Summary
Intravital multiphoton imaging of the tumor milieu allows for the dissection of intricate and dynamic biological processes in situ. Herein, we present a step-by-step protocol for setting up an experimental cancer imaging model that has been optimized for solid tumors such as breast cancer and melanoma implanted in the flanks of mice. This protocol can be utilized for dissecting tumor-immune cell dynamics in vivo or other tumor-specific biological questions.
For complete details on the use of this protocol for intravital imaging of breast cancer, please refer to Tikoo et al. (2021a), and for intravital imaging of melanoma, please refer to Tikoo et al. (2021b).
Subject areas: Cancer, Immunology, Microscopy
Graphical abstract
Highlights
-
•
Detailed protocol for setting up high-resolution intravital imaging of murine tumors
-
•
3D printing of custom stage inserts for tumor stabilization
-
•
Procedures for cannulation of blood vessels
-
•
Surgical preparation and tissue stabilization for imaging tumor milieu in vivo
Intravital multiphoton imaging of the tumor milieu allows for the dissection of intricate and dynamic biological processes in situ. Herein, we present a step-by-step protocol for setting up an experimental cancer imaging model that has been optimized for solid tumors such as breast cancer and melanoma implanted within the flank of mice. This protocol can be utilized for dissecting tumor-immune cell dynamics in vivo or other tumor-specific biological questions.
Before you begin
Experimental design considerations
For the successful implementation of this intravital imaging protocol, several factors associated with the experimental design and setup need to be considered. These factors, either individually or in combination, can dramatically alter a study’s outcome or adversely impact the quality and reproducibility of the data generated. We have highlighted some of these factors below. Researchers with adequate experience in setting up intravital imaging models can directly proceed to the section on the generation of the stage inserts.
-
1.Choice of tumor model
-
a.Breast Cancer: The choice of tumor model relies on the scientific question being pursued (Fantozzi and Christofori, 2006). In the case of mammary tumors, employing genetically modified mouse models such as MMTV-PyMT or MMTV-Neu allows for the spontaneous generation of tumors akin to the human condition. In these model systems, unique oncogenes are overexpressed under tissue-specific promoters (Fantozzi and Christofori, 2006). To visualize immune cells, these mice strains need to be crossed to specific immune cell reporter mice on an identical genetic background. Conversely, fluorescent immune cells can be purified and adoptively transferred before imaging. Please note that MMTV-PyMT mice develop multifocal mammary adenocarcinomas (Guy et al., 1992), wherein the presence of multiple tumors within the ipsilateral mammary fat pads can lead to complications during surgical preparation for imaging, especially the increased risk of damaging patent vasculature. Therefore, the potential risk posed by these complications should be taken into consideration before commencing imaging.Alternatively, implantation of mammary tumors can be performed using syngeneic or xenograft cells. While xenograft models allow for the implantation of well-characterized human mammary tumor cell lines such as MDA-MB-231, the need for implantation into immunocompromised mice, for example, NOD/SCID mice, curtails analysis of the role of adaptive immune cells. Nonetheless, xenograft models are useful for studying innate immune cells as well as the tumor cell behavior and physiology during cancer progression or drug therapy.To gain a detailed understanding of anti-tumor immune responses in breast cancer, syngeneic tumor cell lines such as E0771 mammary adenocarcinoma (compatible with mice on C57BL/6 background) or 4T1 (compatible with mice on BALB/c background) can be utilized. Sub-clones of E0771 and 4T1 with varying metastatic dissemination characteristics are also available for deciphering the mechanisms of tumor cell metastasis (Johnstone et al., 2015). Syngeneic tumor cells can be conveniently implanted into the mammary fat pad to generate tumors with reproducible size and growth dynamics. In our laboratory, the transplantation of E0771 mammary adenocarcinoma cells into the inguinal mammary fat pad of young, virgin female mice is the method of choice (detailed in-depth below) (Tikoo et al., 2021a). Using this methodology, we regularly obtain a >98% success rate in implantation with similar tumor immune profiles and tumor growth rates obtained between replicate experiments.
-
b.Melanoma: For visualizing cellular dynamics within melanoma, researchers can employ various models, including genetically engineered mouse (GEM) melanoma models with or without ultraviolet radiation (UVR) exposure, syngeneic models, or patient-derived xenograft models (Patton et al., 2021, Pérez-Guijarro et al., 2017). GEM melanoma models harbor one or more driver mutations and have been instrumental in deciphering various facets of melanoma cell biology. GEM melanoma models also have an advantage in that they mimic physiological growth rates of melanoma development. Recent and emerging advances in genetic engineering will hopefully lead to the development of GEM melanoma models with inducible and temporally controlled inactivation of genes leading to the generation of more physiological models of melanoma (Patton et al., 2021, Pérez-Guijarro et al., 2017). Conversely, syngeneic mouse models have long been used to understand melanoma cell biology mainly due to their low cost, ease of usage, and fast turnaround times. One of the most commonly used syngeneic cell lines for generating melanoma tumors are the B16 lines (Overwijk and Restifo, 2001), particularly the B16-F10 cells (Guo et al., 2018, Guo et al., 2020, Tikoo et al., 2021b). Our cell line of choice is the recently generated amel-B16-F10 melanoma cell line which is compatible with the intravital imaging modality due to its lack of melanin pigmentation (Tikoo et al., 2021b). Additionally, patient-derived xenografts are an excellent choice for understanding melanoma cell biology as they capture the tumor heterogeneity and provide a spectrum of genetic diversity. However, patient-derived tumors have to be implanted into an immunocompromised mouse, therefore, limiting the understanding generated of the role of immune system (Patton et al., 2021).
-
a.
-
2.
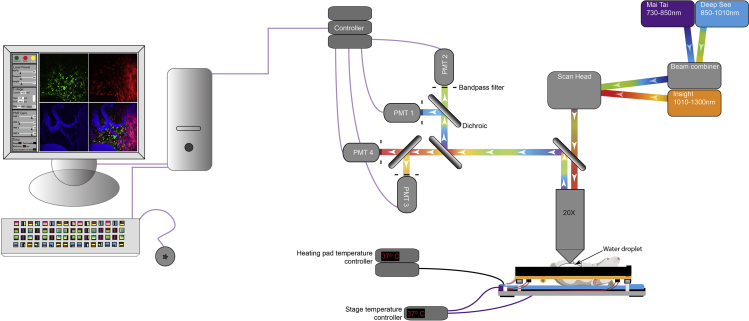
Choice of microscopic equipment: Compared to confocal microscopy, multiphoton microscopy permits deeper tissue imaging with less phototoxicity (Jain et al., 2016). We employ multiphoton imaging modality using the upright LaVision BioTec TriMScope equipped with a 20× (NA 0.95) Olympus water immersion objective (Figure 1), but several other commercial multiphoton systems are also readily available. Our setup has been configured to include up to eight external, non-descanned, dual-channel reflection/fluorescence detectors, and three diode-pumped, wideband, mode-locked Ti-Sapphire femtosecond lasers (Mai Tai®, 730–850 nm; Mai Tai Deep See™, 850–1010 nm, and Insight Deep See™, 1010–1300 nm; supplied by Spectra-Physics, Newport, USA) (Figure 1). For most imaging studies, we use 4–5 detectors to visualize spectrally distinct fluorophores with minimal overlap.
-
3.
Visualization of tumor vasculature: Most tumor imaging studies employ visualization of tumor vasculature (Table 1). The advent of transgenic reporter mice strains, such as mT/mG and Tie2-GFP, which feature fluorescently labeled blood vessels, has enhanced the quality of data that can be obtained without having to inject a plasma dye during the imaging session. Briefly, mT/mG is a double fluorescent, Cre reporter strain wherein all cells constitutively express membrane-targeted, tandem dimer tomato under the control of ROSA26 promoter. In these mice, Cre-mediated excision of mTomato results in expression of the membrane-targeted GFP within the Cre expressing cells (Muzumdar et al., 2007). However, in the absence of Cre activity, blood vessels can be easily distinguished from the surrounding tissue due to the brighter mTomato fluorescent signal. Tie2-GFP transgenic mice were developed by Motoike and co-workers wherein the Tie2 promoter and enhancer elements were utilized to drive the expression of GFP in vascular endothelial cells (Motoike et al., 2000). In the event where these transgenic reporter mice strains are unavailable or the transgenic fluorophores conflict with a given experimental setup, various vascular dyes can be used to visualize blood vessels (Table 1). A good vascular dye is crucial for imaging the behavior of tumor and immune cells at the endothelial interface. Our dye of choice is Evans blue conjugated to bovine serum albumin, referred to as Evans blue-BSA (preparation detailed in Materials). The conjugation of Evans blue to BSA enables the vascular dye to be retained within the vasculature for extended periods of time when imaging inflamed tissues. Experimental designs requiring the analysis of vascular leakage may prefer to use unconjugated Evans blue for imaging. The single-photon emission peak of Evans blue is 680 nm, and the excitation peaks are at 470 and 540 nm. These excitation and emission properties allow for simultaneous visualization of Evans blue-BSA with other commonly used fluorophores such as CFP, GFP, RFP, and mCherry. We regularly use two-photon excitation wavelength range from 890–940 nm to excite Evans blue-BSA, a wavelength range suitable for the excitation of CFP, GFP and YFP. In most cases, we employ the Insight Deep See™ for the excitation of RFP and mCherry (Figure 7B).
Figure 1.
Schematic representation of the intravital tumor imaging setup
Layout of the intravital imaging setup used for imaging tumors.
Table 1.
List of various methodologies utilized for visualizing blood vessels
| Labeling methodology | Advantages | Disadvantages |
|---|---|---|
| mT/mG transgenic reporter mice | Endogenous labeling of all cells (Muzumdar et al., 2007). Blood vessels are marked by high fluorescence intensity of the mTomato signal (Li et al., 2012). If crossed to a Cre reporter strain for example, CD11c-Cre, both blood vessels (red) and dendritic cells (green) could be visualized (Stoler-Barak et al., 2014). It can also be used in conjunction with inducible Cre reporters to temporally regulate transgene expression. | Not useful for studying the vascular and perivascular structures at high resolutions as mTom+ tumor-infiltrating cells could lead to the loss of endothelial cell resolution. |
| Tie2-GFP transgenic reporter mice | Excellent model for labeling blood vessel networks (De Palma et al., 2005). | Highlights other cell types such as Tie2+ macrophages (Pucci et al., 2009, De Palma et al., 2005). GFP expression in blood vessels may interfere with the use of other transgenic reporter strains. |
| Evans blue-BSA | Cheap, readily excited at several wavelengths with emission fluorescence in the far-red spectral range (Tikoo et al., 2021). | Vascular leakage can result in the loss of resolution. |
| Quantum dots | Can be used for a variety of purposes including highlighting blood vasculature and vascular leakage (Vennin et al., 2017). | Expensive. |
| Fluorescently labeled dextrans | Available in a range of different molecular weights. Fluorescently labeled dextrans of varying molecular weights can be employed to visualize vascular permeability. (10,000-2,000,000 Da) (Pai et al., 2014). | Expensive, vascular leakage compromises resolution. |
| Fluorescently labeled lectins | Binds to blood vessels (Robertson et al., 2015). | Toxic at high doses. |
| Fluorescently labeled anti-CD31 antibody | Specifically labels endothelial cells (Robertson et al., 2015). | Expensive. |
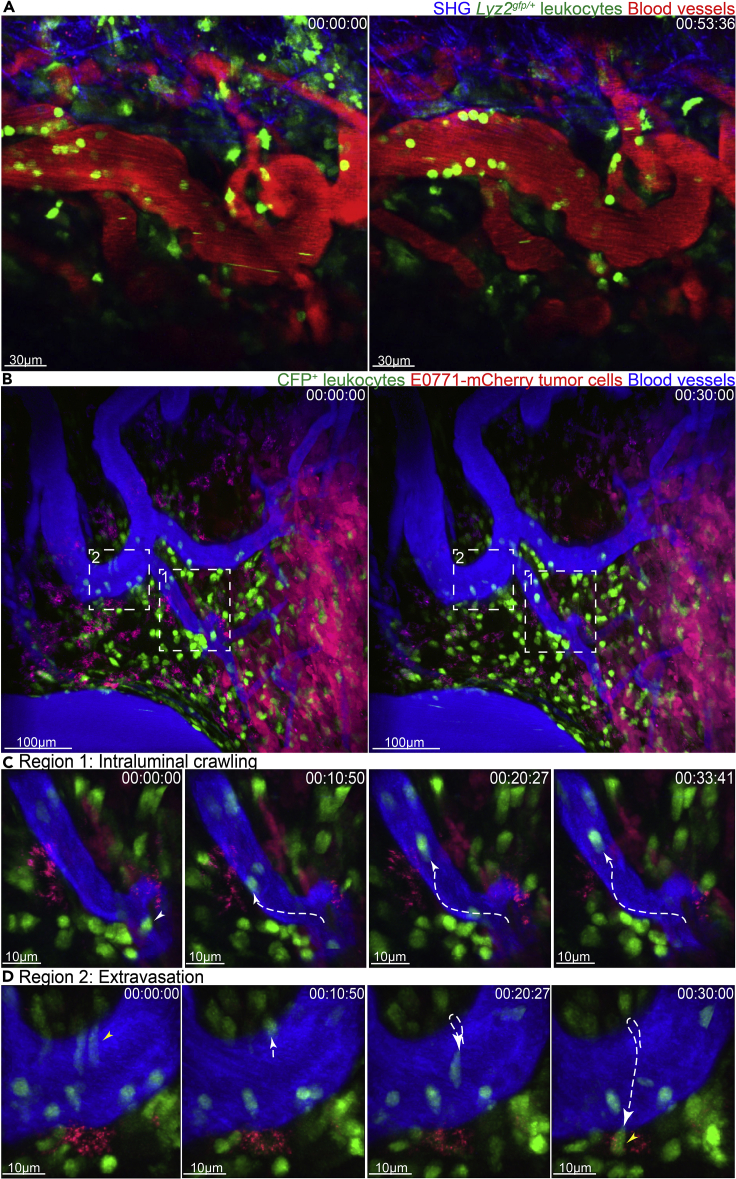
Figure 7.
