Abstract
The popular recreational drug MDMA (3,4-methylenedioxy-methamphetamine) has a documented potential as a psychopharmacological clinical and research tool. This is due to its unique ability to promote reprocessing of traumatic memories, empathetic and pro-social states. Although it is established that MDMA exerts its behavioural effects via the serotonin transporter (SERT), the ligand-protein molecular interplay remains elusive. In order to shed light on the binding of MDMA and its primary congeneric entactogens (MDA, MBDB and MDAI), we first combined induced fit with Monte Carlo simulations. The computed interaction energies of the models correlated well with experimental activities (adjR2 = 0.78). Then we carried out ‘ensemble binding space docking’ on trajectories generated by interpolation of experimentally derived structures of the hSERT from the outward-open, and the occluded, to the inward-open states. This approach revealed low-energy alternative binding modes, suggesting high occupancy of the central site, yet considerable MDMA mobility within it, favouring the paroxetine-like orientation. Finally, we designed a pharmacophore that may be used to recognise hSERT-mediated serotonin releasers and uptake inhibitors of diverse chemical structure, identifying their active conformations and interacting residues. We conclude that the conserved amine-Asp98 ionic and edge-to-face π-π interactions are crucial to the mode of action of MDMA on the hSERT, underscoring the contributions of Tyr95 and gating residues Phe341, Tyr176 and Phe335. Amenable to experimental testing, our modelling may aid the rational design of novel entactogenic compounds and contribute to the understanding of an action mechanism, common and typical of psychotropic agents.
Keywords: Entactogen, Psychoactive, MDA, MDAI, Symporter
Entactogen, Psychoactive, MDA, MDAI, Symporter.
1. Introduction
Due to its rare and reliable ability to produce pro-social states, reduce fear responses, promote introspection, empathy and beneficial emotional processing, 3,4-methylenedioxy-methamphetamine, MDMA or ‘ecstasy’ is the drug with arguably the most potential to usher in a new progressive era in psychedelic science (Sáez-Briones and Hernández, 2013; Dunlap et al., 2018; Curry et al., 2018; Inserra et al., 2021). Proposed to be under the broader category of ‘psychedelics’ (Dunlap et al., 2018), the drug class ‘entactogen’ was coined to classify MDMA, MBDB (3,4-methylenedioxy-N-methyl-ethylphenylethylamine), MDA (3,4-methylenedioxyamphetamine) and MDAI (5,6-methylenedioxy-2-amino-indane), based on early psychiatric use, animal behavioural and human subjective effects, as well as for its departure from the classical SAR (Structure-Activity Relationship) of hallucinogens (Nichols, 1986).
Presently, the number of controlled and peer-reviewed preclinical and clinical trials in phase II and III (54 completed, 22 active) reflects the therapeutic interest in MDMA (https://www.clinicaltrials.gov/; Inserra et al., 2021). Its most common medical use is to treat anxiety disorders, particularly for treatment-resistant post-traumatic stress disorder (PTSD) (Dunlap et al., 2018) as it is proposed to be: “superior to current pharmacotherapies” by the FDA (Inserra et al., 2021; Mitchell et al., 2021). Moreover, MDMA has a recognized potential to improve social interactions in autism spectrum disorders, social anxiety, schizophrenia and, as a research tool to study the neurochemistry of social behaviours, social bonding and empathy, even in invertebrate species (Heifets and Malenka, 2016; Edsinger and Dolen,2018).
The fundamental effects of MDMA include the reuptake inhibition of dopamine, serotonin, i.e. 5-hydroxytryptamine (5HT) and norepinephrine as well as the reversal of the flux of these monoamines into the extracellular space by binding its presynaptic transporters (DAT, SERT and NET), acting as a substrate. Both mechanism of action are prototypical for drugs of psychiatric use such as SSRIs antidepressants that selectively inhibit the reuptake of 5HT and drugs of abuse such as Methamphetamine that acts as a substrate-type monoamine releaser (Henry et al., 2006; Rothman and Baumann, 2003).
The 5HT transporter (SERT) is a twelve transmembrane segments (TM1-12) neurotransmitter Na+ symporter (NSS) that utilizes the electrochemical gradient of Na+ and Cl− to internalize 5HT back to the presynaptic neuron, which initially occupies the central binding site i.e. S1, comprised by sub-sites A, B and C. NSSs undergo a conformational transition from an outward-open into an inward-open state subsequently releasing the neurotransmitter to the cytoplasm (Navratna and Gouaux, 2019). Indeed, the role of the SERT in the mechanism of action of MDMA is preponderant since the entactogenic effects of this drug are mediated by the release of 5HT (Nichols, 1986; Johnson et al., 1986; Tancer and Johanson, 2007; Sáez-Briones and Hernández, 2013) and neuroimaging reveals that SERT density is decreased in brain regions of chronic MDMA users (Müller et al., 2019). Importantly, there is evidence that the compound-induced substrate translocation (5HT release) and macromolecular conformational changes associated to uptake inhibition critically depend on the initial binding mode of the SERT ligands (Sandtner et al., 2016). Although low homology models have provided hints into the molecular interplay between MDMA and the SERT (Gabrielsen et al., 2012; Sandtner et al., 2016), their binding features remain largely unidentified. The paroxetine-bound X-ray (Coleman et al., 2016) and the more recent Cryo-EM structures of the hSERT (Coleman et al., 2020) offer an invaluable opportunity to ascertain critical MDMA/hSERT interactions to a degree of certainty, due to the close structural similarity between MDMA and the selective serotonin reuptake inhibitor (SSRI) paroxetine.
The purpose of this study was twofold:
-
1)
To gain insight into the binding of MDMA and its primary derivatives on the hSERT, at an atomistic level. This was achieved through models based on experimentally and computationally derived structures by combining up-to-date computational methods, e.g. by applying the concept of ‘ensemble binding space’ an approach used in property space analysis (Vistoli et al., 2017).
