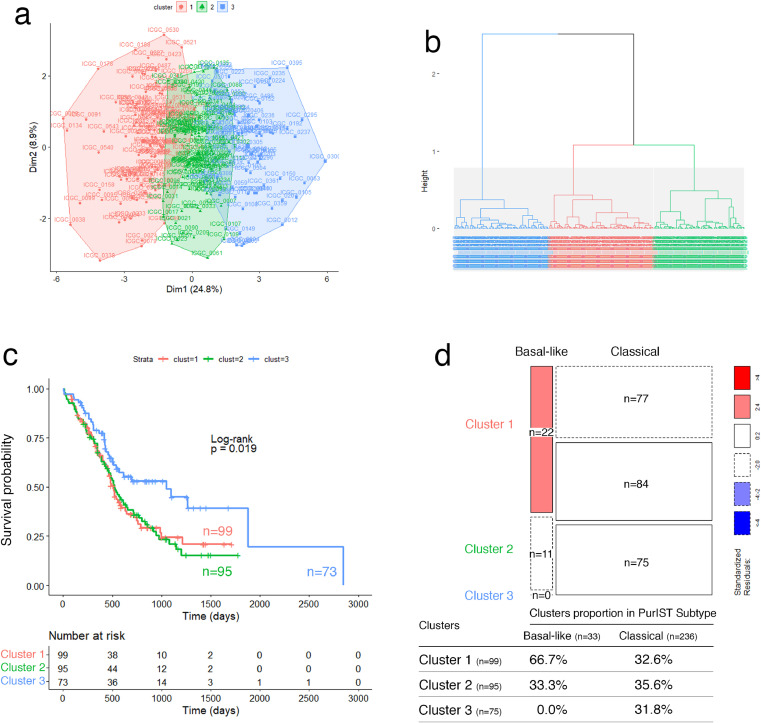
Fig. 3.
Validation of the glyco-signature on ICGC patient's cohort (microarray). (a) Two-dimension representation of the HCPC analysis results and (b) cluster dendrogram related to ICGC affymetrix transcriptomic data (n = 269) using 19 GT genes. (c) Survival curves estimated by using the Kaplan-Meier method and comparing OS probabilities between clusters with the log-rank test (p-value = 0.0019). Clusters 1 and 2 have shorter median OS of 515 and 517 days, respectively compared to cluster 3 with median OS of 1048 days. (d) Mosaic plot showing cross-link between basal-like/classical subtypes according to PurIST classifier and identified clusters 1, 2 and 3 through GT gene prognostic markers. Box height reflects the number of tumors classified in each cluster and box width represents proportion of basal-like/classical subtypes (p-value = 5.424e-05, Chi-squared test).