Significance
Cardiac injury in mammals is associated with a significant and irreversible loss of heart muscle cells (cardiomyocytes). The loss of cardiomyocytes eventually causes heart failure, leading to organ replacement or death. The importance of this study is that we have made the discovery that new cardiomyocytes can be regenerated in the heart via a protein called Sfrp2. This discovery potentially opens up a new avenue for the treatment of cardiac injury.
Keywords: cell differentiation, cell lineage, heart injuries/pathology, myocytes, Wnt signaling pathway
Abstract
Cardiomyogenesis, the process by which the body generates cardiomyocytes, is poorly understood. We have recently shown that Sfrp2 promotes cardiomyogenesis in vitro. The objective of this study was to determine if Sfrp2 would similarly promote cardiomyogenesis in vivo. To test this hypothesis, we tracked multipotent cKit(+) cells in response to Sfrp2 treatment. In control adult mice, multipotent cKit(+) cells typically differentiated into endothelial cells but not cardiomyocytes. In contrast, Sfrp2 switched the fate of these cells. Following Sfrp2 injection, multipotent cKit(+) cells differentiated solely into cardiomyocytes. Sfrp2-derived cardiomyocytes integrated into the myocardium and exhibited identical physiological properties to preexisting native cardiomyocytes. The ability of Sfrp2 to promote cardiomyogenesis was further supported by tracking EdU-labeled cells. In addition, Sfrp2 did not promote the formation of new cardiomyocytes when the cKit(+) cell population was selectively ablated in vivo using a diphtheria toxin receptor–diphtheria toxin model. Notably, Sfrp2-induced cardiomyogenesis was associated with significant functional improvements in a cardiac injury model. In summary, our study further demonstrates the importance of Sfrp2 in cardiomyogenesis.
The mammalian heart lacks the capacity to repair itself following injury. Specialized muscle cells called cardiomyocytes are not replaced in significant numbers, leading to a progressive loss of function and eventual death from heart failure (1–3). Replacing these lost cardiomyocytes requires an understanding of how cardiomyocytes are generated (cardiomyogenesis). Insights into cardiomyogenesis have been provided by inducible pluripotent stem cells (iPSCs) as these cells can be robustly differentiated into cardiomyocytes. In the iPSC model of cardiomyocyte differentiation, the pluripotent iPSCs are first converted into multipotent cardiac progenitors (4–7). Inhibition of β-catenin signaling drives the differentiation of these multipotent cardiac progenitors into cardiomyocytes (4–7). How Wnt/β-catenin signaling is inactivated during cardiomyocyte development in vivo is unclear. We have found a potentially important role for Sfrp2. Lineage-tracing studies indicate that cardiac Sca-1(+) and cKit(+) cells typically differentiate into endothelial cells (8, 9). Analogous to iPSC-derived multipotent cardiac progenitors, we have found that Sfrp2 redirects the fate of Sca-1(+) and cKit(+) cells to cardiomyocyte-like cells (10, 11). Our data suggest that Sfrp2 drives cardiomyocyte differentiation by modulating the competition between canonical Wnt and noncanonical Wnts. Canonical Wnts activate β-catenin and inhibit cardiac specification (12–14). In contrast, noncanonical Wnts inhibit β-catenin and promotes differentiation into cardiomyocytes (15). In the absence of Sfrp2, canonical Wnts out-compete noncanonical Wnts for binding to Fzd receptors. The addition of Sfrp2 upsets this balance as it appears to preferentially bind, and sequester, canonical Wnts such as Wnt3a and Wnt6. This leaves noncanonical Wnts free to bind to Fzd receptors, activating noncanonical pathways, resulting in β-catenin inhibition and cardiac differentiation (10, 11). While our studies have outlined an important role for Sfrp2 in regulating cardiomyocyte differentiation in vitro, we have not demonstrated a role for Sfrp2 in cardiomyogenesis in vivo.
In this study, we wanted to determine if Sfrp2 promoted cardiomyogenesis in vivo. To test this hypothesis, we tracked the fate of the multipotent cKit(+) cell population in vivo in response to Sfrp2 as cKit(+) cells do not differentiate into cardiomyocytes in vivo (8). In our study, in response to Sfrp2, cKit(+) cells were able to develop into cardiomyocytes that exhibited functionally identical properties to preexisting native cardiomyocytes. In addition, we demonstrated the loss of Sfrp2 effects when this cell population was selectively ablated in vivo. Notably, Sfrp2-induced cardiomyogenesis was associated with significant improvements in cardiac function following myocardial infarction (MI). These findings demonstrate that Sfrp2 may play an important role in cardiomyogenesis in vivo and may have clinical relevance for the treatment of MI.
Results
Sfrp2 Induces Cardiomyocyte Differentiation in Lineage-Committed Cells.
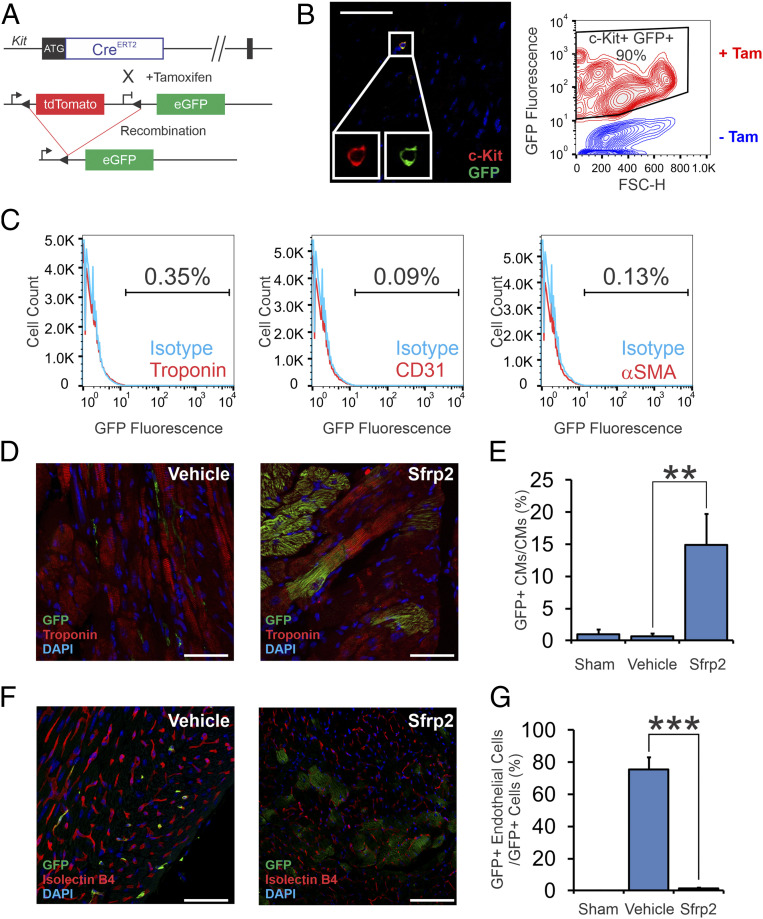
