Significance
Meiotic recombination plays a fundamental role in shaping genetic diversity in eukaryotes. Extensive variation in crossover rate exists between populations and species. The identity of modifier loci and their roles in genome evolution remain incompletely understood. We explored natural variation in Arabidopsis crossover and identified SNI1 as the causal gene underlying a major modifier locus. To date, SNI1 had no known role in crossover. SNI1 is a component of the SMC5/6 complex that is closely related to cohesin and condensin. Arabidopsis sni1 and other SMC5/6 mutants show similar effects on the interference-independent crossover pathway. Hence, our findings demonstrate that the SMC5/6 complex, which is known for its role in DNA damage repair, is also important for control of meiotic crossover.
Keywords: meiosis, crossover, SNI1, SMC5/6, Arabidopsis
Abstract
The frequency and distribution of meiotic crossovers are tightly controlled; however, variation in this process can be observed both within and between species. Using crosses of two natural Arabidopsis thaliana accessions, Col and Ler, we mapped a crossover modifier locus to semidominant polymorphisms in SUPPRESSOR OF NPR1-1 INDUCIBLE 1 (SNI1), which encodes a component of the SMC5/6 complex. The sni1 mutant exhibits a modified pattern of recombination across the genome with crossovers elevated in chromosome distal regions but reduced in pericentromeres. Mutations in SNI1 result in reduced crossover interference and can partially restore the fertility of a Class I crossover pathway mutant, which suggests that the protein affects noninterfering crossover repair. Therefore, we tested genetic interactions between SNI1 and both RECQ4 and FANCM DNA helicases, which showed that additional Class II crossovers observed in the sni1 mutant are FANCM independent. Furthermore, genetic analysis of other SMC5/6 mutants confirms the observations of crossover redistribution made for SNI1. The study reveals the importance of the SMC5/6 complex in ensuring the proper progress of meiotic recombination in plants.
Sexual reproduction is widespread in eukaryotes and promotes genetic variation within the populations. At the heart of this process is the meiotic cell division, in which crossover recombination shuffles genetic information between homologous chromosomes (1). Meiotic recombination initiates from the programmed DNA double-strand breaks (DSBs), which, following resection, form single-stranded DNA filaments (2, 3). These filaments can invade the complementary DNA sequences of a homologous chromosome, leading to the formation of interhomolog joint molecules (JMs) (3). During meiosis, the “ZMM” proteins stabilize interhomolog JMs and promote Class I crossover repair (2, 3), which is the major crossover pathway in most species. The Class I pathway exhibits interference, meaning that crossovers are placed further apart on the chromosome than expected at random (2–4). As an alternative to crossover repair, JMs can be repaired as non-crossovers (NCOs), which is promoted via DNA helicases including BLM/SGS1/RECQ4 (2, 3). In many eukaryotes, the number of DSBs exceeds the final number of crossovers, and DNA helicases play a key role in limiting crossovers by disassembly of meiotic recombination intermediates and promoting their repair as NCOs (5–7). For example, Arabidopsis thaliana recq4a recq4b loss-of-function mutants show a twofold crossover increase due to a failure in NCO formation (7, 8). In budding yeast, the STRUCTURAL MAINTENANCE OF THE CHROMOSOME 5/6 (SMC5/6) complex plays a dual role in DNA helicase control by providing SUMO activity via its NSE2/MMS21 subunit, which stimulates the anti-recombination function of RECQ4, and also by limiting heteroduplex extension, thus reducing helicase substrates (9, 10). JMs may also be repaired via structure-specific endonucleases, including MUS81, which can result in NCOs or Class II non-interfering crossovers (2–4).
The frequency and chromosomal distribution of crossovers are tightly controlled, as high crossover numbers may break down favorable combinations of alleles, potentially reducing fitness (11). However, significant variation in crossover frequency is also observed between individuals, sexes, populations, and species (12–14). Genetic factors that modify crossover levels or patterns have the potential to profoundly influence selective responses during adaptation (13, 15, 16). For example, high variation across the major histocompatibility complex, which encodes cell surface proteins essential for the human adaptive immune system, is achieved by meiotic recombination hotspots within the locus (17, 18). Similarly, crossover hotspots were observed in a subset of plant NLR resistance genes (19).
Genetic loci that act in trans to control variation in meiotic recombination levels or patterns have been described in plants and animals. For example, the PRDM9 zinc finger KRAB-SET domain protein has been shown to recruit DSB machinery to specific DNA sequence motifs at recombination hotspot in humans and mice (20, 21). Genetic variation in PRDM9 governs crossover hotspot location within and between vertebrate species (22–24). A sequence polymorphism in E3 SUMO and ubiquitin ligases related to human RNF212 and HEI10/CCNB1IP1 has been identified that controls crossover frequency in vertebrates, invertebrates, fungi, and plants (25–31). HEI10 in plants acts in the Class I pathway and is a dosage-sensitive regulator of crossover number (30). Variation in the meiosis-specific α-kleisin cohesin subunit REC8 influences the crossover rate in some mammals and plants, which acts to tether chromatin loops to the chromosome axis during prophase I (27, 28, 32, 33). Genetic variation in Arabidopsis also identified TAF4b as a recombination modifier locus, which encodes a general transcription factor paralog expressed specifically during meiosis (34). In this study, we further investigated variation in crossover frequency in A. thaliana and identified SNI1, a component of the SMC5/6 complex, as a factor preventing errors in meiotic recombination in plants.
Results
Genetic Mapping of the Col/Ler Recombination Quantitative Trait Locus rQTL4.
Segregation of linked reporters expressing fluorescent proteins in seed or pollen (fluorescent-tagged lines, FTLs) can be used to measure crossover frequency within defined chromosomal intervals (35–38). In our previous work, we used a Col-420×LLCLL F2 population (in which chromosomes 1, 2, 4, and 5 were Col/Ler segregating) to map quantitative trait loci responsible for variation in crossover frequency between the Arabidopsis Col and Ler accessions (30, 39). We identified two significant rQTLs located on chromosomes 1 and 4, exhibiting logarithm of the (base 10) odds ratio (LOD) scores of 40.1 and 53.5, which explained 23.3% and 33.6% of the variance in crossover frequency, respectively (30). rQTL1 was identified as HEI10, encoding an E3 ubiquitin/SUMO ligase acting in the Class I crossover pathway (30). In this study, we sought to identify the causal locus for rQTL4.
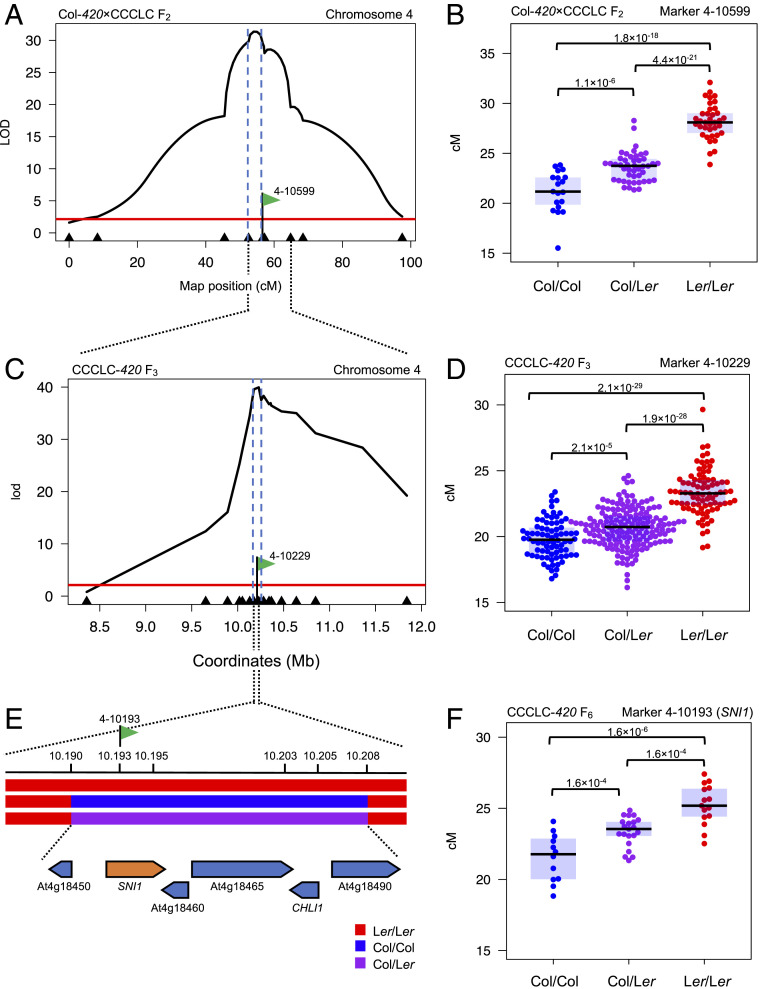
We constructed a Col-420×CCCLC (39) F2 population (n = 102) and genotyped all individuals for 11 markers spanning the previously identified rQTL4 region (30). This revealed a strong rQTL association between markers 4-9652 and 4-10366 with an LOD score of 31.72 (Fig. 1A and SI Appendix, Table S1). rQTL4 is semi-dominant with the Col allele associating with reduced 420 crossover frequency (marker 4-10599; Fig. 1B). We selected a single F2 individual heterozygous over rQTL4 and fixed for rQTL1 and generated an F3 population (n = 2,280), which was genotyped at markers 4-8358, 4-10847, and 4-11840 to identify recombinants within the rQTL4 interval. We identified 325 recombinants, which were measured for 420 crossover frequency and genotyped using 16 markers, which narrowed the credible interval to a 53-kb region containing 26 genes (Fig. 1 C and D and SI Appendix, Table S2). We developed an F4 population from an rQTL4 heterozygous F3 individual (n = 152) (SI Appendix, Fig. S1A and Table S3) and genotyped using four internal markers. This allowed us to identify one individual fixed for Ler upstream of the interval, which showed 420 crossover frequency equivalent to the Col/Ler heterozygous rQTL4 level (mean = 21.31 cM). We used the progeny of this individual to generate a population of 1,056 F5 and genotyped them to find recombinants within the rQTL4 interval. We identified an individual (H-27) that was Col/Ler heterozygous for a 19.5 kb encompassing six genes, with the remainder of the region fixed for one parent (Fig. 1E). The progeny of this plant (n = 48) showed variation in recombination frequency similar to that observed in the original Col-420×CCCLC F2 population (Fig. 1F and SI Appendix, Table S4). Genotyping within the 19.5-kb interval showed that plants fixed for Col allele present a 420 crossover frequency at 21.52 cM, whereas heterozygotes showed 23.35 cM and Ler/Ler homozygotes 25.23 cM on average. Mean 420 crossover frequency was statistically different between the groups, with the greatest difference between the Col/Col and Ler/Ler groups (Welch t test; P = 1.57 × 10−6). Therefore, we conclude that rQTL4 corresponds to one of the six identified genes in the 19.5-kb interval.
