Abstract
A viral outbreak is a global challenge that affects public health and safety. The coronavirus disease 2019 (COVID-19) has been spreading globally, affecting millions of people worldwide, and led to significant loss of lives and deterioration of the global economy. The current adverse effects caused by the COVID-19 pandemic demands finding new detection methods for future viral outbreaks. The environment's transmission pathways include and are not limited to air, surface water, and wastewater environments. The wastewater surveillance, known as wastewater-based epidemiology (WBE), can potentially monitor viral outbreaks and provide a complementary clinical testing method. Another investigated outbreak surveillance technique that has not been yet implemented in a sufficient number of studies is the surveillance of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in the air. Artificial intelligence (AI) and its related machine learning (ML) and deep learning (DL) technologies are currently emerging techniques for detecting viral outbreaks using global data. To date, there are no reports that illustrate the potential of using WBE with AI to detect viral outbreaks. This study investigates the transmission pathways of SARS-CoV-2 in the environment and provides current updates on the surveillance of viral outbreaks using WBE, viral air sampling, and AI. It also proposes a novel framework based on an ensemble of ML and DL algorithms to provide a beneficial supportive tool for decision-makers. The framework exploits available data from reliable sources to discover meaningful insights and knowledge that allows researchers and practitioners to build efficient methods and protocols that accurately monitor and detect viral outbreaks. The proposed framework could provide early detection of viruses, forecast risk maps and vulnerable areas, and estimate the number of infected citizens.
Keywords: SARS-CoV-2, COVID-19, Wastewater based-epidemiology, Viral air surveillance, Artificial intelligence, Artificial neural networks, Machine learning, Deep learning, Reinforcement Learning
Graphical abstract
1. Introduction
On 31 December 2019, the World Health Organization (WHO) office in China was informed by the Wuhan Municipal Health Commission of pneumonia cases of unknown etiology (Mostafa et al., 2021; World Health Organization, 2020c). On 9 January 2020, the Chinese Center for Disease Control and Prevention (China CDC) declared that a novel coronavirus (2019-nCoV) was detected in 15 cases who were suffering from pneumonia (European Centre for Disease Prevention and Control, 2020). On 11 March 2020, the WHO declared 2019-nCoV (later named COVID-19 or Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) a global pandemic after achieving a widespread in over 110 countries resulting in 118,319 confirmed cases (World Health Organization, 2020a).
On the 25th of May 2021, the total number of COVID-19 confirmed cases reached 167.2 million, while the total number of deaths exceeded 3.4 million people (World Health Organization, 2020b). The confirmed cases occurred globally, as follows: 39.6% in America, 18.3% in South-East Asia, 32.4% in Europe, 5.9% in Eastern Mediterranean, 2.1% in Africa, and 1.7% in Western Pacific, respectively (World Health Organization, 2020b). The quick spread of the disease resulted in a global lockdown to stop the virus spread (Al Huraimel et al., 2020; Lau et al., 2020). However, this action has adversely affected the economy, where the International Monetary Fund (IMF) anticipated a 3.5% recession in the global economy in 2020 due to COVID-19, which outweighs the 2008's recession (International Monetary Fund, 2020; Wang and Su, 2020). However, the world output is expected to increase by 5.5% and 4.2% in 2021 and 2022, respectively (International Monetary Fund, 2021). It may be caused by reducing people and goods' movement, especially in countries with the largest world economies (Baldwin and Mauro, 2020; Fernandes, 2020; Mostafa et al., 2021).
According to the International Civil Aviation Organization (ICAO), international air travel has dropped from 4.5 billion passengers taking flights in 2019 to 1.8 billion in 2020, leading to a drop of more than 50%. Hence, this staggered financial losses of more than $370 billion (Economic Development, 2021). Also, the sea freight volumes have decreased by 11% in the United States and by 4% in the EU27 between April and June 2020 compared to June 2008 (International Transport Forum, 2020). It is essential to find a way to provide an early warning for any future pandemic to avoid a deterioration in the health and economic sectors, as happened during the COVID-19 pandemic. The three main approaches that are applied to provide an early warning include sewage-based epidemiology, air biosensors, and computer techniques, such as artificial intelligence (AI) and its related Machine Learning (ML) technologies (Orive et al., 2020; Ribeiro et al., 2020a; Vaishya et al., 2020).
Wastewater-based epidemiology (WBE) is a commonly used approach to provide quantitative and qualitative information about inhabitants' usage of drugs within a given wastewater catchment (Orive et al., 2020). Recently, this approach was proposed in the infectious diseases field (Al Huraimel et al., 2020; Orive et al., 2020). During the SARS-CoV-1 outbreak, between 16 and 37% of the patients suffered from diarrhea (Amoah et al., 2020; Yeo et al., 2020). Also, during the current COVID-19 pandemic, between 2 and 35% of the patients suffered from the same symptom (Wang et al., 2020a). Therefore, the study detected viral ribonucleic acid (RNA) in feces and sewage (Amoah et al., 2020; Pan et al., 2020). Many researchers have also detected infectious coronavirus virions in feces (Xiao et al., 2020). A recent study detected SARS-CoV-2 RNA in wastewater samples before reporting any case, which means that monitoring the virus could be possible before documenting it via the health surveillance system (Medema et al., 2020; Sherchan et al., 2020). In another study, researchers collected sewage samples in Paris during the pandemic, which showed a positive relationship between the SARS-CoV-2 genome units and the number of fatal cases registered regionally and nationally (Wurtzer et al., 2020).
Air biosensors are used as analytical devices to detect the respiratory virus by converting biological material, such as microorganisms, antibodies, tissues, biomimetic, enzymes, or cell receptors, into measurable signals (Ribeiro et al., 2020a). The biological materials immobilize on a transducer surface, which produces a biochemical response by interacting with the analyte in the solution (Nguyen et al., 2019). The transducer then converts this biochemical response to a quantifiable signal measured using a digital detection module (Nguyen et al., 2019). The four main categories of biosensors are electrochemical biosensors, piezoelectric biosensors, thermal biosensors, and optical biosensors (Bukkitgar et al., 2020; Samson et al., 2020; Suleman et al., 2021). The types of biosensors used for the detection of emerging infectious diseases (EIDs) depends on two main factors: (1) the characteristics of the analyte (i.e., concentration, structure, and size); and (2) the matrix in which the analyte exists (i.e., liquid, air) (Pejcic et al., 2006). Different detected viruses include the influenza virus using electrochemical bio/immunosensor (Saylan et al., 2019). Also, the Middle respiratory syndrome coronavirus (MERS) was detected using optical bio/immunosensor (Ravina et al., 2020; Santiago, 2020) and SARS-CoV-2 using piezoelectric immunosensor or thermal biosensor (Lee et al., 2018; Woo et al., 2020).
AI and its related ML technologies are computer techniques and algorithms that turn the available data into valuable insights and knowledge to detect and diagnose the infection (Allam et al., 2020). It can also track the virus; thus, control its spread in real-time (Vaishya et al., 2020). For the current COVID-19 outbreak, an AI-driven algorithm detected and warned about it on 31 December 2019, seven days before the WHO's official notice (Allam et al., 2020). Similarly, a company called "Metabiota (USA)" which is concerned about epidemics monitoring, used a predictive tool to detect and warn some countries like Japan, Thailand, Taiwan, and South Korea of the coronavirus outbreak in their countries seven days before it was officially announced (Heilweil, 2020).
Neural networks was used as a tool for the prediction of the number of COVID-19 cases (Wieczorek et al., 2020a; Wieczorek et al., 2020b). The results were compared to the actual data obtained in different regions. For instance, in their first study, Wieczorek et al. (2020a) developed a complex artificial neural networks model composed of multiple layers to precisely predict the COVID-19 cases count and spread (Wieczorek et al., 2020a). In addition, this model considered the geographical conditions, i.e. location, latitude, and longitude, as input data. The training data was extracted from the actual number of cases obtained from each region over two weeks before developing the model. Although it is difficult to obtain accurate predictions using limited data, this model maintained a high accuracy of spread estimation. Moreover, in the second or further study, the authors introduced new techniques to their model to make it adaptive, i.e. adjust itself automatically according to the new real-time data and pattern (Wieczorek et al., 2020b). Fortunately, the observed overall prediction accuracy was as high as 88%, hitting the ceiling of 99% for some specific regions. Another factor considered by the authors was the time of prediction. Their concern was to minimize this time for a possible fast prediction. Thus, NAdam training model was utilized as it offers the shortest training time while guaranteeing the highest possible efficiency based on extensive tests conducted by the authors.
Machine learning was also used as a detection tool of the disease itself upon evaluating various diagnostic sources such as X-ray, computed tomography (CT), and ultra-sound images (El-Rashidy et al., 2021). Convolutional neural network (CNN) based models have been commonly used in several studies for detection and prediction purposes (Jaiswal et al., 2020; Jin et al., 2020; Ozturk et al., 2020; Sedik et al., 2020). Furthermore, ML and deep learning (DL) models can help fast drug repurposing or even drug discovery for a novel disease, such as in COVID-19 (Ivanov et al., 2020; Jain et al., 2021; Mohanty et al., 2020; Zhou et al., 2020). These examples make it clear that utilizing ML and DL in real-world applications such as detecting individual cases, predicting numbers of cases, spread modeling, and drug development is a must and not a luxurious action.
This comprehensive study aims to review and analyze the primarily used techniques for viral early detection, focusing on the detection of the SARS-CoV-2 virus. The investigated methods include WBE, air biosensors, and artificial intelligence. This paper investigates the literature and analyzes the most commonly applied WBE techniques and the state-of-art technologies used for air virus surveillance. Moreover, it reviews the applicability and potential of the AI and ML algorithms for viral outbreaks. Based on the existing literature reports, a novel framework combining WBE and AI/ML technologies has been proposed in this paper. This framework exploits an ensemble of ML and DL techniques that can help predict and forecast future viral outbreaks, in addition to recommending a set of actions to mitigate the spread of the pandemic. Finally, some remarks and recommendations are provided for applying the framework proposed in this work and the best AI technologies. Additionally, the importance of collaboration between researchers/scientific community and the stakeholders to address future challenges in viral outbreaks is also discussed. The proposed framework can be tested within a collaborative project using datasets collected from different organizations, and the results can be disseminated in a separate study.
2. Modes of environmental transmission
The COVID-19 pandemic is caused by SARS-CoV-2 (Foladori et al., 2020). Coronaviruses have single-stranded RNA (Dabbish et al., 2021; Yeo et al., 2020) and usually infect humans and other animals' respiratory, gastrointestinal, and nervous systems (Chen et al., 2020b). During the past decades, coronaviruses have caused many outbreaks. In 2002, the SARS-CoV started, and in 2012, the Middle East respiratory syndrome (MERS-CoV) took away many lives. The symptoms for SARS-CoV-2 are milder than the symptoms for SARS-CoV and MERS-CoV. However, SARS-CoV-2 is considered to be more infectious than the SARS-CoV variant (Chen et al., 2020a).
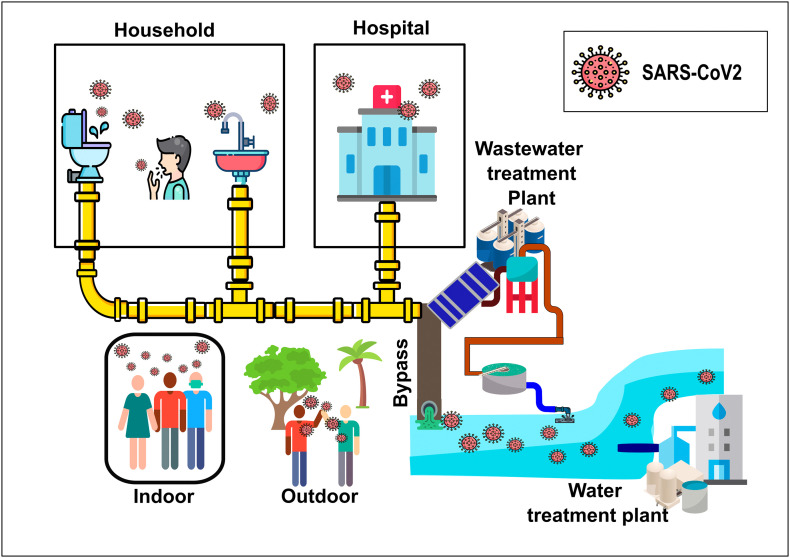
As SARS-CoV-2 is spreading at a faster rate, the mutations of the virus are also observed at a very high rate. To date, more than 1000 mutations for the SARS-CoV-2 have been recorded (Chen et al., 2020a). For the SARS-CoV-2 to infect someone, the virus has to interact with the spike glycoprotein (S protein) and the host angiotensin-converting enzyme 2 (ACE2) receptor while entering the host cells (Sasaki et al., 2021; Walls et al., 2020). The SARS-CoV-2 is becoming more and more infectious as it evolves because the new mutations increase the free binding energy with the ACE2 receptor (Chen et al., 2020a). In the United States alone, a study was done by Wang et al. (2021) showed that the SARS-CoV-2 has four sub-strains and 11 major mutations. The mutation 23403A>G-(D614G) is the second-highest strain found in the United States and is the most dominant strain worldwide (Guruprasad, 2021; Wang et al., 2021; Yurkovetskiy et al., 2020). Toyoshima et al. (2020) found that the two variants, which are ORF1ab 4715 L and S614G, are associated with high fatality rates in 28 countries. The conventional transmission routes for SARS-CoV-2 are respiratory droplets, direct and indirect contact with contaminated surfaces. However, there can be other possible transmission ways depending on the virus load and the surrounding environmental conditions. These possible transmission paths are as follows: (i) stool and sewage, (ii) natural water, and (iii) air environments, which can be indoor and outdoor. Fig. 1 explains how SARS-CoV-2 can be transmitted through different pathways.
Fig. 1.
Transmission pathways of SARS-CoV-2 in the environment.
2.1. SARS-CoV-2 in stool and sewage
The National Institute for Public Health and the Environment (RIVM), a Dutch research institute, detected the SARS-CoV-2 in wastewater treatment plants in the Netherlands. The SARS-CoV-2 was detected in wastewater from Amsterdam Schiphol airport and in a wastewater treatment plant in Kaatsheuvel using molecular Polymerase chain reaction (PCR) methods (National Institute for Public Health and the Environment, The Netherlands, 2020b). Usually, the RNA polymerase gene is amplified to detect the existence of SARS-CoV-2 using conventional PCR techniques (Lau et al., 2003). In a research study conducted by the RIVM, the SARS-CoV-2 virus was detected in 66% of stool samples taken from patients (National Institute for Public Health and the Environment, The Netherlands, 2020a). Wastewater monitoring is a useful approach for detecting viral infections among populations. The RIVM has used this approach to detect norovirus, poliovirus, and measles viruses in wastewater. The RNA of the SARS-CoV-2 can be detected in wastewater. These studies show that SARS-CoV-2 detection in wastewater indicates that the population is infected, however, they do not confirm that the virus particles present in the wastewater are contagious.
A study conducted by Wang et al. (2005) showed that SARS-CoV could survive in wastewater for 14 days at 4oC and for 2 days at 20oC. During these times, the SARS-CoV can be infectious by fecal-oral transmission. Moreover, the MERS-CoV RNA was detected in the feces of 14.6% of the patients. The MERS-CoV can remain viable in sewers at lower temperatures than the SARS-CoV-2 (Yeo et al., 2020). These results confirm that the SARS-CoV-2 and MERS-CoV can be infectious by fecal-oral transmission. In fact, the SARS-CoV-2 belongs to the same coronavirus family as SARS-CoV and MERS-CoV. It is very likely possible that SARS-CoV-2 can also be infectious by fecal-oral transmission. A study done by Zaneti et al. (2021) showed that wastewater treatment plants could be a transmission pathway for the SARS-CoV-2. However, more research is needed to assess the viability of the SARS-CoV-2 under different conditions. There is a high risk for transmission of SARS-CoV-2 through the air when wastewater operators clean the screens manually or when wastewater treatment tanks are not covered. The risk of transmission of SARS-CoV-2 for wastewater treatment workers increases as the population infected increases. The reason is that the lower the infected population is, the higher the dilution of the virus in wastewater (Zaneti et al., 2021).
2.2. SARS-CoV-2 in natural water
Since fecal-oral can be a pathway for SARS-CoV-2 transmission, surface water can be a potential transmission way for SARS-CoV-2, especially in places where there are inadequate sewage facilities and drinking water is contaminated with fecal sludge (Al Huraimel et al., 2020). SARS-CoV-2 pollutes the water bodies by several pathways. One way can be through bypassing the untreated wastewater, which is contaminated by SARS-CoV-2, to rivers, lakes, or other water streams. Another way can be via the treated wastewater effluents contaminated with SARS-CoV-2 due to the inefficient removal of viruses from the wastewater (Bhowmick et al., 2020). The most likely pathway is the discharge of raw sewage directly into water bodies without treatment in areas with low basic sanitation (Al Huraimel et al., 2020). The SARS-CoV-2 has to be active before reaching the water bodies so that the water can be considered as a transmission pathway for the virus. As previously mentioned, the activity of the virus depends on the viral load and the surrounding environmental conditions (Wang et al., 2005). Therefore, the water bodies can be considered as a potential transmission pathway for SARS-CoV-2 when the prevailing environmental conditions can favorably maintain the activity of the infectious virus.
A study conducted on urban rivers of Quito, Ecuador, where wastewater was discharged directly to river streams without any treatment, evaluated the presence of SARS-CoV-2 in river streams. The samples were taken from three different locations along the Quito rivers during the peak of SARS-CoV-2 cases in Ecuador on June 5th, 2020. The results show that the SARS-CoV-2 nucleocapsid protein gene was present in the three sampled locations along the Quito rivers (Guerrero-Latorre et al., 2020). However, it is not confirmed yet that natural water is a transmission pathway for SARS-CoV-2. For SARS-CoV-2 to be transmissible via water, the virus needs to get concentrated enough to persist for long times in water bodies (Bilal et al., 2020). Moreover, the existing activated sludge wastewater treatment plants and drinking water treatment plants need to be more efficient in removing viruses, including the SARS-CoV-2. Treatments like ultraviolet (UV) disinfection and chlorination are expected to kill the SARS-CoV-2 present in contaminated water (Al Huraimel et al., 2020; Pecson et al., 2020). Chlorine doses for removal of bacteria and viruses (including SARS-CoV-2) will change depending on the contact time, chlorine demand, water characteristics, and discharge requirements. However, the chlorine dose often varies between 5 and 20 mg/L (Environmental Protection Agency, 1999). Disinfection using UV radiation is sufficient to inactivate the SARS-CoV-2. Published data suggests that UV doses less than 5 mJ/cm2 are enough to ensure that the effluent water stream is free from coronaviruses (Pecson et al., 2020).
