Abstract
Islets represent an important site of direct action of the hormone ghrelin, with expression of the ghrelin receptor (growth hormone secretagogue receptor; GHSR) having been localized variably to alpha cells, beta cells, and/or somatostatin (SST)-secreting delta cells. To our knowledge, GHSR expression by pancreatic polypeptide (PP)-expressing gamma cells has not been specifically investigated. Here, histochemical analyses of Ghsr-IRES-Cre × Cre-dependent ROSA26-yellow fluorescent protein (YFP) reporter mice showed 85% of GHSR-expressing islet cells coexpress PP, 50% coexpress SST, and 47% coexpress PP + SST. Analysis of single-cell transcriptomic data from mouse pancreas revealed 95% of Ghsr-expressing cells coexpress Ppy, 100% coexpress Sst, and 95% coexpress Ppy + Sst. This expression was restricted to gamma-cell and delta-cell clusters. Analysis of several single-cell human pancreatic transcriptome data sets revealed 59% of GHSR-expressing cells coexpress PPY, 95% coexpress SST, and 57% coexpress PPY + SST. This expression was prominent in delta-cell and beta-cell clusters, also occurring in other clusters including gamma cells and alpha cells. GHSR expression levels were upregulated by type 2 diabetes mellitus in beta cells. In mice, plasma PP positively correlated with fat mass and with plasma levels of the endogenous GHSR antagonist/inverse agonist LEAP2. Plasma PP also elevated on LEAP2 and synthetic GHSR antagonist administration. These data suggest that in addition to delta cells, beta cells, and alpha cells, PP-expressing pancreatic cells likely represent important direct targets for LEAP2 and/or ghrelin both in mice and humans.
Keywords: ghrelin, LEAP2, pancreatic polypeptide, GHSR, islet
The hormone ghrelin is produced primarily in pancreatic islets during the fetal period and in the stomach afterward (1-3). Although ghrelin’s roles during development are unclear, administration of ghrelin or administration of either of 2 other ghrelin gene products, desacyl-ghrelin or obestatin, all have been shown to promote beta-cell survival, beta-cell mass, and increase islet size (4-6). Ghrelin also exhibits well-characterized orexigenic and glucoregulatory actions that are mediated by ghrelin receptors (GHSRs; growth hormone secretagogue receptors), including those in the central nervous system and peripheral tissues (1, 7). For instance, ghrelin’s glucoregulatory actions—emphasized by blood glucose elevation on its administration and by hypoglycemia in calorically restricted ghrelin-knockout (KO) mice—include engagement of central nervous system–expressed GHSRs, GH release from pituitary somatotrophs, and potentially vasoconstrictor effects on GHSR-expressing islet arterioles (7, 8). Ghrelin’s reported glucoregulatory actions also include both direct and indirect effects of ghrelin on islets to modulate secretion of 3 of the major islet-derived hormones—specifically, to inhibit insulin release, modulate glucagon release, and promote somatostatin (SST) release (1, 7, 9, 10). Specifically, ghrelin-induced attenuation of glucose-mediated intracellular calcium increases in isolated rat and mouse beta cells (11, 12), the abolishment of this phenomenon by exposure to the GHSR antagonist [D-Lys3]-GHRP-6 (11), and its rescue in isolated beta cells from mice with selective rat insulin promoter–directed (Ins-Cre-driven) GHSR expression (13) are evidence of direct ghrelin action on beta cells. Additional evidence of a direct effect of ghrelin on beta cells comes from a quantitative reverse transcriptase–polymerase chain reaction analysis demonstrating Ghsr expression by 2 immortalized mouse beta-cell lines and a histochemistry study demonstrating a high degree of colocalized insulin-immunoreactivity and green fluorescent protein (GFP) expression in a GHSR-IRES-tauGFP reporter line (14). Also, ghrelin hyperpolarizes isolated C57BL/6N mouse beta cells and decreases glucose-stimulated insulin secretion from them (12). Moreover, although not entirely aligned with the aforementioned findings, mouse insulin promoter–selective (MIP-Cre-driven) GHSR deletion reduces fasting blood glucose and plasma insulin, reduces glucose-stimulated insulin secretion, and improves insulin sensitivity during insulin tolerance testing (14). Furthermore, Ghsr messenger RNA (mRNA) expression in and ghrelin-induced glucagon secretion from immortalized alpha-cell lines (7, 15) and colocalization of GHSR-immunoreactivity (IR) with glucagon-IR in rat and mouse islets (8, 16) suggest that ghrelin also may act directly on alpha cells. However, bulk sequencing transcriptomic profiling studies have failed to detect Ghsr expression by pools of isolated beta cells and show low or undetectable expression by pools of isolated alpha cells (9, 10). These studies instead reveal high Ghsr expression by delta cells and functionally demonstrate indirect effects of ghrelin on glucagon and/or insulin secretion via stimulating SST release from delta cells (9, 10).
Regulation by ghrelin of the fourth major islet-derived hormone, pancreatic polypeptide (PP), has been described in only a few studies. For instance, ghrelin was shown to dose-dependently inhibit PP release from mouse and rat islets (17, 18). In contrast, one study in humans demonstrated that administered ghrelin acutely increases plasma PP, whereas a second human study showed no effect of administered ghrelin on plasma PP during a meal tolerance test (19, 20). To our knowledge, localization of GHSRs to PP-expressing gamma cells has not been directly investigated. Here, we aimed to further characterize islet GHSR expression with a focus on PP-expressing gamma cells.
Materials and Methods
Animals
Ghsr-IRES-Cre mice (21) were crossed to ROSA26-YFP reporter mice (B6.129X1-Gt[ROSA]26Sortm1[EYFP]Cos/J) (Jackson Laboratory; stock No. 006148) to generate mice with YFP-labeled GHSR-expressing cells for immunohistochemistry (performed on 8- to 9-week-old mice). Injection studies were performed on 8- to 10-week-old male C57BL/6N mice or 8- to 10-week-old male ghrelin-KO mice (22, 23) and wild-type littermates on a C57BL/6N genetic background. Housing conditions were 22 °C to 24 °C under a 12-hour dark-light cycle and free access to water and Teklad Global Diet (2016) (Envigo). All procedures were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Diet-induced Obesity Mouse Model
Plasma samples used here were the same as those published previously (24) and were taken from 16-week-old mice that had been exposed for 12 weeks to either standard chow or a high-fat diet (Envigo Teklad TD88137) since age 4 weeks. The reported ghrelin, LEAP2, and fat mass values are the same as those for 10 of the 11 diet-induced obese mice (1 sample was excluded because of an insufficient remaining sample to perform a PP assay) and 10 of the 10 lean mice described in Fig. 1H and 1I and Supplementary Fig. 3A (25). Samples from these mice, which had been collected into ice-cold, EDTA-coated microfuge tubes, centrifuged at 4 °C at 1500g for 15 minutes, and stored at –80 °C, were newly assayed here for PP.
Figure 1.
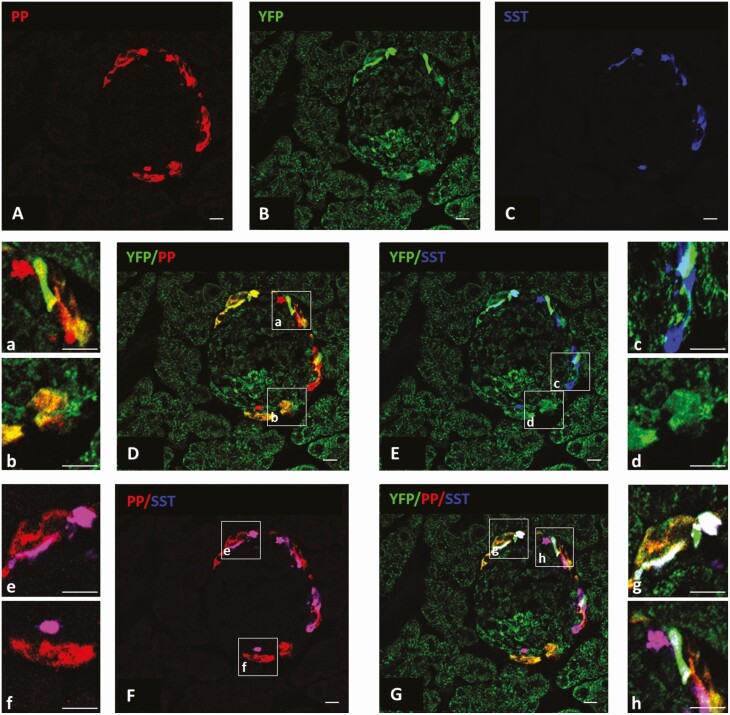
Yellow fluorescent protein (YFP), pancreatic polypeptide (PP), and somatostatin (SST) expression in mouse islets. A to G, a to h, Photomicrographs demonstrating YFP, PP, and SST expression within a single representative mouse islet. A, PP-immunoreactivity (IR) (red); B, YFP-IR (as a reporter for growth hormone secretagogue receptor [GHSR]; green); C, SST-IR (blue); D, YFP-IR (green) and PP-IR (red); E, YFP-IR (green) and SST-IR (blue); F, PP-IR (red) and SST-IR (blue); and G, YFP-IR (green), PP-IR (red), and SST-IR (blue). The boxed areas in D to G are magnified in a to h to emphasize examples of double-labeled cells and triple-labeled cells. Double-labeled cells include YFP-IR + PP-IR (yellow), YFP-IR + SST-IR (cyan), and PP-IR + SST-IR (magenta). Triple-labeled cells include YFP-IR + PP-IR + SST-IR (white). n = 3 mice per group. Scale bars = 50 µm.
