Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) is characterized by an important heterogeneity, reflected by different clinical outcomes and chemoresistance. During carcinogenesis, tumor cells display aberrant glycosylated structures, synthetized by deregulated glycosyltransferases, supporting the tumor progression. In this study, we aimed to determine whether PDAC could be stratified through their glycosyltransferase expression profiles better than the current binary classification (basal-like and classical) in order to improve detection of patients with poor prognosis.
Methods
Bioinformatic analysis of 169 glycosyltransferase RNA sequencing data were performed for 74 patient-derived xenografts (PDX) of resected and unresectable tumors. The Australian cohort of International Cancer Genome Consortium and the microarray dataset from Puleo patient's cohort were used as independent validation datasets.
Findings
New PDAC stratification based on glycosyltransferase expression profile allowed to distinguish different groups of patients with distinct clinical outcome (p-value = 0.007). A combination of 19 glycosyltransferases differentially expressed in PDX defined a glyco-signature, whose prognostic value was validated on datasets including resected whole tumor tissues. The glyco-signature was able to discriminate three clusters of PDAC patients on the validation cohorts, two clusters displaying a short overall survival compared to one cluster having a better prognosis. Both poor prognostic clusters having different glyco-profiles in Puleo patient's cohort were correlated with stroma activated or desmoplastic subtypes corresponding to distinct microenvironment features (p-value < 0.0001). Besides, differential expression and enrichment analyses revealed deregulated functional pathways specific to different clusters.
Interpretation
This study identifies a glyco-signature relevant for a prognostic use, potentially applicable to resected and unresectable PDAC. Furthermore, it provides new potential therapeutic targets.
Funding
This work was supported by INCa (Grants number 2018-078 and 2018-079), Fondation ARC (Grant number ARCPJA32020070002326), Cancéropôle PACA, DGOS (labelization SIRIC, Grant number 6038), Amidex Foundation and Ligue Nationale Contre le Cancer and by institutional fundings from INSERM and the Aix-Marseille Université.
Keywords: Glyco-signature, Glycosyltransferases, Pancreatic ductal adenocarcinoma, Prognosis, Tumor stratification, Transcriptomics
Research in Context.
Evidence before this study
Pancreatic ductal adenocarcinoma (PDAC) is characterized by tumor heterogeneity limiting the adapted patient care and contributing to its poor prognosis related to late diagnosis and chemoresistance. Previous studies have proposed a large-scale molecular classification of PDAC, describing basal-like and classical prognostic subtypes. To refine our ability in deciphering PDAC heterogeneity, we focused on actors of glycosylation pathways. This process highly deregulated in PDAC, is widely involved in tumor properties acquisition, impacting each mechanistic step of carcinogenesis.
Added value of this study
We show that PDAC stratification through glycosyltransferase (GT) gene expression profile is significantly associated with patient survival as an independent prognostic factor. This led to identification of a glyco-signature funded on 19 GT genes that allows the stratification of PDAC with specific molecular profiles and clinical features of patients driving tumor aggressiveness. In addition, the GT gene expression profiles were correlated with the expression of Lewis blood group antigens synthetized by many GT included in the glyco-signature. This signature is able to better describe PDAC than the current classification by splitting PDAC classical subtype into two clusters with poor clinical outcome and another one with better prognosis. The two poor pronostic clusters, defined by the glyco-signature on the validation cohorts, are associated with distinct microenvironment features suggesting that the glycosylation pathways of tumor cells can reflect molecular diversity at stromal level.
Implication of all the available evidence
The glyco-signature, potentially applicable to resected and unresectable PDAC, could be used to predict patient outcome.
Alt-text: Unlabelled box
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) ranks seventh among the causes of cancer deaths [1,2] and is in path to become the second cause of global cancer mortality by 2030 [3]. The 5-year survival rate is < 7% and varies depending on the cancer staging at the time of diagnosis [2,4]. Despite advances in pancreatic cancer research field, PDAC is still characterized by a resistance to chemotherapies and presents a disparity in the response to treatments and survival depending mainly on the surgical management of the patient [5], [6], [7]. The main cause of disparities in clinical evolution and resistance to conventional chemotherapies between patients is related to a significant molecular heterogeneity between their tumors. Therefore, stratification of PDAC appears as a favored strategy to improve patient care and allow the best clinical decision for precision medicine. Several studies have analyzed gene expression of tumor and/or stroma from primary and metastatic PDAC by using mRNA expression microarray and RNA sequencing (RNA-seq); many molecular subtypes of PDAC have been proposed with potential prognostic relevance [8], [9], [10], [11], [12], [13]. Importantly, two clusters of PDAC corresponding to classical and basal-like subtypes are often highlighted with differences in overall survival (OS) and morphological patterns, constituting a well-established consensus.
Glycosylation in eukaryotic cells is the major post-translational modification of macromolecules and participate in the maturation and acquisition of their functions in the cell [14]. This process involves mainly sequential actions of different families of glycosylation enzymes such as glycosyltransferases (GT) and glycosidases, whose expression and function are tightly regulated in each cell. These glyco-enzymes are able to produce numerous and various glycan chains from monosaccharides to form glycoproteins bearing mainly N- and O-linked glycans, glycolipids and proteoglycans. Glycosylation deregulation is one of the important mechanisms contributing to tumor heterogeneity and is now widely accepted as one of the hallmarks of cancer [15]. A large number of studies have described significant changes in glycan chains of glycoconjugates and glycosylation processes in cancers [16,17]. The appearance of aberrant glycosylated structures on tumor cells, but also on cells from the microenvironment, seems to be essential for malignant transformation, tumor growth, cell signaling, as well as cell adhesion and metastatic dissemination [18,19]. Upstream, deregulation of this process occurs partly at the gene expression level of GT: a deep modification of GT gene expression generates aberrant glycosylated antigens and deregulates the whole glycome of cancer cells contributing to their aggressive phenotype [15], [16], [17], [18], [19], [20].
In PDAC, the most common changes in glycan structures affect glycosylated Lewis blood group antigens [20], [21], [22]. The sialyl-Lewisx and sialyl-Lewisa antigens, whose biosynthesis is dependent from α1,3/4-fucosyltransferases (FUT) and α2,3-sialyltransferases (ST3GAL), are preferentially expressed on the surface of tumor cells, which, in the blood or lymphatic circulation, can be recognized by selectins, expressed at the membrane of endothelial cells and could thus promote the formation of metastases [23,24]. The expression of truncated O-glycans such as Tn and sialyl-Tn antigens, described in many cancers, was also observed in PDAC [25,26]. The expression of sialyl-Tn antigen is dependent on many GT such as polypeptide N-acetylgalactosaminyltransferase (GALNT) and α2,6-sialyltransferase (ST6GALNAC) families, but also on deregulated expression of chaperone and GT involved in the elongation of O-glycans such as COSMC, core 1 synthase glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase 1 (C1GALT1) or β1,6-N-acetylglucosaminyltransferases (GCNT1 or GCNT3). These truncated O-glycans could promote tumor growth and metastatic behavior [27].
In a previous study, glycoprotein metabolism has been highlighted as one of the top biological processes significantly deregulated in PDAC [28] suggesting that relevant subtypes could be identified according to their aberrant glycosylation processes. In this study, we aimed to determine whether PDAC could be stratified on the basis of their GT gene expression profiles involved in the biosynthesis of glycoconjugates. By using bioinformatic analysis of RNA-seq data focused on 169 GT genes from patient-derived xenografts (PDX), we have identified a combination of 19 GT, which was able to discriminate two clusters of PDAC on PaCaOmics patient's cohort and three clusters on validation cohorts associated with specific molecular profiles and clinical features of patients. These GT genes were validated on public databases as a prognostic glyco-signature, which could allow best patient care in the future and also highlight new potential targets for diagnosis, prognosis and therapies of different PDAC.
2. Methods
2.1. PaCaOmics patient's cohort and PDX
The PaCaOmics patient's cohort was described in many previous studies [28], [29], [30], [31], [32]. Briefly, three expert clinical centers collaborated on the PaCaOmics project to establish a pancreatic cancer cohort. It includes PDAC samples collected from echoendoscopic ultrasound-guided fine-needle (EUS-FNA) biopsies for patients with unresectable tumors (25 patients), samples obtained from surgical specimens for patients undergoing surgery (40 patients) and samples from patients with carcinomatosis or liver metastasis obtained during explorative laparotomy (11 patients). The consent forms of informed patient were collected in a central database according to ethic principles. Outliers were excluded after a histological selection and 74 patients suffering exclusively with pancreatic adenocarcinoma were retained for this study. The tumor samples of these patients were used to generate PDX. PDX model allows to amplify tumor samples from patients with resected and unresectable tumors; PDX model from the PaCaOmics patient's cohort is representative of patient tissue and faithfully recapitulates patient tumors histological characteristics [32]. It is also possible to discriminate in silico the data from RNA-sequencing between the human tumor cells and mouse stroma cells by assigning each sequence to the human or mouse genome. The protocol to generate PDX was described in detail in previous studies [28], [29], [30], [31], [32]. Briefly, PDAC samples obtained from resected and unresectable tumors were mixed with Matrigel and implanted with a trocar in the subcutaneous upper flank of an anesthetized NMRI-nude mouse. Once xenografted tumors reached 1 cm3, they were removed and passed to NMRI-nude mice for 3 cycles of xenografts. Then, tumors were isolated for RNA extraction and Tissue Microarray construction.
