Abstract
Aptamers are short, single-stranded DNA or RNA oligonucleotide molecules that function as synthetic analogs of antibodies and bind to a target molecule with high specificity. Aptamer affinity entirely depends on its tertiary structure and charge distribution. Therefore, length and structure optimization are essential for increasing aptamer specificity and affinity. Here, we present a general optimization procedure for finding the most populated atomistic structures of DNA aptamers. Based on the existed aptamer LC-18 for lung adenocarcinoma, a new truncated LC-18 (LC-18t) aptamer LC-18t was developed. A three-dimensional (3D) shape of LC-18t was reported based on small-angle X-ray scattering (SAXS) experiments and molecular modeling by fragment molecular orbital or molecular dynamic methods. Molecular simulations revealed an ensemble of possible aptamer conformations in solution that were in close agreement with measured SAXS data. The aptamer LC-18t had stronger binding to cancerous cells in lung tumor tissues and shared the binding site with the original larger aptamer. The suggested approach reveals 3D shapes of aptamers and helps in designing better affinity probes.
Keywords: aptamer, oligonucleotide, tertiary structure, spatial structure, lung adenocarcinoma, small-angle X-ray scattering, SAXS, molecular dynamics, fragment molecular orbital, molecular simulations
Graphical abstract

The article describes a general optimization procedure of the DNA aptamer toward lung cancer. The truncated aptamer LC-18t had stronger binding to cancerous cells and shared the binding site with the original larger aptamer. The suggested approach reveals 3D shapes of oligonucleotides and helps in designing better affinity probes.
Introduction
Nucleic acids (DNA and RNA) hold key information about human development, physiology, and evolution.1 Along with the critical function of encoding proteins, the biological importance of short single-stranded (ss)RNA and ssDNA has been widely described.2, 3, 4, 5, 6 Short ss-oligonucleotides can easily form complexes, enabling ribozymes (small ssRNAs) to catalyze biochemical reactions,7 regulate gene expression,8 and participate in protein synthesis.9
Recent progress in molecular biology and bionanotechnology has made ssDNA a useful tool with various applications in biology and medicine. Long ssDNA structures are created in an approach called DNA origami,10 which has revolutionized the field and led to breakthroughs in many areas of biotechnology and sensing, electronics, and the food and pharmaceutical industry.
An important feature of oligonucleotides is mimicking antibodies.3,11 In vitro evolution of nucleic acid fragments enables aptamers to be selected from large random-sequence libraries of short ssDNA or ssRNA.12,13 Similar to antibodies, aptamers bind to their targets with high affinity and selectivity, because of the ability of oligonucleotides to fold into complex 3D shapes,3,14,15 some of which can exhibit specific binding to their target.
The molecular structure of aptamers determines their function. The former, in turn, depends on the local molecular environment, temperature, pH, solvated ions, and other factors. Obtaining the structure is challenging;16, 17, 18, 19, 20 however, atomistic computer simulations, such as molecular dynamics (MD), can generate plausible molecular models that represent multiple minima that a molecule can adopt in solution.21, 22, 23 Ranking these structures, that is, predicting which of them are more probable, is a very difficult computational task because of the tiny energy differences between these isomers, which necessitates the use of expensive computational methods for a reliable ranking.
Some methods are available for modeling oligonucleotides.24,25 Based on simulations, one can predict molecular structure and optimize existing oligonucleotides for specific purposes.26, 27, 28, 29, 30 Having a molecular structure is needed for molecular docking,31 that is, predicting a binding site.
Models obtained by theoretical methods can be verified experimentally, for instance, by comparing with structures from X-ray crystallography, which requires crystallizing the sample. Thus far, however, obtaining crystals of aptamers has been impossible, and obtaining a crystal structure of their protein complexes is laborious and challenging.16, 17, 18,32
Nuclear magnetic resonance (NMR)20,33 can also be used to obtain molecular structures of aptamers and their complexes. However, NMR has certain limitations for molecules larger than 30 kDa, because the analysis and interpretation of NMR data are challenging.20,34 Some structures of oligonucleotide-protein complexes have been successfully obtained experimentally by using X-ray crystallography and NMR.35, 36, 37, 38
Small-angle X-ray scattering (SAXS)39 is a powerful biophysical method for studying the overall shape and structural transitions of biological macromolecules in solution at nanometer resolution. It is a well-established method for structural investigation of proteins, DNA/RNA, and their complexes, suitable for characterization of aptamers in solution.14,40, 41, 42, 43, 44 SAXS has several advantages over the other methods of structural analysis: it does not require complicated sample preparation, and it features fast data collection and processing. A key advantage of this technology is the ability to measure samples in solution under quasi-physiological conditions, whereas the solution parameters such as temperature, pH, and buffer composition are easily adjustable.45
The promising approach to determine aptamers’ shape in solution is a combination of SAXS data together with MD simulations. Applicability of such an approach was successfully demonstrated in several papers for both proteins46,47 and nucleic acids.48 Simulations were used to sample possible conformations that molecules adopt in the solution, and ensembles of such structures were re-weighted to fit into the SAXS experimental curves.
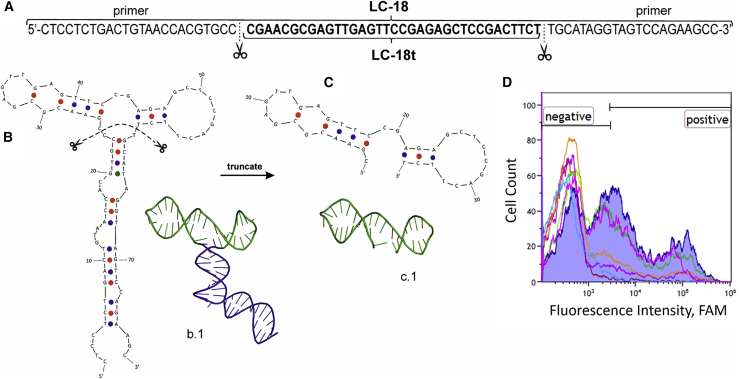
Aptamer LC-18 can bind to lung adenocarcinoma cells,49 tissues,50 and blood plasma51 with high specificity. LC-18 is made of a sequence of 80 nucleotides, including two constant 20-nucleotide primers on each side (Figure 1A). There is a strong need to reduce the size of this aptamer for enhancing its binding properties and making it cheaper to synthesize. In this work, a much-truncated LC-18 (LC-18t) is proposed, demonstrating binding properties alike to those of its predecessor LC-18. To obtain the molecular structure, a combination of theoretical methods, namely, DNA folding tools, quantum-chemical calculations, and MD simulations, is used. The simulated structures are compared with the experimental SAXS shape. In addition, the efficacy of the new aptamer is verified experimentally for cancerous cells.
Figure 1.
