing - review & editing. Robin J. Johnson: Data curation, Writing - review & editing. Carolyn J. Mattingly: Project administration, Funding acquisition, Writing - review & editing.
Declaration of competing interest
The authors declare no conflicts of interest with respect to financial interests, research, authorship, and/or publication of this article.
Acknowledgments
Acknowledgements
We thank Roy McMorran for system administration support.
Funding
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) grant numbers ES014065, ES025128, ES023788 and ES019604. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2021.08.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aherrera A., Olmedo P., Grau-Perez M., Tanda S., Goessler W., Jarmul S., Chen R., Cohen J.E., Rule A.M., Navas-Acien A. The association of e-cigarette use with exposure to nickel and chromium: A preliminary study of non-invasive biomarkers. Environ. Res. 2017;159:313–320. doi: 10.1016/j.envres.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Alhaddad H., Wong W., Sari A.T., Crotty Alexander L.E., Sari Y. Effects of 3-Month Exposure to E-Cigarette Aerosols on Glutamatergic Receptors and Transporters in Mesolimbic Brain Regions of Female C57BL/6 Mice. Toxics. 2020;8(4):95. doi: 10.3390/toxics8040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini R., Sahli M., Ganai S. Cigarette smoking and cognitive function among older adults living in the community. Neuropsychol. Dev. Cogn. B Aging Neuropsychol. Cogn. 2020:1–16. doi: 10.1080/13825585.2020.1806199. [DOI] [PubMed] [Google Scholar]
- Azimi P., Keshavarz Z., Lahaie Luna M., Cedeno Laurent J.G., Vallarino J., Christiani D.C., Allen J.G. An unrecognized hazard in E-cigarette vapor: preliminary quantification of methylglyoxal formation from propylene glycol in E-cigarettes. Int. J. Environ. Res. Public Health. 2021;18(2):385. doi: 10.3390/ijerph18020385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakumar P., Kaur J. Is nicotine a key player or spectator in the induction and progression of cardiovascular disorders? Pharmacol. Res. 2009;60(5):361–368. doi: 10.1016/j.phrs.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Barnes P.J. Cellular and molecular mechanisms of asthma and COPD. Clin. Sci. (Lond) 2017;131(13):1541–1558. doi: 10.1042/CS20160487. [DOI] [PubMed] [Google Scholar]
- Canistro D., Vivarelli F., Cirillo S., Babot Marquillas C., Buschini A., Lazzaretti M., Marchi L., Cardenia V., Rodriguez-Estrada M.T., Lodovici M., Cipriani C., Lorenzini A., Croco E., Marchionni S., Franchi P., Lucarini M., Longo V., Della Croce C.M., Vornoli A., Colacci A., Vaccari M., Sapone A., Paolini M. E-cigarettes induce toxicological effects that can raise the cancer risk. Sci. Rep. 2017;7(1):2028. doi: 10.1038/s41598-017-02317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp P.W., Jaspers I. Electronic cigarettes: their constituents and potential links to asthma. Curr. Allergy Asthma Rep. 2017;17(11):79. doi: 10.1007/s11882-017-0747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.J., Brauer M., Burnett R., Anderson H.R., Frostad J., Estep K., Balakrishnan K., Brunekreef B., Dandona L., Dandona R., Feigin V., Freedman G., Hubbell B., Jobling A., Kan H., Knibbs L., Liu Y., Martin R., Morawska L., Pope C.A., Shin H., Straif K., Shaddick G., Thomas M., van Dingenen R., van Donkelaar A., Vos T., Murray C.J.L., Forouzanfar M.H. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015. Lancet. 2017;389(10082):1907–1918. doi: 10.1016/S0140-6736(17)30505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa D.L. Historical highlights of air pollution toxicology. Toxicol. Sci. 2018;164(1):5–8. doi: 10.1093/toxsci/kfy117. [DOI] [PubMed] [Google Scholar]
- Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., Wiegers J., Wiegers T.C., Mattingly C.J. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49(D1):D1138–D1143. doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Murphy C.G., Saraceni-Richards C.A., Rosenstein M.C., Wiegers T.C., Mattingly C.J. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37(Database issue):D786–D792. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Wiegers T.C., Grondin C.J., Johnson R.J., Sciaky D., Wiegers J., Mattingly C.J. Leveraging the comparative toxicogenomics database to fill in knowledge gaps for environmental health: a test case for air pollution-induced cardiovascular disease. Toxicol. Sci. 2020;177(2):392–404. doi: 10.1093/toxsci/kfaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Wiegers T.C., Wiegers J., Johnson R.J., Sciaky D., Grondin C.J., Mattingly C.J. Chemical-induced phenotypes at CTD help inform the predisease state and construct adverse outcome pathways. Toxicol. Sci. 2018;165(1):145–156. doi: 10.1093/toxsci/kfy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot L.E.S., van der Veen T.A., Martinez F.O., Hamann J., Lutter R., Melgert B.N. Oxidative stress and macrophages: driving forces behind exacerbations of asthma and chronic obstructive pulmonary disease? Am. J. Physiol. Lung Cell Mol. Physiol. 2019;316(2):L369–L384. doi: 10.1152/ajplung.00456.2018. [DOI] [PubMed] [Google Scholar]