2.2. Ethic statements
This work is a data mining of PaCaOmics data previously published [28], [29], [30], [31], [32]. The PaCaOmics study was registered at www.clinicaltrials.gov with registration number NCT01692873. PDAC samples were collected from January 2012 to December 2015. The study was approved by the local ethics committee (Comité de protection des personnes Sud Méditerranée I) following patient informed consent. All experimental procedures on animals were approved by the ethical committee for animal experimentation and French Ministry of Higher Education and Research (APAFIS# 9562-2016051914513578). All experimental protocols were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
2.3. RNA-seq analysis and gene selection
RNA libraries were generated from 74 PDX samples, then RNA-seq data were acquired and processed as previously described [29]. Analyses were performed on genes encoding main GT families involved in glycosylation machinery from human species-specific RNA reads. Therefore a gene set of 211 GT genes was extracted from GlycoGene DataBase (GGDB; https://acgg.asia/ggdb2/) and based on GT gene group section of Hugo Gene Nomenclature Committee (HGNC: https://www.genenames.org/data/genegroup/#!/group/424). Among available genes in our RNA-seq dataset, those belonging to UDP glucuronosyltransferase family were excluded as their expression values were close to 0. Analysis was performed on a list of 169 genes (supplementary Table S1). Unsupervised hierarchical clustering was performed using Euclidean distance algorithm and Ward.D2’s method for linkage. Heatmap representations of 74 PDX focusing on 169 GT genes were generated using “ComplexHeatmap” package on RStudio version 1.2.5033. Relative expression between samples for each gene is depicted in a color gradation and intensities varying from red to blue for upregulated and downregulated genes, respectively.
2.4. Hierarchical clustering on principal component (HCPC) analysis and glyco-signature definition
HCPC analyses were performed using ‘FactoMineR’ and ‘Factoextra’ packages of R programming language. To extract essential information and get rid of noise, only components that explain at least 5% of variances were retained for further analysis. Hierarchical clustering was then performed on PCA, using Euclidean distance algorithm and Ward's method for linkage. The link between the cluster variable and the quantitative variable corresponding to the GT genes was described with the square correlation coefficient of the F-test in a one-way analysis of variance. A total of 19 GT genes showing the most significant p-values were selected as the most relevant GT markers in cluster definition constituting the glyco-signature (for complete HCPC statistics, see supplementary Table S1). In addition, the statistical significance of the 19 GT gene expression between clusters was assessed by using Kruskal-Wallis test.
2.5. Validation on public datasets
The Australian cohort of International Cancer Genome Consortium (ICGC) was used as external validation cohort for the glyco-signature [10]. Both RNA-seq and gene microarray expression datasets, including 91 and 269 resected pancreatic tumors, respectively, were downloaded from ICGC Data Portal (data repositories, release 20 on http://dcc.icgc.org/). The Australian ICGC RNA-seq and microarray cohorts were profiled on Illumina sequencing platform and Illumina microarrays, respectively, from frozen samples. Gene expression raw values were normalized and gene-wise centered. Note that among the 19 GT markers, GALNT4 gene was not part of ICGC gene microarray expression dataset. The glyco-signature was validated on a second independent affymetrix transcriptomic dataset, obtained from the Puleo et al. [12] multi-centric cohort including 309 resected primary PDAC. This cohort was profiled on Affymetrix HG-U219 microarrays from paraffin-embedded samples. Transcriptomic profiles were normalized using robust multi-array average and batch effects were corrected. In both affymetrix transcriptomic datasets, multiple probes are used to target the same gene. The probe showing the highest variance between samples was selected, since it was considered as the one providing the most amount of information.
2.6. Comparison with previously established classification
Basal-like/classical subtyping for each dataset was performed using Purity Independent Subtyping of Tumors (PurIST), an algorithm of a Single Sample Classifier (SSC) developed by Rashid et al. [33]. Proportions of basal-like and classical subtype PDAC in clusters were determined for ICGC RNA-seq dataset and the association between both classifications was assessed using Fisher's exact test from “stats” package of R language. Concerning the ICGC microarray dataset and Puleo patient's cohort, Pearson's chi-squared test from “stats” package of R language was used to determine the significance of subtype distribution in clusters obtained through analysis of GT gene expression. Similarly, proportions of subtypes based on microenvironment features proposed by the authors in Puleo patient's cohort were assessed in clusters, using Pearson's chi-squared test. Standardized residuals for Pearson's chi-squared tests were extracted using “questionr” R package and represented on mosaic plots.
2.7. Survival analyses
Survival analysis based on multivariate Cox proportional hazard regression model was performed using the “survival” package by including multiple covariates (clustering, surgical resection and disease stage) with a confidence interval of CI = 95%. The follow-up starting point of survival time was defined at the diagnostic time. Concerning the Puleo's cohort and the ICGC affymetrix dataset, analyses were performed on the available survival data of 288 and 267 patients, respectively. Overall survival was defined as the time from diagnosis (in PaCaOmics patient's cohort) or surgery (in validation cohorts) to death resulting from any cause. Survival curves were estimated using the Kaplan-Meier method and OS were compared with the log-rank test. Multivariate Cox regression analyses and Kaplan-Meier curves were computed using the “survival” and “survminer” R packages.
2.8. Differential expression and functional analysis
Differential expression analyses comparing pairwise clusters were performed using “DESeq2” and “limma” R packages for RNA-seq and microarray datasets, respectively. Adjusted p-value < 0.05 and |log2(fold change)| > 1 were defined as cut-off criteria. Differentially expressed genes were represented on volcano-plots and Venn diagrams for comparisons between 2 clusters and between 3 clusters, respectively.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed for upregulated and downregulated genes using “ClusterProfiler” R package.
2.9. Statistical analysis
All the statistical analyses performed in this study have been described in each subsection, respectively. HCPC analyses were performed using Euclidean distance algorithm and Ward's method for linkage from the “FactoMineR” and “Factoextra” packages of R language. The link between the cluster and the GT gene variables was assessed using the F-test of a one-way analysis of variance. Moreover, associations between classifications were assessed using either the Pearson's chi-squared test or the Fisher's exact test, both from “stats” package of R language, depending on the sample size. The correlation analysis between GT genes of the glyco-signature and the PAMG was performed using the Pearson correlation test. Survival analysis based on multivariate Cox proportional hazard regression model was performed using the “survival” package by including multiple covariates with a confidence interval of CI = 95%. Overall survivals were compared using the log-rank test.
2.10. Role of the funding source
The funders did not have any role in the study design, the data collection, the data analyses, the result interpretation or the writing of the manuscript.
