Abstract
Congenital heart disease (CHD) is the most common birth defect. Fetal screening ultrasound provides five views of the heart that together can detect 90% of complex CHD, but in practice, sensitivity is as low as 30%. Here, using 107,823 images from 1,326 retrospective echocardiograms and screening ultrasounds from 18- to 24-week fetuses, we trained an ensemble of neural networks to identify recommended cardiac views and distinguish between normal hearts and complex CHD. We also used segmentation models to calculate standard fetal cardiothoracic measurements. In an internal test set of 4,108 fetal surveys (0.9% CHD, >4.4 million images), the model achieved an area under the curve (AUC) of 0.99, 95% sensitivity (95% confidence interval (CI), 84–99%), 96% specificity (95% CI, 95–97%) and 100% negative predictive value in distinguishing normal from abnormal hearts. Model sensitivity was comparable to that of clinicians and remained robust on outside-hospital and lower-quality images. The model’s decisions were based on clinically relevant features. Cardiac measurements correlated with reported measures for normal and abnormal hearts. Applied to guideline-recommended imaging, ensemble learning models could significantly improve detection of fetal CHD, a critical and global diagnostic challenge.
CHD, the most common birth defect1, can be asymptomatic in fetal life but cause substantial morbidity and mortality after birth1–3. Compared to postnatal diagnosis, fetal diagnosis can improve neonatal outcomes and surgical and/or interventional planning4–6 and could enable in utero therapies7,8. Distinguishing normal fetal hearts from complex CHD requiring referral to a fetal cardiologist is therefore a critical and universal need. Low sensitivity in this task can limit palliation options, worsen postnatal outcomes and hamper research on in utero therapies, while low specificity can cause unnecessary additional testing and referrals.
A fetal survey (fetal screening ultrasound) is recommended for every pregnancy worldwide9,10 in the second trimester and generally includes five clinically recommended cardiac views (Fig. 1a) that together could allow clinicians to diagnose up to 90% of complex CHD11,12. In practice, however, detection is often as low as 30%1,13,14, even where ultrasound is universal9,10,14. Specificity is also suboptimal, as low as 40–50%1.
Fig. 1 |. Overview of the ensemble model.
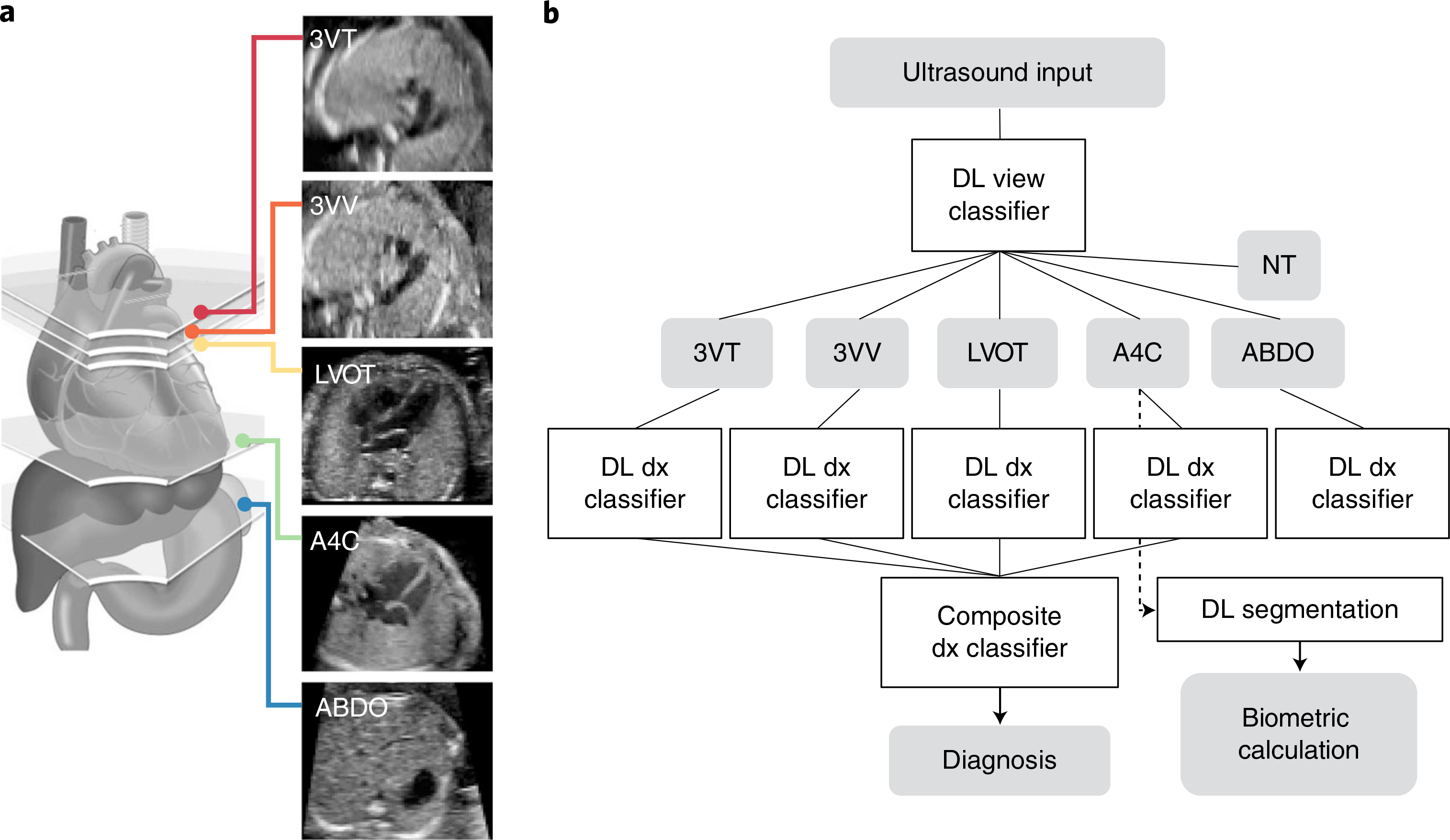
a, Guidelines recommend that the indicated five axial views be used to detect CHD. The illustration was adapted with permission from yagel et. al.45. b, Schematic of the overall model, which is an ensemble of the components shown. From a fetal ultrasound, a DL classifier detects the five screening views (‘DL view classifier’). Subsequent DL classifiers for each view detect whether the view is normal or abnormal (‘DL dx classifiers’). These per-image, per-view classifications are fed into a rule-based classifier (detailed in Extended Data Fig. 1c) to create a composite diagnostic decision as to whether the fetal heart is normal or abnormal (‘composite dx classifier’) (the abdomen view was not included in the composite diagnostic classifier because, clinically, the abdomen view does not commonly contribute to diagnosis; see Methods for further details). A4C views were also passed to a segmentation model to extract fetal cardiac biometrics. NT, non-target; dx, diagnosis.
