Significance
Faithful inheritance of chromosomes during meiosis depends on the formation and repair of double-strand DNA breaks (DSBs), which are generated through the activity of a topoisomerase-like protein known as Spo11. Spo11 exhibits strong conservation throughout eukaryotes, presumably reflecting constraints imposed by its biochemical activity, but auxiliary proteins that collaborate with Spo11 to promote and regulate DSB formation are less well conserved. Here we investigate a cohort of proteins comprising a complex required for meiotic DSB formation in Caenorhabditis elegans, providing evidence for both conservation with and divergence from homologous complexes in other organisms. This work highlights the evolutionary malleability of protein complexes that serve essential, yet auxiliary, roles in fundamental biological processes that are central to reproduction.
Keywords: meiosis, C. elegans, DNA double-strand breaks, meiotic recombination, Mei4
Abstract
Meiotic recombination plays dual roles in the evolution and stable inheritance of genomes: Recombination promotes genetic diversity by reassorting variants, and it establishes temporary connections between pairs of homologous chromosomes that ensure their future segregation. Meiotic recombination is initiated by generation of double-strand DNA breaks (DSBs) by the conserved topoisomerase-like protein Spo11. Despite strong conservation of Spo11 across eukaryotic kingdoms, auxiliary complexes that interact with Spo11 complexes to promote DSB formation are poorly conserved. Here, we identify DSB-3 as a DSB-promoting protein in the nematode Caenorhabditis elegans. Mutants lacking DSB-3 are proficient for homolog pairing and synapsis but fail to form crossovers. Lack of crossovers in dsb-3 mutants reflects a requirement for DSB-3 in meiotic DSB formation. DSB-3 concentrates in meiotic nuclei with timing similar to DSB-1 and DSB-2 (predicted homologs of yeast/mammalian Rec114/REC114), and DSB-1, DSB-2, and DSB-3 are interdependent for this localization. Bioinformatics analysis and interactions among the DSB proteins support the identity of DSB-3 as a homolog of MEI4 in conserved DSB-promoting complexes. This identification is reinforced by colocalization of pairwise combinations of DSB-1, DSB-2, and DSB-3 foci in structured illumination microscopy images of spread nuclei. However, unlike yeast Rec114, DSB-1 can interact directly with SPO-11, and in contrast to mouse REC114 and MEI4, DSB-1, DSB-2, and DSB-3 are not concentrated predominantly at meiotic chromosome axes. We speculate that variations in the meiotic program that have coevolved with distinct reproductive strategies in diverse organisms may contribute to and/or enable diversification of essential components of the meiotic machinery.
Meiotic recombination is important for two reasons. It promotes genetic diversity by reassorting traits, and it creates temporary attachments between pairs of homologous chromosomes that are necessary for their future segregation at the meiosis I division. Recombination is initiated by the programmed introduction of DNA double-strand breaks (DSBs) (1). Some DSBs are repaired by a mechanism that leads to the formation of crossovers (COs) between homolog pairs, and the remaining DSBs are repaired as non-CO products, thereby restoring genome integrity. Although DSBs are required for CO formation, they may lead to genomic instability if they are not repaired or are repaired erroneously. Thus, DSB formation in meiotic cells is governed by regulatory and surveillance mechanisms that function to ensure that enough DSBs are created to guarantee a CO on each homolog pair while limiting excess DSBs that may endanger the genome (2). Without appropriate DSB formation and repair, COs may fail to form between homologs during meiotic prophase, resulting in unattached homologs (univalents) that missegregate during the meiotic divisions, leading to aneuploidy in the resulting progeny.
Meiotic DSB formation is catalyzed by Spo11, a topoisomerase-like protein homologous to the catalytic A subunit of archaeal class VI topoisomerases that is well conserved across eukaryotic kingdoms (3–6). The mechanism of DNA breakage involves formation of a covalent linkage between the Spo11 protein and DNA, analogous to a key intermediate in the topisomerase reaction (1). Despite identification of structural and mechanistic conservation between Spo11 and TopVIA more than 20 y ago, however, counterparts of the archaeal TopVIB subunit that partner with Spo11 in “Spo11 core complexes” were not recognized until much later, reflecting substantial divergence both from TopVIB and among their eukaryotic orthologs (7–9).
DSB formation also depends on multiple additional factors that play critical roles in determining the location, timing, levels, and regulation of DSB formation (2). Several of these auxiliary DSB-promoting factors, including Rec114, Mei4, and Mer2, were originally discovered through genetic screens in Saccharomyces cerevisiae designed to identify genes required for initiation of recombination (10–14) and similar screens in Schizosaccharomyces pombe (15, 16). In contrast to the high level of conservation observed for Spo11, but similar to the other subunits of the Spo11 core complex, many auxiliary DSB protein such Rec114, Mei4, and Mer2 are poorly conserved at the primary sequence level (1). Indeed, apart from limited homology detected between S. cerevisiae Rec114 and S. pombe Rec7 (17–19), high levels of sequence divergence had prevented identification of Rec114, Mei4, and Mer2 homologs outside of budding yeast until nonstandard bioinformatics approaches were applied (20, 21). Homologs of Rec114 and Mei4 that are required for meiotic recombination have now been identified in several species, including Mus musculus (21–23), S. pombe (17, 18, 24), and Arabidopsis thaliana (25, 26). Proteins discovered independently based on roles in meiotic recombination in the ascomycete Sordaria macrospora (Asy1) and the nematode Caenorhabditis elegans (DSB-1 and DSB-2) were also subsequently identified as putative Rec114 homologs (20, 27, 28), but Mei4 homologs were not yet identified in these organisms.
Several studies have established that DSB auxiliary factors Rec114 and Mei4 work closely together with each other and with Mer2 to promote meiotic DSB formation. Physical interactions among these proteins and their orthologs have been demonstrated for several organisms (19, 21, 29–32), and coimmunoprecipitation experiments in M. musculus have further confirmed that these proteins interact with one another in vivo in a meiotic context (23). Recent biochemical analyses have shown that Rec114 and Mei4 together form individual complexes with a stoichiometry of two Rec114 molecules for every one Mei4 molecule and have further suggested that these complexes may self-assemble into large molecular condensates on chromatin during meiotic progression (33). In both S. cerevisiae and M. musculus, all three proteins have been reported to localize together in foci on meiotic prophase chromosomes (19, 23, 29, 32). Further, mouse REC114 and MEI4 and the Mer2 homolog IHO1 all localize predominantly at the meiotic chromosome axis (23, 32), contributing to the idea that they act as an intermediary between chromosome organization and DSB formation. Consistent with this view, chromatin immunoprecipitation experiments in both S. cerevisiae and S. pombe have shown that these proteins interact with both axis-enriched DNA sequences and with DSB sites (31, 34–36). Additionally, S. cerevisiae Rec114 and Mei4 interact with the Rec102 and Rec104 subunits that together comprise the TopVIB-like component of the Spo11 core complex (9, 19). Together these findings implicate Rec114–Mei4 in recruiting Spo11 to the meiotic chromosome axis.
C. elegans DSB-1 and DSB-2, while clearly implicated in meiotic DSB formation, were difficult to recognize as Rec114 homologs owing to high sequence divergence (20, 27, 28). Further, C. elegans differs from yeast and mice regarding the relationships between DSB formation and meiotic chromosome organization. Whereas DSB-dependent recombination intermediates are required to trigger assembly of the synaptonemal complex (SC) between homologous chromosomes in yeast and mice, C. elegans can achieve full synapsis between aligned homologs even in the absence of DSB formation (6). Thus, there are substantial differences in the cellular environments in which DSB-promoting complexes have evolved and function in different organisms.
In our current work, we identify DSB-3 as a protein that partners with DSB-1 and DSB-2 to promote SPO-11–dependent meiotic DSB formation in C. elegans. We demonstrate a requirement for DSB-3 in promoting the DSBs needed for CO formation, and we show that DSB-3 becomes concentrated in germ cell nuclei during the time when DSBs are formed, in a manner that is interdependent with DSB-1 and DSB-2. Through a combination of bioinformatics, interaction data, and colocalization analyses, we identify DSB-3 as a likely Mei4 homolog and establish DSB-1–DSB-2–DSB-3 as functional counterpart of the Rec114-Mei4 complex. Despite homology and a shared role in promoting DSB formation, we find that C. elegans DSB-1, DSB-2, and DSB-3 are distributed broadly on chromatin rather than becoming concentrated preferentially on chromosome axes as observed for mouse REC114–MEI4 complexes. This work highlights the evolutionary malleability of protein complexes that serve essential, yet auxiliary, roles in meiotic recombination. Rapid diversification of such proteins may reflect a relaxation of constraints enabled by changes in another aspect of the reproductive program, or alternatively, they may reflect a capacity of alterations in such proteins to have an immediate impact on reproductive success.
Results
Identification of dsb-3 as a Gene Required for the Formation of Meiotic COs.
