Graphical abstract
Keywords: Ato1, Carboxylic acids, Plasma membrane transport, Yeast, Transporter engineering
Highlights
-
•
Substrate specificity of Ato1 was engineered by altering its pore size.
-
•
The L219A and F98A substitutions enable succinic acid transport activity of Ato1.
-
•
Ato1 E144A substitution causes dominant negative organic acid hypersensitivity.
-
•
Organic acid hypersensitivity is caused by the hyperactive ATO1 transporter alleles.
-
•
First report of a fully functional bacterial transporter (SatP) in yeast.
Abstract
Aiming at improving the transport of biotechnologically relevant carboxylic acids in engineered microbial cell factories, the focus of this work was to study plasma membrane transporters belonging to the Acetate Uptake Transporter (AceTr) family. Ato1 and SatP, members of this family from Saccharomyces cerevisiae and Escherichia coli, respectively, are the main acetate transporters in these species. The analysis of conserved amino acid residues within AceTr family members combined with the study of Ato1 3D model based on SatP, was the rationale for selection of site-directed mutagenesis targets. The library of Ato1-GFP mutant alleles was functionally analysed in the S. cerevisiae IMX1000 strain which shows residual growth in all carboxylic acids tested. A gain of function phenotype was found for mutations in the residues F98 and L219 located at the central constrictive site of the pore, enabling cells to grow on lactic and on succinic acid. This phenotype was associated with an increased transport activity for these substrates. A dominant negative acetic acid hypersensitivity was induced in S. cerevisiae cells expressing the E144A mutant, which was associated with an increased acetic acid uptake. By utilizing computer-assisted 3D-modelling tools we highlight structural features that explain the acquired traits found in the analysed Ato1 mutants. Additionally, we achieved the proper expression of the Escherichia coli SatP, a homologue of Ato1, in S. cerevisiae. To our knowledge, this constitutes the first report of a fully functional bacterial plasma membrane transporter protein in yeast cells.
1. Introduction
Saccharomyces cerevisiae is a common cell factory for organic acid production due to its tolerance to acidic conditions and extensive knowledge of its molecular genetics, physiology and genomics, making it a very attractive platform for metabolic engineering [1]. One of the major bottlenecks for the efficient and cost-effective bioproduction is the export of organic acids through the microbial plasma membrane [2]. So far four native organic acid plasma membrane transporters were functionally characterized in S. cerevisiae [3]: the lactate transporter Jen1 [4], TC #2.A.1.12.2, the acetate transporter Ato1 (previously known as Ady2) [5], TC #2.A.96.1.4, the acetic acid channel Fps1 [6], TC #1.A.8.5.1, and the sorbate/benzoate pump Pdr12 [7], [8], TC #3.A.1.205.3.
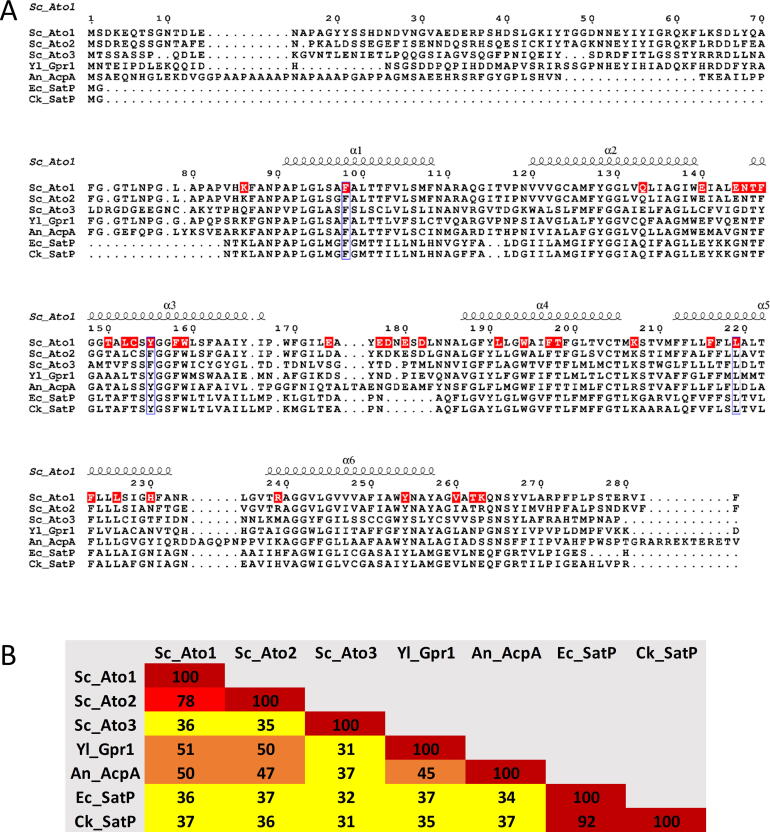
Ato1 and its homologues Ato2 and Ato3 from S. cerevisiae belong to the Acetate Uptake Transporter (AceTr) family [3] and share 78% and 36% of identity with Ato1 respectively, although only Ato1 has been assigned as a monocarboxylate transporter [5], [9], [10]. Recently, specific mutants in Ato2 (L218S) and Ato3 (F95S) selected by adaptive laboratory evolution displayed improved transport activity for monocarboxylates [11]. Members of the AceTr family, distinguishable by the NPAPLGL(M/S) signature motif, are present in a wide range of organisms, namely bacteria, archaea, and fungi [10]. In fungi they are detected in 97% of available genomes and presumably play an important role in spore germination [10], [12], [13]. Although first identified as the main transporter responsible for the uptake of acetic acid into the cytosol [5], subsequent studies demonstrated that Ato1 could transport organic acids in both directions, as it was involved in the export of lactic acid from S. cerevisiae cells engineered for lactic acid production [9], [14]. The export capabilities of AceTr members make Ato1 an interesting engineering target for organic acid production in yeast cell factories.
Acetate is the main substrate of functionally characterized transporters from the AceTr family [5], [10], [13], [15], [16], however, members of this family can also transport other organic acids. Ato1 shows transport activity for other monocarboxylates along acetate, such as formate, propionate, lactate, and pyruvate [9], [17], [18] while the bacterial homologue SatP, besides acetate and lactate, can also transport dicarboxylates such as succinate [16], [19]. Ato1 mutants L219V and A252G selected by adaptive laboratory evolution [18] display improved lactate transport efficiency [11]. Equivalent amino acid substitutions L131V and A164G also increased the lactate transport activity of SatP in E. coli [16]. The crystal structures of bacterial homologues SatP_Ec [19] and SatP_Ck [20] depict hexameric anion channels in their closed state. Each monomer features 6 transmembrane helices and four acetate binding sites lined up along the pore: S1 at the cytoplasmic vestibule, S2 and S3 within the main pore and S4 at the periplasmic vestibule [20]. The structure of the SatP_Ec monomer resembles an hourglass-like shape, with a constrictive site in the pore centre defined by three conserved hydrophobic residues F17, Y72 and L131, thus termed FLY [19]. These studies provided vital information regarding the physiological function, phylogeny, and structure of transporters from the AceTr family. However, more investigation is required to unveil the details of substrate translocation [19] including the ability of Ato1 (L219V & A252G) and SatP_Ec (L131V & A164G) mutants to transport lactic acid [16], [18]. Still puzzling is the severe acetic acid sensitivity observed in Yarrowia lipolytica associated with mutations in GPR1, a member of the AceTr family [21]. The effect of GPR1d alleles was dominant negative since the co-presence of wild-type GPR1 could not alleviate sensitive phenotypes [22]. The sequenced GPR1d alleles contained mutations that led to single amino acid substitutions G62S, G63D, L65Q and G248D [23] and were able to trans-dominantly induce acetic acid sensitivity in S. cerevisiae [24]. Furthermore, single amino acid substitutions A70V, F71V, T74I, A88V, E144G, N145D, M211K, and F212S within Ato1 and N75D, G147D, and S265L within Ato2 were discovered to produce the same hypersensitive effect in S. cerevisiae [24], showing that induction of acetic acid sensitivity is not exclusive for GPR1 mutants. While specific single amino acid substitutions in Gpr1, Ato1 and Ato2 were able to cause such drastic inhibitory effects, surprisingly, knockout of GPR1 in Y. lipolytica or triple knockout of ATO1, ATO2 and ATO3 in S. cerevisiae did not cause any phenotypic changes [25].
In this study, we designed a site-directed mutagenesis approach aiming at deciphering the structure–function relationship of Ato1 in S. cerevisiae. The rational design of mutations in the FLY constrictive site, altering the diameter of the pore, had an impact on transporter specificity and activity. Additionally, we discovered that the cells hypersensitivity to organic acids is a phenotype associated with mutants which possess increased transport activity. Finally, we show that the codon optimized SatP_Ec protein was properly expressed in S. cerevisiae cells and was able to mediate the uptake of organic acids. This is an achievement that, to our knowledge, constitutes the first report of a fully functional bacterial plasma membrane transporter in yeast cells.
2. Material and methods
2.1. Yeast strains, maintenance, and plasmids
The S. cerevisiae IMX1000 strain [26] was used for characterization of ATO1 and SatP_Ec alleles in terms of growth, organic acid uptake, and localization. This strain lacks 25 genes encoding known or putative organic acid transporters and in our experimental conditions presents residual growth on acetic, lactic, succinic, malic and fumaric acid.
The S. cerevisiae CEN.PK 113-5D strain [27] was transformed with ATO1 and SatP_Ec alleles to assess their dominant negative effect on growth on organic acids. The yeast strains were maintained on solid synthetic medium containing Difco Yeast Nitrogen Base (0.67%, w/v), glucose (2%, w/v) and agar (2%, w/v), supplemented with adequate requirements for prototrophic growth, or on solid YPD medium containing yeast extract (1%, w/v), peptone (2%, w/v), glucose (2%, w/v) and agar (2%, w/v). Plasmids used in this study are listed in Table 1. The p416GPD plasmid [28] with glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter was used as a vector for constitutive expression of ATO1 and SatP_Ec alleles in S. cerevisiae strains. The p416GPD contained URA3 marker gene for auxotrophic complementation.
Table 1.