-
2)
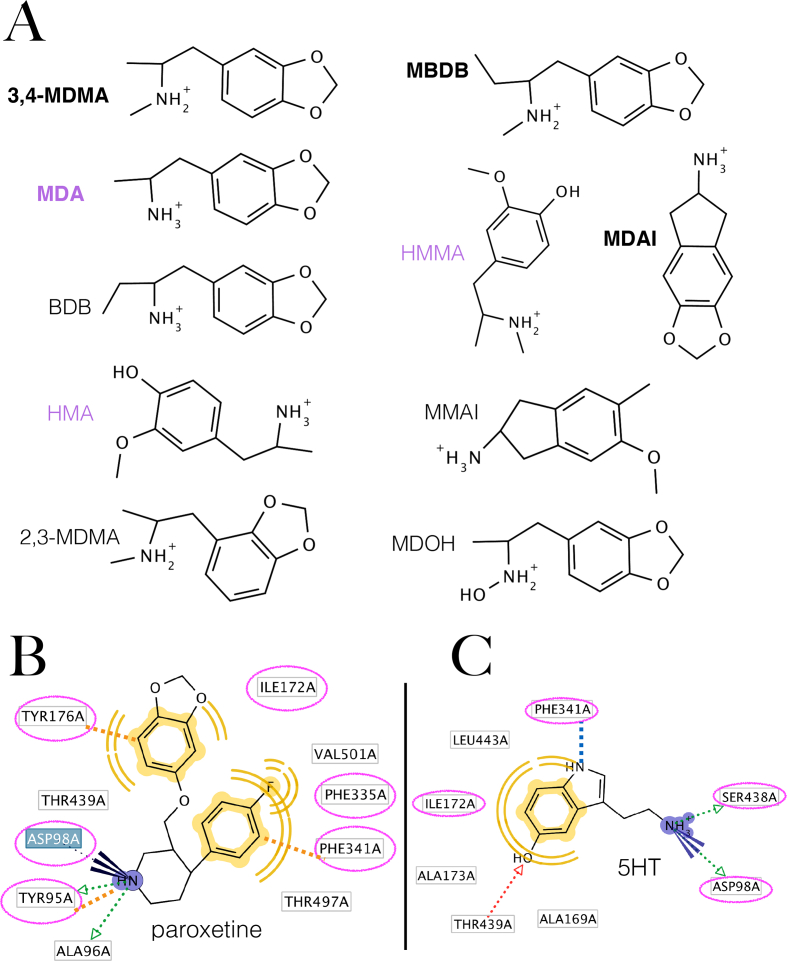
To derive a useful ensemble of the minimal steric and electronic features of MDMA analogues necessary to ensure interaction with the hSERT, i.e. a 3D pharmacophore. To this end, the molecular features of the most essential psychoactive MDMA congeners were combined. Ten compounds were chosen from the literature for their simplicity and relative homogeneity comprising the major metabolites of this drug: 3,4-methylenedioxy-amphetamine (MDA), 4-hydroxy-3-methoxy-amphetamine (HMA) and 3,4-dihydroxy-methamphetamine (HMMA) (in purple Figure 1A) and well-studied entactogens (Figure 1A) (Sáez-Briones and Hernández, 2013; Dunlap et al., 2018).
Figure 1.
Molecules chosen to generate the pharmacophore and 2D representation of the binding mode of paroxetine and serotonin (A) Selected SERT active, MDMA congeneric compounds to design the structure-based pharmacophore. Molecules in bold are the first described entactogens (Nichols, 1986) and compounds in purple are primary metabolites of MDMA. (B) Pharmacophore of paroxetine to the hSERT calculated from its intact observed complex PDB: 6VRH. Computed residues that also bind MDMA (vide infraFigure 3B) are in circles in magenta. (C) Pharmacophore of serotonin on the hSERT generated in the present work. Discontinuous blue line indicates a cation-π interaction, dotted green arrows represent Hbond (donors), blue rays represent an ionic interaction, yellow moieties and waves represent hydrophobic interactions and discontinuous orange lines indicate a cation-π with Y95 and two edge-to-face π-π interactions with Y176 and F341.
The pharmacophore of the intact paroxetine molecule bound to the hSERT is shown for comparison (Figure 1B). The binding features of 5HT, previously elucidated via induced fit and microsecond-long MD simulations by Hellsberg et al. (2019) were reproduced satisfactorily with our induced fit/Monte Carlo approach (Figure 1C).
2. Methods
2.1. Induced fit docking
The atomic coordinates of the hSERT were retrieved from the Protein Data Bank (https://www.rcsb.org/) Protein and ligands were prepared for docking with Chimera1.15 (Pettersen et al., 2004) adding hydrogen atoms, optimizing Hbonds, removing clashes, adding missing sidechains from a rotamer library. Atomic partial charges were assigned under AMBER14SB or MMFF94. Protonation states were verified under the Cresset rules for a pH of 7 in Flare 4.0 (Cresset Biomolecular Discovery Ltd). Flexible-ligand, rigid-receptor dockings were carried out with AutoDock Vina1.1.2 in Chimera1.15 (Pettersen et al., 2004) or in LigandScout 4.4.5 (Inte:Ligand GmbH). The active site was defined either by a grid of 20 × 20 × 20 Å from the ether scaffold group of paroxetine, or by LigandScout 4.4 default environment of approximate diameter of 27Å. Induced fit binding was simulated by successive cycles of either adjustment of bond angles and torsions in Flare 4.0 (Cresset Biomolecular Discovery Ltd), rotamer replacement from the Dunbrack rotamer library (Dunbrack, 2002) and/or energy MMFF94 minimizations in LigandScout 4.4.5 or in Chimera1.15 under AMBER14SB.
2.2. Monte Carlo simulations, interpolation of structures and ensemble docking
Independent conformational search simulations by random perturbation of the torsional angles were carried out in VEGA ZZ 3.0.5 using the Boltzmann jump Monte Carlo method in AMMP 2.4.1(c) 1993–2014 at a temperature of 1000K, with a torsion RMSD of 60° to generate significantly different conformations at each step, followed by 20 minimization steps. This method allows upward jumps in energy to explore the conformational landscape and employs the Metropolis criterion to accept or reject perturbed conformations.
Since we focused on elucidating the most favourable binding modes at the central site of the hSERT rather than on the transport mechanism of the ligand, as an alternative to the costly long MD simulations we used the independent structure interpolation method in Chimera1.15. Trajectories comprising 80 energy-minimized intermediates between the structures: 5I71, 6VRH, 6DZV and 6DZZ were generated at a sinusoidal rate and one every ten was used to dock the ligand to sample a precise and smooth conformational landscape. A docking grid of 30 × 30 × 30Å from the carboxylate of Asp98 was used. The ‘binding space’ of the whole sample (N = 80) was parameterized by calculating the score mean (Sum of scores/N), score range (|Scoremax|–|Scoremin|) and score sensitivity (range/# of active torsions), as proposed by Vistoli et al. (2017).
2.3. Pharmacophore modelling and alignment
Structure-based ‘apo’ and ‘holo’ pharmacophore modelling was carried out with LigandScout 4.4.5 (Inte:Ligand GmbH) (Wolber and Langer, 2005). We first calculated the ‘apo’ side grids to generate the so called ‘apo’ pharmacophore of the entire central site of the hSERT. For an adequate comparison with our combined pharmacophore of carefully selected ligands, it had previously been energy-minimized in the presence of MDMA at the paroxetine location. Then, the pharmacophores were individually generated for each MDMA derivative, based on the latest hSERT structure (PDB:6VRH), via the same induced fit docking protocol i.e. accounting for ligand mobility and ligand-induced Prot-Lig conformational rearrangements within the binding environment. LigandScout 4.4.populates each molecule with diverse conformations after ranking molecules according to their flexibility, it projects pharmacophore features on these molecules and all their conformations. Two top ranked molecules are chosen, i.e. the least flexible and all of their conformations are aligned employing the Inte:Ligand's molecular alignment algorithm. If at least 3 common chemical features can be identified throughout the whole alignment the feature pharmacophore combination is considered to be successful.
2.4. Field and shape properties of ligands
Field points, MEPs, vdWs and hydrophobics were calculated in Flare 4.0 (Cresset Biomolecular Discovery Ltd) with ‘Molecular Field Technology’ by Cresset™ that redefines the charge towards multipole electron distribution akin to a quantum orbital description under the second generation Extended Electron Distribution (XED) force field, unlike conventional monopole atom-centred (point-charge) force fields (Cheeseright et al., 2006; Marshall, 2013). Field points are three-dimensional molecular descriptors as extrema of positive and negative electrostatic, ‘shape’ van der Waals (vdW), and ‘hydrophobic’ fields (a density function correlated with steric bulk and hydrophobicity) (Cheeseright et al., 2006).
