Significance
Abundant surface glycoconjugates play key roles in the infectious cycle of protozoan parasites including Leishmania. Through defining biosynthetic pathways, we identified a fucosyltransferase FUT1 localized to the parasite mitochondrion, an atypical compartment for glycosyltransferases. FUT1 was essential for normal growth, requiring both mitochondrial localization and enzymatic activity. Loss of FUT1 in a single rare segregant (fut1s) showed extensive mitochondrial defects. Enzymatic tests showed FUT1 could fucosylate glycan and peptide substrates in vitro, although as yet the native substrate(s) are unknown. Trypanosomatid mitochondrial FUT1s may offer a facile system in the future for probing mitochondrial glycosylation in a setting uncomplicated by multiple isoforms targeted to diverse compartments, and its essentiality renders it an attractive chemotherapeutic target for these deadly parasites.
Keywords: trypanosomatid protozoan parasites, glycobiology, fucose, glycosyltransferase, chemotherapy
Abstract
Glycoconjugates play major roles in the infectious cycle of the trypanosomatid parasite Leishmania. While GDP-Fucose synthesis is essential, fucosylated glycoconjugates have not been reported in Leishmania major [H. Guo et al., J. Biol. Chem. 292, 10696–10708 (2017)]. Four predicted fucosyltransferases appear conventionally targeted to the secretory pathway; SCA1/2 play a role in side-chain modifications of lipophosphoglycan, while gene deletion studies here showed that FUT2 and SCAL were not essential. Unlike most eukaryotic glycosyltransferases, the predicted α 1–2 fucosyltransferase encoded by FUT1 localized to the mitochondrion. A quantitative “plasmid segregation” assay, expressing FUT1 from the multicopy episomal pXNG vector in a chromosomal null ∆fut1− background, established that FUT1 is essential. Similarly, “plasmid shuffling” confirmed that both enzymatic activity and mitochondrial localization were required for viability, comparing import-blocked or catalytically inactive enzymes, respectively. Enzymatic assays of tagged proteins expressed in vivo or of purified recombinant FUT1 showed it had a broad fucosyltransferase activity including glycan and peptide substrates. Unexpectedly, a single rare ∆fut1− segregant (∆fut1s) was obtained in rich media, which showed severe growth defects accompanied by mitochondrial dysfunction and loss, all of which were restored upon FUT1 reexpression. Thus, FUT1 along with the similar Trypanosoma brucei enzyme TbFUT1 [G. Bandini et al., bioRxiv, https://www.biorxiv.org/content/10.1101/726117v2 (2021)] joins the eukaryotic O-GlcNAc transferase isoform as one of the few glycosyltransferases acting within the mitochondrion. Trypanosomatid mitochondrial FUT1s may offer a facile system for probing mitochondrial glycosylation in a simple setting, and their essentiality for normal growth and mitochondrial function renders it an attractive target for chemotherapy of these serious human pathogens.
Leishmania is a widespread human pathogen, with more than 1.7 billion people at risk, several hundred million infected, and with 12 million showing active disease ranging from mild cutaneous lesions to severe disfiguring or lethal outcomes (1–3). The Leishmania infectious cycle alternates between the extracellular promastigote in the midgut of sand flies and intracellular amastigote residing within macrophages of the mammalian host, where it survives and proliferates in highly hostile environments. These parasites have evolved specific mechanisms enabling them to endure adverse conditions, including a dense cell surface glycocalyx composed of lipophosphoglycan (LPG), glycosylphosphatidylinositol (GPI)-anchored proteins (GP63 and GP46), glycosylinositolphospholipids (GIPLs), and secreted glycoconjugates such as proteophosphoglycan (PPG) and secreted acid phosphatase (sAP) (reviewed in refs. 4–7). One prominent feature of LPG, PPGs, and sAPs is the presence of disaccharide phosphate repeating units ([6Gal(β)1,4)Man(α1)-PO4]), also termed phosphoglycan or PG repeats.
Our laboratory has focused on both forward and reverse genetic approaches to map out glycoconjugate synthesis in Leishmania, emphasizing genes impacting the glycocalyx (8, 9) as well as ether lipids and sphingolipids (10, 11). These studies have provided powerful tools leading to new insights on the requirements for LPG and related phosphoglycan-bearing molecules in both parasite stages within the mammalian and sand fly hosts (5, 6, 12–18).
In several Leishmania species, modifications of the dominant PG repeats of LPG and PPG play key roles in the insect stages in mediating both the attachment and release of promastigotes and metacyclics, respectively, from the sand fly midgut via binding to midgut receptors there (19). In Leishmania major strain FV1 (LmjF), β1–3 galactosyl modifications of the PG repeating units enable replicating promastigotes to bind to the midgut lectin PpGalec, while addition of d-arabinopyranose (d-Arap) to the side-chain galactosyl residues block this interaction and allow release of parasites for subsequent transmission (19–21). We used forward genetic analysis to identify a large family of LPG side chain galactosyltransferases (SCG1-7) and d-arabinopyranosyltransferases (SCA1/2) mediating these modifications (21–26).
As d-Arap is relatively uncommon in nature (27), we were motivated to explore its synthetic pathway. Previously, we identified two genes showing strong homology to the bifunctional Bacteroides protein FKP mediating synthesis of GDP-l-Fucose (GDP-Fuc) through successive kinase and pyrophosphorylase steps (28, 29). Many enzymes using l-Fucose will also accept d-Arap (which differ only by the 6-methyl group), and assays of the two recombinant Leishmania proteins showed that indeed one could synthesize both GDP-d-Arap and GDP-Fuc (AFKP; LmjF.16.0480), while the second could only synthesize GDP- Fuc [FKP; LmjF.16.0440 (28)]. These data were consistent with studies showing the presence of both GDP-Fuc and GDP-Arap in Leishmania (30). Correspondingly, genetic studies showed that knockouts of AFKP completely abrogated GDP-Arap and arabinosylated LPG synthesis, while knockouts of FKP showed little effect (28). However, we were unable to knock out both genes simultaneously, suggesting an unanticipated role for GDP-Fuc. That the essential role of A/FKPs depended on GDP-Fucose was established when GDP-Fuc but not GDP-Arap was provided through expression of the two de novo GDP-Fucose enzymes from Trypanosoma brucei, GDP-mannose 4,6-dehydratase (GMD) and GDP-Fuc synthetase, also known as GDP-4-dehydro-6-deoxy-d-mannose epimerase/reductase (GMER) (31). Similarly, the loss of de novo GDP-fucose synthesis was also lethal in T. brucei, which lacks the FKP salvage pathway (31).
The essentiality of GDP-Fuc was unexpected since there are few reports of fucosylated molecules in Leishmania. One is a “fucose mannose ligand” from Leishmania donovani (32) for which a definitive structure is lacking. Several proteins were predicted to be fucosylated from mass-spectrometric (MS) proteome studies of L. donovani, albeit without experimental confirmation (33). More recently, human erythropoietin expressed in Leishmania tarentolae was shown to bear fucosylated glycans (34). While untested, it seemed unlikely that any of these putative fucosylations would be essential for growth in culture. Together, these data suggest that any putative essential Fuc-glycoconjugate predicted from genetic deletion studies must be relatively rare and/or cryptic.
To better understand the fucose requirement in Leishmania, and guide efforts to identify the essential fucose conjugate, we surveyed candidate fucosyltransferases (FUTs). Unexpectedly, FUT1 localizes within the parasite mitochondrion, an uncommon observation for glycosyltransferases.
Results
Database Mining for FUTs/Arabinopyranosyltransferase (AraTs) Candidates in L. major.
Using a diverse collection of FUTs compiled from GenBank or CAZY databases (35), we searched the predicted L. major proteome for proteins showing significant similarities and/or conserved motifs. Two new candidates (FUT1, LmjF01.0100; FUT2, LmjF.02.0330) as well as three previously described genes emerged (SCA1, LmjF02.0220; SCA2, LmjF02.0180; SCAL, LmjF34.0510; Table 1). Of these, only FUT1 was conserved in all trypanosomes, while FUT2 occurred in Trypanosoma cruzi.
Table 1.
Trypanosomatid l-fucose/d-arabinosylpyranose conjugates, precursors, and candidate transferases
| Leishmania major | Trypanosoma brucei | Trypanosoma cruzi | |
| Fuco-conjugates | Unknown | Unknown | GP72 |
| GDP-Fucose | + | + | + |
| d-Arap conjugates | LPG | Not described | Not described |
| GDP-Arap | + | Not found | Not found |
| α1,2 FUT (CAZY GT11) | LmjF.01.0100 (LmjFUT1) | TTb927.9.3600 (TbFUT1) | TcCLB.506893.90 TcCLB.508501.240 |
| α1,3/4 FUT (CAZY GT10) | LmjF.02.0330 (FUT2) | Not present | Not present |
| Fuc/ArapT (CAZY GT79) | LmjF.02.0220 (SCA1) | Not present | Not present |
| LmjF.02.0180 (SCA2) | |||
| LmjF.34.0510 (SCAL) |
SCA1/2.
The closely related SCA1/2 (side-chain arabinosyltransferase) genes (>99% identity) had been identified functionally and enzymatically as Golgi α1,2 d-Arap transferases modifying Gal-substituted PG repeating units of LPG in L. major during metacyclogenesis (23, 26). Interestingly, when provided with high levels of external l-Fucose, the LPG PG repeating units can become fucosylated, likely mediated by these two enzymes (36). Since Leishmania require neither galactosylated nor arabinosylated LPG for growth in culture or metacyclogenesis (14, 37), their study was deprioritized here.
SCAL.