2.3. SARS-CoV-2 in the air environment (indoor and outdoor)
According to the WHO, the primary method for SARS-CoV-2 spread is direct contact with an infected person (Sohrabi et al., 2020). However, airborne transmission of SARS-CoV-2 is a possible pathway since the other coronaviruses like MERS-CoV and SARS-CoV were transmitted via airborne droplets (Hadei et al., 2020; Morawska and Cao, 2020). Several studies were conducted to assess the likelihood of the spreading of the SARS-CoV-2 via aerosol transmission. A study done by Faridi et al. (2020) did not detect any positive SARS-CoV-2 readings from the samples taken 2 to 5 m away from patients' beds at the Imam Khomeini Hospital complex in Tehran, Iran. Another study conducted at the SARS-CoV-2 center in Singapore showed negative results for the aerosol transmission of the SARS-CoV-2 (Ong et al., 2020a).
On the other hand, at the University of Nebraska Medical Center (USA), thirteen SARS-CoV-2 patients were examined for viral shedding by collecting air samples from each individual. Viral contamination was detected in all samples showing that airborne transmission can be a pathway for SARS-CoV-2 spreading (Santarpia et al., 2020). Moreover, Kenarkoohi et al. (2020) examined the aerosol dispersion of SARS-CoV-2 in a hospital in Iran that treats patients diagnosed with SARS-CoV-2. Two viral RNA air samples were tested and detected positive out of six samples collected from the intensive care unit. Kenarkoohi et al. (2020) indicated that indoor environments can be a potential place for the aerosol transmission of the SARS-CoV-2. Aerosols transmission can be through coughing, sneezing, deep breathing, or talking. As the loudness of voice increases during talking, the rate of particle emission increases; therefore, the risk of aerosol transmission becomes higher (Santarpia et al., 2020).
In outdoor environments, apart from the human-to-human transmission, the SARS-CoV-2 can be transmitted through respiratory droplets of an infected person; while talking, sneezing, breathing, or coughing (Freeman and Eykelbosh, 2020). Particles emitted from the infected person can settle quickly if their size is larger than 5 μm; however, if the particle size is smaller than 5 μm, it can stay suspended in the air. The aerosol transmission of large particles (> 5 μm) is limited to a distance of 1 to 2 m. On the other hand, the small particles (< 5 μm) can travel longer distances and get inhaled by another person (Bourouiba, 2020). The risk of infection by aerosol droplets in outdoor environments depends on the aerosols particle size and the virus's ability to contain aerosols to be concentrated enough to cause an infection when inhaled by another person (Freeman and Eykelbosh, 2020). In general, outdoor environments have a lower risk of SARS-CoV-2 transmission than indoor environments (Weed and Foad, 2020).
2.4. Indirect effects on the air quality due to SARS-CoV-2 (indoor and outdoor)
Air pollution has been increasing worldwide, affecting public health (Amin, 2019). However, with the SARS-CoV-2 pandemic, people stayed in their homes, and businesses started to decrease their workload. As a result, air quality improved, and air pollution had reduced; however, this effect is expected to be only temporary (Zambrano-Monserrate et al., 2020). Mostafa et al. (2021) conducted a study to see how the lockdown measures for combating SARS-CoV-2 affected Egypt's air quality, focusing on the two most populous cities Cairo and Alexandria, between the 15th of March and the 1st of May. They compared noise levels, nitrogen dioxide, carbon monoxide, greenhouse gases, and ozone levels before the pandemic (2015–2019) and during the pandemic (2020). The results showed a 75% decrease in the noise level, a 5% decrease in carbon monoxide emissions leading to an average concentration of 120 ppb. Greenhouse gases have decreased by 4%, while a 15% and 33% reduction in nitrogen dioxide was achieved lead to a concentration reduction estimated by 1.5 × 1015 molecules/cm2 over Cairo and Alexandria. On the other hand, the results showed a 2% increase in the ozone levels over Cairo and Alexandria.
Another study conducted by Zambrano-Monserrate et al. (2020) also showed decreased greenhouse gas emissions, nitrogen dioxide emissions, and noise levels during the pandemic. The decrease of nitrogen dioxide emissions and the increase of ozone levels during the pandemic was also documented by Sicard et al. (2020) and Siciliano et al. (2020). Also, it was found that as a result of the lockdown in China, NO2 and PM2.5 emissions in Wuhan city were reduced by 22.8 μg/m3 and 1.4 μg/m3, respectively (Zambrano-Monserrate et al., 2020). As a conclusion of that study, a decrease in PM2.5 and NO2 emissions were observed during the pandemic in several countries, including Italy, Spain, China, Germany, France, and Egypt (Mostafa et al., 2021; Zambrano-Monserrate et al., 2020).
It is well-known that indoor air quality has a significant effect on human health. Each year, 3.8 million people die from diseases related to indoor air pollution (Nwanaji-Enwerem et al., 2020). The sources of indoor air pollution include, but are not limited to, cooking, cleaning, candle burning, smoking, insulation, and personal care products (Habre et al., 2014). During the pandemic, people stayed at their homes most of the time, which led to an increase in indoor air pollution. In houses, individuals were aware of the risk of SARS-CoV-2, increasing the usage of cleaning products, especially disinfectants with hazardous chemicals, which negatively affected indoor air quality in households (Nwanaji-Enwerem et al., 2020). The components of household cleaning products are terpenes, sodium hypochlorite, acetic acid, or ammonia. These chemicals can undergo various reactions leading to an additional pollution source at homes (Weschler and Carslaw, 2018). Furthermore, the prolonged stay at home increased the indoor pollutants associated with cooking (Nwanaji-Enwerem et al., 2020). These pollutants include nitrogen dioxide, nitric oxide, polycyclic aromatic hydrocarbons, acrolein, and nitrous acid (Weschler and Carslaw, 2018).
3. Wastewater-based epidemiology and surveillance
Surveillance and monitoring programs for COVID-19 at wastewater treatment plants are encouraged to be established to assess its abundance (Naddeo and Liu, 2020). Wastewater provides an opportunity for virus surveillance since the viruses excreted by patients can be traced in a wastewater treatment plant from a plant that serves a population. Additionally, this technique can be adequate to estimate the number of infected people during the early stage of the outbreak (Ahmed et al., 2021; Mallapaty, 2020; Wu et al., 2020b). The early tracing of the SARS-CoV-2 can help the health entities and politicians to adopt policies that can ensure the persistence of healthcare systems against its collapse and overwhelming (Al Huraimel et al., 2020). Wastewater plants usually serve large populations with more than one million inhabitants (Mallapaty, 2020). Hence, wastewater surveillance can act as a supplementary approach to clinical testing campaigns to estimate the spread of SARS-CoV-2 in a community (Wu et al., 2020b).
WBE can provide more opportunities than clinical testing, especially for developing countries with limited resources and a lack of clinical testing equipment. However, it still cannot take over the clinical testing, which is still the optimum choice for the infected patients' determinations (Al Huraimel et al., 2020). Additionally, clinical testing is challenging, especially for large populations where testing is time and labor-intensive (Mao et al., 2020). Besides that, WBE can estimate the total infected citizens, including those asymptomatic and presymptomatic (Lodder and Husman, 2020; Mallapaty, 2020). One of the advantages of the WBE is that it can detect viruses at a very low level, which is essential during the beginning of an outbreak or the end of the stationary phase after the healthcare systems interventions (Ahmed et al., 2020).
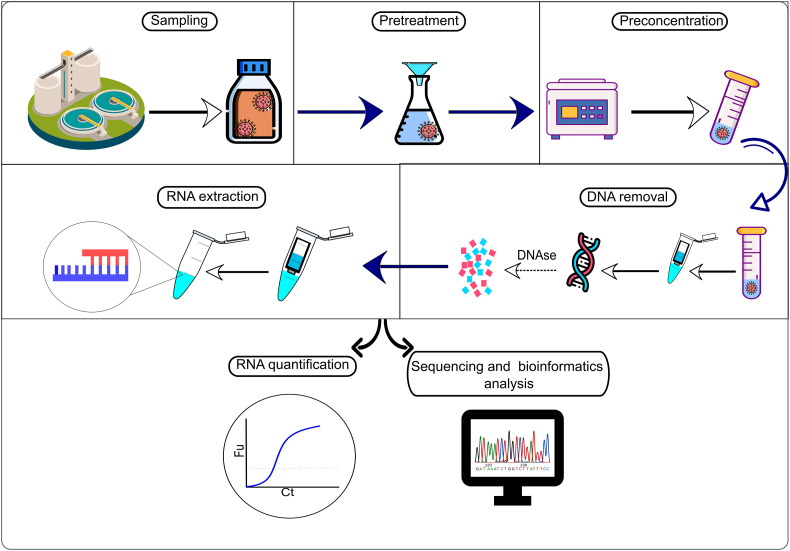
It can be noticed that most of the viral detection techniques are molecular-based. Additionally, the time required, costs, and sensitivity of those techniques varies significantly. The abundance of coronavirus in a population is tracked by wastewater samples determined by the amount of viral RNA excreted in feces per capita. Thereafter, this number can be extrapolated to determine the number of infected people using the concentrations of viral RNA in wastewater (Mallapaty, 2020). To determine the RNA concentration, the sampling preparation process demonstrated in Fig. 2.
Fig. 2.
Sample preparation and analysis process for the SARS-CoV-2's RNA in wastewater.
The WBE has several challenges, especially when building quantitative predictions from the viral RNA, which leads to significant inaccuracies in estimating infected cases. These uncertainties are due but not limited to (i) the complexity of wastewater matrices and the dilute nature of biomarkers in wastewater; (ii) inabilities in pinpointing the suitable sample locations; (iii) non-adequacy of current sampling techniques; and (iv) the need for effective concentrating virus methods (Al Huraimel et al., 2020).
Table 1 demonstrates a summary of the commonly used methods used for viral detection in wastewater. The following subsections present these methods in more detail, along with the testing challenges. Besides, Table 2 illustrates the various approaches used by different countries to detect viruses in wastewater.
Table 1.
A summary of the commonly used methods for viral detection in wastewater.
| Method | Sensitivity | Time required | LOD (SARS-COV2) |
Specificity | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|---|
| RT-PCR | 89-95% | 2 h | 10 copies/μL | 93% |
|
|
(Asif et al., 2021; Nassir et al., 2020; Pilevar et al., 2020; Rabiee et al., 2020; Russo et al., 2020) |
| NASBA | Up to 95% | 60-90 mins | 1 copy/reaction | 98.9% |
|
|
(Fakruddin et al., 2012; Lahrich et al., 2021; Matovu et al., 2010; Chantratita et al., 2004) |
| Biosensors | Up to 96.7% | 15 mins | 0.22 pM | Up to 100% |
|
|
(Choi, 2020; Ejeian et al., 2018; Lahrich et al., 2021; Qiu et al., 2020) |
| Flow cytometry (FCM) | NA | 15-45 mins | NA | NA |
|
|
(Brown et al., 2015; Brussaard, 2004; Ma et al., 2013) |
| ELISA | 20-80% | 2 h | 1 ng/mL | >98% |
|
|
(Feng et al., 2020; Pilevar et al., 2020; Sakamoto et al., 2018; Saville et al., 2001; Streeck et al., 2020) |
| PFGE | NA | 24-26 h | NA | NA |
|
|
(Meays et al., 2004) |
Table 2.
Viral detection and quantification in wastewater and sludge by various countries.
| .Country | Sample source | Sample type (grab or composite) |
Sampling pretreatment | Concentration method | Sample volume (mL) | Concentration (copies/L) | Recovery (%) | Positive percentage (%) |
Detection technique | Sampled virus | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Australia | Raw sewage | Composite |
|
|
100 | 0-120 | NA | 22% | RT–qPCR | SARS-CoV-2 | (Ahmed et al., 2020) |
| Canada | Primary sludge | Composite |
|
|
250 | 1.7 × 103 -3.8 × 105 | 9.3 ± 4.9% | 90% | RT-qPCR | SARS-CoV-2 | (D'Aoust et al., 2021) |
| Chile | Raw sewage | Grab | NA |
|
36 | 4.4 × 103-2.3 × 107 | 47% | 81% | RT-qPCR | polyomavirus JC | (Levican et al., 2019) |
| Czech Republic | Raw sewage | Composite |
|
|
500 | NA | 35.5±13% | 11-27% | RT-PCR | SARS-CoV-2 | (Mlejnkova et al., 2020) |
| Ecuador | River water | Grab |
|
|
2000 | 2.07 × 105- 3.19 × 106 |
NA | 100% | RT-qPCR | SARS-CoV-2 | (Guerrero-Latorre et al., 2020) |
| Egypt | Raw sewage | Grab | NA |
|
5000 | 3.9 × 104 to 3.3 × 108 | NA | 94% | RT-PCR | Pepper mild mottle virus | (Hamza et al., 2019) |
| Egypt | Raw sewage | Grab | NA |
|
5000 | 1.5 × 104 and 1.5 × 107 | NA | 94% | RT-qPCR | Adenovirus | (Hamza et al., 2019) |
| France | Raw sewage | Composite | NA |
|
11 | 5 × 104 - 3 × 106 | NA | 100% | RT-qPCR | SARS-CoV-2 | (Wurtzer et al., 2020) |
| Germany | Raw sewage | Composite |
|
|
45 | 3 × 103 - 2 × 104 | NA | 100% | RT-qPCR | SARS-CoV-2 | (Westhaus et al., 2021) |
| India | Raw sewage | Composite |
|
|
50 | 3.5 × 102 | 57% | 0-100% | RT-PCR | SARS-CoV-2 | (Kumar et al., 2020b) |
| Italy | Raw sewage | Composite |
|
|
250 | 2.4 × 104 - 5.6 × 104 | 2.04 ± 0.70% | 45-65% | RT-qPCR | SARS-CoV-2 | (La Rosa et al., 2021) |
| Italy | Raw sewage | Composite |
|
|
250 | NA | NA | 100% | RT-PCR | SARS-CoV-2 | (La Rosa et al., 2020) |
| Japan | Raw sewage | Grab | NA |
|
100 | 1.2 × 104 -3.5 × 104 | 45% | 47% | RT-qPCR | SARS-CoV-2 | (Hata et al., 2020) |
| Netherlands | Raw sewage | Composite |
|
|
250 | 7.9 × 105 - 2.2× 106 | 73±13% | NA | RT-qPCR | SARS-CoV-2 | (Medema et al., 2020) |
| Spain | Raw sewage | Grab |
|
|
200 | 0-5× 105 | NA | 11% | RT-PCR | SARS-CoV-2 | (Randazzo et al., 2020) |
| Spain | Raw sewage | Composite |
|
|
100 | NA | NA | 100% | RT-qPCR | SARS-CoV-2 | (Balboa et al., 2020) |
| UAE | Raw sewage | Composite |
|
|
250 | 2.86 × 102 -2.90 × 104 | NA | 85% | RT-qPCR | SARS-CoV-2 | (Hasan et al., 2021) |
| USA | Primary sewage sludge | Composite |
|
|
40 | 1.7 × 106 - 4.6 × 108 | NA | 20-60% | RT–qPCR | SARS-CoV-2 | (Peccia et al., 2020) |
| USA | Raw sewage | Composite and grab |
|
|
1000 | 0-7.5 × 103 | 54-56% | 29% (2/7) | RT-PCR | SARS-CoV-2 | (Sherchan et al., 2020) |
3.1. Real-time reverse transcriptase - polymerase-chain-reaction testing (RT-qPCR)
The Reverse Transcription Polymerase Chain Reaction (RT-PCR) is a technique that combines reverse transcription of RNA into DNA and amplifies a specific DNA part with a polymerase chain reaction (Freeman et al., 1999). Real-time PCR (RT-qPCR) is mainly used to quantify the amount of a specified RNA that can be performed by fluorescence monitoring of amplification reactions.
The RT-qPCR was proved to be a sensibly and robust method for the early detection of SARS-CoV-2 during outbreaks (Randazzo et al., 2020). The genetic materials of SARS-CoV-2 have been detected in different wastewater samples worldwide, including Australia, China, France, Germany, Italy, India, Japan, Netherlands, Spain, United Arab Emirates, and the United States of America (Ahmed et al., 2020; Arora et al., 2020; Balboa et al., 2021; Fongaro et al., 2020; Haramoto et al., 2020; Hasan et al., 2021; Kumar et al., 2020b; La Rosa et al., 2020; Rimoldi et al., 2020; Westhaus et al., 2021; Wu et al., 2020a; Wu et al., 2020b; Wurtzer et al., 2020). The RT-PCR was used as a WBE tool and detected viral RNA on several occasions where the number of confirmed cases was low, as in the Netherlands and Spain (Lodder and Husman, 2020; Randazzo et al., 2020). It illustrates the surveillance system's sensitivity and its capabilities as an early warning technique for viral outbreaks.
Kumar et al. (2020b) investigated the presence of the SARS-CoV-2 by using RT-qPCR in India at the Old Pirana wastewater treatment plant, which receives an influent from a hospital that accommodates COVID-19 patients. It was noticed that the quantity of the viral RNA in the wastewater corresponded to the number of declared infected patients at the hospitals. The viral concentration in the influent was ranging between 5.6 × 101 and 3.5 × 102 copies L−1. Gonzalez et al. (2020) studied the frequency of detections and concentrations for twenty-one weeks in south-eastern Virginia, the USA using RT-qPCR. During this period, the concentration range was between 101 and 104 copies per 100 mL in samples where viral RNA was detected. The normalized loading rate fluctuations at the WWTP were aligned with the number of known outbreaks in the study.
3.2. Nucleic acid sequence-based amplification (NASBA)
Nucleic Acid Sequence Based Amplification (NASBA) is another method that can be used for virus detection in wastewater. The NASBA technique relies mainly on the detection of the RNA. NASBA uses several enzymes to amplify the numerous target nucleic acid sequences of the SARS-Cov-2. In contrast to the other nucleic acid amplification methods, NASBA can amplify directly from an RNA without the need for a reverse transcription step (Farkas et al., 2020). The three used enzymes, T7 RNA polymerase, reverse transcriptase, and RNase H, can amplify one strand template of RNA (Hryniszyn et al., 2013). NASBA has the potential to detect a low concentration of DNA or RNA sequences during 15–60 min and at a temperature of 37–65 °C (Farkas et al., 2020).
NASBA was used by Jean et al. (2001) to determine the concentration of Hepatitis A Virus (HAV) in wastewater from the Saint-Nicolas wastewater treatment plant in Canada. Even though there was significant bacterial contamination in the wastewater (2 × 105 fecal coliforms/mL), the results showed high HAV detection. Hence, the bacterial presence doesn't affect the analysis. NASBA was able to detect 0.4 ng of target RNA/mL compared with 4 ng/mL for RT-PCR, indicating NASBA's strong potential (Jean et al., 2001). However, it is noteworthy to mention that several reasons limit the adoption of NASBA as a tool for viral detection in wastewater. This technique has difficulties in producing reliable and quantitative results. Moreover, its costs are relatively higher than the PCR test in terms of viral detection in water and the environment (Walker et al., 2017).