Fig. 1.
Fine-mapping of the rQTL4 recombination modifier locus. (A) LOD scores for markers associated with 420 crossover frequency from Col-420×CCCLC F2 population. The x axis corresponds to the chromosome 4 genetic map with markers used for mapping indicated by arrowheads. The red line indicates the 95% significance threshold, and blue dashed lines mark the credible interval. The marker 4–10599 is labeled with a green flag. (B) Effect plots showing 420 crossover frequency for Col/Col, Col/Ler, and Ler/Ler individuals for the marker 4-10599 in the F2 population. Each dot represents one individual. P values were estimated by Welch t test. (C) As in A but showing the Col-420×CCCLC F3 population. The x axis corresponds to chromosome 4 physical positions (Mb). The marker 4-10229 is labeled with a green flag. (D) As in B but for the marker 4-10229 in the F3 population. (E) The genotypes of progeny of the H-27 recombinant plant, which has only a 19.5-kb region segregating for Col/Ler and the reminder of the genome being fixed. Gene models are shown beneath the genotypes. The marker 4-10193 is labeled with a green flag. (F) As in B but for the marker 4-10193 in the progeny of the H-27 F5 recombinant.
Variation in SNI1 Underlies rQTL4.
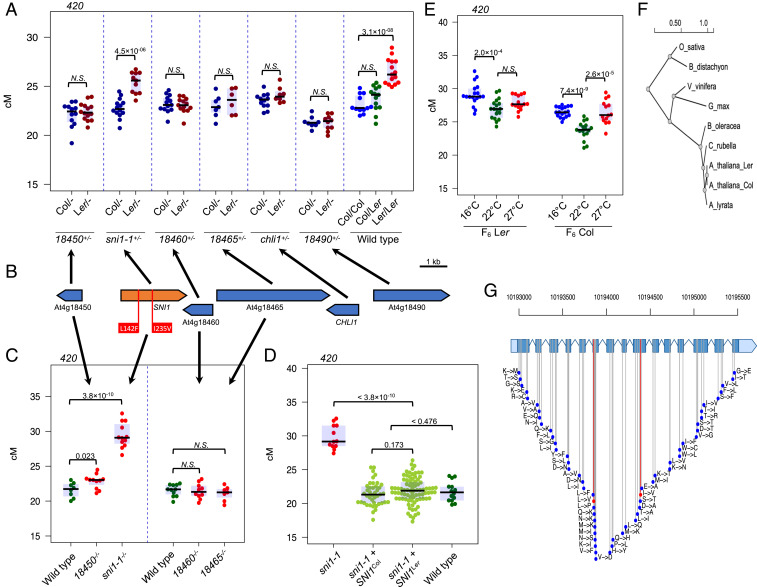
To determine which of the candidate genes corresponds to rQTL4, we performed an allelism test by crossing transfer-DNA mutants in all six genes with F6 individuals fixed for Col/Col and Ler/Ler over the rQTL4 interval. Mutation of the rQTL4 causal gene should result in a significant difference in recombination frequency of plants obtained from crosses with F6Col/Col and F6Ler/Ler plants. We observed that only a mutant in SNI1 (At4g18470) showed a significant difference in 420 crossover frequency between the two crosses, strongly suggesting that this gene corresponds to rQTL4 (Fig. 2 A and B and SI Appendix, Table S5). In addition, SNI1 was the only candidate gene which showed nonsynonymous substitutions in the coding sequence when Col and Ler accessions were compared (Fig. 2B).
Fig. 2.
SNI1 corresponds to rQTL4. (A) 420 crossover frequency for F1 plants obtained from the cross between mutant lines for the six genes and F6 individuals fixed for Col/Col and Ler/Ler over the rQTL4 interval. Measurements for the wild-type F6 individuals for Col/Col, Col/Ler, and Ler/Ler were used as a control. Each dot represents one individual in A, C, D, and E. Significance in A, C, and D was assessed by Welch t test. (B) The genes within the rQTL4 19.5-kb Col/Ler segregating interval with non-synonymous substitutions indicated. (C) 420 crossover frequency for the mutants of genes located within rQTL4 locus. (D) Complementation of the sni1 mutation with SNI1Col and SNI1Ler alleles restores wild-type 420 crossover frequency in T1 generation. (E) F6 lines carrying SNI1Ler are less responsive to temperature stress than the SNI1Col as measured in the 420 interval. (G) Location of all non-synonymous substitution sites in the SNI1 gene observed within 1,135 A. thaliana accessions (46). The two substitutions differing Col and Ler alleles are indicated in red. (F) Phylogenetic tree of SNI1 orthologs identified by basic local alignment search tool (BLAST) searches in selected eukaryotic genomes. The phylogenetic tree was constructed using maximum likelihood method.
We measured 420 crossover frequency in independently isolated homozygous mutations in the candidate genes (Fig. 2C). This assay was not possible for CHLI1 (At4g18480), which has a very strong dwarf mutant phenotype (40), and for At4g18490 due to the FTL reporter silencing. From the four remaining genes, a sni1-1 loss-of-function allele (41–43) showed a significant increase in crossovers (29.41 cM, n = 12) when compared to the wild type (21.58 cM, n = 8) (Fig. 2C and SI Appendix, Table S6). We next performed a complementation experiment in which Col and Ler SNI1 alleles were transformed into the sni1-1 mutant background. This showed that both Col and Ler SNI1 variants can complement the sni1-1 elevated crossover phenotype, demonstrating that the effect on 420 crossover rate in sni1-1 is due to the nonsense mutation at the SNI1 locus and not a secondary mutation (Fig. 2D and SI Appendix, Table S7). We observed that the SNI1Ler transformants were higher recombining than SNI1Col transformants, although the difference was not statistically significant.
Next, we measured whether the Col and Ler SNI1 alleles lead to differences in messenger RNA (mRNA) expression by performing qRT-PCR from closed flower buds, comparing rQTL4 F6 individuals (the progeny of individual H-27). We did not observe a significant difference in expression between the Col and Ler SNI1 alleles, indicating that the variation observed most likely does not act via mRNA expression level (SI Appendix, Fig. S1 B and C and Tables S8 and S9). Consistently, transformation of additional SNI1 copies did not change 420 crossover rate (SI Appendix, Fig. S1D and Table S10).
We investigated how the two alleles might modify recombination rate in different environmental conditions. To this end, we tested the effect of both alleles on recombination at different temperatures, since temperature influences the frequency of recombination (44, 45). We took advantage of the fact that F6Col and F6Ler differ only in the 19.5-kb interval containing SNI1. We observed that the 420 crossover frequency was less responsive to thermal conditions in F6 carrying SNI1Ler than in F6 SNI1Col plants (Fig. 2E and SI Appendix, Table S11). This suggests that variability within the SNI1 gene may be important in modulating meiotic recombination under varying environmental conditions.