SCAL (SCA1/2-like) was identified with 31% identity to the SCA1/2 proteins, which together constitute CAZY family 79 (26, 35). The lack of any related genes in T. brucei and T. cruzi is consistent with the absence of GDP-d-Arap in both (30) (Table 1). We were able to knock out both alleles of SCAL without difficulty and the mutant showed no appreciable growth defect in the culture (SI Appendix, Fig. S1), suggesting that this gene is not essential in vitro, or is redundant with other proteins.
FUT2.
The predicted FUT2 protein was identified by similarity to the CAZY family GT10, which comprises α1,3- or 1,4-fucosyltransferases (35). Strong homology of the 1,604-aa FUT2 protein was seen with PFAM family 00852 (α1,3 or 1,4 fucosyltransferases) from residues 1049–1159, along with other motifs characteristic of this family (38, 39). Using a plasmid segregation test (described further below for FUT1), we were able to knock out both alleles of FUT2 without difficulty (SI Appendix, Fig. S2), suggesting that this gene is not essential during in vitro culture, or is redundant with other proteins.
FUT1.
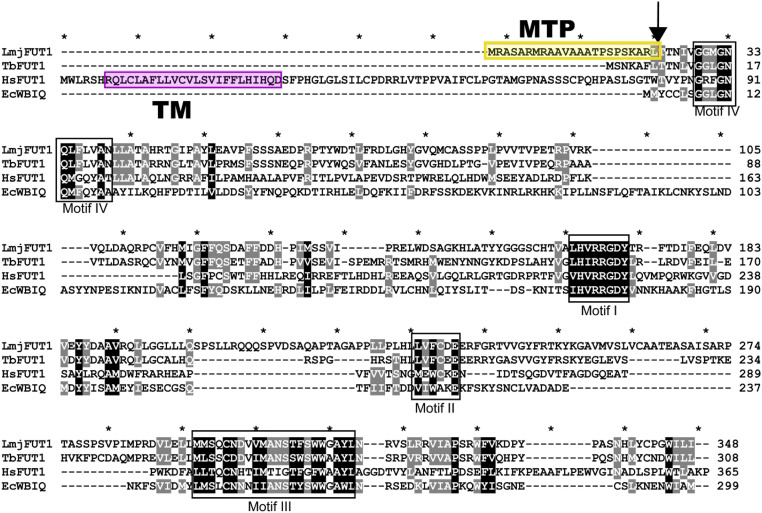
The predicted FUT1 protein showed relationships to the CAZY GT11 family comprised mostly of α1,2 fucosyltransferases (35). The annotated LmjFUT1 predicted a protein of 348 amino acids, exhibiting four characteristic motifs of the GT11 family (Fig. 1), of which motif I (HVRRGDY) has been implicated in the binding of GDP-fucose (40, 41). While SCA1, SCA2, SCAL, and FUT2 resembled typical eukaryotic fucosyltransferases predicted to be type II membrane proteins within the secretory pathway (39, 42), no transmembrane domain was predicted for FUT1. Instead, FUT1 displayed a predicted mitochondrial targeting peptide (MTP) by two algorithms (88%; TargetP and MitoProt) (43, 44). For L. major, the predicted N-terminal MTP comprised the N-terminal 23 amino acids upstream of motif IV, rich in Arg and hydrophobic amino acids (Fig. 1). The LmjFUT1 mitochondrial localization and its requirement for viability were established experimentally below.
Fig. 1.
Comparison of mitochondrial Leishmania major FUT1 to fucosyltransferases from Trypanosoma brucei (TbFUT1), bacteria (EcWBIQ from E. coli O127), and mammals (human HsFUT1). Identical or highly conserved residues are highlighted in black or gray. Conserved fucosyltransferase motifs are marked by black boxes (40, 41). The predicted mitochondrial targeting peptide (MTP) is shaded in yellow, and cleavage site is marked by an arrow. The predicated transmembrane domain (TM) for HsFUT1 is shaded in pink.
Generation of Chromosomal Null Mutants (∆fut1−) in the Presence of Ectopically Expressed FUT1.
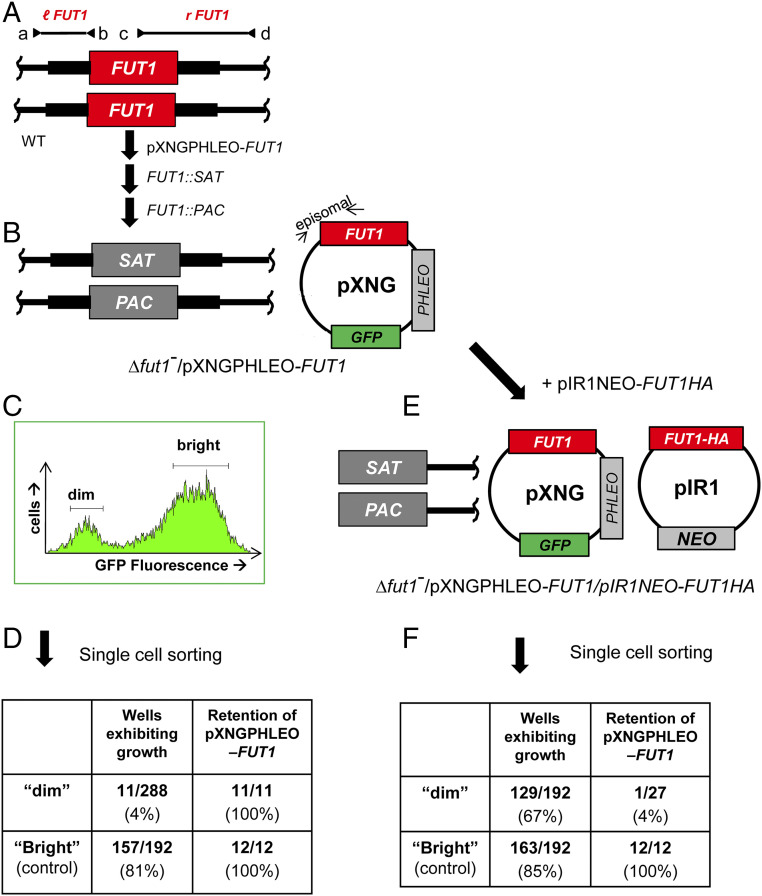
Anticipating that FUT1 could encode an essential fucosyltransferase, we employed a plasmid segregation-based strategy, in which a counter selectable ectopic copy of FUT1 was introduced prior to ablation of the chromosomal FUT1 alleles (45). Wild-type (WT) L. major parasites were first transfected with an episomal pXNGPHLEO-FUT1 construct, followed by successive inactivation of the two chromosomal FUT1 alleles using SAT and PAC targeting constructs. PCR tests confirmed that the marker replacements occurred as planned and the chromosomal alleles of FUT1 were precisely deleted, with retention of the episomal FUT1 (Fig. 2 A and B and SI Appendix, Fig. S3). For simplicity, chromosomal deletions are hereafter referred to as ∆fut1−, with this line termed ∆fut1−/+pXNGPHLEO-FUT1.
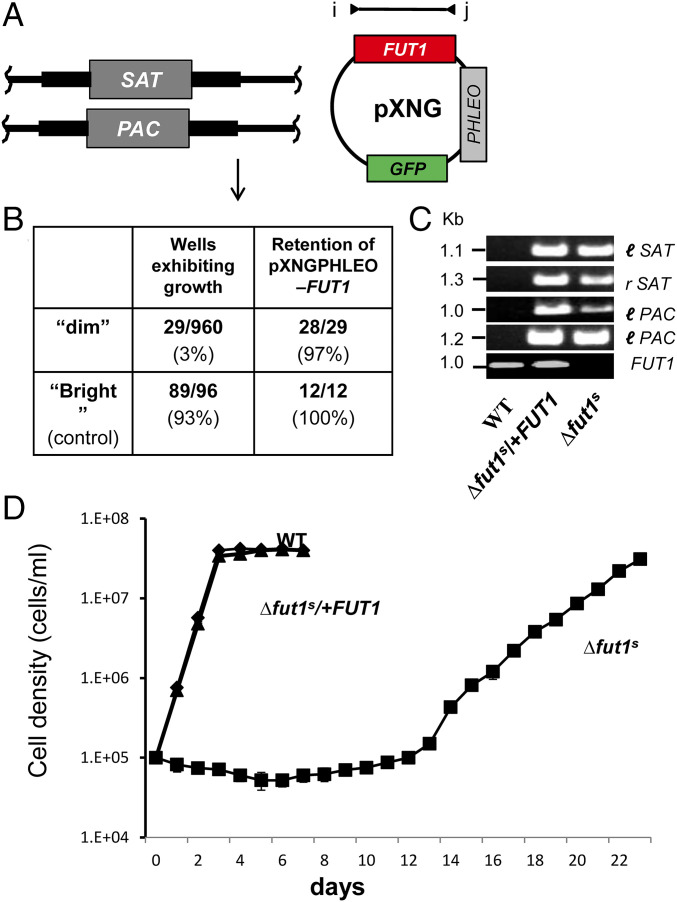
Fig. 2.
