Abstract
Metabonomics in inflammatory bowel disease (IBD) characterizes the effector molecules of biological systems and thus aims to describe the molecular phenotype, generate insight into the pathology, and predict disease course and response to treatment. Nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS), and integrated NMR and MS platforms coupled with multivariate analyses have been applied to create such metabolic profiles. Recent advances have identified quiescent ulcerative colitis as a distinct molecular phenotype and demonstrated metabonomics as a promising clinical tool for predicting relapse and response to treatment with biologics as well as fecal microbiome transplantation, thus facilitating much needed precision medicine. However, understanding this complex research field and how it translates into clinical settings is a challenge. This review aims to describe the current workflow, analytical strategies, and associated bioinformatics, and translate current IBD metabonomic knowledge into new potential clinically applicable treatment strategies, and outline future key translational perspectives.
Keywords: Crohn's disease, Metabolic profiling, Metabolomics, Spectrometry, Spectroscopy, Ulcerative colitis
1. Introduction
Inflammatory bowel disease (IBD) is a condition of recurrent or chronic intestinal inflammation giving rise to diarrhea, bloody stools, abdominal discomfort, and anorexia. IBD is a global disease with increasing incidence in developing and Westernized countries, although a plateau seems to have been reached in high-income countries [1]. IBD is believed to be a multifactorial disease and the consequence of a disturbed immunologic response to environmental and microbial components in genetically susceptible individuals [2]. Ulcerative colitis (UC) [3] and Crohn's disease (CD) [4] are the two main entities of IBD, and although they share a range of characteristics, they are two distinct diseases requiring exact differentiation and phenotyping for tailored treatment and precision medicine [5]. Predicting disease course and treatment response is the very essence of precision medicine but also a challenging task because the dissection of genotype-to-phenotype relations is not straightforward owing to the wide range of downstream regulatory processes that take place from the genome to the metabolome and the concomitant interactions with the exposome and microbiome (Fig. 1A). Thus, the complexity of the network increases toward the metabolome, which is the entire set of intermediates and end products of metabolism, that is, the life-sustaining chemical processes of the biological system. The metabolome is accordingly highly reflective of the actual (patho-)physiologic phenotype and can be characterized by metabolic profiling, or metabonomics, which is the study of metabolic changes in response to internal and external stimuli in an integrated biological system [6]. Metabonomics is used interchangeably with metabolomics, however, metabolomics aims to characterize and quantify all the small molecules in complex biological samples, whereas, metabonomics aims to measure the global, dynamic metabolic response of living systems to biological stimuli or disease. Metabonomics consequently focus on understanding systemic changes through time in complex multicellular systems, which is in line with most studies performed within the field of IBD. Nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS) are usually applied in IBD metabonomic studies to generate extensive targeted or untargeted metabolic profiles. These complex, multivariate data subsequently need to be coupled with bioinformatic tools to extract meaningful information. The ongoing development of metabonomic technologies and advances in bioinformatics have resulted in a steady increase in more sophisticated IBD studies over the past two decades. Recent excellent reviews provide comprehensive and descriptive overviews of currently available IBD metabonomic studies in relation to the microbiome [7], volatome [8], sample type [9], and metabolites [10] by addressing the contribution of the metabolome to the IBD pathogenesis, but one of the major challenges is to translate these findings into new tools of clinical relevance. The vast majority of IBD metabonomic studies focus on IBD diagnostics and differentiation between UC and CD [11], [12], [13], [14], [15], [16], [17], and this has resulted in conflicting outcomes because apparent diagnostic and differential power seems to be lost once the metabonomic data are corrected for potential confounders such as surgery and medication [14]. A more consistent finding is the identification of quiescent UC as a distinct metabonomic phenotype compared with patients with active disease and healthy control individuals [18], [19], [20], [21]. Furthermore, recent studies have also demonstrated metabonomics as a potential clinical tool for predicting relapse [22] and response to treatment with biologics [23] as well as fecal microbiome transplantation (FMT) [24], thus facilitating proactive medical care.
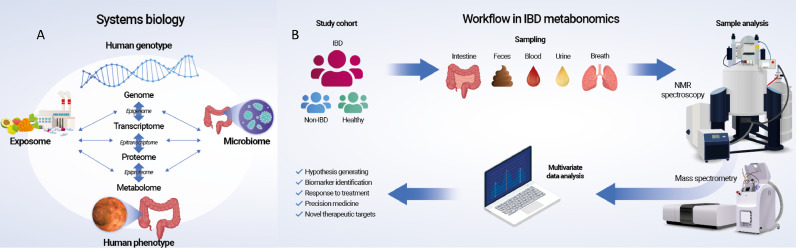
Fig. 1.
Systems biology and common workflow in inflammatory bowel disease (IBD) metabonomics.
With these recent promising studies within the field of IBD metabonomics the objective of this review is to provide an up-to-date overview of currently available analytical technologies and their associated applicability in metabonomic studies; to introduce human IBD metabonomic studies as a tool for molecular phenotyping, prediction of disease course, and treatment response; and finally, to outline future perspectives of importance in the field. This review consequently highlights the importance of metabonomics as an essential tool for the development of future personalized medicine.
