Abstract
Converging clinical and preclinical evidence demonstrates that depressive phenotypes are associated with synaptic dysfunction and dendritic simplification in cortico-limbic glutamatergic areas. On the other hand, the rapid antidepressant effect of acute ketamine is consistently reported to occur together with the rescue of dendritic atrophy and reduction of spine number induced by chronic stress in the hippocampus and prefrontal cortex of animal models of depression. Nevertheless, the molecular mechanisms underlying these morphological alterations remain largely unknown.
Here, we found that miR-9-5p levels were selectively reduced in the hippocampus of rats vulnerable to Chronic Mild Stress (CMS), while acute subanesthetic ketamine restored its levels to basal condition in just 24h; miR-9-5p expression inversely correlated with the anhedonic phenotype. A decrease of miR-9-5p was reproduced in an in vitro model of stress, based on primary hippocampal neurons incubated with the stress hormone corticosterone. In both CMS animals and primary neurons, decreased miR-9-5p levels were associated with dendritic simplification, while treatment with ketamine completely rescued the changes.
In vitro modulation of miR-9-5p expression showed a direct role of miR-9-5p in regulating dendritic length and spine density in mature primary hippocampal neurons. Among the putative target genes tested, Rest and Sirt1 were validated as biological targets in primary neuronal cultures. Moreover, in line with miR-9-5p changes, REST protein expression levels were remarkably increased in both CMS vulnerable animals and corticosterone-treated neurons, while ketamine completely abolished this alteration. Finally, the shortening of dendritic length in corticosterone-treated neurons was shown to be partly rescued by miR-9-5p overexpression and dependent on REST protein expression.
Overall, our data unveiled the functional role of miR-9-5p in the remodeling of dendritic arbor induced by stress/corticosterone in vulnerable animals and its rescue by acute antidepressant treatment with ketamine.
Keywords: miR-9-5p, Stress, CORT, Dendrite morphology, Ketamine, REST
Highlights
-
•
miR-9-5p is reduced in the hippocampus of stress-vulnerable rats.
-
•
miR-9-5p is reduced in primary neurons treated with corticosterone.
-
•
Acute ketamine restores stress-induced morphological changes and miR-9-5p levels.
-
•
miR-9-5p directly modulates dendritic length and spine number in primary neurons.
-
•
REST is a validated target of miR-9-5p.
1. Introduction
Although depression is a widespread debilitating illness with severe socio economic impact, the underlying pathophysiology remains poorly understood. A key role has been attributed to the interaction between genetic background and environmental events, with stress representing the major recognized environmental risk factor for the onset of depressive disorders (Keers and Uher, 2012; Krishnan and Nestler, 2008; Popoli et al., 2011). Indeed, while the response to stressful events is physiologically required to cope with the environment, it can either induce pro-adaptive outcomes in resilient subjects thus improving adaptation or, when dysregulated, exert maladaptive effects in vulnerable individuals (Han and Nestler, 2017; McEwen et al., 2015; Price and Duman, 2020).
Clinical studies on depressed subjects and preclinical models of depression converged in reporting a significant association between depressive phenotype and neuroplasticity alterations in glutamatergic cortico-limbic brain regions, including the hippocampus (HPC) and the prefrontal cortex (PFC) (Duman et al., 2019; Duman and Li, 2012; Pittenger and Duman, 2008; Sanacora et al., 2012). Post-mortem and fMRI studies on depressed patients showed volume reductions and connectivity impairments in both HPC and PFC, which significantly correlated with illness duration (Drevets et al., 2008; McKinnon et al., 2009; O'Connor and Agius, 2015; Sheline et al., 2019). Concomitantly, in stress-based animal models of depression, reduced dendritic length and number of spines were observed in the same brain areas altered in depressed subjects, highlighting a role for stress in causing neuronal atrophy and decreased synaptic function (Krishnan and Nestler, 2008; McEwen et al., 2016; Qiao et al., 2016; Sanacora et al., 2012). On the other hand, antidepressant treatment has been consistently reported to block or reverse dendritic simplification associated with depressive-like phenotype, thus supporting the hypothesis that atrophy and loss of neurons are contributing factors to depression and other stress-related psychiatric disorders (Duman and Li, 2012; Sanacora et al., 2012). Accordingly, the rapid antidepressant effect of acute sub-anesthetic ketamine was consistently demonstrated to restore both dendritic atrophy induced by chronic stress in the HPC and PFC of animal models and hippocampal subfield volume alterations in depressed patients (Kavalali and Monteggia, 2012; Li et al., 2011; Moda-Sava et al., 2019; Tornese et al., 2019; Zhou et al., 2020).
MicroRNAs (miRNAs) have emerged as key modulators of complex patterns of gene/protein expression changes in the brain, where they have a crucial role in the regulation of neuroplasticity, neurogenesis, and neuronal differentiation (Mingardi et al., 2018; Rajman and Schratt, 2017). Although much remains to be uncovered regarding their mechanism of action, recent studies showed that miRNAs are involved in the pathophysiology of mood disorders and in the action of psychotropic drugs (Dwivedi, 2016; Fiori et al., 2018; Mouillet-Richard et al., 2012; O'Connor et al., 2016). The peculiar ability of miRNAs to fine-tune the expression of hundreds of genes simultaneously makes them potential candidates as fast regulators of cellular excitability and morphology. However, to date, only few studies analyzed the involvement of miRNAs in the mechanisms underlying the rapid antidepressant effect of ketamine (O'Connor et al., 2013; Wan et al., 2018; Yang et al., 2014). In particular, the possible involvement of miRNAs in the fast dendritic remodeling of cortico-limbic pyramidal neurons induced by ketamine has never been assessed before.
In the present work, we aimed at unveiling a putative role of miRNAs in the morphological changes of hippocampal pyramidal neurons induced by Chronic Mild Stress (CMS) and ketamine in stress vulnerable adult male rats. We chose to focus our analysis on the HPC because we have recently shown that CMS induces in this brain area, selectively in animals vulnerable to stress (and not resilient ones), impairments of glutamate/GABA presynaptic release, Brain-Derived Neurotrophic Factor (BDNF) mRNA trafficking in dendrites and dendritic shortening, while acute ketamine largely restores cellular/molecular maladaptive changes in only 24h. In the present study, we selected 10 miRNAs, based on their well-known role in the modulation of synaptic plasticity and neuronal morphology, and measured their expression levels in the HPC of rats subjected to CMS and acute ketamine. Selected miRNAs were: miR-9-5p (Dajas-bailador et al., 2012; Giusti et al., 2014; Gu et al., 2018; Xue et al., 2016), miR-16-5p (Burak et al., 2018), miR-29a-5p (Volpicelli et al., 2019; Zou et al., 2015), miR-34a-5p (Andolina et al., 2016; Hu et al., 2020; Yi et al., 2020), miR-124-5p (Gu et al., 2016; Li and Ling, 2017; Xue et al., 2016), miR-132-5p (Jasińska et al., 2016; Pathania et al., 2012), miR-134-5p (Fan et al., 2018; Jimenez-Mateos et al., 2015; Schratt et al., 2006), miR-135a-5p (Hu et al., 2014; Issler et al., 2014; Wang et al., 2020), miR-137-5p (Smrt et al., 2010; Yan et al., 2019), miR-210-5p (Hu et al., 2016; Watts et al., 2021).
Among the analyzed miRNAs, using an integrated in vivo and in vitro approach, we found that miR-9-5p directly controlled neuronal morphology and that the simplification of pyramidal neurons dendritic arbor induced by chronic stress and corticosterone in the hippocampus was associated with reduced miR-9-5p levels. Importantly, we also observed that the rescue of the stress-dependent dendritic shortening induced by acute ketamine involved an increase of miR-9-5p levels.
2. Materials and methods
2.1. Animals
Experiments were performed in accordance with the European Communities Council Directive 2010/63/UE and approved by the Italian legislation on animal experimentation (Decreto Legislativo 26/2014, authorization N 308/2015-PR). A total of 72 Sprague-Dawley male rats weighing 150–175 g (Charles River, Calco, Italy) were housed two per cage at 20–22 °C, 12 h light/dark cycle (light on 7:00 a.m. off 7:00 p.m.), water and food ad libitum, except when required for Chronic Mild Stress (CMS).
2.2. Chronic mild stress and sucrose preference test
After 7 days of acclimatization, CMS was performed as previously reported (Elhussiny et al., 2021; Tornese et al., 2019). Sucrose preference test was used to assess anhedonic phenotype and to divide stressed animals in CMS resilient (CMS-R) and vulnerable (CMS-V), applying a cut-off of preference at 55% (Elhussiny et al., 2021; Tornese et al., 2019). A detailed description of CMS protocol and sucrose preference test can be found in Supplementary Materials.
2.3. Drug treatment, sacrifice, and sampling
After 5 weeks of CMS, half of the CMS-V rats were intraperitoneally injected with a single sub-anesthetic dose (10 mg/kg) of racemic ketamine (MSD Animal Health, Milan, Italy), while the remaining animals received physiological solution (0.9% w/v NaCl) (Elhussiny et al., 2021; Tornese et al., 2019). All the animals were sacrificed by beheading 24 h after ketamine injection. 10 animals/group were randomly selected for RNA extraction/quantitative Real-Time PCR (qPCR) and in-situ hybridization studies: brains were rapidly extracted, and right/left hemispheres were randomly assigned to HPC dissection (qPCR) or fixation in 4% paraformaldehyde (PFA) solution (in-situ hybridization). The HPC of the remaining 8 animals/group were rapidly dissected on ice and assigned to Western blotting experiments. HPC were snap-frozen and kept at −80 °C.
2.4. Primary hippocampal cultures
Primary hippocampal cultures were prepared as described in (Bonini et al., 2015; Musazzi et al., 2014). Briefly, hippocampi at embryonic day 16.5 were dissociated mechanically in 1x Hank's Balanced Salt Solution (HBSS) and neurons were resuspended in Neurobasal™ medium supplemented with B27 (Gibco™, Thermo Fisher Scientific) containing 30 U/ml Penicillin, 30 mg/ml Streptomycin (Sigma-Aldrich), 0.75 mM Glutamax (Gibco™, Thermo Fisher Scientific) and 0.75 mM L-Glutamine (Gibco™, Thermo Fisher Scientific). Half of the medium was replaced with an astrocyte-conditioned medium once a week for a maximum of 4 weeks.
2.5. HEK293T cell cultures
HEK293T cells were cultured as previously reported (Filippini et al., 2017) (see Supplementary Materials for more details).
2.6. RNA isolation, reverse transcription and qPCR
Total RNA was extracted from HPC and primary neuronal cultures using Tri-Reagent (Sigma-Aldrich, Milan, Italy) and Direct-zol RNA MiniPrep (Zymo Research, Freiburg, Germany), according to manufacturer's instructions (Tornese et al., 2019). Reverse transcription was carried out using miRCURY® LNA® RT kit (Exiqon) or Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) (ThermoFisher Scientific), respectively for miRNA or mRNA analysis. qPCR was performed using iTaq Universal SYBR Green supermix (Bio-Rad Laboratories). The full list of primers used for qPCR is reported in Supplementary Table 1 (primers for miRNAs, miRCURY® LNA® miRNA PCR assay, Exiqon) and Supplementary Table 2 (primers for mRNAs).
The relative expression of miRNAs and mRNAs was calculated using the comparative Ct (ΔΔCt) method and is expressed as Fold Change. The mean of SNORD68 and RNU1A1 was used as a control reference for miRNAs, while the geometric mean of GAPDH, ACTIN-β and HPRT was used as a control reference for mRNAs.
2.7. In-situ hybridization
Brain hemispheres were fixed in 4% PFA solution for 24 h and placed in 30% sucrose. Coronal slices (40 μm) were cut and stored in cryo-protectant sectioning buffer (30% ethylene glycol, 30% glycerol and 0.05 M phosphate buffer) at −20 °C. In situ hybridization was performed on free-floating sections as previously reported (La Via et al., 2013; Tornese et al., 2019) (see Supplementary Materials for more details).
2.8. DNA constructs
Vectors to overexpress and downregulate miR-9-5p were designed starting from TWEEN-Lenti vector (7970 bp), that contains the Green Fluorescent Protein (GFP) coding sequence under the control of hPGK promoter (Climent et al., 2015). An additional CMV promoter is present in the vector for gene expression studies.
