Significance
Forest decline due to climate change is increasing worldwide. Accurate forecasting of forest dynamics requires a mechanistic understanding of carbon allocation strategies that can link molecular process regulation to whole-tree responses. However, dedicated transdisciplinary investigations spanning these scales are lacking. Here we used a unique experimental platform to manipulate the whole-tree carbon balance and combine metabolic flux measurements with transcriptional information to reveal tree resource allocation priorities under reduced carbon supply. In contrast to current understanding and model implementations, we show that trees prioritize carbon allocation to storage and mobilization of storage to support respiration rather than growth to ensure immediate and future survival under negative carbon balance that may occur during critical periods of stress.
Keywords: carbon allocation, carbon starvation, isotopic labeling, nonstructural carbohydrate storage, transcriptional regulation
Abstract
Climate change is expected to pose a global threat to forest health by intensifying extreme events like drought and insect attacks. Carbon allocation is a fundamental process that determines the adaptive responses of long-lived late-maturing organisms like trees to such stresses. However, our mechanistic understanding of how trees coordinate and set allocation priorities among different sinks (e.g., growth and storage) under severe source limitation remains limited. Using flux measurements, isotopic tracing, targeted metabolomics, and transcriptomics, we investigated how limitation of source supply influences sink activity, particularly growth and carbon storage, and their relative regulation in Norway spruce (Picea abies) clones. During photosynthetic deprivation, absolute rates of respiration, growth, and allocation to storage all decline. When trees approach neutral carbon balance, i.e., daytime net carbon gain equals nighttime carbon loss, genes encoding major enzymes of metabolic pathways remain relatively unaffected. However, under negative carbon balance, photosynthesis and growth are down-regulated while sucrose and starch biosynthesis pathways are up-regulated, indicating that trees prioritize carbon allocation to storage over growth. Moreover, trees under negative carbon balance actively increase the turnover rate of starch, lipids, and amino acids, most likely to support respiration and mitigate stress. Our study provides molecular evidence that trees faced with severe photosynthetic limitation strategically regulate storage allocation and consumption at the expense of growth. Understanding such allocation strategies is crucial for predicting how trees may respond to extreme events involving steep declines in photosynthesis, like severe drought, or defoliation by heat waves, late frost, or insect attack.
Trees are sessile organisms that cannot escape from increasing environmental stresses or insect outbreaks via migration but must acclimate and adapt anatomically, morphologically, and physiologically (1, 2). A key component of these responses is the ability to optimize resource allocation (3). A core resource, carbon, is assimilated via photosynthesis (source activity) and partitioned among different functions (sinks), including growth, maintenance, storage, defense, and the production of root exudates or volatile compounds (4, 5). However, despite a long history of research on plant carbon allocation (3, 6), we still lack a mechanistic understanding of how trees allocate carbon to grow and survive in a changing environment.
Under normal conditions when source activity is greater than demand for growth and respiration, excess photosynthates can be stored as nonstructural carbohydrates (NSC), a process termed “accumulation” by Chapin et al. (7). However, under stressful environmental conditions, like severe drought, heat waves, late frost, or insect attack, source activity via photosynthesis is reduced by prolonged stomatal closure or leaf loss while demand for respiration, osmoregulation, repair, and defense continues or even increases. This can lead to situations when carbohydrate supply from current photosynthesis cannot meet carbon demand for maintenance metabolism (i.e., negative carbon balance), thus hampering physiological function and survival (8). Under these conditions, trees increasingly depend on the mobilization of stored carbon sources, including NSCs like soluble sugars and starch as well as lipids and proteins (5, 9). Despite the important role of NSC storage in trees, our current understanding assumes that NSC storage is a low-priority sink that accumulates during periods of positive carbon balance, and that can be completely depleted to fuel respiration or growth under negative carbon balance (reviewed in refs. 10–13). This widely held assumption, however, may not be valid if trees prioritize allocation to NSC storage over growth to ensure survival under reduced carbon supply. This conservative allocation strategy. termed “reserve formation” (4), may occur due to the up-regulation of storage and/or the down-regulation of growth (9).
Identifying plant allocation strategies requires investigations that combine ecophysiological approaches with molecular tools (5, 9). Molecular investigations on allocation strategies are often limited to herbaceous model plants like Arabidopsis and show that the supply of respiratory substrates (i.e., carbohydrates) is coordinated to meet current demand for respiration products (e.g., ATP) (14), while ensuring that daytime build-up and nighttime breakdown of starch are tightly regulated to avoid early morning depletion of reserves that could limit growth (15–17). These studies provide compelling insights into plant storage regulation acting over periods ranging from hours to days. However, it remains uncertain whether strategies used by short-lived, fast-cycling herbaceous plants can serve as a model for allocation strategies in long-lived species that reach sexual maturity after many years. The need to survive for decades to reach reproductive maturity combined with high risk of exposure to periodic stresses on seasonal to interannual timescales may require trees to manage their reserves differently than an annual model species such as Arabidopsis.