The dsb-3(me6ts) allele was isolated in a genetic screen for mutants exhibiting a high incidence of males among the progeny of self-fertilizing hermaphrodites, i.e., the “Him” phenotype, which is indicative of errors in segregation of X chromosomes during meiosis. me6ts mutant hermaphrodites exhibit temperature-sensitive meiotic defects affecting both autosomes and X chromosomes (Table 1 and Fig. 1A). Whereas inviable embryos and males (XO) are rare among the self-progeny of wild-type hermaphrodites (XX) raised at either 20 or 25 °C or dsb-3(me6ts) mutant hermaphrodites raised at 20 °C, dsb-3(me6ts) mutant hermaphrodites raised at the nonpermissive temperature of 25 °C produce 28% inviable embryos, and 17% of their surviving progeny are males. Further, DAPI (4′,6-diamidino-2-phenylindole) staining of chromosomes in oocytes at diakinesis, the last stage of meiotic prophase, revealed a defect in chiasma formation in the dsb-3(me6ts) mutant, reflecting an underlying defect in CO formation (see below). Whereas wild-type oocyte nuclei consistently exhibit six pairs of homologous chromosomes connected by chiasmata (bivalents), oocyte nuclei in the dsb-3(me6ts) mutant exhibited a mixture of bivalents and unattached (achiasmate) chromosomes (univalents), with the incidence of univalents increasing with time (Table 1).
Table 1.
Quantitation of embryo viability, male frequency, and diakinesis karyotypes
| Average DAPI bodies/nucleus ± SD (no. of oocytes) | |||||||
| Strain | Genotype | Incubated temperature, °C | Average brood size ± SD (no. of broods) | Dead eggs, % | Males, % | 24 h after L4 | 48 h after L4 |
| N2 | WT | 20 | 264 ± 27 (7) | 0.4 | 0 | 6.0 ± 0.2 (92) | 5.9 ± 0.3 (102) |
| N2 | WT | 25 | 291 ± 22 (7) | 2 | 0 | 5.9 ± 0.3 (110) | 5.9 ± 0.4 (99) |
| AV913 | dsb-3(me6ts) IV | 20 | 274 ± 27 (7) | 0.1 | 0 | 6.0 ± 0.2 (144) | 5.9 ± 0.3 (111) |
| AV913 | dsb-3(me6ts) IV | 25 | 198 ± 47 (7) | 28.2 | 17.3 | 7.0 ± 1.4 (108) | 9.9 ± 1.3 (99) |
| AV1095 | dsb-3(me115) IV | 20 | 156 ± 77 (16) | 99.4 | 24.9 | 11.6 ± 0.6 (150) | |
| AV1029 | meSi7[sun1p::dsb-3::gfp::sun1 3′UTR unc-119+] II; dsb-3(me115) IV | 20 | 150 ± 23 (8) | 5 | 3.7 | 6.1 ± 0.4 (207) | |
| AV1102 | dsb-3(me125)[3xflag::dsb-3] dsb-1(me124)[3xha::dsb-1] IV | 20 | 243 ± 34 (7) | 2.1 | 2.5 | 6.0 ± 0.2 (213) | |
| AV1115 | dsb-2(me132)[3xha::dsb-2] II | 20 | 193 ± 61 (8) | 2.2 | 0.5 | 6.0 ± 0.2 (150) | |
Strains used in and/or created for this study were evaluated for the indicated parameters. Numbers of DAPI bodies were evaluated for worms fixed and stained at 24 h after L4 stage. Analysis of AV913 and corresponding controls were conducted at two temperatures based on the temperature-sensitive nature of the dsb-3(me6ts) mutant. UTR, untranslated region; WT, wild type.
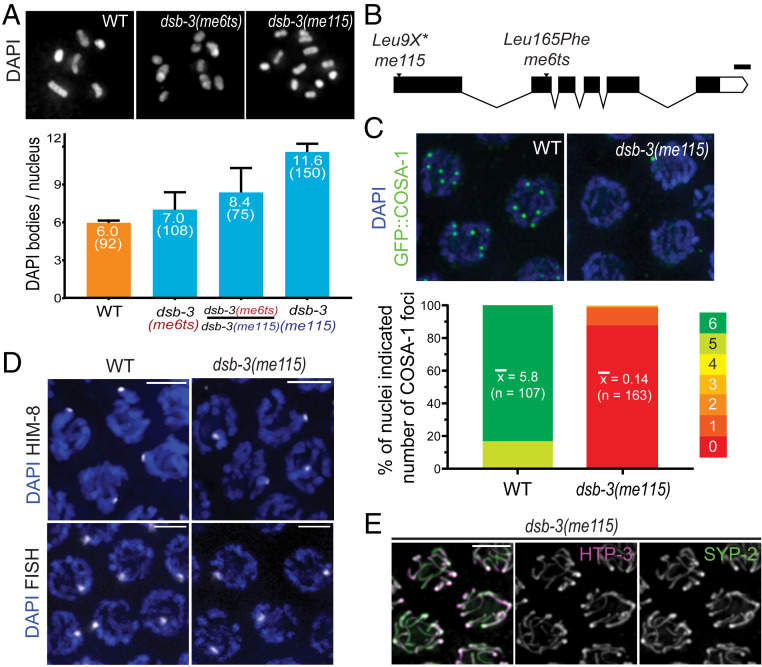
Fig. 1.
Identification of dsb-3 as a gene required for meiotic CO formation. (A, Top) Representative images of diakinesis-stage oocyte nuclei from adult worms fixed at 1 d after L4. (Left) Wild-type (WT) nucleus with six DAPI bodies corresponding to six pairs of homologs connected by chiasmata (bivalents). (Middle) dsb-3(me6ts) nucleus with nine DAPI bodies (three bivalents and six univalents). (Right) dsb-3(me115) nucleus with 12 DAPI bodies (all univalents). (A, Bottom) Quantification of DAPI bodies/ nucleus; error bars indicate standard deviation, and numbers in parentheses indicate the numbers of nuclei assayed. For all pairwise comparisons, Mann–Whitney P values were <0.0001. Assays for WT and dsb-3(me115) homozygotes were performed at 20 °C; assays for dsb-3(me6ts) homozygotes and dsb-3(me6ts)/dsb-3(me115) heterozygotes were performed at 25 °C. (B) Schematic showing the dsb-3 gene structure, with the positions and nature of mutations used in this work; white boxes represent untranslated region sequences, black boxes represent exons, and lines indicate introns. (Scale bar: 100 bp.) (C, Top) Immunofluorescence images of GFP::COSA-1 foci, which correspond to the single CO site on each homolog pair in late pachytene nuclei. WT nuclei have six foci, while foci are reduced or absent in the dsb-3(me115) mutant. (C, Bottom) Stacked bar graphs showing the distribution of GFP::COSA-1 foci counts in nuclei from WT and dsb-3(me115) mutants. Mean numbers of foci per nucleus are indicated, with the numbers of nuclei assayed in parentheses; Mann–Whitney P < 0.0001. (D) Homolog pairing assayed by immunofluorescence of X-chromosome pairing center binding protein HIM-8 (Top) or FISH detecting a 1-Mbp segment of chromosome II (Bottom) in pachytene nuclei of whole-mount gonads. A single focus is observed in each nucleus, indicating successful pairing. (Scale bars: 3.2 µm.) (E) Immunofluorescence image of SC components in late pachytene nuclei in a whole-mount gonad from the dsb-3(me115) mutant. Axis protein HTP-3 and SC central region protein SYP-2 colocalize in continuous stretches between chromosome pairs, indicating successful synapsis. (Scale bar: 3.2 µm.)
Mapping and sequencing identified a missense mutation at genomic position IV: 7758710 (WS279) as the likely causal mutation responsible for the dsb-3(me6ts) mutant phenotype (see SI Appendix); this mutation results in a Leu165Phe substitution in the previously uncharacterized protein C46A5.5. CRISPR-Cas9 genome editing was used to introduce multiple stop codons early into the first exon of C46A5.5, thereby creating the null allele me115 (Fig. 1B). me115 fails to complement dsb-3(me6ts) (Fig. 1A), confirming the identity of C46A5.5 as dsb-3.
Analysis of the dsb-3(me115) null mutant indicates that the DSB-3 protein is required for the formation of meiotic COs between all six pairs of homologous chromosomes. dsb-3(me115) mutant hermaphrodites produced 99% inviable embryos, and 25% of their surviving progeny were male, reflecting missegregation of autosomes and X chromosomes (Table 1). Diakinesis oocytes of dsb-3(me115) mutant hermaphrodites exhibited an average of 11.6 ± 0.6 DAPI-stained bodies, indicating of a lack of chiasmata connecting all six homolog pairs (Fig. 1A). A severe defect in CO formation in the dsb-3(me115) mutant was also revealed using green fluorescent protein (GFP)::COSA-1 as a cytological marker of CO-designated sites in late pachytene nuclei (37) (Fig. 1C). Whereas six GFP::COSA-1 foci (one per homolog pair) were consistently observed in late pachytene nuclei of control worms, GFP::COSA-1 foci were absent from most late pachytene nuclei in the dsb-3(me115) mutant.
