Abstract
Schizophrenia (SCZ) and bipolar disorder (BP) share a number of features. For example, multiple transcriptome analyses have reported molecular alterations common to both diagnoses, findings supported by the considerable overlap in the genetic risk for each disorder. These molecular similarities may underlie certain clinical features that are frequently present in both disorders. Indeed, many individuals with BP exhibit psychosis, and some individuals with SCZ have prominent mood symptoms that warrant the diagnosis of schizoaffective disorder (SA). To explore the potential relationships between molecular alterations and certain clinical features among subjects with these diagnoses, we analyzed RNA sequencing data from the dorsolateral prefrontal and anterior cingulate cortices, provided by the CommonMind Consortium, in subjects from the University of Pittsburgh Brain Tissue Donation Program. Relative to unaffected comparison subjects, in each brain region, robust differential gene expression was present only in SCZ, including a lower expression of genes involved in mitochondrial function and an elevated expression of immune-related genes. However, correlation analyses showed that BP subjects had similar, although less pronounced, gene expression alterations. Comparisons across subgroups of subjects revealed that the similarities between SCZ and BP subjects were principally due to the BP subjects with psychosis. Moreover, the gene expression profile in BP subjects with psychosis was more similar to “pure” SCZ and SA subjects than to BP subjects without psychosis. Together, these analyses suggest that similarities in gene expression between SCZ and BP are at least partially related to the presence of psychosis in some BP subjects.
Keywords: RNA sequencing, human, postmortem, cortex
Introduction
Schizophrenia (SCZ) and bipolar disorder (BP) are recognized as separate diagnostic entities; however, both are highly heterogeneous in symptom presentation and share certain brain abnormalities. For example, both disorders have been associated with lower gray matter volumes in multiple brain regions, including the dorsolateral prefrontal (DLPFC) and anterior cingulate (ACC) cortices.1–3 RNA sequencing studies of total gray matter have reported gene expression alterations in both DLPFC and ACC from SCZ and BP subjects,4 and a microarray analysis of DLPFC gray matter found shared transcriptional alterations in SCZ and BP.5 Moreover, a meta-analysis of transcriptome data from multiple brain regions in subjects with various neuropsychiatric disorders found the strongest overlap in transcriptional dysregulation between SCZ and BP.6 Furthermore, these structural and transcriptome findings are supported by reports of considerable overlap in the genetic risk for SCZ and BP.7
These similarities between SCZ and BP may be reflected, at least in part, in shared clinical features between these disorders. For example, psychotic features like those present in SCZ are seen in a subset of BP subjects. In addition, some subjects with the psychotic features of SCZ can exhibit prominent mood symptoms, warranting a diagnosis of schizoaffective disorder (SA). The presence of psychotic features or mood symptoms has also been associated with differences in genomic and transcriptomic findings across subsets of individuals with SCZ and BP. For example, the polygenic risk score for SCZ is higher in BP subjects with psychosis (BP + P) than in BP subjects without psychosis (BP − P).8 Moreover, a microarray study of DLPFC pyramidal neurons identified a distinct molecular signature in subjects diagnosed with SA disorder relative to those with “pure” SCZ.9
In concert, these findings suggest that shared transcriptional alterations in the DLPFC and ACC of SCZ and BP subjects might be related to the presence of psychotic features and/or the severity of mood symptoms. To explore this idea, previously published RNA sequencing data from the DLPFC and unpublished data from the ACC, obtained from tissue samples provided by the University of Pittsburgh to the CommonMind Consortium, were analyzed. First, a differential expression analysis was performed in each brain region to compare 82 unaffected comparison (C), 57 SCZ, and 35 BP subjects and then to assess similarities in differentially expressed genes (DEGs) across brain regions and diagnostic groups. Next, BP subjects were divided into those with or without psychosis and compared to all SCZ subjects to determine potential alterations in gene expression associated with psychosis. Finally, SCZ subjects were divided into those without (“pure” SCZ) or with prominent mood features (SA) and compared to BP subjects to assess potential differences in gene expression related to mood alterations (supplementary figure 1).
Methods
Data Generation
RNA sequencing data used in these analyses were from the University of Pittsburgh (Pitt) subjects in the CommonMind Consortium (https://www.nimhgenetics.org/available_data/commonmind/) data set. Published analyses of DLPFC data revealed institution as a major source of variance; 10 therefore, only Pitt samples were used in this study to avoid confounding effects of site. Furthermore, 35 of the 45 CommonMind Consortium BP subjects were from the Pitt cohort.
Postmortem Brain Samples
All subjects used in this study were obtained during autopsies conducted at the Allegheny County Office of the Medical Examiner (Pittsburgh, PA) after consent was obtained from the next-of-kin. An independent committee of experienced research clinicians made consensus Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, diagnoses, or confirmed the absence of any diagnoses, using information obtained from medical records and structured diagnostic interviews conducted with the decedent’s family members. Each SCZ and BP subject was matched for sex and as closely as possible for age to one C subject. Subject pairs were processed together to reduce potential batch effects. Ten C subjects were paired to both a SCZ and BP subject; therefore, these C subjects were sequenced twice. For those subjects, analyses were conducted using data from 5 randomly selected C samples from the SCZ pairing and 5 from the BP pairing. All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research and Clinical Training Involving Decedents and Institutional Review Board for Biomedical Research.
