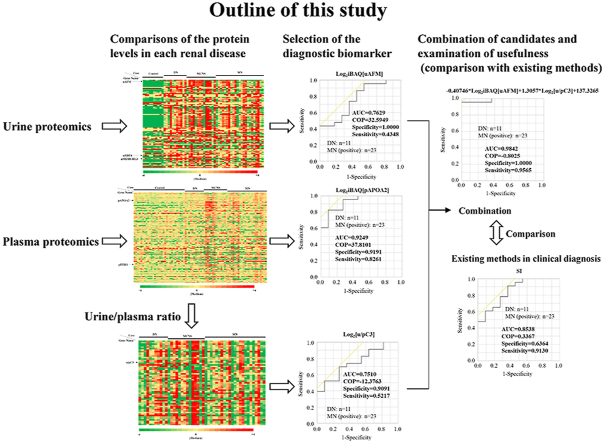
Abstract
The choice of treatment for primary nephrotic syndrome depends on the pathologic type of the disorder. Renal biopsy is necessary for a definitive diagnosis, but it is burdensome for the patients, and can be avoided if tests could be performed using urine or plasma. In this study, we analyzed 100 urinary proteins, 141 plasma proteins, and 57 urine/plasma ratios in cases of diabetic nephropathy (DN; n = 11), minimal change nephrotic syndrome (MCNS; n = 14), and membranous nephropathy (MN; n = 23). We found that the combination of urinary retinol-binding protein 4 and SH3 domain-binding glutamic acid-rich-like protein 3 could distinguish between MCNS and DN, with an area under the curve (AUC) of 0.9740. On the other hand, a selectivity index (SI) based on serotransferrin and immunoglobulin G, which is often used in clinical practice, distinguished them with an AUC of 0.9091. Similarly, the combination of urinary afamin and complement C3 urine/plasma ratio could distinguish between MN and DN with an AUC of 0.9842, while SI distinguished them with an AUC of 0.8538. Evidently, the candidates identified in this study were superior to the SI method. Thus, the aim was to test these biomarkers for accurate diagnosis and to greatly reduce the burden on patients.
Keywords: Urinary proteome, Biomarker, Diabetic nephropathy, Minimal change nephrotic syndrome, Membranous nephropathy
Graphical abstract
Highlights
-
•
Renal biopsy is necessary for a definitive diagnosis of primary nephrotic syndrome.
-
•
Renal biopsy is a heavy burden for patients, and a less burdensome test is desired.
-
•
We discovered at least 6 diagnostic biomarkers using urinary and plasma proteomics.
-
•
Combination allowed to judge the disease more accurately than selectivity index.
1. Introduction
Primary nephrotic syndrome is a group of renal disorders with massive glomerular proteinuria, hypoalbuminemia, and edema, without systemic disease. The main types of disease are minimal change nephrotic syndrome (MCNS) [1], membranous nephropathy (MN) [2], focal segmental glomerulosclerosis, and membranous proliferative glomerulonephritis. It occurs in both children and elderly people—MCNS occurs mainly in children and MN in the elderly. On the other hand, secondary nephrotic syndrome is caused by other underlying diseases, the most common being diabetic nephropathy (DN) caused by diabetes mellitus [3].
It has been reported that about half of the kidney diseases in patients with diabetes are non-diabetic renal diseases, such as MN, immunoglobulin (Ig) A nephropathy, and MCNS [4]. Steroid treatment is required for such cases, which may be delayed if the case is inaccurately diagnosed as DN. Renal biopsy can help in diagnosis, prognosis, and treatment plan, but its use is limited, as it is an invasive procedure. The burden on the patients will be greatly reduced if tests can be performed using urine or plasma that can be collected relatively easily.
In the past, the discovery of biomarkers has been mainly carried out using proteomics technology [5]. Proteomics can identify invaluable disease-specific biomarkers by analyzing the global protein profiling in body fluids, such as urine and blood [6,7]. In fact, in many patients with renal disease, several biomarkers associated with the pathophysiology of the disease have been identified by urinary and plasma proteomics analyses, some of which have been put to practical use [[8], [9], [10]]. The urinary excretion of albumin (uALB) and β2-microglobulin (uB2M) is actually used as a marker for glomerular and tubular damage, respectively.
Few studies have performed quantitative proteomics analyses of urine and plasma simultaneously. In these analyses, it was not clear whether the level of urinary protein was different in each case due to a difference in the level of plasma protein or an increased level of secreted protein caused by renal injury. In this study, we performed label-free quantitative proteomics analyses of both urine and plasma for 11 DN, 14 MCNS, and 23 MN cases. The urine/plasma ratio, in addition to urinary or plasma levels, was evaluated for marker candidates that could be used in the diagnosis of each renal disease. Furthermore, it was compared with the selectivity index (SI), uALB, and uB2M to verify the usefulness of these candidates.