To obtain the overexpression of miR-9-5p, oligonucleotides were designed specific to the sequence of miR-9-5p and annealed in vitro to produce a dsRNA. The CMV promoter was substituted with the Polymerase III specific promoter H1 using ClaI and XbaI (Fermentas, Thermo Fisher Scientific) to obtain Twl-PGKgfp-H1 (7595 bp, Twl-H1). After digestion with XhoI and XbaI, ds-miR-9-5p oligonucleotides were cloned in Twl-H1 using T4 DNA Ligase (Thermo Fisher Scientific). The vector obtained is Twl-PGKgfp-H1-miR-9-5p (7650 bp, Twl-H1-miR-9-5p).
The downregulation vector was generated by cloning a sequence containing 3 repeats of a complementary sequence to miR-9-5p interspersed with spacer nucleotides, acting as a sponge for endogenous miR-9-5p (ds-sponge-miR-9-5p) (Giusti et al., 2014). Red Fluorescent Protein (RFP) was cloned under the control of CMV promoter using XhoI and XbaI to obtain Twl-PGKgfp-CMVrfp (Twl-GFP/RFP). ds-sponge-miR-9-5p was inserted within the 3′UTR of GFP, digesting Twl-GFP/RFP with SalI. The vector obtained is Twl-PGKgfp-CMVrfp-sponge-miR-9-5p (8761 bp, Twl-sponge-miR-9). RFP expression was used as control for transfection, while GFP expression was dependent on the presence of endogenous miR-9-5p binding to the sponge. The empty vectors Twl-H1 and Twl-GFP/RFP were used as controls. The list of the oligonucleotide sequences used in these experiments is reported in Supplementary Table 3. Vectors to downregulate REST protein were based on Twl-H1: after digestion with XhoI and XbaI, two sequences complementary to REST 3′UTR were cloned in Twl-H1 using T4 DNA Ligase (shREST-1: GTGTAATCTACAATACCATTT for Twl-H1-shREST-1; shREST-2: GCGCTAAGAAGTTCTTTG for Twl-H1-shREST-2).
pRRLSIN.cPPT.PGK-GFP.WPRE (Addgene plasmid #12252: PGK-GFP) was used for morphological analysis. This plasmid allows the expression of enhanced GFP (EGFP) under the control of hPGK promoter.
2.9. Transfection of neuronal cultures
Neuronal cultures were transfected at DIV7 (PGK-GFP) or DIV11 (miRNA expression vectors, REST silencing vectors, miR-9-5p miRCURY LNA miRNA mimic 5′-UCUUUGGUUAUCUAGCUGUAUGA-3’; ID: YM00471434 and negative control 5′-TAACACGTCTATACGCCCA-3’; ID: YI00199006, Exiqon) using Magnetofection™ Technology (Oz Biosciences, Marseille, France), according to the manufacturer's protocol (La Via et al., 2013). Briefly, the vectors (PGK-GFP, miRNA expression vectors and REST silencing vectors) and miR-9-5p LNA mimic/negative control were incubated with NeuroMag magnetic beads in the ratio of 1 μg DNA:3.5 μl NeuroMag or 5–25 pmol LNA/RNA:3.5 μl NeuroMag, respectively, in 100 μl of Neurobasal free medium for 20 min, and added drop-by-drop to the cells. Then, the cells were incubated on a magnetic plate for 15 min at 37 °C. After 1 h, a complete change of medium was performed and cells were kept at 37 °C under a 5% CO2 humid atmosphere. 72 h post-transfection, neuronal cells were fixed in 4% PFA for 20 min at RT and mounted on coverslip using SlowFade™ Diamond Antifade Mountant (ThermoFisher, USA) for confocal imaging (miRNA expression vectors, REST silencing vectors), or mechanically harvested in 100 μL of RIPA for protein purification (miR-9-5p LNA miRNA mimic and negative control).
Neuronal cultures transfected with PGK-GFP were kept undisturbed except for the weekly change of medium until DIV17, when they were used for live imaging acquisition and drug treatments.
2.10. In vitro corticosterone and ketamine treatments
DIV14-17 hippocampal neurons were incubated with 200 nM corticosterone (CORT) (or 0.2% dimethyl sulfoxide, DMSO) (Sigma-Aldrich) twice daily (9.00 a.m. and 17.00 p.m.) for 3 consecutive days. Half of the medium was replaced every morning with an astrocyte-conditioned medium. Then, 1 μM ketamine (MSD Animal Health, Milan, Italy) (Cavalleri et al., 2018) was added to the cells (KET treatment group) for 4 h.
2.11. Confocal imaging analysis
Images of fluorescently labeled cells (DIV17 or DIV20) were captured using a confocal microscope LSM880 Upright (Carl Zeiss, Jena, Germany) with a 20× objective at a resolution of 3964 (x/y) pixel, and represent maximum intensity projections of 5 consecutive optical sections taken at an 8 μm interval (for miRNAs expression vectors and REST silencing vectors experiments) or 8 optical sections at 0.96 μm interval (for PGK-GFP vector experiments). Total dendritic length and number of branches were measured using Simple Neurite Tracer Fiji Plugin (Schindelin et al., 2012). The number of spines was manually counted, and spine density was expressed as number of spines/10 μm of dendritic segment (spines of 3 secondary dendrites from a minimum of 15 cells were analyzed).
In PGK-GFP vector experiments, images were acquired at 2 timepoints: at DIV17 (T0) in living cells kept in a humid chamber at 37 °C and 5% CO2, and at DIV21 after CORT and ketamine treatments (T1). The difference between T1 and T0 measurements within the same cell was calculated and expressed as Δ dendritic length T1-T0 (μm). A minimum of 10 cells were acquired for each condition.
2.12. Bioinformatic analysis of miR-9-5p targets and luciferase assay
TargetScan, mirDB and microT-CDS were used to perform target predictions for miR-9-5p (Chen and Wang, 2020; Lewis et al., 2005; Reczko et al., 2012). Given the large number of predicted targets, the following criteria were used to select the targets for reporter gene assays: (i) presence of at least one conserved 8mer, 7mer-m8, or 7mer-A1 site, (ii) reported importance for brain function and plasticity in literature.
For preliminary validation, sequences spanning 30 bp before and after the miR-9-5p binding site within the 3′ untranslated region (3′ UTR) of selected target genes were cloned into a pmirGLO Dual-Luciferase expression vector (Promega) (Rossi et al., 2014) (the primers used are listed in Supplementary Table 4). For final validation, longer 3’ UTR fragments (GSK-3β 839 bp, REST, 1825 bp, SIRT1 403 bp) were amplified (primers listed in Supplementary Table 4), cloned in pmirGLO vector (Promega) and validated by sequencing.
Dual Luciferase® Reporter assay (E1910, Promega) was used according to the manufacturer's protocol. HEK293T cells were plated at 1 x 104 cells/well on 96-well plates in 1x DMEM the day before transfection and co-transfected with pmirGLO construct vectors and miR-9-5p mimics (Dharmacon) or negative control (Dharmacon) using DharmaFECT Duo (0.12 μL/well; Dharmacon). 48h post-transfection, Firefly luciferase activity was measured by reading luminescence (0.1 s) with an EnSight™ Multimode Plate Reader (PerkinElmer). Firefly luciferase signal was normalized on Renilla signal and expressed as RLU. All transfections were performed in triplicate and the experiment was repeated at least 3 times.
2.13. Western blotting
BCA protein concentration assay (Sigma–Aldrich, St. Louis, MO, USA) was used for protein quantitation. Western blotting was performed as previously described (Bonini et al., 2016; Musazzi et al., 2017). Primary antibodies used were: GSK-3β (1:1000, cod. #05–412, Abcam), REST (1:500, cod. ab202962, Abcam), SIRT1 (1:500, cod. ab189494, Abcam). Antibodies against GAPDH (1:8000, cod. Mab374, Millipore) or α-TUBULIN (1:20000, cod. ab7291, Abcam) were used as internal controls. Secondary antibodies used were: anti-mouse (IRDye 926–32210; IRDye 926–68020, LI-COR) or anti-rabbit (IRDye 926–32211, LI-COR), both diluted 1:2000 in TBS-T (see Supplementary Table 5 for details). Signals were detected using Odyssey infrared imaging system (LI-COR Biosciences) and quantified with Image Studio software (version 5.2, LI-COR Biosciences) (Bonini et al., 2016; Musazzi et al., 2017).
2.14. Statistical analysis
Data were shown as mean ± standard error of mean (SEM). Statistical analysis of the data was performed using GraphPad Prism 8 (GraphPad Software Inc., USA). Unpaired Student's t-test followed by Welch's correction was applied in Luciferase assay experiments and in the analysis of morphological changes after miR-9-5p modulation. Correlation between sucrose preference and miR-9-5p expression in CMS rats was analyzed by Pearson's correlation. For all the other experiments, one-way ANOVA followed by Tukey's post-hoc test or Newman-Keuls Multiple Comparison was used. Statistical significance was assumed at p < 0.05.
3. Results
3.1. Acute ketamine rescued the reduction of miR-9-5p in the hippocampus of chronic mild stress-vulnerable animals; miR-9-5p levels inversely correlated with the depressive-like phenotype
In a previous study, we showed that acute ketamine (10 mg/kg) completely rescued in just 24h anhedonic behavior in the sucrose preference test, as well as dendritic retraction and simplification of hippocampal pyramidal neurons, induced by 5 weeks of CMS selectively in stress-vulnerable rats (CMS-V), compared to resilient (CMS-R) and control animals (Tornese et al., 2019). To identify whether these morphological changes were accompanied by alterations in selected miRNAs, we measured by qPCR the expression levels of the 10 selected miRNAs in the HPC of rats subjected to 5 weeks of CMS and acute ketamine.
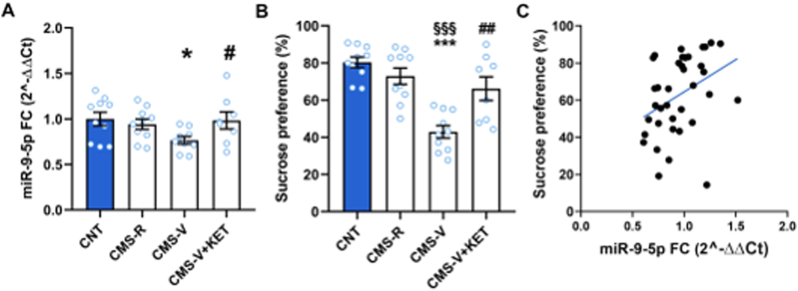
Significant differences were revealed among the experimental groups in the expression of 6 miRNAs (Table 1). Overall, we observed that the levels of several miRNAs were significantly reduced as a result of stress exposure. In more details, miR-134-5p and 135a-5p were significantly decreased in all stressed rats, including CMS-R, CMS-V and CMS-V treated with ketamine (see Table 1 for detailed statistics). However, miR-135a-5p levels were significantly lower in CMS-V compared to CMS-R, and acute ketamine restored the expression to CMS-R levels. Differently, miR-132-5p was significantly reduced only in CMS-R, miR-210-5p selectively in CMS-V, while miR-124-5p was reduced in all CMS-V animals, including those treated with ketamine. Unlike all the other miRNAs, miR-9-5p was selectively reduced only in CMS-V (and not in CMS-R), while acute ketamine completely rescued this reduction to control levels (Fig. 1A). Intriguingly, changes in miR-9-5p expression mirrored alterations of sucrose preference (one-way ANOVA, F3,34 = 15.64, p < 0.001, Tukey's post-hoc test: CMS-V vs CNT p < 0.001, CMS-V vs CMS-R p < 0.001, CMS-V + KET vs CMS-V p < 0.001; Fig. 1B), and significantly correlated with the behavioral phenotype across animals (Pearson r, r = 0.3677, p < 0.05; Fig. 1C). Thus, lower miR-9-5p levels correlated with worse anhedonic phenotype, while higher miR-9-5p expression was associated with higher sucrose preference, suggesting an involvement of miR-9-5p in the regulation of depressive-like behavior.