Pharmacologic Studies
[D-Lys3]-GHRP-6 (200 nmol/30 g body weight [BW]) (D-Lys; Abcam) or vehicle (water) was administered intraperitoneally twice daily at 0630 and 1730 for 4 consecutive days to C57BL/6N mice. Following injection 2 on day 4, mice were fasted overnight (12 hours). Immediately following the fast, mice received a final [D-Lys3]-GHRP-6 or vehicle injection, 2 hours after which blood was collected. In another C57BL/6N cohort, LEAP2 (Peptide International; catalog No. PLP-4405-s) (72 nmol/kg) or vehicle (10% dimethyl sulfoxide in phosphate-buffered saline [PBS]) was delivered intraperitoneally 30 minutes prior to blood collection. In a third C57BL/6N cohort, ghrelin (2 mg/kg BW) (Innovagen AB) or vehicle (saline) was delivered subcutaneously 30 minutes prior to blood collection, as had been done previously in (26). Similarly, ghrelin (2 mg/kg BW) or vehicle was delivered subcutaneously to ghrelin-KO and wild-type littermates, 30 minutes prior to blood collection.
Blood Collection and Plasma Hormone Analysis
Blood glucose was measured in blood obtained from tail snips using a Bayer Contour blood glucose monitoring system. Blood from tail snips also was collected into ice-cold EDTA-coated microcentrifuge tubes. The samples were immediately centrifuged at 4 °C at 1500g for 15 minutes, after which plasma was stored at –80 °C until plasma PP analysis. For some studies, separate samples of blood from the tail snips were collected into ice-cold EDTA-coated microcentrifuge tubes that contained the protease inhibitor p-hydroxymercuribenzoic acid (Sigma Aldrich; final concentration 1 mM). They were immediately centrifuged at 4 °C at 1500g for 15 minutes after which HCl was added (1:10) to achieve a final concentration of 0.1 N to stabilize the acylated form of ghrelin. They were centrifuged again and stored at –80 °C until plasma ghrelin analysis. Plasma PP was determined using a mouse PP enzyme-linked immunosorbent assay kit (Cloud Clone Corp; catalog No. CEB265Mu; range, 24.69-2000 pg/mL, sensitivity < 9.16 pg/mL). Plasma ghrelin was determined using a rat/mouse ghrelin (active) enzyme-linked immunosorbent assay kit (EMD Millipore; catalog No. EZRGRA-90K). End point calorimetric assays were performed with a BioTek PowerWave XS Microplate spectrophotometer and Gen5Biotek software (Thermo Fisher Scientific).
Immunohistochemistry
Formalin-fixed organs from ROSA26-YFP reporter mice carrying the Ghsr-IRES-Cre gene were processed as described previously (21). Pancreas sections 8-µm thick were cut at 150-μm intervals, and brains were sectioned at 25-µm thickness into 5 equal series prior to mounting on glass slides. Antibody details are provided in Supplementary Table 1 (25). No YFP immunolabeling was detected in islets from ROSA26-YFP reporter mice lacking Ghsr-IRES-Cre, indicating a lack of YFP expression without Cre activity (Supplementary Fig. 1) (25). Anti-YFP, anti-insulin, antiglucagon, and anti-SST antibodies were previously validated (27-31). The anti-PP antibody was newly validated here in pancreatic sections adjacent to those used in the main immunohistochemistry experiments. This involved dividing the primary antibody solution in two. In one half, 5X excess PP-blocking peptide (Pancreatic Polypeptide Blocking Peptide, MyBioSource Inc; catalog No. MBS426352) by antibody weight was added (16.5 µg/mL blocking peptide; 3.3 µg/mL antibody). In the other half, an equal volume of PBS was added instead. One set of pancreas sections from 3 different mice was incubated with primary antibody solution + PP-blocking peptide. A second set of pancreas sections from the same 3 mice was incubated with primary antibody solution + PBS. No PP immunoreactivity was observed in islets incubated with primary antibody solution + PP blocking peptide; PP immunoreactivity was observed in islets incubated with primary antibody solution + PBS (Supplementary Fig. 2) (25).
Image Acquisition and Analysis
Fluorescent digital images were taken using 20× and 40× objectives of a Leica TCS SP5 confocal microscope (UT Southwestern Live Cell Imaging Core). Laser intensity, offset, and gain were adjusted to improve signal-to-background ratio. National Institutes of Health ImageJ (http://rsbweb.nih.gov/ij/) software was used to help identify single-, double-, and triple-labeled cells: Images were split into separate channels, edited to highlight the cell edges using the Find Edges plugin, followed by merging 2 or 3 channels (to detect double or triple colocalization) with 4', 6-diamidino-2-phenylindole (DAPI) to manually count colocalized cells. Colocalization criteria included visualization of overlapping immunoreactivity within a single cell, whereby a cell was identified as having a DAPI-stained nucleus with surrounding cell edges. For quantitative analysis, 10 islets from each of 3 mice, the brains of which were first shown to exhibit YFP expression patterns matching the best previously-reported cases (21) (see Supplementary Fig. 3) (25), were examined.
Single-Cell Transcriptome Analyses
Mouse and human single-cell RNA sequencing data were taken from Baron et al (32) (GSE84133) (1 female + 2 male nondiabetic donors; 1 female donor with type 2 diabetes mellitus [T2DM]); Muraro et al (33) (GSE85241) (1 female + 3 male nondiabetic donors); Grün et al (34) (GSE81076) (5 donors with or without T2DM, numbers of each and sexes unspecified); Lawlor et al (35) (GSE86473) (2 female + 3 male nondiabetic donors; 2 female + 1 male donors with T2DM); and Segerstolpe et al (36) (E-MTAB-5061) (1 female + 5 male nondiabetic donors; 2 female + 2 male donors with T2DM) (32, 35, 36).
For GSE81076 and GSE85241, 3′ adaptors (2) were trimmed with Cutadapt 1.16 using ‘AAAAAA’ and mapped with RNA STAR 2.7.5b86 to human GRCh38 genome. Read 1, containing cell barcode (8 nt) and unique molecular identifier (4 nt) was split using fastxtrimmer (http://hannonlab.cshl.edu/fastx_toolkit/) and mapped back to data generated from Read 2 using Fgbio 1.1.0 (http://fulcrumgenomics.github.io/fgbio/). Gene-level unique molecular identifier count was performed using Dropseq tools 2.3.0 (https://github.com/broadinstitute/Drop-seq/) with gene model from Ensembl V100. For GSE86473 and E-MTAB-5061, reads were mapped to human GRCh38 genome and gene-level expression counted using STAR 2.5.0a with Ensembl V100 gene model.
For both mouse and human data sets, counts were analyzed using Seurat v3.1 [https://satijalab.org/seurat/; (37, 38)]. Cells with low reads at less than 1000 (GSE86473, E-MTAB-5061, GSE85241) or less than 500 (GSE81076) were removed. Cells with reads greater than 3 000 000 (GSE86473), greater than 1 000 000 (E-MTAB-5061), greater than 20 000 (GSE81076), or greater than 40 000 (GSE85241) likely represented doublets and were removed. Data were log normalized, scaled, and subjected to principal component analyses. Samples were integrated by identifying anchors to harmonize log-normalized expression across data sets. Notably, clustering of the human data sets was performed to realign the data sets, which had been gathered from multiple sources and collected using differing technologies, to form a single cohesive data set. Differential expression analysis was performed using Wilcoxon rank sum-test to identify marker genes for each cluster. For clusters with no identified canonical marker genes, the top 2 significant genes identified by differential expression analysis were used instead. Using the human data set, a subset of cells expressing a minimum of one transcript read from GHSR was created and reclustered with Seurat for separate analysis.
Differential expression analysis using EdgeR (39) was performed with 8167 cells from 14 nondiabetic donors (4 females + 10 males) and 2362 cells from 8 T2DM donors (5 females + 3 males) (32, 35, 36). Counts were normalized and analyzed using generalized linear model with quasi-likelihood F tests controlling for study, donor, and age. Genes with a false discovery rate (FDR) of less than 0.1 were identified as differentially expressed. In each cluster, only genes expressed in 25% or more of cells within that cluster were included in the analysis. A separate differential expression analysis was also performed for GHSR using the same methodology even though it was expressed in less than 25% of the cells in each cluster.
Statistical Analyses
For functional experiments, significant differences between the groups were determined using a paired t test or 2-way analysis of variance followed by post hoc Sidak multiple comparison testing, and correlations were determined using Pearson correlation coefficient calculations (GraphPad Prism 7.0; GraphPad Software Inc). Data are expressed as mean ± SEM. P values less than .05 were considered statistically significant.
