Abstract
Long-bone fracture is a common injury and its healing process at the fracture site involves several overlapping phases, including inflammation, migration of mesenchymal progenitors into the fracture site, endochondral ossification, angiogenesis and finally bone remodelling. Increasing evidence shows that small noncoding RNAs are important regulators of chondrogenesis, osteogenesis and fracture healing. MicroRNAs are small single-stranded, non-coding RNA-molecules intervening in most physiological and biological processes, including fracture healing. Angiogenin-cleaved 5′ tRNA halves, also called as tiRNAs (stress-induced RNAs) have been shown to repress protein translation. In order to gain further understanding on the role of small noncoding RNAs in fracture healing, genome wide expression profiles of tiRNAs, miRNAs and mRNAs were followed up to 14 days after fracture in callus tissue of an in vivo mouse model with closed tibial fracture and, compared to intact bone and articular cartilage at 2 months of age. Total tiRNA expression level in cartilage was only approximately one third of that observed in control D0 bone. In callus tissue, 11 mature 5′end tiRNAs out of 191 tiRNAs were highly expressed, and seven of them were differentially expressed during fracture healing. When comparing the control tissues, 25 miRNAs characteristic to bone and 29 miRNAs characteristic to cartilage tissue homeostasis were identified. Further, a total of 54 out of 806 miRNAs and 5420 out of 18,700 mRNAs were differentially expressed (DE) in callus tissue during fracture healing and, in comparison to control bone. They were associated to gene ontology processes related to mesenchymal tissue development and differentiation. A total of 581 miRNA-mRNA interactions were identified for these 54 DE miRNAs by literature searches in PubMed, thereby linking by Spearman correlation analysis 14 downregulated and 28 upregulated miRNAs to 164 negatively correlating and 168 positively correlating miRNA-mRNA pairs with chondrogenic and osteogenic phases of fracture healing. These data indicated that tiRNAs and miRNAs were differentially expressed in fracture callus tissue, suggesting them important physiological functions during fracture healing. Hence, the data provided by this study may contribute to future clinical applications, such as potential use as biomarkers or as tools in the development of novel therapeutic approaches for fracture healing.
Keywords: Fracture healing, Bone, Cartilage, tiRNA, miRNA
Highlights
-
•
Content of mature tiRNAs is higher in callus tissue compared to diaphyseal bone.
-
•
11 tiRNAs were highly expressed in callus tissue, incl. 7 differentially expressed.
-
•
54 miRNAs were differentially expressed in callus tissue during fracture healing.
-
•
Validated mRNA targets for 54 DE miRNAs were searched manually in PubMed.
-
•
Integrated expression analysis identified 581 miRNA-target mRNA pairs in callus.
1. Introduction
Fracture healing is a complex process with activation of hundreds of molecules involved in sequential spatially and temporally overlapping regenerative phases, aiming at a full recovery. Secondary healing can be divided into four steps. Fracture healing begins with hematoma formation, followed by formation of soft fibrocartilaginous callus which will then be replaced by hard bony callus. Removal of callus tissue and eventual bone remodelling of the hard callus finalizes the healing process. Fracture repair takes place via inflammation, differentiation of multipotent mesenchymal stromal cells (MSCs) into chondrocytes, osteogenesis, angiogenesis, and osteoclastogenesis.
Immediately after fracture trauma, inflammatory response is activated and factors such as tumor necrosis factor alfa (TNF-α), and interleukins 1α, 1β, 6, 11 and 18 are upregulated (Gerstenfeld et al., 2003; Rundle et al., 2006; Einhorn et al., 1995). In mice, those cytokines are expressed during the first three days after fracture and again after day 14 (Dimitriou et al., 2005). The inflammatory response triggers bone repair as well as angiogenesis (Einhorn and Gerstenfeld, 2015). Following the inflammatory phase, MSCs will be recruited to the fracture site, where they start differentiating into chondrocytes. Chondrogenesis is triggered by expression of a combination of SRY-Box Transcription Factors SOX-5, SOX-6 and SOX-9 (Ikeda et al., 2004). Differentiating chondrocytes secrete cartilage specific extracellular matrix components, e.g., type II collagen and aggrecan core protein. Ultimately, they undergo terminal differentiation and express type X collagen (Liu et al., 2017). Tissue remodelling continues by activation of matrix metalloproteinases, including MMP13 which facilitates the invasion of blood vessels to the injury site. Blood supply from periosteal and medullary circulation is necessary for bone formation in order to provide oxygen and nutrients. Angiogenesis is mainly regulated through angiopoietin and vascular endothelial growth factor (VEGF) signaling pathways (Tsiridis et al., 2007). Furthermore, VEGF is an important regulator for osteogenesis (Street et al., 2002), the expression of which is thought to be triggered by inflammatory cells and MSCs (Hankenson et al., 2011).
Endochondral ossification takes place via the actions of bone forming osteoblasts when the soft callus tissue is replaced by a more stable mineralized hard callus. Osteoblast differentiation is triggered mainly by three transcription factors: Runt-related transcription factor 2 (RUNX2), osterix (SP7) and activating transcription factor 4 (ATF4) (Rutkovskiy et al., 2016). It has also been suggested that hypertrophic chondrocytes could directly transform into osteoblastic cells during endochondral ossification (Hinton et al., 2016). Intervention of bone resorbing osteoclasts is also required to achieve a complete healing of the fracture site (Wang and Yeung, 2017). Osteoblasts release receptor activator of nuclear factor kappa-Β ligand (RANKL) and colony stimulating factor 1 (CSF-1), which are necessary for osteoclast survival, expansion and differentiation (Raggatt and Partridge, 2010).
Gene expression is largely regulated by non-coding RNAs (Guttman and Rinn, 2012). Small non-coding RNAs include several RNA classes such as tRNA derived fragments (tsRNA), microRNAs (miRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNA), piwi-interacting RNA (piRNA). In last decades, next generation sequencing (NGS) and bioinformatics have been used to unveil the function of diverse non-coding RNAs, especially their role on regulation of gene expression.
tsRNA is a novel class of small, 15–36 nt long non-coding and single-stranded RNA. They were reported to derive from precursors or mature tRNA (Li et al., 2018). For long, they were considered to be degradation products of tRNA. Recent studies have shown that tRNA fragments are, in fact, important regulators of cellular homeostasis, and have essential roles in a wide range of physiological and pathological processes by inhibiting global translation of proteins with multiple different mechanisms (Kim, 2019). Under cellular stress, angiogenin cleaves tRNA into 28–36 nt long tiRNAs 3′-tiRNA and 5′-tiRNA halves (tRNA derived stress-induced RNA halves). 5′-tiRNA halves have been shown to be responsible for the assembly of stress granules and induce translational inhibition (Saikia and Hatzoglou, 2015; Emara et al., 2010). The expression of 5′-tiRNAs was also associated with reduced apoptosis and cell survival (Blanco et al., 2014).
MicroRNAs (miRNAs) are an important group of RNA molecules regulating practically all biological processes in the body. They are small single-stranded, non-coding RNA molecules of about 22 nucleotides that mostly bind to the 3′ untranslated region of mRNAs inducing translational repression or degradation of their target mRNA (Bartel, 2004). MicroRNAs are associated with pathological conditions such osteoarthritis (Nugent, 2016), cancer (Nugent, 2014) and tissue regeneration (Nguyen et al., 2015; Michell-Robinson et al., 2015; Dreifke et al., 2015) and are also shown to have a role in bone and cartilage development (Huang et al., 2017; Razmara et al., 2019). Several miRNAs such as miR-140-3p and miR-214 have been found to be differentially expressed (DE) during fracture healing (Nugent, 2017).
The aim of this study was to use a genome wide approach to characterize the expression profiles of tiRNAs, miRNAs and mRNAs associated with secondary fracture healing with the emphasis on chondrogenesis and osteogenesis. This is the first study reporting induced expression of tiRNAs in fracture callus tissue. Target prediction analysis has been typically applied to evaluate the transcriptional impact of miRNA expression profiles. In this study, we searched among published data the verified targets in any tissues or conditions of the differentially expressed miRNAs and performed Spearman's correlation analysis for these miRNA – mRNA pairs to provide physiological verification in callus tissues instead of algorithm-based target prediction analysis.
2. Material and methods
2.1. Animals
Fracture healing experiments were carried out using two months (2 M, 70–74 days) old C57Bl/6N male mice (n = 75). In addition, some skeletal tissue samples were dissected from 10 days (P10) old mice (n = 2). The study plan and procedures were approved by the National Project Authorization Board (project license ESAVI/6129/04.10.03/2011). Animal care was done in accordance with the EU Directive 2010/63/EU and national legislation following the 3R's principles. Mice were maintained in the Central Animal Laboratory of the University of Turku and they received a RM3 (E) soya free rodent diet, SDS, UK (Puolakkainen et al., 2017). Standard closed fracture in mouse tibia was induced by an impact device under isoflurane anaesthesia (250-400 ml/min 2.5%), pain relief was administered by subcutaneous injections of buprenorphine (0.05 mg/kg) and carprofen (5 mg/kg) as previously described (Puolakkainen et al., 2017; Hiltunen et al., 1993). A steel rod was inserted to the medullary canal of the tibia to support the fracture site during healing. Samples were collected at D0 (non-operated control, day zero), and at D5, D7, D10, D14 and D25 after fracture.
2.2. Macroscopic and microscopic evaluation
Skin was removed before X-ray imaging (Faxitron X-ray MX-20). For histological analysis, fat and excess muscle tissue were also removed and the limbs were fixed in fresh 4% paraformaldehyde overnight. Samples were decalcified in 10% Na2-EDTA, 0.1 M phosphate buffer, pH 7.0, embedded in paraffin, cut into 5 μm thick sections, processed and histologically stained with haematoxylin and eosin or with Safranin-O (Kiviranta et al., 1985). Images were captured and digitalized using a Pannoramic 250 Slide Scanner (3DHISTECH, Budapest, Hungary).
2.3. RNA samples
Callus and other tissue samples were dissected under a stereomicroscope. Tibial diaphyseal bone was dissected from non-operated mice. In addition, knee epiphyseal cartilage and tibial diaphyseal bones at P10, and hip articular cartilage and proximal epiphyseal cartilage and proximal metaphyseal bone of the tibiae at 2 M were collected for comparison and validation studies. Dissected tissues were snap frozen in liquid nitrogen and stored at −85 °C until RNA isolation.
Tissue samples were pulverized in liquid nitrogen and homogenized in TRIsure (Bioline) for RNA isolation (Heinonen et al., 2017). RNA concentration was measured using Nanodrop One (Thermo Fisher Scientific). RNA integrity was analysed by Tapestation (Agilent) or equivalent equipment at Turku Bioscience Centre, Turku, Finland.
2.4. Small RNA-Seq analysis
The small RNA libraries were prepared with TruSeq Small RNA Sample Preparation Kit (Illumina, USA) with multiplexing adapters, following the kit user guide (Rev. E). PCR was used to amplify cDNA with primers containing unique six base index sequences distinguishing different samples from one another. Samples were subjected to 6% (w/v) non-denaturing polyacrylamide gel electrophoresis. Fragments between 145 and 160 bp were excised from the gel, and nucleotide quantity was measured with Qubit fluorometer (Thermo Fisher Scientific). Library pools (8−16 samples per pool) were loaded to MiSeq V3 flow cell in 12 pM concentration 10% of PhiX to increase signal integrity. SmallRNA libraries were sequenced using Illumina MiSeq reagent kit V3 150 cycles and 36 bp reads with single-end chemistry.