2. Materials and methods
2.1. Materials
Urine samples from five male and five female healthy control donors were purchased from Lee BioSolutions Inc. (St. Louis, MO, USA), and the normal plasma control samples (n = 30) were obtained from George King Bio-Medical, Inc. (Overland Park, KS).
2.2. Clinical samples
Patient characteristics are summarized in Table 1. Among them, 5, 12, and 12 patients with DN, MCNS, and MN, respectively, were nephrotic syndrome which is defined by a urinary protein level exceeding 3.5 g per day. On the other hand, anti-M-type phospholipase A2 receptor (PLA2R) antibodies in 10 of the 23 MN patients were measured and three of them were positive. The uB2M was evaluated using the latex agglutination method. This study was approved by the institutional review board of Yamagata University School of Medicine. All procedures were conducted in accordance with the principles of the Declaration of Helsinki. Informed consent was obtained from all participants.
Table 1.
Characteristics of DN, MCNS, and MN patients.
| Characteristic | DN (N = 11) | MCNS (N = 14) | MN (N = 23) | P-Value |
|---|---|---|---|---|
| Age (yr) | 57.4 ± 13.7 | 48.1 ± 20.7 | 63.8 ± 10.3 | 0.0623 |
| Male | 7 (63.6) | 6 (42.9) | 13 (56.5) | 0.5571 |
| Serum albumin (g/dL) | 3.6 (2.4–3.8) | 1.7 (1.3–2.1) | 2.4 (2.0–3.6) | 0.0018 |
| Serum Cr (mg/dL) | 1.7 (1.0–3.1) | 0.7 (0.6–1.2) | 0.7 (0.6–1.1) | 0.0009 |
| eGFR (mL/min/1.73 m2) | 29.0 (16.9–46.9) | 80.2 (47.0–103.9) | 75.4 (52.1–85.8) | 0.0004 |
| uB2M (mg/L) | 1.7 (0.4–5.4) | 0.4 (0.1–0.5) | 0.2 (0.1–0.3) | 0.0962 |
| Urinary protein/Cr (g/gCr) | 4.0 (2.3–7.8) | 8.5 (5.1–10.0) | 4.5 (3.0–6.9) | 0.1226 |
Data were presented as mean ± standard deviation, number (%), or median (first quartile-third quartile). Cr: creatinine, eGFR: estimated glomerular filtration rate. The P-value for “male” was calculated using Pearson's chi-square test and the P-values for the other items were calculated using Kruskal-Wallis test. P-values <0.05 are shown in bold.
2.3. Quantification of uALB by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis
Urinary samples were diluted and boiled in 62.5 mM Tris buffer, pH 6.8, containing 2% SDS, 15% glycerol, 5% 2-mercaptoethanol, and 0.02% bromphenol blue. Sample were electrophoresed on a 12% polyacrylamide gel containing 0.1% SDS and stained with Coomassie Brilliant Blue R-250. Stained gels were scanned by the gel documentation system AE-6932GXCF (ATTO, Tokyo, Japan) and quantified by densitometry using Image J software (National Institutes of Health, Bethesda, MD). The band corresponding to uALB was quantified based on the relative intensity to the standard protein.
2.4. Sample preparation, peptide identification, and quantification
Six microliters of urine or 2 μL of plasma were reductively alkylated, digested with trypsin, and desalted using C-Tip (Nikkyo Technos, Tokyo Japan) as described previously [11]. The desalted peptide solution was analyzed by nanoflow liquid chromatography-tandem mass spectrometry using the EASY nLC 1000 system (Thermo Scientific, Hudson, NH, USA) connected to a quadrupole orbitrap mass spectrometer (Q-Exactive, Thermo Scientific) equipped with a nanoelectrospray emitter. The mass spectrometer was operated in data-dependent mode to automatically switch between mass spectrometry and tandem mass spectrometry acquisition. The full-scan spectra (m/z range 350-1800) were acquired. The 10 most intense ions were sequentially isolated and fragmented by higher-energy C-trap dissociation at a normalized collision energy of 28%.
Raw file reads were searched against the Swiss-Prot human database (20,431 sequences), using Proteome Discoverer (version 1.4, Thermo Scientific) with the Sequest HT and Mascot (version 2.6, Matrix Science, Tokyo, Japan) search engines. Precursor and fragment mass tolerances were set to 5 ppm and 0.02 Da, respectively. The enzyme was set to trypsin with maximum missed cleavage sites of 2. Fixed modification for carbamidomethyl cysteine and variable modifications for oxidation of methionine and deamidation of asparagine and glutamine were set. The results were filtered using Percolator with a false discovery rate of 1%. The peak area of each identified peptide was estimated using Proteome Discoverer. The intensity of unique peptides was used to calculate the protein intensity. The intensity-based absolute quantification (iBAQ) algorithm was used for calculating the protein quantification value [12].