Conifers dominate many terrestrial ecosystems in the Northern Hemisphere and are of high ecological and economic importance. However, the critical role of conifers is threatened by climate change as many coniferous species are not adapted to drier, warmer conditions. A mechanistic understanding of carbon allocation priorities under severe photosynthetic deprivation is needed to better predict how conifers may respond to extreme events like drought, heat waves, late frost, or insect attack. In our study, we manipulated the carbon balance of Norway spruce (Picea abies) by progressively reducing atmospheric CO2 concentrations ([CO2]) from 400 ppm (control) to 120 ppm, which is under neutral carbon balance (Materials and Methods), and further to 50 ppm, at which trees go into negative carbon balance. This approach directly imposes source constraints without concomitant sink limitation to prevent oversupply (photosynthetic intake ≤ respiratory losses) and potentially resulting build-up of photosynthetic products and derivatives via accumulative storage, thereby allowing inferences on reserve formation (18). Tree saplings were grown in specially designed flow-through chambers coupled to a [CO2] manipulation system for 8 wks. To assess the overall carbon balance, the changes in [CO2] between inflow and outflow were measured and integrated over the 16-h photoperiod (daytime balance) and the 24-h period (whole-day balance). The fate of carbon fixed after the start of the experiment was tracked by using CO2 that was isotopically distinct (ca. −38‰) from the air under which the plants had grown previously (ca. −9‰). We monitored nighttime respiration, growth, and carbon storage by using metabolomic and isotopic measurements and investigated molecular regulation via transcriptomic analysis. Specifically, we hypothesized that, with declining carbon availability, Norway spruce saplings tend to 1) mitigate losses of carbon through up-regulation of photosynthetic pathways (e.g., light and dark reaction) and down-regulation of respiratory pathways (e.g., glycolysis and tricarboxylic acid cycle), 2) preferentially allocate available carbon to NSC reserves over growth by up-regulation of storage biosynthesis pathways while down-regulating growth-related pathways, and 3) increasingly rely on alternative carbon reserves (lipids and proteins) under negative carbon balance to ensure respiratory maintenance.
Results and Discussion
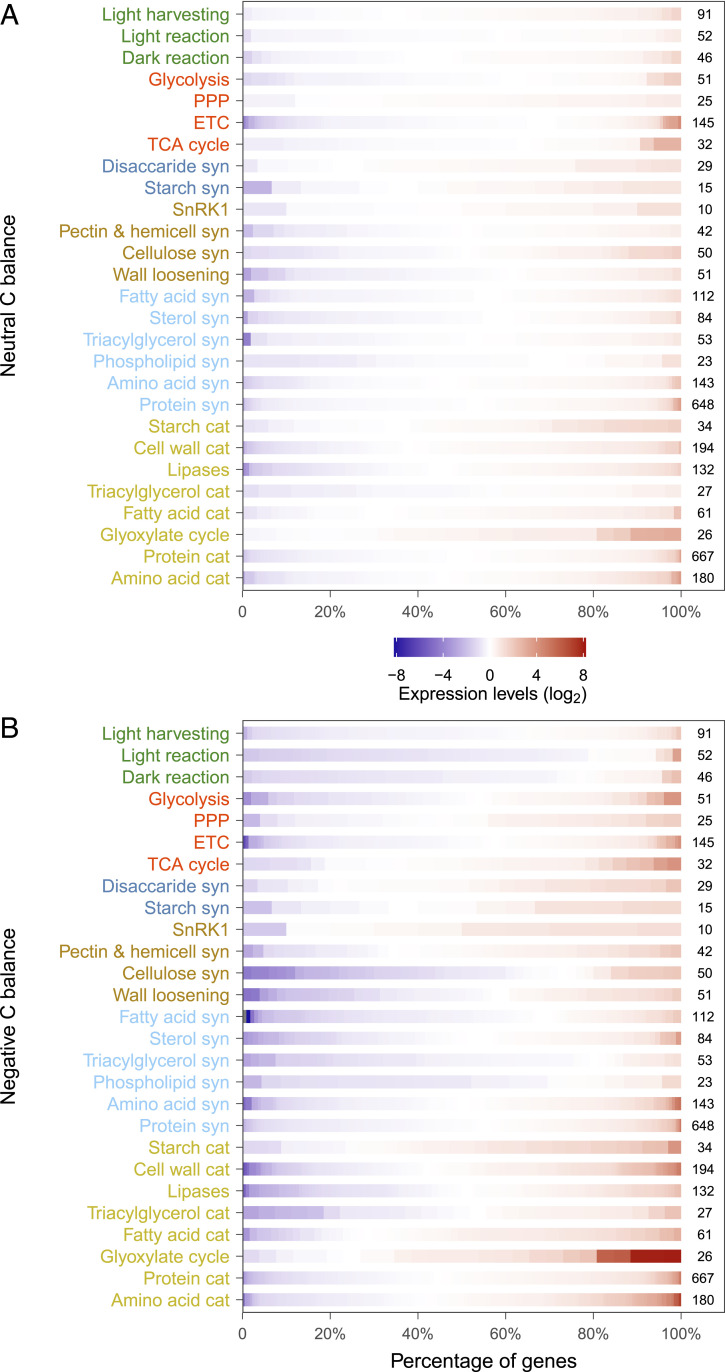
Our results show that with declining carbon, supply trees strategically coordinate storage allocation and consumption to maintain respiratory metabolism, while photosynthesis and growth are down-regulated. Under neutral (photosynthetic intake = respiratory losses) and negative carbon balance (photosynthetic intake < respiratory losses), daytime net assimilation strongly decreased, leading to reduced incorporation of newly assimilated carbon into water-soluble metabolites (Fig. 1) and a decline in concentrations of soluble sugars and starch (Fig. 2 A–D). However, contrary to our first hypothesis, transcript analysis shows that the expression levels of most genes involved in photosynthetic pathways in young leaves were not significantly affected under neutral carbon balance and were even down-regulated under negative carbon balance (Fig. 3 and SI Appendix, Fig. S1). These results suggest that spruce trees may have a limited capacity to up-regulate the whole photosynthetic machinery under neutral or negative carbon balance, possibly as a result of other biochemical constraints (19). Since the initial light-harvesting capacity, for example, is likely to have far exceeded what was needed under negative carbon balance, a general down-regulation of the light-dependent reactions would reduce oxidative damage from redundant light excitation energy (20) and unnecessary maintenance cost (21). A similar molecular mechanism has been used to explain the biochemical (i.e., nonstomatal) limitation of photosynthesis that often occurs under stress such as severe drought (22–24). However, the gene encoding the small subunit of ribulose bisphosphate carboxylase (Rubisco), which catalyzes the first step of CO2 fixation, was up-regulated (SI Appendix, Fig. S1), indicating an attempt to maximize carbon capture under these conditions.