Intravital imaging of tumors
(A) Time lapse intravital imaging of amelanotic melanoma tumors in lyz2gfp/+ mice highlighting the behavior of GFP+ neutrophils.
(B) Time lapse intravital imaging of mCherry-expressing E0771 mammary tumors in Macblue mice highlighting the intraluminal crawling behavior of CFP+ leukocytes (pseudo-colored green).
(C) Leukocyte crawling (white tracks) can be observed in the blood vessels.
(D) Extravasation of CFP+ leukocyte (yellow tracks) can be visualized.
Blood vessels are highlighted by injection of Evans blue-BSA. 00:00:00 represents hr:min:sec. Scale bars have been provided in individual images.
An additional benefit of using Evans blue-BSA is that it acts as an internal control for the quality of the skin flap preparation. Extensive leakage of the vascular dye indicates tissue damage inflicted during the surgical preparation. Caution should be observed during the surgical preparation of the tumor to minimize potential injury to the patent vasculature.
-
4.
Choice of fluorophores: Prior to setting up an imaging model, care should be taken to choose a set of fluorophores with minimal spectral overlap. This ensures relative ease in identifying various cell subsets during imaging and analysis. Availability of fluorescent transgenic reporter strains (Table 2) and their intercrosses should be considered for the experimental design. Another important consideration before choosing the experimental fluorophores is the filter setup available for the multiphoton microscope. Nonetheless, in scenarios wherein spectral overlap cannot be avoided, for example, between fluorophores such as GFP and YFP or CFP and GFP, the extent of bleed-through between channels can be minimized at the time of acquisition by fine-tuning the excitation wavelengths (in the case of tunable lasers) and optimizing the filter setup. Various imaging software, for example, FIJI (Schindelin et al., 2012) and ImageJ (Schneider et al., 2012, Collins, 2007, Rueden et al., 2017), also allow for correcting spectral overlaps post-acquisition.
Note: This protocol can also be employed to carry out label-free intravital imaging using Simultaneous Label-free Autofluorescence Multiharmonic (SLAM) microscopy. SLAM microscopy is an optical imaging platform that enables the detection of Nicotinamide adenine dinucleotide (NADH) from 3-photon autofluorescence imaging (3PAF), Flavin adenine dinucleotide (FAD) from 2-photon autofluorescence imaging (2PAF), detection of non-centrosymmetric structures from second-harmonic generation (SHG), and interfacial features from third-harmonic generation (THG) (You et al., 2018). SLAM microscopy has recently been shown to be capable of simultaneously visualizing leukocytes, red blood cells, tumor cells, adipocytes, collagen fibers, and microvasculature with sufficient resolution owing to the simultaneous excitation of autofluorescence and multi-harmonic processes (You et al., 2018).
Table 2.
List of various fluorescent transgenic reporter mice
| Transgenic reporter mice | Abbreviation | Commonly used for |
|---|---|---|
| cKit-GFP (Berrozpe et al., 2006) | cKit BAC-GFP | GFP expressed under the control of c-kit locus. Can be utilized for imaging mast cells in vivo (Tong et al., 2015, Tikoo et al., 2018). |
| mT/mG (Muzumdar et al., 2007) | mT/mG | mTomato is expressed in all cells (Li et al., 2012). Cre mediated recombination switches expression from mTom to mGFP. |
| Tie2-GFP (Motoike et al., 2000) | Tie2GFP | GFP expression on vascular endothelial cells. Aids in visualizing blood vessels (De Palma et al., 2005). |
| CD11c-YFP (Lindquist et al., 2004) | CD11cYFP | DCs can be visualized by virtue of their expression of eYFP (Ng et al., 2008, Boissonnas et al., 2013, Boldajipour et al., 2016). |
| CXCR6-GFP (Unutmaz et al., 2000) | CXCR6GFP | GFP is knocked into the CXCR6 gene locus. Can be utilized for imaging dermal γδ T cells (Sumaria et al., 2011, Gray et al., 2011), dendritic epidermal T cells (Gray et al., 2011), dermal ILC-2 (Roediger et al., 2013), invariant NKT cells (iNKT cells) (Wong et al., 2011) and tumor infiltrating T cells (Matsumura et al., 2008). |
| MacBlue (Sauter et al., 2014) | Csf1rCFP | Csf1r promoter drives the expression of gal4-VP16, which in turn drives a co-integrated gal4-responsive UAS-ECFP cassette. Monocytes, macrophages, and other CD11b cells express CFP in these mice with highest transgene expression observed in monocytes (Tikoo et al., 2021, Laviron et al., 2019). |
| MacGreen (Sasmono et al., 2003) | Csf1rGFP | Monocytes, macrophages, and other CD11b cells express GFP (Jain and Weninger, 2013, Laviron et al., 2019). |
| CX3CR1-GFP (Jung et al., 2000) | CX3CR1EGFP | Monocytes and macrophages express GFP. Used for in vivo imaging of tumor infiltrating myeloid cells (Laviron et al., 2019, Lee et al., 2018) |
| LyzM-GFP (Faust et al., 2000) | Lyz2EGFP | GFP knock-in in the lysozyme M locus. Neutrophils, monocytes and macrophages express GFP. Care should be taken while breeding so as to use only heterozygous transgenic mice in experiments as the strain is a knock-in (Tikoo et al., 2021, McDonald et al., 2010, Laviron et al., 2019, Lee et al., 2018) . |
-
5.
Choice of anesthesia: Before surgical preparation of mice, we administer via intraperitoneal injection a combined dose of injectable anesthetics, ketamine/xylazine (100/10 mg/kg) to provide deep surgical plane anesthesia. Anesthetized mice are monitored every 20–40 min for adequate depth of anesthesia. Repeated half doses of the anesthetics are administered post-monitoring leading to successful imaging of the tumor tissue for up to a period of five hours. Alternatively, inhalational anesthetics such as isoflurane can be employed to maintain mice under surgical plane anesthesia. Care should be taken to ensure that mice are carefully monitored to avoid discomfort, hypoxemia, and hypercapnia due to the incorrect dosing of anesthetics. In case of excessive movement observed for tissues being imaged, additional drugs such as acepromazine can be used in combination with the anesthetics. Care should be taken when using these drugs as they can potentiate the effects of anesthetics.
Note: Protocols utilizing injectable as well as inhalational anesthetics must be approved by the institutional safety committee to avoid any risks to the operator. The use of anesthetics raises concerns regarding the health and safety of research staff. These concerns should be properly addressed before commencing the experiments.
Optional: In scenarios wherein researchers would like to image continuously without several pauses required for monitoring mice or minimize movement artefacts due to repeated monitoring, and anesthetic injections, automated physiological monitoring systems such as the PhysioSuite from Kent Scientific Corporation can be employed. Additionally, venous cannulation can be used to administer anesthetic top-ups. Care should be taken with the dosing of anesthetics when administering directly into the venous circulation.
Ketamine- Ketamine is a controlled substance (S8, Australia; Schedule III, USA; Class B, UK) and may require relevant licenses to hold and use. Therefore, it is crucial that large volumes of the drug are stored in a safe and secure place with limited access and rigorous monitoring of the usage. As an example, we store large volumes in fireproof safety lockers with a single individual holding access to the drug and regular monitoring of the usage performed by an independent safety and compliance officer. The drug is dispensed to individual (approved) researchers depending on their experimental usage. Similarly, possession and use of xylazine, a prescription drug (S4, Australia), might require approval by relevant authorities.
Isoflurane- The use of an inhalational anesthetic is dependent on the availability of equipment capable of providing a constant flow of the vaporized anesthetic. As tumor imaging is performed over an extended period of time, it is essential that the excess vented anesthetic is sequestered using chemical scrubbers. As most imaging setups employ a chamber for maintaining humidity and preventing interference from extraneous light sources, even small leaks in the anesthetic setup can result in the build-up of anesthetic vapor. Similarly, the use of inhalational anesthetics requires a constant supply of oxygen gas which is highly flammable. Therefore, it is crucial that before proceeding with experiments, the institutional safety officers approve the use of inhalational anesthetics within the facility. Alternatively, an advanced system can be set up wherein the operation of the microscope and monitoring of the mice can be performed remotely (usually in an antechamber).
-
6.
Duration of imaging: Employing this methodology, we have been able to image continuously for durations of more than three hours. Imaging sessions lasting several hours are also feasible with this protocol. However, we do pause the imaging at 20–30 min intervals to check for the depth of anesthesia via palpebral reflex and hindlimb withdrawal. Repeated half doses of ketamine/xylazine are administered intraperitoneally when required. These pauses should be taken into consideration when time-lapse videos are merged, particularly during image analysis. The duration of imaging and the time interval between individual imaging frames depends on the cellular or molecular processes under investigation. Usually, analysis of slower cellular processes such as tumor cell migration or cell cycle dynamics requires imaging over extended periods of time, albeit with large time intervals between individual acquisitions. For example, tumor cell migration dynamics can be studied by imaging for more than 2 h, with data acquisition occurring every 2 min. To study the dynamic behavior of immune cells, for example, intravascular crawling of monocytes, data is acquired for 1 h at 30-s intervals or less. The precise time setup depends on several parameters, including the resolution of the image, the volume of the z stack, the intensity of fluorophores, and the biological process being studied. For most data acquisition, it is crucial that the frame rate is kept constant during the imaging period. Moreover, when comparing datasets from multiple experiments, the frame rate should be kept constant between experiments. To ascertain the frame rate for acquisition, it is recommended that there be a partial overlap between the cell body of a cell being tracked in subsequent frames. In scenarios wherein, cells are migrating at very high speeds, for example, rolling cells within the vascular lumen, the cell body should be clearly distinguishable between the acquisition frames to generate reliable data. If sudden jumps (sudden disappearance followed by reappearance) are observed for the cells being tracked, the dataset should be excluded. Please note that once the parameters for a given study are optimized, they should be kept constant between experiments.
-
7.
Image Analysis: The data generated from multiphoton microscopy can either be analyzed employing commercial software packages, for example, Volocity (Quorum Technologies) and Imaris (Oxford Instruments), or open-source image analysis platforms such as Image J (Schneider et al., 2012, Collins, 2007, Rueden et al., 2017) or FIJI (Schindelin et al., 2012).
Generation of stage inserts
Timing: 8–12 h
Tissue movement introduces artefacts during high-resolution imaging thus interfering with post-acquisition image analysis. Therefore, time-lapse imaging of tumors, similar to other tissues, requires external stabilization. Imaging of melanoma and breast tumors entails surgical exposure of the tumor, thereby providing a large vascularized surface for imaging. Nonetheless, the tumor shape and size can make immobilization of the sample challenging. To achieve stable imaging of the tumor tissue, we have modified our previously described ear skin-imaging model (Li et al., 2012) (Figure 2A). To stabilize and superfuse the tumor tissue, we have employed 3D printing technology to generate stage inserts of variable heights to accommodate varying tumor volumes (Figure 2B), as detailed below.
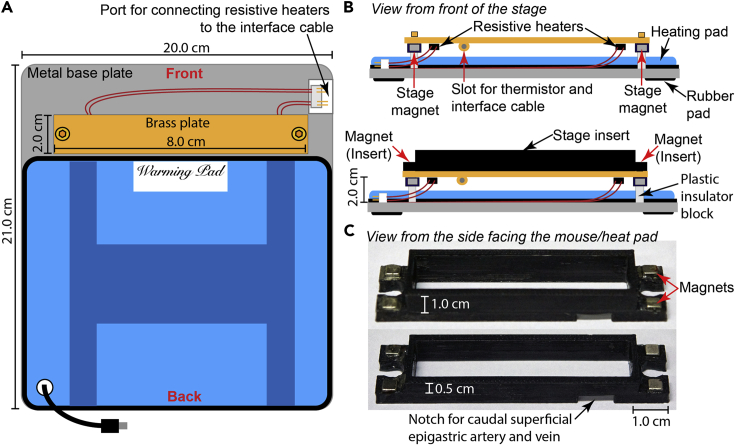
Figure 2.
