Significance
ABCB4 is an ATP-binding cassette transporter expressed in hepatocytes and is essential for the formation of bile by translocating phosphatidylcholine into canaliculi. Mutations in ABCB4 can cause inherited diseases leading to cholestasis. Similarly, commonly used drugs interfering with ABCB4 function can cause drug-induced liver injury. We present cryogenic electron microscopy structures that reveal how ABCB4 recruits phosphatidylcholine from the lipid bilayer and translocates it into bile canaliculi and how this process is inhibited by the antifungal drug posaconazole.
Keywords: ABC transporter, phosphatidylcholine, membrane transport, hepatocyte, cryo-EM
Abstract
ABCB4 is expressed in hepatocytes and translocates phosphatidylcholine into bile canaliculi. The mechanism of specific lipid recruitment from the canalicular membrane, which is essential to mitigate the cytotoxicity of bile salts, is poorly understood. We present cryogenic electron microscopy structures of human ABCB4 in three distinct functional conformations. An apo-inward structure reveals how phospholipid can be recruited from the inner leaflet of the membrane without flipping its orientation. An occluded structure reveals a single phospholipid molecule in a central cavity. Its choline moiety is stabilized by cation-π interactions with an essential tryptophan residue, rationalizing the specificity of ABCB4 for phosphatidylcholine. In an inhibitor-bound structure, a posaconazole molecule blocks phospholipids from reaching the central cavity. Using a proteoliposome-based translocation assay with fluorescently labeled phosphatidylcholine analogs, we recapitulated the substrate specificity of ABCB4 in vitro and confirmed the role of the key tryptophan residue. Our results provide a structural basis for understanding an essential translocation step in the generation of bile and its sensitivity to azole drugs.
One of the essential functions of the liver is the generation of bile, which is produced in canaliculi wedged between adjacent hepatocytes. Its primary constituents are mixed micelles formed by bile salts, phospholipids, and cholesterol. These are transported into bile canaliculi by three distinct ATP-binding cassette (ABC) transporters. Among them, ABCB4 (also known as MDR3) catalyzes the transport of phospholipids (1, 2), with a high selectivity for phosphatidylcholine (PC). The dysfunction of ABCB4 is linked to inherited or acquired liver disease, including progressive familial intrahepatic cholestasis type 3 (PFIC3) (3, 4). Furthermore, therapeutic agents including azole compounds widely used in antifungal therapy can inhibit ABCB4 and cause drug-induced liver injury (DILI) in susceptible patients (5). One such azole, posaconazole, has been shown to interfere with ABCB4 activity, resulting in decreased PC secretion and causing increased toxicity of bile (5, 6).
Results and Discussion
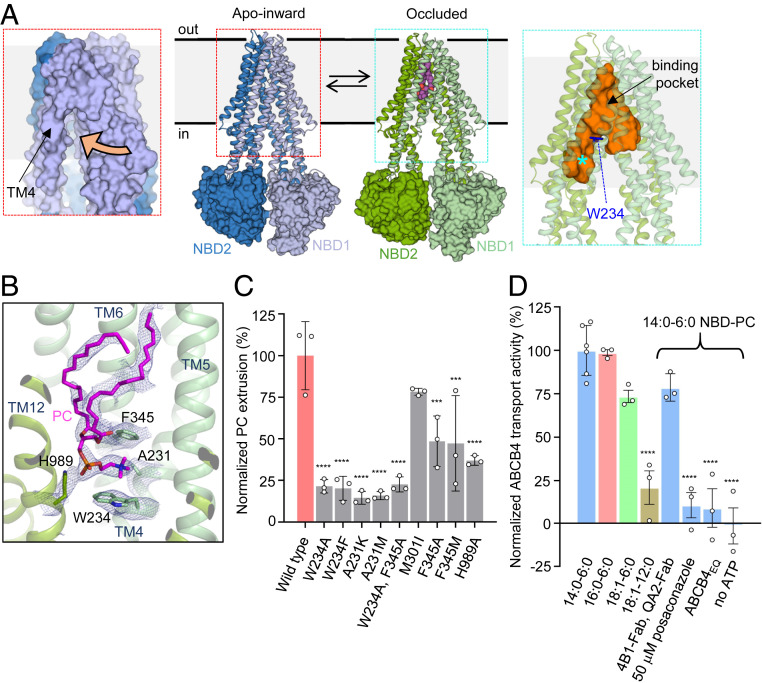
Although ABCB4-related cholestatic diseases have been extensively studied in genetic mouse models (7–10) and in cellular systems (11), little is known about the mechanism of ABCB4 or the structural basis of its inhibition by drugs. The structure of a catalytically inactive variant of human ABCB4 revealed a closed conformation of the nucleotide-binding domains (NBDs) in a prehydrolytic state and a collapsed translocation pathway between the transmembrane domains (TMDs) (12). To visualize wild-type ABCB4, we used phage display mutagenesis employing an epitope masking strategy (13, 14) to generate two antibody fragments (4B1-Fab and QA2-Fab) that bind to two independent epitopes at the interface of the NBDs and the TMDs of ABCB4 (SI Appendix, Fig. S1A). We collected cryogenic electron microscopy (cryo-EM) data of nanodisc-reconstituted ABCB4 bound to 4B1-Fab and QA2-Fab (SI Appendix, Figs. S2A) and found that the transporter adopted two distinct conformations that likely represent states in equilibrium (Fig. 1A). The two EM maps had overall resolutions of 3.6 Å and 4.2 Å and featured side chain resolution for the TMDs (SI Appendix, Fig. S3), allowing for de novo model building. In contrast, the resolution was lower for the NBDs. The ABCB4-Fab interfaces are identical in the two conformational states (SI Appendix, Fig. S4), ruling out conformational bias introduced by the presence of the antibody fragments. The structural differences between the two observed ABCB4 conformations are functionally relevant: We termed the first state “apo-inward” because it featured a side opening to the inner leaflet of the bilayer (Fig. 1A). The second state was termed “occluded” because the kinked conformations of transmembrane helices TM4 and TM10 formed a Y-shaped, occluded cavity of ∼6,500 Å3 at the center of the transporter (Fig. 1A). Whereas the TM helix arrangement in the occluded state resembled structures of the multidrug transporter ABCB1 in substrate- or inhibitor-bound states (15, 16), to our knowledge, the apo-inward-conformation has not previously been observed in human ABC transporters. When compared to the ATP-bound, closed conformation of the ABCB4EQ variant (12), shifts in TM4, TM5, and TM8 are evident, whereas the remaining TM helices superimposed closely (SI Appendix, Fig. S5). The most pronounced shift was that of TM4, which creates a side opening to the lipid bilayer (Fig. 1A).