Comparison of the Col and Ler SNI1 alleles revealed the existence of two nonsynonymous substitutions: L142F and I235V. The first of these substitutions is also present in Ct-1 accession, which was previously used in crosses with Col-420 to map rQTL (SI Appendix, Fig. S2A) (37). As Ct-1×Col-420 mapping population did not reveal rQTL located on chromosome 4, we concluded that the second substitution, I235V, is likely responsible for the SNI1 recombination phenotype. The V235 (Ler) variant exists in 379 out of 1,135 A. thaliana accessions (46). The I235 (Col) allele was considered as ancestral, as it is conserved across Iberian relict accessions and in SNI1 homologs of Arabidopsis lyrata, Arabis alpina, and Camelina sativa (Fig. 2F and SI Appendix, Fig. S3). In general, SNI1 shows high sequence variation across A. thaliana accessions, with more than 30 different alleles and 59 nonsynonymous substitution sites (Fig. 2G and SI Appendix, Fig. S2 B and C) (42).
Mutation of SNI1 Causes Remodeling of the Meiotic Crossover Landscape.
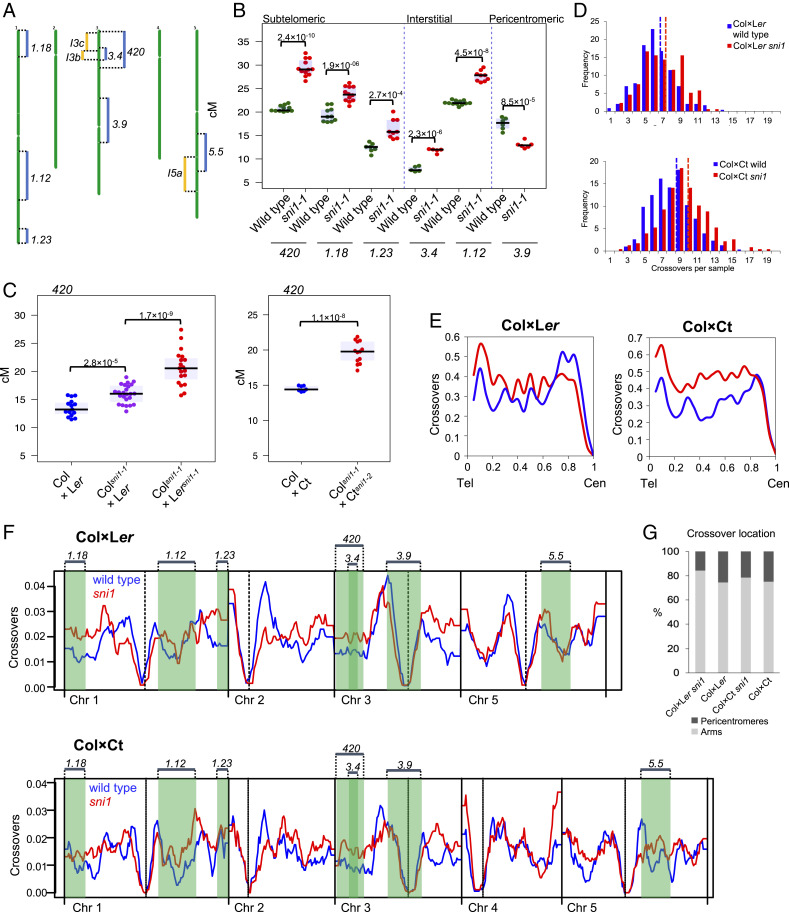
To further investigate crossover frequency in the mutant, we crossed sni1-1 with five different FTL reporter lines positioned in varying chromosomal locations [Col Traffic Lines, CTLs (38)]. In all cases, we observed significant changes in sni1-1 crossover rates, though the extent and direction of those changes varied according to the region (Fig. 3 A and B and SI Appendix, Table S12). Specifically, we observed crossover increases ranging from 22.6% (interval CTL1.18) to 50.4% (interval CTL3.4) in distal chromosomal regions (Fig. 3 A and B). Interestingly, we observed a 25.7% reduction in crossover frequency, from 17.5 to 13.0 cM, in the pericentromeric interval CTL3.9 (Fig. 3 A and B). These observations are consistent with a global redistribution of crossover frequency in sni1-1.
Fig. 3.
The sni1 mutants exhibit elevated crossover levels in chromosome arms and subtelomeric regions but reductions in pericentromeres. (A) Ideogram of A. thaliana chromosomes showing the positions of fluorescent reporter intervals for seed- (blue) and pollen-based (yellow) systems. (B) Crossover frequency in the seed-based intervals as measured in wild type (green) and sni1-1 (red) F2 siblings. (C) 420 crossover frequency (cM) in Col×Ler, Colsni1-1×Ler, and Colsni1-1×Lersni1-1 F1 hybrids (Left) and Col×Ct and Colsni1-1×Ctsni1-2 F1 hybrids (Right). Significance in B and C was assessed by Welch t tests; each dot represents one individual. (D) Histograms presenting the number of crossovers per individual in Col×Ler (blue) and Colsni1-1×Lersni1-1 (red) (Top) and Col×Ct (blue) and Colsni1-1×Ctsni1-2 (red) (Bottom) F2 GBS populations. The mean crossover number of each population is denoted by a vertical dashed line. (E) Crossover frequency along the proportional (scaled) length of the chromosomes from telomeres (Tel) to centromeres (Cen) in Col×Ler (blue) and Colsni1-1×Lersni1-1 (red) F2 populations (Left) and Col×Ct (blue) and Colsni1-1×Ctsni1-2 (red) (Right). Chromosome 4 in Col×Ler populations was excluded from the analyses in D through F to avoid potential bias from the 587-kb Colsni1-1 introgressed region in the Lersni1-1 parent. (F) Crossover frequency over the chromosomes in Col×Ler (blue) and Colsni1-1×Lersni1-1 (red) F2 populations (Top) and Col×Ct (blue) and Colsni1-1×Ctsni1-2 (red) (Bottom) F2 populations. Crossovers were tallied into 300-kb windows, divided by the number of F2 individuals, and a rolling mean plotted along the chromosomes. Cen positions are denoted by vertical dashed lines and TEL positions by vertical solid lines. (G) Crossover location in wild type and sni1 F2 GBS populations as divided into chromosome arms and pericentromeres. The pericentromeres were defined as regions with higher-than-average DNA methylation, which surround the centromeres (88).
To explore the impact of sni1-1 on crossovers genome-wide, we used genotyping-by-sequencing (GBS) of F2 individuals. To this end, we backcrossed (6×) sni1-1 into the Ler accession and produced Colsni1-1×Lersni1-1 F1 hybrids. We also used CRISPR-Cas9 mutagenesis to generate a sni1-2 allele de novo in the Ct accession (SI Appendix, Fig. S2 C and D) and produced Colsni1-1×Ctsni1-2 F1 hybrids. 420 crossover frequency was elevated in both hybrids in the sni1 background compared to wild-type controls (53.2% for Col×Ler; 35.4% for Col×Ct), indicating that the effect is not sensitive to interhomolog polymorphisms (Fig. 3C and SI Appendix, Tables S13 and S14). Interestingly, Col×Ler F1 plants in which only the SNI1Ler allele was active showed significantly higher 420 crossovers than wild-type hybrids (15.98 cM versus 13.50 cM), which is consistent with Ler SNI1 increasing crossover repair. We sequenced genomic DNA from 174 and 229 F2 individuals from each population and identified 1,194 and 2,260 crossovers per Colsni1-1×Lersni1-1 and Colsni1-1×Ctsni1-2 population, respectively, using the TIGER pipeline (47). As a control, we compared to crossovers mapped in wild-type F2 populations generated from Col×Ler and Col×Ct F1 hybrids (8, 48). To eliminate potential effects of sni1-1 introgression in the Colsni1-1×Lersni1-1, chromosome 4 was removed from the analysis. Total crossover number per individual was slightly higher in sni1 than in wild type (9.55 versus 8.18 for Col×Ct, Mann–Whitney–Wilcoxon test P = 1.41 × 10−10 and 6.86 versus 6.25 crossovers per F2 for Col×Ler, P = 6.03×10−3; Fig. 3D). We observed a significant change in the recombination landscape in the sni1 mutants for both hybrids, with elevated crossovers in the chromosome arms and less in the pericentromeres (Fig. 3 E–G).
To exclude a possibility that the crossover estimates are affected by aneuploidy we tested for evidence of copy number variation in our GBS sequencing data. We calculated sequence coverage separately for each chromosome in 241 Colsni1-1×Ctsni1-2 and 305 Col×Ct F2 plants. None of the tested samples showed a coverage deviation larger than 10% from the total sample coverage, consistent with minimal aneuploidy present in these populations (SI Appendix, Table S15).