Aptamer optimization
(A) Sequences of LC-18 and LC-18t aptamers, schematic secondary (1) and tertiary (2) structures of LC-18 (B), and LC-18t (C) aptamers. Replacement analyses demonstrating that the long and short aptamers have the same binding site. Flow cytometry binding histogram where the blue curve corresponds to the long FAM-labeled LC-18 aptamer binding to patient-derived cells of adenocarcinoma tissues at the 100-nM concentration; the pink, green, orange, purple, and light-blue curves indicate the portion of cells bound with non-labeled LC-18 after incubation with 2-, 5-, 10-, 100-, and 1,000-fold excess of LC-18t. The red curve describes intact lung adenocarcinoma cells (D).
Results
Improvement of the previously developed aptamer
The length of the LC-18 aptamer selected from the ssDNA library is 80 nucleotides, including primer regions for amplification. Smaller patterns are usually more specific to the target and are easier to synthesize. The long LC-18 aptamer was truncated to 35 nucleotides in accordance with the predicted secondary and tertiary structures (Figures 1B and 1C). Aptamer LC-18 has the sequence 5′-CTCCTCTGACTGTAACCACGTGCCCGAACGCGAGTTGAGTTCCGAGAGCTCCGACTTCTTGCATAGGTAGTCCAGAAGCC-3′, whereas LC-18t has the sequence 5′-CGAACGCGAGTTGAGTTCCGAGAGCTCCGACTTCT-3′ (Figure 1A).
Binding properties of the new truncated aptamer
To demonstrate that both aptamers LC-18t and LC-18 have the same binding site, replacement analyses were performed. The assay was made by using flow cytometry. Lung cancer cells derived from a patient with lung adenocarcinoma were preincubated with masking RNA and then with 100 nM of the original aptamer LC-18. Applying a higher concentration of the truncated aptamer replaced the original long aptamer in a dose-dependent manner (Figure 1D). Low concentrations (200 and 500 nM) of LC-18t did not replace the parent aptamer (bound with cells at 100 nM concentration). Only 1 μM (a 10-times excess) of LC-18t replaced 58% of LC-18 bound with the cells after preincubation with LC-18t at a concentration of 100 nM. Increasing the concentration of the truncated aptamer to 10 μM resulted in a 68% replacement and to 100 μM in an almost complete replacement of LC-18. The dissociation constant (KD) value for LC-18t appeared to be much lower (19.4 nM) than for LC-18 (149.2 nM) (Figure S5). This means that the binding energy for the LC-18t aptamer is much higher than for LC-18. Therefore, a high concentration of the LC-18 aptamer was needed to replace the LC-18t aptamer (Figure 1D). To demonstrate that the truncated aptamer binds directly to cancer cells in a patient’s tissues in the same manner as its ancestor aptamer LC-18, thin paraffin sections of lung adenocarcinoma tissues (Figure 2A1) were stained. Microscopic analyses revealed a co-localization of LC-18t (Figures 2A3 and 2A4) with anti-carcinoembryonic antigen (CEA) antibodies that are used to stain lung tumor cells (Figure 2A2). Antibodies and aptamers stain the same cells with similar fluorescence intensity (Figures 2A5 and 2A6) but with slightly different localization. Flow cytometry demonstrates that LC-18t binds to patient-derived lung tumor cells but does not bind with healthy lung and inflammation tissues. Oligonucleotide (AG)40, taken as a non-specific control, did not bind to lung cancer cells (Figure 2B).
Figure 2.
LC-18t recognition of the lung cancer cells
Verification of LC-18t targeting lung cancer cells by histological analyses (A) and flow cytometry (B). (A1) Laser-scanning imaging of lung cancer tissue sections stained with hematoxylin and eosin. (A2 and A3) A co-stained adjacent tissue section with Alexa Fluor 405-labeled anti-CEA antibodies (A2) and Cy-5-labeled aptamer LC-18t (A3). (A4) An overlay of (A2) and (A3). (A5) A 3D representation (A6). (B) Histograms of fluorescence intensity with a 40× magnification. Flow cytometry histograms indicate binding of FAM-labeled LC-18t to lung adenocarcinoma cells (the purple curve), cells from the lung with inflammation (red curve), and relatively healthy lung cells (the blue curve). The black curve corresponds to reference intact cells. Binding of lung tumor cells with the control (AG)40 oligonucleotide is shown in brown.
Ab initio SAXS modeling
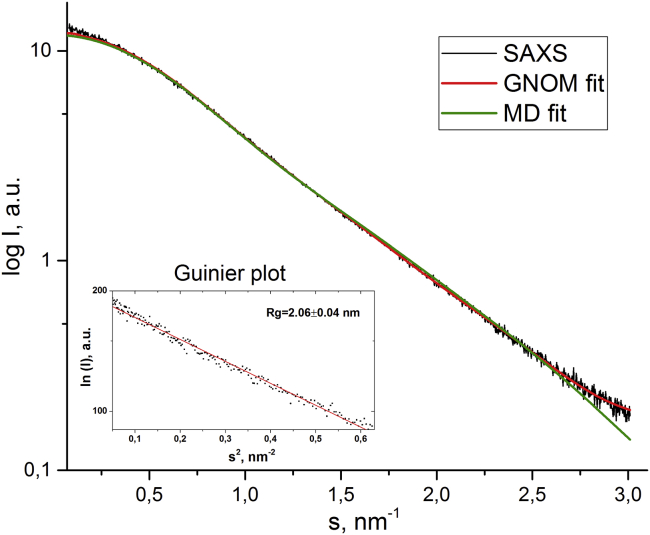
The size-exclusion chromatography (SEC) with SAXS (SEC-SAXS) data clearly indicates a monodisperse solution without any signs of aggregates or oligomeric constituents (see Figure S4). The data were processed for the aptamer LC-18t, and the geometrical parameters were derived. The radius of gyration (Rg) = 2.06 ± 0.04 nm was obtained from the slope of the linear approximation of the Guinier region.39
The maximum dimension (Dmax) = 6.7 nm was found as the largest value r with the nonzero value of the pair distance distribution (p(r)) function, which represents the maximum size of the molecule (Figure 3A). Figure 3B shows the most probable 3D molecular shape based on the obtained p(r) function according to DAMMIN.
Figure 3.
Determination of the aptamer shape based on the small-angle X-ray scattering (SAXS)
(A) Pair distance distribution (p(r)) function. Maximum r value for the p(r). (B) Bead molecular model obtained from the p(r).
