Significance
RAS proteins control many aspects of cellular biology in response to extracellular stimuli. These essential nodes of signal transduction interact with effector proteins that contain conserved RAS-binding domains. Mutations in RAS proteins are a common cause of cancer; hence, understanding the function of their downstream effectors is important for the development of novel therapies. In this study, we characterize the interaction between RAS and the effector protein Sin1, a critical component of the mTORC2, through biochemical and structural approaches. We discover that Sin1 contains an atypical RAS-binding domain and assess the role of this interaction in cells and mice. Moreover, we find that this interaction is dispensable for mTORC2 activity and assembly, suggesting an alternative function for Sin1 in cells.
Keywords: Sin1, mTORC2, KRAS, RAS, RBD
Abstract
RAS proteins are molecular switches that interact with effector proteins when bound to guanosine triphosphate, stimulating downstream signaling in response to multiple stimuli. Although several canonical downstream effectors have been extensively studied and tested as potential targets for RAS-driven cancers, many of these remain poorly characterized. In this study, we undertook a biochemical and structural approach to further study the role of Sin1 as a RAS effector. Sin1 interacted predominantly with KRAS isoform 4A in cells through an atypical RAS-binding domain that we have characterized by X-ray crystallography. Despite the essential role of Sin1 in the assembly and activity of mTORC2, we find that the interaction with RAS is not required for these functions. Cells and mice expressing a mutant of Sin1 that is unable to bind RAS are proficient for activation and assembly of mTORC2. Our results suggest that Sin1 is a bona fide RAS effector that regulates downstream signaling in an mTORC2-independent manner.
Rat Sarcoma virus (RAS) guanosine triphosphate hydrolases (GTPases) are molecular switches that propagate downstream signaling through the engagement of molecular effectors that exhibit high affinity toward the GTP-bound conformation (1). This is achieved by the presence of RAS-associating (RA) or RAS-binding domains (RBD) that promote the interaction between the RAS GTPase and the molecular effector (2). In the case of HRAS, KRAS, and NRAS, which are well-defined oncoproteins, a significant number of effectors that contain such domains have been characterized, including Raf and PI3K kinases, RalGDS, and others (1, 3). These effectors play important roles downstream of GTP-bound RAS by regulating cellular processes including proliferation, survival, metabolism, and differentiation (4). Moreover, RAS effectors are especially attractive therapeutically, and preclinical and clinical studies suggest they are potential targets for the treatment of RAS mutant cancers and developmental disorders (5, 6). Hence, identifying and characterizing effector proteins that act downstream of GTP-bound RAS proteins is critical to understand the molecular and cellular effects of RAS proteins in normal physiology as well as in the context of RAS-driven diseases.
Stress-activated MAP kinase–interacting protein 1 (Sin1) is a highly conserved protein composed of four distinct structural domains that include the N-terminal (NT), conserved region in the middle (CRIM), RBD, and pleckstrin homology (PH) domains (7). Human Sin1 orthologs have been identified in many model organisms, including yeast (Schizosaccharomyces pombe and Saccharomyces cerevisiae), slime mold (Dictyostelium discoideum), roundworms (Caenorhabditis elegans), fruit flies (Drosophila melanogaster) and mammals (8–10). Due to the presence of the RBD, Sin1 is suggested to be an effector of RAS in various model organisms (11). For instance, a screening in S. cerevisiae revealed that the phenotype associated with expression of an activated mutant form of RAS2 (the ortholog of RAS) can be rescued with rat Sin1 complementary DNA (cDNA) (12). In Dictyostelium, interaction between Sin1 and RAS orthologs was found to be important for chemotaxis and the synthesis and relay of cyclic AMP (13). Consistent with reports in other species, human Sin1 also interacts with GTP-loaded RAS proteins in vitro (14). However, Sin1 has been better characterized as a core component of the target of rapamycin complex 2 (TORC2), after AVO1, the Sin1 ortholog in budding yeast, was purified in association with this complex (15). Later studies showed that Sin1 is an essential constituent of mammalian TORC2 (mTORC2) and this appears to be conserved in many model organisms (16–18).
TORC2 is a large serine/threonine kinase complex that phosphorylates a subset of AGC family protein kinases at their hydrophobic and turn motifs, which are required for full activation (19–22); these include AKT, PKC, and SGK (23). Given the importance of these substrates in regulating metabolism, survival, and cytoskeletal remodeling, TORC2 is often considered a master regulator of these cellular functions. However, the molecular cues that regulate the activity and/or localization of the complex remain largely unknown (24) and it has been suggested that mTORC2 might be constitutively active, at least when localized at the plasma membrane (25). In mammalian cells, TORC2 is formed by the catalytic subunit mTOR, a serine/threonine kinase, and the additional proteins mLST8, Deptor, Rictor, Protor-1, and Sin1 (22, 26).
Recent cryoEM reconstruction of the yeast and mammalian TORC2 suggests that the NT domain of Sin1 is required for association with TORC2, while other regions of the protein remain unresolved in such structures (27–30). While Sin1 appears to be dispensable for TORC2 assembly and activity, its CRIM domain adopts a ubiquitin-like fold with an acidic loop required for the interaction with TORC2 substrates, including AKT, PKC, and SGK (31). No structural or biochemical studies to date have addressed the role of the Sin1 RBD in the regulation of TORC2. In Dictyostelium, binding of Sin1 to the small GTPases RAS and Rap1 has been proposed to control TORC2 activity in response to chemoattractants (32). A recent study in human cells, explored the proximal proteome of oncogenic RAS using proximity-dependent biotin labeling. Among well-known interactors of RAS proteins, the authors identified mTORC2 and proposed that oncogenic RAS can regulate this complex to sustain cell and tumor growth (33). Despite these studies, additional molecular analysis is required to understand whether RAS can modulate mTORC2 activity through interaction with the Sin1 RBD.
In this study, we have identified the RAS family members that interact with Sin1 in vitro and in cells. While in vitro the Sin1 RBD interacts with many RAS family proteins at high affinity through anti-parallel β-sheets, in cells the interaction is largely restricted to KRAS4A due to the presence of the RAS C-terminal hypervariable (HVR) region. Moreover, we identified a unique RAS binding module in Sin1 that extends beyond the canonical RBD toward the PH domain. Cross-linking mass spectrometry (XL-MS) indicated that this atypical RBD folds in a conformation auto-inhibited by the PH domain. The atypical RBD forms many contacts with residues of the RAS switch II and effector domains and deletion of this region abrogated interaction with KRAS4A in cells. Strikingly, deletion of this region does not impair basal TORC2 activity or assembly in mammalian cells and was compatible with normal development in mice. Our results highlight the TORC2-independent role of the Sin1-RAS interaction and suggest alternative functions of Sin1 in the context of RAS signaling.
Results
Sin1 Preferentially Interacts with KRAS in Cells in a GTP-Dependent Manner.
In order to test the ability of Sin1 to bind different RAS isoforms in cells, we performed pull-down assays using tagged versions of WT (mostly GDP-bound) or G12V mutant (mostly GTP-bound) NRAS, HRAS, and KRAS. Sin1 preferentially interacted with GTP-bound KRAS (Fig. 1A). The selectivity for KRAS was confirmed in a larger panel of GTP-loaded RAS-related GTPases (Fig. 1B and SI Appendix, Fig. S1A). The GTP requirement for the interaction was also confirmed in vitro, when recombinant Sin1 RBD preferentially interacted with KRAS loaded with the nonhydrolysable GTP analog GTPγS, but not GDP (Fig. 1C). Moreover, the KRAS dominant negative S17N mutation, that only binds GDP (34), prevented interaction with Sin1 in cells (SI Appendix, Fig. S1B). Consistent with oncogenic KRAS mutations existing largely in the GTP-bound form (1), Sin1 interacted preferentially with the cancer-associated variants G12V, G12D, G12C, G13D, Q61L and Q61R (SI Appendix, Fig. S1C). At least five recognized isoforms of Sin1 result from multiple exon skipping during splicing (35) and each of these isoforms retained the ability to interact with KRAS, although binding with isoform 5 was clearly diminished since only part of the RBD is preserved (SI Appendix, Fig. S1D).