We compared Col×Ler and Col×Ct crossover maps using 500-kb windows and observed significant correlation between crossover distributions (Spearman Rho = 0.564; SI Appendix, Fig. S4). When the wild-type maps were compared to sni1, lower correlations were observed (Rho = 0.508 and Rho = 0.339 for Col×Ler and Col×Ct, respectively; SI Appendix, Fig. S4). Similarly, Colsni1-1×Lersni1-1 and Colsni1×Ctsni1-2 maps showed low correlation (Rho = 0.355). Therefore, differences in crossover distribution between both hybrids are exacerbated in the sni1 background.
Mutation of SNI1 Results in Reduced Crossover Interference.
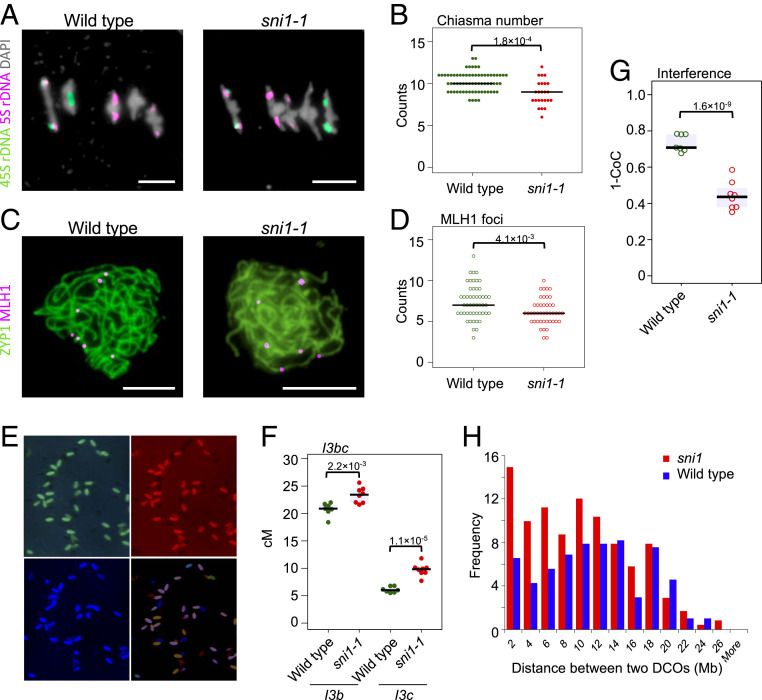
We investigated crossover frequency in sni1-1 using cytological approaches. DAPI staining and fluorescent in situ hybridization (FISH) of metaphase I chromosome spreads were used to count chiasmata on individual chromosome pairs (49). This indicated a small but significant reduction in chiasma numbers in the sni1-1 mutant compared to the wild type (10.2 versus 8.9; Fig. 4 A and B and SI Appendix, Tables S16 and S17), which contrasts with the small overall increase observed via sequencing (Fig. 3D). This may be due to closely spaced crossovers being underestimated by chiasmata counts. Also, we analyzed Class I crossovers via MLH1 immunostaining on pachytene-stage male meiocytes (50) (Fig. 4 C and D and SI Appendix, Table S17). While 7.4 MLH1 foci were detected in Col-420 line (n = 50), 6.2 foci were observed in sni1-1 (n = 44), showing a significant reduction. This suggests that Class I/Class II crossover ratio is changed in sni1-1 and interfering Class I crossovers decrease. Therefore, we tested whether crossover interference is affected in sni1-1, by crossing sni1-1 with the three color FTL I3bc line (51). This allowed us to measure male-specific crossover frequency in two adjacent intervals, I3b and I3c, to measure the number of double crossovers (DCOs) and calculate interference (35, 36). We observed a significant reduction in crossover interference, measured as a coexistence of coincident (CoC), in the sni1-1 mutant when compared to wild type (Fig. 4 E–G and SI Appendix, Table S18).
Fig. 4.
The sni1-1 mutant shows weakening of crossover interference. (A) Representative images of DAPI-stained bivalents at metaphase I in wild type (Col) and sni1-1. FISH probes against 45S (green) and 5S (red) ribosomal DNA were used to identify chromosomes, allowing for chiasma counting depending on chromosome morphology. (Scale bar, 5 μm.) (B) Quantification of chiasma count data from wild type and sni1-1. Significance was assessed by Mann–Whitney–Wilcoxon (MWW) tests. (C) Representative images of pachytene stage male meiocytes from wild type (Col-420) or sni1-1 stained for MLH1 (red) and ZYP1 (green). (Scale bar, 5 μm.) (D) Quantification of MLH1 foci on pachytene stage meiotic chromosomes in wild type and sni1. Significance was assessed by MWW tests. (E) Microphotographs of segregating I3bc pollen grains as seen in three fluorescent channels. Composite image was also shown. (F) I3b and I3c genetic distances in wild type and sni1-1. Each dot represents measurements from four to 10 pooled individuals. Significance was assessed by Welch t tests. (G) Crossover interference (1–CoC) between the I3b and I3c intervals. (H) Histograms of cis-DCO distances in Col×Ct (blue) and Colsni1-1×Ctsni1-2 (red) calculated in 2-Mb windows. Frequency was scaled to the number of F2 individuals in each population.
For our Col×Ct and Colsni1-1×Ctsni1-2 F2 GBS data, we have sufficient crossover events (2,478 and 2,260, respectively) to perform cis-DCO analysis by filtering for parental–heterozygous–parental genotype transitions (52, 53) and measure DCO distances. We compared the distribution of DCO distances between the wild type and sni1 within each F2 population (Fig. 4H) and observed a significantly closer DCOs in sni1 (Kolmogorov–Smirnov test, P = 0.028). Along with the observed decrease in MLH1 foci number, the reduction in crossover interference may be apparent and result from a change in the balance of Class I versus Class II crossover repair in sni1 mutants.
Meiotic Recombination Phenotypes of sni1 Are Likely Independent of Systemic Acquired Resistance.
SNI1 was identified in a genetic screen for regulators of the salicylic acid (SA)–mediated defense response (41), and sni1 mutations have been found to suppress systemic acquired resistance (SAR) (42, 43). To test whether the effect of sni1 on meiotic recombination is linked to its role in SAR, we investigated how SA, the major trigger in SAR, affects crossover frequency. To this end, we treated flowering Colsni1-1-420 and wild-type Col-420 with SA by spraying flowering plants (1 mM of SA solution on a weekly basis). This treatment induced PR1 expression, indicating successful triggering of SAR (SI Appendix, Fig. S5 A and B and Tables S19 and S20). We did not observe any change in 420 crossover frequency, either in wild type or sni1-1. We measured crossover frequency in I5a (Col background) and LTL5.5 (Ler background) chromosome intervals in SA-treated plants and also did not observe any change when compared to nontreated control (SI Appendix, Fig. S5 C and D and Tables S21 and S22). Altogether, our data indicate that SA does not affect meiotic recombination in these regions.
Mutations in some DNA repair genes partially suppress the retarded growth of sni1-1 (42, 54). To see their effect on the meiotic phenotype of sni1, we created the double mutants sni1-1 rad51, sni1-1 brca2a, sni1-1 brca2b, and sni1-1 atr in the background of the reporter line 420. The sni1-1 mutation was not able to suppress the sterility phenotype of rad51 (SI Appendix, Fig. S6A) (however, see ref. 55). We measured 420 crossover frequency in sni1-1 brca2a and sni1-1 brca2b and observed no suppression of recombination (SI Appendix, Fig. S6B and Table S23). Surprisingly, we observed a further increase of 420 recombination frequencies in the sni1-1 atr mutant, which together with no improvement in the fertility of the double mutant compared to sni1-1 suggests that the meiotic recombination phenotype of sni1-1 is independent of ATR (SI Appendix, Fig. S6 C and D and Tables S24 and S35).
Recent studies have shown that the immunity-related phenotypes of sni1 are dependent on EDS1 (56). In the double-homozygous mutant sni1 eds1-2, plant growth is partially restored and transcript accumulation of PR1 is abrogated (56). Therefore, we crossed the sni1-1-420 line with eds1-2 to test whether the meiotic phenotype of sni1 are dependent on EDS1. 420 crossover frequency was not changed in eds1-2 background when compared to wild type, and sni1-1 eds1-2 double-homozygous mutants were not statistically different from the sni1-1 single mutant (SI Appendix, Fig. S5E and Table S25). This indicates that unlike immunity-related phenotypes, the meiotic recombination phenotype of sni1 is independent of EDS1.
Meiotic Recombination Phenotypes of sni1.