Plasmid segregation tests show FUT1 is essential. (A) FUT1 locus and transfection. The figure shows the WT FUT1 locus, with the ORF as a central red box and the flanking sequences used for homologous replacement by SAT or PAC drug resistance cassettes (FUT1::SAT or FUT1::PAC, respectively) shown as heavier flanking lines. The location of “outside” primers used to establish planned replacements (a/b or c/d; to confirm replacements SAT or PAC specific primers were substituted for b or c; SI Appendix, Fig. S3) are shown. WT L. major was first transfected with pXNGPHLEO-FUT1, and successively with the FUT1::SAT and FUT::PAC constructs. (B) The chromosomal null Δfut1− mutant obtained in the presence of episomal FUT1. The genotype of this line is FUT::∆SAT/FUT1::∆PAC/ +pXNGPHLEO-FUT1, abbreviated as Δfut1−/+pXNGPHLEO-FUT1. PCR confirmation tests of the predicted genotype are found in SI Appendix, Fig. S3. pXNG additionally expresses GFP (45). (C) Plasmid segregation tests of FUT1 essentiality. Δfut1−/+pXNGPHLEO-FUT1 was grown for 24 h in the absence of phleomycin, and analyzed by GFP flow cytometry. The two gates used for subsequent quantitation and/or sorting of parasites are shown; dim or weakly fluorescent (2–10 FU) and bright or strongly fluorescent (100–1,000 FU). (D) Results of sorting experiment shown in C. Single cells from both GFP dim or bright populations sorted into 96-well plates containing M199 medium, and incubated for 2 wk, at which time growth was assessed. The numbers sorted and their growth and properties are shown, with bright cells mostly surviving and dim cells mostly not. All 11 survivors from the dim and 12 tested from the bright population sort were confirmed to retain of pXNGPHLEO-FUT1 by growth in media containing phleomycin. (E) Generation of line required for plasmid shuffling. Δfut1−/+pXNGPHLEO-FUT1 was transfected with pIR1NEO-FUT1-HA expressing C-terminally tagged FUT1. PCR tests confirming the predicted genotype Δfut1−/+pXNGPHLEO-FUT1/+pIR1NEO-FUT1-HA are shown in SI Appendix, Fig. S3. This line was then grown 24 h in the absence of phleomycin and subjected to cell sorting and growth tests as described in C. (F) Results of sorting experiment described in E. In this experiment, both dim and bright cells mostly survived (67% and 85%, respectively). Twenty-seven of the dim cells were tested, of which 26 had lost pXNGPHLEO-FUT1 by PCR tests, thereby representing the desired Δfut1−/+pIR1NEO-FUT1-HA.
For plasmid segregation tests, parasites were grown briefly (24 h) in the absence of phleomycin to allow loss of pXNGPHLEO. Parasites were then analyzed for GFP expression by flow cytometry, as a measure of pXNG copy number (45). Two populations emerged: a large population of “bright” cells showing strong fluorescence bearing high copy numbers of pXNGPHLEO-FUT1(>200 fluorescent units [FU]); and a smaller population of “dim” cells, exhibiting control/background fluorescence levels (2–20 FU), lacking pXNG-FUT1 completely, or bearing only a few copies (Fig. 2C).
Fluorescence-activated cell sorting was then used to recover single cells into individual wells of a 96-well microtiter plate, containing M199 medium without phleomycin. For the bright cell population, 157/192 cells inoculated with single bright parasites grew out (81%), representing the “cloning/plating” efficiency of cells subjected to this protocol. In contrast, growth was seen in only 11/288 of the dim parasites tested similarly (4%; Fig. 2D). The 11 survivors still retained pXNGPHLEO-FUT1 as judged by their ability to grow out in the presence of phleomycin and expression of GFP, likely arising by imperfect sorting or other technical factors as seen previously (45). After correction for the plating/cloning efficiency, we estimated that FUT1-null mutants were not obtained from 236 cells tested. This greatly exceeds the resolution of classic “failed to recover deletion” protocols and suggests that, under these conditions, FUT1 is essential.
FUT1 Is Localized in Mitochondria.
To confirm the surprising prediction of mitochondrial localization, we expressed a C-terminal HA-tagged FUT1 protein using the pIR1NEO vector as an episome; this construct was introduced into the ∆fut1−/+pXNGPHLEO-FUT1 line, yielding ∆fut1–/+pXNGPHLEO-FUT1/pIR1NEO-FUT1-HA (Fig. 2E). We then used “plasmid shuffling” to select for a line that had lost the untagged pXNG-FUT1 construct but maintained tagged pIR-FUT1-HA. Following single-cell sorting, 85% of the bright-cell wells grew out (Fig. 2F). Similarly, 67% of the dim cells also grew out, and subsequent tests showed that 96% (26/27) tested lacked GFP and were phleomycin sensitive, while retaining tagged FUT1-HA, yielding ∆fut1−/+ pIR1NEO-FUT1-HA (Fig. 2F). These data confirmed the functionality of FUT1-HA.
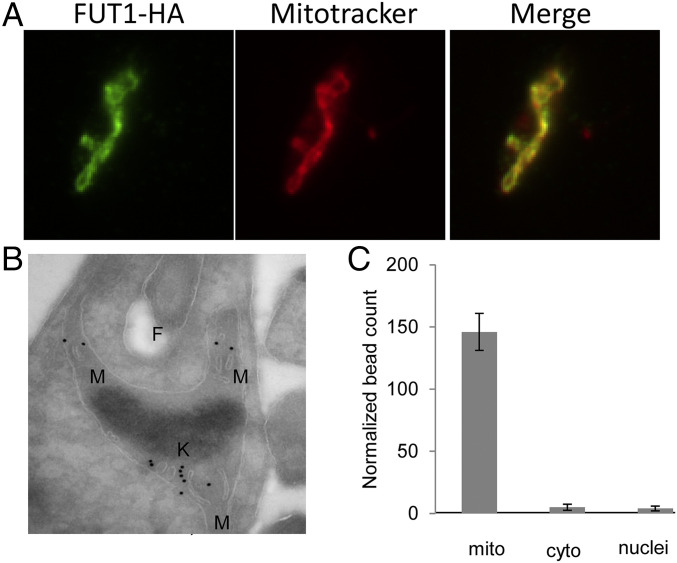
Immunofluorescence analysis (IFA) was used first to assess localization of HA-tagged FUT1 in the ∆fut1−/+pIR1NEO-FUT1-HA line. Anti-HA antibody was used to stain the parasites after labeling with mitochondrial marker MitoTracker Red, which revealed complete colocalization (Pearson’s correlation coefficient = 0.97 ± 0.03; Fig. 3A).
Fig. 3.
LmjFUT1 is localized to the parasite mitochondrion. (A) Indirect immunofluorescence of parasites lines of Δfut1−/+pIR1NEO-FUT1-HA incubated with rabbit anti-HA antibody and visualized with goat anti-rabbit Alex 488-conjugated antibody (leftmost image, green), or MitoTracker Red CMXRos (central image, red), and merged (rightmost image, yellow) are shown. Colocalization by Pearson’s correlation coefficient was 0.97. (B) Cryo-immuno-EM of Δfut1−/+pIR1NEO-FUT1-HA. FUT1-HA was visualized using rabbit anti-HA followed by gold-bead–conjugated mouse anti-rabbit IgG. F, flagellar pocket; K, kinetoplast DNA network; M, mitochondrion. Controls in which the primary anti-HA antibody was omitted yielded no bead counts. (C) Quantitation of cryo-immuno-EM anti-HA bead labeling to FUT-HA in cellular compartments. The data shown are the bead counts and SD, taken from three different experiments comprising three sets of 10 sections each, and normalized to the relative compartment area.
Cryo-immunoelectron microscopy (cryo-immuno-EM) using rabbit anti-HA sera followed by gold-bead–conjugated anti-rabbit sera showed binding primarily to the mitochondrion, generally within the lumen (Fig. 3B). Consistently, no beads were evident with the primary sera omitted. Quantitation of bead density showed more than 94% binding to the mitochondrion, far greater than seen to other compartments (Fig. 3C). Similar results were obtained with C-terminal HA-tagged FUT1 expressed in WT cells. In contrast, expression of an N-terminally HA-tagged FUT1 in WT parasites failed to give any signal in Western blotting or IFA using anti-HA antibody, consistent with the prediction that N-terminal FUT1 functions as a mitochondrial targeting peptide, which would be expected to be cleaved off during import.
We tested deletions of the extended LmjFUT1 MTP for mitochondrial localization and/or function (SI Appendix, Fig. S4). A 16-aa truncation (MSKAR) was fully functional and mitochondrially targeted, a 17-aa truncation (MKAR) was partially functional and mitochondrially targeted, and a 18-aa truncation (MAR) was not functional but partially mitochondrially targeted (SI Appendix, Fig. S4). These data suggest first that LmjFUT1 may possess a cryptic MTP resembling the N terminus of TbFUT1, which we show elsewhere is also localized to the mitochondrion (46). These data further suggest that in trypanosomatids critical residues of the Motif IV region may extend functionally further toward the N terminus than in other species.
Recombinant FUT1 Is an Active Fucosyltransferase.
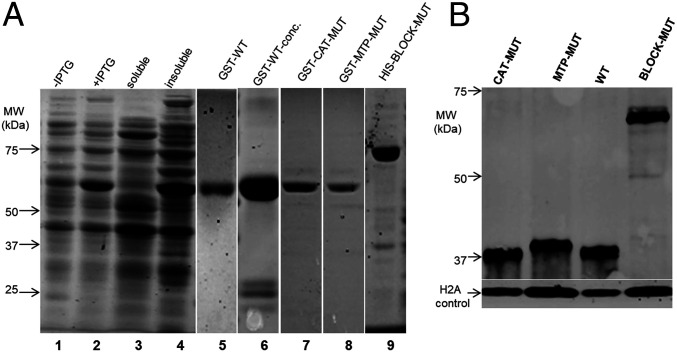
When expressed in Escherichia coli, most FUT1 was insoluble, despite trials with several protein fusion expression systems, a common observation for eukaryotic glycosyltransferases (47) (Fig. 4A, lanes 1–4). The best results were often GST-FUT1 fusions, yielding a small fraction of soluble recombinant protein (<5%), which could be purified by GST affinity chromatography (Fig. 4A, lanes 5 and 6). The fusion protein had an apparent molecular weight of 66 kDa similar to the theoretical value (64,960 Da), and the major impurity appeared to be GST (Fig. 4A, lane 6). All preparations tended to be unstable, and assays were performed on freshly purified material including an active acceptor control (below). Attempts to cleave the GST tag from the fusion protein using factor Xa yielded little free FUT1, and thus the fusion protein was used for enzymatic characterizations.