Fig. 1.
Sin1 interacts with the KRAS splice variant 4A. (A) Immunoblot of the pull-down assay between Flag-Sin1 and GST-tagged RAS isoforms in 293T cells. Both RAS WT, which is mostly GDP loaded, and G12V mutant, which is mostly GTP loaded, were used. WCL: whole-cell lysate; PD: pull-down. (B) Dendrogram depicting evolutionary relationship of different members of the RAS GTPase family and their ability to bind to Sin1. The binding was determined by coimmunoprecipitation as shown in SI Appendix, Fig. S1A. (C) In vitro pull-down assay using recombinant GST or GST-Sin1 RBD (279-353) and KRAS (1-165) loaded with GDP or GTPγS. (D) Immunoblot of the pull-down assay between Flag-Sin1 and GST-tagged KRAS splice variants 4A and 4B in 293T cells. Both KRAS WT, which is mostly GDP loaded, and G12V mutant, which is mostly GTP loaded, were used. WCL: whole-cell lysate; PD: pull-down. (E) Isothermal titration calorimetry plots depicting the calculated KD for the interaction between RAS G domains (1 to 169) and Sin1 RBD (279 to 353). (F) Immunoblot of the pull-down assay between Flag-Sin1 and GST-tagged RAS chimeras in 293T cells. All variants contain the G12V mutation. Chimeras were generated by fusing the G domain of HRAS or NRAS (1 to 169) to the KRAS4A HVR. WCL: whole cell-lysate; PD: pull-down.
Sin1 Selectivity toward the KRAS Isoform 4A Requires the Hypervariable Region.
The KRAS gene is comprised of five exons that give rise to two different isoforms, KRAS 4A and KRAS 4B, through alternative splicing of the fourth exon. These isoforms differ in their C-terminal HVR and, as a consequence, offer different properties to the protein, including posttranslational modifications, folding, and subcellular localization (36). Our pull-down assays showed that Sin1 preferentially interacted with the KRAS4A isoform in cells (Fig. 1D). Next, we used isothermal titration calorimetry (ITC) to determine the interaction affinity between the Sin1 RBD and G-domains (1-169) of NRAS, HRAS, KRAS4A, and KRAS4B bound to GMPPNP, a nonhydrolysable GTP analog. All interactions exhibited a low micromolar binding constant (KD in the range of 3.7–5.5 μM) suggesting that in vitro, the binding to Sin1 RBD was similar across the G-domain of four RAS isoforms (Fig. 1E).
Therefore, these results indicate that the selectivity of Sin1 toward KRAS4A in cells is likely dependent on the HVR. We hypothesized that the HVR provides an exclusive conformation that restricts the interaction of the KRAS4A G-domain in cells. To evaluate this possibility, we generated two chimeric constructs that are composed of the G-domain of either NRAS or HRAS with the KRAS4A HVR. Consistent with our prediction, the presence of the KRAS4A HVR in either HRAS or NRAS G-domains enabled the interaction with Sin1 (Fig. 1F). Similarly, when the KRAS4B HVR was fused to the KRAS4A G-domain, interaction with Sin1 was strongly impaired. (SI Appendix, Fig. S1E). Our results suggest that, despite the ability of RAS G-domains to interact with Sin1 in vitro, the KRAS4A isoform preferentially binds Sin1 in cells as a result of its HVR.
Structural Characterization of the KRAS-Sin1 RBD Interaction.
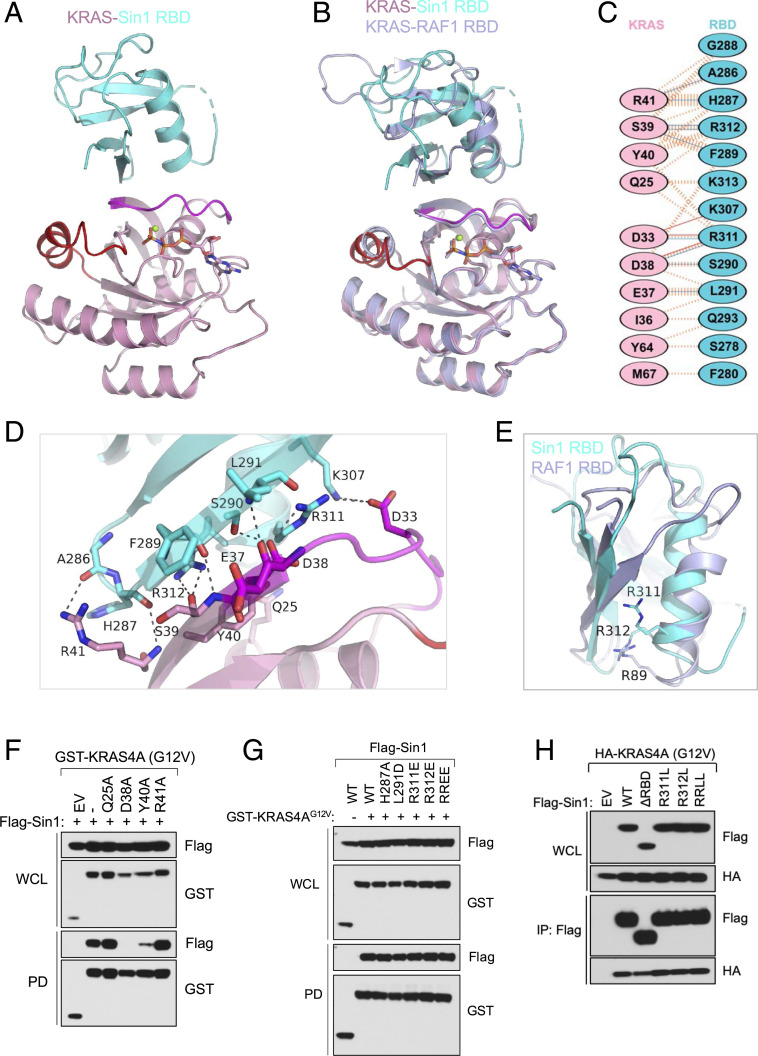
In order to gain structural insights into the interaction between KRAS and Sin1, we employed X-ray crystallography to solve the structure of the G-domain of KRAS (1-169) bound to the GTP analog GMPPNP in complex with Sin1 RBD (275-361). Although the in vitro binding affinity was similar between the two KRAS isoforms, the 4B isoform harboring the Q61R activating mutation yielded crystals with superior diffraction patterns to those of WT KRAS isoforms. In the KRAS4B Q61R - Sin1 RBD complex (hereafter referred to as the KRAS-Sin1 RBD complex), the KRAS-Sin1 RBD interaction interface resembles the RAS-RAF1 RBD interaction interface where RBD and KRAS interact mainly via β-strands and form an extended β-sheet structure (Fig. 2 A–B). This interface contains nine hydrogen bonds and three-salt-bridge interactions between KRAS and Sin1 RBD residues (Fig. 2C). KRAS residues mainly present in the switch I region D33, E37, D38, S39, Y40, R41 form key interactions with Sin1 RBD residues F289, S290, L291, K307 located on β2 strand and residues R311, R312 on α1 helix via main-chain and side-chain atoms (Fig. 2D).
Fig. 2.