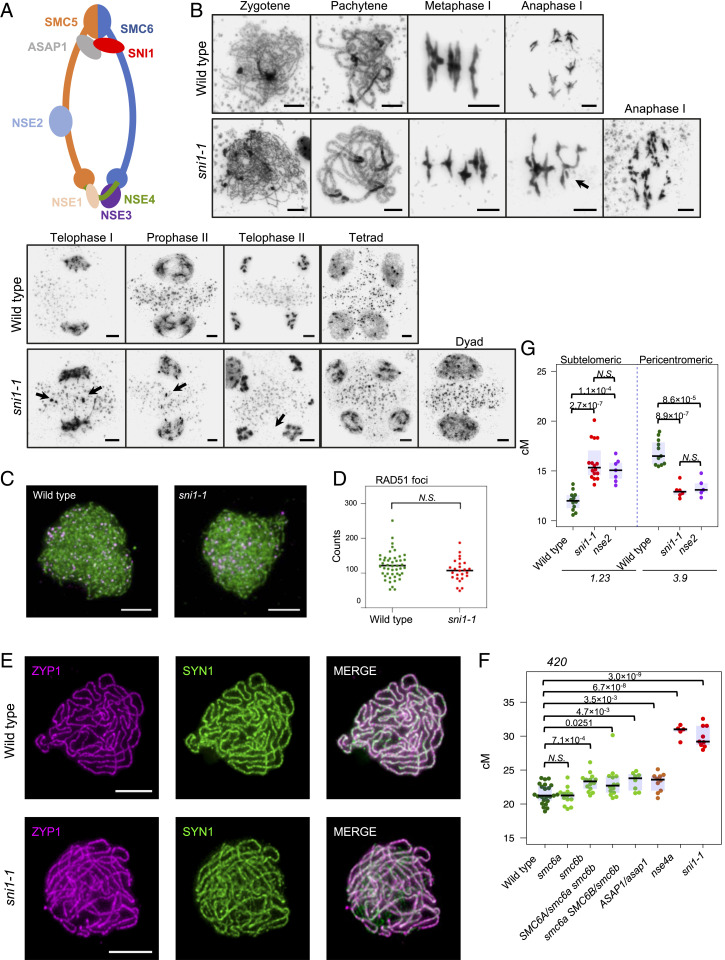
Recent studies have revealed that SNI1 is a component of the SMC5/6 complex and is a homolog of NSE6 (Fig. 5A) (42). The role of SMC5/6 in timely resolution of meiotic recombination intermediates has been described in budding yeast (9, 10, 57). Consistently, we observed anaphase I bridges, chromosome fragments, and micronuclei formation in sni1-1 meiocytes (Fig. 5B), which was also reported for Arabidopsis nse4 and nse2 mutants (58, 59). A single chromosome fragment was observed in one metaphase I cell (out of 24), while all of the analyzed anaphase I cells (n = 15) showed some degree of chromosome fragmentation. Dyads were formed in at least 10% of the meiocytes, which led to the production of unreduced gametes (Fig. 5B), which is also typical for SMC5/6 mutants (59). We confirmed these abnormalities also in the sni1-2 mutant allele (SI Appendix, Fig. S7). Concurrently with the submission of this work, another sni1 allele has been characterized and displayed a similar but stronger phenotype (55). Unreduced gametes may potentially lead to the formation of polyploid seeds, which in turn would affect the crossover frequency measurements. However, only 3.7% (n = 2,292) pollen grains were unreduced in sni1-1 (SI Appendix, Table S26), and we did not observe larger seeds in sni1-1 progeny, which is an indicator of polyploidy. Moreover, seed-based crossover measurements repeatedly returned 3:1 Mendelian segregation ratios of fluorescent reporters (green to non-green and red to non-red seeds), as expected for the diploid plants.
Fig. 5.
The meiotic phenotype of the sni1 mutant is similar to SMC5/6 complex mutants. (A) Schematic representation of SMC5/6 complex. The interface of the protein interaction of the SNI1–ASAP1 subcomplex (gray–red) is unknown in plants. (B) Cytological characterization of the sni1 mutant in comparison to wild type (Col). The stages of meiotic progression were labeled. Chromosome fragments and micronuclei observed in the sni1 mutant are indicated by arrows. (Scale bar, 5 μm.) (C) Representative images of ASY1 (green) and RAD51 (magenta) coimmunostaining on wild type (Col-420) and sni1-1 male meiocytes at zygonema. (D) Quantification of RAD51 foci number per cell in wild type and sni1-1. (E) Representative images of ZYP1 (magenta) and SYN1 (green) coimmunostaining on wild type (Col-420) and sni1-1 male meiocytes at pachynema. (Scale bar, 5 μm.) (F) 420 crossover frequency in SMC5/6 complex mutants. (G) CTL1.23 and CTL3.9 genetic distances (cM) in wild type, sni1-1, and nse2-2. Significance in D through F was assessed by Welch t test; each dot represents one individual.
We investigated the formation of meiotic DSBs in the sni1-1 mutant by scoring RAD51 foci at early prophase I chromosomes and did not observe a significant change when compared to wild type (Fig. 5 C and D and SI Appendix, Table S27). To ensure that sni1-1 does not produce SPO11-independent DSBs that can be repaired by crossovers, we created a sni1-1 spo11-1 double mutant. This double mutant was sterile, showing that sni1-1 mutation is not able to restore fertility in spo11-1 (SI Appendix, Fig. S8). In prophase I, we found no cells at pachytene stage neither in spo11-1 (n = 300) nor sni1-1 spo11-1 (n = 300) (SI Appendix, Fig. S8), which indicates that there is no synapsis in these mutants. Consequently, 10 univalents were observed for both spo11-1 and sni1-1 spo11-1 at metaphase I (SI Appendix, Fig. S8 and Table S28). In two out of 43 metaphase I cells, we observed single bivalents in sni1-1 spo11, which, however, did not differ significantly from the results reported for the single spo11-1 mutant (60). Interestingly, chromosome fragments were detected in 18.6% of sni1-1 spo11-1 cells (n = 43), while this was not observed in spo11-1. As immunostaining with RAD51 antibodies did not show an increase in the number of DSBs in the early stages of sni1-1 mutant prophase (Fig. 5 C and D), these chromosomal fragments likely arose as a consequence of chromosome condensation problems.
Together, these results indicate that DSB formation proceeds normally in sni1-1, and the mutant does not produce SPO11-independent DSBs that could be repaired as crossovers. In addition, we analyzed the synaptonemal complex component ZYP1 (ZIP1 homolog) and meiotic cohesin SYN1 (REC8 homolog) in wild-type and sni1-1 (Fig. 5E). This revealed that synapsis proceeds normally in the sni1-1 mutant without apparent cohesion failures.
Mutants of Other Components of the SMC5/6 Complex Show Similar Changes in Crossover as sni1-1.
We next measured crossover frequency in mutants of other SMC5/6 complex components and compared them to sni1-1 and wild type. In A. thaliana, homologs of all SMC5/6 complex subunits have been identified and are either embryo lethal or show strongly disturbed development which results in partial or complete sterility (42, 61–63). SMC6 is encoded by two genes in the Arabidopsis genome, SMC6A and SMC6B, which are partially functionally redundant (62). NSE4 is encoded by two functionally non-redundant copies, NSE4A and NSE4B, in which expression of NSE4B is significantly lower than NSE4A, and nse4a single mutants show fertility defects (63). We crossed mutants of NSE4A, ASAP1 (homolog of NSE5), and a combination of mutants for SMC6A and SMC6B subunits with the Col-420 reporter line to assess crossover frequency. Crossovers were dramatically elevated in the nse4a mutant (30.71 cM; Fig. 5F and SI Appendix, Table S29). As the asap1 is almost sterile and smc6a smc6b double mutants are lethal in the homozygous state, which precludes measurement of recombination, we tested the effect of their decreased expression. ASAP1/asap1 heterozygotes showed a significant crossover increase from 21.06 to 23.22 cM (Fig. 5F and SI Appendix, Table S29). Out of the smc6 mutants tested, smc6a did not show any effect on crossover frequency, while smc6b (62) showed significant increases (to 24.13 cM). We also constructed smc6a smc6b sesquimutants (homozygous for one mutation and heterozygous for the other) and observed similar increases in 420 crossover frequency as the single smc6b mutant (22.16 cM and 23.35 cM, respectively). Thus, mutants of SMC5/6 components show a consistent change in crossover frequency with that observed in the sni1-1 mutant.
Besides SNI1, mutants of NSE2 are also viable in A. thaliana and produce enough seeds to assess crossover frequency using FTL reporters. NSE2 is located on chromosome 3, within the 420 interval; therefore, it was not possible to test recombination using this FTL. Instead, we crossed nse2-2 with CTL1.23 and CTL3.9, which represent subtelomeric and pericentromeric intervals, respectively (Fig. 3A). The nse2 mutant shows significant changes in both intervals when compared to the wild type (Fig. 5G and SI Appendix, Table S30) (CTL1.23: wild-type 12.0 cM, nse2 15.1 cM; CTL3.9: wild-type 16.9 cM, nse2 13.4 cM). The nse2 changes had the same direction and magnitude as observed in sni1-1 (CTL1.23: sni1-1 15.9 cM; CTL3.9: sni1-1 13.0 cM). Altogether, our results are consistent with a common role of SMC5/6 complex in ensuring proper progress of meiotic crossover.
