Abstract
Ultraviolet (UV) B irradiation of keratinocytes results in the formation of the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) which is a high-affinity ligand for the aryl hydrocarbon receptor (AHR). The resulting activation of AHR signaling induces the expression of cytochrome P450 (CYP) 1A1 which subsequently metabolizes FICZ. Importantly, FICZ is also a nanomolar photosensitizer for UVA radiation. Here, we assess whether a manipulation of the AHR-CYP1A1 axis in human epidermal keratinocytes affects FICZ/UVA-induced phototoxic effects and whether this interaction might be mechanistically relevant for the phototoxicity of the BRAF inhibitor vemurafenib. Treatment of keratinocytes with an AHR agonist enhanced the CYP1A1-catalyzed metabolism of FICZ and thus prevented UVA photosensitization, whereas an inhibition of either AHR signaling or CYP1A1 enzyme activity resulted in an accumulation of FICZ and a sensitization to UVA-induced oxidative stress and apoptosis. Exposure of keratinocytes to vemurafenib resulted in the same outcome. Specifically, CYP phenotyping revealed that vemurafenib is primarily metabolized by CYP1A1 and to a lesser degree by CYP2J2 and CYP3A4. Hence, vemurafenib sensitized keratinocytes to UVA-induced apoptosis by interfering with the CYP1A1-mediated oxidative metabolism of FICZ. In contrast to this pro-apoptotic effect, a treatment of UVB-damaged keratinocytes with vemurafenib suppressed apoptosis, a process which might contribute to the skin carcinogenicity of the drug. Our results provide insight into the mechanisms responsible for the photosensitizing properties of vemurafenib and deliver novel information about its metabolism which might be relevant regarding potential drug-drug interactions. The data emphasize that the AHR-CYP1A1 axis contributes to the pathogenesis of cutaneous adverse drug reactions.
Keywords: Adverse drug reactions, Apoptosis, Aryl hydrocarbon receptor, Cytochrome P450, Phototoxicity, Ultraviolet radiation
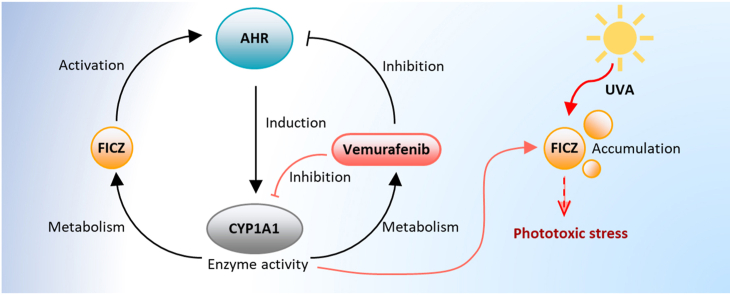
Graphical abstract
1. Introduction
Exposure of the skin to ultraviolet (UV) radiation causes a variety of acute (e.g. sunburn) and chronic (e.g. aging, cancer) adverse health effects [[1], [2], [3]]. The majority of these effects is triggered by UV-induced DNA damage and associated signaling responses. Whereas UVA radiation (315–400 nm) penetrates deep into the skin and even reaches dermal fibroblasts, UVB rays (280–315 nm) are nearly completely absorbed by macromolecular structures, in particular the DNA, of epidermal cells [[1], [2], [3]]. UVB irradiation leads to the formation of mutagenic cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone (6–4) photoproducts [4], whereas UVA rays are poorly absorbed by DNA and exert their harmful effects, i.e. oxidative damage of cellular macromolecules, mainly through an excitation of endogenous photosensitizers, such as protoporphyrin IX and riboflavin, and the resulting formation of reactive oxygen species (ROS) [5]. In addition, various oral drugs, including antibiotics, statins and non-steroidal anti-inflammatory drugs, have been identified as potent photosensitizers. Accordingly, cutaneous phototoxicity is a common side effect of drug treatment [6,7]. A prominent example for a phototoxic drug is vemurafenib (PLX 4032, Zelboraf®), a protein kinase inhibitor approved for the treatment of advanced melanoma and Erdheim-Chester disease with BRAF V600 gain-of-function mutations [[8], [9], [10]]. An enhanced sensitivity to UVA radiation affects 35%–63% of the melanoma patients and is one of the most common side effects of vemurafenib treatment [11,12].
Besides DNA, tryptophan is a potent chromophore for UVB radiation. In keratinocytes, absorption of UVB rays by cytosolic tryptophan leads to the formation of 6-formylindolo[3,2-b]carbazole (FICZ) and other tryptophan photoproducts [[13], [14], [15], [16]]. FICZ is a high-affinity ligand of the aryl hydrocarbon receptor (AHR), a key regulator in xenobiotic metabolism and immunity [[17], [18], [19]]. Upon its activation, AHR shuttles into the nucleus and induces the expression of target genes, for instance encoding cytochrome P450 (CYP) 1A1 and CYP1B1. Importantly, CYP1A1 rapidly monohydroxylates FICZ to enable its detoxification by sulfotransferases and other conjugating enzymes [20,21], thereby ensuring a transient AHR activation by the tryptophan photoproduct [22]. In concert with other signaling pathways, AHR orchestrates UVB stress responses in epidermal cells [2,23].
However, FICZ does not only serve as AHR agonist but is also a potent photosensitizer for UVA radiation [24]. In fact, the Wondrak laboratory has shown that UVA irradiation of FICZ-treated keratinocytes results in ROS generation and associated DNA damage [24]. In a follow-up work, the group proposed to harness the UVA-sensitizing properties of FICZ for the photodynamic elimination of skin cancer [25]. Other investigators, however, suggested that FICZ may contribute to UV-induced skin carcinogenesis by photochemically damaging DNA repair enzymes [26].
Here, we investigated whether a modulation of AHR signaling and/or CYP1A1 enzyme activity stabilizes intracellular FICZ levels and sensitizes keratinocytes to UVA-induced apoptosis. In addition, we assessed whether vemurafenib enhances UVA-induced keratinocyte apoptosis by interfering with the CYP1A1-catalyzed metabolism of FICZ. We performed enzyme activity assays and a detailed CYP phenotyping to identify CYP isoforms involved in the metabolism of vemurafenib. Finally, we analyzed the impact of vemurafenib on the apoptotic clearance of UVB-irradiated keratinocytes harboring mutagenic DNA photoproducts.
2. Results
2.1. FICZ sensitizes human keratinocytes to UVA-induced apoptosis
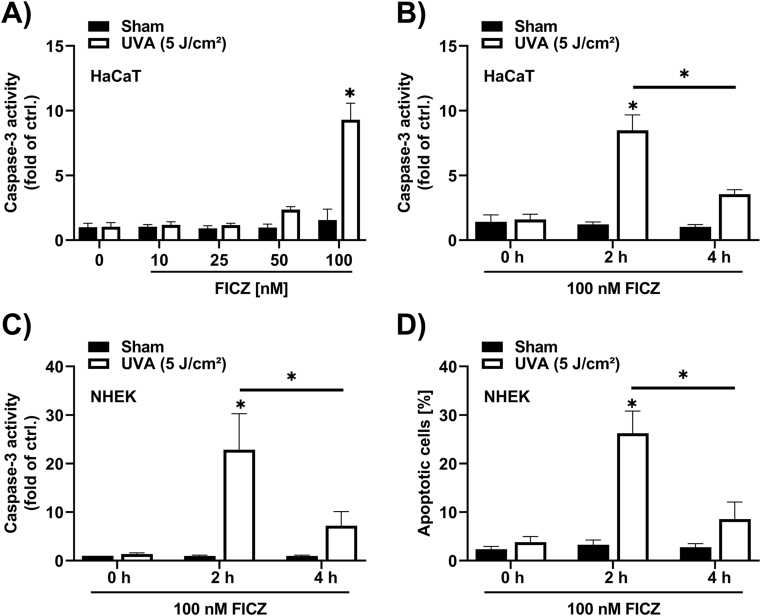
Treatment of HaCaT keratinocytes with FICZ for 30 min and a subsequent exposure to a low dose of 5 J/cm2 UVA radiation resulted in a dose-dependent induction of caspase-3 activity (Fig. 1A). While an exposure to FICZ or UVA radiation alone had no effect, we observed a slight increase of caspase-3 activity in cells pretreated with 50 nM FICZ. A pretreatment with 100 nM FICZ resulted in a nearly 10-fold induction of caspase-3 activity (Fig. 1A). A pretreatment of HaCaT cells or normal human epidermal keratinocytes (NHEKs) with 100 nM FICZ for 2 h strongly increased caspase-3 activity upon UVA irradiation, whereas a FICZ pretreatment for 4 h did not significantly enhance UVA-induced apoptosis (Fig. 1B and C). This observation was confirmed by Nicoletti staining (Fig. 1D), indicating that over a period of 4 h FICZ is being metabolized.
Fig. 1.
FICZ sensitizes keratinocytes to UVA-induced apoptosis.
(A) Caspase-3 activity in HaCaT keratinocytes pretreated with 10 nM, 25 nM, 50 nM and 100 nM FICZ or 0.1% DMSO for 30 min prior to sham or UVA radiation with 5 J/cm2. Cells were lysed for Caspase-3 activity measurement 4 h after UVA radiation (n = 3). (B–D) Cell death measurements of keratinocytes treated with 100 nM FICZ or 0.1% DMSO for 2 h and 4 h before cells were exposed to 0 or 5 J/cm2 UVA radiation. Measurement of Caspase-3 activity in HaCaT cells (B) (n = 3) and NHEKs (C) (n = 3) were carried out 4 h after UVA/sham exposure. Analysis of dead cells (subG1 fraction) by using the Nicoletti assay 24 h post UVA radiation of NHEKs (D) (n = 8). For statistical analysis a 2-way ANOVA (post hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to DMSO sham).
