Abstract
Background
Panicle is a harvesting organ of rice, and its morphology and development are closely associated with grain yield. The current study was carried on a mutant screened through an EMS (ethyl-methane sulphonate) mutagenized population of a Japonica cultivar Kitaake (WT).
Results
A mutant, named as asp-lsl (aberrant spikelet-long sterile lemma), showed a significant decrease in plant height, number of tillers, thousand-grains weight, seed setting rate, spikelet length, kernel length and effective number of grains per panicle as compared to WT. Asp-lsl showed a pleiotropic phenotype coupled with the obvious presence of a long sterile lemma. Cross-sections of lemma showed an increase in the cell volume rather than the number of cells. Genetic segregation analysis revealed its phenotypic trait is controlled by a single recessive nuclear gene. Primary and fine mapping indicated that candidate gene controlling the phenotype of asp-lsl was located in an interval of 212 kb on the short arm of chromosome 8 between RM22445 and RM22453. Further sequencing and indels markers analysis revealed LOC_Os08g06480 harbors a single base substitution (G→A), resulting in a change of 521st amino acid(Gly→Glu. The homology comparison and phylogenetic tree analysis revealed mutation was occurred in a highly conserved domain and had a high degree of similarity in Arabidopsis, corn, and sorghum. The CRISPR/Cas9 mutant line of ASP-LSL produced a similar phenotype as that of asp-lsl. Subcellular localization of ASP-LSL revealed that its protein is localized in the nucleus. Relative expression analysis revealed ASP-LSL was preferentially expressed in panicle, stem, and leaves. The endogenous contents of GA, CTK, and IAA were found significantly decreased in asp-lsl as compared to WT.
Conclusions
Current study presents the novel phenotype of asp-lsl and also validate the previously reported function of OsREL2 (ROMOSA ENHANCER LOCI2), / ASP1(ABERRANT SPIKELET AND PANICLE 1).
Keywords: Rice, Aberrant spikelet, Long sterile lemma, Pleiotropic phenotype, Gene cloning, Relative expression
Background
Rice is the world’s main food crop, and its yield is essential for food security. Spikelet development is one of the most important traits for yield. Rice is considered a model crop for monocotyledons due to its unique structural unit of spikelet compared to dicotyledons. Grasses spikelet consists of two sterile glumes (rudimentary glumes), empty glumes, and a pair of fertile glumes that are further called lemma and palea [1, 2]. According to previous studies, spikelet development can be roughly divided into the following eight stages: the first stage (Sp1) includes the formation of the incomplete rudimentary glume primordia. From second to fourth stage (Sp2-Sp4), glume primordium differentiates into lemma and palea primordium. The formation of lodicule and stamen occur at the fifth (Sp5) and sixth (Sp6) stage of spikelet development, respectively. The seventh (Sp7) and eighth (Sp8) stages involve the formation of carpel primordium and development of ovule and pollen, respectively [3]. So far, numerous genes have been cloned that function at different stages of spikelet development. Mutants with abnormal spikelet development showing hindrance at various stages can be used as important materials for studying molecular mechanism.
In terms of floral organs development, there are certain differences between monocotyledonous and dicotyledonous plants. However, homologous sequence alignment and transgene analysis showed that ABCDE model of dicotyledons applies to monocotyledons to a certain extent. Hence, above-mentioned model can also be applied to understand developmental mode of rice floral organs [4–11]. According to ABCDE model, so far, five types of MADS-box genes have been cloned and divided based on their function. These floral genes e.g., A, B, C, D and E regulate different independent or coordinated actions for a final phenotype of floret.
According to ABCDE model of rice, contrary to Arabidopsis, class A genes are mainly expressed in the outer two whorls of floral organs e.g., OsMADS14 [12, 13], OsMADS15 and OsMADS18 [14] are all AP1 (ACITVATOR PROTEIN 1)-like genes [15, 16]. Exceptionally, OsMADS14 expresses in spikelet meristem (SM), inner lemma, palea, pistil, and stamen at maturity [17]. OsMADS14 affected the flowering period, and its overexpression leads to early flowering in rice [18]. OsMADS15 normally expressed in lemma and lodicules, and mutants of OsMADS15 exhibited elongation of inner and outer glumes [19]. OsMADS18 has transcripts in all floral tissues and affected the flowering period. It’s overexpression caused early flowering but RNAi-silencing did not affect the phenotype compared with its wild type (WT) [20].
In rice, class B genes include e.g., OsMADS2, OsMADS4, and OsMADS16/SUPERWOMAN1 (SPW1) [21]. These are also conserved in angiosperms. OsMADS2 and OsMADS4 are homologous to the Arabidopsis PI (PSEUDOGENE) gene, and OsMADS16 is homologous gene of Arabidopsis AP3 gene [22, 23]. The transgenic plants of OsMADS2 obtained by gene silencing showed abnormal elongation of lodicule [24]. OsMADS2 played a key role in the development of rice panicle by controlling the function of lodicule and stamens [25]. Loss of function of OsMADS4 triggered lodicules to exhibit a palea-like morphology, and stamens exhibited a carpel-like morphology. Similarly, loss of function of OsMADS16 caused the stamens and lodicules to transform into carpels and palea-like structures, respectively [26].
In rice, class C genes include e.g. OsMADS3 [5], OsMADS58 [27] and DROOPING LEAF (DL) [28]. Among them, OsMADS3 and OsMADS58 are homologous genes of Arabidopsis AG gene[29], and DL is homologous to Arabidopsis CRC (CRABSCLAW). OsMADS3 normally expressed in carpels and stamens [30], which mainly controls the development of stamens [31] and inhibits lodicule development. Loss of function of OsMADS3 showed decisive loss of floral meristem (FM), and stamens turn into a pulp-like structure. OsMADS58 mainly controls the development of FM and its RNAi transgenic plant produced cup-shaped carpel [32]. DL regulates the development of FM, and a mutation in dl transformed the carpel into stamens [27].
In rice, class D genes mainly refer to OsMADS13, which is the STK (SERINE/THREONINE KINAS) gene of Arabidopsis and homologous to FBP7 (F-BOX PROTEIN 67)/FBP11 of petunia. OsMADS13 is only expressed in the carpel and mainly concentrated in the ovule. [30–32].
In rice, E genes include several types of genes e.g., SEP (SEPALLATA), LOFSEP (LOSS OF TRANSCRIPTION FACTOR OF SEP), and AGL6 (AGAMOUS-LIKE 6) [33, 34]: OsMADS7 (OsMADS45) and OsMADS8 (OsMADS24) are homologous to Arabidopsis SEP [35, 36]; while, LOFSEP include OsMADS1/LEAFY HULL STERILE 1 (LHS1) [37, 38]. OsMADS5 (OsM5) and OsMADS34/PANlCLE PHYTOMER2 (PAP2) are included in AGL6 and mainly refers to OsMADS6/MOSAIC FLRALORGANS 1 (MFO1) [39]. In rice, the LHS1 regulates the development of lemma and lemma coupled with the development of FM. Its ectopic expression can lead to the elongation of the spikelet [38, 40–42]. OsMADS6 regulates the development of FM and floral organs and interacts with DL to jointly regulate the development of palea, making palea to a lemma-like structure [43]. OsMADS34 has pleiotropic effect and its mutation not only turns the SM abnormal but also transformed the rudimentary glume into an elongated palea [37, 44].