Fig. 1.
Carbon fluxes under declining carbon balance. (A) Carbon (C) uptake, shown as aboveground daytime net assimilation and whole-day net C gain (daytime assimilation minus nighttime respiration). (B) Substrate availability, shown as the proportion of recently assimilated (solid fill) and older stored (diagonal fill) water-soluble C in aboveground organs (previous-year and current-year needles and branches). (C) Allocation of recently assimilated C (solid fill) and older stored (diagonal fill) into sink activities (aboveground nighttime respiration and growth). (D) Changes in sink activity of nighttime respiration and whole-tree biomass growth, expressed as changes to control tree sink activity. All flux and metabolic data were taken between week 3 and week 5. The percentage of new versus old C and sink activities were significantly different (P < 0.05; Tukey’s honestly significant difference, HSD) between the low C availability treatments (neutral and negative C balance) and control (positive C balance).
Fig. 2.
Temporal dynamics of carbon storage under declining carbon balance. Changes of storage compounds in (Left) current-year-young and (Right) previous-year-old leaves. (A and B) Soluble sugars (sum of glucose, sucrose, and fructose). (C and D) Starch. (E and F) Lipids. (G and H) Amino acids (sum of the common amino acids; SI Appendix, Figs. S3–S5). All data are expressed as concentrations (mg/g). Values are the means of four individual chambers (n = 4); error bars represent ± SE. Significant differences between the low carbon (C) availability treatments (neutral and negative C balance) and control (positive C balance) are calculated and indicated by filling of symbols (P < 0.05, Tukey’s HSD).
Fig. 3.
Transcriptional regulation of source and sink activities under declining carbon balance. Changes in the gene expression patterns for current-year-young leaves under neutral (A) and negative carbon (C) balance (B), relative to control (positive C balance) at week 4. Figures showing expression of individual genes for individual pathways at higher resolution can be found in SI Appendix, Figs. S1 and S2. Each bar is composed of a number of genes (small boxes) with different expression patterns, starting from the repressed genes (blue) on the left side to the induced ones (red) on the right side. The number of genes identified for each pathway is listed at the end of the bar. The pathways are grouped based on their functions and indicated by the colors of axis labels: photosynthesis (green), respiration (red), NSC synthesis (dark blue), growth (brown), alternative substrate synthesis (light blue), and storage catabolism (yellow). Note that the width of nested genes of each pathway is adjusted so as to compare the overall expression patterns of different pathways. Full information about the sequence ontology is given in SI Appendix, with a detailed description of the gene expression in SI Appendix. Abbreviations: Cat, catabolism; ETC, electron transport chain; PPP, pentose phosphate pathway; Syn, synthesis; and TCA, trichloroacetic acid.
Although nighttime respiration was limited by substrate availability under neutral carbon balance, isotopic analysis showed that CO2 emitted via nighttime respiration was composed of a higher proportion of newly assimilated carbon than recently grown biomass (Figs. 1 and 2 A–D). This indicates that newly assimilated carbon was preferentially allocated to meet the demands of respiration rather than growth under neutral carbon balance. However, despite the use of a depleted 13CO2 source (δ13C equal −38‰, newly assimilated CO2), respired CO2 was slightly enriched in δ13C under negative carbon balance as compared to the pretreatment period (Fig. 1). This suggests that negative carbon balance forced trees to rely on the degradation of compounds stored prior to the experiment, likely 13C-enriched substrates such as starch (25), cell wall polysaccharides (26), malate (25), or aspartate (27). The utilization of newly assimilated and older stored carbon for respiration appears to be achieved through regulation of respiratory pathways, which were sustained and even up-regulated under neutral and negative carbon balance, respectively (Fig. 3). Therefore, in contrast to our hypothesis that low substrate availability may signal a down-regulation of respiratory demands, trees met respiratory demands under neutral and negative carbon balance, most likely for maintenance or for stress mitigation (e.g., oxidative stress) (28).