2.2. AHR activation decreases the UVA-photosensitizing effect of FICZ
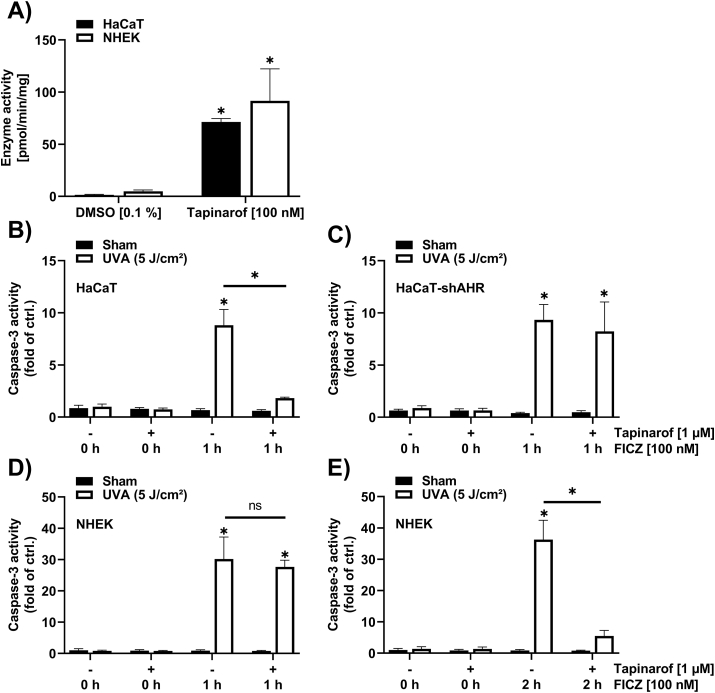
Given that FICZ is monohydroxylated by CYP1A1, we next stimulated keratinocytes with the AHR agonist tapinarof [27] to induce CYP1A1 expression and enzyme activity and analyze its impact on FICZ/UVA-induced caspase-3 activity. Treatment of keratinocytes with 100 nM tapinarof for 24 h induced the deethylation of 7-ethoxyresorufin (Fig. 2A), a reaction that is catalyzed by CYP1A isoforms. As human keratinocytes do not express CYP1A2 [28,29], the observed formation of resorufin was specifically catalyzed by CYP1A1. Hence, a pretreatment of keratinocytes with tapinarof should accelerate FICZ clearance and reduce FICZ/UVA-triggered apoptosis in an AHR-dependent manner. AHR-proficient and AHR-knockdown HaCaT keratinocytes were treated for 24 h with 1 μM tapinarof or solvent. Next, cells were treated for 1 h with 100 nM FICZ and subsequently exposed to 5 J/cm2 UVA radiation. Exposure to tapinarof, FICZ and UVA radiation alone did not affect caspase-3 activity (Fig. 2B and C). We observed a strong induction of caspase-3 activity in AHR-proficient (HaCaT-EV) and AHR-knockdown (HaCaT-shAHR) cells that were pretreated with DMSO and subsequently exposed to FICZ and UVA radiation. As expected, the FICZ/UVA-triggered induction of caspase-3 activity was nearly completely absent in tapinarof-treated AHR-proficient HaCaT cells but not in the HaCaT-shAHR keratinocytes (Fig. 2B and C), providing evidence that the photosensitizing property of FICZ can be manipulated by activating AHR. The same experimental setup was used to confirm this result in NHEKs. However, upon treatment of the tapinarof- and solvent-preexposed cells for 1 h with FICZ prior to UVA irradiation, we noted no significant difference in the FICZ/UVA-induced caspase-3 activity (Fig. 2D). After elongating the incubation with FICZ for another hour, the FICZ/UVA-dependent caspase-3 activity decreased (Fig. 2E), suggesting that FICZ metabolism in NHEKs occurred slower.
Fig. 2.
Treatment of keratinocytes with tapinarof increases CYP1A1 enzyme activity and decreases FICZ/UVA-induced caspase-3 activity in an AHR-dependent manner.
(A) CYP1A1 enzyme activity measurement in HaCaT cells and NHEKs treated for 24 h with 100 nM tapinarof (n = 3). (B–E) Caspase-3 assay performed in AHR-proficient HaCaT cells (b); (n = 3), AHR-knockdown HaCaT cells (C) (n = 3) and NHEKs (D, E) (n = 3). Keratinocytes were treated for 24 h with the specified concentrations of tapinarof, before 100 nM FICZ were added as indicated. Afterwards, cells were irradiated with 5 J/cm2 UVA and 4 h later, caspase-3 activity was determined. For statistical analysis a 2-way ANOVA (post hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to DMSO sham).
2.3. AHR inhibition enhances FICZ/UVA-induced apoptosis and oxidative stress
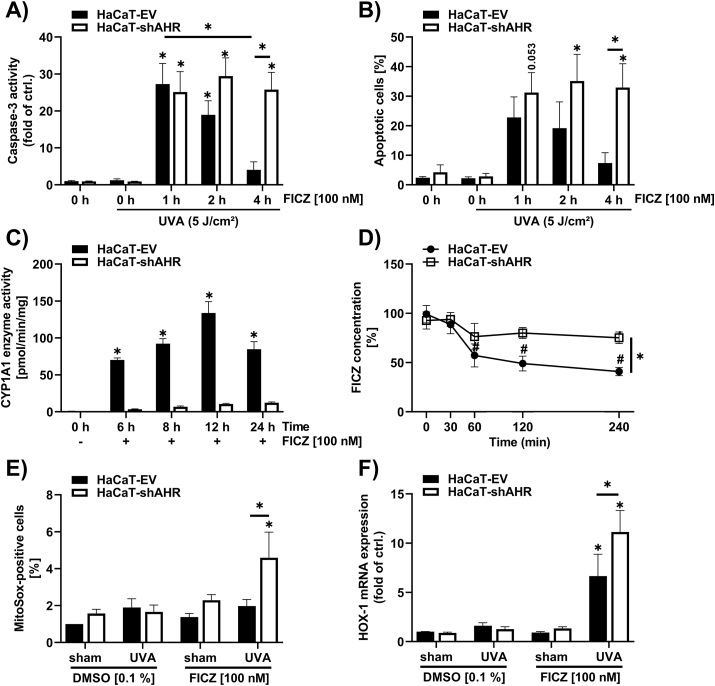
To confirm a critical role of AHR in modulating the FICZ/UVA-induced apoptotic response, HaCaT-EV and HaCaT-shAHR keratinocytes were pretreated for 1, 2 and 4 h with 100 nM FICZ and subsequently exposed to 5 J/cm2 UVA radiation. Notably, FICZ treatment alone did not induce any detectable caspase-3 activity (Supplementary Fig. S1). Compared to the solvent controls, a FICZ pretreatment for 1 h resulted in an increase of the FICZ/UVA-induced apoptosis in both cell-lines (Fig. 3A and B). An elongation of the FICZ preincubation resulted in a decline of FICZ/UVA-triggered apoptosis in HaCaT-EV but not in HaCaT-shAHR keratinocytes (Fig. 3A and B). These results confirmed that by activating AHR FICZ initiated its own CYP1A1-mediated metabolism. As expected, FICZ treatment resulted in an induction of CYP1A1-mediated EROD activity in AHR-proficient but not in AHR-knockdown HaCaT keratinocytes (Fig. 3C). Accordingly, we observed a faster metabolic clearance of exogenously added FICZ in HaCaT-EV compared to HaCaT-shAHR cells (Fig. 3D). After 4 h, ~60% of the applied FICZ was metabolized in HaCaT-EV cells, whereas the FICZ level in HaCaT-shAHR cells declined only for ~25%. When the two cell-lines were treated with 100 nM FICZ for 4 h and subsequently irradiated with UVA, we observed a formation of superoxide, which was paralleled by a transcriptional upregulation of heme oxygenase-1 (HOX-1), an established marker for oxidative stress [30], in the HaCaT-shAHR cells (Fig. 3E and F). However, a lower induction of HOX-1 was also observed in the HaCaT-EV cells. These results provided evidence that a modulation of the AHR-CYP1A1 axis directly affects the metabolism of the UVA-photosensitizer FICZ.
Fig. 3.
Genetic inhibition of AHR in keratinocytes increases FICZ/UVA-induced apoptosis and oxidative stress.
(A, B) Determination of apoptotic AHR-proficient and AHR-knockdown HaCaT keratinocytes exposed to 100 nM FICZ for 1, 2 and 4 h, respectively, followed by irradiation with 0 or 5 J/cm2 UVA. Cells were lysed 4 h after UVA irradiation for determination of caspase-3 activity, (A) (n = 6), for Nicoletti staining, keratinocytes were harvested 24 h after UVA exposure (B) (n = 3). (C) EROD assay to analyze CYP1A1 enzyme activity was performed in HaCaT-EV and HaCaT-shAHR keratinocytes exposed to 100 nM FICZ for different time points (n = 3). (D) HPLC analysis was conducted in HaCaT-EV and HaCaT-shAHR keratinocytes treated with 100 nM FICZ for 30, 60, 120 and 240 min, respectively (n = 3). (E) ROS measurement by analyzing MitoSox positive cells. Cell were exposed to 100 nM FICZ for 4 h and subsequently irradiated with 5 J/cm2 UVA. One hour later, cells were trypsinized and assayed by flow cytometry (n = 4). (F) Gene expression rate of HOX-1 in AHR-proficient and AHR-knockdown HaCaT cells. Keratinocytes were treated for 4 h with FICZ and subsequently irradiated with UVA. After 6 h, cells were harvested for qPCR analyses. HOX-1 transcript levels were normalized to GAPDH (n = 4). For statistical analysis a 2-way ANOVA (post hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to HaCaT-EV keratinocytes exposed to DMSO sham). In addition, for (B, D-F) a Sidak 2-way multiple comparison test was performed to compare HaCaT-EV with HaCaT-shAHR keratinocytes (*, p ˂ 0.05).