The objective of this study was to determine if Sfrp2 promoted cardiomyogenesis in vivo. We used cKit(+) cells for the study of the potential cardiomyogenic actions of Sfrp2 since, analogous to iPSC-derived cardiac progenitors, cKit(+) cells appear to be multipotent. Lineage-tracing studies show that cKit(+) cells typically differentiate into endothelial cells (8). Using high-throughput RNA sequencing (RNA-seq), we found that Sfrp2 switched the fate of these cells and induced cKit(+) cells to express cardiomyocyte-specific messenger RNAs in vivo (10) and suggested that Sfrp2 may drive cKit(+) cells to differentiate into cardiomyocytes in vivo. To test this hypothesis, in this study, we tracked the fate of cKit(+) cells in response to Sfrp2 with a cKit(+) lineage-tracing model (16). In this lineage tracing model, a tamoxifen-inducible Cre (CreERT2) was targeted to the cKit locus (Fig. 1A). The cKitCreERT2 mice were crossed with the double-fluorescent R26mT/mG reporter mouse line to obtain cKitCreERT2/mTmG mice, in which a tamoxifen-inducible Cre is specifically expressed in cKit(+) cells and manifests as enhanced green fluorescent protein (eGFP) expression following tamoxifen administration. This model has been used to track mast cells as well as cKit(+) progenitor cells in the olfactory system (17, 18). The C57BL6 cKitCreERT2/mTmG mice were injected for 14 consecutive days with a low dose of tamoxifen (0.5 mg/mouse/day) to induce Cre expression and to label cKit(+) cells with eGFP. This dosing schedule was developed (19) to prevent the transient cardiotoxicity that can occur in response to tamoxifen treatment/Cre expression (20). Immunocytochemistry and fluorescence-activated single cell sorting (FACS) analysis indicated that cKit(+) cells were efficiently labeled with green fluorescent protein (GFP) (Fig. 1B). Notably for our study, immediately following tamoxifen administration, no eGFP labeling was observed in cardiac cells such as endothelial, smooth muscle, or cardiomyocytes (Fig. 1C).
Fig. 1.
Sfrp2 induces cardiomyogenesis in vivo. (A) Description of the cKit(+) lineage tracing mouse. cKitCreERT2/mTeG mice, where the cKit promoter drives the expression of a tamoxifen-inducible Cre, were used. This transgenic mouse that carries a Cre-ERT2 expression cassette inserted into the ATG start codon of the endogenous cKit locus was generated by homologous recombination. Upon tamoxifen treatment, Cre-mediated recombination at the LoxP sites allows expression of eGFP exclusively in cKit(+) cells. (B) Heart tissue was analyzed for eGFP expression in cKit(+) cells by confocal microscopy (Left) and flow cytometry (Right). (Left) Heart tissue from tamoxifen-treated mice was stained with cKit and GFP antibodies and then visualized by confocal microscopy. (Scale bar, 100 μm for representative confocal microscopy images.) Images are from three separate animals. (Right) Heart tissue from vehicle or tamoxifen-treated mice was digested by collagenase, and the resulting single cell suspension was analyzed for cKit and eGFP expression by flow cytometry. CKit(+) cells represented <0.001% of the total population in the heart. Considering the rarity of these cells, a contour plot was used to visualize what is a very small population. (C) Hearts were digested with collagenase. The resulting single cell suspension was analyzed for eGFP expression in cardiomyocytes, endothelial cells, and smooth muscle cells via flow cytometry. (D) Expression of eGFP [cKit(+) cells and cells derived thereof] and the cardiac marker cardiac troponin-T was analyzed by confocal microscopy. Representative confocal images are shown. (Scale bar, 50 μm.) (E) Quantification of D. n = 7 for vehicle-treated group, and n = 10 for Sfrp2-treated group; ANOVA with Bonferroni post hoc tests. Significance shown: **P < 0.01. (F) Cardiac tissue was analyzed for coexpression of GFP and the endothelial cell–specific stain isolectin B4. Representative confocal images are shown. (Scale bar, 50 μm [vehicle] and 100 μm [Sfrp2].) Individual channels are shown in SI Appendix, Fig. S1B. (G) Quantification of F. n = 7 for vehicle-treated group, and n = 10 for Sfrp2-treated group; ANOVA with Bonferroni post hoc tests. Significance shown: ***P<0.001.
Following verification of our lineage tracing model, mice received tamoxifen for 14 d to induce recombination. At the end of the tamoxifen treatment, the mice were left for 4 d and then subjected to MI by permanent ligation of the left anterior descending artery (LAD). Two days after MI, the point at which cKit(+) cells normally begin to lose their cardiomyocyte lineage gene expression, mice received vehicle or Sfrp2 via direct cardiac injection (11). Tissue was analyzed for eGFP expression 2 mo following injury by confocal microscopy. In agreement with previous reports, there were no eGFP+ cardiomyocytes in control animals (Fig. 1 D and E). In contrast, eGFP+ cardiomyocytes were observed in the Sfrp2 group (Fig. 1 D and E). The effect of Sfrp2 was strong: ∼15% of the cardiomyocytes in the infarct border zone expressed eGFP (Fig. 1D, quantification in Fig. 1E and SI Appendix, Fig. S1A).
Lineage tracing experiments indicate that cardiac injury induces cKit(+) to differentiate into endothelial cells (8, 21). Consequently, we wanted to determine if the typical differentiation process of cKit(+) cells was affected by Sfrp2. As expected, in control mice, following cardiac injury, cKit(+) cells differentiated into endothelial cells (Fig. 1F, with quantification in Fig. 1G. Individual channels are shown in SI Appendix, Fig. S1B). While in control animals cKit(+) cells differentiated into endothelial cells, the lineage tracing showed that most endothelial cells were not derived from cKit(+) cells. In contrast to the control group, there were no cKit(+) derived endothelial cells in the Sfrp2 group (Fig. 1F, with quantification in Fig. 1G. Individual channels are shown in SI Appendix, Fig. S1B). Despite disappearance of cKit(+) derived endothelial cells, GFP+ cardiomyocytes (cardiomyocytes derived from the action of Sfrp2) were supported by de novo vascularization (Fig. 1F). Indeed, the number of endothelial cells was not significantly different between the control and Sfrp2 groups. Further analysis of cKit(+) differentiation indicated that cKit(+) cells did not differentiate into smooth muscle cells in either the control or Sfrp2 groups (SI Appendix, Fig. S2).
Sfrp2-Derived Cardiomyocytes Integrate with the Myocardium and Are Physiologically Normal.