Table 1.
miRNAs expression changes in the HPC of rats induced by CMS and ketamine.
| CNT | CMS-R | CMS-V | CMS-V + KET | ANOVA | |
|---|---|---|---|---|---|
| miR-9-5p | 1 ± 0.22 | 0.94 ± 0.17 | 0.76 ± 0.12* | 1.03 ± 0.24# | F3, 31 = 3,676; p<0.05 |
| miR-16-5p | 1 ± 0.20 | 0.83 ± 0.16 | 0.83 ± 0.20 | 0.80 ± 0.08 | F3, 31 = 2,183; p = 0.11 |
| miR-29a-5p | 1 ± 0.18 | 0.96 ± 0.21 | 0.98 ± 0.15 | 0.97 ± 0.17 | F3, 33 = 0,066; p = 0.98 |
| miR-34a-5p | 1 ± 0.22 | 0.85 ± 0.28 | 0.68 ± 0.18 | 0.77 ± 0.18 | F3, 30 = 2,783; p = 0.06 |
| miR-124-5p | 1 ± 0.17 | 0.88 ± 0.11 | 0.81 ± 0.13* | 0.79 ± 0.16* | F3, 30 = 3,669; p<0.05 |
| miR-132-5p | 1 ± 0.09 | 0.80 ± 0.13** | 0.85 ± 0.12 | 0.89 ± 0.12 | F3, 29 = 4,241; p<0.01 |
| miR-134-5p | 1 ± 0.29 | 0.66 ± 0.07** | 0.79 ± 0.13* | 0.74 ± 0.19* | F3, 30 = 5,153; p<0.01 |
| miR-135a-5p | 1 ± 0.12 | 0.77 ± 0.10** | 0.62 ± 0.10*** | 0.76 ± 0.15** | F3, 24 = 10,96; p<0.001 |
| miR-137-5p | 1 ± 0.23 | 0.85 ± 0.10 | 0.83 ± 0.11 | 0.80 ± 0.14 | F3, 31 = 2,674; p = 0.06 |
| miR-210-5p | 1 ± 0.18 | 0.82 ± 0.08 | 0.79 ± 0.12* | 0.83 ± 0.21 | F3, 32 = 3,218; p<0.05 |
Expression of selected miRNAs in the HPC of rats after CMS and acute ketamine. n = 6–10 (CNT), n = 7–10 (CMS-R), n = 7–10 (CMS-V), n = 7–10 (CMS-V + KET). Newman-Keuls Multiple Comparison Test: *p < 0.05, **p < 0.01, ***p < 0.001 vs CNT; #p < 0.05 vs CMS-V. Data are reported as means ± S.E.M.
Fig. 1.
miR-9-5p expression and sucrose preference measure in CMS rats. (A) qPCR analysis of miR-9-5p expression in the HPC. Tukey's post-hoc test: *p < 0.05 vs CNT; #p < 0.05 vs CMS-V; n = 7–10. Data are reported as means ± S.E.M. (B) Sucrose preference test of CNT and CMS rats at Day +35 of CMS, 24 h after KET/VEH treatment n = CNT 10; CMS-R 10; CMS-V 10; CMS-V + KET 8. Tukey's post-hoc test: ***p < 0.001 vs CNT, ##p < 0.01 vs CMS-V, §§§p < 0.001 vs CMS-R. Data are reported as means ± S.E.M. (C) Correlation between miR-9-5p HPC expression and the sucrose preference in CMS rats. Pearson's r, r = 0.3677, p < 0.05.
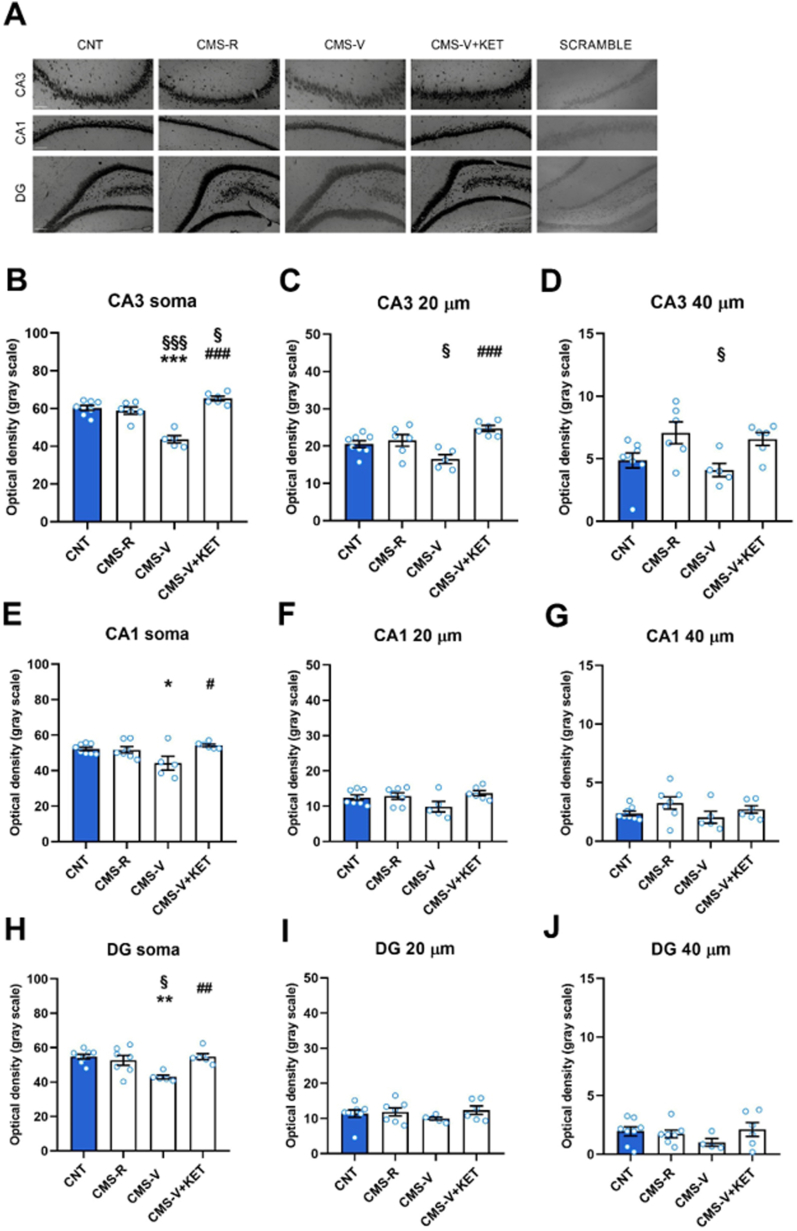
We then used in situ hybridization to further investigate miR-9-5p differential expression in the HPC within CA3, CA1 and DG sub-regions (Fig. 2). Significant differences among groups were found in the somatic expression of miR-9-5p in CA3 (one-way ANOVA, F3,21 = 30.78, p < 0.001), CA1 (one-way ANOVA, F3,22 = 4.47, p < 0.05), and DG (one-way ANOVA, F3,22 = 6.868, p < 0.01). Accordingly, miR-9-5p was selectively reduced in CA3, CA1 and DG of CMS-V animals compared to controls and CMS-R, and ketamine completely rescued the reduction (Tukey's post-hoc test; CA3: CMS-V vs CNT p < 0.001, CMS-V vs CMS-R p < 0.001, CMS-V + KET vs CMS-V p < 0.001, CMS-V + KET vs CMS-R p < 0.05; CA1: CMS-V vs CNT p < 0.05, CMS-V + KET vs CMS-V p < 0.05; DG: CMS-V vs CNT p < 0.01, CMS-V vs CMS-R p < 0.05, CMS-V + KET vs CMS-V p < 0.01). Of note, in CA3 where, specifically, in our previous study we found a reduction of pyramidal neuron apical dendritic length and complexity (Tornese et al., 2019), significant differences were revealed in miR-9-5p expression also along proximal dendrites (20 μm from soma, one-way ANOVA, F3,21 = 7.720, p < 0.01). Multiple comparisons highlighted a significant reduction of miR-9-5p signal in CMS-V compared to CMS-R (not significant compared to control), while ketamine recovered this change (Tukey's post-hoc test; CMS-V vs CNT p = 0.0969, CMS-V vs CMS-R p < 0.05, CMS-V + KET vs CMS-V p < 0.001). miR-9-5p signal in the CA3 of CMS-V rats was significantly weaker compared to CMS-R also at 40 μm from soma (Tukey's post-hoc test; CMS-V vs CMS-R p < 0.05).
Fig. 2.
miR-9-5p expression changes induced by CMS and ketamine in the sub-regions of the HPC of rats. (A) Representative images of miR-9-5p in situ hybridization signal in CA3, CA1 and DG regions of HPC in CNT, CMS-V, CMS-R and CMS-V + KET rats. Scramble probe was used as a negative control. Scale bar 100 μm. (B–D) Expression of miR-9-5p measured as optical density in the soma, proximal (20 μm) and distal (40 μm) dendrites of neurons in CA3. (E–G) Expression of miR-9-5p measured as optical density in the soma, proximal (20 μm) and distal (40 μm) dendrites of neurons in CA1. (H–J) Expression of miR-9-5p measured as optical density in the soma, proximal (20 μm) and distal (40 μm) dendrites of neurons in DG. n = 3–4/group, 2-3 slices/rat. Tukey's post-hoc test: *p < 0.05, **p < 0.01, ***p < 0.001 vs CNT; #p < 0.05, ##p < 0.01, ###p < 0.001 vs CMS-V; §p < 0.05, §§§p < 0.001 vs CMS-R. Data are reported as means ± S.E.M.
3.2. Acute ketamine rescued dendritic retraction and miR-9-5p reduction induced by repeated in vitro corticosterone treatment of primary hippocampal neurons
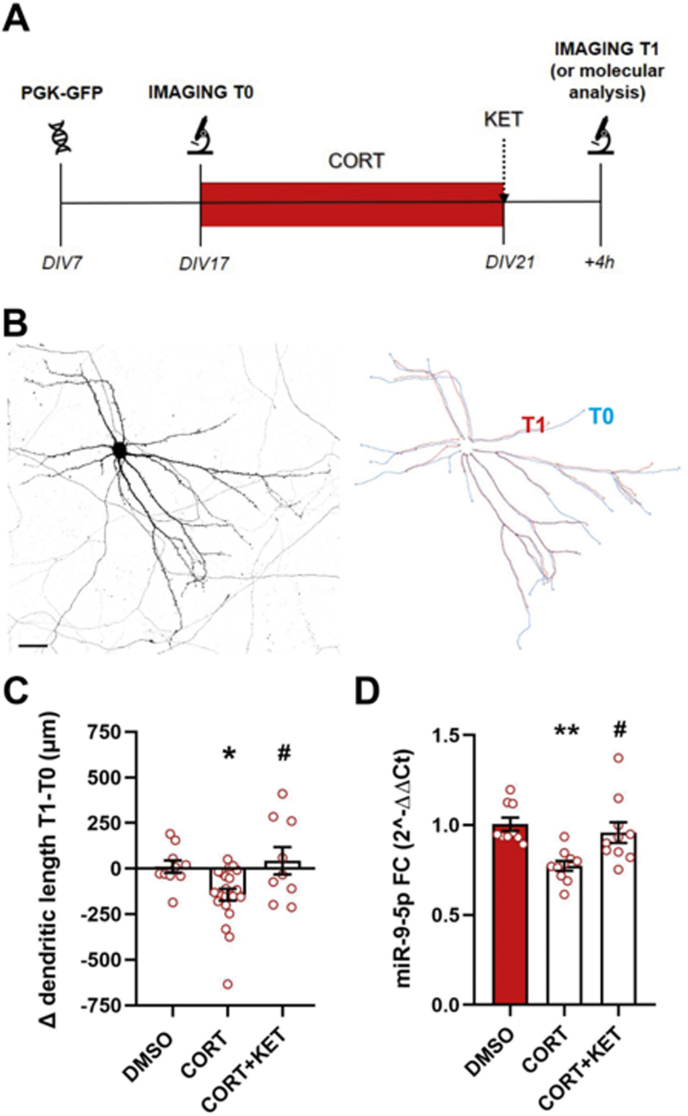
We have previously reported that 5 weeks of CMS induced a significant increase in the serum levels of the stress hormone corticosterone selectively in CMS-V rats (independently of ketamine treatment), but not in CMS-R (Tornese et al., 2019). Thus, to model in a simplified system the neuronal effects of corticosterone (and ketamine) in vulnerable animals and to test their direct action on dendritic remodeling, we designed an in vitro model based on repeated doses of corticosterone (plus acute ketamine) in primary hippocampal neurons (Fig. 3A). Changes of pyramidal neuron total dendritic length were measured before and after corticosterone and ketamine treatments (Fig. 3B). As reported in Fig. 3C, both conditions affected dendritic morphology of primary hippocampal pyramidal neurons (one-way ANOVA, F2,38 = 5.595, p < 0.01). Specifically, repeated corticosterone treatment significantly reduced total dendritic length of pyramidal neurons (Tukey's post-hoc test; CORT vs DMSO p < 0.05), while the incubation of corticosterone-treated cells with ketamine for 4 h fully recovered dendritic retraction to control levels (Tukey's post-hoc test; CORT + KET vs CORT p < 0.05). We then asked whether morphological changes were accompanied by alterations of miR-9-5p expression levels, and qPCR analysis revealed its significant reduction after repeated treatment with corticosterone, that was completely rescued by ketamine (one-way ANOVA, F3,34 = 5.997, p < 0.01; Tukey's post-hoc test; CORT vs DMSO p < 0.01, CORT + KET vs CORT p < 0.05) (Fig. 3D).