2.4. Vemurafenib attenuates FICZ metabolism by interfering with CYP1A1 enzyme activity
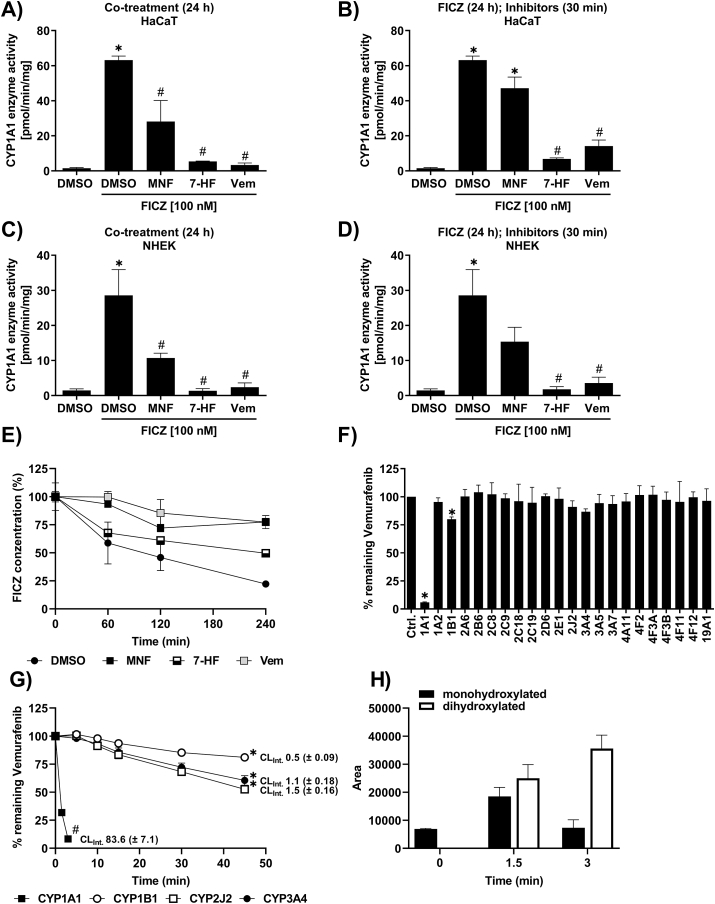
As previously reported, vemurafenib is capable of antagonizing the canonical AHR signaling pathway [31,32], indicating that the phototoxic properties of the BRAF inhibitor (BRAFi) may be related to an interference with the CYP1A1-mediated clearance of FICZ. Therefore, we next assessed whether vemurafenib treatment disturbs CYP1A1 enzyme activity and associated FICZ metabolism. HaCaT cells and NHEKs were treated for 24 h with 100 nM FICZ alone and in combination with vemurafenib, the AHR antagonist 3′-methoxy-4‘-nitroflavone (MNF) [33] or the CYP1A isoform inhibitor 7-hydroxyflavone (7-HF) [34]. FICZ treatment increased CYP1A enzyme activity, an effect which was attenuated by all inhibitors tested (Fig. 4A and B). To investigate whether vemurafenib inhibits CYP1A1 activity directly or indirectly, i.e. by antagonizing AHR and downstream CYP1A1 expression, we pretreated the keratinocytes for 23.5 h with FICZ and subsequently co-exposed the cells to the different inhibitors for just 30 min before the EROD measurement. Importantly, FICZ-induced EROD activity was still reduced in the samples co-treated for 30 min with the BRAFi (Fig. 4C and D), strongly indicating that vemurafenib is a CYP1A1 inhibitor. Noteworthy, in concentrations roughly mirroring or even exceeding plasma levels in patients [35,36], neither dabrafenib nor encorafenib, two additional BRAFi approved for the treatment of advanced melanoma, had an impact on FICZ-induced EROD activity (Supplementary Figs. S2A and b). A co-treatment with vemurafenib reduced the time-dependent metabolic clearance of FICZ in HaCaT keratinocytes (Fig. 4E), supporting our hypothesis that the interference of the BRAFi with CYP1A1 activity is of pathophysiological relevance.
Fig. 4.
Vemurafenib attenuates FICZ metabolism by interfering with CYP1A1 enzyme activity.
a)-d) Identification of CYP1A1 enzyme activity in HaCaT cells and NHEKs treated with different substances (10 μM MNF, 10 μM 7-HF, 25 μM (NHEKs) or 40 μM (HaCaT) Vemurafenib). HaCaT cells (A) (n = 3) and NHEKs (C) (n = 3) were co-exposed for 24 h with one of the three different compounds and 100 nM FICZ. HaCaT cells (B) (n = 3) and NHEKs (D) (n = 3) were treated for 24 h with 100 nM FICZ and MNF, 7-HF or vemurafenib was added 30 min prior to EROD measurement. For statistical analysis a 1-way ANOVA (post-hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to DMSO; #, p ˂ 0.05 relative to DMSO FICZ treatment). (E) FICZ concentration in the culture medium of HaCaT cells was determined by HPLC analysis. Cells were treated for 4 h with 10 μM MNF, 10 μM 7-HF, 40 μM vemurafenib or solvent and subsequently 100 nM FICZ was added for the indicated time (n = 2). (F–H) CYP phenotyping for vemurafenib was conducted using 22 recombinant human CYP isoforms overexpressed in insect cells. (F) Microsomal preparations of each CYP isoform (10 pmol/ml) were incubated for 10 min with 0.5 μM vemurafenib. Subsequently, vemurafenib was analyzed by LC/MS (n = 3) For statistical analysis a 1-way ANOVA (uncorrected Fisher's LSD test) was performed and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to control)). (G) 10 pmol/ml CYP1A1, CYP1B1, CYP2J2 and CYP3A4 were incubated up to 45 min with 0.5 μM vemurafenib in order to determine the intrinsic clearance (CLINT) (n = 3). For statistical analysis a 1-way ANOVA (post-hoc: Tukey test) was performed and data are represented as mean ± SEM. (*, p ˂ 0.05 for 15, 30 and 45 min relative to 0 min; #, p ˂ 0.05 for 1.5 and 3 min relative to 0 min). (H) Identification and formation of CYP1A1-derived monohydroxylated and a dihydroxylated vemurafenib (n = 3).
To clarify whether vemurafenib is a substrate of CYP1A1, we investigated the vemurafenib metabolism in more detail using 22 human recombinant CYP isoforms overexpressed in insect cells. Already after 10 min of incubation with 0.5 μM vemurafenib and 10 pmol/ml of the respective enzymes, we observed a considerable depletion of the drug to ~6% of the control level in microsomes expressing CYP1A1 (Fig. 4F). The CYP phenotyping further indicated a minor involvement of CYP1B1, CYP2J2 and CYP3A4. Hence, we next determined the intrinsic clearance (CLINT) rate for vemurafenib of these four CYP isoforms. These analyses confirmed a major role of CYP1A1 for the metabolic clearance of vemurafenib. Already 3 min after incubation, more than 90% of the drug were metabolized by CYP1A1. According to the U.S. Food and Drug Administration (FDA), vemurafenib is a substrate of CYP3A4 [9]. Hence, we were surprised to see that with a CLINT of 83.6 μl/min/pmol CYP1A1 was 76-times more efficient in metabolizing vemurafenib than CYP3A4 (CLINT = 1.1 μl/min/pmol) (Fig. 4G). In addition, we found that CYP2J2 (CLINT = 1.5 μl/min/pmol) was capable to metabolize vemurafenib as efficient as CYP3A4. In contrast to the CYP phenotyping (Fig. 4F), we could not confirm a noteworthy contribution of CYP1B1 (CLINT = 0.5 μl/min/pmol) (Fig. 4G). According to our experience, such variations may occur, in particular when analyzing compounds which are sparingly soluble in aqueous systems. The rapid metabolic breakdown of vemurafenib by CYP1A1 was paralleled by the formation of a monohydroxylated and a dihydroxylated metabolite (Fig. 4H). The dihydroxylated vemurafenib metabolite was generated specifically by CYP1A1 but not by CYP2J2 and CYP3A4 (data not shown). Taken together, these data strongly indicate that vemurafenib is an excellent CYP1A1 substrate at low concentrations and on the other hand a CYP1A1 inhibitor at higher concentrations. Hence, vemurafenib probably interferes with the oxidative metabolism of FICZ.
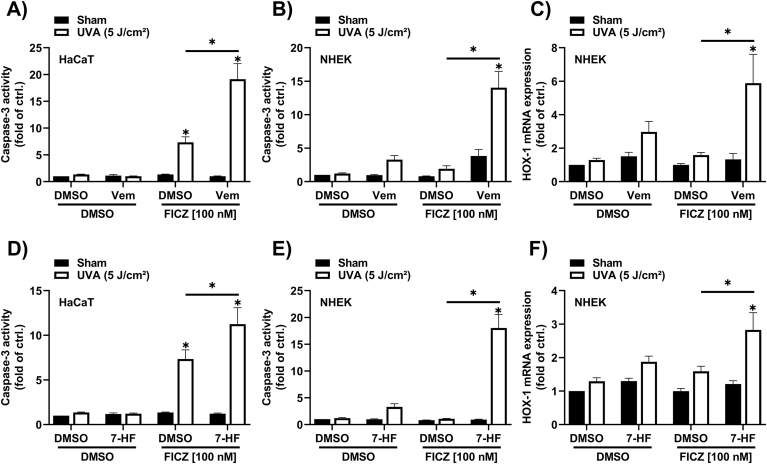
2.5. Vemurafenib enhances FICZ/UVA-induced apoptosis
To assess whether vemurafenib enhances FICZ/UVA-induced apoptosis, keratinocytes were pretreated with 100 nM FICZ for 2 h and 4 h, respectively. Subsequently, cells were exposed to 0 and 5 J/cm2 UVA radiation and immediately treated for 4 h with vemurafenib, 7-HF or solvent. The measurement of caspase-3 activity revealed an increase in the samples treated with FICZ/UVA and vemurafenib (Fig. 5A and B), which was accompanied by a transcriptional induction of the oxidative stress marker HOX-1 (Fig. 5C). Similar results were observed in cells treated with the CYP1A inhibitor 7-HF (Fig. 5D–F), but not in cells treated with the other BRAFi dabrafenib and encorafenib (Supplementary Figs. S2C–E). These results provide evidence that vemurafenib interferes with the metabolic clearance of FICZ and thereby elevates keratinocyte apoptosis in response to UVA irradiation.