Molecular modeling
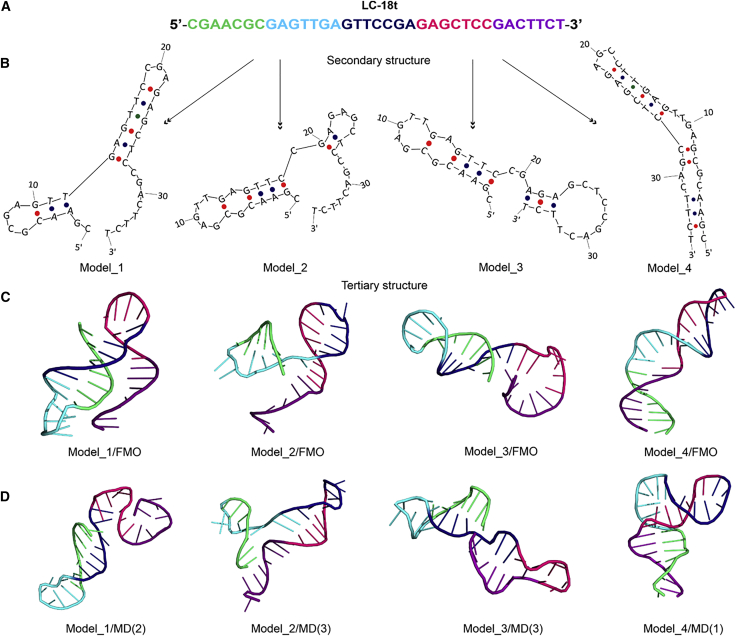
The aptamer LC-18t consists of 35 nucleotides (Figure 4A). To predict the initial structure of LC-18t, OligoAnalyzer25 and MFold24,52,53 were used. Simulations were carried out under the conditions of the SAXS experiments: in the presence of ions Na+ 146 mM and Mg2+ 0.5 mM at 5°C and 20°C. At 5°C, MFold yielded 7 possible models, whereas 17 different conformers were predicted at the same temperature by the OligoAnalyzer server. The MFold and OligoAnalyzer results at 20°C for the LC-18t aptamer include 5 and 11 models, respectively. Further refinement is needed for a reliable determination of the structure in simulations. To this end, four models were chosen (Figure 4B). Model1, Model2, and Model3 are proposed by both OligoAnalyzer and MFold as very probable secondary structures at 5°C, and Model1 has the lowest energy. In addition, Model1 and Model3 are in the top 5 thermodynamically stable structures predicted at 20°C. Model4 is included in the top 10 suggested structures at 5°C and 20°C. The corresponding tertiary structures of Model1, Model2, Model3, and Model4 were prepared by using the Avogadro program and optimized with the fragment molecular orbital (FMO)-density-functional tight-binding (DFTB)3/polarizable continuum model (PCM) method (Figure 4C). The ranking of structures according to their energy based on the FMO method combined with DFTB is different from the MFold and OligoAnalyzer results. According to FMO, the lowest structure is Model4, and the order of structures is Model4 (most probable, lowest energy), Model1, Model3, and Model2 (least probable, highest energy).
Figure 4.
Molecular modeling of the aptamer structure
(A) Sequences of LC18 and LC18t aptamers, (B) schematic structures of LC-18t aptamer models, (C) molecular structures after geometry optimizations with FMO-DFTB3/PCM, and (D) molecular structures after molecular dynamics (MD) simulations (the number in parentheses identifies the cluster). The 3′ end is in purple and pink, the central part is dark blue, and the 5′ end is in cyan and green.
Molecular structures obtained from MD simulations are shown in Figures S1 and 4D for selected clusters numbered 1–3 in parentheses (a cluster means a distinct conformer within the same MD trajectory). Model1 and Model2 with a ss segment at the 3′ end are similar to the models predicted by OligoAnalyzer and MFold as probable structures. The structure relaxation in MD resulted in significant structural changes in single strands for both Model1 and Model2, because single strands of the aptamers are very flexible, whereas the double-helical chains of aptamers are quite rigid. Although some changes took place in MD in the structures of Model1 and Model2, the secondary structure remained the same.
In contrast to Model1 and Model2, Model3 lacks long ss parts, and its structure can be represented as two hairpins at the 3′ and 5′ ends connected by C19 and G20 nucleotides (Figure 4B). As shown in Figures 4C and 4D, Model3 did not change much in the MD simulations. Model3 features preservation of the secondary structure in the 5′ end hairpin (Figure 4D), whereas the 3′ end hairpin shows some variation in it.
Specifically, in Model3 a shifting of the nucleotides relative to each other in the 3′ end hairpin (Figure S2) is observed. This shifting results in a decrease in the number of complementary base pairs, but at the same time, a large number of hydrogen bonds are formed between new pairs of nucleotides.
For Model4, substantial changes in molecule geometry after MD are caused by the presence of the bulge and internal loop in the middle part of the stem. FMO optimization predicts that this part has a linear structure, but in MD, it is bent.
SAXS validation of the obtained models
The three most commonly used programs CRYSOL, WAXSiS, and PEPSI-SAXS were applied to the models obtained in the FMO and MD simulations in order to independently validate them against the SAXS data. The values of the χ2 deviations between experimental and simulated SAXS profiles are summarized in Table 1.
Table 1.
Deviations of χ2 of the experimental SAXS curve for LC-18t from the simulated curves based on FMO and MD calculations
| LC-18t modela | CRYSOL | WAXSiS | PEPSI-SAXS |
|---|---|---|---|
| Model1/FMO | 20.05 | 23.18 | 22.26 |
| Model1/MD(1) | 17.56 | 20.21 | 18.79 |
| Model1/MD(2) | 3.28 | 3.14 | 3.29 |
| Model2/FMO | 132.13 | 160.20 | 108.18 |
| Model2/MD(1) | 17.85 | 24.08 | 25.38 |
| Model2/MD(2) | 17.90 | 27.22 | 4.70 |
| Model2/MD(3) | 8.47 | 15.12 | 7.82 |
| Model2/MD(4) | 19.08 | 35.77 | 15.76 |
| Model3/FMO | 2.59 | 5.99 | 10.63 |
| Model3/MD(1) | 7.04 | 10.23 | 8.26 |
| Model3/MD(2) | 7.51 | 4.51 | 14.11 |
| Model3/MD(3) | 2.62 | 3.50 | 4.47 |
| Model3/MD(4) | 7.06 | 9.37 | 1.78 |
| Model3/MD(5) | 9.46 | 11.94 | 5.77 |
| Model4/FMO | 2.11 | 4.15 | 5.59 |
| Model4/MD(1) | 25.32 | 29.8 | 13.12 |
| Model4/MD(2) | 29.25 | 30.17 | 28.78 |
The number after MD identifies the cluster (a group of structures).