List of plasmids used in this study.
| Plasmid | Characteristics | Reference |
|---|---|---|
| p416GPD | p416; Glyceraldehyde-3-phosphate dehydrogenase promoter | [28] |
| pAto1 | p416GPD derivative; constitutive expression of Ato1 | [9] |
| pL219A | pAto1 derivative; L219A substitution in Ato1 | This study |
| pAto1-GFP | p416GPD derivative; constitutive expression of Ato1-GFP | [10] |
| pK86A-GFP | pAto1-GFP derivative; K86A substitution in Ato1 | This study |
| pF98A-GFP | pAto1-GFP derivative; F98A substitution in Ato1 | This study |
| pQ133A-GFP | pAto1-GFP derivative; Q133A substitution in Ato1 | This study |
| pE140A-GFP | pAto1-GFP derivative; E140A substitution in Ato1 | This study |
| pE144A-GFP | pAto1-GFP derivative; E144A substitution in Ato1 | This study |
| pE144A/F98A-GFP | pAto1-GFP derivative; E144A and F98A substitutions in Ato1 | This study |
| pE144A/Y155A-GFP | pAto1-GFP derivative; E144A and Y155A substitutions in Ato1 | This study |
| pE144A/L219A-GFP | pAto1-GFP derivative; E144A and L219A substitutions in Ato1 | This study |
| pN145A-GFP | pAto1-GFP derivative; N145A substitution in Ato1 | This study |
| pT146A-GFP | pAto1-GFP derivative; T146A substitution in Ato1 | This study |
| pF147A-GFP | pAto1-GFP derivative; F147A substitution in Ato1 | This study |
| pT150A-GFP | pAto1-GFP derivative; T150A substitution in Ato1 | This study |
| pL152A-GFP | pAto1-GFP derivative; L152A substitution in Ato1 | This study |
| pC153A-GFP | pAto1-GFP derivative; C153A substitution in Ato1 | This study |
| pY155A-GFP | pAto1-GFP derivative; Y155A substitution in Ato1 | This study |
| pY155S-GFP | pAto1-GFP derivative; Y155Asubstitution in Ato1 | This study |
| pY155F-GFP | pAto1-GFP derivative; Y155F substitution in Ato1 | This study |
| pF158A-GFP | pAto1-GFP derivative; F158A substitution in Ato1 | This study |
| pW159A-GFP | pAto1-GFP derivative; W159A substitution in Ato1 | This study |
| pE174A-GFP | pAto1-GFP derivative; E174A substitution in Ato1 | This study |
| pE177A-GFP | pAto1-GFP derivative; E177A substitution in Ato1 | This study |
| pD178A-GFP | pAto1-GFP derivative; D178A substitution in Ato1 | This study |
| pE180A-GFP | pAto1-GFP derivative; E180A substitution in Ato1 | This study |
| pD182A-GFP | pAto1-GFP derivative; D182A substitution in Ato1 | This study |
| pL191A-GFP | pAto1-GFP derivative; L191A substitution in Ato1 | This study |
| pW194A-GFP | pAto1-GFP derivative; W194A substitution in Ato1 | This study |
| pF197A-GFP | pAto1-GFP derivative; F197A substitution in Ato1 | This study |
| pT198A-GFP | pAto1-GFP derivative; T198A substitution in Ato1 | This study |
| pK207A-GFP | pAto1-GFP derivative; K207A substitution in Ato1 | This study |
| pF216A-GFP | pAto1-GFP derivative; F216A substitution in Ato1 | This study |
| pL219A-GFP | pAto1-GFP derivative; L219A substitution in Ato1 | This study |
| pL219V-GFP | pAto1-GFP derivative; L219V substitution in Ato1 | This study |
| pL219G-GFP | pAto1-GFP derivative; L219G substitution in Ato1 | This study |
| pL219S-GFP | pAto1-GFP derivative; L219S substitution in Ato1 | This study |
| pL219P-GFP | pAto1-GFP derivative; L219P substitution in Ato1 | This study |
| pL219W-GFP | pAto1-GFP derivative; L219W substitution in Ato1 | This study |
| pF223A-GFP | pAto1-GFP derivative; F223A substitution in Ato1 | This study |
| pL226A-GFP | pAto1-GFP derivative; L226A substitution in Ato1 | This study |
| pH230A-GFP | pAto1-GFP derivative; H230A substitution in Ato1 | This study |
| pR239A-GFP | pAto1-GFP derivative; R239A substitution in Ato1 | This study |
| pY254A-GFP | pAto1-GFP derivative; Y254A substitution in Ato1 | This study |
| pV260A-GFP | pAto1-GFP derivative; V260A substitution in Ato1 | This study |
| pT262A-GFP | pAto1-GFP derivative; T262A substitution in Ato1 | This study |
| pK263A-GFP | pAto1-GFP derivative; K263A substitution in Ato1 | This study |
| pSatP | p416GPD derivative; constitutive expression of SatP_Ec | This study |
| pSatP-L131A | pSatP derivative; L131A substitution in SatP_Ec | This study |
2.2. Spot assays
Spot assays were performed by first cultivating the yeast cells in liquid synthetic medium containing Difco Yeast Nitrogen Base (0.67%, w/v) and glucose (2%, w/v), supplemented with adequate requirements for prototrophic growth. Cells were cultivated until mid-exponential phase and diluted to an OD640 nm of 0.2. Three 1:10 serial dilutions were made and 3 μL of each suspension were spotted on solid synthetic media containing Difco Yeast Nitrogen Base (0.67%, w/v) and agar (2%, w/v), supplemented with adequate requirements for prototrophic growth. Carbon sources used on solid synthetic media were the following: acetic acid (0.5%, w/v, pH 6.0), lactic acid (0.5%, w/v, pH 5.0, unless specified otherwise), succinic acid (0.5%, w/v, pH 5.0, unless specified otherwise), malic acid (0.5%, w/v, pH 5.0), fumaric acid (0.5%, w/v, pH 5.0), or glucose (2%, w/v) as control. Yeast cells were cultivated at 18° C for 10 days.
2.3. Alignment and protein topology prediction
To identify conserved amino acid residues within the AceTr family members that are possibly significant for transporter function, protein sequences of AceTr members from S. cerevisiae: Ato1 (KZV12856.1), Ato2 (KZV08629.1), and Ato3 (KZV12628.1), as well as protein sequences of AceTr members from other organisms that were characterized as acetate transporters: Gpr1 (XP_502188.1), AcpA (Q5B2K4.1), SatP_Ec (AAC73121.1), and SatP_Ck (QQC78103.1) were aligned using the PSI/TM-Coffee server for transmembrane proteins [29]. All protein sequences were retrieved from the National Center for Biotechnology Information (NCBI). The aligned sequences were visualised using the ESPript program [30]. Topology of Ato1 was predicted and visualised using the Protter program [31].
2.4. 3D modelling and HOLE radius prediction of wild-type Ato1 and its mutants
The Ato1 protein sequence was analysed using the HHpred method within the MPI Bioinformatics Toolkit [32], detecting SatP_Ck [20] as the closest homologue with available crystal structure. By selecting SatP_Ck crystal structure as a template and using the Modeller homology modelling program [33], the Ato1 3D structure was obtained. Visual analyses of Ato1 3D structure were performed using the Free Maestro version molecular modelling software as previously reported [34].
Using the 3D structures obtained from HHpred, the radius throughout the transporter pore was calculated with the HOLE program (2.2.005 Linux) [35]. The values of the radiuses were displayed in a graph with the X coordinate being the length of the proteins in a determined vector. The Van der Waals radii file used for the predictions was the hard core radii that is supplied by HOLE. The pore predictions were displayed in Visual Molecular Dynamics program (VMD, 1.9.3) along with the 3D structures [36]. The pore predictions were displayed in Visual Molecular Dynamics program (VMD, 1.9.3) along with the 3D structures [36] as described by Wu et al. [37].
2.5. Heterologous expression of SatP_Ec in S. cerevisiae
The synthetic SatP_Ec gene, optimized for expression in S. cerevisiae, was purchased in the form of pUC18-SatP from GenScript (Piscataway, NJ, USA). The codon optimized SatP_Ec gene was then PCR-amplified from the pUC18-SatP template using the Accuzyme Mix (Bioline, London, UK) and primers SatP_resc_fw and SatP_resc_rew (Table 2). The PCR-amplified SatP_Ec sequence was cut by BamHI and XhoI restriction enzymes (Thermofisher Scientific, Waltham, MA, USA) and cloned into the p416GPD vector, which was digested by the same enzymes under the same conditions, deriving the pSatP.
Table 2.
List of primers used in this study.
| Name | Sequence |
|---|---|
| K86A fw | CCTGCTCCAGTGCACGCTTTTGCTAATCCTGCGC |
| K86A rev | GCGCAGGATTAGCAAAAGCGTGCACTGGAGCAGG |
| F98A fw | CTTAGGTCTTTCAGCCGCCGCGTTGACGACATTTG |
| F98A rev | CAAATGTCGTCAACGCGGCGGCTGAAAGACCTAAG |
| Q133A fw | GTGGTTTGGTGGCTTTGATTGCTGGTATTTG |
| Q133A rev | CAAATACCAGCAATCAAAGCCACCAAACCAC |