Fig. 3.
Analysis of dendritic morphology and miR-9-5p expression in primary hippocampal cultures treated with corticosterone and ketamine. (A) Timing of primary hippocampal cultures in vitro treatment with CORT and KET. (B) Representative image of a hippocampal neuron used for morphological study and reconstruction of the dendritic tree performed using plug-in Fiji at T0 (blue line) and T1 (red line). (C) The difference between T1 and T0 measurements within the same cell is reported as Δ dendritic length T1-T0 (μm), for vehicle, CORT and CORT plus KET treatment. Tukey's post-hoc test: *p < 0.05 vs DMSO, #p < 0.05 vs CORT; n = 10 (DMSO), n = 22 (CORT), n = 10 (CORT + KET). (D) miR-9-5p expression in primary hippocampal neurons treated with CORT and KET. Tukey's post-hoc test: **p < 0.01 vs DMSO, #p < 0.05 vs CORT; n = 9–10. Scale bar 50 μm.. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. miR-9-5p controls dendritic morphology and spine density in primary hippocampal neurons
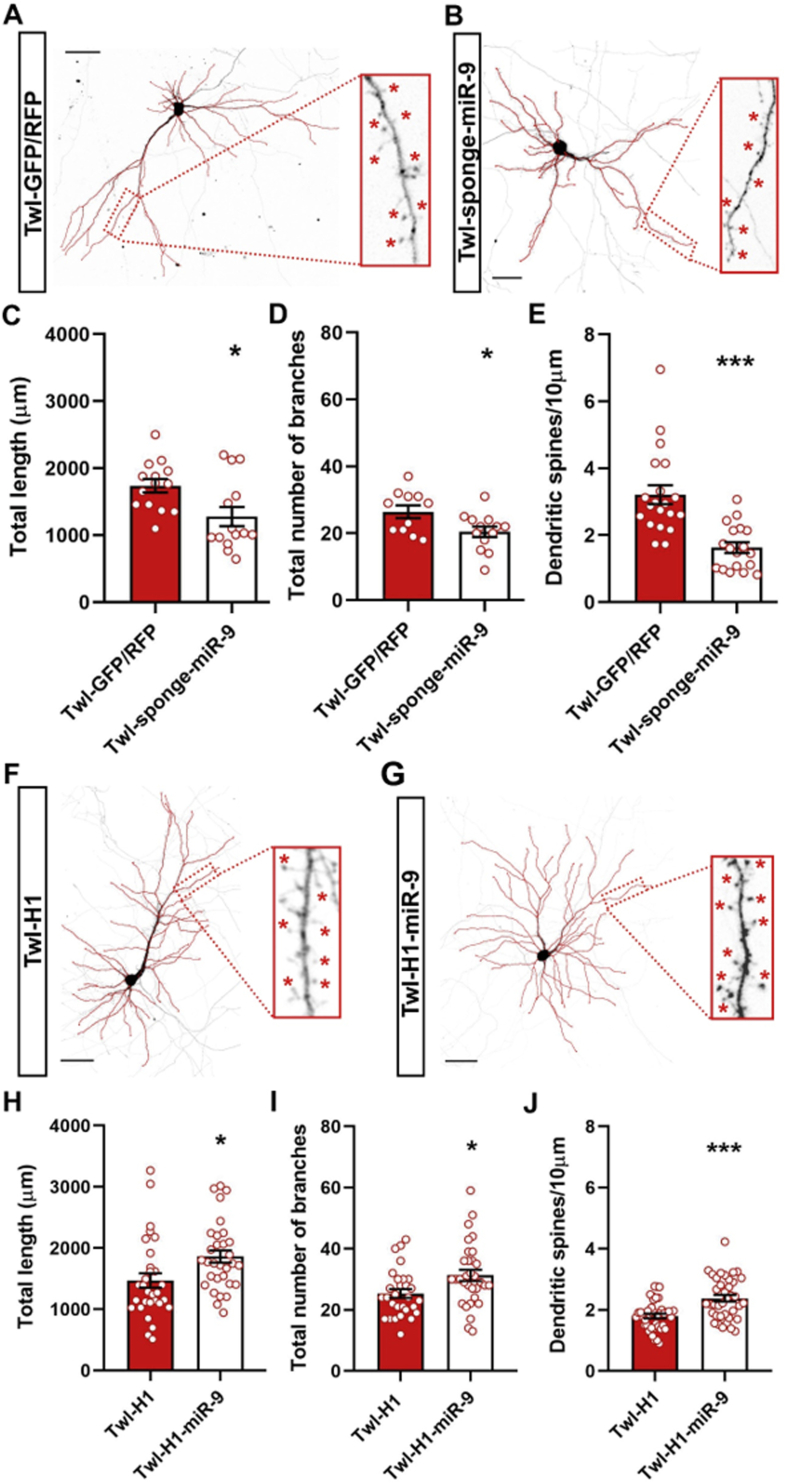
miR-9-5p controls the number of spines and the length and branching of dendrites (Giusti et al., 2014; Xue et al., 2016). Here, having shown parallel changes in dendritic remodeling and miR-9-5p expression, we asked whether miR-9-5p regulated dendritic morphology in mature primary hippocampal cultures. To this end, vectors specifically designed to overexpress or downregulate miR-9-5p were first tested in HEK293T cells (Supplementary Fig. 1 and Supplementary Materials), and then transfected in DIV11 hippocampal neurons. Morphological analysis of pyramidal neurons 72 h post-transfection (DIV14) showed that miR-9-5p downregulation decreased total dendritic length (Student's t-test; p < 0.05), total number of branches (p < 0.05) and dendritic spine density (p < 0.001) (Fig. 4C–E). On the other end, over-expression of miR-9-5p levels produced a significant increase in total dendritic length (Student's t-test; p < 0.05), total number of branches (p < 0.05) and spine density (p < 0.001) of hippocampal neurons (Fig. 4H–J).
Fig. 4.
miR-9-5p modulation in primary neurons changes dendritic morphology and spine density. (A–B) Representative reconstruction using plug-in Fiji of the dendritic tree and spines of primary hippocampal neurons transfected with Twl-GFP/RFP and Twl-sponge-miR-9-5p, respectively. Scale bar 50 μm. Morphological analysis of primary hippocampal neurons after 72 h of miR-9-5p downregulation: (C) total dendritic length; n = 15, (D) total number of branches; n = 15, (E) dendritic spine density; n = 20. Unpaired Student's t-test with Welch's correction: *p < 0.05, ***p < 0.001 vs Twl-GFP/RFP. (F–G) Representative reconstruction using plug-in Fiji of the dendritic tree and spines of primary hippocampal neurons transfected with Twl-H1 and Twl-H1-miR-9-5p, respectively. Scale bar 50 μm. Morphological analysis of primary hippocampal neurons after 72 h of miR-9-5p overexpression: (H) total dendritic length; n = 30, (I) total number of branches; n = 30, (J) dendritic spine density; n = 40. Unpaired Student's t-test with Welch's correction: *p < 0.05, ***p < 0.001 vs Twl-H1. Dendritic length, number of branches and spine density were measured using ImageJ or the plug-in Fiji.
3.4. Bioinformatic analysis of miR-9-5p target genes and in vitro validation of selected targets
The bioinformatic prediction tools TargetScan, mirDB and microT-CDS were used to identify miR-9-5p putative target genes (Supplementary information). Among the most significant genes, we selected those that presented at least one conserved 8mer, 7mer-m8, or 7mer-A1 binding site for miR-9-5p and with reported relevance in the control of brain function and plasticity (selected target genes with relative previous evidence are shown in Table 2).
Table 2.
Bioinformatic selection of miR-9-5p predicted targets.
| GENE | SYMBOL | REPRESENTATIVE TRANSCRIPT | PREDICTION DATABASES | BINDING POSITION ON THE 3′UTR | LITERATURE EVIDENCE |
|---|---|---|---|---|---|
| Dopamine Receptor D2 | DRD2 | ENSMUST00000075764.6 | A | 285–292 |
-Stress response: Żurawek et al., (2013) -Depression and antidepressant response: He et al., 2019, Pecina et al., 2017, Wang et al., 2012 |
| ELAV like RNA binding protein 1 | ELAVL1 | ENSMUST00000098950.4 | A, B, C | 788–795 |
-Stress response: He et al. (2019) -Depression and antidepressant response: He et al. (2019) |
| Glycogen Synthase Kinase 3, β | GSK3β | ENSMUST00000023507.7 | A | 3565–3571 |
-Stress response: Aceto et al., (2020) -Depression and antidepressant response: Duman and Aghajanian, 2014, Liu et al., 2013, Zarate and Machado-Vieira, 2016 |
| Neurotrophic Receptor Tyrosine Kinase 3 | NTRK3 | ENSMUST00000039431.8 | A | 1741–1749 |
-Stress response: Farhang et al., 2014, Xu et al., 2015 -Depression and antidepressant response: Tamasi et al. (2014) |
| RE-1 Silencing Transcription Factor | REST | ENSMUST00000080359.6 | A, C | 1749-1755; 3207–3213 |
-Stress response: Mampay et al., (2019); Soga et al., (2021) -Depression and antidepressant response: Soga et al., (2021) -Morphology: Giusti et al., (2014) |
| Sirtuin 1 | SIRT1 | ENSMUST00000120239.2 | A, B, C | 335–342 |
-Stress response: Kim et al., 2016, Liu et al., 2019, Shen et al., 2019 -Depression and antidepressant response: Abe-Higuchi et al., (2016); Nan et al. (2020) -Morphology: Codocedo et al. (2012); Michan et al., (2010) |
| Sortilin 1 | SORT1 | ENSMUST00000102632.5 | A, B, C | 3274–3280 |
-Stress response: Bai et al., 2016, Ruan et al., 2016 -Depression and antidepressant response: Yang et al. (2020) |
Putative targets of miR-9-5p were bioinformatically identified using TargetScan (A), MirDB (B) and microT-CDS (C). Genes relevant to our study were selected based on their recognized role in synaptic plasticity, response to stress, depression, and antidepressant drugs response.
Luciferase assay in HEK293T cells was used to test the functional interaction between miR-9-5p and its binding sites within mRNA 3′UTRs of each selected putative target gene, cloned in the pmirGLO Dual-Luciferase expression vector. Student's t-test revealed that miR-9-5p mimics significantly decreased the firefly luciferase activity only in cells transfected with plasmids including miR-9-5p target sites within GSK-3β (p < 0.01), REST (p < 0.05) and SIRT1 (p < 0.05) 3′UTRs (Fig. 5). No changes were observed for the other genes.
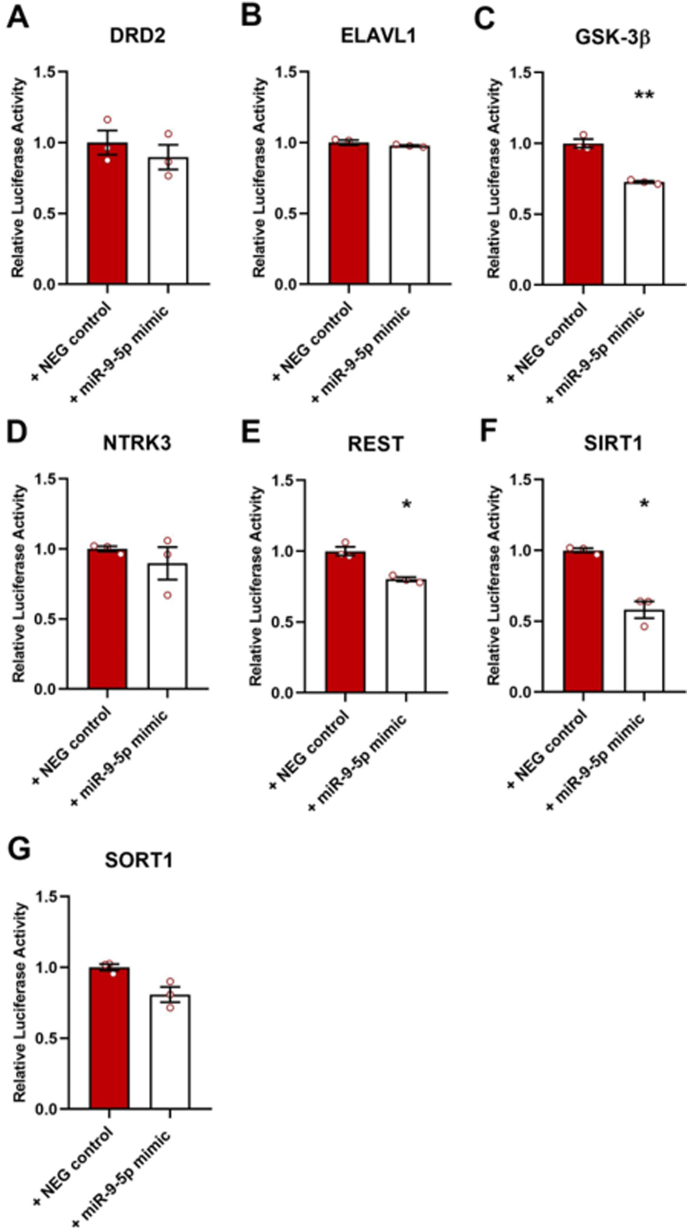
Fig. 5.