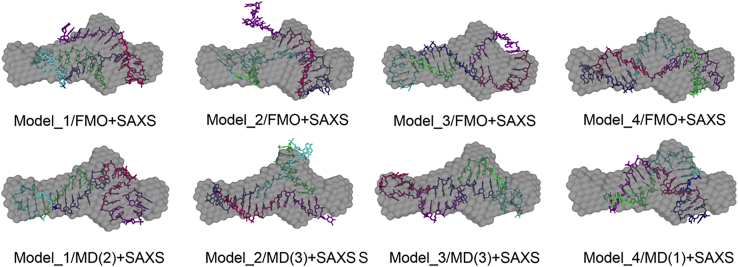
A comparison of the SAXS bead model with LC-18t structures obtained in the FMO optimization and MD simulations is presented in Figure 5. According to Table 1, Model4 after FMO optimization has the best fit with the SAXS bead model (the deviations are the smallest), followed by Model3. In MD, Model1/MD(2) and Model3/MD(3) have the best χ2 values. Note that the labeling of the FMO and MD structures as Modeli may be to some extent misleading because it means only that the particular model was used as a starting structure, and optimization and MD can actually lead to another model (in particular, Model1 and Model2 changed substantially in MD). A detailed comparison of Model3/MD(3) with SAXS is shown in Figure 6.
Figure 5.
Fitting of possible aptamer conformations with experimental SAXS data
Comparison of the SAXS model from the experiment (gray beads) with structures of LC-18t (colored) obtained with FMO optimizations and MD simulations (the number in parentheses identifies the cluster number), for clusters with a small deviation χ2.
Figure 6.
Comparison of experimental and theoretical SAXS curves
Experimental (black) and theoretical SAXS curves based on the bead model structure (red) and MD simulations for Model3/MD(3) (green), with the deviation χ2 = 2.615. The Guinier plot is inserted, demonstrating the calculated gyration radius of the molecule and linearity of the Guinier region, which indicates the monodispersity of the solution.
None of the theoretical models fills the whole SAXS volume (Figure 5). In the real solution, the solvated aptamer molecules are present in multiple conformational states occupying a whole ensemble of conformations, with the relative population ratio governed by the minimum of thermodynamical free energy. A SAXS experiment thus corresponds to an average model over all the possible conformations in solution.
In all calculations in this work, a single conformation is used. In FMO, there is just 1 solute molecule, whereas in MD, there is an infinite set of molecules under periodic boundary conditions; but all solute molecule images are in the same conformation (at a given time).
The most plausible rationalization is that several conformers (models) are relevant; that is, they have a similar energy and occur with a comparable probability. Clearly, one conformer should describe the bulge (the central, thick part in the SAXS volume), whereas another must stretch sideways to the full length. It is physically impossible for 1 conformer to do both (assuming that the SAXS profile is to be trusted). However, it is conceivable that some volume appearing empty for models in Figure 5 may be partially occupied by minor deviations in the structure of the same conformer caused by a local rotation of flexible groups.
Under the assumption that multiple isomers are relevant to fill the SAXS volume, the χ2 values for individual conformers are not very meaningful. However, one can clearly rule out some models. For example, Model2 sticks out of the volume, especially for FMO. A mixture of Model1, Model2, and Model3 could fill the whole SAXS volume. Assigning weights to models is possible by using Boltzmann factors, but it requires a very accurate estimate of the energy to be meaningful.
Discussion
Aptamers are used as clinical reagents for targeted therapeutics and drug delivery, diagnostics and biosensing, and environmental sensors for food and water analysis.4 Successful development of aptamer-based approaches requires information about the aptamer spatial structure. In this work, a new aptamer, LC-18t, has been designed with a much smaller size than its predecessor, LC-18. The new truncated aptamer has been demonstrated to be as efficient as the original one. Histological analyses and flow cytometry analyses have been conducted on lung cancer tissue.
The rational design of highly efficient aptamers depends on knowing their molecular structure. In this work, it is shown that molecular simulations can predict structures from SAXS experiments. OligoAnalyzer and MFold tools have been used to generate a set of several likely models. However, as further atomistic MD and FMO simulations have shown, these models do not accurately predict the relative occupation probability of the conformers in solution.
It has been shown that both geometry optimizations with a quantum-mechanical method and MD with force fields deliver structures that have small deviations from the experimental SAXS curves. With the use of these methods, one can obtain a reliable set of molecular structures. By comparing the calculated and experimentally measured molecular shapes, it has become clear that no single atomistic model can physically fill the bead shape perfectly: When the central bulge is formed, the remaining tails are too short (a crouching pose), leaving empty space at the ends; when tails are widespread, there is no bulge (a stretching pose).
It has been hypothesized that at least several conformers (molecular models) coexist in solution in some ratio so that a scan in SAXS reveals them in an averaged way. Nevertheless, some single calculated models show very good agreement with SAXS, validating the theoretical methods. The best agreement is found for Model3/MD(3). To compare the SAXS curve obtained from simulations with the experimental data, three commonly used programs, CRYSOL, WAXSIS, and PEPSI-SAXS, were applied. Even though all three programs identified Model3 as the closest to the SAXS bead model, the PEPSI-SAXS results differ considerably from the other two programs.
On the basis of presented results for atomistic modeling of aptamer structures, the following protocol is suggested: (1) use SAXS to determine the shape of the aptamer in an experiment, (2) do the initial design of molecular models by using OligoAnalyzer or MFold, (3) perform molecular modeling by using computational methods such as FMO or MD, and (4) compare atomic structure from simulations with the measured SAXS curve using CRYSOL or WAXSIS programs.
Although the proposed procedure to determine the structure is applied to an aptamer in this work, it could also be employed for aptamer-target complexes in future works. And by obtaining a reliable atomistic structure from simulations, one can rationalize the binding of an aptamer to a target, such as a protein. This can be very helpful in designing new, more efficient aptamers based on insight gained from molecular simulations.
Materials and methods
Aptamers LC-18 5′-CTCCTCTGACTGTAACCACGTGCCCGAACGCGAGTTGAGTTCCGAGAGCTCCGACTTCTTGCATAGGTAGTCCAGAAGCC-3′, LC-18t 5′-CGAACGCGAGTTGAGTTCCGAGAGCTCCGACTTCT-3′, and non-specific oligonucleotide (AG)40 5′-CTCCTCTGACTGTAACCACG(AG)40GCATAGGTAGTCCAGAAGCC-3′ were synthesized, labeled, and purified by Integrated DNA Technologies (USA). Aptamers were ordered unlabeled and labeled with Cy-5 or FAM fluorophores.
Flow cytometry
All experiments with human tissues were carried out in accordance with the approved guidelines and principles expressed in the Declaration of Helsinki. The study was approved by the Local Committee on Ethics of the Krasnoyarsk Regional Clinical Cancer Center, named after A. I. Kryzhanovsky No. 6 from March 22, 2016, in Krasnoyarsk, Russia. The patients in this study signed their consent.
Tumor tissues were collected from the patients with lung adenocarcinoma during the complete surgical resection of their tumor. The tissues were immediately placed in ice-cold DMEM (Capricorn Scientific, Germany). Flow cytometry experiments were performed within 2–3 h of tissue collection. Lung cancer tissues, lung inflammation tissues, and relatively healthy tissues distant from the tumor were washed with Dulbecco’s phosphate-buffered saline (DPBS), minced by a blade, pipetted, and filtered through 70 μm filters to obtain a homogeneous solution. The cell suspension was washed three times with DPBS by centrifugation at 3,000 × g for 5 min. Cells (0.5 million in each sample) were preincubated on a shaker with yeast RNA (1 ng μL−1) for 30 min and then with 100 nM of FAM-labeled aptamer LC-18t, LC-18, or (AG)40 oligonucleotide as a control for 30 min at 25°C.
