Significance
Multistep reactions, such as degradation of proteins by the ubiquitin–proteasome system, require that the different components will be colocalized to an assembly that allows catalysis of all steps in an efficient and processive manner. In the absence of colocalization, floating of individual components around the cell would slow the cascade reactions. We demonstrated that such assemblies are generated in the nucleus both under basal and stressed conditions and play a role in efficient degradation of nuclear proteins, unassembled subunits of the proteasome, and unfolded proteins, suggesting they are part of the protein quality-control mechanism and help in preventing cellular damage. The findings suggest that other multicomponent pathways can also be assembled into condensates to allow their efficient activity.
Keywords: LLPS condensates, ubiquitin, proteasome, p62, protein degradation
Abstract
Degradation of a protein by the ubiquitin–proteasome system (UPS) is a multistep process catalyzed by sequential reactions. Initially, ubiquitin is conjugated to the substrate in a process mediated by concerted activity of three enzymes; the last of them—a ubiquitin ligase (E3)—belongs to a family of several hundred members, each recognizing a few specific substrates. This is followed by repeated addition of ubiquitin moieties to the previously conjugated one to generate a ubiquitin chain that serves as a recognition element for the proteasome, which then degrades the substrate. Ubiquitin is recycled via the activity of deubiquitinating enzymes (DUBs). It stands to reason that efficiency of such a complex process would depend on colocalization of the different components in an assembly that allows the reactions to be carried out sequentially and processively. Here we describe nuclear condensates that are dynamic in their composition. They contain p62 as an essential component. These assemblies are generated by liquid–liquid phase separation (LLPS) and also contain ubiquitinated targets, 26S proteasome, the three conjugating enzymes, and DUBs. Under basal conditions, they serve as efficient centers for proteolysis of nuclear proteins (e.g., c-Myc) and unassembled subunits of the proteasome, suggesting they are involved in cellular protein quality control. Supporting this notion is the finding that such foci are also involved in degradation of misfolded proteins induced by heat and oxidative stresses, following recruitment of heat shock proteins and their associated ubiquitin ligase CHIP.
An important, yet unsolved question is how cells recruit the appropriate reactants involved in biochemical reactions, to a defined location at a specific time to increase the reaction efficiency and processivity. This is particularly relevant for sequential transformations along cascades that are catalyzed by different enzymes and complexes, such as degradation of a protein by the ubiquitin–proteasome system (UPS). Separation of the interior milieu of membrane-bound organelles (e.g., nucleus, Golgi apparatus, endoplasmic reticulum, mitochondria, and lysosomes) from the surrounding environment enables the cells to control the spatial distribution of specific reactions and their components. However, this compartmentalization makes it difficult to allow exchange of components necessary for providing high specificity, for example. In addition, cells contain supramolecular assemblies that are not bound by surrounding membranes and can be found both in the cytoplasm (such as the centriole, P bodies, and stress granules) and the nucleus (such as nucleoli, promyelocytic leukemia [PML] bodies, Cajal bodies, and nuclear speckles) (1, 2).
Emerging evidence shows that many membraneless biomolecular condensates are formed by liquid–liquid phase separation (LLPS) (3). These condensates provide a different environment from the surrounding cytoplasm or nucleoplasm and probably have an important role in the concentration of biomolecules, thus allowing efficient multistep processes to occur. In addition, it allows for rapid exchange of specific components—such as ubiquitin ligases—to occur and thus allowing accommodation to changing pathophysiological conditions and the requirement for high specificity of different processes. Recently, it was shown that p62, also known as Sequestosome-1 (SQSTM-1), forms spherical liquid-like condensates (bodies) both in the cytosol and in a cell-free system (4). Formation of these condensates is mediated by the ability of the protein to oligomerize into helical filaments through its N-terminal PB1 (Phox and Bem1) domain, and to bind ubiquitin (Ub) chains attached to target substrates via its C-terminal UBA (ubiquitin-associated) domain (5). The efficiency of p62 condensate formation was shown to be strongly dependent on the length of the Ub chains attached to the substrates (6).
Cytosolic p62 condensates play an important role in maintaining protein homeostasis and quality control by their ability to sequester ubiquitinated proteins and shuttle them for degradation by the autophagy–lysosome system. This is mediated via direct interaction of the p62’s LIR motif with the autophagosome-incorporated LC3 (7). In addition to its role in autophagy, p62 was shown to deliver cytosolic ubiquitinated substrates to the proteasome, by direct interaction of its PB1 domain with the 19S Rpn1 and Rpn10 proteasomal subunits (8).
p62 is widely distributed in the cell, and its nucleocytoplasmic shuttling is mediated by two nuclear localization signals (NLS1 and NLS2) and one nuclear export signal (NES) (9). In addition to its cytosolic functions, p62 forms nuclear bodies that exist both as separate and as hybrid structures by interacting with PML or Cajal bodies (10). Since autophagy occurs exclusively in the cytosol, the role and biological significance of nuclear p62 have remained elusive.
In the present study we show that p62 is involved in the formation of nuclear LLP-separated foci, which recruit components of the UPS and protein substrates, resulting in their efficient degradation under both basal and stressed conditions.
Results
Formation of Nuclear p62-Containing Condensates Is Mediated by LLPS and Depends on the Presence of Ubiquitin Adducts.
p62 shuttles continuously between the nucleus and the cytoplasm, and a fraction of the protein is found in foci (9). Formation of cytosolic p62 foci is mediated by LLPS and characterized by dynamic exchange of p62 between liquid droplets and the surrounding environment (4, 6). As for nuclear foci, the mechanism of their formation and functions are still unknown. To study the nature of nuclear p62 foci, we stained HeLa cells with an anti-p62 antibody. While under basal conditions, two-dimensional (2D) analysis demonstrated the presence of a few foci (Fig. 1A, Left column, and SI Appendix, Fig. S1A), three-dimensional (3D) reconstruction of nuclei from several cell lines enabled the detection of numerous foci (Fig. 1B, dimethyl sulfoxide [DMSO]/control panels, and Movie S1). Following treatment with Leptomycin B (LMB), an inhibitor of nuclear protein export, we observed accumulation of p62 in the nucleus that resulted in an increase in the number and size of p62 foci (Fig. 1A, Right column, and Fig. 1B, LMB panels; SI Appendix, Fig. S1B and Movie S2). To study the mechanism of nuclear p62 condensates formation and dynamics, we used CRISPR/Cas9 to generate p62 knockout (KO) HeLa cells. This cell was transfected (where indicated) with a cDNA coding for a p62 mutant that lacks its nuclear exit signal (FLAG- or GFP-tagged p62ΔNES) which resulted in concentration of p62 in the nucleus. The purpose of this approach was to avoid treatment with LMB that would have resulted in nuclear accumulation of different NES-harboring cellular proteins (SI Appendix, Fig. S1A).