Results
Immunohistologic Evaluation of Growth Hormone Secretagogue Receptor Reporter Mice
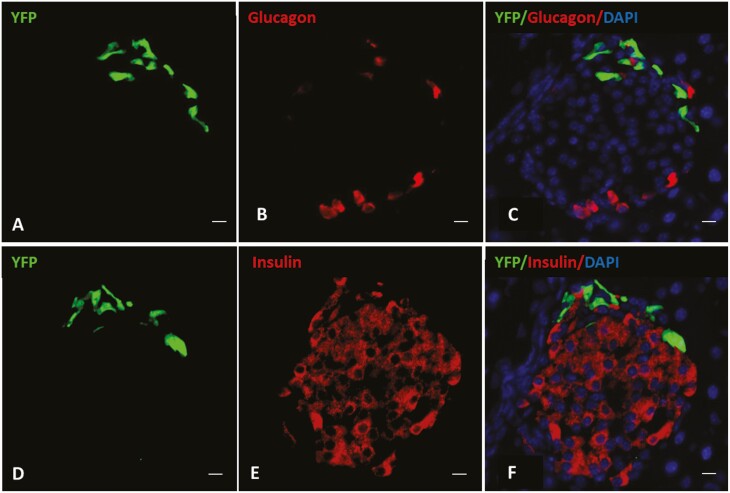
GHSR-expressing islet cells were identified as YFP-IR cells in Cre recombinase transgene-containing + YFP transgene-containing progeny of crosses of Ghsr-IRES-Cre mice × Cre-dependent ROSA26-YFP reporter mice. Nearly all pancreatic islets examined included YFP-IR cells (Fig. 1). YFP-IR cells were limited to the periphery of the islets and were sparsely distributed (see Fig. 1). YFP-IR most frequently colocalized with PP-IR, followed by SST-IR (Table 1, see Fig. 1). Nearly half of YFP-IR cells expressed both PP-IR and SST-IR (see Table 1, Fig. 1). The YFP-IR cells can be further subdivided as follows: 37.9 ± 2.8% contained PP-IR without SST-IR, 3.1 ± 0.9% contained SST-IR without PP-IR, and 12.2 ± 1.6% contained neither SST-IR nor PP-IR. No YFP-IR cells with insulin-IR or glucagon-IR were observed (Fig. 2).
Table 1.
Coexpression of YFP, pancreatic polypeptide, and somatostatin within pancreatic islets of a transgenic mouse model expressing YFP under the direction of the growth hormone secretagogue receptor promoter
| % of cells with YFP immunoreactivity coexpressing | ||
|---|---|---|
| PP immunoreactivity | SST immunoreactivity | PP and SST immunoreactivity |
| 84.7 ± 2.9 | 49.8 ± 2.9 | 46.7 ± 3.0 |
| % of cells with PP immunoreactivity coexpressing | ||
| YFP immunoreactivity | SST immunoreactivity | YFP and SST immunoreactivity |
| 74.9 ± 4.7 | 61.1 ± 3.3 | 40.2 ± 2.3 |
| % of cells with SST immunoreactivity coexpressing | ||
| YFP immunoreactivity | PP immunoreactivity | YFP and PP immunoreactivity |
| 58.3 ± 4.0 | 82.5 ± 4.4 | 55.5 ± 3.7 |
Data are reported as the mean percentage ± SEM for 3 animals at 4 different levels for each animal, each separated by at least 150 μm.
Abbreviations: PP, pancreatic polypeptide; SST, somatostatin.
Figure 2.
YFP, glucagon, and insulin expression in mouse islets. A to C, Photomicrographs demonstrating YFP and glucagon expression + 4', 6-diamidino-2-phenylindole (DAPI) (nuclear) staining within a single representative islet. A, YFP-immunoreactivity (IR) (green); B, glucagon-IR (red); C, merged images of YFP-IR (green) + glucagon-IR (red) + DAPI (blue). D to F, Photomicrographs demonstrating YFP and insulin expression + DAPI (nuclear) staining within a different representative islet. D, YFP-IR (green); E, insulin-IR (red); F, merged images of YFP-IR (green) + insulin-IR (red) + DAPI (blue). n = 3 mice per group. Scale bars = 50 µm.
Single-Cell Transcriptomic Data From Mouse Pancreas
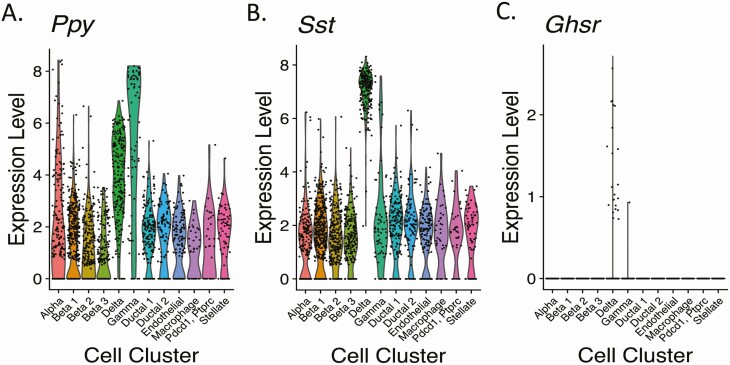
Mouse pancreatic cell type–specific Ghsr mRNA expression was investigated by reanalyzing previously published data (32) in which a droplet-based (inDrop), single-cell RNA sequencing method was implemented to determine the transcriptomes of 1886 individual pancreatic cells from 2 mouse strains (C57BL/6 and ICR). Cells with similar transcriptomes were grouped into 12 distinct cell clusters (Table 2, Fig. 3; Supplementary Fig. 4A) (25), including the 4 traditional islet endocrine cell types (alpha cells [marker = Gcg, which encodes glucagon], 3 distinct beta-cell clusters [markers = Ins1 and Ins2, which encode insulin], delta cells [marker = Sst], and gamma cells [marker = Ppy, which encodes PP]), 2 ductal-cell clusters (marker = Krt19), an endothelial-cell cluster (markers = Pecam1 and Plvap) (36), a macrophage cluster (marker = Cd86) (32), an undefined cluster labeled based on its highest differentially expressed genes (markers = Pdcd1, Ptprc), and a stellate-cell cluster (markers = Pdgfrb and Sparc) (32, 36). Ppy transcript was present in all clusters, yet its expression levels were highest in gamma cells and delta cells (Table 2, Fig. 3A; Supplementary Fig. 4B) in (25). While Table 2 demonstrates that a slightly greater percentage of delta cells (96.9%) express Ppy than gamma cells (91.4%), the expression of Ppy was higher in a greater percentage of gamma cells than delta cells (Fig. 3A). Sst expression also was detectable in all clusters, yet Sst expression was highest in delta cells and only the delta-cell cluster included Sst expression in 100% of its included cells (see Table 2, Fig. 3B; Supplementary Fig. 4C) (25).
Table 2.
Single-cell mouse islet transcriptomic analysis from Baron et al (32) showing gene expression profiles of Ghsr, Ppy, Sst, and Ghrl in 12 different identified cell clusters
| Cluster ID | No. and (%) of total cells | No. and (%) of cells within each cluster expressing the following transcripts: | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ppy | Sst | Ghsr | Ppy + Ghsr | Sst + Ghsr | Ppy + Sst + Ghsr | Ppy + Sst | Ghrl | Ghrl + Ghsr | ||
| Alpha | 195 (10.3) | 149 (76.4) | 147 (75.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 117 (60.0) | 8 (4.1) | 0 (0.0) |
| Beta 1 | 415 (22.0) | 274 (66.0) | 329 (79.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 224 (54.0) | 9 (2.2) | 0 (0.0) |
| Beta 2 | 243 (12.9) | 179 (73.7) | 217 (89.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 163 (67.1) | 7 (2.9) | 0 (0.0) |
| Beta 3 | 205 (10.9) | 118 (57.6) | 181 (88.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 108 (52.7) | 7 (3.4) | 0 (0.0) |
| Delta | 222 (11.8) | 215 (96.9) | 222 (100.0) | 22 (9.9) | 21 (9.5) | 22 (9.9) | 21 (9.5) | 215 (96.9) | 6 (2.7) | 0 (0.0) |
| Gamma | 70 (3.7) | 64 (91.4) | 62 (88.6) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 58 (82.9) | 1 (1.4) | 0 (0.0) |
| Ductal 1 | 168 (8.9) | 144 (85.7) | 144 (85.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 123 (73.2) | 32 (19.1) | 0 (0.0) |
| Ductal 2 | 98 (5.2) | 76 (77.6) | 85 (86.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 68 (69.4) | 23 (23.5) | 0 (0.0) |
| Endothelial | 135 (7.2) | 96 (71.1) | 109 (80.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 85 (63.0) | 7 (5.2) | 0 (0.0) |
| Macrophage | 37 (2.0) | 23 (62.2) | 29 (78.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 20 (54.1) | 1 (2.7) | 0 (0.0) |
| Pdcd1/Ptprc | 31 (1.6) | 19 (61.3) | 26 (83.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 18 (58.1) | 2 (6.5) | 0 (0.0) |
| Stellate | 67 (3.6) | 53 (79.1) | 53 (79.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 46 (68.7) | 2 (3.0) | 0 (0.0) |
Abbreviations: ID, identification; Ppy, pancreatic polypeptide; Sst, somatostatin.
Figure 3.
Expression levels of messenger RNAs encoding pancreatic polypeptide (Ppy), somatostatin (Sst), and growth hormone secretagogue receptor (Ghsr) in 12 different cell clusters from mouse pancreas. Data are presented as violin plots whereby each black dot represents an instance of detectable expression of the transcript within a cell and the corresponding level of expression and the colored regions represent the probability density of the data at different expression levels.
Overall, Ghsr mRNA expression was observed in 1.2% (n = 23/1886) of cells and was restricted to delta-cell and gamma-cell clusters (see Table 2, Fig. 3C; Supplementary Fig. 4D) (25). Within the delta-cell cluster, 9.9% of cells expressed Ghsr. Within the gamma-cell cluster, 1 of 70 (1.4%) cells expressed Ghsr. This single Ghsr-expressing gamma-cell coexpressed Ppy and Sst. For the entire mouse data set, all but one Ghsr-expressing cell coexpressed Ppy (n = 22/23). All but one Ghsr-expressing, Sst-expressing delta cell coexpressed Ppy (n = 21/22). All cells expressing Ghsr + Ppy also expressed Sst (see Table 2). Thus, these data demonstrate that the majority of mouse islet cells expressing Ghsr coexpress Ppy + Sst.