2. Commonly applied workflow, analytical strategies, and multivariate analyses
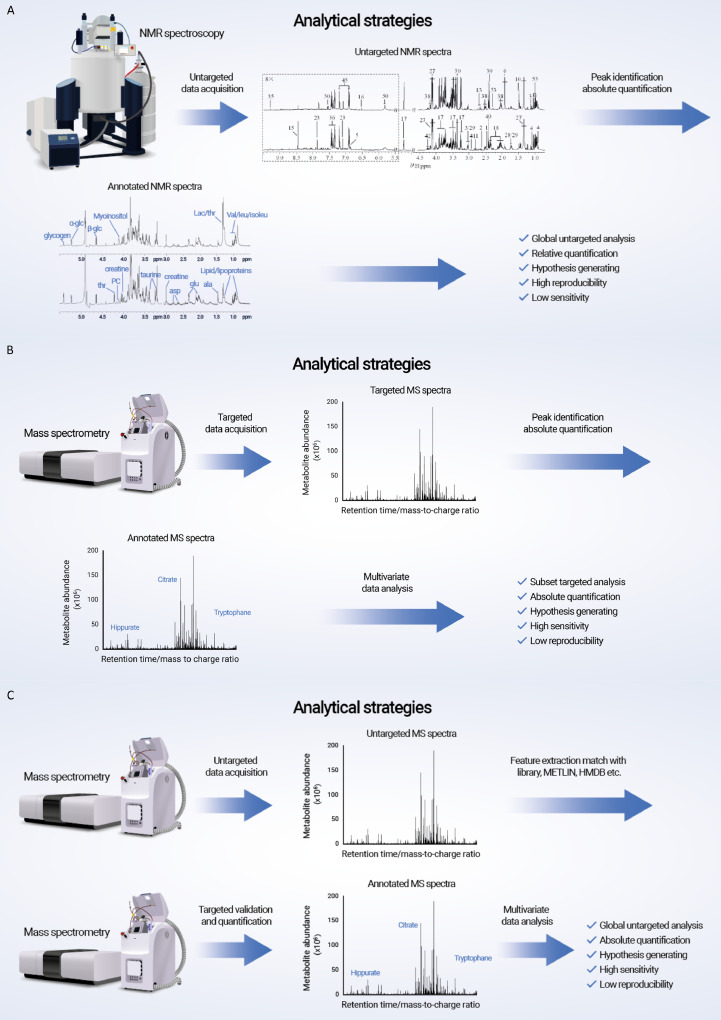
The total estimated number of endogenous human metabolites is approximately 6·500 [25]. However, because the metabolome is the most downstream of omics from drugs, ingested food, and their metabolites, as well as metabolites generated from the interactions between human and symbiotic gut microbiota, the number of metabolites potentially exceeds ∼100·000 [26]. The concentrations of these metabolites range from micromole to millimole (e.g., amino acids and tricarboxylic acid [TCA] cycle intermediates) and from picomole to femtomole (e.g., hormones). The vast number of metabolites with different chemical and physical properties and large dynamic ranges makes the identification and quantification of metabolites challenging tasks, especially because they also need to be associated with disease perturbations such as IBD. Metabonomic studies consequently require large cohorts of patients with IBD and matched control individuals with carefully recorded demographics and clinical details, planning of the sampling type and often successive sampling to identify metabolic trajectories, as well as an integrated approach of different analytical platforms and subsequent carefully performed multivariate analyses (Fig. 1B) [27]. The analytical strategies applied depend on the scientific question because nuclear magnetic resonance spectroscopy (NMR)–based metabonomics is used to identify and semi-quantify metabolites in an untargeted approach (Fig. 2A), whereas mass spectrometry (MS) is ideal for absolute quantification (Fig. 2B and C).
Fig. 2.
Analytical strategies. (A) Nuclear magnetic resonance (NMR), (B) targeted, and (C) untargeted mass spectrometry (MS).
2.1. Nuclear magnetic resonance spectroscopy
NMR employs 1H for the detection of metabolites because protons are the most abundant nuclei present in biological samples [28]. Because the intensity of the 1H NMR spectrum is directly proportional to the number of protons in a molecule, high-resolution NMR spectroscopy is able to quantify the metabolites in an untargeted approach. The resonance of a proton in an NMR spectrum represents its chemical environment and consequently carries molecular structural information that allows identification of the metabolites [28]. The NMR technique additionally permits spectral editing so that the different properties of the metabolite signals can be split up without separating the sample itself; spin-spin relaxation time–edited NMR spectra only carry information on mobile molecules, whereas diffusion-edited NMR spectra attenuate signals from smaller molecules and selectively display only bounded molecules such as lipoprotein lipids [29]. In addition, NMR-based metabonomics is a robust and reliable technique with high reproducibility that requires minimal or no sample preparation, is nondestructive, has a low running cost and high throughput, and is virtually the only technique able to analyze intact tissue [30]. However, NMR-based metabonomics suffers from low intrinsic sensitivity and heavy signal overlap [29]. Although an increasing number of scans and spectral editing/stable isotope labeling strategies to some extent ameliorate these drawbacks, the number of metabolites that can be detected is limited.
2.2. Mass spectrometry and integration with NMR spectroscopy
MS is another commonly applied technology for the investigation of metabonomics that is typically coupled with separation techniques such as liquid chromatography (LC), gas chromatography (GC), and capillary electrophoresis (CE) [31]. MS-based metabonomics can be divided into targeted and untargeted approaches. The targeted approach selectively quantifies specific metabolites, a class of metabolites, or metabolites in a pathway such as bile acids or amino-containing metabolites [32,33]. It requires the addition of internal standards for identification and quantification purposes. The advantage of this approach is that the MS conditions can be optimized for ideal separation and detection, resulting in identification of metabolites in the picomole range, but it requires a priori knowledge and valid hypotheses regarding the samples of interest. For discovery purposes, untargeted MS analysis is employed, which typically generates ∼10·000 features depending on the profiling method and analytical platform. The challenge associated with the untargeted approach is the identification of metabolites. The workflow involves conducting additional MS/MS fragmentation experiments for the unknown metabolites, putative metabolite identification by checking the fragmentation pattern against metabolite databases such as the Human Metabolome Database (HMDB) and Metlin (metabolite and chemical entity database), metabolite verification by spiking the standard compounds, and further quantification [34]. In case the standard molecule is unavailable, synthesis of the compound is necessary for final confirmation. Recently, a high coverage of metabolites employing pseudo-targeted profiling approaches has shown enhanced metabolite identification power, but the ability to discover new metabolites is still limited [35]. Compared with NMR-based metabonomics, the sensitivity of MS is superior [36]. However, more care should be taken when conducting MS experiments because the reproducibility of MS is intrinsically low, and quality control strategies are necessary to obtain reproducible results. Because metabolites are complex and diverse with different chemical properties and concentrations, integrated NMR/MS-based analytical platforms and the combination of targeted and untargeted approaches are necessary. The choice of analytical platforms depends on the scientific questions asked and whether plausible hypotheses are available. An LC/MS-based targeted approach is the first choice when validating a hypothesis in which a known metabolite needs to be detected because an MS-based targeted approach has the most sensitivity. For exploratory purposes, NMR- and MS-based untargeted approaches can be combined for untargeted metabolic profiling studies.