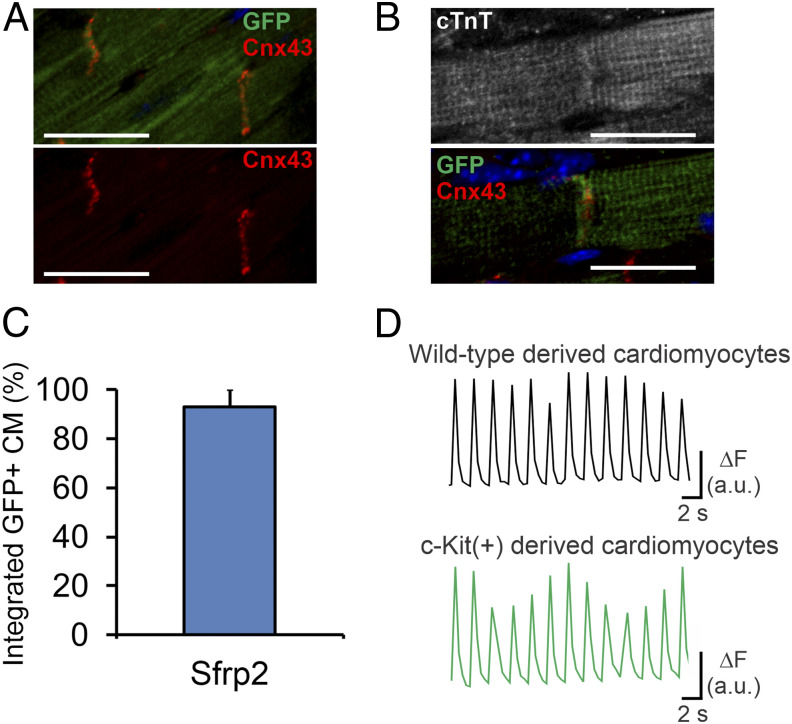
Notably, Sfrp2-derived eGFP+ cardiomyocytes expressed connexin-43 in structures that indicate gap junctions, suggesting integration into the myocardium. Gap junctions were observed between adjacent Sfrp2-derived eGFP+ cardiomyocytes (Fig. 2A) as well as between Sfrp2-derived eGFP+ cardiomyocytes and preexisting (eGFP−) cardiomyocytes (Fig. 2B). A significant majority of the Sfrp2-derived eGFP+ cardiomyocytes were apparently functionally integrated (Fig. 2C).
Fig. 2.
Cardiomyocytes generated via Sfrp2 are physiologically normal and integrate with the myocardium. Following tamoxifen administration to activate the Cre, cKitCreERT2/mTeG mice were subjected to MI. Two days after injury, mice received either vehicle or Sfrp2. Two months after injury, cardiac tissue was analyzed. Sfrp2-derived (eGFP+) cardiomyocytes formed gap junctions with (A) adjacent Sfrp2-derived (eGFP+) cardiomyocytes as well as (B) preexisting (eGFP−) cardiomyocytes, as shown by connexin-43 staining. (Scale bar, 50 μm in A or 25 μm in B.) (C) Quantification of the percentage of Sfrp2-derived (eGFP+) cardiomyocytes, which show apparent integration. n = 7 individual animals. (D) EC coupling in eGFP− and eGFP+ cardiomyocytes. Representative examples of calcium transients obtained from Fura-2–loaded wild-type (eGFP−) and Sfrp2-derived (GFP+) cardiomyocytes during pacing at 0.5 Hz with electric field stimulation.
We then investigated whether Sfrp2-induced eGFP+ cardiomyocytes exhibited the physiological characteristics of isolated mature wild-type cardiomyocytes by measuring excitation-contraction (EC) coupling 4 wk following Sfrp2 treatment. To evaluate calcium signaling and contractility, we used wide field imaging during pacing with electric field stimulation. This revealed that rod-shaped Sfrp2-derived eGFP+ cardiomyocytes exhibited normal sarcomeres, rapid and large calcium transients, and concurrent contractions in response to depolarization (Fig. 2D). The effect was identical to preexisting cardiomyocytes (Fig. 2D). The proportion of eGFP− and eGFP+ cardiomyocytes that showed successful electrical pacing was identical in both groups (∼90%).
Further analysis by RNA-seq indicated that preexisting cardiomyocytes and eGFP+ cells isolated from the post-MI heart 12 d after Sfrp2 treatment were functionally identical as expression levels of cardiomyocyte-specific genes were broadly similar in the two groups (SI Appendix, Fig. S3).
Fate-Mapping with EdU.
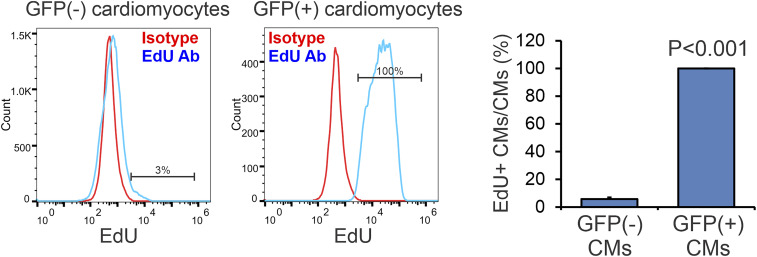
To provide an alternative method to fate-mapping cells, we labeled the eGFP+ cKit(+) cells with the thymidine analog EdU. The number of Sfrp2-derived eGFP+ cardiomyocytes (∼15%) is vastly greater than the cKit(+) population in the heart (<0.1% of all cells in the heart), indicating that the cells must undergo a proliferative phase before differentiating into cardiomyocytes. Thus, the addition of the synthetic thymidine analog EdU during this phase should mark all Sfrp2-derived cardiomyocytes. Consequently, we injected EdU into mice for 10 d immediately post-MI. Following the end of EdU labeling, we waited a further 6 wk after which we measured EdU incorporation in both eGFP(−) and eGFP(+) cardiomyocytes. Approximately 3% of eGFP(−) cardiomyocytes were labeled with EdU (Fig. 3). As expected, 100% of the eGFP(+) cardiomyocytes, i.e., cardiomyocytes derived from a cKit(+) origin, were labeled with EdU (Fig. 3).
Fig. 3.
Fate-mapping with EdU. Mice were subjected to MI and for the following 10 d received EdU by daily i.p. injection. Six weeks post-MI, eGFP(−) and eGFP(+) cardiomyocytes were analyzed for EdU incorporation. n = 3 for vehicle-treated group, and n = 4 for Sfrp2-treated group; t test, P value shown.
Sfrp2 Does Not Produce Cardiomyocytes in the Absence of Multipotent cKit(+) Cells.