We also generated double sni1-1 nse2 and sni1-1 nse4a mutants, which both showed severe developmental abnormalities and did not flower (SI Appendix, Fig. S9). This indicates that the lack of several components of the SMC5/6 complex leads to loss of its function.
Modification of the Crossover Landscape in sni1-1 Is Independent of FANCM.
In budding yeast, the smc5 sgs1 and nse4 sgs1 double mutants are synthetically sick (10). Similarly, when we combined sni1 with recq4a recq4b (orthologous to sgs1), the triple-mutant plants died early after germination (SI Appendix, Fig. S10A), and the same was observed for sni1 RECQ4A/recq4a recq4b mutant. This suggests that RECQ4 helicases are crucial for somatic DNA repair during replication in the absence of SNI1. As the sni1-1 recq4a recq4b triple mutant never enters flowering stage, we were not able to assess crossover frequency. The lethality of sni1 recq4a recq4b triple mutants resembles the synthetic lethality of mus81 recq4a recq4b triple mutants (7, 64). We therefore crossed sni1-1 with mus81 and observed that the resulting double mutant is also synthetically lethal (SI Appendix, Fig. S10B). We propose that SMC5/6-SNI1 prevents formation of complex JMs, which if unrepaired in the absence of RECQ4 or MUS81 cause severe defects in DNA replication and cell division.
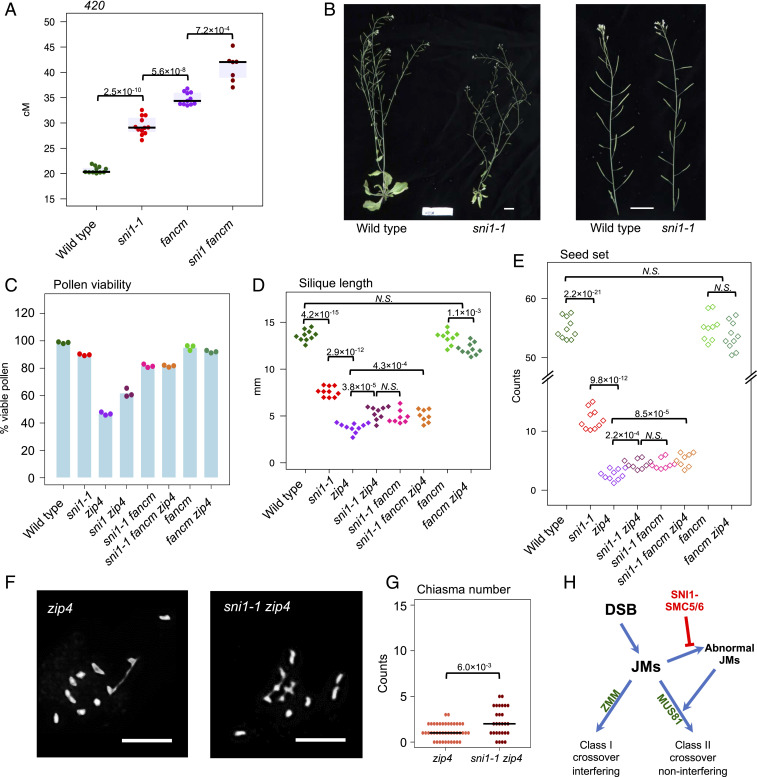
To test the genetic interactions with FANCM, we crossed fancm mutants with sni1-1. The resulting sni1-1 fancm double mutant did not suppress the developmental phenotypes of sni1-1 (SI Appendix, Fig. S10A). In A. thaliana inbred lines, the null fancm mutation leads to a dramatic increase in crossover frequency via the Class II pathway (65). We compared 420 crossover recombination in sni1-1 and fancm single mutants to the double mutant and observed an additive effect of the two mutations which manifests in further elevation of crossover rate from 29.41 cM in sni1-1 and 34.78 cM in fancm to 40.93 cM in sni1-1 fancm (Fig. 6A and SI Appendix, Table S31). This shows that the increase in crossovers observed in sni1-1 mutants is likely not due to mis-regulation of FANCM.
Fig. 6.
SNI1 affects Class II crossover repair in FANCM-independent manner. (A) 420 crossover frequency in wild type, sni1, fancm, and sni1 fancm plants. Significance was assessed by Welch t test; each dot represents one individual. (B) Reduced size and fertility of sni1 mutant plant in comparison with wild type. Whole plants (Left) and primary inflorescences (Right) are shown. (Scale bar, 2 cm.) (C–E) Fertility assays in sni1, zip4, and fancm mutants and their combinations as assessed via pollen viability (C), silique length (D), and seed set (E). Significance was assessed by Welch t test. (F) Representative images of DAPI-stained chromosomes at metaphase I in zip4 and sni1 zip4. (Scale bar, 5 µm.) (G) Quantification of chiasma count data from zip4 and sni1 zip4. Significance was assessed by Mann–Whitney–Wilcoxon tests. (H) Model of SNI1 role in meiotic crossover formation. SNI1, together with SMC5/6 complex, prevents formation of abnormal JMs, which require MUS81 endonuclease for their successful repair.
The sni1-1 Mutation Partially Restores Crossover Formation to zip4 Mutants via Elevated Class II Crossovers.
If the increase in 420 crossover frequency in sni1-1 is due to a higher number of Class II crossovers, then we would expect sni1-1 to partially restore fertility to Class I pathway mutants (2). Therefore, we crossed sni1-1 with the fancm zip4 double mutant and analyzed pollen viability, silique length, and seed set in the resulting double- and triple-mutant lines (Fig. 6 C–E and SI Appendix, Fig. S10 C and D and Tables S32–S35). For all the traits analyzed, sni1-1 exhibits reduced values compared to wild type, showing that the fertility is decreased in this mutant (Fig. 6 B–E and SI Appendix, Fig. S10 C and D). However, the double sni1-1 zip4 mutant showed significantly higher fertility than the zip4 single mutant, supporting the hypothesis that crossover elevation observed in the sni1-1 mutant results from additional Class II events (Fig. 6 C–E and SI Appendix, Tables S32–S35). Interestingly, we found that the fancm mutation is not able to completely restore fertility in the zip4 mutant when combined with the sni1-1 mutation (Fig. 6 C–E and SI Appendix, Fig. S10 C and D). This is likely the result of other meiotic abnormalities observed in the absence of SNI1, including anaphase I bridges and chromosome fragments (Fig. 5B). Also, the sni1-1 fancm double mutant showed similar fertility to sni1-1 fancm zip4, which is much lower than that of the wild-type fancm zip4 but also sni1-1 (Fig. 6 C–E). To confirm that the partial restoration of fertility in sni1-1 zip4 is due to higher crossover numbers, we counted bivalents and chiasmata in metaphase I. While the average number of bivalents per cell in zip4 was 1.2, we observed 2.2 bivalents in sni1-1 zip4 (Fig. 6 F and G and SI Appendix, Table S36). This result is further consistent with the sni1-1 mutant exhibiting elevated Class II crossovers.
Discussion
Our genetic mapping revealed SNI1, a functional homolog of the NSE6 component of the SMC5/6 complex, as a factor ensuring proper course of meiotic recombination in plants. SMC5/6 plays multiple roles in somatic cells during DNA damage repair, replication fork restarting, and telomere maintenance (66). For these reasons, the complex is essential across eukaryotes with most subunits being highly conserved. However, NSE6, together with NSE5, forms a module which in yeast is not permanently bound to the SMC5/6 complex (67, 68). Therefore, the absence of NSE6 is tolerated in many organisms including fission and budding yeast, vertebrate cells, and plants (42, 67, 69, 70). Moreover, SNI1 is poorly conserved at the sequence level, though it shows structural similarities among different organisms, including several armadillo repeats (42). These features indicate that SNI1/NSE6 is more susceptible to variation, which may explain identification of this gene as a modifier of meiotic crossover. Our data suggest that the I235V substitution in SNI1 is casual to rQTL4, although it is unclear how this change affects protein functionality as evidenced by altered recombination frequency (Fig. 1) and different responses to environmental changes (Fig. 2E).