Two reasons for this gap between possible and commonplace CHD detection are (1) inadequate expertise in interpretation and/or (2) inadequate acquisition of diagnostic-quality images15,16. Causes of inadequate imaging include poor acoustic windows, fetal motion and the small size of the fetal heart. Furthermore, a fetal survey includes thousands of image frames spanning multiple structures per single video ‘sweep’, so the diagnostic frames of interest for CHD may be only a handful and are thus easily missed. Finally, the prevalence of CHD in the population (~0.8–1%) is low enough that non-experts see it only rarely and may discount or overlook image acquisition and interpretation can bridge the diagnosis gap; however, these small, single-center programs are difficult to sustain and scale.19
Deep learning (DL) is a state-of-the-art type of machine learning useful in image analysis20–24. DL has been applied to adult cardiac ultrasound25,26, besting clinicians on view classification on small, downsampled datasets27. DL can be used to classify images or to segment structures within images; several DL models can be used together in an ensemble fashion. We hypothesized that DL could improve ultrasound analysis for CHD.
Results
To test whether DL can improve fetal CHD detection, using multimodal imaging and experts in fetal cardiology, we implemented an ensemble of neural networks (Fig. 1b) to (1) identify the five diagnostic-quality, guideline-recommended cardiac views (Fig. 1a) from among all images in a fetal ultrasound (survey or echocardiogram), (2) use these views to provide classification of normal heart versus any of 16 complex CHD lesions (Table 1) and (3) calculate the cardiothoracic ratio (CTR), cardiac axis (CA) and fractional area change (FAC) for each cardiac chamber (Extended Data Fig. 2).
Table 1 |.
Demographics of training and test sets
| Training set (UCSF) | Holdout test sets (UCSF) | Holdout test set (Boston Children’s Hospital) | Holdout test set (UCSF) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Mixed fetal echocardiograms and fetal surveys | Fetal echocardiograms (FETAL-125) and corresponding surveys (OB-125) | Fetal echocardiograms (BCH-400) | Fetal surveys from population, including OB-125 (OB-4000) | |||||||||
|
|
|
|
|
|||||||||
| No. studies | Maternal age (mean ± s.d. (range)) | Gest. age (mean ± s.d. (range)) | No. studies | Maternal age (mean ± s.d. (range)) | Gest. age (mean ± s.d. (range)) | No. studies | Maternal age (mean ± s.d. (range)) | Gest. age (mean ± s.d. (range)) | No. studies | Maternal age (mean ± s.d. (range)) | Gest. age (mean ± s.d. (range)) | |
|
| ||||||||||||
| Normal | 926 | 35 ± 6 (14–51) | 21 ± 1 (18–24) | 88 | 35 ± 6 (17–47) | 21 ± 1 (18–24) | 32 | 33 ± 4 (22–43) | 22 ± 1 (19–23) | 4,071 | 34 ± 5 (13–52) | 20 ± 1 (18–24) |
| TOF | 83 | 32 ± 7 (16–43) | 21 ± 2 (18–24) | 6 | 37 ± 8 (27–48) | 20 ± 1 (19–20) | 74 | 34 ± 5 (18–47) | 21 ± 2 (18–24) | 6 | 37 ± 8 (27–48) | 20 ± 1 (19–20) |
| HLHS | 110 | 32 ± 6 (16–46) | 21 ± 2 (18–24) | 8 | 33 ± 6 (25–41) | 21 ± 2 (18–23) | 66 | 31 ± 5 (21–42) | 21 ± 2 (18–24) | 8 | 33 ± 6 (25–41) | 21 ± 2 (18–23) |
| Isolated aortic stenosis | 9 | 31 ± 5 (24–36) | 21 ± 2 (19–24) | 1 | - | - | 12 | 34 ± 5 (24–43) | 22 ± 2 (19–24) | 1 | - | - |
| Atrioventricular septal defect | 37 | 35 ± 7 (18–44) | 21 ± 2 (18–24) | 4 | 36 ± 3 (–) | 19 ± 2 (–) | 28 | 32 ± 8 (18–44) | 21 ± 2 (18–24) | 4 | 36 ± 3 (–) | 19 ± 2 (–) |
| d-transposltlon of the great arteries | 17 | 35 ± 4 (28–41) | 21 ± 1 (18–24) | 2 | 29 ± 8 (–) | 22 ± 1 (–) | 17 | 32 ± 6 (18–40) | 22 ± 2 (19–24) | 2 | 29 ± 8 (–) | 22 ± 1 (–) |
| l-transposltlon of the great arteries | 1 | - | - | 1 | - | - | 5 | 31 ± 7 (23–39) | 22 ± 1 (21–24) | 1 | - | - |
| Ebstein's anomaly/ tricuspid regurgitation | 24 | 32 ± 6 (19–40) | 21 ± 2 (18–24) | 3 | 29 ± 2 (–) | 21 ± 3 (–) | 12 | 32 ± 6 (21–39) | 21 ± 2 (18–24) | 3 | 29 ± 2 (–) | 21 ± 3 (–) |
| Single ventricle | 20 | 34 ± 5 (25–42) | 21 ± 2 (18–24) | 2 | 31 ± 4 (–) | 23 ± 1 (–) | 29 | 33 ± 5 (21–46) | 21 ± 2 (18–24) | 2 | 31 ± 4 (–) | 23 ± 1 (–) |
| Left-atrial isomerism | 6 | 29 ± 7 (16–36) | 20 ± 1 (19–21) | 1 | - | - | 9 | 34 ± 3 (31–40) | 21 ± 2 (18–24) | 1 | - | - |