Setup of imaging stage
(A) Schematic representation of the tumor imaging stage highlighting the organization and dimensions of various elements.
(B) Side view of the tumor imaging stage.
(C) Images of stage inserts used for stabilizing and perfusing the tumor.
Please note: The height of the stage appears shorter than the equivalent length; this effect is due to the angle of the object within the image.
-
8.
We designed the 3D printed stage inserts compatible with our imaging stage setup (Figures 2A and 2B) using Google Sketchup v8 and exported as STL files for G-code generation. Due to the localization of the tumors in the flank, a large number of angiogenic blood vessels arise from the cranial or caudal superficial epigastric artery and vein. Therefore, the stage inserts need to be printed with a small notch to accommodate the neoangiogenic vasculature (Figure 2C). A template of the design is available as Data S1 and S2.
-
9.
MakerBot Desktop was used to generate printer-specific G-code for 3D printing on a MakerBot Replicator 2× using Acrylonitrile-Butadiene Styrene (ABS).
-
10.
To stabilize the inserts, we embedded 4 mm3 neodymium magnets in the insert, which engages with magnets embedded underneath the stage (Figure 2B and 2C).
Using our methodology, the stage platform along with the tumor is maintained at 37°C throughout the imaging period utilizing the feedback system (Temperature controller TC-344C) (Li et al., 2012). In a scenario wherein, the stage cannot be constructed, researchers can 3D-print a stage with a raised platform (with embedded magnets) and superfuse the inserts with warm saline to maintain the tissue temperature at 37°C.
Note: The use of 3D printed inserts detailed above provides researchers with a unique opportunity for utilizing tumor models wherein the localization of tumor cannot be pre-emptively determined; for example, in the case of GEM models of melanoma and MMTV-PyMT mice. These inserts can be printed in less than an hour and can be generated to varying heights and shapes. Notches for maintaining patent vasculature can be created by gently filing the base of the insert to accomodate the angiogenic vasculature. Although we prefer to retain a large chamber cavity in the inserts (inserts compatible with most tumor models), other researchers may prefer to use inserts with limited space around the tumors to enhance stability. The use of 3D printed stage inserts also allows researchers to utilize their pre-existing imaging setups, thereby reducing the setup cost. These stage inserts also provide a unique access point for adding drugs, dyes, and small molecule inhibitors within the tumor milieu, bypassing the systemic circulation.
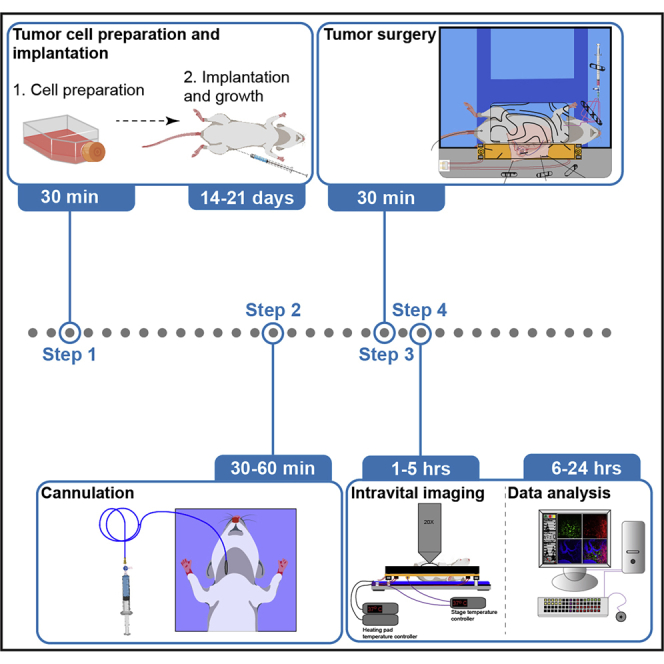
Preparation of tumor cells
Timing: 45 min
For syngeneic or xenograft models, follow the protocol detailed below. For imaging of spontaneous tumors generated in mice, for example, in the MMTV-PyMT model, proceed to the section on step by step method details.
This protocol has been specifically used for:
-
•
B16-F10 and amel-B16-F10 cells and their fluorescent-tagged versions for implanting tumors in the flanks of C57Bl/6 mice.
-
•
E0771 cells and their fluorescent-tagged versions for implanting tumors in the mammary fat pads of C57Bl/6 mice.
-
•
4T1 cells and their fluorescent-tagged versions for implanting tumors in the mammary fat pads of BALB/c mice.
For xenograft models, we recommend rendering the tumor cells fluorescent for imaging purposes. Fluorescent tumor cells can be generated by standard retroviral or lentiviral transduction methods.
-
11.
Maintain the cells in DMEM, high glucose, GlutaMAX™ + 10% FCS + sodium pyruvate, and routinely passage at optimal confluency of 60%–80%.
CRITICAL: Routinely test the tumor cell lines for mycoplasma and other microbial contaminants in accordance with institutional policies.
-
12.
On the day of the injection, wash the tumor cells growing at 70%–80% confluency three times with 1× PBS. We regularly use one TC150 cm2 tissue culture flask at the above-mentioned confluency to prepare cells for injecting up to 15 mice.
CRITICAL: It is crucial to use cells that are metabolically healthy and not overgrown or stressed.
-
13.
Detach the tumor cells by incubating with 0.05% Trypsin EDTA for 30 s–1 min followed by gently tapping the cell culture flask to dislodge the adherent cells.
-
14.
Arrest trypsinization by adding 10–15 mL of FBS containing growth media.
CRITICAL: Care should be taken not to incubate the cells with Trypsin for extended periods of time as this reduces the viability of tumor cells.
-
15.
Collect the cells via centrifugation at 524×g for 5 min.
-
16.Resuspend the resulting pellet in 1× PBS. Count and subsequently adjust the cell suspension to a concentration of 2.5–5.0 million viable tumor cells/mL.
-
a.Assess the cell viability by trypan blue exclusion. Estimate the cell numbers using the Neubauer chamber.
-
b.Cell viability of >95% is deemed acceptable for most experiments.
-
a.
Note: When estimating cell numbers, only viable cell count needs to be taken into consideration.
-
17.
Filter the cell suspension through a 100 μm cell strainer to remove large cell clumps, which can result in variability in the cell number injected. Transfer the cell suspension into an Eppendorf tube and place on ice until injection. Troubleshooting problem 1
Tumor cell implantation
Timing: 20–30 min
-
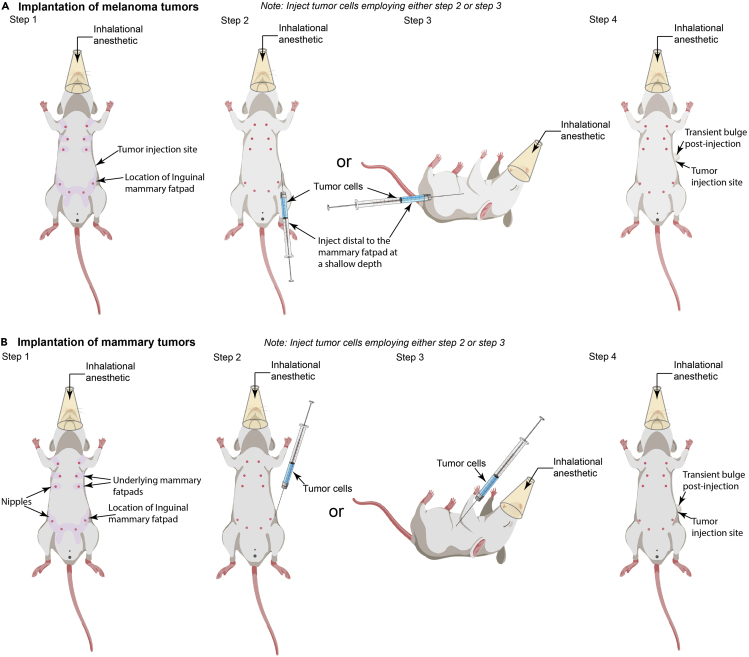
18.Using a 1 mL syringe equipped with a 25G needle, inject 0.1 mL of the tumor cell suspension into the left flank of the anesthetized mice (Figure 3).
 CRITICAL: We inject tumor cells only within the left flank of the mice due to the design of the stage inserts. In case the researchers require tumor injection in the right flank, please ensure that the notch for blood vessels is etched based on the new orientation for tumor imaging. In brief, for successfully imaging the tumors implanted within the right flank, the mouse needs to be rotated 180 degrees on the stage (Figure 6, step 2); therefore, the notch for caudal blood vessels needs to be placed on the cis-end of the insert (Figure 2C, left cis side of the insert rather the presently depicted right).
CRITICAL: We inject tumor cells only within the left flank of the mice due to the design of the stage inserts. In case the researchers require tumor injection in the right flank, please ensure that the notch for blood vessels is etched based on the new orientation for tumor imaging. In brief, for successfully imaging the tumors implanted within the right flank, the mouse needs to be rotated 180 degrees on the stage (Figure 6, step 2); therefore, the notch for caudal blood vessels needs to be placed on the cis-end of the insert (Figure 2C, left cis side of the insert rather the presently depicted right).-
a.Melanoma cell implantation: Inject melanoma cells subcutaneously into the flank of male or female anesthetized mice (Figure 3A, step 2 or 3). We typically inject 0.25–0.5 million B16-F10 melanoma cells in mice using the methodology detailed below. This number of tumor cells generates palpable tumors within ten days, with 0.9 cm3 tumors (end-point criteria) obtained within approximately two to three weeks.
 CRITICAL: During tumor cell implantation, it is crucial to ensure that the needle stays within the subcutaneous tissue and does not enter the peritoneum. We recommend keeping the angle of injection very shallow to achieve this. For successful injection, the syringe should be parallel to the abdominal skin (Figure 3A, step 2), with the cells being injected as soon as the skin is punctured (marked by the loss of resistance to needle penetration). Alternatively, the mice can be positioned on their side, and the syringe can be placed parallel to the chest with the needle facing the forelimb. The injection is performed distal from the hind limb (Figure 3A, step 3). Post-injection, a slight bulge in the skin signifies successful implantation of tumor cells (Figure 3A, step 4). Care should be taken to inject the tumor cells distal to the mammary fat pad at a shallow depth to prevent cell deposition in the mammary fat pad.Note: For injecting melanoma cells in male mice, the general guidelines stated above should be followed, wherein the melanoma cells are injected away from the hind limbs as soon as the skin is punctured. The absence of nipples in male mice creates difficulty in identifying the key landmarks. Therefore, we recommend using female mice for setting up the model. Once sufficiently experienced, researchers can easily perform this procedure on male mice. Also, please note that the male mice are prone to fighting; therefore, adequate observation or separation of individual mice may be required to ensure that the growing tumor is not impacted.
CRITICAL: During tumor cell implantation, it is crucial to ensure that the needle stays within the subcutaneous tissue and does not enter the peritoneum. We recommend keeping the angle of injection very shallow to achieve this. For successful injection, the syringe should be parallel to the abdominal skin (Figure 3A, step 2), with the cells being injected as soon as the skin is punctured (marked by the loss of resistance to needle penetration). Alternatively, the mice can be positioned on their side, and the syringe can be placed parallel to the chest with the needle facing the forelimb. The injection is performed distal from the hind limb (Figure 3A, step 3). Post-injection, a slight bulge in the skin signifies successful implantation of tumor cells (Figure 3A, step 4). Care should be taken to inject the tumor cells distal to the mammary fat pad at a shallow depth to prevent cell deposition in the mammary fat pad.Note: For injecting melanoma cells in male mice, the general guidelines stated above should be followed, wherein the melanoma cells are injected away from the hind limbs as soon as the skin is punctured. The absence of nipples in male mice creates difficulty in identifying the key landmarks. Therefore, we recommend using female mice for setting up the model. Once sufficiently experienced, researchers can easily perform this procedure on male mice. Also, please note that the male mice are prone to fighting; therefore, adequate observation or separation of individual mice may be required to ensure that the growing tumor is not impacted. -
b.Breast cancer cell implantation: For generating mammary tumors, inject the cells into the mammary fat pad of a virgin female mouse (8–12 weeks of age) (Figure 3B, step 2 or 3). We typically inject 0.25–0.5 million E0771 tumor cells in the inguinal mammary fat pad of mice using the methodology detailed below. This number of tumor cells generates palpable tumors within ten days, with 0.9 cm3 tumors (end-point criteria) obtained within approximately two to three weeks.