Previously, variation in REC8, a meiosis-specific component of cohesin, has been implicated as causative for differences in genome-wide recombination rates in even-toed ungulates (27, 28, 71) and plants (32, 33). Our work reveals that the SMC5/6 complex, similar to the cohesion complex (SMC1/SMC3), plays a role in crossover formation. Cohesin is a component of the chromosome axis, which holds sister chromatids together and organizes them into multiple chromatin loops (72). Anchoring chromatin loops to the axis by REC8-cohesin results in local exclusion of the recombination machinery and promoting of intersister repair of DSBs (73–75). The role of SMC5/6 in meiotic crossover formation remains incompletely characterized; however, it is known that the complex can also affect chromatin structure (76). Similarly to cohesin, the SMC5/6 complex can hold two sister chromatids inside its ring, aligning them and stimulating homologous recombination (77). Epistatic interactions between mutants of SMC5/6 and cohesin suggest that the two complexes act in related pathways in somatic cells (78). Moreover, SMC5/6-dependent SUMOylation of the SCC1 subunit of cohesin, a somatic counterpart of REC8, is required for DNA damage-induced sister chromatid cohesion (79–81). It will be interesting to investigate whether these relationships between the cohesion and SMC5/6 complexes are observed for meiotic recombination. Our data indicate that the SNI1 component of SMC5/6 complex is not required for the proper formation of the meiotic chromosome axis and sister chromatid cohesion (Fig. 5E).
In the sni1-1 mutant, we observed no additional DSBs that could be repaired as crossovers (Fig. 5 C and D and SI Appendix, Fig. S8). Therefore, the increase in crossover number is likely due to the formation of abnormal JMs. The elevation of crossover frequency in chromosome distal regions (Fig. 3), seeming drop of crossover interference (Fig. 4 E–H) and partial restoration of the Class I zip4 mutant fertility (Fig. 6 C–G), indicate that some of these abnormal JMs are repaired as Class II crossovers. Furthermore, our genetic assays show that these crossovers are largely independent from FANCM helicase activity (Fig. 6A) but likely depend on MUS81 endonuclease (SI Appendix, Fig. S10B). Together, this is consistent with the SMC5/6-SNI1 complex preventing formation of inappropriate JMs (Fig. 6H).
We propose that natural variation in SNI1 alters the function of the SMC5/6 complex and may have biological significance in modulation of meiotic recombination under varying environmental conditions (Fig. 2E). SMC5/6-SNI1 may affect meiotic DSB repair pathway choice, ultimately leading to changes in the crossover landscape.
Materials and Methods
Plant Material.
All the lines used in the study are described in SI Appendix, Table S37. Genotyping of mutant lines was performed using oligonucleotides as described in SI Appendix, Table S38. Plants were grown in controlled environment chambers at 22 °C with long day 16-/8-h light/dark photoperiods, 60% humidity, and 150-µmol light intensity. Prior to germination seeds were kept for 3 d in the dark at 4 °C to stratify germination.
Measurement of Crossovers Using Seed-Based Systems.
Pictures of seed were acquired using epifluorescent stereomicroscope Lumar version 12 (Zeiss) equipped with a charge-coupled device (CCD) camera in brightfield, ultraviolet (UV) through a dsRed filter, and UV through a green fluorescent protein filter. The CellProfiler program was used to identify seed boundaries in micrographs and to assign a dsRed and eGFP fluorescence intensity value to each seed object (37, 82). Histograms of seed fluorescence were used to classify fluorescent and non-fluorescent seeds for each color. The genetic distance is calculated as cM = 100 × (1–(1−2(NG+NR)/NT)1/2), where NG is the number of green alone seeds, NR is the number of red alone seeds, and NT is the total number of seeds analyzed.
rQTL Mapping.
Genomic DNA was extracted using cetrimonium bromide and genotyped using PCR amplification of Col/Ler simple sequence length polymorphism, cleaved amplified polymorphic sequences (CAPS), or derived CAPS markers (SI Appendix, Table S39). R statistical package rQTL was used to performed one-dimensional QTL mapping (83). The Haley–Knott regression algorithm using 0.1-cM steps for rQTL4 fine mapping was implemented. To fit models with multiple QTLs, the fitqtl function with Haley–Knott regression was used. LOD score significance thresholds were established from 10,000 permutations for each mapping population.
Generation of CRISPR-Cas9 Mutant of SNI1 in Ct Background for GBS.
To obtain a sni1 mutant in Ct background (sni1-2), a single guide RNA (gRNA) targeted within the first exon of the gene was designed. Agrobacterium-based transformation was performed using a vector containing the gRNA (SI Appendix, Table S40) under the U6 promoter and a ICU2::Cas9 transgene. Transformants were genotyped in PCR with primers flanking the SNI1 gRNA target site, and Sanger sequencing was performed to detect deletions. Mutants with heritable deletions causing a frame shift in SNI1 were identified and selfed. M2 plants were screened for individuals not carrying the CRISPR-Cas9 construct. The resulting sni1-2 mutant is described in SI Appendix, Fig. S2 C and D.
Backcrossing of sni1-1 (Col-0) Mutant to Ler Background for GBS.
To obtain sni1-1 mutant in Ler background, the sni1-1 (Col) allele was crossed to Ler-0. F1 plants obtained were backcrossed six times to Ler as recurrent female parent, and each generation was checked to carry sni1-1 mutation by genotyping (SI Appendix, Table S30). The genotype of the BC6 line was verified using 39 previously described Col/Ler indel markers (37).
Complementation of sni1-1 Mutation.
A DNA fragment containing SNI1 was amplified from Col or Ler genomic DNA using primers described in SI Appendix, Table S40. The PCR products were cloned into the pFGC-pcoCas9 binary vector using one step cloning kit (Vazyme). These vectors were transformed into Col-420 and sni1-1-420 hemizygous plants using Agrobacterium strain GV3101 and floral dipping.
Cytological Analysis.
Chromosome spreads and FISH were conducted following the previously described protocol (84). Immunolocalizations were performed as previously described (85). The primary antibodies were anti-AtZYP1 (rat; 1/500 dilution), anti-MLH1 (rabbit; 1/500 dilution), anti-ASY1 (rabbit, 1/500), anti-RAD51 (rat; 1/500), and anti-SYN1 (rabbit; 1/500). Secondary antibodies were anti-rat IgG FITC-conjugated (Agrisera) and anti-rabbit IgG Alexa Fluor 555 conjugated (Molecular Probes) for immunolocalizations to detect ZYP1+MLH1 and ASY1+RAD51 and anti-rat Alexa Fluor 555-conjugated (Molecular Probes) and anti-rabbit FITC-conjugated (Sigma-Aldrich) for the immunolocalization to detect ZYP1+SYN1. For scoring RAD51 and MLH1 foci, blind analyses were applied as a protection against bias. Slides were examined on an Olympus BX61 epifluorescence microscope equipped with a CCD Olympus DP71 camera and analyze using analySIS software (Soft Imaging System).
Fertility Assays.
Pollen viability was assessed from 1,500 pollen grains collected from open flowers of three plants per genotype, using Alexander staining (86). Seed set and silique length were assessed from five fruits, located at positions 6 through 10 of the main stem, in ten plants per genotype.
Additional Experimental Procedures.
A detailed description of qRT-PCR analysis, DNA extraction, and library preparation for GBS, GBS bioinformatics analysis, Pollen-based measurements of genetic distance, and crossover interference can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Gregory Copenhaver (University of North Carolina), Avi Levy (The Weizmann Institute), and Scott Poethig (University of Pennsylvania) for FTLs (CTLs); Erik Wijnker, Jose van der Belt, and Joost Keurentjes (University of Wageningen) for the CCCLC chromosome substitution line; Xinnian Dong (Duke University) for the sni1-1 mutant line; Jane Parker and Raphael Mercier (Max Planck Institute for Plant Breeding Research) for the Col-eds1-2 and fancm-1 mutant lines, respectively; Chris Franklin (University of Birmingham) for ZYP1, MLH1, ASY1, RAD51, and SYN1 antibodies; and Ales Pecinka (Institute of Experimental Botany, CAS) for critical reading of the manuscript. This work was supported by the Polish National Science Center Grants 2016/21/B/NZ2/01757 and 2016/22/E/NZ2/00455 to P.A.Z. and the Foundation for Polish Science Grant (POIR.04.04.00-00-5C0F/17-00) to P.A.Z. N.F.-J. is a PhD fellow funded by the trainee teaching staff program of Spanish Ministry of Education (FPU16/02772). Work in the Henderson group was supported by grants from the European Research Council (SynthHotSpot Consolidator Award) and Biotechnology and Biological Sciences Research Council Grants BB/L006847/1 and BB/M004937/1. M.P. acknowledges the support of the European Union (Marie Curie Innovative Training Networks, Meiotic Control of Recombination in Crops [MEICOM] 765212, and European Cooperation in Science and Technology [COST] Action CA 16212 “INDEPTH”).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021970118/-/DCSupplemental.
Data Availability
All data are publicly available. Sequencing data for sni1 Col×Ct GBS libraries have been deposited under ArrayExpress accession E-MTAB-9413 (87).