| Right-atrial isomerism | 15 | 32 ± 5 (23–39) | 20 ± 2 (18–23) | 2 | 24 ± 8 (–) | 19 ± 1 (–) | 19 | 30 ± 4 (24–37) | 21 ± 2 (18–24) | 2 | 24 ± 8 (–) | 19 ± 1 (–) |
| Aortic coarctation | 21 | 32 ± 5 (20–42) | 21 ± 2 (18–24) | 1 | - | - | 41 | 33 ± 5 (16–42) | 22 ± 2 (18–24) | 1 | - | - |
| Total anomalous pulmonary venous return | 4 | 33 ± 9 (–) | 20 ± 2 (–) | 1 | - | - | 2 | 31 ± 0 (–) | 20 ± 3 (–) | 1 | - | - |
| Truncus arteriosus | 10 | 28 ± 8 (18–38) | 21 ± 2 (19–24) | 1 | - | - | 11 | 34 ± 5 (26–44) | 20 ± 2 (18–23) | 1 | - | - |
| Tricuspid atresia | 9 | 31 ± 6 (23–38) | 21 ± 2 (18–24) | 1 | - | - | 18 | 32 ± 5 (22–40) | 21 ± 2 (18–24) | 1 | - | - |
| Pulmonary atresia with intact ventricular septum | 18 | 31 ± 6 (19–40) | 21 ± 2 (18–24) | 2 | 29 ± 4 (–) | 20 ± 1 (–) | 19 | 32 ± 5 (25–41) | 21 ± 2 (18–24) | 2 | 29 ± 4 (–) | 20 ± 1 (–) |
| Double-outlet right ventricle | 16 | 32 ± 6 (20–42) | 22 ± 2 (19–24) | 1 | - | - | 29 | 31 ± 5 (19–41) | 21 ± 2 (18–24) | 1 | - | - |
| Total | 1,326 | 34 ± 6 (14–51) | 21 ± 2 (18–24) | 125 | 33 ± 6 (17–48) | 21 ± 2 (18–24) | 423 | 33 ± 5 (16–47) | 21 ± 2 (18–24) | 4,108 | 34 ± 5 (13–52) | 20 ± 1 (18–24) |
In small groups where there is an en dash (–), information was withheld to protect patient privacy. The TWINS-10 dataset is not shown (Methods). Maternal age is shown in years; gestational age is shown in weeks. Gest., gestational.
To train the various components in the ensemble, up to 107,823 images from up to 1,326 studies were used. Five test datasets independent from the training dataset were used for evaluating model performance: (1) FETAL-125, which comprises images from axial sweeps from 125 fetal echocardiograms from the University of California, San Fransisco (UCSF) (30% CHD, 19,822 images); (2) OB-125, which comprises all images from the 125 fetal surveys corresponding to the same patients in FETAL-125 (30% CHD, 329,405 images); (3) OB-4000, which comprises all images from 4,108 fetal surveys with 0.9% CHD, similar to the general population prevalence of 0.8–1% (4,473,852 images; includes OB-125); (4) BCH-400, which comprises images from axial sweeps of 423 fetal echocardiograms highly enriched for CHD from Boston Children’s Hospital (BCH), an external medical center (92% CHD, 44,512 images); and (5) TWINS-10, which includes ten sets of twins. Training and test sets are further described in Table 1, the Methods and Supplementary Table 1. Prediction times per image averaged 3 ms for classification and 50 ms for segmentation on a standard laptop (Methods).
View classification.
Identifying the five views of the heart recommended in fetal CHD screening11 (three-vessel trachea (3VT), three-vessel view (3VV), left-ventricular outflow tract (LVOT), axial four chamber (A4C) and abdomen (ABDO)) was a prerequisite for diagnosis. We therefore trained a convolutional neural network28 (Extended Data Fig. 1a) view classifier (‘DL view classifier’, Fig. 1b) to pick the five screening views from fetal ultrasounds, for which any image that was not one of the five guideline-recommended views was classified as ‘non-target’ (for example, head, foot, placenta). Training data were multimodal, including both fetal echocardiograms, which naturally contain more and higher-quality views of the heart, and fetal surveys, offering a full range of non-target images. Notably, only views of sufficient quality to be used for diagnosis (as deemed by expert labelers, Methods) were used to train the view classifier.
On normal studies in the FETAL-125 test set, the F score (the harmonic mean of precision and recall) for view classification was 0.93, (AUC range, 0.94–0.98, Fig. 2a,b). The network’s classification decision on a particular image is determined by the probability of the image belonging to each of the possible classes; by default, the image is assigned to the class with the highest probability. For fetal view classification, as demonstrated for adults27, the mean probability for correct predictions was significantly higher than that for incorrect predictions (P value, Mann–Whitney U test, <1 × 10−300) (Fig. 2c).