2.5. Calculations
Serotransferrin (TF) was calculated as the iBAQ values, as described above. IgG was calculated as the sum of iBAQ values of each of the IgG subtypes, including IgGs1 to 4. Fractional excretion of TF and IgG was calculated using plasma protein levels instead of serum levels. The SI was calculated as the clearance of IgG divided by the clearance of TF using the following formula: ([urinary IgG]*[plasma TF])/([plasma IgG]*[urinary TF]) [13].
2.6. Statistical analyses
Before the analyses, the iBAQ value of each protein was corrected by the level per mL and log2 transformation. The smallest value among the quantified proteins was applied to missing values.
Statistical analyses were performed using JMP software (version 12.2.0; SAS Institute, Cray, NC, USA). Correlation for the reproducibility test was analyzed using Pearson's coefficient and comparisons between each disease group were analyzed using Mann-Whitney or Kruskal-Wallis test. P-values for categorical variables were calculated by Pearson's chi-square test. The receiver operating characteristic (ROC) curve and area under the curve (AUC) were applied to evaluate the diagnostic accuracy. Principal component (PC) analysis was performed to classify the candidates based on their PC. Statistical significance was set at p < 0.05.
3. Results
3.1. Search for diagnostic biomarker from urine
We identified 5488 unique trypsin-digested peptides (Table S1) and 484 urinary proteins, using at least two unique peptides (Table S2). Of these, 100 proteins were analyzed because these proteins were detected in more than half of the total cases. The log2 transformed-iBAQ value of uALB was significantly correlated with the log2 transformed value quantified using the electrophoretic band intensity as an index, and was 39.004 at a concentration of 1 mg/mL (Fig. S1A-C). On the other hand, the log2 transformed-iBAQ value of uB2M was also significantly correlated with the log2 transformed value evaluated using the latex agglutination method, and was 29.393 at a concentration of 1 mg/L (Fig. S1D). The reproducibility test was performed using data from an independent trypsin digestion on a different day. As a result of 6 trials, the Pearson coefficients (R) were between 0.6519 and 0.9787, and the median value was 0.8431. All P-values were less than 0.0001 (Table S3). These results confirmed that the quantitative reproducibility of proteomic analysis in this study was very high.
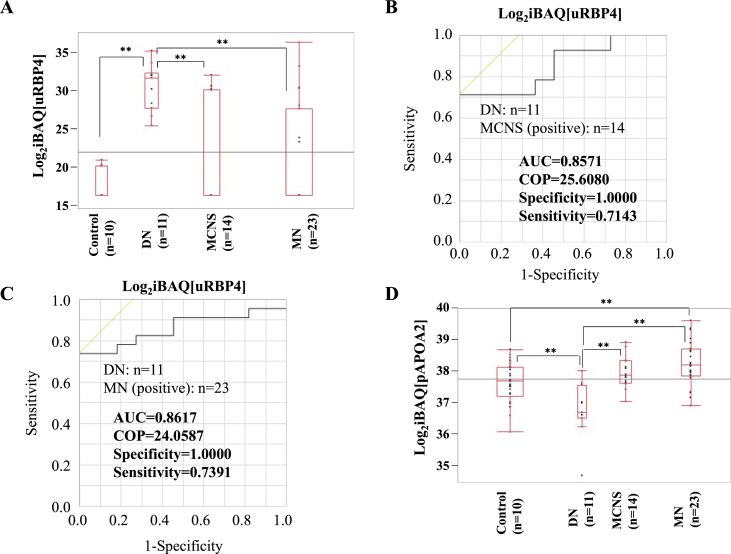
We compared the protein levels in healthy controls, DN, MCNS, and MN (Fig. S2A). Urinary afamin (uAFM) level in MN was the second highest after MCNS and significantly higher than that in DN (Fig. S2B). In contrast, urinary retinol-binding protein 4 (uRBP4) and SH3 domain-binding glutamic acid-rich-like protein 3 (uSH3BGRL3) levels in DN were the highest and significantly higher than those in MCNS and MN (Fig. 1A and S2C). We selected 23 urinary candidates that could distinguish DN from MCNS based on a P-value of less than 0.05 and a difference of more than double (Fig. S2D); we calculated the AUC using ROC curve analysis (Table S4) and compared with uALB and uB2M that are the glomerular and tubular damage biomarkers, respectively. The AUCs for uRBP4 and uSH3BGRL3 were both 0.8571 (Fig. 1B and S2E, and Table 2). On the other hand, the AUCs for uB2M and uALB were 0.7500 and 0.7727, respectively (Fig. S2F and G, and Table 2). We also selected 14 candidates that could distinguish DN from MN (Fig. S2H) and calculated the AUCs (Table S5). The AUCs for uAFM and uRBP4 were 0.7629 and 0.8617, respectively (Fig. 1C, Fig. S2I, and Table 2). On the other hand, the AUCs for uB2M and uALB were 0.7136 and 0.5178, respectively (Fig. S2J and K, and Table 2). These results indicated that uRBP4 could be more accurately diagnosed between DN and MCNS and between DN and MN than uALB and uB2M.