Since pairing and assembly of the SC between homologs are prerequisites for CO formation during C. elegans meiosis, we evaluated whether these features were impaired in the dsb-3(me115) mutant. We assessed pairing using fluorescence in situ hybridization (FISH) for a 1-Mbp segment of chromosome II and immunostaining for HIM-8, a C2H2 zinc-finger DNA-binding protein that concentrates on the chromosome X pairing center (38, 39), demonstrating that dsb-3(me115) mutants are proficient for homolog paring (Fig. 1D). Further, immunostaining for the axial element protein HTP-3 and the SC central region protein SYP-2 (40, 41) revealed fully synapsed chromosomes in early pachytene nuclei in the dsb-3(me115) mutant, indicating successful SC assembly (Fig. 1E). Together these data indicate that the DSB-3 protein is dispensable for pairing and synapsis and point to a role for this protein in the DNA events of recombination.
During C. elegans meiosis, failure to form CO recombination intermediates between one or more chromosome pairs prolongs the early pachytene stage of meiotic prophase, reflecting the operation of a “CO assurance” checkpoint (27, 28, 42). Consistent with the observed deficit of interhomolog COs, dsb-3(me115) mutant gonads display an extended zone of nuclei exhibiting phosphorylation of nuclear envelope protein SUN-1, a marker of CO assurance checkpoint activation (SI Appendix, Fig. S1A).
DSB-3 Is Required for Meiotic DSB Formation.
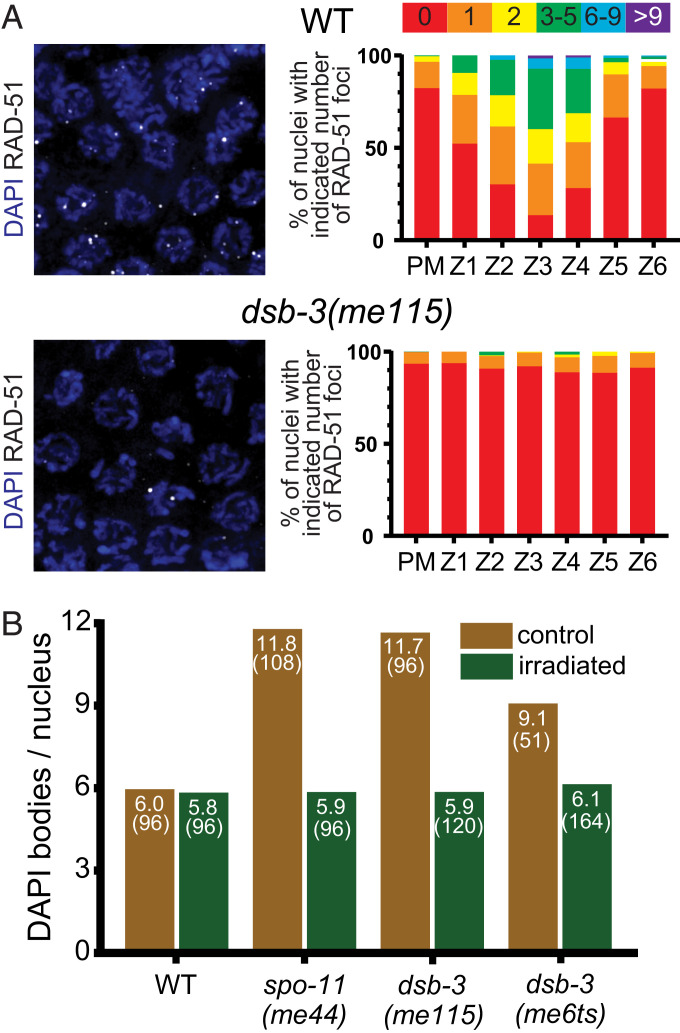
Meiotic recombination is initiated through the formation of DSBs by the conserved topoisomerase-like protein SPO-11 (3, 6). Following formation, DSBs are processed to enable the loading of the DNA strand-exchange protein RAD-51 (43, 44). RAD-51 foci thus mark the sites of recombination intermediates that can be assayed as a proxy for successful initiation of meiotic recombination (41, 45). We observed a strong decrease in the number of RAD-51 foci in dsb-3(me115) mutants relative to wild type (Fig. 2A), suggesting that fewer DSBs are being created in these mutants or that there is a failure to load RAD-51 at DSB sites.
Fig. 2.
DSB-3 is required for meiotic DSB formation. (A, Left) Immunofluorescence images of RAD-51 foci in early pachytene nuclei. RAD-51 foci mark sites of processed DSBs and are strongly reduced in the dsb-3(me115) mutant. (A, Right) Quantification of RAD-51 foci in whole-mount gonads (three or more gonads per genotype). Gonads were divided into seven zones: the premeiotic zone (PM) and six consecutive equal-sized zones encompassing the region of the gonad from the transition zone (where nuclei enter meiotic prophase) to the end of the pachytene stage. (B) Rescue of chiasma formation in dsb-3 mutants by γ-irradiation-induced DNA breaks. Graph showing average numbers of DAPI bodies in diakinesis-stage oocytes of worms exposed to 5 krad of γ irradiation and unirradiated age-matched controls. For WT, spo-11, and dsb-3(me115), worms raised at 20 °C were irradiated at 18 h after L4 and fixed and stained 21 h after irradiation; for dsb-3(me6ts), worms raised at 25 °C were irradiated at 24 h after L4 and fixed and stained 18 h after irradiation.
To determine whether fewer endogenous DSBs was the defect responsible for the deficit of RAD-51 foci, we introduced ectopic DSBs using γ-irradiation to test whether such breaks are sufficient to restore CO formation, as observed previously for other DSB-defective mutants such as dsb-1, dsb-2, and spo-11 (6, 27, 28). Young adult dsb-3(me115) and dsb-3(me6ts) hermaphrodites [alongside wild-type and spo-11(me44) controls] were exposed to 5,000 rad of γ-irradiation and subsequently assayed for CO formation through DAPI staining of chromosomes in oocyte nuclei. We observed a full rescue of normal DAPI-body counts after irradiation (Fig. 2B), suggesting that the dsb-3 mutants are specifically defective in DSB formation.
DSB-3 Is Concentrated in DSB-Competent Nuclei and Is Interdependent with DSB-1 and DSB-2.
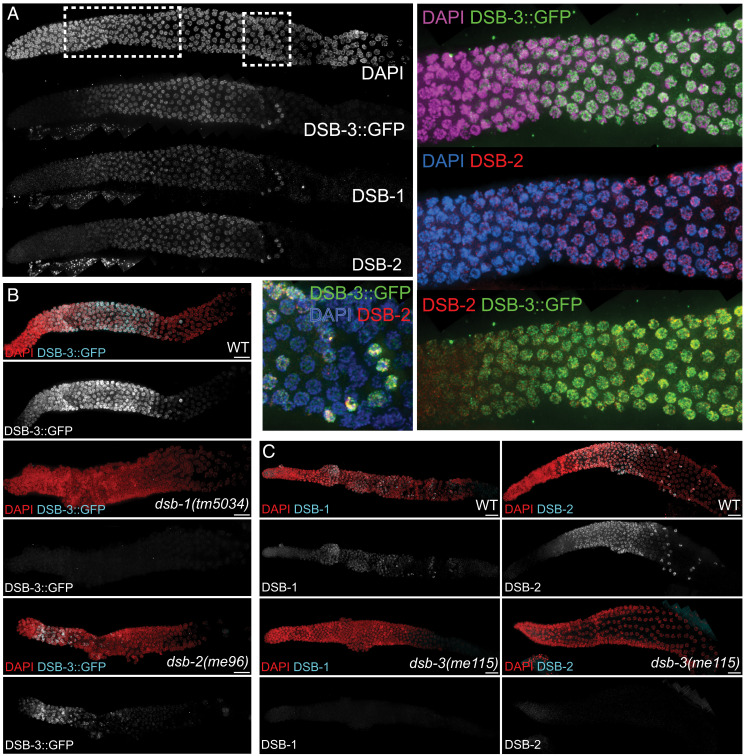
Consistent with its role in promoting DSB formation, DSB-3 localizes to germ cell nuclei during the time when meiotic DSBs are formed. To assess DSB-3 localization, we generated a transgenic strain that expresses a DSB-3::GFP fusion protein in the germ line in the dsb-3(me115) null mutant background (meSi7[sun-1p::dsb-3::gfp::sun13′UTR] II; dsb-3(me115) IV); based on assessment of progeny viability and DAPI bodies in diakinesis oocytes, we infer that this DSB-3::GFP fusion protein is largely functional in promoting meiotic recombination (Table 1). Immunolocalization experiments in whole-mount dissected gonads show that DSB-3::GFP becomes concentrated in germ cell nuclei within the transition zone, soon after entry into meiotic prophase (Fig. 3A). The DSB-3::GFP immunofluorescence signal is strongest in early pachytene nuclei, then declines sharply in midpachytene, albeit with a few outlier nuclei in the late pachytene region of the gonad retaining a strong DSB-3 signal. This pattern of appearance and disappearance of DSB-3 from germ cell nuclei is similar to the patterns observed for the DSB promoting proteins DSB-1 and DSB-2 (27, 28) (Fig. 3A) and corresponds to the timing when nuclei are competent for meiotic DSB formation.