3. Results and discussion
3.1. Binding mode overlap between MDMA and experimentally-derived paroxetine on the hSERT
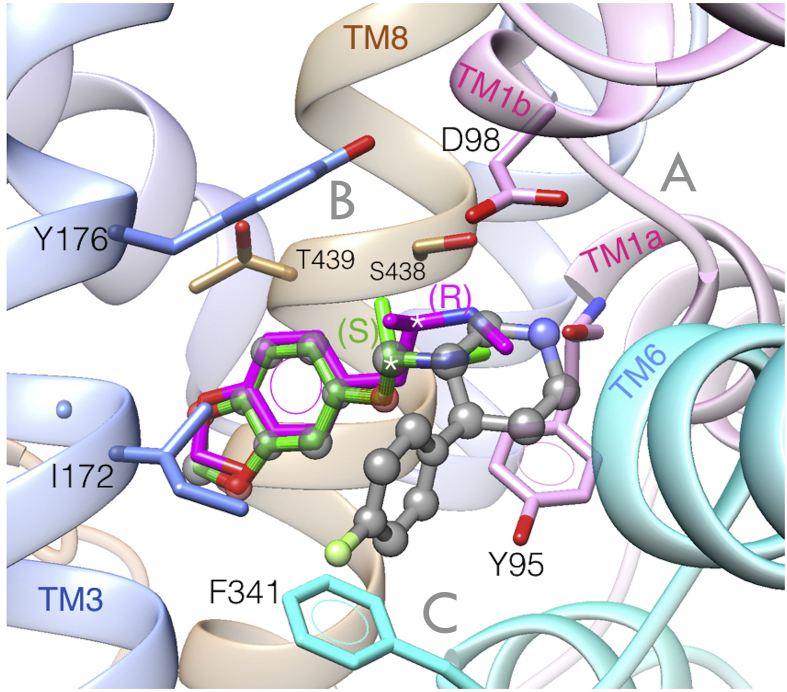
As an preliminary logical approach to delineate the binding of MDMA on the hSERT, we carried out a simple docking of (R)- and (S)-MDMA to the orthosteric central site S1 of the hSERT and the lowest energy solutions indicate that both enantiomers conspicuously overlap paroxetine (Figure 2). Unsurprisingly, the methylammonium of (R)-MDMA forms a salt bridge with Asp98, a conserved feature for substrate and uptake inhibitors, observed in all ligand-bound structures of NSSs (Sandtner et al., 2016; Navratna and Gouaux, 2019). This amine also forms an Hbond with the backbone of Tyr95 at sub-site A, while their benzodioxole ring forms an edge-to-face π-π interaction with Tyr176 of TM3. Additionally, (R)-MDMA may form an Hbond with the sidechain of S438 at sub-site B, i.e. on TM8. The benzodioxole of MDMA lodges between the side chains Ile172, Tyr176 and Thr439, but unlike paroxetine, neither isomer interacts with sub-site C, i.e. under the ‘Lock and key binding model’ (Figure 2). However, this binding pose may only reflect the initial encounter complex based on tentative collisions due to long-range electrostatic recognition events (Du et al., 2016). Hereafter, we delineate the chemical space of dextro-isomers, as (R)-MDMA is the most promising enantiomer for clinical use in terms of safety because it seems to promote social behaviour without producing hyperthermia or neurotoxicity in mice, possibly due to lower dopamine release (Curry et al., 2018; Inserra et al., 2021).
Figure 2.
Top docking solutions of (R)-MDMA (in magenta) and (S)-MDMA (in green) at the central site of the human 5HT transporter (hSERT) superimposed to the Cryo-EM structure bound to paroxetine (in translucent grey) PDB: 6VRH. Of note, the 1,3-benzodioxol moieties of the MDMA isomers overlap exactly on the benzodioxol of paroxetine and can distinguished by its grey coloured translucent atom balls. The asterisks are at the chiral centre of each isomer. Sub-sites A, B and C are in grey.
MDMA simultaneously docked the orthosteric S1 and the allosteric site S2 at the extracellular vestibule on the hSERT structure in lieu of the two molecules of (S)-citalopram (PDB: 5I73) (supplementary figure S1). Because the binding to the orthosteric site determines the mode of action of MDMA and its homologues (Sandtner et al., 2016) and due to the structural similarity of MDMA with paroxetine, this study is focused on the orthosteric central binding site S1.
Next, to uncover the most plausible interactions and evaluate their stability while accounting for local and global protein flexibility, we combined induced fit modelling with Monte Carlo (MC) simulations on the recognition MDMA/hSERT complex.
3.2. MDMA double bonds with Tyr95 on TM1 and with Tyr176 on TM3, residues involved in substrate recognition
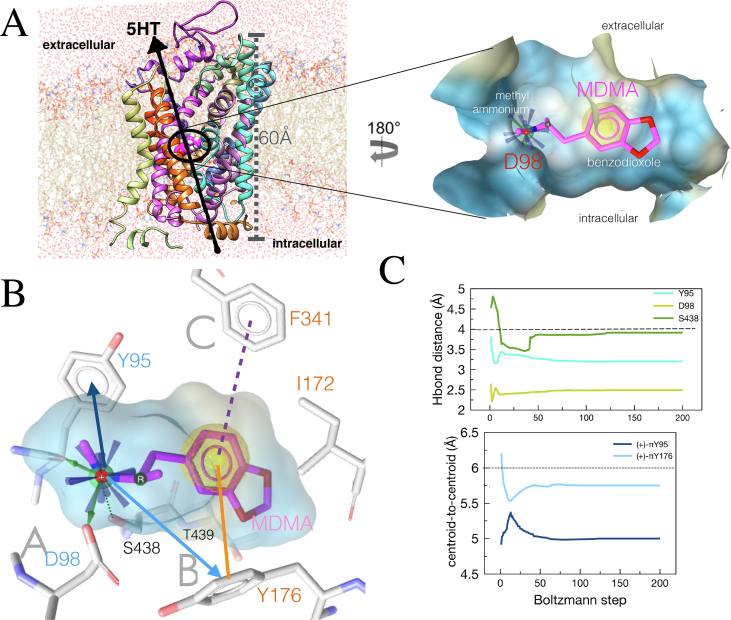
MC simulations show in Figure 3A the relative position of this drug on the hSERT embedded in a lipid bilayer and the insert illustrates the polarity of its binding cavity upon induced fit.
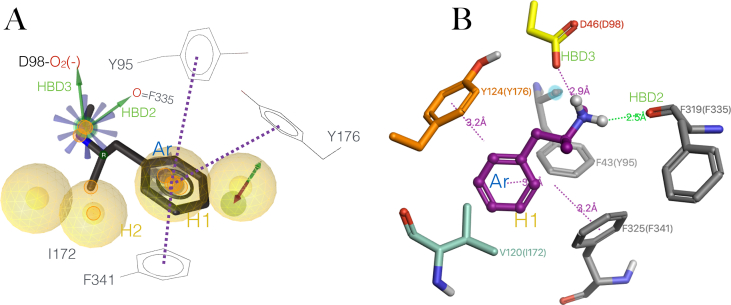
Figure 3.