-
a.
CRITICAL: During tumor cell implantation, it is crucial to ensure that the needle stays within the mammary fat pad and does not enter the peritoneum. We recommend keeping the angle of injection very shallow to achieve this. For successful injection into the mammary fat pad, the syringe should be parallel to the abdominal skin (Figure 3, step 2), with the cells being injected as soon as the skin is punctured (marked by the loss of resistance to needle penetration). Alternatively, the mice can be positioned on their side, and the syringe can be placed parallel to the chest with the needle facing the hind limb. The injection is performed just above the hind limb under the skin (Figure 3, step 3). Post-injection, a slight bulge in the skin signifies successful implantation of tumor cells in the mammary fat pad (Figure 3, step 2).
-
19.
Rotate the syringe while withdrawing the needle to seal the puncture wound and to prevent any loss of cell suspension.
-
20.
Allow the tumor to grow for 2–4 weeks with continuous monitoring.
-
21.
Image the tumor once it reaches a volume of 0.5–0.9 cm3. In our experience, imaging tumors at this particular size is optimal for studying immune responses. However, larger or smaller tumors can also be imaged successfully depending upon the biological question being pursued. Troubleshooting problem 2
Note: Animal monitoring should be carried out in accordance with the approved institutional ethics protocols. Caution should be observed while maintaining tumor-bearing mice. Animals with tumors that begin to show signs of ulceration should be removed and not included in the imaging experiments. Relevant ethics guidelines should always be followed during tumor growth in mice. Undue stress or inflammation can alter the tumor milieu and can result in variability in the data. Special consideration should be given to the well-being of the tumor-bearing mice in accordance with the approved ethics guidelines.
Figure 3.
Methodology for implantation of tumors
(A) Schematic representation of the methodology used to orthotopically implant melanoma in mice.
(B) Illustration of the methodology used to transplant mammary tumors in mice.
Please note, the changes in the direction of tumor injection.
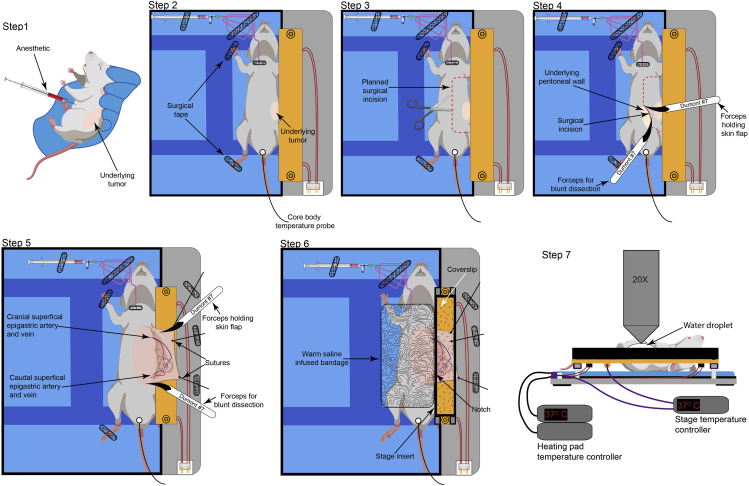
Figure 6.
Surgical preparation of tumors for intravital imaging
Schematic representation of the methodology used to surgically prepare the tumors in mice for intravital imaging. The secure placement of cannula and tubing is also been depicted.
Immune cell preparation and labeling
For experiments wherein labeled immune cells need to be tracked in vivo, several purification and labeling methodologies can be employed depending on the immune cell population being evaluated. The exogenous administration of immune cells can also be used when comparing two immune cell populations under identical conditions. For example, utilizing ex vivo expanded antigen-specific effector CD8+ T cells, we were able to highlight the ability of effector T cells to adhere to inflamed vasculature in the absence of Gi signaling (Shulman et al., 2011). Similarly, we employed this strategy to highlight the importance of Granzyme B in the transmigration of effector T cells during sterile injury (Prakash et al., 2014).
-
22.
Immune cell preparation- The precise methodology used for immune cell preparation is dependent on the immune cell population being studied and the scientific question being pursued. For example, neutrophils can be purified from the bone marrow of mice utilizing the magnetic sorting methodology (Miltenyi Biotec, Neutrophil isolation kit) (Ng et al., 2011) or Percoll gradient (Lämmermann et al., 2013). The number of highly purified neutrophils obtained using the magnetic sorting methodology (2×107 purified neutrophils) corresponds to 2% of total blood neutrophils when administered intravenously and would represent a small fraction of events being studied within the inflamed tissues (Ng et al., 2011). Nonetheless, the purification methodologies mentioned above would be adequate to study migration dynamics of neutrophils when injected directly into the tissue such as the dermis (Ng et al., 2011). In scenarios wherein neutrophil recruitment needs to be studied, it is best to utilize either the Lyz2gfp/+ mice (Lämmermann et al., 2013, McDonald et al., 2010, Peters et al., 2008, Casanova-Acebes et al., 2018) or their bone marrow for adoptive transfer (Abtin et al., 2014).
Similarly, as described above, ex vivo antigen primed and expanded effector T cells can be utilized for studies (Shulman et al., 2011, Prakash et al., 2014). However, the ability of T cells to respond to tumor-derived antigens provides an opportunity wherein endogenously fluorescent T cells can be transferred into mice bearing tumor cells expressing cognate antigen (Mrass et al., 2006, Mrass et al., 2008, Breart et al., 2008, Cazaux et al., 2019, Park et al., 2019). This methodology allows for effector T cells to infiltrate tumor tissue over extended period of time thereby permitting robust tracking in vivo.
-
23.
Immune cell labeling- Several commercially available dyes can be utilized for labeling immune cells with minimal cytotoxicity. However, due to the possibility of dilution during cell division, transfer of dyes between cell populations, or photobleaching during longitudinal imaging, it is highly recommended that endogenously fluorescent cells are utilized wherever possible. A brief list of commonly used fluorescent reporter mice strains is presented in Table 2. Alternatively, labeling can be performed utilizing DNA transfection (Mrass et al., 2006) or retroviral transduction (Obeidy et al., 2020).
Nonetheless, commonly used labeling dyes such as CFSE and CMTMR can be utilized for labeling immune cells as per the manufacturer’s protocol (please see the detailed staining protocol for CFSE and CMTMR) prior to adoptive transfer. To avoid labeling-induced artifacts confounding the study, we recommend that wherever possible, researchers should swap the label between the populations (Lämmermann et al., 2013).
Therefore, we strongly advise that prior to tumor imaging, researchers optimize the immune cell preparation and labeling. An optimization experiment is essential, wherein all steps required for the proposed experiment are performed, but the tumor is harvested, fixed, sectioned and imaged using a confocal microscope to ascertain the abundance and distribution of cells being tracked, their labeling intensity, and spectral separation between fluorophores (when two or more fluorophores are being employed). As intravital imaging focuses on smaller regions (usually less than 500×500 μm), the use of confocal microscopy, especially the tiling function, can help researchers assess the parameters that need to be further optimized before intravital imaging can be successfully undertaken.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | GIBCO/Life Technologies | Cat. No. 11960-044 |
| Sodium pyruvate | Sigma Aldrich | Cat. No. P5280-25G |
| Trypsin EDTA | GIBCO/Life Technologies | Cat. No. 25300-054 |
| Penicillin-Streptomycin | GIBCO/Life Technologies | Cat. No. 15140-122 |
| HBSS | GIBCO/Life Technologies | Cat. No. 14175-095 |
| PBS | GIBCO/Life Technologies | Cat. No. 10010-023 |
| Evans Blue | Sigma Aldrich | E2129-10G |
| Bovine Serum Albumin, Fraction V | Sigma Aldrich | Cat. no.10735078001 |
| Experimental models: cell lines | ||
| mCherry expressing E0771 breast adenocarcinoma cells | Available from authors | Available from authors |
| B16-F10 amelanotic melanoma cells | Available from authors | Tikoo et al., 2021 |
| Experimental models: organisms/strains | ||
| Mouse: Female MacBlue mice on C57Bl/6 background (Age: 8–12-weeks) | Sauter et al. 2014 | JAX Stock no-0260515587942 |
| Mouse: Male or female Lyz2gfp on C57Bl/6 background (Age: 8–12 weeks) | Faust et al., 2000 | N/A |
| Software and algorithms | ||
| Volocity | Quorum Technologies | Quorum Technologies |
| Imaris | Oxford Instruments | Oxford Instruments |
| Other | ||
| RightTemp® Temperature Monitor and Homeothermic Warming Control Module. | Kent Scientific Corporation | RightTemp |
| Interface Cable CC-28 with DIN connector | Warner Instruments | Cat. No. 640106 |
| Resistive heater blocks RH-2 | Warner Instruments | Cat. No. 640274 |
| Temperature Controller TC-344C | Warner Instruments | Order No. 64-2401 |
| Dumont #7 – Fine Forceps | Fine Science Tools | Cat. No. 11274-20 |
| Student Vannas Spring Scissors | Fine Science Tools | Cat. No. 91500-09 |
| Fine Scissors | Fine Science Tools | Cat. No. 14040-10 |
| Adson Forceps | Fine Science Tools | Cat. No. 11006-12 |
| S&T Vascular Clamp- 7mm | Fine Science Tools | Cat. No. 00396-01 |
| S&T Clamp Applying Forceps | Fine Science Tools | Cat. No. 00071-14 |
| Sutures (Dysilk, cat. No. SA4040) | Dysilk | Cat. No. SA4040 |
| Surgical Microscope | Leica | Model no. KL1500 LCD |
| PE-10 tubing | Becton Dickinson (BD) | Cat. No- BD427400 |
| 3-way stopcock | BD | Cat. No. 04301433200 |
| P10 Pipette | Rainin | Cat. No. 17008649 |
| P20 Pipette | Rainin | Cat. No. 17008650 |
| P200 Pipette | Rainin | Cat. No. 17008652 |
| P1000 Pipette | Rainin | Cat. No. 17008653 |
| Pipette gun | Drummond Scientific | Cat. No. 4-000-220 |
| Kimwipes | Liv-Wipe | Cat. No. LIVWIPRFL |
| Kleenex | Kleenex | Cat. No. 4720 |
| Cover glass - 22 × 22 mm | ProSciTech | Cat. No. G422 |
| Cover glass - 22 × 50 mm | ProSciTech | Cat. No. G417 |
| Water bath | Thermomix 1442D | Cat. No. 6003231 |
| TC25 Tissue Culture Flask | Corning | Cat. No. COR430639 |
| TC75 Tissue Culture Flask | Corning | Cat. No. COR430641U |
| TC150 Tissue Culture Flask | Falcon | Cat. No. FAL355001 |
| Falcon Tubes (15 mL) | Falcon | Cat. No. 430791 |
| Falcon Tubes (50 mL) | Falcon | Cat. No. 352070 |
| Eppendorf tubes | Sarstedt | Cat. No. 72-690-001 |
| 10 μL pipette tips | Interpath Services | Cat. No. 24100 |
| 20 μL pipette tips | Interpath Services | Cat. No. 24500 |
| 200 μL pipette tips | Interpath Services | Cat. No. 24700 |
| 1000 μL pipette tips | Interpath Services | Cat. No. 24800 |
| 1 mL Disposable pipettes | Copan | Cat. No. 201C |
| 3 mL Disposable pipettes | Copan | Cat. No. 200C |
| 25G needles | BD | Cat. No. 301807 |
| 1 mL Syringes | BD | Cat. No. 302100 |
| Surgical tape | Various | N/A |
| Surgical Glue | Leukosan | N/A |
| Vacuum grease | Dow Corning; Sigma Aldrich | Cat. No. Z273554 |
| Veet hair removal cream | Reckitt Benckiser | N/A |
| Dialysis cassettes | ThermoFischer Scientific | Cat. no. 66807 |
| 0.22 μm Syringe Filter | Sigma Aldrich | Cat. No. SLGV033RS |
| 0.45 μm Syringe Filter | Sigma Aldrich | Cat. No. SLHV033RS |
| 100 μm Cell Strainer | Corning | Cat No. 352360 |
Materials and equipments
Timing: 6–8 h
Preparation of Evans blue conjugated to Bovine serum albumin (BSA).
-
•
Prepare the BSA solution by solubilizing 1 gm of BSA in 10 mL of sterile saline. Filter the resultant solution using a 0.22 μm syringe filter.
-
•
To conjugate Evans blue to BSA, add 1 mg of Evans blue to 10 mL of 100 mg/mL BSA solution in a 15 mL falcon tube.
-
•
Wrap the tube in aluminum foil, and incubate the mixture at 22°C–24°C for 2 h.
-
•
Post-incubation, filter the solution using a 0.45 μm syringe filter followed by two rounds of filtration through a 0.22 μm syringe filter.
-
•
Place the filtered solution in a dialysis cassette and dialyze against 2 liters of sterile, refrigerated saline for 2–4hrs at 4°C to remove any unconjugated Evans blue.