Structural analyses of the KRAS-Sin1 RBD complex and the impact of point mutations on the KRAS-Sin1 interaction. (A) The overall structure of the KRAS-Sin1 RBD complex is shown in cartoon representation. GMPPNP and Mg2+ (green) are shown as sticks and spheres, respectively. Sin1 RBD is colored cyan, and KRAS is colored pink, with switch I and II regions highlighted in magenta and red, respectively. (B) Superposition of KRAS-Sin1 RBD structure with KRAS-RAF1 RBD structure (light blue, PDB: 6VJJ) shows conformational differences in the RBD of Sin1 and RAF1. The structural superposition was carried out by aligning C-α atoms of KRAS in both structures. (C) Schematic representation of the KRAS-Sin1 RBD interaction interface, as identified by PDBSum (http://www.ebi.ac.uk/pdbsum/). The interactions are colored using the following notations: hydrogen bonds, solid blue lines; salt bridges, solid red lines; nonbonded contacts, striped orange lines (width of the striped line is proportional to the number of atomic contacts). (D) Enlarged view of the KRAS-Sin1 RBD interaction interface formed by residues mainly present in KRAS switch I region (magenta), and β-2 strand and α-1 helix of Sin1 RBD. Dashed black lines indicate intermolecular hydrogen bonds and salt bridges. (E) Structural superposition of Sin1 RBD (cyan) with RAF1 RBD structure (light blue) highlighting conformational differences around R89 of RAF1 RBD. (F) Pull-down assay in 293T cells using Flag-Sin1 and GST-tagged KRAS4A mutants. WCL: whole-cell lysate; PD: pull down. (G) Pull-down assay in 293T cells using GST-tagged KRAS4A and Flag-Sin1 mutants. RREE: R311E, R312E; WCL: whole-cell lysate; PD: pull down. (H) Pull-down assay in 293T cells using GST-tagged KRAS4A and Flag-Sin1 mutants, including deletion of the RBD domain (ΔRBD). RRLL: R311L, R312L; WCL: whole-cell lysate; PD: pull-down.
The overall fold of Sin1 RBD resembles the ubiquitin-fold seen in the RAF1 RBD; however, insertions and deletions are present in loops connecting secondary structural elements and the conformation of these loops differs significantly between Sin1 and the RAF1 RBD domains. Interestingly, unlike the KRAS-RAF1 RBD complex, the helix present in the Sin1 RBD is shifted away from the interface in the KRAS-Sin1 RBD complex. As a result, the last helical turn as seen in the RAF1 RBD, which contains a key residue R89, is missing in the Sin1 RBD (Fig. 2E). However, the guanidinium group present in the sidechain of R312 in the Sin1 RBD extends toward KRAS and forms an interaction similar to that formed by R89 in the KRAS-RAF1 RBD complex. This helical shift in the KRAS-Sin1 RBD complex likely underlies the relatively weaker affinity of the KRAS-Sin1 RBD interaction compared to that of KRAS-RAF1 RBD.
To identify KRAS and Sin1 RBD residues that play an important role in the KRAS-Sin1 RBD interaction, we introduced point-mutations at key KRAS and Sin1 RBD residues. We found that D38A and Y40A mutations impair Sin1 interaction with KRAS in vitro and in cells (Fig. 2F and SI Appendix, Fig. S2A). In vitro ITC binding assays confirmed that KRAS D38A mutation caused complete loss of binding, whereas Y40A resulted in a 12-fold reduction in binding affinity. KRAS R41A exhibited a modest 2-fold decrease in binding affinity and, interestingly, KRAS Q25A mutation resulted in slightly stronger binding between KRAS and the Sin1 RBD (SI Appendix, Fig. S2A).
Similarly, mutation of key residues in the Sin1 RBD demonstrated that R312 and L291 play pivotal roles in the KRAS-Sin1 RBD interaction as charge-reversal mutation of these two residues (R312E and L291D) fully prevented binding between KRAS and Sin1 RBD in vitro (SI Appendix, Fig. S2B). R311E and H287A mutations in the Sin1 RBD had no effect on KRAS-Sin1 RBD interaction. However, when H287A, L291D, R312E or RR311EE mutations were introduced into full-length Sin1, interaction with KRAS4A was not disrupted in cells (Fig. 2G). To further evaluate whether other mutations in that interface could disrupt the interaction with KRAS4A in cells, we generated the Sin1 variants R311L, R312L, RR311LL, and Δ279–353 (ΔRBD). None of these mutations disrupted the interaction with KRAS4A in cells, even when the entire RBD was deleted (Fig. 2H).
Sin1 Contains an Atypical RBD.
Based on our previous results, we hypothesized that a secondary contact motif must be responsible for maintaining the interaction between KRAS and Sin1 in the absence of the RBD. We have recently described protein domain mapping using yeast 2 hybrid-next generation sequencing (DoMY-Seq), an unbiased approach for determining protein–protein interaction domains using yeast two-hybrid (Y2H) screening coupled to next-generation sequencing (NGS) (37). By creating a library of random and overlapping fragments derived from the Sin1 open reading frame that are cloned into a Y2H prey vector, the KRAS interacting peptides are positively selected in drop out auxotrophic media when mated to the yeast containing the KRAS bait. Using NGS, we can then determine the relative position of each peptide within the Sin1 sequence and experimentally determine the KRAS-interacting domain (Fig. 3A). Using DoMY-Seq, we found that the interaction between KRAS and Sin1 was not restricted to the previously described RBD (259-353). Instead, the interaction extended across a larger protein domain that comprised amino acids 279 to 390 (Fig. 3B). We referred to this sequence as the atypical RBD (aRBD), because it extended toward the PH domain of Sin1 and likely contained additional structural elements required for the interaction with KRAS. Importantly, deletion of this aRBD disrupted the interaction between Sin1 and KRAS4A in cells, in contrast to the previously described RBD. In contrast, deletion of the extended region of the aRBD (364-390) or a proximal region encoded by exon 9 (284-376) was not sufficient to abolish binding to KRAS4A (Fig. 3C).
Fig. 3.
Sin1 contains an atypical RBD for proper interaction with KRAS4A. (A) Overview of the DoMY-Seq pipeline used to determine protein–protein interacting motifs. A prey library of fragments generated from Sin1 cDNA is mated with yeast containing the KRAS bait. Selection of interacting peptides is enriched by auxotrophic dropout media, and surviving cells are sequenced by NGS to determine the identity of the sequence. (B) Integrative Genomics Viewer (IGV) plots from DoMY-Seq results depicting KRAS-interacting motif of Sin1. Control media (−LT) is depleted of Leucine and Tryptophane. Dropout mediums (−LTH and −LTHA) are depleted of Leucine, Tryptophane, Histidine, and Adenine, respectively. Based on the plateau of the peaks, we determined this motif to be comprised between Sin1 residues 279 to 390. (C) Immunoblot of the pull-down assay between GST-tagged KRAS4AG12V and Flag-Sin1 mutants in 293T cells. Deletions tested in Sin1 included ΔaRBD (279 to 390), 364 to 390, ΔExon 9 (284 to 376), and the point mutant R311E, S209A. WCL: whole-cell lysate; PD: pull down. (D) In vitro pull-down assay using recombinant GST control, GST-Sin1 RBD (279 to 353), GST-Sin1 aRBD (279 to 390), GST-Sin1 (360 to 390), and KRAS (1 to 165) loaded with GDP or GTPγS. (E) Overlap of the crystal structures of Sin1 PH domain (PDB: 3VOQ; light green) and AKT PH domain (PDB: 1H10; dark green). Unlike AKT PH domain, Sin1 PH domain contains an N-terminal extension that folds as an alpha helix (orange) and is conserved in lower organisms.
In vitro, the interaction between Sin1 aRBD and GTPγS loaded KRAS4A was similar to that of Sin1 RBD. The extended sequence from the aRBD (364-390) exhibited low affinity against KRAS4A, since we were unable to detect the interaction in our pull-down assays. (Fig. 3D). A secondary structure prediction of this extended sequence revealed the presence of a putative α-helix (370-376) that was highly conserved in lower organisms (Fig. 3E). A previously published crystal structure of the Sin1 PH domain contained part of this sequence and, consistent with our predictions, it was folded as an α-helix (38). Importantly, when we compared the Sin1 PH domain with the canonical PH domain crystal structure of AKT (39), we found that the Sin1 370–376 α-helix did not resemble any of the typical elements found in this domain, suggesting the possibility of an alternative function (Fig. 3E).