| E140A fw | CTGGTATTTGGGCTATAGCTTTGGAAAATAC |
| E140 rev | GTATTTTCCAAAGCTATAGCCCAAATACCAG |
| E144A fw | GAGATAGCTTTGGCTAATACTTTTGGTGGTAC |
| E144A rev | GTACCACCAAAAGTATTAGCCAAAGCTATCTC |
| N145A fw | GAGATAGCTTTGGAAGCTACTTTTGGTGGTAC |
| N145A rev | GTACCACCAAAAGTAGCTTCCAAAGCTATCTC |
| T146A fw | GATAGCTTTGGAAAATGCTTTTGGTGGTACCG |
| T146A rev | CGGTACCACCAAAAGCATTTTCCAAAGCTATC |
| F147A fw | TTTGGAAAATACTGCTGGTGGTACCGCATTA |
| F147A rev | TAATGCGGTACCACCAGCAGTATTTTCCAAA |
| T150A fw | AATACTTTTGGTGGTGCTGCATTATGTTCTTA |
| T150 rev | TAAGAACATAATGCAGCACCACCAAAAGTATT |
| L152A fw | TTGGTGGTACCGCAGCTTGTTCTTACGGTG |
| L152A rev | CACCGTAAGAACAAGCTGCGGTACCACCAA |
| C153A fw | GGTGGTACCGCATTAGCTTCTTACGGTGGGTTTTG |
| C153A rev | CAAAACCCACCGTAAGAAGCTAATGCGGTACCACC |
| Y155A fw | CATTATGTTCTGCTGGTGGGTTTTGGTTGAG |
| Y155A rev | CTCAACCAAAACCCACCAGCAGAACATAATG |
| Y155S fw | CATTATGTTCTTCTGGTGGGTTTTGGTTGAG |
| Y155S rev | CTCAACCAAAACCCACCAGAAGAACATAATG |
| Y155F fw | CATTATGTTCTTTCGGTGGGTTTTGGTTGAG |
| Y155F rev | CTCAACCAAAACCCACCGAAAGAACATAATG |
| F158A fw | GTTCTTACGGTGGGGCTTGGTTGAGTTTCGCTG |
| F158A rev | CAGCGAAACTCAACCAAGCCCCACCGTAAGAAC |
| W159A fw | CTTACGGTGGGTTTGCTTTGAGTTTCGCTGC |
| W159A rev | GCAGCGAAACTCAAAGCAAACCCACCGTAAG |
| E174A fw | GTTTGGTATCTTGGCTGCTTACGAAGACAATG |
| E174A rev | CATTGTCTTCGTAAGCAGCCAAGATACCAAAC |
| E177A fw | TCTTGGAAGCTTACGCTGACAATGAATCTG |
| E177A rev | CAGATTCATTGTCAGCGTAAGCTTCCAAGA |
| D178A fw | GGAAGCTTACGAAGCTAATGAATCTGATTTG |
| D178A rev | CAAATCAGATTCATTAGCTTCGTAAGCTTCC |
| E180A fw | CTTACGAAGACAATGCTTCTGATTTGAATAATG |
| E180A rev | CATTATTCAAATCAGAAGCATTGTCTTCGTAAG |
| D182A fw | GAAGACAATGAATCTGCTTTGAATAATGCTTTAG |
| D182A rev | CTAAAGCATTATTCAAAGCAGATTCATTGTCTTC |
| L191A fw | GCTTTAGGATTTTATGCTTTGGGGTGGGCCATC |
| L191A rev | GATGGCCCACCCCAAAGCATAAAATCCTAAAGC |
| W194A fw | TTATTTGTTGGGGGCTGCCATCTTTACGTTTG |
| W194A rev | CAAACGTAAAGATGGCAGCCCCCAACAAATAA |
| F197A fw | GTTGGGGTGGGCCATCGCTACGTTTGGTTTAAC |
| F197A rev | GTTAAACCAAACGTAGCGATGGCCCACCCCAAC |
| T198A fw | GGGTGGGCCATCTTTGCTTTTGGTTTAACCG |
| T198A rev | CGGTTAAACCAAAAGCAAAGATGGCCCACCC |
| K207A fw | CCGTTTGTACCATGGCTTCCACTGTTATGTTCTTTTTG |
| K207 rev | CAAAAAGAACATAACAGTGGAAGCCATGGTACAAACGG |
| F216A fw | GTTATGTTCTTTTTGTTGGCTTTCTTACTAGCATTAAC |
| F216A rev | GTTAATGCTAGTAAGAAAGCCAACAAAAAGAACATAAC |
| L219A fw | GTTGTTCTTCTTAGCTGCATTAACTTTCCTACTG |
| L219A rev | CAGTAGGAAAGTTAATGCAGCTAAGAAGAACAAC |
| L219V fw | GTTGTTCTTCTTAGTTGCATTAACTTTCCTACTG |
| L219V rev | CAGTAGGAAAGTTAATGCAACTAAGAAGAACAAC |
| L219G fw | GTTGTTCTTCTTAGGTGCATTAACTTTCCTACTG |
| L219G rev | CAGTAGGAAAGTTAATGCACCTAAGAAGAACAAC |
| L219S fw | GTTGTTCTTCTTATCTGCATTAACTTTCCTACTG |
| L219S rev | CAGTAGGAAAGTTAATGCAGATAAGAAGAACAAC |
| L219P fw | GTTGTTCTTCTTACCAGCATTAACTTTCCTACTG |
| L219P rev | CAGTAGGAAAGTTAATGCTGGTAAGAAGAACAAC |
| L219W fw | GTTGTTCTTCTTATGGGCATTAACTTTCCTACTG |
| L219W rev | CAGTAGGAAAGTTAATGCCCATAAGAAGAACAAC |
| F223A fw | CTTACTAGCATTAACTGCTCTACTGTTGTCTATTGG |
| F223A rev | CCAATAGACAACAGTAGAGCAGTTAATGCTAGTAAG |
| L226A fw | CATTAACTTTCCTACTGGCTTCTATTGGTCACTTTG |
| L226A rev | CAAAGTGACCAATAGAAGCCAGTAGGAAAGTTAATG |
| H230A fw | CTGTTGTCTATTGGTGCTTTTGCTAATAGACTTGG |
| H230 rev | CCAAGTCTATTAGCAAAAGCACCAATAGACAACAG |
| R239A fw | GACTTGGTGTCACAGCTGCTGGTGGTGTCCTGG |
| R239A rev | CCAGGACACCACCAGCAGCTGTGACACCAAGTC |
| Y254A fw | CTTTCATTGCTTGGGCTAACGCATATGCAGG |
| Y254A rev | CCTGCATATGCGTTAGCCCAAGCAATGAAAG |
| V260A fw | CGCATATGCAGGTGTCGCTACAAAGCAGAA |
| V260 rev | TTCTGCTTTGTAGCGACACCTGCATATGCG |
| T262A fw | CGCATATGCAGGTGTTGCTGCTAAGCAGAATTC |
| T262A rev | GAATTCTGCTTAGCAGCAACACCTGCATATGCG |
| K263A fw | GCAGGTGTTGCTACAGCTCAGAATTCATATGTACTG |
| K263A rev | CAGTACATATGAATTCTGAGCTGTAGCAACACCTGC |
| SatP_resc fw | AAAAGGATCCATGGGTAACACCAAATTAGCC |
| SatP_resc rev | TTTTCTCGAGCTAATGACTTTCACCGATAGGCAAG |
| L131A fw | CGTTTTCTTTTCTGCTACAGTTTTGTTCGCTTTG |
| L131A rev | CAAAGCGAACAAAACTGTAGCAGAAAAGAAAACG |
2.6. Transport assays
Yeast cells were grown overnight in a liquid synthetic medium containing Difco Yeast Nitrogen Base (0.67%, w/v) and glucose (2%, w/v), supplemented with adequate requirements for prototrophic growth. Cells were grown until mid-exponential phase, harvested by centrifugation, washed twice in ice-cold deionized water, and incubated in liquid synthetic media containing lactic acid (0.5%, w/v, pH 5.0) or glycerol (6%, w/v) to alleviate glucose repression. After 4 h, cells were harvested by centrifugation, washed twice in ice-cold deionized water, and resuspended in ice-cold deionized water to a final concentration of about 15 mg dry wt./mL. Yeast cell suspension (30 μL) was transferred into microtubes containing 0.1 M potassium phosphate buffer (60 μL) with a pH of 6.0, unless specified otherwise. After 2 min of incubation at 26 °C, the reaction was started by adding 10 μL of an aqueous solution of the labelled acid (pH 6.0, unless specified otherwise), incubated at 26 °C for 30 s (acetic acid) or 60 s (lactic and succinic acids), and stopped by adding cold 100 mM non-labelled acid (100 μL), of equivalent pH. Microtubes were centrifuged for 5 min at 13000 rpm, the supernatant was discarded, and the cell pellet was resuspended in ice-cold deionized water (1 mL) and centrifuged again for 5 min at 13000 rpm. The supernatant was discarded, and the cell pellet was resuspended in 1 mL of scintillation liquid (Opti-Phase HiSafe II; LKB FSA Laboratory Supplies, Loughborough, UK). Radioactivity was measured in a PerkinElmer Tri-Carb 4810TR liquid scintillation spectrophotometer with disintegrations per minute correction. The following radioactive labelled substrates were utilized: DL-[U-14C] lactic acid (specific activity [S.a.] 13,000 dpm/nmol), sodium salt (PerkinElmer, Massachusetts, USA), [1-14C] acetic acid (S.a. 13,000 dpm/nmol), sodium salt (GE Healthcare, London, UK) and [1-14C] succinic acid (Specific activity of 6000 dpm/nmol), sodium salt (Moravek Biochemicals, California, USA). The transport kinetics best fitting the experimental initial uptake rates and the kinetic parameters, were determined by a computer-assisted non-linear regression analysis (GraphPAD Software version 4.00, San Diego, CA, USA).
2.7. Site directed mutagenesis of Ato1 (Ady2) and SatP_Ec
Site-directed mutagenesis was performed according to Soares-Silva et al. [38], using pAto1, pAto1-GFP and pSatP as templates (Table 1), and primers that confer respective mutations (Table 2).
2.8. Epifluorescence microscopy
Cells were grown overnight in a liquid synthetic medium containing Difco Yeast Nitrogen Base (0.67%, w/v) and glucose (2%, w/v), supplemented with adequate requirements for prototrophic growth. Cells were then washed twice in deionized water and incubated for 4 h in lactic acid (0.5% w/v, pH 5.0). Microscopy analysis was performed by using the Olympus BX63 fluorescence microscope and the cellSens image analysis software.
3. Results
3.1. Screening of critical residues for structure and function of Ato1
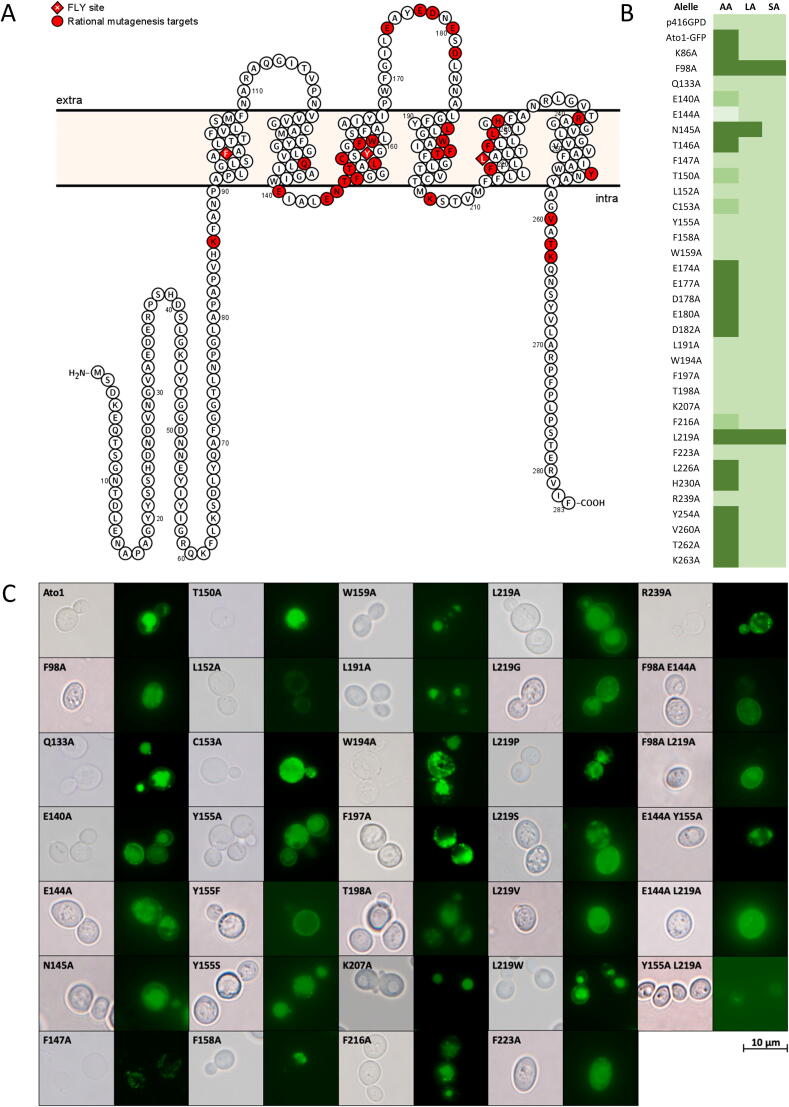
To identify critical residues for the structure and function of S. cerevisiae Ato1, a rational site-directed mutagenesis approach was performed. In the first round, a total of 34 mutations (Fig. 1A –red spots, Table 1) were generated by substituting selected residues with an alanine in the ATO1-GFP template cloned in the p416GPD plasmid [10]. These amino acid residues were selected as mutagenesis targets by combining the already published data (see Introduction) with data obtained from primary sequence analysis tools, namely multiple sequence alignment of AceTr homologues functionally characterized as acetate transporters (Fig. S1). Furthermore, we also took in consideration data obtained from secondary structure-based tools for protein topology prediction allowing the identification of residue location in transmembrane segments, loops or N and C termini (Fig. 1A) as well as from computer-assisted three-dimensional modelling tools (Fig. 3A).