2.5. Quantitative real-time PCR analysis for miRNAs
Reverse transcription was performed using miScript II RT Kit (Qiagen, cat# 218161) using 500 ng of total RNA. Samples were incubated at 37 °C for 1 h followed by an inactivation step at 95 °C for 5 min.
Quantitative PCR analysis was performed using miScript SYBR Green PCR Kit (Qiagen, cat#: 218073). Primers for mmu-miR-98-5p, mmu-let-7d-3p, mmu-let-7 g-5p, Hs_RNU6-2_11, SNORD68, mmu-miR-140-3p, mmu-miR-150-5p, mmu-miR-92a-3p, mmu-miR-148a-3p, mmu-miR-214-3p and mmu-miR-340-5p were purchased from Qiagen. Quantitative PCR was performed in triplicates from three or four biological replicates using cDNA corresponding to 10 ng of template total RNA. PCR was performed using CFX384 ™ instrument (Bio-Rad) with the cycling conditions 95 °C for 15 min, 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s; the last three steps were repeated for 40 cycles.
2.6. RNA-Seq analysis
RNA-Seq analysis for genome wide expression of mRNAs was carried out in the Finnish Microarray and Sequencing Centre's analysis service. Library preparation was done according to Illumina TruSeq® Stranded mRNA Sample Preparation Kit and Guide (part #15031047) for HS protocol using 300 ng of total RNA as a starting material. The final product was purified and enriched by qPCR with an average library fragments in the range of 200–700 bp. All libraries (32 samples) were pooled and run in two lanes using the Illumina HiSeq3000 instrument with single-end sequencing chemistry and 50 bp read length followed by 8 bp index run.
2.7. Quantitative real-time PCR analysis for mRNAs
Expression profiles of Sox5, Col2a1, Acan, Runx2, Sp7 and Bmp2 in callus samples were validated as markers for progression of chondrogenesis and osteogenesis by qRT-rtPCR (Jeyakumar et al., 2017; Jensen et al., 2010). Reverse transcription was performed using SensiFAST ™ cDNA Synthesis kit (Bioline) with 500 ng of total RNA. The qPCR step reaction was performed in triplicates using DyNAmo Flash SYBR Green qPCR kit (ThermoFisher Scientific, cat#F-415L) and 10 ng of total RNA as a template with primers listed in Supplemental File 1: STable 1. PCR was performed using CFX384 ™ instrument (Bio-Rad) with the cycling conditions 95 °C for 15 min, 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. The last three steps were repeated for 40 cycles. ΔCT was calculated using the geometric mean of Tubb_5 and Actb gene expression.
2.8. Bioinformatics and statistical analysis
2.8.1. NGS data analysis
The raw sequences from RNA-Seq and small RNA-Seq analyses were subjected to quality control using FastQC.
Messenger RNA reads were mapped to the UCSC mouse genome (mm10) using STAR version 2.4.2a and aligned using subreads package (v. 1.5.0) in R version 3.0.1.
MicroRNA and tiRNA reads were trimmed using FASTX-Toolkit version 0.0.13 for Linux (32Bit). Then, small sequences were mapped to the reference genome (mm10) using Bowtie version 1.2.2 and aligned using miRDeep2 (Friedlander et al., 2012).
For detection of tiRNAs, SPORTS1.1 (Shi et al., 2018) was used to map small reads to tRNA sequences extracted from mm10 UCSC genome files originating from GtRNAdb using SPORTS default settings. Then, using a pre-compiled Perl script from SPORTS1.1, the original locations of all sequences were identified and analysed whether they originated from 5′end, or 3′end or 3′ having a CCA tail (=3′ CCA end) of tRNA. Then, reads mapping to tiRNA sequences (28 to 36 nt long) were extracted from SPORTS output text file using R. FPM (fragment per million) was calculated after raw counts normalization using SPORTS output in R.
Normalization and differential expression analysis of miRNA, tiRNA and mRNA data was performed using DESeq2 version 1.20.0 (Love et al., 2014) from Rstudio version 1.1.453 for MacOSX 10.6 (64Bit). Raw data was filtered to a minimum raw count of 10 for each gene. Hierarchical gene and sample cluster analysis was performed from log2 transformed normalized data using Pheatmap and genefilter. Principal component analysis (PCA) plot was generated using ggplot2 from log2 transformed normalized data (Supplemental File 1: S.Fig. 1–3). DESeq2 corrects P-values using Benjamini-Hochberg multiple testing adjustment procedure (Benjamini and Hochberg, 1995). DESeq2 reports a value called ‘baseMean’ corresponding to the average of normalized count values over all samples, dividing by size factors (https//:support.illumina.com → DESeq2 Result files). It is referred to in our study in order to filter the results. The PCA and heatmap plots indicate clustering of samples together and show the reproducibility of the experiment.
Only mature miRNA expression was analysed in this study. Seed sequence information was used to associate the miRNAs to their target mRNAs. TargetScanMouse v.7.1 was used to search for the predicted targets to selected miRNAs with baseMean > 100, adjusted P-value < 0.05 that were differentially expressed with absolute value of log2 fold change (log2FC) > 2 (Agarwal et al., 2015). Gene enrichment for pathway analysis was studied using GAGE in R (Luo et al., 2009). Correlation analysis between mRNA and miRNA expression levels was performed in R with a Spearman correlation test, using the variance of each biological sample across all samples. Same correlation analysis was performed to compare NGS data against qPCR differential expression analysis.
2.8.2. qRT-rtPCR data
The gene expression is reported as 2^−ΔΔCt compared to intact bone used as a control. Data was normalized against appropriate reference genes suggested by GeNorm and NormFinder. Details of the reference gene analysis and novel normalization miRNAs are presented in Supplemental File 2 (S.Fig. 6).
Group comparison significance was evaluated using one-way ANOVA test with Tukey post hoc test from ΔCt values.
P-values below 0.05 were considered as significant. Data are presented as box plots with IQR (interquartile range, median and mean). Analyses were performed using IBM SPSS Statistics version 23.
2.9. Key resources table
| Reagents | Source | Identifier |
|---|---|---|
| Buprenorphine (Temgesic 0,3 mg/ml) - dose 0.05–0.1 mg/kg, SC, IP | Reckitt Benckiser Healthcare, Slough, UK | N/A |
| Carprofen (Rymadyl 50 mg/ml) - dose 5 mg/kg SC | Pfizer, New York, NY | N/A |
| Isoflurane (Attane Vet 1000 mg/g) - induction 4–5%, maintenance 1–2,5% | Piramal Critical Care, Northumberland, UK | N/A |
| Trisure | Bioline | Cat# BIO-38032 |
| QPCR reagents | ||
| SensiFAST ™ cDNA Synthesis kit | Bioline | Cat# BIO-65054 |
| MiScript II RT Kit | Qiagen | cat# 218161 |
| MiScript SYBR Green PCR Kit | Qiagen | Cat# 218073 |
| mmu-miR-98-5p | Qiagen | cat# MS00001463 |
| mmu-let-7d-3p | Qiagen | cat# MS00005859 |
| mmu-let-7g-5p | Qiagen | cat# MS00010983 |
| Hs_RNU6-2_11 | Qiagen | cat# MS00033740 |
| SNORD68 | Qiagen | cat# MS00033712 |
| mmu-miR-140-3p | Qiagen | cat# MS00006048 |
| mmu-miR-150-5p | Qiagen | cat# MS00001673 |
| mmu-miR-92a-3p | Qiagen | cat# MS00005971 |
| mmu-miR-148a-3p | Qiagen | cat# MS00001652 |
| mmu-miR-214-3p | Qiagen | cat# MS00032571 |
| mmu-miR-340-5p | Qiagen | cat# MS00032732 |
| Instruments | ||
| Faxitron MX-20 Cabinet X-Ray System | Material resource | RRID:SCR_019878 |
| Pannoramic Slide scanner | Material resource | N/A |
| Pannoramic Viewer | Material resource | RRID:SCR_014424 |
| Nanodrop one Spectrophotometer | Material resource | N/A |