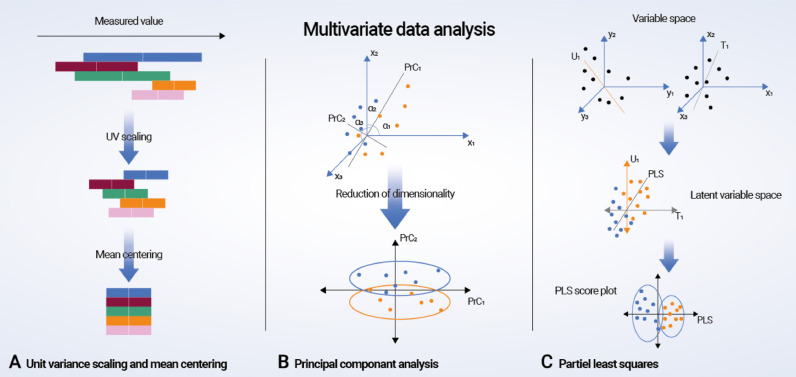
Both 1H NMR- and MS-based metabonomic studies contain high-density information, and series of multivariate statistical analyses such as principal component analysis (PCA), partial least squares/projections to latent structures discriminant analysis (PLS-DA), and orthogonal signal correction projections to latent structures discriminant analysis (O-PLS-DA) are often applied (Fig. 3; Box 1) [37,38]. These methods apply dimensional reduction strategies to complex spectral data in order to decode the changes relating to the biological perturbations of interest. However, despite the current technological and bioinformatic advances, it is essential to acknowledge the limitations of metabonomics, as none of the existing technologies can identify the entire metabolome. Furthermore, the metabolome is an ever-changing and dynamic structure due to the continuous influence of up-stream omics and the environment, and the results from metabonomics studies consequently need to be interpreted under these conditions and with caution.
Fig. 3.
Multivariate data analysis. (A) Preprocessing of metabonomic data using unit variance scaling and mean centering. (B) Principal component analysis (PCA, D2): a virtual matrix X is created with a number of observations (n spectra) and K variables (metabolites). A variable space is constructed with as many dimensions as variables; each variable is characterized by one coordinate axis, but for illustrative purposes, only three variable/metabolite axes (x1, x2, and x3) are displayed. Each observation from the matrix X is then placed in the K-dimensional variable space and mean centered around the value zero, creating a cloud of data points. PCA in this space results in lines or principal components (PrCs) that approximate the variables as closely as possible by reducing correlated variables to a smaller set of uncorrelated variables (PrCs). If the first two PrCs account for 80% of the total variation, only 20% of the total variation will be lost by this two-dimensional reduction. The observation points are projected onto the two-dimensional plane, resulting in new coordinate values for each observation (known as scores), and this makes it possible to produce a PrC score plot. Understanding the scores is possible by calculating loadings, which are given by cosines of the angles α1, α2, and α3 to each PrC. Loading values define the way in which the original variables are linearly combined to form the new variables or PrCs and unravel the degree of correlation and in what manner (positive or negative correlation) the original variables contribute to the scores. (C) Partial least squares (PLS) regression: mean-centered and scaled fabricated x and y data sets are demonstrated as a cloud of points in each variable space. Only three variable axes are displayed, respectively (x1, x2, x3 and y1, y2, y3). PLS regression analysis results in linear arrangements of the original x and y variables, creating new or “latent” variables (T1 and U2). These latent variables, or x and y scores, are in essence principally identical to PrCs. However, the PLS regression model strengthens the relationship between the x and y axes because the iterative algorithm used exchange scores between the two data sets and consequently defines the latent variables in the x data set that have high covariance with those in the y data set. Covariance is required in each dimension, and once it is identified in one dimension, the x data set is decomposed at the same time as the predicted y data set is created. Thus, PLS regression concurrently projects the x and y variables onto a common subspace (TU) so that there is a close relationship between the position of one observation on the x plane and its corresponding position on the y plane. This creates a PLS regression component for the first modeled dimension T1 and U1. The second PLS regression component is created in a similar fashion, and the subsequent PLS data can be illustrated as score and loading plots.
Box 1.
Multivariate data analysis.
| Principal component analysis (PCA) is an unsupervised data analysis method that requires two conditions: (1) each principal component (PrC) is the best explanation of the original variables (x data) in the data set, and (2) each PrC is orthogonal to every other PrC. Data analysis results are visualized using the PrC score and loadings plots. PCA gives an overview of the structure of the data set and is helpful for identifying inherent clusters and separations within the data set and possible outliers. Subsequent supervised analysis methods, such as partial least squares/projections to latent structures discriminant analysis (PLS-DA) and orthogonal signal correction projections to latent structures discriminant analysis (O-PLS-DA), are necessary to extract important biomarkers. In PCA, only x data are considered, whereas partial least squares are an extension of PCA and considers both x and y data sets. Here the y data set can be class membership (PLS-DA) or other biological descriptions (PLS). It is regarded as a supervised modeling method when the multivariate data analysis considers the y data set. The supervised modeling satisfies dimensional reduction (PCA) and correlation with the response matrix (y data) at the same time, generating metabolic information relating to biological events of interest. The purpose of applying orthogonal signal correction is to remove data that are unrelated to the response matrix, such as instrument instability or other irrelevant confounding factors. However, this supervised method increases the risk of overfitting the model, and therefore subsequent model validation procedures are essential when performing supervised data analyses. |
3. Metabonomics characterizes IBD phenotypes
3.1. Metabonomic remission and quiescent disease
Some of the earliest work in IBD metabonomics used NMR spectroscopy on colonic biopsies and differentiated between patients with active UC, patients with active CD, and control individuals with an impressive sensitivity and specificity ranging from 96% to 100% [18]. However, when colonic samples from patients with IBD and quiescent disease had to be validated in the established classifier, only 82% could be classified unambiguously, and 42% of those (primarily patients with UC) were assigned as abnormal or inflamed despite endoscopically and histologically confirmed quiescent disease [18]. This observation was substantiated in a methodologically similar study with NMR spectroscopy on colonic biopsies and isolated colonocytes from UC patients and control individuals; patients with active UC and control individuals were easily differentiated, but 36% of endoscopically assessed quiescent UC patients were classified as inflamed [19]. These 36% were characterized by having a flare-up within 6 months either prior to or after sample collection. Sharma et al. [39] used paired colonic biopsies from uninvolved and involved colonic mucosa from the same UC patients and showed that the metabolic profile of seemingly macroscopically normal mucosa was indistinguishable from that of inflamed colonic tissue. This observation suggests that involvement of the colonic mucosa is far more extensive at the molecular level and might represent a preclinical inflammation of more oral, i.e., proximal, extension of lesions over time. These studies consequently raise two interesting questions: can metabonomics be used to predict imminent flares and thus support proactive personalized medical care, and moreover, is metabonomics-based remission an actual concept and a new potential treatment target? Indeed, integrated metabonomic and transcriptomic analyses have identified quiescent UC as a distinct phenotype as compared with control individuals [40,41]. Intriguingly, these integrated classification models also differentiate between early and late disease onset (above or below 25 years of age) and moreover they differentiate between patients with and without glucocorticoid dependency. This is of utmost importance because early identification of patients with glucocorticoid dependency might lead to more optimized and personalized treatment in terms of timely introduction of immunomodulators and/or biologics.