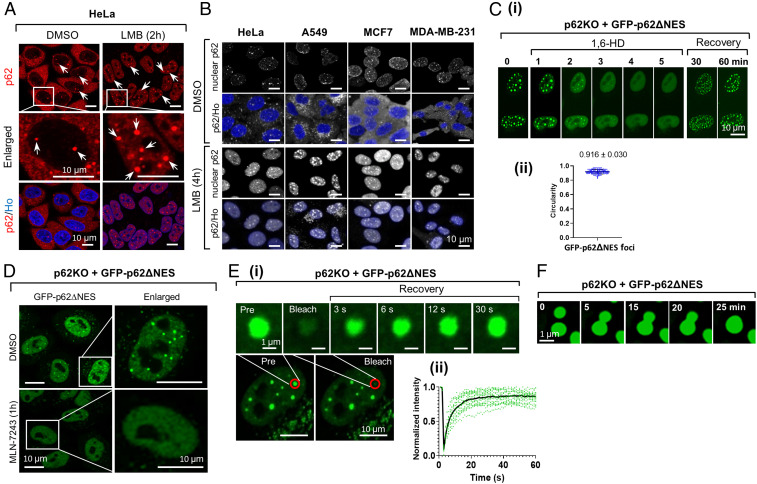
Fig. 1.
Nuclear p62 condensates are formed via LLPS. (A) DMSO (Left) or LMB-treated (Right) HeLa cells were stained with a p62 antibody and Hoechst (Ho) and imaged using 2D confocal microscopy. Arrows point to nuclear p62 foci. (B) The indicated cells were treated with either DMSO (Upper two lines) or LMB (Lower two lines), and stained with anti-p62 and Ho. For highlighting nuclear p62, the cytosolic p62 was hidden by the software when only p62 was stained. (C) Time-lapse imaging of GFP-p62∆NES condensates disruption and recovery following 1,6-HD treatment. (D) Live cell images of the indicated cell treated with the E1 inhibitor MLN-7243. (E) (i) FRAP (Lower, red line surrounded circles) of p62 foci. Time-lapse images (Upper) were taken from bleached cells; (ii) quantification of fluorescence taken from 18 p62 condensates. (F) Time-lapse images of fusion of p62 condensates.
First, we wanted to show that the nuclear foci are indeed phase-separated condensates. We treated GFP-p62ΔNES-transfected p62KO cells with 1,6-hexanediol (1,6-HD), an aliphatic alcohol that disrupts weak hydrophobic protein–protein interactions and destabilizes liquid droplets (11). The treatment resulted in fast disappearance of nuclear p62 foci (t1/2 of ∼2 min) in both p62ΔNES-reconstituted cells (Fig. 1C, i, first six panels, and Movie S3), and in wild-type (WT) HeLa cells (SI Appendix, Fig. S1C, i). The foci recovered completely within 30 to 60 min after removal of 1,6-HD (Fig. 1C, i, seventh and eighth panels). Measurement of the circularity of the foci, showed that they are ball shaped (0.916 ± 0.030; Fig. 1C, ii). Similar data were also obtained in WT HeLa cells (SI Appendix, Fig. S1C, ii).
LLPS of cytosolic p62 is known to be mediated by the interaction of its UBA domain with polyubiquitinated targets. To check whether formation of nuclear p62 condensates is Ub chain dependent, we treated GFP-p62ΔNES-reconstituted p62KO (p62KO + p62ΔNES) cells with MLN-7243, a selective ubiquitin-activating enzyme (UBA1/E1) inhibitor (11). We first showed that the inhibitor almost completely Ub conjugates formation in cells (SI Appendix, Fig. S1D). As can be seen in Fig. 1D, MLN-7243 treatment inhibited almost completely the formation of nuclear p62 foci. Similarly, addition of cycloheximide (CHX), a protein synthesis inhibitor, to LMB-pretreated HeLa cells, resulted in a significant decrease in Ub conjugates (SI Appendix, Fig. S1E, i), a decrease which was specifically inhibited by the proteasome inhibitor MG132. Specifically, we observed a decrease in K48-based Ub chains (SI Appendix, Fig. S1E, ii, first column) with a concomitant decrease in the number and size of nuclear p62 condensates (SI Appendix, Fig. S1E, ii, second column). The disappearance of the K48-based conjugates was also inhibited by MG132 (SI Appendix, Fig. S1E, ii, compare third line to second line from the Top). Importantly, CHX did not affect p62 protein level in the LMB-treated cells (SI Appendix, Fig. S1E, i, Middle), suggesting that its effect on the disappearance of p62 foci is indirect and attributed to the loss of Ub conjugates. In agreement with the effect of CHX on p62 foci disappearance rather than on the quantity of the protein, is the observation that it causes an even distribution of the protein in the nucleus (SI Appendix, Fig. S1E, ii, second column, Middle). Immune staining of p62KO + p62ΔNES cells with antibodies that recognize specific linkages in Ub chains revealed that besides K48-based chains (SI Appendix, Fig. S1E, ii, S1F, i–iii), p62 condensates also contain K63-based chains (SI Appendix, Fig. S1G).
As for the dynamics of p62 condensates, fluorescence recovery after photobleaching (FRAP) of p62KO cells reconstituted with GFP-p62ΔNES, showed a rapid recovery of the foci completed ∼30 s after their bleaching (Fig. 1E). This finding suggests that the foci are not intranuclear inclusion bodies or aggregates, which either have longer recovery time or do not recover at all (12). Interestingly, the recovery of nuclear p62 foci was much faster than that of their cytosolic counterparts (as reported in ref. 4), suggesting that they differ from one another, probably physically. Additionally, we detected fusion of small foci into larger ones (Fig. 1F and Movies S4 and S5).
Components of the UPS Accumulate in p62 Condensates.
In order to study a possible role for p62 nuclear condensates in proteolysis, we searched for components of the UPS. p62 is known to deliver ubiquitinated proteins to the proteasome by direct interaction of its PB1 domain with the 19S Rpn1 and Rpn10 proteasomal subunits (8). Thus, we tested whether the proteasome is recruited to these condensates. HeLa cells were treated with LMB, and the proteasome was detected by immunofluorescent (IF) staining, using initially an anti-α6 (a 20S subunit) antibody. As can be seen in Fig. 2A, the LMB-induced p62 condensates (note that the p62 was endogenous), also contain endogenous proteasome. To further dissect this finding, we overexpressed FLAG-tagged p62ΔNES in p62KO cells. As can be seen in Fig. 2B and C and D, i–iv and Movie S6, the nuclear proteasome is located in the periphery of the condensates, forming an outer shell-like structure around p62, which generates either an inner shell or is distributed in the core of the condensate (Fig. 2D, v–viii and Movie S6). This inner shell surrounds the Ub conjugates, which constitute the core of the condensate (Fig. 2D, v–viii). However, no shell-like structure formation was detected in the p62KO cells that were not transfected with p62, and the nuclear proteasome was mostly evenly distributed (Fig. 2B, Upper Left), suggesting that p62 is essential in recruiting the proteasome to the condensates. Importantly, we found that subunits from the two subcomplexes of the 26S proteasome—the 20S (catalytic particle [CP]; α6) and the 19S (regulatory particle [RP]; Rpn1)—accumulate around the p62 condensates (Fig. 2E), suggesting it is the intact 26S proteasome complex that is recruited.
