Significance
Proteins embedded in membranes experience anisotropic forces from their surroundings. Mechanosensors are activated upon changes in the membrane tension and are the basis of important biological functionalities. However, the molecular mechanisms of mechanosensation are only vaguely understood. Hydrophobic mismatch between protein and membrane, amphiphilic sliding helices, or local membrane bending are examples for different concepts explaining mechanosensation in which one or more of these concepts could be implemented in a particular protein. Here, we show that the mechanosensitive channel of small conductance is gated by the extrusion of lipids from hydrophobic pockets independent of the membrane. A cryoelectron microscopy map at 2.3-Å resolution reveals the so-far-neglected role of aliphatic chains at the periplasmatic side in stabilizing the open state.
Keywords: cryoelectron microscopy, delipidation, force-from-lipid principle, lipid–protein interaction, MscS
Abstract
The mechanosensitive channel of small conductance (MscS) protects bacteria against hypoosmotic shock. It can sense the tension in the surrounding membrane and releases solutes if the pressure in the cell is getting too high. The membrane contacts MscS at sensor paddles, but lipids also leave the membrane and move along grooves between the paddles to reside as far as 15 Å away from the membrane in hydrophobic pockets. One sensing model suggests that a higher tension pulls lipids from the grooves back to the membrane, which triggers gating. However, it is still unclear to what degree this model accounts for sensing and what contribution the direct interaction of the membrane with the channel has. Here, we show that MscS opens when it is sufficiently delipidated by incubation with the detergent dodecyl-β-maltoside or the branched detergent lauryl maltose neopentyl glycol. After addition of detergent-solubilized lipids, it closes again. These results support the model that lipid extrusion causes gating: Lipids are slowly removed from the grooves and pockets by the incubation with detergent, which triggers opening. Addition of lipids in micelles allows lipids to migrate back into the pockets, which closes the channel even in the absence of a membrane. Based on the distribution of the aliphatic chains in the open and closed conformation, we propose that during gating, lipids leave the complex on the cytosolic leaflet at the height of highest lateral tension, while on the periplasmic side, lipids flow into gaps, which open between transmembrane helices.
Mechanosensitive (MS) channels in bacteria provide protection against hypoosmotic shock (1–3). This is an important stress response mechanism because bacteria are frequently exposed to sudden changes in their environment. The MS channel of small conductance (MscS) is often present in bacteria together with several larger paralogs (4, 5). MscS-like channels are not only found in bacteria but also in archaea and in some higher organisms including plants (6).
MscS from Escherichia coli is a well-studied model system for mechanosensation and can directly sense the tension in the membrane as a measure of the osmotic pressure in the cell (7). Many unrelated classes of MS channels detect forces from the membrane and thus follow this force-from-lipid principle for sensing (8–11). However, different membrane and lipid properties may be important to enforce the principle (12–15). MscS is a particularly interesting example, as its molecular structure provides a unique interface for the membrane, which is partly shaped by hydrophobic cavities extending beyond the plane of the membrane.
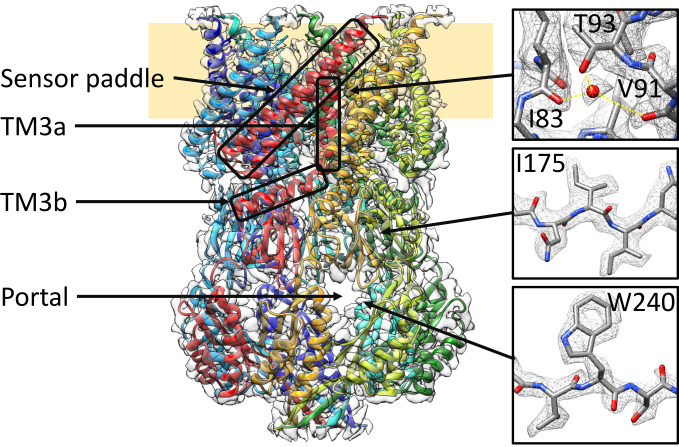
MscS assembles to a homoheptameric complex with a large cytosolic domain and a conical-shaped membrane domain (16) (Fig. 1). The cytosolic domain consists of a vestibule that covers the cytosolic entrance of the pore and is accessible through side portals. Three transmembrane (TM) helices of each subunit contribute to the membrane domain. Helices TM1 and TM2 form an antiparallel bundle connected by a cytosolic loop that reaches, as the so-called sensor paddle, into the membrane. The TM3a helices of the seven subunits establish the pore and, after a kink, connect as TM3b helices to the cytosolic vestibule.
Fig. 1.
Structure of open MscS solubilized in LMNG. MscS consists of seven identical subunits, shown in different colors, and is organized in a membrane domain (Top) and a cytosolic domain (Bottom). In the membrane domain, TM1 and TM2 form the sensor paddle, TM3a the pore, and TM3b connects the two domains. The cytosolic domain shapes a vestibule that can be accessed through portals. Some close-ups of the densities are shown on the right together with the atonic model (oxygen: red, nitrogen: blue, carbon: gray). Yellow lines indicate the coordination of the water. The sample was obtained under condition 2b (see Results).
The paddles are tilted relative to the central axis so that the interface to the membrane is skewed. The tension sensing could be based on local bending of the membrane attached to this interface (12). Cryoelectron microscopy (cryo-EM) structures of closed MscS embedded into membrane nanodiscs showed that the paddles are not completely engulfed in the membrane and that they project into the cytosol (17–19). The cytosolic hydrophobic grooves between the paddles allow lipids from the membrane to penetrate the complex and move along the paddles into pockets between the TM3a and TM3b helices and the paddle loops. Thus, the paddle loops are in contact with lipids but not with the membrane. This is evident from structural data but also from fluorescence quenching and molecular dynamics (MD) simulations (20, 21). As sensing mechanisms, it was proposed that lipid molecules are pulled out of the cytosolic grooves and pockets at high membrane tensions triggering gating (20). Mutational data support this model: R59 that is located in the cytosolic loop forms a salt bridge to a lipid headgroup and with a mutation, which weakens this interaction; R59L, a strong gain-of-function (GOF) phenotype, was observed (21). This agrees with the model because, in this mutant, it should be easier to remove the lipid from the pocket, which will result in gating at lower tension.
Electrophysiological studies showed that MscS is not only found in closed and open states but can also enter adaptive states: If tension is kept on the patch, MscS switches to a nonconducting state in a time-dependent manner (22–24). Part of this nonconducting population can directly be reactivated by a greater stimulus, termed “desensitized,” while the other part cannot and requires minutes of recovery without tension before it can be reactivated, termed “inactivated” (25). Thus, beside the open and closed state, MscS shows two different states of adaptation, which should have different structural properties. Recently, a cryo-EM structure of a nonconductive state of MscS under membrane tension, termed desensitized, was presented with different positioning of the paddles (19). This structure relates to the inactivated state described by Akitake et al. (25) as deduced from the behavior of the mutant G113A. Other studies did not distinguish between desensitization and inactivation in adaptation (26).