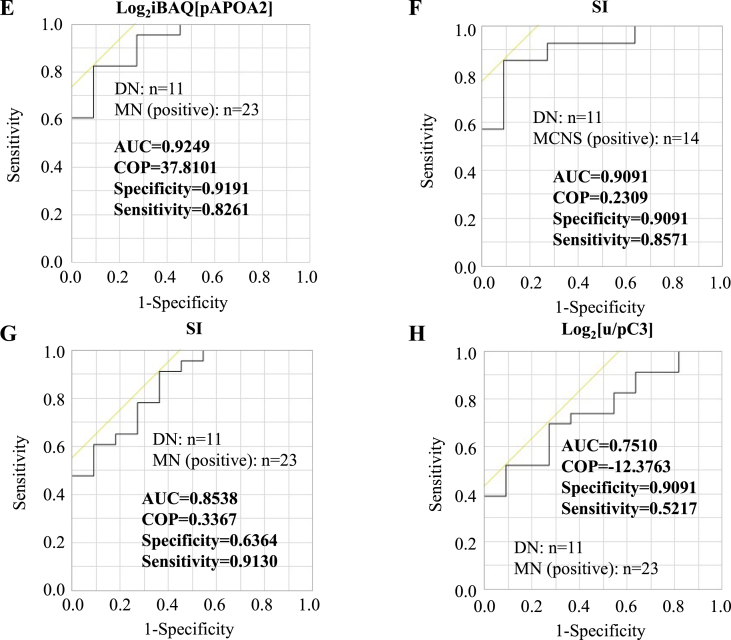
Fig. 1.
Diagnostic marker candidates from the urine, the plasma or the ratio of urinary/plasma protein. (A) The iBAQ logarithm of uRBP4 in each case is shown as box-and whisker plots denoting the median, interquartile range, and the minimum and maximum data points. ROC curve analysis for uRBP4 to discriminate between DN and MCNS (B) or MN (C). (D) The iBAQ logarithm of pAPOA2 in each case is shown as box-and whisker plots. ROC curve analysis for pAPOA2 to discriminate between DN and MN (E), for the SI to discriminate between DN and MCNS (F) or MN (G), and for the u/pC3 to discriminate between DN and MN (H). ** indicates the significant difference in each of the two groups (p < 0.01). COP: cut off point.
Table 2.
ROC curve analysis of various candidates to discriminate between DN and MCNS or MN.
| Candidates | AUC | COP | Specificity | Sensitivity |
|---|---|---|---|---|
| DN vs. MCNS | ||||
| (Set MCNS as positive) | ||||
| SI | 0.9091 | 0.2309 | 0.9091 | 0.8571 |
| uALB | 0.7727 | 930.71 (mg/dL) | 0.9091 | 0.7143 |
| uB2M | 0.7500 | 0.52 (mg/L) | 0.7000 | 0.8182 |
| uRBP4 | 0.8571 | 25.6080 | 1.0000 | 0.7143 |
| uSH3BGRL3 | 0.8571 | 23.1388 | 1.0000 | 0.7143 |
| pITIH1 | 0.9675 | 36.7477 | 0.9091 | 0.9286 |
| uRBP4, uSH3BGRL3 | 0.9740 | −1.6567 | 1.0000 | 0.9286 |
| (04208*[uRBP4] +0.6821*[uSH3BGRL3]-27.9476) | ||||
| uRBP4, pITIH1 | 0.9935 | −1.4938 | 1.0000 | 0.9286 |
| (04238*[uRBP4]-9.4159*[pITIH1] +333.8091) | ||||
| DN vs. MN | ||||
| (Set MN as positive) | ||||
| SI | 0.8538 | 0.3367 | 0.6364 | 0.9130 |
| uALB | 0.5178 | 507.26 (mg/dL) | 0.7273 | 0.4348 |
| uB2M | 0.7136 | 0.39 (mg/L) | 0.7000 | 0.8182 |
| uAFM | 0.7629 | 32.5949 | 1.0000 | 0.4348 |
| uRBP4 | 0.8617 | 24.0587 | 1.0000 | 0.7391 |
| pAPOA2 | 0.9249 | 37.8101 | 0.9091 | 0.8261 |
| u/pC3 | 0.7510 | −12.3763 | 0.9091 | 0.5217 |
| uAFM, uRBP4 | 0.9763 | −1.8943 | 1.0000 | 0.9130 |
| (-1.3560*[uAFM] +0.6013*[uRBP4] +24.4380) | ||||
| uAFM, u/pC3 | 0.9842 | −0.8025 | 1.0000 | 0.9565 |
| (-4.0746*[uAFM] +1.3057*[u/pC3] +137.3265) | ||||
SI: selectivity index; ([urinary IgG]*[plasma TF])/([plasma IgG]* [urinary TF]). The uALB and uB2M are biomarkers for glomerular and tubular disorders, respectively. Those that are superior to SI, uALB, and uB2M are shown in bold. The same values as the best among these biomarkers are underlined. COP: cut off point.