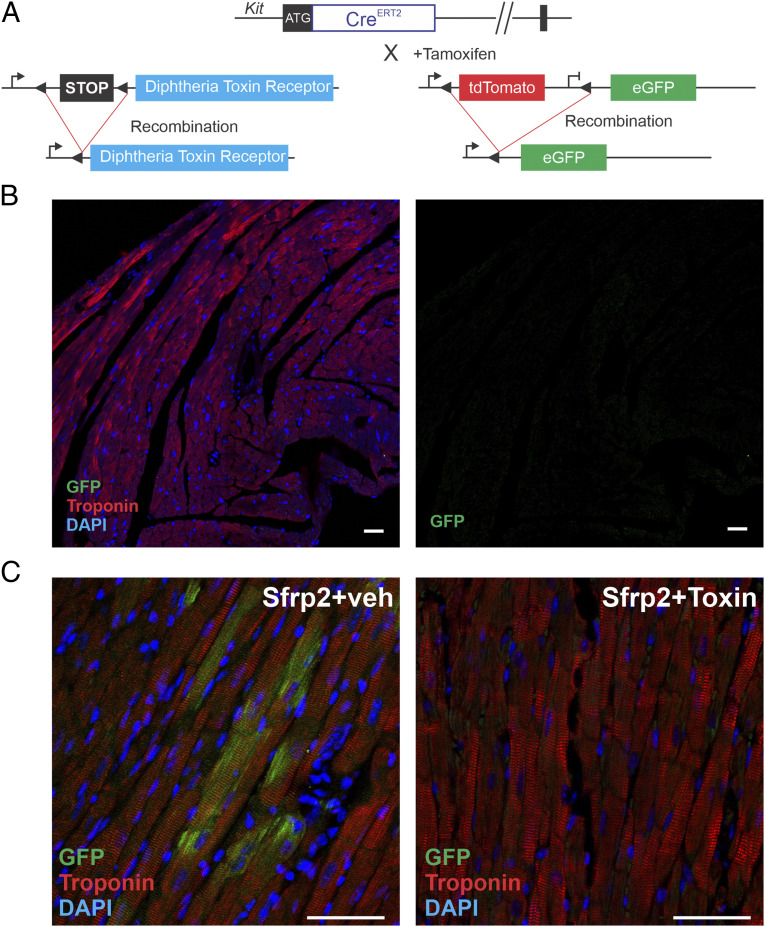
To further demonstrate that eGFP+ cardiomyocytes were not an artifact, we genetically ablated the cKit(+) population from the heart. In the absence of cKit(+) cells, Sfrp2 should not induce eGFP+ cardiomyocyte formation. We ablated the cKit(+) cells from the heart using a diphtheria toxin receptor (DTR)–diphtheria toxin model. The DTR is required to transport diphtheria toxin into a cell. However, mice do not express the DTR and as a result, they are unaffected by diphtheria toxin administration. Expression of DTR allows uptake of diphtheria toxin, leading to cell death. Crucially, adjacent cells not expressing DTR are unaffected by the diphtheria toxin. This model has been successfully employed in vivo to selectively ablate a number of different cell types (22–24).
To selectively ablate cKit(+) cells, we employed the Gt(ROSA)26Sortm1(HBEGF)Awai/J mouse line where DTR expression is activated following expression of Cre. These mice were crossed with the cKitCreERT2/mTmG line (Fig. 4A). Tamoxifen treatment in the progeny results in DTR expression specifically in cKit(+) cells. Administration of diphtheria toxin ablates only those cells expressing DTR, thus selectively removing cKit(+) cells. Diphtheria toxin administration had no effect on gross morphology of heart, kidney, spleen, lung, and liver. We next assessed eGFP staining in the myocardium post–diphtheria toxin administration. No eGFP expression was observed in the preinjury heart (Fig. 4B).
Fig. 4.
Genetic ablation prevents Sfrp2-induced cardiomyogenesis. (A) cKitCreERT2/mTmG mice were crossed with a DTR strain to selectively ablate cKit(+) cells in the heart. Mice (cKitCreERT2/mTmG/DTR) were injected with tamoxifen (0.5 mg/mouse) for 14 consecutive days. In the final 7 d of tamoxifen treatment, dipththeria toxin (100 ng/mouse) was injected daily to ablate cKit(+) positive cells. Four days later, treatment mice were subjected to MI. Two days later, mice were injected with Sfrp2 (0.5 µg) or vehicle at the infarct border zone. (B) cKitCreERT2/mTmG/DTR mice were injected with tamoxifen (0.5 mg/mouse) for 14 consecutive days. In the final 7 d of tamoxifen treatment, dipththeria toxin (100 ng/mouse) was injected daily to ablate cKit positive cells. Heart tissue was analyzed for cardiac troponin-T and eGFP expression by confocal microscopy. n = 3. (C) Representative confocal images of 2 mo post-MI hearts. Colocalization of eGFP and the cardiac marker cardiac troponin-T was determined by confocal microscopy. (Scale bar, 50 μm.) n = 5 per group.
Following these studies, mice received vehicle or diphtheria toxin to ablate the cKit(+) population. Mice then underwent MI and 2 d later were treated with Sfrp2. Two months following MI, mice were killed. In the absence of diphtheria toxin, Sfrp2 promoted the formation of eGFP+ cardiomyocytes (Fig. 4C). In contrast, no eGFP+ cardiomyocytes were observed in the tamoxifen/diphtheria toxin–treated group (Fig. 4C). These data supported our hypothesis that Sfrp2 promotes cardiomyogenesis.
Sfrp2-Induced Cardiomyogenesis Improves Cardiac Function in Mice Following Cardiac Injury.