Genes such as OsMADS34 and G1 can regulate the characteristics of rice glume development, and its mutations caused the glume to exhibit the phenotype of a lemma-like structures [45]. MULTI-FLORET SPIKELET2 (MFS2) encodes a MYB transcription factor and controls the identity of palea. MFS2 is a transcriptional repressor (TRP) and interacts with TOPLESS (TPL)-related proteins and forms a complex; as a result number of floral organs were increased in mfs2 [46]. OsREL2 (ROMOSA ENHANCER LOCI2), orthologous to Arabidopsis TPL, showed a pleiotropic effect including long sterile lemma and defects in panicle heading [47]. Similarly, OsLIS-L1 (LISSENCEPHALY TYPE-1-LIKE-1) encodes a protein with WD (Trp-Asp) domain repeats and revealed semi-dwarfism and defects in fertility [48]. ASP1 (ABERRANT SPIKELET AND PANICLE 1) encodes a TPL protein and control pleiotropic phenotype, including change in the meristem, phyllotaxy, and branching meristem by the intervention of auxin [49].
In this study, we have screened a mutant created by EMS mutagenized population of a wild type cultivar Kitaake (WT). Phenotype, genetic, and expression analysis revealed this gene is homologous to already reported REL2 and ASP1. Although similar phenotypes have already been reported in asp1, but presence of some different phenotypes especially long sterile lemma encouraged us to study further. Our findings not only validate pleiotropic effect of ASP1 and OsREL2 but also present some more phenotypic defects e.g., dwarfism, spikelet shortness and long sterile lemma, that we are reporting for first time.
Results
The phenotypic observation and measurement of agronomic traits revealed pleiotropic effect of ASP-LSL
Data of agronomic traits of asp-lsl and WT (Kitaake) were measured and results revealed that plant height, length of panicle, grain length, number of seeds per panicle, thousand-grain weight, seed setting rate and length of sterile lemma were significantly different in asp-lsl. Compared with its WT, the asp-lsl has reduced plant height by 23.2 %, panicle length by 21.1 %, seed setting rate by 29.2 %, 1000-kernel weight by 28.9 %, spikelet length by 34.4 %, and kernel length by 22.3 %. The most prominent spikelet variation of asp-lsl mutant was its increased and widened sterile lemma, as compared to its WT (Fig. 1). The glume length and width were 4.7 and 2.7 times increased in asp-lsl than that of WT, respectively. Protective glumes accounted ~ 1.9 % of the thousand-grain weight in asp-lsl. After removing glumes, compared with its WT, asp-lsl caryopsis became slender and thousand-grain weight decreased by 33.1 % (as shown in Table 1). It indicates that changes in grain type of asp-lsl led to the decrease in thousand-grain weight, and ultimately decrease in yield.
Fig. 1.
The phenotype of WT and asp-lsl. A The phenotype of WT (left) and asp-lsl (right). B The panicle of WT (up) and asp-lsl (up). C Spikelet of WT (left) and asp-lsl (right). D Sterile lemma of WT (left) and asp-lsl (right) (E) Horizontal view of mature grain of WT (up) and asp-lsl (down). F Horizontal view of kernel of WT (up) and asp-lsl (down). G Vertical view mature grain of WT (up) and asp-lsl (down). H Vertical view of kernel of WT (up) and asp-lsl (down). asp-lsl (down)
Table 1.
Measurement of agronomic traits of asp-lsl and WT
| Material | PMD | PH (cm)** | PL (cm)* | SR (%) ** | GL (mm)** | GW (mm)* | GL-WR* |
|---|---|---|---|---|---|---|---|
| KTK | 89 ± 1 | 62.3 ± 1.52 | 13.3 ± 1.42 | 80.4 ± 0.04 | 7.1 ± 0.04 | 3.49 ± 0.03 | 2.04 ± 0.14 |
| asp-lsl | 96 ± 1 | 47.8 ± 1.11 | 10.5 ± 1.37 | 56.8 ± 0.05 | 8.77 ± 0.04 | 2.72 ± 0.02 | 3.22 ± 0.16 |
PMD Plant maturity days, PH Plant height, PL Plant length, SR seed setting rate, GL Grain length, GW Grain weight, GL/GW Grain length/Grain width.
*indicates the significance at 0.05 level and
**indicates the significance at 0.01 level
asp-lsl showed delayed germination and decreased seed viability
Compared with the WT seeds, the asp-lsl seeds were slender but not filled properly. Observation and statistics of germination record revealed not only the emergence rate of asp-lsl was lower but the germination was also significantly delayed. The germination rate of asp-lsl on the third day of germination was only 8 %, which was significantly lower than 72.3 % of its WT. The overall germination rate of asp-lsl was 69 % that was significantly lower than WT (88.3 %). Results of germination assay revealed that seed viability was significantly decreased in asp-lsl (Fig. 2).
Fig. 2.
Germination and viability analysis of WT and asp-lsl. A The seeds germination of WT (up) and asp-lsl (down). B The grains of WT (up) and asp-lsl (up). C The seed viability of WT (left) and asp-lsl (right)
Analysis of inter-node length of stem
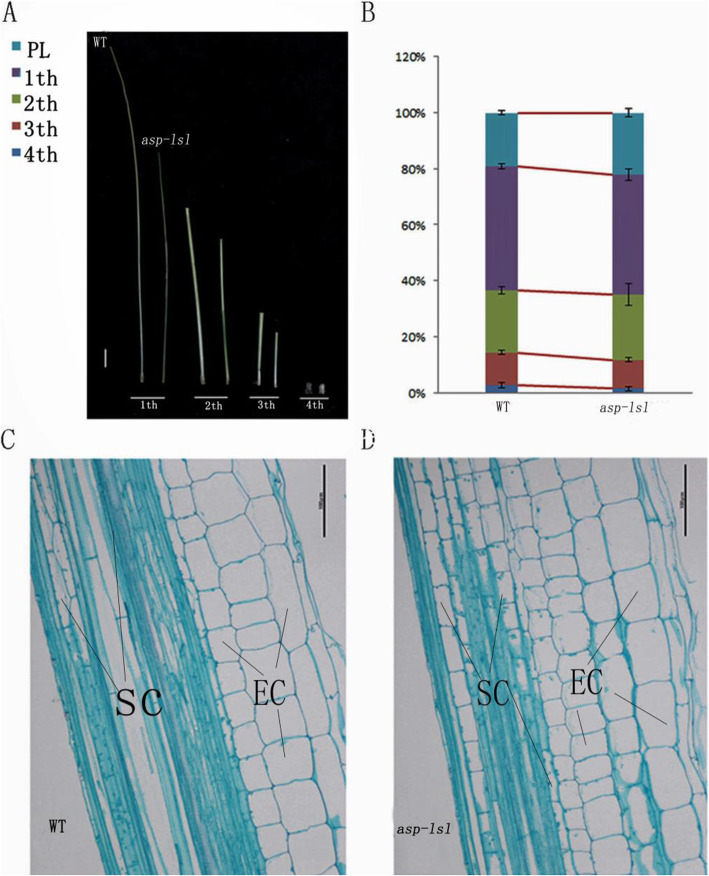
To further analyze of dwarfing of asp-lsl, we measured each stem internode at the maturity stage. Comparison of results showed the first, second, third, and fourth internodes of asp-lsl were shorter in length than those of WT (Fig. 3A). The relative change in length of the first, second, third, and fourth internode between asp-lsl is given in Fig. 3B. Among them, the difference in the first internode was more significant (Table 2). Analysis of the internode length of asp-lsl and WT revealed that dwarfing was mainly caused by the shortening of internodal distance.