-
•
Transfer the cassette containing the labeled Evans blue solution to a cell culture hood. Using a sterile syringe, aseptically withdraw the Evans blue-BSA solution from the dialysis cassette.
-
•
Filter the solution through a 0.22 μm syringe filter, aseptically aliquot into 100 μL aliquots, and store at −20°C.
Note: Wherever possible, all steps should be performed in cell culture hood (BSL-2). Dialysis can be set up in the cell culture hood using sterile refrigerated saline and subsequently moved to a cold room.
CRITICAL: As Evans blue-BSA solution is used for intravenous administration during imaging, one should take utmost care to avoid any microbial contaminations. Wherever possible, aseptic methodology should be followed during the preparation of this solution.
Alternatives: A large array of fluorescently labeled dextran dyes are available for use from commercial vendors; see Table 1.
Step-by-step method details
Animal housing and ethics statements
All mice were housed in individually ventilated cages maintained at 22°C–26°C, 30%–70% relative humidity, and a 12 h light-dark cycle. Sterilized corncob was used as bedding, with sterilized toilet rolls, tissues, and parts of cardboard boxes being provided as environmental enrichment. All mice used were within the age range of 8–12 weeks. For breast cancer studies, only female mice were used, while mice from both sexes were used for imaging melanoma tumors. All experimental techniques, methodology, and experiments were approved by the relevant ethics committee.
Monitoring mice under anesthesia
Post-administration of anesthetics, the mice need to be monitored throughout the duration of imaging. For this purpose, physiological monitoring using PhysioSuite (Kent Scientific Corporation) can be used. Alternatively, the depth of anesthesia can also be monitored by checking for hind limb and palpebral reflex. For testing the palpebral reflex, a sterile cotton tip is used to gently touch the periocular skin to generate a reflex. The absence of reflex indicates deep anesthesia. Similarly, the hind limb can also be gently pinched; if no reflex (hindlimb jerk or withdrawal) is observed, the mouse is under anesthesia.
During the anesthetic period, special attention should be paid to the breathing of the mouse, which should neither be too fast nor labored. Breathing under anesthesia should be deep, rhythmic, and distress-free. Moreover, the testing for palpebral and hindlimb reflexes should not affect the breathing rate. Wherever possible, we suggest that researchers utilize the help of more experienced researchers or veterinary staff to ensure that proper monitoring of anesthesia is being performed and the mice are not in any kind of distress.
Preparation prior to surgery for intravital imaging
Timing: 15 min
The below-detailed steps allow the researchers to prepare mice for cannulation, surgery, and imaging.
-
1.
Assess the tumor-bearing mouse for any visible signs of distress.
-
2.Anesthetize the mouse according to its body weight.
-
a.We administer injectable anesthesia, ketamine/xylazine (100/10 mg/kg) combined dose to provide deep surgical-plane anesthesia.
-
b.Administer repeated half doses upon monitoring during imaging session to enable extended periods of anesthesia. See detailed above.
-
a.
-
3.
Once the mouse is completely under anesthesia (as assessed via palpebral reflex and hindlimb withdrawal), carefully remove the abdominal hair using an electric shaver. We remove 80%–90% of hair by shaving until very little hair remains around the poorly accessible regions such as the hindlimbs.
CRITICAL: While using the electric shaver, take care to avoid damaging the skin overlying the tumor to avoid any accidental incision or injury. The shape of the underlying tumor may necessitate multiple attempts to remove the hair.
-
4.Post-shaving, apply a depilatory cream such as Nair (Church and Dwight) or Veet (Reckitt Benckiser) to the whole abdomen to remove the remaining hair.
-
a.After about 1–2 min (depending on the depilatory cream), clean the hair removal cream with a wet tissue. Thoroughly clean the skin and tap-dry with dry tissues. We recommend using soft tissues so as to not cause any abrasions on the skin. Lastly, swab the tissue with moist, lint-free paper. This step prevents the transfer of auto-fluorescent hair to the tumor region during surgical preparation.
-
b.Post-depilation, quickly place the mouse on its back on a thermal heating pad to maintain its core body temperature.
-
a.
-
5.
Next, proceed with cannulation as described below. If no cannulation is required, please proceed directly to the section on surgical preparation for intravital imaging.
CRITICAL: Do not leave the hair removal creams in contact with the skin for too long as this may irritate and inflame the skin. These steps should be carried out as quickly as possible to minimize the reduction in core body temperature. Alternatively, perform the procedure on a temperature-regulated heat pad or 24 h prior to surgery.
Cannulation
Employ this procedure when repeat injections need to be performed to administer drugs, dyes, or cells. If cannulation is not required, proceed to the surgical preparation for intravital imaging.
For cannulation, we use tail vein, jugular vein, or carotid artery cannulation, depending on the experimental requirements. Both tail vein and jugular vein cannulations transfer the administered agents into the venous circulation and can be used in most scenarios. Carotid artery cannulation is useful for administering cells into the aorta, a methodology utilized when the pulmonary vascular bed needs to be bypassed to avoid sequestration of the adoptively transferred cells in the lungs immediately after administration. As cells are injected into the aorta, they pass through organs (including the tumor mass) before entering the pulmonary vascular bed.
Tail vein cannulation
Timing: 10 min
This procedure can be performed by a researcher with routine mouse-handling experience. It is the method of choice for administering dyes and drugs.
-
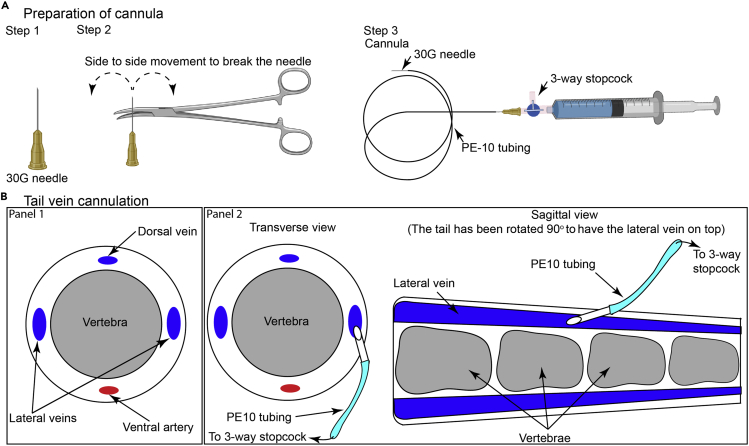
6.Using an artery/toothed-forceps, move the needle of an insulin syringe side-to-side till it breaks from the syringe.
-
a.Insert the broken/blunt end of the needle into PE10 tubing to create a cannula.
-
b.Insert a needle connected to a 3-way stopcock on the other end of the PE10 tubing for administering drugs, dyes, or cells (Figure 4A). Commercially available intravenous cannula can also be used for this purpose.
-
a.
-
7.Anesthetize the mouse (if not previously anesthetized).
-
a.Disinfect the tail by swabbing with 70% w/v ethanol before placing it on its side.
-
b.Warm the tail under a heat lamp for 1–2 min. Lateral veins are the best for cannulation (Figure 4B, Panel 1).
-
a.
Note: Care should be taken while heating the tail of the mouse. Radiant heat should be applied as per the best working practices within the facility. Wherever possible, the mouse should be covered with paper towels (Teri Wipers, Halyard), and only the tail should be exposed to the radiant heat directly. This methodology prevents damage to sensitive tissues such as the eyes and ears. After 1–2 min, researchers can clearly observe the lateral veins within the tail.
-
8.Once the blood vessel appears to be dilated (increased diameter and redness), perform the following steps:
-
a.Lift the tail firmly, reorient and position the needle of the cannula on top of the distal end of the tail parallel to the vessel.
-
b.Gently tilt the needle to an angle of 10°–15° and insert the bevel (facing up) into the blood vessel (Figure 4B, Panel 2).
-
c.Draw the syringe attached to the cannula to see if blood enters the tubing. If yes, then the needle is placed correctly; if not, then repeat the process carefully in another region of the tail.
-
a.
Note: Hold the needle attached to the cannula with artery/toothed forceps using the dominant hand.
-
9.Immobilize the needle and tubing on the tail using surgical tape. Flush 50–100 μL of saline to prevent blood coagulation within the needle.
-
a.If required, inject drugs and cells now via the cannula using the 3-way stopcock.
-
b.Take care so that no more than 10% of the blood volume (approximately 100 μL) is injected at any given time. We usually wait 30–40 min between any two intravenous administrations to avoid fluid overload.
-
a.
Note: In all of our cannulation procedures, we prefer to use a 3-way stopcock to allow for the availability of multiple injection ports for simultaneous injection of dyes and cells. Additionally, this methodology provides surrogate ports in case a port is blocked.
Figure 4.
Cannulation of blood vessels
(A) Illustration of the methodology used to generate the tail vein cannula.
(B) Schematic representation highlighting the methodology for cannulation of tail vein, adapted from The Laboratory Mouse (Shimizu, 2004).
Jugular vein cannulation
Timing: 20–30 min
This procedure requires expertise and is best suited for researchers with experience in small animal surgery. It is most useful for injecting large number of cells in a relatively small volume (to avoid fluid overload/hypervolemia). It is best used for the adoptive transfer of immune cell populations such as neutrophils, monocytes, and effector T cells.
-
10.
Remove hair from approximately 1–2 cm region of skin on the ventral side of the neck using depilatory creams. Perform all the subsequent steps under a surgical microscope.
-
11.Make a superficial skin incision (approximately 1 cm) using a pair of fine-tipped surgical scissors right of the midline of the neck (left to the trachea).
-
a.Gently tease the skin apart to highlight the underlying tissue (Figure 5A, step1).
-
b.Using fine forceps, gently tease the underlying fascia and the surrounding tissue whilst avoiding injury. At this point, the jugular vein becomes visible as a large blood vessel, and care should be taken to avoid any injury.
-
c.Immobilize the skin tissue using a surgical suture (Figure 5A, step 1).
-
d.Tape the other end of the suture onto the heating pad or the metal base plate of the imaging stage.
-
a.
CRITICAL: It is crucial at this stage to make sure that the fascia is well separated to provide access to the jugular vein, but precaution should be taken to avoid any injury to the fascia and the surrounding tissue which have deep-seated veins and may lead to bleeding.
-
12.Gently tease the tissue between the common carotid artery, vagus nerve, and jugular vein.
-
a.Using ultrafine forceps (preferably Dumont #7 forceps), separate the jugular vein and the vagus nerve from the underlying tissue.
-
b.Gently lift the vein using forceps. At this point, no underlying tissue should be displaced, indicating a proper surgical preparation (Figure 5A, step 2).
-
a.
CRITICAL: It is crucial that the vagus nerve and the carotid artery are well separated to avoid any nerve damage when the vein is ligated, but it should be done with utmost care to avoid any injury to the vein, which could lead to extensive bleeding.
-
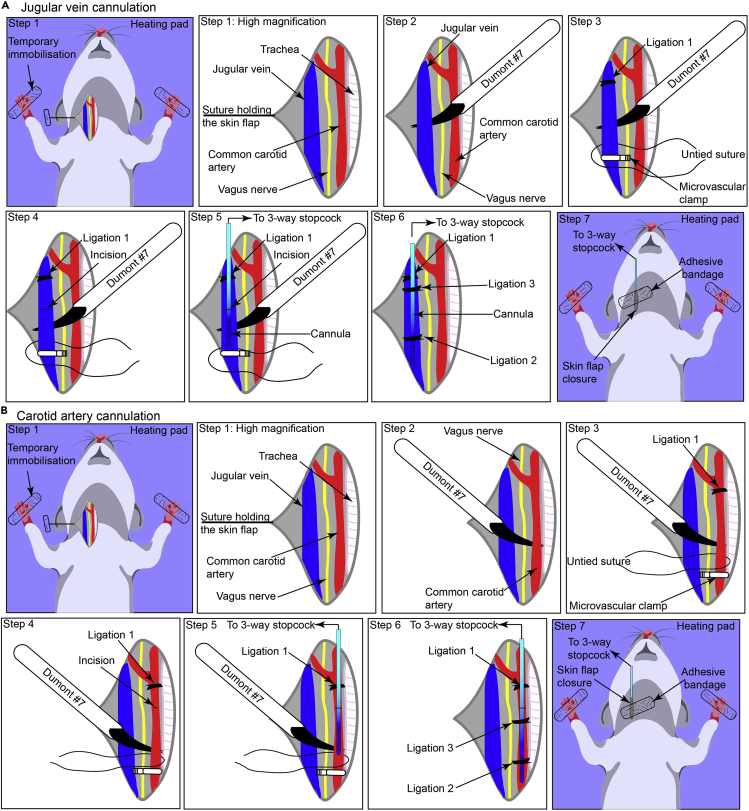
13.