Our results provide evidence for a switch in allocation priorities from growth to storage under neutral and even negative carbon balance. NSCs resulted from accumulation that had occurred prior to the experiment (i.e., under normal conditions) were mobilized to support growth and respiration in the early phases of carbon deprivation (Fig. 1C), as reflected by their initial reductions (Fig. 2 A–D). However, under prolonged carbon deprivation, NSC storage is prioritized overgrowth, and this is supported by multiple lines of evidence. First, concentrations of soluble sugars remained relatively constant between weeks 4 and 8 (Fig. 2 A–D), indicating that trees under neutral and negative carbon balance succeeded in maintaining low but functional levels of NSC. Second, these levels were maintained with continuous input from recently assimilated carbon and remobilization of older stored carbon (Fig. 1B). Third, transcriptomic analysis showed that the expression of genes encoding disaccharide (e.g., sucrose synthase) and starch biosynthesis was up-regulated under negative carbon balance (Fig. 3 and SI Appendix, Fig. S1), a finding that supports “reserve formation” that results from up-regulation of storage.
To ensure substrate supply for NSC biosynthesis, processes related to growth were strongly down-regulated under negative carbon balance. Reducing carbon supply strongly decreased biomass accumulation (Fig. 1C), accompanied by a down-regulation of genes encoding cellulose and lipid biosynthesis, as well as cell wall loosening—an important process involved in leaf expansion (29) (Fig. 3 and SI Appendix, Figs. S1 and S2). Negative carbon balance also induced expression of genes involved in the SnRK1 signaling pathway that has been shown to repress energy-consuming biosynthetic actions (e.g., growth and defense) under sugar and energy starvation conditions (30, 31). These results also support the idea of “reserve formation” that growth was down-regulated by the trees to divert carbon to synthesis of storage compounds. Similar processes may explain the observed shift in allocation toward NSC storage rather than growth during or after shading (32), defoliation (33), and drought (34). The up-regulation of storage compound synthesis and down-regulation of sink activities like growth in spruce trees under negative carbon balance have also been observed in Arabidopsis during extended nighttime periods (15–17).
Our results have strong implications for understanding how trees avoid death in the face of severe photosynthetic deprivation that may occur during extreme events causing prolonged stomatal closure or defoliation, including severe drought, heat waves, late frost, or insect attack. Trees under normal conditions store substantial amounts of NSC when carbon supply exceeds carbon demands for growth and other metabolic processes such as defense and reproduction, and these NSC can be mobilized to buffer shortages in the early phases of photosynthetic deprivation. However, as photosynthetic deprivation progresses and NSCs are reduced to levels at which tree survival is at risk, trees preferentially allocate available carbon to NSC storage rather than to growth (i.e., reserve formation). These NSCs are likely reserved to ensure survival especially under herbivore and pathogen attack (35–37), which often co-occurs with climate extremes such as drought and heat (38, 39). For example, preferential allocation of limited carbon to carbohydrate storage rather than to growth may allow trees to produce osmoregulatory compounds or defenses such as terpenes and phenolics when necessary. Further research is needed to show exactly where on the gradient of carbon supply allocation priority shifts.
The observed changes in NSCs may result from changes in the rates of both biosynthesis and degradation under neutral and negative carbon balance. Transcriptomic analysis showed that genes encoding enzymes involved in starch degradation, such as β- and α-amylases, were significantly up-regulated (Fig. 3 and SI Appendix, Fig. S1), demonstrating that in spruce trees, like in Arabidopsis (40, 41), starch degradation is up-regulated to maintain sugar levels to cope with environmental stress. Furthermore, processes related to catabolism of cell wall polysaccharides were also up-regulated under negative carbon balance (Fig. 3 and SI Appendix, Fig. S1). In addition, concentrations of total fatty acids strongly decreased in old leaves under negative carbon balance (Fig. 2F), which is supported by the transcriptomic analysis showing that fatty acid catabolism was up-regulated (Fig. 3). More strikingly, the glyoxylate pathway, which converts lipid-derived substrates (i.e., acetyl-CoA) into malate and succinate for the synthesis of carbohydrates, was strongly induced under negative carbon balance (Fig. 3 and SI Appendix, Fig. S2). Such regulation likely occurred earlier and to a greater extent in mature leaves than in developing leaves, given that concentrations of total fatty acids had not yet been significantly affected in the latter (Fig. 2 E and F). Together, these results are consistent with our third hypothesis that autolysis of polysaccharides and lipids provides alternative substrates for respiration in spruce trees under negative carbon balance, similar to what has been observed in Arabidopsis (42, 43).