Fig. 3.
Observation and analysis of internodal distance of WT and asp-lsl. A The comparison of each respective internode of WT and asp-lsl, from left to right: first, second, third and fourth internode of WT and asp-lsl, respectively. bar=5cm (B)The column showing comparative length of four internodes at maturity. C WT transverse sections of the first internode, bar=50μm. D asp-lsltransverse sections of the first internode, where SC: silicide cell, EC: epidermis cell and bar=50μm
Table 2.
Analysis of internodal distance of asp-lsl and WT,
| Internode length | WT (cm) | Proportion(%) | MT(cm) | Proportion(%) |
|---|---|---|---|---|
| 1st | 31.7 ± 0.6 | 54.9 | 23.4 ± 3.3** | 53.5 |
| 2nd | 15.7 ± 0.86 | 27.2 | 12.77 ± 2.10 | 30.9 |
| 3rd | 8.4 ± 0.44 | 14.6 | 5.63 ± 2.61 | 13.6 |
| 4th | 1.9 ± 0.57 | 3.3 | 0.8 ± 0.23 | 2.0 |
**indicates the significance at 0.01 level
Spikelet morphology and fertility observation of asp-lsl
The spikelet of asp-lsl has a similar floret structure to WT except a longer sterile lemma (Fig. 4A and B). Pollen viability was checked using KI-I2 staining, and results revealed no significant changes in fertility between WT and asp-lsl (Fig. 4C and D). To further look down minor changes in lemma morphology, paraffin cross-sections were prepared (Fig. 4E and F). The results of lemma morphology observation revealed the presence of larger cell volume. However, there was no significant change in the number of cells of sterile lemma. So, larger lemma was produced as result of the change in cells volume rather than the number of cells.
Fig. 4.
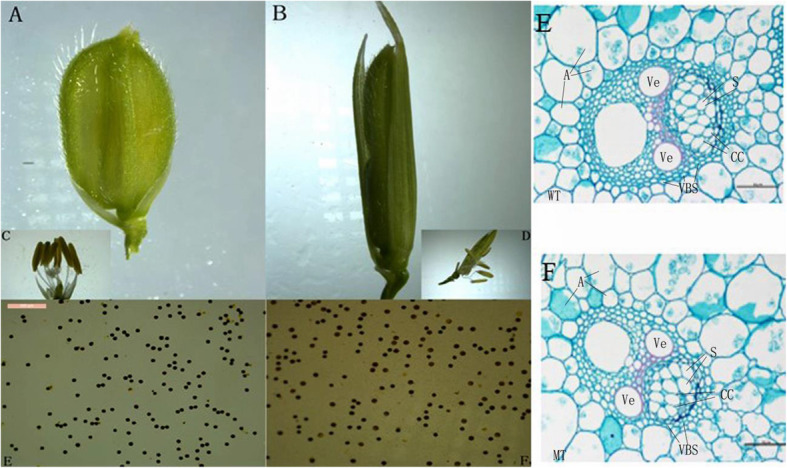
The sterile lemma morphology and Potassium Iodide staining of WT and asp-lsl (A) The sterile lemma of WT. B The sterile lemma of asp-lsl (C) Pollen grains of WT (D) Pollen grains of asp-lsl E) The paraffin sections of WT (F) and asp-lsl. Where A: amyloplast, Ve: vessel, VBS: vascular bundle sheath and CC: companion cells
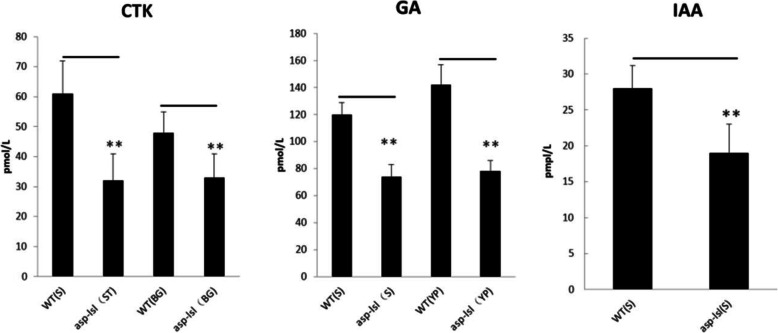
asp-lsl showed significant changes GA, CTK, and IAA phytohormones
In order to compare changes in the shape of asp-lsl architecture, we determined the relevant hormones e.g., GA (Gibberellins), IAA (Indole acetic acid), and CTK (cytokinin) content in stem and panicle. The results showed GA, CTK, and IAA of asp-lsl had significant changes as compared with its WT (Fig. 5). GA, CTK, and IAA of asp-lsl were found significantly lower in stem and young panicle than that of WT.
Fig. 5.
Endogenous hormone contents of WT and asp-lsl. S: stem, YP: young panicle, BG: before grouting, MS: mature stage
A single recessive gene controls the phenotype in asp-lsl
Using Yixiang 1B and KTK as male parents and asp-lsl as female parent, were crossed to get two mapping populations (asp-lsl *YXB and asp-lsl *KTK). Both populations were observed for phenotypes in F1 and F2. The results showed that the phenotype of all F1 plants were consistent with the WT phenotype. F1 were selfed to get F2 progeny; in later one, plants appeared with the both (WT and asp-lsl) type of phenotype. χ2 (Chi-square) test of segregation ratio between of WT vs. asp-lsl was in accordance with the Mendelian ratio of 3:1 (Table 3). The phenotypic segregation ratio of both F2 population can validate that mutant phenotype is controlled by a single recessive gene.
Table 3.
The genetic segregation ratio of ASP-LSL
| Cross | F1 | F2 | χ2(3:1) | ||
|---|---|---|---|---|---|
| Total No. of plants | No. of mutant type plants | No. of wild type plants | |||
| asp-lsl /KTK | Normal | 570 | 128 | 442 | 1.17a |
| asp-lsl /YXB | Normal | 670 | 174 | 496 | 0.67a |
aindicates the significance at 0.05 level
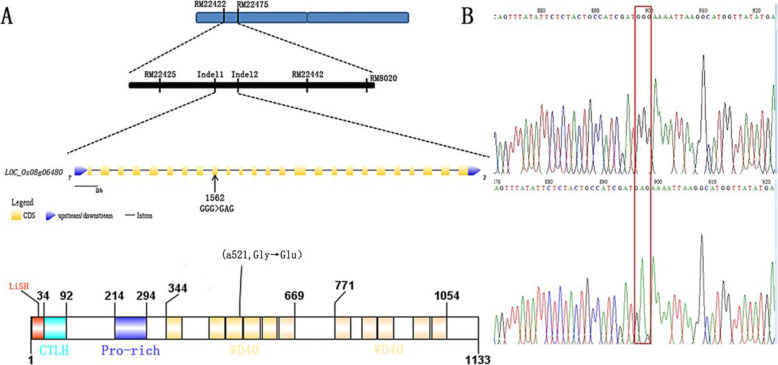
Primary and fine mapping of ASP-LSL
An F2 segregated population, constructed from the cross of asp-lsl and Yixiang 1B was used for gene mapping. Primarily, 456 pairs of SSR markers were used for primary gene mapping and locus was initially positioned between RM22418 and RM22529 (Fig. 6). In order to further locate, different Indel markers were applied and the interval was narrowed down to a region of 212 kb between Indel-1 and Indel-2. Twenty-six candidate genes were present in an interval of 212 kb. Analyzing all genes in this interval, it was found that the LOC_Os08g06480 contains an SNP. Using WT and ASP-LSL cDNA as templates, the gene was amplified and sequenced. An SNP was detected that changed cDNA (1562nd nucleotide) of LOC_Os08g06480 from G (guanine) → A (adenine). As a result of single frame-shift mutation, 521th amino acid was changed from G (glycine) → E (glutamic acid). So, we regarded the ASP-LSL gene as a candidate gene for the given phenotype of asp-lsl. To verify the mutation, 20 single plants were selected from the WT and asp-lsl from the F2 segregated population and verified using Tetra-Primer ARMS PCR.