Fig. 5.
Vemurafenib stimulates FICZ/UVA-induced caspase-3 activity in keratinocytes.
(A, D) FICZ/UVA-induced caspase-3 activity in HaCaT cells exposed to 40 μM vemurafenib and 10 μM 7-HF. Keratinocytes were pretreated for 4 h with vemurafenib or 7-HF. Next, FICZ was added for another 2 h before cells were irradiated with 5 J/cm2 UVA radiation. Four hours later cells were lyzed for caspase-3 activity assay (n = 6). (B, E) Caspase-3 activity and (C, F) gene expression level of HOX-1 was determined in NHEKs sequentially exposed to vemurafenib or 7-HF (4 h), 100 nM FICZ (4 h) and UVA radiation. Four hours after irradiation, cells were lysed for caspase-3 activity assay (n = 8); for qRT-PCR analysis cells were lyzed after 6 h (n = 8). For statistical analysis a 2-way ANOVA (post-hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to DMSO sham).
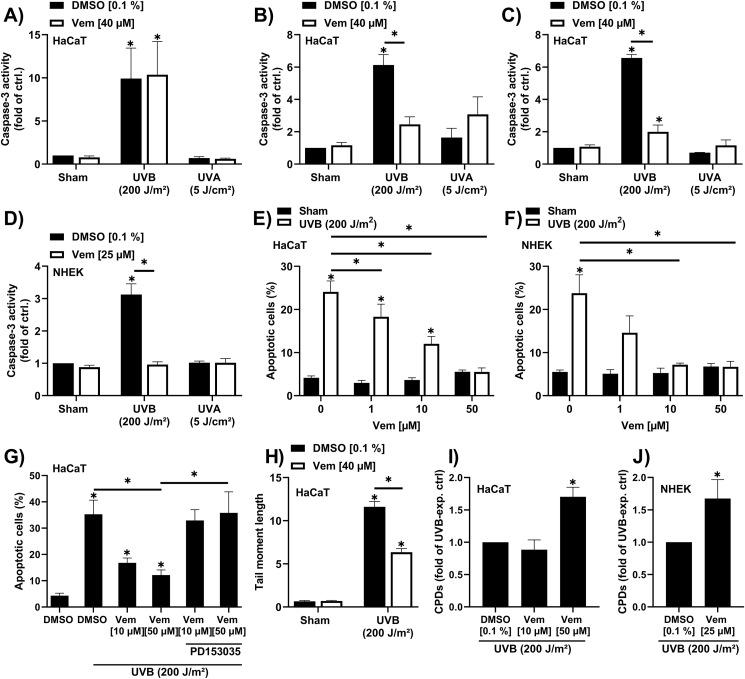
2.6. Vemurafenib inhibits UVB-induced apoptosis and CPD removal independently from phototoxicity
The proapoptotic effect of vemurafenib in FICZ/UVA-exposed keratinocytes is to some extent contradictory to the previously reported anti-apoptotic properties of the BRAFi in UVB-irradiated keratinocytes and cutaneous squamous cell carcinoma (SCC) cells [37]. Thus, we next investigated whether vemurafenib also affects UVB-induced apoptosis in our keratinocyte cultures. Irradiation of HaCaT keratinocytes with 200 J/m2 UVB enhanced caspase-3 activity, whereas an exposure to a low dose of UVA (5 J/cm2) had no effect on apoptosis (Fig. 6A). A 4 h pretreatment with 40 μM vemurafenib did neither affect the UVB-induced caspase-3 activity nor sensitize the cells to UVA-induced apoptosis (Fig. 6A). However, when HaCaT keratinocytes were pretreated for 4 h with vemurafenib, UVB-irradiated through PBS and subsequently exposed to vemurafenib for another 16 h, a pronounced decline in caspase-3 activity was observed (Fig. 6B). Surprisingly, we observed the same decrease in UVB-induced caspase-3 activity in cells that were only treated with vemurafenib post UVB exposure (Fig. 6C), indicating that this anti-apoptotic effect occurred independently from photochemistry. This result was confirmed in NHEKs treated with vemurafenib immediately after UVB exposure (Fig. 6D). Further analyses of the subG1 fraction confirmed a dose-dependent anti-apoptotic effect of vemurafenib when added to UVB-irradiated HaCaT cells and NHEKs (Fig. 6E and F). In melanoma patients, vemurafenib treatment stimulates the EGF receptor (EGFR) [38], which is known to inhibit keratinocyte apoptosis [39]. In fact, a co-exposure of UVB-irradiated HaCaT keratinocytes with the EGFR inhibitor PD153035 reversed the anti-apoptotic effect of vemurafenib (Fig. 6G), suggesting an involvement of the receptor tyrosine kinase. A major driver of UVB-induced apoptosis are DNA double-strand breaks (DSBs) [40]. Comet assay analyses revealed that a treatment of UVB-irradiated HaCaT keratinocytes with vemurafenib signficantly reduced DSB formation (Fig. 6H). Given that UVB-induced DSBs mainly result from collapsing replication forks in dividing CPD-positive keratinocytes [41,42] and vemurafenib reversibly inhibits keratinocyte proliferation (Supplementary Figs. S3A and B), we next investigated the impact of vemurafenib on CPD content. Indeed, treatment of HaCaT keratinocytes and NHEKs with vemurafenib immediately after exposure to 200 J/m2 UVB resulted in an accumulation of CPDs compared to solvent controls (Fig. 6H and I). These data indicate that vemurafenib, when applied to keratinocytes directly after UVB irradiation, inhibits the proliferation-related formation of DSBs and the associated apoptotic clearance of CPD-positive cells.
Fig. 6.
Vemurafenib affects apoptosis and CPD removal independently from phototoxicity.
(A) Caspase-3 activity was determined in HaCaT cells pretreated for 4 h with 40 μM vemurafenib and irradiated with either 200 J/m2 UVB (n = 4), 5 J/cm2 UVA (n = 3) or sham (n = 7). 16 h later caspase-3 activity was measured. (B) HaCaT keratinoyctes were pretreated with 40 μM vemurafenib for 4 h, were irradiated with 200 J/m2 UVB (n = 3), 5 J/cm2 UVA (n = 3) = or sham (n = 6) and post treated with solvent control or vemurafenib (40 μM) before cells were lysed for caspase-3 activity assay. (C, D) Caspase-3 activity measurement in HaCaT keratinocytes (n = 4) and NHEKs (n = 7) irradiated with 200 J/m2 UVB, 5 J/cm2 UVA or sham, prior to a 16 h treatment with the indicated vemurafenib concentrations. (E, F) Percentage of apoptotic HaCaT keratinocytes (n ≥ 4) and NHEKs (n = 4) exposed to 200 J/m2 UVB radiation or sham and a posttreatment of different vemurafenib concentrations for 24 h. (G) Rate of apoptosis was determined in HaCaT keratinocytes unirradiated or irradiated with 200 J/m2 UVB and subsequently treated with vemurafenib for 24 h. In addition, a combined treatment with vemurafenib and PD153035 was conducted for 24 h after UVB radiation or sham treatment (n = 4). (H) DNA-double strand breaks were analyzed in HaCaT cells that were unirradiated or exposed to UVB radiation prior to a treatment with DMSO or vemurafenib for 16 h (n = 3). (I, J) CPD-content was identified in HaCaT keratinocytes (n = 4) and NHEKs (n = 7) irradiated with UVB or sham and post treated with DMSO or the indicated vemurafenib concentrations. For statistical analysis a 2-way ANOVA (post hoc: Tukey test) was performed for all experiments and data are represented as mean ± SEM. (*, p ˂ 0.05 relative to DMSO sham).
3. Discussion
Sensitization of keratinocytes to UVA radiation by the endogenous tryptophan photoproduct FICZ results in the generation of oxidative stress and associated macromolecular damage, which may disrupt skin homeostasis and contribute to extrinsic aging and carcinogenesis [24,26]. Here, we demonstrate that a manipulation of the AHR-CYP1A1 axis in human keratinocytes has a direct effect on the UVA phototoxicity of FICZ and associated apoptosis, a process that might be relevant for drug-induced cutaneous adverse effects. Specifically, we found that an activation of AHR signaling and the associated induction of CYP1A1 activity results in a rapid clearance of FICZ, whereas an inhibition of this axis leads to an accumulation of FICZ and an enhanced susceptibility to UVA-induced apoptosis.
Although FICZ was identified by mass spectrometry in human skin [43], the concentration range of cutaneous FICZ levels is quite enigmatic. However, as UVB irradiation of human volunteers induces the expression of AHR target genes in the skin [44,45] and sulfated FICZ metabolites are present in human urine [21], it is likely that FICZ is formed in sufficient amounts to activate AHR signaling in vivo. Moreover, UVB radiation-independent sources exist, such as skin-colonizing yeasts, which may elevate cutaneous FICZ level [46,47]. We assume that in healthy skin FICZ levels are rather low and even upon transient inhibition of the AHR-CYP1A1 axis may not reach UVA-reactive concentrations. In contrast, a longer-lasting inhibition of CYP1A1 expression or activity may result in an accumulation of pathophysiologically relevant amounts of FICZ. This might be also true for other tryptophan photoproducts with comparable properties. For example, it is well-known that a UVB irradiation of tryptophan leads to the formation of N-formylkynurenine [[48], [49], [50]], which is a micromolar photosensitizer for UVA radiation [49] and a precursor for low-affinity AHR ligands, such as kynurenine and kynurenic acid [51]. However, to the best of our knowledge, N-formylkynurenine is not a substrate of CYP1 isoforms.