Tissue Processing and RNA Sequencing
Details of tissue processing and RNA sequencing have been reported.10 Briefly, blocks containing DLPFC (area 9/46) or ACC (area 24) were cut on a cryostat and total gray matter collected.11 Total RNA from 50 mg of tissue was isolated at the Mount Sinai Medical Center and ribosome depletion-based RNA sequencing was performed. After quality control analysis and mapping to a reference human genome (GRCh38), samples with a minimum of 50 million mapped reads were retained for downstream analysis.
Normalization of Gene Expression, Covariate Adjustment, and Statistics
The limma package12 in R was used for voom normalization and count per million (CPM) determination for each of 58 347 ENSEMBL genes. Genes with at least 1 CPM in >50% of the samples were retained for downstream analysis, resulting in 18 480 unique genes. Initial principal component analysis of the normalized data detected 4 outlier samples (3 C and 1 SCZ), all from the ACC, which were removed from subsequent analyses. For the subjects included for analyses (supplementary table 1), sex distribution, mean age, and mean postmortem interval (PMI) did not differ across any subject groups, although pH was marginally different across subject groups in the ACC samples (tables 1 and 2). RNA integrity (RIN) values were obtained following RNA isolation from the tissue samples used in the RNAseq analyses. In both DLPFC and ACC samples, mean RIN was lower in the SZ and BP subjects relative to C subjects (table 1).
Table 1.
Summary of subject characteristics for samples retained after QC and PCA analysis
| DLPFC samples | C | SCZ | BP | Statistics (χ 2/ANOVA) |
|---|---|---|---|---|
| Number | 82 | 57 | 35 | |
| Sex | 59 M, 23 F | 44 M, 13 F | 20 M, 15 F | χ 2 = 4.33, P = .12 |
| Age (years) | 48.1 ± 14.2 | 48.1 ± 13.0 | 45.5 ± 12.2 | F(2,171) = 0.52, P = .60 |
| PMI (hours) | 19 ± 5.4 | 20.0 ± 8.4 | 20.5 ± 7.0 | F(2,171) = 0.71, P = .50 |
| RIN | 8.5 ± 0.4 | 8.2 ± 0.6* | 8.2 ± 0.5* | F(2,171) = 9.82, P = 9.0 × 10−5 |
| pH | 6.7 ± 0.3 | 6.6 ± 0.3 | 6.6 ± 0.3 | F(2,171) = 3.0, P = .052 |
| ACC samples | ||||
| Number | 77 | 57 | 32 | |
| Sex | 54 M, 23 F | 44 M, 13 F | 19 M, 13 F | χ 2 = 2.52, P = .28 |
| Age (years) | 48.0 + 14.5 | 47.4 + 13.2 | 45.5 + 12.5 | F(2,163) = 0.37, P = .69 |
| PMI (hours) | 18.9 + 5.3 | 19.3 + 8.0 | 20.5 + 6.7 | F(2,163) = 0.69, P = .50 |
| RIN | 7.9 + 0.6 | 7.6 + 0.7* | 7.5 + 0.8* | F(2,163) = 3.83, P = .02 |
| pH | 6.7 ± 0.3 | 6.6 ± 0.3* | 6.6 ± 0.3* | F(2,163) = 4.0, P = .02 |
Note: Values are mean (standard deviation).
DLPFC, dorsolateral prefrontal; SCZ, schizophrenia; BP, bipolar disorder; ANOVA, analysis of variance; PMI, postmortem interval; RIN, RNA integrity number obtained for samples isolated for RNA sequencing; ACC, anterior cingulated; SA, schizoaffective disorder; BP + P, bipolar disorder subjects with psychosis; BP – P, bipolar disorder subjects without psychosis; QC, quality control; PCA, principal component analysis.
*Significantly different from C subjects.
Table 2.
Summary of subject characteristics for samples retained after QC and PCA analysis
| DLPFC samples | C | SA | “pure” SCZ | BP − P | BP + P | Statistics (χ 2/ANOVA) |
|---|---|---|---|---|---|---|
| Number | 82 | 20 | 37 | 24 | 11 | |
| Sex | 59 M, 23 F | 12 M, 8 F | 32 M, 5 F | 14 M, 10 F | 6 M, 5 F | χ 2 = 8.77, P = .07 |
| Age (years) | 48.1 ± 14.2 | 44.6 ± 12.2 | 50.0 ± 13.2 | 45.7 ± 12.7 | 45.3 ± 11.7 | F(4,169) = 0.80, P = .53 |
| PMI (hours) | 19 ± 5.4 | 20.3 ± 7.7 | 19.9 ± 7.7 | 20.2 ± 7.4 | 21.1 ± 6.4 | F(4,169) = 0.39, P = .81 |
| RIN | 8.5 ± 0.4 | 8.2 ± 0.7* | 8.2 ± 0.6* | 8.3 ± 0.5* | 8.1 ± 0.6* | F(4,169) = 5.00, P = 7.0 × 10−4 |
| pH | 6.7 ± 0.3 | 6.6 ± 0.3 | 6.6 ± 0.3 | 6.7 ± 0.3 | 6.5 ± 0.2 | F(4,169) = 2.19, P = .07 |
| ACC samples | ||||||
| Number | 77 | 20 | 37 | 24 | 11 | |
| Sex | 54 M, 23 F | 11 M, 9 F | 32 M, 5 F | 13 M, 9 F | 6 M, 4 F | χ 2 = 8.63, P = .07 |
| Age (years) | 48.0 ± 14.5 | 43.9 ± 12.2 | 49.3 ± 13.4 | 45.5 ± 12.9 | 45.6 ± 12.2 | F(4,161) = 0.70, P = .60 |
| PMI (hours) | 18.9 ± 5.3 | 19.0 ± 8.6 | 19.5 ± 7.8 | 20.7 ± 7.2 | 20.1 ± 5.8 | F(4,161) = 0.37, P = .83 |
| RIN | 7.9 ± 0.6 | 7.7 ± 0.6 | 7.6 ± 0.7 | 7.5 ± 0.9 | 7.7 ± 0.6 | F(4,161) = 2.03, P = .09 |
| pH | 6.7 ± 0.3 | 6.6 ± 0.3* | 6.6 ± 0.3 | 6.7 ± 0.3 | 6.5 ± 0.2* | F(4,161) = 2.70, P = .03 |
Note: Abbreviations are explained in the first footnote to table 1.