Fig. 1.
Structure of wild-type ABCB4 and functional analysis. (A) Structures of apo-inward and occluded ABCB4 (Center), with TMDs shown as ribbons and NBDs as surfaces. The N- and carboxyl-terminal halves of ABCB4 are colored differently. Bound phosphatidylcholine is shown as purple spheres. (Left) Close-up of the apo-inward conformation shown as a surface representation. The side opening to the inner leaflet of the membrane is indicated by an orange arrow. (Right) Close-up of the occluded conformation. The central cavity, generated with HOLLOW (47), is shown as an orange surface. The phosphatidylcholine binding pocket is located above the residue W234, shown as blue sticks. The asterisk depicts the area termed the access tunnel in the related ABCB1 protein (16). (B) Binding pocket of PC molecule in occluded conformation. Bound PC and interacting residues are shown as sticks; EM density is shown as blue mesh. (C) Normalized PC extrusion rates from HEK293T cells expressing wild-type or mutant ABCB4. The results are normalized to the amount of ABCB4 extracted from the membranes relative to wild-type ABCB4. Means of measurements from at least three biological replicates are shown; error bars represent SDs. Statistical significance was determined using Dunnett’s test. Differences between wild-type and mutants were depicted as *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (D) Normalized in vitro transport activity of wild-type ABCB4. Proteoliposomes contained liver polar lipid extract and cholesterol (4:1 wt/wt) supplemented with 0.3% (wt/wt) NBD-PC. Numbers refer to the alkyl tail lengths and the number of double bonds. Data were normalized to the highest transport rate. Means of measurements from at least three independent proteoliposomes-based samples are shown; error bars represent SDs. Statistical significance was determined using Dunnett’s test. Differences between 14:0 and 6:0 NBD-PC condition and other samples were depicted as *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
In the occluded conformation, a distinctive EM density feature in the central cavity matched the shape of a phospholipid and was modeled as a PC molecule (Fig. 1B). While the density for the headgroup was well defined, that of the alkyl chains was less well resolved. This may be due to flexibility within the binding pocket or reflect a mixture of fatty acid tails of the bound PC molecules. The alkyl chains of the fatty acid moieties fill the hydrophobic end of the cavity in the vicinity of extracellular loops ECL3 and ECL4. The occluded cavity containing bound PC extends further toward the outer membrane boundary than in any of the structures of ABCB1 (16). As a consequence, the phospholipid bound to ABCB4 is shifted almost to the level of the outer leaflet but without having been flipped and with the headgroup at the level of the center of the lipid bilayer. The choline moiety is wedged between two aromatic side chains, W234 from TM4 and F345 from TM6 (Fig. 1B). This arrangement allows for strong cation-π interactions (17), which suggests these may strongly contribute to PC recognition. This is notably reminiscent of choline recognition by acetylcholine receptors, where similar aromatic cages were observed (18–20). The phosphate group of bound PC interacts with the side chain of H989 (Fig. 1B). Because there is no positively charged residue nearby, the negative charge of the phosphate group may be partially transferred to the histidine side chain.
To investigate the functional relevance of these structural observations, we tested the activity of ABCB4 variants in HEK293 cells (Fig. 1C). While mutating W234 and/or F345 did not fully abolish activity, it reduced phospholipid extrusion to ∼20%. The activity of the ABCB4 variant H989A was reduced to ∼40%. Mutating A231 to lysine or methionine significantly reduced ABCB4 activity, likely due to steric clashes introduced by these mutations (Fig. 1C). In addition to the cellular assay, we developed an in vitro assay of ABCB4-mediated PC transport (SI Appendix, Fig. S6) based on a previously described method (21). Fluorescent phosphatidylcholine analogs (PCfluo) carrying a nitrobenzoxadiazole (NBD) group attached to one of the fatty acid chains were coreconstituted with ABCB4 in proteoliposomes. The fluorophore can be quenched by dithionite, but only for lipids located in the outer liposome leaflet given that dithionite cannot permeate the bilayer (21, 22). We determined that ABCB4-mediated PCfluo transport was ATP-dependent and inhibited by posaconazole (Fig. 1D). The two Fab fragments used for structural studies had no significant effect on PCfluo transport, thus they do not inhibit reconstituted ABCB4. The specificity of ABCB4 is faithfully recapitulated in vitro, as translocation was observed for PCfluo but not for the equivalently NBD-labeled phosphatidylethanolamine (PEfluo, SI Appendix, Fig. S6C). We found that a PCfluo analog containing longer alkyl tails (18:1–12:0) resulted in a reduced transport rate (Fig. 1D). While the NBD moiety is likely to influence the process, we conclude that PC molecules can only be translocated as long as they fit the binding pocket, which can be rationalized by the limited space of the cavity.
Our results provide a molecular description for the observed substrate specificity of ABCB4. The binding of lipids with headgroups larger than choline (e.g., phosphatidylinositol) would be impeded due to steric clashes. Lipids without a net positive charge in the alcohol entity (e.g., phosphatidylserine or phosphatidylglycerol) would not benefit from the cation-π interactions provided by W234 and F345, weakening any potential binding. The specificity of PC over PE is less obvious. Given that phosphatidylethanolamine (PE) is a poor substrate of ABCB4 both in vivo [very low concentration in bile (23)] and in vitro (SI Appendix, Fig. S6C), we surmise that despite being compatible in size and containing a positive charge if protonated, its interaction with the aromatic cage of ABCB4 must be weak. While we cannot exclude other effects, the most straightforward explanation is that unlike the trimethylamine group of choline, the primary amine group of ethanolamine can be deprotonated as the lipid travels from the inner leaflet to the binding pocket of ABCB4. Deprotonation is possible despite the pKa of 9.6 (24) given that the headgroup has to enter the hydrophobic cavity of the access tunnel, where deprotonation would lead to an uncharged amine group and thus a lower energetic penalty while traveling through the low-dielectric region of the protein. A similar effect is responsible for keeping aspartate and glutamate side chains of the c-ring of ATP synthase protonated when they are exposed to the hydrophobic section of the lipid bilayer (25).