3.2. Search for diagnostic biomarker from plasma
In addition to urinary samples, we identified 4175 unique trypsin-digested peptides (Table S6) and 223 plasma proteins (Table S7), of which 141 proteins were analyzed. We compared the protein levels in normal plasma controls, DN, MCNS, and MN groups (Fig. S3A). Plasma apolipoprotein A-II (pAPOA2) level in MN was the highest and significantly higher than that in DN, while that in DN was the lowest (Fig. 1D). In contrast, plasma inter-alpha-trypsin inhibitor heavy chain H1 (pITIH1) level in MCNS was the highest and significantly higher than that in DN (Fig. S3B).
We selected 24 candidates that could distinguish DN from MCNS (Fig. S3C) and calculated the AUC (Table S4). The AUC for pITIH1 was 0.9675 (Fig. S3D and Table 2). We also selected 8 candidates that could distinguish DN from MN (Fig. S3E) and calculated the AUC (Table S5). The AUC for pAPOA2 was 0.9249 (Fig. 1E and Table 2). These results indicated that pITIH1 and pAPOA2 were suitable candidates for distinguishing DN from MCNS or MN, respectively.
3.3. Search for diagnostic biomarker from the ratio of urinary to plasma proteins
SI, which is based on the ratio between IgG and TF clearance, was calculated for DN, MCNS, and MN. Most DN cases had SIs above 0.2, significantly higher than MCNS and MN, while most MCNS cases had SIs less than 0.2 (Fig. S4A). We found that the AUCs of SI for diagnosis between DN and MCNS, and DN and MN were 0.9091 and 0.8538, respectively (Fig. 1F and G, and Table 2). These results indicated that proteinuria in DN was nonselective and that in MCNS was selective to moderately selective, which was consistent with the previous reports [13].
We compared the urinary to plasma protein ratios in DN, MCNS, and MN groups (Fig. S4B-E). The urinary to plasma complement C3 ratio (u/pC3) in DN was the highest and significantly higher than that in MN (Fig. S4C). We selected 21 and 9 candidates that could distinguish DN from MCNS (Fig. S4D) and DN from MN (Fig. S4E), respectively; the AUC was calculated (Tables S4 and S5), which was 0.7510 for the latter u/pC3 (Fig. 1H and Table 2).
3.4. Selection of the best diagnostic biomarker from candidates
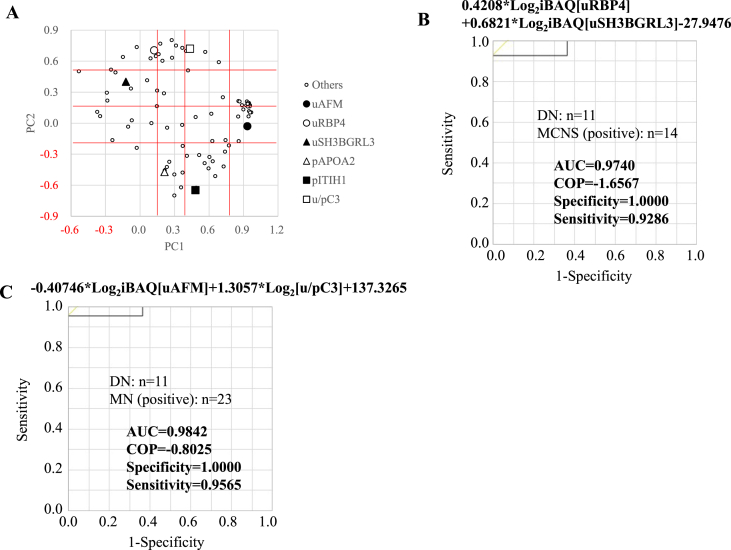
Of the candidates identified and analyzed in this study, pITIH1 and pAPOA2 suited the best to distinguish between DN and MCNS, and DN and MN, respectively, owing to their highest AUC. It was then examined whether combining the two markers would increase the discrimination accuracy. Since it was considered that the markers to be combined should have different properties, PC analysis of the candidates was performed. There were 68 and 31 candidates to distinguish between DN and MCNS, and DN and MN, respectively. Excluding duplication, PC analysis was performed on 83 candidates (Fig. 2A).