| Illumina TruSeq Small RNA Sample Preparation Kit | Resource | N/A |
| Thermo Fisher Qubit fluorimeter | Material resource | RRID:SCR_018095 |
| Illumina MiSeq System | Resource | RRID:SCR_016379 |
| Bio-Rad CFX384 Real-Time Detection System | Material resource | RRID:SCR_018057 |
| Illumina HiSeq3000 instrument | Resource | N/A |
| Bioinformatics analysis | ||
| FastQC | Software resource | RRID:SCR_014583 |
| STAR | Software resource | RRID:SCR_004463 |
| Subreads | Software resource | http://subread.sourceforge.net/ |
| FASTX-Toolkit | Software resource | RRID:SCR_005534 |
| Bowtie | Software resource | RRID:SCR_005476 |
| miRDeep2 | Software resource | RRID:SCR_010829 |
| SPORTS1.1 | Software resource | https://github.com/junchaoshi/sports1.1 |
| DESeq2 | Software resource | RRID:SCR_015687 |
| ggplot2 | Software resource | RRID:SCR_014601 |
| pheatmap | Software resource | RRID:SCR_016418 |
| genefilter | Software resource | https://bioconductor.org/packages/release/bioc/html/genefilter.html |
| TargetScan | Software resource | RRID:SCR_010845 |
| geNORM | Software resource | RRID:SCR_006763 |
| NormFinder | Software resource | RRID:SCR_003387 |
| IBM SPSS Statistics | Software resource | RRID:SCR_019096 |
3. Results
3.1. Characterization of the fracture healing model
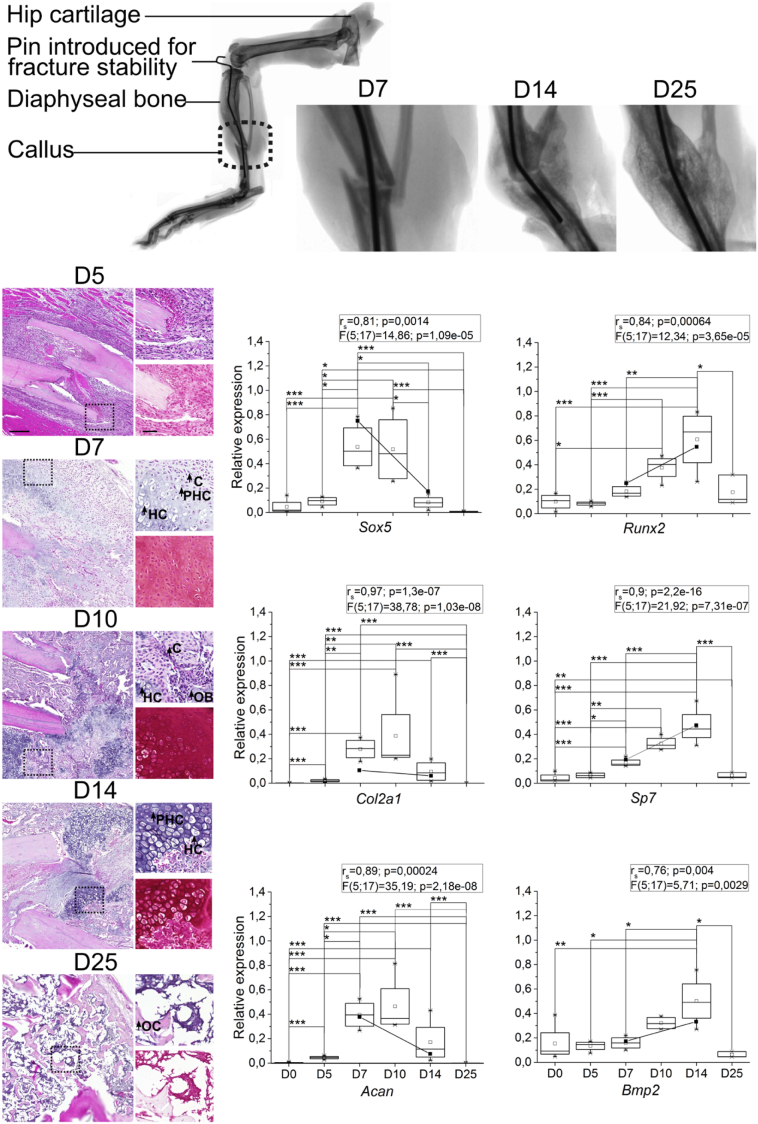
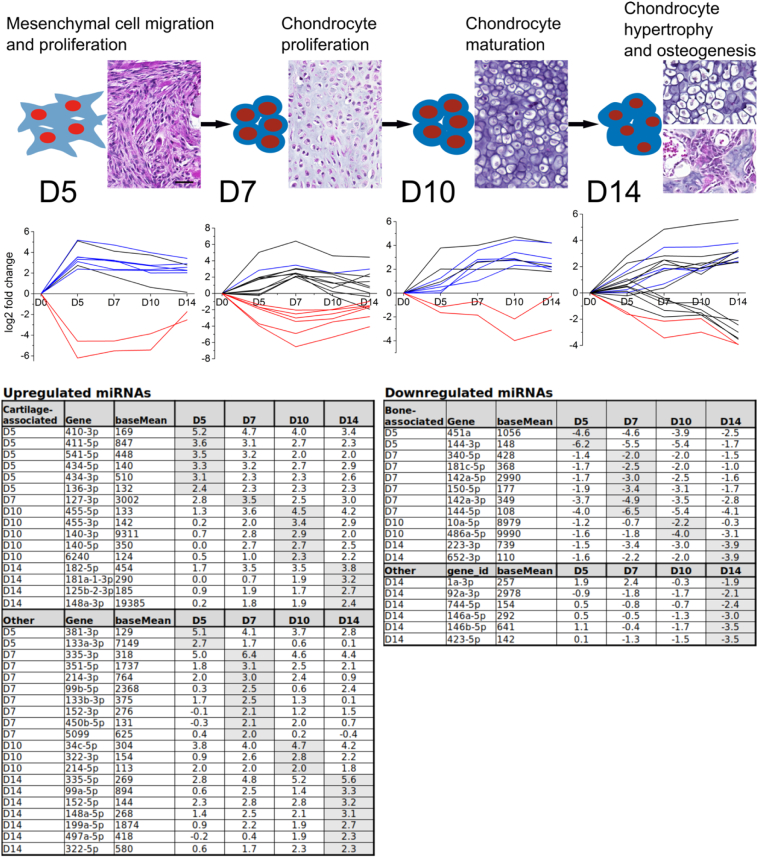
To study the genome wide RNA expression profiles involved in fracture healing, a tibial fracture was induced using an impact device and the progression of fraction healing was followed for 25 days. Macroscopic evaluation of the callus and the appropriate positioning of the steel rod in the bone marrow was performed by X-ray imaging (Fig. 1). Tissue sections stained were with haematoxylin and eosin for evaluation of the morphological changes or with Safranin O for proteoglycans (Fig. 1). At D5, MSC recruitment was observed at the fracture site. At D7, round chondrocytes embedded in proteoglycan-rich matrix were observed. By D10, chondrogenesis had reached its most active stage with many of the chondrocytes being already hypertrophic and the matrix rich of proteoglycans. At D14, hypertrophy had progressed, and osteogenesis was active, as indicated by the presence of bony regions in the callus. By D25, cartilaginous matrix with chondrocytes had disappeared, and the whole callus tissue consisted of bone.
Fig. 1.
Characterization of the fracture healing phases of mouse closed fracture model. (Top panel) X-ray images of operated hind limbs at days 7 (D7), 14 (D14) and 25 (D25) after fracture. (Left panels) Callus tissue morphology; tissue sections were stained with haematoxylin and eosin (higher magnification right top) and Safranin O (higher magnification, right bottom). Scale bar (left) equals to 200 μm, magnified images scale bar (right) equals to 50 μm. The magnified areas are highlighted in the left panels by dashed line boxes. C: round chondrocytes; HC: hypertrophic chondrocytes; PHC: prehypertrophic chondrocytes, OB: osteoblast, OC: osteocyte. (Right panels) Relative expression of selected mRNAs used as markers of chondrogenesis: Acan, Col2a1 and Sox5 and as markers of osteogenesis: Runx2, Sp7 and Bmp2 in callus tissue at D5, D7, D10, D14 and D25. Diaphysis samples were used as controls (D0). Data was normalized against Tubb_5 and Actb. The boxplot indicates the interquartile range (IQR) and median, □ represents the mean. Inter-group comparison was performed using one-way ANOVA test, *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001. * above the box charts represent the inter-group one-way ANOVA test P-value. F (degree of freedom 1; degree of freedom 2) = ratio of variances; P-value. D5–D14: n = 4; D25: n = 3; 2 M diaphyseal bone control (D0): n = 4. Correlation coefficient (rs: Spearman coefficient) and statistical significance as P-values between qRT-rtPCR data and NGS data of each mRNA are presented above each panel.
Also, at the molecular level, the chondrogenic differentiation was most active at D7 and D10, as indicated by the expression of chondrogenic marker genes Sox5, Acan and Col2a1 (Fig. 1). Expression of osteogenic marker genes Runx2, Sp7 and Bmp2 gradually increased throughout D5–D10 with the highest expression at D14. By D25, the expression of all marker genes had returned to the control bone level. Characterization at both histological and molecular levels indicated that our data covers periods of endochondral ossification from early chondrogenesis at D5 to ongoing osteogenesis and remodelling at D14. To identify the changes in RNA expression during fracture healing process, we performed NGS analysis of tiRNAs and miRNAs and RNA-Seq analysis of mRNAs in callus samples at different time points during fracture healing compared to D0 bone.
3.2. Increased quantity of mature 5′ end tiRNAs in fracture healing callus
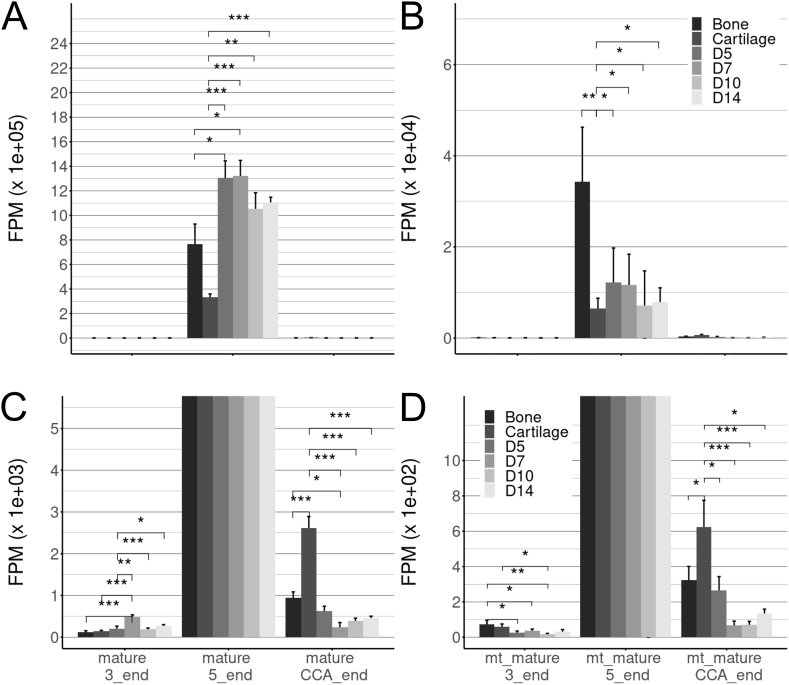
The impact of fracture healing process on tiRNAs was studied in callus tissues due to their described roles in several human diseases and traumatic conditions. The levels of 28-36 nucleotide long tRNA halves (stress-induced RNAs, tiRNA) were analysed in NGS data. A total of 191 tiRNAs were identified. Mature tiRNA reads, from SPORTS output, were averaged within biological replicates and collapsed whether they originated from 5′end, 3′end or 3′ CCA addition end of cellular and mitochondrial tRNA (Fig. 2). Overall expression of 5′derived tiRNAs was expressed at appr. 6500-fold higher than that of 3′end derived tiRNAs. In cartilage, the normalized tiRNA reads originating from mature 5′end were only approximately one third and from mature 3′end about a half of that compared to D0 bone.
Fig. 2.
Increased quantity of 5' tiRNAs in fracture callus tissue. Mean ± SD of normalized tiRNA reads (FPM, fragment per million) was calculated in intact diaphyseal bone and hip articular cartilage and in D5, D7, D10 and D14 callus samples. (A) Mature tiRNAs originating from 3′end, 5′end and 3′ CCA end. One way ANOVA test for mature 3′end: [F(5, 15)=14.36, p = 2.9e-05], 5′end: [F(5, 15)=10.34, p = 0.000192] and CCA end: [F(5, 15)=25.45, p = 7.9e-07]. (B) Mature mitochondrial tiRNAs originating from 3′end, 5′end or 3′ CCA end. (C) Zoom of the panel A for better vizualization of 3’ and CCA end mature tiRNAs. (D) Zoom of the panel B for better vizualization of 3’ and CCA end mature mitochondrial tiRNAs. One way ANOVA test for mt_mature 3′end: [F(5, 15)=6.631, p = 0.00191] 5′end: [F(5, 15)=6.379, p = 0.00229] and CCA end: [F(5, 15)=8.397, p = 0.000588]. SD = sqrt(sum variances). Statistically significant comparisons are marked, *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001. Samples are D0 control bone (n = 4), hip articular cartilage (n = 3), callus samples at D5 (n = 3), D7 (n = 5), D10 (n = 4), and D14 (n = 2).
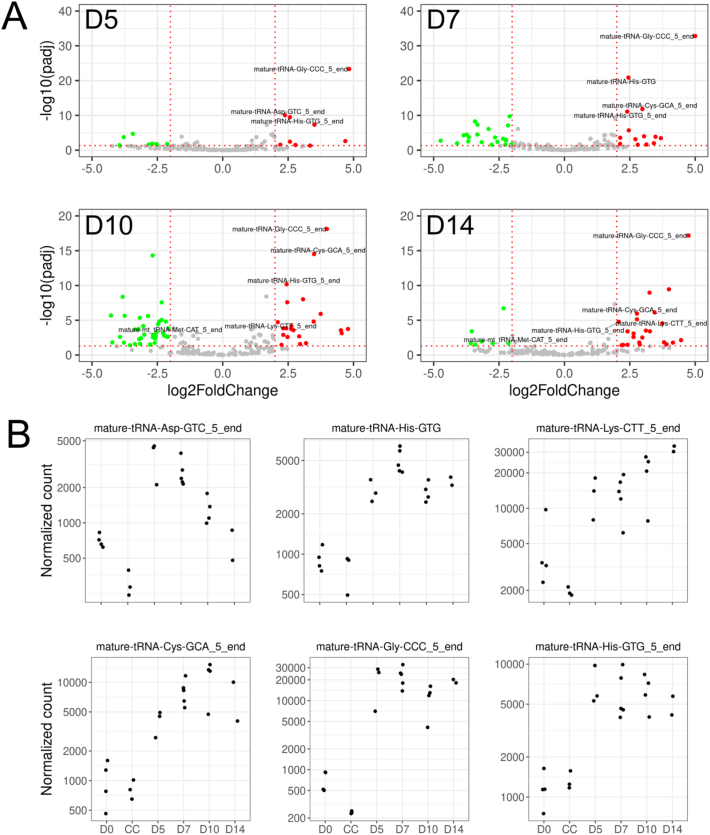
Proportionally the normalized counts of both mature 3′ and 5′end tiRNAs were increased by appr. 15–50% in callus tissue, as compared to D0 bone although mature 5′end originated tiRNAs prevailed in bone and callus tissues. Also, normalized reads of mature 3’ end fragments slightly increased while reads of all mitochondrial fragments were lower in callus tissue when compared to bone. Major changes in 191 tiRNA levels are visualized as volcano plots at D5, D7, D10 and D14 (Fig. 3A, full data in Supplemental File 3).