These phenotypic findings have also been observed with MS-based lipidomic [21] (i.e., a subset of the metabolome) and metabonomic studies [20] and may discriminate between patients with flares of UC, patients with quiescent UC, and control individuals in multivariate modeling. Here 220 different lipids and 177 metabolites were identified, respectively, but the highest significant variation was observed within the sphingolipids, phosphatidylcholine (PC), lysophosphatidylcholine (LPC), and a range of amino acids. The changes were also registered in patients with quiescent UC, albeit to a lesser extent, but correlated with previous MS-based lipidomic studies on mucus from patients with UC and CD and control individuals, in whom reduced levels of PC and LPC in both flaring and quiescent UC, but not in CD, were found [42]. These aforementioned findings have led to a phase III placebo-controlled trial of a modified release of PC, which showed significant improvement of the activity in mesalazine-refractory UC [43]. The low levels of PC and LPC in quiescent disease also indicate that PC treatment might be beneficial for sustained clinical remission, but this needs to be elucidated.
In recent years it has gradually become evident that the main risk factors for disease flares, colectomy, and colorectal cancer in UC are sustained wound formation and inflammation and that medication improving mucosal healing modifies the disease course favorably [44]. Thus, histologic remission has become the new treatment target. However, with the identification of a potential metabonomic remission in UC, large-scale prospective studies based on metabonomics and intestinal biopsies are warranted to validate whether metabonomic remission is associated with an even better long-term state of remission and consequently a disease-modifying course [45].
3.2. Serum metabonomics and a proatherogenic lipid profile
Williams et al. [46] were the first to demonstrate significant differences in the metabolic profiles of serum in patients with quiescent UC and CD and control individuals. The major discriminatory metabolites were low levels of low-density lipoprotein (LDL), high-density lipoprotein (HDL), unsaturated lipids, choline, alanine, acetate, and isoleucine, whereas very low-density lipoprotein (VLDL), glucose, lactate, and N-acetylglycoprotein were increased. The patients were evaluated based on clinical activity scores, that is, the Simple Clinical Colitis Activity Index (SCCAI) [47] or the Harvey Bradshaw Index (HBI) [48] for UC and CD, respectively. Subclinical inflammatory processes might explain the observed differences at clinically quiescent disease, because no endoscopic or indirect measures of inflammation were provided (e.g., increased levels of C-reactive protein or calprotectin). All subsequent and methodologically similar studies, that is, NMR-based studies, have been unable to differentiate between patients with UC and CD [49], [50], [51], [52], [53], although several of the metabolic perturbations have been replicated, albeit in patients with flaring IBD. Especially the proatherogenic lipid profile with increased VLDL [46,51,53] and decreased HDL [46,53] is repeatedly found in patients with active IBD and seems to be the consequence of increased transit times, malabsorption, and the inflammatory setting. In this environment, tumor necrosis factor alpha (TNF-α) stimulates the release of free fatty acids from adipocytes and the production of triglycerides in the liver and inhibits the hydrolysis of triglycerides in VLDL, leading to an increase in VLDL and decrease in HDL [54]. Moreover, in a longitudinal study monitoring the metabolic trajectory during induction treatment of IBD patients with infliximab, the proatherogenic lipid profile was ameliorated in the patients with IBD brought into remission, which was not the case for nonresponders [53]. This is of immediate clinical importance because an increased incidence of cardiovascular morbidity is found among patients with IBD [55,56], making patients with frequent flares or chronically active disease potential candidates for concomitant statin treatment and lifestyle intervention. This kind of intervention is currently not recommended in any international IBD treatment algorithms, but as the proatherogenic nature of the inflammation is further underlined by the low levels of choline [46,51] and glycerophosphocholine (GPC) [51,53] and an increased level of trimethylamine-N-oxide (TMAO) [57], all of which are correlated with cardiovascular disease [58,59] this might change in a foreseeable future.
3.3. Serum metabonomics and fatigued quiescent IBD patients
Fatigue is one of the most disabling [60] and prevalent extraintestinal manifestations of IBD and is registered in 80% of patients with active IBD and 50% of patients with quiescent IBD [61]. Despite these numbers, surprisingly little is known about fatigue in IBD, and the etiology still remains elusive. However, a recent exploratory lipidomic study associated fatigue and clinically quiescent IBD in patients with significantly decreased levels of PCs, LPCs, plasmanyls, sphingomyelins, phosphatidylethanolamines (PAs), phosphatidylinositols (PIs), phosphatidylserines (PSs), and eicosanoids [62]. Furthermore, a prevailing gut–brain-axis link hypothesis has animated a prospective multiomic study including quiescent IBD patients with and without fatigue; fatigued quiescent IBD patients were identified with lower biodiversity and a reduced abundance of Faecalibacterium, Rominococcus, and Alistipes and a higher abundance of Coprobacillus, which correlated with decreased levels of tryptophan and branched-chain amino acids (BCAAs) when compared with nonfatigued patients with quiescent IBD [63]. The proteome data revealed no significant differences in inflammatory cytokines (e.g., interleukin 6 [IL-6]) and thus no overt systemic inflammation. The authors consequently speculate whether a local proinflammatory environment exists in the intestines that is associated with fatigue despite clinical and endoscopic remission. This is in line with the previously described state of metabonomic remission, and whether the imbalance in tryptophan and BCAAs might play a pivotal part in the pathophysiology of fatigue in patients with quiescent IBD because these metabolites are known to modify mood and depressive symptoms [64,65]. This classical hypothesis-generating metabonomic study obviously merits further investigation because these findings have relevance beyond IBD owing to the high prevalence of fatigue throughout a wide range of chronic inflammatory diseases. A placebo-controlled crossover trail to determine the effect of oral 5-hydroxytryptophan on fatigue in patients with IBD is currently being performed and results are eagerly awaited (ClinicalTrials.gov; identifier: NCT03574948).
4. IBD metabonomics predicts disease course
Prediction of disease course in terms of progression and severity is essential to proactive precision medicine. Today the disease course is monitored by clinical scores, imaging, endoscopy, calprotectin, and blood samples resulting in active rather than proactive management, which calls for identification of predictive biomarkers.