Fig. 2.
The usefulness of each combination of the two candidates as a diagnostic marker and comparison with that of SI. (A) Factor scores of the observations are plotted on the first two components. PC1 is classified into four categories: less than 0.15, 0.15 or more and less than 0.4, 0.4 or more and less than 0.8, and 0.8 or more. PC2 is also classified into four categories: -0.2 or less, −0.2 or more and less than 0.15, 0.15 or more and less than 0.5, and 0.5 or more. ROC curve analyses for the combination of uRBP4 and uSH3BGRL3 to discriminate between DN and MCNS (B) and the combination of uAFM and u/pC3 to discriminate between DN and MN (C).
PC 1 was divided into 4 groups so that the number of numerically classified candidates was approximately the same. PC2 was also divided into 4 groups; the combination of PC1 and PC2 was classified into a maximum of 16 groups, and the candidate with the highest AUC was selected for each group. Finally, 11 candidates were selected to distinguish between DN and MCNS, as well as DN and MN (bold letters in Tables S4 and S5).
We performed discriminant analysis using 2 of these candidates to select the best combination and nominal logistic regression analysis to estimate the parameters for discrimination of renal disease. The uRBP combined with uSH3BGRL3 (Fig. 2B and S5A and B, and Table 2) or pITIH1 (Fig. S5C-E and Table 2) were optimal for discriminating DN and MCNS, and the AUCs were 0.9740 and 0.9935, respectively. Similarly, uAFM combined with u/pC3 (Fig. 2C and S5F and G, and Table 2) or uRBP4 (Fig. S5H-J and Table 2) were optimal for distinguishing DN from MN, with the AUCs of 0.9842 and 0.9763, respectively. These combinations of the candidates were better than SI at discriminating DN and MCNS or MN.
3.5. Correlation of patient characteristics and each candidate
We compared each candidate with patient characteristics (Table S8). The uAFM and u/pC3 showed significant negative correlation with serum albumin levels. The uRBP4 in DN and pITIH1 in MCNS also showed significant negative correlation with serum albumin, but uRBP4 in MCNS and MN, and pITIH1 in DN did not. In addition, uRBP4 and u/pC3 in MN showed significant positive correlation with serum creatinine levels, but those in DN did not. Furthermore, uSH3BGRL3 level in DN showed significant positive correlation with estimated glomerular filtration rate, while that in MCNS did not. These results suggested that uRBP4, uSH3BGRL3, pITIH1, and u/pC3 could more likely be disease-discriminating markers because their characteristics differed depending on the disease.
3.6. Effects of other confounding factors
About half patients with DN and MN were nephrotic syndrome, and the other half were not, while most patients with MCNS were nephrotic syndrome. To evaluate the usefulness of each candidate for distinguishing DN from MN in patients without extreme symptoms such as nephrotic syndrome, we compared it in patients except for nephrotic syndrome. As a result, AUCs of all candidates were superior to SI (Table S9). On the other hand, each combination of the two candidates were better than SI at discriminating DN and MCNS or MN even in patients with nephrotic syndrome (Table S10).
In the clinical practice, plasma anti-PLA2R autoantibodies are used to diagnose MN. This study also measured anti-PLA2R autoantibodies in 10 of the 23 MN patients and found that three of them were positive. To evaluate the usefulness of each candidate for distinguishing DN from MN patients without anti-PLA2R autoantibodies, we compared it. Each combination of the two candidates were better than SI at discriminating DN and MN patients even without anti-PLA2R autoantibodies (Table S11).
4. Discussion
The number of patients with diabetes is increasing, and the number of patients with non-diabetic renal diseases such as MN and MCNS is also increasing [4]. In such cases, steroid treatment is required, but if DN is inaccurately diagnosed, steroid treatment may be delayed. Urinary and plasma proteomics was performed to search for potential markers for the differential diagnosis of DN and MCNS or MN and compared with SI, uALB, and uB2M that have already been put into practical use. The combination of uRBP4 and uSH3BGRL3 could distinguish between DN and MCNS with excellent AUC, sensitivity, and specificity as compared with these markers (Figs. 1F and 2B, and S2F and G). The combination of uAFM and uRBP4 or u/pC3 could also distinguish between DN and MN with excellent AUC, sensitivity, and specificity (Figs. 1G and 2C, S2 J and K, and S5J).