Fig. 2 |. Performance of the view detection step of the ensemble model.
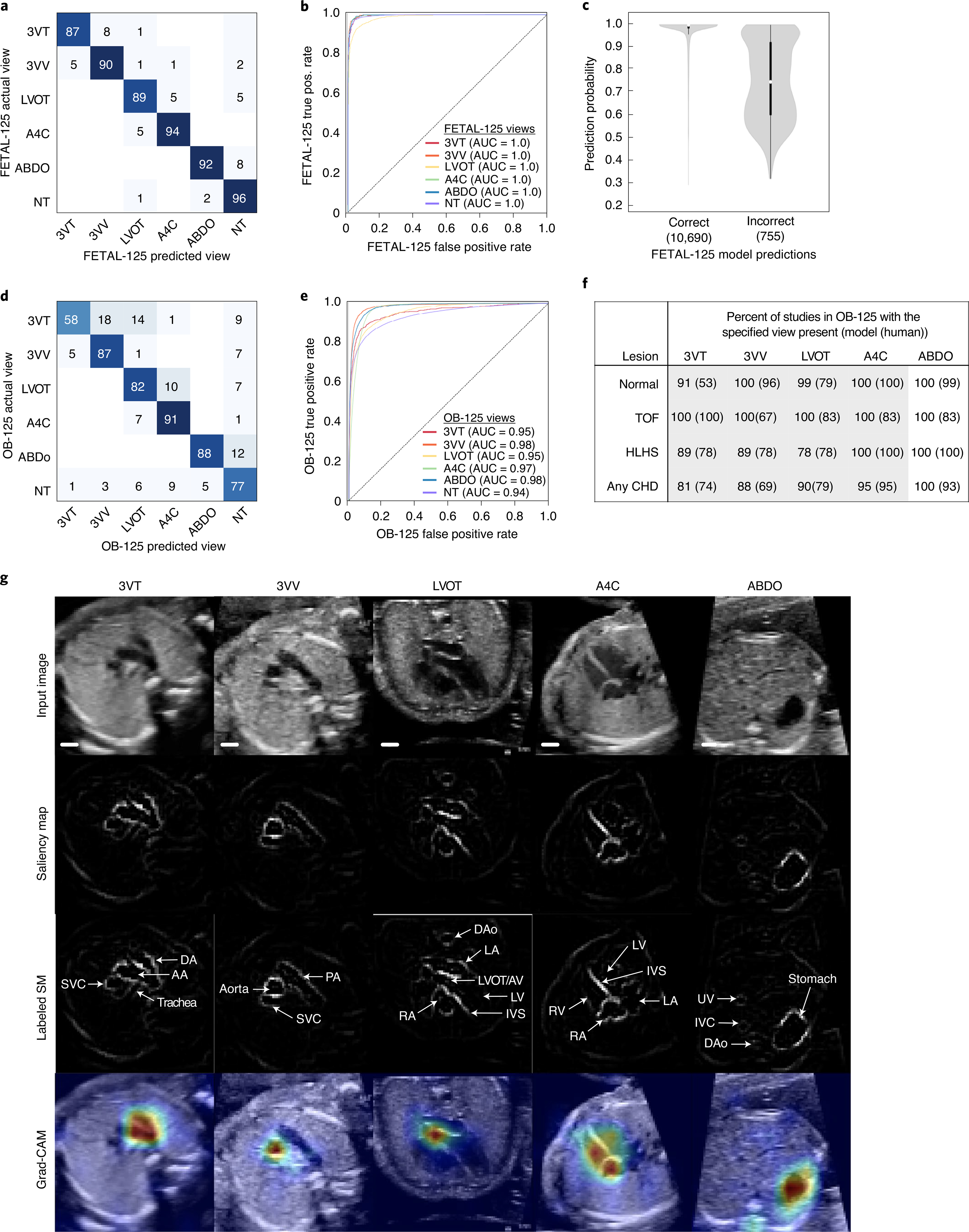
Normalized confusion matrix (a) and ROC curve (b) showing classifier performance on normal hearts from the FETAL-125 test set. Pos., positive. c, Violin plots showing prediction probabilities for this test set, by correctness. In violin plots, white dots signify medians, the thick black line signifies first to third quartiles. Numbers in parentheses below the x axis indicate the number of independent images in each violin plot. For correctly predicted images, the minimum, first quartile, median, third quartile and maximum prediction probabilities are 0.29, 0.98, 1.0, 1.0 and 1.0, respectively. For incorrectly predicted images, the minimum, first quartile, median, third quartile and maximum prediction probabilities are 0.32, 0.60, 0.75, 0.92 and 1.0, respectively. Normalized confusion matrix (d) and ROC curve (e) showing classifier performance on normal hearts from the OB-125 test set. f, Percent of fetal surveys from the OB-125 test set with model-detected views (compared to human-detected views shown in parentheses). Gray shading indicates views with AUC ≥ 75 for normal versus abnormal prediction from Fig. 3a,d. g, One example test image is shown per view (top row), with a corresponding saliency map (unlabeled, second row; labeled, third row). Fourth row, Grad-CAM for the example images. Scale bars indicate 5 mm. SM, saliency map; DA, ductal arch; AA, aortic arch; SVC, superior vena cava; PA, pulmonary artery; TV, tricuspid valve; AV, aortic valve; MV, mitral valve; IVS, interventricular septum; IAS, interatrial septum (foramen ovale); RA, right atrium; RV, right ventricle; LA, left atrium; DAo, descending aorta; LV, left ventricle; UV, umbilical vein; IVC, inferior vena cava.