The use of amino acids as alternative substrates to fuel respiration under negative carbon balance has been also reported for Arabidopsis (44) but not for trees. Under negative carbon balance, the concentrations of total amino acids significantly increased in spruce needles (Fig. 2 G and H). These changes were accompanied by a concurrent up-regulation of pathways that yield metabolites (e.g., pyruvate, acetyl-CoA, malate, and succinate) to support the tricarboxylic acid cycle or glyoxylate pathway (Fig. 3 and SI Appendix, Fig. S2) (44). Concentrations of aspartate, asparagine, proline, glutamate, and glutamine all significantly increased under negative carbon balance (SI Appendix, Figs. S3 and S4), and there was a strong up-regulation of genes encoding proline dehydrogenase and asparagine synthetase, which catalyze conversion of proline into glutamate and of aspartate and glutamine to asparagine and glutamate, respectively (Fig. 3 and SI Appendix, Fig. S2). Furthermore, concentrations of most amino acids remained high in old senescent leaves (Fig. 2 G and H), likely because these amino acids act as osmoprotectants under negative carbon balance, similar to their roles during abiotic stresses such as drought, salinity, or extreme temperatures (41, 45, 46). The increase in amino acids may help compensate for decreased soluble sugars in maintaining osmotic potential under neutral and negative carbon balance.
In conclusion, by combining data from metabolite fluxes and pools with isotopic measurements and transcriptomics, our study shows that under negative carbon balance, Norway spruce trees 1) down-regulate photosynthetic pathways, in particular those related to the light reactions, likely to reduce oxidative damage and associated maintenance costs, while sustaining respiratory pathways; 2) up-regulate NSC biosynthesis pathways while growth-related pathways are down-regulated, to preferentially allocate available carbon to storage over growth; abd 3) induce cell wall polysaccharide degradation and lipid catabolism as well as amino acid metabolism to provide energy and substrates required for survival under prolonged negative carbon balance. Our study provides insights on how trees strategically regulate carbon source–sink relationships to prioritize allocation to NSC storage and mobilization of storage compounds to support respiration over growth when faced with severe photosynthetic limitations. Understanding these strategies is of critical importance for assessing how trees respond to extreme events, particularly those associated with reductions in carbon supply, such as severe drought (47) or defoliation from heat waves, late frost, or insect attack (48).
Materials and Methods
Plant Material and Growth Conditions.
The experiment was conducted in 12 glass chambers built in the greenhouse of the Max Planck Institute for Biogeochemistry in Jena; refer to Hartmann, Ziegler, Kolle, and Trumbore (49) for more details. Control of [CO2] of air entering the 12 glass chambers was achieved by scrubbing all CO2 (δ13C equal −9‰) from the ambient air and readding CO2 (−38‰) to the CO2-free air stream via mass flow controllers to achieve desired concentrations. Twelve chambers were divided into three [CO2] treatments, thus yielding four replicates for each of the three treatments: 1) 400 ppm maintained throughout the experiment as control; 2) immediate reduction from 400 ppm to 280 ppm, which was maintained for 2 wks, then further reduced to 120 ppm to reach neutral carbon balance (i.e., whole-day, aboveground carbon gain was 0, maintained for 6 wk); 3) immediate reduction from 400 ppm to 170 ppm, which was maintained for 2 wks, then further reduced to 50 ppm to reach negative carbon balance (i.e., whole-day aboveground carbon gain was negative, maintained for 6 wk) (50). This experimental design is intended to reduce carbon supply in which metabolic demands for carbon are not met by current photosynthate, leading to declines in the NSC pool. Norway spruce clones were acquired from the Forestry Research Institute of Sweden. One spruce seedling (ca. 7-y-old seedlings) was placed into each glass chamber in May 2016. Roots were grown in a mixture of sand and a slow-releasing inorganic fertilizer (Osmocote Start, Everris International B.V., Netherlands) in an airtight pot separated from the aboveground (50).
Measurements of Flux and Biomass.
Aboveground CO2 flux was measured using a Picarro cavity ring-down spectrometer 2131-i (Picarro Inc. Santa Clara) and calculated by the differences between the [CO2] of air entering ([CO2]in) the chambers measured once per hour and the [CO2] of air leaving ([CO2]out) the chambers measured once every two hours (SI Appendix, Methods). Daytime net assimilation and nighttime respiration were computed depending on the duration over which each occurred (16 and 8 h, respectively). To estimate biomass increment over time, each pot with the seedling was placed on a balance (resolution: 0.1 g; PCB 10000-1, KERN & SOHN GmbH, Germany) inside the growth chamber, and the weight was continuously recorded. To minimize interference from transpiration and watering, ca. 2 cm of sand from the bottom was constantly immersed into water, and the upper sand was wetted by capillary action. The bottom of the pot was connected to a separate water reservoir where the water level was kept constant using a water level sensor. All flux and biomass data were averaged over ca. w wks between week 3 and week 5.
Analysis of Metabolites.
Current-year-young and previous-year-old organs (leaves) were collected from each chamber before the experiment (week 0), in the middle of the experiment (week 4), and at the end of the experiment (week 8). Leaves and branches were separated and homogenized in liquid nitrogen, freeze-dried, and then ground to a fine powder. Soluble sugars were extracted with distilled water at 65 °C and determined with High-Performance Liquid Chromatography-Pulsed Amperometric Detection (HPLC-PAD). Starch in the remaining pellet was digested with a-amylase and amyloglucosidase (Sigma-Aldrich) and determined with HPLC-PAD (51). Total concentrations of fatty acids were determined by a NMR-based fat analyzer (ORACLE; CEM, Matthews). Amino acids were extracted by methanol and diluted by water containing 13C, 15N labeled amino acids, and then analyzed by an HPLC coupled to a tandem mass spectrometer (Triple Quad 6500+, API-Sciex) (52).