Fig. 3.
Differential expression of tiRNAs in callus tissue. (A) Volcano plots visualize the expression levels of a total of 191 annotated tiRNAs with average normalized read counts (baseMean) above 1000. Green dots represent downregulated genes (Padjusted-value < 0.05 and log2FC < −2), red dots represent upregulated genes (Padjusted-value < 0.05 and log2FC > 2) and grey dots represent other genes (X-axis, absolute value of log2FC > 2 and Padjusted-value > 0.05). Horizontal dotted lines represent Padjusted-value threshold (0.05) and vertical dotted lines represent log2FC thresholds (−2 and 2). (B) Distribution of normalized expression levels of six differentially expressed mature tiRNAs with average normalized read counts (baseMean) > 1000, log2 fold change > 2 and Padjusted-value < 0.05 during fracture healing. Samples are D0 control bone (n = 4), and callus samples at D5 (n = 3), D7 (n = 5), D10 (n = 4), and D14 (n = 2).
Eleven out of 191 mature tiRNAs were expressed at a distinctly higher level than the others with baseMean > 1000. Out of these seven tiRNAs were differentially expressed meeting the filtering criteria of baseMean > 1000, adjusted P-value < 0.05 and absolute value of log2FC > 2 (Supplemental File 3). The high baseMean filtering was set in order to select only highly expressed tiRNAs possibly serving better as biomarker candidates. Levels of six mature tiRNAs (tRNA-Gly-CCC_5_end, tRNA-Asp-GTC_5_end, tRNA-His-GTG_5_end, tRNA-Lys-CTT_5_end, tRNA-Cys-GCA_5_end and tRNA-His-GTG) were induced (Fig. 3B). In addition, level of one mitochondrial tiRNA, i.e., tRNA-Met-CAT_5_end was reduced, as compared to D0 bone, with log2FC varying between −1.5 and −2.3 at D5–D14. Two other 5′end mature tiRNAs tRNA-Val-AAC_5_end and tRNA-Val-CAC_5_end were expressed at levels with baseMean of over 350,000 reads, but their log2FC was 1.86 and 1.92. and adjusted Padjusted-value < 1e−04. Mature-tRNA-Glu-CTC_5_end and mature-tRNA-Gly-GCC_5_end were stably expressed with baseMean of 15,341 and 153,810 reads, respectively (Supplemental File 3).
3.3. Bone and cartilage tissues express distinct miRNA populations
Bone and cartilage tissues were included in the NGS analysis in order to identify miRNAs that are characteristic to the homeostatic stage of these tissues and how they behave during fracture healing. Abundant bone and cartilage miRNA populations were identified by comparing miRNAs in hip articular cartilage and tibial diaphyseal bone at 2 M. Altogether, 683 miRNAs were identified in the NGS data of 2 M bone and cartilage samples. Heatmap analysis was performed for the DE miRNAs between bone and cartilage tissue with baseMean > 100, absolute value of log2FC > 2 and adjusted P-value < 0.05. A total of 29 miRNAs were highly expressed in cartilage and lower in bone, while 25 miRNAs were highly expressed in bone and lower in cartilage (Fig. 4, Supplemental File 3). In the later analysis, these miRNAs are referred to as cartilage or bone associated miRNAs.
Fig. 4.
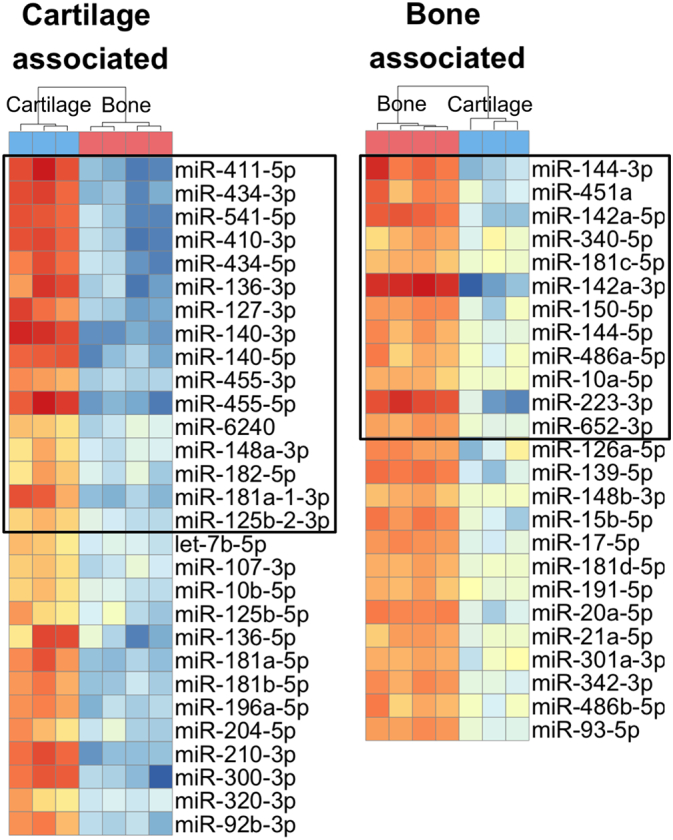
Bone and cartilage associated miRNA populations. Heatmaps indicate the occurrence of differentially expressed miRNAs between cartilage and bone. Abundant miRNAs in 2 M hip articular cartilage (n = 3) and in 2 M control tibial diaphyseal bone D0 (n = 4). MiRNAs with average normalized read counts (baseMean) above 100 are presented, with absolute log2FC > 2 and Padjusted-value < 0.05. Black frames delineate miRNAs which during fracture healing were differentially expressed (Fig. 5).
3.4. In fracture callus, differentially expressed bone associated miRNAs were suppressed while differentially expressed cartilage associated miRNAs were induced
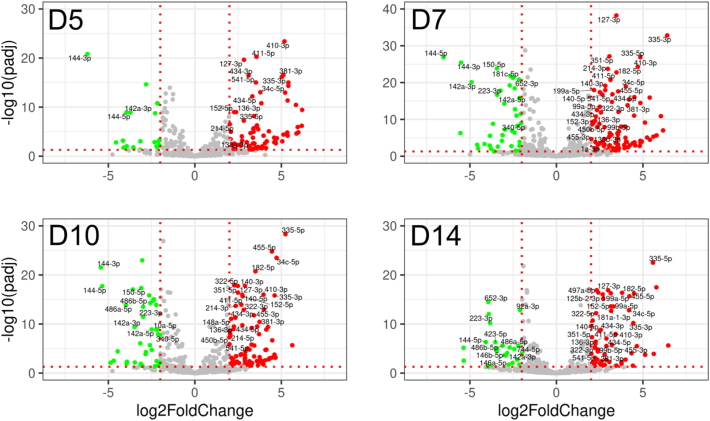
In order to show the putative interactions between miRNAs and mRNAs, the differential expression of miRNAs in callus tissue during fracture healing was first explored. Altogether, 806 mature miRNAs were identified in the NGS data of callus and control tissue samples. In comparison to D0 bone, 54 miRNAs were differentially expressed with baseMean over 100, absolute log2FC > 2 and adjusted P-value < 0.05 (Supplemental File 3). Their expression levels are visualized in volcano plots at D5, D7, D10 and D14 (Fig. 5).
Fig. 5.
Differential expression of miRNAs visualized by volcano plots. A total of 806 annotated miRNAs with average normalized read counts (baseMean) above 100 are presented in the volcano plots. Green dots represent downregulated genes (Padjusted-value < 0.05 and log2FC < −2), red dots represent upregulated genes (Padjusted-value < 0.05 and log2FC > 2) and grey dots represent other genes (X-axis, absolute value of log2FC > 2 and Padjusted-value > 0.05). Horizontal dotted lines represent Padjusted-value threshold (0.05) and vertical dotted lines represent log2FC thresholds (−2 and 2). Samples are D0 bone (n = 4), and callus samples at D5 (n = 3), D7 (n = 5), D10 (n = 4), and D14 (n = 2). For detailed list of genes, see Supplemental File 3.
Differential expression of miRNAs visualized by volcano plots. A total of 806 annotated miRNAs with average normalized read counts (baseMean) above 100 are presented in the volcano plots. Green dots represent downregulated genes (Padjusted-value < 0.05 and log2FC < −2), red dots represent upregulated genes (Padjusted-value < 0.05 and log2FC > 2) and grey dots represent other genes (X-axis, absolute value of log2FC > 2 and Padjusted-value > 0.05). Horizontal dotted lines represent Padjusted-value threshold (0.05) and vertical dotted lines represent log2FC thresholds (−2 and 2). Samples are D0 bone (n = 4), and callus samples at D5 (n = 3), D7 (n = 5), D10 (n = 4), and D14 (n = 2). For detailed list of genes, see Supplemental File 3.
Further, expression profiles of the DE miRNAs were divided into clusters based on time point of maximal absolute value of log2FC against D0 bone (Fig. 6). Thirty-five miRNAs were upregulated in comparison to D0 bone, including 16 cartilage associated miRNAs. Nineteen miRNAs were downregulated, as compared to D0 bone, including all 12 bone-associated DE miRNAs (Fig. 4, Fig. 6). In addition, 26 other miRNAs were differentially expressed during fracture healing. Majority of them were upregulated at D5, D7 and D10 while five miRNAs were downregulated at D10 and D14.
Fig. 6.
Expression profiles of differentially expressed miRNAs during fracture healing. MiRNA clusters were formed based on the date with maximum absolute value of log2 fold change against D0 at D5, D7, D10 and D14. Phases of fracture healing at D5, D7, D10 and D14 are also indicated on top. Red lines indicate bone associated and blue lines cartilage associated (see Fig. 4), and black lines other differentially expressed MiRNAs with average normalized read counts (baseMean) > 100 with an absolute value of log2FC > 2 against 2 M diaphyseal bone (D0) baseline and Padjusted-value < 0.05 are presented. Log2FC values are presented in tables for each of the 54 differentially expressed miRNAs, divided in upregulated and downregulated groups. Differential expression was calculated in R using DESeq2 against D0 (2M control diaphyseal bone). D0: n = 4, D5: n = 3, D7: n = 5, D10: n = 4, D14: n = 2. Scale bar equals to 50 μm.