Overall, our results indicate the presence of an atypical RBD in Sin1 that is required for KRAS4A interaction in cells. Moreover, the results highlight the importance of deleting the full domain when developing Sin1 mutants unable to bind KRAS4A.
The Sin1 PH Domain Occludes the RBD by Adopting an Autoinhibited Conformation.
In order to better understand the molecular determinants of aRBD and KRAS binding, we decided to crystallize a Sin1 fragment (275-510) containing both RBD and PH domains in complex with KRAS. However, when determining the binding affinity between the Sin1 RBD-PH recombinant protein and KRAS we noticed that, compared to Sin1 RBD, the KD had increased significantly to ∼65 μM (Sin1 RBD ∼5.4 μM) (SI Appendix, Fig. S3A). Given that PH domains have been previously shown to block functional domains until they interact with their lipid ligands (40, 41), we hypothesized that the lower affinity of the Sin1 RBD-PH toward KRAS was likely due to steric hindrance by the PH domain. Hence, we undertook XL-MS experiments using recombinant Sin1 RBD-PH (275-510) and the crosslinker Bis[Sulfosuccinimidyl] glutarate (BS2G), which allows for amino-reactive crosslinks of 7.7 Å. Our results showed major crosslinks between the C-terminal region of the Sin1 PH domain and the RBD, specifically around aa 290 to 320 (SI Appendix, Fig. S3B). Given the monomeric nature of our recombinant protein, we concluded that these links are intramolecular and support the idea of an autoinhibited conformation. Using our XL-MS data, our RBD crystal structure, and the previously described PH domain crystal structure, we modeled the potential autoinhibited Sin1 RBD-PH structure (SI Appendix, Fig. S3C). Modeled structure of Sin1 RBD-PH suggests that these two domains likely interact via residues present on the α1 helix of RBD and β5-β8 strands of PH domain. Structural superposition of modeled Sin1 RBD-PH structure with the crystal structure of KRAS-Sin1 RBD complex shows that the PH domain partially blocks the KRAS interacting interface on Sin1 RBD (SI Appendix, Fig. S3D).
Structure of the Sin1 RBD-PH Domain in Complex with KRAS.
To gain structural insights into how the linker region between the RBD and PH domains of Sin1 interacts with KRAS, we attempted to solve the structure of the KRAS/Sin1 RBD-PH complex. However, because of the low micromolar affinity (KD ∼65 μM) between KRAS and Sin1 RBD-PH (SI Appendix, Fig. S3A), it was not possible to form a stable complex for crystallization. Interestingly, as we described before, the KRAS Q25A mutation increased the binding affinity to the Sin1 RBD in vitro and in cells (Fig. 2D and SI Appendix, Fig. S2A). Indeed, ITC confirmed that KRAS Q25A exhibited a six-fold higher affinity for Sin1 RBD-PH (KD∼10.5 μM), which was sufficient to obtain a stable complex for crystallography (SI Appendix, Fig. S4A). Using GMPPNP-bound KRAS-Q25A, we were able to crystallize and solve the structure of the KRAS Q25A/Sin1 RBD-PH complex (hereafter referred to as KRAS/Sin1 RBD-PH complex) at a resolution of 2.3 Å (SI Appendix, Table S1).
The overall structure of the KRAS/Sin1 RBD-PH complex shows that the Sin1 linker region and C-terminal end of PH domain interact extensively with residues in the switch II region of KRAS (Fig. 4 A–B). As predicted, the linker region contains a helix formed by residues 370 to 379. There is another small helix in the linker region (near the RBD) formed by residues 357 to 361, but it is not involved in the KRAS-Sin1 interaction (SI Appendix, Fig. S4B). The PH domain structure in the KRAS/Sin1 RBD-PH complex resembles the previously solved structure of the isolated Sin1 PH domain (38), except that in our structure, residues at the C-terminal end form an additional helix (residues 492 to 500) which points away from KRAS. Unlike the KRAS/Sin1-RBD interface, all the interactions formed between KRAS and Sin1 linker and PH domain are mainly van der Waals and hydrophobic (Fig. 4C). Sin1 linker residues F363, I369, M377, L378, S380 and H381 interact with KRAS switch I residues I36 and R41, and switch II residues E63, Y64, M67 and Q70. Sin1 PH domain residues Y484, S487, and S488 interact with A66, E62 and S65 residues present in the switch II region of KRAS Fig. 4C). Structural comparison between KRAS-Sin1 RBD and KRAS/Sin1 RBD-PH shows that a small sidechain containing amino acid in KRAS (Q25A mutation) causes conformational changes in the loop preceding the α1 helix in the Sin1 RBD, resulting in the formation of additional interactions at the interface (SI Appendix, Fig. S4C). These additional interactions likely contribute to the 6-fold higher affinity between KRAS-Q25A and Sin1-RBD-PH domains. Calculation of the solvent-accessible area shows that 560 Å2 of RBD and 435 Å2 of linker and PH domain are buried when Sin1 RBD-PH forms a complex with KRAS (Fig. 4D). Mapping the interaction interface between KRAS and Sin1 RBD-PH domains suggests that both switch regions contribute significantly to KRAS-Sin1 interaction. The interaction interface formed by the KRAS switch II region in this complex is much more extensive than any other KRAS-effector complexes solved to date, including RAS-PI3Kγ or RAS-NORE1 complex (42, 43).
Fig. 4.
Structure of the KRAS/Sin1 RBD-PH complex shows additional interactions formed by KRAS switch II region and Sin1 linker region and PH domain. (A and B) The overall structure of the complex formed by GMPPNP-bound KRAS and Sin1 RBD-PH domains shown in (A) cartoon and (B) surface representations. Panel A has the same color coding as in Fig. 2A, and Sin1 linker and PH domain are colored orange and green, respectively. (C) Schematic representation of the KRAS/Sin1 linker-PH interaction interface, as identified by PDBSum. The interactions are colored using the same notation as defined in Fig 2C. (D) Enlarged view of the KRAS/Sin1 linker-PH interaction interface formed by residues mainly present in the KRAS switch II region (red) and Sin1 linker region (orange) and PH domain (green). (D) The Top and Bottom panels show the region (colored gray) of Sin1 RBD-PH and KRAS that are involved in the KRAS-Sin1 complex formation.
A Sin1 Mutant Unable to Bind RAS Does Not Affect mTORC2 Activity or Assembly.