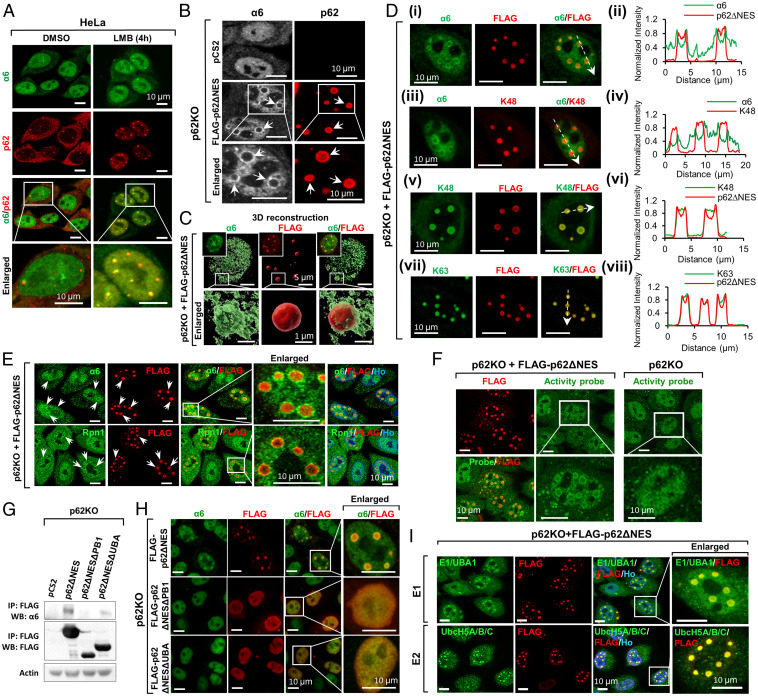
Fig. 2.
p62 condensates recruit components of the UPS. (A) Colocalization of the α6 subunit of the proteasome with p62 foci in cells treated with DMSO or LMB. (B) α6 is recruited to p62 foci (arrows) in p62KO cells transfected with p62∆NES. (C) Three-dimensional reconstruction of α6 and p62 in the nucleus. Squares in the Upper Left corner of the three Upper panels represent photographs of the entire nucleus. (D) Colocalization profiles of α6, K48- and K63-based Ub chains, and FLAG-p62∆NES: (i, iii, v, and vii) fluorescent images; (ii, iv, vi, and viii) intensity tracings (along the dashed arrows in i, iii, v, and vii) of α6, p62, and K48 and K63-based chains, respectively. (E) Colocalization of α6 (green, Upper) and Rpn1, the 19S subunit (green, Lower) with p62 condensates (arrows). (F) Proteasome activity demonstrated in p62 foci, using a specific activity probe. (G) Co-IP of nuclear (WT) and mutant (ΔPB1 and ΔUBA) p62s with the α6 subunit. (H) IF of colocalization of nuclear (WT) and mutant p62s, with the α6 subunit. (I) Nuclear p62 foci colocalize with E1/UBA1 and E2s (UbcH5A/B/C).
To study whether the proteasome recruited to the p62 condensates is proteolytically active, we used a fluorescent proteasome activity probe (13). We found that in p62KO + p62ΔNES cells, the fluorescent probe was also detected around the p62 condensates (Fig. 2F), strongly suggesting the proteasome that surrounds the condensates is active.
To identify which domain of p62 is responsible for the recruitment of the proteasome, we constructed two p62ΔNES mutants, which lacked the PB1 or the UBA domain (p62ΔNESΔPB1 or p62ΔNESΔUBA, respectively). We found that while p62ΔNES and p62ΔNESΔUBA did interact with the proteasome, the deletion of PB1 abrogated this interaction (Fig. 2G). Furthermore, we found that the proteasome did not form the shell-like structures in cells reconstituted with either p62ΔNESΔPB1 or p62ΔNESΔUBA (Fig. 2H, second and third lines). That is in agreement with the role of the UBA domain in recruiting Ub chains (SI Appendix, Fig. S2A, i and ii), and the role of the PB1 domain (and the Lys7 that resides within it) (14) in oligomerizing p62, which is essential for foci formation (SI Appendix, Fig. S2B).
In addition, we identified the first (E1/UBA1), second (E2, ubiquitin-conjugating protein) (Fig. 2I), and third (E3, ubiquitin ligase) (see below) components in the conjugation cascade in the foci, raising the possibility that while conjugation occurs initially in the nucleoplasm, once the seed of conjugates is formed, it continues more efficiently in the foci. Deubiquitinating enzymes (DUBs) were also identified in the foci (SI Appendix, Fig. S2C), which appear therefore to contain all the elements required for protein degradation and Ub recycling.
Nuclear p62 Condensates Act as Active Proteolytic Sites that Accelerate the Proteasomal Degradation of Nuclear Proteins.
The finding that an active 26S proteasome is recruited to p62 condensates that contain ubiquitinated proteins, raised the obvious question of whether these foci act as proteolytic sites. To answer this question, we used a model fusion protein—NLS-GFP-CL1—in which the NLS directs GFP to the nucleus, and the CL1-degron destabilizes the protein and renders it an efficient proteasomal substrate (15). As can be seen in Fig. 3A, i, Upper line, besides the presence of NLS-GFP-CL1 in the nuclei of p62KO cells, we detected a significant accumulation of the protein in the nucleoli. Reconstitution of NLS-GFP-CL1–transfected p62KO cells with p62ΔNES, changed its distribution: the protein was recruited into p62 foci, which was accompanied by a significant reduction in its quantity in nucleoli and the nucleoplasm, most probably due to degradation in the p62 foci (Fig. 3A, i, Lower line and Fig. 3A, ii). Indeed, as can be seen in Fig. 3B, i and ii, and Movie S7, the presence of nuclear p62 (either in WT or in p62KO + p62ΔNES HeLa cells) stimulates significantly the degradation of the fusion protein. That is in comparison to cells that lack p62. In correlation with this finding, we observed that the level of the fusion protein in p62KO cells was higher than in cells that express p62 (Fig. 3B, i). Of note is that p62ΔNES was stable (Fig. 3B, iii). As expected, treatment of cells with MG132 abolished the degradation of nuclear GFP-CL1 (Fig. 3B, i). Similar findings were observed microscopically (Fig. 3C).