3.3. Biosensors
Biosensors are portable analytical instruments that can detect biological pathogens (i.e., SARS-CoV-2) and proteins with measurable physicochemical properties by converting biological reactions into measurable electrical signals (Goode et al., 2015; Mehrotra, 2016). Given that viral detection, the biological molecule could be the isolated microorganism, the viral nucleic acid, viral protein particles, or the host's antibodies against the virus. A biochemical response is obtained specific to the viral particles or their genome. The transducer captures the recognition response converting it into a quantifiable signal computed by a digital detector, pursued by analyzing and presenting the detected signal via a processor (Samson et al., 2020; Yang et al., 2020). Biological responses can be transduced to measurable signals once they interacted with their specific targets (Neethirajan et al., 2017). Aptamer-based biosensors (Aptasensor) have a single-stranded DNA that can bind to the target protein or DNA with high accuracy, producing a detectable signal once it gets bound (Farkas et al., 2020).
Biosensors have been successfully used to detect norovirus in aquatic samples. The aptasensors used are based on electrochemical, fluorescence, colorimetric, and surface plasmon resonance detection platforms (Farkas et al., 2020; Weerathunge et al., 2019). Generally, biosensors are resistant to environmental inhibitors, allowing high recovery of it and allowing low detection limits (Schilling et al., 2018). Moreover, the biosensors can detect viruses in a short period of 7–16 min (Liu and Zhu, 2005). Furthermore, the interaction between molecules can be observed in real-time (Liu and Zhu, 2005). Qiu et al. (2020) succeeded in using a dual-function plasmonic biosensor that combines plasmonic photothermal effect and surface plasmon resonance to detect SARS-CoV-2 RNA. The biosensor showed high accuracy with a low detection limit of 0.22 pM. Nevertheless, considering the necessity of rapid detection of the infection status within a population and the viral presence in the environment, biosensors can serve as a potential tool in the surveillance and detecting SARS-CoV-2 (Lahrich et al., 2021).
3.4. Nano(bio)sensors and quantum dots
The development of nano(bio)sensors is essential due to their unique physicochemical and optical properties of materials of small sizes (Suleman et al., 2021). These properties give the nano-sensors high accuracy and sensitivity besides the fast detection of viral infections. In addition, merging the nano-sized sensors with the electrochemical detection methods offers the advantage of reduced viral detection time and the ability to detect low concentrations (Bukkitgar et al., 2020). This combination is considered a cost-effective method for fast detecting of coronavirus-induced infections (Lim and Bonanni, 2020).
In order to prepare the nano-sensors, a surface modification step of the parent nano-particles should be done. One method of surface modification or functionalization is the loading or doping with other active materials having high sensitivity to viruses and pathogens (Shetti et al., 2021). In this regard, various nano-sensors were developed, including nano-spherical particles, nano-islands and nano-wires of gold, and graphene nano-particles to detect various viral infections, including SARS-CoV-2 (Antiochia, 2020; Asif et al., 2020). For instance, Au fiber Bragg grating FBG probe decorated with graphene oxide nanoparticles was used as a sensor to rapidly detect COVID-19 from patients' saliva (Samavati et al., 2020). Additionally, Vadlamani et al. (2020) prepared a low-cost and highly sensitive sensor consisting of nano‑cobalt supported on titania nano-tubes to detect the SARS-CoV-2 virus (Vadlamani et al., 2020). Fortunately, this detection method was relatively quick because of the use of an electrochemical sensor that detects the spike protein on the virus's surface. In a recent study, a new dual-functional plasmonic biosensor containing two-dimensional gold nano-islands decorated with DNA receptors was tested as a promising COVID-19 virus analyzer (Qiu et al., 2020). It is worth noting that this analytical device combines both the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction.
In addition, other forms of carbon-based nano-sensors are deemed to be excellent candidates for viral detection, such as carbon nano-tubes and dots (Wang and Dai, 2015), in addition to biochars (Spanu et al., 2020). This is owed to their excellent electrochemical and optical sensing characteristics. From an economic and environmental point of view, biomass-derived from waste matrices (e.g., agricultural residues) has received much attention to prepare these carbonaceous materials (Bhat et al., 2020). Moreover, there are other types of bio-sensors such as electro-spun nano-fibers, 1-D carbon nano-tubes, and quantum dots that are small in size and have high activity and accuracy towards viral detection (Castillo-Henriquez et al., 2020; Shetti et al., 2021).
Quantum dots (QDs) are considered very promising sensors as they are highly selective (Castillo-Henriquez et al., 2020). Besides, they can detect a diverse range of substances/compounds in a wide range of clinical conditions rapidly and without the requirement of further laboratory tests. The fluorescence and photo-electrochemical features of the developed QDs enable them to detect various microRNAs, DNAs, and proteins which are evidence of their ability to detect a wide variety of viruses (Ma et al., 2018). Therefore, they were successfully used for the detection of circulating cell-free miRNAs in lung carcinogenesis (Singh et al., 2018), plant viruses (Hong and Lee, 2018), and most recently, the COVID-19 virus (Manivannan and Ponnuchamy, 2020). They were used not only for the detection but also for combatting the virus by hampering the genomic replication of the viral RNA.
3.5. Nucleic acid staining with fluorescent dyes (NASFD)
Highly fluorescent nucleic acid dyes can be used to detect the SARS-CoV-2 virus in wastewater, wherein the wastewater samples pass via a filter with a pore size of 0.22 μm (Maestre-Carballa et al., 2019). After that, the nucleic acids in the SARS-CoV-2 virus particles get stained by a fluorescent dye that allows the formation of fluorescent dots with dimensions more significant than the actual virus particles. These fluorescent dots appear after the excitation of the fluorescent dye bounded with the nucleic acids. Thus, the fluorescent dots will be counted as SARS-CoV-2 virus particles by viewing the sample under an epifluorescence microscope (Corpuz et al., 2020).
One of the most common dyes that have been used widely in recent research is SYBR Green I, whose blue emission is observed with the help of an epifluorescence microscope (Tonkrongjun et al., 2019). Wu and Liu (2009) used SYBR Green I to count viruses from influent, effluent, and sludge samples. Also, Otawa et al. (2007) adopted a direct count method with SYBR Green I as a staining agent to count the number of virus particles in mixed liquor-activated sludge samples.
One of the pros of using this method is that counting stained virus particles even under lower magnifications is possible; thus, the need for transmission electron microscopy (TEM) instrumentation can be avoided (Ortmann and Suttle, 2009). Also, there is no need to cultivate the samples in the laboratories since the virus particles can still be counted using this method. Moreover, it will be easier to differentiate the virus particles with nucleic acids from virus-like particles without nucleic acids through DNase treatment (Forterre et al., 2013).
3.6. Flow cytometry (FCM)
Another method employed to count SARS-CoV-2 virus in wastewater samples is by combining fluorescent nucleic acid-specific dyes with flow cytometry (FCM). Firstly, in the FCM unit, wastewater samples are diluted with the buffer solutions, stained with the fluorescent dyes (e.g., propidium iodide, thiazole orange, SYBR Green I, SYBR Green II, and SYBR Gold), and injected into the flow cytometer. After that, the SARS-CoV-2 virus particles enter a stream in a single file under the hydrodynamics effect of the surrounding sheath fluid. Then, the individual particles intersect with a monochromatic light beam, usually from an argon-ion laser. Finally, each particle's interactions with the incident laser beam produce scattered and fluorescence particles that can be collected by detectors and get analyzed as scatter and fluorescence intensity, respectively (Corpuz et al., 2020).
Furthermore, in the FCM method, the wastewater samples are stained with a fluorescent dye that binds selectively either to the DNA or RNA. Hence, the intensity of the DNA/dye and RNA/dye complexes' fluorescence correlates with the sample's DNA/RNA content (Adan et al., 2017). The positives about using FCM are its high accuracy and quantification speed (Brown et al., 2019). The FCM approach was used by Huang et al. (2016) to quantify the virus particles in activated sludge and effluent samples from three wastewater reclamation plants. According to another study done by Brown et al. (2019) on determining the number of virus particles from activated sludge samples using FCM, it was shown that the FCM method has a higher sensitivity than that of epifluorescence microscopy (EFM). Consequently, it can be said that FCM has a higher sensitivity and quantification speed in counting virus particles compared to the EFM method.
3.7. In situ fluorescence
A different method that can only be performed in specialized laboratories is fluorescence-based analysis. The conceptual design of a fluorescence instrument has been presented by Pollard (Pollard, 2012) that in-situ and online monitoring of viruses' numbers in varying water matrices, including effluent wastewater, can be done. The proposed fluorescence device has an inline filter to remove any bacteria present in the wastewater sample. After this filter, a mixing coil is placed, where DNase and the fluorescent probe SYBR-Gold are added to the sample to form the DNA/RNA-SYBR viral complex. After that, this mixture is directed to reverse osmosis (RO) filter to concentrate the viruses. The RO concentrate, stained viral particles, passes across a unit to measure the fluorescence signal. While the permeate becomes the background fluorescence that is subtracted from the sample viral fluorescence.
Despite that the proposed device is still under the conceptual stage, this method's tests have shown that the excitation-emission matrix (EEM) fluorescence intensity has a linear correlation (r2 = 0.97) with the viral count.
3.8. Immunofluorescence assay (IFA)
Immunofluorescence (IFA) is a method that can be used to quantify and analyze the infectivity of SARS-CoV-2 virus. In this method, an infected cell culture sample is adsorbed on a microscopic slide. Next, the fixed sample is incubated sequentially with a specific antibody and a fluorescent chemical-conjugated secondary antibody that recognizes the former to detect the viral protein antigen. Finally, the fluorophore-conjugated antibody is fluoresced under optical excitation, where the antigen-antibody complex appears as a fluorescent particle under a fluorescent microscope (Corpuz et al., 2020; Im et al., 2019).
Calgua et al. (2011) used this method successfully to quantify HAdV and JCPyV in raw sewage inflowing into wastewater treatment plants. The results showed that IFA has one higher order of magnitude in sensitivity compared with other cell culture methods used in this study. Schlindwein et al. (2010) used the IFA method to verify human adenovirus 2 (HAdV 2) viability in an activated sludge and effluent samples from a wastewater treatment plant.
3.9. Enzyme-linked immunosorbent assay (ELISA)
The enzyme-linked immunosorbent assay (ELISA) is a method utilized to detect the presence of microbial antigens in various matrices. ELISA depends on the antigen-binding principle to its specific antibody, causing a change in color or fluorescence due to the resultant enzyme activity.
This method works as follows: firstly, antigen-binding at a specific antibody is immobilized on a surface, usually in a set of 96-well microtiter plates. Secondly, a second enzyme-linked antibody, which is specified for the same antigen, is utilized to form an antibody-antigen-antibody sandwich. Then, the enzyme-coupled antibody is reacted with a substrate that alters its color when modified by the enzyme. Finally, the color change or fluorescence is correlated with the probed antigens' concentration in the sample (Corpuz et al., 2020). This method was utilized successfully by Atabakhsh et al. (2020) to detect rotavirus in influent and effluent samples of wastewater treatment plants. In that study, ELISA methods helped determine the removal efficiencies of rotavirus from urban wastewater treatment systems.
3.10. Pulsed-field gel electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) is a method that uses a pulsating electric field that enables the separation of high molecular weight DNA fragments of the SARS-CoV-2 virus according to their molecular sizes (Corpuz et al., 2020; Nassonova, 2008). In this method, two separate electrodes are used to generate alternating electric fields that cause reorientation of the molecules periodically to align to the imposed electric field (Le et al., 2017). Molecular sizes and DNA molecules' charges affect their ability to re-orient themselves and respond to the set modulated electric fields. Smaller DNA molecules usually take a shorter time to re-orient themselves by migrating through the gel matrix pores towards the new anodes; while, larger molecules take a longer time. Also, the larger DNA molecules, which migrate slower than the set pulse time, tend to migrate as one band via the gel matrix (Le et al., 2017; Lopez-Canovas et al., 2019). Furthermore, the band pattern formed by a viral community works as its fingerprint, where the number of the formed bands estimates the number of different viruses (diversity) in a sample (Corpuz et al., 2020).
The PFGE method was applied by Otawa et al. (2007) to estimate the diversity of viruses in activated sludge samples from 14 different wastewater treatment plants. The results showed that the prevailing sizes of viral DNAs in the samples were in the range of 40–70 kb. The PFGE method was also able to demonstrate the mutual similarity in the detected viral species from the different activated sludge wastewater treatment plants based on the similarity of the band patterns. Another study was done by Wu and Liu (2009), where they used PFGE to determine the virus diversities in influent, primary settling tank, effluent, and sludge samples from a municipal wastewater treatment plant. The results revealed that the activated sludge, anaerobic digestion sludge, and effluent had the largest number of bands; besides, there were similar band patterns between the influent and primary settling tank samples. The results also showed that the dominant sizes of viral DNAs were between the ranges of 30–80 kb and 200–350 kb.
3.11. Transmission electron microscopy (TEM)
Optical microscopes cannot detect viruses. However, Electron microscopes can identify and determine viruses (Corpuz et al., 2020). Transmission electron microscopy is one of the earliest techniques to identify and classify viruses based on morphology (Roingeard et al., 2019). The working principle of a TEM depends on negative staining, where the SARS-CoV-2 viral particles are adsorbed on a pre-treated electron transparent sample support grid (Corpuz et al., 2020). Usually uranyl acetate and phosphotungstic acid, are then used for staining. The samples are then analyzed using the electron microscope. The negative staining allows SARS-CoV-2 viral counting and viral size and structure determinations (Laue, 2010).
García-Fontana et al. (2020) visualized viral particles from Grenada, Spain's Los Vados wastewater treatment plant. The TEM analysis showed differences depending on the sampling location. For example, a wide diversity of Eukarya viral particles and bacteriophages were detected in the influent. Similarly, the same observations were noticed in the WWTP effluent and sludge. The examination of wastewater samples by TEM also elaborated the diversity of viral morphologies with filamentous viral particles. Moreover, the TEM examination showed that 300 nm length of viral filamentous particles are typical for tobamoviruses (Bacnik et al., 2020).
The advantages of using the TEM to determine viruses during an outbreak rely on the fact that viruses' morphologies do not change after a mutation of the nucleic acid. On the other hand, the TEM is highly selective to host-specific infectious viruses, leading to a lower count than the real one (Brown et al., 2015; Corpuz et al., 2020). Additionally, the TEM is challenging to be used for investigating high numbers of samples since it requires experienced personnel (Barreto-Vieira and Barth, 2015; Corpuz et al., 2020).
3.12. Testing challenges in wastewater
The testing for SARS-CoV-2 in wastewater is not fully standardized yet and still lacks a fully standardized protocol for accurate detection and determination (Sherchan et al., 2020). Al Huraimel et al. (2020) illustrated that quantitative determination of infected citizens by SARS-CoV-2 from wastewater is a challenging process due to several reasons such as (i) the amount of viral RNA existing in feces, (ii) the lack of representative samples along the periods, (iii) the detection ability of kits at low concentrations, (iv) the variability of viral RNA excretion rates between inhabitation, (v) the vague links between the predictions of the number of cases and the amount of detection RNA concentration in wastewater, (vi) the necessity to evolve envelop virus concentrating methods (Ahmed et al., 2020; Mallapaty, 2020).
4. Coronaviruses air sampling and detection
4.1. Coronaviruses air sampling
An essential step in microbial air sampling is to eliminate any damage that could affect the microbial cell by limiting the microbial stress (e.g., long sampling time and dryness effect) during the sampling procedure. Furthermore, preserving the cell viability is not as important as ensuring the nucleic acid integrity, especially for RNA viruses, as the RNA possesses high instability (Verreault et al., 2008). Maintaining optimum sampling conditions (sampling volume, flow rate, and time) also plays a major role in precise sampling and targeted virus detection. Multiple factors can affect the indoor sampling of SARS-like viruses, including air movement, ventilation, humidity, and temperature (Guo et al., 2020). For hospital air sampling, patients' numbers inside the room, occupancy, distances from the patient's bed, and activity during sampling could influence the results (Cox et al., 2020). Therefore, surrounding factors such as sampling environmental conditions and sampler type should be assessed upon standardizing a protocol for SARS-CoV-2 air sampling (Rahmani et al., 2020).
Bioaerosols samplers are classified into four categories, as follows: liquid impingers, filters, impactors, and cyclones (Borges et al., 2021; Sung et al., 2017). For indoor air sampling, biosamplers are usually located 40–50 cm (Dubuis and Duchaine, 2021) and up to a distance of 1.5–2 m (Dumont-Leblond et al., 2021; Dumont-Leblond et al., 2020) from the resident for collecting the optimum and representative air sampling volume.
Liquid impinge-type biosamplers collect/absorb the bioaerosols into a liquid medium. Most of the recent studies have recommended using the biosampler for air sampling of SARS like viruses (Rahmani et al., 2020). The biosampler is normally equipped with a high flow rate vacuum pump ranging from 10 to 60 L/min to collect the air samples (Rahmani et al., 2020). Due to the use of a high flow rate vacuum pump, this sampling tool may be more appropriate for outdoor air sampling rather than for collecting samples from indoor environments. However, Faridi et al. (2020) have applied this impinger technique for the sampling of SARS-Cov-2 in an indoor environment (Imam Khomeini Hospital Complex) after replacing the high volume vacuum pump by an average flow-rate pump, although the limited flow rate capacity negatively impacted the overall effectiveness of the viral detection. It has been reported that liquid impingers are the most widely used tool for sampling viruses in the air because they help to maintain the viability and integrity of the viral particles during sampling (Rahmani et al., 2020). Additionally, the viral analyses can be conducted directly without the need for viral extraction from the medium and/or a filter surface (Pan et al., 2019; Rahmani et al., 2020).
Moreover, filters are extensively used for viral air sampling because of the high efficiency in capturing virus-containing aerosols having a particle size ˂ 500 nm. The four basic filtration mechanisms are interception, diffusion, inertial impaction, and electrostatic attraction. Different materials for the membrane filters have been tested in sampling airborne viruses, such as polytetrafluoroethylene (PTFE), cellulose, tightly packed cotton, gelatin, Teflon, and polycarbonate (PC) (Pan et al., 2019; Rahmani et al., 2020). Gelatin and PTFE are the most widely used filters for air sampling of SARS-like viruses (Booth et al., 2005; Fabian et al., 2009). The advantage of using the gelatin filter over the other commonly used filters is their ability to dissolve in the liquid medium that facilitates the enumeration of microbial or viral particles in cell cultures without any harmful impact on the viability of the viruses (Pan et al., 2019). Besides, gelatin filters have been commonly used in modern air sampling instruments, such as MD-8 Airscan sampler (Rahmani et al., 2020). Zhao et al. (2014) have reported that the MD-8 Airscan sampler with a gelatin filter enhanced the efficiency of collecting RNA viruses, i.e., ~ 100%. Similarly, PTFE filter has been recommended by the National Institute for Occupational Safety and Health (NIOSH) for sampling airborne viruses (Rahmani et al., 2020), owing to its unique structure, which offers an advantage of easy elution of the target viruses from the membrane without any interaction between them (Lindsley et al., 2017).