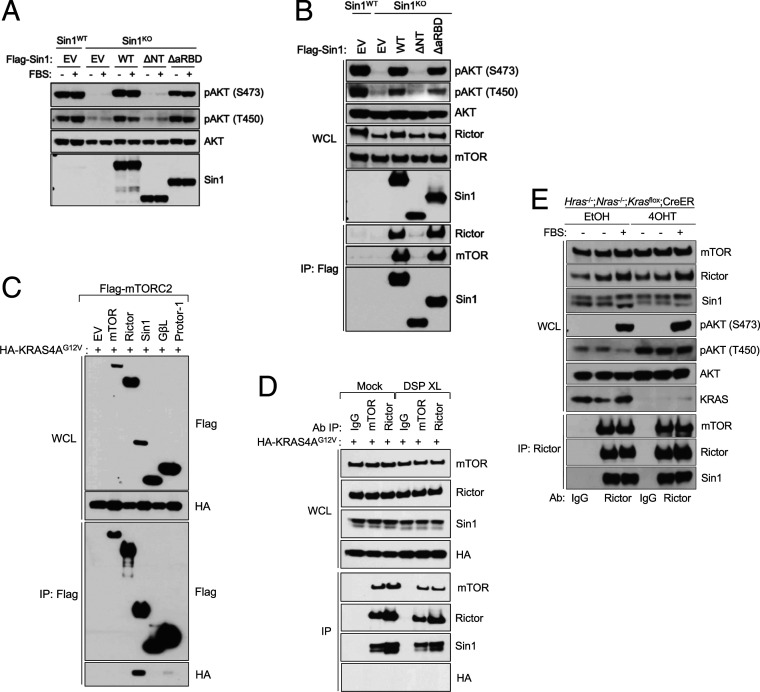
To determine the role of the KRAS-Sin1 interaction on the activity and assembly of mTORC2, we generated two separation-of-function Sin1 mutants. The first, Sin1 ΔaRBD, is unable to bind KRAS; the second, Sin1 ΔNT, was generated by deleting the NT domain (1-192) and is unable to bind mTORC2 (44). To test the function of these mutants, we generated Sin1 knockout 293T cells using CRISPR/Cas9, so that we could reconstitute with the aforementioned Sin1 mutants. Sin1 knockout cells lost phosphorylation of AKT at both hydrophobic (S473) and turn (T450) motifs, two robust readouts of mTORC2 activity (19, 20), under basal conditions and upon growth factor stimulation (SI Appendix, Fig. S5A). Importantly, the absence of Sin1 did not affect the stability of the complex, because the mTOR-Rictor interaction was preserved in Sin1 knockout cells (SI Appendix, Fig. S5B). Overexpression of WT Sin1 or the ΔaRBD mutant was sufficient to rescue the AKT phosphorylation in these cells to the levels observed in parental Sin1 WT cells. In contrast, the Sin1 ΔNT mutant failed to rescue mTORC2 activity, given its inability to bind the complex (Fig. 5A). Similar results were obtained when looking at PKCα phosphorylation, another substrate of mTORC2 (SI Appendix, Fig. S5C) (45). Consistent with these results, both Sin1 WT and ΔaRBD associated with mTORC2 while the ΔNT mutant did not, as assessed by immunoprecipitation of either Sin1 or Rictor (Fig. 5B and SI Appendix, Fig. S5D). In addition, the Sin1 ΔaRBD mutant did not exhibit any changes in mTORC2 or MAPK signaling upon growth factor stimulation (SI Appendix, Fig. S5 E–F).
Fig. 5.
mTORC2 basal activity is not regulated by KRAS4A. (A) Immunoblot of phosphorylated AKT in 293T Sin1 KO cells reconstituted with Sin1 WT, ΔNT, and ΔaRBD plasmids. Cells were starved overnight in serum-free media and stimulated with fetal bovine serum (FBS) for 15 min. (B) Flag immunoprecipitation in 293T Sin1 KO cells reconstituted with Flag-Sin1 WT, ΔNT, and ΔRBD plasmids using 0.3% CHAPS lysis buffer. WCL: whole-cell lysate; IP: immunoprecipitation. (C) Immunoblot of the coimmunoprecipitation between HA-KRAS4AG12V and Flag-tagged components of the mTORC2 (mTOR, Rictor, Sin1, GβL, and Protor-1) using 0.3% CHAPS lysis buffer. WCL: whole-cell lysate; IP: immunoprecipitation. (D) Endogenous immunoprecipitation of mTOR or Rictor in 293T cells transfected with HA-KRAS4AG12V. Cells were either mock-treated or crosslinked with DSP for 40 min and quenched with 25 mM Tris-HCl for 20 min. WCL: whole-cell lysate; IP: immunoprecipitation. (E) Immunoblot analysis of lysates from Hras−/−; Nras−/−; Krasflox; CreER MEFs treated with tamoxifen (4-OHT) to activate Cre recombinase and delete the floxed Kras allele. Cells were serum starved overnight and treated for 15 min with FBS.
These results prompted us to speculate that KRAS does not interact directly with mTORC2. Hence, we assessed the interaction between KRAS4A (G12V) and mTORC2 by coimmunoprecipitation. First, when the different main components of the complex (namely mTOR, Rictor, Sin1, GβL, and Protor-1) were coexpressed with KRAS4A (G12V), we only saw interaction with Sin1 (Fig. 5C). Because these experiments are performed in the presence of a zwitterionic detergent that preserves the integrity of the complex (19, 46), our results indicate that KRAS4A (G12V) is unable to associate directly with mTORC2; otherwise, we should have detected KRAS4A (G12V) in the pull-downs for each mTORC2 component. Second, we assessed whether KRAS4A (G12V) had the ability to associate with endogenous mTORC2. We immunoprecipitated endogenous mTORC2 from mammalian cells using a Rictor antibody, or both mTORC1/2 with an mTOR antibody. We were unable to detect the ectopic KRAS4A (G12V) protein, even when cells were crosslinked with the cell-permeable bifunctional crosslinker dithiobis(succinimidyl propionate) (DSP), which has been successfully used in the past to detect weak interactors of mTORC1 (47) (Fig. 5D). We also knocked out KRAS in mammalian cells using CRISPR/Cas9 (SI Appendix, Fig. S6A). Stimulation of these cells with growth factors did not affects the levels of AKT phosphorylation at the hydrophobic or turn motifs (SI Appendix, Fig. S6B). In addition, we did not observe any changes in the formation of the mTORC2 complex, as assessed in Rictor immunoprecipitates (SI Appendix, Fig. S6C).
We further analyzed the effect of KRAS loss in RASless mouse embryonic fibroblasts (MEFs). These cells are derived from mice that are knockout for the Nras and Hras genes and contain homozygous floxed alleles for Kras and a ubiquitous Cre recombinase that is activated in response to tamoxifen (48). The RASless MEFs are a unique model, because they allow the effect of Kras to be tested without the possibility of redundancy with other RAS isoforms; when treated with tamoxifen, recombination of the Kras allele occurs and cells undergo cell cycle arrest. We used RASless MEFs to assess the effect of RAS deletion on mTORC2 activity and complex formation. We found that, upon recombination of the Kras allele, no changes were observed in AKT hydrophobic motif phosphorylation or mTORC2 assembly (Fig. 5E). Altogether, these results indicate that the absence of KRAS in cells does not affect mTORC2 function.
Sin1 ΔaRBD Mice Are Healthy and Exhibit Normal mTORC2 Function.
Mice lacking Sin1 are embryonically lethal due to mTORC2 inhibition during development (17). We leveraged this phenotype to assess whether germline deletion of the Sin1 aRBD could affect mTORC2 signaling. The mouse Sin1 aRBD is encoded by exons 6 to 9 of the Mapkap1 gene, which spans ∼62 kb. Using CRISPR/Cas9, we engineered mouse embryonic stem cells to introduce a fusion between exons 6 and 9 that gives rise to an in-frame deletion of the aRBD (SI Appendix, Fig. S7A). Mice with homozygous deletion of Sin1 aRBD were born at expected Mendelian rates and without any obvious morphological or histological abnormalities (Fig. 6A and SI Appendix, Fig. S7B). Sanger sequencing of genomic DNA isolated from homozygous Sin1 ΔaRBD mice confirmed the presence of the in-frame deletion (Fig. 6B). Consistent with previous reports, Sin1 knockout mice exhibited embryonic lethality (17) (Fig. 6A); in these embryos we observed underdevelopment of the placenta architecture, which exhibited fewer obvious maternal spaces and compaction of the trophoblast-lined cords, similar to Rictor and mLST8 knockout mice (49, 50). In contrast, the placenta of homozygous Sin1 ΔaRBD embryos was normal (Fig. 6C). Next, we harvested several tissues from WT, heterozygous, and homozygous Sin1 ΔaRBD mice and analyzed the phosphorylation of AKT at its hydrophobic motif (S473) as a surrogate of mTORC2 activity. The levels of phosphorylated AKT were similar between compound mutant and WT mice in all the tissues analyzed, which included brain, heart, kidney, liver, lung, and spleen (Fig. 6D). Consistent with these results, immunoprecipitation of endogenous mTORC2 using an antibody against Rictor showed intact assembly of mTORC2 in homozygous Sin1 ΔaRBD mice (Fig. 6E and SI Appendix, Fig. S7C). Next, we generated Sin1 ΔaRBD WT, heterozygous, and homozygous primary MEFs from E13.5 embryos. Similar to the analysis performed in the tissues derived from adult mice, we did not observe differences in AKT or PKCα phosphorylation, even when cells were stimulated with growth factors (Fig. 6F and SI Appendix, Fig. S7 D–E). Immunoprecipitation of Rictor in these fibroblasts showed that Sin1 ΔaRBD protein associated with mTORC2 to the same extent as WT Sin1. Similarly, no differences in the association of mTOR and Rictor were observed (Fig. 6G). Overall, the results obtained from these mice confirm that the Sin1 interaction with KRAS is dispensable for proper mTORC2 function and assembly.