Several structural studies of MscS resulted in closed (16–19, 27–29) or open (20, 30–32) conformations. The questions arise how the sample conditions during these structural studies determined a certain conformation and if we can learn something about the tension sensing from this information. In this study, we show that the reversible lipid removal and addition is decisive if MscS is in an open or a closed conformation. Structurally, we find a network of elongated densities in the hydrophobic pockets and the pore, caused by the aliphatic chains of lipids and detergent.
Results
Experimental Design Producing Closed and Open States in Solubilized MscS.
The degree of delipidation of a membrane protein depends on the exact conditions of detergent incubation: Higher detergent concentrations cause a stronger delipidation (33, 34). A long incubation time promotes lipid removal as well as a higher incubation temperature (35). An important factor is also what kind of detergent is used; for example, octyl glucose neopentyl glycol and Cymal-5 effectively remove lipids from membrane proteins (34, 36, 37).
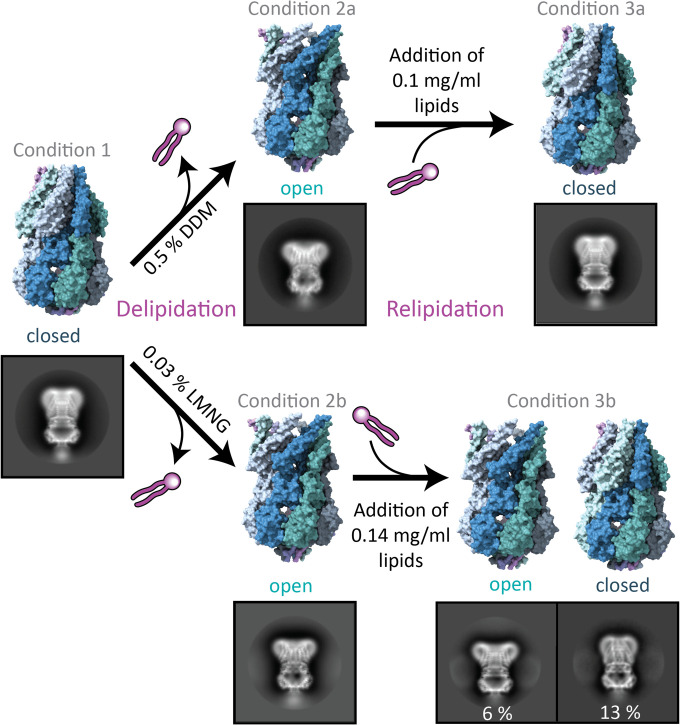
We solubilized MscS in 1.5% dodecyl-β-maltoside (DDM) for 1 h at 4 °C and then purified it in a two-step protocol, using an Ni-NTA immobilized metal affinity chromatography (IMAC) in the presence of 0.05% DDM followed by a size exclusion chromatography (SEC) in the presence of 0.03% DDM (condition 1). To determine the conformational state of MscS, we obtained a cryo-EM map by single-particle analysis to a resolution of 3.9 Å and found MscS in a closed conformation (Fig. 2 and SI Appendix, Fig. S1). The assignment of the conformational state was confirmed by comparison to reference structures (19, 30) and was based on root mean square deviation values between the Cα-atoms of the N-terminal region (residues ≤ G113), where most of the conformational rearrangements occur (SI Appendix, Table S1). Then, we increased the DDM concentration in the wash buffer on the IMAC from 0.05 to 0.5% and incubated overnight at 4 °C. The DDM concentration of 0.03% DDM in the SEC buffer was maintained. Under this condition, we acquired a structure of MscS at 3.1-Å resolution in an open conformation (condition 2a). In a last step, we took the same sample that produced the open structure and added back lipids as mixed micelles (0.1 mg/mL final concentration azolectin from a 10 mg/mL stock solution in 0.5% DDM) and incubated it for 30 min at room temperature before preparing grids. Again, a closed conformation was obtained at 3.1-Å resolution (condition 3a). Thus, solubilized MscS can reversibly change back from the open to the closed state when more lipids are present.
Fig. 2.
Experimental strategy to obtain solubilized MscS in open and closed conformations. MscS solubilized in DDM produced a closed conformation (condition 1), while washing with higher DDM concentrations during the purification led to an open conformation (condition 2a). Addition of lipids back to the same sample again produced a closed MscS (condition 3a). Exchange of DDM to LMNG during purification resulted in an open MscS (condition 2b), while addition of lipids partially closed MscS (condition 3b). Percentages refer to the number of particles in side-view 2D classes in the open or closed conformation relative to the total number of particles. The ratio of these percentages is an indication for the conformational distribution in the sample. MscS structures are shown in surface representation with different colors for the subunits. Examples of 2D class images are shown for all conditions in which the edge has a length of 27.2 nm.
Thin-layer chromatography (TLC) confirmed that under condition 1, higher amounts of lipid were associated with MscS than under condition 2a (SI Appendix, Fig. S2). Earlier, we found by SEC online with inductively coupled plasma mass spectrometry that about three phospholipid molecules per subunit (corresponding to 21 lipids per complex) are present in the MscS and that lipids were still detectable in protein-free micelles in the same sample solution (38). This experiment was performed under our less delipidating standard purification conditions with 1.0% DDM in the solubilization buffer. It agrees with three resolved lipids per subunit in the closed structure in nanodiscs, which are perhaps less dynamic than other lipids in the complex (17). In contrast, under the more stringent conditions used for crystallization (which produced open conformations in crystals), only five lipids were left in the whole MscS complex, detected by native mass spectrometry (20). This is within an error consistent with one resolved lipid per subunit that we found in the open detergent-solubilized MscS structures (see Lipid and Detergent Molecules within the MscS Complex).
