Significance
Biologics, and especially antibodies, are promising therapeutics. Antibodies are expected to show higher subtype selectivity and less off-target activity than small molecules. G protein–coupled receptors (GPCRs) being the main drug targets, there is a need for antibodies modulating these receptors. Here, we describe the first single-domain antibody fully activating a GPCR. This nanobody activates the homodimeric metabotropic glutamate receptor type 4 (mGlu4), an interesting target for the treatment of Parkinson's disease or pain. Using modeling tools, we show this nanobody acts by stabilizing the active form of the binding domain. It does not activate heterodimeric mGlu receptors containing the mGlu4 subunit. These data revealed that nanobodies can be useful tools to specifically control mGlu receptor subtypes.
Keywords: G protein–coupled receptor, single-domain antibody, activation mechanism, agonist
Abstract
There is growing interest in developing biologics due to their high target selectivity. The G protein–coupled homo- and heterodimeric metabotropic glutamate (mGlu) receptors regulate many synapses and are promising targets for the treatment of numerous brain diseases. Although subtype-selective allosteric small molecules have been reported, their effects on the recently discovered heterodimeric receptors are often not known. Here, we describe a nanobody that specifically and fully activates homodimeric human mGlu4 receptors. Molecular modeling and mutagenesis studies revealed that the nanobody acts by stabilizing the closed active state of the glutamate binding domain by interacting with both lobes. In contrast, this nanobody does not activate the heterodimeric mGlu2-4 but acts as a pure positive allosteric modulator. These data further reveal how an antibody can fully activate a class C receptor and bring further evidence that nanobodies represent an alternative way to specifically control mGlu receptor subtypes.
There is more and more interest in developing antibodies as possible pharmacological and even therapeutic agents (1, 2). The single-domain antibodies, also named nanobodies, are prone for such activities, as their short variable loops can interact in surface cavities that can vary between the conformational states of a protein (1). As such, nanobodies regulating drug targets have already been reported (3, 4). Among the main drug targets are the G protein–coupled receptors (GPCRs) that play important roles in cell–cell communication (5).
Among the large GPCR family, class C receptors are those activated by the main neurotransmitters, glutamate and γ-amino butyric acid (GABA), as well as by Ca2+ ions, sweet and umami compounds (6). There are eight genes encoding metabotropic glutamate (mGlu) receptor subunits. Although only considered as disulfide-linked homodimers, the complexity of the mGlu receptor family increased with the description of heterodimeric entities made of two different mGlu subunits (7). Indeed, the postsynaptic mGlu1 and -5 on one side and presynaptic mGlu2, -3, -4, -7, and -8 subunits on another side can form heterodimeric entities in recombinant cells, leading to the possible existence of 16 additional mGlu subtypes (7, 8). Among these, the mGlu2-4 heterodimer retained much attention. It was found to display a specific pharmacological profile and was identified in transfected neurons as well as in both cortico-striatal (9) and lateral perforant path (10) terminals. Identifying the various heterodimeric mGlu receptors in the brain and their possible roles is essential.
We recently reported nanobodies acting as positive allosteric modulators (PAMs) specific to the mGlu2 homodimer (11). Among the mGlu subtypes, the mGlu4 containing receptors, and especially mGlu4 homodimers, are of interest for the treatment of Parkinson’s disease (12–14) and pain (15, 16). In the present study, we report the identification of DN45, a nanobody that specifically and fully activates human mGlu4 homodimers. We show that this nanobody acts by stabilizing the closed active state of the glutamate binding Venus flytrap domain (VFT). However, this nanobody was unable to directly activate the mGlu2-4 heterodimer, acting instead as a pure PAM. These data reveal a way a nanobody can activate a class C GPCR and confirm the potential of using nanobodies to discriminate between homo- and heterodimeric receptors.
Results
DN45 Is Specific to the Human mGlu4 Receptor.
To identify nanobodies targeting mGlu4, HEK293 cells transiently expressing either the human or the rat mGlu4 receptor were injected into a llama. Genes encoding VHH domains were amplified by RT-PCR from the total RNA of peripheral blood mononuclear cells and used to create a phage display library. This library was depleted on controlled Human Embryonic Kidney (HEK293) cells and enriched by two cycles of positive selection on human mGlu4 transfected cells in the presence of an excess of anti-HEK293 nanobodies (17). Nanobody containing Escherichia coli supernatants were screened by flow cytometry.