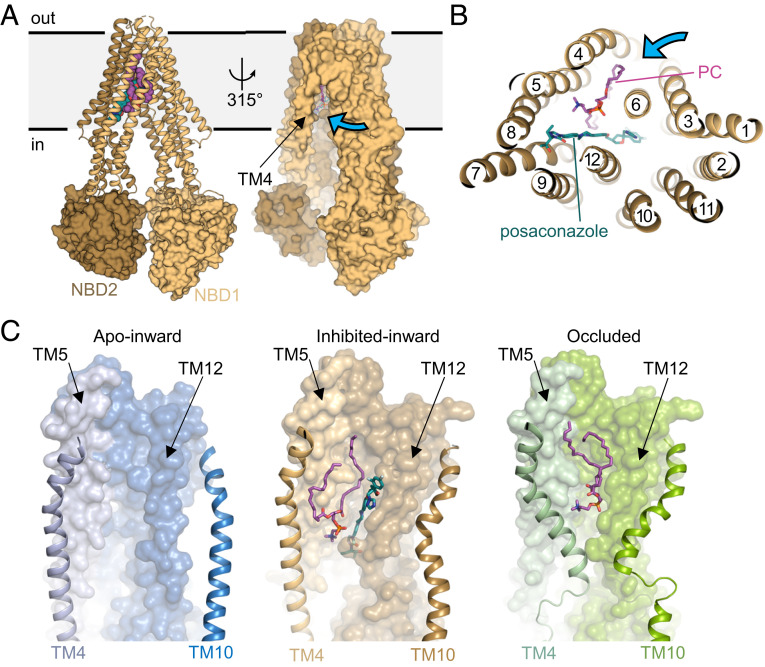
To elucidate the molecular basis of azole inhibition, we determined the structure of the catalytically inactive variant of ABCB4 (ABCB4EQ) in a complex with 4B1-Fab and in the presence of posaconazole at an overall resolution of 3.8 Å (SI Appendix, Figs. S1C, S2B, and S7). ABCB4EQ was chosen due to its lower dynamics and flexibility compared with the wild-type protein. The structure revealed an inward-open conformation with a wider domain separation compared to the other structures (Fig. 2A). The central cavity observed in the occluded state is open to the cytosolic side and to the inner leaflet of the membrane (Fig. 2 A and B). We observed two distinct EM density features in the cavity: The first matched a posaconazole molecule approximately at the position where phospholipid is observed in the occluded conformation (SI Appendix, Fig. S8). The second matched a phospholipid and was therefore built as a PC molecule. It is proximal to the side opening to the membrane, with the choline moiety positioned to form cation-π interaction with W234 (SI Appendix, Fig. S8C). Whereas one of the alkyl tails extends into the binding cavity, the headgroup and the second alkyl tail of bound PC are closer to the side opening to the membrane than in the occluded conformation (Fig. 2C). The posaconazole-bound ABCB4 structure therefore not only reveals the structure of an intermediate state of PC recruitment from the lipid bilayer but also how antifungal azoles inhibit ABCB4, which may help develop antifungal agents with a lower potential for causing liver damage. Bound posaconazole does not interfere with PC advancing to an intermediate location but prevents it from fully entering the binding pocket. The inhibitor also prevents ABCB4 from adopting an occluded conformation as it interferes with the required kinking of TM4 and TM10 (Fig. 2C).
Fig. 2.
Structure of posaconazole-inhibited ABCB4. (A, Left) TMDs are shown as ribbons and NBDs as surfaces. The N- and carboxyl-terminal halves of ABCB4 are colored differently. Posaconazole and PC are shown in sphere representation and colored green and purple, respectively. (Right) Surface representation. The blue arrow shows the side opening to the membrane. (B) Horizontal slice through a ribbon representation, with posaconazole and PC shown as green and purple sticks. (C) Close-up views of the TMDs in distinct states. Bound PC and posaconazole are shown in sticks and colored as in B. The TM helices 4 and 10 undergoing conformational changes are shown as ribbons. Posaconazole prevents bound PC from reaching the central pocket and TM4 from adopting a kinked conformation.
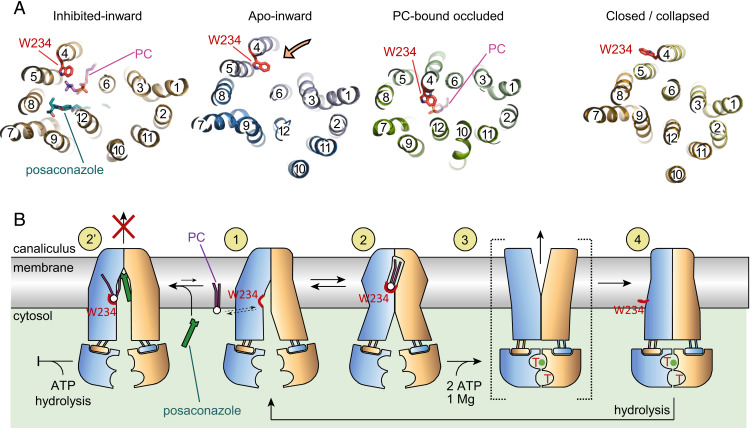
A comparison of the available ABCB4 structures provides the context for the conformational changes associated with PC translocation (Fig. 3A and SI Appendix, Fig. S9). The tryptophan residue W234 plays a key role and adopts three distinct conformations during the transport cycle. In the apo-inward state, it is positioned near the side opening to the bilayer, where it may contribute to the recruitment of choline-containing phospholipids. This premise is supported by the structure of posaconazole-inhibited ABCB4, where a bound lipid molecule was observed in a loading position. In the occluded conformation, W234 is located in the central cavity, where it participates in strong cation-π interactions with the choline group, ensuring specificity for PC. In the ATP-bound state (12), W234 is no longer at the center of the transporter but has swung outwards, facing the lipids and eliminating the choline binding pocket.