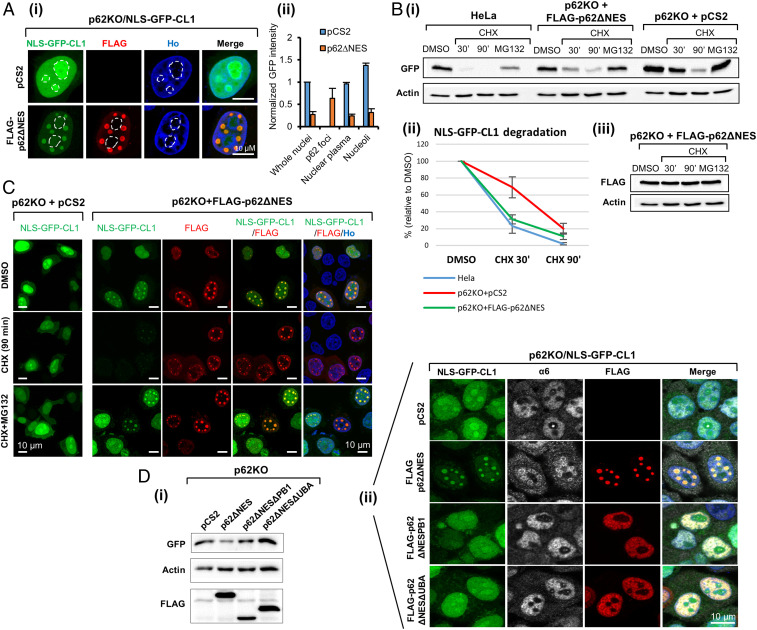
Fig. 3.
p62 phase separation accelerates the degradation of NLS-GFP-CL1. (A) (i) Distribution of stably expressed NLS-GFP-CL1 in p62KO cells without (Upper) or with (Lower) FLAG-p62∆NES. Dashed circles mark nucleoli; (ii) GFP intensity in the nuclei, p62 foci, nucleoplasm, and nucleoli as determined in 40 cells and normalized artificially to the intensity in whole nuclei in p62KO cells. (B) (i) Degradation (as determined during CHX chase and Western blotting) of NLS-GFP-CL1 in HeLa and p62KO cells transfected with either p62ΔNES or an empty vector; (ii) quantification of degradation data from three independent experiments; (iii) p62ΔNES stability in the experiment presented under B, i. (C) Degradation of NLS-GFP-CL1 occurs in p62 foci and is mediated by the proteasome. (D) The ΔPB1 and ΔUBA mutants of p62ΔNES do not support degradation of NLS-GFP-CL1 (i) and p62 foci formation (ii).
Since phase separation of p62 is mediated by its PB1 and UBA domains, we tested whether deletion of each of them—which prevents formation of p62 condensates—affects the degradation of nuclear GFP-CL1. We found that reconstitution of NLS-GFP-CL1–expressing p62KO cells with p62ΔNES, results in a decrease in the level of nuclear GFP-CL1 (Fig. 3D, i and ii, Left column); compare the Upper Top to the second from Top) with its recruitment to p62 condensates. There, it is surrounded by the recruited proteasome (Fig. 3D, ii, second line, second panel from Left). Such effect on condensation of the substrate could not be observed in the absence of either PB1 or UBA domains (Fig. 3D, i and ii, third and fourth lines).
p62 Accelerates the Degradation of Unassembled Proteasomal Subunits.
The proteasome is widely distributed in the cell and is predominantly located in the nucleus (16). It is composed of 33 distinct subunits, which are arranged in two subcomplexes—a 20S (CP) and a 19S (RP). The assembly of the proteasome is a complicated process, and an excess of unassembled proteasomal subunits is degraded by the UPS, following their ubiquitination (17, 18).
First, we studied whether p62 condensates play a role in recruiting these unincorporated subunits (as an example for proteins the cell does not need anymore). We stably overexpressed the β4-GFP 20S proteasomal subunit in p62KO cells. The expressed subunit was distributed throughout the cell with predominant nuclear localization (Fig. 4 A, Upper Left). Reconstitution of β4-GFP–expressing cells with p62ΔNES (Fig. 4 A, Lower line, two Left panels; and Fig. 4 B, Upper line) resulted in accumulation of β4-GFP in p62 condensates. Additionally, we found that p62ΔNES directly interacts with β4-GFP (Fig. 4C). Importantly, staining of the endogenous proteasome with anti-α6 antibody in p62ΔNES-expressing cells (Fig. 4 A, Lower line, α6), revealed a different distribution pattern from that of the overexpressed β4-GFP in the p62 condensates: only the outer margin of the accumulated β4-GFP colocalized with the endogenous proteasome (which surrounds the condensates; see also Fig. 2 B–E). This finding suggests that besides the fraction of GFP-β4 that has been incorporated into the proteasome, a significant portion of the overexpressed subunit has remained free/unincorporated, and localized to the core of the condensate. Since the proteasome is a stable complex with a half-life of approximately 1 wk, it was reasonable to assume that following CHX treatment, the unincorporated β4-GFP subunits will be degraded, while the subunits that are incorporated into the proteasome will remain stable. To test this hypothesis, we treated β4-GFP/p62ΔNES–transfected p62KO cells with CHX. Immunostaining revealed that following CHX treatment, only the inner part of the accumulated β4-GFP in the p62 condensates was degraded (Fig. 4B, second line, second panel from Left; SI Appendix, Fig. S3A, second line; and Movie S8), while its outer part remained stable and displayed a similar distribution to that of the endogenous proteasome. Further confirming these findings is the experiment depicted in Fig. 4D, i and ii. p62KO + p62ΔNES cells expressing GFP-β4 were treated with CHX. Western blot analysis revealed that a fraction of the GFP-β4 disappeared, but left behind a stable portion. When the cell lysates from the same experiment were now treated with anti-α6 antibody, and the entire proteasome was precipitated, the quantity of GFP-β4 in the CHX-treated and nontreated cells was identical, strongly suggesting that indeed, the β4 subunit is divided between two pools—a free unincorporated pool, which is unstable, and a stable proteasome-incorporated pool. In addition, treatment of cells with both MG132 and CHX prevented the degradation of β4-GFP (Fig. 4B and SI Appendix, Fig. S3A, compare Lower to Middle lines), confirming its proteasome-dependent degradation. Similar results were obtained for Rpn11-GFP, a 19S RP subunit (Fig. 4E and Movie S9).
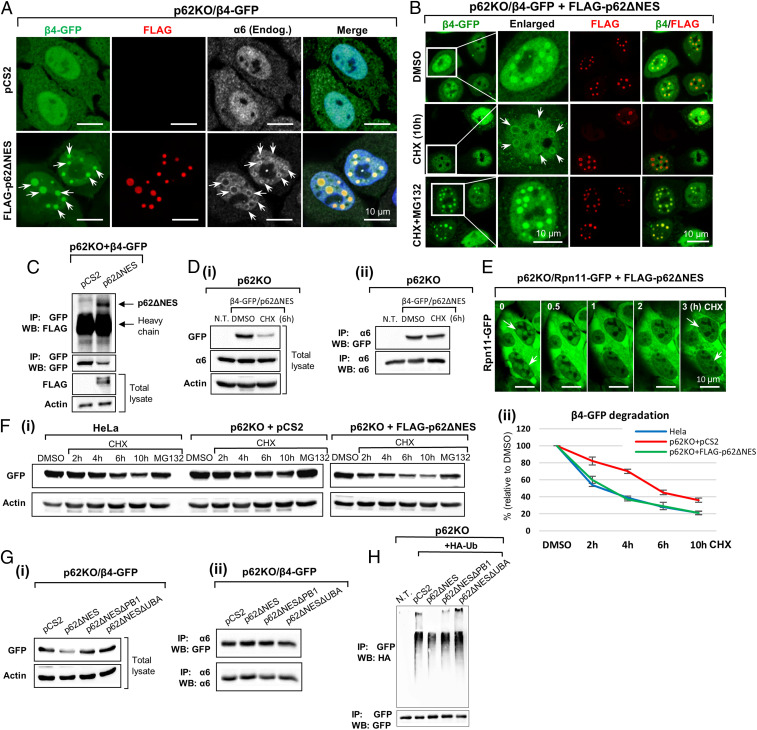
Fig. 4.