3. Results
3.1. Expression profiles of GT genes predict overall survival of PDAC patients
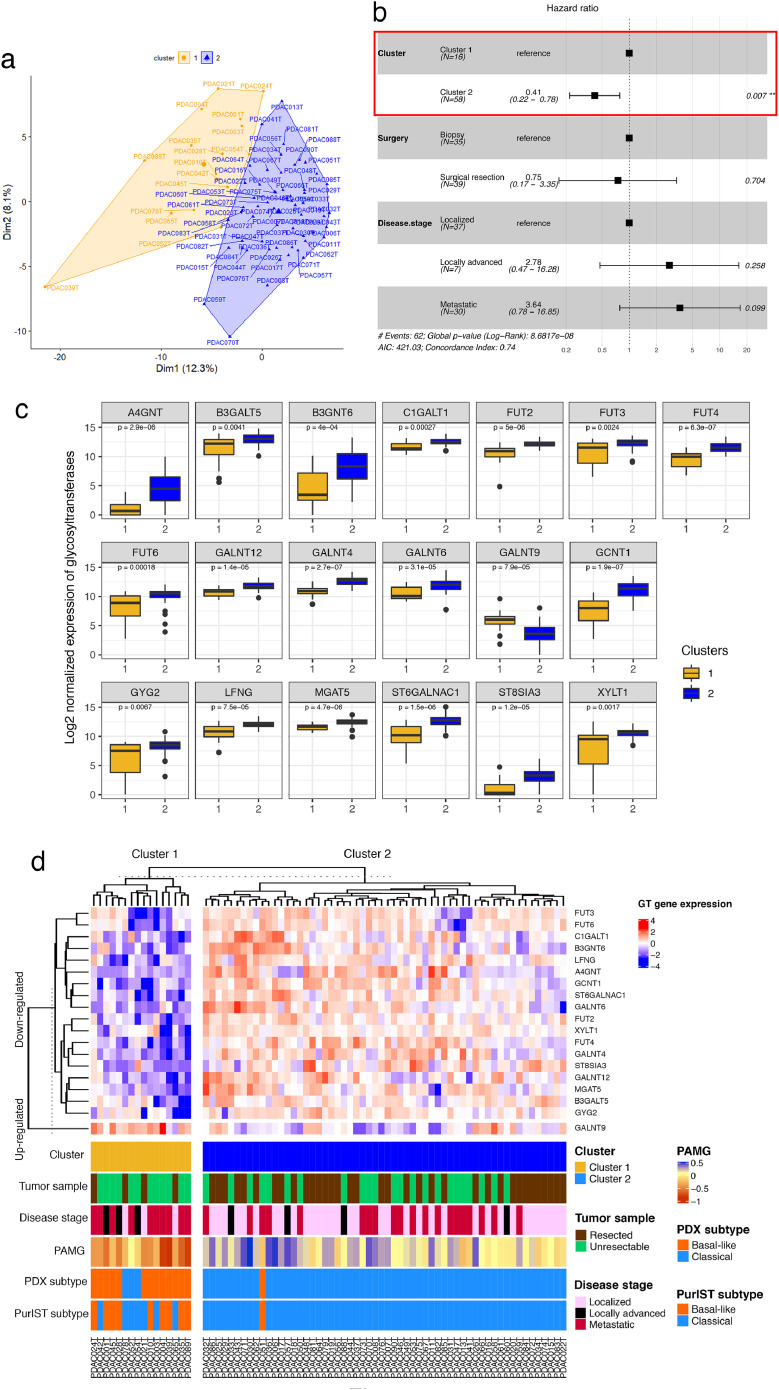
RNA-seq data of 169 GT genes revealed significant differences in gene expression between tumor cells from 74 PDX (Supplementary Fig. S1a) highlighting the tumor heterogeneity in GT gene expression. Subsets of PDAC were distinguishable with a high expression of specific genes and low expression of others. To deeply explore these PDAC glyco-profiles, we performed a principal component analysis (PCA), followed by a hierarchical clustering (HCPC). Two different clusters of PDAC were identified on PaCaOmics patient's cohort (Fig. 1a; Supplementary Fig. S1b, c) based on a specific GT gene expression profile (Supplementary Table S1). Cluster 1 and cluster 2 included, respectively 21.6% (n = 16/74) and 78.4% (n = 58/74) of PDAC patients. Interestingly, specific GT gene expression profiles defined for each cluster were related to prognosis in a multivariate analysis including surgical resection and disease stage corresponding to the most relevant factors impacting survival, predicting at best patient prognosis. PDAC stratification through GT gene expression profile was significantly associated to patient OS as an independent prognostic factor with a hazard ratio HR = 0.41 (p-value = 0.007 [95% CI: 0.22–0.78]) (Fig. 1b) indicating that PDAC patients in cluster 2 have a best survival compared to patients in cluster 1. This result was shown with OS analysis of clusters 1 and 2, considering separately resected, locally advanced and metastatic origin samples (Supplementary Fig. S2). This prognostic value, independent of other factors, suggests that GT gene expression profiles have a significant impact on the survival of patients.
Fig. 1.
Prognostic stratification of PDAC through their GT gene expression profile (a, b) and clinical features of patients with their PDAC molecular profiles (c, d). (a) Biplot of the HCPC analysis result based on RNA-seq data of 74 PDAC and 169 GT genes. (b) Forest plot of the multivariate survival analysis including PDAC clustering, surgical resection and the disease stage. (c) Boxplot showing Log2 normalized expression of 19 GT genes stratified by clusters 1 and 2. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). Kruskal-Wallis p-value was used for the statistical significance between clusters. (d) Heatmap of the two clusters based on transcription signature of 19 GT genes. Clinical characteristics and molecular classifications of each patient tumor were shown in the corresponding annotation. Color bars indicate the tumor sample type, disease stage, PAMG and basal-like/classical subtyping based on previously established consensus [30] and PurIST classifier.
3.2. Identification of GT genes as prognostic markers
To define GT genes with a major impact on the survival, the top GT genes whose expressions contributed significantly to cluster definition, were extracted from HCPC analysis. This results in a selection of 19 GT genes as the most relevant markers to stratify PDAC (Table 1; Supplementary Table S1). Among these genes, several functional paralogues of fucosyltransferases including FUT2, FUT3, FUT4, FUT6, and of N-acetygalactosaminyltransferases GALNT4, GALNT6 and GALNT12 were significantly downregulated in cluster 1 compared to cluster 2 while GALNT9 was upregulated in cluster 1 (Fig. 1c, d; Supplementary Fig. S1d, Table 1; Supplementary Table S1). Our results also showed a significant downregulation of A4GNT, B3GALT5, B3GNT6, C1GALT1, GCNT1, GYG2, LFNG, MGAT5, ST6GALNAC1, ST8SIA3 and XYLT in cluster 1 compared to cluster 2 (Fig. 1c, d; Supplementary Fig. S1d).
Table 1.
List of 19 GT genes as prognostic markers.
| Gene symbol* | Gene name* | Glycan synthesis / Function |
|---|---|---|
| GALNT9 | Polypeptide N-acetylgalactosaminyltransferase 9 | Transfers a GalNAc residue to serine or threonine residue of an acceptor core protein. Initiation of mucin-type O- linked protein glycosylation |
| A4GNT | Alpha-1,4-N-acetylglucosaminyltransferase | Transfers an alpha-1,4-GlcNAc residue onto core 2 branched O-glycans. Biosynthesis of mucin-type O-glycan |
| B3GALT5 | Beta-1,3-galactosyltransferase 5 | Transfers a beta-1,3-Gal residue to GlcNAc-based acceptors such as the core 3 O-glycan. Biosynthesis of type 1 Lewis antigens |
| B3GNT6 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 | Transfers a beta-1,3-GlcNAc residue to GalNAc-serine or -threonine. Biosynthesis of mucin-type core 3 O-glycan |
| C1GALT1 | Core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1 | Transfers a beta-1,3-Gal residue to O-linked GalNAc residue onto protein. Biosynthesis of mucin-type core 1 O-glycan |
| FUT2 | Fucosyltransferase 2 | Transfers an alpha-1,2-Fuc residue to terminal Gal-based acceptors. Lewis and ABO blood group antigen biosynthesis |
| FUT3 | Fucosyltransferase 3 (Lewis blood group) | Transfers an alpha-1,3- or alpha-1,4-Fuc residue to GlcNAc-based acceptors. Last step of Lewis blood group antigen biosynthesis |
| FUT4 | Fucosyltransferase 4 | Transfers an alpha-1,3-Fuc residue to GlcNAc-based acceptors. Lewis x (CD15) antigen biosynthesis |
| FUT6 | Fucosyltransferase 6 | Transfers an alpha-1,3-Fuc residue to GlcNAc of alpha-2,3 sialylated substrates. Sialyl-Lewis x antigen biosynthesis |
| GALNT4 | Polypeptide N-acetylgalactosaminyltransferase 4 | Transfers a GalNAc residue to serine or threonine residue of an acceptor core protein. Initiation of mucin-type O- linked protein glycosylation |
| GALNT6 | Polypeptide N-acetylgalactosaminyltransferase 6 | Transfers a GalNAc residue to serine or threonine residue of an acceptor core protein. Initiation of mucin-type O- linked protein glycosylation |
| GALNT12 | Polypeptide N-acetylgalactosaminyltransferase 12 | Transfers a GalNAc residue to serine or threonine residue of an acceptor core protein. Initiation of mucin-type O- linked protein glycosylation |
| GCNT1 | Glucosaminyl (N-acetyl) transferase 1 | Transfers a beta-1,6-GlcNAc residue onto mucin-type core 1 O-glycan. Biosynthesis of mucin-type core 2 branched O-glycan |
| GYG2 | Glycogenin 2 | Self-glucosylation (Glc). Initiation reaction of glycogen biosynthesis |
| LFNG | LFNG O-fucosylpeptide | Transfers a beta-1,3-GlcNAc residue to O-linked fucose residue onto Notch molecules. Regulation of Notch molecules activity (Notch signaling pathway) |
| MGAT5 | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase | Transfers a beta-1,6-GlcNAc residue to mannose of biantennary N-linked glycan present onto glycoproteins. Biosynthesis of tri- and tetra-antennary complexe N-glycans |
| ST6GALNAC1 | ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | Transfers an alpha-2,6-NeuAc residue to O-linked GalNAc residues onto protein. Biosynthesis of cancer-associated sialyl-Tn antigen |
| ST8SIA3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | Transfers an alpha-2,8-NeuAc residue to terminal NeuAc of glycolipids and N-linked glycan of glycoproteins. Biosynthesis of polysialic acid chains |
| XYLT1 | Xylosyltransferase 1 | Transfers a Xyl residue to a serine residue of an acceptor core protein. Biosynthesis of glycosaminoglycan chains |
according to the HGNC. GALNT9 is up-regulated in cluster 1 and down-regulated in cluster 2; the 18 other GT are down-regulated in cluster 1 and up-regulated in cluster 2.