Induced fit binding mode of the MDMA/hSERT complex. (A) The hSERT embedded in a DPPC bilayer with explicit solvent by MemProtMD (Newport et al., 2019), MDMA in magenta is docked perpendicular to the y-axis of the transporter, and the arrow indicates the drug-induced reverse transport of serotonin. The insert shows the binding cavity coloured by Aggregated Lipophilicity/Hydrophobicity (non-polar regions in yellow). (B) Induced fit binding mode of the MDMA/hSERT complex. Interactions observed during Monte Carlo (MC) simulations: Cation-π interactions are in blue arrows. Green arrows represent Hbonds. Blue rays represent a positive ionisable moiety. The yellow sphere represents hydrophobic interactions. The line in orange and the discontinuous purple line represent π-π interactions. Sub-sites are in grey letters. (C) A typical trajectory of MC simulations on the MDMA/hSERT system. The paroxetine molecule was replaced by a molecule of MDMA (PDB: 6VRH) and subjected to stochastic conformational search with the Boltzmann jump method. Limits (cut-off) distances to define an interaction are shown in discontinuous black lines.
The results along the MC trajectories show that, in an optimized, developed complex, the methylated ammonium moiety of MDMA may interact with four residues (Figure 3B), including the ‘tripod grip’ conserved in the D. melanogaster dopamine transporter dDAT, comprising Tyr95 and Asp98 in sub-site A and Phe341 from sub-site C (Navratna and Gouaux, 2019). The donor-to-acceptor distances along the Boltzmann energy jumps indicate a strong salt bridge with Asp98, a moderate Hbond with the backbone of Tyr95 and an additional weak Hbond with the sidechain of Ser438. Interestingly, this moiety is also capable of forming two cation-π interactions with Tyr95 and Tyr176 (blue lines in Figures 3B and 3C) and or an edge-to-face π-π interaction via its benzol ring with the latter and with Phe341. Finally, Thr439, Ile172, Tyr176 and Phe341 contribute to accommodate the half hydrophobic half polar benzodioxole tail group of MDMA at the orthosteric S1 (see interactions in common with the intact paroxetine structure in Figure 1B and the verified in silico model of 5HT in Figure 1C).
The double bonding with Tyr95 may be crucial for the mechanism of action of MDMA as this residue mediates substrate recognition and it is implicated in reuptake inhibition binding, particularly in the stereoselectivity of citalopram (Henry et al., 2006). Similarly, the simultaneous ionic, cation-π and π-π stacking with Asp98 and Tyr176 respectively, may play a role in the mode of action of MDMA, since the Hbond between these two residues is also decisive for substrate recognition (Navratna and Gouaux, 2019).
To support our induce fit modelling methods and docking energy calculations, we next tested the hypothesis of whether the computed intermolecular energies of a set of entactogenic compounds could correlate to their experimental efficacy activities.
3.3. Quantitative correlation between experimental data and induced fit models of MDMA metabolites with the hSERT
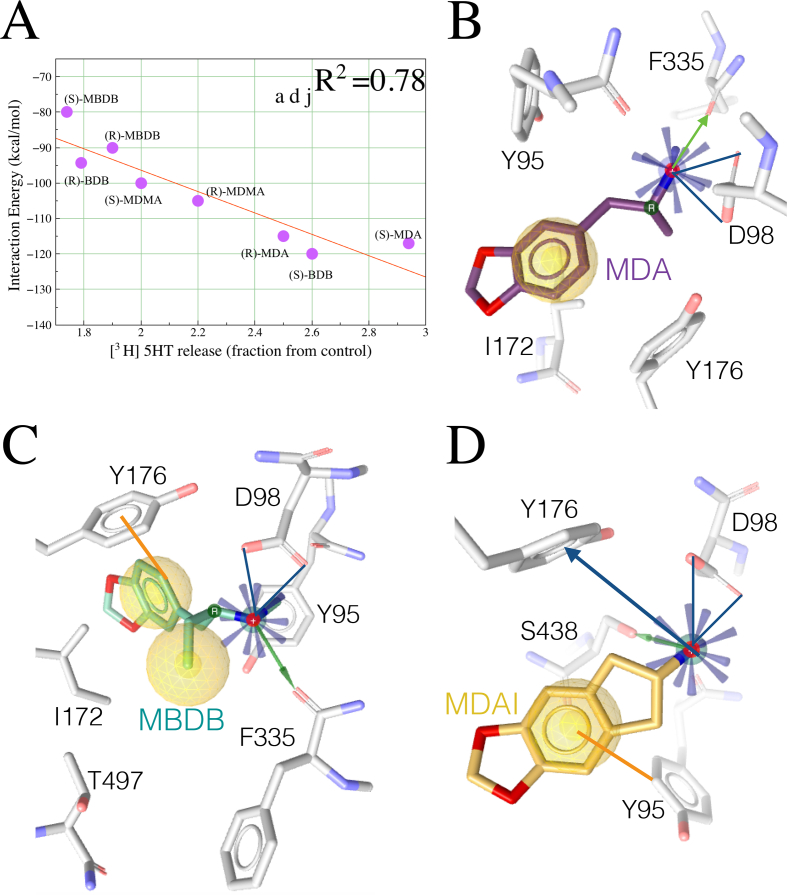
Indeed there was a clear trend of the Coulombic and vdW terms towards partially explaining the efficacy of tritiated neurotransmitter release in rat brain slices (Johnson et al., 1986). To avoid the risk of chance correlation, we calculated the coefficient of determination, which was adjusted for the number of independent explanatory terms relative to the number of data points (adjR2) (Figure 4A). The correlation of the in silico interaction energies with the experimental data was possible despite interspecies variations (92% identities between the hSERT and the rSERT), because the MDMA active site of the human and rat SERT (within 5.5Å) are identical.
Figure 4.
Correlation between experimental and computed data and induced fit models of MDMA derivatives. (A) Linear correlation: y(x) = a∗x + b between experimental evidence and computed results of the main MDMA congeneric entactogens. Binding modes resulting from Monte Carlo simulations of (B) MDA, a primary metabolite of this MDMA, and entactogens (C) MBDB and (D) MDAI Cation-π interactions are in blue arrows. Green arrows represent Hbonds. Blue rays represent a positive ionisable moiety. Yellow spheres represent hydrophobic interactions. Lines in orange represent π-π interactions.
Next, we proceeded to analyse the binding modes of the (R)-enantiomers of the main derivatives of MDMA. In contrast to MDMA, MDA and MBDB optimizes the salt bridge with Asp98, allowing their primary and secondary amine to Hbond with F335 of sub-site C at TM6, at the expense, in the case of MDA, of loosing π-π interactions. On the contrary MBDB shares this key interaction with Tyr176, favourably accommodating its ethyl side chain. Ile172 seems to anchor the benzodioxol moieties of MDMA, MDA and MBDB, but not MDAI, similarly than in the ibogaine-bound complex (Figures 4B and 4C). Nevertheless, the bulkier mutant I172M recognizes multiple inhibitors by significantly decreasing their potency, sparing substrates like 5HT and MDMA (Henry et al., 2006). The shorter molecule of the entactogen MDAI also forms a strong salt bridge with Asp98 concomitant to an edge-to-face π-π with Tyr95, a cation-π with Tyr176 and an Hbond with Ser438 (Figure 4D).
These models may be of clinical relevance, e.g. MDA showed a stronger ionic interaction and greater energy than MDMA, which correlated with its higher in vitro activity may contribute to explain why the human subjective effects of this metabolite last longer that those of MDMA, as it is believed that this is attributed to its mechanism of action rather than to its pharmacokinetics (Baggott et al., 2019).