References
- 1.Barton N. H., Charlesworth B., Why sex and recombination? Science 281, 1986–1990 (1998). [PubMed] [Google Scholar]
- 2.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villeneuve A. M., Hillers K. J., Whence meiosis? Cell 106, 647–650 (2001). [DOI] [PubMed] [Google Scholar]
- 5.De Muyt A., et al., BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakharyevich K., Tang S., Ma Y., Hunter N., Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Séguéla-Arnaud M., et al., Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. U.S.A. 112, 4713–4718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serra H., et al., Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 2437–2442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xaver M., Huang L., Chen D., Klein F., Smc5/6-Mms21 prevents and eliminates inappropriate recombination intermediates in meiosis. PLoS Genet. 9, e1004067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copsey A., et al., Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions. PLoS Genet. 9, e1004071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto S. P., The evolutionary enigma of sex. Am. Nat. 174 (suppl. 1), S1–S14 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Coop G., Przeworski M., An evolutionary view of human recombination. Nat. Rev. Genet. 8, 23–34 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Smukowski C. S., Noor M. A. F., Recombination rate variation in closely related species. Heredity 107, 496–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M., Modification of linkage intensity by natural selection. Genetics 57, 625–641 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman M. W., Otto S. P., Christiansen F. B., Population genetic perspectives on the evolution of recombination. Annu. Rev. Genet. 30, 261–295 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Cullen M., Perfetto S. P., Klitz W., Nelson G., Carrington M., High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71, 759–776 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauppi L., Sajantila A., Jeffreys A. J., Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum. Mol. Genet. 12, 33–40 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Choi K., et al., Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 12, e1006179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baudat F., et al., PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinch A. G., et al., The landscape of recombination in African Americans. Nature 476, 170–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers S., Freeman C., Auton A., Donnelly P., McVean G., A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40, 1124–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Hinch A. G., et al., Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science 363, eaau8861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auton A., et al., A fine-scale chimpanzee genetic map from population sequencing. Science 336, 193–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong A., et al., Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319, 1398–1401 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Qiao H., et al., Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston S. E., Bérénos C., Slate J., Pemberton J. M., Conserved genetic architecture underlying individual recombination rate variation in a wild population of soay sheep (Ovis aries). Genetics 203, 583–598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandor C., et al., Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 8, e1002854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L., et al., Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genet. 11, e1005387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziolkowski P. A., et al., Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao H. B. D. P., et al., A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright K. M., et al., Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol. Biol. Evol. 32, 944–955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreissig S., et al., Natural variation in meiotic recombination rate shapes introgression patterns in intraspecific hybrids between wild and domesticated barley. New Phytol. 228, 1852–1863 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Lawrence E. J., et al., Natural variation in TBP-ASSOCIATED FACTOR 4b controls meiotic crossover and germline transcription in Arabidopsis. Curr. Biol. 29, 2676–2686.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Berchowitz L. E., Copenhaver G. P., Fluorescent Arabidopsis tetrads: A visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 3, 41–50 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Yelina N. E., et al., High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat. Protoc. 8, 2119–2134 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Ziolkowski P. A., et al., Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. eLife 4, e03708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G., Rossidivito G., Hu T., Berlyand Y., Poethig R. S., Traffic lines: New tools for genetic analysis in Arabidopsis thaliana. Genetics 200, 35–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wijnker E., et al., Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat. Genet. 44, 467–470 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Koncz C., et al., Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 9, 1337–1346 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Zhang Y., Clarke J. D., Li Y., Dong X., Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Yan S., et al., Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 52, 602–610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durrant W. E., Wang S., Dong X., Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. U.S.A. 104, 4223–4227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd A., Morgan C., H Franklin F. C., Bomblies K., Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208, 1409–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modliszewski J. L., et al., Elevated temperature increases meiotic crossover frequency via the interfering (type I) pathway in Arabidopsis thaliana. PLoS Genet. 14, e1007384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso-Blanco C.et al.; 1001 Genomes Consortium , 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowan B. A., Patel V., Weigel D., Schneeberger K., Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 5, 385–398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackwell A. R., et al., MSH2 shapes the meiotic crossover landscape in relation to interhomolog polymorphism in Arabidopsis. EMBO J. 39, e104858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Moran E., Armstrong S. J., Santos J. L., Franklin F. C. H., Jones G. H., Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162, 1415–1422 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chelysheva L., et al., An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129, 143–153 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Francis K. E., et al., Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 3913–3918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drouaud J., et al., Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16, 106–114 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H., et al., Linked-read sequencing of gametes allows efficient genome-wide analysis of meiotic recombination. Nat. Commun. 10, 4310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., Durrant W. E., Song J., Spivey N. W., Dong X., Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 107, 22716–22721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H., et al., RAD51 supports DMC1 by inhibiting the SMC5/6 complex during meiosis. Plant Cell 51, 10.1093/plcell/koab136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez E., et al., DNA damage as a consequence of NLR activation. PLoS Genet. 14, e1007235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lilienthal I., Kanno T., Sjögren C., Inhibition of the Smc5/6 complex during meiosis perturbs joint molecule formation and resolution without significantly changing crossover or non-crossover levels. PLoS Genet. 9, e1003898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zelkowski M., et al., Arabidopsis NSE4 proteins act in somatic nuclei and meiosis to ensure plant viability and fertility. Front Plant Sci 10, 774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu M., et al., SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol. 14, 153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grelon M., Vezon D., Gendrot G., Pelletier G., AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert V., SMC proteins and their multiple functions in higher plants. Cytogenet. Genome Res. 124, 202–214 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Watanabe K., et al., The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688–2699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Díaz M., et al., The SMC5/6 complex subunit NSE4A is involved in DNA damage repair and seed development. Plant Cell 31, 1579–1597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartung F., Suer S., Bergmann T., Puchta H., The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34, 4438–4448 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crismani W., et al., FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Diaz M., Pecinka A., Scaffolding for repair: Understanding molecular functions of the SMC5/6 complex. Genes (Basel) 9, 36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bustard D. E., et al., During replication stress, non-SMC element 5 (NSE5) is required for Smc5/6 protein complex functionality at stalled forks. J. Biol. Chem. 287, 11374–11383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leung G. P., Lee L., Schmidt T. I., Shirahige K., Kobor M. S., Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J. Biol. Chem. 286, 26250–26257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R. III, Boddy M. N., The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 26, 1617–1630 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Räschle M., et al., DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnston S. E., Huisman J., Pemberton J. M., A genomic region containing REC8 and RNF212B is associated with individual recombination rate variation in a wild population of red deer (Cervus elaphus). G3 (Bethesda) 8, 2265–2276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klein F., et al., A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Hong S., et al., The logic and mechanism of homologous recombination partner choice. Mol. Cell 51, 440–453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoon S. W., et al., Meiotic prophase roles of Rec8 in crossover recombination and chromosome structure. Nucleic Acids Res. 44, 9296–9314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambing C., et al., Interacting genomic landscapes of REC8-cohesin, chromatin and meiotic recombination in Arabidopsis thaliana. Plant Cell 32, 1218–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palecek J. J., SMC5/6: Multifunctional player in replication. Genes (Basel) 10, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kegel A., Sjögren C., The Smc5/6 complex: More than repair? Cold Spring Harb. Symp. Quant. Biol. 75, 179–187 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Potts P. R., Porteus M. H., Yu H., Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu N., et al., Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 26, 1473–1485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Almedawar S., Colomina N., Bermúdez-López M., Pociño-Merino I., Torres-Rosell J., A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 22, 1576–1581 (2012). [DOI] [PubMed] [Google Scholar]
- 81.McAleenan A., et al., SUMOylation of the α-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr. Biol. 22, 1564–1575 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Lamprecht M. R., Sabatini D. M., Carpenter A. E., CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Arends D., Prins P., Jansen R. C., Broman K. W., R/qtl: High-throughput multiple QTL mapping. Bioinformatics 26, 2990–2992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez-Garcia M., Pradillo M., “Functional analysis of Arabidopsis ARGONAUTEs in meiosis and DNA repair – Plant argonaute proteins: Methods and protocols” in Methods in Molecular Biology, Carbonell A., Ed. (Springer, New York, 2017), pp. 145–158. [DOI] [PubMed] [Google Scholar]
- 85.Varas J., Pradillo M., “Immunolabeling protocols for studying meiosis in plant mutants defective for nuclear envelope components – The LINC complex: Methods and protocols” in Methods in Molecular Biology, Gundersen G. G., Worman H. J., Eds. (Springer, New York, 2018), pp. 237–247. [DOI] [PubMed] [Google Scholar]
- 86.Alexander M. P., Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122 (1969). [DOI] [PubMed] [Google Scholar]
- 87.Zhu L., Szymanska-Lejman M., Ziolkowski P. A., Identifying crossover locations in Arabidopsis thaliana sni1 Col-0 x Ct-1 F2 populations using genotyping-by-sequencing. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9413/. Deposited 30 July 2020.
- 88.Choi K., et al., Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 532–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available. Sequencing data for sni1 Col×Ct GBS libraries have been deposited under ArrayExpress accession E-MTAB-9413 (87).