Analysis of miR-9-5p interaction with predicted target genes using Luciferase assay. Relative Luciferase Activity was measured in HEK293T cells co-transfected with specific dual-reporter vectors containing an approximately 70 bp-long sequence of the 3′UTR of (A) DRD2, (B) ELAVL1, (C) GSK-3β, (D) NTRK3, (E) REST, (F) SIRT1, (G) SORT1 and with specific mimics for miR-9-5p or negative control. Unpaired Student's t-test with Welch's correction: *p < 0.05, **p < 0.01 vs NEG control. n = 3; all experiments were repeated 3 times.
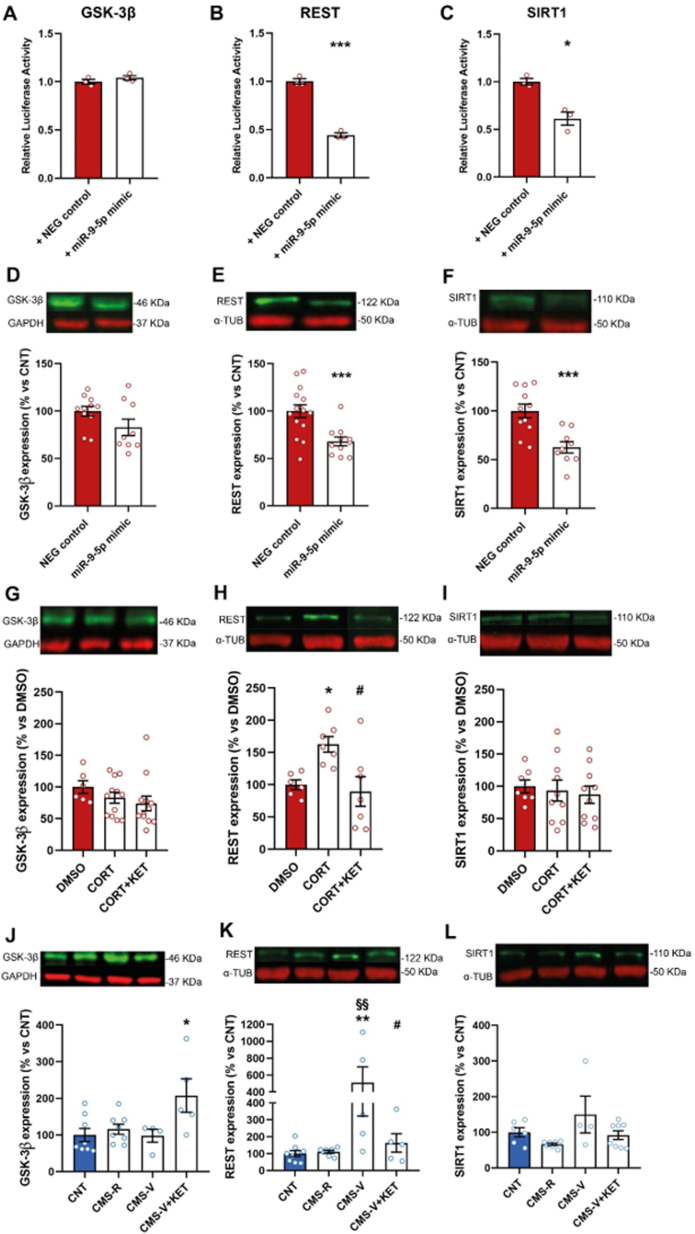
To further validate the interaction of miR-9-5p with these three predicted targets in a more complex biological context, a longer segment of GSK-3β, REST and SIRT1 3′UTRs containing miR-9-5p binding site was cloned in the pmirGLO vector. As shown in Fig. 6, miR-9-5p mimics suppressed firefly luciferase activity in cells transfected with the reporter vectors containing REST 3′UTR (Fig. 6B; Student's t-test; p < 0.001) and SIRT1 3′UTR (Fig. 6C; Student's t-test, p < 0.05), but not in cells carrying GSK-3β 3′UTR (Fig. 6A; Student's t-test, p = 0.2994). Accordingly, the measure of GSK-3β, REST and SIRT1 protein expression levels in primary hippocampal cultures treated with miR-9-5p mimics revealed a reduction of REST (Fig. 6E; Student's t-test, p < 0.001) and SIRT1 (Fig. 6F; Student's t-test, p < 0.001), while no significant differences were found for GSK-3β (Fig. 6D; Student's t-test, p = 0.11). Overall, these data validated REST and SIRT1 as biological target genes of miR-9-5p, while GSK-3β was not confirmed.
Fig. 6.
Validation of miR-9-5p target genes. (A,B,C) Relative Luciferase Activity for GSK-3β, REST and SIRT1, long 3′ UTR fragments. Unpaired Student's t-test: *p < 0.05, ***p < 0.001 vs NEG control. n = 3; all experiments were repeated 3 times. (D,E,F) Analysis of GSK-3β, REST and SIRT1 expression in primary hippocampal neurons treated with miR-9-5p mimics or negative control. Unpaired Student's t-test: ***p < 0.001 vs NEG control. n = 9–14. (G,H,I) Analysis of GSK-3β, REST and SIRT1 expression in primary hippocampal neurons treated with corticosterone and ketamine. One-way ANOVA followed by Tukey's post-hoc test: *p < 0.05 vs DMSO; #p < 0.05 vs CORT. n = 6–12. (J,K,L) Analysis of GSK-3β, REST and SIRT1 protein expression in the HPC of CMS rats. One-way ANOVA followed by Tukey's post-hoc test: *p < 0.05, **p < 0.01 vs CNT; #p < 0.05 vs CMS-V; §§p < 0.01 vs CMS-R. n = 4–8.
3.5. Alterations of REST protein levels in primary hippocampal neurons and in the HPC of CMS rats were in line with miR-9-5p changes
To evaluate whether the expression levels of the validated miR-9-5p target genes were changed in primary neurons treated with corticosterone/ketamine and in CMS rats, we measured the levels of GSK-3β, REST and SIRT1 in both systems.
In primary hippocampal neurons, we found a significant treatment effect only in REST protein expression levels (one-way ANOVA, F2,17 = 6,143, p < 0.01), and no changes in SIRT1 (one-way ANOVA, F2,24 = 0,1957, p = 0.8236) or GSK-3β (one-way ANOVA, F3,32 = 0,9883, p = 0.4107) (Fig. 6G–I). Repeated corticosterone administration strongly increased REST levels, and ketamine completely rescued the change (Tukey's post-hoc test: CORT vs DMSO p < 0.05, CORT + KET vs CORT p < 0.05).
In the HPC of CMS rats, significant differences among groups were revealed in the protein expression levels of GSK-3β (one-way ANOVA, F3,21 = 4.080, p < 0.05) and REST (one-way ANOVA, F3,21 = 5,796, p < 0.01), while SIRT1 levels were unchanged (one-way ANOVA, F3,21 = 2.771, p = 0.0669) (Fig. 6J-L). As regarding GSK-3β, we found only a significant increase in CMS-V rats treated with ketamine compared to control (Tukey's post-hoc test, CMS-V + KET vs CNT p < 0.05). Instead, in line with changes of miR-9-5p levels, REST protein was remarkably increased selectively in CMS-V animals and ketamine completely reversed this change to control levels (Tukey's post-hoc test: CMS-V vs CNT p < 0.01, CMS-V + KET vs CMS-V p < 0.05).
We also measured GSK-3β, REST and SIRT1 mRNA levels (Supplementary Fig. 2), and found no significant changes for GSK-3β (one-way ANOVA, F3,24 = 2.190, p = 0.1154) and REST (one-way ANOVA, F3,21 = 2.345, p = 0.1020), while SIRT1 mRNA levels were significantly increased in CMS-V compared to both CNT and CMS-R (one-way ANOVA, F3,23 = 5.011, p < 0.01; Tukey's post-hoc test: CMS-V vs CNT p < 0.05, CMS-V vs CMS-R p < 0.05); ketamine only partly rescued the increase, since CMS-V + KET group is not significantly different from either controls or CMS-V (Tukey's post-hoc test: CMS-V + KET vs CNT p = 0.2346, CMS-V + KET vs CMS-V p = 0.7643).
3.6. Dendritic shortening induced by corticosterone in primary cultures is partly rescued by miR-9-5p overexpression and dependent on REST protein expression
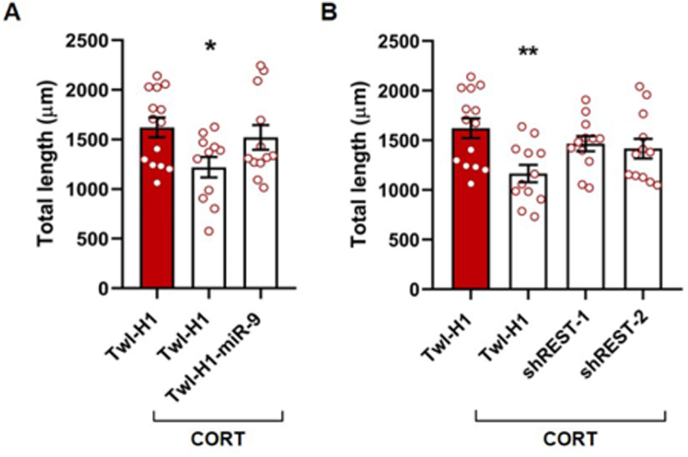
Finally, to provide further evidence that miR-9-5p plays a role in stress-induced dendritic remodeling, primary hippocampal neurons were transfected with miR-9-5p overexpression vector (or empty control) before being exposed to repeated treatment with corticosterone as before. While in line with the results in Fig. 3, corticosterone induced a reduction of total dendritic length (one-way ANOVA, F2,34 = 3.509, p = 0.0412; Tukey's post-hoc test: Twl-H1+CORT vs Twl-H1+DMSO p < 0.05) (Fig. 7A), miR-9-5p overexpression partly prevented the effects of corticosterone (Tukey's post-hoc test: Twl-H1-miR-9+CORT vs Twl-H1+CORT p = 0.11; Twl-H1-miR-9+CORT vs Twl-H1+DMSO p = 0.78).
Fig. 7.
Analysis of preventive effect of miR-9-5p overexpression and REST downregulation on dendritic morphology changes induced by CORT treatment. (A) Total dendritic length analysis of primary hippocampal neurons transfected with Twl-H1 or Twl-H1-miR-9 and treated with repeated CORT (or DMSO). one-way ANOVA followed by Tukey's post-hoc test: *p < 0.05 vs Twl-H1+DMSO. n = 12–14. (B) Total dendritic length analysis of primary hippocampal neurons transfected with Twl-H1, Twl-H1-shREST-1 or Twl-H1-shREST-2 and treated with repeated CORT (or DMSO). one-way ANOVA followed by Tukey's post-hoc test: **p < 0.01 vs Twl-H1+DMSO. n = 12–14. Dendritic length was measured using plug-in Fiji.
We finally asked whether REST, which we demonstrated to be a functional target of miR-9-5p, was implicated in corticosterone-dependent dendritic shortening. To this end, we transfected primary hippocampal neurons with two different REST silencing vectors and then treated cultures with corticosterone. Downregulation of REST prevented, at least in part, the reduction of dendritic length induced by corticosterone, as demonstrated by the observation that no difference was found between corticosterone-treated cells with REST downregulation and controls (one-way ANOVA, F3,46 = 4.303, p = 0.0093; Tukey's post-hoc test: Twl-H1+CORT vs Twl-H1+DMSO p < 0.01; Twl-H1-shREST-1+CORT vs Twl-H1+DMSO p = 0.62; Twl-H1-shREST-2+CORT vs Twl-H1+DMSO p = 0.39) (Fig. 7B).
Overall, these data suggest that the reduction of miR-9-5p levels and the related increase of REST protein expression are key molecular effectors of dendritic remodeling induced by stress.