We also tested what effect a different detergent has on the conformational state of MscS (conditions 2b and 3b). For this, MscS solubilized in 1.0% DDM was exchanged with the branched detergent lauryl maltose neopentyl glycol (LMNG) during IMAC (0.03% LMNG) and SEC (0.02% LMNG). Because neopentyl glycol detergents, like LMNG, have been shown to have a stronger delipidating effect (34), we predicted a further stabilization of the open conformation. This produced an open conformation of MscS at 2.3-Å resolution described in more detail in the next section. Both open structures, obtained in DDM (condition 2a) and LMNG (condition 2b), agree well with each other (SI Appendix, Fig. S3) and the open crystal structures (20, 31). Addition of a similar amount of lipids back to the sample under condition 3a (0.14 mg/mL 1,2-di-(9,10-dibromo)stearoyl-sn-glycero-3-phosphoethanolamine [BrPE]; Materials and Methods) resulted in a mixture of twice as many closed as open MscS (condition 3b). The relipidation in LMNG was not further optimized but showed that the addition of lipids was sufficient to close most of the channels. Three-dimensional reconstruction of the closed population under condition 3b produced a map of 3.7-Å resolution while, for the open conformation, a resolution of 2.7 Å was obtained. In conclusion, the different detergents have a similar effect of reversibly switching the conformation of MscS dependent on the lipid content, but the exact conditions vary.
Structure of Open MscS Solubilized in LMNG at 2.3-Å Resolution.
Using the detergent LMNG instead of DDM produced a structure of the open conformation to a high resolution (condition 2b; Fig. 1 and SI Appendix, Fig. S4), which allowed us to identify structural water. These are predominantly found in the cytosolic vestibule, in contrast to the membrane domain, where hardly any water is resolved. However, an interesting exception is a water molecule at the loop between TM2 and TM3a, which is presumably important for the coupling between tension sensing (from the senor paddle) and gating (pore helices). This water is coordinated by the carbonyl groups of residues I83 and V91 and the hydroxy group of T93 (Fig. 1). We expected that mutating T93 to T93A destabilizes the water by providing one less hydrogen bond. In electrophysiological measurements, this mutation showed a GOF phenotype as the PL:PS ratio increased from 1.58 ± 0.13 (wild-type [WT]) to 1.85 ± 0.16 (T93A; Student’s unpaired t test against WT: P = 0.0004) (SI Appendix, Fig. S5B). The PL:PS ratio is the ratio of pressures required to open MscL (mechanosensitive channel of large conductance) WT (as internal standard) relative to MscS and thus provides a measure of the tension threshold for opening MscS (39). In addition, dwell times of 10 ± 15 ms (SI Appendix, Fig. S5 D and E) are short in comparison to 150 ms for WT (40). The phenotype of this mutation suggested that the coupling between the sensor paddle and the pore helices is enhanced. Therefore, this water must somewhat restrain opening but then, once open, prevents a fast return to the closed state. Mutation to T93V showed two different types of openings: Activations with a short dwell time were observed at PL:PS = 1.92 ± 0.15 (P = 0.0002), and activations with steady dwell time at PL:PS = 1.26 ± 0.09 (P < 0.0001) (SI Appendix, Fig. S5C). The dwell times for T93V could not be reliably quantified with our data but only qualitatively assessed (SI Appendix, Fig. S5F). Earlier, it had been shown that a T93R mutation also caused a GOF phenotype (41), while a T93W mutation had little effect (42). However, the indole nitrogen in the Trp side chain also supports hydrogen bonding.
On the cytosolic side of the TM3b helices, water channels connect the inside of the vestibule with the cytosol (SI Appendix, Fig. S6). These channels are probably irrelevant for conductance or equilibration between the vestibule and the cytosol because they are much smaller than the large cytosolic portals, which provide rapid exchange of water and ions between the vestibule and the cytosol (43). Instead, it is more likely that the channels allow TM3b to move relative to the rigid β-domain as they become smaller in the closed conformation. A prominent residue at the vestibule entrance of these channels is the conserved residue N117, which is implicated in adaptation (44).
Lipid and Detergent Molecules within the MscS Complex.
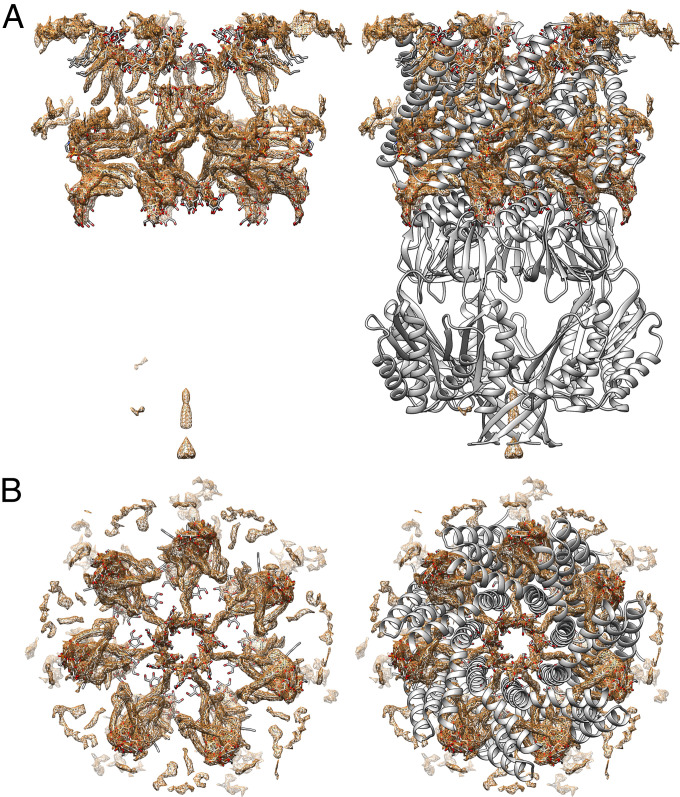
In addition to the protein and the water molecules, a network of elongated densities in the pockets and the pore reveals the position of the aliphatic chains of the detergents and the residual lipids (Fig. 3). In general, the complex could contain lipids from the membrane, DDM from the solubilization, and LMNG from the detergent exchange (condition 2b, SI Appendix, Fig. S7). We found all three types of molecules and modeled one lipid, five LMNG, and three DDM molecules per subunit of MscS (SI Appendix, Figs. S8 and S9). The lipid headgroup of the only lipid is electrostatically bound to R59 as in the closed structure (Fig. 4A). In contrast to the closed structure, one fatty acid chain is bent, and the other was resolved as C14 fatty acid.
Fig. 3.
Difference map of open MscS in LMNG (condition 2b). (A) Difference map after subtraction of protein density showing elongated densities (brown). The interpretation of these densities as lipids and detergent molecules is shown in stick representation (oxygen: red, nitrogen: blue, carbon: gray). Close-ups are shown in Fig. 4. On the right side a model of the protein in cartoon representation is also shown (gray). (B) View along the symmetry axis from the cytosol. On the right side with the protein model, the view was cut in the height of TM3b for clarity.