Gal: Galactose; Fuc: Fucose; Glc: Glucose; Xyl: Xylose; GalNAc: N-acetylgalactosamine; GlcNAc: N-acetylglucosamine; NeuAc: N-acetylneuraminic acid or sialic acid.
3.3. Clinical features of patients and their PDAC molecular profiles
The HCPC analysis allowed the identification of two prognostic clusters of PDAC with distinct clinical features of patients from PaCaOmics cohort. Unresectable PDAC represented 75% (n = 12/16) in cluster 1 whereas cluster 2 included 60% (n = 35/58) of patients with resected tumors (Fig. 1d). Among the unresectable PDAC, 25% and 17% were locally advanced tumors in clusters 1 and 2, respectively. Moreover, cluster 1 contains 63% (n = 10/16) of metastatic tumors compared to cluster 2 with only 34% (n = 20/58) of metastatic tumors (Fig. 1d). The classification of PDAC into different transcriptomic subtypes using PurIST classifier showed that 90% of basal-like subtypes were classified in cluster 1 and inversely cluster 2 regrouped 89% of classical subtype (p-value < 0.0001, Fisher's exact test). However, the composition of cluster 1 is heterogeneous with 56% basal-like vs 44% classical subtypes and 81% basal-like vs 19% classical subtypes according to PurIST classifier and stratification proposed by Nicolle et al. [30], respectively. On the other hand, cluster 2 contains around 2% of PDAC basal-like subtype whatever the classifier used. Then, the cluster 1 regroups basal-like PDAC subtypes and classical PDAC subtypes of patients with short survival for the majority of them (Supplementary Table S1). This idea could be refined by the Pancreatic Adenocarcinoma Molecular Gradient (PAMG) [29], a more precise stratification of tumors represented by a gradient from pure basal-like to pure classical. Indeed, the PAMG confirms cluster 1 mixed composition including tumors with intermediate gradient, although it does contain the top scored basal-like tumors. Moreover, the comparison of the glyco-signature to the PAMG showed that all the 19 genes were significantly correlated to the PAMG. Interestingly, only GALNT9 gene expression (upregulated in cluster 1) correlated negatively with the PAMG (Pearson correlation coefficient R = -0.47, p-value = 2.01e-05) indicating its association with basal-like phenotype in similar way as Vimentin (VIM), a marker of mesenchymal differentiation and aggressive carcinoma. However, other 18 GT gene expression (upregulated in cluster 2) correlates positively with the PAMG (Pearson correlation coefficient 0.36 < R < 0.71 and p-value < 0.01) confirming their association with classical phenotype like GATA6 expression (a marker of classical phenotype) (Supplementary Fig. S3; Supplementary Table S3). These combined features allow a precise tumor characterization, which is essential to dissect molecular diversity. Therefore, the glyco-signature of 19 GT genes was found relevant to stratify PDAC according to their aggressiveness.
In order to assess whether specific GT gene expression profiles defined for each cluster could have an impact on the formation of glycosylated antigens, immunohistochemical staining using antibodies against Lewis blood group antigens and GT was performed on PDX-TMA. As shown in Supplementary Fig. S4, a large majority of PDAC from cluster 1 displayed a weak or negative staining for Lewisa, Lewisb, Lewisx and sialyl-Tn antigens compared to cluster 2. Although many GT are involved in the formation of these glycosylated antigens, these results are consistent with the transcriptomic data of several GT genes included in the glyco-signature (Supplementary Fig. S5a–f). Interestingly, all negative tissues for sialyl-Lewisa expression, except one case, are included in cluster 1 (Supplementary Fig. S5a). The simultaneous downregulation of FUT2, FUT3 B3GALT5 gene expression in cluster 1 also affects the biosynthesis of Lewisb antigen (Supplementary Fig. S5c). This is also the case for the expression of truncated O-glycans sialyl-Tn antigens whose biosynthesis is dependent on the level of expression of ST6GALNAC1 and C1GALT1 which can compete for the same glycan chain as substrate (Supplementary Fig. S5f-g).
3.4. Validation of the glyco-signature prognostic value on independent cohorts
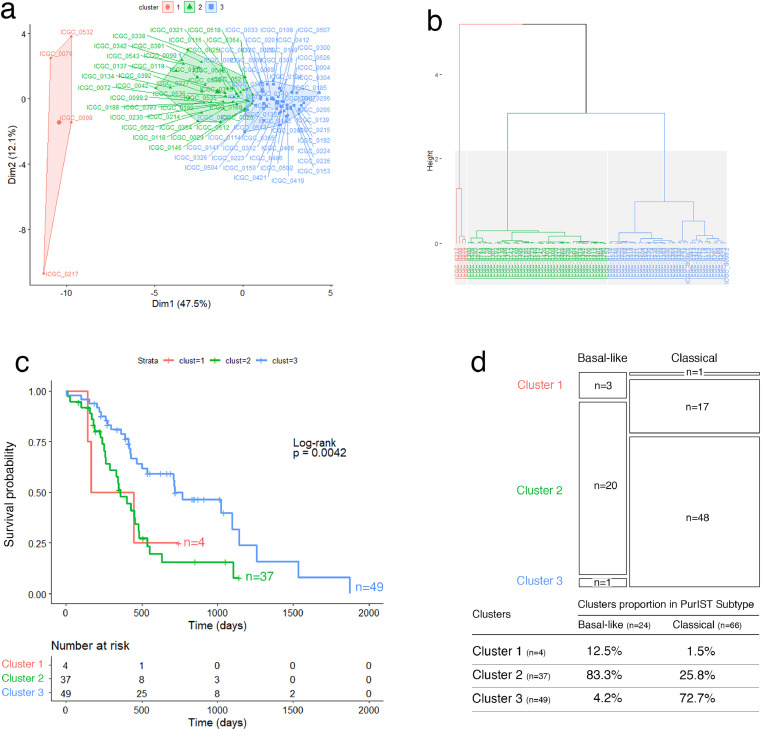
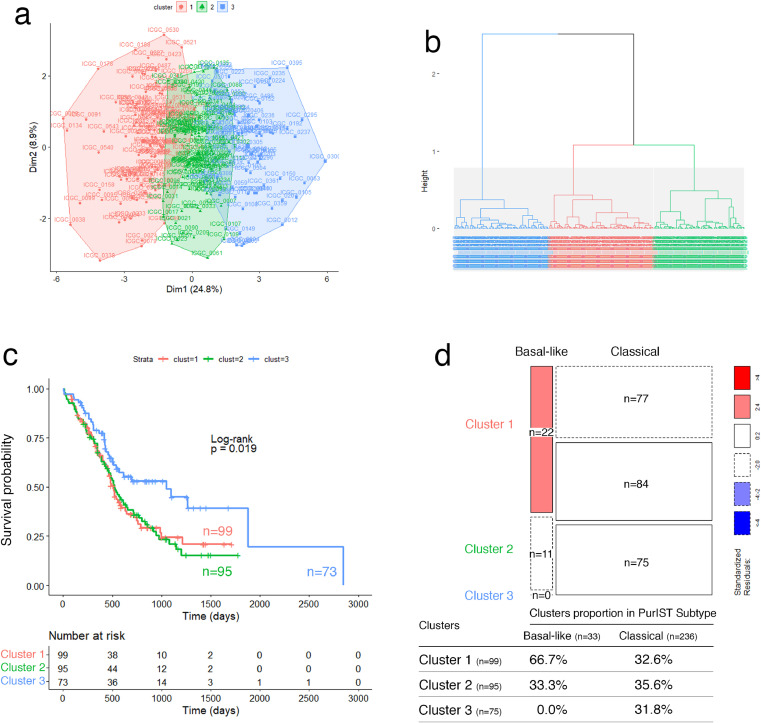
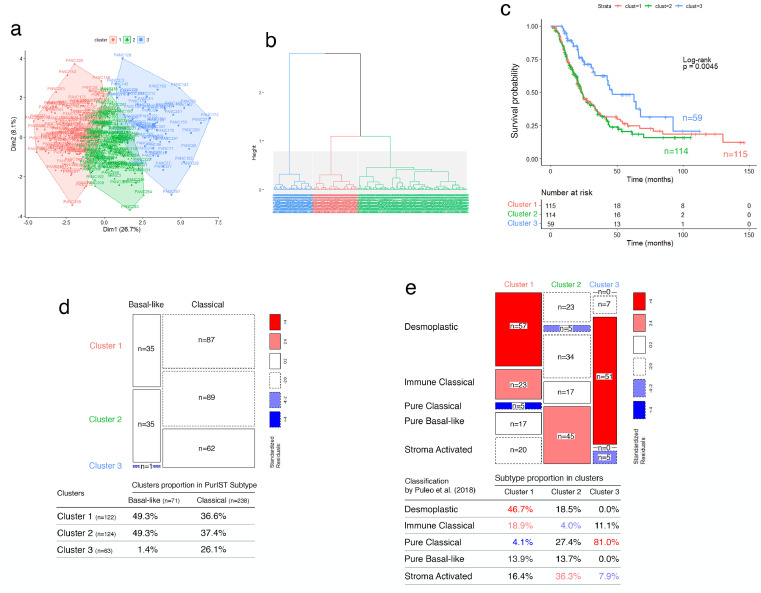
To validate the prognostic accuracy of the proposed glyco-signature, we applied it on three independent external datasets of resected PDAC tumors from ICGC (RNA-seq and microarray) and Puleo patient's cohorts. Note that the glyco-signature was determined on PDX, considering exclusively the epithelial compartment of tumors, unlike the validation performed on resected whole tumor tissues, including stromal component of tumors. In this context, HCPC analyses identified systematically three clusters in the three validation cohorts, through the 19 GT genes, with significant differences in OS. Clusters 1 and 2 were associated with poor prognosis compared to the cluster 3 which showed about a double median OS (Figs. 2a–c; 3a–c; 4a–c; Supplementary Fig. S6 a, b, e, f, i, j). Furthermore, statistical analyses showed that the three clusters were significantly associated with basal-like/classical subtyping (ICGC patient's cohort (RNA-seq): p-value < 0.0001, Fisher's exact test; ICGC patient's cohort (microarray): p-value = 5.424e-05, Chi-squared test; Puleo patient's cohort (microarray): p-value = 3.597e-05, Chi-squared test). While the overwhelming majority of tumors in cluster 3 had a classical subtype, both clusters 1 and 2 included most of the basal-like subtype PDAC but contained also a large proportion of classical subtype PDAC having a poor prognosis (Figs. 2d, 3d; 4d; Supplementary Fig. S6 c, d, g, h, k, l, m–o). Besides, expression differences of the 19 GT genes in the 3 clusters showed globally a trend of gradual expression from cluster 1 to cluster 3, except for GALNT9, which is downregulated in cluster 3 (Supplementary Fig. S7). This is consistent with what was previously shown in PaCaOmics patient's cohort. More importantly, the expression of B3GALT5, FUT3, C1GALT1, GALNT4, GALNT12 and ST6GALNAC1 were strongly downregulated in cluster 1 compared to cluster 2 in the three validation cohorts.