Values are mean (standard deviation).
*Significantly different from C subjects
Because brain region was identified as a primary driver of variance in the aggregated data (accounting for 33.7% of the variance), each brain region was analyzed separately. For differential gene expression, a basic linear regression model was used along with the precision weights obtained during voom normalization. Covariates included in the final model were sex, age, PMI, RIN, and 5 ancestry-related factors. These covariates explained 33% of the variance in the original analysis of the DLPFC by the CommonMind Consortium10 and are covariates commonly included in postmortem studies. The same statistical model was used to analyze subjects stratified into 5 groups (supplementary figure 1): C subjects, “pure” SCZ, SA, BP + P or BP − P subjects (table 2). All BP + P subjects had bipolar type 1 (BP1), whereas BP − P subjects were a combination of BP1 (n = 10), bipolar type 2 (n = 8), and bipolar not otherwise specified (n = 6).
Statistical significance for differential expression (DE) was determined using the Benjamini–Hochberg procedure13 with a false discovery rate (FDR) of 5%. Pathway analysis on over 500 canonical pathways was performed on the DEGs with INGENUITY analysis software using all filtered genes as background. Pathway significance was determined using Fisher’s Exact test. Cohen’s d effect sizes were calculated for each of the 18 480 genes within each diagnostic group and region and used to compare the overall transcriptional profile between groups. Use of Cohen’s d effect size is dependent upon the t-statistics obtained in the DE analysis, accounts for the differences in sample sizes among groups and yields results highly similar to correlation of log2 fold change. Pearson correlation coefficients were calculated for each comparison (eg, between the SCZ effect sizes for DLPFC and ACC). Analyzing the correlation between such a large number of observations results in statistically significant correlations for even very small correlation coefficients.14 Thus, the potential strength of correlation coefficients was inferred by designating them as weak (r < .2), moderate (.2 < r < .4), strong (.4 < r > .6), or very strong (r > .6).
Antipsychotic-Exposed Monkeys
Briefly, a cohort of 34 Rhesus macaque monkeys were randomly assigned to receive clozapine (n = 9), haloperidol (n = 17), or vehicle (n = 8) daily for 6 months. Details of these antipsychotic-exposed monkeys have been reported.10 DLPFC tissue (A46) was used for RNA sequencing and differential expression analysis was performed as for the human data.
Results
Differential Gene Expression Is More Robust in SCZ than in BP Subjects
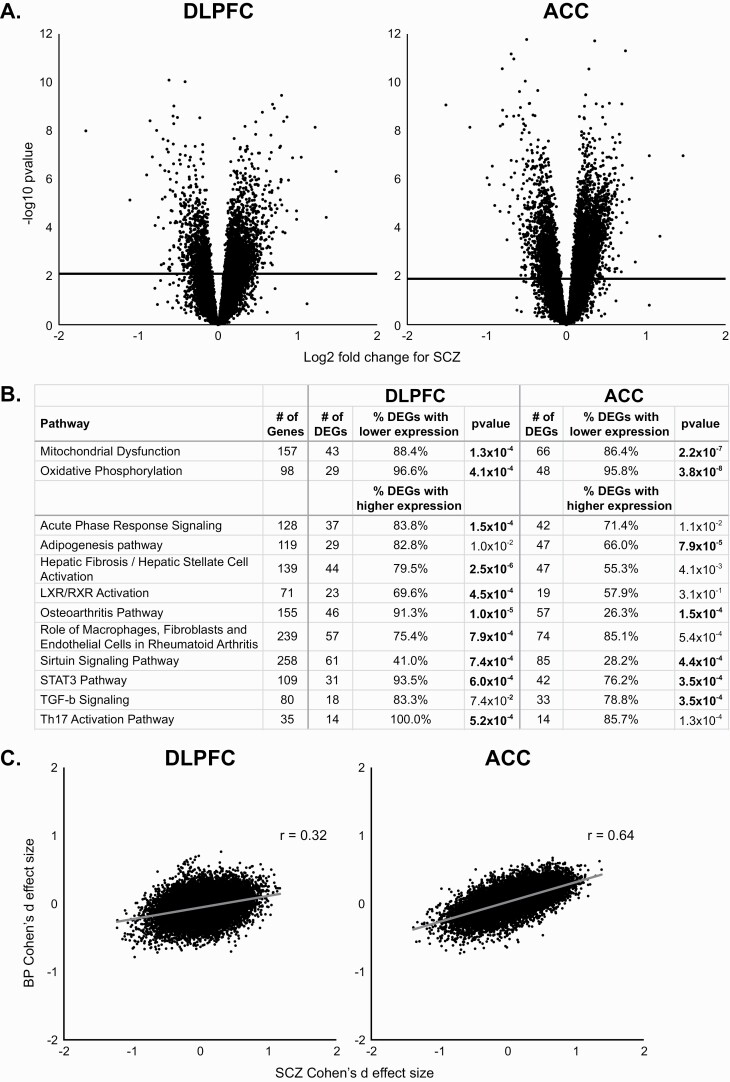
Initial analysis of variance (ANOVA) of the DLPFC data identified 2936 DEGs in 57 SCZ relative to 82 C subjects (figure 1A; see supplementary table 2 for top DEGs; supplementary figure 2); 1057 (35%) of the DEGs had lower expression in SCZ subjects. In contrast, no DEGs were detected between 35 BP and 82 C subjects, although 417 genes were differentially expressed between SCZ and BP subjects; of these, 323 were among the DEGs detected between SCZ and C subjects. In the initial ANOVA of the ACC data, 4712 DEGs were detected in SCZ relative to C subjects (figure 1A; see supplementary table 3 for top DEGs; supplementary figure 2); 1783 (38%) of the DEGs had lower expression in SCZ. Again, no DEGs were detected in BP subjects relative to C subjects and only 18 genes were differentially expressed between SCZ and BP subjects, all of which were among the DEGs detected between SCZ and C subjects.