DN is thought to be caused by a combination of glycation-induced CD59 inactivation and hyperglycemia-induced complement activation that increases the deposition of membrane attack complexes in the kidney [14]. Glomerular C3 mRNA and protein were shown to be positively correlated with DN [15], with urinary C3 levels in DN being significantly higher than those in diabetes without renal involvement [16]. The u/pC3 in DN was the highest and was significantly higher than that in MN (Fig. S4C). These results suggest that u/pC3 is a promising diagnostic marker for DN but not for diabetes.
RBP4, which is readily filtered through the glomerulus and almost reabsorbed in the renal proximal tubules, has been reported as a urinary marker of tubular injury in early DN [10]. The uRBP4 and serum albumin had a significant negative correlation in DN but not in MCNS and MN (Table S8). In addition, the SI of DN was significantly higher than that of MCNS and MN (Fig. S4A). These results suggested that tubular dysfunction occurred in DN, plasma proteins were non-selectively leaked into the urine, and uRBP4 may be an indicator.
SH3BGRL3 was reported to be a binding partner of the epidermal growth factor receptor [17], which inhibits tumor necrosis factor α-induced apoptosis and promotes cell survival [18]. The uSH3BGRL3 in DN showed significant positive correlation with estimated glomerular filtration rate, whereas uRBP4 was negatively correlated (Table S8). The uRBP4 and uSH3BGRL3 in DN were significantly higher than those in MN and MCNS (Fig. 1A and S2C), and the AUC, specificity, and sensitivity for distinguishing DN and MCNS were all the same (Fig. 1B and S2E), but the false positive and false negative cases were different from each other. Therefore, it was expected that DN and MCNS could be discriminated more accurately by combining these two markers. In fact, the combination improved AUC, sensitivity, and specificity (Fig. 2B).
In contrast, pediatric idiopathic nephrotic syndrome has been shown to be associated with the upregulation of uAFM [19]. The uAFM in MN was the second highest after MCNS, which was significantly higher than that in DN (Fig. S2B), and negatively correlated with serum albumin (Table S8). These results suggest that uAFM can be a promising marker for determining whether diabetic renal disease is DN or MCNS/MN.
In this study, by combining the two candidates, DN and MCNS or MN could be distinguished with excellent AUC, sensitivity, and specificity that are superior to SI, uALB, and uB2M. However, there are two limitations to our study: (1) the number of patients was too small because of one institution study; therefore, it is necessary to increase the number of patients and verify them in the future to determine whether the clinical course is consistent with the behavior of the candidate. (2) Due to the characteristics of the clinical department, DN patients were often targeted for diabetic patients with apparent renal failure, and the DN group used in this study had more patients with impaired renal function than the MCNS and MN groups (Table 1); therefore, the possibility that DN-specific markers are renal function markers cannot be ruled out and should be validated using the urine of DN patients with comparable renal function to patients in the MCNS and MN groups.
Although the renal function of the MCNS and MN patients used in this study was closer to normal than that of the DN patients, the MCNS-specific or MN-specific markers showed remarkable differences, so at least these markers are likely to be disease-specific and useful markers. Because MCNS-specific or MN-specific proteins are useful in diagnosing MCNS or MN complications in diabetic patients, the practical application of the diagnostic markers identified in this study may greatly reduce the burden on patients and make them more accurate diagnosis may be possible.
Author contributions
AA collected samples, performed experiments, and edited the paper; TO performed experiments and wrote the paper; KI, KK, NS, and SW collected samples, assisted the experiments, and edited the paper; MW and TK created the research project and edited the paper.
Declaration of competing interest
None declared.
Acknowledgements
We would like to thank Ms. Emiko Nishidate for her help in sample management and assistance in experiments. We would like to thank Editage (www.editage.com) for English language editing. This study was supported, in part, by a Grant-in-Aid for Scientific Research of Japan Society for the Promotion of Science, (C) (21K08271 to TK).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101102.