Analysis of Isotopes.
The use of an isotopic tracer (δ13C equal −38‰) allowed us to partition allocation of newly assimilated versus stored carbon to different pools. All δ13C values were reported relative to the international Vienna Pee Dee Belemnite (SI Appendix, Methods). The δ13C signature of CO2 of air entering (δ13Cin) and leaving the chamber (δ13Cout) was measured with Fourier transform infrared spectroscopy (FTIR, Ecotech Pty Ltd., Knoxfield, Victoria, Australia) using otherwise the same protocol of the flux measurements. The δ13C signature of nighttime respired CO2 was calculated by using the mass balance:
| [1] |
We calculated carbon isotopic discrimination (∆) under 400 ppm and 120 ppm [CO2] treatment using equation (53):
| [2] |
where ξ is defined as
| [3] |
To reduce interferences from daytime respiration of recently assimilated and old stored substrates, we selected periods of high daytime net assimilation (i.e., above ca. 20 ppm and ca. 10 ppm [CO2] for 400 ppm and 120 ppm [CO2], respectively). Our calculations showed that trees under 400 ppm and 120 ppm [CO2] treatment had a similar ∆ of ca. 22‰, which was 5‰ higher than trees grown under ambient conditions prior to the experiment (17‰). In addition, we assume a similar ∆ between 120 ppm and 50 ppm [CO2].
We measured bulk δ13C with an isotope-ratio mass spectrometry (Thermo Finnigan GmbH, Germany). An aliquot of hot water extract was pipetted into a tin cup and dried at 40 °C for subsequent measurement of the δ13C of water-soluble carbon, which is a reliable proxy for the δ13C of soluble sugars, as previously shown in needles of Pinus sylvestris (54). The proportion of newly assimilated carbon in the corresponding carbon pool, Pnew, was calculated using the equation
| [4] |
where δ13CS is the δ13C signature of the corresponding carbon pool measured around week 4, δ13CB is the background δ13C signature of the corresponding carbon pool measured at week 0 (i.e., pre-label), and δ13Cfull is the theoretical 13C signature of the corresponding carbon pool that is fully labeled, accounting for the difference in isotopic signature between the air used to flush the chambers before (−9‰) and during the experiment (−38‰), as well as an increase of 5‰ in ∆. For example, δ13CB of biomass was ca. −26‰; thus, δ13Cfull is assumed to be ca. −60‰ under the new labeling environment. To compare the proportion of new versus old carbon between aboveground respiration, water-soluble carbon, and biomass growth, δ13C was measured and weighted across current-year-young and previous-year-mature leaves and branches.
Transcriptome Analysis.
Developing leaves collected from each chamber at week 4 (i.e., four replicates per treatment) were ground to fine powder in liquid nitrogen in a mortar. Total RNA was extracted with an Invitrap Spin Plant RNA Mini Kit and (STRATEC Molecular GmbH, Berlin, DE) and RNase-Free DNase Set (Qiagen GmbH, Hilden, DE). Transcriptome sequencing was carried out on an Illumina HiSeq2500 Genome Analyzer platform using poly(A)-enriched RNA (Max Planck Genome Center Cologne). On average, each sample yielded 36 million high-quality paired-end reads of ∼150 base pairs each (bp). Quality control, trimming, and de novo assembly of the pooled reads were achieved using CLC Genomics Workbench version 11.0 (https://digitalinsights.qiagen.com). The assembly was carried out with the following settings: nucleotide mismatch cost = 2; insertion costs = 3; deletion costs = 3; length fraction = 0.3; and similarity = 0.9 as previously described by Vogel, Badapanda, Knorr, and Vilcinskas (55). The sequences in the assembly were annotated using Blast 2GO software suite version 5 (56) using standard settings (Blast x homology searches against National Center for Biotechnology Information and functional annotations using Gene Ontology (GO) terms [http://www.geneontology.org], InterPro terms [InterProScan, EBI] and enzyme classification [EC]). The dataset of ∼450 million reads of 150 bp in length was assembled into 29,696 contigs. The length of the contigs ranges from 350 to 16,560 bp, with an average length of 1,380 bp. Approximately 23,900 contigs were annotated. Differential gene expression was analyzed by mapping the reads of each individual sample to the pooled de novo–assembled transcriptome using CLC Genomics Workbench with the following settings: mismatch cost = 2; insertion cost = 3, deletion cost = 3; length fraction = 0.8; similarity fraction = 0.8; and maximum number of hits for a read = 10. Expression profiles for individual metabolic processes were generated using GOterms, EC numbers, and interPro terms and were manually curated based on standard plant biochemical texts (57). Heat maps for gene expression profiles were rendered using Morpheus (https://clue.io/morpheus).
Statistical Analyses.
For analysis of flux, metabolic, and isotopic data, the normality and homogeneity of variances were checked with Shapiro–Wilk test and Levene test, respectively, and were log-transformed as needed. Significant differences between the treatments were tested using Tukey’s honest significance test (P < 0.05). For data expressed as the percentage of control, SEs were propagated according to error propagation rules. All statistical analysis was conducted in R (version 3.3.2) (58). Statistical differences in gene expression were calculated for each gene between the treatments using the Reads Per Kilobase Million (RPKM) algorithm to obtain correct estimates for relative expression levels (biases in the sequence datasets and different transcript sizes were corrected).