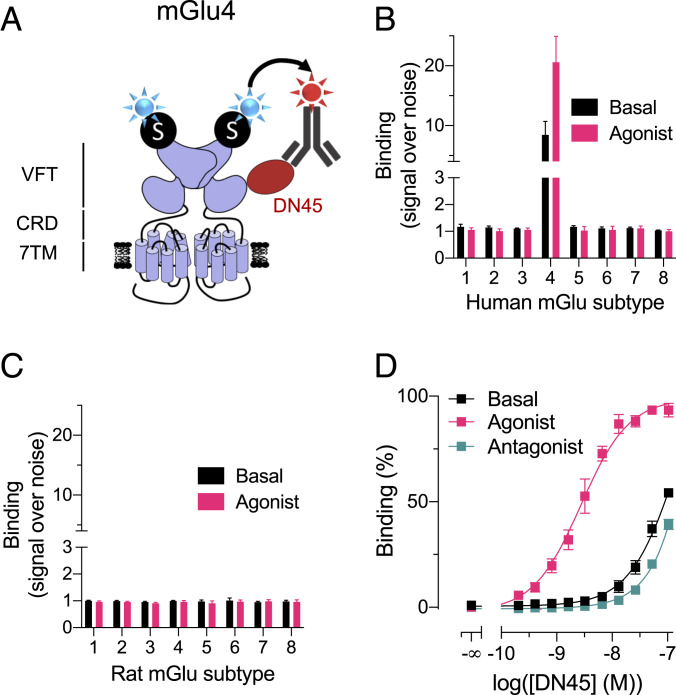
Among the selected clones, nanobody DN45 was further characterized. We performed a binding assay based on time-resolved fluorescence energy transfer (TR-FRET) between the SNAP-tagged receptor and an anti-c-Myc-antibody-d2 interacting with the c-Myc-tag inserted at the carboxyl-terminal end of the DN45 sequence (Fig. 1A). The nanobody binds to human mGlu4 exclusively when tested at 100 nM, and no binding was observed on any other human mGlu receptor (Fig. 1B) nor on the eight rat mGlu receptors (Fig. 1C). To avoid constitutive receptor activation by ambient glutamate produced by the cultured HEK293 cells, excitatory amino acid transporter 3 was cotransfected with the indicated receptors such that receptors were essentially in an inactive state under basal condition. Activating any of the mGlu receptors with an agonist did not allow binding of DN45 either, except on the human mGlu4, in which a higher signal was observed (Fig. 1 B and C). Indeed, DN45 had a preferred high binding affinity (KD = 3.1 nM) for the active conformation of the human mGlu4 receptor and a low binding affinity for the basal and antagonist-induced conformation (Fig. 1D and SI Appendix, Table S1).
Fig. 1.
DN45 is selective for the human mGlu4 receptor and preferentially binds to the active receptor. (A) Cartoon representing a TR-FRET–based binding assay. SNAP-mGlu receptors were labeled with 100 nM BG-Lumi4-Tb (blue star). Then, 100 nM DN45 containing a c-Myc epitope was labeled with 200 nM anti-c-Myc-antibody (gray) coupled to d2 (red star). (B) Specific binding of DN45 to human mGlu4 receptor without (black) and with (pink) a saturating concentration of agonist (mGlu group I: 1 µM quisqualic acid; group II: 100 nM LY379268; group III: 10 µM L-AP4), represented by an increase of signal compared to an irrelevant nanobody containing the c-Myc sequence. (C) No specific binding was observed between the nanobody and any rat mGlu receptors in the absence (black) or presence (pink) of agonist. (D) Binding of an increasing concentration of DN45 on hmGlu4 under basal condition (black) in the presence of the agonist L-AP4 (10 µM) or in the presence of the antagonist LY341495 (100 µM). Data in B–D are mean ± SEM of three individual experiments (SI Appendix, Table S1).
DN45 Is a Full Agonist of the mGlu4 Receptor.
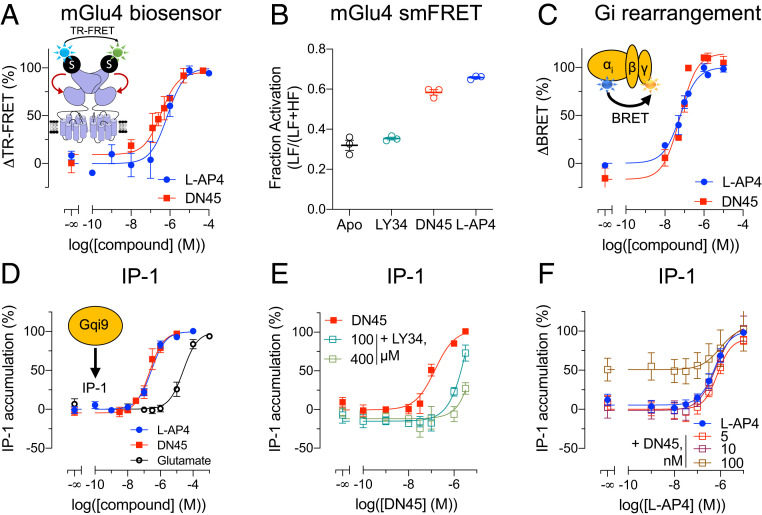
In agreement with the higher binding affinity of DN45 for the active form of the hmGlu4 receptor, DN45 alone stabilizes the active conformation of the VFT dimer as revealed using a TR-FRET–based conformational biosensor (Fig. 2A) (7, 18). This biosensor measures the distance variation between the N-terminally inserted SNAP tags of the subunits such that a high TR-FRET is measured in the basal state (after random labeling with the SNAP substrates O6-benzylguanine [BG]-Lumi4-Tb and BG-Green), while a lower TR-FRET is observed upon the reorientation of the VFTs upon activation (18). DN45 displays a potency and efficacy not significantly different from the group-III mGlu receptor agonist L-AP4 (Fig. 2A and SI Appendix, Table S2). Subsequently, we setup a single-molecule FRET (smFRET) approach to compare the agonist effect of L-AP4 and DN45 at the single-molecule level in an environment without ambient glutamate (Fig. 2B and SI Appendix, Fig. S1 and Table S3). There was no difference between the apo-state and the antagonist-stabilized state. However, both L-AP4 and DN45 acted as full agonists, as illustrated by the similar increase in the proportion of molecules in the low FRET, active conformation. DN45 also activated the natural Gi protein of the mGlu4 receptor with the same potency as L-AP4, as revealed with a Gi bioluminescence resonance energy transfer (BRET) sensor (Fig. 2C and SI Appendix, Fig. S2 and Table S2). The agonist activity of DN45 was further confirmed by measuring the accumulation of IP-1 upon activation of the chimeric G protein Gqi9 (Fig. 2D). DN45 potency and efficacy were similar to those obtained with L-AP4, both being more potent than glutamate (Fig. 2D and SI Appendix, Table S2). In the presence of LY341495, the potency of DN45 was decreased (Fig. 2E). Of note, the L-AP4 potency for accumulation of IP-1 was not significantly increased in the presence of nonsaturating DN45 concentrations (Fig. 2F and SI Appendix, Table S2).