For the validation of NGS data, expression levels of mmu-miR-150-5p, mmu-miR-340-5p, mmu-miR-92a-3p, mmu-miR-140-3p, mmu-miR-214-3p and mmu-miR-148a-3p were analysed by qRT-rtPCR in D0, D5, D7, D10, D14 and D25 callus samples (Supplemental File 2: S.Fig. 7). These miRNAs were chosen for the validation analysis to represent various roles in chondrogenesis, osteogenesis or fracture healing, either suggested by target prediction by TargetScanMouse, or on the basis of previously published data (Tuddenham et al., 2006; Wang et al., 2013; Tian et al., 2017). A strong correlation (rs above 0.7) was observed between the expression profiles from NGS and qRT-rtPCR analyses. The correlation of mmu-miR-92a-3p expression profile, with a high variability of expression observed between biological replicates, however, did not reach statistical significance even though the profiles were quite similar. The statistical significance was observed only at D14 between expression values by NGS and qRT-rtPCR analyses.
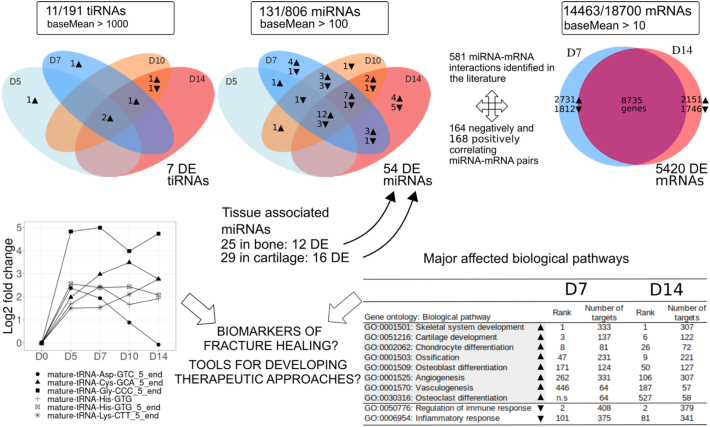
3.5. Integrative analysis of miRNA and target mRNA expression levels delineates a population of miRNA – mRNA pairs characteristic to chondrogenic and osteogenic phases
RNA-Seq analysis at D0, and at D7 and D14 samples, representing the chondrogenic and osteogenic phases of fracture healing, respectively, was carried out to characterize how fracture healing affected mRNA expression. A total of 18,700 genes were identified in the analysis. After data normalization, 14,463 genes were identified with a baseMean level 10 or higher, with adjusted P-value < 0.05. Out of these, 5420 were differentially expressed with an absolute value of log2FC > 2 (Fig. 7, Supplemental File 4). At D7, 1807 mRNAs were downregulated and 2731 upregulated, as compared to D0 bone. At D14, 1760 mRNAs were downregulated and 2179 upregulated. The top 10 biological processes that were suppressed were mostly related to immune response, while the top induced biological processes were related to organ and skeletal tissue development and chondrocyte differentiation (Supplemental File 1: S.Fig. 4). Volcano plots are presented to list log2FC in the expression levels of 247 mRNAs that were strictly related to three significant and important fracture healing associated biological processes, i.e., skeletal system development, cartilage development and ossification (Supplemental File 1: S.Fig. 5, Supplemental File 4).
Fig. 7.
Schematic summary of the genome wide expression analysis of tiRNAs, miRNAs and mRNAs during early fracture healing in mice. Cutoff for the presented values, average normalized read count (baseMean) for tiRNAs was >1000, for miRNAs > 100 and for mRNAs > 10. Values in boxes indicate the number of genes with expression level above the baseMean cutoff, and total number of identified genes. Number of bone and cartilage associated miRNAs are presented. Differential expression (DE) (with an absolute value of log2FC > 2 against 2 M diaphyseal bone (D0) baseline and Padjusted-value < 0.05) is illustrated at D5, D7, D10 and D14 for tiRNAs and miRNAs and at D7 and D14 for mRNAs. The gene ontology analysis was based on mRNA expression data at D7 and D14. ▼: downregulation; ▲: upregulation. Rank associated with GO terms have P-value < 0.05. Grey color highlights the upregulated biological GO pathways.
Differentially expressed mRNAs were sub-divided into ten gene ontology categories: skeletal system development, cartilage development, chondrocyte differentiation, ossification, osteoblast differentiation, angiogenesis, vasculogenesis, osteoclast differentiation, regulation of immune response and inflammatory response (Supplemental File 4). Selected GO categories were all related to the main steps involved in fracture healing (Einhorn and Gerstenfeld, 2015).
Integrative analysis was carried out by first searching for verified mRNA targets for the 54 DE in PubMed in human and rodent species (Supplemental File 5). Altogether, 581 putative interactions for the 54 DE miRNAs were identified in the data published by the end of June 2020. Majority of the targets were verified in various cancer tissues and cells, but some studies in skeletal tissues and mesenchymal cell lines were also found.
Spearman correlation analysis was carried out in individual callus samples (n = 10) between the expression levels of 54 DE miRNAs (NGS data) and their reported verified target mRNAs (RNA-Seq data) (Supplemental File 5). Correlation analysis for downregulated and upregulated miRNAs and their targets resulted in identification of 164 negatively correlating and 168 positively correlating miRNA - mRNA pairs during fracture healing (Table 1, Table 2).
Table 1.
Downregulated miRNAs during fracture healing and their significantly correlated mRNAs. Reported verified target mRNAs in human and rodent species for the 54 DE miRNAs (listed in Fig. 6) were searched in PubMed (data published by June 2020). Spearman correlation test in R was carried out between miRNA and target mRNA expression levels in individual samples. miRNAs that were downregulated compared to D0 bone and their target mRNA with negative correlation in the expression levels are listed here. No correlation was found between any of the published targets for downregulated miRNAs miR-1a-3p, miR-146a-5p, 146b-5p and miR-744-5p. Entire list of the validated target search, with references and correlation coefficient values between miRNAs and target mRNAs is presented in Supplemental File 5.
Downregulated miRNAs during fracture healing and their significantly correlated mRNAs. Reported verified target mRNAs in human and rodent species for the 54 DE miRNAs (listed in Fig. 6) were searched in PubMed (data published by June 2020). Spearman correlation test in R was carried out between miRNA and target mRNA expression levels in individual samples. miRNAs that were downregulated compared to D0 bone and their target mRNA with negative correlation in the expression levels are listed here. No correlation was found between any of the published targets for downregulated miRNAs miR-1a-3p, miR-146a-5p, 146b-5p and miR-744-5p. Entire list of the validated target search, with references and correlation coefficient values between miRNAs and target mRNAs is presented in Supplemental File 5.
| Cluster | miRNA |
Targets with negative correlation |
Targets with positive correlation |
|---|---|---|---|
| 14 downregulated miRNAs ↔ 59 negatively correlating and 61 positively correlating mRNAs | |||
| D5 | miR-144-3pa | Hif1a, Pbx3, Fn1, App, Fzd4, Smad4, Fosb, Ctbp2, Hoxa7, Tie2 | Map3k8, Smarca4, Atg4a, Klf3, Ezh2, Sgk3, Arid1a |
| D5 | miR-451aa | Osr1, Cav1, Trim66, Mif, Tbx1, Oxtr | Psmb8, Ywhaz, Atf2, Il6r |
| D7 | miR-142a-3pa | Fam98a, Fzd7, Rab3a, Adam9, Il6, Ctnnb1, Nr2f6 | Tgfbr1, Hmga1, Hmgb1, Peli1 |
| D7 | miR-142a-5pa | Cyr61, Socs1, Ghr | Becn1, Pten |
| D7 | miR-144-5pa | Smad1 | Cdc25a, Atf2, Cdkl1 |
| D7 | miR-150-5pa | Socs1, Rab9a, Mmp14, Slc2a1, Elk1 | Gsk3b, Sfpi1 |
| D7 | miR-181c-5pa | Il1a, Tnf | |
| D7 | miR-340-5pa | Hif1a, Fmod, Yap1, Arg1, Stat3, Nrp1, Ctnnb1 | Neurod4, Ddx58, Nfkb1, Cdk1, Nfe2l2, Anxa3, Oas2, Pik3c3, Rap1a, Atf1, Ḱpna4, Mapk14, Vav3, Rock1 |
| D10 | miR-10a-5pa | Ccn2, Hoxa1 | |
| D10 | miR-486a-5pa | Dock1, Cemip, Nrp2 | Pik3r1, Pten |
| D14 | miR-223-3pa | Nf2, Lif, Fat1, Ctnnd1, Skil, Zeb1, Cdh6, Smad3, Fam5c, Rasa1, Il17rd | Inpp5p, Arid1a, Fbxo30, Foxo3, Nlrp3, Syne1, Il1b, Stat4 |
| D14 | miR-652-3pa | Cdh1 | |
| D14 | miR-92a-3p | Snhg14 | Atg4a, Fam220a, Cdh1, Notch1, Pten |
| D14 | miR-423-5p | Tnip2, Cdkn1a, Igf2bp1 | Nacc1, Rab11b, Mybl2, Stmn1, Gstm1, Srf, Nlrx1 |
Bone-associated DE miRNA, see Fig. 4.
Table 2.
Upregulated miRNAs during fracture healing and their significantly correlated mRNAs. Reported verified target mRNAs in human and rodent species for the 54 DE miRNAs (listed in Fig. 6) were searched in PubMed (data published by June 2020). Spearman correlation test in R was carried out between miRNA and target mRNA expression levels in individual samples. miRNAs that were downregulated compared to D0 bone and their target mRNA with negative correlation in the expression levels are listed here. No correlation was found between any of the published targets for upregulated 133b-3p, 181a-1-3p, and miR-497a-3p. No targets have been verified in the literature for miR-133b-3p, miR-434-5p, miR-5099 and miR-322-3p. Entire list of the validated target search, with references and correlation coefficient values between miRNAs and target mRNAs is presented in Supplemental File 5.
Upregulated miRNAs during fracture healing and their significantly correlated mRNAs. Reported verified target mRNAs in human and rodent species for the 54 DE miRNAs (listed in Fig. 6) were searched in PubMed (data published by June 2020). Spearman correlation test in R was carried out between miRNA and target mRNA expression levels in individual samples. miRNAs that were downregulated compared to D0 bone and their target mRNA with negative correlation in the expression levels are listed here. No correlation was found between any of the published targets for upregulated 133b-3p, 181a-1-3p, and miR-497a-3p. No targets have been verified in the literature for miR-133b-3p, miR-434-5p, miR-5099 and miR-322-3p. Entire list of the validated target search, with references and correlation coefficient values between miRNAs and target mRNAs is presented in Supplemental File 5.