The binding level was measured on an FC-500 Flow Cytometer (Beckman Coulter, USA). Replacement analyses were performed to prove that aptamers share the same binding site. Higher concentrations of LC-18t (0.2, 0.5, 1, 10, and 100 μM) were used to replace LC-18 from the cells.
Determination of KDs of aptamers
Apparent KDs for the aptamers LC-18 or LC-18t were determined using flow cytometry. Patient-derived lung cancer cells were incubated with 2 nМ, 6 nM, 10 nМ, 20 nM, 30 nМ, 60 nМ, 80 nM, 100 nМ, 150 nМ, and 300 nМ of FAM-labeled aptamers. The data were analyzed with Kaluza 1.2 software. Apparent KDs for the aptamers LC-18 or LC-18t were determined from the regression affinity curves corresponding to the percentage of bound lung cancer cells versus aptamer concentrations as half from maximum binding.
In accordance with the Michaelis-Menten equation, under conditions of excess aptamer concentration:
where [R0] is LC cell concentration, [L0] is aptamer concentration; [RLinf] is the steady-state equilibrium concentration of the LC cells and aptamer complex; Kd is aptamer’s KD; and [RLinf]/[R0] is the steady fraction of bound cells.
A hyperbolic regression model was plotted in accordance with the equation F ([L0]) = [L0]/([L0] + KD) from the dataset of serial measurements [RLinf]/[R0] depending on the value [L0] to determine the KD value.
Histological tissue staining
Lung cancer tissue pieces were frozen in liquid nitrogen and sliced into 5 μm sections by a Microm HM525 Cryostat and placed on polylysine-coated glass slides. Tissue sections were incubated with 10% bovine serum albumin (Sigma-Aldrich, USA) for 30 min, followed by incubation with a primary anti-CEA antibody (2 ng μL−1) in a humidified atmosphere for 1 h and then with a secondary antibody labeled with Alexa Fluor 405 (2 ng μL−1) in a humidified atmosphere for 1 h and then washed three times with DPBS.
Nonspecific binding of the aptamers was blocked by yeast RNA (1 ng μL−1; Sigma-Aldrich, USA) incubated for 30 min. Then 50 nM of Cy-5-labeled LC-18t aptamer was added. The mixture was incubated for 30 min in a humidified atmosphere and washed three times with DPBS. The Bio Mount mounting medium (Bio-Optica, Italy) was used to fix the sections. The tissue sections were analyzed by laser-scanning fluorescence microscopy using a Carl Zeiss LSM800 system.
SAXS
SAXS measurements were performed at the P12 BioSAXS beamline of the European Molecular Biology Laboratory (EMBL) at the Petra III storage ring (German Synchrotron Research Centre [DESY], Hamburg, Germany).54 The data were collected on a photon-counting Pilatus-6M detector at a sample-to-detector distance of 3.0 m and a wavelength λ of 0.12 nm covering the momentum vector range 0.2 < s < 7.8 nm−1 (s = 4πsinθ/λ, where 2θ is the scattering angle). The measurements were performed in two regimes: (1) a high-throughput “batch” mode using a robotic sample changer at several concentration points for a further extrapolation to infinite dilution and (2) a combined in-line use of SEC-SAXS. The latter allows one to address on stream the sample monodispersity as well as conformational heterogeneity.
In the batch mode, the scattering patterns from the dilution series were collected at four different concentrations: 8, 4, 2, and 1 mg/mL, each in a volume of 50 μL. The extrapolation of the scattering signal to zero concentration allows one to minimize the influence of interparticle interactions at low angles, thus effectively eliminating the impact of the structure factor. Strictly speaking, this regime does not guarantee the monodispersity of the solution, and the data should be treated cautiously. However, if a single biologically relevant model fits the SAXS data, it may indicate the monodispersity of the sample and confirm the feasibility of the derived SAXS parameters.
The SEC-SAXS mode allows one to directly validate the monodispersity of the solution and, for example, to collect the SAXS data from different oligomeric states separately.55 The largest available on-site column Superdex 75 Increase 10/300 GL was utilized to confirm the sample monodispersity. The flow rate 0.4 mL/min with an exposition period of 1 frame per second was used, resulting in profiles of a total of 3,600 individual SAXS items.
The initial SEC-SAXS data-processing steps were done in CHROMIXS.56 The obtained 1D data were processed by using standard procedures.45,57,58 The program PRIMUS from the ATSAS package59 was used to derive the overall SAXS parameters: the Rg, intraparticle Dmax, and Porod volume (Vp). The p(r) function was calculated by using the indirect Fourier transform by GNOM.60 The p(r) function represents a histogram of distances between all possible pairs of atoms within a particle, weighted by their individual scattering contrasts. The molecular mass of the solute was calculated by comparison of the forward scattering intensity (I(0)) of the sample with that of the scattering from a reference bovine serum albumin solution and also from the excluded solute volume calculated with DATPOROD.
The spatial shape of the biomolecule was obtained by an ab initio method of molecular envelope determination using the DAMMIN program.61 In the case of ab initio reconstruction, the scattering particle is approximated by uniformly scattering “dummy” pseudo atoms in order to reconstruct the overall molecular envelope.61 Since SAXS is an intrinsically ambiguous method, multiple models were reconstructed and cross validated to choose the most representative one for further analysis.62 This model was used as a guide for molecular modeling calculations.
The scattering from the obtained atomistic models was simulated and back validated against the experimental data by CRYSOL,63 WAXSiS,64 and PEPSI-SAXS.65 CRYSOL uses a multipole expansion of the scattering amplitudes to calculate the spherically averaged scattering pattern and approximates the hydration shell by a border layer of a given thickness and density. WAXSiS is based on explicit-solvent, all-atom MD simulations providing a realistic model for both the hydration layer and the excluded solvent. PEPSI-SAXS is also based on a multipole expansion principle, whereas its run time is reduced even more than that of CRYSOL because of the introduction of several additional speed-up features. The SAXS dataset is available at the SASBDB;66 accession code: SASDK36.
Secondary structure prediction
Based on the sequence, the secondary structure of the LC-18t aptamer was predicted by using the online software OligoAnalyzer25 3.1 and MFold.24,52,53 The simulations were done with 146 mM Na+ and 0.5 mM Mg2+ at 5°C and 20°C.
Details of theoretical calculations
Two kinds of computational methods were used to make separate predictions of the structures of aptamers: (1) geometry optimizations using a quantum-mechanical method, third-order DFTB3 with the 3ob-3-1 set of parameters67 and (2) MD with force fields. A comparison of the DFTB and MD structures with SAXS experiments reveals deviations for each method.