Fig. 2.
Validation of the glyco-signature on ICGC patient's cohort (RNA-seq). (a) Two-dimension representation of the HCPC analysis results and (b) cluster dendrogram related to ICGC RNA-seq data (n = 91) using 19 GT genes. (c) Survival curves estimated by using the Kaplan-Meier method and comparing OS probabilities between clusters with the log-rank test (p-value = 0.0042). Cluster 1 and 2 have shorter median OS of 304 and 359 days, respectively compared to cluster 3 with median OS of 719 days. (d) Mosaic plot showing cross-link between basal-like/classical subtypes and clusters 1, 2 and 3 identified through GT gene prognostic markers. Box height reflects the number of tumors classified in each cluster and box width represents proportion of basal-like/classical subtypes (p-value < 0.0001, Fisher's exact test).
Fig. 3.
Validation of the glyco-signature on ICGC patient's cohort (microarray). (a) Two-dimension representation of the HCPC analysis results and (b) cluster dendrogram related to ICGC affymetrix transcriptomic data (n = 269) using 19 GT genes. (c) Survival curves estimated by using the Kaplan-Meier method and comparing OS probabilities between clusters with the log-rank test (p-value = 0.0019). Clusters 1 and 2 have shorter median OS of 515 and 517 days, respectively compared to cluster 3 with median OS of 1048 days. (d) Mosaic plot showing cross-link between basal-like/classical subtypes according to PurIST classifier and identified clusters 1, 2 and 3 through GT gene prognostic markers. Box height reflects the number of tumors classified in each cluster and box width represents proportion of basal-like/classical subtypes (p-value = 5.424e-05, Chi-squared test).
Fig. 4.
Validation of the glyco-signature on Puleo patient's cohort. (a) Two-dimension representation of the HCPC analysis results and (b) cluster dendrogram related to affymetrix transcriptomic data (n = 309) using 19 GT genes. (c) Survival curves estimated by using the Kaplan-Meier method and comparing OS probabilities between clusters with the log-rank test (p-value = 0.0045). Cluster 1 and 2 have shorter median OS of 23.8 and 22.9 months, respectively compared to cluster 3 with median OS of 46.4 months. (d) Mosaic plot showing cross-link between basal-like/classical subtypes according to PurIST classifier and identified clusters 1, 2 and 3 through GT gene prognostic markers. Box height reflects the number of tumors classified in each cluster and box width represents proportion of basal-like/classical subtypes (p-value = 3.597e-05, Chi-squared test). (e) Mosaic plot showing the microenvironment subtype proportion in clusters 1, 2 and 3 through GT gene prognostic markers, related to microenvironment-based classification of PDAC tumors, proposed by Puleo et al. [12]. Box width reflects the number of tumors classified in each cluster and box height represents proportion of microenvironment subtypes (p-value < 0.0001, Chi-squared test). For both mosaic plots, the standardized residuals, with an absolute value greater than 2.0, indicate boxes contributing to significant chi-square test statistic. The color range from red to blue indicates whether observed frequency is significantly higher or lower, respectively than the expected frequency (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
These validation analyses on expression datasets, obtained from diverse technological platforms (RNA-seq or microarray) allowed to assess the glyco-signature accuracy on stratifying PDAC samples independently of the technological provenance of generated expression data and increased its robustness.
Tumor subtyping based on the epithelial compartment was not sufficient to explain the two poor prognostic clusters including simultaneously basal-like and classical subtype PDAC. Therefore, we assessed whether identified clusters through GT gene expression could be associated with specific microenvironment features as suggested by the Puleo et al. study [12]. Interestingly, the stromal-driven subtype composition was different between the two poor prognostic clusters on the Puleo patient's cohort. Cluster 1 includes predominantly desmoplastic and immune classical microenvironment (46.7% and 18.9%, respectively), whereas cluster 2 is significantly uncorrelated with immune classical subtype and is mainly enriched in stroma-activated microenvironment (36.3%) (p-value < 0.0001, Pearson's chi-squared test) (Fig. 4e). Cluster 3, which includes PDAC patients with a better prognosis, was characterized by a predominant pure classical microenvironment (81%). These particular distributions highlight the importance of microenvironment features in prognosis related to specific GT gene expression profiles. Beyond the established basal-like/classical classification, the glyco-signature was then able to discriminate two clusters with similar poor prognosis but differing by their microenvironment features.
3.5. Identification of functional pathways
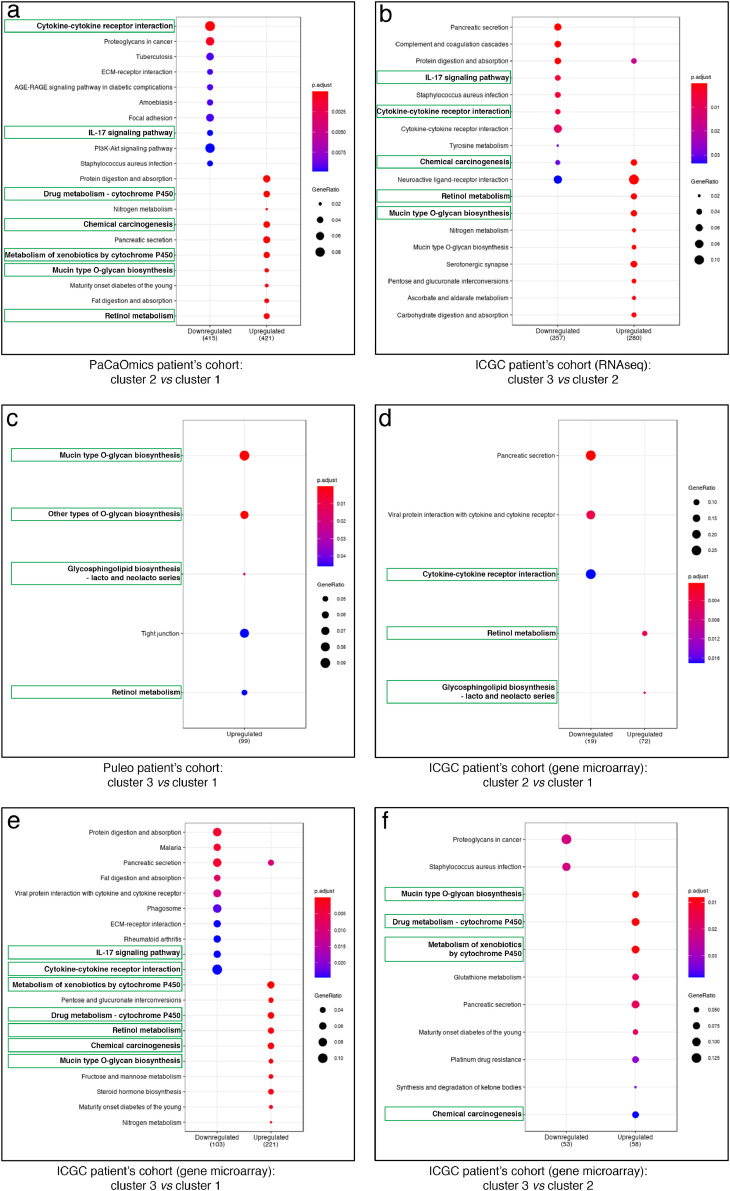
In order to better characterize the tumor biology and decipher global transcription patterns related to each cluster, a pairwise differential expression analysis was performed for each dataset allowing us to highlight lists of differentially expressed genes used further for the KEGG pathway analyses. Results showed the specificity and relevance of glycosylation process in the determination of the prognostic clusters. ‘Mucin type O-glycan biosynthesis’ was an enriched pathway for upregulated genes in clusters with better prognosis (Fig. 5; Supplementary Figs. S8–10; Supplementary Tables S1-2 for the complete enriched biological processes and associated statistics). A large majority of the 19 GT gene markers is part of this pathway. In particular the glycosylation pathways involving GALNT4, GALNT6, GALNT12, GCNT1 and ST6GALNAC1 or B3GALT5, FUT2 and FUT3 were upregulated in cluster 3 and/or 2 vs cluster 2 and/or 1 in ICGC and Puleo patient's cohorts (Supplementary Table S1). Similarly, a core of intrinsically connected pathways including ‘Retinol metabolism’, ‘Chemical carcinogenesis’ as well as ‘Drug metabolism – cytochrome P450’ and ‘Metabolism of xenobiotics by cytochrome P450’ were systematically identified as a KEGG pathway network of upregulated genes in clusters with better prognosis (Fig. 5; Supplementary Figs. S8–10; Supplementary Tables S1-2). A focused analysis on redundant genes composing these pathways and identified across different datasets allowed us to bring out the most relevant ones. In this way, ADH1C, CYP2C18, CYP2S1, CYP3A5, GSTA1, GSTA2 and UGT2B17 were found as the major contact points within the network linking these pathways (Supplementary Tables S1-2). In particular CYP3A5 involved in these pathways is upregulated in cluster 2 vs cluster 1 in PaCaOmics patient's cohort and in cluster 3 and 2 vs cluster 1 in ICGC and Puleo patient's cohorts (Supplementary Table S1).