Ligate the anterior region of the vein using the instrument tie methodology (with two opposing throws) using a surgical silk suture (Dynek Dysilk 4/0 suture) (Figure 5A, step 3, Ligation 1).
-
14.
Clamp the distal end using a microvascular clamp attached to the S&T clamp applying forceps (Figure 5A, step 3). Leave a single suture untied at the distal end.
CRITICAL: To ligate the vein, cut appropriately sized sutures (approximately 10 cm in length) and pass under the vein which has been gently lifted by inserting forceps underneath (Figure 5A, step 3). Take care to avoid excessive abrasion with the sutures or forceps to prevent any tear or bleeding.
-
15.
Using a pair of Vannas scissors, make a small incision on the vein. A small amount of blood retained in the lumen of the clamped vein will leak from the incision site (Figure 5A, step 4).
CRITICAL: To make a precise incision on the vein, we recommend lifting the vein from the top using Dumont#7 forceps, positioning the Vannas scissors 30° to 45° to the forceps so that just a small part of the vein comes in contact with the scissors. Make a single, sharp cut to avoid excessive damage to the vein.
-
16.Insert a cannula (without the needle) (PE10®, BD) connected to a 3-way stopcock containing saline into the vein via the incision (Figure 5A, step 5).
-
a.Immobilize the cannula in the vein by ligating with a surgical suture employing instrument tie methodology with two opposing throws (Figure 5A, step 6, Ligation 2).
- b.
-
a.
CRITICAL: This part of the procedure requires gentle manipulations and dexterity. The tip of the cannula should be cut with a scalpel blade to provide a sharp 45° bevel. Before inserting the cannula, confirm that the cannula or the connecting stopcock has no air entrapped and the stopcock is in a locked position.
-
17.Remove the microvascular clamp using the S&T clamp forceps.
-
a.Trim the excess suture thread.
-
b.Placing the skin flap on the underlying tissue, seal the incision site using surgical glue.
-
c.The researchers can also attach the cannula onto the skin surface using surgical glue followed by an adhesive bandage. These additional steps prevent the accidental removal of cannula either during the surgical preparation of the tumor or during the transfer of the stage to the microscope.
-
d.Flush 50–100 μL of saline to prevent blood coagulation within the cannula.
-
e.Inject drugs and cells as required via the cannula using the 3-way stopcock, taking care that no more than 10% of the blood volume is injected at any given time to avoid any changes in the hemodynamic parameters (Figure 5A, step 7).
-
a.
Note: We usually lightly tape the syringe, 3-way stopcock, and the tubing to the stage or heat pad so as to prevent any accidental damage, bending, or removal. This further helps during the surgical preparation of the tumor as the tubing is securely placed away from the surgical setup (Figure 6).
Figure 5.
Cannulation of blood vessels
(A) Schematic representation depicting the stepwise methodology used for cannulating jugular vein.
(B) Illustration depicting the stepwise methodology used for cannulating carotid artery.
Pause point: Take brief pauses (<30 sec) between steps 10 to 17 to prepare for the upcoming step, change surgical instruments, and check ligations or cannula.
Carotid artery cannulation
Timing: 30–45 min
This procedure is best suited for researchers with extensive experience in small animal surgery. It is very useful for injecting large number of cells in experiments wherein the pulmonary vascular bed needs to be bypassed to avoid sequestration of cells immediately post-transfer.
-
18.
Perform steps 10 and 11 as described for jugular vein cannulation (Figure 5B, step 1).
-
19.Gently tease the tissue between the trachea and the jugular vein to reveal the presence of the common carotid artery which is easily detected by rhythmic pulsation and the vagus nerve which runs parallel to the common carotid artery.
-
a.Using ultrafine forceps (preferably Dumont #7 forceps), separate the common carotid artery and the vagus nerve. Isolate the carotid artery from the underlying tissue.
-
b.Gently lift the artery using the forceps. At this point, no underlying tissue should be displaced, thus indicating a good surgical preparation (Figure 5B, step 2).
-
a.
CRITICAL: It is crucial that the vagus nerve and carotid artery are well separated to avoid any nerve damage when the artery is ligated, but it should be done with utmost care to avoid any injury to the artery, which would lead to excessive bleeding.
-
20.
Ligate the anterior region of the common carotid artery prior to the partition into internal and external carotid arteries using the instrument tie methodology (with two opposing throws) employing a silk surgical suture (Dynek Dysilk 4/0 suture) (Figure 5B, step 3, Ligation 1).
-
21.
Clamp the distal end using microvascular clamps attached to the S&T clamp applying forceps.
CRITICAL: To ligate the artery, cut appropriately sized sutures (approximately 10 cm in length) and pass under the artery which has been gently lifted by inserting forceps underneath. Care should be taken to avoid excessive abrasion with the sutures or forceps that may lead to bleeding.
-
22.
Using Vannas scissors, make a small incision in the carotid artery. Small amounts of blood retained in the lumen of the clamped artery will leak from the incision site, and the artery will collapse (Figure 5B, step 4). Troubleshooting problem 4
CRITICAL: To make a precise incision on the artery, we recommend lifting the artery from the top using Dumont#7 forceps, positioning the Vannas scissors 45° to the forceps so that just a small part of the artery comes in contact with the scissors. Make a single, sharp cut to avoid excessive damage to the artery.
- 23.
CRITICAL: This part of the procedure requires gentle manipulation and dexterity. We advise pulling the cannula tube at the tip to make it as thin as possible without generating any kinks in the cannula. The tip of the cannula should then be cut with a scalpel blade to provide a sharp 30° to 45° bevel. Before inserting the cannula, confirm that the cannula or the connecting stopcock has no air entrapped and the stopcock is in a locked position.
-
24.Remove the microvascular clamp using the S&T clamp forceps.
-
a.Trim the excess suture thread.
-
b.Placing the skin flap on the underlying tissue, seal the incision site using a surgical glue.
-
c.The researchers can also attach the cannula onto the skin surface using surgical glue followed by an adhesive bandage. These additional steps prevent accidental removal of the cannula during either tumor surgery or transfer of the stage to the microscope.
-
d.Flush 50–100 μL of saline to prevent blood coagulation within the cannula.
-
e.Inject drugs and cells as required via the cannula using the 3-way stopcock, taking care that no more than 10% of the blood volume is injected at any given time to avoid any changes in the hemodynamic parameters (Figure 5B, step 7).
-
a.
Note: We usually lightly tape the syringe, 3-way stopcock, and the tubing to the stage or heat pad so as to prevent any accidental damage, bending, or removal. This further helps during the tumor surgery as the tubing is securely placed away from the surgical setup (Figure 6).
Pause point: Take brief pauses (<30 sec) between steps 18–24 to prepare for the upcoming step, change surgical instruments, and check the ligations or cannula.
Surgical preparation for intravital imaging
Timing: 30 min
This procedure requires expertise and is best suited for researchers with experience in small animal surgery. These steps allow for access to the underlying mammary or melanoma tumor for intravital imaging.
-
25.
Place the mouse on its back (Figure 6, step 2), on the thermal-controlled surgical board. Gently restrain the limbs using surgical tape to avoid excessive movement during surgery.
-
26.
Insert a rectal probe into the mouse to continuously monitor the core body temperature using a thermal regulator set at maintaining the core body temperature at 37°C (Figure 6, step 2).
-
27.
Make a lateral incision from the lower abdomen to the neck area. Exercise appropriate caution so as to not puncture the peritoneum (Figure 6, step 3).
-
28.
Using blunt dissection, lift the skin from the underlying peritoneal wall (Figure 6, step 3).
-
29.
Make angled incisions pointing towards the limbs (Figure 6, step 4).
-
30.
Gently pull the skin back. Using blunt dissection, separate the skin from the underlying peritoneal wall to create a skin flap, thus exposing the tumor.
CRITICAL: The tumor is vascularized by newly formed angiogenic blood vessels originating from both cranial and caudal superficial epigastric artery and vein. Take utmost care to avoid injury to these blood vessels.
-
31.Remove the fascia connecting the skin to the peritoneal wall very carefully and with a lot of precision using the curved edge of the Dumont #7 forceps and a pair of surgical scissors if required (Figure 6, step 4).
- a.
-
b.Furthermore, remove any non-vascularized fascia, connective or adipose tissue from the surface of the tumor to improve the quality of imaging.
CRITICAL: If the fascia connecting the skin flap to the peritoneal wall is not adequately removed, these connections may result in lateral movement observed during tumor imaging caused due to breathing, with the inhalation and exhalation of the breath changing the tension within the skin flap.
CRITICAL: Remove any residual fat (for mammary tumor) or fascia (for melanoma) from the tumor surface as this may hamper the imaging. Gently lift the non-vascularized fascia or adipose tissue and cut from the underlying tumor. The presence of excess fascia, connective tissue, or mammary fat is occasionally observed, especially in smaller tumors wherein the surrounding tissue has not yet been replaced by the tumor mass.
-
32.Once the tumor has been surgically prepared, apply hydrophobic ointment (such as vacuum grease) to the base of an appropriately sized stage insert and gently place it onto the stage to generate a watertight seal.
-
a.Superfuse the tumor with saline solution at 37°C.
-
b.Place a coverslip on top of the stage insert. Alternatively, image the tumor in the superfused chamber using a water-dipping objective. Troubleshooting problem 7
-
a.
-
33.
Cover the exposed tissue with a sterile cotton-gauze sheet. Keep the tissue warm and perfused with saline at 37°C.
-
34.
To prevent the drying of the animal’s eyes during the imaging session, apply a lubricating ophthalmic ointment before imaging is commenced.
-
35.
Remove or relax the temporary restraint on the limbs to allow for a normal posture.
-
36.
Scan the large exposed tumor to find a suitable region for imaging.
-
37.
While the tumor is being imaged, inject vascular dyes, drugs, or cells to study the real-time dynamics of immune cells in vivo.
-
38.
Clean the microscope stage by wiping it down with 70% ethanol.
Note: All steps should be carried out using sterile surgical instruments. Sterile, surgical-grade cotton-gauze should be used in this procedure. Consider administration of analgesics during the surgical procedure as per the institutional ethics protocols and experimental design.
Intravital imaging
Timing: 1–5 h
For operating the intravital microscope, please follow the specific guidelines of the manufacturer. We suggest turning the microscope on, including all the laser components, and detectors to allow sufficient time for the lasers and optics to warm up and stabilize. This should be done before the researcher starts preparing the mouse for surgery.
-
39.
Clean the microscope stage by wiping it down with 70% ethanol.
-
40.
Following the surgical preparation of the tumor, transfer the imaging stage (Figure 6, step 6) onto the microscope stage.
-
41.Once the mouse is placed under the microscope, scan the tissue using the eyepiece for the region of interest.
-
a.Use the endogenous fluorescence of transgenic immune cells or tumor cells to determine an appropriate region of interest (ROI).
-
b.Usually, regions with blood vessels are chosen to study leukocyte trafficking. However, the choice of the region to be imaged depends on the scientific query being pursued.
-
c.Take care to image regions that reflect the overall biology of the tissue and to avoid any biases being introduced based on selective imaging.
-
a.
-
42.
Once an ROI is selected, switch the imaging to the multiphoton mode.
-
43.
At this point, imaging is controlled by computer-assisted software. The authors of this article employ IMSpector (LaVision). The imaging is performed as described previously (Li et al., 2012). Troubleshooting Problem 8
-
44.
Post-completion of the imaging session, euthanize the mice in accordance with the approved ethical guidelines.
-
45.
Save and export the data.
Expected outcomes
Using the methodology detailed above, we regularly generate longitudinal imaging datasets as detailed in Figure 7. We have been successfully able to use this methodology for imaging mammary tumors and fatpad (Tikoo et al., 2021a), and melanoma (Tikoo et al., 2021b). We have detailed the most common steps for data processing and analysis below.
Quantification and statistical analysis
-
1.Store and export the data collected by the IMSpector software (LaVision) as ome.tif files containing the metadata for the imaging sessions.
-
a.The files can be imported into various image processing software programs such as Image J, Volocity, and Imaris. Depending on the experimental query, we employ both Volocity and Imaris software for data processing and analyses.
-
b.The caveat of using commercial software is the prohibitive licensing costs associated with them; therefore, non-commercial image analysis software, for example, Image J or FIJI, can also be utilized for post-acquisition image analysis. Nonetheless, the incorporation of a large number of features, ease of performing multiple tasks, and the ability to process multiple datasets provided by commercial software are of immense value to researchers.
-
c.To identify the best software for analysis, we often consider the ease of use, list of features, stability of the analysis platform, batch processing capabilities, compatibility with acquired data files from various imaging instruments, ease of employing server-based licenses, and the cost of licensing.
-
a.
-
2.To highlight some of the most commonly used features, we will utilize Volocity and Imaris software as an example. A detailed list of all the features and capabilities of the software can be found at Volocity and Imaris websites. Outlined below is a brief overview of the processes performed utilizing these software:
-
a.Visualizing the data set: Post-acquisition, import the data files into either Volocity or Imaris. Files imported into Volocity and Imaris can be viewed in different modes.