Single-Cell Transcriptomic Data From Human Pancreatic Cells
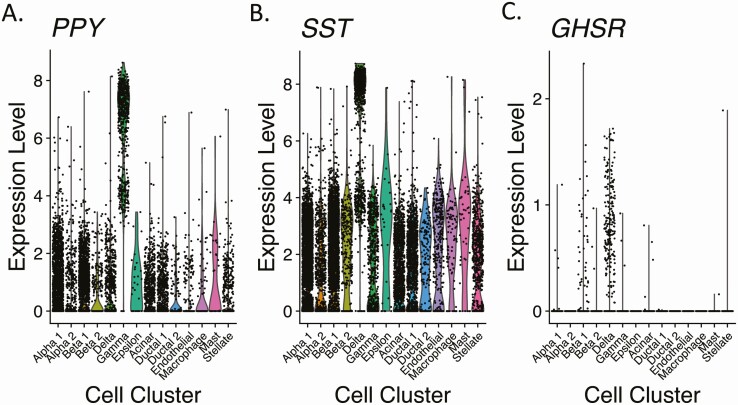
Single-cell RNA sequencing data from human pancreatic cells were obtained from Baron et al (32) and 4 other studies (33-36). Data were from nondiabetic donors, T2DM donors, and donors of unknown health status. After removing low-quality cells and doublets, this combined database consisted of 14 011 cells (8167 from nondiabetic donors, 2362 from T2DM donors, and 3482 from donors of unknown health status). Fourteen distinct cell clusters were identified (Table 3, Fig. 4; Supplementary Fig. 5A) (25), including the 4 traditional islet endocrine cell types (2 distinct alpha-cell clusters [marker = GCG], 2 distinct beta-cell clusters [marker = INS], delta cells [marker = SST], and gamma cells [marker = PPY], a fifth islet endocrine cell type [epsilon-cells [marker = GHRL, which encodes ghrelin], an acinar-cell cluster [marker = CPA1], 2 ductal-cell clusters [marker = KRT19], an endothelial-cell cluster [marker = VWF], a macrophage cluster [marker = SDS], a mast-cell cluster [marker = TPSAB1], and a stellate-cell cluster [marker = PDGFRA]). Like the mouse data, PPY and SST expression within human pancreas was rather indiscriminate, although cells with the highest PPY and SST expression levels were concentrated within gamma-cell and delta-cell clusters, respectively (see Table 3, Fig. 4A and 4B). While there was a relatively similar percentage of cells within the gamma-cell clusters of human pancreas and mouse pancreas that expressed SST (84.8% and 88.6%, respectively), the delta-cell clusters of human and mouse pancreata were characterized by obvious differences in PPY coexpression (40.5% of cells in humans vs 96.9% of cells in mice; see Tables 2 and 3).
Table 3.
Single-cell human islet transcriptomic analysis showing gene expression profiles of GHSR, PPY, SST, and GHRL in 14 different identified cell clusters
| Cluster ID | No. and (%) of total cells | No. and (%) of cells within each cluster expressing the following transcripts: | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PPY | SST | GHSR | PPY + GHSR | SST + GHSR | PPY + SST + GHSR | PPY + SST | GHRL | GHRL + GHSR | ||
| Alpha 1 | 3472 (24.8) | 1509 (43.5) | 2704 (77.9) | 6 (0.2) | 5 (0.1) | 5 (0.1) | 4 (0.1) | 1314 (37.9) | 139 (4.0) | 2 (0.06) |
| Alpha 2 | 878 (6.3) | 250 (28.5) | 536 (61.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 197 (22.4) | 19 (2.2) | 0 (0.0) |
| Beta 1 | 3197 (22.8) | 1139 (35.6) | 2602 (81.4) | 45 (1.4) | 24 (0.8) | 33 (1.0) | 18 (0.6) | 1069 (33.4) | 98 (3.1) | 1 (0.0) |
| Beta 2 | 301 (2.2) | 109 (36.2) | 251 (83.4) | 3 (1.0) | 1 (0.3) | 3 (1.0) | 1 (0.3) | 107 (35.6) | 8 (2.7) | 0 (0.0) |
| Delta | 954 (6.8) | 386 (40.5) | 944 (99.0) | 211 (22.1) | 126 (13.2) | 211 (22.1) | 126 (13.2) | 385 (40.4) | 28 (2.9) | 6 (0.6) |
| Gamma | 607 (4.3) | 604 (99.5) | 515 (84.8) | 3 (0.5) | 3 (0.5) | 3 (0.5) | 3 (0.5) | 513 (84.5) | 42 (6.9) | 0 (0.0) |
| Epsilon | 29 (0.2) | 18 (62.1) | 24 (82.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 16 (55.2) | 29 (100.0) | 0 (0.0) |
| Acinar | 1696 (12.1) | 479 (28.2) | 1071 (63.2) | 5 (0.3) | 4 (0.2) | 4 (0.2) | 4 (0.2) | 403 (23.8) | 37 (2.2) | 1 (0.06) |
| Ductal 1 | 1524 (10.9) | 489 (32.1) | 1127 (74.0) | 1 (0.07) | 1 (0.07) | 1 (0.07) | 1 (0.07) | 420 (27.6) | 28 (1.8) | 0 (0.0) |
| Ductal 2 | 155 (1.1) | 56 (36.1) | 120 (77.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 47 (30.2) | 3 (1.9) | 0 (0.0) |
| Endothelial | 337 (2.4) | 84 (24.9) | 262 (77.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 78 (23.2) | 12 (3.6) | 0 (0.0) |
| Macrophage | 90 (0.6) | 30 (33.3) | 76 (84.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 30 (33.3) | 3 (3.3) | 0 (0.0) |
| Mast | 45 (0.3) | 18 (40.0) | 37 (82.2) | 1 (2.2) | 0 (0.0) | 1 (2.2) | 0 (0.0) | 15 (33.3) | 0 (0.0) | 0 (0.0) |
| Stellate | 726 (5.2) | 209 (28.8) | 550 (75.8) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 195 (26.9) | 16 (2.2) | 0 (0.0) |
Abbreviations: ID, identification; PPY, pancreatic polypeptide; SST, somatostatin.
Figure 4.
Expression levels of messenger RNAs encoding pancreatic polypeptide (PPY), somatostatin (SST), and growth hormone secretagogue receptor (GHSR) in 14 different cell clusters from human pancreas. Data are presented as violin plots whereby each black dot represents an instance of detectable expression of the transcript within a cell and the corresponding level of expression and the colored regions represent the probability density of the data at different expression levels.
GHSR was detected in a slightly higher percentage of cells in the human data set (2.0%; (n = 276/14 011; see Table 3) than in the mouse data set. In humans, GHSR expression was observed in all 4 traditional islet endocrine cell types and in acinar-cell, ductal 1-cell, mast-cell, and stellate-cell clusters (see Table 3, Fig. 4C). The percentage of cells with GHSR expression was highest in the delta-cell cluster (22.1%), followed by mast cells, beta cells, gamma cells, acinar cells, alpha cells, stellate cells, and ductal cells. This distribution also was apparent on reclustering only the data from GHSR-expressing cells (raw read count ≥ 1; Supplementary Fig. 6) (25). This reclustering identified 3 distinct clusters of GHSR-expressing cells (clusters 0, 1, and 2; Supplementary Fig. 6A) (25). These clusters appear to be driven by cell type as they were not defined wholly by the original cell clusters, the original study from which the data were derived, or the expression levels of GHSR, PPY, SST, or INS (Supplementary Fig. 6B-6G) (25). PPY was coexpressed in 59.4% (n = 164/276) of GHSR-expressing cells, and 95.7% (n = 157/164) of those also expressed SST (see Table 3). PPY expression was low and scattered among clusters 0, 1, and 2; SST expression was more prevalent and higher in clusters 0 and 2; high INS expression was concentrated in cluster 1 (see Supplementary Fig. 6E-6G) (25).
Ghrelin Expression
The ghrelin gene encodes ghrelin, desacyl-ghrelin, and obestatin. In mice, Ghrl was observed in all cell clusters—most notably, the 2 ductal-cell clusters (see Table 2). In none of these cell clusters were Ghrl expression levels particularly high (Supplementary Fig. 7A) (25). No Ghrl-expressing cell in the mouse data set coexpressed Ghsr (see Table 2), nor was a distinct Ghrl-expressing epsilon-cell cluster identified.
In humans, GHRL expression occurred in all but one cluster, including an epsilon-cell cluster, as reported by Baron et al (32). All epsilon cells expressed GHRL, whereas the other clusters included only a minority of GHRL-expressing cells (see Table 3). Epsilon cells all had relatively high GHRL expression levels compared to most other GHRL-expressing human pancreatic cells (Supplementary Fig. 7B) (25). No epsilon cell coexpressed GHSR, while only occasional GHRL-expressing cells in the other clusters coexpressed GHSR (see Table 3).
Effects of Type 2 Diabetes Mellitus on Gene Expression
Within the human pancreatic data set, T2DM was associated with several changes in gene expression levels (Supplementary Fig. 8 and Supplementary Table 2) (25). In most clusters, more genes were upregulated than downregulated in the presence of T2DM. Exceptions included the endothelial-cell and alpha 1-cell clusters, which showed more downregulated genes. The ductal 1-cell and beta 1-cell clusters showed the highest changes in gene expression in T2DM.