To accelerate the DFTB calculations, they were combined with the FMO method68,69 at the two-body level FMO2, whereas the solvent (water) was described by a PCM.70. In FMO, each nucleotide in LC-18t was treated as a fragment (35 fragments total); the fragmentation was performed by using the Facio71 program. The modeling of the aptamers was performed with Avogadro72 and fully optimized with FMO-DFTB/PCM using the GAMESS program.73
MD simulations were conducted by using GROMACS 2019.8 software.74 The aptamer was solvated in a 10 × 10 × 10-nm periodic cubic box of water. Then the whole system was neutralized with Na+ ions, and additional Na+ and Cl− ions were added up to total 0.1 M concentration of salt. The Amber14sb75 force field for the aptamer and counterions as well as the TIP3P model76 for water was used. MD simulations of 200 ns were performed with the NPT (at constant number of particles N, pressure P, and temperature T) ensemble at 310 K and 1 atmosphere (atm) using the velocity-rescaling thermostat with a 1-ps time constant77 and at 1 bar pressure using the Parrinello-Rahman barostat with a 5-ps time constant78 and 4.5 × 10−5 bar−1 compressibility. Hydrogen-containing bond lengths were constrained to their equilibrium values in the force field by using the LINCS method,79 which permitted using a 2-fs time step for the trajectory integration. A particle mesh Ewald (PME) method80 was used to sum periodic electrostatic interactions with a real-space cutoff of 1.2 nm, and Lenard-Jones interactions were treated by using a cut-off scheme with the same 1.2-nm threshold. In addition, MD simulations with the same set-up were performed but using an Amber14sb_parmbsc181 set of force-field parameters for the aptamer, which is specifically tuned for DNA simulations. However, it shows much less conformational flexibility for the aptamers (see Figure S6). Thus, we considered only Amber14sb trajectories in the clustering.
Clusters of structures and their centers were computed by using the quality threshold algorithm82 implemented in VMD.83 Root-mean-square deviation (RMSD) of phosphorus atoms was used as a metric function with a cutoff of 0.4 nm. At most, five clusters were extracted for each aptamer.
Computation of scattering curves from molecular simulations
Scattering curves can be computed by using molecular structures obtained in simulations and validated against experimental SAXS curves. Historically, the first and the most straightforward approach to generate SAXS patterns from a given model is to explicitly compute the scattering contributions from all atoms of a model using the Debye equation.84 Although it gives a mathematically exact solution, this approach has two main drawbacks: (1) the hydration layer is difficult to model explicitly, and thus it is hard to get its scattering contribution, and (2) use of the Debye formula requires significant computational resources for big molecules. Therefore, we employed several popular approaches that work around these problems utilizing spherical harmonics expansion along with other performance improvements. Specifically, we used three programs: CRYSOL,62 WAXSiS,63 and PEPSI-SAXS.64
In CRYSOL, the parameters were maximum angle = 3 nm−1, number of points = 256, solvent density = 334 e/nm3, hydration shell contrast = 0.03 e/A3, maximum order of harmonics = 25, and order of the Fibonacci grid = 17. In WAXSiS the parameters were maximal angle = 3 nm−1, number of points = 101, solvent density = 334 e/nm3, and envelope distance = 0.7 nm. In PEPSI-SAXS, the parameters were hydration shell = 5%, use of explicit hydrogens, no smearing, and automatic background.
Acknowledgments
The research was performed using equipment of the Shared Core Facilities of Molecular and Cell Technologies at Krasnoyarsk State Medical University. The synchrotron SAXS data were collected at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). A.S.K. is grateful to Aptamerlab LLC for the assistance in aptamer design and 3D structure analyses. We thank Ivan Lapin for his help with microscopic analyses. Microscopic analyses using Carl Zeiss LSM 800 were carried out at the Center for Bioassay, Nanotechnology and Nanomaterials Safety (“Biotest-Nano”) (Multiple-Access Center, Tomsk State University, Tomsk, Russia). D.M. also thanks the CSC-IT Center in Espoo, Finland, for providing computational resources. The study was supported by a grant from the Russian Science Foundation (project number 21-73-20240) for A.S.K. R.V.M aknowledges Russian Foundation for Basic Research (project number 19-32-90266) for funding. D.G.F. acknowledges financial support by JSPS KAKENHI, grant number 19H02682. D.S.M. acknowledges financial support by BMBF grant number 16QK10A (SAS-BSOFT). Y.A.’s work at Argonne National Laboratory was supported by the US Department of Energy, Office of Science, under contract DE-AC02-06CH11357. D.M. received funding as a part of BioExcel CoE (https://bioexcel.eu/), a project funded by the European Union contracts H2020-INFRAEDI-02-2018-823830 and H2020-EINFRA-2015-1-675728. V.M. thanks Russian Foundation for Basic Research (project number 19-03-00043) for funding.
Author contributions
D.M., V.M., I.A.S., P.V.A., D.G.F., Y.A., and F.N.T. performed molecular simulation of the aptamers. R.V.M., D.S.M., V.N.Z., and A.E.S. performed SAXS experiments. G.S.Z., O.S.K., T.N.Z., A.V.K., R.A.Z., and A.S.K. performed biological experiments. D.V.V. performed mathematical analyses. M.V.B. and A.S.K. conceived of the general idea and provided supervision. D.M., V.M., I.A.S., P.V.A., M.V.B., D.G.F., Y.A., F.N.T., R.V.M., D.S.M., G.S.Z., O.S.K., T.N.Z., and A.S.K. wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.015.
Contributor Information
Yuri Alexeev, Email: yuri@alcf.anl.gov.
Anna S. Kichkailo, Email: azamay@krasgmu.ru.