Fig. 6.
Gene mapping of ASP-LSL (A) Primary gene mapping of locus on short arm chromosome 8, Yellow line showed the structure of candidate gene, the Protein structure of candidate gene. B The chromatogram of WT and asp-lsl showing SNP
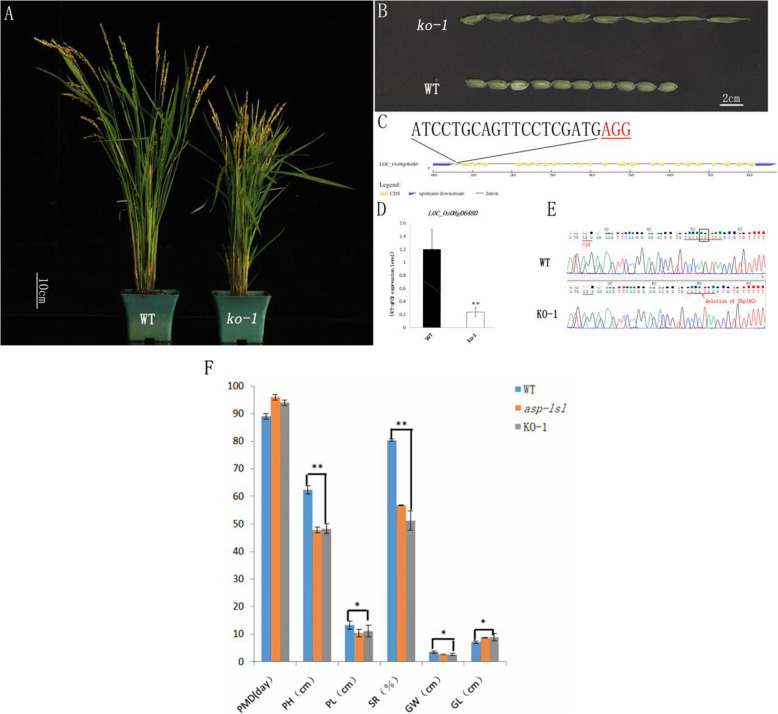
Knock out of ASP-LSL plants showed asp-lsl phenotype
In order to verify, the ASP-LSL gene is responsible for the above-mentioned mutant phenotype (Fig. 7A). A CRISPR/Cas9 vector targeted at the first exon (ATCCTGCAGTTCCTCGATGAGG) of ASP-LSL. The knock-out line-1 (ko-1) showed an increased length of sterile lemma, short plant height coupled with phenotype of asp-lsl (Fig. 7B). Meanwhile, we analyzed gene expression by RT-qPCR, and results showed a significant decrease in the expression ASP-LSL in knock-out lines (Fig. 7D). The chromatogram of ko-1 showed a 2 bp deletion as compared to its WT (Fig. 7E). Number of days of maturity, plant height, panicle length, seed setting rate, grain width and grain length have been significantly decreased in ko-1 as compared to WT (Fig. 7F).
Fig. 7.
Phenotype of WT and knockout plant. (A) The phenotype of WT and ko-1 (B) The phenotype of WT and ko-1 floret. (C) The Targeted sequence of the first exon of ASP-LSL gene. (D) RT-qPCR analysis of ASP-LSL gene in WT and ko-1 (E) Chromatogram (sequencing peaks) of WT and ko-1(F) Agronomic traits comparison of WT, KO, and asp-lsl. Where PMD: Plant maturity days, PH: Plant height, PL: Panicle length SR: seed setting rate, GW: Grain weight GL: Grain length
Phylogenetic analysis of ASP-LSL
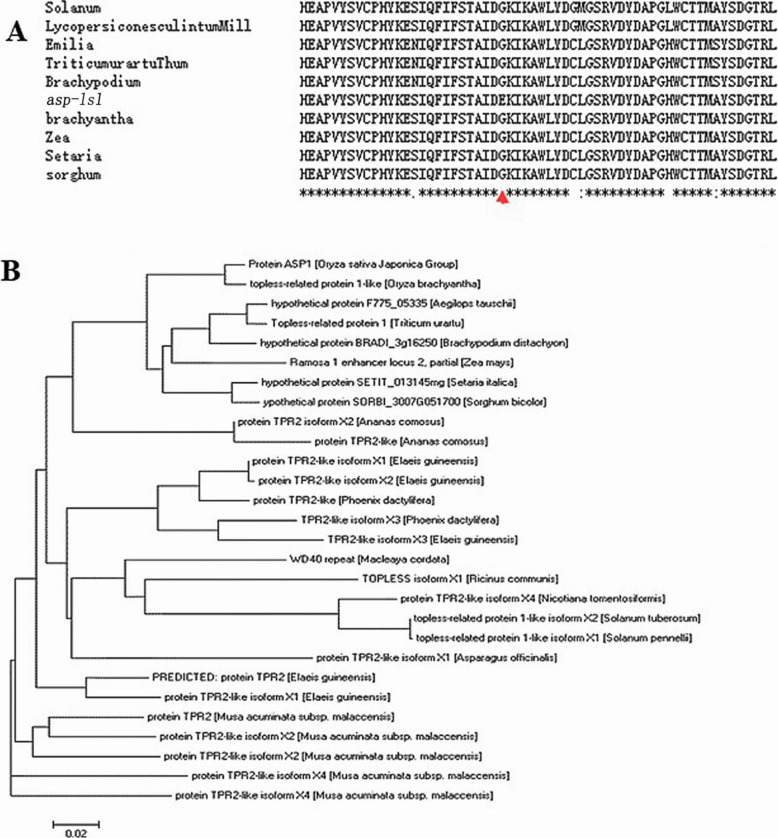
A complete comparison of amino acid sequence of ASP-LSL was used for construction of a phylogenetic tree at threshold of 70-99 %. As shown in the figure, all the reported mutations were occurred in a highly conserved action site, similar as in the mutant asp-lsl (Fig. 8A). The ASP-LSL protein has highest homology with a TOPLESS- protein in higher plants (Fig. 8B).
Fig. 8.