Fig. 1.
Differential gene expression in subjects with schizophrenia (SCZ) and bipolar disorder (BP). (A) Volcano plots showing the log2 fold change in SCZ subjects for dorsolateral prefrontal (DLPFC; left) and anterior cingulate (ACC; right). Solid horizontal line represents the Benjamini–Hochberg-corrected P value of .05. (B) Results of INGENUITY analysis software for genes differentially expressed in SCZ from DLPFC and ACC. Bolded P values represent differences from C subjects at a 5% FDR level of significance. In the mitochondrial dysfunction and oxidative phosphorylation pathways, both of which are involved in mitochondrial function, most differentially expressed genes (DEGs) had a lower expression in SCZ. In most of the other pathways, many of which are involved in immune responses, the majority of DEGs had a higher expression in SCZ. (C) Correlations between the SCZ and BP Cohen’s d effect sizes for DLPFC (left) and ACC samples (right).
Pathway analysis of the DEGs in the DLPFC and ACC from SCZ subjects identified several pathways enriched for the SCZ DEGs at a 5% FDR (figure 1B). These included pathways for mitochondrial function (which predominantly had genes with lower expression in SCZ) and several pathways involved in immune-related functions (which predominantly had genes with elevated expression in SCZ). These pathways were affected to similar degrees in both cortical regions, suggesting that the diagnosis-related gene expression alterations were similar between regions. Indeed, the Cohen’s d effect sizes were very strongly correlated between the DLPFC and ACC (r = .77) in SCZ subjects.
The absence of DEGs in BP subjects could be due to the smaller sample size relative to the SZ subjects. To evaluate this possibility, a secondary analysis of the DLPFC samples was performed using all 82 C subjects and 33 pairs of SCZ and BP subjects matched perfectly for sex and as closely as possible for age. Results revealed 1077 DEGs in SCZ but none in BP subjects, suggesting that smaller sample size does not account for the absence of DEGs in BP subjects. However, because altered gene expression in BP might be of lower magnitude relative to SCZ, overall similarities in gene expression patterns between SCZ and BP subjects were assessed in each brain region using the correlation between Cohen’s d effect sizes. The correlation (figure 1C) between SCZ and BP effect sizes of the expression level of each gene was moderate in DLPFC (r = .32) but very strong in ACC (r = .64).
In concert, these findings suggest that SCZ is associated with robust alterations in gene expression that are similar in DLPFC and ACC. In contrast, gene expression alterations are much more modest in BP subjects and are more similar to SCZ in the ACC than in the DLPFC.