Fig. 1.
Screening of S. cerevisiae Ato1 mutants obtained by site-directed mutagenesis. A – The predicted Ato1 topology showcasing the locations of 34 amino acid residues (highlighted in red) which were substituted by alanine. The residues which form the central constrictive site (FLY) are diamond-shaped and represented by white characters. B – Heatmap of the 34 Ato1-GFP mutants expressed in S. cerevisiae IMX1000 based on their effect on growth on acetic, lactic, and succinic acids as sole carbon sources. C – Localization of GFP-tagged Ato1 and mutants mentioned in B that presented an altered growth profile, as well as Y155S, Y155F, L219G, L219P, L219S, L219V, L219W, F98A E144A, F98A L219A, E144A Y155S, E144A L219A, Y155A L219A mutants. Cells were collected at mid-exponential growth phase from YNB glucose (2% w/v), washed and derepressed for 4 h in YNB lactic acid (0.5% w/v), and observed by epifluorescence microscopy. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
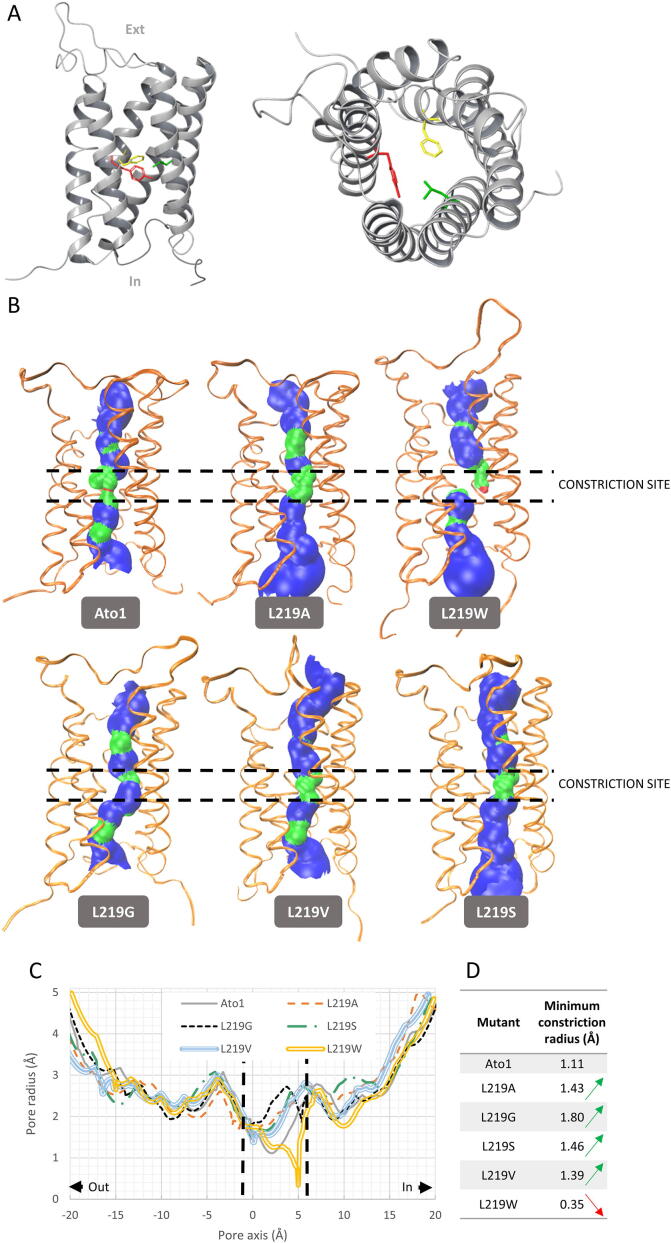
Fig. 3.
Predicted 3D structure model of Ato1 obtained by homology threading using SatP_Ck crystal structure (PDB Entry: 5YS3) as template. A – Ato1 consists of six transmembrane α-helices with both termini at the intracellular side. The residues F98 (yellow), Y155 (red), and L219 (green) in the 3D structure of the Ato1 (transversal and extracellular view) form the central and narrowest hydrophobic constriction of the anion pathway. In-intracellular; Ext-extracellular. B – Analysis of the effect of the mutations L219A, L219G, L219S, L219V and L219W on the protein 3D structure using HOLE software (see Material and Methods). C – Prediction of the pore constriction along the axis of the native Ato1 and of mutations L219A, L219G, L219S, L219V and L219W using HOLE software. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
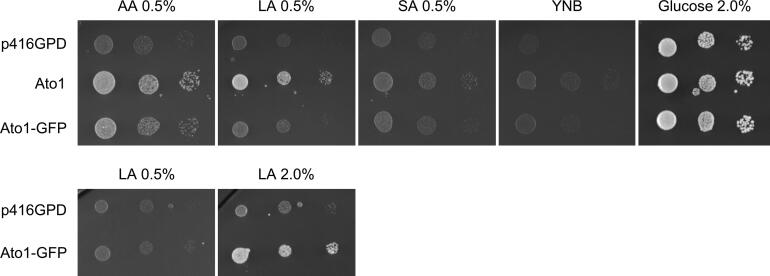
The phenotype of Ato1-GFP mutants was evaluated by growth on acetic, lactic or succinic acids as sole carbon and energy sources (Fig. 1B). S. cerevisiae IMX1000 cells transformed with the plasmid harbouring the Ato1-GFP construct displayed a reduction on acetic acid and lactic acid uptake of 20–30% and 35–50% respectively, which did not affect growth on YNB acetic acid (0.5%) whereas cells require higher concentrations of lactic acid (2%) to fully recover growth on this acid in comparison with cells harbouring the untagged Ato1 (Fig. S2A). No growth is found on succinate, confirming the inability of Ato1 and Ato1-GFP to transport succinate.
The phenotype of the mutants could be assigned into distinct categories. The mutants Q133A, E140A, F147A, T150A, L152A, C153A, Y155A, F158A, W159A, L191A, W194A, F197A, T198A, K207A, F216A, F223A, and R239A presented reduced growth on acetic acid (Fig. 1B). To distinguish whether this phenotype was due to an alteration in the transporter activity or to a deficient protein localization at the plasma membrane, we analysed the expression and localization of each of these mutant alleles by fluorescence microscopy (Fig. 1C). The mutant transporters E140A and Y155A displayed proper localization at the plasma membrane, whereas the remaining mutants from this group showed absence of GFP labelling at the plasma membrane which is an indication of deficient expression, protein misfolding and/or improper trafficking. The N145A mutant displayed increased ability to grow on lactic acid whereas the F98A and L219A mutants presented increased growth both on lactic and on succinic acid, all showing localization at the plasma membrane. Cells expressing the mutant E144A, properly located at the plasma membrane, displayed even lower ability to grow on acetic acid than the strain transformed with the empty vector, which is a sign of hypersensitivity to this acid as previously reported [24]. Finally, no changes compared to the wild-type were found for K86A, T146A, L226A, H230A, Y254A, V260A, T262A, and K263A, an indication that no detectable functional alteration occurred in these mutants which were properly located at the plasma membrane (not shown).
3.2. The F98-Y155-L219 constrictive site is essential for substrate specificity and transport activity
The Y155A mutant, although properly localized at the plasma membrane, showed only residual growth on acetic acid, similar to cells transformed with p416GPD vector (Fig. 2A). The same residual growth was observed in the Y155S mutant, which is associated with a very low expression level and protein mislocalization evaluated by fluorescence microscopy (Fig. 1C). On the other hand, substitution by another aromatic amino acid (Y155F) enabled growth (Fig. 2A) and has an expression profile similar to the wild-type Ato1 (Fig. 1C).
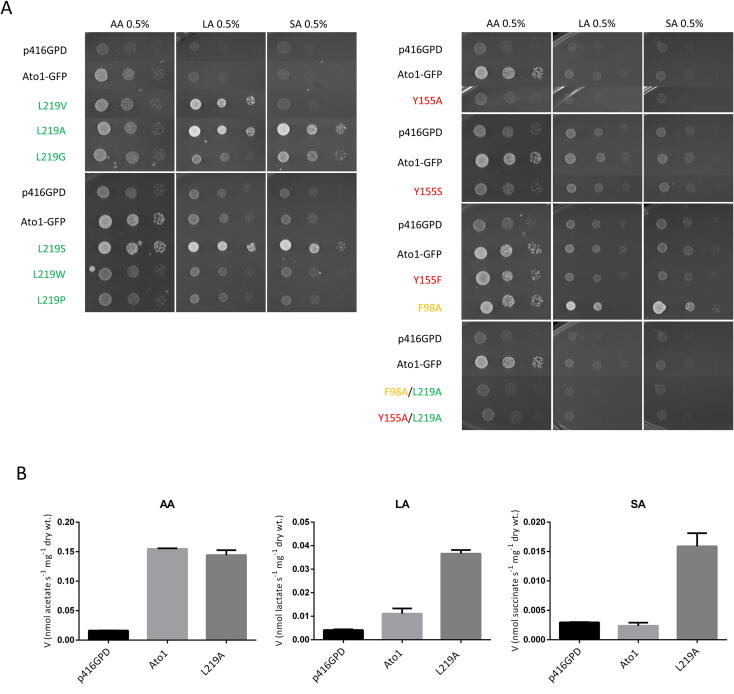
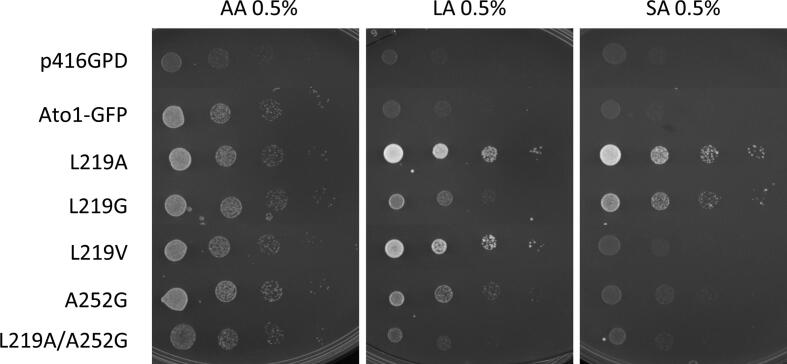
Fig. 2.
Functional analysis of the Ato1 residues F98-Y155-L219. A – Spot assay of S. cerevisiae IMX1000 cells expressing Ato1-GFP mutants in the residues F98 (yellow), Y155 (red), and L219 (green) grown on YNB media containing acetic acid (0.5%, pH 6.0) (AA), lactic acid (0.5%, pH 5.0) (LA), or succinic acid (0.5%, pH 5.0) (SA) at 18 °C for 10 days. B – Uptake of acetic acid (1.0 mM, pH 6.0), lactic acid (3.0 mM, pH 6.0) and succinic acid (2.0 mM, pH 6.0) in S. cerevisiae IMX1000 cells transformed with the plasmids p416GPD, pAto1, and pL219A. Cells were collected at mid-exponential growth phase from YNB-glucose (2% w/v), washed and derepressed for 4 h in YNB glycerol (6% w/v). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The substitution F98A enabled increased growth on lactic and succinic acids, while keeping growth on acetic acid similar to the wild-type (Fig. 2A). The same growth pattern was observed for L219A mutant, a phenotype further confirmed by measuring the uptake for labelled acetic, lactic and succinic acid (Fig. 2B), providing evidence for the role of these amino acid residues on transporter specificity and activity. According to our findings, the presence of an aromatic residue at position F98 is not essential for proper function of Ato1, whereas an aromatic residue in position Y155 seems to be essential for protein function. Interestingly, the double substitution F98A/L219A (Fig. 2A), despite being expressed at the plasma membrane (Fig. 1C), showed only residual growth on acetic acid and did not allow increased growth on lactic or succinic acids. The double mutant Y155A/L219A, with a very low expression level and protein mislocalization (Fig. 1C), also presented only residual growth on all organic acids tested (Fig. 2A).