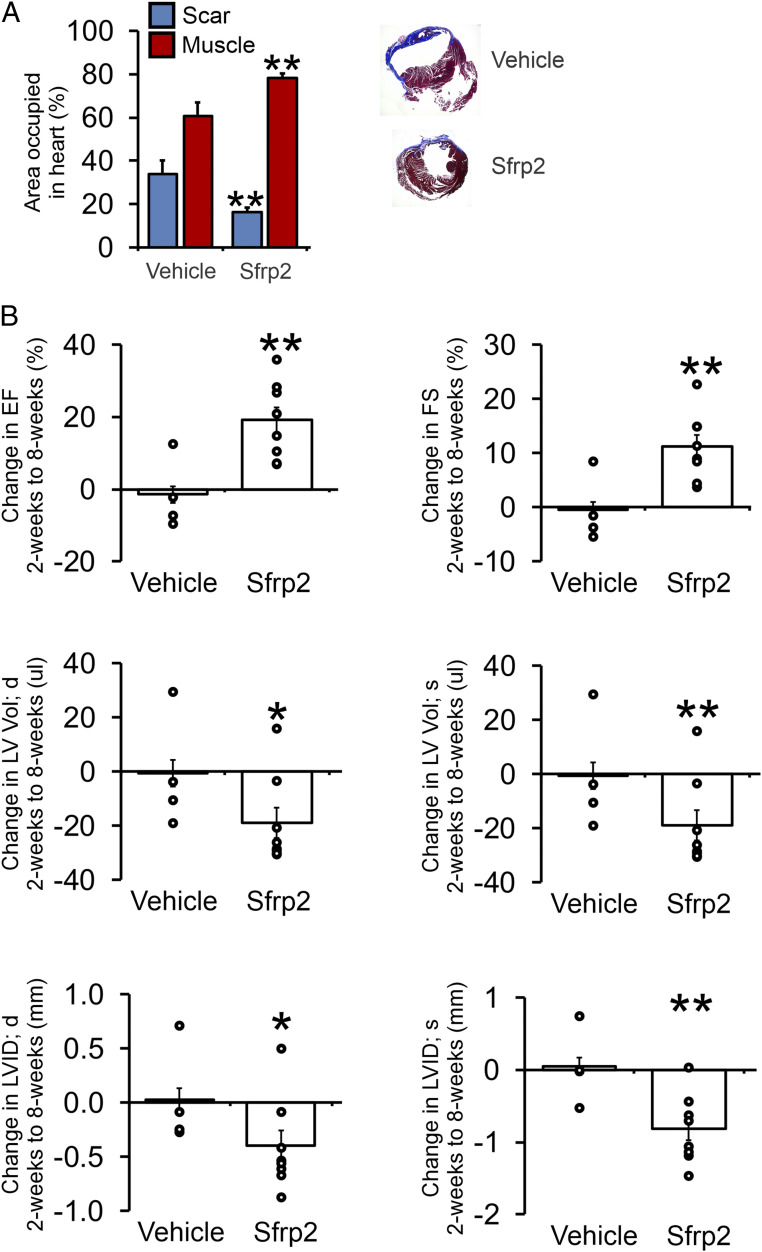
We next wanted to determine if Sfrp2-induced cardiomyogenesis influenced cardiac function following cardiac injury. Mice (cKitCreERT2/mTmG) received tamoxifen for 14 d to induce recombination. At the end of the tamoxifen treatment, the mice were left for 4 d and then subjected to MI by permanent ligation of the LAD. Two days after MI, mice received vehicle or Sfrp2 via direct cardiac injection. Two months after MI, tissue sections 0.5 mm and 1 mm below the injection site were analyzed for the areas occupied by scar tissue and cardiac muscle. When compared to the control group, Sfrp2 was associated with reduced scar size (Fig. 5A). Corresponding with reduced scar size, Sfrp2 increased the cardiac muscle area (Fig. 5A).
Fig. 5.
Sfrp2-induced cardiomyogenesis is associated with improved therapeutic outcomes. Mice (cKitCreERT2/mTmG) were injected with tamoxifen (0.5 mg/mouse) for 14 consecutive days. Four days after tamoxifen treatment, mice were subjected to MI. Two days after injury, mice were injected with Sfrp2 (0.5 µg) or PBS at the infarct border zone. (A) Analysis of Masson’s trichrome staining for cardiac muscle (red) and fibrosis (blue). Sections were taken at 0.5 mm and 1 mm below the Sfrp2/vehicle injection point. n = 4 (vehicle) or 8 (Sfrp2); t test, **P < 0.01. Representative Masson’s trichrome–stained cardiac sections are shown. (B) Cardiac function was assessed by echocardiography immediately prior to injury, 2 wk after injury, and finally 8 wk after injury. Raw cardiac function data are provided in SI Appendix, Table S1. Comparisons were made between 2 and 8 wk postinjury in the control and Sfrp2 groups. The graphs show the comparison for each animal (open circle) as well as the average value in each group (bar). n = 4 (vehicle) or 8 (Sfrp2). t tests were carried out between the control and Sfrp2 groups; *P < 0.05, **P < 0.01.
Cardiac function was assessed immediately prior to injury and then 2 and 8 wk after injury. Cardiomyocyte differentiation is typically a long process, taking >4 wk to generate mature cardiomyocytes (25, 26). Cardiac function 2 wk after injury, prior to the development of mature cardiomyocytes, was broadly similar between the control and Sfrp2 groups (SI Appendix, Table S1). By 8 wk post-MI, by which point mature cardiomyocytes had developed, the control and Sfrp2 groups had markedly diverged. In the control group, there were further decreases in cardiac function (SI Appendix, Table S1). In contrast, the Sfrp2 group showed significantly improved cardiac function (SI Appendix, Table S1). To further evaluate the effect of Sfrp2, the change in cardiac function between 2 and 8 wk post-MI was calculated for each animal. Further underlying the beneficial effect of Sfrp2, while the control mice typically demonstrated worsening cardiac function, all of the mice in the Sfrp2 group showed improvements in ejection fraction and fractional shortening (Fig. 5B). Further analysis of the cardiac function data showed that Sfrp2-induced cardiomyogenesis had a stronger effect on systolic measurements (Fig. 5B), indicating improved cardiac contractility.
Discussion
Cardiomyocyte differentiation remains a poorly understood process. It has been proposed that Wnt/β-catenin signaling has a biphasic function during cardiac development (27). Early cardiac specification requires Wnt/β-catenin activation, whereas differentiation requires inhibition of this pathway (27). Crescent, a member of the Sfrp family, and Dkk1 promote cardiac development in chicken and Xenopus embryos via inhibition of the Wnt/β-catenin pathway (28, 29). Similarly, β-catenin genetic ablation specifically in the mouse endoderm results in the formation of multiple ectopic hearts (30), and the development of the heart from the posterior mesoderm requires Wnt inhibition (28, 29). Important insights into cardiomyogenesis have also been provided by iPSCs as these cells can be robustly differentiated into cardiomyocytes. In the iPSC model of cardiomyocyte differentiation, pluripotent iPSCs are first converted into multipotent cardiac progenitors (4–7). Generation of multipotent cardiac progenitors requires Wnt/β-catenin activation (4–7). The status of the Wnt/β-catenin signaling pathway determines the subsequent fate of these multipotent cardiac progenitors. Inactivation of Wnt/β-catenin signaling drives the differentiation of the iPSC-derived multipotent cardiac progenitors into cardiomyocytes (4–7). In contrast, if Wnt/β-catenin signaling is not inhibited, these iPSC-derived multipotent cardiac progenitors will differentiate into a restricted number of alternative cell types (31). How Wnt/β-catenin signaling is inactivated during cardiomyocyte development in vivo is unclear. Endogenous Wnt/β-catenin inhibitors include proteins in the Dikkopf and secreted frizzled receptor-protein (Sfrp) families. Dikkopf-1 has been implicated in cardiomyocyte development from multipotent cardiac progenitors (29, 32). Our findings described previously as well as previous work in our laboratory suggest that Sfrp2 plays an important role in the switch from Wnt/β-catenin activation to Wnt/β-catenin inactivation. In this respect, it is interesting that we found Sfrp2 was apparently sufficient to completely switch the fate of cKit(+) cells. Under standard conditions, cKit(+) cells differentiated into endothelial cells. In contrast, Sfrp2 redirected the fate of these cells to cardiomyocytes. Switching the fate of cells from endothelial cell differentiation to cardiomyocyte differentiation further suggests that Sfrp2 is influencing the Wnt/β-catenin pathway, as Wnt/β-catenin activation is believed to be important for endothelial cell differentiation (33–35). Our studies (10, 11) suggest that Sfrp2 promotes cardiomyocyte differentiation by modulating the competing actions of canonical and noncanonical Wnts. Canonical Wnts inhibit cardiomyocyte activation via β-catenin activation (12–14). In contrast, noncanonical Wnts promote differentiation into cardiomyocytes via the inhibition of β-catenin (15). What we have discovered is that in the absence of Sfrp2, canonical Wnts out-compete noncanonical Wnts for binding to Fzd receptors such as Fzd5. Sfrp2 appears to preferentially bind, and sequester, canonical Wnts such as Wnt3a and Wnt6. This tips the balance in favor of the noncanonical Wnts which are now free to bind to Fzd receptors. Once bound to Fzd receptors, these noncanonical Wnts inhibit β-catenin via the planar cell polarity pathway and JNK (10, 11). These studies outlined an important role for Sfrp2 in regulating cardiomyocyte differentiation in vitro. The importance of the current study is the demonstration that Sfrp2 promotes cardiomyocyte differentiation in vivo. Of note, our genome profiling of cKit(+) cells isolated from the heart revealed that Sfrp2 induced the expression of genes involved in noncanonical Wnt signaling (10). While this suggests that the outlined mechanism is also important in vivo, future studies with genetic knockout models will be needed to fully address this question. Another important question to address is the origin of the cKit(+) cells, as our models are unable to distinguish between resident cKit(+) cells and cKit(+) cells that entered the heart following injury. Such studies would provide useful information regarding the role of Wnt inhibitors in cardiac development, for example, by potentially acting as chemoattractants to localize cells to the correct environment for cardiomyocyte differentiation.