We then tested the view classifier on OB-125 (Fig. 2d,e). When diagnostic-quality target views were present, the view classifier found them with 90% sensitivity (95% CI, 90%) and 78% specificity (95% CI, 77–78%). Using only images with prediction probabilities at or above the first quartile, sensitivity and specificity increased to 96% and 92% (95% CI, 96% and 92–93%). Recommended views were not always present in each fetal survey and were more commonly present in normal studies (Fig. 2f). The view classifier’s greatest confusion was between 3VT and 3VV (Fig. 2d), adjacent views that often also cause clinical uncertainty11,16,29.
To validate that the view classifier used clinically relevant features, we performed both saliency mapping and gradient-weighted class activation mapping (Grad-CAM) experiments27,30 on test images to show the pixels (saliency mapping) or region (Grad-CAM) most important to the classifier in making its decision. Both experiments show that the view classifier makes its decisions based on clinically relevant image features (Fig. 2g).
Classification of normal versus complex CHD.
We trained the same convolutional neural network architecture used above to classify normal hearts versus CHD for each of the five view classes (binary ‘DL dx classifiers,’ Fig. 1b). On FETAL-125, the AUC ranged from 0.72 (ABDO) to 0.88 (3VV and A4C; Fig. 3a). Across all test datasets, AUCs for the ABDO view reflected the clinical finding that the abdomen view is the least useful for CHD diagnosis. For each heart, we arrived at a composite diagnostic decision of normal heart versus CHD by applying a rule-based classifier (‘composite dx classifier’ Fig. 1b) to the per-image, per-view predictions that is essentially a weighted average (Methods and Extended Data Fig. 1c).
Fig. 3 |. Performance of the diagnostic steps of the ensemble model.
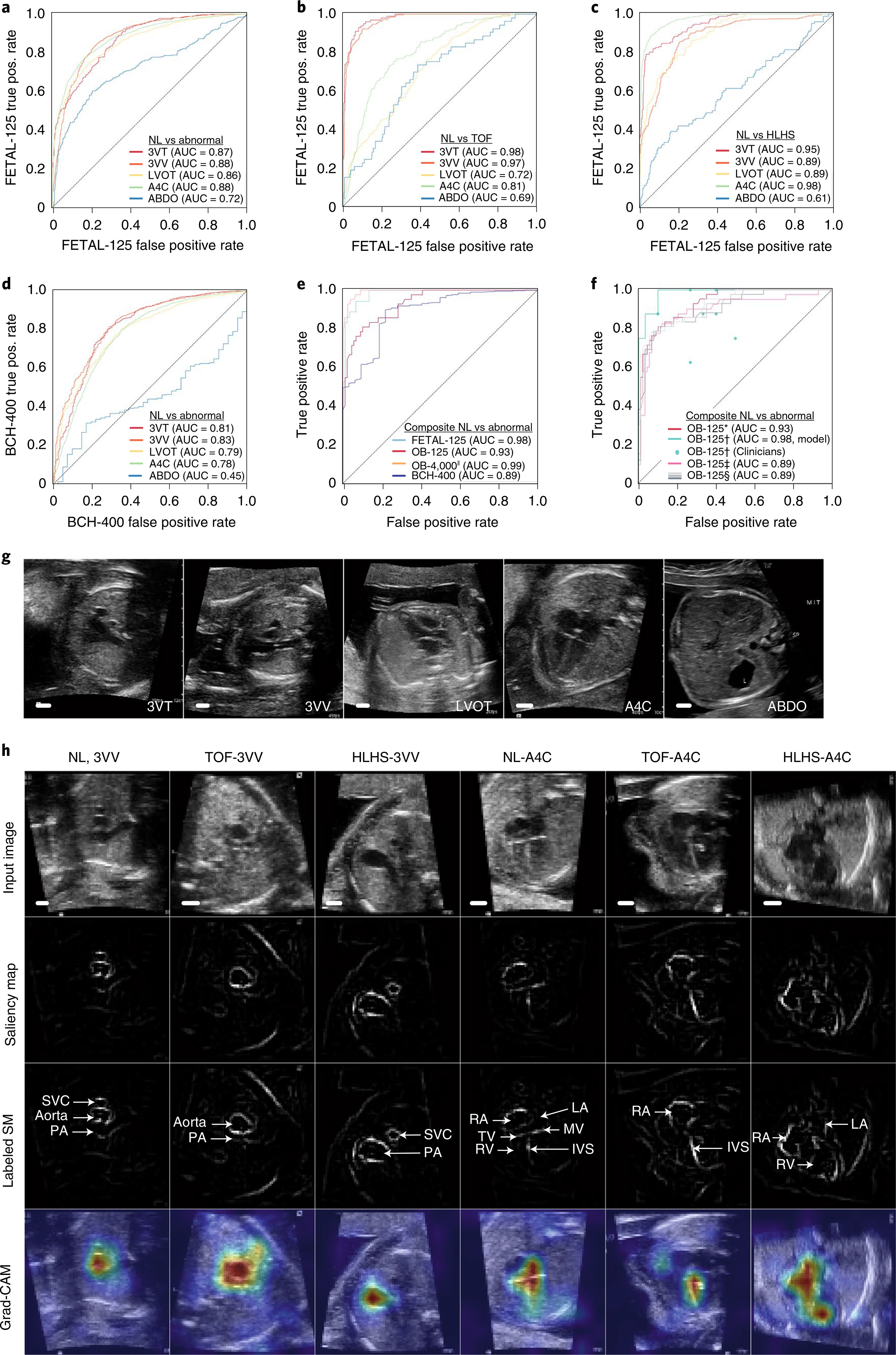
ROC curves showing the model’s ability to distinguish normal hearts versus any CHD lesion mentioned in Table 1 (a), normal heart (NL) versus TOF (b) and NL versus HLHS (c) for each of the five views in the FETAL-125 test dataset. d, ROC curve for prediction of per-view normal versus abnormal hearts from external data (BCH-400 test set). e, ROC curves for composite (per-heart) prediction of normal versus abnormal hearts for each of the four test datasets. ‘OB-4000ll’ indicates the high-confidence target images from the OB-4000 test set (images with view-prediction probability at or above the first quartile). f, ROC curve for composite (per-heart) prediction of normal heart versus CHD for different testing scenarios for OB-125. OB-125*, all possible images present. OB-125†, only five images present, one image per view (teal line is model performance; teal dots denote clinician performance). OB-125‡, low-quality images. OB-125§, 6.5% of views scrambled to simulate error in view classification (average of three replicates). g, Example of images given to both the model and clinicians for determination of normal versus abnormal hearts in a head-to-head comparison. h, Top row, one example test image is shown for normal heart, TOF and HLHS; 3VV and A4C views are shown. Second row, corresponding unlabeled saliency map. Third row, labeled saliency map. Fourth row, Grad-CAM provides a heatmap of regions of the image most important to the model in prediction. In 3VV, the relative sizes of the aorta and pulmonary artery distinguish these lesions from normal hearts; and in A4C, the angled intraventricular septum and enlarged right heart distinguish TOF and HLHS, respectively, from normal hearts. Scale bars indicate 5 mm.