Although filters are very effective in collecting virus-containing particles, they also have some drawbacks, including the possibility of viral destruction in certain conditions, such as sampling of large air volumes (Pan et al., 2019; Rahmani et al., 2020). High volume sample pumps require a longer sampling time, resulting in filter dryness and thus causing damage to the viruses (Rahmani et al., 2020). Other drawbacks that were reported for gelatin filters include: (i) dissolution of gelatin filters may occur in case of sampling at high relative humidity or if high volume sample pumps are used, (ii) desiccation of viruses may take place in the case of sampling at low relative humidity (Fabian et al., 2009; Pan et al., 2019; Rahmani et al., 2020). Impactors like the Andersen 6-stage sampler and the slit sampler collect airborne particles into a liquid medium, where a vacuum pump is used to withdraw the aerosol. The collection media is then recovered, and aliquots are used for viral isolation (Pan et al., 2019). The use of a high-resolution slit-sampler in the collection of SARS coronaviruses was also addressed in a previous study (Booth et al., 2005). However, negative results were reported for all the viral cultures, although 2 out of the 10 samples were tested positive using RT-PCR. This could be attributed to the low concentrations of the viruses in the air or that these instruments are inappropriate for sampling aerosolized viruses (Pan et al., 2019).
Cyclone sampler exerts centrifugal forces on particles in order to be separated from the air flow. This sampler has been used for sampling influenza A virus (IAV) and SARS-like viruses (Pan et al., 2019; Rahmani et al., 2020). The NIOSH has developed a multistage cyclone sampler that operates at 3.5 L/min. The first stage consists of a 15 mL tube that captures aerosol particles larger than 4 μm. The second stage consists of a 1.5 mL tube that captures aerosol particles between 1 and 4 μm. While the third stage consists of a polytetrafluoroethylene (PTFE) filter that collects aerosol particles smaller than 1 μm (Chia et al., 2020). However, the cyclone sampler was not efficiently successful in collecting SARS-CoV-2 viruses present in indoor air of a hospital complex (Chia et al., 2020). It was reported that physical damage might have occurred to the viruses due to the impaction forces created during the sampling process (Blachere et al., 2009; Brown et al., 2013; Cao et al., 2011; Pan et al., 2019).
Table 3 summarizes the commonly used air samplers for coronaviruses sampling and the limitations of each sampler. Several precautions must be taken in consideration during the selection step of the air sampler for both indoor and outdoor environments. For example, it is recommended not to operate the cyclone sampler with an airflow rate greater than 200 L/min to avoid the degradation of viral RNA in the collected samples (Borges et al., 2021). Additionally, it is recommended not to use high-volume sample pumps with filter samplers to evade viral dehydration (Robotto et al., 2021).
Table 3.
Summary of the commonly used air samplers for corona viruses sampling.
| Sampler | Target virus | Sampling time | Sampling flow rate (sampling volume) | Limitations | References |
|---|---|---|---|---|---|
| Filter: MD-8 airscan sampler |
MERS-CoV |
20 min |
|
|
(Kim et al., 2016) |
| Filter: MD-8 airscan sampling device (Sartorius) and sterile gelatin filters |
MERS-CoV |
20 min |
50 L/min (1000 L) |
(Azhar et al., 2014) |
|
| Filter: High-efficiency particulate air (HEPA) filters installed in the pipeline connecting sampler and vacuum pump |
SARS coronaviruses |
4 h |
4 L/min (960 L) |
(Agranovski et al., 2004) |
|
| Filter: PTFE membrane filter with a pore size of 0.3 μm in a closed-face, 3-piece disposable plastic cassette attached to a personal sample pump |
SARS coronaviruses |
10.5 – 13 h |
2 L/min (1260–1560 L) |
(Booth et al., 2005) |
|
| Filter: MD-8 airscan sampler |
SARS-CoV-2 |
15 min |
100 L/min (1500 L) |
(Ong et al., 2020a) |
|
| Filter: Sterilized gelatin filters with pore size 3 μm placed in styrene filter cassette |
SARS-CoV-2 |
1 h |
5 L/min (300 L) |
|
(Liu et al., 2020) |
| Impinger: attached to a personal sample pump with average flow |
SARS-CoV-2 |
1 h |
1 L/min (60 L) |
|
(Faridi et al., 2020) |
| Impactor: High-resolution slit-sampler system |
SARS coronaviruses |
18 min |
30 L/min (540 L) |
|
(Booth et al., 2005) |
| Cyclone: SASS 2300 wetted wall cyclone sampler |
SARS-CoV-2 |
30 min |
|
|
(Borges et al., 2021; Guo et al., 2020) |
| Cyclone Bioaerosol Sampler |
SARS-CoV-2 |
4 h |
3.5 L/min (840 L) |
(Chia et al., 2020) |
|
| Cyclone and filter: Cyclone sampler and 37-mm filter cassettes and 0.3-μm polytetrafluoroethylene filters | SARS-CoV-2 | 4 h | 100 L/min (400 L) | (Ong et al., 2020a) |
4.2. Coronaviruses detection techniques
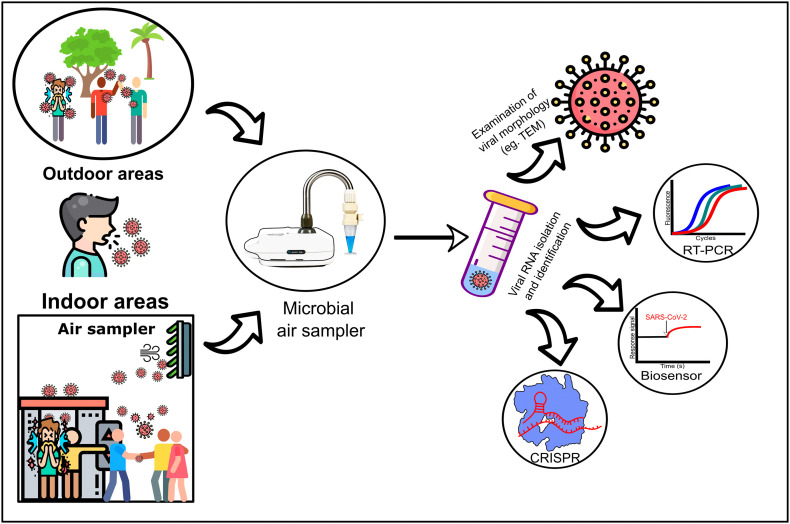
Following the sampling phase, the viral RNA is extracted from the sample matrices using the viral RNA/DNA isolation kit and stored in an elution buffer for further analysis (Azhar et al., 2014; Faridi et al., 2020). Once the viral genomic material is extracted, several examinations and identification techniques are performed, as demonstrated in Fig. 3 (Corman et al., 2020; Drosten et al., 2002; Shetti et al., 2021). Nowadays, multiple identification techniques are modified to detect viruses in air samples with high efficiency as shown in Table 4 . Moreover, Cheng et al. (2020) studied SARS-CoV-2 air sampling in an isolation room and highlighted that positive environmental air samples were detected only if the patient's viral load within the clinical samples exceeded 1000 copies/mL. They also reported that the SARS-CoV-2 viral load ranging from 110 to 9400 copies/mL resulted in a positive SARS-CoV-2 in the environmental air samples (Cheng et al., 2020), with a distance ≥2 m between the air sample and the patient (Lednicky et al., 2020).
Fig. 3.
SARS-CoV-2 air survaillence, sampling, and identification.
Table 4.
Summary of the commonly used techniques for viral detection in air.
| Method | Mechanism of detection | Time required* | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| RT-qPCR | Nucleic acid amplification | 2 h |
|
|
(Guo et al., 2020; Ong et al., 2020b) |
| RT-LAMP | Nucleic acid amplification | 20 min |
|
|
(Park et al., 2020; Thi et al., 2020; Yu et al., 2020; Zhu et al., 2020) |
| TEM | Viral morphological examination | Few min |
|
|
(Agranovski et al., 2004; Kim et al., 2016) |
| CRISPR-Cas | Nucleic acid identification –gene editing | 40 min |
|
|
(Abduljalil, 2020; Broughton et al., 2020; Jia et al., 2020) |
| Biosensors | Measuring biological response | 15 min |
|
|
(Iravani, 2020; Kim et al., 2020; Qiu et al., 2020; Samson et al., 2020) |
4.3. Reverse transcriptase real-time polymerase chain reaction testing (RT-qPCR)
The gold standard technique for SARS-CoV-2 detection in air samples is RT-qPCR owing to its high sensitivity and accuracy (Lednicky et al., 2020; Lei et al., 2020; Rahmani et al., 2020), as previously mentioned. Although RT-qPCR can precisely identify SARS-CoV-2 in several clinical specimens including, blood, saliva, sputum, stool, and urine (Peng et al., 2020; Wang et al., 2020b), various studies have proved the effectiveness of RT-qPCR in detecting SARS-CoV-2 in the air as well. This technique can predict thousands of viral copies present in 1 mL of liquid aerosol within the proper limit of detection (LOD) (Ong et al., 2020b).
Therefore, a key step in the air monitoring system is to enrich the viral concentrations, which are distributed in the environment, into smaller volumes of liquid to exceed the LOD, in conjunction with increasing the sample volume. Guo et al. (2020) have examined the indoor environment within hospitals using RT-qPCR, and they detected the presence of SARS-CoV-2 in patients' rooms, around air outlets, and in physician's office areas. A large sample volume (~ 9000 L) was collected using the SASS 2300 wetted wall cyclone sampler to achieve positive results (Guo et al., 2020). The advantages of using RT-qPCR are the high detection sensitivity, ease of use, relative rapid results, and the simultaneous diagnosis of multiple respiratory agents.
4.4. Reverse transcription-loop-mediated isothermal amplification (RT-LAMP)
Another technique for SARS-CoV-2 identification based on nucleic acid amplification is the reverse transcription-loop-mediated isothermal amplification (RT-LAMP), which offers an advantage of accurate and sensitive detection of SARS-CoV-2 in clinical samples (Yu et al., 2020). This novel technique is used to overcome the constraints of qPCR as long processing time. Unlike RT-qPCR, RT-LAMP is processed under the isothermal condition at constant temperature (65 °C), thus reducing the time consumption as temperature changes; therefore, the reaction lasts only 20 min (Park et al., 2020). Additionally, the LOD of RT-LAPM is only 80 copies of RNA/mL of samples, which is considered lower than the limit detected by qPCR (Huang et al., 2020). The concept of nucleic acid detection relies on magnesium pyrophosphate production upon amplification resulting in turbidity that could be seen by the naked eye and quantified using a turbidimeter (Hong et al., 2004).
Based on the LAMP technique, Zhu et al. (2020) have developed nanoparticle-based biosensors (NBS) to achieve the benefits of rapid and precise detection of SARS-CoV-2. In RT-LAMP-NBS one-step assay, the LAMP primers are designed to detect and amplify the opening reading frame fragment 1a/b sequence and nucleoprotein sequence of the SARS-CoV-2 genome in one tube. The NBS detects and analyzes the results. With the aid of this biosensor, it can predict very low concentrations of SARS-CoV-2 (12 copies) per reaction (Zhu et al., 2020). Therefore, it overcomes non-specific amplification of other non-SARS-CoV-2 genomic sequences, increasing the specificity and reducing false-positive outcomes. RT-LAMP was widely used for SARS-CoV-2 detection in clinical samples. Nevertheless, it has not been applied yet for SARS-CoV-2 detection in air samples (Thi et al., 2020).
4.5. Transmission electron microscope (TEM)
As previously discussed in an earlier section, TEM is beneficially used to recognize and characterize viruses along with various matrices. However, viral identification in air samples using TEM is challenged by the viral concentration enrichment within the sample to reach a detectable limit (Richert-Poggeler et al., 2018). Subsequently, previous studies on different coronaviruses relied primarily on large sample volumes. Kim et al. (2016) have reported MERS-CoV existence, and Agranovski et al. (2004) had positive results for SARS-CoV in air samples when the volume of the sample in both studies exceeded 1000 L (Agranovski et al., 2004; Kim et al., 2016). Another pre-requisite step to increase the viral load in the collected sample is through viral cultures. The tubing used to interconnect the sampler with the vacuum pump are fitted with HEPA (high-efficiency particulate air) filters. Such filters are inoculated in a flask with confluent monolayers of Vero cells (viral host cells); after that, the flask is incubated at 37 °C for 60 min. The inoculated cell cultures are then preserved at the proper culture medium, and finally, the culture supernatant is examined under the TEM to identify the virus (Agranovski et al., 2004).
4.6. Clustered regularly interspaced short palindromic repeats (CRISPR) – Cas (CRISPR associated) technique
Introduction of modern gene-editing CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) - Cas (CRISPR associated) technique was extremely vital in the field of viral detection (Bayat et al., 2018). This technology targets viral RNA degradation; thus, it controls viral replication within the host cell and reduces transmission (Brouns et al., 2008). CRISPR-Cas system was discovered firstly as part of natural bacterial and archaeal immune responses against viruses. Afterward, scientists repurposed its use for gene-editing in mammalian cells (Jinek et al., 2012). Recently, the CRISPR-Cas system is a potential tool for COVID-19 molecular diagnosis and therapeutic approaches (Kumar et al., 2020c; Zuo et al., 2017).
Applying the CRISPR-Cas system for SARS-CoV-2 detection improved the sensitivity, specificity, and detection time (30–60 min) over traditional PCR techniques (Jia et al., 2020). Broughton et al. (2020) applied CRISPR-Cas12-based lateral flow assay to diagnose SARS-CoV-2 infection. They took swabs from the nasopharyngeal or oropharyngeal in a consistent period (around 40 min). Whereas, Hajian et al. (2019) combined CRISPR-Cas technology with FET to develop a new biological sensor. The biosensor is composed of a CRISPR-chip connected to graphene-based FET. With the advantage of skipping the amplification process, it managed to detect a 1.7 femtomolar concentration of nucleic acid in only 15 min (Hajian et al., 2019).
4.7. Biosensors for SARS-CoV-2 detection
Several biosensors were developed to detect coronaviruses such as MERS and SARS-CoV-1 to diagnose the infection rapidly. Most of the biosensors were designed based on optical systems. Moreover, the piezoelectric mechanism was used for SARS-CoV-1 detection, compared to an electrochemical biosensor for MERS identification. Huang et al. (2009) introduced a new biosensor relying on a fiber-optical mechanism to detect SARS-CoV-1 nucleocapsid protein (Huang et al., 2009). Similarly, Roh and Jo (2011) utilized the same biorecognition molecule to detect 0.1 pg/mL of SARS-CoV-1 through a quantum dots-coupled RNA aptamer chip (Roh and Jo, 2011). However, nanoparticle-based biosensors were used widely in MERS detection. Teengam et al. (2017) created pyrrolidinyl peptide nucleic acid probes via nanoparticle aggregation to develop an optical biosensor to determine MERS. These nucleic acid probes were synthesized to effectively hybridize with the complementary sequence of MERS RNA (Teengam et al., 2017). Moreover, Layqah and Eissa (2019) utilized gold nanoparticles to modify carbon electrodes carrying MERS antigen, then quantifying the specific antibodies in the samples volumetrically (Layqah and Eissa, 2019). Since the biorecognition material, whether it is a nucleic acid, antigen, antibody, or PCR product, is typically similar across the species, the previously developed biosensors can be modified to determine SARS-CoV-2.
A recognition target is identified to design a biosensor specific for SARS-CoV-2 detection, such as any region of SARS-CoV-2 RNA, its proteins, viral proteases, or human immunoglobulins as a part of the human immune response towards the virus (Escors et al., 2001; Huang et al., 2009; Tian et al., 2020; Xu et al., 2020). The recognition method is then chosen either through nucleic acid hybridization, receptor-ligand interaction, antigen-antibody binding, or enzymatic reaction. An additional component included for the environmental sampling of SARS-CoV-2 directly from the ambient air is an air sampler. Given the fact that the airborne SARS-CoV-2 concentration in the air is relatively low (103–104 of viral RNA copies per 1 m3 of air during the COVID-19 pandemic), a complex air sampling procedure is essential. Accordingly, this implements point-of-care risk assessment, which boosts viral concentration to a detectable limit (Yang et al., 2011). Therefore, a proper aerosol to hydrosol enrichment capacity equivalent to 105–106 folds (obtained from 103 to 104 of SARS-CoV-2 genome copies per 1 m3 of air) is necessary (Kim et al., 2020). Since the particles carrying the SARA-CoV-2 virus are loaded with other compounds (e.g., proteins, salts, organic and inorganic substances, in addition to other viruses), the viral-containing particles are larger than the size of the virus. Therefore, the sampler used for airborne viral detection should be designed with filters of larger pore sizes than the actual viral size (25–400 nm) (Hogan et al., 2005).
A novel biosensor was introduced by Qiu et al. (2020) after studying the RNA sequences of both SARS-CoV viruses. They designed a specific oligonucleotide merging the sequencing of both SARS-CoV-1 and SARS-CoV-2 and measured the hybridization effect via dual plasmonic-photothermal mechanisms. The biosensor effectively detected 0.22 pM concentrations of SARS-CoV-2 (Qiu et al., 2020). Seo et al. (2020) have developed a sensor-based technique for the detection of SARS-CoV-2. The sensor was designed based on field-effect transistor (FET) biosensing ability, in which a FET graphene film is coated with a specific antibody targeting SARS-CoV-2 spike protein (Seo et al., 2020). This method provided high accuracy and sensitivity in detecting low concentrations of SARS-CoV-2 in nasopharyngeal swabs of infected patients without the need for sample pretreatment. FET biosensor was proved to detect 1 fg/mL concentration of SARS-CoV-2 spike proteins if it dissolved in a phosphate buffer saline and a concentration of 100 fg/mL. Also, if it is present in clinical transport medium (Samson et al., 2020). Although it has not been applied, it is considered a potential method for the sensitive detection of SARS-CoV-2 in the air. Extensive research is invested nowadays to reduce the drawbacks and improve these biosensors' performance for an efficient and more reliable diagnosis of SARS-CoV-2 in different matrices.