Another feature of our study is the use of a cell ablation model. In the first instance, we employed standard lineage tracing. Genetic lineage tracing is carried out by combining an inducible Cre expressed in the cell type of interest with a Cre-activated reporter expressing a fluorescent protein. In essence, lineage tracing is a “pulse-chase” experiment where cells are marked only when the Cre is activated. In our study, a tamoxifen-inducible Cre marked cKit(+) cells with the fluorescent protein GFP. Using this model, we found that Sfrp2 gave rise to GFP+ cardiomyocytes. This finding suggested that Sfrp2 was promoting cKit(+) differentiation into cardiomyocytes. We verified the findings of the lineage tracing by ablating the cKit(+) cell population in a DTR–diphtheria toxin model. The significant advantage of this DTR cell ablation model is that it definitively ties the presence of the fluorescent reporter to the progeny of the marked cell. In standard lineage tracing, the presence of the fluorescent reporter is taken to mean that the fluorescent cell developed from the initially marked cell. However, random events could potentially activate the fluorescent reporter in any cell type during the duration of the study. These random events would give rise to “false positives”: the appearance of differentiation from the initially marked cell when in reality there was none. In a cell ablation model, random activation of a fluorescent reporter would be obvious. We lost all fluorescent events with the DTR model, indicating that Sfrp2 was definitively inducing cKit(+) to differentiate into cardiomyocytes.
Finally, there are potential clinical implications of our study. Our observation that Sfrp2-induced cardiomyogenesis promoted functional recovery in the infarcted heart suggests that Sfrp2, or small molecules that inhibit Wnt, could be delivered into patients following MI to regenerate heart muscle from cells within the scar tissue. Our findings may also prove applicable in other settings where tissue is lost postinjury. Following stroke, damage to the brain could be repaired through molecular activation of resident cells (36) that reside in the tissue.
In summary, we have identified that Sfrp2 plays an important role in cardiomyocyte differentiation in vivo. This finding may aid the development of therapies to replace cardiomyocytes that are lost following injury.
Materials and Methods
Study Design.
Sample size.
Sample size was selected based on previous studies in the laboratory. The number was not altered during the course of the study.
Rules for stopping data collection.
Death before the end of the study period stopped data collection.
Data inclusion/exclusion criteria.
All data were included and no data were excluded. This was decided before the study was undertaken.
Outliers.
No data were removed. Thus, there were no outlier tests.
Selection of end points.
The primary end point was determined prior to the study.
Replicates.
Animal numbers can be found in each figure legend.
Research objectives.
The hypothesis tested was that Sfrp2 induces cKit(+) cells to differentiate into cardiomyocytes.
Research subjects or units of investigation.
Animal lineage tracing models (see Animal Models).
Experimental design.
This was a laboratory experiment. Vehicle or Sfrp2 was injected into mice 2 d after MI. See below for further details [MI (Acute LAD Coronary Artery Ligation) and Sfrp2 Injection].
Randomization.
Animals of the correct genotype were randomly selected. Randomization was performed blinded (see Blinding).
Blinding.
Three separate blinded investigators carried out the staining and counting protocols described. Investigators were blinded with respect to animal identifiers and group assignments. Values were averaged between the three blinded investigators. The animal surgeon who carried out the mice procedures did not know the animal genotype or the identity of the chemicals given to the animals. All compounds used in this study were colorless and odorless; thus, there was no way to differentiate between vehicle, Sfrp2, or diphtheria toxin. For any given experiment, all groups were subjected to surgery at the same time; thus, there was no bias in how the groups were treated with respect to surgeries.
Animal Models.
Mice were maintained at the Duke University animal facility following the local guidelines. Animal procedures were approved by the Duke University Institutional Animal Care and Use Committee prior to starting the experiments.
C57BL/6:cKit CreERT2: R26mT-mG/+ (cKitCreERT2/mTmG) mice were used where the cKit promoter drives the expression of a tamoxifen-inducible Cre (Fig. 1A). This transgenic mouse carries a Cre-ERT2 expression cassette inserted into the start codon of the endogenous cKit locus (16). Upon tamoxifen treatment, Cre-mediated recombination at the Locus of Crossover in P1 (LoxP) sites allows expression of eGFP exclusively in cKit(+) cells. cKit CreERT2/mTmG mice were then crossed with a DTR strain (C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J; Jackson Labs) to selectively ablate cKit cells. Mice were maintained on a C57BL6 background. Eight-week-old mice received 0.5 mg/day 4-OH tamoxifen (Sigma) diluted in sunflower oil (Sigma) by intraperitoneal (i.p.) administration for 14 consecutive days in order to induce Cre expression and recombination. In the final 7 d of tamoxifen treatment diphtheria toxin (100 ng/mouse, Sigma) was injected daily to ablate cKit positive cells. One week after the last 4-OH tamoxifen dose, mice were subjected to permanent ligation of the LAD.
MI (Acute LAD Coronary Artery Ligation) and Sfrp2 Injection.