Alignment and phylogenetic tree of homologus proteins of ASP-LSL (A) Alignment analysis of the amino acid sequence of ASP-LSL (B) Phylogentic tree of homologus proteins of ASP-LSL
Subcellular localization of ASP-LSL
We constructed ASP-LSL-YFP and empty YFP vectors and transformed them into rice protoplasts. Compared with WT, nuclear colocalization of ASP-LSL revealed yellow fluorescence that overlaps with the red and formed an orange fluorescence as a nuclear localization signal (Fig. 9). Results of subcellular localization showed ASP-LSL is a nuclear protein.
Fig. 9.
Subcellular localization of ASP-LSL
Relative expression of floral development-related genes in ASP-LSL
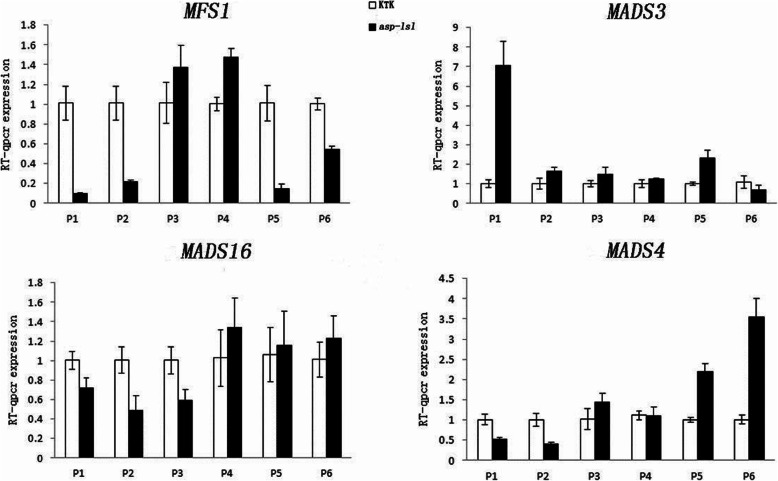
Development of asp-lsl was different than that of WT, to know which related genes were involved in expressing the different phenotype of asp-lsl. Young panicle at different growth periods were selected to detect the expression of different panicle development related genes. Different MADS genes e.g., MFS1, OsMADS3, OsMADS4 and OsMADS16 showed a significant change in asp-lsl as compared to WT. Quantitative expression of floral development-related genes showed different expression levels at different stages. The expression level of MFS1 in the mutants was higher than that of WT at P3 and P4 stages (Fig. 10). OsMADS4 and OsMADS16 had higher expression levels in asp-lsl at the later stage of young panicle development, while OsMADS3 showed the highest expression level at P1-P5. The sequence of primers used for relative expression analysis is given in Table 4.
Fig. 10.
Relative expression analysis of different floral development-related genes in WT and asp-lsl
Table 4.
The primers used for the quantitative relative expression analysis of genes related to floret development
| Primer | Sequence 5’ →3’ |
|---|---|
| OsMADS1-F | TCTTGGTGAGGATTTGGGC |
| OsMADS1-R | CCTTGCTCTTCAGATCAAACAG |
| OsMADS3-F | AACGCAAACAGTAGGACCATAGTG |
| OsMADS3-R | CCCCTCTCATTCTCAACAACC |
| OsMADS4-F | AGCACAAGATGTTGGCTTTTAGGG |
| OsMADS13-F | ATGGGGAGGGGCAGGATTGAG |
| OsMADS13-R | TGCGCCTTCTTGTACCTGTCA |
| OsMADS16-F | CGAGGCGTACGAGACTCTGC |
| OsMADS16-R | ACCACGCGGAAGGCGAACAT |
| DL-F | CCCATCTGCTTACAACCGCTT |
| DL-R | GTTGGAGGTGGAAACCGTCG |
| MFS1-F | CGGCTCGTGATCTCGACACGTAC |
| MFS1-R | CACAGCCGGACCAGTGCTCTC |
Discussion
ASP-LSL encodes a single recessive nuclear transactional repressor gene
In this study, an aberrant spikelet mutant was obtained by EMS mutagenesis of a wild cultivar, Kitaake. Through the inspection and comparison of agronomic traits, asp-lsl showed multiple defects including, the structure of spikelet and major agronomic yield traits e.g., plant height, number of tillers, seed setting rate, 1000-kernel weight, and number of effective grains per panicle were significantly compromised. Segregation and gene mapping analysis revealed its phenotypes were controlled by a single recessive nuclear. LOC_Os08g06480 encodes a TPR/TPL nuclear transcriptional co-repressor. Transcriptional co-repressors have been already reported to play their role in controlling the spikelet, seed size and weight in rice and Arabidopsis [33, 50]. Phylogenetic analysis revealed that mutation was occurred in highly conserved site and have been reported in maize, sorghum and rice [1, 49]. This site can be targeted by molecular breeder to solve the specific problem of germination in hybrid rice [51]. Protein of this site may have specific binding activity to some other genes that regulate different traits including the development of lemma [48]. In recent years, many genes controlling flower development in rice have been cloned. Most of them are homologous to Arabidopsis and have a certain degree of conservation. These findings provide a basis for studying more about the regulation mechanism of rice flower development, without disturbing other major agronomic yield traits [51, 52]. In grasses, these homologous proteins can interact with ETHYLENE RESPONSE FACTOR (ERF), Jasmonic acid (JA), and IAA [47, 49]. Endogenous measurement of phytohormone contents suggested that ASP-LSL is sensitive to IAA that has an extensive role in plant growth and development activities. ASP-LSL also showed the action of IAA in regard to transition meristem fate and phyllotaxy during the vegetative phase [49, 53, 54]. The different phenotype of ASP-LSL could be a result of its molecular interaction with other spikelet development and auxin signaling pathway genes.
ASP-LSL controls the novel phenotype in japonica cv. Kitaake
According to National Rice Data Center (ricedata.cn), ASP-LSL locus has already been reported in different articles. All the previous materials that were identified and reported so far belong to Nipponbare and Dongjin. In asp-lsl, site of mutation (521th amino acid Gly to Glu) is also novel, and this mutation has not been reported before. Moreover, all previous studies used T-DNA mutations, while we used EMS as a source of mutagenesis. Moreover, asp-lsl showed different salient features as compared to other mutants, which are given below.
OsREL2 produced pleiotropic phenotype e.g., a greater number of secondary branches than primary branches and reduced number of floral organs etc., as compared to WT. T-DNA insertion mutant, osrel2 also showed phenotype defects in panicle development. Contrary to OsREL2, ASP-LSL has an extra-long sterile lemma that bypasses the whole length of its spikelet. While in osrel2 sterile lemma merely passes the half of the length of spikelet [47].
OsLIS-L1 was a single recessive gene, reported to regulate pleiotropic phenotype and considered essential for fertility and internode elongation. OsLIS-L1 encodes a protein that contains a WD40 motif, which is necessary for human brain development. OsLIS-L1 was highly expressed in rice stems and panicles, but its expression in panicles showed dynamic changes at different developmental stages. Two independent alleles, oslis-1-1 and oslis-1-2 both showed reduced male gamete fertility, semi-dwarfism and shorter length of panicle. OsLIS-L1 plays an important role in the elongation of the inverted internodes and the formation of male gametophytes [48]. The apparent difference of OsLIS-L1 and ASP-LSL is the presence of a long sterile lemma in a later one.