We have previously reported that vemurafenib binds to AHR and inhibits downstream CYP1A1 gene expression [31]. Here we demonstrate that vemurafenib is a substrate for CYP1A1 and interferes with the metabolic degradation of FICZ via inhibition. The resulting accumulation of the potent UVA photosensitizer may contribute to the clinically well-documented and frequently occurring phototoxic side effects of a vemurafenib therapy [11,12]. Importantly, exposure of vemurafenib to UVA and UVB radiation in chemico does not lead to ROS formation [52], implying that vemurafenib interacts with cellular molecules and structures in order to unleash its UVA-photosensitizing properties. Given that cancer patients are treated for several month with vemurafenib, it is tempting to speculate that the cutaneous FICZ level reach UVA-photosensitizing concentrations. Whether at a certain point the accumulating FICZ level might be high enough to override the vemurafenib-mediated inhibition of the canonical AHR pathways and to induce CYP1A1 expression is not known. It is tempting to speculate, whether a topical treatment of the affected skin areas with non-toxic high-affinity AHR ligands may help to reestablish CYP1A1 expression and enforce FICZ clearance. An obvious candidate drug is the bacteria-derived AHR agonist tapinarof which has successfully passed clinical testing for the treatment of psoriasis and atopic dermatitis [53,54].
According to the Zelboraf® approval summary of the FDA [9], vemurafenib is primarily metabolized by CYP3A4. This stands in stark contrast to the results of our in vitro metabolism studies, identifying CYP1A1 as being ~76-times more efficient in clearing vemurafenib than CYP3A4. This discrepancy might be due to the fact that the majority of metabolism studies conducted by the pharmaceutical industry encompasses CYP1A2, but not CYP1A1. The same might be true for CYP2J2, which exhibits a comparable intrinsic clearance rate for vemurafenib as CYP3A4. Along the same line, neither CYP1A1 nor CYP2J2 are part of the CYP panel recommended by the guidance documents for the assessment of drug interactions from the FDA [55] and the European Medicines Agency [56]. Interestingly, a co-application of the CYP3A4 inhibitor itraconazole had only moderate effects on the steady state level of vemurafenib in patients [57], and both the expression of hepatic CYP1A1 and the pharmacokinetics of vemurafenib exhibit a remarkable interindividual variability [58,59]. However, in combination with our previous findings [31], our current data imply that by interfering with AHR and CYP1A1, vemurafenib does not only enforce its own accumulation, but also interferes with the metabolism of other CYP1A1 substrates, in particular co-applied drugs. In fact, CYP1A1 is involved in the metabolism of various clinical drugs, including the antiemetic granisetron [58], the antihypertensive drug riociguat [58], and the anticancer drugs dacarbazine [60], erlotinib [61] and imiquimod [28].
A detrimental consequence of the UVA photosensitization by vemurafenib is the oxidative stress-related disturbance of nucleotide excision repair [62,63]. Our data, however, clearly show that vemurafenib inhibits the apoptotic clearance of CPD-positive keratinocytes independently from phototoxic stress. Given that UVB-induced keratinocyte apoptosis is mainly triggered by DNA double-strand breaks, which occur when CPD-positive cells start to replicate their genetic material, an inhibition of cell proliferation might be causative for the observed anti-apoptotic effects. The vemurafenib-mediated inhibition of proliferation was reversible by washing out the drug which may resemble declining drug levels in patients. However, in concert with the accumulated CPDs, this process may not only cause an elevated apoptotic response but also increase the risk for mutagenesis and, possibly, skin carcinogenesis. In fact, the development of SCC is a common adverse side effect of vemurafenib monotherapy that occurs in 19%–26% of the treated patients [11]. Unlike UV-induced photocarcinogenesis, vemurafenib therapy-associated SCCs develop rapidly, within the first three month of treatment, and do not only occur in highly sun-exposed but also in low sun-exposed areas of the skin [11,64]. The pathogenesis of these SCCs may involve a stimulation of mitogenic MEK/ERK signal transduction in BRAF wild-type keratinocytes which probably promotes the proliferation of latent mutant keratinocytes present in the epidermis [11]. A co-treatment with a MEK inhibitor (i.e. the current clinical routine for treating advanced melanoma) does not completely abrogate the development of vemurafenib-associated SCC [11,65], supporting the idea that additional pathomechanisms exist. Results from in vitro studies on human keratinocytes indicated that the activation of mitogenic MEK/ERK signaling occurred only in response to low doses of vemurafenib, i.e. 2 μM or less [66]. We assume that at higher drug concentrations, vemurafenib may promote tumor development by protecting mutated keratinocytes against apoptosis and enhancing genomic instability. A further elucidation of the mechanisms is particularly relevant for patients with Erdheim-Chester disease, which are still treated with vemurafenib and frequently suffer from secondary skin cancers [10].
We have previously reported that an inhibition of AHR signaling attenuates UVB-induced skin carcinogenesis in mice [67]. Specifically, AHR represses nucleotide excision repair and apoptosis in UVB-irradiated keratinocytes [67,68]. Accordingly, AHR inhibition accelerated the removal of DNA-damaged keratinocytes by enforcing DSB-triggered apoptosis [67]. As we have previously identified vemurafenib as an AHR antagonist [31], the anti-apoptotic effect of the BRAFi observed in UVB-exposed keratinocytes was unexpected and urgently requires further investigation. In accordance with literature [38,69], our data suggest that a stimulation of EGFR may be causative for the anti-apoptotic property of vemurafenib.
A limitation of our study is the applied concentration range of vemurafenib. The median maximum plasma concentration at steady state determined in melanoma patients receiving 960 mg vemurafenib b.i.d. is 56.7 ± 21.8 μg/ml, which corresponds to 115.7 ± 44.5 μM [59]. As mentioned above, the authors noted a high interindividual pharmacokinetic variability with vemurafenib plasma concentrations ranging from 13 μg/ml (26.5 μM) to 109.8 μg/ml (224.1 μM) [59]. With more than 99%, vemurafenib exhibits a very high plasma protein binding [9], indicating that the concentrations used in this study exceeded clinically relevant concentrations, i.e. the free fraction. However, it should not be underestimated that also during in vitro testing, considerable amounts of vemurafenib will bind to albumin and other proteins present in FBS-supplemented culture medium or even to cell culture plastics [70]. The HIV-1 protease inhibitor lopinavir, for instance, exhibits a comparably high plasma protein binding of 98.9%. A determination of the free fraction of lopinavir in culture medium supplemented with 10% FBS revealed that 96.1% of the drug was bound to serum proteins [71]. However, the potential clinical relevance of the identified mechanisms for the phototoxicity of vemurafenib is strengthened by the fact that a treatment of keratinocytes with dabrafenib and encorafenib neither affected FICZ-induced CYP1A1 enzyme activity nor enhanced FICZ/UVA-induced apoptosis. Importantly, a monotherapy with these BRAFi is less often (≤5%) associated with an elevated UVA photosensitivity than vemurafenib [65].
Taken together, a modulation of the CYP1A1-catalyzed metabolism of the endogenous photosensitizer FICZ might be a relevant molecular mechanism responsible for the UVA phototoxicity of vemurafenib. Moreover, vemurafenib treatment inhibited the apoptotic clearance of UVB-irradiated CPD-positive keratinocytes, a mechanism which might contribute to therapy-associated skin carcinogenesis. As vemurafenib is currently in clinical testing for other BRAF V600-mutant malignancies, for instance in combination with rituximab for the treatment of hairy-cell leukemia [72] and as single agent for the treatment of non-small-cell lung cancer [73], a further elucidation of the identified molecular mechanisms is required.
4. Material and methods
4.1. Cell culture, UV irradiation and treatment
HaCaT keratinocytes were cultured in DMEM (PAN Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (PAN Biotech) and 1% antibiotics/antimycotics (PAN Biotech). The generation of HaCaT-EV and HaCaT-shAHR keratinocytes has been previously described [16]. The culture medium of HaCaT-EV and HaCaT-shAHR keratinocytes was supplemented with G418 (Carl Roth, Karlsruhe, Germany). Normal human epidermal keratinocytes were obtained from PromoCell (Heidelberg, Germany) and cultured in Keratinocyte Growth Medium 2 (PromoCell). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. UV exposure of cells was carried out by using the BS-02 irradiation chamber (Opsytec Dr. Gröbel, Ettlingen Germany) equipped with individually controllable UVA Actinic and UVB bulbs and respective sensors. For both, UV and sham irradiation, cell culture medium was replaced by PBS. For cell treatment, 6-formylindolo[3,2-b]carbazole (Biomol, Hamburg, Germany), tapinarof, dabrafenib, encorafenib (MedChem Express, Monmouth Junction, NJ, USA), 3’-methoxy-4′-nitroflavone (a kind gift from I. Meyer, Symrise AG, Holzminden, Germany), 7-hydroxyflavone (Sigma-Aldrich, Munich, Germany) and vemurafenib (Selleck Chemicals, Houston, TX, USA) were dissolved in dimethyl sulfoxide.
4.2. Caspase-3 activity assay
Caspase-3 activity was determined by using the Caspase-3 Fluorometric assay Kit (PromoCell) according to the manufacturer's instructions.
4.3. Nicoletti assay
The analysis of DNA content via Nicoletti staining and flow cytometry was carried out as described previously [68].
4.4. 7-Ethoxyresorufin deethylase (EROD) assay
Measurement of EROD activity in living keratinocyte monolayers was carried out as described previously [74].