Fig. 5.
KEGG enrichment pathway analysis of prognostic clusters. Dot-plot representations of the top 10 downregulated (left) and upregulated (right) enriched pathways comparing different clusters as follows: (a) cluster 2 vs cluster 1 in PaCaOmics patient's cohort, (b) cluster 3 vs cluster 2 in ICGC patient's cohort (RNA-seq), (c) cluster 3 vs cluster 1 in Puleo patient's cohort, and (d) cluster 2 vs cluster 1, (e) cluster 3 vs cluster 1, (f) cluster 2 vs cluster 1 in ICGC patient's cohort (gene microarray). Dot color gradient corresponds to p-adjusted values of the enrichment score while circle size is proportional to gene ratio in the corresponding pathway. Green squares highlight the best common enriched pathways between clusters. For the complete enriched biological processes and associated statistics, see Supplementary Table S2 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Moreover, most of the enriched pathways of downregulated genes in clusters having the best prognosis were related to inflammatory process or immune system such as ‘Cytokine-cytokine receptor interaction’ and ‘IL-17 signaling pathway’ (Fig. 5; Supplementary Figs. S8–10; Supplementary Tables S1-2). At the gene level, IL6 is common to several downregulated pathways in clusters with the higher OS across multiple comparisons of different datasets (Supplementary Tables S1-2). These microenvironment features were also found by analyzing the murine stroma compartment between clusters 1 and 2 of PDX samples in PaCaOmics patient's cohort. In particular, the ‘Cytokine- cytokine receptor interaction’ pathway is downregulated in murine stroma of cluster 2 compared to cluster 1. Although the microenvironment of tumor cells is specific to the host (tumors from murine xenografts or patient tissues), these data show that common pathways can be deregulated between both samples (Supplementary Fig. S11).
4. Discussion
PDAC heterogeneity remains a significant limitation in adapted patient care, contributing to its poor prognosis related to late diagnosis and chemoresistance. The approaches to overcome phenotypic heterogeneity of PDAC were based on deciphering and characterizing molecular profiles of tumors to define the most homogeneous subtypes sharing common features [8], [9], [10], [11], [12], [13]. In our study, we explored PDAC heterogeneity by investigating GT expression profiles, which contribute to the formation of aberrant glycans acquired during cancer development [16], [17], [18]. We focused on genes involved in glycan biosynthesis to provide insights on mechanisms driving disrupted cellular functions and tumor aggressiveness in PDAC. Analysis of PDX samples from resected and unresectable tumors based on GT gene expression profile allowed the identification of two PDAC clusters with different patient prognosis. However, the small number of patients with unresectable PDAC remains a limitation of the study and further investigation is needed on more large cohorts to determine if the glyco-signature is potentially applicable to these patients. The identified glyco-signature of 19 GT was applied on the validation cohorts of resected whole tumor tissues, including the stromal component. In this validation, the use of expression datasets obtained from diverse technology (RNA-seq and microarray) increases its robustness as well as the different types of sample (frozen samples and paraffin-embedded tissues). The specificity of the glyco-signature is related to its identification on the epithelial compartment of PDX samples, before its validation on whole resected tumor tissues. Although the combination of 19 GT genes is important to stratify PDAC, further investigation is needed to identify if this combination is reducible while retaining the same prognostic value. This glyco-signature was able to discriminate more specifically two separate PDAC clusters with poor prognosis of patients and one having a better outcome; these three clusters were associated with distinct microenvironment compositions as suggested by results obtained through stratification based on stromal features proposed by Puleo et al. [12]. Moreover, the composition of these two poor prognosis PDAC clusters is mixed including basal-like subtype and classical subtype associated to short survival of patients; whereas the cluster including patients with longer OS contained mainly PDAC of classical subtype. Recently, Chan-Seng-Yue et al. [34] have identified two subtypes of basal-like tumors. In parallel, single-cell RNA-seq studies showed the intra-tumor coexistence of basal-like and classical phenotypes [35]. These findings highlight the challenge associated with the binary classification of PDAC tumors. Glyco-profile analysis also tends to detect aggressive PDAC. Indeed, the expression of B3GALT5, FUT3 and ST6GALNAC1 genes was downregulated in cluster 1 compared to cluster 2, whatever the validation cohort used. The ability of these three GT (B3GALT5, FUT3 and ST6GALNAC1) in association with microenvironment markers to discriminate both poor prognosis clusters requires further investigations to be used as biomarkers in order to improve prognostic accuracy and treatment efficacy of different PDAC patients.