We wondered if the gain of function for lactate and succinate transport found in the L219A mutant would be related with the side-chain size of the amino acid residue at this position. We performed a new set of mutations by changing the pore size of the narrowest constriction of Ato1 to assess if the activity and specificity of the transporter for lactate or succinate would be altered. The substitutions L219A, L219G and L219S allowed growth on lactic and succinic acids, however, the substitution L219V only allowed cells to grow on lactic acid (Fig. 2A). These results pointed to the existence of a correlation between the side-chain size of the amino acid at the position 219 and the ability to transport higher molecular weight anions such as: Leucine (acetate) > Valine (acetate and lactate) > Serine > Alanine > Glycine (acetate, lactate, succinate). To further confirm this hypothesis, we performed substitutions L219W and L219P that presented absence of growth on carboxylic acids. Protein localization was determined for all the substitutions in the residue 219. (Fig. 2A). The loss of function of the mutant L219P is associated with its improper localization (Fig. 1C), however the L219W was expressed and properly localized at the plasma membrane (Fig. 1C), demonstrating that an increased side-chain size at this position decreased the transport capacity. Pacheco et al. [9] demonstrated that lactate is a substrate of Ato1, however its transport activity is not sufficient to allow growth on this acid in the conditions here tested (Fig. 2A). Thus, in Ato1, the L129A/G/S/V substitutions increased the capacity of transporting lactate, whereas the L129A/G/S also changed the specificity, enabling the transport of succinate. All of these substitutions were properly localized at the plasma membrane (Fig. 1C)
To determine the effect of the mutations L219A, L219G, L219S, L219V and L219W on the protein structure, we obtained a 3D model of Ato1 and mutated proteins, using the crystal structure of the bacterial homolog SatP_Ck (PDB 5YS3) as a template. Additionally, the pore size of the predicted structures was analysed using the HOLE software (see Material and Methods). The pore radius profile varied significantly in the different mutants, in particular at the narrowest constriction formed by the sidechains of the FLY constrictive site, located in the middle of the protein. As observed in Fig. 3B and C, the native Ato1 3D model predicts a radius of 1.11 Å whereas mutations L219A/G/S/V result in an increased pore radius in the narrowest constrictive site from 1.39 to 1.8 Å. Conversly, the mutation L219W presents a decreased pore radius (0.35 Å) at the constrictive site, that results from the blockage of the pore by the aromatic ring of tryptophan, as observed in the three-dimensional representation of the protein pore (Fig. 3B and C). With this analysis, it was possible to verify the close association between the side-chain size of the amino acid residue at the position 219 and the substrate specificity of Ato1 transporter.
3.3. Acetic acid hypersensitivity is associated with a hyperactive transporter
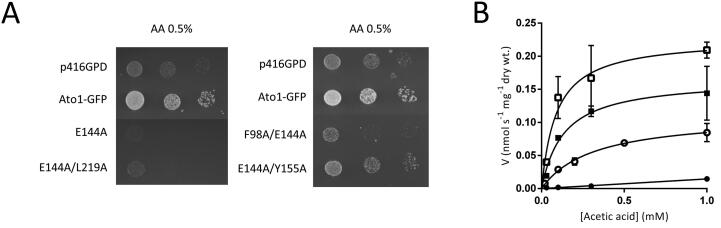
The E144A substitution in Ato1, allowing proper localization at the plasma membrane (Fig. 1C), causes acetic acid hypersensitivity of S. cerevisiae (Fig. 4A), similar to the E144G mutation previously described by Gentsch et al. [24]. Surprisingly, when analysing the kinetics of acetic acid transport of cells expressing the E144A mutant, we observed a much higher activity by the mutant than the wild-type Ato1 (Fig. 4B) suggesting that hypersensitivity is caused by the overactivity of the transporter. The Vmax for acetic acid uptake estimated for the E144A mutant was 0.17 nmol.s−1.mg dry weight−1, which represents a 1.5-fold increase over 0.11 nmol.s−1.mg dry weight−1 estimated for the wild-type strain. The affinity for acetic acid transport increased in the mutant E144A, with a Km value of 0.13 mM whereas the value estimated for the cells expressing the wild-type allele is 0.31 mM. We wondered whether the cells' acetic acid hypersensitivity could be associated with a toxic effect of the protein in the membrane and not caused directly by its activity. To test this hypothesis, we generated the E144A/Y155A double mutant because we previously demonstrated that the single Y155A substitution abolishes acetic acid transport activity of Ato1, while still allowing its proper localization. The double mutant E144A/Y155A was not functional, presenting improper localization (Fig. 1C) and inability to grow on acetic acid (Fig. 4A). On the other hand, the double mutations E144A/L219A and F98A/E144A presented acetic acid hypersensitivity (Fig. 4A) and displayed a high expression level at the plasma membrane (Fig. 1C), revealing the cis-dominant effect of the E144A over the other functional alleles.
Fig. 4.
Functional analysis of S. cerevisiae IMX 1000 cells expressing Ato1-GFP, Ato1-GFP mutants (E144A, E144A/L219A, F98A/E144A and E144A/Y155A) and SatP. A - Growth of Ato1-GFP E144A, E144A/L219A, F98A/E144A and E144A/Y155A mutant cells spotted on YNB media containing acetic acid (0.5% w/v), pH 6.0) (AA) at 18 °C for 10 days. B - Initial uptake rates of radiolabelled 14C-acetic acid as a function of the acid concentration in cells transformed with the plasmids p416GPD (full circles), pAto1-GFP (open circles), pE144A-GFP (full squares), pSatP (empty squares). Cells were collected at mid-exponential growth phase from YNB-glucose (2% w/v), washed and derepressed for 4 h in YNB lactic acid (0.5% w/v, pH 5.0).
3.4. Engineering a hyperactive lactate and succinate Ato1 transporter
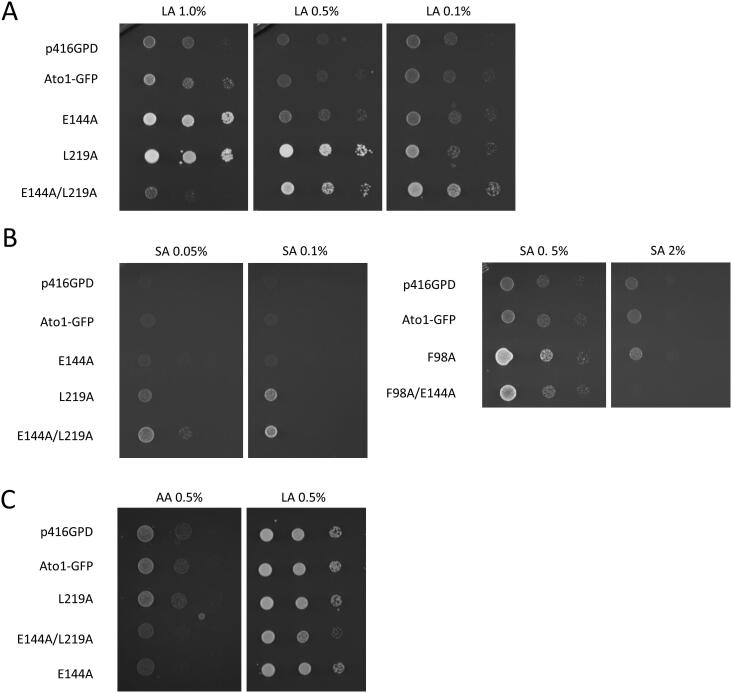
The E144A substitution did not cause significant alterations in cell growth on lactic acid 0.5% or succinic acid 0.5% at pH 5.0 when compared to wild-type Ato1 (Fig. 1A). Since hypersensitivity was associated with increased transport activity, we investigated whether the hypersensitive phenotype could be observed for other acids by combining the E144A and L219A substitutions. The double mutant E144A/L219A displayed hypersensitivity to 1.0% lactic acid (pH 5.0), while the respective single mutants displayed increased growth in these conditions (Fig. 5A). The observed loss of growth of the double mutant did not correspond to a loss of function, since the same mutant grew well on 0.5% lactic acid (pH 5.0) (Fig. 5A). In fact, at a lower concentration of lactic acid (0.1%), the double mutant exhibited better growth performance than the respective single mutants (Fig. 5A). We observed similar concentration-dependent behaviour of the double mutant E144A/L219A on varying concentrations of succinic acid (Fig. 5B). These results suggest that hyperactive transport can be advantageous for cell growth, as long as the acid metabolism can cope with the increased uptake rates. Next, we questioned whether the simultaneous co-expression of the wild-type Ato1 and the hyperactive mutant would affect the hypersensitive phenotypes observed. If the expression of hyperactive organic acid transporter was the cause of hypersensitive phenotypes, the same phenotypes would also occur in strains that co-express wild-type and hyperactive Ato1. The S. cerevisiae CEN.PK 113-5D strain can utilize both acetic and lactic acid as sole carbon and energy sources (Fig. 5C). This strain displayed impaired growth on acetic acid when expressing the mutant Ato1 transporter E144A (Fig. 5C), even though the wild-type ATO1 gene is present in the genome of the cell. When expressing the E144A/L219A mutant, the S. cerevisiae CEN.PK 113-5D strain displayed impaired growth on acetic and lactic acids (Fig. 5C). The observed trans-dominant negative behaviour of E144A allele for acetic acid sensitivity is in accordance with a previous report by Gentsch et al. [24].
Fig. 5.
Analysis of the hypersensitivity phenotype caused by Ato1-GFP E144A mutation. A – Spot assay of S. cerevisiae IMX1000 transformed with Ato1-GFP mutant alleles in the residues E144, L219 and E144/L219, grown on YNB media containing 1%, 0.5% and 0.1% of lactic acid, pH 5.0 (LA) at 18 °C for 10 days. B – Spot assay of S. cerevisiae IMX1000 transformed with Ato1-GFP mutant alleles in the residues E144A, L219A, E144/L219A and F98A/E144A, grown on YNB containing 0.05%, 0.1%, 0.5% and 2.0% succinic acid, pH 5.0 (SA) at 18 °C for 10 days. C – Spot assay of S. cerevisiae CEN.PK 113-5D transformed with Ato1-GFP mutants in the residues E144, L219, E144/L219, grown on YNB media containing acetic acid (0.5%, pH 6.0) (AA) or lactic acid (0.5%, pH 5.0) (LA) at 18 °C for 10 days.