ASP1 encodes a transcriptional co-repressor, homologous to TPL of Arabidopsis and REL2 in maize. Multiple phenotypic defects in asp1 mutants were occurring, possibly due to the activation of multiple genes involved in meristem function. Aps1 showed disorganized phyllotaxy and transition from branch to meristem formation was also compromised. The mutant with the loss of function of ASP1 also revealed the bleaching of branches and spikelets. [55] A salient and novel feature of ASP-LSL with respect to ASP1 is also the presence of long sterile lemma. In asp1 the sterile lemma length reaches up merely to half of spikelet.
All of above three genes were located at the same gene locus as that of ASP-LSL, but mutant used in the current study contain conserved structure of the mutation site, a different source of germplasm, and the mode of mutagenesis was also different. Current study not only analyzes the biological function of this gene in more detail but also provide new germplasm resources for mutant bank pool. Current research was focused on the functional and phenotypic analysis of ASP-LSL. In the future, more in-depth work on the interaction and molecular regulation of ASP-LSL would be carried out.
Conclusions
Current study reports the novel phenotype of asp-lsl that showed a significant decrease in plant height, number of tillers, thousand-grains weight, seed setting rate, spikelet length, kernel length and effective number of grains per panicle as compared to its WT. Asp-lsl showed delayed germination, deceased seed viability and shorter internodal distance than that of WT. Segregation and gene mapping analysis revealed its phenotypes were controlled by a single recessive nuclear. Subcellular localization of ASP-LSL revealed that its protein is localized in the nucleus. The knockout line of ASP-LSL produced a similar phenotype as that of asp-lsl. In the Hybrid seed production, spike germination is a prominent factor that affects the overall yield and milling quality of rice. Molecular breeders can manipulate the genes related to the pathway of long sterile lemma to solve the problem of spike germination with minimal damage to the other major agronomic traits.
Methods
Materials
A wild type (WT) japonica variety (Kitaake) was mutagenized by ethyl-methane sulfonate (EMS), and a mutant showing long sterile lemma coupled with other phenotypic defects was selected from M2 population and named as asp-lsl. Its phenotypic traits were found to be inherited stably after multiple generations of selfing. Two indica varieties Yixiang 1B and Kitaake were used as male parents and crossed with asp-lsl to construct F2 segregating populations. The materials used in experiment were planted under field conditions alternately at the experimental sites of Rice Research Institute, Sichuan Agricultural University in Lingshui (N18.47°, E110.04°), Hainan and Chongzhou (N30.67°, E103.67°), Sichuan, China. The sampling of material and experimental data was recorded with the permission and regular legislation of Rice Research Institute, Sichuan Agricultural University, China.
Measurement of agronomic traits
At the maturity period of mutant and WT, 10 plants were randomly selected and data of plant height, ear length, grain length, grain width, thousand-grain weight, seed setting rate, inner and outer glume length and number of first branches, etc. were measured. The data were finally represented as an average of 10 individual plants for each of the corresponding trait index.
Histology observations and identification of pollen fertility
Under normal field growth conditions, several primary differentiation stages of panicles were used for histology observation. The different tissue parts of mutant and WT panicles, lemma and glume at panicle elongation, grain filling and fully mature stage were fixed in FAA fixative at 4℃. After rinsing with series of 50 %, 70 %, and 80 % ethanol, and pictures were taken using a scanning electron microscope (FEI QUANTA 450). Fresh spikelets of mutant and WT with their pollens were observed under a microscope using potassium iodide staining.
Dynamic observation of growth and development
To explore the difference between mutant and WT, a seed germination test was carried out in the laboratory. The specific parameters that were followed are given below.
Indoor germination test
One hundred healthy seeds of WT and mutant with their three biological repetitions were germinated at 37℃ in a light incubator. Germination rate was observed after 3 and 7 days, respectively.
Seed vigor examination
In order to verify the accuracy of the germination test results, 100 seeds were selected and cut down longitudinally and finally soaked in a 0.1 % tri-phenyltetrazolium chloride (TTC) solution for 3 h at 35 °C. After dyeing, TTC solution was poured out, and seeds were washed with clean water twice, and the stained number of embryos were counted.
Observation of seedling emergence time in an incubator
Fifty white seeds (stained) of WT and mutant were selected and sown in plastic pots using the soil culture method. Pots were kept under continuous observations, and emergence time from one-leaf to the five-leaves stage was recorded.
Observation of root growth parameters
The WT and mutant plants were grown in a nutrient solution for 7 and 14 days were selected for root parameters observation. At the same time, the change of hypocotyl length was also measured in the absence of light at 32℃ in order to determine the relationship between asp-lsl and IAA. To explore its relationship with hormone transport and signal response, IAA solution was sprayed on the mutant plants grown for three days on culture. Whereas only distilled water was used as control treatment.
Construction of the genetic map
For hundred fifty-six pairs of SSR markers constructed that were evenly distributed on all chromosomes of rice, were selected as primers, and the DNA of individual F2 populations was used as a template for PCR amplification. Bulk segregant analysis (BSA) was used to construct WT and mutant mixed DNA pools to screen out polymorphic differences through linkage markers. For confirmation of mutant phenotype, DNA of F2 progeny of single plant was used as DNA template. During primary gene mapping, the sequence of polymorphic loci was acquired from the biological database RGP (http://rgp.dna.affrc.go.jp/) and NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared it with the sequence of model indica (9311) and japonica (nipponbare) varieties. SSR markers were designed using public databases e.g., RIS (http://rice.genomics.org.cn/rice/index2.jsp) and RGP (http://rgp.dna.affrc.go.jp/). To further fine map, indels markers were constructed and applied to the mutant DNA template. Chengdu Kinco Biological Co., Ltd synthesized the primers.
PCR amplification
The specific quantity of ingredients used in PCR amplification was as follows; DNA (2 µl), F-Primer (1 µl), R-Primer (1 µl), dNTP (10mM) mixture (0.3 µl), Taq(5U/µl)0.2 µl, 10×Buffer mg2+ 2 µl and ddH2O (13.5 µl). The specific PCR amplification procedure was as follows: 95 °C pre-denaturation for 5 min; 95 °C denaturation for 30 s, 56 °C annealing for 30 s, 72 °C extension for 1 min, 34 cycles; 72 °C extension for 7 min, 4 °C storage for 1 min. The PCR products were separated on 3 % agarose gel electrophoresis and photographed under a Gel Documentation System (Bio-Rad Gel Doc 2000).
Sequence analysis
Primer premier 6.0 and lingo 7.0 software were used primer designing and sequence analysis. As the target gene ASP-LSL has a relatively bigger size, we used the segmented amplification method and divided the gene into four fragments for easier amplification. PCR product was processed with Tiangen DNA recovery kit that is used for purification and separation. Recovered products were sent to Sichuan Qingke Biological Company (Ltd.) for gene sequencing.