| Cluster | miRNA |
Targets with negative correlation |
Targets with positive correlation |
|---|---|---|---|
| 28 upregulated miRNAs ↔ 105 negatively correlating and 107 positively correlating mRNAs | |||
| D5 | miR-136-3pa | Pten | |
| D5 | miR-410-3pa | Cxcr5, Fmr1, Yy1, Pten | Stat3, Tgfbr2, Snai1 |
| D5 | miR-411-5pa | Txnip, Vasp, Grb2 | |
| D5 | miR-434-3pa | Eif5a1 | |
| D5 | miR-541-5pa | Cdk6 | |
| D5 | miR-133a-3p | Met | |
| D5 | miR-381-3p | Ube2c, Cdk6, Map3k8, Cxcr4 | Ets2, Fgf7, Lrp6 |
| D7 | miR-127-3pa | Kif3b | Bag5 |
| D7 | miR-99b-5p | Fzd8 | |
| D7 | miR-152-3p | Atg12, Fasl, Brd4, Dnmt1 | Klf4, Slc2a1, Foxf1a, Klf4 |
| D7 | miR-214-3p | Nlrc5, Hmga1, St6gal1, Atg12, Ezh2, Ezh1, Pim1, Stat6, Cadm1, Pten | Ctnnb1, Pdk1, Abcb1, C1qtnf9, Irs1, Runx3, Foxp2, Mmp2, Plcb1, Ctrp9, Pdk1 |
| D7 | miR-335-3p | Kdm4c, Plaur | |
| D7 | miR-351-5p | Mapk13 | Sirt6 |
| D7 | miR-450b-5p | Yap1, Kif26b | |
| D10 | miR-140-3pa | Cxcl12, Trpm2, Cd38, Tnfa, Jak1, Sirt1, Atp1b, Myb, Mcf2l, Atp8a1, Bcl2 | Klf4, Pthlh, Masp1, Acan, Col4a1, Pycr1, Tgfb3, Trps1 |
| D10 | miR-140-5pa | Tnf, Nfe2l2, Map3k11, Stat1, Bloc1s2, Creb1, Birc5, Glul, Hdac4, Pin1, Adam10, Igf1r, Hmgb1, Hmgn5, Tlr4 | Vegfa, Sept2, Pak4, Six1, Smurf1, Meg3, Plod1, Sox4, Thy1, Ndrg3, Frs2, Sox9, Cemip, Fgfrl1, Muc1, Runx3, Hoxa11, Snai2, Pdgfra, Smad3, Yes1, Foxo1, Zeb1 |
| D10 | miR-455-3pa | Fam83f | Runx2, Dusp3, Zeb1 |
| D10 | miR-455-5pa | Lgals9, Jak1, Myd88, Dnmt1, Kdm6b | Rab31, S1pr1, Socs3, Runx2, Rab18 |
| D10 | miR-34c-5p | Gucy1b3, Flot2, Sp1, Etv6, Atg4b, Ccl22, Gucy1b3 | Bmf |
| D10 | miR-214-5p | Rock1, Klf5, Cxcr5, E2f2, Ciz1 | Wasl, Dpysl5, Tm4sf1, Col4a1 |
| D14 | miR-148a-3pa | Dnmt1, Ikbkb, Mcl1, Kdm6b, Snhg4 | Runx3 |
| D14 | miR-182-5pa | Rab27a, Flot1, Foxo3, Cdkn1b, Bcl2l12, Sesn2, Cfl1, Creb1, Pten | Bnip3, Igf1, Pthlh, Reck, Smad4, Cd2ap, Hoxa9, Rnd3, Gli2, Dll4, Efnb1 |
| D14 | miR-99a-5p | Hoxa1 | |
| D14 | miR-148a-5p | Serpinh1, Tlr3 | |
| D14 | miR-152-5p | Txnip | Smad3, Fbxl7, Pik3ca |
| D14 | miR-199a-5p | Ccr7, Mst1, Ccnb1, Slit1, Nfkb1, Rela, Ccn2, Rock1, March8, Fkbp5, Sirt1 | Fzd6, Hif1a, Zeb1, Osgin2, Tgfb2, Pik3ca, Ece1, Pias3, Pvrl1, Srgap2, Vegfa, Arhgap12, Map4k3, Mafb |
| D14 | miR-322-5p | Fam3b, Nfkb1 | |
| D14 | miR-335-5p | Ldhb, Bcl2l2, Dancr, Aplnr, Tnfsf11 | |
Cartilage-associated DE miRNA, see Fig. 4.
4. Discussion
In this study, the genome wide expression profiles of tiRNAs, miRNAs and mRNAs were investigated during chondrogenic and early osteogenic phases of secondary fracture healing of a standard closed fracture where the healing occurs via callus formation. Also, expression levels of tiRNAs in normal 2 M bone and cartilage tissues were compared, and bone and cartilage homeostasis associated miRNA populations were identified.
During fracture healing, a correlation analysis of the expression levels of 54 DE miRNA and their reported target mRNAs (identified by PubMed search) was performed. These findings are summarized in Fig. 7.
4.1. TiRNAs associated with fracture healing as a stress response
The role of tRNA fragments in regulation of cellular functions have emerged as a hot topic in recent years. This is the first study to describe the expression profiles of tiRNAs in callus tissue during fracture healing. An overall increase in tiRNA expression was observed in callus tissues compared with healthy bone or cartilage tissues. In bone, 5′end tiRNAs were expressed at about five times higher level compared to cartilage. Several unique features of the cartilage may contribute to this difference, including poor cell proliferation rate and hypoxic nature of the avascular mature cartilage. Also, angiogenin which cleaves tRNA into tiRNA fragments was expressed at a slightly lower level in cartilage, as compared to bone.
Expression levels of 11 mature 5′end tiRNAs stood out from the remaining 191 tiRNAs due to their much higher expression levels. Six 5′end derived mature tiRNAs were upregulated and presented distinct profiles related to different phases of fracture healing.
Peaking of tRNA-Asp-GTC_5′end expression was emphasized to D5, a phase with mesenchymal cell migration and proliferation, tRNA-His-GTG expression emphasized to D7 associating with chondrogenesis while tRNA-Lys-CTT_5′end peaked at D14, a phase with active osteogenesis. The expression of tRNA-Cys-GCA_5′end, tRNA-Gly-CCC_5′end and tRNA-His-GTG_5′end tiRNAs remained high throughout all the main phases of fracture healing at D5-D14. In addition, two other abundant 5′end tiRNAs mature-tRNA-Val-AAC_5′end and mature-tRNA-Val-CAC_5′end reached slightly lower level of log2FC differential expression peaking at D5.
TiRNAs originating from the 5′end have been reported to affect gene regulation by translational activation, inhibition and reprogramming, and to modulate stem cell pluripotency in mESCs (Kim, 2019; Krishna et al., 2019; Advani and Ivanov, 2019). The present study indicated that in fracture callus, ten 5′end tiRNAs are abundant, and six of them were significantly upregulated with log2FC over 2 tiRNAs, as compared to healthy bone, suggesting them a special role in regulation of fracture healing. On the other hand, two highly abundant mature 5′end tiRNAs tRNA-Glu-CTC_5′end and tRNA-Gly-GCC_5′end, were stably expressed and may therefore have role in general homeostasis of these tissues.
More detailed investigations are required to clarify the relevance and functions of tiRNAs in fracture healing follow-up as well as their potential value as biomarkers.
4.2. MiRNAs associated to cartilage and bone homeostasis
Comparison of control bone and hip articular cartilage at two months resulted in identification of 29 highly abundant miRNAs in articular cartilage, and 25 highly abundant miRNAs in bone, which suggests the importance of these miRNAs in the maintenance of homeostasis in these tissues. Several of the miRNAs enriched in cartilage tissue, such as miR-140, miR-181a, miR-204 and miR-455-3p, have been previously shown to be important in regulation of chondrogenesis (Razmara et al., 2019). Many of them (miR-125b-5p, 140p-5p, miR-204-5p, 320-3p) have been reported to inhibit osteogenesis or vascularization, cell growth and invasion (miR-127-3p, miR-136-3p), thereby supporting the specific characteristics of the cartilage tissue which lacks blood vessels and nerves. Further, mature chondrocytes have very low proliferation rate (Dreier, 2010). In a similar manner as the cartilage associated miRNAs mentioned above have been reported to have a role in cartilage development, the bone associated miRNAs, such as miR-15b, miR-17-5p, miR-20a, miR-21 and miR-93, have been reported to have a role in the regulation of osteogenesis (Huang et al., 2017). In bone tissue, several miRNAs are also related to regulation of inflammation, e.g., miR-142a-3p, and miR-142a-5p (Talebi et al., 2017). MiR-142a-5p was recently reported to promote osteoblast differentiation via targeting nuclear factor 1A and to promote osteoclast differentiation of bone marrow-derived macrophages via the PTEN/PI3K/AKT/FoxO1 pathway (Yuan et al., 2021; Lou et al., 2019). MiR-142-3p was reported to be a negative regulator of osteoclastogenesis in monocytes, macrophages and dendritic cells, and its FGD5-AS1-mediated reduction was shown to induce expression of its target SOCS6 mediating signals to NF-kappa pathway to inhibit lipopolysaccharide induced experimental periodontitis (Fordham et al., 2016; Chen et al., 2019a).
In addition, a few of the bone-associated miRNAs, e.g., miR-150-5p, miR-223-3p, miR-93-5p have been detected in matrix particles or circulating exosomes (Kelch et al., 2017; Legrand et al., 2020; Lu et al., 2019; Mäkitie et al., 2018; Perez-Sanchez et al., 2018; Hosokawa et al., 2017) and may therefore be important in cell-cell communication and systemic regulation of body functions.
4.3. Differential expression during fracture healing
When evaluating the in vivo data, it is important to keep in mind that callus is a complex multicellular tissue where the healing phases in parallel cells and regions proceed partially non-synchronized through inflammation, chondrogenesis, vascularization, osteogenesis, and shifting gradually to osteoclastogenesis and finally reach bone remodelling. Out of the 54 DE miRNAs, 16 miRNAs were cartilage-associated and 12 bone-associated. Altogether, 18 miRNAs were expressed in callus tissue at a lower level than in bone, including all DE bone-associated miRNAs. Interestingly, all cartilage-associated DE miRNAs were expressed at a higher level in callus tissue. In order to find out how these DE miRNAs related to different phases of fracture healing, they were divided into four clusters according to the date after fracture with the highest absolute log2FC in the expression profile.
4.4. Impact of miRNA downregulation on fracture healing
In the present study, 18 miRNAs were downregulated after fracture. Twelve of them were bone associated miRNAs. Reduced expression of a particular miRNA in callus tissue is expected to allow the expression of its target gene(s) necessary to drive fracture healing. The majority of these miRNAs were found to regulate genes important for osteoblast differentiation, chondrogenesis, vascularization or bone remodelling. For example, miR-150-5p was expressed at a lower level at D7 and correlated negatively with its verified targets Mmp14 and Slc2a1. Mmp14 codes for a membrane bound metalloproteinase MT1-MMP which is important in extracellular matrix remodelling. It is associated to terminal differentiation of chondrocytes, and during fracture healing has been earlier reported to be induced during chondrogenic phase (Lehmann et al., 2005). Slc2a1 codes for glucose transporter GLUT1, which is important for the cellular uptake of glucose and thus in skeletal tissue development as well as for all glucose dependent cellular processes in chondrocytes (Lee et al., 2018). Similarly, negative correlation was observed between miR-340-5p and Ctnnb1 that codes for beta-catenin. In the early stages of fracture healing, beta-catenin regulates multipotent mesenchymal cells to differentiate either into osteoblasts or chondrocytes and once the cells have committed to osteoblastic lineage, it supports positively their osteoblastic phenotype (Chen et al., 2007). Hypoxia-inducible factor 1-alpha (HIF1A) inhibits miR-340-5p thereby enhancing Ctnnb1 expression and osteogenesis under hypoxic conditions (Du et al., 2017). MiR-144-3p was downregulated in callus tissues and negatively correlated with its target Hif1a, thereby allowing translation of HIF1A during fracture healing. Downregulation of miR-340-5p has been shown to have a role in promoting cell proliferation and suppressing chondrocyte apoptosis in osteoarthritis by inhibiting extracellular signal-regulated kinase (ERK) signaling via fibromodulin (Zhang et al., 2018). In the present data, a negative correlation was observed between miR-340-5p and fibromodulin gene (Fmod), suggesting a similar role during fracture healing. miR-340 gene originates from the intronic region Rnf130 (Ring Finger Protein 130 gene), a gene involved in protein ubiquitination, which was also downregulated during fracture. miR-340 expression correlated with its host gene Rnf130 (rs = 0.7, p = 0.031) suggesting that expression of miR-340 is regulated by the expression of its host gene as has been shown before for intragenic miRNAs (Gulyaeva and Kushlinskiy, 2016).