Fig. 6.
Proficient mTORC2 function in Sin1 ΔaRBD mice. (A) Mendelian rates obtained when interbreeding heterozygous Sin1ΔaRBD and Sin1KO mice. Pups were genotyped at P21, and the resulting genotype is shown as green (wild type), orange (heterozygous), and red (homozygous). χ2 test was used to compare with expected Mendelian rates. (B) Chromatogram obtained from Sanger sequencing the region encoding for the in-frame deletion of Sin1 ΔaRBD. (C) (Upper) Representative hematoxylin and eosin (H&E)-stained sections of the placental labyrinth demonstrate appropriately developed architecture in the WT and Sin1 ΔaRBD with abundant maternal-blood–filled spaces surrounding trophoblast-lined cords that contain fetal vessels but an underdeveloped architecture in the Sin1 knockout with fewer obvious maternal spaces and compaction of the trophoblast-lined cords. (Lower) Representative sections of immunohistochemistry (IHC) staining for Vimentin shows positive finely trabeculated fetal endothelium in the WT and Sin1ΔaRBD placenta while highlighting the underdevelopment and compaction in the complete knockout (scale bar, 200 μm). (D) Immunoblot analysis of AKT phosphorylation in lysates obtained from different organs of Sin1 ΔaRBD WT, heterozygous, or homozygous mice. Each lane represents an independent animal. HSP90 antibody was used to normalize. (E) Immunoprecipitation of endogenous Rictor in brain lysates obtained from Sin1 ΔaRBD WT, heterozygous, or homozygous mice. Each lane represents an independent animal. Note the change in Sin1 molecular weight as a result of the ΔaRBD deletion. IP: immunoprecipitation. (F) Western blot analysis of lysates isolated from Sin1 KO, Sin1 ΔaRBD WT, heterozygous, or homozygous MEFs. (G) Immunoprecipitation of endogenous Rictor in lysates obtained from Sin1 ΔaRBD WT, heterozygous, or homozygous primary MEFs. WCL: whole-cell lysate; IP: immunoprecipitation.
Discussion
Sin1 is a multidomain protein necessary for the proper function of mTORC2 (7, 16–18). Through its N-terminal domain, Sin1 is inserted into this large complex, while the acidic ubiquitin fold of the CRIM domain provides critical interactions with canonical mTORC2 substrates (29, 31). Hence, depletion of Sin1 in mammalian cells and model organisms results in mTORC2 inhibition (16–18). Other regions of the protein, such as the RBD and PH domains of Sin1 remain less studied, despite the fact that they are highly conserved in many eukaryotes (8, 9). In the particular case of the RBD, sequence homology to the RAF kinase RBD suggests a potential interaction with RAS GTPases. Analysis of such interaction in yeast, slime mold, mice, and humans have demonstrated the association between Sin1 and RAS orthologs (11–14). Our data also supports this interaction, both in vitro using recombinant proteins and in cells by means of coimmunoprecipitation assays. The interaction is direct, mediated by the G-domain, and requires the presence of GTP, similar to other RAS effectors such as RAF, PI3K or RalGDS (1, 2). However, despite the ability to interact with all RAS isoforms, Sin1 shows preferential binding to the KRAS4A isoform when expressed in cells. This selectivity is afforded by the RAS HVR, which despite not interacting directly with Sin1, likely positions the G-domain in the correct orientation for interaction. Indeed, computational modeling has shown that the KRAS4A HVR affects the orientation of the protein at the plasma membrane (51). Moreover, other RAS effectors, namely RAF kinases, have been shown to preferentially interact with different RAS isoforms depending on their HVR, even though the interaction occurs with the RAS G-domain (52).
Given that expression of the KRAS4A isoform is restricted to specific tissues and enriched in stem cells and embryonic tissues (53–55), future work will be required to assess whether the KRAS4A-Sin1 interaction plays a role in these cellular lineages. In our pull-down experiments, we have found that endogenous Sin1 is particularly resistant to interacting with ectopically expressed GTP-loaded KRAS4A. A plausible explanation for this observation is that Sin1 appears to adopt an auto-inhibited conformation in which the PH domain folds back to occlude the RBD, as suggested by our XL-MS experiments. Previous reports have demonstrated that the Sin1 PH domain can associate with phosphatidylinositols that can act as second messengers of mitogenic signaling (14, 56). Therefore, we hypothesize that interaction with such lipids can relieve the autoinhibitory conformation to allow RAS interaction in vivo. In fact, many other PH domain-containing proteins exhibit a similar mechanism of activation, where the PH domain blocks the catalytic domain unless it interacts with lipids (57, 58). Alternatively, the presence of additional molecular triggering events, such as posttranslational modifications, could facilitate the open conformation (59).
In this study, we also show that the KRAS binding interface of Sin1 is longer than previously described; it not only includes the prototypical RBD but also expands toward the PH domain. We identified this so-called aRBD using an unbiased Y2H approach and have confirmed the key interacting residues by X-ray crystallography. Interestingly, we find that many of the interactions mediated by the RBD-PH linker occur around the KRAS switch II region. In the last two decades, structures of RAS in complex with various effectors such as RAF, PI3Kγ, RalGDS, RASSF5, PLCε, Byr2, and Grb14 have been solved (42, 43, 60, 61, 62–64). Even though these effectors share low sequence similarity in the RA/RBD, their interaction mode with RAS is well conserved and involves residues mainly present in the switch I region of RAS. In these structures, interaction involving the switch II region of KRAS is either absent or limited to a couple of polar interactions formed mainly by switch II residues E63 and Y64. The helical region located at the N terminus of the RBD in the HRAS-RASSF5 complex and the kinase domain in the HRAS-PI3Kγ complex have been observed to form polar and hydrophobic interaction with 2–3 residues in switch II (42, 43). In the KRAS/Sin1 RBD-PH structure, the interaction interface formed by the switch II region of KRAS and linker-PH domain of Sin1 is much more extensive than observed in any of the RAS-effector complexes characterized so far. This structural feature suggests that the interaction interface formed by the switch II region of RAS plays an important role in RAS-Sin1 interaction. Considering that most of the structural studies on RAS-effector complexes have been limited to RA/RBD domains, it is possible that in other RAS-effector complexes, the switch II region of KRAS may also interact with other neighboring domains of RA/RBD and thereby contribute to RAS-effector interactions.
Among the structures of RAS-effector complexes solved so far, the structure of HRAS in complex with Grb14, a member of the Grb7-10-14 family of cytoplasmic adaptor proteins, includes both RA and PH domain (64). However, unlike the KRAS/Sin1 RBD-PH complex, the linker region and PH domain of Grb14 are located far from the RAS-Grb14 interface in the HRAS/Grb14 RA-PH complex. In the later structure, RAS interaction with Grb14 is limited to the RA domain, and it has been suggested that this keeps the PH domain free to bind to headgroups of membrane phosphoinositide via basic residues in the β1-β2 loop. In the KRAS/Sin1 RBD-PH complex, the β1-β2 loop of the PH domain is fully accessible and is located on the opposite end from the KRAS/Sin1 RBD PH interface. Thus, our structural work suggests that the Sin1 PH domain may be able to interact with lipid headgroup even when Sin1 is in complex with RAS.