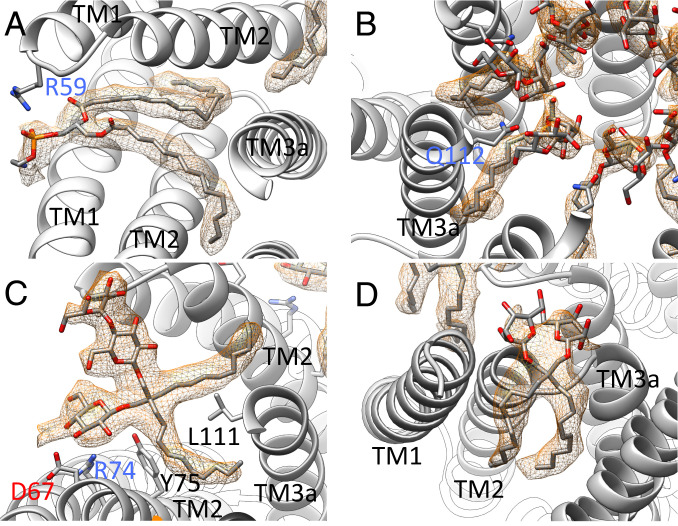
Fig. 4.
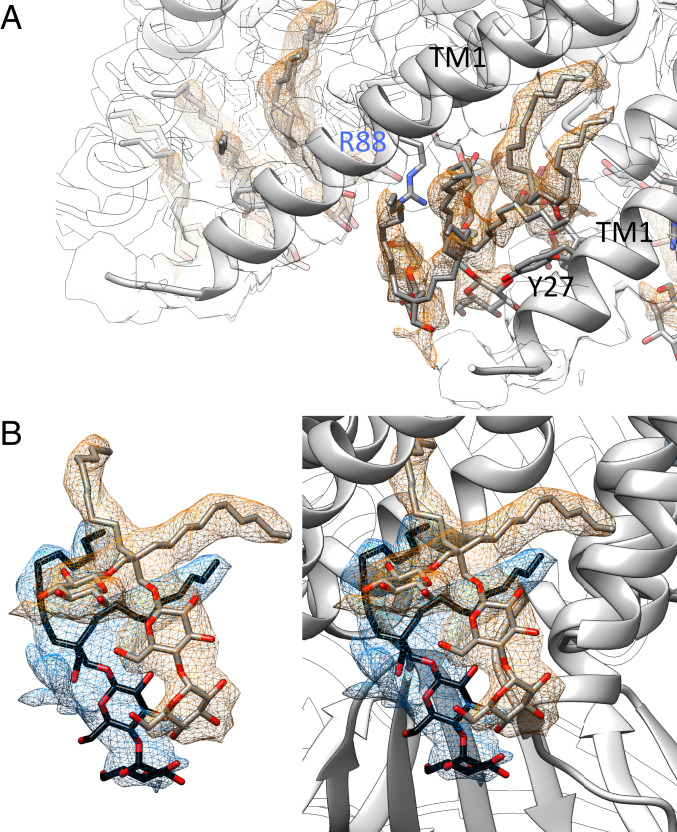
Selected ligands in the structure of open MscS in LMNG. (A) One lipid, modeled as DOPE, is resolved in the structure at the paddle loop. A salt bridge is formed with R59. The fatty acid chains are bent because the TM3a helices restricting the space in the open conformation (B) DDM molecules are located in the pore entering the gaps between the TM3a helices. The headgroup is coordinated by Q112. (C) An LMNG molecule close to the paddle loop and the lipid. This LMNG molecule, together with a second, cover a large hydrophobic opening toward the cytosol. One maltose unit is well resolved, while in the second unit, only one sugar ring is resolved. (D) One LMNG molecule per subunit of the LMNG stack on the periplasmic side. This LMNG is close to the TM2–TM3a loop and relates to a well-resolved lipid at this loop in the closed structure. The headgroups are not well resolved, and only one sugar ring each is shown.
In the open structure, the TM3a pore helices move outwards and provide space for intercalating aliphatic chains (Fig. 4B), which emerge from the inside of the pore. We modeled these as parts of DDM molecules (1 DDM per subunit) with their headgroups coordinated by Q112 at the cytosolic exit of the pore. DDM molecules may take the place of lipids inside the pore, which move with their aliphatic chains into the gaps between the TM3a helices during opening, as suggested by recent MD simulations (19). Another set of DDM molecules in the pore face the periplasmic exit with their headgroups, where they are coordinated by Q92 in the loop between TM2 and TM3a. A third DDM molecule per subunit is lying parallel to the lipid in the pocket, and its hydrophobic tail extends into the gaps between the pore helices.
Short hydrophobic chains and branching of the densities are easily recognizable structural signatures that identified LMNG molecules within the complex (Fig. 4 C and D). Interestingly, the LMNG molecules stack: Two LMNG molecules are stacked at the paddle loop close to the resolved lipid, and three are stacked on the periplasmic side between the TM1 helices (Fig. 5). Perhaps such an LMNG stacking is the reason for the stabilizing effect of LMNG reported for many membrane proteins (45, 46). Between the TM1 helices, the hydrophilic headgroups of LMNG face inwards to the pore funnel, while the hydrophobic tails face outwards to the micellar belt (Fig. 5A). The middle of the three LMNG molecules is clamped between Y27 in TM1 and R88 in the TM2–TM3a loop. R88 forms a salt bridge with a lipid close to this loop in the closed structure of MscS in nanodiscs (17, 18). It is plausible that lipids have similar positions to the LMNG stack when MscS is embedded in the membrane and R88 moves with the lipid outwards during opening. The periplasmic ends of the TM1 helices separate from each other so that lipids from the periplasmic leaflet can move into the gap and can exchange with the bound lipid on R88.
Fig. 5.
Stacking of LMNG molecules. (A) On the periplasmic side, three LMNG molecules stack on each other in between the TM1 helices. The best-resolved one is at the loop between TM2 and TM3a (see Fig. 4D). The middle one is clamped by R88 and Y27. The headgroups face toward the water-filled funnel in the center, while the hydrophobic tails face outwards into the micelle. The protein density is shown as an outline at the same threshold. (B) Two LMNG molecules stack on the cytosolic side covering a hydrophobic opening. The image on the right side also shows the context of the protein structure.
The structures of the DDM-solubilized samples also showed lipid and detergent densities. Comparing the two closed structures (condition 1 and condition 3a), the sample with the added lipids (condition 3a) produced an overall better resolved structure, which may be due to more complete lipidation but also to the larger number of particles in the dataset (SI Appendix, Table S2). Here, a lipid molecule is resolved at the periplasmic entrance of the pore coordinated to R88 as in the structure of the closed MscS reconstituted into nanodiscs with a native-like lipid environment (17). Close to this lipid, a DDM molecule is resolved in the pore with its headgroup bound to Q92, the carbonyl group of G87, and the phosphate of the lipid (SI Appendix, Fig. S10). In the pockets at the paddle loops, some density is present but could not be modeled as lipids or detergent. The open conformation under condition 2a showed a lipid molecule in this region with similarly bent tails as under condition 2b in LMNG but with a slightly changed position of the headgroup (SI Appendix, Fig. S11A). Regions that are occupied with LMNG under condition 2b show DDM molecules under condition 2a but with less resolution. For example, two DDM molecules are located close to the hinge between TM2 and TM3a (SI Appendix, Fig. S11B) and close to the paddle loop (SI Appendix, Fig. S11C).