Supplemental information
▪▪▪
References
- 1.Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- 2.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Dunn M.R., Jimenez R.M., Chaput J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1:0076. [Google Scholar]
- 4.Chen C., Zeng Z., Liu Z., Xia R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018;5:63. doi: 10.1038/s41438-018-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onoa B., Tinoco I., Jr. RNA folding and unfolding. Curr. Opin. Struct. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Rich R.R., Fleisher T.A., Shearer W.T., Schroeder H.W., Frew A.J., Weyand C.M. Third Edition. Elsevier; 2008. Clinical Immunology: Principles and Practice. [Google Scholar]
- 7.Doudna J.A., Cech T.R. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 8.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore P.B., Steitz T.A. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 10.Wang P., Meyer T.A., Pan V., Dutta P.K., Ke Y. The Beauty and Utility of DNA Origami. Chem. 2017;2:359–382. [Google Scholar]
- 11.Bauer M., Strom M., Hammond D.S., Shigdar S. Anything You Can Do, I Can Do Better: Can Aptamers Replace Antibodies in Clinical Diagnostic Applications? Molecules. 2019;24:4377. doi: 10.3390/molecules24234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 14.Tomilin F.N., Moryachkov R., Shchugoreva I., Zabluda V.N., Peters G., Platunov M., Spiridonova V., Melnichuk A., Atrokhova A., Zamay S.S. Four steps for revealing and adjusting the 3D structure of aptamers in solution by small-angle X-ray scattering and computer simulation. Anal. Bioanal. Chem. 2019;411:6723–6732. doi: 10.1007/s00216-019-02045-0. [DOI] [PubMed] [Google Scholar]
- 15.Lupu L., Wiegand P., Hüttmann N., Rawer S., Kleinekofort W., Shugureva I., Kichkailo A.S., Tomilin F.N., Lazarev A., Berezovski M.V., Przybylski M. Molecular Epitope Determination of Aptamer Complexes of the Multidomain Protein C-Met by Proteolytic Affinity-Mass Spectrometry. ChemMedChem. 2020;15:363–369. doi: 10.1002/cmdc.201900489. [DOI] [PubMed] [Google Scholar]
- 16.Adrian M., Heddi B., Phan A.T. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Fürtig B., Richter C., Wöhnert J., Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 18.Mao X., Marky L.A., Gmeiner W.H. NMR structure of the thrombin-binding DNA aptamer stabilized by Sr2+ J. Biomol. Struct. Dyn. 2004;22:25–33. doi: 10.1080/07391102.2004.10506977. [DOI] [PubMed] [Google Scholar]
- 19.van Buuren B.N.M., Schleucher J., Wittmann V., Griesinger C., Schwalbe H., Wijmenga S.S. NMR spectroscopic determination of the solution structure of a branched nucleic acid from residual dipolar couplings by using isotopically labeled nucleotides. Angew. Chem. Int. Ed. Engl. 2004;43:187–192. doi: 10.1002/anie.200351632. [DOI] [PubMed] [Google Scholar]
- 20.van der Werf R.M., Tessari M., Wijmenga S.S. Nucleic acid helix structure determination from NMR proton chemical shifts. J. Biomol. NMR. 2013;56:95–112. doi: 10.1007/s10858-013-9725-y. [DOI] [PubMed] [Google Scholar]
- 21.Gong Z., Zhao Y., Chen C., Xiao Y. Role of ligand binding in structural organization of add A-riboswitch aptamer: a molecular dynamics simulation. J. Biomol. Struct. Dyn. 2011;29:403–416. doi: 10.1080/07391102.2011.10507394. [DOI] [PubMed] [Google Scholar]
- 22.Rhinehardt K.L., Srinivas G., Mohan R.V. Molecular dynamics simulation analysis of anti-MUC1 aptamer and mucin 1 peptide binding. J. Phys. Chem. B. 2015;119:6571–6583. doi: 10.1021/acs.jpcb.5b02483. [DOI] [PubMed] [Google Scholar]
- 23.Jeevanandam J., Tan K.X., Danquah M.K., Guo H., Turgeson A. Advancing Aptamers as Molecular Probes for Cancer Theranostic Applications-The Role of Molecular Dynamics Simulation. Biotechnol. J. 2020;15:e1900368. doi: 10.1002/biot.201900368. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owczarzy R., Tataurov A.V., Wu Y., Manthey J.A., McQuisten K.A., Almabrazi H.G., Pedersen K.F., Lin Y., Garretson J., McEntaggart N.O. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiridonova V.A., Novikova T.M., Sizov V.A., Shashkovskaya V.S., Titaeva E.V., Dobrovolsky A.B., Zharikova E.B., Mazurov A.V. DNA Aptamers to Thrombin Exosite I. Structure-Function Relationships and Antithrombotic Effects. Biochemistry (Mosc.) 2019;84:1521–1528. doi: 10.1134/S0006297919120113. [DOI] [PubMed] [Google Scholar]
- 27.Carothers J.M., Oestreich S.C., Szostak J.W. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J. Am. Chem. Soc. 2006;128:7929–7937. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gevertz J., Gan H.H., Schlick T. In vitro RNA random pools are not structurally diverse: a computational analysis. RNA. 2005;11:853–863. doi: 10.1261/rna.7271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chushak Y., Stone M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009;37:e87. doi: 10.1093/nar/gkp408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai J.J.P., Ng K.-L. World Scientific; 2017. Computational Methods with Applications in Bioinformatics Analysis. [Google Scholar]
- 31.Vorobyeva M., Vorobjev P., Venyaminova A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules. 2016;21:1613. doi: 10.3390/molecules21121613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y., Fujii S., Hiroaki H., Sakata T., Tanaka T., Uesugi S., Tomita K., Kyogoku Y. A′-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC) Nucleic Acids Res. 1999;27:949–955. doi: 10.1093/nar/27.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leupin W., Wagner G., Denny W.A., Wüthrich K. Assignment of the 13C nuclear magnetic resonance spectrum of a short DNA-duplex with 1H-detected two-dimensional heteronuclear correlation spectroscopy. Nucleic Acids Res. 1987;15:267–275. doi: 10.1093/nar/15.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin T.G., Bharat T.A.M., Joerger A.C., Bai X.C., Praetorius F., Fersht A.R., Dietz H., Scheres S.H.W. Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. USA. 2016;113:E7456–E7463. doi: 10.1073/pnas.1612720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruigrok V.J.B., Levisson M., Hekelaar J., Smidt H., Dijkstra B.W., van der Oost J. Characterization of aptamer-protein complexes by X-ray crystallography and alternative approaches. Int. J. Mol. Sci. 2012;13:10537–10552. doi: 10.3390/ijms130810537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012;40:8119–8128. doi: 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J.H., Canny M.D., De Erkenez A., Krilleke D., Ng Y.S., Shima D.T., Pardi A., Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Convery M.A., Rowsell S., Stonehouse N.J., Ellington A.D., Hirao I., Murray J.B., Peabody D.S., Phillips S.E.V., Stockley P.G. Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution. Nat. Struct. Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 39.Guinier A. La diffraction des rayons X aux très petits angles : application à l’étude de phénomènes ultramicroscopiques. Ann. Phys. 1939;11:161–237. [Google Scholar]
- 40.Hammel M. Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS) Eur. Biophys. J. 2012;41:789–799. doi: 10.1007/s00249-012-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambo R.P., Tainer J.A. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu. Rev. Biophys. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 42.Viéville J.M.P., Barluenga S., Winssinger N., Delsuc M.A. Duplex formation and secondary structure of γ-PNA observed by NMR and CD. Biophys. Chem. 2016;210:9–13. doi: 10.1016/j.bpc.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Preus S., Wilhelmsson L.M. Advances in quantitative FRET-based methods for studying nucleic acids. ChemBioChem. 2012;13:1990–2001. doi: 10.1002/cbic.201200400. [DOI] [PubMed] [Google Scholar]