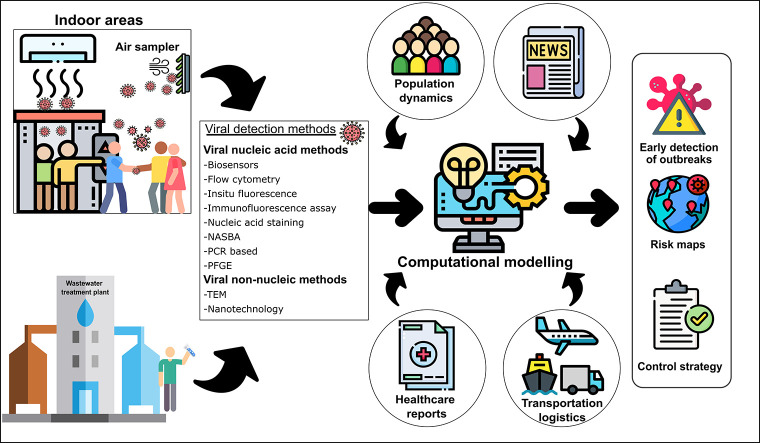
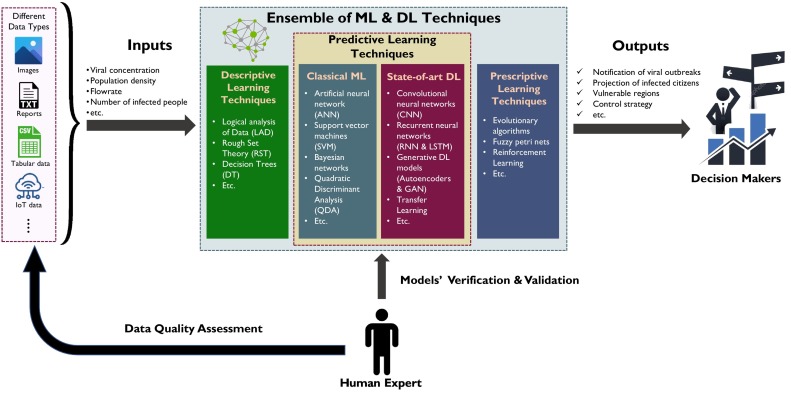
5. Role of computational modeling in viral outbreaks
Besides theory, experiment, and simulation, computational modeling has quickly evolved to become one of the main pillars of science (Chang and Makatsoris, 2001). The advancement in computational modeling tools and technologies and their versatile adaptation in many fields has led to a significant quality improvement of the executed tasks. Among those tasks are the early detection of disease vector outbreaks and the risk map assessments (Agbehadji et al., 2020; Simsek and Kantarci, 2020). The availability of massive global data (health and environmental) led to the advancement in computing capacities and the emergence of technologies such as Big Data, AI, ML, Internet of Things (IoT), and Cloud Computing (Allam et al., 2020). Notably, this allowed massive amounts of data collected from different sources to be retrieved and analyzed in real-time to provide informative insights and predictions, as presented in Fig. 4 . Hence, the availability of predictive modeling techniques should be expected to contribute remarkably to detect future viral outbreaks using the current available data, including environmental and health datasets. Currently, the detection of outbreaks using computational methods such as AI is focused mainly on identifying the virus spread between humans using data collected from infected humans. Using the same technology, the AI can also detect outbreaks using WBE.
Fig. 4.
Computational modeling strategies for viral outbreak management.
5.1. Artificial intelligence and machine learning for detection of outbreaks of infected persons
AI and its related ML technologies can accurately track and survey the virus spread, alarming the patients and healthcare authorities, based on the availability of representative data (Ahmad et al., 2020; Kumar et al., 2020a). These technologies can contribute to providing fast, reliable, and cost-effective tools to help manage viral outbreaks. Moreover, they can rapidly identify high-risk patients and provide risk maps and valuable insights to control infection in real-time. Table 5 summarizes the computational modeling methods, mainly AI/ML techniques used recently in different jurisdictions for forecasting the status of the SARS-CoV-2 pandemic.
Table 5.
A summary of different computation methods used for SARS-CoV-2 forecasting.
| Studied location | Forecasting Method | Input | Validation/Error method | Results | Reference |
|---|---|---|---|---|---|
| Brazil |
|
|
|
|
(Ribeiro et al., 2020b) |
| Canada |
|
|
|
|
(Chimmula and Zhang, 2020) |
| China |
|
|
|
|
(Al-Qaness et al., 2020) |
| India USA UK |
|
|
|
|
(Prasanth et al., 2021) |
| Italy |
|
|
|
|
(Chintalapudi et al., 2020) |
| Italy Spain France China Australia USA |
|
|
|
|
(Zeroual et al., 2020) |
| Mexico |
|
|
|
|
(Muhammad et al., 2021) |
| Ukraine |
|
|
|
|
(Chumachenko et al., 2020) |
| 24 Countries and 24 States |
|
|
|
|
(Wieczorek et al., 2020b) |
| 12 countries |
|
|
|
|
(Peng and Nagata, 2020) |
Moreover, advanced ML techniques such as deep learning (DL) networks are used to detect and predict the SARS-CoV-2 pandemic. A deep learning network (nCOVnet) has also been proposed for the fast detection of the pandemic using chest X-ray images (Panwar et al., 2020). As a transfer learning approach, the authors of another study (Nayak et al., 2021) compared the performance of different pre-trained convolutional neural network (CNN) models such as AlexNet (Krizhevsky et al., 2012), VGG-16 (Simonyan and Zisserman, 2014), SqueezeNet (Iandola et al., 2016) and ResNet-34 (He et al., 2016) for detecting the pandemic using chest X-ray images. Another approach was recently proposed by Zheng et al. (2020) to predict the probability of the pandemic infection. This approach exploits the 3D chest CT volumes through the segmentation of the 3D lung region using pretrained U-Net (Ronneberger et al., 2015) architecture; after that, the segmented lung is fed into a 3D DL network for prediction.
Outside China, the AI-driven algorithms were able to detect the SARS-CoV-2 even before the WHO was informed. Consequently, several alarms were triggered to warn travelers of the risks of traveling to specific locations. On the other hand, Chinese companies did not trigger those alarms, indicating that the data-sharing restriction might have led to these severe consequences (Allam et al., 2020). BlueDot was one of the companies with a successful history of detecting outbreaks (Dananjayan and Raj, 2020). It had previously detected the outbreak of the Zika virus in Florida in addition to spotting the COVID-19 pandemic wave 9 days before the WHO released the alerting statement of the emergence of coronavirus (Bogoch et al., 2016). BlueDot used the available data from news reports, airline flights, and previous outbreaks in predicting the SARS-CoV-2 emerging from Chinese regions. In recent work, eight of ten cities that could be potentially affected by the new coronavirus were successfully predicted (Bogoch et al., 2020). Metabiota is another company that successfully used a similar approach by using Big Data analytics to exploit flight data to forecast infections, several days before the first reported case, in countries at risks such as Japan, Thailand, Taiwan, and South Korea (Allam et al., 2020). Companies such as BlueDot (Netherlands), Metabiota (USA), and others that can exploit the breakthrough of AI technologies and big data analytics can help the healthcare sectors, governments, and businesses minimize the risk resulting from unexpected viral outbreaks. Accordingly, various AI techniques, big data analytics, and their applicability in contact tracing, detection of viral outbreaks, and recommending control strategies are discussed in the following subsections.
5.2. Contact tracing
As the SARS-CoV-2 is spread mainly from person to person through droplets, discharges the nose, or contact transmission, therefore, contact tracing can be an appropriate method for monitoring and control the spread of the outbreak (World Health Organization, 2020a). The contact tracing process relies on using infected patients' data to forecast and predict the virus spread. Multiple COVID-19 infected countries used digital contact tracing, mainly smartphone applications synergized with different technologies such as Bluetooth, Global Positioning System (GPS), Social media, contact details, network-based APIs, mobile tracking data, card transaction data, and system physical address (Lalmuanawma et al., 2020). Such data from different sources change over time and grows quickly; hence, it is called “big data” (Devakunchari, 2014). Many platforms can handle big data challenges in terms of volume, velocity, and variety. Storm (Apache Storm), S4 (Neumeyer et al., 2010), Kafka (Apache Kafka), Apache Spark (Apache Spark™ - Unified Analytics Engine for Big Data), and Flink (Apache Flink: Stateful Computations over Data Streams) can handle data stream processing that can process large volumes of data in a relatively short time span. The digitalization of contract tracing and the exploitation of big data analytics are superior by their virtual real-time fast abilities compared to the non-digital process. The collected individual personal data through digital applications are then used by ML and AI techniques to track citizens who are vulnerable to the viral outbreak due to their recent contact with infected persons. Lalmuanawma et al. (2020) listed over 36 countries that have successfully used digital contact tracing by using either centralized, decentralized, or hybrid techniques.
Digital contact tracing is limited mainly by privacy and control over data and data security breaches. According to Sohrabi et al. (2020), several contact tracing applications, including those produced by countries, violated privacy laws and were reported to be unsafe. Several countries already have their contact tracing applications to track the spread of viruses; however, to fight the viral spread, a global action should be performed for a better tracing. Wen et al. (2020) have analyzed the documentation of 41 released contracted tracing applications for COVID-19 contacts tracing. The results showed that several applications could expose the identity information to enable fingerprinting and tracking specifying users. This addresses issues regarding the question of who should be capable and has the right to trace people. Confidential and publically available data should be addressed appropriately. Restrictions of using or sharing data would also be a matter of concern.
5.3. Accurate detection of outbreaks: motivations to combine WBE and AI
Water is a vital resource for society, the economy, and the environment (Abdeldayem et al., 2020), especially during a pandemic where hygiene is critical for spreading the virus. During the last decade, the water sector has been navigating parallelly into two transformations (Garrido-Baserba et al., 2020). The first transformation is the fourth water revolution, “Water 4.0,” which aims to more sustainable and rational water resources management (Sedlak, 2014). The second one is the digitalization of the water sector and its empowerment with AI and Big Data to initiate new functionalities in the water and wastewater management sectors (Poch et al., 2020; Ramírez Calderón et al., 2020). In the context of viral detection, AI-based tools that exploit the data collected from smart city sources have the potential to monitor and control viral outbreaks.
AI has been conventionally used for controlling state variables in wastewater treatment plants or outbreaks detection from Big Data (Allam et al., 2020; Ramírez Calderón et al., 2020; Sundui et al., 2021). As previously mentioned, WBE can provide great opportunities to detect viral outbreaks, especially for countries with limited resources to carry out clinical tests. However, upon conducting an extensive search, it was found that no approach in the scientific literature combines AI with WBE for the detection of viral outbreaks. Hence, there is a clear need to fill this gap and propose new frameworks for such purposes. In response to this need, this paper proposes a framework for using AI to predict viral outbreaks using WBE. The framework is presented in the following subsection.
5.4. The proposed WBE-AI framework
The proposed framework depends on the concept of data-driven modeling that exploits an ensemble of diversified types of ML methods; predictive, descriptive, and prescriptive. This is because no single method can work perfectly in all situations and all types of data. These methods use various algorithms from statistical learning, AI, and combinatorial optimization. It is essential to combine human expertise with additional knowledge extracted by the AI/ML methods (Ragab et al., 2019). The role of human expertise is central to the framework proposed in this work. First, the expert quantifies the data quality by using an assessment technique consistent with his/her subjective knowledge. This is to provide a clear assessment of the data uncertainty. The second task is to perform the verification and validation procedures to ensure the trustworthiness of the resulting predictive/descriptive/prescriptive models. This is based on his/her subjective knowledge and enables models to be updated with minimal efforts from human experts.
For the case of viral outbreaks, the inputs to the framework consist of different types of data, including images, reports, tabular data, IoT, and others. These data are processed, and a number of parameters are then extracted, such as the concentration of viruses in wastewater samples, the official number of infected people in the region of the wastewater plant, population density, and wastewater flow/generation rate. Those parameters are potentially able to complement each other and contribute effectively to build the model. Other parameters are also considered, such as the external disturbances that influence the WBE operations, e.g., change of water consumption patterns, delayed load peaks, and the shift between industrial and residential flows. The outputs of the framework resemble the notification of an outbreak, projection of infected citizens and mortality, vulnerable regions of the outbreaks, and others. Moreover, the framework provides the decision-maker with control strategies that should be followed to minimize any expected risks. The proposed framework is illustrated in Fig. 5 , while the framework's building blocks are also presented in more detail in the following sections.
Fig. 5.
Proposed framework for viral outbreak detection and decision making.
5.4.1. Predictive machine learning and deep learning methods
The general concept of predictive ML methods is to learn specific properties from a training dataset to build a model capable of making predictions (Alpaydin, 2014). Predictive methods can be divided into two groups: regression and pattern classification. Machine learning regression methods are based on analyzing existing relationships between the variables and trends in the data to make predictions about numerical variables, e.g., the number of infected citizens. In contrast to regression methods, the task of pattern classification is to assign discrete class labels to particular observations as outcomes of a prediction. To go back to the above example: a pattern classification task could predict a high or low infection. The most common ML predictive methods are the support vector machine (SVM) (Bishop, 2006), Bayesian networks (Jensen and Nielsen, 2007), Quadratic discriminant analysis (QDA) (Friedman, 1989), and the artificial neural network (ANN) (Sundui et al., 2021).
Although traditional ML methods have been proven to be effective for predictive analysis, these methods cannot be successfully applied in some cases. Therefore, advanced DL methods such as convolutional neural networks (CNN) (LeCun et al., 1989), recurrent neural networks (RNN) (Hochreiter and Schmidhuber, 1997; Medsker and Jain, 2001), stacked automatic encoders (SAE) (Kingma et al., 2014), deep belief networks (DBN) (Hinton, 2009) should be added to the list to ensure building accurate and representative predictive models. Although DL algorithms are powerful, the functions they create during the training procedure are sophisticated and opaque. Humans who would like to use these models should have the basic ability to understand and trust them before making important decisions.
5.4.2. Descriptive machine learning methods
Descriptive machine learning methods do precisely what the name implies. They describe or summarize raw data and make it interpretable by humans (Novak et al., 2009). The main advantage of descriptive methods over predictive ones is that they enable and allow decision-makers to analyze past behaviors and understand how they might influence future outcomes. Among the descriptive ML methods, the decision trees (DT) (Quinlan, 1986), rough set theory (RST) (Pawlak, 1998), and Logical Analysis of Data (LAD) (Crama et al., 1988) are the most popular ones.
These descriptive methods represent a cornerstone in the framework since decision-makers always seek accurate predictions and meaningful insights that help them take the right actions very rapidly. In the proposed framework, the decision trees are suggested as one of the most widely used and practical descriptive methods and is quite popular and ranked as number 1 in the pre-eminent paper entitled”Top 10 Algorithms in Data Mining” (Wu et al., 2008). It is a flow-chart-like structure, represented as sets of “if” and “then” rules that are readable (interpretable) by humans. LAD is a ML classification technique based on the theories of Boolean algebra and combinatorial optimization (Boros et al., 2000). The fundamental concept of LAD is the extraction of interpretable patterns from the data to discover hidden knowledge from a set of training observations. The extracted patterns are represented as sets of “if” and “then” rules that are easy to interpret (Ragab et al., 2018).
5.4.3. Prescriptive machine learning methods
Prescriptive learning methods aim to recommend actions, control strategies, and policies for optimizing a target process. This is an advantageous characteristic over the predictive and descriptive methods reported in the literature. Providing the decision-makers with recommended control strategies through the proposed framework can help in minimizing the spread of the viral outbreak. This can be done through predicting and analyzing the current situation through predictive and descriptive methods and then select from many prescriptions the best one that mitigates the outbreak of the infection. Several prescriptive ML techniques can be adopted and used in the framework, such as deep reinforcement learning (RL) (Mousavi et al., 2018), fuzzy petri nets (Liu et al., 2017), and evolutionary-based methods (Yu and Gen, 2010).
6. Discussions: remarks, recommendations, and future research perspectives
Several approaches for the detection of viral outbreaks have been presented in this article. However, there are still many aspects that require further attention to obtain a more accurate viral outbreaks surveillance approach. This prioritized need of viral outbreaks surveillance tools is attributed to the fact that the world has been in several dangerous situations over the last two decades, primarily due to the spread of several infectious diseases that could even last for the coming few years. Therefore, the different scientific research communities (biologists, computer scientists, doctors, engineers, statisticians, etc.) should strongly collaborate and identify the existing knowledge gaps in outbreaks surveillance tools and develop tools and techniques that will rapidly and efficiently protect the safety of communities and its people. With the ongoing COVID-19 pandemic, which has left behind many deserted city centers worldwide, we must ask ourselves an important question concerning the future vitality of this pandemic. The authors are deeply convinced that the strength of any city center is that it is alive, with its residents, workers, students, restaurants, shops, schools, shows, museums, lively streets, green spaces, and public squares. In this line of progressive research, AI and related digitalization technologies can play a major role in achieving that goal and making cities more inclusive and sustainable. In this section, several remarks are discussed on the existing viral detection tools and recommendations on the exploitation of advanced AI technologies are provided in order to achieve reliable and robust outbreak monitoring and control strategies.
Viral air surveillance is a complicated process demanding advanced technologies and governmental approvals for widespread applications. The development of these technologies requires proper standardization techniques, relevant calibration, and efficient reporting methods. Furthermore, air sampling methodology design must consider the choice of the sampling location, sample treatment before analysis, boosting the viral load concentration, physical and biological viability of the virus after sampling. Nevertheless, efforts have to be exerted to select the air sampling method compatible with the downstream analytical procedures. Further research is required to design novel biosensors for viral detection in air samples, in addition to the identification in clinical samples.
This study shows that the WBE can have great potential for the detection of viral outbreaks. However, some challenges are still faced while applying it and need to be further addressed. Firstly, viral source concentration should be accurately identified from exogenous sources to avoid overestimating or underestimating the real number of patients. Secondly, it is challenging to account for temporal fluctuations needed to estimate the accurate population size. The standardization of methods should be considered to measure the microbial factors in wastewater. A standardized approach used by all stakeholders is not provided yet. Hence, most of the current approaches are resulting from diligent efforts by researchers. Therefore, it would be fundamentally essential to have standardized methods for future research in WBE. Besides, existing data analytics techniques need to be further improved to avoid inaccuracies related to a false positive or false negative. Moreover, the cross-validation of data and interdisciplinary collaborations between different agencies would be vital for better WBE.
The ML and DL have recently contributed to the improvement of screening, prediction, and forecasting tools for viral outbreaks with reasonable accuracy and reliability. However, the urgency associated with outbreaks mandates top-notch performance in terms of precisely predicting and screening, which has not yet been achieved. Hence, the continuous upgrading of the current models is mandatory for better forecasting in the future. An important aspect that needs to be addressed is the data privacy associated with specific ML that requires private data as an input for viral outbreak detection, especially for technologies such as contact tracing. The use of ML and DL for detecting outbreaks using environmental parameters such as WBE is essential. To the authors' current knowledge, no employed ML/DL is used for early outbreak detection using WBE. Consequently, using these AI technologies for viral detection using WBE needs to be explored because it has a vast potential in improving viral outbreak detection and mapping.
As previously mentioned, several attempts were done to predict how the viral spread took place. Consequently, the number of cases was very close to the observed ones, with a deviation of only 2 to 10 cases over the actual reported cases (De Simone and Piangerelli, 2020; Namasudra et al., 2021). As an additional real-world application of ML usage, diagnosis of the cases proved its wide range coverage and multiple capabilities (Ahmad et al., 2021; Fan et al., 2020). For example, Liang et al. (2021) proposed a deep learning framework to analyze COVID-19 cases using medical images to increase testing sensitivity. Fortunately, the developed CNN model showed high detection accuracy after being trained by multiple X-ray and computed tomography images. The statistical indices of the developed CNN were calculated and had the values of F1 score > 97% and specificity >99%. In another study, deep CNN was developed and utilized to detect COVID-19 using medical images (Gaur et al., 2021). In this case, the overall model accuracy was around 92%, with a sensitivity of >94%. These techniques were applied in other studies where their accuracy and statistical indices were relatively high (Pham, 2020; Shah et al., 2020).