Discussion
The membrane directly exerts forces on MscS, which can be described as bulk properties of the membrane. For example, it was recently established that the stiffness of the membrane is important for tension sensing of MscS (15). However, tension sensing of MscS cannot be solely understood on the level of membrane properties because lipid molecules leave the membrane and penetrate the MscS complex. Lipid molecules are found in pockets close to TM3b, far away from the plane of the membrane and even in the pore (17–21), which is not in direct contact with the surrounding membrane. Based on these observations, a tension-sensing model of MscS was proposed, which puts the extraction of these lipids at high tension center stage for triggering gating (20). But how important is this process, really, in comparison to the direct interaction between membrane and paddles?
Our experiments show that MscS without a membrane environment can be switched between open and closed conformations dependent on the amount of associated lipids. In this context, the conformational state in past structures can be understood. MscS solubilized in DDM was found in the open (20, 31, 32) and closed (18) states. Our structures of MscS under the different conditions show that these lipids are located within the pockets. Thus, these artificial conditions of detergent-solubilized MscS can separate the effect of the membrane from the effect of the lipids in the pockets. Although our experiments stress the importance of the lipids in the pockets, the extrusion model for tension sensing must be refined. While with the opening transition, the volume of the pockets decreases on the cytosolic side in agreement with the model (20), the largest conformational changes are seen on the periplasmic side: The N-terminal ends of the TM1 helices at the periplasmic surface of the membrane increase the covered diameter from about 30 Å in the closed state to 65 Å in the open state (SI Appendix, Fig. S12B). If that would be a simple area expansion of the protein, then that would be a strong driving force for tension sensing. However, the situation is more complicated because gaps between the TM1 helices open, and a new interface between the membrane and the bulk water phase in the funnel appears. Our results suggest that lipids from the periplasmic leaflet flow into these gaps, with their headgroups facing the water phase and thus avoiding an unfavorable exposure of the hydrophobic core. It seems likely that lipid molecules take similar positions as the LMNG molecules in our structure (Fig. 6B and SI Appendix, Fig. S8B). Consequently, lipid headgroups would be located at the height of the hydrophobic membrane core facing the hydrophilic surface between the N-terminal end of TM1 and the TM2-TM3a loop.
Fig. 6.
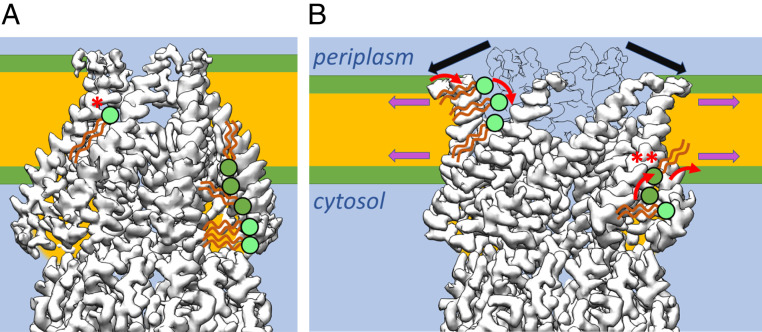
Scheme of lipid interaction with MscS during gating. (A) MscS in the closed and (B) open conformation schematically together with the membrane and interacting lipids. For clarity, only on the left side of MscS examples for interacting lipids from the periplasmic leaflet are shown (for each conformation). On the right side of MscS interacting lipids from the cytosolic leaflet are exemplified. Lipids that were directly seen in the structures or were suggested by the positions of LMNG are shown in light green. (A) In the closed conformation, a lipid is locked between two adjacent sensor paddles in the middle of the membrane with its headgroup facing the funnel (*). There is no pathway for its charged headgroup to cross to the cytosolic leaflet, and the pathway to the periplasmic leaflet is blocked by TM1 helices. On the cytosolic side, large hydrophobic pockets are filled with lipids from the cytosolic leaflet. (B) Additional tension in the membrane leads to thinning of the membrane and is highest at the onset of the hydrophobic chains (magenta arrows). Our model proposes that lipids in the pockets in this area (**) will initiate the conformational change in MscS. The N-terminal domains at the periplasmic interface are not completely resolved, but this region of MscS shows a substantial outward movement (black arrows; an outline of the closed conformation is shown for comparison), which leads to the separation of the TM1 helices. Now, the bulk water phase in the funnel would be exposed to the membrane core. To prevent that from happening, lipid head groups must move from the periplasmic leaflet into these gaps and equilibrate with the lipid trapped in the closed state. The amphipathic N-terminal domains may also sense tension directly in the membrane in the periplasmic leaflet.
MscS under native conditions embedded in the membrane has a different occupation of the pockets in the open conformation than in our structures because only lipids are present but no detergent. Zhang et al. recently established a protocol to produce tension in a native membrane environment around MscS by extracting lipids from nanodiscs with β-cyclodextrin (19). They found open MscS to be dynamic and unstable, while continued tension settled MscS in a novel desensitized conformation. We suggest that our high-resolution open structure was only obtained because of the stabilization by the detergent. Especially, it is difficult to imagine how the openings of the hydrophobic pockets toward the cytosol close to the paddle loops (SI Appendix, Fig. S13) are completely shielded by lipid headgroups toward the bulk water phase and at the same time accommodate all their hydrophobic chains within the pockets. Close to TM3b, less space is available for long hydrophobic chains than in the closed structure. These space requirements are satisfied by the shorter hydrophobic chains of detergents, as in our LMNG structure, in which two stacked LMNG molecules with their large sugar headgroups and short hydrophobic chains fill the cytosolic pockets. In a native membrane without detergent, asymmetric flexible adjustments of the paddle positions probably avoid these large hydrophobic openings. However, one lipid is resolved in this region in our open MscS structures next to the LMNG stack with bent hydrophobic chains. It is coordinated to R59, which shows a strong phenotype when mutated to R59L as mentioned in the Introduction. It may surprise that this lipid is observed in the closed as well as the open state as if it is static, but this is perhaps also due to the nonnative condition. The reason why we didn’t observe the desensitized state as described by Zhang et al. is perhaps explained by the fact that this lipid was not removed by our detergent treatments.