- 44.1st International Conference “Aptamers in Russia 2019.”. Mol. Ther. Nucleic Acids. 2019;17:1–24. [Google Scholar]
- 45.Da Vela S., Svergun D.I. Methods, development and applications of small-angle X-ray scattering to characterize biological macromolecules in solution. Curr. Res. Struct. Biol. 2020;2:164–170. doi: 10.1016/j.crstbi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen A.H., Wang Y., Bottaro S., Grudinin S., Arleth L., Lindorff-Larsen K. Combining molecular dynamics simulations with small-angle X-ray and neutron scattering data to study multi-domain proteins in solution. PLoS Comput. Biol. 2020;16:e1007870. doi: 10.1371/journal.pcbi.1007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Björling A., Niebling S., Marcellini M., van der Spoel D., Westenhoff S. Deciphering solution scattering data with experimentally guided molecular dynamics simulations. J. Chem. Theory Comput. 2015;11:780–787. doi: 10.1021/ct5009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernetti M., Hall K.B., Bussi G. Reweighting of molecular simulations with explicit-solvent SAXS restraints elucidates ion-dependent RNA ensembles. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab459. Published online June 9, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zamay G.S., Kolovskaya O.S., Zamay T.N., Glazyrin Y.E., Krat A.V., Zubkova O., Spivak E., Wehbe M., Gargaun A., Muharemagic D. Aptamers Selected to Postoperative Lung Adenocarcinoma Detect Circulating Tumor Cells in Human Blood. Mol. Ther. 2015;23:1486–1496. doi: 10.1038/mt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamay T.N., Zamay G.S., Kolovskaya O.S., Zukov R.A., Petrova M.M., Gargaun A., Berezovski M.V., Kichkailo A.S. Current and prospective protein biomarkers of lung cancer. Cancers (Basel) 2017;9:155. doi: 10.3390/cancers9110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zamay G.S., Zamay T.N., Kolovskii V.A., Shabanov A.V., Glazyrin Y.E., Veprintsev D.V., Krat A.V., Zamay S.S., Kolovskaya O.S., Gargaun A. Electrochemical aptasensor for lung cancer-related protein detection in crude blood plasma samples. Sci. Rep. 2016;6:34350. doi: 10.1038/srep34350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker M., Jacobson A.B. Using reliability information to annotate RNA secondary structures. RNA. 1998;4:669–679. doi: 10.1017/s1355838298980116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waugh A., Gendron P., Altman R., Brown J.W., Case D., Gautheret D., Harvey S.C., Leontis N., Westbrook J., Westhof E. RNAML: a standard syntax for exchanging RNA information. RNA. 2002;8:707–717. doi: 10.1017/s1355838202028017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanchet C.E., Spilotros A., Schwemmer F., Graewert M.A., Kikhney A., Jeffries C.M., Franke D., Mark D., Zengerle R., Cipriani F. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J. Appl. Cryst. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graewert M.A., Da Vela S., Gräwert T.W., Molodenskiy D.S., Blanchet C.E., Svergun D.I., Jeffries C.M. Adding size exclusion chromatography (SEC) and light scattering (LS) devices to obtain high-quality small angle X-ray scattering (SAXS) data. Crystals. 2020;10:1–18. [Google Scholar]
- 56.Panjkovich A., Svergun D.I. CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics. 2018;34:1944–1946. doi: 10.1093/bioinformatics/btx846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mertens H.D.T., Svergun D.I. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 58.Gräwert T., Svergun D. Application of small-angle X-ray scattering in studies of biological macromolecules. In: Myers R.A., editor. Encyclopedia of Analytical Chemistry. Wiley Online Library; 2021. [Google Scholar]
- 59.Manalastas-Cantos K., Konarev P.V., Hajizadeh N.R., Kikhney A.G., Petoukhov M.V., Molodenskiy D.S., Panjkovich A., Mertens H.D.T., Gruzinov A., Borges C. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Cryst. 2021;54:343–355. doi: 10.1107/S1600576720013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 61.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svergun D., Barberato C., Koch M.H. CRYSOL - A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 64.Knight C.J., Hub J.S. WAXSiS: a web server for the calculation of SAXS/WAXS curves based on explicit-solvent molecular dynamics. Nucleic Acids Res. 2015;43:W225–W230. doi: 10.1093/nar/gkv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grudinin S., Garkavenko M., Kazennov A. Pepsi-SAXS: an adaptive method for rapid and accurate computation of small-angle X-ray scattering profiles. Acta Crystallogr. D Struct. Biol. 2017;73:449–464. doi: 10.1107/S2059798317005745. [DOI] [PubMed] [Google Scholar]
- 66.Kikhney A.G., Borges C.R., Molodenskiy D.S., Jeffries C.M., Svergun D.I. SASBDB: Towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaus M., Lu X., Elstner M., Cui Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theory Comput. 2014;10:1518–1537. doi: 10.1021/ct401002w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fedorov D.G. The fragment molecular orbital method: theoretical development, implementation in GAMESS and applications. WIREs Comput. Mol. Sci. 2017;7:e1322. [Google Scholar]
- 69.Nishimoto Y., Fedorov D.G., Irle S. Density-functional tight-binding combined with the fragment molecular orbital method. J. Chem. Theory Comput. 2014;10:4801–4812. doi: 10.1021/ct500489d. [DOI] [PubMed] [Google Scholar]
- 70.Nishimoto Y., Fedorov D.G. The fragment molecular orbital method combined with density-functional tight-binding and the polarizable continuum model. Phys. Chem. Chem. Phys. 2016;18:22047–22061. doi: 10.1039/c6cp02186g. [DOI] [PubMed] [Google Scholar]
- 71.Suenaga M. Facio: New Computational Chemistry Environment for PC GAMESS. J. Comput. Chem. 2005;4:25–32. [Google Scholar]
- 72.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barca G.M.J., Bertoni C., Carrington L., Datta D., De Silva N., Deustua J.E., Fedorov D.G., Gour J.R., Gunina A.O., Guidez E. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020;152:154102. doi: 10.1063/5.0005188. [DOI] [PubMed] [Google Scholar]
- 74.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex. 2015;1–2:19–25. [Google Scholar]
- 75.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 77.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 78.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Physiol. 1981;52:7182. [Google Scholar]
- 79.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. 3 LINCS: a linear constraint solver for molecular simulations. J. Comp. Chem. 1997;18:1463–1472. [Google Scholar]
- 80.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577. [Google Scholar]
- 81.Ivani I., Dans P.D., Noy A., Pérez A., Faustino I., Hospital A., Walther J., Andrio P., Goñi R., Balaceanu A. Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2016;13:55–58. doi: 10.1038/nmeth.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heyer L.J., Kruglyak S., Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Debye P. Zerstreuung von Röntgenstrahlen. Ann. Phys. 1915;351:809–823. [Google Scholar]
- 83.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
▪▪▪