Using this approach, we achieved AUCs of 0.98, 0.93, 0.99 and 0.89 in distinguishing normal from abnormal hearts on FETAL-125, OB-125, OB-4000 and BCH-400, respectively (Fig. 3e) (to achieve this AUC for OB-4000, only images with view-prediction probabilities above the first quartile were used in the composite diagnostic classifier). This allowed a sensitivity of 95% (95% CI, 83–99%), a specificity of 96% (95% CI, 95–97%), a positive predictive value of 20% (95% CI, 17–23%) and a negative predictive value of 100% in OB-4000. Performance on these and additional testing scenarios discussed below are summarized in Fig. 3f and Supplementary Table 2; notably, we compared testing scenarios for OB-125, where all images (regardless of view-prediction probability) are used for diagnosis, where only true positive views are used, where only images with view-prediction probabilities above the first quartile are used and where a portion of true positive views are purposefully scrambled to mimic view misclassification. Overall, model sensitivity on fetal echocardiograms rivaled that cited across several papers31–33 (P value, 0.3, assuming normal distribution of measures in the literature). More importantly, model sensitivity and specificity on fetal surveys were significantly better than reported performance1,13,14,32 (P values, 0.002 and 0.04, respectively).
While the clinician’s interpretation of the entire ultrasound study served as our gold-standard label for testing and training, we also wished to compare model performance on fetal surveys (OB-125) directly against that of clinicians on a task-for-task basis by giving each the following test: one full-resolution image per view, with only five images in total per heart (Fig. 3g). This test was chosen both to make the task feasible for humans and, given the potential variation in image acquisition protocols across different institutions, to simulate a ‘lean protocol’ in which only minimal recommended views are acquired. Thirty-eight of the 125 fetal surveys (30%) in OB-125 contained all five views. On this test, the model achieved 88% sensitivity (95% CI, 47–100%) and 90% specificity (95% CI, 73–98%). Clinicians (n = 7) achieved an average sensitivity of 86% (95% CI, 82–90%) and a specificity of 68% (95% CI, 64–72%) (Fig. 3f). The model was comparable to clinicians (P = 0.3) in sensitivity and superior to them (P = 0.04) in specificity.
To validate that the model generalizes beyond the medical center where it was trained34, we tested it on fetal echocardiograms from an unaffiliated, geographically remote medical center with a high prevalence of CHD (BCH-400; Table 1). AUCs for view detection on normal hearts ranged from 0.95 to 0.99 (not shown). The AUC for composite classification of normal versus abnormal hearts was 0.89, despite a high prevalence of abnormal hearts in this test set (Fig. 3e and Supplementary Table 2).
Multifetal pregnancies have a higher risk of CHD than the general population1. Therefore, a CHD detection model applicable to ultrasounds of twins and other multiple pregnancies would be useful. Based on saliency mapping and Grad-CAM experiments (Figs. 2g and 3h), we hypothesized that our model could perform adequately on surveys of twins. We used our model to predict views and diagnoses for ten sets of twins (TWINS-10 test set; n = 20 fetuses), including those with tetralogy of Fallot (TOF) and hypoplastic left heart syndrome (HLHS). Sensitivity and specificity were 100% and 72% (Supplementary Table 2).
Models should be robust to minor variation in image quality to be useful for a range of patients and medical centers. We therefore assessed model performance on images within OB-125 that expert clinicians did not label as high-quality views but that the model did classify as target views (Fig. 2d,f). We inspected these ‘false positive’ images directly and analyzed their prediction probabilities. Of images with probability ≥0.9, two-thirds (66%) were in fact target views but of lower quality (for example, slightly off axis, heavily shadowed) than ones chosen by experts, and most (59%) of these low-quality target views had probabilities ≥0.9 (Extended Data Fig. 3). Therefore, the model can appropriately detect target views of lower quality. We submitted these lower-quality target images for diagnostic prediction and found a sensitivity of 95% (95% CI, 83–99%) and a specificity of 39% (95% CI, 28–50%). Thus, the ensemble model can make use of suboptimal images in fetal surveys to detect complex CHD, albeit with lower specificity.