Deletion of the Sin1 aRBD yielded a mutant that is unable to interact with KRAS, yet when expressed in Sin1 knockout cells we show that it completely rescues mTORC2 activity. The truncated Sin1 (ΔaRBD) incorporates into the mTORC2 complex, as it can be immunopurified with other components of the complex, including mTOR and Rictor. In contrast, a mutant that lacks the N-terminal domain (ΔNT) is unable to associate with the complex and rescue the activity in Sin1 knockout cells. This observation had been previously noted, because expression of ectopic Sin1 containing large N-terminal tags cannot associate with mTORC2 (16). Moreover, as cryo-EM analyses of mTORC2 provide further insights and increased resolution of this large complex, our data appear to be consistent with a model in which the Sin1 N-terminal domain, but not RBD or aRBD, is required for interaction with and activation of mTORC2 (29, 30). The 3.2 Å structure of the human mTORC2 complex has shown that the N-terminal domain (residues 2–137) of Sin1 integrates into the Rictor fold and connects Rictor with mLST8, thus playing a direct role in stabilizing the mTORC2 complex. Sin1 uses mSLT8 as a platform to position the substrate recruiting CRIM domain in the mTORC2 complex. Even though the construct used in the cryoEM study included full-length Sin1 protein, the RBD and PH domains were not visible in the cryoEM map, which leaves the possibility that these two domains do not interact directly with mTORC2.
The rescue of Sin1 ΔaRBD in cells was also confirmed by generating a novel mouse germline knock-in allele that endogenously expresses this mutant. Sin1 homozygous knockout mice are embryonic lethal (17) due to a defect in the placental architecture that resembles the phenotype seen in Rictor knockout mice (50), while Sin1 homozygous ΔaRBD mice are normal and fertile. In fact, tissues and embryonic fibroblasts derived from ΔaRBD mice reveal normal mTORC2 activation and assembly.
Other groups have shown that RAS proteins can modulate the activity of TORC2 in Dictyostelium and human cells, by interacting directly with both Sin1 and mTOR (11, 32, 33, 65). In the present study, we have not detected interaction between KRAS and components of mTORC2 other than Sin1. Although we have not determined interaction with mTOR using recombinant proteins, coimmunoprecipitation assays have failed to reveal this interaction, even when the mTORC2 was crosslinked or in the presence of zwitterionic detergents known to preserve the integrity of the complex (46). Additionally, deletion of KRAS in human cells did not impact the activation of the complex, even upon growth factor stimulation. However, it is important to highlight that some of the previous studies have used oncogenic variants of RAS, which could explain the differences observed here. It is possible that oncogenic RAS might modulate mTORC2 allosterically, indirectly, or that different subcellular pools of mTORC2 are regulated differently (25). Because oncogenic KRAS also promotes PI3K activation and, hence, translocation of AKT to the plasma membrane (66), caution is certainly warranted when using phosphorylated AKT as a surrogate of mTORC2 activity in these experiments.
Our results open the possibility that the interaction between KRAS4A and Sin1 has an mTORC2-independent function. Sin1 was initially identified as a component of the stress-activated MAPK; it interacts with both JNK kinases and MEKK2 (67, 68). Despite these findings, little progress has been made in elucidating the role of Sin1 in response to stress. In addition, these experiments do not address the contribution of specific Sin1 domains, so it is not possible to distinguish any phenotype from an mTORC2-dependent or independent mechanism. Our separation-of-function Sin1 mutants provide a robust and reliable tool to address this issue. For instance, in future experiments, it would be interesting to assess if Sin1 ΔaRBD mutant cells and mice respond differentially to certain types of cellular stress or to KRAS oncogenic transformation. In the latter, the KRAS4A isoform has been shown to be required for lung cancer initiation (69), providing an excellent model to test the potential contribution of Sin1.
In summary, our work has provided biochemical and structural insights into the interaction between KRAS and Sin1 and revealed that mTORC2 activity and assembly are not affected by Sin1 mutants that are unable to bind RAS, in cultured cells and mice. These results contribute to our overall understanding of mTORC2 regulation and support a potential role for Sin1-KRAS interaction independently of mTORC2.
Materials and Methods
HEK293T cells were purchased from the American Type Culture Collection and used at low passages. KRAS and Sin1 KO clones were generated using CRISPR/Cas9 technology. Primary Sin1ΔaRBD MEFs were derived from E13.5 embryos as previously described (70). All proteins used for crystallization and biophysical assay were expressed in Escherichia coli. All RAS proteins were purified as outlined in Kopra et al. (71). Briefly, the expressed proteins of the form His6-TEV-target or His6-MBP-TEV-target were purified from clarified lysates by immobilized metal affinity chromatography, treated with His6-TEV protease to release the target protein, and the target protein separated from other components of the tobacco etch virus (TEV) protease reaction by a second round of IMAC. Proteins were further purified by gel-filtration chromatography.
Crystals of KRAS Q61R complexed with Sin1 RBD and KRAS Q25A complexed with Sin1 (RBD-PH) diffracted to a resolution of 2.70 and 2.35 Å, respectively. Crystallographic datasets were integrated and scaled using X-ray detector software (XDS) (72). Crystal parameters and data collection statistics are summarized in SI Appendix, Table S1. The structure of KRAS in complex with Sin1 RBD was solved by molecular replacement using the program Phaser as implemented in the Phenix suite of programs (73), with a protein-only structure of GMPPNP-bound KRAS present in KRAS-RAF1(RBD) complex (Protein Data Bank ID: 6VJJ) as a search model. The initial solution obtained from molecular replacement was refined using the program Phenix.refine within the Phenix suite of programs (73). The model was further improved using iterative cycles of manual model building in Coot (74) and refinement with Phenix.refine. Molecular docking of Sin1 RBD and PH domains was carried out using the HADDOCK (75). ITC experiments were performed with MicroCal PEAQ-ITC calorimeter at 25 °C using GDP/GMPPNP-bound RAS proteins exchanged as previously described (76). Crosslinking mass spectrometry was performed as previously described (77). For details regarding all materials and methods, reference SI Appendix.
Supplementary Material
Acknowledgments
We thank Allan Balmain for scientific insights and feedback, Bernie Suter from Next Interactions for help with Sin1 DoMY-Seq, Cathy Tournier for providing Sin1 knockout mice, and Alexandra Newton for the Sin1 knockout MEFs. We also thank Bill Gillette, John-Paul Denson, Jennifer Mehalko, Rosemilia Reyes, Vanessa Wall, Jose Sanchez Hernandez, Nitya Ramakrishnan, Shelley Perkins, Mukul Sherekar, Stephanie Widmeyer, Troy Taylor, Xiaoying Ye, and Kelly Snead of the Protein Expression Laboratory (Frederick National Laboratory for Cancer Research) for their help in cloning, expressing, and purifying recombinant proteins. We are grateful to the staff of 24-ID-C/E beamline at the Advanced Photon Source, Argonne National Laboratory, for their help with data collection. We are thankful to Timothy Tran for his help with the crystallographic studies. Funding was provided by the NIH/NCI Grant No. R35CA197709-01 (to F.M.). P.C. work is supported by the NCI K99/R00 Pathway to Independence Award (Award No. K99CA245122). Mass spectrometry experiments were supported by the Adelson Medical Research Foundation and the University of California, San Francisco Program for Breakthrough Biomedical Research. Part of this work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from NIH Grant No. P30 GM124165. The Eiger 16M detector on 24-ID-E is funded by an NIH Office of Research Infrastructure Programs High-End Instrumentation grant (Grant No. S10OD021527). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was funded in part with federal funds from the National Cancer Institute, NIH Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Footnotes
Competing interest statement: F.M. is a consultant for Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Pfizer, Daiichi Sankyo, Amgen, PMV Pharma, OPNA-IO, and Quanta Therapeutics and has received research grants from Boehringer-Ingelheim and is a consultant for and cofounder of BridgeBio Pharma. P.C. is a cofounder and advisory board member of Venthera.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103261118/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank and are available under Accession Nos. 7LC2 (KRAS Q61R–Sin1 RBD complex) and 7LC1 (KRAS Q25A–Sin1 RBD-PH complex).