Fig. 4.
Expression of WT or mutant FUT1s in E. coli or within Leishmania. (A) Expression and purification of recombinant GST-FUT1 fusion proteins from E. coli visualized following SDS-PAGE. Lanes 1–6 are GST-FUT1 (WT). Lane 1: before-induction whole-cell lysates; lane 2: postinduction whole-cell lysates; lane 3: soluble fraction; lane 4: insoluble fraction; lane 5: elution; lane 6: concentrated eluted protein; lane 7, purified GST-CAT-MUT; lane 8, purified GST-MTP-MUT; and lane 9, purified HIS-BLOCK-MUT. Molecular weight markers are shown on the left. The images from lanes 1–4 and 5–9 are from separate experiments. (B) Western blot analysis of C-terminal HA-tagged FUT1s expressed in Leishmania. Lysates from parasites expressing the indicated HA-tagged FUT1s in a ∆fut1−/+pXNGPHLEO-FUT1 background are shown (the presence of untagged WT FUT1 was required as none of the mutants were viable in its absence; Fig. 6). Western blots were performed with anti-HA to visualize the tagged FUT1 expression, and anti–L. major H2A as a loading control.
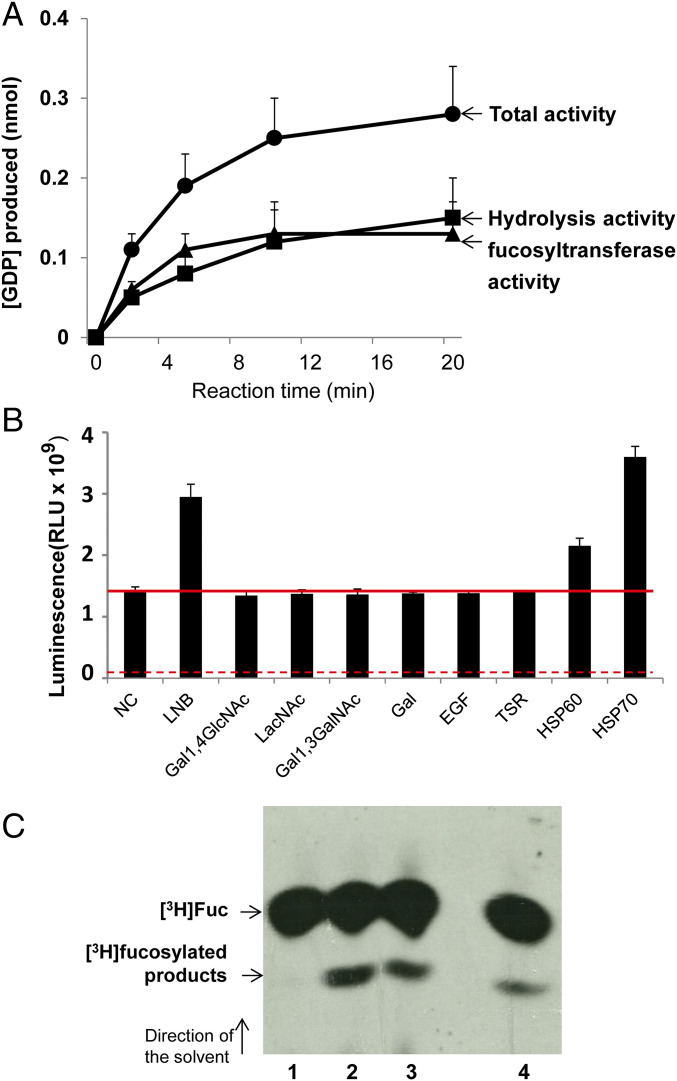
To assay recombinant GST-FUT1, we used an indirect assay that measures GDP arising from metabolism of highly purified GDP-Fuc, through coupling to ATP synthesis and measurement by a luciferase/luciferin reaction (48). Initially, recombinant GST-FUT1 was incubated with GDP-Fuc as donor and β-d-Gal-(1→3)-d-GlcNAc (lac-N-biose [LNB]) as acceptor. A reaction time course established the product to increase linearly for the first 6 min and to plateau by 20 min (Fig. 5A).
Fig. 5.
LmjFUT1 is a fucosyltransferase. (A) Time course of GDP production with GST-FUT1 in the presence or absence of GDP-Fucose and 1 mM LNB. Solid circles, incubation in the presence of GDP-Fucose + LNB (“total activity”); solid squares, incubation with GDP-Fucose alone (hydrolysis activity); solid triangles, difference between presence and absence of LNB (acceptor-dependent fucosyltransferase activity). (B) GST-FUT1 was incubated with GDP-Fucose in the presence of various acceptors, and the luminescence from GDP produced was measured after 5 min of incubation. Lane 1, NC, no acceptor (hydrolysis only); lane 2, LNB; lane 3, Gal1,4GlcNAc; lane 4, LacNAc; lane 5, Gal1,3GalNAc; lane 6; galactose; lane 7, EGF, EGF-like repeat; lane 8, thrombospondin like repeat; lane 9, mHSP60 peptide; and lane 10, mHSP70 peptide. The red horizontal line represents the hydrolysis activity (lane 1). The solid red line shows activity in the absence of acceptor or with inactive acceptors; the dashed red line marks the background seen in the absence of enzyme (about 7 × 107 RLU under these conditions vs. 1.3 × 109 RLU in the presence of enzyme). (C) Fucosyltransferase activity assayed in L. major in vivo. Whole-cell lysates were incubated with anti-HA agarose beads, which were washed, and then incubated with 1 μCi GDP-[3H]Fuc and 1 mM LNB as acceptor. Reaction products were separated by HPTLC and detected by fluorography as shown. Lane 1, WT; lane 2, Δfut1−/+pIR1NEO-LmjFUT1-HA; lane 3, Δfut1−/+pIR1NEO-TbFUT1; lane 4, WT/+pIR1NEO-Lmj-FUT1-HA. Elsewhere, we show that the level of free [3H]Fuc is considerably lower in the absence of enzyme than in its presence (46).
In the absence of the acceptor, GDP-Fuc was hydrolyzed and an increase in GDP could be detected (Fig. 5A, curve labeled “hydrolysis activity”). Such acceptor-independent activity has been described previously for other FUTs in the absence or with suboptimal substrates (48, 49). In the presence of LNB acceptor, GDP formation increased approximately twofold (Fig. 5B, curve labeled “total activity”). Acceptor-dependent fucosyltransferase activity was calculated from the difference between these two curves (Fig. 5A; curve labeled “fucosyltransferase activity”). Initial rate data were calculated from a 5-min reaction, and established that the optimum conditions for the coupled reaction were 37 °C, pH 6, with LNB as an acceptor, yielding a specific activity of recombinant GST-FUT1 of 59 ± 5.4 nmol⋅min−1⋅mg−1 protein (Fig. 6E). In the presence of a good acceptor hydrolysis activity might decrease, which would be undetectable in this assay. LmjFUT1 lacked activity with the glycan acceptors N-acetyllactosamine (LacNAc), β-galactose, and Galβ1,3GalNAc (Fig. 5B). Overall, the activity seen with the substrates tested resembled that seen for TbFUT1, where a wider range was tested (46).
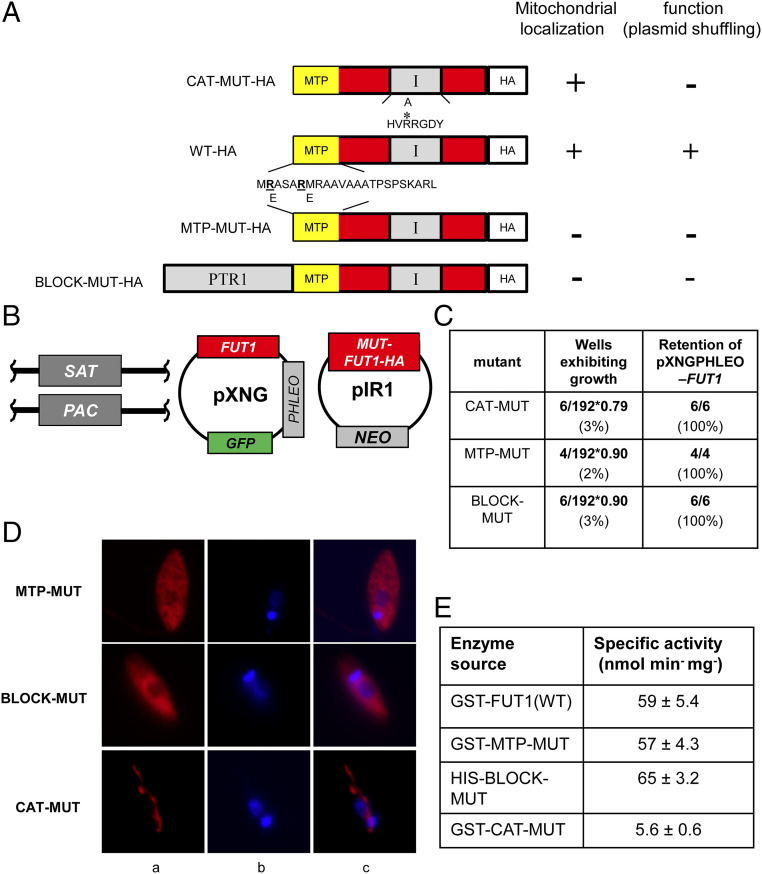
Fig. 6.