As expected, INS expression levels were downregulated in beta 1-cells while GCG expression levels were upregulated in alpha 1-cells in the setting of T2DM (see Supplementary Table 2) (25)). PPY expression levels were downregulated in the gamma-cell cluster while SST expression levels were not significantly changed in the delta-cell cluster in T2DM donors (see Supplementary Table 2) (25). GHRL expression levels were unaffected by T2DM in the epsilon-cell cluster (data not shown).
Notably, GHSR expression levels were not significantly changed by T2DM in the delta-cell cluster (log2 fold change [FC] = –0.295; P = 9.96 × 10–1; FDR = 1), although in the beta 1-cell cluster, they were upregulated (log2FC = 0.677; P = 1.14 × 10–29; FDR = 2.09 × 10–27).
Effects of Growth Hormone Secretagogue Receptor Ligands on Plasma Pancreatic Polypeptide
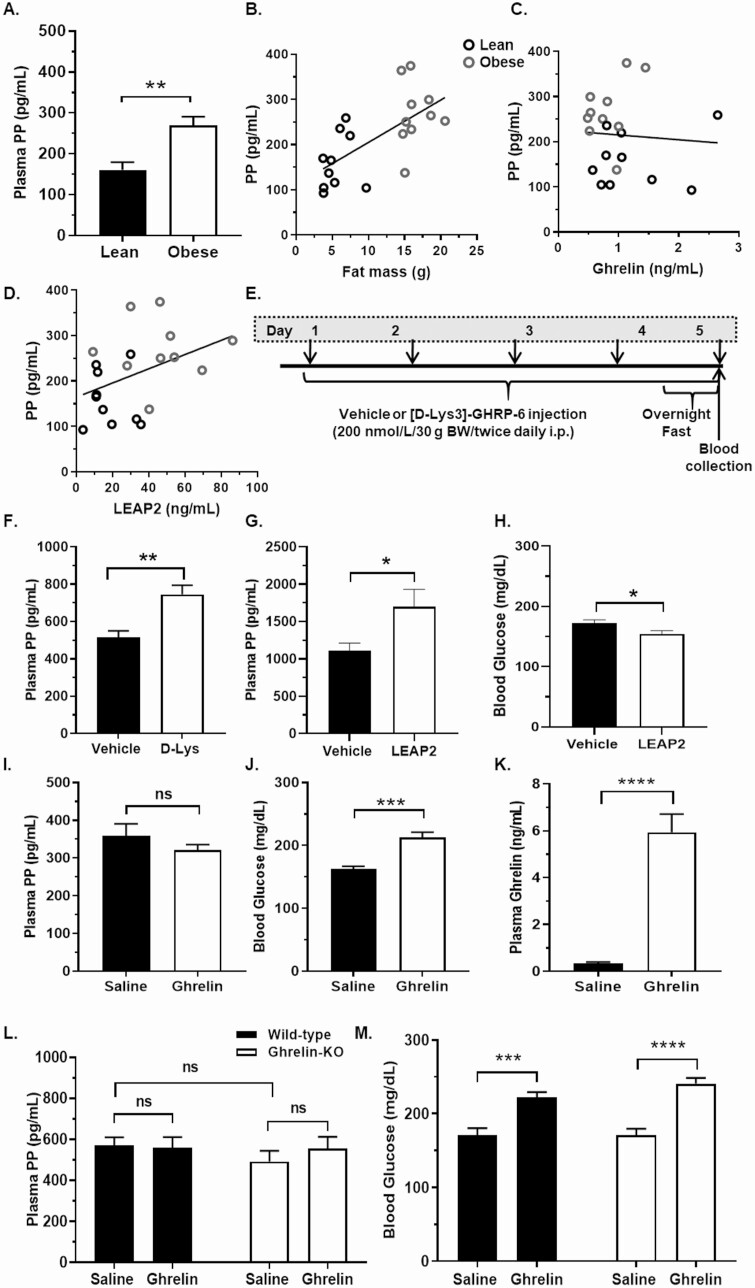
We also assessed GHSR-induced modulation of plasma PP. As plasma levels of ghrelin and the endogenous GHSR antagonist/inverse agonist LEAP2 are oppositely regulated in diet-induced obesity (24), we first determined how diet-induced obesity affects plasma PP. Plasma PP was higher in diet-induced obese mice than in lean controls (Fig. 5A). Moreover, PP was positively correlated with fat mass (Fig. 5B). Although plasma ghrelin did not correlate with PP, LEAP2 and PP were positively correlated (Fig. 5C and 5D).
Figure 5.
Effects of growth hormone secretagogue receptor (GHSR) agonism and antagonism on plasma pancreatic polypeptide (PP). A, Plasma PP in C57BL6/N mice fed for 12 weeks either chow (lean) or a high-fat diet (obese). Scatter plots showing correlations between plasma PP and B, vs corresponding fat mass; C, plasma acyl-ghrelin; and D, plasma LEAP2 in lean and obese mice, with the lines indicating the linear regression fits for the data points (B, Pearson correlation coefficient, r2 = 0.435, P = .002; C, Pearson correlation coefficient, r2 = 0.005, P = .761; D, Pearson correlation coefficient, r2 = 0.170, P = .049) n = 10 mice per group. E, Schematic of [D-Lys3]-GHRP-6 (D-Lys) vs vehicle (water) administration protocol, whereby down-facing arrows indicate the administration schedule (every 12 hours on days 1-4, and then once on day 5 following a 12-hour overnight fast). F, Plasma PP in overnight-fasted C57BL/6N mice 2 hours following the final D-Lys vs vehicle injection, on day 5. n = 6 to 11 mice per group. G, Plasma PP, and H, blood glucose in C57BL/6N mice 30 minutes following LEAP2 (72 nmol/kg, intraperitoneally) vs vehicle (10% dimethyl sulfoxide in phosphate-buffered saline) injection. n = 12 to 16 mice per group. I, Plasma PP; J, blood glucose; and K, plasma ghrelin in C57BL/6N mice 30 minutes following ghrelin (2 mg/kg, subcutaneously) vs saline injection. n = 8 to 10 mice per group. L, Plasma PP, and M, blood glucose in ghrelin-knockout (KO) and wild-type littermates 30 minutes following ghrelin (2 mg/kg, subcutaneously) vs saline injection. n = 10 mice per group. ns, nonsignificant. *P < .05, **P < .01, ***P < .001, and ****P < .0001. Data were analyzed by A and F to K, t test; B to D, Pearson correlation; and L and M, 2-way analysis of variance followed by post hoc Sidak multiple comparison tests.
Administration of [D-Lys3]-GHRP-6 increased plasma PP in overnight-fasted C57BL/6N mice (by 44.5%; Fig. 5E and 5F). Similarly, LEAP2 administration increased plasma PP in ad lib–fed mice (by 52.7%; Fig. 5G) and decreased blood glucose (Fig. 5H). Ghrelin administration did not significantly reduce plasma PP in ad lib–fed C57BL/6N mice (Fig. 5I), although it expectedly (18) raised blood glucose (Fig. 5J) and plasma ghrelin (Fig. 5I-5K). Furthermore, plasma PP was unaffected by ghrelin deletion in saline-administered ad lib–fed mice (Fig. 5L). Nor did ghrelin administration affect plasma PP in ad lib–fed ghrelin-KO mice or wild-type littermates (see Fig. 5L). Blood glucose levels in saline-administered ad lib–fed ghrelin-KO mice and ad lib–fed wild-type littermates were equivalent, and ghrelin raised blood glucose in both genotypes (Fig. 5M).
Discussion
We report for the first time high GHSR + PP coexpression within the pancreas. In particular, 84.7% of YFP-IR islet cells in GHSR reporter mice coexpressed PP-IR. The next highest amount of GHSR coexpression was with SST. Specifically, 49.8% of YFP-IR islet cells coexpressed SST-IR (of which the vast majority also coexpressed PP-IR). No YFP-IR islet cells coexpressed insulin-IR or glucagon-IR. Single-cell transcriptomic data from mouse pancreas also demonstrated a highly overlapping pattern of mRNAs encoding GHSR, PP, and SST: A total of 95.7% of Ghsr-expressing cells coexpressed Ppy and 100% of Ghsr-expressing cells coexpressed Sst, with most Ghsr-expressing cells (95.7%) expressing both Ppy and Sst. In contrast, the immunohistologic and single-cell transcriptomic analyses differed in the percentages of PP-expressing cells and SST-expressing cells that coexpress GHSR: The percentage of PP-expressing cells that coexpress GHSR was 74.9% by immunohistologic analysis of reporter mice vs 1.6% by transcriptomic analysis; the percentage of SST-expressing cells that coexpress GHSR was 58.3% by immunohistologic analysis of reporter mice vs 1.5% by transcriptomic analysis. A high percentage of GHSR-expressing cells coexpressing SST was also revealed by single-cell transcriptomic analysis of human pancreas (94.6%). However, in contrast to the high percentages (95.7%) of Ghsr-expressing cells that coexpress Ppy or both Ppy + Sst as revealed by single-cell transcriptomic analysis of mouse pancreas, the percentages of GHSR-expressing cells that coexpress PPY or both PPY and SST were somewhat lower (59.4% and 56.9%, respectively) in human pancreas.
In general, the variable expression of GHSR within different cells of the same cell clusters was expected based on the well-characterized phenomenon of heterogeneity within different islet cell populations (40, 41). However, the high coexpression of GHSR + PP within islets was unexpected. Most studies investigating a relationship between the ghrelin system and islets have centered on insulin, glucagon, and SST, which are undoubtedly better understood and characterized in terms of their actions, including their effects on blood glucose regulation and/or BW regulation (7, 9, 10). Other than a few papers investigating ghrelin’s effects on PP release (see the second paragraph of this work), the interaction of the ghrelin system with PP has not been a major area of study.