Fig. 3.
Structural changes of ABCB4 during transport cycle. (A) Horizontal slices through the TMDs of available ABCB4 structures, shown from the inside of the cell. TM helices are shown as ribbons and numbered. Bound PC and posaconazole and the side chain of W234 are shown as sticks. The orange arrow shows the side entrance from the inner leaflet of the membrane. (B) Proposed structure-based reaction mechanism. The N- and carboxyl-terminal halves of ABCB4 are colored yellow and blue. The red arch depicts the choline binding site including the side chain of W234. Numbers in yellow circles depict functionally relevant conformational states: States 1, 2, and 2′ represent structures from this study, state 4 is based on PDB: 6s7p (12), and state 3 is based on a putative outward-open conformation (12).
The structures described here provide detailed snapshots of three of the functional states (states 1, 2, and 2′) of ABCB4 involved in lipid recruitment and binding (Fig. 3B). Taken together with previous structural data, we propose a mechanistic framework for how ABCB4 performs its functions and how its distinctive specificity is built into its structure (Fig. 3B). The three structures reveal the early, ATP-independent steps of ABCB4-catalyzed phosphatidylcholine translocation (states 1 and 2). The subsequent steps require an outward-open, ATP-bound state (state 3) as modeled previously and a collapsed state (state 4), both of which feature dimerized NBDs (12). PC extrusion and resetting of the transporter require the binding (state 3) and hydrolysis (state 4 to state 1) of ATP.
Conclusions
ABCB4 was originally identified as a sister protein to the multidrug transporter ABCB1 (P-glycoprotein), and the two proteins share remarkably high sequence similarity (86%) (26). Our results suggest that rather than a credit card swipe mechanism (27), ABCB4 employs an alternating access mechanism similar to that proposed for ABCB1, but with a central pocket that harbors a phospholipid molecule rather than a drug. While ABCB1 was reported to display in vitro transport activity of fluorescently labeled phospholipids containing very short acyl chains (21, 28), its physiological role is to transport drugs, not phospholipids. The difference in shape and size of the substrate-binding cavities in the occluded states can rationalize the distinct functions. The cavity in ABCB4 is larger and provides space for alkyl chains of phospholipids that would not fit the cavity of ABCB1 (15, 16). ABCB4 could in principle interact with, be inhibited by, and transport several ABCB1 substrates, but this moonlighting activity is likely not of physiological relevance given its exclusive expression in the canalicular membrane. In conclusion, our results provide a structural framework for understanding the specific function and inhibition of ABCB4 and contribute to a better understanding of a key function of the liver.
Materials and Methods
Protein Expression and Purification.
Wild-type human ABCB4 (UniProt ID P21439-2) and the catalytically inactive variant ABCB4EQ (E558Q, E1200Q) were expressed and purified as described (12). Briefly, the proteins were expressed as fusion constructs with a 3C cleavable carboxyl-terminal eYFP/rho-1D4 tag from a stable cell line generated in a tetracycline-inducible Flp-In T-Rex system (Thermo Fisher Scientific). Cells were adapted and grown in EX-Cell 293 serum-free media (Sigma) supplemented with 2% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 μg mL−1 streptomycin, 100 units mL−1 penicillin (Gibco), and 6 mM L-glutamine (Gibco) at 37 °C, 6% CO2, and 80% humidity. Cell expression was induced by adding tetracycline (Sigma) to a concentration of 0.50 to 0.75 μg⋅mL−1. Induced cells were maintained for 48 h under the same conditions, followed by harvesting and storing at −80 °C.
All purification steps were performed at 4 °C or on ice, whenever possible. The cell pellet was resuspended (4:1 vol/wt) in working buffer (25 mM Hepes [pH 7.4], 150 mM NaCl, 20% glycerol) supplemented with cOmplete EDTA-free protease inhibitor tablets (Roche) and DNaseI (Roche) and dounce homogenized. A mixture of n-dodecyl-β-D-maltopyranoside (DDM, Anatrace) and cholesteryl hemisuccinate (CHS, Anatrace) was added to a final concentration (wt/vol) of 1% and 0.2%, respectively. The protein extraction lasted 2 h, followed by centrifugation at 100,000×g for 40 min. The supernatant was applied to pre-equilibrated Sepharose-coupled rho-1D4 antibody (University of British Columbia) resin (1D4 resin) and incubated for 2 h. The resin was washed four times with 10 column volumes (CVs) of the running buffer (the working buffer supplemented with 0.02% DDM and 0.004% CHS). To elute the protein, resin was incubated for at least 1 h with 3 CV of the running buffer supplemented with His-tagged 3C protease (1:10 wt/wt protease to estimated protein). The solution was passed through equilibrated Ni-NTA resin to capture the protease. The eluted protein in detergent was measured at 280 nm absorbance using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) to determine the concentration.
Proteoliposome and Nanodisc Reconstitution.
A mixture of Liver Polar Lipid Extract (LPL, Avanti Polar Lipids) and cholesterol (CLR, Avanti Polar) (4:1 wt/wt) was prepared as described (29). The mixture was used for both liposome and nanodisc generation. For experiments involving fluorescently labeled lipids, 0.3% (wt/wt) NBD-lipid (Avanti Polar) was added at the initial step where LPL and CLR were mixed.
Proteoliposomes were prepared as described (29). In short, the LPL:CLR mixture was frozen and thawed at least 3 times, followed by extruding 11 times through a 400 nm polycarbonate filter (Whatman). Large unilamellar vesicles were destabilized with 0.23% (vol/vol) Triton X-100 and mixed with detergent-solubilized protein at 100:1 (wt/wt) ratio. Prewashed Bio-Beads SM-2 (Bio-Rad) were added to the solution to uptake the detergent. Formed proteoliposomes were passed through a gravity column, spun, and resuspended in HBS (25 mM Hepes [pH 7.4], 150 mM NaCl) to a final lipid concentration of 10 mg⋅mL−1. The reconstitution efficiency was determined by silver stained gel densitometry analyzed in ImageJ (30).