Gene Expression Profiles Are Similar Between SCZ Subjects and BP Subjects With Psychosis
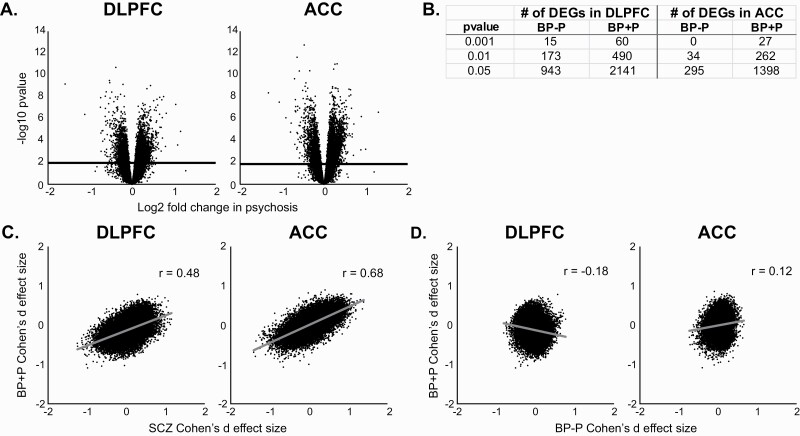
The positive correlations between the SCZ and BP effect sizes might be related to a history of psychosis in some BP subjects. Therefore, a cross-diagnosis analysis of psychosis was performed by stratifying all the subjects into those with or without psychosis (ie, SCZ and BP + P versus C and BP − P subjects; table 2). This analysis revealed that psychosis was associated with 4076 DEGs (42.6% with lower expression) in the DLPFC and 5244 DEGs (40.1% with lower expression) in the ACC (figure 2A). After including diagnosis (ie, C, SCZ, or BP) as a covariate, 3722 DEGs were identified in DLPFC and 3420 DEGs were identified in ACC, suggesting that the presence of psychosis is associated with differential gene expression, regardless of diagnosis.
Fig. 2.
Association of psychosis with differential gene expression. (A) Volcano plots showing the log2 fold change in subjects with psychosis for dorsolateral prefrontal (DLPFC; left) and anterior cingulate (ACC) samples (right). Solid horizontal line represents the Benjamini–Hochberg-corrected P value of .05. (B) Number of differentially expressed genes in bipolar disorder subjects with psychosis (BP + P) versus bipolar disorder subjects without psychosis (BP − P) at various P-value thresholds in DLPFC and ACC. (C) Correlations between Cohen’s d effect sizes for schizophrenia and BP + P subjects for DLPFC (left) and ACC samples (right). (D) Correlations between Cohen’s d effect sizes for BP − P and BP + P subjects for DLPFC (left) and ACC samples (right).
To explore more fully the effect of psychosis specifically in BP, BP + P (n = 11), and BP − P subjects (n = 24) were compared to C subjects. Although no DEGs were detected in either BP group relative to C subjects, at more relaxed statistical cutoffs (based on non-FDR corrected P values), many more DEGs were detected in the BP + P (figure 2B; supplementary figure 2) compared to the BP − P group, which had more than twice as many subjects. Furthermore, the SCZ and BP + P effect sizes were strongly correlated (r = .48) in DLPFC and very strongly correlated (r = .67) in ACC (figure 2C), whereas the correlations between SCZ and BP − P effect sizes were weak (r = .01) in DLPFC and moderate (r = .30) in ACC. Moreover, the correlations between the 2 BP groups in the DLFPC (r = −.18) and ACC (r = .12) were weak (figure 2D) and much smaller than the correlations between SCZ and BP+P (figure 2C).
Because all the BP + P subjects were diagnosed with BP1, the correlation between BP + P and SCZ might be related to the severity of the manic episodes in BP1 subjects versus the mixture of BP subtypes in the BP − P subjects. To address this issue, correlations of effect sizes were compared between BP1 subjects with (BP1 + P) or without (BP1 − P) psychosis and SCZ subjects. In DLPFC, correlations were strong between BP1 + P and SCZ (r = .48), moderate between BP1 + P and BP1 − P (r = .29), and weak between SCZ and BP1 − P (r = .01). In ACC, the correlations were similarly very strong (r = .68) between BP1 + P and SCZ subjects, but weak (r = .11) between BP1 + P and BP1 − P subjects and moderate (r = .21) between SCZ and BP1 − P subjects. Together, these data suggest that BP + P subjects have a gene expression profile more like SCZ than BP − P subjects and that the severity of manic episodes among BP subjects is unlikely a major contributor to these similarities.