As with view classification above, we performed several analyses to determine whether the diagnostic classifications were based on clinically relevant image features. We trained a set of per-view binary classifiers for each of the two most common lesions in our dataset (TOF and HLHS) and examined receiver operating characteristic (ROC) curves, saliency maps and Grad-CAM experiments. For TOF, AUCs were highest for the two views from which TOF is most easily clinically appreciable: 3VT and 3VV (Fig. 3b). For HLHS, 3VT, 3VV, LVOT and A4C are all abnormal, consistent with higher AUC values in Fig. 3c. Saliency mapping and Grad-CAM highlighted pixels and image regions relevant to distinguishing these lesions from normal hearts (Fig. 3h). In clinical practice, reported sensitivity in detecting TOF and HLHS is as low as 50% and 30%, respectively35. With our model, sensitivity is 71% for TOF and 89% for HLHS (specificities of 89% and 92%; Supplementary Table 2). Examples of diagnostic misclassifications can be found in Extended Data Fig. 4.
Segmentation for fetal biometrics.
Biometric measurements aid in fetal CHD screening and diagnosis11. We therefore trained a modified U-Net36 (Extended Data Fig. 1b and Methods) to find cardiothoracic structures in A4C images and used these segmented structures to calculate the CTR, CA and FAC for each cardiac chamber (Table 2 and Fig. 4). Normal, TOF and HLHS hearts from UCSF were represented in training and testing.
Table 2 |.
Fetal biometrics calculated from pixel-level segmentation of anatomic structures
| CTR | CA (°) | FAC | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||||||||
| Left ventricle | Right ventricle | Left atrium | Right atrium | |||||||||||||||
|
|
|
|
|
|
|
|||||||||||||
| Label | Prediction | P valuea | Label | Prediction | P valuea | Label | Prediction | P valuea | Label | Prediction | P valuea | Label | Prediction | P valuea | Label | Prediction | P valuea | |
|
| ||||||||||||||||||
| Normal | 0.53 ± 0.03 | 0.52 ± 0.03 | 0.2 | 46 ± 6 | 38 ± 6 | 0.003 | 0.47 ± 0.10 | 0.44 ± 0.06 | 0.3 | 0.47 ± 0.11 | 0.52 ± 0.13 | 0.2 | 0.52 ± 0.13 | 0.55 ± 0.12 | 0.5 | 0.49 ± 0.08 | 0.48 ± 0.10 | 0.5 |
| TOF | 0.53 ± 0.07 | 0.50 ± 0.04 | 0.3 | 65 ± 14 | 63 ± 16 | 0.5 | 0.53 ± 0.29 | 0.66 ± 0.21 | 0.5 | 0.42 ± 0.15 | 0.30 ± 0.26 | 0.3 | 0.30 ± 26 | 0.69 ± 0.22 | 0.1 | 0.42 ± 0.09 | 0.67 ± 0.28 | 0.1 |
| HLHS | 0.55 ± 0.07 | 0.52 ± 0.08 | 0.2 | 59 ± 18 | 49 ± 6 | 0.2 | 0.37 ± 0.21 | 0.75 ± 0.09 | 0.03 | 0.35 ± 0.10 | 0.36 ± 0.10 | 0.06 | 0.36 ± 0.10 | 0.68 ± 0.12 | 0.03 | 0.41 ± 0.08 | 0.55 ± 0.14 | 0.1 |
| P valueb | 0.5 | 0.5 | 0.04 | 0.007 | 0.7 | 0.02 | 0.2 | 0.08 | 0.09 | 0.2 | 0.2 | 0.3 | ||||||
Mann-Whitney Utest.
Kruskal-Wallistest
Fig. 4 |. Analysis of fetal cardiac structure and function measurements based on segmentation provided by the ensemble model.
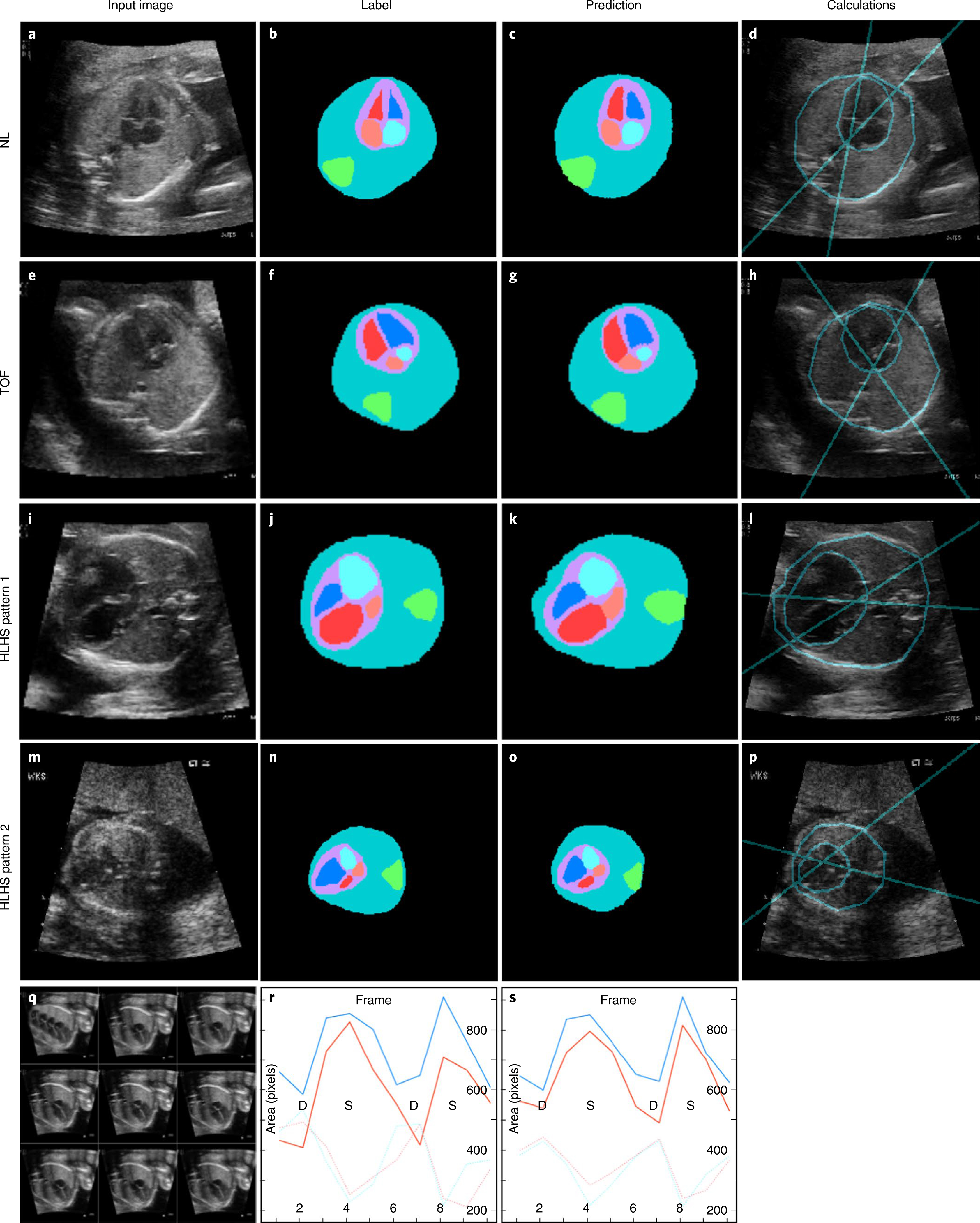
a,s Example input image, ground-truth label of anatomic structures, prediction of anatomic structures and calculations of the CTR and CA for a normal heart (a–d), TOF (e–h) and HLHS (i–p). Segmentation of an image series (q) allows plots of chamber area over time (label, r; prediction, s) and identification of image frames in ventricular systole (S) and diastole (D) for FAC calculation. Scale bars indicate 5 mm. Teal, thorax; green, spine; purple, heart; red, left ventricle; pink, left atrium; blue, right ventricle; light blue, right atrium.