Both mitochondrial localization and fucosyltransferase activity are required for the essential function of FUT1. (A) Depiction of mutant LmjFUT1-HA designed to block mitochondrial import or catalysis. The predicted MTP is shown in yellow, the catalytic motif I is shown in gray, and the HA tag is shown in white, with the remainder of FUT1 in red. CAT-MUT-HA has an R297A substitution; MTP-MUT-HA replaces two Glu residues in the MTP with Arg; BLOCK-MUT-HA has the cytoplasmic protein PTR1 fused to the N terminus. The results of mitochondrial localization tests (D) and plasmid shuffling tests (C) are summarized on the Right of the illustrations. (B) Scheme of plasmid shuffling to test the function of mutant FUT1s. HA-tagged mutant FUT1s were expressed from pIR1NEO in the Δfut1−/+pXNGPHLEO-FUT1 line. Growth in the absence of phleomycin and FACS sorting of dim and bright cells was performed as in Fig. 2. (C) Plasmid shuffling tests of FUT1 mutants. Δfut1−pXNG-FUT1/pIR-MUT-HA lines was grown for 24 h in the absence of phleomycin, and analyzed by GFP flow cytometry. In all tests, survival of bright (control) cells was 79–90%. The results show that few dim cells yielded growth, and all of those arose from incomplete sorting (retention of pXNGPHLEO-FUT1). (D) Indirect immunofluorescence of HA-tagged FUT1 expressed from pIRNEO in the Δfut1−/+pXNGPHLEO-FUT1 background. Column a, anti-HA (red); column b, DNA (Hoechst, blue); and column c, merge of columns a and b. (E) Acceptor-dependent specific activity of purified recombinant FUT1 proteins assayed by GDP formation in the presence of GDP-Fucose and LNB. The average and SD of three preparations are shown.
We confirmed the fucosyltransferase activity of FUT1 using enzyme expressed within Leishmania. Whole-cell lysates of WT or ∆fut1−/pIRNEO-FUT1-HA were applied to anti-HA-agarose beads, which were washed and then incubated with GDP-[3H]Fuc and LNB. With both cell lines, a new product was observed, similar to that seen with a control expressing TbFUT1, shown elsewhere to be to Fucα1,2Galβ1,3GlcNAc (46). As expected, no products other than [3H]Fuc were found with WT lacking tagged FUT1, again attributed to hydrolysis of the GDP-Fuc substrate (Fig. 5 A and C). As the ∆fut1−/pIR1NEO-TbFUT1 mutant was obtained by plasmid shuffling in a ∆fut1− background (Materials and Methods), these data proved this heterologous enzyme could satisfy the Leishmania FUT1 requirement (Fig. 5C, lane 3).
Two peptide substrates for mammalian protein-O-transferases 1 and 2, corresponding to the epidermal growth factor-like repeats (EGFR) or thrombospondin-like repeats (TSR) [kindly provided by R. Haltiwanger, University of Georgia, Athens, GA (50, 51)] lacked activity with LmjFUT1 (Fig. 5B). We synthesized peptides bearing 5 aa on either side of the proposed fucosylation sites of the Leishmania infantum mitochondrial HSP60 or HSP70 (33), which both showed significant acceptor-dependent fucosyltransferase activity (Fig. 5B). To confirm this unexpected activity against a peptide substrate, we incubated LmjFUT1 with the mHSP70 peptide and GDP-Fucose, and analyzed the product by higher-energy collisional dissociation (HCD) MS; fucosylation of the central target serine residue was clearly evident (SI Appendix, Fig. S5). MS methods for detecting peptide fucosylation vary greatly in their efficiency (52, 53), and the GDP-Glo assay may provide a better quantitative view of the efficiency of the peptide substrates. Thus, LmjFUT1 could recognize a diverse spectrum of acceptors in vitro, including both glycan and peptide substrates.
Mitochondrial Localization Is Required for the Essential Role(s) of FUT1.
While FUT1 was found primarily in the mitochondrion (Fig. 3), occasionally the predominant distribution of a protein may be misleading about its true site of function (54). Thus, we asked whether mitochondrial localization was required for FUT1 function, by testing mutants where mitochondrial targeting was compromised (Fig. 6). Guided by characteristic features of MTPs (55), we replaced the first two Arg residues with Glu, predicted to reduce the probability of mitochondrial import from 88 to 3% (MTP-MUT, Fig. 6A). Similarly, we fused the cytosolic protein PTR1 to the N terminus of FUT1 (BLOCK-MUT), anticipating that the large 30-kDa PTR1 would block MTP recognition (Fig. 6A). Both modifications were made with the C-terminally HA-tagged FUT1 expressed in pIR1NEO, rendering them suitable for immunodetection and functional tests (Fig. 6B). Plasmid shuffling tests showed that neither MTP-MUT nor BLOCK-MUT could substitute for WT-FUT1, as only 2–3% of the dim cells survived (vs. 90–99% for bright cells; Fig. 6C). Tests showed that the few surviving dim cells had retained the WT FUT1 plasmid and thus were not true segregants (Fig. 6C).
Immunofluorescence analysis of the tagged proteins showed that, unlike the HA-tagged WT FUT1, HA-tagged MTP-MUT and BLOCK-MUT proteins were now localized to the cytoplasm (Fig. 6D). Two essential controls were performed to validate the conclusion above. First, Western blot analysis showed that despite their cytosolic localization, expression of the MTP-MUT or BLOCK-MUT proteins was comparable to that of the WT FUT1 in Leishmania, relative to an H2A loading control (SI Appendix, Fig. 4B). Consistent with the cytosolic localization and lack of MTP cleavage, the size of MTP-MUT-HA was slightly greater than WT-HA, about 40 kDa, in good agreement with the predicted uncut protein (39.5 kDa; Fig. 4B). Second, we expressed and purified tagged MTP-MUT and BLOCK-MUT proteins in E. coli, and showed that their specific FUT activity was the same as WT (Fig. 6E). Thus, the failure of the mutants to rescue the chromosomal ∆fut1− mutant did not arise because of a lack of activity, instead rising solely from their failure to enter the mitochondrion.
Fucosyltransferase Activity Is Required for the Essential Role(s) of FUT1.
We next tested whether the essential function of FUT1 required fucosyltransferase activity (56). We mutated the first arginine to alanine (R297A) in the FUT1 motif I (Fig. 1B), conserved in many fucosyltransferase families (39) and mutation of which causes 97–100% loss of in catalytic activity (40, 57). A C-terminal HA-tagged catalytic FUT1 mutant (CAT) was expressed using the pIR1NEO vector, and immunofluorescence assays showed it remained in the mitochondrion like WT (Fig. 6D). However, when subjected to plasmid shuffling only 3% of the dim cells grew out, vs. 79% for bright cells (Fig. 6C). Subsequent tests of the dim survivors showed they still retained the WT FUT1 plasmid (Fig. 6C).
These data suggested that CAT mutant was unable to support Leishmania growth. Again, two controls strongly support this interpretation. First, Western blot analysis showed that the level of CAT mutant expression within Leishmania was comparable to that of a tagged WT FUT1, relative to an H2A loading control (Fig. 4B). As expected, the size of CAT-HA was similar to WT FUT1-HA, indicating that it was similarly cleaved during import (Fig. 4B). Second, we expressed and purified the CAT mutant protein from E. coli, and assayed FUT activity. While the behavior and yield during purification of this enzyme were similar to the WT and MTP mutants, the CAT-MUT only retained less than 10% of WT activity, very close to the background of this assay (Fig. 6E).
Collectively, these data argue that expression of an enzymatically active mitochondrion fucosyltransferase is required for Leishmania viability.
Recovery of a Rare ∆fut1s Mutant by Plasmid Segregation in Rich Medium.
The segregation tests above yielding no FUT1-null cells were performed in standard Leishmania M199 media (Fig. 2). Reasoning that a richer media might spare mitochondrial deficiencies arising from FUT1 loss, we repeated this experiment several times using a richer medium, Schneider insect medium. In multiple experiments collectively screening nearly 1,000 cells, few dim cells survived as before (Fig. 7B), even after extended incubations. However, in just one experiment a dim clone was recovered which completely lacked FUT1 by PCR tests (Fig. 7 A–C). When both WT and the sole fut1-null mutant (here termed ∆fut1s) were inoculated into fresh media, WT cells doubled after about 8 h, while ∆fut1s increased slowly in cell number for the first 2 wk, after which it grew somewhat more rapidly and eventually reached similar cell density as WT (Fig. 7D). This pattern of very slow initial growth repeated in subsequent passages, although after extended passage the growth increased somewhat. The growth defect of primary ∆fut1s could be fully restored by FUT1 reexpression (Fig. 7D), as could all other defects described below.
Fig. 7.
Recovery of a single FUT1-null mutant segregant (∆fut1s). (A) Starting cell line for plasmid segregation in rich culture medium. Δfut1−/+pXNGPHLEO-FUT1 cells were grown and subjected to single-cell sorting in Schneider’s media. (B) Control bright cells showed a typically high frequency of wells supporting growth (93%), while few dim cells survived as in M199 medium (Fig. 2). Of the 29 survivors, 28 retained the pXNGPHLEO plasmid, while 1 designated ∆fut1s did not. (C) PCR tests of ∆fut1s, WT and a ∆fut1s /pXNGPHLEO-FUT1 “addback.” Primer sets l-SAT, r-SAT, l-PAC, and r-PAC confirm ∆fut1s lacks chromosomal FUT1 (SI Appendix, Fig. S3) and FUT1 ORF primers (l, 3955; j, 3956) confirm the absence of FUT1 sequences in ∆fut1s and its presence in the addback. (D) Growth of WT, ∆fut1s, and ∆fut1s/+pXNGPHLEO-FUT1 in M199 medium. Parasites were inoculated at a density of 105/mL and growth was followed by Coulter counting.
∆fut1s Showed Ultrastructural and Functional Mitochondrial Changes.