Effects of Mood Do Not Account for Differential Gene Expression in SCZ
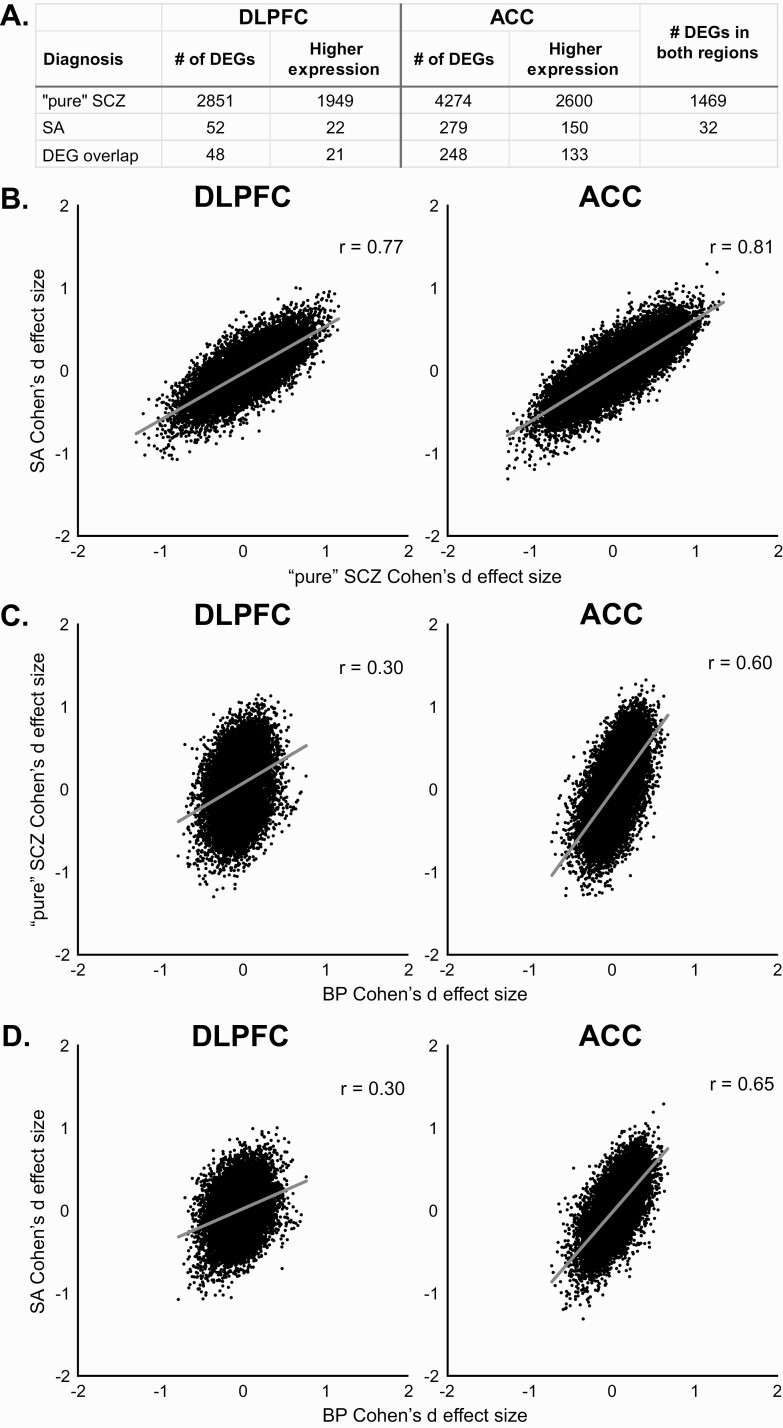
The correlated gene expression between the SCZ and BP subject groups might also be attributable to the pronounced mood alterations in the subset of subjects with SA disorder, especially in the ACC, a core cortical hub in mood regulation. Therefore, the SA and BP subjects were combined and compared to the C and “pure” SCZ subjects. Using this approach, no “mood-related” DEGs were detected in either region, even after statistically adjusting for diagnosis. Comparisons of SA (n = 20), “pure” SCZ (n = 37), and C subjects revealed DEGs in both SA and “pure” SCZ relative to C subjects (figure 3A; supplementary figure 2) with considerable overlap between diagnoses and regions for specific DEGs. The smaller number of DEGs in the SA subjects is likely due, in part, to the lower sample size in the SA group. Indeed, the correlation of the effect sizes between “pure” SCZ and SA (figure 3B) was very strong in both DLPFC (r = .77) and ACC (r = .81). Although no DEGs were detected between “pure” SCZ and SA subjects at a 5% FDR, 526 genes in DLPFC and 422 genes in ACC showed differences in expression between these groups at a more relaxed statistical cutoff (P < .05). The correlations between BP subjects and “pure” SCZ or SA subjects were similar within a region, although both correlations were stronger in ACC than DLPFC (figures 3C and D).
Fig. 3.
Association of mood symptoms with differential gene expression. (A) Number of differentially expressed genes (DEGs) detected in dorsolateral prefrontal (DLPFC) or anterior cingulate (ACC) from “pure” schizophrenia (SCZ) or schizoaffective disorder (SA) subjects. (B) Correlations between Cohen’s d effect sizes for “pure” SCZ and SA subjects from DLPFC (left) and ACC (right). (C) Correlations between Cohen’s d effect sizes for BP and “pure” SCZ subjects from DLPFC (left) and ACC (right). (D) Correlations between Cohen’s d effect sizes for BP and SA subjects from DLPFC (left) and ACC (right).
In concert, these findings suggest that gene expression alterations are robust, and strongly correlated between “pure” SCZ and SA subjects in both DLPFC and ACC, and that the correlated gene expression between BP and SCZ subjects is not primarily related to the pronounced mood disturbances in the subset of SCZ subjects with SA disorder.