4.5. HPLC analysis
To determine FICZ concentration in cell culture medium, 600 μl medium were removed and centrifuged for 5 min at 21.000×g. Subsequently, 500 μl of the supernatant was taken and mixed with 500 μl of acetonitrile before samples were analyzed by Reverse Phase-HPLC (Shimadzu HPLC system interfaced with the LabSolution software). FICZ was injected onto a Kromasil Eternity C18 column (length = 250 nm, internal diameter = 4 mm, particle size = 5 μm) at 45 °C. The aqueous mobile phase consisted of 0.1% formic acid and the organic phase was acetonitrile with 0.1% acid formic. The flow rate was 1 ml/min. FICZ was eluted by the following discontinuous gradient at a flow rate of 1 ml/min: the initial concentration of 50% in solvent B increase to 80% over 15 min and this was followed by a decrease to 50% over the next minute, and the initial conditions were then maintained for 9 min. The FICZ was monitored by fluorescence emission (at 525 nm) after excitation at 390 nm. The FICZ was quantified by integration of the peak fluorescence area, employing a respective calibration curve.
4.6. Quantitative real-time PCR
Isolation of total RNA, cDNA synthesis and quantitative real-time PCR was carried out as described previously [74]. Oligonucleotides used in this work were: 5′-CCCCAGGCACCAGGGCGTGAT-3′ and 5′-GGTCATCTTCTCGCGGTTGGCCTTGGGGT-3′ for β-actin, and 5′-GCCATGAACTTTGTCCGGTG-3′ and 5′-GGATGTGCTTTTCGTTGGGG– 3′ for HOX-1.
4.7. ROS measurement
ROS production was measured by using the fluorogenic dye MitoSox (Red Mitochondiral Superoxide Indicator, Thermo Fisher Scientific) and a BD FACS Canto II flow cytometer (BD Biosciences, San Jose, CA, USA). Data analysis was performed using the FlowJo software (Tree Star).
4.8. Incubation of insect microsomes with vemurafenib
Microsomal preparations of 22 heterologously expressed human CYP isoforms were incubated with vemurafenib and prepared for subsequent LC-MS analysis as previously described [28].
4.9. LC–MS analysis
Incubations were analyzed using reversed-phase HPLC with gradient elution using 10 mM ammonium formate and acetonitrile containing 0.1% formic acid as solvents. The HPLC was coupled to a high-resolution mass spectrometer Q-Exactive® (Thermo Scientific, Bremen Germany) operated in full-scan mode. Selectivity of the analytes was gained by extracting a very narrow mass range in the order of 10–20 ppm of the exact mass of the analyte.
4.10. Cell viability assay
Cell viability was analyzed by using CellTiter-Blue Cell Viability Assay (Promega, Mannheim, Germany) according to the manufacturer's instructions.
4.11. Cell proliferation assay
Cell division was analyzed using the CellTrace Cell Proliferation Kit (Invitrogen, Carlsbad, CA, USA) and a FACS Canto II Flow Cytometer (BD, Franklin Lakes, NJ, USA).
4.12. Comet assay
The analysis of DNA double-strand breaks via Comet assay was carried out as described previously [67].
4.13. Southwestern slot–blot analysis
Isolation of DNA and Southwestern slot–blot-based quantification of CPDs was conducted as described previously [67]. The anti-CPD monoclonal antibody (clone TDM-2) was purchased from Cosmo Bio (Carlsbad, CA, USA).
4.14. Statistical analysis
All experiments were performed independently at least three times unless otherwise noted. Data are presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using Graph Pad Prism 8.4.2 (GraphPad Prism Software, California, USA). A one-factor analysis of variance (ANOVA) was performed to compare means of more than two samples. In the case of multiple factor dependence, a two-factor ANOVA was performed. A value of p ≤ 0.05 was considered statistically significant.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgment
We thank Petra Boukamp and Ellen Fritsche for providing cell-lines and Imke Meyer for generously donating MNF. We thank Volker Lambertz and Frederick Hartung for technical support. VG was supported by the EU program Erasmus+. This work was supported by grants of the Jürgen Manchot Foundation (to KMR and NCS).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102110.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Young A.R., Claveau J., Rossi A.B. Ultraviolet radiation and the skin: photobiology and sunscreen photoprotection. J. Am. Acad. Dermatol. 2017;76(3S1):S100–S109. doi: 10.1016/j.jaad.2016.09.038. [DOI] [PubMed] [Google Scholar]
- 2.Bernard J.J., Gallo R.L., Krutmann J. Photoimmunology: how ultraviolet radiation affects the immune system. Nat. Rev. Immunol. 2019;19(11):688–701. doi: 10.1038/s41577-019-0185-9. [DOI] [PubMed] [Google Scholar]
- 3.Krutmann J., Morita A., Chung J.H. Sun exposure: what molecular photodermatology tells us about its good and bad sides. J. Invest. Dermatol. 2012;132(3 Pt 2):976–984. doi: 10.1038/jid.2011.394. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J., Douki T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018;17(12):1816–1841. doi: 10.1039/c7pp00395a. [DOI] [PubMed] [Google Scholar]
- 5.Wondrak G.T., Jacobson M.K., Jacobson E.L. Endogenous UVA-photosensitizers: mediators of skin photodamage and novel targets for skin photoprotection. Photochem. Photobiol. Sci. 2006;5(2):215–237. doi: 10.1039/b504573h. [DOI] [PubMed] [Google Scholar]
- 6.Blakely K.M., Drucker A.M., Rosen C.F. Drug-induced photosensitivity-an update: culprit drugs, prevention and management. Drug Saf. 2019;42(7):827–847. doi: 10.1007/s40264-019-00806-5. [DOI] [PubMed] [Google Scholar]
- 7.Lembo S., Raimondo A., Conti V., Venturini M. Photosensitivity and cancer immune-targeted therapies. Photodermatol. Photoimmunol. Photomed. 2020;36(3):172–178. doi: 10.1111/phpp.12533. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O'Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim G., McKee A.E., Ning Y.M., Hazarika M., Theoret M., Johnson J.R., Xu Q.C., Tang S., Sridhara R., Jiang X., He K., Roscoe D., McGuinn W.D., Helms W.S., Russell A.M., Miksinski S.P., Zirkelbach J.F., Earp J., Liu Q., Ibrahim A., Justice R., Pazdur R. FDA approval summary: vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin. Canc. Res. 2014;20(19):4994–5000. doi: 10.1158/1078-0432.CCR-14-0776. [DOI] [PubMed] [Google Scholar]
- 10.Oneal P.A., Kwitkowski V., Luo L., Shen Y.L., Subramaniam S., Shord S., Goldberg K.B., McKee A.E., Kaminskas E., Farrell A., Pazdur R. FDA approval summary: vemurafenib for the treatment of patients with erdheim-chester disease with the BRAFV600 mutation. Oncol. 2018;23(12):1520–1524. doi: 10.1634/theoncologist.2018-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacouture M.E., Duvic M., Hauschild A., Prieto V.G., Robert C., Schadendorf D., Kim C.C., McCormack C.J., Myskowski P.L., Spleiss O., Trunzer K., Su F., Nelson B., Nolop K.B., Grippo J.F., Lee R.J., Klimek M.J., Troy J.L., Joe A.K. Analysis of dermatologic events in vemurafenib-treated patients with melanoma. Oncol. 2013;18(3):314–322. doi: 10.1634/theoncologist.2012-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dummer R., Rinderknecht J., Goldinger S.M. Ultraviolet A and photosensitivity during vemurafenib therapy. N. Engl. J. Med. 2012;366(5):480–481. doi: 10.1056/NEJMc1113752. [DOI] [PubMed] [Google Scholar]
- 13.Rannug A., Rannug U., Rosenkranz H.S., Winqvist L., Westerholm R., Agurell E., Grafstrom A.K. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 1987;262(32):15422–15427. [PubMed] [Google Scholar]
- 14.Helferich W.G., Denison M.S. Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol. Pharmacol. 1991;40(5):674–678. [PubMed] [Google Scholar]
- 15.Rannug U., Rannug A., Sjoberg U., Li H., Westerholm R., Bergman J. Structure elucidation of two tryptophan-derived, high affinity Ah receptor ligands. Chem. Biol. 1995;2(12):841–845. doi: 10.1016/1074-5521(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 16.Fritsche E., Schafer C., Calles C., Bernsmann T., Bernshausen T., Wurm M., Hubenthal U., Cline J.E., Hajimiragha H., Schroeder P., Klotz L.O., Rannug A., Furst P., Hanenberg H., Abel J., Krutmann J. Lightning up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc. Natl. Acad. Sci. U.S.A. 2007;104(21):8851–8856. doi: 10.1073/pnas.0701764104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel C.F.A., Van Winkle L.S., Esser C., Haarmann-Stemmann T. The aryl hydrocarbon receptor as a target of environmental stressors - implications for pollution mediated stress and inflammatory responses. Redox Biol. 2020:101530. doi: 10.1016/j.redox.2020.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avilla M.N., Malecki K.M.C., Hahn M.E., Wilson R.H., Bradfield C.A. The ah receptor: adaptive metabolism, ligand diversity, and the xenokine model. Chem. Res. Toxicol. 2020;33(4):860–879. doi: 10.1021/acs.chemrestox.9b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothhammer V., Quintana F.J. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 2019;19(3):184–197. doi: 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- 20.Bergander L., Wincent E., Rannug A., Foroozesh M., Alworth W., Rannug U. Metabolic fate of the Ah receptor ligand 6-formylindolo[3,2-b]carbazole. Chem. Biol. Interact. 2004;149(2–3):151–164. doi: 10.1016/j.cbi.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Wincent E., Amini N., Luecke S., Glatt H., Bergman J., Crescenzi C., Rannug A., Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J. Biol. Chem. 2009;284(5):2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 22.Wei Y.D., Bergander L., Rannug U., Rannug A. Regulation of CYP1A1 transcription via the metabolism of the tryptophan-derived 6-formylindolo[3,2-b]carbazole. Arch. Biochem. Biophys. 2000;383(1):99–107. doi: 10.1006/abbi.2000.2037. [DOI] [PubMed] [Google Scholar]