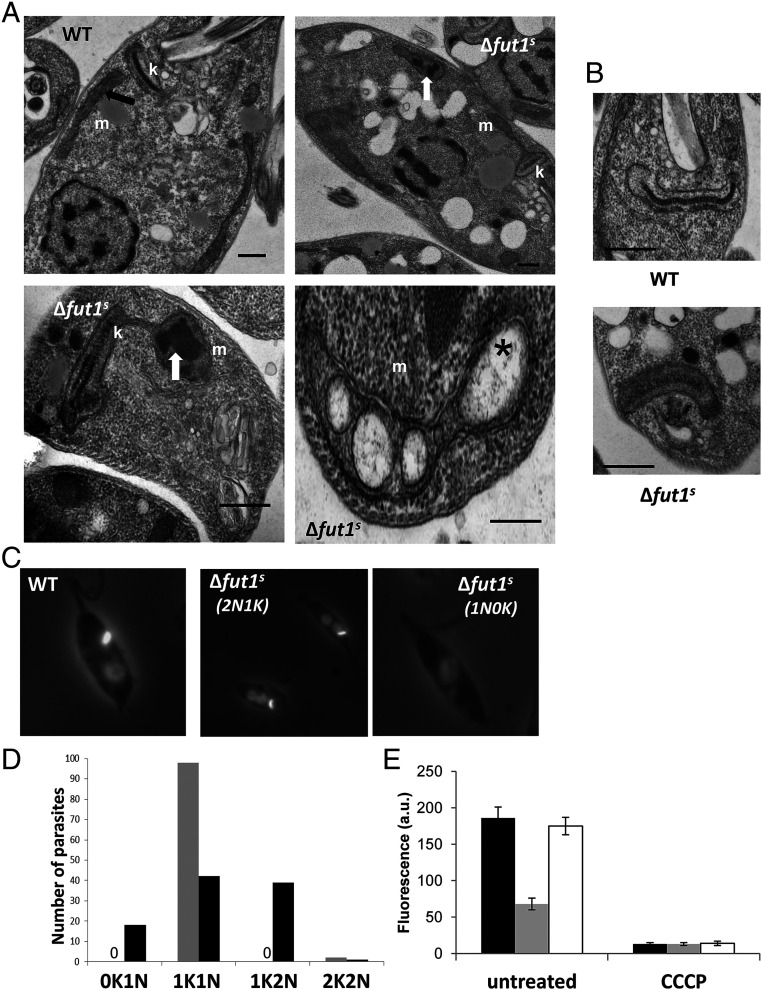
Transmission EM of ∆fut1s revealed changes in the ultrastructure of mitochondria, with swelling and bloated cristae (Fig. 8A) often containing dark electron-dense aggregates, similar to that seen previously in disrupted cristae associated with dysfunctional mitochondria (58). The kDNA network showed alterations in size, decreasing 80% in length while increasing 12% in thickness relative to WT (Fig. 8B and SI Appendix, Fig. S6). DAPI staining of the nucleus and kinetoplast DNA network revealed that while WT parasites mostly showed a typical one kinetoplast–one nuclei (1K1N) pattern (59), only 42% of ∆fut1s were 1K1N, with 39% 1K2N, and remarkably, 18% lost the kDNA network entirely (0K1N; Fig. 8 C and D). Few abnormalities were observed in the ultrastructure of other organelles such as the nucleus, endoplasmic reticulum, flagellum, and acidocalcisomes (Fig. 8 A and B).
Fig. 8.
∆fut1s shows multiple mitochondrial abnormalities. (A) transmission EM of WT (subpanel a) and ∆fut1s (subpanels b–d). m, mitochondria; k, kinetoplast; black arrow, normal mitochondrial cristae; white arrows, aggregates inside mitochondria; star, bloated cristae. (Scale bars: 500 nm.) (B) Ultrastructural analysis of kDNA in WT and ∆fut1s. While WT cell presents typical compact kDNA structure, most mutant cells show a “looser” kDNA network that is wider with decreased length (see SI Appendix, Fig. S6 for quantitation). (Scale bar: 500 nm.) (C) Loss of kDNA network in ∆fut1s. WT or ∆fut1s were stained with DAPI to visualize the kDNA network (small bright spot) and nucleus (dimmer large circle). A typical one kinetoplast/one nucleus (1K/1N) pattern is shown for WT in the leftmost panel; the central panel shows a ∆fut1s cell with a 1K/2N pattern, and the rightmost panel shows a ∆fut1s cell lacking kDNA (0K/1N). (D) Quantitation of kDNA/nucleus patterns seen in WT (gray bars) vs. ∆fut1s (dark bars). A “0” indicates no cells of that pattern. (E) Mitochondrial potential assessed by staining with TMRE. Parasites were incubated with 100 nM TMRE and analyzed by flow cytometry, with the signal on the FL-2 channel expressed in arbitrary units (a.u.). The left portion shows cells incubated for 15 min; the right portion shows cells further incubated with 300 µM cyanide m-chlorophenylhydrazone (CCCP) for 60 min. Lines tested are as follows: WT, black bar; ∆fut1s, gray bar; and ∆fut1s/+pXNGPHLEO-FUT1, white bar.
We assessed the mitochondrial membrane potential (∆Ψm) of ∆fut1s using tetramethylrhodamine ethyl ester (TMRE), a cationic lipophilic dye whose accumulation depends on mitochondrial membrane potential. Logarithmic phase ∆fut1s incubated with TMRE showed only 36% of the WT fluorescence (Fig. 8E), which returned to normal upon restoration of FUT1 (Fig. 8E, panel labeled “untreated”). As a control, cells were treated with the protonophore carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (CCCP) to induce loss of mitochondrial membrane potential, which yielded greatly decreased TMRE fluorescence (Fig. 8E, panel labeled “CCCP”). Thus, loss of FUT1 results in a profound mitochondrial dysfunction.
Discussion
Previous studies have shown that GDP-Fucose synthesis is essential in trypanosomatids, yet for Leishmania and Trypanosoma brucei fucosylated molecules that might account for this requirement remained unknown. We reasoned that identification of essential fucosyltransferase(s) and characterization of their substrate specificities would better inform our search for the postulated essential fuco-conjugates. Given the structural similarity of l-Fucose and d-arabinopyranose and the tendency of enzymes using either substrate (or their activated GDP derivatives) to accept the other, we searched in Leishmania genomes for proteins showing relationship to diverse collection of prokaryotic and eukaryotic fucosyl- or d-arabinosyl transferases (Table 1). Four candidates were predicted to be targeted to the secretory pathway, as typical for most parasite glycosyltransferases. SCA1 and SCA2 encoded transferases mediating developmentally regulated side-chain d-arabinopyranosylation of LPG within the Golgi apparatus, and unlikely to be essential given that LPG is not (14). The role(s) of SCAL and FUT2 are unknown (Table 1); however, both genes could be readily deleted with little apparent consequence in vitro (SI Appendix, Figs. S1 and S2). Among possible roles could be d-Arap modification of GIPLs (60), especially as GIPLs are not required for survival (11).
FUT1, in contrast, was conserved in both African and South American trypanosomes, and uniquely, was predicted and then experimentally confirmed to be targeted exclusively to the parasite mitochondrion, most likely within the lumen (Figs. 1 and 3). By plasmid segregation tests, FUT1 appeared to be essential for normal growth (Fig. 2). As mitochondrial glycosyltransferases are uncommon, we used a “plasmid shuffle” assay to test whether mitochondrial localization was required by the parasite, examining FUT1s with mutated or blocked MTPs. Disruption of mitochondrial targeting was found as expected, but these were unable to satisfy the WT FUT1 requirement (Fig. 6). We similarly explored whether the FUT1 catalytic activity was required for growth, mutating conserved putative active site residues, and again, the mutant enzyme could not satisfy the FUT1 requirement (Fig. 6). Importantly, all mutant FUT1 proteins were expressed at levels comparable to WT in vivo (Fig. 4B), and in vitro assays of recombinant enzymes showed similar activities for the WT and mitochondrial targeting mutants, while the catalytic mutant showed near-background activity (Fig. 6E). These data established that both mitochondrial localization and catalytic activity were required for essential FUT1 function.
One advantage of the plasmid segregation test for essentiality is that it allows tests of many more events than typical transfection-based approaches (45). In standard media, we tested more than 200 events (Fig. 7), and once FUT1’s mitochondrial role was recognized, we repeated these studies in richer media potentially sparing mitochondrial function. However, despite testing collectively more than 1,000 estimated events, we were able to obtain only a single mutant completely lacking FUT1 entirely (∆fut1s; Figs. 7 and 8). Notably, ∆fut1s displayed exactly the phenotypes expected for a dysfunctional mitochondrial mutant, including slow growth, structural alterations of cristae, mitochondrial inclusions and abnormalities, alterations and even loss of the kinetoplast DNA network, and greatly decreased mitochondrial membrane potential (Figs. 7 and 8). Importantly, restoration of FUT1 expression (∆fut1s/+FUT1) completely reversed these defects back to WT. Nonetheless, the question remains as to why only a single ∆fut1s could be obtained; one explanation is that it bears a second site compensating mutation, albeit one unable to restore normal mitochondrial function. For example, in trypanosomes, compensating mutations in the ATP synthase gamma-subunit are able to rescue kDNA deficiency (61). Future studies will address these questions.