Anesthetized 10-wk-old mice were intubated prior to left thoracotomy and exposure of the left ventricle of the heart. Mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg) by i.p. injection. Ophthalmic ointment was applied to both eyes to prevent corneal desiccation during the procedure. Following washing of the skin with iodophore/alcohol, an endotracheal intubation was performed, and the mouse was connected to a ventilator (model 683, Harvard Apparatus, tidal volume 0.7 to 1 mL, respiratory rate 120 breaths per minute) via an intravenous catheter (20 GA 1 IN) as the cannula under direct laryngoscopy. The chest cavity was opened between the fourth and the fifth rib in the intercostals muscle, the heart externalized, and a 7–0 nylon suture placed through the myocardium into the anterolateral left ventricular (LV) wall, corresponding to the course of the LAD. The suture was positioned approximately midway between the apex and base and a ligature made. Before ligation, left coronary artery entrapment was confirmed by upward traction. The suture was completely tied off (MI), and the apex of the LV was observed for evidence of myocardial blanching, indicating interruption in coronary flow. The wound was then closed using 7–0 nylon and the chest cavity was subsequently closed in layers with 5–0 monofilament suture. Negative pressure was re-established at closure, and the animal was gradually removed from the respirator. Following the resumption of spontaneous respiration, the endotracheal tube was removed and the animal allowed to recover on a deltaphase isothermal pad set at 37 °C. The animals remained in a supervised setting until fully conscious and then were returned to their cages and given standard chow and water. Postoperative analgesia was used: bupivacaine locally + buprenorphine subcutaneous twice daily for 5 d.
Two days following MI, mice underwent a second left thoracotomy followed by intracardiac injection of a total of 0.5 µg of recombinant mouse Sfrp2 (R&D Systems) or normal saline (vehicle) at 3 to 5 sites within the infarct border zone. Direct cardiac injection of the Wnt inhibitor Sfrp2 significantly reduces the possibility of Sfrp2 affecting organs other than the heart.
All skin sutures were removed by 7 d postoperation, if the sutures remained.
Serial Echocardiography.
High-resolution two-dimensional echocardiography was performed pre-MI and at 2 wk and 8 wk post-MI. At each time point, the following information was acquired: fractional shortening, ejection fraction, LV mass, LV end-diastolic diameter, LV end-systolic diameter, LV end-diastolic volume, and LV end-systolic volume.
Immunohistochemistry.
Sixteen heart sections were immunostained with GFP (abcam, ab6556), cardiac troponin (abcam, ab105439), isolectin β4 (Life Technologies, I21413), alpha smooth muscle actin (Sigma, A5228), or connexin 43 (Sigma, C6219) antibodies as appropriate in a blinded manner. The entire peri-infarct region in each heart slice was analyzed by confocal imaging using an LSM 510 Meta DuoScan microscope (Zeiss), and processed and positive cells were quantified using Zeiss Zen and cell profiler software. All secondary antibodies were Alexa-Fluor (Invitrogen) and were used according to standard techniques. Note: as samples were paraffin embedded, there is no fluorescence from GFP or tdTomato. Antibodies are needed for the detection of the fluorescent proteins.
Heart Tissue for EdU Analysis.
Mice were subjected to MI. One day later, mice received EdU injections by i.p. [48 mg/kg (37)] daily for 10 d post-MI. Six-week post-MI hearts were homogenized as described above. Cardiomyocytes were collected by centrifugation at 300 × g for 1 min. All other cells were collected by centrifugation at 400 × g for 7 min. Pellets were combined and fixed with 2% vol/vol paraformaldehyde for 15 min. Cells were incubated with a BV421-GFP antibody (BD Biosciences 566040) for 1 h at 4 °C in Perm/Wash buffer (BD Biosciences). Following washing in Perm/Wash buffer, cells were incubated with a 647-EdU antibody according to the manufacturer’s instructions (BD Biosciences 647 EdU Click Proliferation Kit 565456). Following washing according to the manufacturer’s instructions, cells were incubated with phycoerythrin-conjugated cardiac troponin-T antibody (Miltenyi Biotech 130-106-746) for 1 h at 4 °C in Perm/Wash buffer (BD Biosciences). After washing in Perm/Wash buffer, cells were analyzed by flow cytometry. Compensation was carried out by FloJo software postacquisition. Gating was based on isotype controls.
Flow cytometry data were compensated and analyzed with FlowJo version 10.
Ex Vivo Analysis: Calcium and Contractility Measurements.
Cardiomyocytes were isolated according to Louch et al. with modifications (38). Prior to harvest, the heart was flushed with Krebs-Henseleit buffer until clear of blood. The heart was then excised, carefully dissecting mediastinal adhesions, atria surgically removed, and ventricles dissociated using standard procedures. Following isolation, dissociated cells were suspended in Tyrode solution containing 140 mM NaCl, 5.4 mM KCl, 1.05 mM MgCl2, 1.8 mM CaCl2, 0.33 mM NaH2PO4, 5 mM Hepes, and 10 mM glucose (pH 7.4 with NaOH) and analyzed with 12 h of isolation.
Dissociated cells were plated on laminin-coated dishes, the cells were allowed to attach, and the dishes were loaded with the calcium indictor Fura-2 (1 μM, Molecular Probes/Invitrogen 20 min) for 20 min, then mounted on a Nikon TE2000 inverted microscope equipped with a Photometrics CoolSnap camera, a xenon arc lamp, and a lambda DG-4 rapid filter changer (Sutter). Dishes were perfused at room temperature with Tyrode solution containing 140 mM NaCl, 5.4 mM KCl, 1.05 mM MgCl2, 1.8 mM CaCl2, 0.33 mM NaH2PO4, 5 mM Hepes, and 10 mM glucose (pH 7.4 with NaOH). Dishes were visually scanned, rod-shaped cardiomyocytes identified, and cells paced at 0.5 and 1 Hz using electrical field stimulation with 5 ms voltage pulses at 150% of threshold (Warner Field Stimulation chamber). Calcium dynamics and contraction were determined for single cells. Images were acquired using a 40× S Plan 1.3 NA objective and image acquisition controlled using Metafluor/Metamorph software (Molecular Devices). Single wavelength fluorescence images were acquired by excitation at 380 nm and emission at 510 nm to increase time resolution (acquisition interval of 38 to 39 ms). This sampling frequency was suboptimal for accurately capturing calcium transient peaks, which resulted in apparent variability in transient amplitudes. The changes in Fura-2 fluorescence intensity (ΔF) shown thus provide a qualitative measure of the calcium signals evoked during EC coupling. For each group (eGFP− and eGFP+), 20 cardiomyocytes were analyzed for electrical pacing.
Masson’s Trichrome: Fibrosis and Muscle Area Measurements.
Hearts of anesthetized (isoflurane, Henry Schein-VetUS) mice 2 mo post-MI were arrested via cold cardioplegic solution (35 mmol/L KCl, 68.6 mmol/L mannitol, 5% dextrose, 1.6 mmol/L bicarbonate, 1,000 units heparin in normal saline). Hearts were excised and fixed overnight at 4 °C in 10% vol/vol neutral buffered formalin, followed by incubation in 30% wt/vol sucrose for a further 24 h at 4 °C. The entire heart from the suture to the apex was cut into a number of 0.5-mm sections using a Zivic Stainless Steel Mouse Heart Matrix (HSHS005-1). These sections were paraffin embedded. Sections taken at 0.5 mm and 1 mm distal to the suture were used for the fibrosis and muscle measurements. For each section, five 10-μm slices were taken and mounted onto a glass slide. Masson’s trichrome staining was then performed according to standard techniques. All slides were stained at the same time to avoid any environmental bias. For each tissue slice total LV, fibrosis and muscle area were calculated as sum of areas in the slices by blinded investigators using ImageJ software. Fibrosis and muscle areas are expressed as a percentage of total LV area.