Genetic segregation analysis and knock out line development
The mutation sites were analyzed by dCAPS molecular marker primers, which were designed on (http://helix.wustl.edu/dcaps/dcaps.html). Twenty plants from F2 segregated population of WT and mutant were taken for DNA extraction, and PCR amplification was performed with a restriction endonuclease. A CRISPR/Cas9 vector targeted at the first exon of ASP-LSL was developed using BG Biotech Kit (BGK03), purchased from Hongzhou Bioge Biotechnology Co., Ltd and followed the protocol given by the manufacturer (http://www.biogle.cn/index/excrispr). ASP-LSL 19-bp PAM sequence was selected, and the Oligo sequence was generated online using BioGe website (www.bioge.cn). Target sites were selected to prevent off-target effects in the BLAST database (http://www.gramene.org/) for specificity. The synthesized oligo was dissolved in 10 ul of water. The following (buffer aneal 18ul, forward Oligo 1 ul, and reverse Oligo 1 ul) ingredients were mixed on ice and heated in a PCR apparatus at 95 °C for 3 min. The temperature was decreased to 20 °C at the rate of 0.2 °C/s. According to the manufacturer’s instruction, oligo dimer was constructed into CRISPR/Cas9 vector, each component (Oligo primer 1 ul, the CRISPR/Cas Vector 2 ul, enzyme mix 1 ul, 6 ul H2O) was evenly mixed on the ice and reacted in the PCR apparatus (20℃) for 60 min after mixing.
The recombinant system was transformed into E. coli according to the reference procedure. 10 ul of the reaction solution was added to 100 ul of competent cells, and then the mixture was mixed and placed in an ice bath for 30 min (do not shake during this period and keep standing strictly). The reaction mixture was gently taken out at 42℃ for 90 s, immediately placed on ice for 2 min, added 500 ul preheated LB liquid medium, and cultured at 37℃/200 RPM for 1 h. An appropriate amount of bacterial liquid was coated on the LB plate containing Kanamycin and placed upside down at 37℃ for overnight culture. The constructed vector was sent to Wuhan Boyuan Biotechnology Co., Ltd for genetic transformation.
Quantitative relative expression analysis
The total RNA of mutant and WT roots, stems, leaves, and spikelets at the young panicle and early flowering stage were extracted using the RNA extraction kit. The purified RNA was reversed transcribed into cDNA. The sequence of a specific gene and its other homologous genes were quantified using primers. The template and reference gene (actin) was used in three biological repeats with two NTC (no template control) repeats. Quantitative expression was analyzed by Bio-Rad CFX Manager V2 that is based on △△CT method.
Bioinformatics analysis
Amino acid sequence homology was compared using NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) tool. Clustal X and MEGA software were used to perform comparison of complete sequence and construction of NJ evolutionary tree.
Subcellular localization
The subcellular localization was predicted online using ChloroP 1.1 and TargetP 1.1 server. The full-length of ASP-LSL cDNA was amplified from its WT using specific primers (F: 5′ATGTCGTCGCTTAGCAGG-3′; R: 5′GACTTCTGGTTTGTTAGCT–3′) with BamHI at 5′-end and a SalI at 3′-end. The target fragments were cloned into pC1300-35 S-eYFP vector at the N-terminus of YELLOW FLUORESCENT PROTEIN (YFP). The constructs were transformed into rice protoplasts and incubated before the examination. YFP fluorescence signals were detected using a Laser Scanning Confocal Microscope (Nikon A1).
Acknowledgements
Not Applicable.
Abbreviations
- A
Amyloplast
- AP1
ACITVATOR PROTEIN 1
- ASP1
ABERRANT SPIKELET AND PANICLE 1
- asp-lsl
Aberrant spikelet-long sterile lemma
- BG
Before grouting
- CC
Companion cells
- CRC
CRABSCLAW
- CTK
Cytokinin
- DL
DROOPING LEAF
- EC
Epidermis cell
- EMS
Ethyl-methane sulphonate
- FBP
F-BOX PROTEIN
- FM
Floral meristem
- GA
Gibberellins
- GL
Grain length
- GW
Grain weight
- IAA
Indole acetic acid
- LHS1
LEAFY HULL STERILE 1
- LOFSEP
LOSS OF TRANSCRIPTION FACTOR OF SEP
- MFO1
MOSAIC FLRALORGANS 1
- MFS2
MULTI-FLORET SPIKELET2
- MS
Mature stage
- OsLIS-L1
LISSENCEPHALY TYPE-1-LIKE-1
- OsREL2
ROMOSA ENHANCER LOCI2
- OsREL2
ROMOSA ENHANCER LOCI2
- PAP2
PANlCLE PHYTOMER2
- PH
Plant height
- PI
PSEUDOGENE
- PL
Panicle length
- PMD
Plant maturity days
- S
Stem
- SC
Silicide cell
- SEP
SEPALLATA
- SM
Spikelet meristem
- SPW1
SUPERWOMAN1
- SR
Seed setting rate
- STK
SERINE/THREONINE KINAS
- TPL
TOPLESS
- VBS
Vascular bundle sheath
- Ve
Vessel
- WT
Wild type
- YP
Young panicle
Authors' contributions
JW, TW, XP designed the study. AA, and XW wrote of manuscript. TW, JS, YF, TZ and YL1 performed experiments and analyses. HZ, AA, YX, YL2, CS, and JW planted materials and carried out analysis for proteomes. AA and XW revised the manuscript content. All authors read and approved the final manuscript.
Funding
We acknowledge grants-in-aid from Department of Science and Technology of Sichuan Province (2021YFYZ0020, 2018JY0144), NSFC (National Natural Science Foundation of China) Grant No. 31771763 and a Breeding Research Project (21ZDYF2186). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tingkai Wu, Asif Ali and Jinhao Wang contributed equally to this work.
References
- 1.Yu Y. Genetic Analysis and Gene Mapping of Rice glume-protecting Lemma Mutant lemma-like sterile lemma (lsl) Chongqing: Southwest University; 2015. [Google Scholar]
- 2.Keming Zhu HT, et al. Advances in molecular genetics of panicle development regulation in rice. Mol Plant Breed. 2015;13(9):2109–19. [Google Scholar]
- 3.Ikeda KSH, Nagato Y. Developmental course of inflorescence and spikelet in rice. Breed Sci. 2004;54:147–56.
- 4.Kater MM, Dreni L, Colombo L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot. 2006;57(13):3433–44. doi: 10.1093/jxb/erl097. [DOI] [PubMed] [Google Scholar]
- 5.Kyozuka JSK. Ectopic expression of OsMADS3, a rice orthoiog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol. 2002;43:130–5. [DOI] [PubMed]
- 6.Pelaz SDGS, Baumann E, Wisman E, et al. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;4(5):200–3. [DOI] [PubMed]
- 7.Preston JC, Kellogg EA. Reconstructing the Evolutionary History of ParalogousAPETALA1/FRUITFULL-Like Genes in Grasses (Poaceae) Genetics. 2006;174(1):421–37. doi: 10.1534/genetics.106.057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinheimer R, Kellogg EA. Evolution of AGL6-like MADS box genes in grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell. 2009;21(9):2591–605. doi: 10.1105/tpc.109.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S. bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell. 2009;21(9):2578–90. doi: 10.1105/tpc.109.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson BE, Hake S. Translational Biology: From Arabidopsis Flowers to Grass Inflorescence Architecture. Plant Physiol. 2009;149(1):38–45. doi: 10.1104/pp.108.129619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang DBWZA. Stamen specification and anther development in rice. Chin Sci Bull. 2009;54:2342–53.