Fig. 2.
DN45 is a full agonist of the hmGlu4 receptor. (A) Measurement of the change in TR-FRET induced upon stimulation with increasing concentrations of L-AP4 (blue) or DN45 (red). (B) Measurement of the fraction of activated receptor by single-molecule FRET in basal/apo state (black) in the presence of LY341495 (green), DN45 (red), or L-AP4 (blue). (C) Measurement of the rearrangement of the Gi protein in a BRET experiment upon stimulation with increasing concentrations of L-AP4 (blue) and DN45 (red). (D) Measurements of the activation of the Gq pathway via transient overexpression of Gqi9 upon stimulation with increasing concentrations of L-AP4 (blue), DN45 (red), or glutamate (black). (E) Measurements of the activation of the Gq pathway via transient overexpression of Gqi9 upon stimulation with increasing concentrations of DN45 alone (red) or in the presence of LY341495 (i.e., 100 or 400 µM). (F) Measurement of the activation of the Gq pathway upon stimulation with increasing concentrations of L-AP4 alone (blue) or L-AP4 with nonsaturating concentrations (i.e., 5, 10, or 100 nM) of DN45. Data in A–F are mean ± SEM of three or more individual experiments. Statistical analysis for A–D was performed using unpaired two-tailed t tests, and the statistical analysis of F was an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (SI Appendix, Tables S2 and S3).
Residues in Lobe 2 of the Human mGlu4 VFT Confer Subtype Selectivity to DN45.
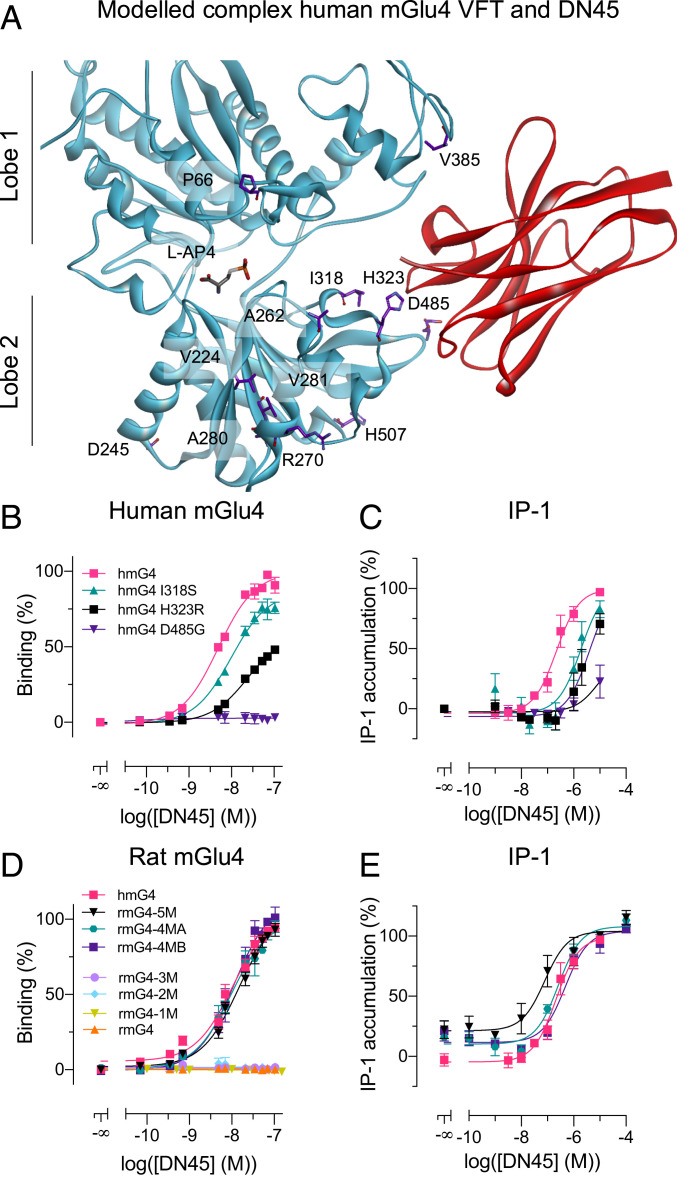
To explain the subtype selectivity of DN45, we substituted any individual residue at the surface of the human mGlu4 VFT into its rat equivalent. We identified 12 residues on the surface of the VFT that are different in the human and rat proteins (Fig. 3A). Among the 12 mutations tested, three (i.e., I318S, H323R, and D485G) affected DN45 binding, with D485G suppressing binding completely (Fig. 3B). Consistent with these binding data, the DN45 agonist effect was largely affected or even suppressed in these human mGlu4 mutants (Fig. 3C), while L-AP4 could still activate them (SI Appendix, Fig. S3A).