The high GHSR + PP coexpression suggests that PP might be regulated by GHSR and might mediate some actions of ghrelin and/or LEAP2. Indeed, here we showed that LEAP2 and [D-Lys3]-GHRP-6 increased plasma PP in fed and fasted conditions, respectively. This suggests that blockade of GHSR activity might counteract the decreased plasma PP associated with fasting (7, 42, 43). However, we did not demonstrate significant change in plasma PP following ghrelin administration in either ad lib–fed wild-type or ghrelin-KO mice, nor were plasma PP levels altered in saline-administered ad lib–fed ghrelin-KO mice as compared to wild-type mice. Perhaps the positive correlation between plasma LEAP2 and PP but absent correlation between plasma ghrelin and PP reflect the findings in the LEAP2 and ghrelin administration studies showing a statistically-significant effect of administered LEAP2 but not ghrelin on plasma PP. Further studies would help clarify discrepancies between the present findings and the previous reports of both ghrelin-inhibited and ghrelin-enhanced PP secretion (17-19). Given the relatively high GHSR + PP coexpression, it would also be worthwhile to investigate whether the ghrelin system’s orexigenic and glucoregulatory actions involve mediation by PP. Indeed, although ghrelin raises food intake and BW, LEAP2 blocks ghrelin-induced food intake (44-46). This is relevant because, similar to those reported actions of LEAP2, peripherally administered PP and transgenic PP overexpression reduce food intake and decrease BW in mice and humans (43, 47-50). Also, although ghrelin increases hepatic gluconeogenesis, decreases insulin sensitivity, and decreases insulin secretion, LEAP2 blocks ghrelin-induced blood glucose elevations and exaggerates falls in blood glucose resulting from chronic caloric restriction (44, 45). This is relevant because, similar to those reported overall actions of LEAP2 on blood glucose, administered PP reduces hepatic glucose production, increases insulin sensitivity, and increases insulin secretion (51, 52). Altogether, the reported metabolic actions of PP coincide with those of LEAP2, which we herein demonstrate increases plasma PP.
Notably, although our study demonstrated that plasma PP both was higher in diet-induced obese C57BL/6N mice than in lean mice and positively correlated with fat mass in those mice, prior studies in rodents correlating plasma PP with BW have proved difficult to find and those in humans do not show consistent results. A study in Sprague-Dawley rats demonstrated that 4 weeks of a high-fat diet increases the percentage of PP-immunoreactive cells from approximately 9% (in standard chow–fed animals) to approximately 15% (53). In contrast, in C57Bl/6J mice, 7 months of a high-fat diet reduced the percentage of PP-immunoreactive cells (54). Corresponding plasma PP levels were not available in either study. Another study described more than double the pancreatic content of PP in ob/ob mice than in lean controls, as well as a loss in the usual postprandial rise in plasma PP in the ob/ob mice (55). In a clinical study, human adult volunteers with obesity and normal glucose tolerance (mean body mass index [BMI] = 34) showed similar overnight fasted plasma PP levels as a cohort of healthy controls (mean BMI = 23) (56). Yet, the same study demonstrated that volunteers with obesity (mean BMI = 43) who had abnormal glucose tolerance exhibited on average 2 times higher plasma PP (56). The obese, glucose-intolerant group also exhibited a higher maximum plasma PP in response to administered secretin than the healthy controls (56). These findings are different from a study comparing children with obesity to lean controls, in which plasma PP was negatively correlated to BMI (57). Thus, our new data demonstrating a positive correlation between plasma PP and fat mass in mice are consistent with some but not all the available rodent and human data in the literature.
The highly coinciding GHSR + PP + SST coexpression, especially in mouse islets, is another interesting finding revealed here. These data might reflect previously reported highly coexpressed GHSR + SST (9, 10) and highly coexpressed PP + SST (58, 59). Although our histochemistry data do not distinguish GHSR-expressing delta cells that also express PP from GHSR-expressing gamma cells that also express SST, the transcriptomic analyses demonstrated Ghsr/GHSR + Ppy/PPY + Sst/SST coexpression in gamma cells and delta cells in mice and humans. To our knowledge, the significance of PP produced by gamma cells vs that produced by other islet cell types (mostly at lower expression levels) has not heretofore been defined.
Also of note, the human pancreas transcriptomic analysis revealed GHSR expression in small percentages of insulin-producing beta cells. This finding, alongside the more prevalent GHSR expression within delta cells, reflects the previously described functional studies attributing ghrelin-inhibited insulin secretion alternatively to direct engagement of GHSR-expressing beta cells or to indirect engagement of beta cells via GHSR-expressing delta cells (9-13). Our observation of occasional GHSR-expressing alpha cells matches that of Adriaenssens and colleagues (10). Despite the small percentage of GHSR-expressing human beta cells and alpha cells revealed here (1.3% of beta cells and 0.14% of alpha cells), this translates to a likely consequential complement of cells given the total number of cells within human beta-cell and alpha-cell clusters. Future studies will help to determine the functional significance of the newly revealed upregulated GHSR expression within beta cells from T2DM donors.
As a penultimate topic of discussion, our single-cell transcriptomic analyses also revealed Ghrl/GHRL expression both in mouse and human data sets. In humans, this included epsilon cells, which are defined by high ghrelin expression but absent expression of insulin-IR, glucagon-IR, SST-IR, or PP-IR (2, 3, 60). GHSR expression was not observed in epsilon cells. Although expectedly (60) no epsilon cells were identified in the adult mouse data set, ghrelin was also observed in all or most other cell clusters in the mouse and human data sets. For most of these other cell clusters, ghrelin-expressing cells represented only very small percentages. Exceptions included the 2 mouse ductal-cell clusters in the mouse data set, of which 19.1% to 23.5% of the cells exhibited ghrelin expression. The low number of epsilon cells in the human data set reflects previous descriptions of far fewer epsilon cells in adulthood than in the fetal period (2, 3, 60-63). For instance, a previous single-cell transcriptomic study of 14 779 adult human islet cells identified only 11 epsilon cells (61). A separate immunohistochemical analysis demonstrated a 30% occupancy of islets by ghrelin-IR cells at gestation week 23, followed by a subsequent decline to only the occasional ghrelin-IR cell per adult islet (63). Our observation of ghrelin expression in cell types other than epsilon cells may be explained by the description of the ghrelin-expressing cell as a multipotent progenitor within the pancreas, which gives rise to significant numbers of alpha cells and gamma cells, rare beta cells, occasional ductal cells, and 5% of exocrine tissue (64).
Some caveats of our study also deserve mention. One caveat relates to the use of the Ghsr-IRES-Cre X Cre-dependent ROSA26-YFP reporter mice to report on GHSR expression in the mouse. In general, GHSR expression has been studied in a variety of species and organs using techniques ranging from various histochemistry analyses (using labeled antisense Ghsr riboprobes, anti-GHSR antiserum samples, binding of tagged-GHSR ligands, or GHSR promoter-driven reporters) to Western blot, reverse transcriptase–polymerase chain reaction, ribonuclease protection assay, and transcriptomic analyses (as described in Mani et al) (65). Among these techniques, only the histochemical techniques offer an in situ anatomical view of the expressed GHSR protein or mRNA. However, techniques such as in situ hybridization histochemistry are often insensitive, tending to underestimate actual gene expression levels, especially of cell surface receptors that, as a group, have relatively low mRNA abundance. Furthermore, cell surface receptors often lack adequate antigenicity to permit the generation of reliable antibodies for use in immunohistochemistry, and the antibodies that are commercially available often fail to show specificity when validated using KO animals, as is the case for many commercially available anti-GHSR antibodies (see Reichenbach [66] and data not shown]. Thus, to avoid some of those limitations, we paired the single-cell transcriptomic analyses with histochemical analyses of reporter mice with YFP expression driven by a Ghsr-IRES-Cre knockin gene. Yet, the Ghsr-IRES-Cre mouse line also has its shortcomings. As described previously, although the pattern of Cre activity within the brains of this Ghsr-IRES-Cre mouse line mostly faithfully reproduces the known GHSR expression pattern as determined using in situ hybridization histochemistry and a GHSR-eGFP transgenic mouse line, 100% agreement among those techniques is lacking and occasional Ghsr-IRES-Cre mice exhibit a somewhat asymmetric pattern (more expression on one side of the brain) or a less extensive bilateral pattern of Cre activity (21, 65, 67). Nonetheless, for the study here, we used only mice in which the brains were first shown to exhibit YFP expression patterns matching the best previously reported cases (21). Also of note, a similar reporter mouse made by inserting an IRES-tauGFP cassette (as opposed to the IRES-Cre cassette) into the endogenous Ghsr locus recently was characterized as having high coexpression of GFP (as a marker of GHSR expression) with insulin-IR and glucagon-IR islet cells; coexpression of GFP with SST or PP was not examined, nor has GFP expression within that mouse model been fully characterized within the brain (14, 68). The reasons underlying the differential islet GHSR expression observed here using the GHSR-IRES-Cre X ROSA26-YFP mice vs that described for the GHSR-IRES-tauGFP line are as of yet unclear.