Unincorporated proteasomal subunits are degraded more efficiently in nuclear p62 condensates. (A) Stably overexpressed β4-GFP is recruited to nuclear p62 foci in a p62ΔNES-dependent manner. The endogenous α6 subunit is localized to the periphery of these foci. (B) Degradation of β4-GFP in p62 foci following addition of CHX. (C) Lysates from cells described under A were immunoprecipitated with an anti-GFP and immunoblotted with anti-FLAG to demonstrate association between FLAG-p62ΔNES and β4-GFP. (D) The β4 proteasomal subunit serves both as a proteolytic substrate (i; Western blot of whole cell lysates) and as an incorporated/integral subunit of the proteasomal complex (ii; IP of whole lysates with anti-α6). (E) The Rpn11 subunit of the 19S subcomplex of the proteasome is degraded in what appears to be p62 foci in cells stably transfected with p62∆NES. Time-lapse images were captured along the CHX treatment (images were taken from Movie S9). (F) (i) p62 stimulates proteasomal degradation of overexpressed β4-GFP; (ii) quantification of degradation data from three independent experiments. (G) (i) Effect of p62∆NES, p62∆NES∆PB1, and p62∆NES∆UBA on the steady-state level of β4-GFP; (ii) p62 and its mutants have no effect on the level of the β4 subunit incorporated into the intact proteasome. (H) Ubiquitination of β4-GFP in the presence of p62ΔNES or its ΔPB1 and ΔUBA species. β4-GFP was immunoprecipitated, and the sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE)–resolved proteins were blotted with anti-HA to detect the conjugated HA-Ub.
Next, we examined the requirement for nuclear p62 in the degradation of unincorporated β4-GFP. We found that the degradation of β4-GFP in p62KO cells was slower compared to both HeLa and p62KO + p62ΔNES cells (Fig. 4F and SI Appendix, Fig. S3B). In agreement with this finding, the steady-state level of β4-GFP was higher in p62KO cells compared with the two other cells (SI Appendix, Fig. S3B). Furthermore, we noted that the level of β4-GFP reached its minimal point 12 h after initiation of the CHX treatment and remained so until the end of the experiment after 20 h (SI Appendix, Fig. S3B), suggesting that the remaining β4-GFP represents the stable proteasome-incorporated fraction. When we reconstituted p62KO cells with either p62ΔNES or its ΔPB1 and ΔUBA mutants, only the intact protein could stimulate degradation of β4-GFP (Fig. 4G, i). Importantly, immunoprecipitation (IP) of the 26S proteasome from p62KO cells and from cells that were reconstituted with p62ΔNES and its ΔUBA and ΔPB1 deletion mutants, revealed no difference in the amount of the proteasome-incorporated β4-GFP subunit (Fig. 4G, ii), suggesting that p62 has no effect on the fate of β4 in the proteasome.
Since p62 recruits substrates through their Ub chains, we examined the ubiquitination state of overexpressed β4-GFP. We found that the subunits were ubiquitinated, and the level of the conjugates decreased in cells that were transfected with p62ΔNES (Fig. 4H), most probably due to their degradation. In contrast, expression of the two p62 mutants did not affect the level of the conjugates, and the p62ΔUBA domain even caused a slight increase in their level. This finding suggests that the UBA domain is more involved in recruitment of ubiquitinated targets, probably due to its higher affinity to Ub conjugates (Fig. 4H). It raises the possibility that the nature of the interaction of p62 with the unincorporated β4-GFP subunit and its recruitment to the p62 condensates as proteolytic substrates (mediated via its UBA domain), is different from its interaction with the intact proteasome (mediated via its PB1 domain) and its resulting recruitment to the condensate (SI Appendix, Fig. S3C [compare to Fig. 2G], SI Appendix, Fig. S2A, and Fig. 6A [see below]).
Fig. 6.
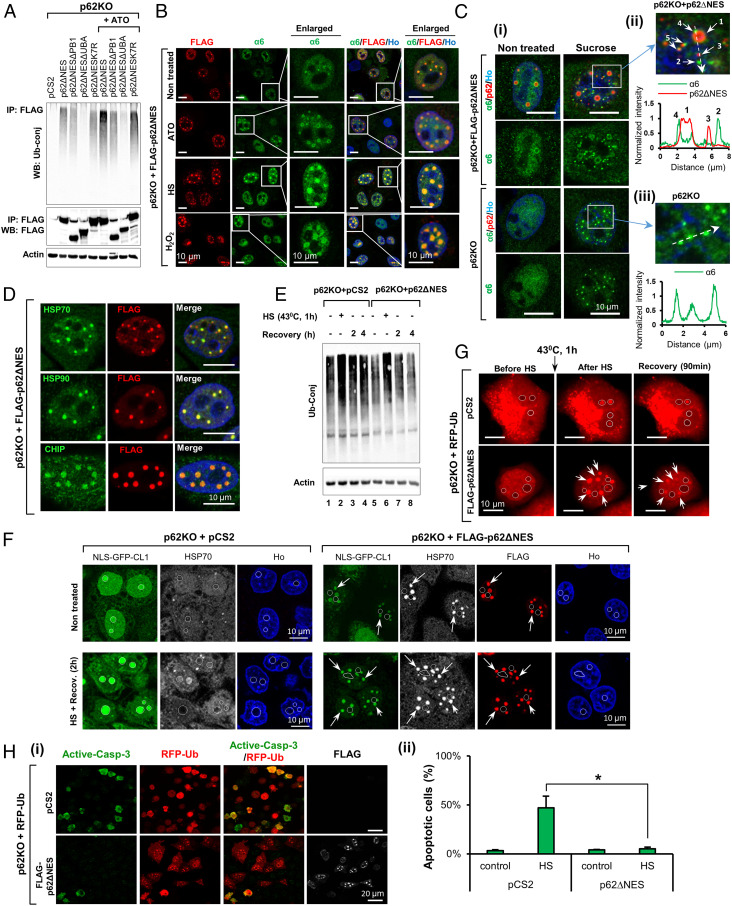
Nuclear p62 condensates are part of the PQC machinery. (A) ATO increases the UBA domain-mediated interaction of p62ΔNES with Ub-conjugated proteins. The precipitated p62 species with which the ubiquitin conjugates interacted, are shown in the Middle panel. (B) ATO, H2O2, and heat stress stimulate formation of p62∆NES condensates and recruitment of the proteasome to these foci. (C) (i) Sucrose treatment induces p62-independent proteasome LLPS and subsequent fusion of the foci with p62ΔNES condensates (that most probably existed prior to the stress, but also were induced by the stress). (ii and iii) Fluorescent images (of the enlarged sections in C, i) and intensity tracings (along the dashed arrows) of α6 and p62 taken from the p62KO cells reconstituted with FLAG-p62ΔNES (ii) and the p62KO cells (iii). Arrows in ii point to different types of foci (see main text). (D) HSP70, HSP90, and CHIP localize in p62∆NES condensates. (E) p62∆NES accelerates the elimination of Ub conjugates induced by HS. (F) p62ΔNES recruits NLS-GFP-CL1 to p62 condensates (arrows) and therefore prevents its HS-induced accumulation in nucleoli (marked by circles). (G) RFP-Ub accumulates in p62 foci following HS. In the absence of p62, it appears in nucleoli. Cells were subjected to HS, and RFP-Ub was visualized immediately after or following 90 min at 37 °C. Arrows point to p62 foci. Circles denote nucleoli. (H) (i) p62ΔNES protects the RFP-Ub-transfected cells from heat-induced apoptosis. Cells were exposed to heat stress and moved to 37 °C for 24 h; (ii) measurements of apoptosis (three independent experiments) were carried out in RFP-Ub–expressing cells in the presence or absence of p62ΔNES (n > 500 cells in each condition; *P < 0.05).