For nanodisc reconstitution, the mixture of LPL:CLR was solubilized in 1% DDM and 0.2% CHS followed by sonication. The translucent lipid mixture was mixed with detergent-solubilized ABCB4 and incubated at ambient temperature for 5 min. Next, membrane scaffold protein (MSP1D1) was added and the solution was diluted with room temperature HBS to decrease the final glycerol concentration to 4%. The mixture was incubated for another 20 min. Note that the molar ratio of the components (protein to MSP to lipids) was 1:5:100. Prewashed Bio-Beads SM-2 (Bio-Rad) (∼0.7 g per 100 μg of the protein in the solution) were added. and the solution was incubated at 4 °C overnight with gentle stirring. The sample was passed through a gravity column, and the supernatant was concentrated on a 100,000 molecular weight cutoff centrifugal filter (Amicon, Merck). The final purification step involved size exclusion chromatography (SEC) running with HBS.
Phage Display.
To facilitate the biopanning selections, wild-type ABCB4 was reconstituted into nanodiscs using biotinylated MSP1D1. The MSP1D1 protein was chemically biotinylated as described (31), using fivefold molar excess of EZ-Link-NHS-PEG4-Biotin (Thermo Fisher Scientific). A pull-down assay on the streptavidin-coated paramagnetic particles (Promega) confirmed the biotinylation efficiency. Fab Library E (13, 32), a phage library expressing Fab-fragments on the surface, was used for biopanning (DNA was kindly provided by S. Koide). All five rounds were performed in a selection buffer (HBS supplemented with 1% bovine serum albumin). In the first round, 200 nM nanodisc-reconstituted ABCB4 was immobilized on magnetic beads, followed by manual biopanning. The beads were washed three times with the selection buffer, where afterward only phage expressing Fab-fragments specific to the immobilized target remained attached. The beads were resuspended and used to infect log-phase Escherichia coli XL-1 Blue cells. Phage were amplified overnight in media with M13-KO7 helper phage (109 pfu⋅mL−1) and ampicillin (100 μg⋅mL−1) and used as an input for the subsequent round. Additional four rounds of biopanning were performed with decreasing target concentrations: 100, 50, 20, and 10 nM, respectively. For these rounds a KingFisher magnetic beads handler (Thermo Fisher Scientific) was used to semiautomate the procedure. In addition, to disassemble the nanodiscs and elute binders specific only to the reconstituted target, the final step of each round included 15 min incubation in 1% Fos-choline-12 (prepared in the selection buffer). The reduction of nonspecific binders was achieved by i) preclearing phage pools from each of 2 to 5 rounds with 100 μL streptavidin and ii) using 1.5 μM nonbiotinylated empty MSP1D1 nanodiscs as soluble competitors in every round.
The phage pools from the fourth and fifth round were screened for individual clones and validated by single-point phage ELISA using 96 well plates (Nunc). The plates were coated with 2 μg⋅mL−1 neutravidin and blocked with selection buffer. Individual phage were amplified by inoculating 400 μL media (supplemented with 109 pfu⋅mL−1 M13-KO7 and 100 μg⋅mL−1 ampicillin) with E. coli XL-1 colonies harboring individual phagemids and incubating overnight in 96 well deep blocks at 37 °C and 280 rpm shaking. The supernatant, which contained the phage particles was collected and tenfold diluted in the selection buffer prior the assay. Nanodisc-reconstituted ABCB4 and empty nanodiscs (control sample) were immobilized on a coated plate at 50 nM concentration for 30 min at ambient temperature, followed by 30 min incubation with diluted phage. Next, 30 min incubation with HRP-conjugated anti-M13 monoclonal antibody (GE Heatlhcare) and an addition of TMB substrate (Thermo Fisher Scientific) detected a signal corresponding to bound phage particles. Specific binders were selected based on the absorbance signal ratio between the immobilized target wells and corresponding background from the empty nanodisc wells. The clones were sequenced at the University of Chicago Comprehensive Cancer Center DNA Sequencing Facility.
Fab Expression and Purification.
The clones were subcloned into pRH2.2 plasmid using an In-Fusion Cloning kit (Takara). E. coli BL21-Gold cells (Agilent) were transformed with the plasmids expressing verified the Fab-fragments. Cells were grown in 1 L media supplemented with 100 μg⋅mL−1 ampicillin to OD600 of 0.8, then induced with 1 mM IPTG and maintained for another 4 h at 37 °C. The cells were harvested and stored at −80 °C. The antigen-binding fragments were purified by Protein A chromatography, followed by ion-exchange chromatography as described previously (33).
To estimate the ABCB4:Fab binding affinity, a multipoint ELISA was conducted. Briefly, the same neutravidin-coated 96 well plates were used to immobilize 50 nM nanodisc-reconstituted ABCB4. Fab-fragments were subsequently diluted threefold (starting from 3 μM) and assayed against the immobilized transporter. The binder detection was done using HRP-conjugated mouse anti-human IgG F(ab′)2 (Jackson) and TMB substrate (Thermo Fisher Scientific). Data were plotted, and the binder with the highest affinity (4B1-Fab, the lowest estimated EC50 value) was selected for epitope-masking phage display.
Epitope-Masking Phage Display.
The epitope-masking phage display (34) was conducted as an epitope-exclusion strategy in isolating another ABCB4-specific binder. The biopanning procedure was similar to the one described above for nanodisc-reconstituted wild-type ABCB4 as an immobilized target; however, it was conducted in the presence of isolated 4B1-Fab fragment to block the immunodominant epitope. The excess of the 4B1-Fab (1.0 μM) was used in every round of biopanning and at every step of the procedure (immobilization, washing, etc.). Subsequent rounds (first manual followed by semiautomated) were performed with decreasing target concentrations: 300, 150, 75, 50, and 25. Similarly, 2.0 μM nonbiotinylated empty MSP1D1 nanodiscs were used as soluble competitors in every round. The phage pools from the fourth and fifth rounds were also screened for individual clones using single-point ELISA as described above; however, from this point on 4B1-Fab was not included in the subsequent steps. The procedures of selecting phage binders, sequencing phagemids, subcloning, expressing and purifying Fab-fragments, and performing multipoint ELISA were repeated. The second QA2-Fab binder was selected and the simultaneous binding of 4B1-Fab and QA2-Fab to nanodisc-reconstituted ABCB4 was confirmed with SEC (SI Appendix, Fig. S2).