All these reported studies represent strong evidence that these techniques should be applied on a large scale in different areas, especially the medical sector and environmental monitoring for viral detection. However, one obstacle for this to happen on a large scale is the lack of data transfer. Collaborations and official agreements between different sectors to provide the needed data to the research community will serve significantly in overcoming this obstacle. Open-source data sets (Shuja et al., 2020), alongside rigorous analysis based on sophisticated models, lead to suitable decisions during hard times. Our proposed framework offers a good way for any viral outbreak, especially COVID-19, surveillance in wastewater or air based on different data sources and forms inspired by the aforementioned techniques.
Besides detecting the viral outbreak, there is an indispensable need for a fast drug discovery process. Drug discovery is an exhausting process due to the very-high dimensional search space of molecules that can reach up to 1060 small drug-like molecules or peptides. As a result, it may take years to develop an effective and certified drug for commercial use.
Fortunately, ML can play a key role in accelerating the drug discovery process, not only tracking the spread of the contagious virus. Prescriptive ML techniques such as RL can be used for training a search agent. This search agent can introduce good candidates of protein sequences according to an evaluation function. As a result, the number of lab experiments and clinical trials needed for a final vaccine can gradually decrease.
The buzz around AI and related ML/DL technologies reflects its enormous disruptive potential based on key tangible benefits over conventional modeling methods. An important question is how to fully and better exploit the available historical data as essential assets to build accurate and versatile outbreak descriptive/predictive/prescriptive models that will help to address the above-mentioned challenges. The aim is to predict and describe how various components and phenomena interact and influence each other to achieve accurate monitoring and control of different pandemic-like scenarios. The spectacular success of advanced AI methods in a wide range of data analytics applications offers them as appropriate/best available candidates. These advanced AI methods, combined with innovative processing techniques and human expertise, have shown great success in large-scale industrial applications where the data have complex and ill-defined distributions (Elhefnawy et al., 2021; Ragab et al., 2019). Over the past few years, AI has been exploited in different cases, driven by advanced and relatively inexpensive online computing infrastructures. The multitude of diversified ML and DL has proven to be promising for many data-driven modeling applications. This is one of the main motivations of the proposed WBE-AI framework.
The proposed framework aims to deliver timely and highly reliable insight into the viruses' presence for accurately monitoring and controlling their outbreaks. Confidently, the proposed framework can help to protect people and communities while maintaining business and operational continuity. Facilities and stakeholders such as hospitals, manufacturing/industrial, and healthcare facilities can benefit from the proposed framework.
Researchers need flexible and generic tools that can be easily adapted to meet the new needs to gain the true rewards and values from existing data. To do so, here are some remarks and recommendations on the proper use and applicability of this framework.
6.1. Hyperparameters optimization and AutoML
In order to get the best performance of the ML or DL models, hyperparameter optimization is highly recommended. For example, in case of the existing ML methods, the optimization algorithm, its learning rate, regularization parameters, etc. are among the hyperparameters that need to be tuned within a search space. In the case of the DL method(s), other hyperparameters should be optimized, such as the number of layers (fully connected or convolutional), the activation function, the number of filters in each convolutional layer in the case of convolutional neural networks, the batch size and the number of epochs for training, etc.
For automating the design of ML models and deep neural networks, automated machine learning tools such as Auto-Keras (Jin et al., 2019), H2OautoML (LeDell and Poirier, 2020), Auto-sklearn (Feurer et al., 2019), Auto-WEKA (Thornton et al., 2013), and Auto-Pytorch (Zimmer et al., 2021), in addition to neural architecture search (NAS) techniques (Wistuba et al., 2019) are commonly used instead of the exhausting task of manual design of such models with numerous hyperparameters. However, these techniques have some limitations due to their highly demanding computational power to obtain the best performing model in a large search space.
6.2. Human-centric and Data-centric AI
In an analogy to the existing practices on continuous software engineering “DevOps” (Bass et al., 2015), the practices for continuous delivery of ML applications are called “MLOps” (Mäkinen et al., 2021). The ML/DL technique used for implementing the predictive/descriptive/prescriptive models is just the first step within the whole cycle of MLOps. There is an urgent need to highlight the importance of data processing techniques for successfully deploying any robust AI products in the commercial market. Using the same AI technique while working on the data engineering process is a potential research direction under the “Data-centric AI” umbrella. One of the reasons behind that is the availability of several sophisticated ML/DL techniques in the absence of representative and informative datasets and the need to monitor and update the deployed models over time based on their performance and upcoming data.
The proposed computational modeling workflow is done under the full supervision of a human expert. The involvement of the human expert within our proposed AI framework is an indispensable need for the emerging “Human-centric AI”. Despite AI technologies' reliability and unprecedented performance, these technologies cannot completely replace human expertise and knowledge. Human expertise is essential due to the following reasons: (i) assess the quality of data, and (ii) select and leverage the suitable ML/DL method that fits the need. However, the combination of “Data-centric AI” & “Human-centric AI” can result in an augmented intelligence that may facilitate the adoption of AI in various complex applications such as the one addressed in this article.
6.3. Transfer learning: from COVID-19 to other viruses
To solve the problem of the insufficiently labeled SARS-COV-2 data, a common and effective method is transfer learning, which realizes the diagnosis of any target virus by simply transferring the information of other viruses to the target COVID-19 case. Moreover, generative learning AI techniques can predict and simulate new virtual viral strains based on the historical viral data. Accordingly, these potential upcoming strains can be confronted and dealt with based on the previously mentioned prescriptive learning techniques that can accelerate the drug discovery process. Furthermore, different simulation and AI techniques can be used to design new adaptive and accurate viral detectors for rapid monitoring and control of potential viral outbreaks. The proposed framework exploits the deep transfer learning methods and combines the advantages of DL and transfer learning and their excellent feature extraction capabilities.
Aside from the use of AI techniques to simulate and design new detectors, the scale-up of producing highly detective nano (bio)sensors, including quantum dots, in the future is a must despite being challenging due to techno-economic considerations and royalty issues. However, they possess some appealing features, such as high sensitivity, which means working with very low concentrations of pathogens, high accuracy, and fast detection. These advantages make them excellent candidates to replace the traditional detection and diagnosis methods currently being used to overcome their drawbacks. Hence, future research should focus on feasibility studies in real environments, scale-up and commercial-scale production of nano-sensors.
Finally, the effects of cross talk and viral defense signaling between the viruses and the host should also be investigated, especially in the newly mutated viruses such as SARS-CoV-2. Such investigations should also address the existing knowledge gaps in detection using the currently applied techniques, identify the effects of transmission in the environment and their spreading abilities.
7. Conclusions
Viral outbreaks are matters of great concern for human survival. Therefore, understanding its transmission means and its early detection is a matter of concern at various levels. WBE has been a growing technique for viral outbreak detection using different monitoring techniques. Besides that, surveillance of viruses in indoor areas has been performed; however, this study shows that the extension and its applicability on higher scales still need further studies. AI and its related ML and DL technologies are promising for viral outbreak detection, risk map formation, and decision support. These technologies augment researchers and decision-makers with multiple perspectives, which can address the outbreaks monitoring and surveillance challenges. Synergizing WBE and AI tools/techniques will significantly contribute to the surveillance of viral outbreaks. In this work, a novel framework has been proposed that combines an ensemble of ML and DL techniques that can be used for different “fit-the-purpose” and by using various types of monitored data sets. This ensemble includes descriptive, predictive and prescriptive techniques that can help decision-makers to accurately detect and forecast future viral outbreaks in addition to recommending versatile control strategies for preventing the spread of the pandemic. Additionally, remarks and recommendations are provided for the successful implementation/development of the proposed framework, including hyperparameter optimization of ML and DL techniques, augmentation of human-centric & data-centric AI, and the exploitation of transfer learning for the detection of current viral outbreaks using the past experiences in this field.
CRediT authorship contribution statement
Omar M. Abdeldayem: Data collection and analysis, Writing - original draft, Visualization, Conceptualization, Methodology, Project administration, Writing - review & editing
Areeg M Dabbish: Writing - original draft, Methodology, Visualization, Data collection and analysis, Writing - review & editing
Mahmoud M. Habashy: Writing - original draft, Methodology, Visualization, Data collection and analysis, Writing - review & editing
Mohamed K. Mostafa: Conceptualization, Methodology, Visualization, Writing - original draft, Resources, Writing - review & editing, Supervision
Mohamed Elhefnawy: Data collection and analysis, Writing - original draft, Visualization
Lobna Amin: Data collection and analysis, Writing - original draft, Visualization
Eslam G. Al-Sakkari: Data collection and analysis, Writing - review & editing
Ahmed Ragab: Conceptualization, Methodology, Writing - review & editing, Supervision
Eldon R. Rene: Conceptualization, Writing - review & editing, Supervision
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors dedicate this paper to the memorial of their colleague Omar Eldaghar who passed away in Uganda while serving the underprivileged communities to have access to basic human needs. We hope that we can carry on with his legacy and ethos in making this world a better place for everyone “Shoulder to shoulder for those who lack basic human needs.”.
E.R. Rene thanks IHE Delft for providing staff time support under the project “Support to Society” to co-work with researchers from Egypt and students of the ERASMUS+ IMETE programme.
Editor: Eakalak Khan
References
- Abdeldayem O.M., Eldaghar O., Mostafa M.K., Habashy M.M., Hassan A.A., Mahmoud H., et al. Mitigation plan and water harvesting of flashflood in arid rural communities using modelling approach: a case study in Afouna village, Egypt. Water. 2020;12:2565. [Google Scholar]
- Abduljalil J.M. Laboratory diagnosis of SARS-CoV-2: available approaches and limitations. New Microbes New Infect. 2020;36 doi: 10.1016/j.nmni.2020.100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adan A., Alizada G., Kiraz Y., Baran Y., Nalbant A. Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- Agbehadji I.E., Awuzie B.O., Ngowi A.B., Millham R.C. Review of big data analytics, artificial intelligence and nature-inspired computing models towards accurate detection of COVID-19 pandemic cases and contact tracing. Int. J. Environ. Res. Public Health. 2020;17:1–16. doi: 10.3390/ijerph17155330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranovski I.E., Safatov A.S., Pyankov O.V., Sergeev A.N., Agafonov A.P., Ignatiev G.M., et al. Monitoring of viable airborne SARS virus in ambient air. Atmos. Environ. 2004;38:3879–3884. doi: 10.1016/j.atmosenv.2004.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A., Garhwal S., Ray S.K., Kumar G., Malebary S.J., Barukab O.M. The number of confirmed cases of covid-19 by using machine learning: methods and challenges. Arch. Comput. Meth. Eng. 2020:1–9. doi: 10.1007/s11831-020-09472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F., Farooq A., Ghani M.U. Deep ensemble model for classification of novel coronavirus in chest x-ray images. Comput. Intell. Neurosci. 2021;2021:8890226. doi: 10.1155/2021/8890226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., et al. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Huraimel K., Alhosani M., Kunhabdulla S., Stietiya M.H. SARS-CoV-2 in the environment: modes of transmission, early detection and potential role of pollutions. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allam Z., Dey G., Jones D. Artificial intelligence (AI) provided early detection of the coronavirus (COVID-19) in China and will influence future urban health policy internationally. AI. 2020;1:156–165. [Google Scholar]
- Alpaydin E. 2014. Introduction to machine learning ethem alpaydin. Introd. to Mach. Learn. [Google Scholar]
- Al-Qaness M.A.A., Ewees A.A., Fan H., Abd El Aziz M. Optimization method for forecasting confirmed cases of COVID-19 in China. J. Clin. Med. 2020;9:1–15. doi: 10.3390/jcm9030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin L. Control of nitrogen oxide emission from vehicular engines: brief perspectives. Austin Environ. Sci. 2019;4:1–2. [Google Scholar]
- Amoah I.D., Kumari S., Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020;143 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiochia R. Nanobiosensors as new diagnostic tools for SARS, MERS and COVID-19: from past to perspectives. Microchim. Acta. 2020;187:639. doi: 10.1007/s00604-020-04615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Nag A., Rajpal A., Tiwari S.B., Sethi J., Sutaria D., et al. 2020. Detection of SARS-CoV-2 RNA in fourteen wastewater treatment systems in Uttarakhand and Rajasthan States of North India. (Published in medRxiv) [Google Scholar]
- Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., et al. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Xu Y., Xiao F., Sun Y. Diagnosis of COVID-19, vitality of emerging technologies and preventive measures. Chem. Eng. J. 2021;423 doi: 10.1016/j.cej.2021.130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabakhsh P., Kargar M., Doosti A. Molecular detection and genotyping of group a rotavirus in two wastewater treatment plants, Iran. Braz. J. Microbiol. 2020;51:197–203. doi: 10.1007/s42770-019-00131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., Hashem A.M., El-Kafrawy S.A., Sohrab S.S., Aburizaiza A.S., Farraj S.A., et al. Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. mBio. 2014;5:1–4. doi: 10.1128/mBio.01450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacnik K., Kutnjak D., Pecman A., Mehle N., Tusek Znidaric M., Gutierrez Aguirre I., et al. Viromics and infectivity analysis reveal the release of infective plant viruses from wastewater into the environment. Water Res. 2020;177 doi: 10.1016/j.watres.2020.115628. [DOI] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. (145268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R., Mauro B.Wd. 2020. Economics in the Time of COVID-19. [Google Scholar]
- Barreto-Vieira D.F., Barth O.M. Negative and positive staining in transmission electron microscopy for virus diagnosis. Microbiology in Agriculture and Human Health. 2015:45–56. [Google Scholar]
- Bass L., Weber I., Zhu L. Pearson Education; New Jersey, USA: 2015. DevOps: A Software Architect's Perspective. [Google Scholar]
- Bayat H., Naderi F., Khan A.H., Memarnejadian A., Rahimpour A. The impact of CRISPR-cas system on antiviral therapy. Adv. Pharm. Bull. 2018;8:591–597. doi: 10.15171/apb.2018.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat V.S., S S., Hegde G. Review—biomass derived carbon materials for electrochemical sensors. J. Electrochem. Soc. 2020;167 [Google Scholar]
- Bhowmick G.D., Dhar D., Nath D., Ghangrekar M.M., Banerjee R., Das S., et al. Coronavirus disease 2019 (COVID-19) outbreak: some serious consequences with urban and rural water cycle. npj Clean Water. 2020;3:1–32. [Google Scholar]
- Bilal M., Nazir M.S., Rasheed T., Parra-Saldivar R., Iqbal H.M.N. Case Studies in Chemical and Environmental Engineering. Vol. 2. 2020. Water matrices as potential source of SARS-CoV-2 transmission – an overview from environmental perspective; pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.M. 2006. Pattern Recognition and Machine Learning. [Google Scholar]
- Blachere F.M., Lindsley W.G., Pearce T.A., Anderson S.E., Fisher M., Khakoo R., et al. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Dis. 2009;48:438–440. doi: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- Bogoch I.I., Brady O.J., Kraemer M.U.G., German M., Creatore M.I., Kulkarni M.A., et al. Anticipating the international spread of zika virus from Brazil. Lancet. 2016;387:335–336. doi: 10.1016/S0140-6736(16)00080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth T.F., Kournikakis B., Spence M., Paton S., Simor A., Bastien N., et al. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 2005;191:1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges J.T., Nakada L.Y.K., Maniero M.G., Guimaraes J.R. SARS-CoV-2: a systematic review of indoor air sampling for virus detection. Environ. Sci. Pollut. Res. 2021;28:40460–40473. doi: 10.1007/s11356-021-13001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros E., Hammer P.L., Ibaraki T., Kogan A., Mayoraz E., Muchnik I. An implementation of logical analysis of data. IEEE Trans. Knowl. Data Eng. 2000;12:292–306. [Google Scholar]
- Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA. 2020;323:1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.S., Gordon T., Price O., Asgharian B. Thoracic and respirable particle definitions for human health risk assessment. Part. Fibre Toxicol. 2013;10:12. doi: 10.1186/1743-8977-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.R., Camezuli S., Davenport R.J., Petelenz-Kurdziel E., Ovreas L., Curtis T.P. Flow cytometric quantification of viruses in activated sludge. Water Res. 2015;68:414–422. doi: 10.1016/j.watres.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Brown M.R., Hands C.L., Coello-Garcia T., Sani B.S., Ott A.I.G., Smith S.J., et al. A flow cytometry method for bacterial quantification and biomass estimates in activated sludge. J. Microbiol. Methods. 2019;160:73–83. doi: 10.1016/j.mimet.2019.03.022. [DOI] [PubMed] [Google Scholar]
- Brussaard C.P.D. Viral control of phytoplankton populations - a review. J. Eukaryot. Microbiol. 2004;51:125–138. doi: 10.1111/j.1550-7408.2004.tb00537.x. [DOI] [PubMed] [Google Scholar]
- Bukkitgar S.D., Shetti N.P., Aminabhavi T.M. Electrochemical investigations for COVID-19 detection-a comparison with other viral detection methods. Chem. Eng. J. 2020;127575 doi: 10.1016/j.cej.2020.127575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B., Barardi C.R., Bofill-Mas S., Rodriguez-Manzano J., Girones R. Detection and quantitation of infectious human adenoviruses and JC polyomaviruses in water by immunofluorescence assay. J. Virol. Methods. 2011;171:1–7. doi: 10.1016/j.jviromet.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Cao G., Noti J.D., Blachere F.M., Lindsley W.G., Beezhold D.H. Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler. J. Environ. Monit. 2011;13:3321–3328. doi: 10.1039/c1em10607d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Henríquez L., Brenes-Acuña M., Castro-Rojas A., Cordero-Salmerón R., Lopretti-Correa M., Vega-Baudrit J.R. Biosensors for the detection of bacterial and viral clinical pathogens. Sensors. 2020;20(23):6926. doi: 10.3390/s20236926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Makatsoris H. Supply chain modeling using simulation. Int. J. Simul. 2001;2:24–30. [Google Scholar]