4. Discussion
Preclinical studies have provided evidence that hippocampal synaptic remodeling and functional connectivity are associated with depression-related behavior in animal models of chronic stress (Duman et al., 2019; Krishnan and Nestler, 2008; McEwen et al., 2016; Qiao et al., 2016), as well as with rapid antidepressant effects of acute ketamine (Deyama and Duman, 2020; Kavalali and Monteggia, 2012; Tornese et al., 2019). Accordingly, we have previously demonstrated that animals vulnerable to the effects of CMS exhibit dendritic retraction and simplification of hippocampal CA3 pyramidal neurons, while acute ketamine, together with the rescue of depressive-like phenotype, completely recovers morphological deficits in just 24 h (Tornese et al., 2019).
In the present work, we investigated miR-9-5p as a putative molecular effector of dendritic remodeling induced by chronic stress and ketamine in the hippocampus. The expression levels of miR-9-5p were selectively lower in the hippocampus of CMS vulnerable animals and in primary neurons repeatedly incubated with the stress hormone corticosterone, compared to controls. Importantly, although previous studies consistently reported dendritic atrophy in brain areas of animal models of stress (Duman et al., 2019; Krishnan and Nestler, 2008; McEwen et al., 2016; Qiao et al., 2016), to our knowledge, here we showed for the first time that in vitro repeated treatment with corticosterone of primary neuronal cultures reproduces the reduction of pyramidal neuron dendritic length induced in vivo by stress. Our data clearly demonstrate that cellular effects of corticosterone are key mediators of both neuronal remodeling and decrease of miR-9-5p levels induced by stress. Of note, CMS resilient animals did not display any alterations in hippocampal miR-9-5p expression, suggesting that the reduction of miR-9-5p levels may be part of the maladaptive response to CMS, and related to anhedonic phenotype. Accordingly, miR-9-5p levels in the hippocampus negatively correlated with the anhedonic phenotype, further confirming the involvement of this miRNA in mechanisms involved in behavioral vulnerability to chronic stress. This idea is also supported by the fact that acute ketamine completely reversed the reduction of miR-9-5p both in the hippocampus of CMS vulnerable animals and in primary neurons. The effect of ketamine was rapid (observed in vivo after 24h, and in vitro after 4 h), suggesting a putative involvement of miR-9-5p in the mechanisms underlying the fast antidepressant effect of ketamine.
Since the changes in miR-9-5p levels occurred together with dendrite remodeling, we took advantage of reporter vectors allowing the simultaneous expression of GFP and overexpression or downregulation of miR-9-5p in primary neuronal cultures, to study the direct effect of miR-9-5p modulation on dendritic arbor. We found that miR-9-5p downregulation in mature hippocampal cultures decreased the length and complexity of pyramidal neuron dendrites, as well as the number of spines, while the overexpression of miR-9-5p exerted opposite effects. miR-9 is an evolutionarily conserved small regulatory RNA, highly enriched in the brain, and it has been consistently reported to play a key role in neurodevelopment (Radhakrishnan and Anand, 2016) and in the regulation of neurogenesis, neuronal morphology and function (Coolen et al., 2013; Dajas-bailador et al., 2012; Giusti et al., 2014). In particular, miR-9-5p was shown to control axonal extension and branching (Dajas-bailador et al., 2012), as well as in vitro and in vivo dendritic development (Giusti et al., 2014). However, miR-9-5p has never been implied before in dendritic remodeling of hippocampal pyramidal neurons associated with depressive behavior and antidepressant effects.
To further establish the molecular mechanism by which miR-9-5p might modulate dendritic remodeling, we predicted its target transcripts by bioinformatic analysis. We focused on selected targets that had previously shown to regulate brain function and plasticity. After biological validation of the interaction between miR-9-5p with its predicted target sequences, two main genes were confirmed: Sirt1 and Rest.
SIRT1 belongs to the family of sirtuins, histone deacetylases associated with transcriptional silencing, aging, cell differentiation, stress response and inflammation (Lu et al., 2018). SIRT1 is abundantly expressed in the brain where, throughout an epigenetic control of plasticity-associated genes, it can regulate synaptic plasticity, memory formation and cognitive function (Gao et al., 2010; Michan et al., 2010). Sirtuins are also known to promote axonal elongation, neurite outgrowth and dendritic branching during neuronal development (Herskovits and Guarente, 2014).
SIRT1 has been consistently reported to play neuroprotective roles in different models of neurodegenerative disorders, including Alzheimer's, Parkinson's, Huntington's and motor neuron diseases among others (Jęśko et al., 2017). More recently, variants near the Sirt1 gene were associated with depression (CONVERGE Consortium, 2015). Nevertheless, in our hands, SIRT1 protein was not significantly modulated by neither stress, nor ketamine treatment, suggesting a limited involvement of SIRT1 in our model.
REST, also known as neuron-restrictive silencer factor (NRSF), is a transcription repressor of more than 2000 neuron-specific target genes, encoding proteins involved in synaptic plasticity, cell death, neurotransmitter receptors, ion channel proteins and adhesion molecules (Mampay and Sheridan, 2019). REST expression is critical for brain development, cell differentiation and cell maturation, but its levels are remarkably downregulated during late stages of development, resulting in acquisition of neuronal phenotype (Zhao et al., 2017).
Although REST expression is low in mature neurons, it increases after specific cellular insults, including ischemia, seizure and neurotoxicity (Mampay and Sheridan, 2019). Moreover, both physical and psychological stress have been consistently shown to upregulate REST levels in brain areas of animal models of stress (Chen et al., 2016; Mou and Zhao, 2016; Soga et al., 2021), as well as in clinical studies (Goswami et al., 2010). During the adult stage, activated REST in mature neurons represses neurotransmission and leads to neuronal death and cognitive disorders (Giusti et al., 2014; Hwang et al., 2017; Pajarillo et al., 2020).
We found a remarkable increase of REST protein in the hippocampus of CMS vulnerable animals and in corticosterone-treated primary neurons, while ketamine treatment restored its expression to control levels in both models. These results are consistent with the concurrent reduction of miR-9-5p measured in both CMS vulnerable animals and in corticosterone-treated primary neurons. At the same time, ketamine, together with recovering the stress-induced morphological alterations, completely rescued miR-9-5p levels and down-regulated REST protein expression to control levels, both in stressed animals and in primary neurons treated with corticosterone.
Despite we did not analyze the effects of miR-9-5p/REST in vivo modulation in the hippocampus of CMS animals, both in vitro overexpression of miR-9-5p and silencing of REST were able to partly prevent dendritic shortening induced by corticosterone treatment of primary neuronal cultures. These results strongly suggest that the reduction of miR-9-5p induced by stress, and the consequent increase of REST protein expression, are key molecular effectors of hippocampal neuron dendritic simplification induced by chronic stress.
In line with our data, a previous study reported that miR-9-5p controls dendritic growth in developing mouse primary hippocampal neurons by targeting REST (Giusti et al., 2014). The transfection of neural precursor cells with shRNA against REST rescued dendritic growth defects in neurons with conditional down-regulation of miR-9-5p, confirming that REST is a major downstream molecular effector of miR-9-5p for the modulation of dendritic morphology. The authors hypothesized that given the abundant miR-9-5p expression in the adult brain and low expression levels of REST, except in areas involved in neurogenesis, miR-9-5p could be a key factor in keeping low REST levels during maturation. This may be necessary to avoid REST-dependent inhibition of plasticity-associated genes that mediate vesicular transport, signal transduction, neurite outgrowth and cell adhesion (Sun et al., 2005). In line with a role of miR-9/REST not only during development but also in mature neuronal populations, we hypothesize that stress-induced downregulation of miR-9-5p in vulnerable animals reduces hippocampal pyramidal neuron dendritic length through mechanisms implying REST upregulation, in turn inhibiting the transcription of genes involved in synaptic plasticity, while acute ketamine, restoring miR-9-5p levels, rescues morphological effects of stress.
5. Conclusion
We have investigated the role of miR-9-5p in the functional and morphological changes induced by chronic stress and ketamine both in the hippocampus of rats subjected to chronic mild stress and in primary hippocampal cultures. We demonstrated that morphological changes induced by chronic corticosterone and rescued by ketamine are accompanied by a decrease of miR-9-5p levels both in the hippocampus of stressed rats and in primary neuronal cultures. This in turn induces an upregulation of REST protein expression that negatively influences neuronal dendritic morphology, presumably by controlling the expression of synaptic plasticity-associated genes. We have to acknowledge that a limitation of our study is the absence of in vivo miR-9-5p/REST expression manipulation that would definitely confirm the involvement in morphological and behavioral effects of chronic stress. Further studies are needed to address this issue.
Funding
This work was supported by research grants from the Italian Ministry of University (PRIN project n. 2015HRE757) and Cariplo Foundation (Prog. 2014–1133 and 2017–0620).
CRediT authorship contribution statement
Jessica Mingardi: Methodology, Investigation, Formal analysis, Writing – original draft. Luca La Via: Methodology, Investigation. Paolo Tornese: Investigation. Giulia Carini: Investigation. Kalevi Trontti: Investigation. Mara Seguini: Investigation. Daniela Tardito: Investigation. Federica Bono: Resources. Chiara Fiorentini: Resources. Leonardo Elia: Resources. Iiris Hovatta: Conceptualization, Resources, Writing – original draft, Funding acquisition. Maurizio Popoli: Conceptualization, Resources, Writing – original draft, Funding acquisition. Laura Musazzi: Conceptualization, Formal analysis, Resources, Writing – original draft, Supervision, Funding acquisition. Alessandro Barbon: Conceptualization, Formal analysis, Resources, Writing – original draft, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100381.
Contributor Information
Laura Musazzi, Email: laura.musazzi@unimib.it.