Fig. 3.
DSB-3 is concentrated in DSB-competent nuclei and is interdependent with DSB-1 and DSB-2. (A) Whole-mount hermaphrodite gonad (from distal tip to end of pachytene) stained with DAPI and antibodies detecting DSB-3::GFP, DSB-1, and DSB-2. Meiotic progression proceeds from left to right in all images. DSB-3::GFP signal is first detected in nuclei within the transition zone (rectangular inset) soon after meiotic prophase entry, is strongest in early pachytene nuclei, and declines sharply in midpachytene, albeit with a few outlier nuclei (square inset) in the late pachytene region retaining a strong DSB-3 signal. This pattern of appearance and disappearance is similar to those observed for DSB-1 and DSB-2. (B) Whole-mount hermaphrodite gonads stained with DAPI and antibody detecting DSB-3::GFP. DSB-3::GFP is not detected in the dsb-1(null) mutant and is strongly reduced and limited to a smaller region of the gonad in the dsb-2(null) mutant. (C) Images of gonads stained for DSB-1 (Left) or DSB-2 (Right), showing that DSB-1 and DSB-2 immunofluorescence signals are not detected in the dsb-3(me115) mutant. (Scale bars: 16.2 µm.)
The DSB-3, DSB-2, and DSB-1 proteins are not only abundant in the same nuclei during early meiotic prophase, but they are also interdependent for this immunolocalization. Previous studies had demonstrated interdependence for DSB-1 and DSB-2 (27, 28), with loss of the DSB-2 immunofluorescence signal in a dsb-1 null mutant (which lacks meiotic DSB-promoting activity) and reduction of DSB-1 immunofluorescence signal in a dsb-2 null mutant (which retains a low residual level of DSB-promoting activity). Similarly, DSB-3::GFP immunofluorescence signal was abolished in dsb-1 null mutant germ lines (Fig. 3B). Likewise, DSB-3::GFP signal was reduced in dsb-2 null mutant germ lines and was restricted to a few rows of nuclei in the transition zone and very early pachytene region (Fig. 3B). Conversely, DSB-1 and DSB-2 immunostaining were lost in dsb-3(me115) mutant germ lines (Fig. 3C). Collectively, these data indicate that DSB-3, DSB-2, and DSB-1 are interdependent for proper localization to germ cell nuclei, indicating that they function together in promoting meiotic DSB formation.
Together, our data demonstrating 1) a similar requirement in promoting DSB formation, 2) concentration in the same nuclei, and 3) interdependence for localization and/or abundance in meiotic nuclei are all consistent with DSB-3 functioning in a complex together with DSB-1 and DSB-2 to promote the formation of SPO-11–dependent meiotic DSBs.
Evidence That DSB-1, DSB-2, and DSB-3 Form a Complex Homologous to the Yeast and Mammalian Rec114–Mei4 Complexes.
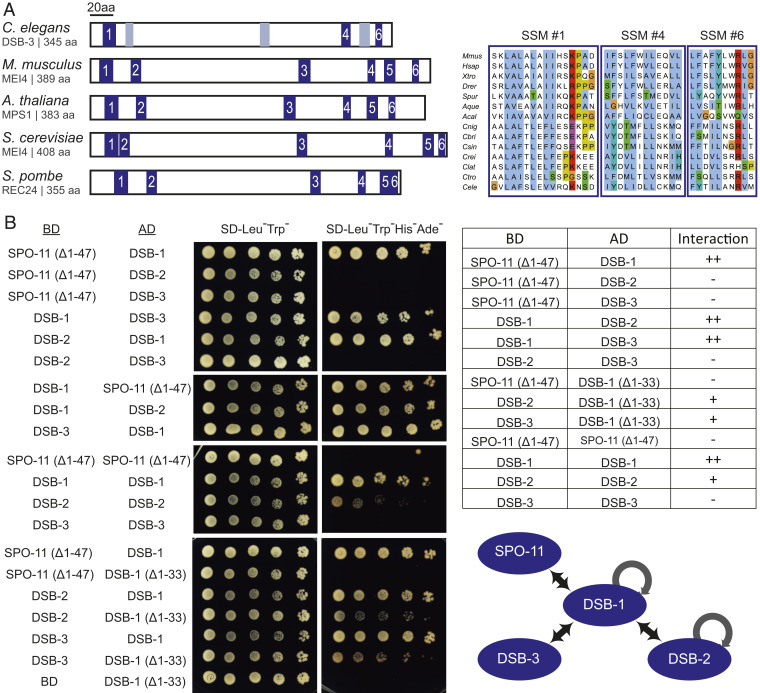
Although the initial PSI-BLAST searches conducted for DSB-1 and DSB-2 had not identified homologs outside of Caenorhabditis (27, 28), DSB-1 and DSB-2 were subsequently identified as likely distant homologs of the Rec114 proteins from fungi and mammals (20). This identification was enabled using an approach involving PSI-BLAST searches initiated using sequence alignments, in combination with scanning for patterns of similarity in predicted secondary structure, to identify short signature motifs (SSMs) in poorly conserved proteins (46). We obtained additional support for the assignment of DSB-1 and DSB-2 as Rec114 homologs using the Phyre2 structure prediction server (47), which identified medium confidence (no. 6 hit, 40.3%) and low confidence (no. 19 hit, 11.9%) alignments between the N-terminal domains of DSB-2 and DSB-1 and the N-terminal domain (where the identified SSMs are located) of the solved structure of mouse Rec114 (48). We therefore used an alignment-driven PSI-BLAST approach similar to that described above to identify DSB-3 as a putative homolog of Mei4 (Materials and Methods, Fig. 4A, and SI Appendix, Fig. S2), which is required for meiotic DSB formation in yeast and mice and forms a complex with Rec114 (21–23, 29, 30, 49). Based on local amino acid composition and relative position in the protein sequence, three of the six SSMs previously identified in Mei4 homologs from diverse species (SSMs no. 1, 4, and 6) are well-supported in the Caenorhabditis DSB-3 orthologs, while the other three SSMs are less well conserved.
Fig. 4.
Evidence that DSB-1, DSB-2, and DSB-3 form a complex homologous to the yeast and mammalian REC114-MEI4 complexes. (A, Left) Schematic depicting the positions of six SSMs (blue boxes) previously defined for Mei4 homologs in diverse species, indicating the three SSMs (1, 4, 6) that are most strongly supported in C. elegans DSB-3; gray boxes indicate positions that potentially correspond to SSMs 2, 3, and 5 based on a multiple sequence alignment (MSA) with vertebrate and marine invertebrate homologs of similar size, but are less well conserved. (A, Right) Aligned sequences of SSMs 1, 4, and 6 from M. musculus, Homo sapiens, Xenopus tropicalis, Danio rerio, Strongylocentrotus purpuratus, Amphimedon queenslandica, Aplysia californica, and the following nematodes of the genus Caenorhabditis: C. nigoni, C. briggsae, C. sinica, C. remanei, C. latens, C. tropicalis, and C. elegans. SSMs are cropped from an MSA generated in JalView (78) using the ClustalX coloring scheme. For additional information, see SI Appendix, Fig. S2 and Supplementary Materials and Methods. (B) Y2H assay revealing interactions among the DSB-1, -2, and -3 proteins and between DSB-1 and SPO-11. Potential interactions between proteins fused with the GAL4 activation domain (AD) and proteins fused with the GAL4 DNA binding domain (BD) were assayed by growth on media lacking histidine and adenine. A construct producing an N-terminal truncation of SPO-11 lacking the first 47 amino acids (Δ1–47) was used for these analyses, as severe auto-activation was observed for full-length SPO-11. Negative controls showing lack of auto-activation for the constructs used are presented in SI Appendix, Fig. S2. A schematic summarizing the identified interactions is shown on the bottom right.
To complement these in silico analyses, we used yeast two-hybrid (Y2H) assays to establish a network of interactions among the DSB-1, DSB-2, and DSB-3 proteins and SPO-11, the protein that catalyzes DSB formation (Fig. 4B and SI Appendix, Fig. S2). Y2H interactions were detected between DSB-1 and DSB-2 and between DSB-1 and DSB-3, consistent with an ability of these proteins to form complexes. Homotypic interactions were also detected both for DSB-1 and for DSB-2. In addition to the interactions detected among the putative Rec114 and Mei4 homologs, DSB-1 also interacted with SPO-11 in the Y2H assay.