- Dinardo P., Rome E.S. Vaping: The new wave of nicotine addiction. Cleve Clin. J. Med. 2019;86(12):789–798. doi: 10.3949/ccjm.86a.19118. [DOI] [PubMed] [Google Scholar]
- Farzal Z., Perry M.F., Yarbrough W.G., Kimple A.J. The Adolescent Vaping Epidemic in the United States-How It Happened and Where We Go From Here. JAMA Otolaryngol Head Neck Surg. 2019;145(10):885–886. doi: 10.1001/jamaoto.2019.2410. [DOI] [PubMed] [Google Scholar]
- Fiordelisi A., Piscitelli P., Trimarco B., Coscioni E., Iaccarino G., Sorriento D. The mechanisms of air pollution and particulate matter in cardiovascular diseases. Heart Fail Rev. 2017;22(3):337–347. doi: 10.1007/s10741-017-9606-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.E., Cooke W.H. Acute effects of electronic cigarettes on arterial pressure and peripheral sympathetic activity in young nonsmokers. Am. J. Physiol. Heart Circ. Physiol. 2021;320(1):H248–H255. doi: 10.1152/ajpheart.00448.2020. [DOI] [PubMed] [Google Scholar]
- Gotts J.E., Jordt S.E., McConnell R., Tarran R. What are the respiratory effects of e-cigarettes? BMJ. 2019;366 doi: 10.1136/bmj.l5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Lynch J., Richardson A., Lorkiewicz P., Srivastava S., Theis W., Shirk G., Hand A., Bhatnagar A., Srivastava S., Conklin D.J. Electronic cigarette solvents, pulmonary irritation, and endothelial dysfunction: role of acetaldehyde and formaldehyde. Am. J. Physiol. Heart Circ. Physiol. 2021;320(4):H1510–H1525. doi: 10.1152/ajpheart.00878.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavuluru R., Han S., Hahn E.J. On the popularity of the USB flash drive-shaped electronic cigarette Juul. Tob Control. 2019;28(1):110–112. doi: 10.1136/tobaccocontrol-2018-054259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B.L., Davis A.P., Rosenstein M.C., Wiegers T.C., Mattingly C.J. Ranking transitive chemical-disease inferences using local network topology in the comparative toxicogenomics database. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0046524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal A.M., Strong D.R., Sussman S., Kirkpatrick M.G., Unger J.B., Barrington-Trimis J.L., Audrain-McGovern J. Psychiatric comorbidity in adolescent electronic and conventional cigarette use. J. Psychiatr. Res. 2016;73:71–78. doi: 10.1016/j.jpsychires.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losacco C., Perillo A. Particulate matter air pollution and respiratory impact on humans and animals. Environ. Sci. Pollut. Res. Int. 2018;25(34):33901–33910. doi: 10.1007/s11356-018-3344-9. [DOI] [PubMed] [Google Scholar]
- Mallock N., Trieu H.L., Macziol M., Malke S., Katz A., Laux P., Henkler-Stephani F., Hahn J., Hutzler C., Luch A. Trendy e-cigarettes enter Europe: chemical characterization of JUUL pods and its aerosols. Arch Toxicol. 2020;94(6):1985–1994. doi: 10.1007/s00204-020-02716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney S., Eversole A., Crabtree M., Soule E., Eissenberg T., Breland A. Acute effects of JUUL and IQOS in cigarette smokers. Tob Control. 2020;30(4):449–452. doi: 10.1136/tobaccocontrol-2019-055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.M., Clapp P.W., Rebuli M.E., Pawlak E.A., Glista-Baker E., Benowitz N.L., Fry R.C., Jaspers I. E-cigarette use results in suppression of immune and inflammatory-response genes in nasal epithelial cells similar to cigarette smoke. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;311(1):L135–L144. doi: 10.1152/ajplung.00170.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumalage T., Lamb T., Friedman M.R., Rahman I. E-cigarette flavored pods induce inflammation, epithelial barrier dysfunction, and DNA damage in lung epithelial cells and monocytes. Sci. Rep. 2019;9(1):19035. doi: 10.1038/s41598-019-51643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, E., Medicine, Health, D. Medicine, H. Board on Population, P. Public Health and S. Committee on the Review of the Health Effects of Electronic Nicotine Delivery (2018). Public Health Consequences of E-Cigarettes. D. L. Eaton, L. Y. Kwan and K. Stratton. Washington (DC), National Academies Press (US). Copyright 2018 by the National Academy of Sciences. All rights reserved.