Next, to provide a more in-depth characterization of the binding of MDMA throughout the transitions of the hSERT by making use of the available structural data, we carried out ‘ensemble binding space docking’. Briefly, this incorporates the statistically confirmed idea that alternative binding modes and the degree and ease of mobility of a ligand within a binding site significantly contributes to the observed affinity (Vistoli et al., 2017). This approach also accounts for the dynamic processes of protein flexibility and ligand mobility by analysing and parameterizing multiple binding poses on various representative protein conformations.
3.4. Binding space analysis of MDMA along the transition of hSERT from the outward-open to the inward-open conformation
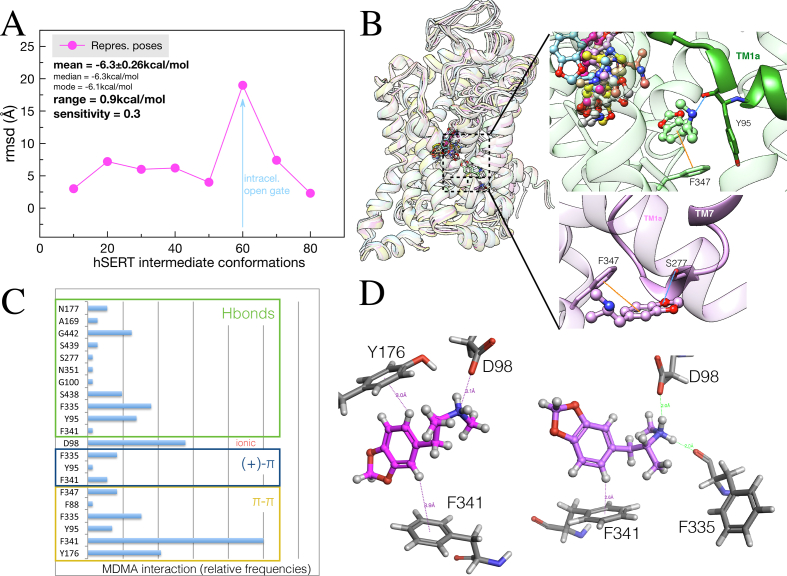
We analyse MDMA binding along the transition from the outward-open and occluded to the inward-open states of the hSERT (see Methods section 2.2 for details). The mean and range scores denote a fair degree of homogeneity in docking energies, which implies that alternative binding modes are low-energy and may play a part in the mechanism of action of MDMA. Therefore every binding pose was clustered irrespective of its ranking order. The sensitivity score encodes the capacity of a given ligand to vary its docking scores by adjusting its own conformation. To visualize the degree of mobility of MDMA within the central site every docking solution was clustered and the rmsd between the most populated representatives were calculated consecutively (Figure 5A).
Figure 5.
Ensemble binding space analysis of MDMA on the hSERT. (A) Parameterized ‘Ensemble binding space docking’ of MDMA on interpolated trajectories between two hSERT outward-open, one occluded and an inward-open structure (PDBs: 5I71, 6VRH, 6DZV and 6DZZ). Data points (in magenta) represent the rmsd values between the most populated representative docking solutions at each intermediate model. The blue arrow represents the migration of MDMA towards the open gate at the cytoplasmic side. (B) Representative MDMA/hSERT binding complexes, the inserts show the interactions of the most populated binding solutions closest to the inward gate. π-π stackings are in orange and in Hbonds in blue. (C) Relative frequencies of the residues interacting electronically with MDMA along the interpolated trajectories, i.e. calculated from representative binding poses from dockings simulations carried out in 1 every 10 intermediate conformations. Every docking solution (N = 80) was clustered into 8 representative biding poses. (D) Two representative binding poses illustrating the most frequent interactions.
In general, the ‘degree of binding space exploration’ i.e. displacement from the binding site (Figures 5A and 5B) and the repertoire of bonding residues (15 in total) (Figure 5C) was not large considering the size of the docking grid. This indicates a high occupancy of the outward central binding site by MDMA with a rich exploration of alternative binding modes e.g. 30% of the observed conformations were ‘flipped’ from tail to head groups, as previously reported (Sandtner et al., 2016) and binding modes parallel to the Y axis of the transporter were also representative.
The frequency plot in Figure 5C represents the bonding contacts of the ‘ensemble binding space’ analysis. This approach allowed capturing MDMA as a substrate albeit partially, moving towards the cytoplasmic side at the open internal gate (outlier in Figure 5A). The inserts in Figure 5B highlight the role of Phe347 in this migration showing MDMA above and below this residue interacting aromatically. In fact, the top three interactions occurred between gating residues Tyr176, Phe335 and Phe341 that allow the formation of the permeation pathway into the cytoplasm (Coleman et al., 2019). While, Phe335 formed an Hbond with the secondary amine of MDMA (Fig. 5D), a residue that blocks the release of reuptake inhibitors such as ibogaine, S-citalopram and paroxetine via its aromatic side chain (Coleman et al., 2019; Möllet et al., 2019; Coleman et al., 2020). Of note, the orientation of Phe335 remains the same for all inhibitor bound structures, with the exception of sertraline, which forms an edge-to-face π-π with its double ring system, a relatively frequent analogous feature with MDMA identified by the binding space analysis (Figure 5 C). Thus, it is likely that this interaction is implicated in the mode of action of MDMA as a 5HT reuptake inhibitor.
Importantly, aromatic bonds were more frequent than Hbonds or ionic interactions and two simultaneous π-π interactions between sub-sites B and C were common (Figure 5C). These results indicate that aromatic interactions may be just as important to the binding of MDMA as the amine-SERT ionic interaction with Asp98 (Sandtner et al., 2016).
Because sodium (two Na+) ions are indispensable for substrate transport and binding 5HT uptake inhibitors (Coleman et al., 2016; Hellsberg et al., 2019), we explored the binding of MDMA in the presence of Na+ by cloning the coordinates of the two Na+ ions from the ts3 hSERT (PDB: 5I71) into our system, followed by 3000 steps of Steepest descend energy minimization before docking, similarly to the procedure carried out by Hellsberg et al. (2019) for 5HT. The binding pose orientation obtained in the presence of the two Na+ and Cl− (Supplementary Fig S2) was that of our model (Figure 3B) with a slight change in the binding mode, involving interactions accounted for in the ‘space binding analysis’ (Figure 5C).
The second goal of this work was to design a 3D pharmacophore from the most basic MDMA congeneric compounds on the hSERT, capable of identifying active molecules. Such structure-based representation of the essential features of hSERT binders may then be helpful to the medicinal chemistry research of agents with desirable psychotropic effects, e.g. antidepressants and pro-social empathogenic compounds. The next sections are dedicated to this end.