Densities within the pore are best explained by DDM molecules, seven from the periplasmic side and seven from the cytosolic side. In our open MscS structure, the hydrophobic chains enter the gaps between the pore helices. The closed electron microscopy structures of MscS reconstituted in nanodiscs showed similar densities for the periplasmic side with a slight rotation around the symmetry axis. Theoretically, it could be that DDM is present in the pore not only in the open but also in the closed, reconstituted state if it is not removed during reconstitution. However, that seems unlikely, at least for the densities from the periplasmic side, as fluorescence quenching experiments showed that residues within the pore, A94 and G101, have contact with lipids (21). Quenching was far lower on the cytosolic side of the pore at the positions L105 and G113. In these fluorescence studies, MscS was presumably in the closed state as it was reconstituted. Consistently, the elongated densities are only present on the periplasmic side of the pore in our closed structures but on both sides in the open structures. Lipids within the pore were suggested to be part of the molecular mechanism for adaptation (14, 47). Thus, our structures may represent the adapted rather than the open state. However, it is difficult to imagine how lipids get into the pore without energetically unfavorable passages of the hydrophobic tails or the hydrophilic head. One possible scenario is that the bent tails of the resolved lipid straighten and enter the gaps between the pore helices instead of the DDM molecules within the pore. Another DDM molecule parallel to the lipid stretches from outside the pore into the gap. In a native environment, this position could be taken by a lipid.
In conclusion, we show that delipidation triggers opening of MscS independently of a membrane and that the presence of detergent stabilizes the open conformation further. A network of elongated densities caused by hydrophobic chains of lipids and detergent molecules is well preserved between different samples in the same conformation, indicating that specific voids are provided by MscS. The least resolved region with respect to hydrophobic chains is in the grooves in which the cytosolic leaflet is attached (SI Appendix, Fig. S9B). We propose that here, the most dynamic lipids are located, and these are the lipids that are easiest removed by increased membrane tension triggering gating or at least initiating the opening transition (Fig. 6B and SI Appendix, Fig. S12A, green arrow). Tension is highest at the onset of the fatty acid chains on both leaflets, and here, also, an increase in tension is seen when the membrane is stretched (48). Thus, this region must be most important for tension sensing (49). Further experiments are required to test this proposition. Recently, it was suggested that the lipid that is typically best resolved in MscS structures bound to R88 is responsible for gating (18, 19). This lipid, which was termed the “hook lipid” (18) or “gatekeeper lipid” (19), is in the closed conformation of MscS with its hydrophobic tails exposed to the center of the membrane and the cytosolic leaflet (Fig. 6 A; marked as *). Because of the anisotropy of the pressure profile in the membrane, this lipid experiences positive pressure in the center of the membrane and reaches the region of highest tension with the ends of the tails (SI Appendix, Fig. S12A). There are good arguments that this lipid has the functional role of a gatekeeper. When lipids were extracted with β-cyclodextrin from MscS reconstituted in nanodiscs, this lipid disappeared from the EM map first, with MscS still in a closed conformation (19). However, for this lipid to leave MscS in the closed conformation, it requires that the charged lipid head group crosses the hydrophobic core of the membrane, which is energetically extremely unfavorable. Our data suggest that lipids enter the gaps between TM1 helices during the opening of MscS from the periplasmic leaflet because the N-terminal ends of these helices move radially outward (Fig. 6B). We think that the hook lipid equilibrates with these periplasmic lipids in the open state and acts in adaptation, probably by blocking the pore. However, that is speculative, and further experiments are required to prove or disprove the role of this lipid in gating or adaptation.
The N-terminal domains preceding TM1 may also sense tension directly because they are exposed to the region of highest tension on the periplasmic leaflet. This domain has been proposed to be an amphipathic helix (19). Such helices could contribute to gating by dragging the TM1 helices outwards together with the flowing of the lipid molecules, which has been suggested as a more general tension sensing mechanism for MS channels (50). Such a contribution is supported by the sensitive LOF phenotype of mutations in the N-terminal domain (51, 52).
Materials and Methods
A detailed description of all materials and methods is given in the SI Appendix.
Expression and Purification of MscS.
Expression and purification of MscS has been described elsewhere (38) with only small modifications. Cells that expressed MscS with a carboxyl-terminal His6-tag were suspended in a buffer containing 1% (condition 2b and 3b) or 1.5% (condition 1, 2a, 3a) DDM (from Glycon) and incubated in total for 60 min on ice. After centrifugation and filtration, MscS was bound to prepacked 0.5 mL Ni-NTA agarose columns and washed with 30 mL washing buffer containing either 0.05% (condition 1) or 0.5% DDM (condition 2a to 3a) or 0.03% LMNG (condition 2b to 3b), 50 mM sodium phosphate buffer pH 7.5, 300 mM NaCl, 10% (wt/vol) glycerol, and 50 mM imidazole. For elution, 300 mM imidazole in washing buffer was used. The peak fraction was further purified by SEC. For the DDM-solubilized samples (condition 1, 2a, and 3a), the samples were injected on a Superdex200 Increase 10/300 column (GE Healthcare) equilibrated with buffer A containing 50 mM Hepes pH 7.5, 150 mM NaCl, and 0.03% DDM. For the LMNG sample, buffer A contained 0.02% LMNG instead of DDM (condition 2b and 3b). Peak fractions were concentrated in 100-kDa spin filters (Amicon) to 5 mg/mL and used for grid preparation. For condition 3a, the sample in condition 2a was supplemented with 0.1 mg/mL azolectin added from a stock solution of 10 mg/mL solved in 0.5% DDM and incubated for 30 min at room temperature before grid preparation. For condition 3b, 0.14 mg/mL BrPE was added (stock 14 mg/mL in 0.5% LMNG) to a sample of MscS that was obtained under condition 2b. It should be noted that neither azolectin nor BrPE are native to E. coli. However, many studies used azolectin with little effect on the function of MscS (15, 53), while the molecular properties of brominated lipids are similar to their unsaturated counterparts (54).
Cryo-EM and Data Analysis.
All MscS samples were frozen on R1.2/1.3 holey carbon coated copper 400 grids (Quantifoil) employing a Vitrobot IV (FEI) freezing device. Data were acquired at the cryo-EM facility in Würzburg on an FEI Titan Krios G3 microscope equipped with a direct Falcon III detector (55). Details for the measurement conditions are given in SI Appendix (SI Appendix, Table S2 and SI Materials and Methods). Motion correction and dose weighting was performed with the program MotionCor2 (56). For further data processing, the workflow in Relion 3.1 was used (57). The contrast transfer functions were determined with the program CTFFind4 (58). Particles were first autopicked with the program crYOLO (59) and then, after two-dimensional (2D) classification, with appropriate class averages as references in Relion (60). The resolution of the final refinements is given according to gold standard at a cutoff of 0.143 as implemented in Relion (61). Coot version 0.9.2 (62) and Phenix realspace.refine version 1.19-4092 (63) were used for model building, followed by validation with MolProbity (64). Images were generated with Chimera 1.14 (65) or ChimeraX (66).