We note that a truncated version of DSB-1 lacking the N-terminal 33 amino acids loses the ability to interact with SPO-11 but retains its ability to associate with DSB-2 and DSB-3. This suggests that the interactions between DSB-1 and SPO-11 and the interactions between DSB-1 and DSB-2 or DSB-3 may be mediated, at least in part, by different parts of the DSB-1 protein.
DSB-3, DSB-2, and DSB-1 Colocalize in Meiotic Nuclei.
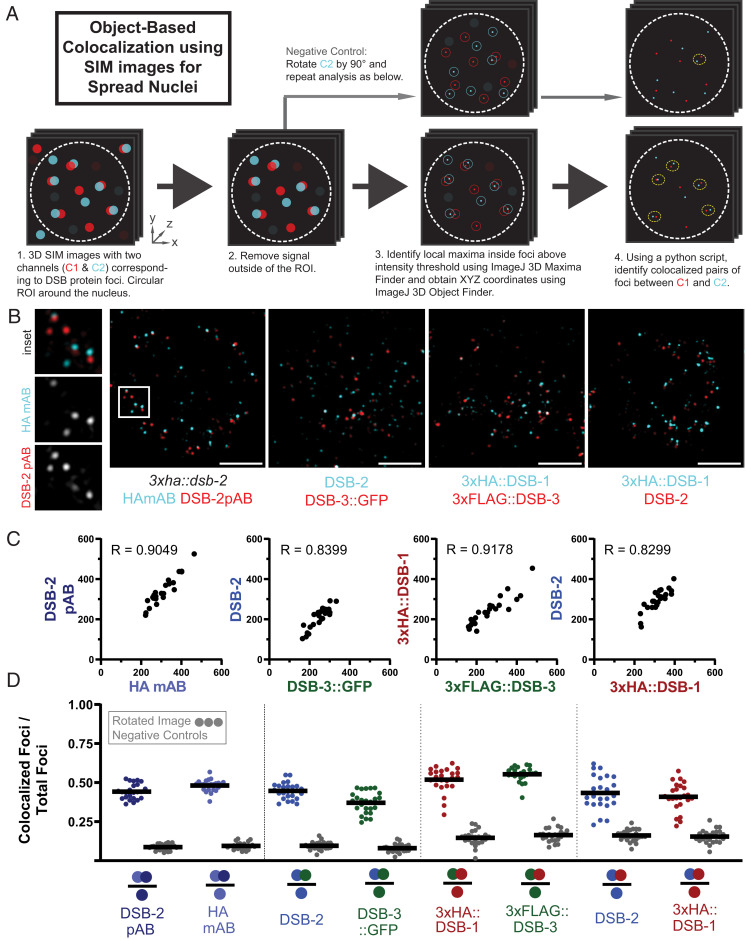
To complement our genetic, bioinformatic, and Y2H evidence that DSB-3, DSB-2, and DSB-1 function together as components of conserved protein complexes to promote DSB formation, we investigated their colocalization using structured illumination microscopy (SIM) on spread preparations of meiotic nuclei (Fig. 5). For most of these analyses, we used a moderate nuclear spreading protocol (50), coupled with SIM imaging to provide improved spatial resolution below the limits of standard light microscopy (51). This approach enabled detection of these proteins as chromosome-associated foci. To facilitate costaining of protein pairs for these colocalization analyses, we created strains expressing hemaggluninin (HA)- or FLAG-tagged versions of the DSB proteins from the endogenous loci (Table 1), and we detected the proteins using indirect immunofluorescence.
Fig. 5.
DSB-3, DSB-2, and DSB-1 colocalize in meiotic nuclei. (A) Schematic summarizing the object-based image analysis pipeline used to assess colocalization of foci in SIM immunofluorescence images of partially spread nuclei (see Materials and Methods and SI Appendix, Fig. S3 for more details). (B) Representative SIM images of individual spread nuclei stained with antibodies targeting the indicated DSB proteins; each panel depicts a single Z slice from a 3D image stack. The following experiments are represented in order from left to right: mouse monoclonal HA and rabbit polyclonal DSB-2 antibodies, both detecting the same 3xHA::DSB-2 tagged protein in a 3xha::dsb-2 II strain; chicken GFP and rabbit DSB-2 antibodies, detecting DSB-3::GFP and DSB-2 in a meSi7[dsb-3::gfp] II; dsb-3(me115) IV strain ; mouse HA and rabbit FLAG antibodies, detecting 3xHA::DSB-1 and 3xFLAG::DSB-3 in a 3xflag::dsb-3 3xha::dsb-1 IV strain; and mouse HA and rabbit DSB-2 antibodies, detecting 3xHA::DSB-1 and DSB-2 in a 3xflag::dsb-3 3xha::dsb-1 IV strain. (Scale bars: 2 µm.) (C) Quantification of DSB protein foci for the indicated pairwise combinations; each data point represents the numbers of foci for the two analyzed channels in a single nucleus. Spearman R values indicate that numbers of the two types of foci are strongly correlated within nuclei. (D) Graphs showing the fraction of foci of a given type (indicated by a single colored circle in the denominator in each schematic below the horizontal axis) that are colocalized with the other type of focus analyzed in that same experiment (colocalizing foci are represented by two colored circles in the numerator). For each pair of focus types analyzed, two sets of experimental analyses (represented by colored data points) and paired negative controls (represented by gray data points) are presented.
We used the image analysis pipeline outlined in Fig. 5A (described in more detail in SI Appendix, Fig. S3) to identify DSB protein foci and to conduct object-based colocalization analyses to assess the degree of colocalization detected for pairwise combinations of the imaged DSB proteins within individual nuclei. As negative controls, we generated virtual nuclei in which the second channel in each combination was rotated by 90° in XY, resulting in virtual composite images in which foci are modified in location but numbers, sizes, and intensity distributions of foci remain unaltered. Collectively, our analyses indicate that DSB-3, DSB-2, and DSB-1 strongly colocalize with each other in meiotic prophase nuclei.
Analyses of all three pairwise combinations of DSB-1, DSB-2, and DSB-3 foci are presented in Fig. 5 B–D and SI Appendix, Fig. S4. For all three pairs, numbers of foci for the two channels detected in each nucleus were strongly correlated, consistent with expectations for components of the same protein complex. Further, substantial colocalization was observed for each pair. For example, 45 ± 5% of DSB-2 foci colocalized with DSB-3::GFP foci, and, conversely, 37 ± 7% of DSB-3::GFP foci colocalized with DSB-2 foci. In contrast, negative control coincidental colocalization values were 10 ± 3% and 8 ± 3%, respectively. Similarly, 52 ± 8% of 3xFLAG::DSB-3 foci colocalized with 3xHA::DSB-1 foci, and, conversely, 55% ± 5% of 3xHA::DSB-1 foci colocalized with 3xFLAG::DSB-3 foci. Likewise, 43 ± 11% of 3xHA::DSB-1 foci colocalized with DSB-2 foci, and, conversely, 41 ± 9% of DSB-2 foci colocalized with 3xHA::DSB-1 foci.
Although substantial colocalization was observed for all pairwise combinations, the fraction of colocalization may seem lower than might be anticipated for proteins comprising the same protein complex. We note, however, that incomplete colocalization was similarly observed for the Rec114 and Mei4 proteins in both budding yeast and mouse meiocytes (29, 32). One possible explanation is that only a subset of these protein molecules occur together in complexes, while other molecules exist separately within the nucleus; however, this explanation is not easily reconciled with the observed interdependence among these proteins. Another possibility is that the observed degree of colocalization reflects limitations on our ability to detect all of the DSB-1, DSB-2, and DSB-3 target molecules that are present, e.g., because of isoforms that lack epitopes or because the complexes and/or their components may be organized in a manner that makes some epitopes inaccessible to detection reagents.
This latter possibility is supported by data from an experiment in which we assessed colocalization for fluorescent foci representing separate epitopes on the same protein, 3xHA::DSB-2, expressed from the endogenous dsb-2 locus. Specifically, we used a mouse monoclonal antibody (mAB) against the HA epitope and rabbit polyclonal (pAB) antibodies raised against the C-terminal 100 amino acids of the DSB-2 protein. The numbers of foci for the two channels detected in each nucleus were strongly correlated (Fig. 5C), as expected for foci representing the same target molecule. However, colocalization was again incomplete, in both directions: 44 ± 5% of DSB-2 pAB foci colocalized with HA mAB foci, and, conversely, 48 ± 2% of HA mAB foci colocalized with DSB-2 pAB foci. This incomplete colocalization of HA mAB and DSB-2 pAB fluorescent signals supports the conclusion that a subset of epitopes on DSB-2 proteins present in the nucleus were not detected in these experiments.