Supplementary Material
Acknowledgments
We thank Waldemar Ziegler, Olaf Kolle, René Schwalbe, Daniel Rzesanke, Agnes Fastnacht, Frank Voigt, and Bernd Schloeffel for their assistance in setting up the experiment and greenhouse work. We thank Savoyane Lambert, Jessica Heublein, and Iris Kuhlmann for their assistance in sample collection and processing, Anett Enke for NSC measurements, Heiko Moossen and Heike Geilmann for isotopic measurements, Katharina Grosser and Anja Worrich for NMR-based lipid analysis, and Michael Reichelt for amino acid analysis. J.G., N.M.v.D., G.G., and S.T. gratefully acknowledge the German Research Foundation for funding iDiv (DFG–FZT 118, 202548816). The study was funded primarily by Max Planck Society.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023297118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Transcriptome data have been deposited in the NCBI database under BioProject accession no. PRJNA751264.
References
- 1.Nicotra A. B., et al., Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Anderegg W. R. L., Trugman A. T., Bowling D. R., Salvucci G., Tuttle S. E., Plant functional traits and climate influence drought intensification and land-atmosphere feedbacks. Proc. Natl. Acad. Sci. U.S.A. 116, 14071–14076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooney H. A., The carbon balance of plants. Annu. Rev. Ecol. Syst. 3, 315–346 (1972). [Google Scholar]
- 4.Körner C., Plant CO2 responses: An issue of definition, time and resource supply. New Phytol. 172, 393–411 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Hartmann H., Trumbore S., Understanding the roles of nonstructural carbohydrates in forest trees - From what we can measure to what we want to know. New Phytol. 211, 386–403 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hartmann H., Bahn M., Carbone M., Richardson A. D., Plant carbon allocation in a changing world - Challenges and progress: Introduction to a Virtual Issue on carbon allocation. New Phytol. 227, 981–988 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Chapin F. S., Schulze E.-D., Mooney H. A., The ecology and economics of storage in plants. Annu. Rev. Ecol. Syst. 21, 423–447 (1990). [Google Scholar]
- 8.McDowell N. G., et al., The interdependence of mechanisms underlying climate-driven vegetation mortality. Trends Ecol. Evol. 26, 523–532 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Dietze M. C., et al., Nonstructural carbon in woody plants. Annu. Rev. Plant Biol. 65, 667–687 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Kozlowski T. T., Carbohydrate sources and sinks in woody plants. Bot. Rev. 58, 107–222 (1992). [Google Scholar]
- 11.Le Roux X., Lacointe A., Escobar-Gutiérrez A., Le Dizès S., Carbon-based models of individual tree growth: A critical appraisal. Ann. For. Sci. 58, 469–506 (2001). [Google Scholar]
- 12.Sala A., Woodruff D. R., Meinzer F. C., Carbon dynamics in trees: Feast or famine? Tree Physiol. 32, 764–775 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Hartmann H., et al., Identifying differences in carbohydrate dynamics of seedlings and mature trees to improve carbon allocation in models for trees and forests. Environ. Exp. Bot. 152, 7–18 (2018). [Google Scholar]
- 14.O’Leary B. M., Asao S., Millar A. H., Atkin O. K., Core principles which explain variation in respiration across biological scales. New Phytol. 222, 670–686 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Smith A. M., Stitt M., Coordination of carbon supply and plant growth. Plant Cell Environ. 30, 1126–1149 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Gibon Y., et al., Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 32, 859–874 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Verbančič J., Lunn J. E., Stitt M., Persson S., Carbon supply and the regulation of cell wall synthesis. Mol. Plant 11, 75–94 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gessler A., Grossiord C., Coordinating supply and demand: Plant carbon allocation strategy ensuring survival in the long run. New Phytol. 222, 5–7 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Fatichi S., Leuzinger S., Körner C., Moving beyond photosynthesis: From carbon source to sink-driven vegetation modeling. New Phytol. 201, 1086–1095 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Gururani M. A., Venkatesh J., Tran L. S., Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 8, 1304–1320 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Bilgin D. D., et al., Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Drake J. E., et al., Stomatal and non-stomatal limitations of photosynthesis for four tree species under drought: A comparison of model formulations. Agric. For. Meteorol. 247, 454–466 (2017). [Google Scholar]
- 23.Galmés J., Medrano H., Flexas J., Photosynthetic limitations in response to water stress and recovery in Mediterranean plants with different growth forms. New Phytol. 175, 81–93 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Flexas J., et al., Decreased Rubisco activity during water stress is not induced by decreased relative water content but related to conditions of low stomatal conductance and chloroplast CO2 concentration. New Phytol. 172, 73–82 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Gessler A., et al., On the metabolic origin of the carbon isotope composition of CO2 evolved from darkened light-acclimated leaves in Ricinus communis. New Phytol. 181, 374–386 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Hartmann H., Gleixner G., Thoma M., Schwab V. F., Carbon isotope fractionation including photosynthetic and post-photosynthetic processes in C3 plants: Low [CO2] matters. Geochim. Cosmochim. Acta 245, 1–15 (2019). [Google Scholar]