Histidine is consistently reported at low levels in patients with IBD, which was also noted by Hisamatsu et al. [66], who in a prospective study of patients with quiescent UC and among 19 investigated metabolites solely identified low levels of plasma histidine to be predictive of relapse within a 1-year period. In this study, remission was based only on clinical scores, and subclinical inflammation might explain the low levels of histidine. However, Probert et al. [67] identified plasma metabolic profiles capable of discriminating between high and low endoscopic activity and corresponding histologic severity, demonstrating for the first time that plasma-based metabonomics can distinguish between mild and severe active UC. The patients were followed prospectively, and a distinct baseline metabolic profile was identified that predicted worsening of symptoms within a period of 6 months (accuracy 74% ± 4%), and again histidine, at low levels, was found to be one of the major predictive metabolites. Kohashi et al. [68] identified 114 metabolites in the sera from UC patients and control individuals. Based on test and validation cohorts (60 UC patients and 60 control individuals in each) and a predefined selection strategy for metabolites holding differential power, a diagnostic model that differentiated between UC and CD was created based on four metabolites: oxalate, 3-hydroxy-butyrate, ribulose, and 1,6-anhydroglucose. Applying the same strategy, a UC assessment model was also established that correlated with the clinical activity score. This model contained two metabolites, p-hydroxybenzoic acid and histidine, and again histidine was found at low levels, and in an inverse correlation with the clinical activity score. In a subsequent prospective monitoring study of patients with UC, patients in remission with succeeding flares had a concomitant increase in the assessment model's value and patients with active disease had a reduction of the value during treatment, suggesting that this model could be applied as a monitoring tool. Thus Kohashi et al. [68] developed potential diagnostic and assessment models for monitoring UC, but it is important to note that treatment with sulfasalazine was found to be positively correlated with high levels of p-hydroxybenzoic acid, which was one of the metabolites holding strong differential power, and that no cohort with non-IBD intestinal inflammation was included in the study. Thus, histidine seems to be a candidate biomarker for predicting disease course, but a major prospective IBD cohort study including clinical, endoscopic, and histologic scores is required to validate its potential.
Prediction of disease course also appears achievable using a multiomics approach on serum samples because it seems possible to identify otherwise quiescent IBD patients who are not in histologic remission and consequently at risk of an imminent flare. An established model of four metabolites, propionyl-l-carnitine, carnitine, sarcosine, and sorbitol, combined with three proteins, IL-10, glial cell line–derived neurotrophic factor, and the T-cell surface CD8 alpha chain, was found to be predictive of relapse within 2 years [22]. An independent validation of these results would translate into an applicable clinical tool and empower proactive treatment strategies.
5. IBD metabonomics predicts treatment response
5.1. Metabonomics predicts response to biologic therapy
A bile acid dysmetabolism characterized by defective deconjugation, transformation, and desulfation has repeatedly been documented in IBD and associated with dysbiosis [11,17,23,69,70]. This results in a relative overabundance of primary and sulfated bile acids and a dysbiosis-induced secondary bile acid deficiency that promotes colonic inflammation. The apparent bile acid dysmetabolism combined with a disturbed lipid profile has been shown to predict response to TNF inhibitor treatment because primary bile acids and increased levels of especially sphingomyelin in feces have been associated with nonresponders [23]. Similarly, butyrate and substrates involved in butyrate synthesis (e.g., acetaldehyde) have been identified as predictive metabolites of clinical remission following biologic therapy in a metabologenomics-based (i.e., microbiome) gene-sequencing and fecal metabolic profiling in silico prediction study [71]. Prior to treatment, a clear dysbiosis was found with reduced biodiversity and a reduction in individual phylotypes such as Coprococcus and Roseburia inulinivorans (short-chain fatty acid [SCFA] producers). After treatment, these differences were almost nonexistent in both responders and nonresponders. The fecal metabolic profile, in contrast, remained significantly disrupted in nonresponders, whereas responders experienced an increase especially in butyric acid (an SCFA). Importantly, already at baseline nonresponders had a more profoundly affected metabolic profile, making it possible to predict drug response. This study ultimately validated the results in an IBD cohort and confirmed that metabolic profiling of fecal samples has the potential to identify patients likely to achieve clinical remission with biologic treatment. Thus fecal-based metabolic profiles of bile acids, lipids, and SCFAs are already available for clinical testing and validation.
Serum-based metabonomics has identified lipid profiles with increased levels of phosphocholine, ceramide, sphingomyelin, and triglycerides in nonresponders to TNF inhibitors, demonstrating that a potential pharmacodynamic explanation for the primary driver of inflammation in anti-TNF nonresponders might be lipid based [23]. Thus, it is likely that the genesis of the dominant inflammatory process in patients with IBD correlates with different metabolic phenotypes in which some are driven by TNF and others are driven by lipid. If this kind of a priori knowledge can be made available in the clinical setting, it would undoubtedly have a significant impact on personalized medicine when choosing between a TNF inhibitor and other regimens from the therapeutic armamentarium.
5.2. Fecal metabonomics predicts fecal microbiota transplantation response
A few studies have indicated that FMT in patients with UC has the ability to induce remission [72], [73], [74]. However, the underlaying microbial processes, mechanisms of action, and predictors of outcome still remain unknown. An exploratory study applied a metabologenomics approach and revealed greater biodiversity before and after FMT in patients achieving remission, with enrichment of Eubacterium hallii and R. inulivorans and increased levels of SCFA biosynthesis and secondary bile acids in treated versus nontreated patients [24], which is in line with changes observed in anti-TNF responders and nonresponders. An essentially similar pediatric study of FMT in patients with UC combining 16S rRNA gene sequencing and metabonomics illustrated that the gut microbial and metabolic profiles, including changes in SCFAs among patients responding to FMT, shifted toward those of the donors. However, the metabolic profile of one patient remained stable and significantly different from those of healthy donors despite successful transfer of microbes. This patient experienced only a partial response to FMT, highlighting the fact that the response to treatment does not depend on single factors such as SCFAs or specific microbiota, but rather on an interplay among multiple factors [75]. Thus, treatment and subsequent reestablishment of the microbiota is not sufficient to achieve remission but have to be paralleled with concomitant changes in the metabolome. The mechanism of action by which FMT and biologics induce these changes is currently unknown and necessitates further investigation of the host–microbiota interaction with detailed in-depth molecular characterization of the mucosa-related microbiota and its interplay with host omics during treatment.
6. Outstanding questions and future key clinical perspectives
Is the potential metabonomics remission associated with an even better long-term state of remission compared to histological remission and consequently a disease modifying course?
The treat-to-target paradigm is currently aimed at mucosal healing, however, increasing evidence support deep mucosal healing with histological remission as the next target. With the existence of a potential metabonomics remission in IBD a future treatment target might be set at the molecular level to achieve better long-term outcomes.