For the DSB-2 - DSB-3::GFP combination we also conducted a colocalization analysis on “superspread” nuclei, in which chromosomes were dispersed over an area 6 to 10 times larger than that of an unperturbed nucleus (52) (SI Appendix, Fig. S5). The numbers of foci detected using this approach were three to five times higher than the numbers observed in our analysis of partial spreads, but a similar degree of colocalization was detected: 55 ± 16% of DSB-2 foci colocalized with DSB-3::GFP foci, and, conversely, 44 ± 10% of DSB-3::GFP foci colocalized with DSB-2 foci. The observation of larger numbers of foci with a comparable degree of colocalization suggests the possibility that groups of DSB protein complexes may be split into smaller cohorts by the superspread procedure.
The Presence and Colocalization of DSB-3, DSB-2, and DSB-1 Are Not Confined to the Meiotic Chromosomal Axis.
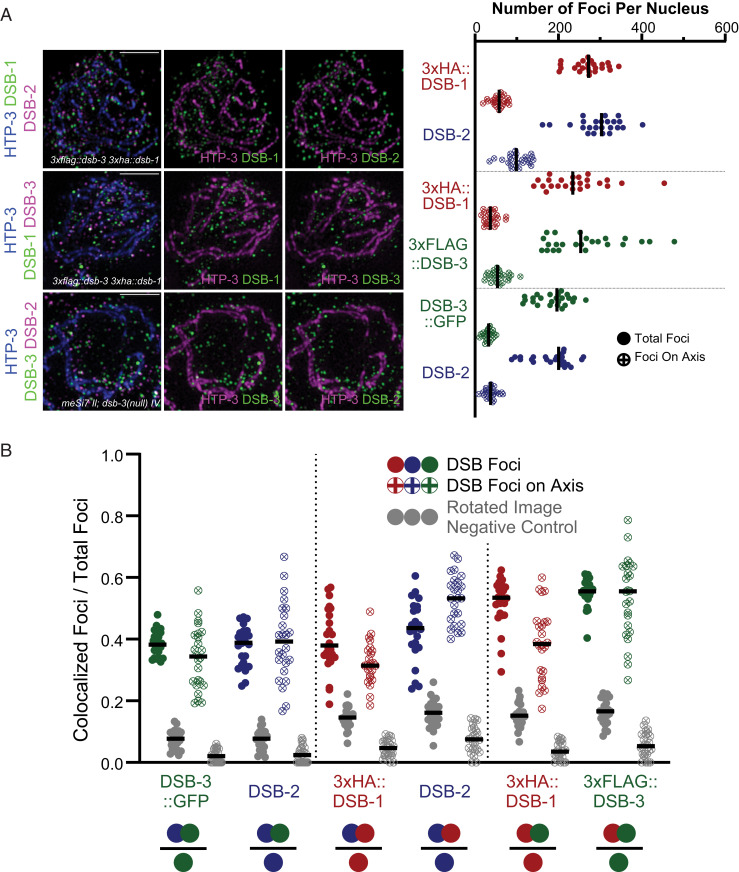
Rec114 and Mei4 are often referred to as “axis-associated” proteins, but the available data reveal a more complex picture. Colocalized immunostaining foci of mouse MEI4 and REC114 occur primarily on the axes of spread meiotic prophase chromosomes (22, 23, 32), but accumulation of REC114 at pseudoautosomal region minisatellite repeats only partially overlaps with axes (53). In S. cerevisiae, axis association has been inferred from chromatin immunoprecipitation experiments showing that peaks of Mei4 and Rec114 enrichment strongly correlate with chromatin immunoprecipitation signals for meiotic axis proteins Hop1 and Red1 (34, 36); however, immunostaining experiments show that a substantial fraction of Mei4 and Rec114 localizes to off-axis chromatin (19, 36). Further, in whole-mount C. elegans gonads immunostained for DSB-1, the majority of DSB-1 signal was detected on off-axis chromatin (28).
Our analysis of spread meiotic nuclei clearly indicates that DSB-1, DSB-2, and DSB-3 foci are not enriched at the chromosome axis during C. elegans meiosis. Rather, most foci occur away from the axis, in the associated chromatin loops (Fig. 6A). To quantify axis association, we first segmented images by creating axis masks for each nucleus that corresponded to the pixels containing the immunofluorescence signal derived from the axis protein HTP-3 (SI Appendix, Fig. S3B), then for each DSB protein tested we identified the subset of foci, termed “axis-associated foci” for which some or all of the pixels coincided with the axis mask. This approach indicated that only 15 to 30% of DSB protein foci detected in our analyses overlapped with chromosome axis signal, indicating that on spread chromosomes from meiotic nuclei, the preponderance of DSB protein foci detected were not associated with the meiotic chromosome axes.
Fig. 6.
The presence and colocalization of DSB-3, DSB-2, and DSB-1 are not confined to the meiotic chromosomal axis. (A, Left) Representative maximum-intensity projections of SIM images, representing the middle third of Z-stacks collected for the depicted spread nuclei. Nuclei were stained with antibodies targeting DSB proteins and the axis component HTP-3. (Scale bars: 2 µm.) (A, Right) Graph showing quantification of numbers of DSB protein foci that colocalize with the axis signal, as well as the total numbers of DSB protein foci identified in the analyzed nuclei. (B) Graphs depicting the fractions of colocalized foci/total foci for DSB protein foci of the indicated types (represented as in Fig. 5), reported for axis-associated foci and for total nuclear foci analyzed within the same datasets. The filled dots represent colocalization fractions for all DSB protein foci of a given type within the nucleus, and dots with crosses represent the colocalization fractions for the subset of DSB protein foci of the indicated type that colocalize with the axis.
We also assessed whether DSB protein foci associated with the chromosome axis might exhibit a higher degree of colocalization with their DSB protein partners relative to the level of colocalization observed for the full set of foci within the nucleus (Fig. 6B). However, this analysis did not reveal any consistent enrichment of colocalization for DSB protein foci that were linked to the chromosome axis. Thus, while components of the meiotic chromosome axis do have roles in promoting and regulating SPO-11–dependent DSB-forming activity in C. elegans meiosis (54–56) these roles do not appear to be mediated by concentrating DSB-1–DSB-2–DSB-3 complexes in close proximity to the chromosome axis.
Discussion
Initiation of meiotic recombination by programmed DSB formation is an ancient and conserved feature of the meiotic program that predates divergence of plants, animals, and fungi. Thus, it is not surprising that Spo11, the protein directly responsible for catalyzing DSB formation, is strongly conserved across kingdoms, given constraints imposed by its requirement to interact with and perform chemistry on DNA. However, many additional proteins required for DSB formation had been identified in the yeast system, but plant and metazoan homologs of these auxiliary DSB proteins had long eluded detection by standard BLAST analyses. The barrier to detection of homologs outside fungi was eventually breached using secondary structure prediction coupled to MAFFT alignment and phylogenomically oriented PSI-BLAST searches (21), which identified characteristic SSMs for putative Mei4 and Rec114 homologs; moreover, the veracity of these predictions was borne out by demonstration of meiotic roles in mouse mutants (21–23).
Auxiliary proteins involved in DSB formation during C. elegans meiosis were identified independently based on analysis of meiotic mutants (refs. 27 and 28 and this work). However, recognition of these proteins as distant homologs of conserved DSB-promoting factors came later, after their functional importance in DSB formation was already established (20). Identification of DSB-1 and DSB-2 as Rec114 homologs was further solidified by alignments of the predicted structure of DSB-1 and DSB-2 with the solved structure of mouse REC114 (ref. 48 and this work). Likewise, our initial identification of C. elegans DSB-3 as a factor important for meiotic DSB formation was similarly based on functional data. The identity of DSB-3 as a putative Mei4/MEI4 ortholog was derived computationally from alignments and collinearity of SSMs among metazoan homologs, and this identification was reinforced by demonstration of Y2H interactions, colocalization, and interdependence with Rec114 homologs DSB-1 and DSB-2. Thus, despite a high degree of divergence at the amino acid sequence level, our data collectively support the conclusion that DSB-1, DSB-2, and DSB-3 together form complexes that are the functional counterpart of Rec114–Mei4 complexes.