Nuclear p62 Stimulates Degradation of c-Myc.
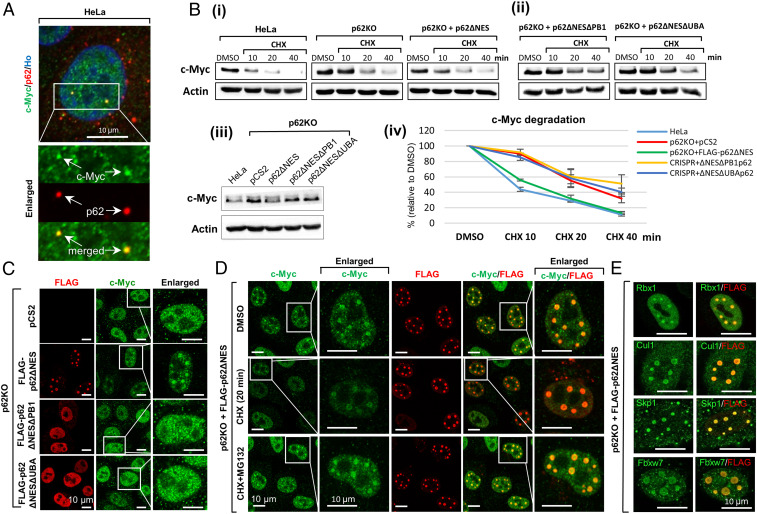
Next, we examined whether nuclear p62 condensates are involved in the degradation of native substrates of the UPS, which in many cases are short-lived nuclear transcription factors and regulatory proteins. One typical representative of this group is the c-Myc oncoprotein, which is rapidly degraded with a half-life of ∼20 to 30 min (19). First, we asked whether endogenous nuclear p62 colocalizes with endogenous c-Myc. IF staining of HeLa cells revealed that the two are indeed colocalized in p62 condensates (Fig. 5A). To further study the significance of this interaction, we examined whether nuclear p62 accelerates the degradation of c-Myc. As can be seen in Fig. 5B, this is indeed the case. The degradation of c-Myc in p62KO cells was slower than in both HeLa and p62KO + p62ΔNES cells (Fig. 5B, i). A slowed degradation was also observed when p62KO cells were reconstituted with the ΔPB1 and ΔUBA domain deletion mutants of nuclear p62 (Fig. 5B, ii). Further corroborating these observations were the higher steady-state levels of c-Myc in these cells (Fig. 5B, iii) and the increased intensity of its staining, as was visualized microscopically (Fig. 5C). Interestingly, it appears that in the absence of p62, c-Myc can be identified in the nucleolus, similarly to our model substrate NLS-GFP-CL1 (Fig. 3 A and C) and is probably stored there. In the presence of p62, the nucleoli are emptied, and the substrates appear in p62 foci, where they are degraded. In addition, we observed that CHX treatment of p62KO + p62ΔNES cells results in a decrease in the level of c-Myc in p62 condensates compared to untreated cells (Fig. 5D). It is noteworthy that when c-Myc is degraded in the condensates, the merge with p62 disappears, and the remaining and stable p62 shows up again in its original red fluorescent emission (Fig. 5D, Middle line). The decrease is due to degradation of the protein by the proteasome, as it is inhibited by MG132 (Fig. 5D, Lower). An important question was whether the condensates also contain SCFFbxw7, the main E3 ligase complex that ubiquitinates c-Myc (19). As can be seen in Fig. 5E, staining with specific antibodies to SCFFbxw7 complex, which is composed of Cullin-1, Skp1, Rbx1, and F-box protein Fbxw7, shows that the entire complex colocalizes with p62 in the condensates. Similar microscopic findings were also shown for c-Jun (SI Appendix, Fig. S5).
Fig. 5.
c-Myc degradation is stimulated in nuclear p62 condensates. (A) Endogenous c-Myc is colocalized with endogenous p62 in HeLa cells. (B) p62 (i) but not its ΔPB1 and ΔUBA deletion mutants (ii) stimulates degradation of c-Myc in the nucleus; (iii) steady-state level of c-Myc was monitored in cells transiently expressing pCS2, p62ΔNES, and its ΔPB1 and ΔUBA mutants; (iv) quantification of degradation data obtained in three independent experiments. (C) Degradation of c-Myc occurs in WT p62ΔNES foci. Note the lower level of c-Myc when WT p62ΔNES is present (compare Right panel in the second line, to Right panels in the first, third, and fourth lines). (D) Proteasome-dependent c-Myc degradation in p62ΔNES condensates as monitored following addition of CHX. (E) Skp1, Cullin 1, and the F-box protein Fbxw7: the components of the SCF E3 ligase complex that ubiquitinates c-Myc, are localized to p62ΔNES condensates.
Nuclear p62 Condensates Act as Part of the Cellular Protein Quality-Control (PQC) Mechanism.