Fig. 3.
Residues in lobe 2 of the human mGlu4 VFT confer subtype selectivity to DN45. (A) Front view of the closed conformation of the VFT of the human mGlu4 receptor, stabilized by DN45 (red) in the best-scored nonconstrained docking followed by the 10-ns molecular dynamics simulation. Residues on the surface of the mGlu4 VFT that are specific for the human ortholog are highlighted in purple (side chains). (B) Binding of DN45 was determined in the TR-FRET binding assay as illustrated in Fig. 1A with human wild-type (hmG4) and mutated mGlu4 receptors (i.e., hmG4 I318S, hmG4 H323R, or hmG4 D485G) in the presence of 1 µM L-AP4. (C) Activation of the Gq pathway by hmG4, hmG4 I318S, hmG4 H323R, and hmG4 D485G with increasing concentrations of DN45. (D) Binding of DN45 to mutated rat mGlu4 receptors rmG4-5M (KD = 22.8 nM), rmG4-4MA (KD = 10.4 nM), and rmG4-4MB (KD = 13.6 nM) is not significantly different from binding to hmG4 (KD = 5.4 nM), and no binding is observed for rmG4-3M, rmG4-2M, rmG4-M1, or rmG4. (E) DN45 activation of the Gq pathway by rmG4-5M, rmG4-4MA, and rmG4-4MB. Data in B–E are mean ± SEM of three or more individual experiments. The KD values and statistical analysis for B–E was performed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test and are presented in SI Appendix, Tables S1 and S2.
Molecular Modeling Identifies DN45-mGlu4 Interaction Site on Both Lobe 2 and Lobe 1.
We built three-dimensional (3D) models of both human mGlu4 (in an active closed conformation) and DN45 and performed docking experiments using ZDOCK (19) without any specific constraint. Our best-scored model using ZRANK (20), which keeps the main interactions with or without L-AP4 within the VFT binding site after a 10-ns dynamics simulation, revealed possible interaction sites of DN45 on the VFT (Fig. 3A). In the presence of bound L-AP4, more interactions between the partners were detected (SI Appendix, Fig. S4 A and B) in agreement with the higher binding affinity in the active state. A time-dependent analysis of the model revealed the mGlu4 residues maintaining interaction with DN45 during the 10-ns molecular dynamics simulations (SI Appendix, Fig. S5). These include the residues that are human-specific: I318, H323, D485 (lobe 2), and also residues on lobe 1 (SI Appendix, Fig. S5). The virtual mutagenesis analysis performed on our 3D model of the mGlu4–DN45 complex is perfectly in line with the evaluated impact of individually substituting the 12 human-specific residues for its rat equivalents (SI Appendix, Fig. S5 and Table S4). In addition, the model revealed other key residues such as L322 (SI Appendix, Fig. S6).
DN45 Binding Was Restored on Rat mGlu4 Mutant Bearing At Least Four Human Residues.
The proposed model was further confirmed by introducing human-specific residues into the rat sequence, restoring DN45 binding and agonist activity on the mutated rat mGlu4 receptor. We considered five human-specific residues based on their proximity to the apparent epitope of DN45 (i.e., I318, H323, D485, V385, and H507 [Fig. 3A]). When the five rat residues were mutated to their human equivalent (rmG4-5M), DN45 bound (Fig. 3D) and activated the receptor (Fig. 3E) with an affinity (KD = 22.8 nM) and potency not different from those measured on the human mGlu4 receptor (SI Appendix, Tables S1 and S2). The substitutions G485D (rmG4-1M), G485D+R323H (rmG4-2M), and G485D+R323H+S318I (rmG4-3M) on the rat mGlu4 receptor were, however, not sufficient to recover binding of DN45 (Fig. 3D). Indeed, the model of rmG4-3M reveals that DN45 is not able to interact with the three mutated residues (i.e., S318I, R323H, and G485D) (SI Appendix, Fig. S4A) because a loop is closing the entrance of the epitope cavity (SI Appendix, Fig. S7). The additional mutation of I385V or Q507H restores the opening of the cavity and allows for interactions between DN45 and the three key human residues (SI Appendix, Fig. S7). This was further supported when binding of DN45 to the rat mGlu4 bearing the three main aforementioned substitutions plus I385V (rmG4-4MB) or Q507H (rmG4-4MA) was recovered (Fig. 3D) and both mutants were activated by DN45 (Fig. 3E). The observed affinities (KD = 13.6 nM and 10.4 nM, respectively) and potencies were not significantly different from those measured for human mGlu4 (Fig. 3 D and E and SI Appendix, Tables S1 and S2). All mutants were functional, as we successfully activated them with L-AP4 (SI Appendix, Fig. S3B).
Taken together, these modeling and mutagenesis data provide a reliable explanation for the species selectivity of DN45 by interacting with residues mainly located on lobe 2 of the VFT.
DN45 Agonist Activity Needs Interaction with Both Lobes of the VFT.