3.5. ‘Unliganded’ and structure-based merged pharmacophore design of primary MDMA derivatives
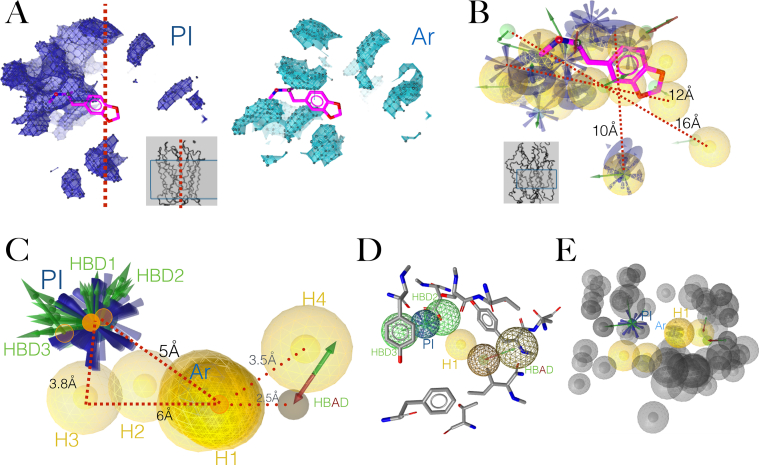
Since positive ionisable (PI) and aromatic (Ar) features seemed to mediate MDMA binding, we calculated the interaction potential of these two features and projected them on the ‘Apo site grids’ of the hSERT/MDMA energy-minimized complex. The localized density of the PI potentials agrees with the ensemble binding space analysis, suggesting that the ‘orthosteric orientation’ of MDMA is favoured, while the Ar densities illustrates the likelihood of the ligand forming simultaneous π- π interactions (Figure 6A). For comparison the unliganded (or ‘Apo’) pharmacophore i.e. a complete description of potential features disregarding the ligand, is shown in Figure 6B. Comprising 45 features, this is an exhaustive and complex, yet static representation of ligand binding potentials.
Figure 6.
Unliganded (‘Apo’) and ‘merged’ designed pharmacophores of the hSERT (A) Regions of the hSERT with positive ionisable (in blue) and aromatic π-π (in cyan) potential interactions. (B) ‘Apo pharmacophore’ of the hSERT. The visualised zones are inside a rectangle in the grey inserts. (C) Designed induced fit structure-based pharmacophore. (PI.- positive ionisable, HBDs.- Hbond donors, Ar.-aromatic interaction, H.-hydrophobic interactions, HBAD.- Hbond donor/acceptor. (D) Tolerance spheres for electrostatic interactions of the pharmacophore in C. (E) Exclusion volumes for the pharmacophore in C.
Instead, our pharmacophore model was obtained after individual induced fit modelling, (10 features). This 3D hypothesis offers a dynamic, clearer, more specific representation of the proposed stereoelectronic properties required to bind the hSERT (Figure 6C). The tolerance spheres for electrostatic interactions are shown in Figure 6D, while Figure 6E shows in grey the exclusion volume spheres i.e. the zones of the binding environment prone to steric clash by a ligand.
3.6. Testing the merged pharmacophore with diverse SERT-active compounds
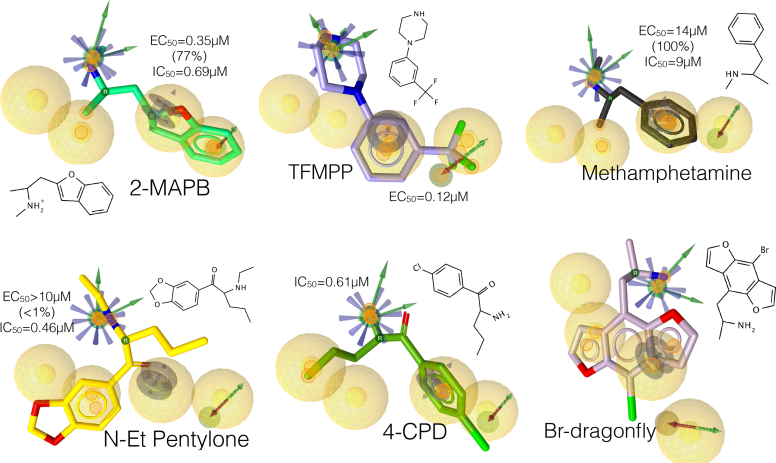
We selected compounds of different chemical class with known hSERT activities (Eshleman et al., 2019), to test our merged pharmacophore hypothesis. Every molecule tested met the minimal chemical and spatial requirements to fit the model (Figures 7, 9B and 9C), suggesting that this designed pharmacophore can recognize both 5HT reuptake inhibitors (IC50) and 5HT releasers (EC50) of certain structural diversity, thereby providing the most likely i) bioactive conformations of a range of active SERT binders, ii) the nature and iii) the location of the interactions with and on the receptor, i.e. the residues and functional groups involved in protein-ligand recognition.
Figure 7.
‘Test’ active drugs at the hSERT aligned to the designed pharmacophore in Figure 6C. Their 2D structure and 5HT release (EC50) and uptake inhibition (IC50) activities are associated to each compound. The small orange circles on the compounds represent the matched feature pairs that aligned to the pharmacophore. To our knowledge, there is no data on Br-dragonfly, although the pharmacophore fit predicts it may be active at the hSERT.
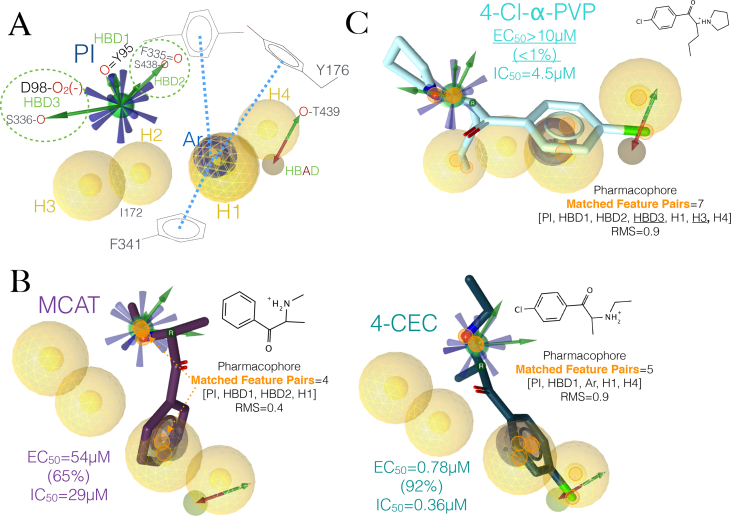
Figure 9.
Molecular determinants of the proposed pharmacophore and ‘test’ molecules aligned to this pharmacophore. (A) Pharmacophore hypothesis associated to the functional groups of the interacting residues. (B) Two hSERT active, 5HT uptake inhibitors and releasers aligned to the pharmacophore in A. (C) A hSERT active, 5HT uptake inhibitor that does not cause the release of 5HT. The EC50 values and those in parenthesis represent the potencies and efficacies of 5HT release, respectively. The IC50 values represent the potencies of 5HT reuptake inhibition. The small orange circles on the compounds represent the matched feature pairs that aligned to the pharmacophore: PI.-positive ionisable, HBD.-Hbond donor, H.-hydrophobic.
Remarkably, the alignment of Methamphetamine with the hSERT 3D merged pharmacophore accurately predicted the crystallographic binding features of this drug (at 3.10Å resolution) to the Drosophila melanogaster dopamine transporter (dDAT). Methamphetamine on the hSERT model (Figure 8A) and on the crystal on dDAT (Figure 8B) has the same exact type of interactions and functional groups involved with the exact number of corresponding residues, despite a sequence identity of 52%. Of note, the pharmacophore alignment also predicted the lack of the backbone-to-amine Hbond with the residue corresponding to Tyr95 (HBD1), highlighted in cyan in Figure 8B. This binding mode may be relevant for substrate-type monoamine releasers in other transporters, since MDMA and Methamphetamine elevate dopamine, norepinephrine and 5HT concentrations through this mechanism, albeit with different potencies (Rothman and Baumann, 2003). For a complete description of interacting residues of the designed pharmacophore see Figure 9A.