References
- 1.Simanshu D. K., Nissley D. V., McCormick F., RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel M., Côté J.-F., Ras GTPases’ interaction with effector domains: Breaking the families’ barrier. Commun. Integr. Biol. 6, e24298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D., RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustelo X. R., Crespo P., Fernández-Pisonero I., Rodríguez-Fdez S., RAS GTPase-dependent pathways in developmental diseases: Old guys, new lads, and current challenges. Curr. Opin. Cell Biol. 55, 42–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castel P., Rauen K. A., McCormick F., The duality of human oncoproteins: Drivers of cancer and congenital disorders. Nat. Rev. Cancer 20, 383–397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore A. R., Rosenberg S. C., McCormick F., Malek S., RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruan C., et al., Sin1-mediated mTOR signaling in cell growth, metabolism and immune response. Natl. Sci. Rev. 6, 1149–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatebe H., Shiozaki K., Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 7, 77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S.-Z., Roberts R. M., The evolution of the Sin1 gene product, a little known protein implicated in stress responses and type I interferon signaling in vertebrates. BMC Evol. Biol. 5, 13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson M. G., et al., Sin1: An evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18, 4210–4221 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith S. F., Collins S. E., Charest P. G., Ras, PI3K and mTORC2 - Three’s a crowd? J. Cell Sci. 133, jcs234930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colicelli J., et al., Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 2913–2917 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Parent C. A., Insall R., Firtel R. A., A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10, 2829–2845 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroder W. A., et al., Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell. Signal. 19, 1279–1289 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Loewith R., et al., Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Frias M. A., et al., mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865–1870 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Jacinto E., et al., SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Yang Q., Inoki K., Ikenoue T., Guan K.-L., Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M., Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Facchinetti V., et al., The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Guan K.-L., mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearce L. R., Komander D., Alessi D. R., The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Gaubitz C., Prouteau M., Kusmider B., Loewith R., TORC2 structure and function. Trends Biochem. Sci. 41, 532–545 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ebner M., Sinkovics B., Szczygieł M., Ribeiro D. W., Yudushkin I., Localization of mTORC2 activity inside cells. J. Cell Biol. 216, 343–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaubitz C., et al., Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol. Cell 58, 977–988 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Karuppasamy M., et al., Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat. Commun. 8, 1729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuttfeld E., et al., Architecture of the human mTORC2 core complex. eLife 7, e33101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., et al., Cryo-EM structure of human mTOR complex 2. Cell Res. 28, 518–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatebe H., et al., Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. eLife 6, e19594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khanna A., et al., The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci. Rep. 6, 25823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalski J. R., et al., The functional proximal proteome of oncogenic Ras includes mTORC2. Mol. Cell 73, 830–844.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nassar N., Singh K., Garcia-Diaz M., Structure of the dominant negative S17N mutant of Ras. Biochemistry 49, 1970–1974 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroder W., Cloonan N., Bushell G., Sculley T., Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene 339, 17–23 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Mo S. P., Coulson J. M., Prior I. A., RAS variant signalling. Biochem. Soc. Trans. 46, 1325–1332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castel P., Holtz-Morris A., Kwon Y., Suter B. P., McCormick F., DoMY-Seq: A yeast two-hybrid-based technique for precision mapping of protein-protein interaction motifs. J. Biol. Chem. 100023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan D., Matsuura Y., Structures of the pleckstrin homology domain of Saccharomyces cerevisiae Avo1 and its human orthologue Sin1, an essential subunit of TOR complex 2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 386–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas C. C., Deak M., Alessi D. R., van Aalten D. M. F., High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 12, 1256–1262 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Drugan J. K., Rogers-Graham K., Gilmer T., Campbell S., Clark G. J., The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J. Biol. Chem. 275, 35021–35027 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Parikh C., et al., Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc. Natl. Acad. Sci. U.S.A. 109, 19368–19373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pacold M. E., et al., Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Stieglitz B., et al., Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 27, 1995–2005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cameron A. J. M., Linch M. D., Saurin A. T., Escribano C., Parker P. J., mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 439, 287–297 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K.-L., Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarbassov D. D., Bulgakova O., Bersimbaev R. I., Shaiken T., “Isolation of the mTOR complexes by affinity purification” in MTOR: Methods and Protocols, Methods in Molecular Biology, Weichhart T., Ed. (Humana Press, 2012), pp. 59–74. [DOI] [PubMed] [Google Scholar]
- 47.Kim D.-H., et al., mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Drosten M., et al., Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiota C., Woo J.-T., Lindner J., Shelton K. D., Magnuson M. A., Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11, 583–589 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Guertin D. A., et al., Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11, 859–871 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Li Z.-L., Buck M., Computational modeling reveals that signaling lipids modulate the orientation of K-Ras4A at the membrane reflecting protein topology. Structure 25, 679–689.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terrell E. M., et al., Distinct binding preferences between Ras and Raf family members and the impact on oncogenic Ras signaling. Mol. Cell 76, 872–884.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pells S., et al., Developmentally-regulated expression of murine K-ras isoforms. Oncogene 15, 1781–1786 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Tsai F. D., et al., K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. U.S.A. 112, 779–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newlaczyl A. U., Coulson J. M., Prior I. A., Quantification of spatiotemporal patterns of Ras isoform expression during development. Sci. Rep. 7, 41297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu P., et al., PtdIns(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov. 5, 1194–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemmon M. A., Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheffzek K., Welti S., Pleckstrin homology (PH) like domains - Versatile modules in protein-protein interaction platforms. FEBS Lett. 586, 2662–2673 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Liu P., et al., Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 15, 1340–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fetics S. K., et al., Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang L., Hofer F., Martin G. S., Kim S. H., Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol. 5, 422–426 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Bunney T. D., et al., Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol. Cell 21, 495–507 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Scheffzek K., et al., The Ras-Byr2RBD complex: Structural basis for Ras effector recognition in yeast. Structure 9, 1043–1050 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Qamra R., Hubbard S. R., Structural basis for the interaction of the adaptor protein grb14 with activated ras. PLoS One 8, e72473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charest P. G., et al., A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev. Cell 18, 737–749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez-Viciana P., et al., Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370, 527–532 (1994). [DOI] [PubMed] [Google Scholar]
- 67.Cheng J., et al., Mip1, an MEKK2-interacting protein, controls MEKK2 dimerization and activation. Mol. Cell. Biol. 25, 5955–5964 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schroder W., Bushell G., Sculley T., The human stress-activated protein kinase-interacting 1 gene encodes JNK-binding proteins. Cell. Signal. 17, 761–767 (2005). [DOI] [PubMed] [Google Scholar]
- 69.To M. D., et al., Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat. Genet. 40, 1240–1244 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castel P., et al., RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science 363, 1226–1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kopra K., et al., Homogeneous dual-parametric-coupled assay for simultaneous nucleotide exchange and KRAS/RAF-RBD interaction monitoring. Anal. Chem. 92, 4971–4979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liebschner D., et al., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Zundert G. C. P., et al., The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Dharmaiah S., et al., Structures of N-terminally processed KRAS provide insight into the role of N-acetylation. Sci. Rep. 9, 10512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGilvray P. T., et al., An ER translocon for multi-pass membrane protein biogenesis. eLife 9, e56889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank and are available under Accession Nos. 7LC2 (KRAS Q61R–Sin1 RBD complex) and 7LC1 (KRAS Q25A–Sin1 RBD-PH complex).