- 12.Bing Wang XW, et al. Research progress on ABC model of flower organ development. Agric Biotechnol Sci. 2003;19(5):1–3. (Abstract in English)
- 13.Moon YH, KHG, Jung JY, et al. Determination of the motif responsible for interaction between the rice APETALA1AGAMOUS-like9 family proteins using a yeast two-hybrid system. Plant Physiol. 1999;120(4):1193–204. [DOI] [PMC free article] [PubMed]
- 14.Dimarcq JL, HD, Meister M, et al. Characterization and transcriptional profilesof a Drosophila gene encoding an inset defensin. Eur J Biochem. 1994;221(1):201–9. [DOI] [PubMed]
- 15.Litt AIVF. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics. 2003;165(2):821–33. [DOI] [PMC free article] [PubMed]
- 16.Litt MLAJA. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Human Genet. 1989;44(3):397–401. [PMC free article] [PubMed]
- 17.Pelucchi N, Fornara F, Favalli C, Masiero S, Lago C, Pè EM, Colombo L, Kater MM. Comparative analysis of rice MADS-box genes expressed during flower development. Sex Plant Reprod. 2002;15(3):113–22. doi: 10.1007/s00497-002-0151-7. [DOI] [Google Scholar]
- 18.Jeon JS, LS, Jung KH, et al. Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Plant Cell. 2000;12:581–92.
- 19.Masiero S, Imbriano C, Ravasio F, Favaro R, Pelucchi N, Gorla MS, Mantovani R, Colombo L, Kater MM. Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J Biol Chem. 2002;277(29):26429–35. doi: 10.1074/jbc.M202546200. [DOI] [PubMed] [Google Scholar]
- 20.Fornara F, Parenicova L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 2004;135(4):2207–19. doi: 10.1104/pp.104.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung YY, KSR, Kang HG, et al. Characterization of two rice MADS-box genes homologous to Globosa. Plant Sci. 1995;109:45–56.
- 22.Moon YH, JJV, Kang HG, et al. Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol Biol. 1999;40:167–77. [DOI] [PubMed]
- 23.Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development. 2003;130(4):705–18. doi: 10.1242/dev.00294. [DOI] [PubMed] [Google Scholar]
- 24.Prasad KVU. Double-stranded RNA interference of a rice PIGLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics. 2003;165(4):2301–5. [DOI] [PMC free article] [PubMed]
- 25.Kang HGJJS, Lee S, et al. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol Biol. 1998;38(6):1021–9. [DOI] [PubMed]
- 26.Shinozuka YKS, Shomura A, et al. Isolation and characterization of rice MADS-box gene homologues and their RFLP mapping. DNA Res. 1999;6(2):123–9. [DOI] [PubMed]
- 27.Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16(2):500–9. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang HG. ea: Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS-box gene in tobacco. Plant Mol Biol. 1995;29(1):1–10. [DOI] [PubMed]
- 29.Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell. 2011;23(8):2850–63. doi: 10.1105/tpc.111.087007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Liang W, Yin C, Zhu L, Zhang D. Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 2011;156(1):263–74. doi: 10.1104/pp.111.172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007;52(4):690–9. doi: 10.1111/j.1365-313X.2007.03272.x. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Dee ZP, WP, Enrico PM, et al. OsMADSl3, a novel rice MADS-box gene expressed during ovule development. Dev Genet. 1999;25:237–44. [DOI] [PubMed]
- 33.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169(4):2209–23. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa) Plant J. 2010;61(5):767–81. doi: 10.1111/j.1365-313X.2009.04101.x. [DOI] [PubMed] [Google Scholar]
- 36.Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 2005;10(9):427–35. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Jeon JS, LS et al. Leafy hull sterile 1 is a homeotic mutation in a rice MADS-box gene affecting rice flower development. Plant Cell. 2000;12:871–84. [DOI] [PMC free article] [PubMed]
- 38.Malcomber ST, Kellogg EA. Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell. 2004;16(7):1692–706. doi: 10.1105/tpc.021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim JMYH, An G, et al. Two rice MADS domain proteins interact with OsMADS1. Plant Mol Biol. 2004;44:513–27. [DOI] [PubMed]
- 40.Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol. 2005;59(1):125–35. doi: 10.1007/s11103-005-2161-y. [DOI] [PubMed] [Google Scholar]
- 41.Prasad K, Parameswaran S, Vijayraghavan U. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 2005;43(6):915–28. doi: 10.1111/j.1365-313X.2005.02504.x. [DOI] [PubMed] [Google Scholar]
- 42.Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol. 2001;211(6):281–90. doi: 10.1007/s004270100153. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH. Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta. 2006;223(5):882–90. doi: 10.1007/s00425-005-0141-8. [DOI] [PubMed] [Google Scholar]
- 44.De Bodt S, Raes J, Van de Peer Y, Theissen G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003;8(10):475–83. doi: 10.1016/j.tplants.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Ling Zhu XC, Kangxi Du. Cloning and expression analysis of long glume mutant gene in rice. Chinese Agric Sci. 2015;48(11):275–7.
- 46.Li Y-F, Zeng X-Q, Li Y, Wang L, Zhuang H, Wang Y, Tang J, Wang H-L, Xiong M, Yang F-Y. MULTI-FLORET SPIKELET 2, a MYB transcription factor, determines spikelet meristem fate and floral organ identity in rice. Plant Physiol. 2020;184(2):988–1003. doi: 10.1104/pp.20.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon Y, Yu S-i, Park J-h, Li Y, Han J-H, Alavilli H, Cho J-I, Kim T-H, Jeon J-S, Lee B-h. OsREL2, a rice TOPLESS homolog functions in axillary meristem development in rice inflorescence. Plant Biotechnology Reports. 2012;6(3):213–24. doi: 10.1007/s11816-011-0214-z. [DOI] [Google Scholar]
- 48.Gao X, Chen Z, Zhang J, Li X, Chen G, Li X, Wu C. OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta. 2012;235(4):713–27. doi: 10.1007/s00425-011-1532-7. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida A, Ohmori Y, Kitano H, Taguchishiobara F, Hirano H. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co‐repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012;70(2):327–39. doi: 10.1111/j.1365-313X.2011.04872.x. [DOI] [PubMed] [Google Scholar]
- 50.Xu D, Lin F, Jiang Y, Ling J, Hettiarachchi C, Tellgrenroth C, Holm M, Wei N, Deng XW. Arabidopsis COP1 SUPPRESSOR 2 Represses COP1 E3 Ubiquitin Ligase Activity through Their Coiled-Coil Domains Association. PLOS Genetics. 2015;11(12):e1005747. [DOI] [PMC free article] [PubMed]
- 51.Yang D, He N, Zheng X, Zhen Y, Xie Z, Cheng C, Huang F. Cloning of long sterile lemma (lsl2), a single recessive gene that regulates spike germination in rice (Oryza sativa L.) BMC plant biology. 2020;20:1–10. doi: 10.1186/s12870-019-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jieya Wang FZ. Research Progress of flower organ mutants and related genes in rice. J Changjiang Univ. 2011;2(8):1-4. (Abstract in English)
- 53.Ikedakawakatsu K, Maekawa M, Izawa T, Itoh JI, Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012;69(1):168–80. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- 54.Ikeda K, Nagasawa N, Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol. 2005;282(2):349–60. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell. 2001;13(11):2471–81. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.