Figure 8.
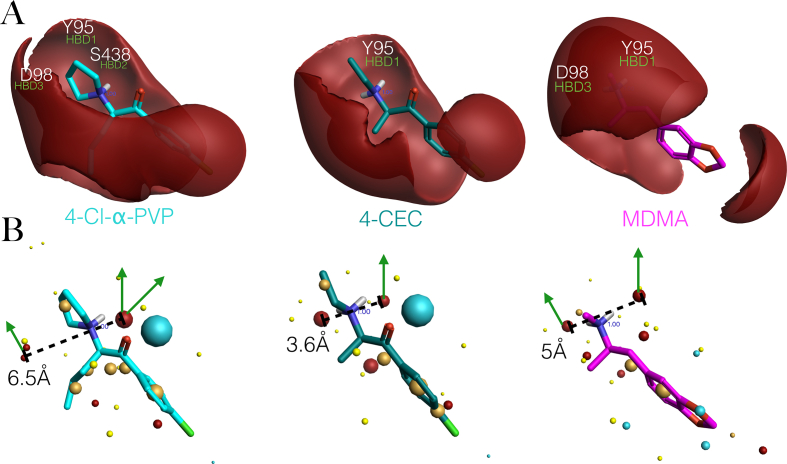
Binding modes of Methamphetamine on the hSERT and on the dDAT (A) Proposed active conformation of Methamphetamine in black, stick representation aligned to the pharmacophore hypothesis based on MDMA congenerics to the hSERT, showing the interacting residues and functional groups. HBDs.- Hbond donors in green arrows, Ar.-aromatic π-π interactions in discontinued purple lines, H.- hydrophobic interactions in yellow spheres. (B) Binding mode of Methamphetamine (in purple) from the X-ray complex with D. melanogaster dopamine transporter (PDB: 4XP6), the residues are numbered from the original structure and in parenthesis the corresponding residues of hSERT upon sequence alignment. Interactions associated to their distances. In purple edge-to-face π-π stackings and a salt bridge with the acidic residue (in yellow), an Hbond in green, some features of the pharmacophore are identified e.g HBD3, HBDS, yet highlighted in cyan the absence of HBD1 with the side chain of residue corresponding to Tyr95.
It is possible that efficient SERT reuptake inhibitors that lack potency and efficacy to induce the reverse flow of 5HT may bind more tightly at the central site than those that have balanced EC50 and IC50 activities. For instance, the psychoactive, ‘second generation bath salt’ 4-Cl-α-PVP, practically inactive in [3H]5HT release assays, has more theoretical favourable non-covalent interactions with the transporter than the structurally related MCAT and 4-CEC, as assessed by the number of computed matched pharmacophore feature pairs (Figures 9B and 9C). Additionally, it can be derived from the model that the reason behind the drastic activity loss of 4-Cl-α-PVP may be two-fold: i) the ring arrangement of its tertiary ammonium allows three potential Hbonds and ii) the occupation of a hydrophobic cavity (H3) by its n-butyl chain is flanked by Tyr95 and Ile172 of TM1a and TM3, respectively.
The calculated positive electrostatic fields and ‘Field points’ of the two cathinones and MDMA (aligned for comparison) in Figures 10A and 10B, respectively can explain the difference in their Hbond donor behaviour on the pharmacophore hypothesis. The more directional, larger but discontinuous positive field of 4-Cl-α-PVP compared with the more compact and smooth field of structurally related 4-CEC, makes the cyclic pyrrolidinyl group of the former, able to Hbond with three residues of the hSERT. In fact, the relative volume and the degree of spread between the +ve Field points of the three molecules are proportional to the number of Hbonds in each case (Figure 10B).
Figure 10.
Computed electrostatic potentials and ‘Point field’ representation of hSERT active compounds. (A) Positive electrostatic fields of each compound with the relative position of the residues with which they form Hbonds according to their alignment to the designed pharmacophore. (B) ‘Field points’ of each molecule in A representing the maxima and minima of steric and electrostatic potentials. The distances between the +ve ‘Field points’ of their ammonium moieties illustrate their ability to form each Hbond (in green arrows).
Of Note, the pharmacophore is limited in that it may only find the ‘high-affinity binding mode’, i.e. being biased towards fitting the amine ionic and Hbonds interactions and not the (inverted) ‘low-affinity binding mode’ estimated by Sandtner et al. (2016). However, this designed pharmacophore agrees with and builds on the theoretical findings of Eshleman et al. (2019).
Taken together, our models provide new insights into a prototypical mechanism of action and a molecule of current relevance. This work may have potential clinical implications as far as it may guide new experiments to improve the design of entactogenic molecules. Perhaps more importantly, the pharmacophore may not only be able to recognise compounds with such properties but also molecules, active at the hSERT that may serve as novel serotonin reuptake inhibitors.
4. Conclusion
Our computed results indicate that, at the outward-open conformation of the hSERT, MDMA occupies the allosteric extracellular site S2 and the central site S1. The orientation of this drug coincides with that of the structurally related paroxetine. Primary congeneric entactogens MDA, MBDB and MDAI essentially retain the binding mode of MDMA, forming an Hbond with Phe335 or Ser438. MDMA may explore alternative low-energy binding modes within the same binding pocket with a relatively small repertoire of bonded residues along interpolated trajectories of the outward-open, and occluded to the inward-open conformations of the hSERT. The calculated frequency of MDMA/hSERT π-π stackings was higher than that of Hbonds indicating a significant interaction with Phe341, Tyr176 and Phe335 (in ranking order), i.e. residues that form the principal permeation pathway. In common with paroxetine, in addition to the highly conserved amine-Asp98 salt bridge, MDMA may concurrently form an Hbond and a cation-π interaction with Tyr95, a residue involved in substrate recognition. This drug may migrate from or into the central site via aromatic interactions with Phe347. A ten feature, structure-based 3D pharmacophore was designed by merging the stereoelectronic properties of selected MDMA metabolites and derivatives upon induced fit to the hSERT. This pharmacophore hypothesis can recognize 5HT reuptake inhibitors and releasers of diverse chemical class. This work may pave the way to the design of more potent hSERT binders with potentially attractive psychoactive effects of interest in the clinic or to neuropharmacological research.
Declarations
Author contribution statement
Ángel A. Islas: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Thomas Scior: Analyzed and interpreted the data; Wrote the paper.
Laura G. Moreno: Analyzed and interpreted the data.
Funding statement
Dr Ángel A. Islas was supported by Consejo Nacional de Ciencia y Tecnología and Benemérita Universidad Autónoma de Puebla.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Ángel A. Islas, Email: angel.islasn@correo.buap.mx.