-
i.While using Volocity, we prefer to use the extended focus mode to visualize the tumor tissue (Figure 7A and Methods video S1). This projection allows for rapid screening of the imaging data, optimization of the signal intensity for individual channels, and identification of any imaging artefacts; for example, a distortion caused by muscle twitch can be observed at timepoint 00:27:28 (hr:min:sec) in the lower half of the Methods video S1.
-
ii.Similarly, in Imaris, under ‘3D view’, images are opened as a maximum intensity projection (MIP) interactive 3D image that can be rotated and zoomed in and out (Figure 7B and Methods video S2). The ‘Slice’ mode allows images of a z stack to be viewed as individual slices within the z plane, whilst, ‘Section’ mode is chosen to view the xyz dimensions of an image stack. For simplicity, we have outlined a few features of the Imaris software below.Note: When utilizing automated analysis, it is highly recommended to normalize/negate the effects of transient movements from the final data sets.Methods video S1. Intravital imagingof amelanotic melanoma tumours highlighting the dynamic behaviour of GFP+ leukocytes, related to Quantification and statistical analysis, Step 3 a,iDownload video file (24.8MB, mp4)Methods video S2. Intravital imaging of E0771 tumours highlighting the dynamic behaviour of monocytes and tumour cells, related to Quantification and statistical analysis, Step 3 a,iiDownload video file (14.9MB, mp4)
-
i.
-
b.Editing the data set: Under the ‘Edit’ menu, various functions are located that can be used to edit the original uploaded data set.
-
i.Time points, slices, and channels can be added or deleted by choosing the corresponding option and uploading the additional files.
-
ii.Images can also be cropped to a specific region of interest (ROI) by choosing the ‘Crop 3D’ option and defining the ROI (Figures 7C and 7D).
-
i.
-
c.Adjusting brightness and contrast: Using the ‘Display Adjustment’ window, the brightness, contrast, and color of each individual channel can be adjusted based on an intensity color scale.
-
d.Correcting drift: Correct the translational drift in the XY plane and rotational drift in relation to stationary reference objects within the same image frame.
-
i.First, create tracks for the reference objects for example, non- migratory cells or blood vessels using the ‘Spots’ or ‘Surface’ function.
-
ii.Once tracks are created, select the ‘Correct Drift’ function, found in the ‘Edit Tracks’ tab, to correct the drift of the whole image frame based on the tracks of the stationary objects created.
-
i.
-
e.3D rendering: Perform the 3D rendering of the objects of interest, such as blood vessels or cells, by creating or reconstructing a surface over the selected objects using the ‘Surface’ function.
-
i.Create surfaces on objects within a chosen channel by following a series of steps.
-
ii.Once the final surface is generated, a properties panel appears that can be used to edit various aspects of the surface such as color and transparency and calculate statistical metrics including surface volume, intensity.
-
i.
-
f.Cell tracking: Using the ‘Spots’ function, track the individual cells and statistically analyze over time.
-
i.Create a spot on each individual cell within a chosen channel by following the series of steps outlined.
-
ii.Create tracks from these spots automatically using an algorithm. Alternatively, draw and track individual spots manually for each time frame of the data set (Figures 7B and 7C).
-
iii.Similar to the ‘Surface’ function, a properties panel enables tracks to be edited and statistically analyzed for parameters such as track distance and velocity.
-
iv.Export all statistical data as an excel spreadsheet file.
-
v.Other than the spot function, researchers can also employ the surface function for tracking cell populations.
-
vi.The choice of the methodology used for tracking cells relies on the quality of the data, the physiology and behavior of the cell population being tracked, and the experimental query being answered. We recommend utilizing the resources provided within the Learning center, IMARIS.
-
i.
-
g.Exporting the data set: Once all the image processing is completed, export all data sets from Imaris as an mp4 movie file. Alternatively, a ‘Snapshot’ function is also available to capture a single image at any given time point.
-
a.
Limitations
One of the limitations of this protocol is its surgical invasiveness. Therefore, it is carried out as an end-point approach, thus limiting its potential in repeated longitudinal imaging (Alexander et al., 2008, Ilina et al., 2018). In a scenario wherein longitudinal imaging would be beneficial, tumors can be implanted utilizing the dorsal skin-fold window chamber methodology and imaged longitudinally (Maeda and DaCosta, 2014, Alexander et al., 2008, Biel et al., 2014). Similarly, poor surgical preparation of the tumor might affect the inflammatory response. However, if performed correctly, the surgery causes minimal change to the inflammatory status of the tissue being imaged. Moreover, the protocol allows for reproducible growth and development of tumors within the mammary fat pad and skin. The surgical preparation provides accessibility to a large region of the growing tumor mass, permitting increased flexibility in selecting the regions of interest.
Troubleshooting
Problem 1
Rapid growth of tumors with minimal vascularization.
Potential solution
The growth rate of tumor in vivo is determined by the number of cells injected. While injection of 1 million cells results in the development of tumors within two weeks, injection of 0.25–0.5 million cells results in highly vascularized tumors within approximately three weeks. The number of tumor cells injected influences the growth rate of the tumors, the degree of vascularization, infiltration of immune cells, and the localization of immune infiltrate. Therefore, it is prudent that researchers optimize these parameters before proceeding with the intravital imaging studies.
Problem 2
Variable rate of tumor growth within a cohort of injected mice.
Potential solution
Once the cell suspension is aspirated into the syringe, injections should be performed within 30 s. Due to their size and density, tumor cells can readily settle within the syringe leading to inaccurate injections. We prefer to aspirate tumor cell suspensions for a single mouse each time to avoid variability in tumor cell numbers injected between mice. The Eppendorf tube with the tumor cell suspensions is thoroughly mixed before each aspiration. Also, the variability introduced due to the characteristics of the tumor cell line being employed should be taken into consideration when ascertaining the experimental group size.
Problem 3
Difficulty in cannulating the vein.
Potential solution
The ligation should be performed such that it provides a large accessible region for cannulating the blood vessel. Similarly, the clamping should be done as far as possible from the ligation site to provide accessibility to the vein for cannulation. Care should be taken that during clamping, no tissue is caught between the clamps as this will lead to imperfect clamping.
Problem 4
Rapid bleeding after the incision is made on the artery
Potential solution
Rapid bleeding is probably caused by the improper use of a vascular clamp. To alleviate the issue, apply a second clamp on the artery to stop excessive bleeding. Readjust the first clamp taking care that during clamping, no tissue is caught between the clamps as this will lead to imperfect clamping. Only the artery should be present within the clamp.
Problem 5
Difficulty in cannulating the artery.
Potential solution
The ligation should be performed just before the artery bifurcates to provide accessibility to as much region as possible for cannulation. Similarly, the clamping should be performed as far away as possible from the ligation site to provide accessibility to the artery for cannulation. Care should be taken such that during clamping, no tissue is caught between the clamps as this will lead to imperfect clamping. Only the artery should be present in the clamp.
Problem 6
Inability to insert the cannula.
Potential solution
To efficiently insert the cannula, make sure that it is well-positioned, preferably 45° to the incision on the artery. It may also be helpful to insert the tip of the forceps into the incision and then insert the cannula in the artery using the lower side of the forceps as a guide. Excessive force will disrupt the artery, resulting in blockage or a tear.
Problem 7
Poor perfusion of the tumor tissue as evidenced by lack of blood flow or non-adherent stationary cells within the vascular lumen.
Potential solution
To ensure good vascular perfusion, ensure that the notch on the insert aligns with the region of the cranial and caudal superficial epigastric artery and vein. Any compression of this vasculature will lead to poor perfusion of the tumor tissue. Within a well perfused tumor tissue, most of the labeled cells within the vascular lumen can be visualized to rapidly flow through with a small number of cells depicting the behavior of rolling or vascular adhesion (see Methods videos S1 and S2). Please ensure that the insert height is 2–3 mm higher than the tumor being imaged and no overt pressure is being applied onto the tumor by either the insert or the coverslip.
Problem 8
Tissue drifts observed during the imaging sessions.
Potential solution
It is not uncommon to observe tissue drifts for the first 20–40 min of the imaging sessions. These drifts are caused by the difference in temperature between the tumor tissue, heated platform, buffer, and the objective. These differences normalize over time and the drifts are stabilized. However, if tissue drifts are observed over extended periods, these may be caused by improper stabilization of the skin flap, fluctuation in stage temperature, or tissue movement due to breathing. To correct for these issues, check that all the sutures are adequately secured, ensure proper connection of electronics and ensure that the fascia between the skin flap has been separated from the peritoneal wall.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rohit Jain (R.Jain@centenary.org.au).
Request for reagents and resources can also be directed to Prof. Wolfgang Weninger (W.Weninger@centenary.org.au; w.weninger@meduniwien.ac.at).
Materials availability
The breast cancer and melanoma cell lines are available from the laboratory upon request. Amelanotic B16-F10 melanoma and mCherry expressing E0771 breast adenocarcinoma cell lines require material transfer agreement (MTA), as per institutional guidelines.
Data and code availability
The sample imaging dataset utilized in this study can be obtained from the authors upon request. Analysis was performed utilizing commercially available software, Volocity (Quorum Technologies) and Imaris (Oxford Instruments).
Acknowledgments
The authors would like to acknowledge the core flow cytometry and imaging facility at the Centenary Institute of Cancer Medicine and Cell Biology (Drs. Adrian Smith and Kristina Jahn). We would like to acknowledge Sydney cytometry for providing the resources needed for generating the 3-D printed stage inserts and thank Dr. Frank Kao and Mr. Steven Allen for their help in generating 3D printed imaging stage inserts. We would also like to acknowledge various funding bodies especially, National Health and Medical Research Council, PdCCRS research scheme from Cancer Australia (CanToo), and The University of Sydney’s internal funding schemes. Schematics have been created using BioRender.
Author contributions
R.J. and S.T. conceptualized and designed the study, standardized the protocol, and carried out the experiments. K.O. assisted with data analysis. S.D. assisted with designing and standardizing of the 3D inserts. L.C. and B.M. assisted with animal ethics. R.J., S.T., and W.W. supervised the study, wrote, and edited the manuscript. All authors have read and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100722.
Contributor Information
Rohit Jain, Email: R.Jain@centenary.org.au.
Wolfgang Weninger, Email: W.Weninger@centenary.org.au.