- 23.Vogeley C., Esser C., Tuting T., Krutmann J., Haarmann-Stemmann T. Role of the aryl hydrocarbon receptor in environmentally induced skin aging and skin carcinogenesis. Int. J. Mol. Sci. 2019;20(23) doi: 10.3390/ijms20236005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park S.L., Justiniano R., Williams J.D., Cabello C.M., Qiao S., Wondrak G.T. The tryptophan-derived endogenous aryl hydrocarbon receptor ligand 6-Formylindolo[3,2-b]Carbazole is a nanomolar UVA photosensitizer in epidermal keratinocytes. J. Invest. Dermatol. 2015;135(6):1649–1658. doi: 10.1038/jid.2014.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justiniano R., de Faria Lopes L., Perer J., Hua A., Park S.L., Jandova J., Baptista M.S., Wondrak G.T. The endogenous tryptophan-derived photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) is a nanomolar photosensitizer that can be harnessed for the photodynamic elimination of skin cancer cells in vitro and in vivo. Photochem. Photobiol. 2021;97(1):180–191. doi: 10.1111/php.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brem R., Macpherson P., Guven M., Karran P. Oxidative stress induced by UVA photoactivation of the tryptophan UVB photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) inhibits nucleotide excision repair in human cells. Sci. Rep. 2017;7(1):4310. doi: 10.1038/s41598-017-04614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S.H., Jayawickreme C., Rickard D.J., Nicodeme E., Bui T., Simmons C., Coquery C.M., Neil J., Pryor W.M., Mayhew D., Rajpal D.K., Creech K., Furst S., Lee J., Wu D., Rastinejad F., Willson T.M., Viviani F., Morris D.C., Moore J.T., Cote-Sierra J. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J. Invest. Dermatol. 2017;137(10):2110–2119. doi: 10.1016/j.jid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Mescher M., Tigges J., Rolfes K.M., Shen A.L., Yee J.S., Vogeley C., Krutmann J., Bradfield C.A., Lang D., Haarmann-Stemmann T. The Toll-like receptor agonist imiquimod is metabolized by aryl hydrocarbon receptor-regulated cytochrome P450 enzymes in human keratinocytes and mouse liver. Arch. Toxicol. 2019;93(7):1917–1926. doi: 10.1007/s00204-019-02488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron J.M., Holler D., Schiffer R., Frankenberg S., Neis M., Merk H.F., Jugert F.K. Expression of multiple cytochrome p450 enzymes and multidrug resistance-associated transport proteins in human skin keratinocytes. J. Invest. Dermatol. 2001;116(4):541–548. doi: 10.1046/j.1523-1747.2001.01298.x. [DOI] [PubMed] [Google Scholar]
- 30.Keyse S.M., Tyrrell R.M. Heme oxygenase is the major 32-kDa stress protein induced in human skin fibroblasts by UVA radiation, hydrogen peroxide, and sodium arsenite. Proc. Natl. Acad. Sci. U. S. A. 1989;86(1):99–103. doi: 10.1073/pnas.86.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawerkamp H.C., Kislat A., Gerber P.A., Pollet M., Rolfes K.M., Soshilov A.A., Denison M.S., Momin A.A., Arold S.T., Datsi A., Braun S.A., Olah P., Lacouture M.E., Krutmann J., Haarmann-Stemmann T., Homey B., Meller S. Vemurafenib acts as an aryl hydrocarbon receptor antagonist: implications for inflammatory cutaneous adverse events. Allergy. 2019;74(12):2437–2448. doi: 10.1111/all.13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corre S., Tardif N., Mouchet N., Leclair H.M., Boussemart L., Gautron A., Bachelot L., Perrot A., Soshilov A., Rogiers A., Rambow F., Dumontet E., Tarte K., Bessede A., Guillemin G.J., Marine J.C., Denison M.S., Gilot D., Galibert M.D. Sustained activation of the Aryl hydrocarbon Receptor transcription factor promotes resistance to BRAF-inhibitors in melanoma. Nat. Commun. 2018;9(1):4775. doi: 10.1038/s41467-018-06951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y.F., Santostefano M., Cunningham B.D., Threadgill M.D., Safe S. Identification of 3'-methoxy-4'-nitroflavone as a pure aryl hydrocarbon (Ah) receptor antagonist and evidence for more than one form of the nuclear Ah receptor in MCF-7 human breast cancer cells. Arch. Biochem. Biophys. 1995;316(1):470–477. doi: 10.1006/abbi.1995.1062. [DOI] [PubMed] [Google Scholar]
- 34.Zhai S., Dai R., Friedman F.K., Vestal R.E. Comparative inhibition of human cytochromes P450 1A1 and 1A2 by flavonoids. Drug Metab. Dispos. 1998;26(10):989–992. [PubMed] [Google Scholar]
- 35.Falchook G.S., Long G.V., Kurzrock R., Kim K.B., Arkenau H.T., Brown M.P., Hamid O., Infante J.R., Millward M., Pavlick A., Chin M.T., O'Day S.J., Blackman S.C., Curtis C.M., Lebowitz P., Ma B., Ouellet D., Kefford R.F. Dose selection, pharmacokinetics, and pharmacodynamics of BRAF inhibitor dabrafenib (GSK2118436) Clin. Canc. Res. 2014;20(17):4449–4458. doi: 10.1158/1078-0432.CCR-14-0887. [DOI] [PubMed] [Google Scholar]
- 36.Delord J.P., Robert C., Nyakas M., McArthur G.A., Kudchakar R., Mahipal A., Yamada Y., Sullivan R., Arance A., Kefford R.F., Carlino M.S., Hidalgo M., Gomez-Roca C., Michel D., Seroutou A., Aslanis V., Caponigro G., Stuart D.D., Moutouh-de Parseval L., Demuth T., Dummer R. Phase I dose-escalation and -expansion study of the BRAF inhibitor encorafenib (LGX818) in metastatic BRAF-mutant melanoma. Clin. Canc. Res. 2017;23(18):5339–5348. doi: 10.1158/1078-0432.CCR-16-2923. [DOI] [PubMed] [Google Scholar]
- 37.Vin H., Ojeda S.S., Ching G., Leung M.L., Chitsazzadeh V., Dwyer D.W., Adelmann C.H., Restrepo M., Richards K.N., Stewart L.R., Du L., Ferguson S.B., Chakravarti D., Ehrenreiter K., Baccarini M., Ruggieri R., Curry J.L., Kim K.B., Ciurea A.M., Duvic M., Prieto V.G., Ullrich S.E., Dalby K.N., Flores E.R., Tsai K.Y. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. Elife. 2013;2 doi: 10.7554/eLife.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girotti M.R., Pedersen M., Sanchez-Laorden B., Viros A., Turajlic S., Niculescu-Duvaz D., Zambon A., Sinclair J., Hayes A., Gore M., Lorigan P., Springer C., Larkin J., Jorgensen C., Marais R. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Canc. Discov. 2013;3(2):158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastore S., Mascia F., Mariani V., Girolomoni G. The epidermal growth factor receptor system in skin repair and inflammation. J. Invest. Dermatol. 2008;128(6):1365–1374. doi: 10.1038/sj.jid.5701184. [DOI] [PubMed] [Google Scholar]
- 40.Roos W.P., Thomas A.D., Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Canc. 2016;16(1):20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 41.Garinis G.A., Mitchell J.R., Moorhouse M.J., Hanada K., de W.H., Vandeputte D., Jans J., Brand K., Smid M., van der Spek P.J., Hoeijmakers J.H., Kanaar R., van der Horst G.T. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005;24(22):3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunkern T.R., Kaina B. Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol. Biol. Cell. 2002;13(1):348–361. doi: 10.1091/mbc.01-05-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schallreuter K.U., Salem M.A., Gibbons N.C., Maitland D.J., Marsch E., Elwary S.M., Healey A.R. Blunted epidermal L-tryptophan metabolism in vitiligo affects immune response and ROS scavenging by Fenton chemistry, part 2: epidermal H2O2/ONOO(-)-mediated stress in vitiligo hampers indoleamine 2,3-dioxygenase and aryl hydrocarbon receptor-mediated immune response signaling. Faseb. J. 2012;26(6):2471–2485. doi: 10.1096/fj.11-201897. [DOI] [PubMed] [Google Scholar]
- 44.Tigges J., Haarmann-Stemmann T., Vogel C.F., Grindel A., Hubenthal U., Brenden H., Grether-Beck S., Vielhaber G., Johncock W., Krutmann J., Fritsche E. The new aryl hydrocarbon receptor antagonist E/Z-2-benzylindene-5,6-dimethoxy-3,3-dimethylindan-1-one protects against UVB-induced signal transduction. J. Invest. Dermatol. 2014;134(2):556–559. doi: 10.1038/jid.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katiyar S.K., Matsui M.S., Mukhtar H. Ultraviolet-B exposure of human skin induces cytochromes P450 1A1 and 1B1. J. Invest. Dermatol. 2000;114(2):328–333. doi: 10.1046/j.1523-1747.2000.00876.x. [DOI] [PubMed] [Google Scholar]