Should IBD patients with chronic active disease or frequent flares be treated with lifestyle intervention and statins due to a proatherogenic lipid profile?
A proatherogenic serum lipid profile characterizes patients with active IBD and correlates with an increased incidence of cardiovascular morbidity. Achieving a state of remission ameliorates the proatherogenic profile underlining the importance of treat-to-target and concomitant statin treatment and lifestyle intervention in patients with frequent flares or chronical activity. The latter is not included in any international IBD treatment guidelines, but with the current knowledge this might change in a foreseeable future.
Is it possible to treat fatigue in quiescent IBD patients?
Fatigue is the most prominent extraintestinal manifestation in IBD with up to 50% of patients in remission being affected, yet little is known about the cause. Metabonomics has, however, identified tryptophane and branched chain amino acids to be potential treatment strategies and current clinical trials with tryptophane is ongoing.
Is histidine and integrated omics able to predict eminent flares prior to manifest clinical symptoms enabling precision medicine?
Predicting disease course is the very essence of precision medicine. Serum-based metabonomics seem to have identified histidine and an integrated metabonomics and proteomics profile able to predict eminent flares enabling proactive medical care if validated in large prospective IBD cohorts.
Can fecal metabonomics predict response to biological and FMT treatment?
Fecal bile acids, lipids, and SCFA profiles have been shown to predict response to biologics and FMT facilitating a priori identification of appropriate candidates. If validated, this will have a tremendous impact on future treatment strategies.
7. Conclusions
This review has provided op-to-date knowledge on the workflow and commonly applied analytical techniques in IBD metabonomics and has shown metabonomics to be a very promising clinical tool for predictive modeling in terms of phenotyping, imminent flares, and response to treatment. Thus, the proatherogenic lipid profile in active IBD and tryptophan deficits in the fatigued phenotype are readily converted into clinically applicable treatment strategies. Moreover, the identified concept of metabonomic remission might in the foreseeable future change our treatment target for patients with UC and potentially alter the natural history of the disease. Finally, real world precision medicine is one step closer in IBD because of the identified predictive metabolites and omics profiles. We anticipate that future standardized protocols, affordable instruments, and user-friendly analysis platforms will become more widely available, and metabonomics will consequently play an increasingly important role alongside other diagnostic and prognostic tools in a clinical setting and provide us with personalized medicine, as well as clues for new therapeutic avenues.
Search strategy
Data for this Review were identified by searches of MEDLINE, Current Contents, PubMed, and references from relevant articles using the search terms “metabolomics”, “metabonomics”, inflammatory bowel disease”, “ulcerative colitis” and “Crohn's disease”. Only articles published in English between 1980 and 2021 were included.
Contributors
Conceptualisation: JTB, OHN; literature search: JTB, OHN; original draft: JTB; writing: JTB, YLW, JBS, OHN; figures and visualization: JTB; review and editing: JTB, YLW, JBS, OHN
Declaration of Competing Interest
The authors have no interests to declare.
Acknowledgements
The authors acknowledge the financial support from Aase og Ejnar Danielsens Fond, Civilingeniør Frode V. Nygaard og Hustrus Fond, Aage og Johanne Louis-Hansens Fond, Colitis-Crohn Foreningen, and Frimodt-Heineke Fonden. The funders were not involved in the study in any way.
References
- 1.Zhao M., Gönczi L., Lakatos P.L., Burisch J. The burden of inflammatory bowel disease in Europe in 2020. J Crohn’s Colitis. 2021 doi: 10.1093/ecco-jcc/jjab029. In press. [DOI] [PubMed] [Google Scholar]
- 2.Chang J.T. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T., Siegmund B., Le Berre C. Ulcerative colitis. Nat Rev Dis Prim. 2020;6:74. doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- 4.Roda G., Chien Ng S., Kotze P.G. Crohn's disease. Nat Rev Dis Prim. 2020;6:22. doi: 10.1038/s41572-020-0156-2. [DOI] [PubMed] [Google Scholar]
- 5.Le Berre C., Ananthakrishnan A.N., Danese S., Singh S., Peyrin-Biroulet L. Ulcerative colitis and Crohn's disease have similar burden and goals for treatment. Clin Gastroenterol Hepatol. 2020;18:14–23. doi: 10.1016/j.cgh.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson J.K., Lindon J.C., Holmes E. Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 7.Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 8.Van Malderen K., De Winter B.Y., De Man J.G., De Schepper H.U., Lamote K. Volatomics in inflammatory bowel disease and irritable bowel syndrome. EBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher K., Catesson A., Griffin J.L., Holmes E., Williams H.R.T. Metabolomic analysis in inflammatory bowel disease: a systematic review. J Crohn's Colitis. 2021;15:813–826. doi: 10.1093/ecco-jcc/jjaa227. [DOI] [PubMed] [Google Scholar]
- 10.Bauset C., Gisbert-Ferrándiz L., Cosín-Roger J. Metabolomics as a promising resource identifying potential biomarkers for inflammatory bowel disease. J Clin Med. 2021;10:622. doi: 10.3390/jcm10040622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzosa E.A., Sirota-Madi A., Avila-Pacheco J. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee T., Clavel T., Smirnov K. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. 2017;66:863–871. doi: 10.1136/gutjnl-2015-309940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santoru M.L., Piras C., Murgia A. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjerrum J.T., Wang Y., Hao F. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122–133. doi: 10.1007/s11306-014-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan S., Jia B., Chao K. UPLC-QTOF-MS-based plasma lipidomic profiling reveals biomarkers for inflammatory bowel disease diagnosis. J Proteome Res. 2020;19:600–609. doi: 10.1021/acs.jproteome.9b00440. [DOI] [PubMed] [Google Scholar]