Having established conservation among the auxiliary complexes that promote the DSB-forming activity of Spo11, our analyses also reveal interesting differences. First, whereas yeast and mice each have only a single Rec114/REC114 ortholog, nematodes in the Caenorhabditis genus each have two paralogs, indicating duplication and divergence in the parental lineage. DSB-1 and DSB-2 are neither identical to nor functionally interchangeable with each other, as DSB formation is strongly reduced in dsb-2 mutants and eliminated in dsb-1 mutants. Interestingly, recent biochemical analyses indicate a 2 Rec114 : 1 Mei4 stoichiometry of the yeast complex (33). The interdependence of DSB-1 and DSB-2 for nuclear enrichment, in combination with the colocalization observed in chromosome spreads (this work), suggests that the C. elegans complexes may typically contain one DSB-1 subunit and one DSB-2 subunit (rather than two identical subunits). However, there is low residual DSB-promoting activity present in dsb-2–null mutants, suggesting that complexes with two DSB-1 subunits may form and be partially functional when DSB-2 is absent. While the data do support functional diversification of the C. elegans Rec114 paralogs, how this diversification came about and/or how and why it persisted remain unknown.
A second apparent distinction between C. elegans DSB-1, DSB-2, and DSB-3 and their mouse counterparts is the observed relationship to meiotic chromosome axes. On mouse meiotic prophase chromosome spreads, REC114 and MEI4 localize predominantly at chromosome axes throughout most of the genome (with the exception of regions with abundant minisatellite repeats) (23, 32, 53). This prominent association with chromosome axes has led to the proposal that a major role for REC114 and MEI4 is to recruit the SPO11 core complex to the axis to activate its DSB-promoting activity specifically in close proximity to the axis, where DSB repair predominantly occurs. In C. elegans, meiosis-specific HORMAD proteins HTP-1 and HTP-3, which are major building blocks of the chromosome axis, are implicated in promoting and regulating meiotic DSB formation (54–56). HTP-3 is strictly required for DSB formation, and HTP-1 is required for normal levels of DSB activity. However, despite a requirement for axis components in DSB formation, DSB-1–DSB-2–DSB-3 complexes are not preferentially enriched adjacent to axes. Interestingly, whereas recruitment of mouse REC114–MEI4 complexes to meiotic chromosomes axes is mediated through a direct interaction between IHO1 (the mouse Mer2 homolog) and axis component HORMAD1 (32), no Caenorhabditis Mer2/IHO1 ortholog has yet been detected.
Given the observed lack of enrichment of DSB-1, DSB-2, and DSB-3 at the axes, the role(s) of C. elegans meiotic HORMAD proteins in promoting DSB formation may not be strictly limited to recruiting the homologs of Rec114 and Mei4 to chromosomes. One possibility is that the C. elegans HORMADs might be involved in “activating” the subset of DSB-1–DSB-2–DSB-3 complexes that do occur in close proximity to the axis, such that only axis-associated complexes are competent for SPO-11 recruitment and/or DSB formation. Alternatively, the role(s) of the C. elegans axis proteins might be indirect; e.g., assembly of the axis might potentially signal successful formation of chromosome structure that is proficient for meiotic DSB repair, thereby licensing the nucleus that it is safe to proceed with DSB formation. The possibility of this signaling scenario is strengthened by prior work demonstrating a role for C. elegans HORMAD proteins in a signaling process that sustains activity of protein kinase CHK-2, a master regulator of multiple processes during meiosis, including nuclear enrichment of DSB-1 and DSB-2 (27, 28, 42, 57).
It is possible that the apparent difference between mice and C. elegans regarding axis enrichment of Rec114–Mei4 complexes may be related to differences in spatial organization of recombination events in the genome and/or in coupling between DSB repair and homolog recognition. Meiotic recombination in mice occurs predominantly within 1- to 2-kb “hotspot” regions, separated by larger (50- to 100-kb) cold regions where the probability of recombination is very low. A similar “local hotspot” distribution of recombination events was not observed in C. elegans (for the portion of the genome studied) (58, 59), suggesting that different constraints are operating to dictate where DSBs may form. Further, there is substantial variation among organisms regarding their relative dependence on different mechanisms that promote pairwise alignment and synapsis between homologous chromosomes. In mouse meiosis, formation of early SPO11-dependent DSB repair intermediates appears to be the main mechanism of homology verification, required to trigger SC assembly and constraining it to occur strictly between aligned homologous chromosomes (60, 61). In contrast, in C. elegans, local cis-acting chromosomal domains known as pairing centers play a primary role in homolog recognition, and these are capable of promoting largely successful pairwise synapsis between homologs even in the absence of recombination (62). We speculate that differences in the constraints governing the genomic locations of DSBs and/or differences in dependence on DSBs for homology verification may have either contributed to, or been enabled by, diversification of meiotic DSB auxiliary protein complexes. We note that high divergence among essential components of key biological processes is a hallmark not only of the meiotic program but of reproduction more generally (1, 63, 64). This likely reflects multiple underlying factors, including the potential for changes in such proteins to have an immediate impact on processes that directly affect fitness by modulating reproductive success.
A key question regarding the role(s) of the Rec114–Mei4 (or DSB-1–DSB-2–DSB-3) complexes is how exactly they are functioning to promote Spo11 activity. It was recently proposed that Rec114–Mei4 complexes function by forming large DNA-dependent biomolecular condensates that promote DSB activity by causing a high local concentration of Spo11 core complexes at presumptive DSB sites held adjacent to the chromosome axis (33). This model was proposed based on 1) a large segment of the Rec114 protein exhibiting a high probability of disorder, 2) the ability of purified Rec114–Mei4 complexes to promote formation of DNA-dependent condensates in vitro, and 3) an ability of Rec114–Mei4 complexes to interact with and recruit the Spo11 core complex. While evaluating potential for in vitro condensation is outside the scope of the current study, we note that some of the above attributes may be shared with the C. elegans DSB-1–DSB-2–DSB-3 complex. First, based on the Y2H data, the DSB-1–DSB-2–DSB-3 complex is expected to be able to interact with SPO-11 complexes. However, in yeast, interactions occur between Rec114 and the Rec102 and Rec104 components (which together correspond to Top6BL) of the Spo11 core complex, whereas in C. elegans a Top6BL homolog has not yet been identified, and DSB-1 can interact directly with SPO-11 itself in the Y2H assay. Second, the DSB-1 and DSB-2 proteins have long segments with predicted protein disorder scores that hover around 0.5 and include short segments scoring >0.5, leaving it ambiguous whether these might represent bona fide disordered regions. Third, the observation that higher numbers of DSB-2 and DSB-3 foci are detected in superspread nuclei than in moderately spread nuclei raises the possibility that these proteins might normally occur in larger groups within intact nuclei, potentially analogous to the condensates proposed to occur during yeast meiosis. In either system, future investigations aiming to test predictions of the condensation model will need to address the challenge of visualizing complex dynamic behavior in vivo.
Materials and Methods
C. elegans Genetics and Genome Engineering.
Worms were cultured at 20 °C using standard conditions (65) unless otherwise noted. A list of strains used is provided in SI Appendix. Established approaches were used to generate inserted transgenes (66) or edited endogenous loci (67, 68) expressing tagged proteins; details are available in SI Appendix. dsb-3(me6ts) was isolated in a screen for mutants exhibiting a high incidence of males (69); mapping and sequence analyses identifying the causal mutation in C46A5.5 are described in SI Appendix.
DAPI Staining of Oocyte Chromosomes and Irradiation Assay.
Numbers of DNA bodies in diakinesis oocytes were assessed in ethanol-fixed adult hermaphrodites stained with DAPI as in ref. 70. This method underestimates the frequency of achiasmate chromosomes, as some univalents lie too close to each other to be resolved unambiguously. To test for rescue of bivalent formation by exogenously derived DSBs, worms were exposed to 5,000 rad (50 Gy) of γ-irradiation using a Cs-137 source, fixed, and stained 18 to 21 h after irradiation, and numbers of DAPI bodies were counted in oocyte nuclei in the −1 to −3 positions.
Bioinformatic Identification of Homology between DSB-3 and Mei4.
PSI-BLAST searches using the MPI BLAST server (71), initiated using an alignment of DSB-3 homologs from diverse roundworm species as the query, identified a putative Brugia malayi DSB-3 homolog. A subsequent round of PSI-BLAST searches, initiated using an alignment with the putative B. malayi homolog as the header sequence and initially focusing on the N-terminal portion of the protein, led to retrieval of plant and animal Mei4 homologs. Similarity in protein lengths and patterns of predicted secondary structure were prioritized over E-value considerations in selection of proteins chosen for the multiple sequence alignment presented in SI Appendix, Fig. S2, which was generated using MAFFT Version 7.0 with gap opening penalty parameter set to 2.0 and offset value parament set to 0.125.
Immunofluorescence.
Immunofluorescence experiments involving whole-mount gonads, partially spread nuclei, or superspread nuclei were conducted as in refs. 52, 55, and 72, with modifications. Antibodies used, details of procedures, and image acquisition and processing are provided in SI Appendix.
Quantification of RAD-51 foci and GFP::COSA-1 foci is described in the Fig. 1 legend.
FISH.
FISH experiments were conducted as in ref. 52, with modifications as detailed in SI Appendix. Peaks of FISH signals were identified using the ImageJ plugin 3D Maxima Finder (73).