TLC.
MscS samples (500 µg), purified under conditions 1 and 2a, were extracted as described earlier (67). A 20 × 20 cm2 silica F254 aluminum plate was dried at 70 °C for 10 min before application of the MscS extracts and lipid standards. A closed tank was lined with filter paper, filled, and equilibrated with chloroform, methanol, and 1 M KCl in a 10:10:3 volume ratio as mobile phase (20). The plate was placed in the tank and left until the mobile phase ascended two-thirds of the plate. After drying the plate, it was stained with 0.05% (wt/vol) primuline in acetone:water (volume ratio 8:2), and spots were imaged under ultraviolet light (68). Then the plate was sprayed with 0.1% ninhydrin (Thermo Fisher Scientific) in acetone:water (volume ratio 8:2) followed by heating at 120 °C until pink spots developed to specifically detect phosphoethanolamine (69).
Patch Clamp Electrophysiology.
Patch clamp experiments were carried out on inside-out patches of MscS expressed in E. coli strain MJF429 (ΔyggB, ΔmscK) as previously described (21). All data were acquired using an HEKA EPC8 amplifier and Patch Master software (HEKA). Measurements were conducted on at least six patches derived from a minimum of two independent sample preparations. Pressure ratios PL:PS for activation of MscL in relation to MscS are given as mean ± SD. The dwell time was analyzed with the software Origin 2019b with a bin size of 5 ms and fitting of a Gaussian distribution to the logarithmic dwell times.
Accession Numbers.
The filtered map and unfiltered half maps are available in the electron microscopy database (EMDB), while the models at the Protein Data Bank (PDB) database are available under the respective accession numbers: EMD-12997, PDB: 7ONL (DDM-solubilized, condition 1), EMD-13003, PDB: 7OO0 (DDM-solubilized, condition 2a), EMD-13006, PDB: 7OO6 (DDM-solubilized with added lipid, condition 3a), EMD-12996, PDB: 7ONJ (LMNG-solubilized, condition 2b), EMD-13008, PDB: 7OOA (LMNG-solubilized with added lipid, open, condition 3b), and EMD-13007, PDB: 7OO8 (LMNG-solubilized with added lipid, closed, condition 3b).
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Project Grant Bo1150/15-1 and DFG Equipment Grant INST 93/903-1 FUGG.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107095118/-/DCSupplemental.
Data Availability
EM maps and PDB models have been deposited in EMDB and PDB [PDB: 7ONL (70), 7OO0 (71), 7OO6 (72), 7ONJ (73), 7OO8 (74), and 7OOA (75); EMDB: EMD-12997 (76), EMD-13003 (77), EMD-13006 (78), EMD-12996 (79), EMD-13007 (80), and EMD-13008 (81)].
References
- 1.Cox C. D., Bavi N., Martinac B., Bacterial mechanosensors. Annu. Rev. Physiol. 80, 71–93 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen T., Rasmussen A., “Bacterial mechanosensitive channels” in Membrane Protein Complexes: Structure and Function. Subcellular Biochemistry, Harris J., Boekema E., Eds. (Springer, 2018), pp. 83–116. [DOI] [PubMed] [Google Scholar]
- 3.Blount P., Iscla I., Life with bacterial mechanosensitive channels, from discovery to physiology to pharmacological target. Microbiol. Mol. Biol. Rev. 84, e00055-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pivetti C. D., et al., Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 67, 66–85 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards M. D., et al., Characterization of three novel mechanosensitive channel activities in Escherichia coli. Channels (Austin) 6, 272–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton E. S., Schlegel A. M., Haswell E. S., United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 66, 113–137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sukharev S., Purification of the small mechanosensitive channel of Escherichia coli (MscS): The subunit structure, conduction, and gating characteristics in liposomes. Biophys. J. 83, 290–298 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brohawn S. G., Su Z., MacKinnon R., Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc. Natl. Acad. Sci. U.S.A. 111, 3614–3619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sukharev S. I., Martinac B., Arshavsky V. Y., Kung C., Two types of mechanosensitive channels in the Escherichia coli cell envelope: Solubilization and functional reconstitution. Biophys. J. 65, 177–183 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syeda R., et al., Piezo1 channels are inherently mechanosensitive. Cell Rep. 17, 1739–1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murthy S. E., et al., OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7, e41844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips R., Ursell T., Wiggins P., Sens P., Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haswell E. S., Phillips R., Rees D. C., Mechanosensitive channels: What can they do and how do they do it? Structure 19, 1356–1369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen T., How do mechanosensitive channels sense membrane tension? Biochem. Soc. Trans. 44, 1019–1025 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Xue F., et al., Membrane stiffness is one of the key determinants of E. coli MscS channel mechanosensitivity. Biochim. Biophys. Acta Biomembr. 1862, 183203 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Bass R. B., Strop P., Barclay M., Rees D. C., Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen T., Flegler V. J., Rasmussen A., Böttcher B., Structure of the mechanosensitive channel MscS embedded in the membrane bilayer. J. Mol. Biol. 431, 3081–3090 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Reddy B., Bavi N., Lu A., Park Y., Perozo E., Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS. eLife 8, e50486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al., Visualization of the mechanosensitive ion channel MscS under membrane tension. Nature 590, 509–514 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Pliotas C., et al., The role of lipids in mechanosensation. Nat. Struct. Mol. Biol. 22, 991–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen T., et al., Interaction of the mechanosensitive channel, MscS, with the membrane bilayer through lipid intercalation into grooves and pockets. J. Mol. Biol. 431, 3339–3352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sukharev S. I., Blount P., Martinac B., Kung C., Mechanosensitive channels of Escherichia coli: The MscL gene, protein, and activities. Annu. Rev. Physiol. 59, 633–657 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Koprowski P., Kubalski A., Voltage-independent adaptation of mechanosensitive channels in Escherichia coli protoplasts. J. Membr. Biol. 164, 253–262 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Levina N., et al., Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: Identification of genes required for MscS activity. EMBO J. 18, 1730–1737 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akitake B., Anishkin A., Liu N., Sukharev S., Straightening and sequential buckling of the pore-lining helices define the gating cycle of MscS. Nat. Struct. Mol. Biol. 14, 1141–1149 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Edwards M. D., Bartlett W., Booth I. R., Pore mutations of the Escherichia coli MscS channel affect desensitization but not ionic preference. Biophys. J. 94, 3003–3013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbacher S., Bass R., Strop P., Rees D. C., Mechanosensitive Ion Channels, Part A (Elsevier, 2007). [Google Scholar]