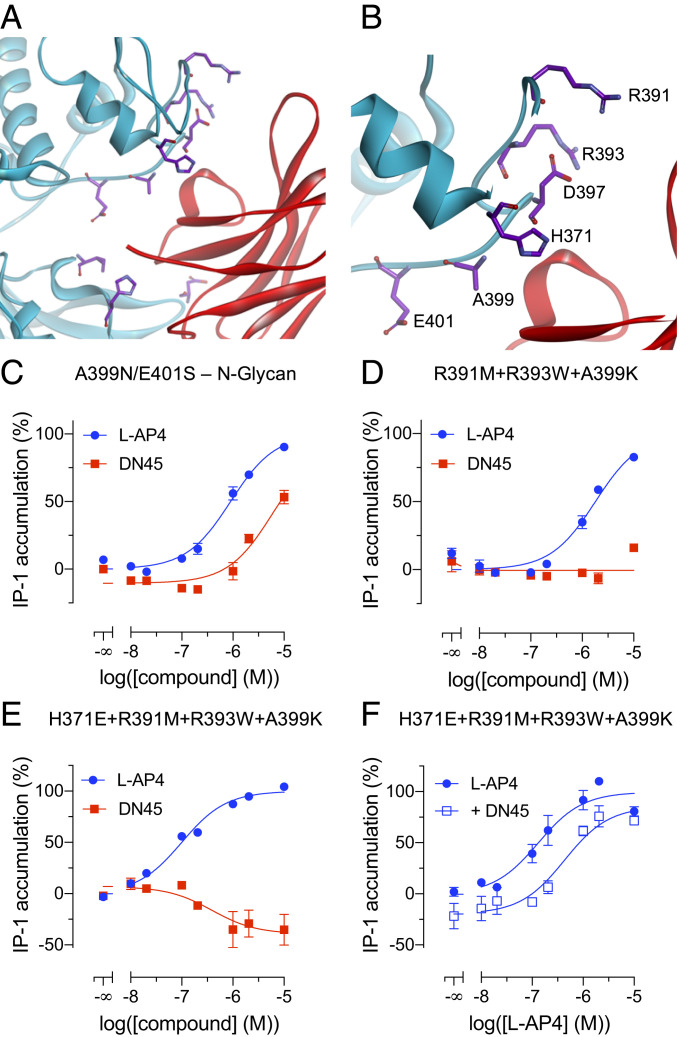
The predictions of interactions between DN45 and lobe 1 of the VFT in a closed active state especially comprise residues H371 to E401 (Fig. 4 A and B and SI Appendix, Fig. S4B). Interactions with this region are expected to stabilize the closed conformation of the VFT, providing an explanation for the agonist activity of DN45 and its increased affinity for the active mGlu4 (Fig. 1D). In a first attempt to prevent the interaction between DN45 and lobe 1 of the mGlu4 VFT, an N-glycosylation site, A399N/E401S, was introduced. This resulted in a large decrease in DN45 agonist potency but did not suppress its agonist activity (Fig. 4C).
Fig. 4.
DN45 agonist activity needs interactions with lobe 1 of the VFT. (A) A front view of the interactions between DN45 and lobe 1 and lobe 2 of the human mGlu4 receptor. Residues on lobe 1 in the loop spanning from H371 to E401 and residues I318, H323, and D485 are highlighted in purple (side chains). (B) A zoomed-in view of the loop comprising residues H371 to E401. (C) IP-1 production by L-AP4 (blue) and DN45 (red) of a human mGlu4 receptor bearing an N-glycosylation site at N399. (D) Effect of L-AP4 (blue) and DN45 (red) on IP-1 production by a human mGlu4 receptor bearing three mutations (i.e., R391M, R393W, and A399K). (E) Effect of L-AP4 (blue) and DN45 (red) on a human mGlu4 receptor bearing four mutations (i.e., H371E, R391M, R393W, and A399K). (F) Effect of L-AP4 alone (filled circles) and in the presence of 100 nM DN45 (open squares) on the activity of the quadruple hmGlu4 receptor mutant. Data of C–F are mean ± SEM of three individual experiments. Statistical analysis of C and F was performed using an unpaired one-tailed t test (SI Appendix, Table S2).
Single-point mutations H371A, K386A, H392A, D397A, E401A, and E403A did not lead to antagonist effects of DN45 on these mutants (SI Appendix, Fig. S8). Therefore, a virtual saturation mutagenesis was performed to predict the impact of multiple mutations of lobe 1 residues on the binding of DN45 and also the impact of these mutations on the overall stability of mGlu4. The best-predicted mutations were H371E, R391M, R393W, and A399K in a loop of lobe 1 (Fig. 4 A and B). The calculated binding energy revealed that the highest destabilizing values were found for the triple mutant R391M+R393W+A399K (4.82 kcal/mol) and quadruple mutant H371E+R391M+R393W+A399K (4.81 kcal/mol).
Experimental data support that this loop is involved in the agonist effect of DN45, as DN45 no longer activated either the triple or the quadruple mutant (Fig. 4 D and E). Of note, the quadruple mutant displays constitutive activity that is reduced by DN45, revealing that DN45 acts as an inverse agonist on this receptor (Fig. 4E). This is confirmed by the significant antagonistic effect of DN45 on this mutant upon activation by L-AP4 (Fig. 4F and SI Appendix, Table S2).
Taken together, these data reveal that DN45 primarily binds to lobe 2 but also to lobe 1 in the active state, an interaction increasing its affinity and required for its agonist activity.
DN45 Acts as a Positive Allosteric Modulator on Heterodimeric mGlu2-4.
It is now recognized that the mGlu4 subunit can associate with other mGlu subunits to form heterodimeric mGlu receptors (7, 8). Among these, the mGlu2-4 heterodimer has retained much attention and has specific pharmacological properties that were used to illustrate its existence in the brain (9, 10).