Alessandro Barbon, Email: alessandro.barbon@unibs.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abe-Higuchi N., Uchida S., Yamagata H., Higuchi F., Hobara T., Hara K., Kobayashi A., Watanabe Y. Hippocampal sirtuin 1 signaling mediates depression-like behavior. Biol. Psychiatr. 2016;80:815–826. doi: 10.1016/j.biopsych.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Aceto G., Colussi C., Leone L., Fusco S., Rinaudo M., Scala F., Green T.A., Laezza F., D'Ascenzo M., Grassi C. Chronic mild stress alters synaptic plasticity in the nucleus accumbens through GSK3β-dependent modulation of Kv4.2 channels. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:8143–8153. doi: 10.1073/pnas.1917423117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolina D., Di Segni M., Bisicchia E., D'Alessandro F., Cestari V., Ventura A., Concepcion C., Puglisi-Allegra S., Ventura R. Effects of lack of microRNA-34 on the neural circuitry underlying the stress response and anxiety. Neuropharmacology. 2016;107:305–316. doi: 10.1016/j.neuropharm.2016.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.Y., Ruan C.S., Yang C.R., Li J, Kang Z-L, Zhou L, Liu D, Zeng Y-Q, Wang T-H, Tian C-F, Liao H, Bobrovskaya L, Zhou X-F. ProBDNF signaling regulates depression-like behaviors in rodents under chronic stress. Neuropsychopharmacology. 2016;41(12):2882–2892. doi: 10.1038/npp.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini D., Filippini A., La Via L., Fiorentini C., Fumagalli F., Colombi M., Barbon A. Chronic glutamate treatment selectively modulates AMPA RNA editing and ADAR expression and activity in primary cortical neurons. RNA Biol. 2015;12:43–53. doi: 10.1080/15476286.2015.1008365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini D., Mora C., Tornese P., Sala N., Filippini A., La Via L., Milanese M., Calza S., Bonanno G., Racagni G., Gennarelli M., Popoli M., Musazzi L., Barbon A. Acute footshock stress induces time-dependent modifications of AMPA/NMDA protein expression and AMPA phosphorylation. Neural Plast. 2016. 2016 doi: 10.1155/2016/7267865. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burak K., Lamoureux L., Boese A., Majer A., Saba R., Niu Y., Frost K., Booth S.A. MicroRNA-16 targets mRNA involved in neurite extension and branching in hippocampal neurons during presymptomatic prion disease. Neurobiol. Dis. 2018;112:1–13. doi: 10.1016/j.nbd.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Cavalleri L., Merlo Pich E., Millan M.J., Chiamulera C., Kunath T., Spano P.F., Collo G. Ketamine enhances structural plasticity in mouse mesencephalic and human iPSC-derived dopaminergic neurons via AMPAR-driven BDNF and mTOR signaling. Mol. Psychiatr. 2018;23:812–823. doi: 10.1038/mp.2017.241. [DOI] [PubMed] [Google Scholar]
- Chen C.-C., Huang C.-C., Hsu K.-S. Chronic social stress affects synaptic maturation of newly generated neurons in the adult mouse dentate gyrus. Int. J. Neuropsychopharmacol. 2016;19 doi: 10.1093/ijnp/pyv097. pyv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climent M., Quintavalle M., Miragoli M., Chen J., Condorelli G., Elia L. TGFβ Triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, Thereby modulating vessel stabilization. Circ. Res. 2015;116:1753–1764. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- Codocedo J.F., Allard C., Godoy J., Varela-Nallar L., Inestrosa N.C. SIRT1 Regulates Dendritic Development in Hippocampal Neurons. PLoS ONE. 2012;7(10):e47073. doi: 10.1371/journal.pone.0047073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M., Katz S., Bally-cuif L. miR-9 : a versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013;7:1–11. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-bailador F., Bonev B., Garcez P., Stanley P., Guillemot F., Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012;15 doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- Deyama S., Duman R.S. Neurotrophic mechanisms underlying the rapid and sustained antidepressant actions of ketamine. Pharmacol. Biochem. Behav. 2020;188:172837. doi: 10.1016/j.pbb.2019.172837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets W.C., Price J.L., Furey M.L. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct. Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R.S., Aghajanian G.K. Neurobiology of Rapid Acting Antidepressants: Role of BDNF and GSK-3β. Neuropsychopharmacology. 2014;39(1) doi: 10.1038/npp.2013.217. 233–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R.S., Li N. A neurotrophic hypothesis of depression: role of synaptogenesis in the actions of NMDA receptor antagonists. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:2475–2484. doi: 10.1098/rstb.2011.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R.S., Sanacora G., Krystal J.H. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron. 2019;102:75–90. doi: 10.1016/j.neuron.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Pathogenetic and therapeutic applications of microRNAs in major depressive disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2016;64:341–348. doi: 10.1016/j.pnpbp.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhussiny M.E.A., Carini G., Mingardi J., Tornese P., Sala N., Bono F., Fiorentini C., La Via L., Popoli M., Musazzi L., Barbon A. Modulation by chronic stress and ketamine of ionotropic AMPA/NMDA and metabotropic glutamate receptors in the rat hippocampus. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2021;104:110033. doi: 10.1016/j.pnpbp.2020.110033. [DOI] [PubMed] [Google Scholar]
- Fan C., Zhu X., Song Q., Wang P., Liu Z., Yu S.Y. MiR-134 modulates chronic stress-induced structural plasticity and depression-like behaviors via downregulation of Limk1/cofilin signaling in rats. Neuropharmacology. 2018;131:364–376. doi: 10.1016/j.neuropharm.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Farhang S., Barar J., Fakhari A., Mesgariabbasi M., Khani S., Omidi Y., Farnam A. Asymmetrical expression of BDNF and NTRK3 genes in frontoparietal cortex of stress-resilient rats in an animal model of depression. Synapse. 2014;68(9):387–393. doi: 10.1002/syn.21746. [DOI] [PubMed] [Google Scholar]
- Filippini A., Bonini D., Lacoux C., Pacini L., Zingariello M., Sancillo L., Bosisio D., Salvi V., Mingardi J., La Via L., Zalfa F., Bagni C., Barbon A. Absence of the Fragile X Mental Retardation Protein results in defects of RNA editing of neuronal mRNAs in mouse. RNA Biol. 2017;14:1580–1591. doi: 10.1080/15476286.2017.1338232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori L.M., Lin R., Ju C., Belzeaux R., Turecki G. Using epigenetic tools to investigate antidepressant response. 2018. 255-272. [DOI] [PubMed]
- Gao J., Wang W.-Y., Mao Y.-W., Gräff J., Guan J.-S., Pan L., Mak G., Kim D., Su S.C., Tsai L.-H. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti S.A., Vogl A.M., Brockmann M.M., Vercelli C.A., Rein M.L., Trümbach D., Wurst W., Cazalla D., Stein V., Deussing J.M., Refojo D. MicroRNA-9 controls dendritic development by targeting REST. Elife 1–22. 2014 doi: 10.7554/eLife.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D.B., May W.L., Stockmeier C.A., Austin M.C. Transcriptional expression of serotonergic regulators in laser-captured microdissected dorsal raphe neurons of subjects with major depressive disorder: sex-specific differences. J. Neurochem. 2010;112:397–409. doi: 10.1111/j.1471-4159.2009.06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Fu C., Lin L., Liu S., Su X., Li A., Wu Q., Jia C., Zhang P., Chen L., Zhu X., Wang X. miR-124 and miR-9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C-GPM6A pathway. J. Cell. Physiol. 2018;233:673–687. doi: 10.1002/jcp.25927. [DOI] [PubMed] [Google Scholar]
- Gu X., Li A., Liu S., Lin L., Xu S., Zhang P., Li S., Li X., Tian B., Zhu X., Wang X. MicroRNA124 regulated neurite elongation by targeting OSBP. Mol. Neurobiol. 2016;53:6388–6396. doi: 10.1007/s12035-015-9540-4. [DOI] [PubMed] [Google Scholar]
- Han M.-H., Nestler E.J. Neural substrates of depression and resilience. Neurotherapeutics. 2017;14:677–686. doi: 10.1007/s13311-017-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., He H., Yang L., Zhang J., Chen K., Duan Z. Functional tag SNPs inside the DRD2 gene as a genetic risk factor for major depressive disorder in the Chinese Han population. Int. J. Clin. Exp. Pathol. 2019;12(2):628–639. [PMC free article] [PubMed] [Google Scholar]
- He Z.X., Song H.F., Liu T.Y., Ma J., Xing Z.K., Yin Y.Y., Liu L., Zhang Y.N., Zhao Y.F., Yu H.L., He X.X., Guo W.X., Zhu X.J. HuR in the Medial Prefrontal Cortex is Critical for Stress-Induced Synaptic Dysfunction and Depressive-Like Symptoms in Mice. Cerebr. Cortex. 2019;29(6):2737–2747. doi: 10.1093/cercor/bhz036. [DOI] [PubMed] [Google Scholar]
- Herskovits A.Z., Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-W., Jiang J.-J., Yan-Gao, Wang R.-Y., Tu G.-J. MicroRNA-210 promotes sensory axon regeneration of adult mice in vivo and in vitro. Neurosci. Lett. 2016;622:61–66. doi: 10.1016/j.neulet.2016.04.034. [DOI] [PubMed] [Google Scholar]
- Hu Yue, Pei W., Hu Ying, Li P., Sun C., Du J., Zhang Y., Miao F., Zhang A., Shen Y., Zhang J. MiR34a regulates neuronal MHC class I molecules and promotes primary hippocampal neuron dendritic growth and branching. Front. Cell. Neurosci. 2020;14 doi: 10.3389/fncel.2020.573208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yu D., Gu Q., Yang Y., Tu K., Zhu J., Li Z. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 2014;5:3263. doi: 10.1038/ncomms4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.-Y., Aromolaran K.A., Zukin R.S. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 2017;18:347–361. doi: 10.1038/nrn.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O., Haramati S., Paul E.D., Maeno H., Navon I., Zwang R., Gil S., Mayberg H.S., Dunlop B.W., Menke A., Awatramani R., Binder E.B., Deneris E.S., Lowry C.A., Chen A. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- Jasińska M., Miłek J., Cymerman I.A., Łęski S., Kaczmarek L., Dziembowska M. miR-132 regulates dendritic spine structure by direct targeting of matrix metalloproteinase 9 mRNA. Mol. Neurobiol. 2016 doi: 10.1007/s12035-015-9383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jęśko H., Wencel P., Strosznajder R.P., Strosznajder J.B. Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem. Res. 2017;42:876–890. doi: 10.1007/s11064-016-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos E.M., Engel T., Merino-Serrais P., Fernaud-Espinosa I., Rodriguez-Alvarez N., Reynolds J., Reschke C.R., Conroy R.M., McKiernan R.C., DeFelipe J., Henshall D.C. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct. Funct. 2015;220:2387–2399. doi: 10.1007/s00429-014-0798-5. [DOI] [PubMed] [Google Scholar]
- Kavalali E.T., Monteggia L.M. Synaptic mechanisms underlying rapid antidepressant action of ketamine. Am. J. Psychiatr. 2012;169:1150–1156. doi: 10.1176/appi.ajp.2012.12040531. [DOI] [PubMed] [Google Scholar]
- Keers R., Uher R. Gene–environment interaction in major depression and antidepressant treatment response. Curr. Psychiatr. Rep. 2012;14:129–137. doi: 10.1007/s11920-011-0251-x. [DOI] [PubMed] [Google Scholar]
- Kim H.D., Hesterman J., Call T., Magazu S., Keeley E., Armenta K., Kronman H., Neve R.L., Nestler E.J., Ferguson D. SIRT1 Mediates Depression-Like Behaviors in the Nucleus Accumbens. J. Neurosci. 2016;36(32):8841–8852. doi: 10.1523/JNEUROSCI.0212-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Nestler E.J. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Via L., Bonini D., Russo I., Orlandi C., Barlati S., Barbon A. Modulation of dendritic AMPA receptor mRNA trafficking by RNA splicing and editing. Nucleic Acids Res. 2013;41:617–631. doi: 10.1093/nar/gks1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that Thousands of human genes are MicroRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li G., Ling S. MiR-124 promotes newborn olfactory bulb neuron dendritic morphogenesis and spine density. J. Mol. Neurosci. 2017;61:159–168. doi: 10.1007/s12031-016-0873-x. [DOI] [PubMed] [Google Scholar]
- Li N., Liu R.-J., Dwyer J.M., Banasr M., Lee B., Son H., Li X.-Y., Aghajanian G., Duman R.S. Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol. Psychiatr. 2011;69:754–761. doi: 10.1016/j.biopsych.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Fuchikami M., Dwyer J.M., Lepack A.E., Duman R.S., Aghajanian G.K. GSK-3 Inhibition Potentiates the Synaptogenic and Antidepressant-Like Effects of Subthreshold Doses of Ketamine. Neuropsychopharmacology. 2013;38(11):2268–2277. doi: 10.1038/npp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Yan H., Zhou D., Cai X., Zhang Y., Li S., Li H., Li S., Zhou D-S., Li X., Zhang C., Sun Y, Dai J., Zhong J., Yao Y-J., Luo X-J., Fang Y., Zhang D., Ma Y., Yue W., Li M., Xiao X. The depression GWAS risk allele predicts smaller cerebellar gray matter volume and reduced SIRT1 mRNA expression in Chinese population. Transl. Psychiatry. 2019;9(1):333. doi: 10.1038/s41398-019-0675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Li J., Zhang H., Zhao X., Yan L.J., Yang X. Role and possible mechanisms of sirt1 in depression. Oxid. Med. Cell. Longev. 2018. 2018 doi: 10.1155/2018/8596903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampay M., Sheridan G.K. REST: an epigenetic regulator of neuronal stress responses in the young and ageing brain. Front. Neuroendocrinol. 2019;53:100744. doi: 10.1016/j.yfrne.2019.04.001. [DOI] [PubMed] [Google Scholar]