Studies on classifications of tumors based on their glyco-gene expression profiles are beginning to emerge [35], [36], [37], [38]. A subtype of patient's tumors with poor prognosis colorectal cancer was identified by a glyco-gene signature in which the loss of GALNT6 gene expression was correlated with invasion and chemoresistance of cancer cells and was highlighted as a prognostic biomarker [39]. Recently, B3GNT3, B4GALNT3, FUT3, FUT6, GCNT3 and MGAT3 were shown to be overexpressed in PDAC [40]. In our study, we bring new insights on how the glycosylation pathways are altered between different PDAC clusters. The initiation step of glycosylation is affected with a downregulation of several polypeptide N-acetylgalactosaminyltransferases, GALNT4/GALNT6/GALNT12, as well as GT involved in sialylation such as ST6GALNAC1, or elongation of O-glycans such as C1GALT1 and GCNT1. Interestingly, the low expression of GALNT6 was associated with poorly differentiated pancreatic tumors and poor clinical outcome of PDAC patients [41]. It was also shown that the loss of C1GALT1 expression contributes to PDAC progression and metastasis formation [42]. These results are consistent with our findings where these two glyco-genes display a lower expression in clusters having the poor outcomes. The same profile is observed with the α1,2-fucosyltransferase FUT2 and several α1,3-fucosyltransferases, FUT3 (having also an α1,4-fucosyltransferase activity)/FUT4/FUT6, which show a concerted and simultaneous deregulation in tumors. This highlights the importance of impaired fucosylated antigen expression during malignant transformation and acquisition of cancer cell phenotype. Among them, the carbohydrate antigen CA19-9 corresponding to sialyl-Lewisa structure remains the only clinically used biomarker to monitor disease relapse [43,44]. Our prognostic signature brings to light deregulation of key GT involved in CA19-9 biosynthesis, such as FUT3 and B3GALT5 [45,23], as important as α1,2-fucosyltransferase FUT1 downregulation and α2,3-sialyltransferase ST3GAL (over)-expression [46]. Moreover, CA19-9 was involved in the development of pancreatitis and PDAC, demonstrating that abnormal glycosylation can play a key role in pancreatic oncogenesis [45]. Interestingly, FUT3 and B3GALT5, but also FUT2 were downregulated in cluster 1 compared to cluster 2, all three associated with poor prognosis of patients, suggesting a decrease of CA19-9 expression but also Lewisb expression in this cluster. Indeed the TMA analyses showed that 37.50% (n = 6/16) of tumors are negative for the expression of sialyl-lewisa in cluster 1 while only 4.25% (n = 2/47) of tumors are negative for the expression of this antigen in cluster 2. We have also observed that 81.25% (n = 13/16) of tumors are negative for the expression of Lewisb in cluster 1 while only 23.40% (n = 11/47) of tumors are negative for the expression of this antigen in cluster 2. The association of CA19-9 and/or Lewisb with FUT3, B3GALT5 and/or FUT2 could be interesting prognostic biomarkers to discriminate PDAC patients with poor prognosis. Further investigations are needed to validate the potential prognostic values of these markers. Other GT genes included in the glyco-signature could also play an important role in PDAC progression. We have observed a downregulation of A4GNT gene expression encoding for the α1,4-N-acetylglucosaminyltransferase in the cluster having a poor outcome. A4GNT is involved in the biosynthesis of O-glycan carrying terminal α1,4 N-acetylglucosamine residues (α1,4GlcNAc) onto mucin 6 (Muc6) (Table 1). A loss of α1,4GlcNAc expression has been shown in gastric tumorigenesis [47]. Interestingly, a decreased of α1,4GlcNAc related with MUC6 expression was observed during tumor progression of PDAC [48]. In recent years, several studies have also shown that glycosylation of the Notch extracellular domain may regulate Notch activity during development in mammals but also in cancer [49]. In particular, Lunatic Fringe (LNFG), which encodes for O-fucosylpeptide β1,3 N-acetylglucosaminyltransferase (Table 1) can modulate Notch signalling pathways through the binding between Notch receptor-Notch ligand [50]. Recently, Zhang et al. [51] have shown an increase of PDAC development in the LFNGflox/flox;KrasLSL-G12D;Pdx1-Cre mouse model where LNFG was deleted. These results are consistent with our findings where this GT displays a lower expression in clusters having the poor outcomes. Interestingly, GYG2, a glyco-gene which encodes for glycogenin-2 (Table 1), was identified in a glucose metabolism-related gene signature from TCGA for predicting the prognosis of clear cell renal cell carcinoma [52]. Although these studies have shown the functionnal importance of down-regulated C1GALT1, GALNT6, LFNG and A4GNT in tumor progression of PDAC, a fully functional validation of other individual GT gene identified in the glyco-signature is needed. Further investigations will bring more insights to better characterize the functional and mechanistic impact of the GT genes in the PDAC progression.
Considering glycosylation involvement in most biological functions, it is widely admitted that its deregulation substantially impairs and remodels the global tumor biology. Therefore, comparing our GT expression profile-based classification to a non-tumor driven subtypes linked to microenvironment features was a relevant way to capture differential mechanisms driving tumor aggressiveness at a larger scale. The prognostic value of the glyco-signature may be explained by its ability to reflect molecular diversity at stromal and immune level, distinguishing the role of microenvironment signals in cancer cell phenotype acquisition. This was confirmed with Puleo patient's cohort, where two poor prognostic clusters identified through glycosylation pattern were significantly related to desmoplastic and stroma-activated subtypes, suggesting that in cluster 1, aggressiveness may be linked to the presence of immune cells, endothelial cells and fibroblasts; whereas cluster 2 was characterized by low immune infiltrates with high level of fibroblasts and endothelial cells. Interestingly, enriched downregulated pathways in clusters with best survival are related to inflammatory process and immune system, confirming the importance of microenvironment signals in tumor aggressiveness characteristic of patients with poor outcome. Proinflammatory cytokine IL-6 is found to be downregulated in clusters with best survival but also differentially expressed between the two poor prognostic clusters in Puleo patient's cohort. It has been shown that inflammation mediated by JAK/STAT pathway and driven by IL-6 signaling is a major cause of chemotherapy resistance and that blocking IL-6 receptor could enhance chemotherapy efficacy [53]. In addition, blockade of IL-6 showed an interesting anti-tumor activity when combined with checkpoint blockade immunotherapy (PD-L1), by modulating immunological features [54]. This might be promising to select patients and propose them the adapted care.
Taken together, enriched upregulated pathways in clusters having the best survival, including ‘Retinol metabolism’, ‘Chemical carcinogenesis’, ‘Drug metabolism – cytochrome P450’ and ‘Metabolism of xenobiotics by cytochrome P450’ form an interconnected network. These results are consistent with the identification of the ‘Drug metabolism – cytochrome P450’ pathway as a functional signature to improve the prognosis and sensitivity to chemotherapeutic agents for PDAC of classical subtype [55]. Among them CYP family and especially CYP3A5 which shows differential expression between clusters is known to be involved in tumor cell autonomous drug detoxification by mediating systemic drug metabolism [56]. This may be indicative of resistance to paclitaxel or tyrosine kinase inhibitors such as erlotinib, or irinotecan involved in Folfirinox protocol, metabolized by CYP3A5 [56], [57], [58]. Therefore, by comparing the two poor prognostic clusters identified in Puleo and ICGC patient's cohorts, we could hypothesize a better sensitivity to these drugs for cluster 1 with downregulated CYP3A5 compared to cluster 2. Further functional investigations are needed to consolidate this hypothesis.
In conclusion, we propose a glyco-signature funded on GT gene expression allowing a better classification of PDAC according to their specific mechanisms driving tumor aggressiveness through the glycosylation pathways in tumors and microenvironment. Although further investigations are needed, this glyco-signature could contribute to guide clinical decision by predicting patient outcomes and identifying sensitivity to specific therapies.
Contributors
E. Mas, J. Iovanna, and N. Dusetti conceived and designed the project. E. Mas supervised the study. Y. Mohamed Abd-El-Halim conducted and performed the study with the help of A. El Kaoutari, F. Silvy and E. Mas and the contribution of M. Bigonnet and J. Roques for material provision and data collection. M. Rubis performed the immunohistochemistry experiments. Y. Mohamed Abd-El-Halim, A. El Kaoutari, F. Silvy, J. Cros, R. Nicolle, J. Iovanna, N. Dusetti, and E. Mas analysed and interpreted the results. Y. Mohamed Abd-El-Halim and E. Mas wrote and illustrated the manuscript. A. El Kaoutari, J. Cros, R. Nicolle, J. Iovanna, and N. Dusetti contributed to discussion and reviewed and edited the manuscript. Y. Mohamed Abd-El-Halim, A. El Kaoutari, J. Iovanna, E. Mas and N. Dusetti have verified the underlying data. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare no competing financial interests
Acknowledgments
Acknowledgments
This work was supported by INCa (Grants Nos. 2018-078 and 2018-079), Fondation ARC (Grant No. ARCPJA32020070002326), Cancéropôle PACA, DGOS (labelization SIRIC, Grant No. 6038), Amidex Foundation and Ligue Nationale Contre le Cancer and by institutional fundings from INSERM (Paris, France) and the Aix-Marseille Université (Marseille, France).
Data sharing statement
Data in this study are available upon reasonable request by sending a message to the corresponding authors.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103541.
Contributor Information
Nelson Dusetti, Email: nelson.dusetti@inserm.fr.