Differential Gene Expression Is Not Associated With Exposure to Antipsychotic Drugs
The similarities in gene expression between SCZ and BP + P subjects might be due to effects of antipsychotic drugs. Indeed, almost all the SCZ subjects were prescribed antipsychotic medications at the time of death. Interestingly, only 4 (36%) of the 11 BP + P subjects were prescribed antipsychotic medications at the time of death, making it unlikely that the similarities between SZ and BP + P subjects were due to these drugs. Furthermore, 7 (29%) of the 24 BP − P subjects were prescribed antipsychotic medications at the time of death, suggesting that the presence of antipsychotic medications cannot account for the gene expression differences between BP + P and BP − P subjects. To further explore any potential contributions of antipsychotic drugs to altered gene expression, DLPFC samples from macaque monkeys chronically exposed to haloperidol, clozapine, or vehicle were subjected to RNA sequencing. As previously reported,10 no DEGs (q < .05) were detected in either the haloperidol- or clozapine-exposed animals. Even at nominal statistical cutoffs (P < .01), only 35 DEGs were detected in the clozapine-exposed animals and only 143 DEGs were detected in the haloperidol-exposed monkeys. Of these transcripts, only PDK4 was also a DEG in SCZ subjects, and PDK4 levels were 17.5% lower in the haloperidol-exposed monkeys but 41.4% higher in the SCZ subjects. Since lower statistical power in the monkey cohort compared to the human cohort could explain the lack of DEGs, correlations between the Cohen’s d effect sizes for DEGs detected in DLPFC data from SCZ subjects and orthologous genes in the antipsychotic-exposed monkeys were determined. In both groups of antipsychotic-exposed monkeys, the effect sizes were only weakly correlated with the human findings (supplementary figure 3). Moreover, combining all the antipsychotic-exposed animals into one group still resulted in a weak (r = −.06) correlation with the human data. Together, these data suggest that treatment with antipsychotic medications is not likely to account for the altered gene expression in SCZ or BP + P subjects.
Discussion
In DLPFC, robust transcriptome alterations were detected in SCZ relative to C subjects, including lower expression of genes involved in mitochondrial function and higher expression of genes involved in inflammation, findings consistent with prior studies of the DLPFC.5,6,15 In ACC, we found a similar pattern of robust transcriptome alterations. These alterations were very strongly correlated between DLPFC and ACC, suggesting that a conserved disease process drives altered gene expression in very different cortical regions in subjects with SCZ. Consistent with this interpretation, studies in 2 independent subject cohorts reported similar alterations in RNA editing in both DLFPC and ACC in SCZ16 and multiple imaging studies have reported smaller gray matter volumes in both regions.1–3
Prior studies have reported considerable overlap in both transcriptome alterations4,6 and genetic risk for SCZ and BP.7,8 However, although we did not detect any DEGs in BP using a rigid statistical cutoff, the effect sizes in SCZ and BP were strongly positively correlated, suggesting that many gene expression alterations are shared by both disorders, albeit with a weaker disease effect in BP. These findings are comparable to a previous meta-analysis showing similarities in gene expression and altered pathways between BP and SCZ.6 Interestingly, the correlations among gene expression alterations were consistently stronger in the ACC than DLPFC. Alterations in mood and affect, which are substantially mediated by the ACC,17 are shared by BP and SCZ, suggesting that gene expression alterations in the ACC may be related to these similarities. Furthermore, the weaker correlations between groups in DLPFC gene expression may be more related to impairments in cognitive processes that are dependent upon the DLPFC and frequently more severe and persistent in SCZ than BP.18
The similar, but weaker, profile of gene expression alterations in BP across both brain regions might be related to other clinical features, such as psychosis, that are shared with SCZ but are present in only a subset of BP subjects. Indeed, analyses of subgroups of subjects revealed that BP + P subjects had a gene expression profile much more comparable to SCZ than to BP − P subjects. This similarity in gene expression alterations between SCZ and BP + P subjects might reflect a shared genetic liability for psychosis as the correlation between SCZ polygenic risk score and the presence of psychotic features in BP is quite strong.8 Furthermore, structural MRI studies show similar alterations in DLPFC and ACC in subjects with affective and nonaffective first-episode psychosis,19 whereas structural features of the ACC differ between BP + P and BP − P subjects.20 However, it should be noted that structural MRI studies of other brain regions (eg, hippocampus) found similar volumetric alterations between pure “SCZ” and SA subjects that were not present in BP + P subjects.21 Additionally, the clinical presentation of psychosis in SCZ and BP can be quite different, suggesting that factors other than shared patterns of gene expression in the DLPFC and ACC likely contribute to diagnosis-associated differences in the content of delusions. Therefore, further studies are needed to determine if psychosis per se is associated with molecular and structural brain alterations.
In contrast, the presence of mood alterations in the subset of SZ subjects with SA disorder does not appear to be related to similar gene expression alterations between SCZ and BP subjects. For example, we did not identify any genes whose expression differed between subjects with (BP and SA) and without (pure “SCZ” and C) pronounced mood alterations. Furthermore, the correlation in gene expression alterations between SA and BP subjects was considerably weaker than between SA and pure “SCZ” subjects, even in the ACC, a cortical region strongly associated with mood regulation. However, the diagnostic challenges in distinguishing SA from SCZ and BP + P must be kept in mind in interpreting these findings.
The potential effects of psychotropic medications need to be considered in interpreting the results of the present study. This issue is challenging to address in postmortem studies, given the absence of SCZ or BP subjects with no lifetime history of pharmacotherapy. However, the inclusion of BP + P subjects who were not on antipsychotics in this study, along with the lack of differential expression in the antipsychotic-exposed monkeys, suggests that the bulk of findings reported here are unlikely to be due to antipsychotic drugs. Furthermore, the prescription of other medications differed substantially across SCZ and BP + P subjects, making it unlikely that any one drug or combination of drugs was responsible for the gene expression alterations shared among subjects with psychosis. For example, only 2 BP − P, 1 BP + P, and 2 SCZ subjects were prescribed lithium at the time of death. However, 59% of SZ subjects and 63% of BP + P subjects, but only 38% of BP − P subjects, were on valproic acid and/or benzodiazepines; thus, we cannot exclude the potential contribution of these medications to the gene expression alterations that were shared among subjects with psychosis.