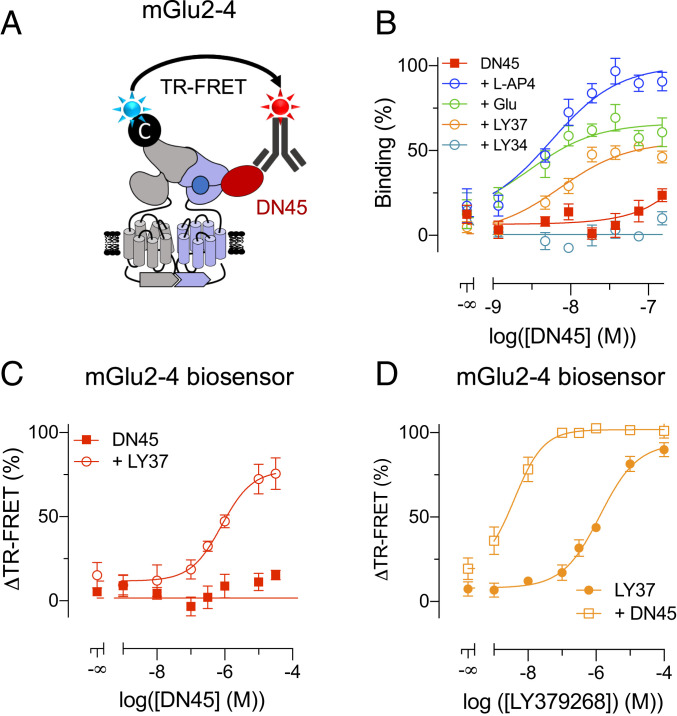
We first examined the binding of DN45 to the mGlu2-4 heterodimer using a combination of mGlu2 N-terminally labeled with CLIP-tag and carboxyl-terminal GABAB1 endoplasmic reticulum retention sequence C1KKXX (21) (CLIP-mGlu2-C1KKXX) and mGlu4 N-terminally tagged with SNAP-tag and carboxyl-terminal GABAB2 endoplasmic reticulum sequence C2KKXX (21) (SNAP-mGlu4-C2KKXX). After labeling CLIP-mGlu2-C1KKXX with Lumi4-Tb, binding of DN45 to the mGlu4 subunit should generate a TR-FRET signal between the Lumi4-Tb on the mGlu2 subunit and the d2 acceptor on an anti-c-Myc-antibody that binds to DN45 (Fig. 5A). A very low signal could be detected under basal condition that almost disappeared in the presence of the competitive antagonist LY341495 (Fig. 5B). However, a large signal could be measured in the presence of the mGlu4 agonist L-AP4, the general mGlu agonist glutamate, or the mGlu2 selective agonist LY379268 (Fig. 5B), revealing binding affinities of 5.0 nM, 3.1 nM, and 7.4 nM, respectively, close to the KD measured on the agonist-occupied mGlu4 homodimers (3.1 nM).
Fig. 5.
DN45 acts as a positive allosteric modulator on heterodimeric mGlu2-4. (A) A cartoon representing a TR-FRET–based assay to measure binding of DN45 to the mGlu2-4 heterodimer using CLIP-mGlu2-C1KKXX and SNAP-mGlu4-C2KKXX. (B) Binding of DN45 to mGlu2-4 alone (red), in the presence of 100 µM mGlu4 antagonist LY341495 (light blue), of 1 mM glutamate (green), of 10 µM LY379268 (orange), or of 10 µM L-AP4 (blue). (C) Activation of the mGlu2-4 heterodimer by DN45 alone (red) or in the presence of the mGlu2 agonist LY379268 (50 nM) measured by a TR-FRET biosensor using CLIP-mGlu2-C1KKXX and SNAP-mGlu4-C2KKXX, labeled with 1 µM BC-Green and 100 nM BG-Lumi4Tb. Values are normalized to the maximal response to LY379268. (D) The response of LY379268 (filled circles) on the TR-FRET biosensor is potentiated by 10 µM DN45 (open squares). Values are normalized to the maximal response to LY379268. Data of B–D are mean ± SEM of three or more individual experiments. Statistical analysis of B was performed using an ordinary one-way ANOVA with Tukey’s multiple comparisons test, and analysis of D was performed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (SI Appendix, Tables S1 and S2).
The effect of DN45 on the conformation of the mGlu2-4 heterodimer was then examined. Using CLIP-mGlu2-C1KKXX labeled with O6-benzylcytosine (BC)-Lumi4Tb and SNAP-mGlu4-C2KKXX labeled with BG-Green, only the heterodimer generates a TR-FRET signal that is largely decreased upon activation (10, 18). As previously reported, the mGlu2 agonist LY379268 activated the heterodimer, while the mGlu4 agonist L-AP4 remains very partial (10, 18). DN45, on the other hand, could not activate the mGlu2-4 by itself (Fig. 5C). However, in the presence of 5% of the maximal effective concentration (EC5) of LY379268 (50 nM), DN45 could activate the receptor, revealing a clear PAM effect (Fig. 5C). Indeed, DN45 largely increased the potency of the mGlu2 agonist LY379268 (Fig. 5D), demonstrating its potent pure PAM effect on the mGlu2-4 heterodimer.