3.5. The bacterial acetate channel SatP_Ec is functional in yeast
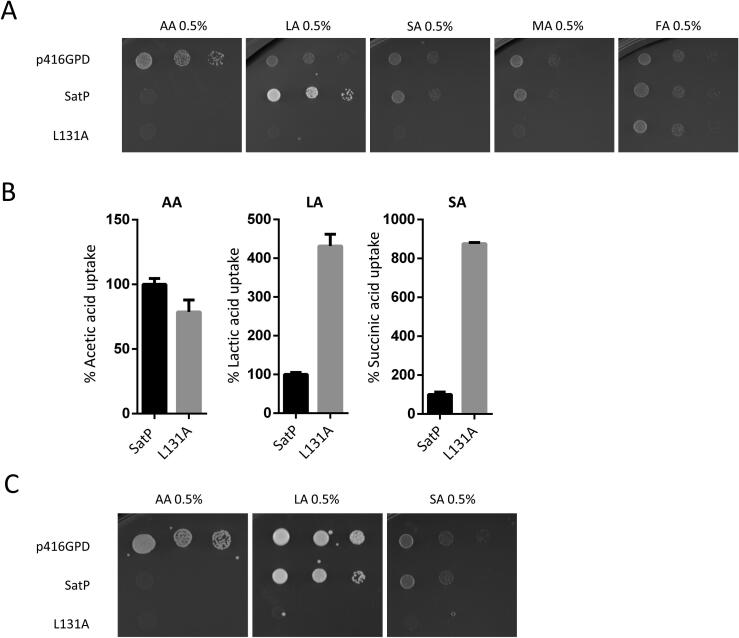
The acetate transporter SatP_Ec shares 36% protein sequence similarity with Ato1 (Fig. S1). In this work, we expressed a codon optimized version of the SatP_Ec in the strain S. cerevisiae IMX1000. The resultant strain is hypersensitive to acetic acid, similar to the strain expressing Ato1 mutant E144A (Fig. 6A). Additionally, SatP_Ec expression allowed the S. cerevisiae IMX1000 strain to grow on medium containing lactic acid (0.5%, pH 5.0) as a sole carbon source (Fig. 6A). Initial uptake rates of acetic acid were increased in cells transformed with SatP_Ec, when compared to cells transformed with the corresponding empty vector (Fig. 4B), further demonstrating that SatP_Ec is functional in S. cerevisiae. The kinetics for acetic acid in S. cerevisiae cells expressing SatP_Ec were a Vmax of 0.23 nmol.s−1.mg (dcw)-1 and a Km value of 0.09 mM (Fig. 4B).
Fig. 6.
Functional analysis of SatP_Ec in S. cerevisiae.A – Spot assay of S. cerevisiae IMX1000 transformed with pSatP and pSatP-L131A plasmids. Cells were grown on YNB media containing acetic acid (0.5%, pH 6.0) (AA), lactic acid (0.5%, pH 5.0) (LA), succinic acid (0.5%, pH 5.0) (SA), malic acid (0.5%, pH 5.0) (MA), or fumaric acid (0.5% pH 5.0) (FA), at 18 °C for 10 days. B – Uptake of acetic acid (1.0 mM, pH 6.0), lactic acid (3.0 mM, pH 5.5) and succinic acid (2.0 mM, pH 5.5) in S. cerevisiae IMX1000 cells transformed with the plasmids pSatP and pSatP-L131A. Cells were collected at mid-exponential growth phase from YNB-glucose 2% (w/v), washed and derepressed for 4 h in YNB glycerol (6%). C – Spot assay of S. cerevisiae CEN.PK 113-5D strain transformed with p416GPD, pSatP and pSatP-L131A on YNB containing acetic acid (0.5%, pH 6.0) (AA), lactic acid (0.5%, pH 5.0) (LA) or succinic acid (0.5%, pH 5.0) (SA), at 18 °C for 10 days.
The central constrictive site (FLY) of SatP_Ec was also engineered by introducing the L131A substitution (equivalent to L219A in Ato1). This substitution caused hypersensitivity not only to acetic, but also to lactic, succinic, and malic acids, indicating that it generates a hyperactive transporter for these acids (Fig. 6A). The same cells did not exhibit any growth differences on fumaric acid compared to the wild-type SatP_Ec or the empty vector (Fig. 6A). The SatP_Ec L131A mutant showed similar uptake of acetic acid and increased uptake of lactic and succinic acids compared to wild-type SatP_Ec (Fig. 6B). Trans-dominant negative hypersensitive phenotypes were present when S. cerevisiae CEN.PK 113-5D strains were transformed with pSatP and pSatP L131A (Fig. 6C).
4. Discussion
4.1. Ato1 residues involved in substrate specificity and activity
Ato1 is a plasma membrane transporter from the model organism S. cerevisiae, and a member of the Acetate Uptake Transporter (AceTr) family, TC #2.A.96. It shows transport activity for industrially relevant organic acids such as acetate and lactate, while its bacterial homologue SatP_Ec also shows transport activity for succinate, which is another biotechnological product of interest. Prior to this work, equivalent amino acid substitutions resulted in an increased lactate transport capacity of Ato1 (L219V and A252G) and SatP_Ec (L131V and A164G), but the structural mechanisms causing these alterations in the transporter function were not unveiled. Recently, a work of Baldi et al. [11] identified amino acid substitutions in Ato2 (L218S) and Ato3 (F95S) which also led to an increased lactic acid transport capacity in S. cerevisiae, proposing the widening of the narrowest constriction site of these two mutants as the reason behind the observed gain-of-function phenotypes. Notably, residues L218 in Ato2 and F95 in Ato3 are equivalent to L219 and F98 in Ato1, respectively.
Also, the reasons underlying acetic acid hypersensitivity of yeast cells expressing specific Ato1 mutants [24] were not yet elucidated. Considering the aforementioned features, and the fact that the recently published crystal structures of SatP_Ck [20] and SatP_Ec [19] allowed a more precise 3D modelling of AceTr homologues, we decided to deepen the knowledge on Ato1 structure–function relationship. Our initial rational design of amino acid substitutions to alanine included 34 single Ato1-GFP mutants (Fig. 1A). The 34 single substitutions led to distinct phenotypes such as reduction or complete loss of Ato1 function, increased Ato1 transport capacity for lactate, gain of function for succinate transport, and hypersensitivity of the yeast cells to acetic acid (Fig. 1B). Among the mutants which presented reduced or lost function, only E140A and Y155A (Fig. 1C) could be detected at the plasma membrane, which points towards the involvement of these residues in substrate translocation through Ato1. The residue equivalent to E140 in SatP_Ck, the E57 residue, interacts with acetate molecules via a water-mediated hydrogen bond network within the S1 binding site [20]. Furthermore, the E57A substitution abolished the acetate transport activity in SatP_Ck [20]. The remaining mutants from this group were not detected in the plasma membrane, probably playing a role in structural stability, folding and trafficking of Ato1, although we cannot completely exclude their potential involvement in substrate translocation. This can be the case for the conserved aromatic residues within the substrate pathway, such as F64 and W76 (equivalent to F147 and W159 in Ato1, respectively) which coordinate the translocation of acetate molecules via anion-π interactions in SatP_Ck [20]. Additionally, the residue Q50, equivalent to Q133 in Ato1, was identified as the key residue responsible for the transition of SatP_Ck from closed into the open state, via molecular dynamics simulation [37]. Moreover, Q50 was found to be essential for acetate transport in SatP_Ck by functioning as a hydrogen bond donor [20].
The F98A and L219A Ato1 mutants display a gain of function phenotype by enabling cell growth on succinic acid as a sole carbon source associated with the gain of transport activity for this acid (Fig. 2). It was previously shown that the L219V and A252G substitutions improve yeast growth on lactic acid [18], but unlike F98A and L219A, they do not enable growth on succinic acid as a sole carbon source (Fig. 2A and S3). Similarly to L219A, the L219S Ato1 mutation also improved cell growth on lactic and succinic acids as sole carbon sources. The equivalent substitution in Ato2 (L218S) improved growth on lactic acid, while growth on succinic acid was not tested [11]. In this work, we show that the size of the residue at the position 219 modulates transporter specificity and activity. By increasing the size of the narrowest constriction in the L219A mutant, the transport of larger molecules, like succinate, becomes possible and the transport of other larger molecules, like lactate, is more efficient (Fig. 2). On the other hand, a larger residue can cause the complete blockage of acetate transport through the pore (Fig. 3), as observed in the L219W mutant that is unable to grow on all tested substrates. It is important to note that the original model of SatP_Ck obtained under the crystallization conditions is based on the closed state of the transporter, and the central constriction is physically too narrow to allow the passage of acetate or other small anions [20]. Additionally, we cannot exclude that other structural features besides the pore size are also contributing to Ato1 substrate specificity and transport activity. This was observed for the substitution Y155A that widens the pore but results in the loss of transporter function (Fig. 1B and 2B). Molecular dynamic simulations predict that the outward movement of SatP_Ck residues F17 and Q50 open the FLY constrictive site enabling the flow of acetate [37], thus the presence of specific interactions is necessary for protein function. Furthermore, excessive modifications of the Ato1 pore, such as in F98A/L219A (Fig. 2A) or L219A/A252G (Fig. S3) double mutants, led towards the complete abolishment of Ato1 transport activity.
4.2. Understanding the dominant negative acetic acid hypersensitivity phenotype
The AceTr members Ato1, SatP_Ec, and Gpr1 are all functionally characterized as acetic acid transporters [5], [10], [16]. In this work, we found that the hypersensitive phenotype exhibited by yeast expressing Ato1 mutant E144A or SatP_Ec is linked to high uptake rates of acetic acid. In S. cerevisiae, the Ato1 mutant E144A displayed significantly higher activity than the wild-type Ato1 and the expression of the bacterial SatP_Ec led to even higher acetic acid uptake rates than the Ato1 E144A mutant (Fig. 4B). By combining the mutations which lead to loss of Ato1 function and to the yeast acetic acid hypersensitivity, we demonstrated that transport activities of Ato1 E144A mutant and SatP_Ec are directly responsible for the observed higher uptake rates of organic acids, causing hypersensitivity of yeast cells to these acids. Additionally, the expression of the mutant transporter Ato1 E144A and SatP_Ec triggered acetic acid hypersensitivity in a dominant negative manner in S. cerevisiae (Fig. 4, Fig. 6, respectively). The dominant negative effect of mutant alleles from the AceTr family on acetate utilization was first observed by Kujau et al. [22], as Y. lypolytica GPR1d mutants were unable to grow on acetate as sole carbon source and the expression of the wild-type GPR1 could not complement the observed lack of growth. At the time, Gpr1 was proposed to function as a glyoxylate pathway regulator, hence the Gpr1 = glyoxylate pathway regulator name. Tzschoppe et al. [21] showed that acetate had a negative effect on growth of Y. lypolytica GPR1d mutants even in the presence of glucose, thus proposing the involvement of Gpr1 in acetic acid stress response. The same type of acetic acid hypersensitivity was later reported for several Ato1 and Ato2 mutants in S. cerevisiae (see Introduction), including the E144G mutant of Ato1 [24]. As we demonstrated in this work, the hypersensitivity phenotype was extended to other carboxylic acids through the engineering of the central constrictive site (FLY), and consequent increase of substrate specificity and activity of Ato1 E144A/L219A as well as of SatP_Ec L131A mutants (Fig. 5, Fig. 6). Cells expressing the Ato1 E144A/L219A double mutant displayed hypersensitivity to lactic and succinic acids in a concentration dependent manner (Fig. 5), as the L219A substitution allowed Ato1 E144A/L219A double mutant to import these acids more efficiently. Accordingly, SatP_Ec L131A mutant enabled significantly higher uptake rates of lactic and succinic acids compared to SatP_Ec and consequently caused the hypersensitivity of yeast cells to these acids (Fig. 6). At lower concentrations of lactic and succinic acids in the culture media, the Ato1 E144A/L219A double mutant expression exhibited a beneficial rather than toxic effect and enabled growth of S. cerevisiae IMX1000 strain on these acids as sole carbon sources even more than the Ato1 L219A allele (Fig. 5). Our results indicate that Ato1 becomes hyperactive upon E144A substitution, while the transport level presented by SatP_Ec in S. cerevisiae is already sufficient to promote a toxic effect when cells are grown on acetic acid (0.5%, pH 6.0). Uptake of organic acids by hyperactive transporter variants from the AceTr family can overwhelm the yeast cells and inhibit their growth, but such uptake can also be advantageous when concentrations of organic acids are low, as it provides the cells with more carbon and energy sources within a given time. This may explain why yeast species typically possess several AceTr homologues in their genomes [10], [39], which may switch on and off depending on their kinetic properties and the acid concentration in the environment, however confirmation of this hypothesis remains to be proved. Still, our findings explain why the organic acid hypersensitivity related with the expression of Ato1 E144A mutant and SatP_Ec displayed dominant negative behaviour and support the idea that the same phenomenon occurred in previously reported Gpr1, Ato1 and Ato2 mutants [22], [24].