Various stress conditions such as oxidation, heat, exposure to heavy metals, and ultraviolet (UV) irradiation, cause damage to proteins. These proteins have to be removed efficiently in order to prevent cytotoxicity and cell death. They are first recognized and ubiquitinated by specific E3s and are subsequently degraded by the proteasome or the autophagic machinery (7). As p62 binds ubiquitinated proteins via its UBA domain, we hypothesized that stress-induced increase in ubiquitination should result in increased formation of p62 condensates. To test this, we subjected HeLa, p62KO, and p62KO + p62ΔNES cells to oxidative [induced by arsenic trioxide (ATO) (20) or H2O2] and heat stress (HS), and monitored formation of Ub conjugates and p62 condensates. We found that the oxidative stress increased the amount of Ub conjugates bound to p62ΔNES (Fig. 6A), which resulted probably from an increase in their general level (SI Appendix, Fig. S4A, i and ii). As expected, the conjugates did not bind to the p62 mutant that lacks the UBA domain (5, 21), and bound less efficiently to the ΔPB1 deletion mutant and to p62K7R, which cannot oligomerize (14) (Fig. 6A). The increased interaction between nuclear p62 and Ub conjugates led, most probably, to an increase in both the number and size of p62 condensates (Fig. 6B, SI Appendix, Fig. S4B, and Movies S10–12) that was also accompanied by an increased recruitment of the proteasome to the foci (Fig. 6B, second and third columns). Importantly, pretreatment of the cells with an E1 inhibitor prior to the induction of the stress, abrogated the formation of the condensates (SI Appendix, Fig. S4C). When we subjected cells to a short, 30 min, osmotic stress by sucrose, we detected two types of proteasome-containing nuclear foci: 1) p62 condensates—the ones that are the main subject of this study—that are positive for both p62 and the proteasome (which is located around the condensate) (Fig. 6C, i, Upper Right, and Fig. 6C, ii, arrow 1); and 2) much smaller foci that are positive only for the proteasome (Fig. 6C, i, Upper Right, and Fig. 6C, ii, arrow 2). Additionally, we detected the appearance of newly formed small p62-only foci (Fig. 6C, i, Upper Right, and Fig. 6C, ii arrow 3). It appears that the small proteasomal foci fuse with either the large p62 condensates (Fig. 6C, ii, arrow 4) or with the small p62 foci (Fig. 6C, ii, arrow 5, SI Appendix, Fig. S4D, i, Upper line, and SI Appendix, Fig. S4D, ii, arrow 1), which might later fuse to one another to form new large functional p62 condensates (as shown in Fig. 1F and Movies S4 and S5). Importantly, it has been shown recently that sucrose treatment induces phase separation of the nuclear proteasome in a RAD23B-dependent manner (11). It appears that these foci are similar or identical to the small proteasome-only foci that we observe, and that importantly, the formation of which appears to be p62 independent. To show that p62 is not involved in sucrose-induced phase separation of the proteasome, we treated p62KO cells with sucrose. We found that the lack of p62 did not prevent phase separation of the nuclear proteasome (Fig. 6C, i, third and fourth lines, and Fig. 6C, iii, SI Appendix, Fig. S4D, i, Lower line, and SI Appendix, Fig. S4D, iii). Additionally, we found that the nature of the condensates is determined by the type of Ub chains involved in their formation. Proteasome-only condensates mostly contain K48-based chains, while p62 condensates contain both K48- and K63-based chains (SI Appendix, Fig. S4E). Thus, it appears that the sucrose-induced condensates, described by Yasuda et al. (11), are formed by a different mechanism and might have a different function(s), thought they can fuse with already existing mature or newly formed p62 condensates.
To prevent aggregation of unfolded/damaged proteins, cells rely on PQC systems, which either refold them (a process mediated by chaperones such as HSP70 and HSP90) or target them for degradation by the proteasome or the autophagic machinery, following their ubiquitination by cochaperone(s)/E3 ligase(s) (such as CHIP) (7). Since p62 is known to interact with the multichaperone Hsc70/HSP70 complex via the cochaperone BAG3 and/or CHIP (7), we stained p62KO + p62ΔNES cells for different components of this PQC system. As can be seen in Fig. 6D, staining of p62 revealed that in the nucleus, it colocalizes with HSP70, HSP90, and CHIP. An important question relates to the possible role of these proteins in eliminating damaged proteins. To address this question, we subjected p62KO and p62KO + p62ΔNES cells to HS. As can be seen in Fig. 6E, the stress induced formation of Ub conjugates that were degraded more efficiently in cells that were reconstituted with p62. Of note is that the steady-state level of the conjugates was lower in the p62-reconstituted cells compared to their p62KO counterparts (compare lane 1 to lane 5 in Fig. 6E) highlighting the role of p62 in eliminating Ub conjugates even under basal conditions. Similar corollary observations were seen using the rapidly degrading proteolytic substrate NLS-GFP-CL1. As can be seen in Fig. 6F (first column), in the absence of p62, following heat stress, the protein accumulates in a large amount in nucleoli and remains stable after the cells recover from the stress. When p62 is reconstituted, NLS-GFP-CL1 appears in p62 condensates and undergoes degradation (Fig. 6F, fourth column; note that the intensity of GFP is decreased significantly in the p62-containing compared to the p62-lacking cells). Similarly, RFP-Ub, some of which might be conjugated to target substrates, also accumulates in nucleoli following HS in the absence of p62. When p62 is present, it is recruited to p62 condensates (Fig. 6G). When the cell recovers, its level in the condensates decreases as it is partially degraded along with its target substrates (22). Since the accumulation of misfolded or aggregated proteins in both the nucleus and nucleolus induces cytotoxicity and cell death (23, 24), we examined whether p62ΔNES protects the cells from HS-induced death. As can be clearly seen in Fig. 6H, the presence of p62 is important in protecting cells from apoptotic death during their recovery after HS.
Discussion
In the present study, we show that both under basal and stress conditions, nuclear p62 forms condensates in a process that is mediated by LLPS. These condensates serve as active proteolytic centers by sequestering the 26S proteasome and ubiquitinated substrates along with the conjugation machinery. We demonstrate that in the nuclear proteolytic p62 foci, the degradation of nuclear proteins, c-Myc for example, is stimulated. Similarly, the degradation of excess of free and unincorporated Rpn11 and β4 proteasomal subunits is also more efficient. Furthermore, we show that p62 condensates play a role in cellular PQC. Induction of formation of unfolded proteins by heat or oxidative stress results in recruitment of chaperones (HSP70 and HSP90) and the chaperone-dependent E3 ligase CHIP to the p62 condensates, with faster disappearance of the stress-induced Ub adducts.
Several problems remain unsolved, however. One relates to the distinct roles of p62 in the cytosol where it is involved in autophagy and in the nucleus where it is involved in proteasomal degradation. It is possible that different signals and adaptors (such as different Ub chains) are involved in these different processes. Functionally, most transcriptional regulators that are tightly regulated and short lived, are residing in the nucleus, and it is possible that the p62 foci that employ the highly specific ubiquitin system evolved mostly in this organelle, while autophagy that is mostly a response to stress and is less specific, evolved in the cytosol. Another one relates to the mechanism of assembly of the foci: What is the signal(s) that alerts cells for inducing foci formation in order to degrade a specific substrate(s)? It is possible that it is initiated by a target substrate on which a short Ub chain was assembled. This short chain is sufficient to recruit p62 (6), which in turn recruits the proteasome. In parallel, elongation of the chain is continued using E1, and the cognate E2 and E3, to generate a Ub chain(s) optimal for proteasomal activity. In that sense, the presence of DUBs that we have shown suggests the notion that the foci serve as efficient proteolytic centers. Another question relates to the composition of the foci and how they are adapted to changing pathophysiological conditions: Do E3s and E2s change with changing conditions and the need to degrade different substrates? A related problem is whether the foci can target, at the same time, different substrates, or are they substrate specific? Also, the role of the K63-based chains in the foci is not clear, although a role for these chains in targeting substrates to the proteasome has been reported (25). Further, it was shown that K63-based short chains can trigger degradation by seeding K48/K63 branched chains that serve as degradation signals (26). Since K63-based chains/target substrates are also involved in signaling, it is possible that the foci serve to sequester them as part of a regulatory mechanism. Most of these questions can be answered using visual and biochemical tools. Thus, uniquely tagging substrates and ligases can tell us whether they all colocalize to the same foci, or whether the condensates are substrate-specific.