The potential influence of other comorbid factors, such as substance use disorder and death by suicide, appear unlikely to account for the gene expression alterations associated with psychosis. For example, 62.5% of BP − P subjects but only 39.7% of SCZ and BP + P subjects had a substance use disorder diagnosis, suggesting that substance abuse is not a likely explanation for the present findings. Moreover, 33.3% of BP − P subjects died by suicide, a similar suicide rate to that of the psychotic subjects (29.4%).
Overall, our data suggest that alterations in gene expression in SCZ are similar in cortical regions that differ substantially in structure and function, consistent with a conserved disease process that is common to at least DLPFC and ACC. In addition, consistent with recent reports of substantial genetic overlap in the risk for SCZ and BP, our transcriptome findings suggest that this overlap may underlie the presence of psychosis in some subjects with BP but not the presence of mood symptoms in some subjects with SCZ. Thus, our findings provide support for the concept that psychosis represents an emergent brain property with a molecular basis that transcends traditional diagnostic boundaries.22,23
Supplementary Material
Acknowledgments
The authors thank Mary Brady for excellent technical assistance and Bernie Devlin and Lambertus Klei for help accessing the data and critical comments on the manuscript. D.A.L. serves as a consultant to Astellas. The authors declare that there are no conflicts of interest in relation to the subject of this study.
Funding
This work was supported by National Institutes of Mental Health grants (MH103204 and MH043784 to D.A.L.). D.A.L. currently receives investigator-initiated research support from Pfizer and Merck.
References
- 1.Takayanagi Y, Sasabayashi D, Takahashi T, et al. . Reduced cortical thickness in schizophrenia and schizotypal disorder. Schizophr Bull. 2020;46(2):387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugihara G, Oishi N, Son S, Kubota M, Takahashi H, Murai T. Distinct patterns of cerebral cortical thinning in schizophrenia: a neuroimaging data-driven approach. Schizophr Bull. 2017;43(4):900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162(12):2233–2245. [DOI] [PubMed] [Google Scholar]
- 4.Ramaker RC, Bowling KM, Lasseigne BN, et al. . Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanz TA, Reinhart V, Sheehan MJ, et al. . Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: a comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major depressive disorder. Transl Psychiatry. 2019;9(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandal MJ, Haney JR, Parikshak NN, et al. . Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018;359(6376):693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrath LM, Cornelis MC, Lee PH, et al. . Genetic predictors of risk and resilience in psychiatric disorders: a cross-disorder genome-wide association study of functional impairment in major depressive disorder, bipolar disorder, and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(8):779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 2018;173(7):1705–1715.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arion D, Corradi JP, Tang S, et al. . Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015;20(11):1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromer M, Roussos P, Sieberts SK, et al. . Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsubomoto M, Kawabata R, Zhu X, et al. . Expression of transcripts selective for GABA neuron subpopulations across the cortical visuospatial working memory network in the healthy state and schizophrenia. Cereb Cortex. 2019;29(8):3540–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 1995;57(1):289–300. [Google Scholar]
- 14.Lin M, Lucas HC, Shmueli G. Research commentary-too big to fail: large samples and the p-value problem. Inf Syst Res. 2013;24(4):906–917. [Google Scholar]
- 15.Hauberg ME, Fullard JF, Zhu L, et al. ; CommonMind Consortium . Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol Psychiatry. 2019;24(11):1685–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breen MS, Dobbyn A, Li Q, et al. ; CommonMind Consortium . Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat Neurosci. 2019;22(9):1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riva-Posse P, Holtzheimer PE, Mayberg HS. Cingulate-mediated depressive symptoms in neurologic disease and therapeutics. Handb Clin Neurol. 2019;166:371–379. [DOI] [PubMed] [Google Scholar]
- 18.Lynham AJ, Hubbard L, Tansey KE, et al. . Examining cognition across the bipolar/schizophrenia diagnostic spectrum. J Psychiatry Neurosci. 2018;43(4):245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo A, Delvecchio G, Altamura AC, Soares JC, Brambilla P. Gray matter differences between affective and non-affective first episode psychosis: a review of Magnetic Resonance Imaging studies. J Affect Disord. 2019;243:564–574. [DOI] [PubMed] [Google Scholar]
- 20.Anticevic A, Savic A, Repovs G, et al. . Ventral anterior cingulate connectivity distinguished nonpsychotic bipolar illness from psychotic bipolar disorder and schizophrenia. Schizophr Bull. 2015;41(1):133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold SJ, Ivleva EI, Gopal TA, et al. . Hippocampal volume is reduced in schizophrenia and schizoaffective disorder but not in psychotic bipolar I disorder demonstrated by both manual tracing and automated parcellation (FreeSurfer). Schizophr Bull. 2015;41(1):233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill SK, Reilly JL, Keefe RS, et al. . Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. Am J Psychiatry. 2013;170(11):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker JT, Holmes AJ, Masters GA, et al. . Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry. 2014;71(2):109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.