ATPase Assays.
The assay was conducted to measure the amount of inorganic phosphate released from the ABCB4-mediated ATP hydrolysis reaction at various conditions (35). Nanodisc-reconstituted wild-type ABCB4 in HBS was used in the concentration range of 0.01 to 0.03 mg⋅mL−1. Fab-fragments were used in at least threefold excess to ensure saturation. Posaconazole (Sigma) was dissolved in dimethyl sulfoxide (DMSO) and sequentially diluted to 100× working stock solutions. Upon addition of posaconazole, each reaction sample contained 1% DMSO. All ATPase reactions were started by addition of 4 mM ATP and 10 mM MgCl2, incubated for 0 to 60 min at 37 °C and stopped by adding 6% SDS. Using GraphPad Prism 8, ATPase rates were calculated from linear regressions, and the IC50 value was determined from the sigmoidal interpolation (four parameter logistic equation) of the data points.
ABCB4 Mutant Generation and PC Extrusion Assay.
The ABCB4 mutants were generated by replacing the gene fragment of interest with corresponding synthetic, high-fidelity, double-stranded gBlocks Gene Fragments (IDT) using T4 DNA ligase (NEB). Positives clones were confirmed by sequencing at Microsynth AG prior to the assay.
The extrusion assays were performed in transiently transfected HEK293T cells as described before with minor modifications (12). Cells were seeded, grown, and maintained as adherent cultures in Dulbecco Modified Eagle Medium (DMEM, gibco) supplemented with 100 μg⋅mL−1 streptomycin, 100 units mL−1 penicillin and 10% FBS at 37 °C under 5% CO2 and humidified condition. At 50 to 80% cell confluency the medium was exchanged with one with lower FBS concentration (2%) and the transfection was done by dropwise adding a DNA:branched polyethylenimine mixture (1:2.5 wt/wt ratio), followed by 24 h expression in similar conditions. Medium was aspirated, cells were washed with prewarmed PBS (Gibco) and fresh DMEM without FBS but containing antibiotics and with 0.02% BSA. After 18 h, the media was collected and spun, and the supernatant was used to extract the lipids, while the cells were collected and harvested. A modified Bligh-Dyer liquid extraction method (36) was used: 0.8 mL supernatant mixed with 4.5 mL 1:2 (vol/vol) chloroform:methanol mixture was vigorously vortexed, followed by addition of 1.5 mL 0.1 M KCl and another mixing. The lowest, organic phase was collected and dried under the argon stream. Dried lipids were resuspended and PC content was analyzed with a Phosphatidylcholine Assay Kit (Sigma). The amount of extruded PC was normalized relative to the expression level of DDM:CHS solubilized, fluorescently labeled construct. Briefly, the cells were mixed with 25 mM Hepes (pH 7.4), 150 mM NaCl, 20% glycerol supplemented with 1% DDM and 0.2% CHS, extracted for 2 h, and spun at 250,000×g. Supernatant was loaded on a TSKgel G3000SWXL column at 0.4 mL⋅min−1 in 25 mM Hepes (pH 7.4), 150 mM NaCl, 0.02% DDM, and 0.004% CHS buffer and analyzed for a fluorescent signal. Results were normalized relative to the wild-type ABCB4.
Proteoliposome-Based Translocation Assay.
ABCB4 proteoliposomes were prepared as described above using LPL:CLR containing 0.3% of respective NBD-lipid. At each step the lipid mixture was prevented from photobleaching by minimizing exposure to light. The translocation reaction mixture was prepared in HBS diluting proteoliposomes to 0.5 mg⋅mL−1 in the presence or absence of 0.05 mM posaconazole or at least threefold molar excess of Fab-fragments. The mixture was prewarmed at 37 °C with gentle shaking, and the reaction was started by addition of ATP and MgCl2 mixture to the final 4 mM and 10 mM concentrations, respectively. The translocation reaction timepoints were collected between 0 and 40 min and stopped by mixing with vanadate (final concentration of 1 mM). Each condition of the translocation reaction was conducted at least in triplicate. Each timepoint sample was transferred to a 10 mm Quartz cuvette (Hellma) with a magnetic stirring bar prior to the measurement of fluorescent signal. Time-based measurements (counts per second) were recorded on the PTI QuantaMaster 800 spectrofluorometer (Horiba) at 470 nm excitation and 540 nm emission wavelengths at constant 22 °C temperature with 35 rpm mixing. Signal was recorded for 129 s until it stabilized and freshly prepared sodium dithionite (Sigma, dissolved in unbuffered Tris) was added to a final concentration of 5 mM. After 360 s Triton X-100 was added to a final concentration of 0.5% and measured for another 100 s.
From each sample measurement, three plateaus were denoted as averages of the last ten data points of each plateau: prior dithionite addition (Bmax), prior Triton X-100 addition and at the end of the measurement (Bmin, background signal). To reduce the background signal, Bmin was subtracted from the data points, and the values were normalized to 100% (SI Appendix, Fig. S7B, panel I). The plateau upon dithionite addition was denoted (panel II). These values from each timepoint of the translocation reaction were converted into differences of quenched NBD-lipids and plotted as a function of reaction time (panel III). The values were converted into moles of the corresponding NBD-lipids and normalized with the moles of active ABCB4 (determined from reconstitution efficiency and assumed 50% orientation distribution; panel IV). The transport rate is an average of at least three slopes of the linear regressions. Interestingly, the activity of the transporter depends on the commercially available batch of LPL lipids used for the preparation of the liposomes; therefore, the conditions were studied in reference to the “basal” activity of each lipid batch. Observed noninhibited transport rates were in a range of 50 to 300 mmol NBD-PC (mol ABCB4)-1 min−1. All data were normalized to the basal activity of 14:0–6:0 NBD-PC lipid sample.