Thomas Scior, Email: thomas.scior@correo.buap.mx.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Baggott M.J., Garrison K.J., Coyle J.R., Galloway G.P., Barnes A.J., Huestis M.A., Mendelson J.E. Effects of the psychedelic amphetamine MDA (3,4-methylenedioxyamphetamine) in healthy volunteers. J. Psychoact. Drugs. 2019 Apr-Jun;51(2):108–117. doi: 10.1080/02791072.2019.1593560. [DOI] [PubMed] [Google Scholar]
- Cheeseright T., Mackey M., Rose S., Vinter A. Molecular field extrema as descriptors of biological activity: definition and validation. J. Chem. Inf. Model. 2006 Mar-Apr;46(2):665–676. doi: 10.1021/ci050357s. [DOI] [PubMed] [Google Scholar]
- Coleman J.A., Green E.M., Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016 Apr 21;532(7599):334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.A., Navratna V., Antermite D., Yang D., Bull J.A., Gouaux E. Chemical and structural investigation of the paroxetine-human serotonin transporter complex. Elife. 2020 Jul 3;9 doi: 10.7554/eLife.56427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry D.W., Young M.B., Tran A.N., Daoud G.E., Howell L.L. Separating the agony from ecstasy: R(-)-3,4-methylenedioxymethamphetamine has prosocial and therapeutic-like effects without signs of neurotoxicity in mice. Neuropharmacology. 2018 Jan;128:196–206. doi: 10.1016/j.neuropharm.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Ji X.L., Liu S.Q. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016 Jan 26;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack R.L., Jr. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002;12(4):431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Dunlap L.E., Andrews A.M., Olson D.E. Dark classics in chemical neuroscience: 3,4-methylenedioxymethamphetamine. ACS Chem. Neurosci. 2018 Oct 17;9(10):2408–2427. doi: 10.1021/acschemneuro.8b00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsinger E., Dölen G. A conserved role for serotonergic neurotransmission in mediating social behavior in Octopus. Curr. Biol. 2018 Oct 8;28(19):3136–3142. doi: 10.1016/j.cub.2018.07.061. e4. [DOI] [PubMed] [Google Scholar]
- Eshleman A.J., Nagarajan S., Wolfrum K.M., Reed J.F., Swanson T.L., Nilsen A., Janowsky A. Structure-activity relationships of bath salt components: substituted cathinones and benzofurans at biogenic amine transporters. Psychopharmacology (Berl) 2019 Mar;236(3):939–952. doi: 10.1007/s00213-018-5059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen M., Ravna A.W., Kristiansen K., Sylte I. Substrate binding and translocation of the serotonin transporter studied by docking and molecular dynamics simulations. J. Mol. Model. 2012 Mar;18(3):1073–1085. doi: 10.1007/s00894-011-1133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifets B.D., Malenka R.C. MDMA as a probe and treatment for social behaviors. Cell. 2016 Jul 14;166(2):269–272. doi: 10.1016/j.cell.2016.06.045. [DOI] [PubMed] [Google Scholar]
- Hellsberg E., Ecker G.F., Stary-Weinzinger A., Forrest L.R. A structural model of the human serotonin transporter in an outward-occluded state. PloS One. 2019 Jun 28;14(6) doi: 10.1371/journal.pone.0217377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry L.K., Field J.R., Adkins E.M., Parnas M.L., Vaughan R.A., Zou M.F., Newman A.H., Blakely R.D. Tyr-95 and Ile-172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J. Biol. Chem. 2006 Jan 27;281(4):2012–2023. doi: 10.1074/jbc.M505055200. [DOI] [PubMed] [Google Scholar]
- Inserra A., De Gregorio D., Gobbi G. Psychedelics in psychiatry: neuroplastic, immunomodulatory, and neurotransmitter mechanisms. Pharmacol. Rev. 2021 Jan;73(1):202–277. doi: 10.1124/pharmrev.120.000056. [DOI] [PubMed] [Google Scholar]
- Johnson M.P., Hoffman A.J., Nichols D.E. Effects of the enantiomers of MDA, MDMA and related analogues on [3H]serotonin and [3H]dopamine release from superfused rat brain slices. Eur. J. Pharmacol. 1986 Dec 16;132(2-3):269–276. doi: 10.1016/0014-2999(86)90615-1. [DOI] [PubMed] [Google Scholar]
- Marshall G.R. Limiting assumptions in molecular modeling: electrostatics. J. Comput. Aided Mol. Des. 2013 Feb;27(2):107–114. doi: 10.1007/s10822-013-9634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.M., Bogenschutz M., Lilienstein A. MDMA-assisted therapy for severe PTSD: a randomized, double-blind, placebo-controlled phase 3 study. Nat. Med. 2021 Jun;27(6):1025–1033. doi: 10.1038/s41591-021-01336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller I.R., Slivacka M., Nielsen A.K., Rasmussen S.G.F., Gether U., Loland C.J., Rand K.D. Conformational dynamics of the human serotonin transporter during substrate and drug binding. Nat. Commun. 2019 Apr 11;10(1):1687. doi: 10.1038/s41467-019-09675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Brändle R., Liechti M.E., Borgwardt S. Neuroimaging of chronic MDMA ("ecstasy") effects: a meta-analysis. Neurosci. Biobehav. Rev. 2019 Jan;96:10–20. doi: 10.1016/j.neubiorev.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Navratna V., Gouaux E. Insights into the mechanism and pharmacology of neurotransmitter sodium symporters. Curr. Opin. Struct. Biol. 2019 Feb;54:161–170. doi: 10.1016/j.sbi.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport T.D., Sansom M.S.P., Stansfeld P.J. The MemProtMD database: a resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res. 2019 Jan 8;47(D1):D390–D397. doi: 10.1093/nar/gky1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D.E. Differences between the mechanism of action of MDMA, MBDB, and the classic hallucinogens. Identification of a new therapeutic class: entactogens. J. Psychoact. Drugs. 1986 Oct-Dec;18(4):305–313. doi: 10.1080/02791072.1986.10472362. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rothman R.B., Baumann M.H. Monoamine transporters and psychostimulant drugs. Eur. J. Pharmacol. 2003 Oct 31;479(1-3):23–40. doi: 10.1016/j.ejphar.2003.08.054. [DOI] [PubMed] [Google Scholar]
- Sáez-Briones P., Hernández A. MDMA (3,4-methylenedioxymethamphetamine) analogues as tools to characterize MDMA-like effects: an approach to understand entactogen pharmacology. Curr. Neuropharmacol. 2013 Sep;11(5):521–534. doi: 10.2174/1570159X11311050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandtner W., Stockner T., Hasenhuetl P.S., Partilla J.S., Seddik A., Zhang Y.W., Cao J., Holy M., Steinkellner T., Rudnick G., Baumann M.H., Ecker G.F., Newman A.H., Sitte H.H. Binding mode selection determines the action of ecstasy homologs at monoamine transporters. Mol. Pharmacol. 2016 Jan;89(1):165–175. doi: 10.1124/mol.115.101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancer M., Johanson C.E. The effects of fluoxetine on the subjective and physiological effects of 3,4-methylenedioxymethamphetamine (MDMA) in humans. Psychopharmacology (Berl) 2007 Jan;189(4):565–573. doi: 10.1007/s00213-006-0576-z. [DOI] [PubMed] [Google Scholar]
- Vistoli G., Mazzolari A., Testa B., Pedretti A. Binding space concept: a new approach to enhance the reliability of docking scores and its application to predicting butyrylcholinesterase hydrolytic activity. J. Chem. Inf. Model. 2017 Jul 24;57(7):1691–1702. doi: 10.1021/acs.jcim.7b00121. [DOI] [PubMed] [Google Scholar]
- Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005 Jan-Feb;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.