Thus, cell biological, genetic manipulation and the severe mitochondrial dysfunction of a ∆fut1s mutant all establish the localization and critical functionality of FUT1 within the parasite mitochondrion. The questions now become what are the natural FUT1 acceptor(s) and products therein, and what role do they play in essential mitochondrial biology? Remarkably, despite knowledge of essential GDP-Fucose synthesis (28, 30, 31), and extensive work characterizing parasite glycoconjugates since the early 1980s, few candidate fucosylated molecules have been found in any Leishmania species, and none in L. major. Several L. donovani proteins exhibited MS/MS signatures suggestive of fucosylation (33). Two were mitochondrial HSPs, peptides of which showed fucosylation activity in the GDP-Glo assay in vitro (Fig. 5B) and confirmed by MS of the HSP70 peptide product (SI Appendix, Fig. S5). Going forward, global or mitochondrial-targeted proteomic screens will be necessary to ascertain whether the in vitro results with peptides extrapolate more generally across the Leishmania proteome.
Similarly, comparisons of WT and ∆fut1s using a variety of antisera or lectins specific for fucoconjugates have proven uninformative so far, as was biorthogonal labeling with 6-alkynl fucose (specific methods are provided in SI Appendix). Failure to detect FUT1-dependent mitochondrial products might signify 1) low abundance below the detection limits, 2) inability of modified fucose derivatives such as 6-alkynyl fucose to be properly transported, activated or resist degradation within the mitochondrion, or 3) further incorporation and/or metabolism of fucose to forms not recognizable by the methods used here. A variety of fucose modifications have been described in prokaryotes, and the internal fucose within the SKP1 pentasaccharide (d-Galα1–3-d-Gal [or Glc] α1–3-l-Fucα1–2-d-Galβ1–3-d-GlcNAcα1-) of several single-celled eukaryotes is not recognized by lectins or antibodies tested here (62). Studies searching for the cellular FUT1-dependent products in T. brucei have been thus far similarly unrequited (46). Future studies will be necessary to resolve the nature of the GDP-Fucose and FUT1-dependent product(s) in trypanosomatids, which genetic and biochemical data strongly predict must nonetheless exist.
We did not deeply explore the glycan acceptor specificity of LmjFUT1, with the best acceptor being LNB and little activity with others tested (Fig. 5B). The presence of significant GDP-Fuc hydrolysis with all substrates tested may be a sign that the native acceptor moiety has yet to be identified (Fig. 5). More extensive studies with TbFUT1 showed a preference for lac-N-biose and its β-methyl glycoside, again typically accompanied by significant background hydrolysis (46). The strong homology between these two species enzymes and the ability of TbFUT1 to satisfy the Leishmania FUT1 requirement (Fig. 5C) (46) suggests that biologically relevant specificities may be similar in both species. While two substrates for mammalian protein O-fucosyltransferase were inactive with LmjFUT1, two peptides predicted to be fucosylated in L. donovani functioned as acceptors, and MS/MS of the one tested confirmed fucosylation (Fig. 5B and SI Appendix, Fig. S5). Peptide substrates have yet to be tested with the TbFUT1, which may or not behave similarly to LmjFUT1. Thus, LmjFUT1 is unusual in its ability to fucosylate a diverse spectrum of substrates in vitro.
While intramitochondrial glycosylation is uncommon, that mediated by the mitochondrial isoform of the mammalian O-GlcNAc transferase has received considerable attention (63, 64). Proteomic surveys have revealed ∼100 O-GlcNAc modified mitochondrial proteins (65–67), mostly encoded by nuclear genes raising the possibility of glycosylation prior to mitochondrial import (68). However, to date no mechanistic studies establish that glycosylated proteins can be imported into mitochondria, while several proteins encoded by mitochondrial DNA are glycosylated (thus rendering cytoplasmic activity unlikely), and mitochondrial transporters able to transport UDP-GlcNAc have been identified (66, 67, 69, 70), in support of the intramitochondrial glycosylation paradigm. Importantly, O-GlcNAc modifications of mitochondrial proteins have been postulated to play key roles in mitochondrial energy metabolism and function (66, 67), and an analogous role could be imagined for intramitochondrial fucosylation in Leishmania. Potentially, trypanosomatid mitochondrial FUT1s may offer a facile system in the future for probing mitochondrial glycosylation in a setting uncomplicated by multiple isoforms targeted to diverse compartments. Moreover, its essentiality for normal growth and mitochondrial function renders FUT1 an attractive target for chemotherapy of trypanosomatid parasites, where numerous current drugs inhibit mitochondrial pathways (71).
Materials and Methods
Detailed protocols are provided in SI Appendix.
Leishmania Culture and Transfection.
L. major strain FV1 was grown in M199 or Schneider’s complete medium with 10% heat-inactivated fetal bovine serum and other supplements, and transfected by electroporation using a high-voltage protocol. Clonal lines were obtained from colonies after plating on semisolid media, and grown thereafter in selective medium for less than 10 passages before use, prior to which selective drug was removed for one passage.
Molecular Constructions and Primers.
Molecular constructs are described in detail in SI Appendix, Table S1, and oligonucleotide primers are described in SI Appendix, Table S2. Molecular constructs were confirmed by restriction mapping, sequencing, and functional testing. Molecular methods were performed as described (29).
Homologous Replacement of Chromosomal FUTs, Often in the Presence of Ectopically Expressed FUTs.
The workflow for gene is shown in Fig. 2 and SI Appendix, Figs. S1 and S2 for FUT1, SCAL1, and FUT2, respectively. All planned replacements and the presence selectable markers were confirmed by PCR, and the presence of the plasmid bearing GFP by PCR and FACS, respectively.
Plasmid Segregation or Shuffling by Single-Cell Sorting.
Δfut1− parasites bearing pXNGPHLEO-FUT1 alone (segregation) or additionally expressing test FUT1 sequences from pIR1NEO (shuffling) were subjected to flow cytometry and single-cell cloning to recover GFP+ cells (bright) or GFP− (dim), the latter representing potential segregants or “shuffles.” All dim cells were tested for retention of the pXNGPHLEO-FUT1 plasmid to eliminate ones arising from imperfect sorting; only one exceptional true negative was recovered from cells grown in Schneiders’ medium, termed fut1s.
Subcellular Localization of FUT1 and Mitochondrial Membrane Potential.
The cellular location of a tagged FUT-HA construct was monitored in fixed cells by immunofluorescence or immuno-EM; negative controls lacking the primary anti-HA antibody showed little if any reactivity. In some experiments, colocalization with the mitochondrion was established in parallel by staining MitoTracker Red. Overall, cellular morphology was monitored by transmission electron microscopy. The mitochondrial membrane potential (∆Ψm) in live Leishmania promastigotes was measured in cells incubated with 100 nM tetramethylrhodamine ethylester perchlorate, followed flow cytometry. Treatment with CCCP was then added as a control.
Protein Expression and Purification.
GST- and His-tagged recombinant FUT1 constructs were prepared and expressed in E. coli, and the tagged proteins purified by affinity chromatography followed by analysis by SDS-PAGE.
Fucosyltransferase Activity Assays.
Enzymatic activity of recombinant LmFUT1 preparations was determined by a GDP-Glo bioluminescent GDP detection assay (Promega), with highly purified GDP-fucose in the presence of various glycan or peptide substrates, for various times as indicated; controls included omission of enzyme and/or acceptor. After termination of the reaction, the GDP detection regent was added and luminescence was recorded after 1 h.
Peptide MS was performed after fucosylation of a mtHSP70 peptide (EWKYVSDAEKE), followed by purification, separation, and analysis by liquid chromatography (LC)-MS with a Q Exactiv Quadrupole-Orbitrap mass spectrometer. The acquisition of the HCD spectra were triggered from the values corresponding to the expected charge states of the doubly and triply charged protonated molecular ions for the unmodified and fucosylated EWKYVSDAEKE peptide (m/z = 692.325, 461.888, 765.354, and 510.572). The unprocessed LC-MS data were analyzed using SKYLINE (version 3.6.9).
Enzymatic assay of HA-tagged FUT1 expressed in Leishmania and bound to anti-HA beads, and resolution of products by high-performance thin-layer chromatography (HPTLC) were performed as described (46).
Supplementary Material
Acknowledgments
We thank Robert Haltiwanger for providing EGFR and TSR substrates, L. Mahal and L. Zhang for anti-Glc-Fuc antibody, Giulia Bandini for discussions and encouragement, Deborah E. Dobson and Lon-Fy Lye for comments on the manuscript, Wandy Beatty of the Molecular Microbiology Imaging Facility for transmission and immuno-electron microscopy, I. C. Almeida and S. Portillo for providing IB4 lectin, and R. R. Townsend, P. E. Gilmore, and R. W. Sprung of the Washington University Proteomics Shared Resource for performing MS analysis. For single-cell sorting, we thank the staff of the high-speed sorter core, Alvin J. Siteman Cancer Center, Washington University Medical School. This work was supported by NIH Grants AI31078 and AI29646 to S.M.B., an administrative supplement to AI31078 to G.P., Berg Postdoctoral Fellowships from the Department of Molecular Microbiology (S.M. and H.G.), and a Wellcome Investigator Award (101842/Z/13/Z) to M.A.J.F.