Images.
Images were processed with CorelDraw. Confocal microscopy images were processed using Zeiss software Axiovision Rel4.8 and Zen.
Statistics.
All statistical analysis was performed using GraphPad. For experiments containing two conditions, a t test was performed. ANOVA was used for experiments with three or more conditions followed by Bonferroni post hoc tests for comparisons between individual groups. Grubb’s test was used to identify outliers. A P value of less than 0.05 was considered significant.
Study Approval.
Animal procedures and experiments were approved by the Duke University Institutional Animal Care and Use Committee prior to starting the experiments.
Supplementary Material
Acknowledgments
We thank John Wong of the Duke Human Vaccine Institute for the use of flow cytometers and the Mandel Cardiovascular Research Center Echocardiography Core for the measurements of cardiac function. We also thank Sophie Dal-Pra for technical assistance. Research conducted in these studies was supported by National Heart, Lung, and Blood Institute Grants RO1 HL35610, HL81744, HL72010, HL73219, and HL081744-07S1, as well as by the Edna and Fred L. Mandel Jr. Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103676118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Raw and processed RNA-seq data can also be found in the NIH Gene Expression Omnibus (GSE90615) (39).
References
- 1.Bergmann O., et al., Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jennings R. B., Reimer K. A., The cell biology of acute myocardial ischemia. Annu. Rev. Med. 42, 225–246 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Cleutjens J. P., Blankesteijn W. M., Daemen M. J., Smits J. F., The infarcted myocardium: Simply dead tissue, or a lively target for therapeutic interventions. Cardiovasc. Res. 44, 232–241 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Später D., Hansson E. M., Zangi L., Chien K. R., How to make a cardiomyocyte. Development 141, 4418–4431 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Jeziorowska D., Korniat A., Salem J. E., Fish K., Hulot J. S., Generating patient-specific induced pluripotent stem cells-derived cardiomyocytes for the treatment of cardiac diseases. Expert Opin. Biol. Ther. 15, 1399–1409 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kleinsorge M., Cyganek L., Subtype-directed differentiation of human iPSCs into atrial and ventricular cardiomyocytes. STAR Protoc 1, 100026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y., Zou J., Differentiation of cardiomyocytes from human pluripotent stem cells in fully chemically defined conditions. STAR Protoc 1, 100015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Berlo J. H., et al., c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509, 337–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vagnozzi R. J., et al., Genetic lineage tracing of Sca-1+ cells reveals endothelial but not myogenic contribution to the murine heart. Circulation 138, 2931–2939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkinson C. P., et al., Insights from molecular signature of in vivo cardiac c-Kit(+) cells following cardiac injury and β-catenin inhibition. J. Mol. Cell. Cardiol. 123, 64–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmeckpeper J., et al., Inhibition of Wnt6 by Sfrp2 regulates adult cardiac progenitor cell differentiation by differential modulation of Wnt pathways. J. Mol. Cell. Cardiol. 85, 215–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen E. D., Tian Y., Morrisey E. E., Wnt signaling: An essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development 135, 789–798 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Oikonomopoulos A., et al., Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3. Circ. Res. 109, 1363–1374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita J. K., et al., Prospective identification of cardiac progenitors by a novel single cell-based cardiomyocyte induction. FASEB J. 19, 1534–1536 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Bisson J. A., Mills B., Paul Helt J. C., Zwaka T. P., Cohen E. D., Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT. Dev. Biol. 398, 80–96 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Klein S., et al., Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat. Commun. 4, 1630 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Goldstein B. J., et al., Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J. Comp. Neurol. 523, 15–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goss G. M., et al., Differentiation potential of individual olfactory c-Kit+ progenitors determined via multicolor lineage tracing. Dev. Neurobiol. 76, 241–251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loffredo F. S., Steinhauser M. L., Gannon J., Lee R. T., Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell 8, 389–398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koitabashi N., et al., Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ. Res. 105, 12–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L., et al., Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 23, 1488–1498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Blijswijk J., et al., Altered lymph node composition in diphtheria toxin receptor-based mouse models to ablate dendritic cells. J. Immunol. 194, 307–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka K., et al., Generation of mouse models for type 1 diabetes by selective depletion of pancreatic beta cells using toxin receptor-mediated cell knockout. Biochem. Biophys. Res. Commun. 436, 400–405 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Golub J. S., et al., Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J. Neurosci. 32, 15093–15105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayawardena T. M., et al., MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 116, 418–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgkinson C. P., et al., Cardiomyocyte maturation requires TLR3 activated nuclear factor kappa B. Stem Cells 36, 1198–1209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gessert S., Kühl M., The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 107, 186–199 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Marvin M. J., Di Rocco G., Gardiner A., Bush S. M., Lassar A. B., Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15, 316–327 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider V. A., Mercola M., Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 15, 304–315 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lickert H., et al., Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev. Cell 3, 171–181 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., et al., Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat. Commun. 10, 2238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bondue A., Blanpain C., Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 107, 1414–1427 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z., et al., Wnt/β-catenin signaling determines the vasculogenic fate of postnatal mesenchymal stem cells. Stem Cells 34, 1576–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masckauchán T. N., Shawber C. J., Funahashi Y., Li C. M., Kitajewski J., Wnt/beta-catenin signaling induces proliferation, survival and interleukin-8 in human endothelial cells. Angiogenesis 8, 43–51 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Yan J., et al., Cyclic stretch induces vascular smooth muscle cells to secrete connective tissue growth factor and promote endothelial progenitor cell differentiation and angiogenesis. Front. Cell Dev. Biol. 8, 606989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan C., Scotting P., Expression of Kit and Etv1 in restricted brain regions supports a brain-cell progenitor as an origin for cranial germinomas. Cancer Genet. 208, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Beigi F., et al., C3orf58, a novel paracrine protein, stimulates cardiomyocyte cell-cycle progression through the PI3K-AKT-CDK7 pathway. Circ. Res. 113, 372–380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louch W. E., Sheehan K. A., Wolska B. M., Methods in cardiomyocyte isolation, culture, and gene transfer. J. Mol. Cell. Cardiol. 51, 288–298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodgkinson C. P., et al., A crucial function for c-Kit(+) in cardiac regeneration: Role of Wnt inhibition. NIH GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90615. Deposited 30 November 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Raw and processed RNA-seq data can also be found in the NIH Gene Expression Omnibus (GSE90615) (39).