- 16.Manfredi M., Conte E., Barberis E. Integrated serum proteins and fatty acids analysis for putative biomarker discovery in inflammatory bowel disease. J Proteom. 2019;195:138–149. doi: 10.1016/j.jprot.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Jansson J., Willing B., Lucio M. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bezabeh T., Somorjai R.L., Smith I.C.P. The use of 1H magnetic resonance spectroscopy in inflammatory bowel diseases: distinguishing ulcerative colitis from Crohn's disease. Am J Gastroenterol. 2001;96:442–448. doi: 10.1111/j.1572-0241.2001.03523.x. [DOI] [PubMed] [Google Scholar]
- 19.Bjerrum J.T., Nielsen O.H., Hao F. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J Proteome Res. 2010;9:954–962. doi: 10.1021/pr9008223. [DOI] [PubMed] [Google Scholar]
- 20.Diab J., Hansen T., Goll R. Mucosal metabolomic profiling and pathway analysis reveal the metabolic signature of ulcerative colitis. Metabolites. 2019;9:291. doi: 10.3390/metabo9120291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diab J., Hansen T., Goll R. Lipidomics in ulcerative colitis reveal alteration in mucosal lipid composition associated with the disease state. Inflamm Bowel Dis. 2019;25:1780–1787. doi: 10.1093/ibd/izz098. [DOI] [PubMed] [Google Scholar]
- 22.Borren N.Z., Plichta D., Joshi A.D. Multi-"-omics" profiling in patients with quiescent inflammatory bowel disease identifies biomarkers predicting relapse. Inflamm Bowel Dis. 2020;26:1524–1532. doi: 10.1093/ibd/izaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding N.S., McDonald J.A.K., Perdones-Montero A. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn's disease. J Crohn's Colitis. 2020;14:1090–1102. doi: 10.1093/ecco-jcc/jjaa039. [DOI] [PubMed] [Google Scholar]
- 24.Paramsothy S., Nielsen S., Kamm M.A. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Panner Selvam M.K., Finelli R., Agarwal A., Henkel R. Proteomics and metabolomics-current and future perspectives in clinical andrology. Andrologia. 2021;53:e13711. doi: 10.1111/and.13711. [DOI] [PubMed] [Google Scholar]
- 26.Wishart D.S., Tzur D., Knox C. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjerrum J.T., Nielsen O.H., Wang Y.L., Olsen J. Technology insight: metabonomics in gastroenterology-basic principles and potential clinical applications. Nat Clin Pract Gastroenterol Hepatol. 2008;5:332–343. doi: 10.1038/ncpgasthep1125. [DOI] [PubMed] [Google Scholar]
- 28.Shi X., Xiao C., Wang Y., Tang H. Gallic acid intake induces alterations to systems metabolism in rats. J Proteome Res. 2013;12:991–1006. doi: 10.1021/pr301041k. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Bollard M.E., Keun H. Spectral editing and pattern recognition methods applied to high-resolution magic-angle spinning 1H nuclear magnetic resonance spectroscopy of liver tissues. Anal Biochem. 2003;323:26–32. doi: 10.1016/j.ab.2003.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Tang H., Holmes E. Biochemical characterization of rat intestine development using high-resolution magic-angle-spinning 1H NMR spectroscopy and multivariate data analysis. J Proteome Res. 2005;4:1324–1329. doi: 10.1021/pr050032r. [DOI] [PubMed] [Google Scholar]
- 31.Theodoridis G., Gika H.G., Wilson I.D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. TrAC Trends Anal Chem. 2008;27:251–260. [Google Scholar]
- 32.Lin H., An Y., Tang H., Wang Y. Alterations of bile acids and gut microbiota in obesity induced by high fat diet in rat model. J Agric Food Chem. 2019;67:3624–3632. doi: 10.1021/acs.jafc.9b00249. [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Zhou L., Lei H. Simultaneous quantification of amino metabolites in multiple metabolic pathways using ultra-high performance liquid chromatography with tandem-mass spectrometry. Sci Rep. 2017;7:1423. doi: 10.1038/s41598-017-01435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courant F., Antignac J.P., Dervilly-Pinel G., Le Bizec B. Basics of mass spectrometry based metabolomics. Proteomics. 2014;14:2369–2388. doi: 10.1002/pmic.201400255. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z., Chen Y., Gao Y. Development of a high-coverage metabolome relative quantitative method for large-scale sample analysis. Anal Chim Acta. 2020;1109:44–52. doi: 10.1016/j.aca.2020.02.049. [DOI] [PubMed] [Google Scholar]
- 36.Gathungu R.M., Kautz R., Kristal B.S., Bird S.S., Vouros P. The integration of LC-MS and NMR for the analysis of low molecular weight trace analytes in complex matrices. Mass Spectrom Rev. 2020;39:35–54. doi: 10.1002/mas.21575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bylesjö M. Extracting meaningful information from metabonomic data using multivariate statistics. Methods Mol Biol. 2015;1277:137–146. doi: 10.1007/978-1-4939-2377-9_11. [DOI] [PubMed] [Google Scholar]
- 38.Bjerrum J.T. Metabonomics: analytical techniques and associated chemometrics at a glance. Methods Mol Biol. 2015;1277:1–14. doi: 10.1007/978-1-4939-2377-9_1. [DOI] [PubMed] [Google Scholar]
- 39.Sharma U., Singh R.R., Ahuja V., Makharia G.K., Jagannathan N.R. Similarity in the metabolic profile in macroscopically involved and un-involved colonic mucosa in patients with inflammatory bowel disease: an in vitro proton (1H) MR spectroscopy study. Magn Reson Imaging. 2010;28:1022–1029. doi: 10.1016/j.mri.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 40.Rantalainen M., Bjerrum J.T., Olsen J., Nielsen O.H., Wang Y. Integrative transcriptomic and metabonomic molecular profiling of colonic mucosal biopsies indicates a unique molecular phenotype for ulcerative colitis. J Proteome Res. 2015;14:479–490. doi: 10.1021/pr500699h. [DOI] [PubMed] [Google Scholar]
- 41.Bjerrum J.T., Rantalainen M., Wang Y., Olsen J., Nielsen O.H. Integration of transcriptomics and metabonomics: improving diagnostics, biomarker identification and phenotyping in ulcerative colitis. Metabolomics. 2014;10:280–290. doi: 10.1007/s11306-013-0580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehehalt R., Wagenblast J., Erben G. Phosphatidylcholine and lysophosphatidylcholine in intestinal mucus of ulcerative colitis patients. A quantitative approach by nanoElectrospray-tandem mass spectrometry. Scand J Gastroenterol. 2004;39:737–742. doi: 10.1080/00365520410006233. [DOI] [PubMed] [Google Scholar]
- 43.Karner M., Kocjan A., Stein J. First multicenter study of modified release phosphatidylcholine “LT-02” in ulcerative colitis: a randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol. 2014;109:1041–1051. doi: 10.1038/ajg.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dal Buono A., Roda G., Argollo M., Peyrin-Biroulet L., Danese S. Histological healing: should it be considered as a new outcome for ulcerative colitis? Expert Opin Biol Ther. 2020;20:407–412. doi: 10.1080/14712598.2020.1701652. [DOI] [PubMed] [Google Scholar]