One can argue that the expression of a nuclear-only p62 may affect the normal proteolytic processes in the cell, for example, by inhibiting cytosolic autophagy. We suggest that this is not the case, as even in an extreme case of p62-deficient cells, autophagy still proceeds and is probably enabled by proteins like NBR1 that have similar functions to that of p62 (27).
Experimental Procedures
Materials (purchased from commercial sources) and methods are described in SI Appendix, Supplementary Experimental Procedures.
Cell Transfection.
HeLa cells or HeLa cells in which p62 was deleted (p62KO) were transiently transfected with the following cDNA constructs, which were cloned into the pCS2 plasmid (Invitrogen): FLAG-p62∆NES, FLAG-p62∆NES∆PB1, FLAG-p62∆NES∆UBA, FLAG-p62∆NESK7R, GFP-p62∆NES, RFP-Ub, and HA-Ub (SI Appendix, Supplementary Experimental Procedures, Cell Culture and Transfection).
Generation of p62KO Cells.
HeLa cells were transfected with a pSpCas9 (BB)-2A-GFP plasmid containing the guide RNA sequences for p62 (forward: CACCGCGCTACACAAGTCGTAGTCT; reverse: AAACAGACTACGACTGTGTAGCGC) targeting the p62 gene at exon 3. Fluorescence-activated cell sorting (FACS) was performed 48 h after transfection, and GFP-expressing cells were selected. Cells were grown for recovery, and single-cell clones derived from the sorted cells were isolated by limiting dilution. PCR was performed to identify the adjacent regions beside the targeted DNA region of sgRNA, a selection method by which clones containing mutations were identified. The sequences that were obtained were compared to the wild-type sequence. Insertion/deletion mutations causing a frame-shift disruption in both alleles were identified as null mutations. Cell lysates were confirmed for the absence of the p62 protein by Western blot analysis. One obtained clone of p62-depleted HeLa cell was denoted a p62KO cell and used throughout the current study.
Proteasome Activity Detection.
To visualize proteasome activity and the distribution of active proteasomes in the nucleus, p62KO cells were transfected with an empty pCS2 vector or with the same vector that contains cDNA coding for FLAG-p62ΔNES. After 24 h, cells were incubated with 2 µM of the cell-permeable fluorescent proteasome activity probe (described in SI Appendix, Supplementary Experimental Procedures, Antibodies and Reagents) for 2 h. Cells were then washed with fresh Dulbecco's Modified Eagle Medium to remove residual unbound probe. After fixation with 4% paraformaldehyde in phosphate buffered saline for 20 min at room temperature, cells were stained with anti-FLAG antibody. Imaging of the probe was performed on a Zeiss LSM700 confocal microscope with a 488-nm laser and a 63× oil immersion objective lens.
Induction of Oxidative, Heat, and Osmotic Stress.
Oxidative stress was induced by treatment of cells with either arsenic trioxide (ATO; 2 µM) or hydrogen peroxide (H2O2; 200 µM) for 2 h. Osmotic stress was induced by incubating cells in a medium containing 0.2 M sucrose for 30 min. For heat stress, cells were heated at 43 °C for 1 h followed by recovery at 37 °C for 2 or 4 h. Cells were processed for analysis as described in SI Appendix, Supplementary Experimental Procedures, Immunofluorescence Microscopy, Live Cell Imaging and Cell Lysates, Immunoprecipitation, and Western Blotting. Time-lapse imaging was carried out using a 40× objective at 4-min intervals for the indicated times. To study heat-induced apoptosis, the recovery time after HS treatment was extended to 24 h.
Supplementary Material
Acknowledgments
A.C. was supported by grants from the Adelson Medical Research Foundation, the Israel Science Foundation, and by a grant from the Albert Sweet Foundation. A.C. is an Israel Cancer Research Fund Professor. A.F. acknowledges support from the Israel Academy of Sciences and Humanities Postdoctoral Fellowship for Foreign Researchers.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107321118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Matera A. G., Izaguire-Sierra M., Praveen K., Rajendra T. K., Nuclear bodies: Random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell 17, 639–647 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyon A. S., Peeples W. B., Rosen M. K., A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun D., Wu R., Zheng J., Li P., Yu L., Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isogai S., et al., Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 286, 31864–31874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaffagnini G., et al., p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308. 10.15252/embj.201798308. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Kaplan V., Livneh I., Avni N., Cohen-Rosenzweig C., Ciechanover A., The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int. J. Biochem. Cell Biol. 79, 403–418 10.1016/j.biocel.2016.07.019. (2016). [DOI] [PubMed] [Google Scholar]
- 8.Seibenhener M. L., et al., Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24, 8055–8068 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pankiv S., et al., Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol. Chem. 285, 5941–5953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Souquere S., Weil D., Pierron G., Comparative ultrastructure of CRM1-nucleolar bodies (CNoBs), intranucleolar bodies (INBs) and hybrid PML/p62 bodies uncovers new facets of nuclear body dynamic and diversity. Nucleus 6, 326–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda S., et al., Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Hinz J., Lehnhardt L., Zakrzewski S., Zhang G., Ignatova Z., Polyglutamine expansion alters the dynamics and molecular architecture of aggregates in dentatorubropallidoluysian atrophy. J. Biol. Chem. 287, 2068–2078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkers C. R., et al., Profiling proteasome activity in tissue with fluorescent probes. Mol. Pharm. 4, 739–748 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Wilson M. I., Gill D. J., Perisic O., Quinn M. T., Williams R. L., PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12, 39–50 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Gilon T., Chomsky O., Kulka R. G., Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17, 2759–2766 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enenkel C., Proteasome dynamics. Biochim. Biophys. Acta 1843, 39–46 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Livneh I., Cohen-Kaplan V., Cohen-Rosenzweig C., Avni N., Ciechanover A., The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 26, 869–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters L. Z., et al., The protein quality control machinery regulates its misassembled proteasome subunits. PLoS Genet. 11, e1005178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welcker M., et al., The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alarifi S., Ali D., Alkahtani S., Siddiqui M. A., Ali B. A., Arsenic trioxide-mediated oxidative stress and genotoxicity in human hepatocellular carcinoma cells. OncoTargets Ther. 6, 75–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen-Kaplan V., et al., p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 113, E7490–E7499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shabek N., Herman-Bachinsky Y., Ciechanover A., Ubiquitin degradation with its substrate, or as a monomer in a ubiquitination-independent mode, provides clues to proteasome regulation. Proc. Natl. Acad. Sci. U.S.A. 106, 11907–11912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon I, et al., Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saudou F., Finkbeiner S., Devys D., Greenberg M. E., Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Saeki Y., et al., Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtake F., Tsuchiya H., Saeki Y., Tanaka K., K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 115, E1401–E1408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirkin V., et al., A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.