Eric Mas, Email: eric.mas@inserm.fr.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rawla P., Sunkara T., Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., Neoptolemos J.P. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 5.Ying H., Dey P., Yao W., Kimmelman A.C., Draetta G.F., Maitra A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 7.Lai E., Puzzoni M., Ziranu P., Pretta A., Impera V., Mariani S. New therapeutic targets in pancreatic cancer. Cancer Treat Rev. 2019;81 doi: 10.1016/j.ctrv.2019.101926. [DOI] [PubMed] [Google Scholar]
- 8.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203. doi: 10.1016/j.ccell.2017.07.007. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puleo F., Nicolle R., Blum Y., Cros J., Marisa L., Demetter P. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155:1999–2013. doi: 10.1053/j.gastro.2018.08.033. e3. [DOI] [PubMed] [Google Scholar]
- 13.Collisson E.A., Bailey P., Chang D.K., Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 14.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munkley J., Elliott D.J. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7:35478–35489. doi: 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stowell S.R., Ju T., Cummings R.D. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510. doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues J.G., Balmaña M., Macedo J.A., Poças J., Fernandes Â., De-Freitas-Junior J.C.M. Glycosylation in cancer: selected roles in tumor progression, immune modulation and metastasis. Cell Immunol. 2018;333:46–57. doi: 10.1016/j.cellimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Pinho S.S., Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 19.Chandler K.B., Costello C.E., Rahimi N. Glycosylation in the tumor microenvironment: implications for tumor angiogenesis and metastasis. Cells. 2019;8:544. doi: 10.3390/cells8060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mereiter S., Balmaña M., Gomes J., Magalhães A., Reis CA. Glycomic approaches for the discovery of targets in gastrointestinal cancer. Front Oncol. 2016;6:55. doi: 10.3389/fonc.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trinchera M., Aronica A., Dall'Olio F. Selectin ligands sialyl-lewis a and sialyl-lewis x in gastrointestinal cancers. Biol (Basel) 2017;6:16. doi: 10.3390/biology6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munkley J. The glycosylation landscape of pancreatic cancer. Oncol Lett. 2019;17:2569–2575. doi: 10.3892/ol.2019.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubert M., Panicot-Dubois L., Crotte C., Sbarra V., Lombardo D., Sadoulet M.O. Peritoneal colonization by human pancreatic cancer cells is inhibited by antisense FUT3 sequence. Int J Cancer. 2000;88:558–565. doi: 10.1002/1097-0215(20001115)88:4<558::aid-ijc7>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 24.Witz I.P. The selectin-selectin ligand axis in tumor progression. Cancer Metastasis Rev. 2008;27:19–30. doi: 10.1007/s10555-007-9101-z. [DOI] [PubMed] [Google Scholar]
- 25.Remmers N., Anderson J.M., Linde E.M., DiMaio D.J., Lazenby A.J., Wandall H.H. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 2013;19:1981–1993. doi: 10.1158/1078-0432.CCR-12-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuessler M.H., Pintado S., Welt S., Real F.X., Xu M., Melamed M.R. Blood group and blood-group-related antigens in normal pancreas and pancreas cancer: enhanced expression of precursor type 1, Tn and sialyl-Tn in pancreas cancer. Int J Cancer. 1991;47:180–187. doi: 10.1002/ijc.2910470204. [DOI] [PubMed] [Google Scholar]
- 27.Radhakrishnan P., Dabelsteen S., Madsen F.B., Francavilla C., Kopp K.L., Steentoft C. Proceedings of the National Academy of Sciences United State of America. Vol. 111. 2014. Immature truncated O-glycophenotype of cancer directly induces oncogenic features; pp. E4066–E4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bian B., Bigonnet M., Gayet O., Loncle C., Maignan A., Gilabert M. Gene expression profiling of patient-derived pancreatic cancer xenografts predicts sensitivity to the BET bromodomain inhibitor JQ1: implications for individualized medicine efforts. EMBO Mol Med. 2017;9:482–497. doi: 10.15252/emmm.201606975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolle R., Blum Y., Duconseil P., Vanbrugghe C., Brandone N., Poizat F. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EbioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juiz N., Elkaoutari A., Bigonnet M., Gayet O., Roques J., Nicolle R. Basal-like and classical cells coexistence in pancreatic cancer revealed by single cell analysis. FASEB J. 2020;34:12214–12228. doi: 10.1096/fj.202000363RR. [DOI] [PubMed] [Google Scholar]
- 32.Duconseil P., Gilabert M., Gayet O., Loncle C., Moutardier V., Turrini O. Transcriptomic analysis predicts survival and sensitivity to anticancer drugs of patients with a pancreatic adenocarcinoma. Am J Pathol. 2015;185:1022–1032. doi: 10.1016/j.ajpath.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 33.Rashid N.U., Peng X.L., Jin C., Moffitt R.A., Volmar K.E., Belt B.A. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clin Cancer Res. 2020;26:82–92. doi: 10.1158/1078-0432.CCR-19-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan-Seng-Yue M., Kim J.C., Wilson G.W., Ng K., Figueroa E.F., O'Kane G.M. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 35.Milde-Langosch K., Karn T., Schmidt M., Zu Eulenburg C., Oliveira-Ferrer L., Wirtz R.M. Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res Treat. 2014;145:295–305. doi: 10.1007/s10549-014-2949-z. [DOI] [PubMed] [Google Scholar]
- 36.Potapenko I.O., Lüders T., Russnes H.G., Helland Å., Sørlie T., Kristensen V.N. Glycan-related gene expression signatures in breast cancer subtypes; relation to survival. Mol Oncol. 2015;9:861–876. doi: 10.1016/j.molonc.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashkani J., Naidoo KJ. Glycosyltransferase gene expression profiles classify cancer types and propose prognostic subtypes. Sci Rep. 2016;6:26451. doi: 10.1038/srep26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagatsuma T., Nagai-Okatani C., Matsuda A., Masugi Y., Imaoka M., Yamazaki K. Discovery of pancreatic ductal adenocarcinoma-related aberrant glycosylations: a multilateral approach of lectin microarray-based tissue glycomic profiling with public transcriptomic datasets. Front Oncol. 2020;10:338. doi: 10.3389/fonc.2020.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noda M., Okayama H., Tachibana K., Sakamoto W., Saito K., Thar Min A.K. Glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin Cancer Res. 2018;24:4468–4481. doi: 10.1158/1078-0432.CCR-17-3533. [DOI] [PubMed] [Google Scholar]
- 40.Gupta R., Leon F., Thompson C.M., Nimmakayala R., Karmakar S., Nallasamy P. Global analysis of human glycosyltransferases reveals novel targets for pancreatic cancer pathogenesis. Br J Cancer. 2020;122:1661–1672. doi: 10.1038/s41416-020-0772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Yamada S., Inenaga S., Imamura T., Wu Y., Wang K.Y. Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br J Cancer. 2011;104:1882–1889. doi: 10.1038/bjc.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chugh S., Barkeer S., Rachagani S., Nimmakayala R.K., Perumal N., Pothuraju R. Disruption of C1galt1 gene promotes development and metastasis of pancreatic adenocarcinomas in mice. Gastroenterology. 2018;155:1608–1624. doi: 10.1053/j.gastro.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azizian A., Rühlmann F., Krause T., Bernhardt M., Jo P., König A. CA19-9 for detecting recurrence of pancreatic cancer. Sci Rep. 2020;10:1332. doi: 10.1038/s41598-020-57930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphris J.L., Chang D.K., Johns A.L., Scarlett C.J., Pajic M., Jones M.D. The prognostic and predictive value of serum CA19.9 in pancreatic cancer. Ann Oncol. 2012;23:1713–1722. doi: 10.1093/annonc/mdr561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engle D.D., Tiriac H., Rivera K.D., Pommier A., Whalen S., Oni T.E. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science. 2019;364:1156–1162. doi: 10.1126/science.aaw3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aubert M., Panicot L., Crotte C., Gibier P., Lombardo D., Sadoulet M.O. Restoration of alpha(1,2) fucosyltransferase activity decreases adhesive and metastatic properties of human pancreatic cancer cells. Cancer Res. 2000;60:1449–1456. [PubMed] [Google Scholar]
- 47.Karasawa F., Shiota A., Goso Y., Kobayashi M., Sato Y., Masumoto J. Essential role of gastric gland mucin in preventing gastric cancer in mice. J Clin Invest. 2012;122:923–934. doi: 10.1172/JCI59087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohya A., Yamanoi K., Shimojo H., Fujii C., Nakayama J. Gastric gland mucin-specific O-glycan expression decreases with tumor progression from precursor lesions to pancreatic cancer. Cancer Sci. 2017;108:1897–1902. doi: 10.1111/cas.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi H., Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 2014;453:235–242. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moloney D.J., Panin V.M., Johnston S.H., Chen J., Shao L., Wilson R. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 51.Zhang S., Chung W.C., Xu K. Lunatic Fringe is a potent tumor suppressor in Kras-initiated pancreatic cancer. Oncogene. 2016;35:2485–2495. doi: 10.1038/onc.2015.306. [DOI] [PubMed] [Google Scholar]
- 52.Wang S., Zhang L., Yu Z., Chai K., Chen J. Identification of a glucose metabolism-related signature for prediction of clinical prognosis in clear cell renal cell carcinoma. J Cancer. 2020;11:4996–5006. doi: 10.7150/jca.45296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long K.B., Tooker G., Tooker E., Luque S.L., Lee J.W., Pan X. IL6 receptor blockade enhances chemotherapy efficacy in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2017;16:1898–1908. doi: 10.1158/1535-7163.MCT-16-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mace T.A., Shakya R., Pitarresi J.R., Swanson B., McQuinn C.W., Loftus S. IL-6 and PD-L1 antibody blockade combination therapy reduces tumor progression in murine models of pancreatic cancer. Gut. 2018;67:320–332. doi: 10.1136/gutjnl-2016-311585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang P., Zhang C., Li W., Zhai B., Jiang X., Reddy S. Identification of a robust functional subpathway signature for pancreatic ductal adenocarcinoma by comprehensive and integrated analyses. Cell Commun Signal. 2020;18:34. doi: 10.1186/s12964-020-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noll E.M., Eisen C., Stenzinger A., Espinet E., Muckenhuber A., Klein C. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22:278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas H. Pancreatic cancer: CYP3A5 contributes to PDAC chemoresistance. Nat Rev Gastroenterol Hepatol. 2016;13:188. doi: 10.1038/nrgastro.2016.36. [DOI] [PubMed] [Google Scholar]
- 58.de Man F.M., Goey A.K.L., van Schaik R.H.N., Mathijssen R.H.J., Bins S. Individualization of irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 2018;57:1229–1254. doi: 10.1007/s40262-018-0644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.