- McEwen B.S., Bowles N.P., Gray J.D., Hill M.N., Hunter R.G., Karatsoreos I.N., Nasca C. Mechanisms of stress in the brain. Nat. Neurosci. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S., Nasca C., Gray J.D. Stress effects on neuronal structure: Hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology. 2016;41:3–23. doi: 10.1038/npp.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon M.C., Yucel K., Nazarov A., MacQueen G.M. A meta-analysis examining clinical predictors of hippocampal volume in patients with major depressive disorder. J. Psychiatry Neurosci. 2009;34:41–54. [PMC free article] [PubMed] [Google Scholar]
- Michan S., Li Y., Chou M.M.-H., Parrella E., Ge H., Long J.M., Allard J.S., Lewis K., Miller M., Xu W., Mervis R.F., Chen J., Guerin K.I., Smith L.E.H., McBurney M.W., Sinclair D.A., Baudry M., de Cabo R., Longo V.D. SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingardi J., Musazzi L., De Petro G., Barbon A. miRNA editing: new insights into the fast control of gene expression in Health and disease. Mol. Neurobiol. 2018;55:7717–7727. doi: 10.1007/s12035-018-0951-x. [DOI] [PubMed] [Google Scholar]
- Moda-Sava R.N., Murdock M.H., Parekh P.K., Fetcho R.N., Huang B.S., Huynh T.N., Witztum J., Shaver D.C., Rosenthal D.L., Alway E.J., Lopez K., Meng Y., Nellissen L., Grosenick L., Milner T.A., Deisseroth K., Bito H., Kasai H., Liston C. Sustained rescue of prefrontal circuit dysfunction by antidepressant-induced spine formation. Science. 2019;364 doi: 10.1126/science.aat8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Zhao X. NRSF and CCR5 established neuron-glia communication during acute and chronic stresses. J. Drug Metabol. Toxicol. 2016;7 doi: 10.4172/2157-7609.1000197. [DOI] [Google Scholar]
- Mouillet-Richard S., Baudry A., Launay J.-M., Kellermann O. MicroRNAs and depression. Neurobiol. Dis. 2012;46:272–278. doi: 10.1016/j.nbd.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Musazzi L., Rimland J., Ieraci A., Racagni G., Domenici E., Popoli M. Pharmacological characterization of BDNF promoters I, II and IV reveals that serotonin and norepinephrine input is sufficient for transcription activation. Int. J. Neuropsychopharmacol. 2014;17:779–791. doi: 10.1017/S1461145713001685. [DOI] [PubMed] [Google Scholar]
- Musazzi L., Tornese P., Sala N., Popoli M. Acute stress is not acute: sustained enhancement of glutamate release after acute stress involves readily releasable pool size and synapsin I activation. Mol. Psychiatr. 2017;22:1226–1227. doi: 10.1038/mp.2016.175. [DOI] [PubMed] [Google Scholar]
- O'Connor R.M., Grenham S., Dinan T.G., Cryan J.F. microRNAs as novel antidepressant targets: converging effects of ketamine and electroconvulsive shock therapy in the rat hippocampus. Int. J. Neuropsychopharmacol. 2013;16:1885–1892. doi: 10.1017/S1461145713000448. [DOI] [PubMed] [Google Scholar]
- O'Connor R.M., Gururajan A., Dinan T.G., Kenny P.J., Cryan J.F. All roads lead to the miRNome: miRNAs have a central role in the molecular pathophysiology of psychiatric disorders. Trends Pharmacol. Sci. 2016;37:1029–1044. doi: 10.1016/j.tips.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Nan Dai, Yuqi C., Zonglin S., Chenglong D., Na L., Fang L., Cong Z., Xiufeng X. Association of a SIRT1 polymorphism with changes of gray matter volume in patients with first-episode medication-naïve major depression. Psychiatry Res.: Neuroimaging. 2020;301:111101. doi: 10.1016/j.pscychresns.2020.111101. [DOI] [PubMed] [Google Scholar]
- O'Connor S., Agius M. A systematic review of structural and functional MRI differences between psychotic and nonpsychotic depression. Psychiatr. Danub. 2015;27(Suppl. 1):S235–S239. [PubMed] [Google Scholar]
- Pajarillo E., Rizor A., Son D.-S., Aschner M., Lee E. The transcription factor REST up-regulates tyrosine hydroxylase and antiapoptotic genes and protects dopaminergic neurons against manganese toxicity. J. Biol. Chem. 2020;295:3040–3054. doi: 10.1074/jbc.RA119.011446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M., Torres-Reveron J., Yan L., Kimura T., Lin T.V., Gordon V., Teng Z.-Q., Zhao X., Fulga T.A., Van Vactor D., Bordey A. miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PloS One. 2012;7 doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecina M., Sikora M., Avery E.T., Heffernan J., Peciña S., Mickey B.J., Zubieta J-K. Striatal dopamine D2/3 receptor-mediated neurotransmission in major depression: Implications for anhedonia, anxiety and treatment response. Eur. Neuropsychopharmacol. 2017;27(10):977–986. doi: 10.1016/j.euroneuro.2017.08.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C., Duman R.S. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- Popoli M., Yan Z., McEwen B.S., Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 2011;13:22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.B., Duman R. Neuroplasticity in cognitive and psychological mechanisms of depression: an integrative model. Mol. Psychiatr. 2020;25:530–543. doi: 10.1038/s41380-019-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Li M.X., Xu C., Chen H. Bin, An S.C., Ma X.M. Dendritic spines in depression: what we learned from animal models. Neural Plast. 2016. 2016;20–24 doi: 10.1155/2016/8056370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan B., Anand A.A.P. Role of miRNA-9 in brain development. J. Exp. Neurosci. 2016;10 doi: 10.4137/JEN.S32843. JEN.S32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman M., Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development. 2017;144:2310–2322. doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- Reczko M., Maragkakis M., Alexiou P., Grosse I., Hatzigeorgiou A.G. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- Rossi M., Kilpinen H., Muona M., Surakka I., Ingle C., Lahtinen J., Hennah W., Ripatti S., Hovatta I. Allele-specific regulation of DISC1 expression by miR-135b-5p. Eur. J. Hum. Genet. 2014;22:840–843. doi: 10.1038/ejhg.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan C-S., Yang C-R., Li J-Y., Luo H-Y., Bobrovskaya L., Zhou X-F. Mice with Sort1 deficiency display normal cognition but elevated anxiety-like behavior. Exp. Neurol. 2016;281:99–108. doi: 10.1016/j.expneurol.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Sanacora G., Treccani G., Popoli M. Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology. 2012;62:63–77. doi: 10.1016/j.neuropharm.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sheline Y.I., Liston C., McEwen B.S. Parsing the Hippocampus in depression: chronic stress, hippocampal volume, and major depressive disorder. Biol. Psychiatr. 2019;85:436–438. doi: 10.1016/j.biopsych.2019.01.011. [DOI] [PubMed] [Google Scholar]
- Shen J., Li Y., Qu C., Xu L., Sun H., Zhang J. The enriched environment ameliorates chronic unpredictable mild stress-induced depressive-like behaviors and cognitive impairment by activating the SIRT1/miR-134 signaling pathway in hippocampus. J. Affect. Disord. 2019;248:81–90. doi: 10.1016/j.jad.2019.01.031. [DOI] [PubMed] [Google Scholar]
- Smrt R.D., Szulwach K.E., Pfeiffer R.L., Li X., Guo W., Pathania M., Teng Z.-Q., Luo Y., Peng J., Bordey A., Jin P., Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin Ligase mind bomb-1. Stem Cell. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T., Nakajima S., Kawaguchi M., Parhar I.S. Repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) in social stress and depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2021;104:110053. doi: 10.1016/j.pnpbp.2020.110053. [DOI] [PubMed] [Google Scholar]
- Sun Y.-M., Greenway D.J., Johnson R., Street M., Belyaev N.D., Deuchars J., Bee T., Wilde S., Buckley N.J. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol. Biol. Cell. 2005;16:5630–5638. doi: 10.1091/mbc.e05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamasi V., Petschner P., Adori C., Kirilly E., Ando R.D., Tothfalusi L., Juhasz G., Bagdy G. Transcriptional Evidence for the Role of Chronic Venlafaxine Treatment in Neurotrophic Signaling and Neuroplasticity Including also Glutatmatergic- and Insulin-Mediated Neuronal Processes. PLoS ONE. 2014;9(11):e113662. doi: 10.1371/journal.pone.0113662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornese P., Sala N., Bonini D., Bonifacino T., La Via L., Milanese M., Treccani G., Seguini M., Ieraci A., Mingardi J., Nyengaard J.R., Calza S., Bonanno G., Wegener G., Barbon A., Popoli M., Musazzi L. Chronic mild stress induces anhedonic behavior and changes in glutamate release, BDNF trafficking and dendrite morphology only in stress vulnerable rats. The rapid restorative action of ketamine. Neurobiol. Stress. 2019;10:100160. doi: 10.1016/j.ynstr.2019.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli F., Speranza L., Pulcrano S., De Gregorio R., Crispino M., De Sanctis C., Leopoldo M., Lacivita E., di Porzio U., Bellenchi G.C., Perrone-Capano C. The microRNA-29a modulates serotonin 5-HT7 receptor expression and its effects on hippocampal neuronal morphology. Mol. Neurobiol. 2019;56:8617–8627. doi: 10.1007/s12035-019-01690-x. [DOI] [PubMed] [Google Scholar]
- Wan Y.-Q., Feng J.-G., Li M., Wang M.-Z., Liu L., Liu X., Duan X.-X., Zhang C.-X., Wang X.-B. Prefrontal cortex miR-29b-3p plays a key role in the antidepressant-like effect of ketamine in rats. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu X., Yu Y., Han Y., Wei J., Collier D., Li T., Ma X. The role of single nucleotide polymorphism of D2 dopamine receptor gene on major depressive disorder and response to antidepressant treatment. Psychiatry Res. 2012;200(2–3):1047–1050. doi: 10.1016/j.psychres.2012.06.024. [DOI] [PubMed] [Google Scholar]
- Wang N., Yang Y., Pang M., Du C., Chen Y., Li S., Tian Z., Feng F., Wang Y., Chen Z., Liu B., Rong L. MicroRNA-135a-5p promotes the functional recovery of spinal cord injury by targeting SP1 and ROCK. Mol. Ther. Nucleic Acids. 2020;22:1063–1077. doi: 10.1016/j.omtn.2020.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts M., Williams G., Lu J., Nithianantharajah J., Claudianos C. MicroRNA-210 regulates dendritic morphology and behavioural flexibility in mice. Mol. Neurobiol. 2021;58:1330–1344. doi: 10.1007/s12035-020-02197-6. [DOI] [PubMed] [Google Scholar]
- Xu Y., Cheng X., Cui X., Wang T., Liu G., Yang R., Wang J., Bo X., Wang S., Zhou W., Zhang Y. Effects of 5-h multimodal stress on the molecules and pathways involved in dendritic morphology and cognitive function. Neurobiol. Learn. Mem. 2015;123:225–238. doi: 10.1016/j.nlm.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Xue Q., Yu C., Wang Y., Liu L., Zhang K., Fang C., Liu F., Bian G., Song B., Yang A., Ju G., Wang J. MiR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.-L., Sun X.-W., Wang Z.-M., Liu P.-P., Mi T.-W., Liu C., Wang Y.-Y., He X.-C., Du H.-Z., Liu C.-M., Teng Z.-Q. MiR-137 deficiency causes anxiety-like behaviors in mice. Front. Mol. Neurosci. 2019;12 doi: 10.3389/fnmol.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yang Q., Wang X., Luo C., Wan Y., Li J., Liu K., Zhou M., Zhang C. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. NeuroMolecular Med. 2014;16:594–605. doi: 10.1007/s12017-014-8312-z. [DOI] [PubMed] [Google Scholar]
- Yang C.R., Zhang X.Y., Liu Y., Du J.Y., Liang R., Yu M, Zhang F.Q., Mu X.F., Li F., Zhou L., Zhou F.H., Meng F.J., Wang S., Ming D., Zhou X.F. Antidepressant Drugs Correct the Imbalance Between proBDNF/p75NTR/Sortilin and Mature BDNF/TrkB in the Brain of Mice with Chronic Stress. Neurotox. Res. 2020;37(1):171–182. doi: 10.1007/s12640-019-00101-2. [DOI] [PubMed] [Google Scholar]
- Yi L.T., Zhu J.X., Dong S.Q., Li C.F., Zhang Q.P., Cheng J., Liu Q. miR-34a induces spine damages via inhibiting synaptotagmin-1 in depression. Neurobiol. Stress. 2020;13:100243. doi: 10.1016/j.ynstr.2020.100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate C.A., Machado-Vieira R. GSK-3: A key regulatory target for ketamine’s rapid antidepressant effects mediated by enhanced AMPA to NMDA throughput. Bipolar Disord. 2016;18(8):702–705. doi: 10.1111/bdi.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhu M., Yu Y., Qiu L., Zhang Y., He L., Zhang J. Brain REST/NRSF is not only a silent repressor but also an active protector. Mol. Neurobiol. 2017;54:541–550. doi: 10.1007/s12035-015-9658-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y.-L., Wu F.-C., Liu W.-J., Zheng W., Wang C.-Y., Zhan Y.-N., Lan X.-F., Ning Y.-P. Volumetric changes in subcortical structures following repeated ketamine treatment in patients with major depressive disorder: a longitudinal analysis. Transl. Psychiatry. 2020;10:264. doi: 10.1038/s41398-020-00945-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Ding Y., Wang K., Xiong E., Peng W., Du F., Zhang Z., Liu J., Gong A. MicroRNA-29A/PTEN pathway modulates neurite outgrowth in PC12 cells. Neuroscience. 2015;291:289–300. doi: 10.1016/j.neuroscience.2015.01.055. [DOI] [PubMed] [Google Scholar]
- Żurawek D., Faron-Górecka A., Kuśmider M., Kolasa M., Gruca P., Papp M., Dziedzicka-Wasylewska M. Mesolimbic dopamine D2 receptor plasticity contributes to stress resilience in rats subjected to chronic mild stress. Psychopharmacology (Berl) 2013;227:583–593. doi: 10.1007/s00213-013-2990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.