Discussion
The mGlu4 receptor has been proposed as a potential drug target for the treatment of Parkinson’s disease (12, 22, 23) or pain (15). In the brain, mGlu4 can be found in a homodimeric form or associated with other mGlu subunits, like mGlu2, in heteromeric complexes. Using ligands with different properties at mGlu4 homo- and heterodimers, evidence has been provided supporting homodimers as the best targets for Parkinson’s disease treatment (13). However, the respective role of both types of mGlu4 containing receptors remains to be clarified (8–10). In the present study, we report an mGlu4 nanobody with selective agonist activity at mGlu4 homodimers and with a pure PAM action on the mGlu2-4 heterodimers.
Nanobodies have become popular in pharmaceutical research. They have a high specificity and affinity and can stabilize specific conformations of their targets. Due to their small size compared to traditional IgG (∼15 versus ∼150 kDa), nanobodies have several advantages like lower immunogenicity, better pharmacokinetics, and the possibility to reach smaller cavities in proteins (3, 4). As such, nanobodies can stabilize specific conformations of their target, being interesting tools for structural studies (1) and innovative pharmacological agents (24). Moreover, some nanobodies can cross the blood–brain barrier and can then be used to target the central nervous system (25). Of course, nanobodies may have a much better subtype selectivity than orthosteric ligands acting at a binding site often conserved in various receptor subtypes activated by the same natural ligand. Most recently described nanobodies with pharmacological action at GPCRs act as antagonists (26, 27), as does the first Food and Drug Administration (FDA)-approved nanobody, caplacizumab (24).
Recently, nanobodies acting as selective PAMs for mGlu2 (11) and mGlu5 (28) homodimers were reported, the former having in vivo activities. Both nanobodies were found to stabilize the agonist-bound active orientation of the two VFTs of the dimers, with the mGlu2 nanobodies acting at the active interface of the VFTs (11), while the mGlu5 one acts on a loop on the top of the mGlu5 VFT (28). Here, we identified the mode of action of DN45 by combining state-of-the-art modeling and docking approaches and mutagenesis. We show that DN45 can stabilize the closed active state of each VFT in the mGlu4 homodimer, as the binding epitope includes residues from both lobe 1 and lobe 2. DN45, like L-AP4, increases the same proportion of mGlu4 in the active conformation, as shown by smFRET in the total absence of glutamate, demonstrating an agonist activity equivalent to that of L-AP4. While the primary binding epitope is located on lobe 2, further interaction between the VFT and DN45 occurs upon spontaneous closure of the VFT. These additional contacts increase the affinity of DN45 and lead to stabilization of the active state of the receptor, as does the agonist. This model is well supported by the observation that preventing contact between lobe 1 and the nanobody through mutations converts the nanobody into an antagonist or an inverse agonist, stabilizing the inactive receptor by preventing VFT closure. Taken together, our data confirm that stabilizing the closed VFTs is sufficient for receptor activation even in the absence of glutamate. Such a mode of action is, then, different from what has been proposed for the mGlu2 and mGlu5 PAM nanobodies and reveals ways for the development of selective agents acting at a specific mGlu subtype.
When tested on the mGlu2-4 heterodimer, DN45 was found unable to activate the receptor on its own. In contrast to the mGlu4 homodimer, on which two nanobodies are likely acting (one per VFT), only one is expected to bind to the heterodimer, on the mGlu4 VFT only. The fact that the nanobody cannot activate such a heterodimer suggests that the mGlu4 VFT may have less tendency to spontaneously close when associated with mGlu2. One may also consider the positive allosteric effect between each VFTs (21). The transient closure of one VFT may facilitate the closure of the other such that binding of a first nanobody may facilitate the binding of the second one in the homodimer, leading to the stabilization of the active state without agonist and then to the full agonist activity. Such an allosteric process is unlikely to occur in the heterodimer in the presence of DN45 alone, since DN45 only acts on one VFT. In agreement with this model, agonist binding in the mGlu2 VFT largely favored DN45 binding on the mGlu4 subunit. Such a model is consistent with the pure PAM effect of DN45 observed on the mGlu2-4 heterodimer.
Such a PAM action of DN45 on mGlu2-4 is also consistent with the effects of group-III agonists acting at the mGlu4 VFT. Indeed, L-AP4 could barely activate the heterodimer, while it largely potentiates the effect of an mGlu2 agonist (10, 29). This supports the idea that the closure of the mGlu2 VFT is the driving force for the activation of the mGlu2-4 heterodimer that is then further stabilized in the active state through the closure of the mGlu4 VFT by DN45.
This model points to a transactivation mechanism within the mGlu2-4 receptor, as the mGlu4 TMD is mainly responsible for G-protein activation in this heterodimer (30). This is then similar to what is well established for the GABAB receptor in which GABA stabilizes the closed state of the GB1 VFT, leading to the G-protein activation by the GB2 7TM (31).
Overall, we describe a nanobody with a full agonist action at a GPCR. Such a tool will be useful to solve the active structure of human mGlu4. We show DN45 acts by stabilizing the closed state of the mGlu4 VFT by a distinct mode of action compared to other nanobodies that enhance the activity of class C GPCRs. Our study, then, provides insight into the activation mechanism of mGlu4 and mGlu2-4 receptors and illustrates how powerful nanobodies can be to decipher the function of these different receptor subtypes.