A second caveat relates to limitations of transcriptomic analyses. Although single-cell sequencing allows for transcriptomic profiling of individual cells, transcripts with low copy numbers are not easily detected using this method (69). As such, Ghsr/GHSR mRNA expression levels, which like other G-protein coupled receptors tend to be low (70-72), could be underrepresented in the transcriptomic data. In general, the biological relevance of low-expressed transcripts that are captured is not necessarily certain. While here we include functional data and cite functional data suggesting key roles for islet-expressed GHSR, it remains unclear for most transcripts if biologically relevant amounts of protein or immunohistochemically detectable amounts of protein depend on high expression levels of its transcript, or whether low expression levels are sufficient. Thus, the single-cell RNA sequencing data are presented here unfiltered, in 3 different ways—in the form of a violin plot, a UMAP plot, and in tabular form. Using all 3 of these methods, the numbers and percentages of cells of a certain cluster expressing a certain transcript are indicated. Also, using the violin and UMAP plots, the expression levels of a certain transcript within individual cells are indicated. If high expression of a transcript is physiologically relevant, the violin and UMAP plots, in particular, clearly delineate those clusters that contain cells with high levels of expression of our genes of interest.
We also chose to present the unfiltered expression values, instead of filtering the transcript expression to reduce potential “noise”, for another reason. As the human transcriptomic data set contains RNA sequencing data from a number of sources that used different sequencing technologies, the median reads per cell differ between the different data sets. Filtering out low-level transcript expression could result in the removal of data from sources with a lower read depth, and not result in filtering of “noise.” Although the level of coexpression of canonical peptide transcripts could possibly represent ambient RNA expression and contamination across cells, we consistently observed overlap of peptide transcript expression within islet cells across different technologies within the human RNA sequencing data sets. This suggests that the pancreatic cells do indeed express transcripts for multiple peptides/hormones; however, this does not necessarily translate into coexpression and secretion of multiple peptides/hormones from single islet cells.
As a third caveat, we acknowledge that this study is mainly descriptive. While the functional analyses performed provide some clues, they do not enable us to make definitive statements regarding the biological significance of GHSR expression within the PP-expressing islet cells. We do hope that these newly reported expression data, together with the available functional analyses, inspire further investigation regarding the metabolic effects of PP-expressing islet cells and the roles of PP in mediating the actions of the ghrelin system.
Acknowledgements
We acknowledge Lavanya Vishvanath at the Touchstone Diabetes Center and Dr Luis Leon Mercado, Division of Hypothalamic Research, Internal Medicine, UT Southwestern, for helpful discussions.
Financial Support: This work was supported by the Diana and Richard C. Strauss Professorship in Biomedical Research; the Mr. and Mrs. Bruce G. Brookshire Professorship in Medicine; the Kent and Jodi Foster Distinguished Chair in Endocrinology, in Honor of Daniel Foster, MD; institutional funds from the University of Texas Southwestern Medical Center and the National Institutes of Health (Nos. R01DK103884 and R01DK103884 to J.M.Z.). G.K.C.D. is funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) doctoral training program. B.Y.H.L. is supported by the BBSRC (project grant No. BB/S017593/1). G.S.H.Y. is supported by the UK Medical Research Council (MRC Metabolic Diseases Unit grant No. MC_UU_00014/1) and the BBSRC (project grant No. BB/S017593/1).
Author Contribution: D.G. designed the study, collected the data, performed the statistical analyses, and helped write the manuscript. B.K.M. contributed to the study design and data collection. K.S. helped with data collection. G.K.C.D., B.Y.H.L., and G.S.H.Y. evaluated the transcriptomic data, helped write the manuscript, and helped secure funding. S.O.L. and N.P.M. generated the mice used in the study. J.M.Z. oversaw all aspects of the study, helped write the manuscript, and secured funding. All authors approved the final version.
Glossary
Abbreviations
- BMI
body mass index
- BW
body weight
- DAPI
4', 6-diamidino-2-phenylindole
- FC
fold change
- FDR
false discovery rate
- GFP
green fluorescent protein
- GH
growth hormone
- Ghrl
ghrelin gene
- GHSR
growth hormone secretagogue receptor
- IR
immunoreactivity
- KO
knockout
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- PP
pancreatic polypeptide
- SST
somatostatin
- T2DM
type 2 diabetes mellitus
- YFP
yellow fluorescent protein
Additional Information
Disclosures: B.K.M. is currently employed by Novo Nordisk Research Center Indianapolis, Inc, although his major contributions to the study occurred while on the faculty of University of Texas Southwestern Medical Center. The other authors have no conflicts of interest to disclose.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Heppner KM, Tong J. Mechanisms in endocrinology: regulation of glucose metabolism by the ghrelin system: multiple players and multiple actions. Eur J Endocrinol. 2014;171(1):R21-R32. [DOI] [PubMed] [Google Scholar]
- 2. Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101(9):2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wierup N, Svensson H, Mulder H, Sundler F. The ghrelin cell: a novel developmentally regulated islet cell in the human pancreas. Regul Pept. 2002;107(1-3):63-69. [DOI] [PubMed] [Google Scholar]
- 4. Granata R, Settanni F, Biancone L, et al. Acylated and unacylated ghrelin promote proliferation and inhibit apoptosis of pancreatic beta-cells and human islets: involvement of 3′,5′-cyclic adenosine monophosphate/protein kinase A, extracellular signal-regulated kinase 1/2, and phosphatidyl inositol 3-kinase/Akt signaling. Endocrinology. 2007;148(2):512-529. [DOI] [PubMed] [Google Scholar]
- 5. Granata R, Settanni F, Gallo D, et al. Obestatin promotes survival of pancreatic beta-cells and human islets and induces expression of genes involved in the regulation of beta-cell mass and function. Diabetes. 2008;57(4):967-979. [DOI] [PubMed] [Google Scholar]
- 6. Granata R, Settanni F, Trovato L, et al. Unacylated as well as acylated ghrelin promotes cell survival and inhibit apoptosis in HIT-T15 pancreatic beta-cells. J Endocrinol Invest. 2006;29(9):RC19-RC22. [DOI] [PubMed] [Google Scholar]
- 7. Mani BK, Shankar K, Zigman JM. Ghrelin’s relationship to blood glucose. Endocrinology. 2019;160(5):1247-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drott CJ, Franzén P, Carlsson PO. Ghrelin in rat pancreatic islets decreases islet blood flow. Am J Physiol Endocrinol Metab. 2019;317(1):E139-E146. [DOI] [PubMed] [Google Scholar]
- 9. DiGruccio MR, Mawla AM, Donaldson CJ, et al. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol Metab. 2016;5(7):449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adriaenssens AE, Svendsen B, Lam BY, et al. Transcriptomic profiling of pancreatic alpha, beta and delta cell populations identifies delta cells as a principal target for ghrelin in mouse islets. Diabetologia. 2016;59(10):2156-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dezaki K, Hosoda H, Kakei M, et al. Endogenous ghrelin in pancreatic islets restricts insulin release by attenuating Ca2+ signaling in beta-cells: implication in the glycemic control in rodents. Diabetes. 2004;53(12):3142-3151. [DOI] [PubMed] [Google Scholar]
- 12. Kaiser J, Krippeit-Drews P, Drews G. Acyl-Ghrelin influences pancreatic β-cell function by interference with KATP channels. Diabetes. 2021;70(2):423-435. [DOI] [PubMed] [Google Scholar]
- 13. Kurashina T, Dezaki K, Yoshida M, et al. The β-cell GHSR and downstream cAMP/TRPM2 signaling account for insulinostatic and glycemic effects of ghrelin. Sci Rep. 2015;5:14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pradhan G, Wu CS, Villarreal D, et al. β-Cell GHS-R regulates insulin secretion and sensitivity. Int J Mol Sci. 2021;22(8):3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wierup N, Yang S, McEvilly RJ, Mulder H, Sundler F. Ghrelin is expressed in a novel endocrine cell type in developing rat islets and inhibits insulin secretion from INS-1 (832/13) cells. J Histochem Cytochem. 2004;52(3):301-310. [DOI] [PubMed] [Google Scholar]
- 16. Kageyama H, Funahashi H, Hirayama M, et al. Morphological analysis of ghrelin and its receptor distribution in the rat pancreas. Regul Pept. 2005;126(1-2):67-71. [DOI] [PubMed] [Google Scholar]
- 17. Kumar R, Salehi A, Rehfeld JF, Höglund P, Lindström E, Håkanson R. Proghrelin peptides: desacyl ghrelin is a powerful inhibitor of acylated ghrelin, likely to impair physiological effects of acyl ghrelin but not of obestatin a study of pancreatic polypeptide secretion from mouse islets. Regul Pept. 2010;164(2-3):65-70. [DOI] [PubMed] [Google Scholar]
- 18. Qader SS, Håkanson R, Rehfeld JF, Lundquist I, Salehi A. Proghrelin-derived peptides influence the secretion of insulin, glucagon, pancreatic polypeptide and somatostatin: a study on isolated islets from mouse and rat pancreas. Regul Pept. 2008;146(1-3):230-237. [DOI] [PubMed] [Google Scholar]
- 19. Arosio M, Ronchi CL, Gebbia C, Cappiello V, Beck-Peccoz P, Peracchi M. Stimulatory effects of ghrelin on circulating somatostatin and pancreatic polypeptide levels. J Clin Endocrinol Metab. 2003;88(2):701-704. [DOI] [PubMed] [Google Scholar]