4.5. Impact of miRNA upregulation on fracture healing
The upregulation of miRNAs is expected to directly reduce its target's translation in a given tissue or increase chromatin accessibility by recruiting chromatin modifying proteins and thereby facilitating expression of another gene. A significant number of upregulated miRNAs in callus tissue were associated with cartilage tissue development (Razmara et al., 2019). The expression of miR-140-3p, miR-148a-3p and miR-214 have been found to be important for chondrogenesis and osteogenesis (Tuddenham et al., 2006; Wang et al., 2013; Tian et al., 2017; Roberto et al., 2018). A negative correlation was observed between miR-214-3p and Ezh2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit). Ezh2 inhibition has been reported to accelerate osteogenic differentiation (Camilleri et al., 2018). In addition, miR-214 gene expression correlated with the expression of its host gene Dnm3os (Dynamin 3 opposite strand gene) and miR-140 with Wwp2 (WW Domain Containing E3 Ubiquitin Protein Ligase 2 gene) which were also upregulated after fracture which is again demonstrating a possible consequence of host gene – miRNA expression pattern, as described above for miR-340.
4.6. Data interpretation
Several aspects influence on the data interpretation. These data describe tiRNA, miRNA and mRNA transcriptomes during fracture healing in two months old fertile but skeletally immature male C57Bl/6 N mice. An important issue in interpreting the data is that osteogenic differentiation potential of mesenchymal progenitor cells is affected by aging (Liu et al., 2015). In addition, the genetic variability between different mouse strains as well as the gender significantly contribute to the process of bone regulation and fracture healing (Manigrasso and O'Connor, 2008; Deng et al., 2020). Hence, age, gender and species contribute to the transcriptome signature and are important aspects while comparing the data between different experiments as well as when applying to the different experimental settings.
Considerable amounts of miRNAs and tiRNAs in circulation originate from erythrocytes. It has been shown that extracellular vesicles are secreted during erythropoiesis, physiological cellular aging, disease conditions, and in response to environmental stressors (Thangaraju et al., 2020). Noncoding RNAs may be absorbed from lysing red blood cells and hence cause contamination in the sample tissue. Although the volume of the blood contamination is most likely very small in comparison to callus tissue volume, we cannot exclude the possibility of minor contamination by erythrocytes. Angiogenesis is an important component of fracture healing and it is also possible that erythrocytes residing in the new vessels contribute - in addition to endogenous synthesis - to the miRNA and tiRNA signature. Some of the miRNAs, such as miR-451, miR-144, and miR-486 and 5′ tiRNAs that are highly expressed in erythrocytes and abundant in circulation, such as tRNA-His-GTG and tRNA-Cys-GCA, tRNA-Lys-CTT were also highly expressed in the present data (Xu et al., 2019; Dhahbi et al., 2013). Supporting the physiological role, these miRNAs were differentially expressed during fracture healing and negatively correlated with the expression levels of their confirmed targets, e.g., miR-144-5p negatively correlated with Smad1, miR-451 with Osr1, and miR-486 with Cemip, thereby connecting these miRNAs to bone formation (Tasca et al., 2018; Karvande et al., 2018; Chen et al., 2019b). In conclusion, the possible physiological role of these miRNAs and tiRNAs in fracture healing may be a combined effect of endogenous synthesis in callus tissue and blood/erythrocyte originated vesicles of vascularized callus tissue.
The data obtained with the current approach provides a general view on the gene expression profiles during fracture healing in vivo. Future studies using, e.g., single cell population sequencing and spatial transcriptomic platforms will allow more localized and differentiation phase specific information on RNA expression and fine-tuning of the miRNA/tiRNA relationships during the cell differentiation in fracture repair. Altogether, these results show that changes in miRNA expression were directly associated with physiological events in vivo and, may play a role in mRNA regulation during fracture healing. Some of these miRNAs (miR-142-5p, miR-199a-5p, miR-223, miR-144 and miR-497) were previously found in a genome wide study to be associated with impaired fracture healing in aging mice by downregulating angiogenesis-, chondrogenesis-, and osteogenesis-related pathways and upregulating osteoclastogenesis (He et al., 2016). We sub-divided here differentially expressed mRNAs into ten related gene ontology categories: skeletal system development, cartilage development, chondrocyte differentiation, ossification, osteoblast differentiation, angiogenesis, vasculogenesis, osteoclast differentiation, regulation of immune response and inflammatory response. Selected GO categories were all related to the main steps involved of fracture healing (Einhorn and Gerstenfeld, 2015).
Whether or not the miRNA expression is linked to a direct regulation of target mRNAs, to their host gene expression, or expression of transcription factors directly targeting miRNA promoters, selected miRNA genes may have value as indicators of fracture healing progression, or even as tools for developing novel pharmaceutical agents promoting fracture healing.
5. Conclusions
Genome wide expression profiles of small RNAs and mRNAs were investigated by next generation sequencing and bioinformatics analyses in callus tissues, cartilage and control intact diaphyseal bone during early secondary fracture healing using an in vivo model of tibial closed fracture in mice.
This is the first study linking differential expression of tiRNAs in callus tissue to fracture healing. The quantities of mature 3′-end and 5′-end total tiRNAs and six individual mature 5′end tiRNAs were increased in comparison to control diaphyseal bone. Also, the quantity of total tiRNAs in cartilage was low, only appr. one third of that observed in bone. A combined literature search for target mRNAs to 54 DE miRNAs in all human and rodent tissues, and correlation analysis of miRNA – target mRNA expression levels in callus tissue resulted in 581 putative interactions, thereby linking 14 downregulated miRNAs and 28 upregulated miRNAs to 164 negatively correlating and 168 positively correlating miRNA-mRNA pairs with chondrogenic and osteogenic phases of fracture healing (Fig. 7). A few new miRNAs, including miR-150-5p and miR-340-5p, were linked to bone homeostasis and formation during fracture healing. Altogether, this study contributes to a better understanding of the molecular mechanisms of fracture healing in vivo and may contribute to the future clinical applications, such as potential use of miRNAs and tiRNAs as biomarkers or as tools in the development of novel therapeutic approaches for fracture healing.
The following are the supplementary data related to this article.
Validation studies of the data.
Validation studies of the data; miRNA normalization genes.
TiRNA and miRNA normalized full data.
RNASeq full data and GO data.
Integrative miRNA-mRNA expression analysis
CRediT authorship contribution statement
Matthieu Bourgery: Software, Validation, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft. Erika Ekholm: Investigation, Writing – review & editing. Katja Fagerlund: Investigation, Writing – review & editing. Ari Hiltunen: Investigation, Writing – review & editing. Tero Puolakkainen: Investigation, Writing – review & editing. Juha-Pekka Pursiheimo: Investigation, Writing – review & editing. Terhi Heino: Investigation, Writing – review & editing. Jorma Määttä: Investigation, Writing – review & editing. Jussi Heinonen: Investigation, Writing – review & editing. Emrah Yatkin: Investigation, Writing – review & editing. Tiina Laitala: Supervision, Funding acquisition, Writing – review & editing. Anna-Marja Säämänen: Investigation, Supervision, Project administration, Resources, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge Merja Lakkisto, Liudmila Shumskaya and Jukka Karhu for their excellent technical assistance, the Central Animal Laboratory of Turku for their excellent staff who took care of our animals, and Noora Kotaja for reviewing the manuscript. Matthieu Bourgery is a member of University of Turku TUDMM doctoral program.
References
- Advani V.M., Ivanov P. Translational control under stress: reshaping the translatome. Bioessays. 2019;41(5) doi: 10.1002/bies.201900009. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. elife. 2015;4 doi: 10.7554/eLife.05005. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57(1):289–300. http://www.jstor.org/stable/2346101 [Internet] [Google Scholar]
- Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–2039. doi: 10.15252/embj.201489282. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri E.T., Dudakovic A., Riester S.M., Galeano-Garces C., Paradise C.R., Bradley E.W. Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development. J. Biol. Chem. 2018;293(49):19001–19011. doi: 10.1074/jbc.RA118.003909. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Whetstone H.C., Lin A.C., Nadesan P., Wei Q., Poon R. Beta-catenin signaling plays a disparate role in different phases of fracture repair: implications for therapy to improve bone healing. PLoS Med. 2007;4(7) doi: 10.1371/journal.pmed.0040249. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lan Z., Li Q., Li Y. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-?B pathway. Artif. Cells Nanomed. Biotechnol. 2019;47(1):2098–2106. doi: 10.1080/21691401.2019.1620256. (Dec) [DOI] [PubMed] [Google Scholar]
- Chen L., Shi K., Andersen T.L., Qiu W., Kassem M. KIAA1199 is a secreted molecule that enhances osteoblastic stem cell migration and recruitment. Cell Death Dis. 2019;10(2):126. doi: 10.1038/s41419-018-1202-9. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Gao X., Sun X., Cui Y., Amra S., Huard J. Gender differences in tibial fractures healing in normal and muscular dystrophic mice. Am. J. Transl. Res. 2020;12(6):2640–2651. [PMC free article] [PubMed] [Google Scholar]
- Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Boffelli D., Mote P. 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriou R., Tsiridis E., Giannoudis P.V. Current concepts of molecular aspects of bone healing. Injury. 2005;36(12):1392–1404. doi: 10.1016/j.injury.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Dreier R. Hypertrophic differentiation of chondrocytes in osteoarthritis: the developmental aspect of degenerative joint disorders. Arthritis Res. Ther. 2010;12(5):216. doi: 10.1186/ar3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreifke M.B., Jayasuriya A.A., Jayasuriya A.C. Current wound healing procedures and potential care. Mater. Sci. Eng. Mater. Biol. Appl. 2015;48:651–662. doi: 10.1016/j.msec.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K., Li Z., Fang X., Cao T., Xu Y. Ferulic acid promotes osteogenesis of bone marrow-derived mesenchymal stem cells by inhibiting microRNA-340 to induce ß-catenin expression through hypoxia. Eur. J. Cell Biol. 2017;96(6):496–503. doi: 10.1016/j.ejcb.2017.07.002. Sep. [DOI] [PubMed] [Google Scholar]
- Einhorn T.A., Gerstenfeld L.C. Fracture healing: mechanisms and interventions. Nat Rev. 2015;11(1):45–54. doi: 10.1038/nrrheum.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhorn T.A., Majeska R.J., Rush E.B., Levine P.M., Horowitz M.C. The expression of cytokine activity by fracture callus. J. Bone Miner. Res. 1995;10(8):1272–1281. doi: 10.1002/jbmr.5650100818. [DOI] [PubMed] [Google Scholar]
- Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham J.B., Guilfoyle K., Naqvi A.R., Nares S. MiR-142-3p is a RANKL-dependent inducer of cell death in osteoclasts. Sci. Rep. 2016;6:24980. doi: 10.1038/srep24980. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenfeld L.C., Cullinane D.M., Barnes G.L., Graves D.T., Einhorn T.A. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J. Cell. Biochem. 2003;88(5):873–884. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- Gulyaeva L.F., Kushlinskiy N.E. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016;14(1):143. doi: 10.1186/s12967-016-0893-x. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankenson K.D., Dishowitz M., Gray C., Schenker M. Angiogenesis in bone regeneration. Injury. 2011;42(6):556–561. doi: 10.1016/j.injury.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Z.-K., Liu J., He Y.-X., Tang T., Li J. Bioinformatics and microarray analysis of miRNAs in aged female mice model implied new molecular mechanisms for impaired fracture healing. Int. J. Mol. Sci. 2016;17(8) doi: 10.3390/ijms17081260. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen J., Zhang F.P., Surmann-Schmitt C., Honkala S., Stock M., Poutanen M. Defects in chondrocyte maturation and secondary ossification in mouse knee joint epiphyses due to snorc deficiency. Osteoarthr. Cartil. 2017;25(7):1132–1142. doi: 10.1016/j.joca.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Hiltunen A., Vuorio E., Aro H.T. A standardized experimental fracture in the mouse tibia. J. Orthop. Res. 1993;11(2):305–312. doi: 10.1002/jor.1100110219. [DOI] [PubMed] [Google Scholar]
- Hinton R.J., Jing Y., Jing J., Feng J.Q. Roles of chondrocytes in endochondral bone formation and fracture repair. J. Dent. Res. 2016 Jan;96(1):23–30. doi: 10.1177/0022034516668321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa K., Kajigaya S., Feng X., Desierto M.J., Fernandez Ibanez M.D.P., Rios O. A plasma microRNA signature as a biomarker for acquired aplastic anemia. Haematologica. 2017;102(1):69–78. doi: 10.3324/haematol.2016.151076. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Geng J., Jiang S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2017;368(2):229–238. doi: 10.1007/s00441-016-2462-2. [DOI] [PubMed] [Google Scholar]
- Ikeda T., Kamekura S., Mabuchi A., Kou I., Seki S., Takato T. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50(11):3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- Jensen E.D., Gopalakrishnan R., Westendorf J.J. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. doi: 10.1002/biof.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyakumar V., Halbwirth F., Niculescu-Morzsa E., Bauer C., Zwickl H., Kern D. Chondrogenic gene expression differences between chondrocytes from osteoarthritic and non-OA trauma joints in a 3D collagen type I hydrogel. Cartilage. 2017;8(2):191–198. doi: 10.1177/1947603516657641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvande A., Kushwaha P., Ahmad N., Adhikary S., Kothari P., Tripathi A.K. Glucose dependent miR-451a expression contributes to parathyroid hormone mediated osteoblast differentiation. Bone. 2018;117:98–115. doi: 10.1016/j.bone.2018.09.007. Dec. [DOI] [PubMed] [Google Scholar]
- Kelch S., Balmayor E.R., Seeliger C., Vester H., Kirschke J.S., van Griensven M. miRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci. Rep. 2017;7(1):15861. doi: 10.1038/s41598-017-16113-x. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K. Transfer RNA-derived small non-coding RNA: dual regulator of protein synthesis. Mol. Cells. 2019;42(10):687–692. doi: 10.14348/molcells.2019.0214. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviranta I., Jurvelin J., Tammi M., Saamanen A.M., Helminen H.J. Microspectrophotometric quantitation of glycosaminoglycans in articular cartilage sections stained with safranin O. Histochemistry. 1985;82(3):249–255. doi: 10.1007/BF00501401. [DOI] [PubMed] [Google Scholar]
- Krishna S., Yim D.G., Lakshmanan V., Tirumalai V., Koh J.L., Park J.E. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019 Jul;20(7) doi: 10.15252/embr.201947789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Abel E.D., Long F. Glucose metabolism induced by bmp signaling is essential for murine skeletal development. Nat. Commun. 2018;9(1):4831–4835. doi: 10.1038/s41467-018-07316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M.A., Millet M., Merle B., Rousseau J.-C., Hemmendinger A., Gineyts E. A signature of circulating miRNAs associated with fibrous dysplasia of bone: the mirDys study. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2020 doi: 10.1002/jbmr.4111. Jun. [DOI] [PubMed] [Google Scholar]
- Lehmann W., Edgar C.M., Wang K., Cho T.-J., Barnes G.L., Kakar S. Tumor necrosis factor alpha (TNF-alpha) coordinately regulates the expression of specific matrix metalloproteinases (MMPS) and angiogenic factors during fracture healing. Bone. 2005;36(2):300–310. doi: 10.1016/j.bone.2004.10.010. Feb. [DOI] [PubMed] [Google Scholar]
- Li S., Xu Z., Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel) 2018;9(5) doi: 10.3390/genes9050246. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Xia X., Li B. Mesenchymal stem cell aging: mechanisms and influences on skeletal and non-skeletal tissues. Exp. Biol. Med. (Maywood). 2015;240(8):1099–1106. doi: 10.1177/1535370215591828. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.F., Samsa W.E., Zhou G., Lefebvre V. Transcriptional control of chondrocyte specification and differentiation. Semin. Cell Dev. Biol. 2017;62:34–49. doi: 10.1016/j.semcdb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., Peng Z., Wang B., Li X., Li X., Zhang X. miR-142-5p promotes the osteoclast differentiation of bone marrow-derived macrophages via PTEN/PI3K/AKT/FoxO1 pathway. J. Bone Miner. Metab. 2019;37(5):815–824. doi: 10.1007/s00774-019-00997-y. Sep. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550–558. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.-B., Chen D.-Z., Chen L., Hu E.-D., Wu J.-L., Li H. Attenuation of experimental autoimmune hepatitis in mice with bone mesenchymal stem cell-derived exosomes carrying MicroRNA-223-3p. Mol. Cells. 2019;42(12):906–918. doi: 10.14348/molcells.2019.2283. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinf. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkitie R.E., Hackl M., Niinimäki R., Kakko S., Grillari J., Mäkitie O. Altered MicroRNA profile in osteoporosis caused by impaired WNT signaling. J. Clin. Endocrinol. Metab. 2018;103(5):1985–1996. doi: 10.1210/jc.2017-02585. May. [DOI] [PubMed] [Google Scholar]
- Manigrasso M.B., O’Connor J.P. Comparison of fracture healing among different inbred mouse strains. Calcif. Tissue Int. 2008;82(6):465–474. doi: 10.1007/s00223-008-9144-3. Jun. [DOI] [PubMed] [Google Scholar]
- Michell-Robinson M.A., Touil H., Healy L.M., Owen D.R., Durafourt B.A., Bar-Or A. Roles of microglia in brain development, tissue maintenance and repair. Brain. 2015;138(Pt 5):1138–1159. doi: 10.1093/brain/awv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.H., Diao H.J., Chew S.Y. MicroRNAs and their potential therapeutic applications in neural tissue engineering. Adv. Drug Deliv. Rev. 2015;88:53–66. doi: 10.1016/j.addr.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag. Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthr. Cartil. 2016;24(4):573–580. doi: 10.1016/j.joca.2015.10.018. Apr. [DOI] [PubMed] [Google Scholar]
- Nugent M. MicroRNAs and fracture healing. Calcif. Tissue Int. 2017;101:355–361. doi: 10.1007/s00223-017-0296-x. [DOI] [PubMed] [Google Scholar]
- Perez-Sanchez C., Font-Ugalde P., Ruiz-Limon P., Lopez-Pedrera C., Castro-Villegas M.C., Abalos-Aguilera M.C. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Hum. Mol. Genet. 2018;27(5):875–890. doi: 10.1093/hmg/ddy008. Mar. [DOI] [PubMed] [Google Scholar]
- Puolakkainen T., Rummukainen P., Lehto J., Ritvos O., Hiltunen A., Saamanen A.M. Soluble activin type IIB receptor improves fracture healing in a closed tibial fracture mouse model. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0180593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggatt L.J., Partridge N.C. Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 2010;285(33):25103–25108. doi: 10.1074/jbc.R109.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmara E., Bitaraf A., Yousefi H., Nguyen T.H., Garshasbi M., Cho W.C. Non-coding RNAs in cartilage development: an updated review. Int. J. Mol. Sci. 2019;20(18) doi: 10.3390/ijms20184475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto V.P., Gavaia P., Nunes M.J., Rodrigues E., Cancela M.L., Tiago D.M. Evidences for a new role of miR-214 in chondrogenesis. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-21735-w. pp. 3704–018-21735-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle C.H., Wang H., Yu H., Chadwick R.B., Davis E.I., Wergedal J.E. Microarray analysis of gene expression during the inflammation and endochondral bone formation stages of rat femur fracture repair. Bone. 2006;38(4):521–529. doi: 10.1016/j.bone.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Rutkovskiy A., Stenslokken K.O., Vaage I.J. Osteoblast differentiation at a glance. Med. Sci. Monit. Basic Res. 2016;22:95–106. doi: 10.12659/msmbr.901142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M., Hatzoglou M. The many virtues of tRNA-derived stress-induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J. Biol. Chem. 2015;290(50):29761–29768. doi: 10.1074/jbc.R115.694661. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ko E.-A., Sanders K.M., Chen Q., Zhou T. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genomics Proteomics Bioinformatics. 2018;16(2):144–151. doi: 10.1016/j.gpb.2018.04.004. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street J., Bao M., deGuzman L., Bunting S., Peale F.V., Jr., Ferrara N. Vascular endothelial growth factor stimulates bone repair by promoting angiogenesis and bone turnover. Proc. Natl. Acad. Sci. U. S. A. 2002;99(15):9656–9661. doi: 10.1073/pnas.152324099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebi F., Ghorbani S., Chan W.F., Boghozian R., Masoumi F., Ghasemi S. MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J. Neuroinflammation. 2017;14(1):55. doi: 10.1186/s12974-017-0832-7. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasca A., Astleford K., Blixt N.C., Jensen E.D., Gopalakrishnan R., Mansky K.C. SMAD1/5 signaling in osteoclasts regulates bone formation via coupling factors. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0203404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraju K., Neerukonda S.N., Katneni U., Buehler P.W. Extracellular vesicles from red blood cells and their evolving roles in health, coagulopathy and therapy. Int. J. Mol. Sci. 2020;22(1) doi: 10.3390/ijms22010153. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Zheng F., Li Z., Wang H., Yuan H., Zhang X. miR-148a-3p regulates adipocyte and osteoblast differentiation by targeting lysine-specific demethylase 6b. Gene. 2017;627:32–39. doi: 10.1016/j.gene.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Tsiridis E., Upadhyay N., Giannoudis P. Molecular aspects of fracture healing: which are the important molecules? Injury. 2007;38(Suppl. 1):S11–S25. doi: 10.1016/j.injury.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M.K., Clark I. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Wang W., Yeung K.W.K. Bone grafts and biomaterials substitutes for bone defect repair: a review. Bioact Mater. 2017;2(4):224–247. doi: 10.1016/j.bioactmat.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19(1):93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- Xu P., Palmer L.E., Lechauve C., Zhao G., Yao Y., Luan J. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood. 2019;133(23):2518–2528. doi: 10.1182/blood.2018854604. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Li M., Feng X., Zhu E., Wang B. miR-142a-5p promoted osteoblast differentiation via targeting nuclear factor IA. J. Cell. Physiol. 2021;236(3):1810–1821. doi: 10.1002/jcp.29963. Mar. [DOI] [PubMed] [Google Scholar]
- Zhang W., Cheng P., Hu W., Yin W., Guo F., Chen A. Downregulated microRNA-340-5p promotes proliferation and inhibits apoptosis of chondrocytes in osteoarthritis mice through inhibiting the extracellular signal-regulated kinase signaling pathway by negatively targeting the FMOD gene. J. Cell. Physiol. 2018;234(1):927–939. doi: 10.1002/jcp.26921. Jan. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation studies of the data.
Validation studies of the data; miRNA normalization genes.
TiRNA and miRNA normalized full data.
RNASeq full data and GO data.
Integrative miRNA-mRNA expression analysis