- 45.Bjerrum J.T., Nielsen O.H. Metabonomics in gastroenterology and hepatology. Int J Mol Sci. 2019;20:3638. doi: 10.3390/ijms20153638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams H.R.T., Willsmore J.D., Cox I.J. Serum metabolic profiling in inflammatory bowel disease. Dig Dis Sci. 2012;57:2157–2165. doi: 10.1007/s10620-012-2127-2. [DOI] [PubMed] [Google Scholar]
- 47.Walmsley R.S., Ayres R.C.S., Pounder R.E., Allan R.N. A simple clinical colitis activity index. Gut. 1998;43:29–32. doi: 10.1136/gut.43.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey R.F., Bradshaw J.M. A simple index of Crohn's-disease activity. Lancet. 1980;1:514. doi: 10.1016/s0140-6736(80)92767-1. (London, England) [DOI] [PubMed] [Google Scholar]
- 49.Schicho R., Shaykhutdinov R., Ngo J. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res. 2012;11:3344–3357. doi: 10.1021/pr300139q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Lin L., Xu Y. 1H NMR-based spectroscopy detects metabolic alterations in serum of patients with early-stage ulcerative colitis. Biochem Biophys Res Commun. 2013;433:547–551. doi: 10.1016/j.bbrc.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Dawiskiba T., Deja S., Mulak A. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol. 2014;20:163–174. doi: 10.3748/wjg.v20.i1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fathi F., Majari-Kasmaee L., Mani-Varnosfaderani A. 1H NMR based metabolic profiling in Crohn's disease by random forest methodology. Magn Reson Chem. 2014;52:370–376. doi: 10.1002/mrc.4074. [DOI] [PubMed] [Google Scholar]
- 53.Bjerrum J.T., Steenholdt C., Ainsworth M. Metabonomics uncovers a reversible proatherogenic lipid profile during infliximab therapy of inflammatory bowel disease. BMC Med. 2017;15:184. doi: 10.1186/s12916-017-0949-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popa C., Netea M.G., van Riel P., van der Meer J.W.M., Stalenhoef A.F.H. The role of TNF-alpha in chronic inflammatory conditions, intermediary metabolism, and cardiovascular risk. J Lipid Res. 2007;48:751–762. doi: 10.1194/jlr.R600021-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Kristensen S.L., Ahlehoff O., Lindhardsen J. Inflammatory bowel disease is associated with an increased risk of hospitalization for heart failure: a Danish nationwide cohort study. Circ Heart Fail. 2014;7:717–722. doi: 10.1161/CIRCHEARTFAILURE.114.001152. [DOI] [PubMed] [Google Scholar]
- 56.Biondi R.B., Salmazo P.S., Bazan S.G.Z. Cardiovascular risk in individuals with inflammatory bowel disease. Clin Exp Gastroenterol. 2020;13:107–113. doi: 10.2147/CEG.S243478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun M., Du B., Shi Y. Combined signature of the fecal microbiome and plasma metabolome in patients with ulcerative colitis. Med Sci Monit. 2019;25:3303–3315. doi: 10.12659/MSM.916009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Syme C., Czajkowski S., Shin J. Glycerophosphocholine metabolites and cardiovascular disease risk factors in adolescents: a cohort study. Circulation. 2016;134:1629–1636. doi: 10.1161/CIRCULATIONAHA.116.022993. [DOI] [PubMed] [Google Scholar]
- 59.Roncal C., Martínez-Aguilar E., Orbe J. Trimethylamine-N-oxide (TMAO) predicts cardiovascular mortality in peripheral artery disease. Sci Rep. 2019;9:15580. doi: 10.1038/s41598-019-52082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Gennep S., Gielen M.E., Rietdijk S.T. Work productivity loss is determined by fatigue and reduced quality of life in employed inflammatory bowel disease patients. Eur J Gastroenterol Hepatol. 2021 doi: 10.1097/MEG.0000000000002178. In press. [DOI] [PubMed] [Google Scholar]
- 61.Borren N.Z., van der Woude C.J., Ananthakrishnan A.N. Fatigue in IBD: epidemiology, pathophysiology and management. Nat Rev Gastroenterol Hepatol. 2019;16:247–259. doi: 10.1038/s41575-018-0091-9. [DOI] [PubMed] [Google Scholar]
- 62.Horta D., Moreno-Torres M., Ramírez-Lázaro M.J. Analysis of the association between fatigue and the plasma lipidomic profile of inflammatory bowel disease patients. J Proteome Res. 2021;20:381–392. doi: 10.1021/acs.jproteome.0c00462. [DOI] [PubMed] [Google Scholar]
- 63.Borren N.Z., Plichta D., Joshi A.D. Alterations in fecal microbiomes and serum metabolomes of fatigued patients with quiescent inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2021;19:519–527. doi: 10.1016/j.cgh.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Jenkins T., Nguyen J., Polglaze K., Bertrand P. Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain axis. Nutrients. 2016;8:56. doi: 10.3390/nu8010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baranyi A., Amouzadeh-Ghadikolai O., von Lewinski D. Branched-chain amino acids as new biomarkers of major depression - a novel neurobiology of mood disorder. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hisamatsu T., Ono N., Imaizumi A. Decreased plasma histidine level predicts risk of relapse in patients with ulcerative colitis in remission. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Probert F., Walsh A., Jagielowicz M. Plasma nuclear magnetic resonance metabolomics discriminates between high and low endoscopic activity and predicts progression in a prospective cohort of patients with ulcerative colitis. J Crohn's Colitis. 2018;12:1326–1337. doi: 10.1093/ecco-jcc/jjy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohashi M., Nishiumi S., Ooi M. A novel gas chromatography mass spectrometry-based serum diagnostic and assessment approach to ulcerative colitis. J Crohns Colitis. 2014;8:1010–1021. doi: 10.1016/j.crohns.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 69.Duboc H., Rajca S., Rainteau D. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 70.Weng Y.J., Gan H.Y., Li X. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J Dig Dis. 2019;20:447–459. doi: 10.1111/1751-2980.12795. [DOI] [PubMed] [Google Scholar]
- 71.Aden K., Rehman A., Waschina S. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel siseases. Gastroenterology. 2019;157:1279–1292. doi: 10.1053/j.gastro.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 72.Moayyedi P., Surette M.G., Kim P.T. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Paramsothy S., Kamm M.A., Kaakoush N.O. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 74.Costello S.P., Hughes P.A., Waters O. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis. JAMA. 2019;321:156. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nusbaum D.J., Sun F., Ren J. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol Ecol. 2018;94:fiy133. doi: 10.1093/femsec/fiy133. [DOI] [PMC free article] [PubMed] [Google Scholar]