- 28.Angiulli G., et al., New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins. eLife 9, e53530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belyy V., Anishkin A., Kamaraju K., Liu N., Sukharev S., The tension-transmitting ‘clutch’ in the mechanosensitive channel MscS. Nat. Struct. Mol. Biol. 17, 451–458 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Wang W., et al., The structure of an open form of an E. coli mechanosensitive channel at 3.45 A resolution. Science 321, 1179–1183 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pliotas C., et al., Conformational state of the MscS mechanosensitive channel in solution revealed by pulsed electron-electron double resonance (PELDOR) spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, E2675–E2682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai J. Y., Poon Y. S., Kaiser J. T., Rees D. C., Open and shut: Crystal structures of the dodecylmaltoside solubilized mechanosensitive channel of small conductance from Escherichia coli and Helicobacter pylori at 4.4 Å and 4.1 Å resolutions. Protein Sci. 22, 502–509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigal N., Lewinson O., Wolf S. G., Bibi E., E. coli multidrug transporter MdfA is a monomer. Biochemistry 46, 5200–5208 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Gupta K., et al., Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc. 13, 1106–1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechara C., et al., A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat. Chem. 7, 255–262 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Laganowsky A., Reading E., Hopper J. T. S., Robinson C. V., Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 8, 639–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilgü H., et al., Variation of the detergent-binding capacity and phospholipid content of membrane proteins when purified in different detergents. Biophys. J. 106, 1660–1670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen T., et al., Tryptophan in the pore of the mechanosensitive channel MscS: assessment of pore conformations by fluorescence spectroscopy. J. Biol. Chem. 285, 5377–5384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blount P., Sukharev S. I., Schroeder M. J., Nagle S. K., Kung C., Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 11652–11657 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards M. D., et al., Pivotal role of the glycine-rich TM3 helix in gating the MscS mechanosensitive channel. Nat. Struct. Mol. Biol. 12, 113–119 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Miller S., et al., Domain organization of the MscS mechanosensitive channel of Escherichia coli. EMBO J. 22, 36–46 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen T., et al., Properties of the mechanosensitive channel MscS pore revealed by tryptophan scanning mutagenesis. Biochemistry 54, 4519–4530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flegler V. J., et al., The MscS-like channel YnaI has a gating mechanism based on flexible pore helices. Proc. Natl. Acad. Sci. U.S.A. 117, 28754–28762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koprowski P., Grajkowski W., Isacoff E. Y., Kubalski A., Genetic screen for potassium leaky small mechanosensitive channels (MscS) in Escherichia coli: Recognition of cytoplasmic β domain as a new gating element. J. Biol. Chem. 286, 877–888 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breyton C., et al., Assemblies of lauryl maltose neopentyl glycol (LMNG) and LMNG-solubilized membrane proteins. Biochim. Biophys. Acta Biomembr. 1861, 939–957 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Lee S., et al., How do branched detergents stabilize GPCRs in micelles? Biochemistry 59, 2125–2134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X. C., Liu Z., Li J., From membrane tension to channel gating: A principal energy transfer mechanism for mechanosensitive channels. Protein Sci. 25, 1954–1964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gullingsrud J., Schulten K., Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 86, 3496–3509 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantor R. S., Lateral pressures in cell membranes: A mechanism for modulation of protein function. J. Phys. Chem. B 101, 1723–1725 (1997). [Google Scholar]
- 50.Bavi N., et al., The role of MscL amphipathic N terminus indicates a blueprint for bilayer-mediated gating of mechanosensitive channels. Nat. Commun. 7, 11984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen A., et al., The role of tryptophan residues in the function and stability of the mechanosensitive channel MscS from Escherichia coli. Biochemistry 46, 10899–10908 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Vásquez V., et al., Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 378, 55–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura T., et al., Differential effects of lipids and lyso-lipids on the mechanosensitivity of the mechanosensitive channels MscL and MscS. Proc. Natl. Acad. Sci. U.S.A. 109, 8770–8775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.East J. M., Lee A. G., Lipid selectivity of the calcium and magnesium ion dependent adenosinetriphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry 21, 4144–4151 (1982). [DOI] [PubMed] [Google Scholar]
- 55.Song B., et al., Capabilities of the Falcon III detector for single-particle structure determination. Ultramicroscopy 203, 145–154 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheres S. H. W. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner T., et al., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres S. H. W., Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen E. F., et al., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Pettersen E. F., et al., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bligh E. G., Dyer W. J., A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 68.Skipski V. P., Thin-layer chromatography of neutral glycosphingolipids. Methods Enzymol. 35, 396–425 (1975). [DOI] [PubMed] [Google Scholar]
- 69.Skipski V. P., Peterson R. F., Barclay M., Quantitative analysis of phospholipids by thin-layer chromatography. Biochem. J. 90, 374–378 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in closed conformation. Protein Data Bank. https://www.rcsb.org/structure/7ONL. Deposited 25 May 2021.
- 71.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in open conformation. Protein Data Bank. https://www.rcsb.org/structure/7OO0. Deposited 26 May 2021.
- 72.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid. Protein Data Bank. https://www.rcsb.org/structure/7OO6. Deposited 26 May 2021.
- 73.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in open conformation. Protein Data Bank. https://www.rcsb.org/structure/7ONJ. Deposited 25 May 2021.
- 74.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in closed conformation with added lipid. Protein Data Bank. https://www.rcsb.org/structure/7OO8. Deposited 26 May 2021.
- 75.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid. Protein Data Bank. https://www.rcsb.org/structure/7OOA. Deposited 27 May 2021.
- 76.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in closed conformation. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-12997. Deposited 25 May 2021.
- 77.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in open conformation. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-13003. Deposited 26 May 2021.
- 78.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-13006. Deposited 26 May 2021.
- 79.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in open conformation. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-12996. Deposited 25 May 2021.
- 80.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in closed conformation with added lipid. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-13007. Deposited 26 May 2021.
- 81.Rasmussen T., Flegler V. J., Boettcher B., Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/entry/EMD-13008. Deposited 27 May 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM maps and PDB models have been deposited in EMDB and PDB [PDB: 7ONL (70), 7OO0 (71), 7OO6 (72), 7ONJ (73), 7OO8 (74), and 7OOA (75); EMDB: EMD-12997 (76), EMD-13003 (77), EMD-13006 (78), EMD-12996 (79), EMD-13007 (80), and EMD-13008 (81)].