Sample Preparation for Cryo-EM.
Both nanodisc-reconstituted ABCB4 samples were prepared as described above, and before the final step of SEC, Fab-fragments were added to the sample. For both wtABCB4:4B1-Fab:QA2-Fab and ABCB4EQ:4B1-Fab, samples were mixed with the corresponding Fabs in 1:2 molar ratio and incubated on ice for 30 min. Samples were loaded on a TSKgel G3000SWXL column at 0.4 mL⋅min−1 in HBS (SI Appendix, Fig. S2), and selected fractions were pooled. The wtABCB4:4B1-Fab:QA2-Fab sample was concentrated on a 100,000 molecular weight cutoff centrifugal filter (Amicon, Merck) to 0.60 mg⋅mL−1 and used for plunge freezing. Similarly, the ABCB4EQ:4B1-Fab sample was concentrated to 0.48 mg⋅mL−1, and posaconazole stock solution was added to a final concentration of 0.05 mM (0.5% DMSO). Samples were applied onto glow discharged Quantifoil R1.2/1.3 carbon/copper Cu 300 mesh grids, followed by plunge freezing in liquid ethane using Vitrobot Mark IV (FEI) at 4 °C and 100% humidity.
Cryo-EM Data Collection and Processing.
The samples were imaged on a Titan Krios 300 kV microscope (Thermo Fisher Scientific) with a Gatan K3 camera and Gatan BioQuantum 1967 energy filter. Data were acquired semiautomatically with EPU (Thermo Fischer Scientific) at a nominal magnification of 130,000 and in superresolution mode (0.33 Å/px). The defocus range was set between −0.6 and −2.4 μm with a 0.2 μm step. Each image stack contained 40 frames with an average dose of 2.0 e−/Å2 per frame (wtABCB4:4B1-Fab:QA2-Fab) or 1.7 e−/Å2 per frame (posaconazole-bound ABCB4EQ:4B1-Fab). We collected a total of 9477 multiframe micrographs of wtABCB4:4B1-Fab:QA2-Fab data set and 23,673 multiframe micrographs (two data sets) of the posaconazole-bound ABCB4EQ:4B1-Fab.
The data processing details are presented in SI Appendix, Figs. S3 and S8. Briefly, multiframe micrographs were imported into Relion 3.1 (37) and were motion-corrected (MotionCor2) (38), dose-weighted, and binned by a factor of 2 to a pixel size of 0.66 Å/px. Contrast transfer function (CTF) parameters were estimated with Gctf (39). Micrographs with estimated resolution lower than 4 Å were excluded from further processing. For each data set, initially, reference-free autopicking with Laplacian-of-Gaussian filtering was performed, followed by several rounds of 2D classifications. The best classes (612,269 particles of wtABCB4:4B1-Fab:QA2-Fab data set and 305,910 particles the posaconazole-bound ABCB4EQ:4B1-Fab) were then used to generate an ab initio model. The initial 3D model was then used as a template for particle autopicking. The particles were threefold binned and used for several rounds of 2D classification followed by two rounds of 3D classification. The particles from the best resolved 3D class were reextracted (unbinned to 0.66 Å/px), 3D refined, and 3D classified without alignment and with masking out the nanodisc and constant domain (Fc) of the Fabs. The wtABCB4:4B1-Fab:QA2-Fab sample was classified into six classes and two of them, the most distinct (apo-inward and occluded), were processed further. The remaining classes presented transition states between these two conformations. The particles of these two classes were 3D refined, Bayesian polished (all frames), and CTF refined. The final 3D refinement and postprocessing (B-factor sharpening) was performed subsequently applying a few different masks: a soft mask around the entire complex; masking out the nanodisc; masking out the nanodisc and Fc; masking out the nanodisc, Fc, and NBDs (TMDs only). The final two maps (with masked out nanodisc and Fc, and masked TMDs only) were at 4.3 Å and 4.2 Å resolution, respectively, for the apo-inward state, and 3.8 Å and 3.6 Å for the occluded state. The posaconazole-bound ABCB4EQ:4B1-Fab sample was processed in a similar way, resulting in a final EM density map at 3.8 Å resolution (masked out nanodisc and Fc). For each conformation, the higher resolution maps were used for de novo model building, whereas the lower resolution maps facilitated rigid body fitting of certain domains.
Model Building and Refinement.
Model building was performed in Coot (40). All three final EM density maps featured well-resolved TMD and Fc regions allowing for de novo model building, except for extracellular loop 1 (ECL1, residues 93 through 105), which is not built in the final models. Another ambiguous region was observed in the PC-bound, occluded map. While the part of the TM4 and TM10 helices occluding the biding pocket have well-defined density (residues 215– through 41 and 854 through 875, respectively), the regions following the kinks in these helices have weaker density (residues 242 through 248 and 876 through 886, respectively), indicating flexibility. In order to correctly build these fragments, the EM density map was low-pass filtered. Less-resolved regions of the NBDs and constant domain of the Fabs were built by rigid body fitting using previous structures (12, 33). A posaconazole molecule and a PC lipid (1,2-dilinoleoyl-sn-glycero-3-phosphocholine) and their geometry were generated from the SMILES codes in eLBOW (41). The lipid with two 18:2 alkyl chains was chosen for a model building, as this fatty acid appears in human bile. Refinement of the coordinates was performed in Phenix (42) and the model validation in MolProbity (43) and presented in SI Appendix, Table S1. Further analyses are presented in SI Appendix, Fig. S10. Histograms and directional FSC plots were generated on the remote 3DFSC Processing Server (44) with a mask excluding the Fc domains. Q-scores for all transporter residues were calculated with the MapQ plug-in (45) in UCSF Chimera (46) and plotted.
Figure Preparation.
Graph preparation, data analysis, and statistical analysis were performed in GraphPad 8 (GraphPad Software). The images of models and EM density maps were prepared in PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger) and UCSF Chimera (46), respectively. The cavity was visualized with HOLLOW (47), and the volume was calculated in 3V (48).