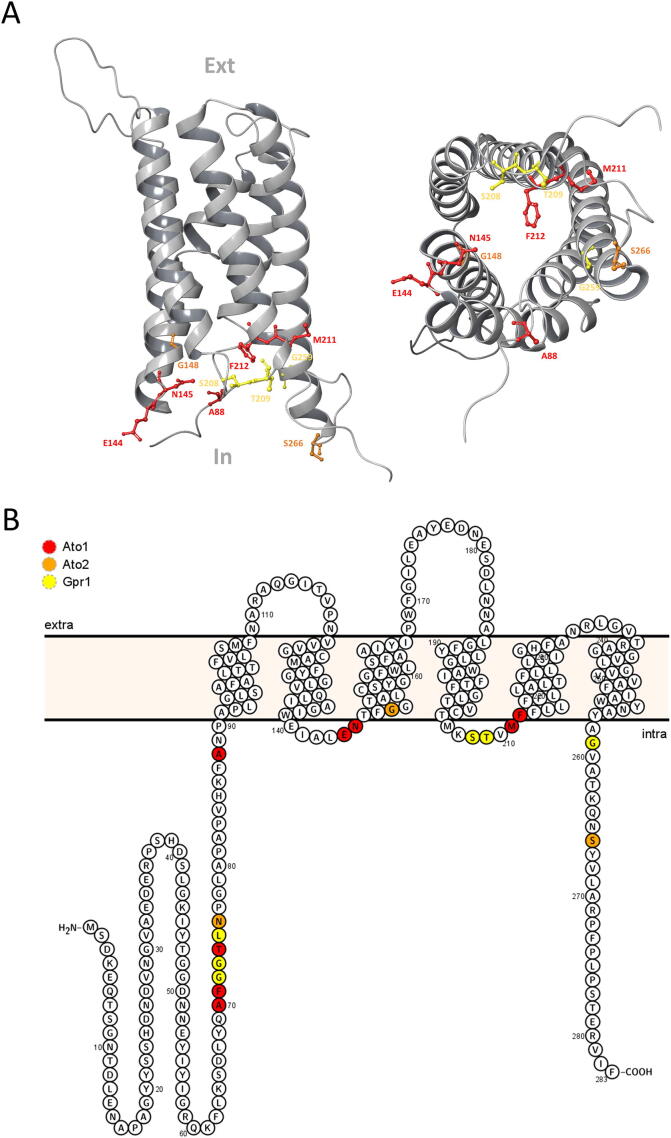
4.3. The regulatory role of Ato1 termini and loops
In an effort to better understand the structural basis of Ato1 mutants displaying overactivity phenotype, we analysed the locations of these amino acid residues within Ato1, including the locations of the equivalent amino acids from Ato2 and Gpr1, which also led to acetic acid hypersensitivity in yeast. According to the predicted 3D model of Ato1 (Fig. 7) these amino acids are only found in the cytosolic termini and cytosolic loops. Cytosolic termini and loops of Ato1, Ato2 and Gpr1 are most probably playing a role in regulating the influx of organic acids from the extracellular medium into the cytosol, possibly by assembling a barrier which slows down or blocks organic acid entry, such as gates. Introduction of mutations which were described as hypersensitive may prevent the assembly of such barrier, and promote the constantly open state of Ato1, Ato2 and Gpr1. The involvement of tails and loops of various transporters in gating function was recently reviewed by Mikros & Diallinas [40]. Alternatively, the cytosolic termini and loops of these transporters may assemble a structure which serves as a binding site for an unknown negative regulator. Being a bacterial transporter, SatP_Ec has shorter termini and loops than its eukaryotic homologs. Moreover, SatP_Ec contains a glycine residue (G61) instead of glutamate at the position which corresponds to the E144 of Ato1, where the E144G substitution caused acetic acid hypersensitivity [24]. These observations might explain why the expression of SatP_Ec causes acetic acid hypersensitivity in yeast, without the requirement of any further mutation. Overall, these data provide strong evidence that the cytosolic termini and loops play an important role in the regulation of transporters from the AceTr family.
Fig. 7.
Structural analysis of amino acids from Ato1 (red), Ato2 (orange), and Gpr1 (yellow) whose substitutions led to acetic acid hypersensitivity of yeast. A – Location of amino acids whose substitutions led to acetic acid hypersensitivity of yeast within a predicted 3D structure model of Ato1 obtained by homology threading using SatP_Ck crystal structure (PDB Entry: 5YS3) as template. In-intracellular; Ext-extracellular. B – Location of amino acids whose substitutions led to acetic acid hypersensitivity of yeast within a visualized predicted topology of Ato1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The toxic effects of organic acids on microbial cell growth are a well-known phenomenon typically caused by the passive diffusion of undissociated form of the acids into the cytosol [17]. Inside the cell, such organic acids predominantly dissociate into protons and anions because their pKa values are lower than the near-neutral cytosolic pH value. The released protons acidify the cytosol and disturb cell homeostasis, while anions may produce specific toxic effects on the metabolism depending on the organic acid in play [41]. In this work, organic acid toxicity occurred due to the hyperactive organic acid uptake via Ato1 E144A mutant and SatP_Ec, rather than passive diffusion, since at the tested pH values the diffusion component of the acids is negligible. Published crystal structures of SatP_Ec and SatP_Ck depict channel-like structures, and in vitro transport studies demonstrated anion conducting activities of SatP_Ec and SatP_Ck, without proton symport [19], [20]. On the other hand, in vivo transport studies of Ato1 and Gpr1 in S. cerevisiae, and SatP_Ec in E. coli, showed that the acetate uptake depends on proton motive force, indicating that these transporters function, directly or indirectly, coupled with protons [5], [10], [16].
With the current knowledge on the mechanism of transport from the AceTr family it is unclear whether hypersensitivity emerged solely from the overwhelming influx of organic acid anions, or organic acid anions and protons combined.
4.4. Functional expression of the bacterial transporter SatP_Ec in yeast
An important achievement of this work is the functional expression of the full amino acid sequence of SatP_Ec in S. cerevisiae without any further modifications besides the use of a codon adapted gene sequence (Fig. 6). To our knowledge this is the first report of a prokaryotic plasma membrane transporter that is functional in a yeast cell. The only evidence of a bacterial transporter expressed in yeast is the araE transporter that could enhance resveratrol accumulation, but without transporting resveratrol directly, possibly by affecting lipid bilayer permeability [42].
SatP_Ec shares 36% of protein sequence similarity with Ato1 and mediates the uptake of acetate, lactate and succinate in vivo [16] and in vitro [19]. Surprisingly, in S. cerevisiae, SatP_Ec presented a different specificity, mediating the uptake of acetic and lactic acids but not of succinic acid. Perhaps the differences in membrane composition, intracellular charge, or intracellular pH between S. cerevisiae and E. coli may have a strong enough impact on the SatP_Ec structure to allow the passage of monocarboxylates and restrict the passage of succinic acid molecules. However, the L131A substitution in the central FLY constrictive site of SatP_Ec enabled the passage of succinic acid, increased the lactic acid uptake rate and triggered malic acid hypersensitivity (Fig. 6A and B) supporting the role of the constrictive FLY site on substrate specificity and activity in this family of transporters.
4.5. Applicability of the engineered Ato1 and SatP_Ec in the biotech industry
The scientific community and the biotech industry are currently focused on the development of robust microbial cell factories for efficient organic acid production from cheap industrial biowastes [43]. Since the recognition that organic acid export over the plasma membrane represents one of the key steps in microbial production of these compounds, organic acid transporters started receiving greater attention in metabolic engineering strategies [43]. Efforts aimed at the identification of biotechnologically relevant membrane transporters and their further improvement by the means of transporter engineering will lead towards more efficient biotechnological processes, however, more structure–function relationship models are needed to boost the emerging field of transporter engineering [2].
An important achievement of this work was the ability to engineer Ato1 and SatP_Ec proteins to transport succinic acid, a biotechnological product of interest. Many efforts have been made to engineer efficient bio-production of succinic acid (for a review, see [44]) and efficient succinic acid exporters are crucial to achieve high titres of this acid by yeast cell factories. Currently, industrial strains were engineered for the expression of transporters SpMae1 and AnDCT-02, both described as anion channels [45] that promote succinate export, allowing the production of high succinic acid titres at low pH [46]. In this work, we analysed ATO1 and SatP_Ec alleles only in terms of organic acid import. Whether Ato1 L219A, Ato1 E144A/L219A, Ato1 F98A/E144A and SatP_Ec L131A can work as efficient succinic acid exporters in yeast cell factories will be a further step of our research. However, as anion channels, these transporters may allow the production of high succinic acid titres similarly to SpMae1 and AnDCT-02. In this study, we showed that S. cerevisiae can be a useful platform for the characterization of codon optimized bacterial transporters. S. cerevisiae “null-platform” strains with deleted native transporters of substrates such as organic acids [26] and sugars [47] are nowadays available and can be easily implemented for screening and characterization of transporters of these substrates via spot assays, radiolabelled substrate uptake assays, and fermentations. Bacteria can inhabit the most diverse environments, often surviving in extreme conditions with limited nutrients. As such, they are a great source of transporter genes that might be useful for yeast-based biotechnology. We hope that our finding will promote the study of other functional bacterial transporters in yeast and bring a contribution to the biotech industry.