- 20. Tong J, Davis HW, Gastaldelli A, D’Alessio D. Ghrelin impairs prandial glucose tolerance and insulin secretion in healthy humans despite increasing GLP-1. J Clin Endocrinol Metab. 2016;101(6):2405-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mani BK, Osborne-Lawrence S, Mequinion M, et al. The role of ghrelin-responsive mediobasal hypothalamic neurons in mediating feeding responses to fasting. Mol Metab. 2017;6(8):882-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez JA, Bruggeman EC, Mani BK, et al. Ghrelin receptor agonist rescues excess neonatal mortality in a Prader-Willi syndrome mouse model. Endocrinology. 2018;159(12):4006-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shankar K, Gupta D, Mani BK, et al. Acyl-ghrelin is permissive for the normal counterregulatory response to insulin-induced hypoglycemia. Diabetes. 2020;69(2):228-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mani BK, Puzziferri N, He Z, et al. LEAP2 changes with body mass and food intake in humans and mice. J Clin Invest. 2019;129(9):3909-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gupta D, Mani BK, Shankar K, et al. High co-expression of the ghrelin and LEAP2 receptor GHSR with pancreatic polypeptide in mouse and human islets. Data for: Extended data set and supplemental materials. UT Southwestern Institutional Repository 2021. Deposited July 9, 2021. https://hdl.handle.net/2152.5/9595 [Google Scholar]
- 26. Chuang JC, Sakata I, Kohno D, et al. Ghrelin directly stimulates glucagon secretion from pancreatic alpha-cells. Mol Endocrinol. 2011;25(9):1600-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chambers AP, Sorrell JE, Haller A, et al. The role of pancreatic preproglucagon in glucose homeostasis in mice. Cell Metab. 2017;25(4):927-934.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jhou TC, Geisler S, Marinelli M, Degarmo BA, Zahm DS. The mesopontine rostromedial tegmental nucleus: a structure targeted by the lateral habenula that projects to the ventral tegmental area of Tsai and substantia nigra compacta. J Comp Neurol. 2009;513(6):566-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Krieken PP, Dicker A, Eriksson M, et al. Kinetics of functional beta cell mass decay in a diphtheria toxin receptor mouse model of diabetes. Sci Rep. 2017;7(1):12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510(1):79-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gautron L, Lee C, Funahashi H, Friedman J, Lee S, Elmquist J. Melanocortin-4 receptor expression in a vago-vagal circuitry involved in postprandial functions. J Comp Neurol. 2010;518(1):6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baron M, Veres A, Wolock SL, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3(4):346-360.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muraro MJ, Dharmadhikari G, Grün D, et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3(4):385-394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grün D, Muraro MJ, Boisset JC, et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 2016;19(2):266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lawlor N, George J, Bolisetty M, et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2017;27(2):208-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Segerstolpe Å, Palasantza A, Eliasson P, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24(4):593-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lun ATL, Chen Y, Smyth GK. It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol. 2016;1418:391-416. [DOI] [PubMed] [Google Scholar]
- 40. Bru-Tari E, Oropeza D, Herrera PL. Cell heterogeneity and paracrine interactions in human islet function: a perspective focused in β-cell regeneration strategies. Front Endocrinol (Lausanne). 2020;11:619150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miranda MA, Macias-Velasco JF, Lawson HA. Pancreatic β-cell heterogeneity in health and diabetes: classes, sources, and subtypes. Am J Physiol Endocrinol Metab. 2021;320(4):E716-E731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adrian TE, Besterman HS, Cooke TJ, Bloom SR, Barnes AJ, Russell RC. Mechanism of pancreatic polypeptide release in man. Lancet. 1977;1(8004):161-163. [DOI] [PubMed] [Google Scholar]
- 43. Batterham RL, Le Roux CW, Cohen MA, et al. Pancreatic polypeptide reduces appetite and food intake in humans. J Clin Endocrinol Metab. 2003;88(8):3989-3992. [DOI] [PubMed] [Google Scholar]
- 44. Ge X, Yang H, Bednarek MA, et al. LEAP2 is an endogenous antagonist of the ghrelin receptor. Cell Metab. 2018;27(2):461-469.e6. [DOI] [PubMed] [Google Scholar]
- 45. Islam MN, Mita Y, Maruyama K, et al. Liver-expressed antimicrobial peptide 2 antagonizes the effect of ghrelin in rodents. J Endocrinol. 2020;244(1):13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. M’Kadmi C, Cabral A, Barrile F, et al. N-Terminal liver-expressed antimicrobial peptide 2 (LEAP2) region exhibits inverse agonist activity toward the ghrelin receptor. J Med Chem. 2019;62(2):965-973. [DOI] [PubMed] [Google Scholar]
- 47. Asakawa A, Inui A, Yuzuriha H, et al. Characterization of the effects of pancreatic polypeptide in the regulation of energy balance. Gastroenterology. 2003;124(5):1325-1336. [DOI] [PubMed] [Google Scholar]
- 48. Sainsbury A, Shi YC, Zhang L, et al. Y4 receptors and pancreatic polypeptide regulate food intake via hypothalamic orexin and brain-derived neurotropic factor dependent pathways. Neuropeptides. 2010;44(3):261-268. [DOI] [PubMed] [Google Scholar]
- 49. Ueno N, Inui A, Iwamoto M, et al. Decreased food intake and body weight in pancreatic polypeptide-overexpressing mice. Gastroenterology. 1999;117(6):1427-1432. [DOI] [PubMed] [Google Scholar]
- 50. Jesudason DR, Monteiro MP, McGowan BM, et al. Low-dose pancreatic polypeptide inhibits food intake in man. Br J Nutr. 2007;97(3):426-429. [DOI] [PubMed] [Google Scholar]
- 51. Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci U S A. 2006;103(7):2334-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim W, Fiori JL, Shin YK, et al. Pancreatic polypeptide inhibits somatostatin secretion. FEBS Lett. 2014;588(17):3233-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruipan Z, Xiangzhi M, Li L, et al. Differential expression and localization of neuropeptide Y peptide in pancreatic islet of diabetic and high fat fed rats. Peptides. 2014;54:33-38. [DOI] [PubMed] [Google Scholar]
- 54. Adeghate E, Christopher Howarth F, Rashed H, Saeed T, Gbewonyo A. The effect of a fat-enriched diet on the pattern of distribution of pancreatic islet cells in the C57BL/6J mice. Ann N Y Acad Sci. 2006;1084:361-370. [DOI] [PubMed] [Google Scholar]
- 55. Jia BQ, Taylor IL. Failure of pancreatic polypeptide release in congenitally obese mice. Gastroenterology. 1984;87(2):338-343. [PubMed] [Google Scholar]
- 56. Glaser B, Zoghlin G, Pienta K, Vinik AI. Pancreatic polypeptide response to secretin in obesity: effects of glucose intolerance. Horm Metab Res. 1988;20(5):288-292. [DOI] [PubMed] [Google Scholar]
- 57. Reinehr T, Enriori PJ, Harz K, Cowley MA, Roth CL. Pancreatic polypeptide in obese children before and after weight loss. Int J Obes (Lond). 2006;30(10):1476-1481. [DOI] [PubMed] [Google Scholar]
- 58. Portela-Gomes GM, Johansson H, Olding L, Grimelius L. Co-localization of neuroendocrine hormones in the human fetal pancreas. Eur J Endocrinol. 1999;141(5):526-533. [DOI] [PubMed] [Google Scholar]
- 59. Katsuta H, Akashi T, Katsuta R, et al. Single pancreatic beta cells co-express multiple islet hormone genes in mice. Diabetologia. 2010;53(1):128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heller RS, Jenny M, Collombat P, et al. Genetic determinants of pancreatic epsilon-cell development. Dev Biol. 2005;286(1):217-224. [DOI] [PubMed] [Google Scholar]
- 61. Dominguez Gutierrez G, Kim J, Lee AH, et al. Gene signature of the human pancreatic ε cell. Endocrinology. 2018;159(12):4023-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mastracci TL, Sussel L. The endocrine pancreas: insights into development, differentiation, and diabetes. Wiley Interdiscip Rev Membr Transp Signal. 2012;1(5):609-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andralojc KM, Mercalli A, Nowak KW, et al. Ghrelin-producing epsilon cells in the developing and adult human pancreas. Diabetologia. 2009;52(3):486-493. [DOI] [PubMed] [Google Scholar]
- 64. Arnes L, Hill JT, Gross S, Magnuson MA, Sussel L. Ghrelin expression in the mouse pancreas defines a unique multipotent progenitor population. PLoS One. 2012;7(12):e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mani BK, Walker AK, Lopez Soto EJ, et al. Neuroanatomical characterization of a growth hormone secretagogue receptor-green fluorescent protein reporter mouse. J Comp Neurol. 2014;522(16):3644-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reichenbach A, Steyn FJ, Sleeman MW, Andrews ZB. Ghrelin receptor expression and colocalization with anterior pituitary hormones using a GHSR-GFP mouse line. Endocrinology. 2012;153(11):5452-5466. [DOI] [PubMed] [Google Scholar]
- 67. Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494(3):528-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jiang H, Betancourt L, Smith RG. Ghrelin amplifies dopamine signaling by cross talk involving formation of growth hormone secretagogue receptor/dopamine receptor subtype 1 heterodimers. Mol Endocrinol. 2006;20(8):1772-1785. [DOI] [PubMed] [Google Scholar]
- 69. Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Howard AD, Feighner SD, Cully DF, et al. A receptor in pituitary and hypothalamus that functions in growth hormone release. Science. 1996;273(5277):974-977. [DOI] [PubMed] [Google Scholar]
- 71. Fredriksson R, Schiöth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67(5):1414-1425. [DOI] [PubMed] [Google Scholar]
- 72. Amisten S, Atanes P, Hawkes R, et al. A comparative analysis of human and mouse islet G-protein coupled receptor expression. Sci Rep. 2017;7:46600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”