Abbreviations
Abbreviations are followed:
- MCNS
minimal change nephrotic syndrome
- MN
membranous nephropathy
- DN
diabetic nephropathy
- Ig
immunoglobulin
- uALB
urinary albumin
- uB2M
urinary β2-microglobulin
- SI
selectivity index
- PLA2R
M-type phospholipase A2 receptor
- SDS
sodium dodecyl sulfate
- iBAQ
intensity-based absolute quantification
- TF
serotransferrin
- ROC;
receiver operating characteristic
- AUC
area under the curve
- PC
principal component
- uAFM
urinary afamin
- uRBP4
urinary retinol-binding protein 4
- uSH3BGRL3
urinary SH3 domain-binding glutamic acid-rich-like protein 3
- pAPOA2
plasma apolipoprotein A-II
- pITIH1
plasma inter-alpha-trypsin inhibitor heavy chain H1, u/pC3; urinary to plasma complement C3 ratio
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Vivarelli M., Massella L., Ruggiero B., Emma F. Minimal change disease. Clin. J. Am. Soc. Nephrol. 2017;12(2):332–345. doi: 10.2215/CJN.05000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couser W.G. Primary membranous nephropathy. Clin. J. Am. Soc. Nephrol. 2017;12(6):983–997. doi: 10.2215/CJN.11761116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umanath K., Lewis J.B. Update on diabetic nephropathy: core curriculum 2018. Am. J. Kidney Dis. 2018;71(6):884–895. doi: 10.1053/j.ajkd.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Liu D., Huang T., Chen N., Xu G., Zhang P., Luo Y. The modern spectrum of biopsy-proven renal disease in Chinese diabetic patients-a retrospective descriptive study. PeerJ. 2018;6 doi: 10.7717/peerj.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amiri-Dashatan N., Koushki M., Abbaszadeh H.A., Rostami-Nejad M., Rezaei-Tavirani M. Proteomics applications in health: biomarker and drug discovery and food industry. Iran. J. Pharm. Res. (IJPR) 2018;17(4):1523–1536. [PMC free article] [PubMed] [Google Scholar]
- 6.Geyer P.E., Holdt L.M., Teupser D., Mann M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017;13(9):942. doi: 10.15252/msb.20156297. Pubmed:28951502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beasley-Green A. Urine proteomics in the era of mass spectrometry. Int. Neurourol. J. 2016;20(Suppl 2):S70–S75. doi: 10.5213/inj.1612720.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schanstra J.P., Mischak H. Proteomic urinary biomarker approach in renal disease: from discovery to implementation. Pediatr. Nephrol. 2015;30(5):713–725. doi: 10.1007/s00467-014-2790-y. [DOI] [PubMed] [Google Scholar]
- 9.Siwy J., Zürbig P., Argiles A., Beige J., Haubitz M., Jankowski J. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol. Dial. Transplant. 2017;32(12):2079–2089. doi: 10.1093/ndt/gfw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiseha T., Tamir Z. Urinary markers of tubular injury in early diabetic nephropathy. Internet J. Nephrol. 2016;2016:4647685. doi: 10.1155/2016/4647685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osaki T., Sugiyama D., Magari Y., Souri M., Ichinose A. Rapid immunochromatographic test for detection of anti-factor XIII A subunit antibodies can diagnose 90 % of cases with autoimmune haemorrhaphilia XIII/13. Thromb. Haemostasis. 2015;113(6):1347–1356. doi: 10.1160/TH14-09-0745. [DOI] [PubMed] [Google Scholar]
- 12.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 13.Laurent J., Philippon C., Lagrue G., Laurent G., Weil B., Rostoker G. Proteinuria selectivity index - prognostic value in lipoid nephrosis and related diseases. Nephron. 1993;65(2):185–189. doi: 10.1159/000187472. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh P., Sahoo R., Vaidya A., Chorev M., Halperin J.A. Role of complement and complement regulatory proteins in the complications of diabetes. Endocr. Rev. 2015;36(3):272–288. doi: 10.1210/er.2014-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woroniecka K.I., Park A.S., Mohtat D., Thomas D.B., Pullman J.M., Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X.Q., Chang D.Y., Chen M., Zhao M.H. Complement activation in patients with diabetic nephropathy. Diabetes Metab. 2019;45(3):248–253. doi: 10.1016/j.diabet.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Chiang C.Y., Pan C.C., Chang H.Y., Lai M.D., Tzai T.S., Tsai Y.S. SH3BGRL3 protein as a potential prognostic biomarker for urothelial carcinoma: a novel binding partner of epidermal growth factor receptor. Clin. Canc. Res. 2015;21(24):5601–5611. doi: 10.1158/1078-0432.CCR-14-3308. [DOI] [PubMed] [Google Scholar]
- 18.Berleth E.S., Henn A.D., Gurtoo H.L., Wollman R., Alderfer J.L., Mihich E. A novel tumor necrosis factor-alpha inhibitory protein, TIP-B1. Int. J. Immunopharm. 2000;22(12):1137–1142. doi: 10.1016/s0192-0561(00)00071-0. [DOI] [PubMed] [Google Scholar]
- 19.Sedic M., Gethings L.A., Vissers J.P., Shockcor J.P., McDonald S., Vasieva O. Label-free mass spectrometric profiling of urinary proteins and metabolites from paediatric idiopathic nephrotic syndrome. Biochem. Biophys. Res. Commun. 2014;452(1):21–26. doi: 10.1016/j.bbrc.2014.08.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.