- 27.Melzer E., O’leary M. H., Anapleurotic CO2 fixation by phosphoenolpyruvate carboxylase in C3 plants. Plant Physiol. 84, 58–60 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couée I., Sulmon C., Gouesbet G., El Amrani A., Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57, 449–459 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Cosgrove D. J., Catalysts of plant cell wall loosening. F1000 Res., 10.12688/f1000research.7180.1. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez M., Parola R., Andreola S., Pereyra C., Martínez-Noël G., TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the “yin-yang” model? Plant Sci. 288, 110220 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Margalha L., Confraria A., Baena-González E., SnRK1 and TOR: Modulating growth-defense trade-offs in plant stress responses. J. Exp. Bot. 70, 2261–2274 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Weber R., Gessler A., Hoch G., High carbon storage in carbon-limited trees. New Phytol. 222, 171–182 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wiley E., Casper B. B., Helliker B. R., Recovery following defoliation involves shifts in allocation that favour storage and reproduction over radial growth in black oak. J. Ecol. 105, 412–424 (2017). [Google Scholar]
- 34.Galiano L., et al., The fate of recently fixed carbon after drought release: Towards unravelling C storage regulation in Tilia platyphyllos and Pinus sylvestris. Plant Cell Environ. 40, 1711–1724 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Goodsman D. W., Lusebrink I., Landhäusser S. M., Erbilgin N., Lieffers V. J., Variation in carbon availability, defense chemistry and susceptibility to fungal invasion along the stems of mature trees. New Phytol. 197, 586–594 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Wiley E., Rogers B. J., Hodgkinson R., Landhäusser S. M., Nonstructural carbohydrate dynamics of lodgepole pine dying from mountain pine beetle attack. New Phytol. 209, 550–562 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Raffa K. F., et al., Defence syndromes in lodgepole - Whitebark pine ecosystems relate to degree of historical exposure to mountain pine beetles. Plant Cell Environ. 40, 1791–1806 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Kautz M., Meddens A. J. H., Hall R. J., Arneth A., Biotic disturbances in Northern Hemisphere forests – A synthesis of recent data, uncertainties and implications for forest monitoring and modelling. Glob. Ecol. Biogeogr. 26, 533–552 (2017). [Google Scholar]
- 39.Seidl R., et al., Forest disturbances under climate change. Nat. Clim. Chang. 7, 395–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stitt M., Zeeman S. C., Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Dong S., Zhang J., Beckles D. M., A pivotal role for starch in the reconfiguration of 14C-partitioning and allocation in Arabidopsis thaliana under short-term abiotic stress. Sci. Rep. 8, 9314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E.-J., Matsumura Y., Soga K., Hoson T., Koizumi N., Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 48, 405–413 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Usadel B., et al., Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146, 1834–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hildebrandt T. M., Nunes Nesi A., Araújo W. L., Braun H. P., Amino acid catabolism in plants. Mol. Plant 8, 1563–1579 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Bown A. W., Shelp B. J., Plant GABA: Not just a metabolite. Trends Plant Sci. 21, 811–813 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Szabados L., Savouré A., Proline: A multifunctional amino acid. Trends Plant Sci. 15, 89–97 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Adams H. D., et al., A multi-species synthesis of physiological mechanisms in drought-induced tree mortality. Nat. Ecol. Evol. 1, 1285–1291 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Huang J., et al., Tree defence and bark beetles in a drying world: Carbon partitioning, functioning and modelling. New Phytol. 225, 26–36 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Hartmann H., Ziegler W., Kolle O., Trumbore S., Thirst beats hunger - Declining hydration during drought prevents carbon starvation in Norway spruce saplings. New Phytol. 200, 340–349 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Huang J., et al., Eyes on the future - Evidence for trade-offs between growth, storage and defense in Norway spruce. New Phytol. 222, 144–158 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Landhäusser S. M., et al., Standardized protocols and procedures can precisely and accurately quantify non-structural carbohydrates. Tree Physiol. 38, 1764–1778 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crocoll C., Mirza N., Reichelt M., Gershenzon J., Halkier B. A., Optimization of engineered production of the glucoraphanin precursor dihomomethionine in Nicotiana benthamiana. Front. Bioeng. Biotechnol. 4, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cernusak L. A., et al., Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 200, 950–965 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Brandes E., et al., Short-term variation in the isotopic composition of organic matter allocated from the leaves to the stem of Pinus sylvestris: Effects of photosynthetic and postphotosynthetic carbon isotope fractionation. Glob. Change Biol. 12, 1922–1939 (2006). [Google Scholar]
- 55.Vogel H., Badapanda C., Knorr E., Vilcinskas A., RNA-sequencing analysis reveals abundant developmental stage-specific and immunity-related genes in the pollen beetle Meligethes aeneus. Insect Mol. Biol. 23, 98–112 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Conesa A., Götz S., Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchanan B. B., Gruissem W., Jones R. L., Biochemistry and molecular biology of plants (John Wiley & Sons Ltd, Chichester, UK, 2nd Ed., 2015). [Google Scholar]
- 58.R Development Core Team , R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna). https://www.r-project.org. (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Transcriptome data have been deposited in the NCBI database under BioProject accession no. PRJNA751264.