Supplementary Material
Acknowledgments
We thank the Scientific Center for Optical and Electron Microscopy (ScopeM) at ETH Zürich for technical support. We thank Julia Kowal for help with cryo-EM data collection and Jeppe A. Olsen for preparing the nanodisc-reconstituted sample for the first round of phage display. We thank Silvia Napolitano for technical support with fluorescence measurements. This research was supported by Swiss National Science Foundation Grant 310030_189111 (to K.P.L.), National Institutes of Health Grant GM117372 (to A.A.K.), and Swiss National Science Foundation Grant 310030_155563 (to B.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106702118/-/DCSupplemental.
Data Availability
Coordinates for atomic models and cryo-EM density maps were deposited at the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMD), respectively: apo-inward-open ABCB4 (PDB ID 7NIU, EMD-12365), phosphatidylcholine-bound, occluded ABCB4 (PDB ID 7NIV, EMD-12366), and posaconazole-bound inward-open (PDB ID 7NIW, EMD-12367).
References
- 1.Elferink R. P. J. O., Tytgat G. N. J., Groen A. K., Hepatic canalicular membrane 1: The role of mdr2 P-glycoprotein in hepatobiliary lipid transport. FASEB J. 11, 19–28 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Linton K. J., Lipid flopping in the liver. Biochem. Soc. Trans. 43, 1003–1010 (2015). [DOI] [PubMed] [Google Scholar]
- 3.de Vree J. M. L., et al., Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc. Natl. Acad. Sci. U.S.A. 95, 282–287 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucena J. F., et al., A multidrug resistance 3 gene mutation causing cholelithiasis, cholestasis of pregnancy, and adulthood biliary cirrhosis. Gastroenterology 124, 1037–1042 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Mahdi Z. M., Synal-Hermanns U., Yoker A., Locher K. P., Stieger B., Role of multidrug resistance protein 3 in antifungal-induced cholestasis. Mol. Pharmacol. 90, 23–34 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Frampton J. E., Scott L. J., Posaconazole: A review of its use in the prophylaxis of invasive fungal infections. Drugs 68, 993–1016 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Nibbering C. P., et al., Regulation of biliary cholesterol secretion is independent of hepatocyte canalicular membrane lipid composition: A study in the diosgenin-fed rat model. J. Hepatol. 35, 164–169 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Henkel C., et al., Changes of the hepatic proteome in murine models for toxically induced fibrogenesis and sclerosing cholangitis. Proteomics 6, 6538–6548 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Karatayli E., Hall R. A., Weber S. N., Dooley S., Lammert F., Effect of alcohol on the interleukin 6-mediated inflammatory response in a new mouse model of acute-on-chronic liver injury. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 298–307 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Sokolović A., et al., Overexpression of insulin like growth factor binding protein 5 reduces liver fibrosis in chronic cholangiopathy. Biochim. Biophys. Acta 1822, 996–1003 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Stieger B., Mahdi Z. M., Model systems for studying the role of canalicular efflux transporters in drug-induced cholestatic liver disease. J. Pharm. Sci. 106, 2295–2301 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Olsen J. A., Alam A., Kowal J., Stieger B., Locher K. P., Structure of the human lipid exporter ABCB4 in a lipid environment. Nat. Struct. Mol. Biol. 27, 62–70 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Fellouse F. A., et al., High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924–940 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Paduch M., et al., Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam A., Kowal J., Broude E., Roninson I., Locher K. P., Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nosol K., et al., Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl. Acad. Sci. U.S.A. 117, 26245–26253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J. C., Dougherty D. A., The cation-π interaction. Chem. Rev. 97, 1303–1324 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Celie P. H. N., et al., Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Zhong W., et al., From ab initio quantum mechanics to molecular neurobiology: A cation-π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 12088–12093 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S., Qu Q., Robertson M. J., Skiniotis G., Kobilka B. K., Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckford P. D. W., Sharom F. J., The reconstituted P-glycoprotein multidrug transporter is a flippase for glucosylceramide and other simple glycosphingolipids. Biochem. J. 389, 517–526 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malvezzi M., et al., Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat. Commun. 4, 2367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman R., Rahman K., Lipid flow in bile formation. Biochim. Biophys. Acta (BBA). Lipids Lipid Metab. 1125, 113–133 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Tsui F. C., Ojcius D. M., Hubbell W. L., The intrinsic pKa values for phosphatidylserine and phosphatidylethanolamine in phosphatidylcholine host bilayers. Biophys. J. 49, 459–468 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vik S. B., Antonio B. J., A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 269, 30364–30369 (1994). [PubMed] [Google Scholar]
- 26.Morita S. Y., Terada T., Molecular mechanisms for biliary phospholipid and drug efflux mediated by ABCB4 and bile salts. BioMed Res. Int. 2014, 954781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen J. P., et al., P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 7, 275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Helvoort A., et al., MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 87, 507–517 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Geertsma E. R., Nik Mahmood N. A. B., Schuurman-Wolters G. K., Poolman B., Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominik P. K., et al., Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure 24, 300–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fellouse F. A., Wiesmann C., Sidhu S. S., Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. U.S.A. 101, 12467–12472 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., et al., Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 576, 315–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S., Ura M., Hoey R. J., Kossiakoff A. A., A new versatile immobilization tag based on the ultra high affinity and reversibility of the calmodulin-calmodulin binding peptide interaction. J. Mol. Biol. 427, 2707–2725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal. Biochem. 168, 1–4. (1988). [DOI] [PubMed] [Google Scholar]
- 36.Sündermann A., Eggers L. F., Schwudke D., “Liquid extraction: Bligh and Dyer” in Encyclopedia of Lipidomics, Wenk M. R., Ed. (Springer Netherlands, 2016), pp. 1–4. [Google Scholar]
- 37.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 1–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams C. J., et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Y. Z., et al., Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pintilie G., et al., Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Ho B. K., Gruswitz F., HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voss N. R., Gerstein M., 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for atomic models and cryo-EM density maps were deposited at the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMD), respectively: apo-inward-open ABCB4 (PDB ID 7NIU, EMD-12365), phosphatidylcholine-bound, occluded ABCB4 (PDB ID 7NIV, EMD-12366), and posaconazole-bound inward-open (PDB ID 7NIW, EMD-12367).