Identification of DSB Protein Foci and Object-Based Colocalization Analysis.
For Figs. 5 and 6, images were analyzed using an object-based colocalization analysis pipeline that combined standard functions available in ImageJ in conjunction with a custom Python script. A detailed description of the colocalization analysis pipeline is presented in SI Appendix, Fig. S3. For these analyses, 32-bit Z-stacks of SIM images of immunofluorescence signals for at least two different antibodies detecting DSB proteins (C1 and C2) were imported into ImageJ (74, 75) with the Fiji distribution (76). The signal maxima for each channel, identified as foci by the image analysis pipeline, were qualitatively compared to the original image to verify accurate identification of foci.
For colocalization analysis of DSB-2 and DSB-3::GFP foci on superspread nuclei the same pipeline was used, except that foci were analyzed within 3.43 μm × 3.43 μm square regions of interest (ROIs) located entirely within the spread (one to three ROIs per nucleus).
Supplementary Material
Acknowledgments
We thank C. Girard and S. Ramakrishnan for assistance with irradiation experiments and generation of sequencing libraries, W. Zhang for early analysis of the me6ts mutant, and David Paul for discussions regarding object-based colocalization. We acknowledge N. Bhatla for creating the Exon-Intron Graphic Maker (http://wormweb.org/) used for Fig. 1B. We thank A. Dernburg and V. Jantsch for antibodies and the Caenorhabditis Genetics Center (funded by NIH Office of Research Infrastructure Programs P40 OD010440) for strains. This work was supported by an American Cancer Society Research Professor Award (RP-15-209-01-DDC) and NIH Grants R01GM53804 and R35GM126964 to A.M.V., by a Blavatnik Family Foundation Fellowship and a Stanford Mason Case Fellowship to A.W.H., by an Austrian Science Fund Erwin Schrödinger Fellowship (J-3676) to A.W., by a Stanford Graduate Fellowship to K.Y., by grants from the Taiwan Ministry of Science and Technology (107-2923-B-002-001-MY4 and MoST 104-2628-B-002-002-MY3) to P.C., and by Grant 1S10OD01227601 from the National Center for Research Resources to the Stanford Cell Sciences Imaging Facility.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109306118/-/DCSupplemental.
Data Availability
The original 32-bit individual nucleus ImageJ files, the segmented axis channel files, the identified peaks, the values used for and the output position files from the 3D Maxima Finder for each nucleus, the custom python script used to identify colocalization, and the resulting spreadsheet files showing colocalization data have been deposited in the BioStudies Database (accession no. S-BSST568) (77). Materials used in this research are available on request from A.M.V. (annev@stanford.edu).
References
- 1.Keeney S., “Spo11 and the formation of DNA double-strand breaks in meiosis” in Recombination and Meiosis, Genome Dynamics and Stability, Egel R., Lankenau D.-H., Eds. (Springer, Berlin, 2008), pp. 81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeney S., Lange J., Mohibullah N., Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 48, 187–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Bergerat A., et al., An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997). [DOI] [PubMed] [Google Scholar]
- 5.McKim K. S., et al., Meiotic synapsis in the absence of recombination. Science 279, 876–878 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Dernburg A. F., et al., Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Robert T., et al., The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351, 943–949 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Vrielynck N., et al., A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Claeys Bouuaert C., et al., Structural and functional characterization of the Spo11 core complex. Nat. Struct. Mol. Biol. 28, 92–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menees T. M., Roeder G. S., MEI4, a yeast gene required for meiotic recombination. Genetics 123, 675–682 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malone R. E., et al., Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128, 79–88 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittman D., Lu W., Malone R. E., Genetic and molecular analysis of REC114, an early meiotic recombination gene in yeast. Curr. Genet. 23, 295–304 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Engebrecht J. A., Voelkel-Meiman K., Roeder G. S., Meiosis-specific RNA splicing in yeast. Cell 66, 1257–1268 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Cool M., Malone R. E., Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol. Cell. Biol. 12, 1248–1256 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponticelli A. S., Smith G. R., Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123, 45–54 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Veaux L. C., Hoagland N. A., Smith G. R., Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130, 251–262 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone R. E., Pittman D. L., Nau J. J., Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol. Gen. Genet. 255, 410–419 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Molnar M., et al., Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157, 519–532 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maleki S., Neale M. J., Arora C., Henderson K. A., Keeney S., Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 116, 471–486 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tessé S., et al., Asy2/Mer2: An evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 31, 1880–1893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R., Bourbon H.-M., de Massy B., Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar R., et al., MEI4—A central player in the regulation of meiotic DNA double-strand break formation in the mouse. J. Cell Sci. 128, 1800–1811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar R., et al., Mouse REC114 is essential for meiotic DNA double-strand break formation and forms a complex with MEI4. Life Sci. Alliance 1, e201800259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonfils S., Rozalén A. E., Smith G. R., Moreno S., Martín-Castellanos C., Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J. Cell Sci. 124, 1328–1338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Muyt A., et al., A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5, e1000654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronceret A., Doutriaux M.-P., Golubovskaya I. N., Pawlowski W. P., PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 106, 20121–20126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosu S., et al., The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9, e1003674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamper E. L., et al., Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9, e1003679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Hooker G. W., Roeder G. S., Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173, 1969–1981 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner S., Kohli J., Ludin K., Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 56, 237–249 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi T., et al., A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47, 722–733 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Stanzione M., et al., Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claeys Bouuaert C., et al., DNA-driven condensation assembles the meiotic DNA break machinery. Nature 592, 144–149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panizza S., et al., Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Sasanuma H., et al., Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 22, 398–410 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carballo J. A., et al., Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet. 9, e1003545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoo R., et al., COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips C. M., et al., HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051–1063 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacQueen A. J., et al., Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123, 1037–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severson A. F., Ling L., van Zuylen V., Meyer B. J., The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23, 1763–1778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colaiácovo M. P., et al., Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Woglar A., et al., Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 9, e1003335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pâques F., Haber J. E., Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung P., Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Alpi A., Pasierbek P., Gartner A., Loidl J., Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112, 6–16 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Bourbon H.-M., Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boekhout M., et al., REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell 74, 1053–1068.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arora C., Kee K., Maleki S., Keeney S., Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13, 549–559 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Woglar A., Villeneuve A. M., Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173, 1678–1691.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gustafsson M. G. L., Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. U.S.A. 102, 13081–13086 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woglar A., et al., Quantitative cytogenetics reveals molecular stoichiometry and longitudinal organization of meiotic chromosome axes and loops. PLoS Biol. 18, e3000817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acquaviva L., et al., Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Couteau F., Zetka M., HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19, 2744–2756 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Perez E., Villeneuve A. M., HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19, 2727–2743 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodyer W., et al., HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14, 263–274 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Kim Y., et al., The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev. Cell 31, 487–502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaur T., Rockman M. V., Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics 196, 137–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernstein M. R., Rockman M. V., Fine-scale crossover rate variation on the Caenorhabditis elegans X Chromosome. G3 (Bethesda) 6, 1767–1776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Romanienko P. J., Camerini-Otero R. D., The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Hillers K. J., Jantsch V., Martinez-Perez E., Yanowitz J. L., “Meiosis.” WormBook: The Online Review of C. elegans Biology (2017). http://www.wormbook.org/chapters/www_meiosis/meiosis.html. Accessed 1 December 2019. [DOI] [PMC free article] [PubMed]
- 63.Swanson W. J., Vacquier V. D., The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Dapper A. L., Payseur B. A., Molecular evolution of the meiotic recombination pathway in mammals. Evolution 73, 2368–2389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frøkjaer-Jensen C., et al., Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paix A., Folkmann A., Rasoloson D., Seydoux G., High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arribere J. A., et al., Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villeneuve A. M., A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136, 887–902 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bessler J. B., Reddy K. C., Hayashi M., Hodgkin J., Villeneuve A. M., A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics 175, 2029–2037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabler F., et al., Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinformatics 72, e108 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Pattabiraman D., Roelens B., Woglar A., Villeneuve A. M., Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13, e1006670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ollion J., Cochennec J., Loll F., Escudé C., Boudier T., TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rueden C. T., et al., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hinman A., Villeneuve A., C. elegans DSB-3 reveals conservation and divergence among protein complexes promoting meiotic double-strand breaks - Colocalization data. BioStudies. https://www.ebi.ac.uk/biostudies/studies/S-BSST568. Accessed 1 August 2021. [Google Scholar]
- 78.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original 32-bit individual nucleus ImageJ files, the segmented axis channel files, the identified peaks, the values used for and the output position files from the 3D Maxima Finder for each nucleus, the custom python script used to identify colocalization, and the resulting spreadsheet files showing colocalization data have been deposited in the BioStudies Database (accession no. S-BSST568) (77). Materials used in this research are available on request from A.M.V. (annev@stanford.edu).