Supplemental information
References
- Abtin A., Jain R., Mitchell A.J., Roediger B., Brzoska A.J., Tikoo S., Cheng Q., Ng L.G., Cavanagh L.L., Von Andrian U.H. Perivascular macrophages mediate neutrophil recruitment during bacterial skin infection. Nat. Immunol. 2014;15:45–53. doi: 10.1038/ni.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander S., Koehl G.E., Hirschberg M., Geissler E.K., Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem. Cell Biol. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- Berrozpe G., Agosti V., Tucker C., Blanpain C., Manova K., Besmer P. A distant upstream locus control region is critical for expression of the Kit receptor gene in mast cells. Mol. Cell. Biol. 2006;26:5850–5860. doi: 10.1128/MCB.01854-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel N.M., Lee J.A., Sorg B.S., Siemann D.W. Limitations of the dorsal skinfold window chamber model in evaluating anti-angiogenic therapy during early phase of angiogenesis. Vasc. Cell. 2014;6:17. doi: 10.1186/2045-824X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonnas A., Licata F., Poupel L., Jacquelin S., Fetler L., Krumeich S., Théry C., Amigorena S., Combadière C. CD8+ tumor-infiltrating T cells are trapped in the tumor-dendritic cell network. Neoplasia. 2013;15:85–94. doi: 10.1593/neo.121572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B., Nelson A., Krummel M.F. Tumor-infiltrating lymphocytes are dynamically desensitized to antigen but are maintained by homeostatic cytokine. JCI Insight. 2016;1:e89289. doi: 10.1172/jci.insight.89289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breart B., Lemaître F., Celli S., Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest. 2008;118:1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Acebes M., Nicolás-Ávila J.A., Li J.L., García-Silva S., Balachander A., Rubio-Ponce A., Weiss L.A., Adrover J.M., Burrows K., A-González N. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med. 2018;215:2778–2795. doi: 10.1084/jem.20181468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazaux M., Grandjean C.L., Lemaître F., Garcia Z., Beck R.J., Milo I., Postat J., Beltman J.B., Cheadle E.J., Bousso P. Single-cell imaging of CAR T cell activity in vivo reveals extensive functional and anatomical heterogeneity. J. Exp. Med. 2019;216:1038–1049. doi: 10.1084/jem.20182375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T.J. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- De Palma M., Venneri M.A., Galli R., Sergi Sergi L., Politi L.S., Sampaolesi M., Naldini L. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell. 2005;8:211–226. doi: 10.1016/j.ccr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fantozzi A., Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212. doi: 10.1186/bcr1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust N., Varas F., Kelly L.M., Heck S., Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- Gray E.E., Suzuki K., Cyster J.G. Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J. Immunol. 2011;186:6091–6095. doi: 10.4049/jimmunol.1100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Beaumont K.A., Sharp D.M., Lui G.Y.L., Weninger W., Haass N.K., Tikoo S. Abrogation of RAB27A expression transiently affects melanoma cell proliferation. Pigment Cell Melanoma Res. 2020;33:889–894. doi: 10.1111/pcmr.12903. [DOI] [PubMed] [Google Scholar]
- Guo D., Lui G.Y.L., Lai S.L., Wilmott J.S., Tikoo S., Jackett L.A., Quek C., Brown D.L., Sharp D.M., Kwan R.Y.Q. RAB27A promotes melanoma cell invasion and metastasis via regulation of pro-invasive exosomes. Int. J. Cancer. 2018;144:3070–3085. doi: 10.1002/ijc.32064. [DOI] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., Muller W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina O., Campanello L., Gritsenko P.G., Vullings M., Wang C., Bult P., Losert W., Friedl P. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model. Mech. 2018;11:dmm034330. doi: 10.1242/dmm.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Tikoo S., Weninger W. Recent advances in microscopic techniques for visualizing leukocytes in vivo. F1000Research. 2016;5:915. doi: 10.12688/f1000research.8127.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Weninger W. Shedding light on cutaneous innate immune responses: the intravital microscopy approach. Immunol. Cell Biol. 2013;91:263–270. doi: 10.1038/icb.2012.76. [DOI] [PubMed] [Google Scholar]
- Johnstone C.N., Smith Y.E., Cao Y., Burrows A.D., Cross R.S., Ling X., Redvers R.P., Doherty J.P., Eckhardt B.L., Natoli A.L. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model. Mech. 2015;8:237–251. doi: 10.1242/dmm.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., Littman D.R. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviron M., Combadière C., Boissonnas A. Tracking Monocytes and Macrophages in Tumors With Live Imaging. Front. Immunol. 2019;10:1201. doi: 10.3389/fimmu.2019.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Park S.A., Zou Y., Seo S.U., Jun C.D., Lee W.J., Hyun Y.M., Cho N.H. Real-Time Monitoring of Cancer Cells in Live Mouse Bone Marrow. Front. Immunol. 2018;9:1681. doi: 10.3389/fimmu.2018.01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.L., Goh C.C., Keeble J.L., Qin J.S., Roediger B., Jain R., Wang Y., Chew W.K., Weninger W., Ng L.G. Intravital multiphoton imaging of immune responses in the mouse ear skin. Nat. Protoc. 2012;7:221–234. doi: 10.1038/nprot.2011.438. [DOI] [PubMed] [Google Scholar]
- Lindquist R.L., Shakhar G., Dudziak D., Wardemann H., Eisenreich T., Dustin M.L., Nussenzweig M.C. Visualizing dendritic cell networks in vivo. Nat. Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- Lämmermann T., Afonso P.V., Angermann B.R., Wang J.M., Kastenmüller W., Parent C.A., Germain R.N. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., DaCosta R.S. Optimization of the dorsal skinfold window chamber model and multi-parametric characterization of tumor-associated vasculature. Intravital. 2014;3:e27935. doi: 10.4161/intv.27935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S., Wang B., Kawashima N., Braunstein S., Badura M., Cameron T.O., Babb J.S., Schneider R.J., Formenti S.C., Dustin M.L. Radiation-induced CXCL16 release by breast cancer cells attracts effector T cells. J. Immunol. 2008;181:3099–3107. doi: 10.4049/jimmunol.181.5.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., Pittman K., Menezes G.B., Hirota S.A., Slaba I., Waterhouse C.C., Beck P.L., Muruve D.A., Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- Motoike T., Loughna S., Perens E., Roman B.L., Liao W., Chau T.C., Richardson C.D., Kawate T., Kuno J., Weinstein B.M. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Mrass P., Kinjyo I., Ng L.G., Reiner S.L., Puré E., Weninger W. CD44 mediates successful interstitial navigation by killer T cells and enables efficient antitumor immunity. Immunity. 2008;29:971–985. doi: 10.1016/j.immuni.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrass P., Takano H., Ng L.G., Daxini S., Lasaro M.O., Iparraguirre A., Cavanagh L.L., Von Andrian U.H., Ertl H.C., Haydon P.G. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J. Exp. Med. 2006;203:2749–2761. doi: 10.1084/jem.20060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Ng L.G., Hsu A., Mandell M.A., Roediger B., Hoeller C., Mrass P., Iparraguirre A., Cavanagh L.L., Triccas J.A., Beverley S.M., Scott P., Weninger W. Migratory dermal dendritic cells act as rapid sensors of protozoan parasites. PLoS Pathogen. 2008;4:e1000222. doi: 10.1371/journal.ppat.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.G., Qin J.S., Roediger B., Wang Y., Jain R., Cavanagh L.L., Smith A.L., Jones C.A., De Veer M., Grimbaldeston M.A. Visualizing the neutrophil response to sterile tissue injury in mouse dermis reveals a three-phase cascade of events. J. Invest. Dermatol. 2011;131:2058–2068. doi: 10.1038/jid.2011.179. [DOI] [PubMed] [Google Scholar]
- Obeidy P., Ju L.A., Oehlers S.H., Zulkhernain N.S., Lee Q., Galeano Niño J.L., Kwan R.Y., Tikoo S., Cavanagh L.L., Mrass P. Partial loss of actin nucleator actin-related protein 2/3 activity triggers blebbing in primary T lymphocytes. Immunol. Cell Biol. 2020;98:93–113. doi: 10.1111/imcb.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overwijk W.W., Restifo N.P. B16 as a mouse model for human melanoma. Curr. Protoc. Immunol. 2001;Chapter 20:Unit 20.1. doi: 10.1002/0471142735.im2001s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai S., Qin J., Cavanagh L., Mitchell A., El-Assaad F., Jain R., Combes V., Hunt N.H., Grau G.E., Weninger W. Real-time imaging reveals the dynamics of leukocyte behaviour during experimental cerebral malaria pathogenesis. PLoS Pathog. 2014;10:e1004236. doi: 10.1371/journal.ppat.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.L., Buzzai A., Rautela J., Hor J.L., Hochheiser K., Effern M., Mcbain N., Wagner T., Edwards J., Mcconville R. Tissue-resident memory CD8 promote melanoma-immune equilibrium in skin. Nature. 2019;565:366–371. doi: 10.1038/s41586-018-0812-9. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Mueller K.L., Adams D.J., Anandasabapathy N., Aplin A.E., Bertolotto C., Bosenberg M., Ceol C.J., Burd C.E., Chi P. Melanoma models for the next generation of therapies. Cancer Cell. 2021;39:610–631. doi: 10.1016/j.ccell.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N.C., Egen J.G., Secundino N., Debrabant A., Kimblin N., Kamhawi S., Lawyer P., Fay M.P., Germain R.N., Sacks D. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science. 2008;321:970–974. doi: 10.1126/science.1159194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash M.D., Munoz M.A., Jain R., Tong P.L., Koskinen A., Regner M., Kleifeld O., Ho B., Olson M., Turner S.J. Granzyme B promotes cytotoxic lymphocyte transmigration via basement membrane remodeling. Immunity. 2014;41:960–972. doi: 10.1016/j.immuni.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Pucci F., Venneri M.A., Biziato D., Nonis A., Moi D., Sica A., Di Serio C., Naldini L., De Palma M. A distinguishing gene signature shared by tumor-infiltrating Tie2-expressing monocytes, blood "resident" monocytes, and embryonic macrophages suggests common functions and developmental relationships. Blood. 2009;114:901–914. doi: 10.1182/blood-2009-01-200931. [DOI] [PubMed] [Google Scholar]
- Pérez-Guijarro E., Day C.P., Merlino G., Zaidi M.R. Genetically engineered mouse models of melanoma. Cancer. 2017;123:2089–2103. doi: 10.1002/cncr.30684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson R.T., Levine S.T., Haynes S.M., Gutierrez P., Baratta J.L., Tan Z., Longmuir K.J. Use of labeled tomato lectin for imaging vasculature structures. Histochem. Cell Biol. 2015;143:225–234. doi: 10.1007/s00418-014-1301-3. [DOI] [PubMed] [Google Scholar]
- Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S., Mitchell A.J., Tay S.S., Jain R., Forbes-Blom E. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C.T., Schindelin J., Hiner M.C., Dezonia B.E., Walter A.E., Arena E.T., Eliceiri K.W. Imagej2: Imagej For The next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono R.T., Oceandy D., Pollard J.W., Tong W., Pavli P., Wainwright B.J., Ostrowski M.C., Himes S.R., Hume D.A. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Sauter K.A., Pridans C., Sehgal A., Bain C.C., Scott C., Moffat L., Rojo R., Stutchfield B.M., Davies C.L., Donaldson D.S. The MacBlue binary transgene (csf1r-gal4VP16/UAS-ECFP) provides a novel marker for visualisation of subsets of monocytes, macrophages and dendritic cells and responsiveness to CSF1 administration. PLoS One. 2014;9:e105429. doi: 10.1371/journal.pone.0105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S. The Laboratory Mouse. 1st Edition. Elsevier Ltd.; 2004. Chapter 32- Routes of Administration. [Google Scholar]
- Shulman Z., Cohen S.J., Roediger B., Kalchenko V., Jain R., Grabovsky V., Klein E., Shinder V., Stoler-Barak L., Feigelson S.W. Transendothelial migration of lymphocytes mediated by intraendothelial vesicle stores rather than by extracellular chemokine depots. Nat Immunol. 2011;13:67–76. doi: 10.1038/ni.2173. [DOI] [PubMed] [Google Scholar]
- Stoler-Barak L., Moussion C., Shezen E., Hatzav M., Sixt M., Alon R. Blood vessels pattern heparan sulfate gradients between their apical and basolateral aspects. PLoS One. 2014;9:e85699. doi: 10.1371/journal.pone.0085699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumaria N., Roediger B., Ng L.G., Qin J., Pinto R., Cavanagh L.L., Shklovskaya E., Fazekas De St Groth B., Triccas J.A., Weninger W. Cutaneous immunosurveillance by self-renewing dermal gammadelta T cells. J. Exp. Med. 2011;208:505–518. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo S., Barki N., Jain R., Zulkhernain N.S., Buhner S., Schemann M., Weninger W. Imaging of mast cells. Immunol. Rev. 2018;282:58–72. doi: 10.1111/imr.12631. [DOI] [PubMed] [Google Scholar]
- Tikoo S., Jain R., Martinez B., Song R., Wielscher M., Rizzetto S., Shaw L.E., Mitchell A.J., Torres-Pacheco M.E., Luciani F. A distinct CD115- erythro-myeloid precursor present at the maternal-embryonic interface and in the bone marrow of adult mice. bioRxiv. 2021 doi: 10.1101/2021.07.24.453629. [DOI] [Google Scholar]
- Tikoo S., Jain R., Tomasetig F., On K., Martinez B., Heu C., Stehle D., Obeidy P., Guo D., Vincent J.N. Amelanotic B16-F10 melanoma compatible with advanced 3-D imaging modalities. J. Invest. Dermatol. 2021;141:2090–2094. doi: 10.1016/j.jid.2021.01.025. [DOI] [PubMed] [Google Scholar]
- Tong P.L., Roediger B., Kolesnikoff N., Biro M., Tay S.S., Jain R., Shaw L.E., Grimbaldeston M.A., Weninger W. The skin immune atlas: three-dimensional analysis of cutaneous leukocyte subsets by multiphoton microscopy. J. Invest. Dermatol. 2015;135:84–93. doi: 10.1038/jid.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unutmaz D., Xiang W., Sunshine M.J., Campbell J., Butcher E., Littman D.R. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. Journal of immunology. 2000;165:3284. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- Vennin C., Chin V.T., Warren S.C., Lucas M.C., Herrmann D., Magenau A., Melenec P., Walters S.N., Del Monte-Nieto G., Conway J.R. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl. Med. 2017;9:eaai8504. doi: 10.1126/scitranslmed.aai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.H., Jenne C.N., Lee W.Y., Léger C., Kubes P. Functional innervation of hepatic iNKT cells is immunosuppressive following stroke. Science. 2011;334:101–105. doi: 10.1126/science.1210301. [DOI] [PubMed] [Google Scholar]
- You S., Tu H., Chaney E.J., Sun Y., Zhao Y., Bower A.J., Liu Y.Z., Marjanovic M., Sinha S., Pu Y., Boppart S.A. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun. 2018;9:2125. doi: 10.1038/s41467-018-04470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sample imaging dataset utilized in this study can be obtained from the authors upon request. Analysis was performed utilizing commercially available software, Volocity (Quorum Technologies) and Imaris (Oxford Instruments).