CRediT authorship contribution statement
Toni Rendulić: Investigation, Validation, Visualization, Writing – original draft. João Alves: Investigation, Visualization. João Azevedo-Silva: Investigation. Isabel Soares-Silva: Investigation, Validation, Writing - review & editing. Margarida Casal: Conceptualization, Investigation, Validation, Funding acquisition, Supervision, Writing – original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the strategic programme UID/BIA/04050/2019 funded by Portuguese funds through the FCT-IP, the project TransAcids (PTDC/BIAMIC/5184/2014) funded by FCT-IP and ERDF by COMPETE 2020-POCI and the project River2Ocean NORTE-01-0145-FEDER-000068, co-financed by the European Regional Development Fund (ERDF), through Programa Operacional Regional do Norte (NORTE 2020) as well as the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Yeastdoc grant agreement No 764927.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
Supplementary Fig. 2.
Supplementary Fig. 3.
References
- 1.Abbott D.A., Zelle R.M., Pronk J.T., Van Maris A.J.A. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: Current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 2.Kell D.B., Swainston N., Pir P., Oliver S.G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015;33(4):237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Casal M., Queirós O., Talaia G., Ribas D., Paiva S. Carboxylic acids plasma membrane transporters in Saccharomyces cerevisiae. Adv Exp Med Biol. 2016;892:229–251. doi: 10.1007/978-3-319-25304-6_9. [DOI] [PubMed] [Google Scholar]
- 4.Casal M., Paiva S., Andrade R.P., Gancedo C., Leão C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J Bacteriol. 1999;181(8):2620–2623. doi: 10.1128/JB.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paiva S., Devaux F., Barbosa S., Jacq C., Casal M. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast. 2004;21(3):201–210. doi: 10.1002/yea.1056. [DOI] [PubMed] [Google Scholar]
- 6.Mollapour M., Piper P.W. Hog1 Mitogen-Activated Protein Kinase Phosphorylation Targets the Yeast Fps1 Aquaglyceroporin for Endocytosis, Thereby Rendering Cells Resistant to Acetic Acid. Mol Cell Biol. 2007;27 doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piper P., Mahé Y., Thompson S., Pandjaitan R., Holyoak C., Egner R. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17 doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holyoak C.D., Bracey D., Piper P.W., Kuchler K., Coote P.J. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy- dependent mechanism. J Bacteriol. 1999;181(15):4644–4652. doi: 10.1128/JB.181.15.4644-4652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacheco A., Talaia G., Sá-Pessoa J., Bessa D., Gonçalves M.J., Moreira R. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Res. 2012;12(3):375–381. doi: 10.1111/j.1567-1364.2012.00790.x. [DOI] [PubMed] [Google Scholar]
- 10.Ribas D., Soares-Silva I., Vieira D., Sousa-Silva M., Sá-Pessoa J., Azevedo-Silva J. The acetate uptake transporter family motif “NPAPLGL(M/S)” is essential for substrate uptake. Fungal Genet Biol. 2019;122:1–10. doi: 10.1016/j.fgb.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Baldi N., de Valk S.C., Sousa-Silva M., Casal M., Soares-Silva I., Mans R. Evolutionary engineering reveals amino acid substitutions in Ato2 and Ato3 that allow improved growth of Saccharomyces cerevisiae on lactic acid. FEMS Yeast Res. 2021;21:1–12. doi: 10.1093/femsyr/foab033. [DOI] [PubMed] [Google Scholar]
- 12.Rabitsch K.P., Tóth A., Gálová M., Schleiffer A., Schaffner G., Aigner E. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11(13):1001–1009. doi: 10.1016/S0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 13.Sá-Pessoa J., Amillis S., Casal M., Diallinas G. Expression and specificity profile of the major acetate transporter AcpA in Aspergillus nidulans. Fungal Genet Biol. 2015;76:93–103. doi: 10.1016/j.fgb.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Turner T.L., Lane S., Jayakody L.N., Zhang G.C., Kim H., Cho W. Deletion of JEN1 and ADY2 reduces lactic acid yield from an engineered Saccharomyces cerevisiae, in xylose medium, expressing a heterologous lactate dehydrogenase. FEMS Yeast Res. 2019:19. doi: 10.1093/femsyr/foz050. [DOI] [PubMed] [Google Scholar]
- 15.Robellet X., Flipphi M., Pégot S., MacCabe A.P., Vélot C. AcpA, a member of the GPR1/FUN34/YaaH membrane protein family, is essential for acetate permease activity in the hyphal fungus Aspergillus nidulans. Biochem J. 2008;412 doi: 10.1042/BJ20080124. [DOI] [PubMed] [Google Scholar]
- 16.Sá-Pessoa J, Paiva S, Ribas D, Silva IJ, Viegas SC, Arraiano CM. SATP (YaaH), a succinate-acetate transporter protein in Escherichia coli. Biochem J. 2013;454 doi: 10.1042/BJ20130412. [DOI] [PubMed] [Google Scholar]
- 17.Casal M., Paiva S., Queirós O., Soares-Silva I. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev. 2008;32(6):974–994. doi: 10.1111/j.1574-6976.2008.00128.x. [DOI] [PubMed] [Google Scholar]
- 18.Kok S., Nijkamp J.F., Oud B., Roque F.C., Ridder D., Daran J.-M. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res. 2012;12(3):359–374. doi: 10.1111/j.1567-1364.2012.00787.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun P., Li J., Zhang X., Guan Z., Xiao Q., Zhao C. Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel. J Biol Chem. 2018;293(50):19492–19500. doi: 10.1074/jbc.RA118.003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu B., Xia B., Zhou Q., Lu Y., He M., Hasegawa K. Succinate-acetate permease from Citrobacter koseri is an anion channel that unidirectionally translocates acetate. Cell Res. 2018;28(6):644–654. doi: 10.1038/s41422-018-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzschoppe K., Augstein A., Bauer R., Kohlwein S.D., Barth G. Trans-dominant mutations in the GPR1 gene cause high sensitivity to acetic acid and ethanol in the yeast Yarrowia lipolytica. Yeast. 1999;15(15):1645–1656. doi: 10.1002/(ISSN)1097-006110.1002/(SICI)1097-0061(199911)15:15<>1.0.CO;2-I10.1002/(SICI)1097-0061(199911)15:15<1645::AID-YEA491>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 22.Kujau M., Weber H., Barth G. Characterization of mutants of the yeast Yarrowia lipolytica defective in acetyl-coenzyme a synthetase. Yeast. 1992;8(3):193–203. doi: 10.1002/yea.320080305. [DOI] [PubMed] [Google Scholar]
- 23.Augstein A., Barth K., Gentsch M., Kohlwein S.D., Barth G. Characterization, localization and functional analysis of Gpr1p, a protein affecting sensitivity to acetic acid in the yeast Yarrowia lipolytica. Microbiology. 2003;149:589–600. doi: 10.1099/mic.0.25917-0. [DOI] [PubMed] [Google Scholar]
- 24.Gentsch M., Kuschel M., Schlegel S., Barth G. Mutations at different sites in members of the Gpr1/Fun34/YaaH protein family cause hypersensitivity to acetic acid in Saccharomyces cerevisiae as well as in Yarrowia lipolytica. FEMS Yeast Res. 2007;7:380–390. doi: 10.1111/j.1567-1364.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 25.Matthäus F, Barth G. The Gpr1/Fun34/YaaH Protein Family in the Nonconventional Yeast Yarrowia lipolytica and the Conventional Yeast Saccharomyces. cerevisiae. 2013:145–163. doi: 10.1007/978-3-642-38320-5_7. [DOI] [Google Scholar]
- 26.Mans R, Hassing EJ, Wijsman M, Giezekamp A, Pronk JT, Daran JM. A CRISPR/Cas9-based exploration into the elusive mechanism for lactate export in Saccharomyces cerevisiae. FEMS Yeast Res. 2017;17(8) doi: 10.1093/femsyr/fox085. [DOI] [PubMed] [Google Scholar]
- 27.Entian K.D., Kötter P. 25 Yeast Genetic Strain and Plasmid Collections. Methods in Microbiology. 2007;36 doi: 10.1016/S0580-9517(06)36025-4. [DOI] [Google Scholar]
- 28.Mumberg D., Müller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156(1):119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 29.Chang J.-M., Di Tommaso P., Taly J.-F., Notredame C. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinf. 2012;13(Suppl 4):S1. doi: 10.1186/1471-2105-13-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omasits U., Ahrens C.H., Müller S., Wollscheid B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30(6):884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann L., Stephens A., Nam S.-Z., Rau D., Kübler J., Lozajic M. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430(15):2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribas D, Sá-Pessoa J, Soares-Silva I, Paiva S, Nygård Y, Ruohonen L. Yeast as a tool to express sugar acid transporters with biotechnological interest. FEMS Yeast Research. 2017;17 doi: 10.1093/femsyr/fox005. [DOI] [PubMed] [Google Scholar]
- 35.Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., Sansom M.S.P. A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14(6):354–360. doi: 10.1016/S0263-7855(97)00009-X. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Wu M., Sun L., Zhou Q., Peng Y., Liu Z., Zhao S. Molecular Mechanism of Acetate Transport through the Acetate Channel SatP. J Chem Inf Model. 2019;59(5):2374–2382. doi: 10.1021/acs.jcim.8b00975. [DOI] [PubMed] [Google Scholar]
- 38.Soares-Silva I., Paiva S., Diallinas G., Casal M. The conserved sequence NXX[S/T]HX[S/T]QDXXXT of the lactate/pyruvate:H+ symporter subfamily defines the function of the substrate translocation pathway. Mol Membr Biol. 2007;24(5-6):464–474. doi: 10.1080/09687680701342669. [DOI] [PubMed] [Google Scholar]
- 39.Alves R., Sousa-Silva M., Vieira D., Soares P., Chebaro Y., Lorenz M.C. Carboxylic Acid Transporters in Candida Pathogenesis. MBio. 2020;11(3) doi: 10.1128/mBio.00156-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikros E., Diallinas G. Tales of tails in transporters. Open Biology. 2019;9(6):190083. doi: 10.1098/rsob.190083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. Weak acid adaptation: The stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology. 2001;147(10):2635–2642. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Halls C., Zhang J., Matsuno M., Zhang Y., Yu O. Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng. 2011;13(5):455–463. doi: 10.1016/j.ymben.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Soares-Silva I., Ribas D., Sousa-Silva M., Azevedo-Silva J., Rendulić T., Casal M. Membrane transporters in the bioproduction of organic acids: State of the art and future perspectives for industrial applications. FEMS Microbiol Lett. 2020;367 doi: 10.1093/femsle/fnaa118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn J.H., Jang Y.S., Lee S.Y. Production of succinic acid by metabolically engineered microorganisms. Curr Opin Biotechnol. 2016;42:54–66. doi: 10.1016/j.copbio.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Darbani B., Stovicek V., van der Hoek S.A., Borodina I. Engineering energetically efficient transport of dicarboxylic acids in yeast Saccharomyces cerevisiae. PNAS. 2019;116(39):19415–19420. doi: 10.1073/pnas.1900287116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jansen MLA, Heijen JJ, Verwaal R. (12) United States Patent. WO/2013/004670, 2013.
- 47.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Letters. 1999;464(3):123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]