- 46.Magiatis P., Pappas P., Gaitanis G., Mexia N., Melliou E., Galanou M., Vlachos C., Stathopoulou K., Skaltsounis A.L., Marselos M., Velegraki A., Denison M.S., Bassukas I.D. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J. Invest. Dermatol. 2013;133(8):2023–2030. doi: 10.1038/jid.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smirnova A., Wincent E., Vikstrom Bergander L., Alsberg T., Bergman J., Rannug A., Rannug U. Evidence for new light-independent pathways for generation of the endogenous aryl hydrocarbon receptor agonist FICZ. Chem. Res. Toxicol. 2016;29(1):75–86. doi: 10.1021/acs.chemrestox.5b00416. [DOI] [PubMed] [Google Scholar]
- 48.Asquith R.S., Rivett D.E. Studies on the photooxidation of tryptophan. Biochim. Biophys. Acta. 1971;252(1):111–116. doi: 10.1016/0304-4165(71)90098-5. [DOI] [PubMed] [Google Scholar]
- 49.Walrant P., Santus R., Grossweiner L.I. Photosensitizing properties of N-formylkynurenine. Photochem. Photobiol. 1975;22(1–2):63–65. doi: 10.1111/j.1751-1097.1975.tb06723.x. [DOI] [PubMed] [Google Scholar]
- 50.Youssef A., von Koschembahr A., Caillat S., Corre S., Galibert M.D., Douki T. 6-Formylindolo[3,2-b]carbazole (FICZ) is a very minor photoproduct of tryptophan at biologically relevant doses of UVB and simulated sunlight. Photochem. Photobiol. 2019;95(1):237–243. doi: 10.1111/php.12950. [DOI] [PubMed] [Google Scholar]
- 51.Platten M., Nollen E.A.A., Rohrig U.F., Fallarino F., Opitz C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019;18(5):379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 52.Heppt M.V., Clanner-Engelshofen B.M., Marsela E., Wessely A., Kammerbauer C., Przybilla B., French L.E., Berking C., Reinholz M. Comparative analysis of the phototoxicity induced by BRAF inhibitors and alleviation through antioxidants. Photodermatol. Photoimmunol. Photomed. 2020;36(2):126–134. doi: 10.1111/phpp.12520. [DOI] [PubMed] [Google Scholar]
- 53.Stein Gold L., Bhatia N., Tallman A.M., Rubenstein D.S. A phase IIb, randomized clinical trial of tapinarof cream for the treatment of plaque psoriasis: secondary efficacy and patient-reported outcomes. J. Am. Acad. Dermatol. 2021;84(3):624–631. doi: 10.1016/j.jaad.2020.04.181. [DOI] [PubMed] [Google Scholar]
- 54.Paller A.S., Gold L.S., Soung J., Tallman A.M., Rubenstein D.S., Gooderham M. Efficacy and patient-reported outcomes from a phase IIb, randomized clinical trial of tapinarof cream for the treatment of adolescents and adults with atopic dermatitis. J. Am. Acad. Dermatol. 2021;84(3):632–638. doi: 10.1016/j.jaad.2020.05.135. [DOI] [PubMed] [Google Scholar]
- 55.U.S. Food and Drug Administration In vitro drug interaction studies — cytochrome P450 enzyme- and transporter-mediated drug interactions guidance for industry. 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions
- 56.European Medicines Agency . 2012. Guideline on the Investigation of Drug Interactions.https://www.ema.europa.eu/en/investigation-drug-interactions [Google Scholar]
- 57.Zhang W., Mathisen M., Goodman G.R., Forbes H., Song Y., Bertran E., Demidov L., Shin S.J. Effect of itraconazole, a potent CYP3A4 inhibitor, on the steady-state pharmacokinetics of vemurafenib in patients with BRAF(V600) mutation-positive malignancies. Clin. Pharmacol. Drug Dev. 2021;10(1):39–45. doi: 10.1002/cpdd.822. [DOI] [PubMed] [Google Scholar]
- 58.Lang D., Radtke M., Bairlein M. Highly variable expression of CYP1A1 in human liver and impact on pharmacokinetics of riociguat and granisetron in humans. Chem. Res. Toxicol. 2019;32(6):1115–1122. doi: 10.1021/acs.chemrestox.8b00413. [DOI] [PubMed] [Google Scholar]
- 59.Funck-Brentano E., Alvarez J.C., Longvert C., Abe E., Beauchet A., Funck-Brentano C., Saiag P. Plasma vemurafenib concentrations in advanced BRAFV600mut melanoma patients: impact on tumour response and tolerance. Ann. Oncol. 2015;26(7):1470–1475. doi: 10.1093/annonc/mdv189. [DOI] [PubMed] [Google Scholar]
- 60.Reid J.M., Kuffel M.J., Miller J.K., Rios R., Ames M.M. Metabolic activation of dacarbazine by human cytochromes P450: the role of CYP1A1, CYP1A2, and CYP2E1. Clin. Canc. Res. 1999;5(8):2192–2197. [PubMed] [Google Scholar]
- 61.Li J., Zhao M., He P., Hidalgo M., Baker S.D. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin. Canc. Res. 2007;13(12):3731–3737. doi: 10.1158/1078-0432.CCR-07-0088. [DOI] [PubMed] [Google Scholar]
- 62.Peacock M., Brem R., Macpherson P., Karran P. DNA repair inhibition by UVA photoactivated fluoroquinolones and vemurafenib. Nucleic Acids Res. 2014;42(22):13714–13722. doi: 10.1093/nar/gku1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimeswenger S., Mann U., Hoeller C., Foedinger D., Jantschitsch C. Vemurafenib impairs the repair of ultraviolet radiation-induced DNA damage. Melanoma Res. 2019;29(2):134–144. doi: 10.1097/CMR.0000000000000546. [DOI] [PubMed] [Google Scholar]
- 64.Anforth R., Menzies A., Byth K., Carlos G., Chou S., Sharma R., Scolyer R.A., Kefford R., Long G.V., Fernandez-Penas P. Factors influencing the development of cutaneous squamous cell carcinoma in patients on BRAF inhibitor therapy. J. Am. Acad. Dermatol. 2015;72(5):809–815 e1. doi: 10.1016/j.jaad.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 65.Daud A., Tsai K. Management of treatment-related adverse events with agents targeting the MAPK pathway in patients with metastatic melanoma. Oncol. 2017;22(7):823–833. doi: 10.1634/theoncologist.2016-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roh M.R., Kim J.M., Lee S.H., Jang H.S., Park K.H., Chung K.Y., Rha S.Y. Low-concentration vemurafenib induces the proliferation and invasion of human HaCaT keratinocytes through mitogen-activated protein kinase pathway activation. J. Dermatol. 2015;42(9):881–888. doi: 10.1111/1346-8138.12950. [DOI] [PubMed] [Google Scholar]
- 67.Pollet M., Shaik S., Mescher M., Frauenstein K., Tigges J., Braun S.A., Sondenheimer K., Kaveh M., Bruhs A., Meller S., Homey B., Schwarz A., Esser C., Douki T., Vogel C.F.A., Krutmann J., Haarmann-Stemmann T. The AHR represses nucleotide excision repair and apoptosis and contributes to UV-induced skin carcinogenesis. Cell Death Differ. 2018;25(10):1823–1836. doi: 10.1038/s41418-018-0160-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frauenstein K., Sydlik U., Tigges J., Majora M., Wiek C., Hanenberg H., Abel J., Esser C., Fritsche E., Krutmann J., Haarmann-Stemmann T. Evidence for a novel anti-apoptotic pathway in human keratinocytes involving the aryl hydrocarbon receptor, E2F1, and checkpoint kinase 1. Cell Death Differ. 2013;20(10):1425–1434. doi: 10.1038/cdd.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gross A., Niemetz-Rahn A., Nonnenmacher A., Tucholski J., Keilholz U., Fusi A. Expression and activity of EGFR in human cutaneous melanoma cell lines and influence of vemurafenib on the EGFR pathway. Targeted Oncol. 2015;10(1):77–84. doi: 10.1007/s11523-014-0318-9. [DOI] [PubMed] [Google Scholar]
- 70.Kisitu J., Hollert H., Fisher C., Leist M. Chemical concentrations in cell culture compartments (C5) - free concentrations. ALTEX. 2020;37(4):693–708. doi: 10.14573/altex.2008251. [DOI] [PubMed] [Google Scholar]
- 71.Hickman D., Vasavanonda S., Nequist G., Colletti L., Kati W.M., Bertz R., Hsu A., Kempf D.J. Estimation of serum-free 50-percent inhibitory concentrations for human immunodeficiency virus protease inhibitors lopinavir and ritonavir. Antimicrob. Agents Chemother. 2004;48(8):2911–2917. doi: 10.1128/AAC.48.8.2911-2917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tiacci E., De Carolis L., Simonetti E., Capponi M., Ambrosetti A., Lucia E., Antolino A., Pulsoni A., Ferrari S., Zinzani P.L., Ascani S., Perriello V.M., Rigacci L., Gaidano G., Della Seta R., Frattarelli N., Falcucci P., Foa R., Visani G., Zaja F., Falini B. Vemurafenib plus rituximab in refractory or relapsed hairy-cell leukemia. N. Engl. J. Med. 2021;384(19):1810–1823. doi: 10.1056/NEJMoa2031298. [DOI] [PubMed] [Google Scholar]
- 73.Mazieres J., Cropet C., Montane L., Barlesi F., Souquet P.J., Quantin X., Dubos-Arvis C., Otto J., Favier L., Avrillon V., Cadranel J., Moro-Sibilot D., Monnet I., Westeel V., Le Treut J., Brain E., Tredaniel J., Jaffro M., Collot S., Ferretti G.R., Tiffon C., Mahier-Ait Oukhatar C., Blay J.Y. Vemurafenib in non-small-cell lung cancer patients with BRAF(V600) and BRAF(nonV600) mutations. Ann. Oncol. 2020;31(2):289–294. doi: 10.1016/j.annonc.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 74.Frauenstein K., Tigges J., Soshilov A.A., Kado S., Raab N., Fritsche E., Haendeler J., Denison M.S., Vogel C.F., Haarmann-Stemmann T. Activation of the aryl hydrocarbon receptor by the widely used Src family kinase inhibitor 4-amino-5-(4-chlorophenyl)-7-(dimethylethyl)pyrazolo[3,4-d]pyrimidine (PP2) Arch. Toxicol. 2015;89(8):1329–1336. doi: 10.1007/s00204-014-1321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.