- Chantratita Wasun, Pongtanapisit Wiroj, Piroj Wantanich, Srichunrasmi Chutatip, Seesuai Somying. Development and comparison of the real-time amplification based methods–NASBA-Beacon, RT-PCR taqman and RT-PCR hybridization probe assays–for the qualitative detection of sars coronavirus. Southeast Asian J. Trop. Med. Public Health. 2004;35:623–629. [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C., Wong S.C., Chan V.W., So S.Y., Chen J.H., Yip C.C., et al. Air and environmental sampling for SARS-CoV-2 around hospitalized patients with coronavirus disease 2019 (COVID-19) Infect. Control Hosp. Epidemiol. 2020;41:1258–1265. doi: 10.1017/ice.2020.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K.…Marimuthu K. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapudi N., Battineni G., Sagaro G.G., Amenta F. COVID-19 outbreak reproduction number estimations and forecasting in Marche, Italy. Int. J. Infect. Dis. 2020;96:327–333. doi: 10.1016/j.ijid.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumachenko D., Chumachenko T., Meniailov I., Pyrohov P., Kuzin I., Rodyna R. Data Stream Mining & Processing. Springer International Publishing; 2020. On-line data processing, simulation and forecasting of the coronavirus disease (COVID-19) propagation in Ukraine based on machine learning approach; pp. 372–382. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Jr., Campiglia P., et al. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mbareche H., Lindsley W.G., Duchaine C. Field sampling of indoor bioaerosols. Aerosol Sci. Technol. 2020;54:572–584. doi: 10.1080/02786826.2019.1688759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crama Y., Hammer P.L., Ibaraki T. Cause-effect relationships and partially defined boolean functions. Ann. Oper. Res. 1988;16:299–325. [Google Scholar]
- Dabbish A.M., Yonis N., Salama M., Essa M.M., Qoronfleh M.W. Inflammatory pathways and potential therapies for COVID-19: a mini review. Eur. J. Inflamm. 2021;19:1–22. [Google Scholar]
- Dananjayan S., Raj G.M. Artificial intelligence during a pandemic: the COVID-19 example. Int. J. Health Plann. Manag. 2020;35:1260–1262. doi: 10.1002/hpm.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A., Piangerelli M. A bayesian approach for monitoring epidemics in presence of undetected cases. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devakunchari R. Analysis on big data over the years. Int. J. Sci. Res. Publ. 2014;4:1–7. [Google Scholar]
- Drosten C., Gottig S., Schilling S., Asper M., Panning M., Schmitz H., et al. Rapid detection and quantification of RNA of ebola and Marburg viruses, Lassa virus, crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuis M.E., Duchaine C. Aerosol production during blood and urine pre-analytical processing and handling in a hospital biochemistry clinical laboratory during the COVID-19 pandemic. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.643724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont-Leblond N., Veillette M., Mubareka S., Yip L., Longtin Y., Jouvet P., et al. Low incidence of airborne SARS-CoV-2 in acute care hospital rooms with optimized ventilation. Emerg. Microbes Infect. 2020;9:2597–2605. doi: 10.1080/22221751.2020.1850184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont-Leblond N., Veillette M., Bherer L., Boissoneault K., Mubareka S., Yip L., et al. Positive no-touch surfaces and undetectable SARS-CoV-2 aerosols in long-term care facilities: an attempt to understand the contributing factors and the importance of timing in air sampling campaigns. Am. J. Infect. Control. 2021;49:701–706. doi: 10.1016/j.ajic.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economic Development . UN News; 2021. Air Travel Down 60 Per Cent, as Airline Industry Losses Top $370 Billion: ICAO. [Google Scholar]
- Ejeian F., Etedali P., Mansouri-Tehrani H.A., Soozanipour A., Low Z.X., Asadnia M., et al. Biosensors for wastewater monitoring: a review. Biosens. Bioelectron. 2018;118:66–79. doi: 10.1016/j.bios.2018.07.019. [DOI] [PubMed] [Google Scholar]
- Elhefnawy M., Ragab A., Ouali M.S. Fault classification in the process industry using polygon generation and deep learning. J. Intell. Manuf. 2021 doi: 10.1007/s10845-021-01742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rashidy N., Abdelrazik S., Abuhmed T., Amer E., Ali F., Hu J.W., et al. Comprehensive survey of using machine learning in the COVID-19 pandemic. Diagnostics. 2021;11(7) doi: 10.3390/diagnostics11071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental Protection Agency . Office of Water Washington, D.C.; 1999. Wastewater Technology Fact Sheet: Chlorine Disinfection. [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Timeline of ECDC's response to COVID-19. 2021. [Google Scholar]
- Fabian P., McDevitt J.J., Houseman E.A., Milton D.K. Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: considerations for a new infectious virus aerosol sampler. Indoor Air. 2009;19:433–441. doi: 10.1111/j.1600-0668.2009.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakruddin M., Mazumdar R.M., Chowdhury A., Mannan K.S.B. Nucleic acid sequence based amplification (NASBA) - prospects and applications. Int. J. Life Sci. Pharma Res. 2012;2:106–121. [Google Scholar]
- Fan Z., Jamil M., Sadiq M.T., Huang X., Yu X. Exploiting multiple optimizers with transfer learning techniques for the identification of COVID-19 patients. J. Healthc. Eng. 2020;2020:8889412. doi: 10.1155/2020/8889412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi S., Niazi S., Sadeghi K., Naddafi K., Yavarian J., Shamsipour M., et al. A field indoor air measurement of SARS-CoV-2 in the patient rooms of the largest hospital in Iran. Sci. Total Environ. 2020;725 doi: 10.1016/j.scitotenv.2020.138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Health. 2020;16:1–6. [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., et al. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Fernandes N. 2020. Economic Effects of Coronavirus Outbreak (COVID-19) on the World Economy. [Google Scholar]
- Feurer M., Klein A., Eggensperger K., Springenberg J.T., Blum M., Hutter F. Automated Machine Learning. Springer; Cham: 2019. Auto-sklearn: efficient and robust automated machine learning; pp. 113–134. [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P.…Rodríguez-Lázaro D. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P., Soler N., Krupovic M., Marguet E., Ackermann H.W. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013;21:1–5. doi: 10.1016/j.tim.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Freeman S., Eykelbosh A. COVID-19 and outdoor safety: considerations for use of outdoor recreational spaces. National Collaborating Centre for Environmental Health. 2020:829. [Google Scholar]
- Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. BioTechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- Friedman J.H. Regularized discriminant analysis. J. Am. Stat. Assoc. 1989;84:165–175. [Google Scholar]
- García-Fontana C., Rodriguez-Sanchez A., Muñoz-Palazon B., Gonzalez-Martinez A., Vela-Cano M., Gonzalez-Lopez J. Profile of the spatial distribution patterns of the human and bacteriophage virome in a wastewater treatment plant located in the south of Spain. Water. 2020;12:2316. [Google Scholar]
- Garrido-Baserba M., Corominas L., Cortes U., Rosso D., Poch M. The fourth-revolution in the water sector encounters the digital revolution. Environ. Sci. Technol. 2020;54:4698–4705. doi: 10.1021/acs.est.9b04251. [DOI] [PubMed] [Google Scholar]
- Gaur L., Bhatia U., Jhanjhi N.Z., Muhammad G., Masud M. Medical image-based detection of COVID-19 using deep convolution neural networks. Multimedia Systems. 2021:1–10. doi: 10.1007/s00530-021-00794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode J.A., Rushworth J.V., Millner P.A. Biosensor regeneration: a review of common techniques and outcomes. Langmuir. 2015;31:6267–6276. doi: 10.1021/la503533g. [DOI] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacres-Granda I., Granda M.G., Freire-Paspuel B., Rios-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020;26:1583–1591. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habre R., Coull B., Moshier E., Godbold J., Grunin A., Nath A., et al. Sources of indoor air pollution in New York City residences of asthmatic children. J. Expo. Sci. Environ. Epidemiol. 2014;24:269–278. doi: 10.1038/jes.2013.74. [DOI] [PubMed] [Google Scholar]
- Hadei M., Hopke P.K., Jonidi A., Shahsavani A. A letter about the airborne transmission of SARS-CoV-2 based on the current evidence. Aerosol Air Qual. Res. 2020;20:911–914. [Google Scholar]
- Hajian R., Balderston S., Tran T., deBoer T., Etienne J., Sandhu M., et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza H., Rizk N.M., Gad M.A., Hamza I.A. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efficiency of the treatment process. Arch. Virol. 2019;164:2707–2713. doi: 10.1007/s00705-019-04383-x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., et al. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Honda R., Hara-Yamamura H., Meuchi Y. 2020. Detection of SARS-CoV-2 in Wastewater in Japan by Multiple Molecular Assays Implication for Wastewater-based Epidemiology (WBE) [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- Heilweil R. Vox; 2020. How AI Is Battling the Coronavirus Outbreak. [Google Scholar]
- Hinton G.E. Deep belief networks. Scholarpedia. 2009;4:5947. [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hogan C.J., Jr., Kettleson E.M., Lee M.H., Ramaswami B., Angenent L.T., Biswas P. Sampling methodologies and dosage assessment techniques for submicrometre and ultrafine virus aerosol particles. J. Appl. Microbiol. 2005;99:1422–1434. doi: 10.1111/j.1365-2672.2005.02720.x. [DOI] [PubMed] [Google Scholar]
- Hong S., Lee C. The current status and future outlook of quantum dot-based biosensors for plant virus detection. Plant Pathol. J. 2018;34:85–92. doi: 10.5423/PPJ.RW.08.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryniszyn A., Skonieczna M., Wiszniowski J. Methods for detection of viruses in water and wastewater. Adv. Microbiol. 2013;03:442–449. [Google Scholar]
- Huang J.C., Chang Y.F., Chen K.H., Su L.C., Lee C.W., Chen C.C., et al. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhao Z., Hernandez D., Jiang S. Near real-time flow cytometry monitoring of bacterial and viral removal efficiencies during water reclamation processes. Water. 2016;8:464. [Google Scholar]
- Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microbiol. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. SqueezeNet: AlexNet-level Accuracy With 50x Fewer Parameters and < 0.5 MB Model Size. arXiv; 2016. <book-title>SqueezeNet: AlexNet-level Accuracy With 50x Fewer Parameters and < 0.5 MB Model Size</book-title>. [Google Scholar]
- Im K., Mareninov S., MFP Diaz, Yong W.H. In: Biobanking: Methods and Protocols, Methods in Molecular Biology. 1897. Yong W.H., editor. 2019. An introduction to performing immunofluorescence staining; pp. 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Monetary Fund . World Economic Outlook Reports. 2020. World economic outlook update: a crisis like no other, an uncertain recovery. [Google Scholar]
- International Monetary Fund . 2021. World Economic Outlook Update. [Google Scholar]
- 2020. International Transport Forum. Unprecedented Impact of COVID-19 on Freight Volumes in Second Quarter. 2021. [Google Scholar]
- Iravani S. Nano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater. Adv. 2020;1:3092–3103. [Google Scholar]
- Ivanov J., Polshakov D., Kato-Weinstein J., Zhou Q., Li Y., Granet R., et al. Quantitative structure-activity relationship machine learning models and their applications for identifying viral 3CLpro- and RdRp-targeting compounds as potential therapeutics for COVID-19 and related viral infections. ACS Omega. 2020;5:27344–27358. doi: 10.1021/acsomega.0c03682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Potschka H., Chandra P.P., Tripathi M., Vohora D. Management of COVID-19 in patients with seizures: mechanisms of action of potential COVID-19 drug treatments and consideration for potential drug-drug interactions with anti-seizure medications. Epilepsy Res. 2021;174 doi: 10.1016/j.eplepsyres.2021.106675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- Jean J., Blais B., Darveau A., Fliss I. Detection of hepatitis a virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 2001;67:5593–5600. doi: 10.1128/AEM.67.12.5593-5600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen F.V., Nielsen T.D. 2007. Bayesian Networks and Decision Graphs. [Google Scholar]
- Jia F., Li X., Zhang C., Tang X. The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein Cell. 2020;11:624–629. doi: 10.1007/s13238-020-00708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Song Q., Auto-keras Hu.X. Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2019. An efficient neural architecture search system; pp. 1946–1956. [Google Scholar]
- Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020;11:5088. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenarkoohi A., Noorimotlagh Z., Falahi S., Amarloei A., Mirzaee S.A., Pakzad I., et al. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Sci. Total Environ. 2020;748 doi: 10.1016/j.scitotenv.2020.141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Chang S.Y., Sung M., Park J.H., Bin Kim H., Lee H., et al. Extensive viable Middle East respiratory syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Clin. Infect. Dis. 2016;63:363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., An S., Hwang J. An integrated system of air sampling and simultaneous enrichment for rapid biosensing of airborne coronavirus and influenza virus. Biosens. Bioelectron. 2020;170 doi: 10.1016/j.bios.2020.112656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D.P., Mohamed S., Rezende D.J., Welling M. Advances in Neural Information Processing Systems. 2014. Semi-supervised learning with deep generative models; pp. 3581–3589. [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. AlexNet. Adv. Neural Inf. Process. Syst. 2012;2012:1–9. [Google Scholar]
- Kumar A., Gupta P.K., Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. <journal-title>Diabetes Metab. Syndr. Clin. Res. Rev.</journal-title>. 2020;14:569–573. doi: 10.1016/j.dsx.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Malik Y.S., Ganesh B., Rahangdale S., Saurabh S., Natesan S., et al. CRISPR-cas system: an approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., et al. SARS-CoV-2 has been circulating in northern Italy since december 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahrich S., Laghrib F., Farahi A., Bakasse M., Saqrane S., El Mhammedi M.A. Review on the contamination of wastewater by COVID-19 virus: impact and treatment. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.142325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L.T., Fung Y.W., Wong F.P., Lin S.S., Wang C.R., Li H.L., et al. A real-time PCR for SARS-coronavirus incorporating target gene pre-amplification. Biochem. Biophys. Res. Commun. 2003;312:1290–1296. doi: 10.1016/j.bbrc.2003.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H., Khosrawipour V., Kocbach P., Mikolajczyk A., Schubert J., Bania J., et al. The positive impact of lockdown in Wuhan on containing the COVID-19 outbreak in China. J. Travel Med. 2020;27:1–7. doi: 10.1093/jtm/taaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue M. Electron microscopy of viruses. Methods Cell Biol. 2010;96:1–20. doi: 10.1016/S0091-679X(10)96001-9. [DOI] [PubMed] [Google Scholar]
- Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T., Chun-Xiao W., Shu-Lin L. Pulsed field gel electrophoresis. Reference module. Life Sci. 2017 doi: 10.1016/B978-0-12-809633-8.06980-6. [DOI] [Google Scholar]
- LeCun Y., Boser B., Denker J.S., Henderson D., Howard R.E., Hubbard W., et al. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989;1:541–551. [Google Scholar]
- LeDell E., Poirier S. Proceedings of the AutoML Workshop at ICML. 2020. H2O automl: scalable automatic machine learning. [Google Scholar]
- Lednicky J.A., Lauzardo M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Ahn J.H., Park S.Y., Kim G.H., Kim J., Kim T.H., et al. Recent advances in AIV biosensors composed of nanobio hybrid material. Micromachines. 2018;9:651. doi: 10.3390/mi9120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Ye F., Liu X., Huang Z., Ling S., Jiang Z., et al. SARS-CoV-2 environmental contamination associated with persistently infected COVID-19 patients. Influenza Other Respir. Viruses. 2020;14:688–699. doi: 10.1111/irv.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican J., Levican A., Ampuero M., Gaggero A. JC polyomavirus circulation in one-year surveillance in wastewater in Santiago, Chile. Infect. Genet. Evol. 2019;71:151–158. doi: 10.1016/j.meegid.2019.03.017. [DOI] [PubMed] [Google Scholar]
- Liang S., Liu H., Gu Y., Guo X., Li H., Li L., et al. Fast automated detection of COVID-19 from medical images using convolutional neural networks. Commun. Biol. 2021;4:35. doi: 10.1038/s42003-020-01535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R.R.X., Bonanni A. The potential of electrochemistry for the detection of coronavirus-induced infections. Trends Anal. Chem. 2020;133 doi: 10.1016/j.trac.2020.116081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley William G., Green Brett J., Blachere Francoise M., Martin Stephen B., Law Brandon F., Jensen Paul A., et al. Manual of Analytical Methods (NMAM) 2017. Sampling and characterization of bioaerosols. [Google Scholar]
- Liu W.T., Zhu L. Environmental microbiology-on-a-chip and its future impacts. Trends Biotechnol. 2005;23:174–179. doi: 10.1016/j.tibtech.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Liu H.C., You J.X., Li Z.W., Tian G. Fuzzy petri nets for knowledge representation and reasoning: a literature review. Eng. Appl. Artif. Intell. 2017;60:45–56. [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Lodder W., Husman A.Md.R. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Canovas L., Martinez Benitez M.B., Herrera Isidron J.A., Flores Soto E. Pulsed field gel electrophoresis: past, present, and future. Anal. Biochem. 2019;573:17–29. doi: 10.1016/j.ab.2019.02.020. [DOI] [PubMed] [Google Scholar]
- Ma L., Mao G., Liu J., Yu H., Gao G., Wang Y. Rapid quantification of bacteria and viruses in influent, settled water, activated sludge and effluent from a wastewater treatment plant using flow cytometry. Water Sci. Technol. 2013;68:1763–1769. doi: 10.2166/wst.2013.426. [DOI] [PubMed] [Google Scholar]
- Ma F., Li C.C., Zhang C.Y. Development of quantum dot-based biosensors: principles and applications. J. Mater. Chem. B. 2018;6:6173–6190. doi: 10.1039/c8tb01869c. [DOI] [PubMed] [Google Scholar]
- Maestre-Carballa L., Lluesma Gomez M., Angla Navarro A., Garcia-Heredia I., Martinez-Hernandez F., Martinez-Garcia M. Insights into the antibiotic resistance dissemination in a wastewater effluent microbiome: bacteria, viruses and vesicles matter. Environ. Microbiol. 2019;21:4582–4596. doi: 10.1111/1462-2920.14758. [DOI] [PubMed] [Google Scholar]
- Mäkinen S., Skogström H., Laaksonen E., Mikkonen T. 2021. Who Needs MLOps: What Data Scientists Seek to Accomplish and How Can MLOps Help? [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Manivannan S., Ponnuchamy K. Quantum dots as a promising agent to combat COVID-19. Appl. Organomet. Chem. 2020;34 doi: 10.1002/aoc.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Matovu E., Mugasa C.M., Ekangu R.A., Deborggraeve S., Lubega G.W., Laurent T., et al. Phase II evaluation of sensitivity and specificity of PCR and NASBA followed by oligochromatography for diagnosis of human african trypanosomiasis in clinical samples from D.R. Congo and Uganda. PLoS Negl. Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meays C.L., Broersma K., Nordin R., Mazumder A. Source tracking fecal bacteria in water: a critical review of current methods. J. Environ. Manag. 2004;73:71–79. doi: 10.1016/j.jenvman.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medsker L.R., Jain L.C. Recurrent neural networks. Des. Applic. 2001:5. [Google Scholar]