Materials and Methods
Information on materials, llama immunization, selection, production, and purification of DN45, mutagenesis, cell culture and transfection, labeling of CLIP and SNAP-tag, the DN45 binding and selectivity assay, the IP-1 accumulation assay, the mGlu4 and mGlu2-4 biosensor assay, the BRET assay, statistical analysis, single-molecule FRET approach, and molecular modeling is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Arpège platform facilities at the Institut de Génomique Fonctionnelle for all fluorescence-based assays. J.-P.P. was supported by la Fondation pour la Recherche Médicale (Grant DEQ20170336747), Cisbio Bioassays (Eidos collaborative team Institut de Genomique Fonctionnelle-Cisbio, Grant 039293), the Fond Unique Interministériel (FUI) of the French government (grant Cell2Lead project), the Agence Nationale pour la Recherche (ANR) (Grant ANR-18-CE11-0004-01), and LabEx MAbImprove (Grant NR-10-LABX-5301). P.R. was supported by the ANR (Grant ANR-15-CE18-0020-01). J.-P.P., L.P., and P.R. were further supported by the Centre National de la Recherche Scientifique and the Institut National de la Santé et de la Recherche Médicale. P.C. and D.N. were supported by the FUI of the French government (FUI, Cell2Lead project). F.A. was supported by the Centre National de la Recherche Scientifique and the Science Ambassador Program from Dassault Système BIOVIA. J.H. was supported by a fellowship from la Région Occitanie and Cisbio Bioassays (TransACT, Grant 156544) and la Ligue contre le cancer (PhD Grant IP/SC-16487). J.F. was supported by la Fondation pour la Recherche Médicale (Grant DEQ20170336747). R.B.Q. was supported by a grant from the ANR (Grant ANR-18-CE11-0004-02). The Centre de Biologie Structurale belongs to the France-BioImaging national infrastructure supported by the ANR (Grant ANR-10-INBS-04, “Investments for the future”).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105848118/-/DCSupplemental.
Data Availability
All data and associated protocols are available in the main paper and the SI Appendix. Materials described in this study are available upon request (to J.-P.P.).
References
- 1.Cromie K. D., Van Heeke G., Boutton C., Nanobodies and their use in GPCR drug discovery. Curr. Top. Med. Chem. 15, 2543–2557 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Gupta A., et al., Increased abundance of opioid receptor heteromers following chronic morphine administration. Sci. Signal. 3, ra54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchings C. J., Koglin M., Olson W. C., Marshall F. H., Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nat. Rev. Drug Discov. 16, 787–810 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hino T., et al., G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 482, 237–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kniazeff J., Prézeau L., Rondard P., Pin J. P., Goudet C., Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol. Ther. 130, 9–25 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Doumazane E., et al., A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 25, 66–77 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Lee J., et al., Defining the homo- and heterodimerization propensities of metabotropic glutamate receptors. Cell Rep. 31, 107605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin S., et al., Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J. Neurosci. 34, 79–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno Delgado D., et al., Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. eLife 6, 1–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholler P., et al., Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear consolidation. Nat. Commun. 8, 1967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charvin D., mGlu4 allosteric modulation for treating Parkinson’s disease. Neuropharmacology 135, 308–315 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Niswender C. M., et al., Development and antiparkinsonian activity of VU0418506, a selective positive allosteric modulator of metabotropic glutamate receptor 4 homomers without activity at mGlu2/4 heteromers. ACS Chem. Neurosci. 7, 1201–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubellini P., Melon C., Dale E., Doller D., Kerkerian-Le Goff L., Distinct effects of mGlu4 receptor positive allosteric modulators at corticostriatal vs. striatopallidal synapses may differentially contribute to their antiparkinsonian action. Neuropharmacology 85, 166–177 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Vilar B., et al., Alleviating pain hypersensitivity through activation of type 4 metabotropic glutamate receptor. J. Neurosci. 33, 18951–18965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zussy C., et al., Dynamic modulation of inflammatory pain-related affective and sensory symptoms by optical control of amygdala metabotropic glutamate receptor 4. Mol. Psychiatry 23, 509–520 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Even-Desrumeaux K., et al., Masked selection: A straightforward and flexible approach for the selection of binders against specific epitopes and differentially expressed proteins by phage display. Mol. Cell. Proteomics 13, 653–665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scholler P., et al., HTS-compatible FRET-based conformational sensors clarify membrane receptor activation. Nat. Chem. Biol. 13, 372–380 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Chen R., Li L., Weng Z., ZDOCK: An initial-stage protein-docking algorithm. Proteins 52, 80–87 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Pierce B., Weng Z., ZRANK: Reranking protein docking predictions with an optimized energy function. Proteins 67, 1078–1086 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kniazeff J., et al., Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Urwyler S., Allosteric modulation of family C G-protein-coupled receptors: From molecular insights to therapeutic perspectives. Pharmacol. Rev. 63, 59–126 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Sebastianutto I., Cenci M. A., mGlu receptors in the treatment of Parkinson’s disease and L-DOPA-induced dyskinesia. Curr. Opin. Pharmacol. 38, 81–89 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Duggan S., Caplacizumab: First global approval. Drugs 78, 1639–1642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T., et al., Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: Application to brain imaging. FASEB J. 26, 3969–3979 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Low S., et al., VHH antibody targeting the chemokine receptor CX3CR1 inhibits progression of atherosclerosis. MAbs 12, 1709322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon C., et al., Synthetic nanobodies as angiotensin receptor blockers. Proc. Natl. Acad. Sci. U.S.A. 117, 20284–20291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehl A., et al., Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitz J., et al., Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron 92, 143–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., et al., Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer. eLife 6, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galvez T., et al., Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J. 20, 2152–2159 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and associated protocols are available in the main paper and the SI Appendix. Materials described in this study are available upon request (to J.-P.P.).