- Olds J.L., Kabbani N. Is nicotine exposure linked to cardiopulmonary vulnerability to COVID-19 in the general population? Febs J. 2020;287(17):3651–3655. doi: 10.1111/febs.15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo P., Goessler W., Tanda S., Grau-Perez M., Jarmul S., Aherrera A., Chen R., Hilpert M., Cohen J.E., Navas-Acien A., Rule A.M. Metal concentrations in e-cigarette liquid and aerosol samples: the contribution of metallic coils. Environ. Health Perspect. 2018;126(2) doi: 10.1289/EHP2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omaiye E.E., McWhirter K.J., Luo W., Pankow J.F., Talbot P. High-nicotine electronic cigarette products: toxicity of JUUL fluids and aerosols correlates strongly with nicotine and some flavor chemical concentrations. Chem. Res. Toxicol. 2019;32(6):1058–1069. doi: 10.1021/acs.chemrestox.8b00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S., Al-Kindi S.G., Brook R.D. Air pollution and cardiovascular disease: JACC state-of-the-art review. J. Am. Coll Cardiol. 2018;72(17):2054–2070. doi: 10.1016/j.jacc.2018.07.099. [DOI] [PubMed] [Google Scholar]
- Ramirez J.E.M., Karim Z.A., Alarabi A.B., Hernandez K.R., Taleb Z.B., Rivera J.O., Khasawneh F.T., Alshbool F.Z. The JUUL e-cigarette elevates the risk of thrombosis and potentiates platelet activation. J. Cardiovasc. Pharmacol. Ther. 2020;25(6):578–586. doi: 10.1177/1074248420941681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P., Liu J., Springer M.L. JUUL and combusted cigarettes comparably impair endothelial function. Tob. Regul. Sci. 2020;6(1):30–37. doi: 10.18001/TRS.6.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidel B., Radicioni G., Clapp P.W., Ford A.A., Abdelwahab S., Rebuli M.E., Haridass P., Alexis N.E., Jaspers I., Kesimer M. E-cigarette use causes a unique innate immune response in the lung, involving increased neutrophilic activation and altered mucin secretion. Am. J. Respir. Crit. Care. Med. 2018;197(4):492–501. doi: 10.1164/rccm.201708-1590OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, S.M., Bitzer, Z.T., Goel, R., Trushin, N., Richie, J.P. (2019). “Free Radical, Carbonyl, and Nicotine Levels Produced by Juul Electronic Cigarettes.” Nicotine Tob Res 21(9): 1274-1278. [DOI] [PMC free article] [PubMed]
- Schwilk E., Zhang L., Smith M.T., Smith A.H., Steinmaus C. Formaldehyde and leukemia: an updated meta-analysis and evaluation of bias. J. Occup. Environ. Med. 2010;52(9):878–886. doi: 10.1097/JOM.0b013e3181ef7e31. [DOI] [PubMed] [Google Scholar]
- Sifat A.E., Vaidya B., Kaisar M.A., Cucullo L., Abbruscato T.J. Nicotine and electronic cigarette (E-Cig) exposure decreases brain glucose utilization in ischemic stroke. J. Neurochem. 2018;147(2):204–221. doi: 10.1111/jnc.14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffritti M., Belpoggi F., Lambertin L., Lauriola M., Padovani M., Maltoni C. Results of long-term experimental studies on the carcinogenicity of formaldehyde and acetaldehyde in rats. Ann. N. Y. Acad. Sci. 2002;982:87–105. doi: 10.1111/j.1749-6632.2002.tb04926.x. [DOI] [PubMed] [Google Scholar]
- Talih S., Salman R., El-Hage R., Karam E., Karaoghlanian N., El-Hellani A., Saliba N., Shihadeh A. Characteristics and toxicant emissions of JUUL electronic cigarettes. Tob. Control. 2019;28(6):678–680. doi: 10.1136/tobaccocontrol-2018-054616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirión-Romero I., Pérez-Padilla R., Zabert G., Barrientos-Gutiérrez I. respiratory impact of electronic cigarettes and “low-risk” tobacco. Rev. Invest. Clin. 2019;71(1):17–27. doi: 10.24875/RIC.18002616. [DOI] [PubMed] [Google Scholar]
- Walley S.C., Wilson K.M., Winickoff J.P., Groner J. A public health crisis: electronic cigarettes, vape, and JUUL. Pediatrics. 2019;143(6):e20182741. doi: 10.1542/peds.2018-2741. [DOI] [PubMed] [Google Scholar]
- Xu F., Cao J., Luo M., Che L., Li W., Ying S., Chen Z., Shen H. Early growth response gene 1 is essential for urban particulate matter-induced inflammation and mucus hyperproduction in airway epithelium. Toxicol. Lett. 2018;294:145–155. doi: 10.1016/j.toxlet.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Yingst J.M., Hrabovsky S., Hobkirk A., Trushin N., Richie J.P., Jr., Foulds J. Nicotine absorption profile among regular users of a pod-based electronic nicotine delivery system. JAMA Netw. Open. 2019;2(11) doi: 10.1001/jamanetworkopen.2019.15494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.