Footnotes
Competing interest statement: One of the reviewers (G.W.H.) and one of the authors (M.A.J.F.) are coauthors on chapter 12 in Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 3, 2015–2017).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108963118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Alvar J.et al.; WHO Leishmaniasis Control Team , Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7, e35671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigott D. M., et al., Global distribution maps of the leishmaniases. eLife 3, e02851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bañuls A. L., et al., Clinical pleiomorphism in human leishmaniases, with special mention of asymptomatic infection. Clin. Microbiol. Infect. 17, 1451–1461 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Naderer T., McConville M. J., Intracellular growth and pathogenesis of Leishmania parasites. Essays Biochem. 51, 81–95 (2011). [DOI] [PubMed] [Google Scholar]
- 5.McConville M. J., Mullin K. A., Ilgoutz S. C., Teasdale R. D., Secretory pathway of trypanosomatid parasites. Microbiol. Mol. Biol. Rev. 66, 122–154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilgoutz S. C., McConville M. J., Function and assembly of the Leishmania surface coat. Int. J. Parasitol. 31, 899–908 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Ilg T., Proteophosphoglycans of Leishmania. Parasitol. Today 16, 489–497 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Beverley S. M., Turco S. J., Lipophosphoglycan (LPG) and the identification of virulence genes in the protozoan parasite Leishmania. Trends Microbiol. 6, 35–40 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Capul A. A., Barron T., Dobson D. E., Turco S. J., Beverley S. M., Two functionally divergent UDP-Gal nucleotide sugar transporters participate in phosphoglycan synthesis in Leishmania major. J. Biol. Chem. 282, 14006–14017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K., Bangs J. D., Beverley S. M., Sphingolipids in parasitic protozoa. Adv. Exp. Med. Biol. 688, 238–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zufferey R., et al., Ether phospholipids and glycosylinositolphospholipids are not required for amastigote virulence or for inhibition of macrophage activation by Leishmania major. J. Biol. Chem. 278, 44708–44718 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Späth G. F., et al., Persistence without pathology in phosphoglycan-deficient Leishmania major. Science 301, 1241–1243 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Turco S. J., Späth G. F., Beverley S. M., Is lipophosphoglycan a virulence factor? A surprising diversity between Leishmania species. Trends Parasitol. 17, 223–226 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Späth G. F., et al., Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc. Natl. Acad. Sci. U.S.A. 97, 9258–9263 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacks D. L., et al., The role of phosphoglycans in Leishmania-sand fly interactions. Proc. Natl. Acad. Sci. U.S.A. 97, 406–411 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lázaro-Souza M., et al., Leishmania infantum lipophosphoglycan-deficient mutants: A tool to study host cell-parasite interplay. Front. Microbiol. 9, 626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco L. H., Beverley S. M., Zamboni D. S., Innate immune activation and subversion of Mammalian functions by Leishmania lipophosphoglycan. J. Parasitol. Res. 2012, 165126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capul A. A., Hickerson S., Barron T., Turco S. J., Beverley S. M., Comparisons of mutants lacking the Golgi UDP-galactose or GDP-mannose transporters establish that phosphoglycans are important for promastigote but not amastigote virulence in Leishmania major. Infect. Immun. 75, 4629–4637 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacks D., Kamhawi S., Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 55, 453–483 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Kamhawi S., et al., A role for insect galectins in parasite survival. Cell 119, 329–341 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Beverley S. M., Dobson D. E., Flypaper for parasites. Cell 119, 311–312 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Dobson D. E., et al., Leishmania major survival in selective Phlebotomus papatasi sand fly vector requires a specific SCG-encoded lipophosphoglycan galactosylation pattern. PLoS Pathog. 6, e1001185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goswami M., Dobson D. E., Beverley S. M., Turco S. J., Demonstration by heterologous expression that the Leishmania SCA1 gene encodes an arabinopyranosyltransferase. Glycobiology 16, 230–236 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Dobson D. E., Scholtes L. D., Myler P. J., Turco S. J., Beverley S. M., Genomic organization and expression of the expanded SCG/L/R gene family of Leishmania major: Internal clusters and telomeric localization of SCGs mediating species-specific LPG modifications. Mol. Biochem. Parasitol. 146, 231–241 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Dobson D. E., et al., Functional identification of galactosyltransferases (SCGs) required for species-specific modifications of the lipophosphoglycan adhesin controlling Leishmania major-sand fly interactions. J. Biol. Chem. 278, 15523–15531 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Dobson D. E., et al., Identification of genes encoding arabinosyltransferases (SCA) mediating developmental modifications of lipophosphoglycan required for sand fly transmission of Leishmania major. J. Biol. Chem. 278, 28840–28848 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Mengeling B. J., Turco S. J., Microbial glycoconjugates. Curr. Opin. Struct. Biol. 8, 572–577 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Guo H., et al., Genetic metabolic complementation establishes a requirement for GDP-fucose in Leishmania. J. Biol. Chem. 292, 10696–10708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyne M. J., Reinap B., Lee M. M., Comstock L. E., Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Turnock D. C., Ferguson M. A., Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Eukaryot. Cell 6, 1450–1463 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnock D. C., Izquierdo L., Ferguson M. A. J., The de novo synthesis of GDP-fucose is essential for flagellar adhesion and cell growth in Trypanosoma brucei. J. Biol. Chem. 282, 28853–28863 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Palatnik de Sousa C. B., et al., The FML (fucose mannose ligand) of Leishmania donovani. A new tool in diagnosis, prognosis, transfusional control and vaccination against human kala-azar. Rev. Soc. Bras. Med. Trop. 29, 153–163 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Rosenzweig D., Smith D., Myler P. J., Olafson R. W., Zilberstein D., Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 8, 1843–1850 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Breitling R., et al., Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 25, 209–218 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson I. B., O’Donnell N., Allen S., Mehlert A., Ferguson M. A., Typing of Leishmania lipophosphoglycans by electrospray mass spectrometry. Mol. Biochem. Parasitol. 100, 207–215 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Späth G. F., Beverley S. M., A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp. Parasitol. 99, 97–103 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Edbrooke M. R., et al., The alpha(1-3)-fucosyltransferases come of age. Biochem. Soc. Trans. 25, 880–886 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Ma B., Simala-Grant J. L., Taylor D. E., Fucosylation in prokaryotes and eukaryotes. Glycobiology 16, 158R–184R (2006). [DOI] [PubMed] [Google Scholar]
- 40.Li M., et al., Identification of a new alpha1,2-fucosyltransferase involved in O-antigen biosynthesis of Escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry 47, 11590–11597 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Duncker I., Mollicone R., Candelier J. J., Breton C., Oriol R., A new superfamily of protein-O-fucosyltransferases, alpha2-fucosyltransferases, and alpha6-fucosyltransferases: Phylogeny and identification of conserved peptide motifs. Glycobiology 13, 1C–5C (2003). [DOI] [PubMed] [Google Scholar]
- 42.Paulson J. C., Colley K. J., Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. J. Biol. Chem. 264, 17615–17618 (1989). [PubMed] [Google Scholar]
- 43.Almagro Armenteros J. J., et al., Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2, e201900429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claros M. G., Vincens P., Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Murta S. M., Vickers T. J., Scott D. A., Beverley S. M., Methylene tetrahydrofolate dehydrogenase/cyclohydrolase and the synthesis of 10-CHO-THF are essential in Leishmania major. Mol. Microbiol. 71, 1386–1401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandini G., et al., An essential, kinetoplastid-specific GDP-Fuc: β-d-Gal α-1,2-fucosyltransferase is located in the mitochondrion of Trypanosoma brucei. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/726117v2 (Accessed 27 July 2021). [DOI] [PMC free article] [PubMed]
- 47.Kim S. G., Kweon D. H., Lee D. H., Park Y. C., Seo J. H., Coexpression of folding accessory proteins for production of active cyclodextrin glycosyltransferase of Bacillus macerans in recombinant Escherichia coli. Protein Expr. Purif. 41, 426–432 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zegzouti H., et al., Homogeneous detection of glycosyltransferase activities with universal bioluminescent assays. Glycobiology 26, 1543 (2016). [Google Scholar]
- 49.Zhang L., et al., Helicobacter hepaticus Hh0072 gene encodes a novel alpha1-3-fucosyltransferase belonging to CAZy GT11 family. Glycobiology 20, 1077–1088 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Y., Koles K., Vorndam W., Haltiwanger R. S., Panin V. M., Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J. Biol. Chem. 281, 9393–9399 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., et al., Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 276, 40338–40345 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Mikesh L. M., et al., The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swearingen K. E., et al., A tandem mass spectrometry sequence database search method for identification of O-fucosylated proteins by mass spectrometry. J. Proteome Res. 18, 652–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huh W. K., et al., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Omura T., Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 123, 1010–1016 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Levine Z. G., et al., Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proc. Natl. Acad. Sci. U.S.A. 118, e2016778118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi T., et al., A sequence motif involved in the donor substrate binding by alpha1,6-fucosyltransferase: The role of the conserved arginine residues. Glycobiology 10, 503–510 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Guler J. L., Kriegova E., Smith T. K., Lukes J., Englund P. T., Mitochondrial fatty acid synthesis is required for normal mitochondrial morphology and function in Trypanosoma brucei. Mol. Microbiol. 67, 1125–1142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ambit A., Woods K. L., Cull B., Coombs G. H., Mottram J. C., Morphological events during the cell cycle of Leishmania major. Eukaryot. Cell 10, 1429–1438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyder M. A., Sul D., Kaneshiro E. S., The fatty acid and monosaccharide compositions of three neutral and three phosphorylated glycolipids isolated from Leishmania donovani promastigotes grown in a chemically defined medium. J. Parasitol. 85, 771–778 (1999). [PubMed] [Google Scholar]
- 61.Dean S., Gould M. K., Dewar C. E., Schnaufer A. C., Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 110, 14741–14746 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teng-umnuay P., et al., The cytoplasmic F-box binding protein SKP1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem. 273, 18242–18249 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A., Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116, 647–654 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Hanover J. A., et al., Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Wulff-Fuentes E., et al., The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sacoman J. L., Dagda R. Y., Burnham-Marusich A. R., Dagda R. K., Berninsone P. M., Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function, and survival in HeLa cells. J. Biol. Chem. 292, 4499–4518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Banerjee P. S., Ma J., Hart G. W., Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burnham-Marusich A. R., Berninsone P. M., Multiple proteins with essential mitochondrial functions have glycosylated isoforms. Mitochondrion 12, 423–427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y., et al., Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma J., et al., O-GlcNAcomic profiling identifies widespread O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29141–29153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fidalgo L. M., Gille L., Mitochondria and trypanosomatids: Targets and drugs. Pharm. Res. 28, 2758–2770 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.