- Mehrotra P. Biosensors and their applications - a review. J. Oral Biol. Craniofac. Res. 2016;6:153–159. doi: 10.1016/j.jobcr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of SARS-Cov-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health. 2020;17:5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S., Harun Ai Rashid M., Mridul M., Mohanty C., Swayamsiddha S. Application of artificial intelligence in COVID-19 drug repurposing. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:1027–1031. doi: 10.1016/j.dsx.2020.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa M.K., Gamal G., Wafiq A. The impact of COVID 19 on air pollution levels and other environmental indicators - a case study of Egypt. J. Environ. Manag. 2021;277 doi: 10.1016/j.jenvman.2020.111496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S.S., Schukat M., Howley E. Lecture Notes in Networks and Systems. Vol. 16. 2018. Deep reinforcement learning: an overview; pp. 426–440. [Google Scholar]
- Muhammad L.J., Algehyne E.A., Usman S.S., Ahmad A., Chakraborty C., Mohammed I.A. Supervised machine learning models for prediction of COVID-19 infection using epidemiology dataset. SN Comput. Sci. 2021;2:11. doi: 10.1007/s42979-020-00394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. [Google Scholar]
- Namasudra S., Dhamodharavadhani S., Rathipriya R. Nonlinear neural network based forecasting model for predicting COVID-19 cases. Neural. Process. Lett. 2021:1–21. doi: 10.1007/s11063-021-10495-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir A.A., Baptiste M.J., Mwikarago I., Habimana M.R., Ndinkabandi J., Murangwa A., et al. medRxiv; 2020. RPA-based Method for the Detection of SARS-COV2. [Google Scholar]
- Nassonova E.S. Pulsed field gel electrophoresis: theory, instruments and application. Cell Tissue Biol. 2008;2:557–565. [Google Scholar]
- National Institute for Public Health and the Environment, The Netherlands . 2020. Coronavirus Monitoring in Sewage Research. 2021. [Google Scholar]
- National Institute for Public Health and the Environment, The Netherlands . 2020. Novel Coronavirus Found in Wastewater. 2021. [Google Scholar]
- Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process. Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neethirajan S., Ahmed S.R., Chand R., Buozis J., Nagy E. Recent advances in biosensor development for foodborne virus detection. Nanotheranostics. 2017;1:272–295. doi: 10.7150/ntno.20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumeyer L., Robbins B., Nair A., Kesari A. Proceedings - IEEE International Conference on Data Mining. ICDM; 2010. S4: distributed stream computing platform; pp. 170–177. [Google Scholar]
- Nguyen H.H., Lee S.H., Lee U.J., Fermin C.D., Kim M. Immobilized enzymes in biosensor applications. Materials. 2019;12:121. doi: 10.3390/ma12010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P.K., Lavrac N., Webb G.I. Supervised descriptive rule discovery: a unifying survey of contrast set, emerging pattern and subgroup mining. J. Mach. Learn. Res. 2009;10 [Google Scholar]
- Nwanaji-Enwerem J.C., Allen J.G., Beamer P.I. Another invisible enemy indoors: COVID-19, human health, the home, and United States indoor air policy. J. Expo. Sci. Environ. Epidemiol. 2020;30:773–775. doi: 10.1038/s41370-020-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Sutjipto S., Chia P.Y., Young B.E., Gum M., et al. Absence of contamination of personal protective equipment (PPE) by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Infect. Control Hospi. Epidemiol. 2020;41:614–616. doi: 10.1017/ice.2020.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732 doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann A.C., Suttle C.A. In: Bacteriophages: Methods and Protocols. MRJ Clokie, Kropinski A.M., editors. Vol. 501. 2009. Determination of virus abundance by epifluorescence microscopy; pp. 87–95. [DOI] [PubMed] [Google Scholar]
- Otawa K., Lee S.H., Yamazoe A., Onuki M., Satoh H., Mino T. Abundance, diversity, and dynamics of viruses on microorganisms in activated sludge processes. Microb. Ecol. 2007;53:143–152. doi: 10.1007/s00248-006-9150-9. [DOI] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Lednicky J.A., Wu C.Y. Collection, particle sizing and detection of airborne viruses. J. Appl. Microbiol. 2019;127:1596–1611. doi: 10.1111/jam.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in x-rays using nCOVnet. Chaos, Solitons and Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak Z. Rough set theory and its applications to data analysis. Cybern. Syst. 1998;29:661–688. [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B., Gerrity D., Bibby K., Drewes J.E., Gerba C., Gersberg R., et al. Editorial perspectives: will SARS-CoV-2 reset public health requirements in the water industry? Integrating lessons of the past and emerging research. Environ. Sci.: Water Res. Technol. 2020;6:1761–1764. [Google Scholar]
- Pejcic B., De Marco R., Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131:1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- Peng Y., Nagata M.H. An empirical overview of nonlinearity and overfitting in machine learning using COVID-19 data. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020;92:1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.D. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020;10:16942. doi: 10.1038/s41598-020-74164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilevar M., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2020;124656 doi: 10.1016/j.jhazmat.2020.124656. [DOI] [PubMed] [Google Scholar]
- Poch M., Garrido-Baserba M., Corominas L., Perello-Moragues A., Monclus H., Cermeron-Romero M., et al. When the fourth water and digital revolution encountered COVID-19. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P.C. Fluorescence instrument for in situ monitoring of viral abundance in water, wastewater and recycled water. J. Virol. Methods. 2012;181:97–102. doi: 10.1016/j.jviromet.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Prasanth S., Singh U., Kumar A., Tikkiwal V.A., Chong P.H.J. Forecasting spread of COVID-19 using google trends: a hybrid GWO-deep learning approach. Chaos Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Quinlan J.R. Induction of decision trees. Mach. Learn. 1986;1:81–106. [Google Scholar]
- Rabiee N., Bagherzadeh M., Ghasemi A., Zare H., Ahmadi S., Fatahi Y., et al. Point-of-use rapid detection of SARS-CoV-2: nanotechnology-enabled solutions for the COVID-19 pandemic. Int. J. Mol. Sci. 2020;21:5126. doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab A., El-Koujok M., Poulin B., Amazouz M., Yacout S. Fault diagnosis in industrial chemical processes using interpretable patterns based on logical analysis of data. Expert Syst. Appl. 2018;95:368–383. [Google Scholar]
- Ragab A., El Koujok M., Ghezzaz H., Amazouz M., Ouali M.-S., Yacout S. Deep understanding in industrial processes by complementing human expertise with interpretable patterns of machine learning. Expert Syst. Appl. 2019;122:388–405. [Google Scholar]
- Rahmani A.R., Leili M., Azarian G., Poormohammadi A. Sampling and detection of corona viruses in air: a mini review. Sci. Total Environ. 2020;740 doi: 10.1016/j.scitotenv.2020.140207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez Calderón O.A., Abdeldayem O.M., Pugazhendhi A., Rene E.R. Current updates and perspectives of biosorption technology: an alternative for the removal of heavy metals from wastewater. Curr. Pollut. Rep. 2020;6:8–27. [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravina Dalal A., Mohan H., Prasad M., Pundir C.S. Detection methods for influenza a H1N1 virus with special reference to biosensors: a review. Biosci. Rep. 2020;40:1–18. doi: 10.1042/BSR20193852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro B.V., TAR Cordeiro, Oliveira e Freitas G.R., Ferreira L.F., Franco D.L. Biosensors for the detection of respiratory viruses: a review. Talanta Open. 2020;2 doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M., da Silva R.G., Mariani V.C., Coelho L.D.S. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Poggeler K.R., Franzke K., Hipp K., Kleespies R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2018;9:3255. doi: 10.3389/fmicb.2018.03255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robotto A., Quaglino P., Lembo D., Morello M., Brizio E., Bardi L., et al. SARS-CoV-2 and indoor/outdoor air samples: a methodological approach to have consistent and comparable results. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard P., Raynal P.I., Eymieux S., Blanchard E. Virus detection by transmission electron microscopy: still useful for diagnosis and a plus for biosafety. Rev. Med. Virol. 2019;29:1–9. doi: 10.1002/rmv.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. In: Medical Image Computing and Computer-assisted Intervention – MICCAI 2015. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Springer International Publishing; Cham: 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Russo A., Minichini C., Starace M., Astorri R., Calo F., Coppola N., et al. Infection and Drug Resistance. Vol. 13. 2020. Current status of laboratory diagnosis for COVID-19: a narrative review; pp. 2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Putalun W., Vimolmangkang S., Phoolcharoen W., Shoyama Y., Tanaka H., et al. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018;72:32–42. doi: 10.1007/s11418-017-1144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati A., Samavati Z., Velashjerdi M., Fauzi Ismail A., Othman M.H.D., Eisaabadi B.G., et al. Sustainable and fast saliva-based COVID-19 virus diagnosis kit using a novel GO-decorated Au/FBG sensor. Chem. Eng. J. 2020;127655 doi: 10.1016/j.cej.2020.127655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R., Navale G.R., Dharne M.S. Biosensors: frontiers in rapid detection of COVID-19. 3 Biotech. 2020;10:385. doi: 10.1007/s13205-020-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J.L., Rivera D.N., Herrera V., Morwitzer M.J., Creager H., Santarpia G.W.…Lowe J.J. Aerosol and surface transmission potential of SARS-CoV-2. MedRxiv. 2020 [Google Scholar]
- Santiago I. Trends and innovations in biosensors for COVID-19 mass testing. ChemBioChem. 2020;21:2880–2889. doi: 10.1002/cbic.202000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville R.D., Constantine N.T., Cleghorn F.R., Jack N., Bartholomew C., Edwards J., et al. Fourth-generation enzyme-linked immunosorbent assay for the simultaneous detection of human immunodeficiency virus antigen and antibody. J. Clin. Microbiol. 2001;39:2518–2524. doi: 10.1128/JCM.39.7.2518-2524.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylan Y., Erdem O., Unal S., Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9:65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K.B., DeGrasse J., Woods J.W. The influence of food matrices on aptamer selection by SELEX (systematic evolution of ligands by exponential enrichment) targeting the norovirus P-domain. Food Chem. 2018;258:129–136. doi: 10.1016/j.foodchem.2018.03.054. [DOI] [PubMed] [Google Scholar]
- Schlindwein A.D., Rigotto C., Simoes C.M., Barardi C.R. Detection of enteric viruses in sewage sludge and treated wastewater effluent. Water Sci. Technol. 2010;61:537–544. doi: 10.2166/wst.2010.845. [DOI] [PubMed] [Google Scholar]
- Sedik A., Iliyasu A.M., Abd El-Rahiem B., Abdel Samea M.E., Abdel-Raheem A., Hammad M., et al. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses. 2020;12 doi: 10.3390/v12070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak D. Yale University Press; New York, USA: 2014. Water 4.0: The Past, Present, and Future of the World's Most Vital Resource. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shah F.M., SKS Joy, Ahmed F., Hossain T., Humaira M., Ami A.S., et al. 2020. A Comprehensive Survey of COVID-19 Detection Using Medical Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetti N.P., Mishra A., Bukkitgar S.D., Basu S., Narang J., Raghava Reddy K., et al. Conventional and nanotechnology-based sensing methods for SARS coronavirus (2019-nCoV) ACS Appl. Bio Mater. 2021;4:1178–1190. doi: 10.1021/acsabm.0c01545. [DOI] [PubMed] [Google Scholar]
- Shuja J., Alanazi E., Alasmary W., Alashaikh A. COVID-19 open source data sets: a comprehensive survey. Appl. Intell. 2020;51:1296–1325. doi: 10.1007/s10489-020-01862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard P., De Marco A., Agathokleous E., Feng Z., Xu X., Paoletti E., et al. Amplified ozone pollution in cities during the COVID-19 lockdown. Sci. Total Environ. 2020;735 doi: 10.1016/j.scitotenv.2020.139542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano B., Dantas G., da Silva C.M., Arbilla G. Increased ozone levels during the COVID-19 lockdown: Analysis for the city of Rio de Janeiro, Brazil. Sci. Total Environment. 2020;737 doi: 10.1016/j.scitotenv.2020.139765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. arXiv; 2014. Very Deep Convolutional Networks for Large-scale Image Recognition. [Google Scholar]
- Simsek M., Kantarci B. Artificial intelligence-empowered mobilization of assessments in COVID-19-like pandemics: a case study for early flattening of the curve. Int. J. Environ. Res. Public Health. 2020;17:3437. doi: 10.3390/ijerph17103437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.D., Shandilya R., Bhargava A., Kumar R., Tiwari R., Chaudhury K., et al. Quantum dot based nano-biosensors for detection of circulating cell free mirnas in lung carcinogenesis: from biology to clinical translation. Front. Genet. 2018;9:616. doi: 10.3389/fgene.2018.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu D., Binda G., Dossi C., Monticelli D. Biochar as an alternative sustainable platform for sensing applications: a review. Microchem. J. 2020:159. [Google Scholar]
- Streeck H., Schulte B., Kummerer B.M., Richter E., Holler T., Fuhrmann C., et al. Infection fatality rate of SARS-CoV2 in a super-spreading event in Germany. Nat. Commun. 2020;11:5829. doi: 10.1038/s41467-020-19509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleman S., Shukla S.K., Malhotra N., Bukkitgar S.D., Shetti N.P., Pilloton R., et al. Point of care detection of COVID-19: advancement in biosensing and diagnostic methods. Chem. Eng. J. 2021;414 doi: 10.1016/j.cej.2021.128759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundui B., Ramirez Calderon O.A., Abdeldayem O.M., Lázaro-Gil J., Rene E.R., Sambuu U. Applications of machine learning algorithms for biological wastewater treatment: updates and perspectives. Clean Techn. Environ. Policy. 2021;23:127–143. [Google Scholar]
- Sung G., Ahn C., Kulkarni A., Shin W.G., Kim T. Highly efficient in-line wet cyclone air sampler for airborne virus detection. J. Mech. Sci. Technol. 2017;31:4363–4369. [Google Scholar]
- Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12:1–13. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C., Hutter F., Hoos H.H., Auto-WEKA Leyton-Brown K. Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2013. Combined selection and hyperparameter optimization of classification algorithms; pp. 847–855. [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkrongjun P., Phetpeng S., Asawutmangkul W., Sotthibandhu S., Kitpipit T., Thanakiatkrai P. Improved STR profiles from improvised explosive device (IED): fluorescence latent DNA detection and direct PCR. Forensic Sci. Int. Genet. 2019;41:168–176. doi: 10.1016/j.fsigen.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamani B.S., Uppal T., Verma S.C., Misra M. 2020. Functionalized TiO2 Nanotube-based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishya R., Javaid M., Khan I.H., Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault D., Moineau S., Duchaine C. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 2008;72:413–444. doi: 10.1128/MMBR.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.I., McQuillan J., Taiwo M., Parks R., Stenton C.A., Morgan H., et al. A highly specific Escherichia coli qPCR and its comparison with existing methods for environmental waters. Water Res. 2017;126:101–110. doi: 10.1016/j.watres.2017.08.032. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(281–292) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dai Z. Carbon nanomaterial-based electrochemical biosensors: an overview. Nanoscale. 2015;7:6420–6431. doi: 10.1039/c5nr00585j. [DOI] [PubMed] [Google Scholar]
- Wang Q., Su M. A preliminary assessment of the impact of COVID-19 on environment - a case study of China. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021;4:228. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed M., Foad A. 2020. Rapid Scoping Review of Evidence of Outdoor Transmission of COVID-19. [Google Scholar]
- Weerathunge P., Ramanathan R., Torok V.A., Hodgson K., Xu Y., Goodacre R., et al. Ultrasensitive colorimetric detection of murine norovirus using nanozyme aptasensor. Anal. Chem. 2019;91:3270–3276. doi: 10.1021/acs.analchem.8b03300. [DOI] [PubMed] [Google Scholar]
- Wen H., Zhao Q., Lin Z., Xuan D., Shroff N. In: Security and Privacy in Communication Networks. Namkee P., et al., editors. Springer Nature; Switzerland: 2020. A study of the privacy of COVID-19 contact tracing apps; pp. 297–317. [Google Scholar]
- Weschler C.J., Carslaw N. Indoor chemistry. Environ. Sci. Technol. 2018;52:2419–2428. doi: 10.1021/acs.est.7b06387. [DOI] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M., Silka J., Polap D., Wozniak M., Damasevicius R. Real-time neural network based predictor for cov19 virus spread. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M., Silka J., Wozniak M. Neural network powered COVID-19 spread forecasting model. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba M., Rawat A., Pedapati T. 2019. A Survey on Neural Architecture Search. [Google Scholar]
- Woo C.H., Jang S., Shin G., Jung G.Y., Lee J.W. Sensitive one-step isothermal detection of pathogen-derived RNAs. MedRxiv. 2020 [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease 2019 (COVID-19): Situation Report – 51. [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease (COVID-19): Weekly Updates. [Google Scholar]
- World Health Organization . 2020. Novel Coronavirus (2019-nCoV): Situation Report-3. [Google Scholar]
- Wu Q., Liu W.T. Determination of virus abundance, diversity and distribution in a municipal wastewater treatment plant. Water Res. 2009;43:1101–1109. doi: 10.1016/j.watres.2008.11.039. [DOI] [PubMed] [Google Scholar]
- Wu X., Kumar V., Quinlan J.R., Ghosh J., Yang Q., Motoda H., et al. Top 10 algorithms in data mining. Knowl. Inf. Syst. 2008;14:1–37. [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Medrxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Elankumaran S., Marr L.C. Concentrations and size distributions of airborne influenza a viruses measured indoors at a health Centre, a day-care Centre and on aeroplanes. J. R. Soc. Interface. 2011;8:1176–1184. doi: 10.1098/rsif.2010.0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Ma Yuanyuan, Stanciu Stefan G., Wu Aiguo. In: Nanobiosensors: From Design to Applications. Wu A., Khan W.S., editors. Wiley-VCH Verlag GmbH & Co. KGaA; 2020. Transduction process-based classification of biosensors; pp. 23–44. [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Gen M. 2010. Introduction to Evolutionary Algorithms. [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (rt-lamp) diagnostic platform. Clin. Chem. 2020;66:975–986. doi: 10.1093/clinchem/hvaa102. (Springer, London (UK)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(739–751) doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano-Monserrate M.A., Ruano M.A., Sanchez-Alcalde L. Indirect effects of COVID-19 on the environment. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneti R.N., Girardi V., Spilki F.R., Mena K., Westphalen A.P.C., da Costa Colares E.R., et al. Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeroual A., Harrou F., Dairi A., Sun Y. Deep learning methods for forecasting COVID-19 time-series data: a comparative study. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Aarnink A.J., Wang W., Fabri T., Groot Koerkamp P.W., de Jong M.C. Airborne virus sampling: efficiencies of samplers and their detection limits for infectious bursal disease virus (IBDV) Ann. Agric. Environ. Med. 2014;21:464–471. doi: 10.5604/12321966.1120585. [DOI] [PubMed] [Google Scholar]
- Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., et al. Deep learning-based detection for COVID-19 from chest CT using weak label. IEEE Trans. Med. Imaging. 2020:1–13. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer L., Lindauer M., Hutter F. Auto-pytorch: multi-Fidelity MetaLearning for efficient and robust AutoDL. IEEE Trans. Pattern Anal. Mach. Intell. 2021;43(9):3079–3090. doi: 10.1109/TPAMI.2021.3067763. [DOI] [PubMed] [Google Scholar]
- Zuo X., Fan C., Chen H.-Y. Biosensing: CRISPR-powered diagnostics. Nat. Biomed. Eng. 2017;1:1–2. [Google Scholar]