Per-class Jaccard similarities measuring overlap of labeled and predicted segmentations are found in Supplementary Table 3. Predictably, Jaccard values were higher for more highly represented pixel classes (for example, background) and were similar to intra-labeler Jaccard values (range, 0.53–0.98; mean, 0.76). Example labels and predictions for segmented structures are shown in Fig. 4.
Normal cardiothoracic circumference ratios range from 0.5 to 0.6 (ref. 1). Mann–Whitney U testing showed no statistical differences among clinically measured and labeled CTRs for normal hearts, nor between labeled and model-predicted CTRs. CTRs for TOF and HLHS hearts were normal, as previously reported1.
A normal CA is 45° ± 20° (ref. 11). Consistent with the literature37, the mean CA value was increased in TOF at 63° ± 16° (range, 54–80°; P value, 0.007). CA values for HLHS were not found in the literature, but the model-predicted CA was 49° ± 2° (range, 33–72°; P value, 0.04).
In addition to the five still-image views, it is best practice to also obtain a video of the A4C view to assess cardiac function1. The FAC quantifies this assessment. From a study measuring 70 normal 18- to 24-week fetuses, the 50th percentile for left- and right-ventricular FAC averaged 0.34 ± 0.01 and 0.33 ± 0.02, respectively38. In our test dataset, the labeled FAC values for normal left ventricle and right ventricle were 0.47 ± 0.10 and 0.47 ± 0.11, respectively, and model predictions were 0.44 ± 0.06 (P value, 0.3) and 0.52 ± 0.13 (P value, 0.2), respectively. Although there are no fetal atrial FAC values established in the literature, model-predicted left-atrium and right-atrium FAC values were statistically indistinguishable from labels at 0.52 ± 0.12 and 0.48 ± 0.10, respectively (P values, 0.5 and 0.5). All measurements are summarized in Table 2 and Extended Data Fig. 2.
In sum, the data show that fetal cardiothoracic biometrics can be derived from image segmentation, showing good agreement between previously reported values and the potential to provide additional metrics not yet benchmarked.
Discussion
With clear benefit to early diagnosis and treatment of CHD and growing research on in utero interventions, the need for accurate, scalable fetal screening for CHD has never been stronger39, while sensitivity and specificity for CHD detection are quite variable at centers and clinics worldwide and in many centers remain quite low1. To address this, we investigated the impact of combining real-world fetal ultrasounds and trusted clinical guidelines with cutting-edge DL to achieve expert-level CHD detection from fetal surveys, one of the most difficult diagnostic challenges in ultrasound. In over 4,000 fetal surveys (over 4 million images), the ensemble model achieved an AUC of 0.99.
DL has been used on various medical tasks21,23,40, but to our knowledge this is the first use of deep learning to approximately double reported community-level sensitivity and specificity on a global diagnostic challenge in a test set of real fetal screening ultrasounds with a CHD prevalence similar to the general population (OB-4000).
The model’s performance and speed allow its integration into clinical practice as software onboard ultrasound machines to improve real-time acquisition and to facilitate telehealth approaches to prenatal care41. As a key benefit, the view classifier could be used on its own to help ensure adequate view acquisition. For retrospectively collected images, the model could be used as standalone software for which a user uploads a study and receives model-chosen views and diagnostic predictions.
To ensure that our model could work robustly in real-world settings, we used two-dimensional ultrasound and standard recommended fetal views rather than specialized or vendor-specific image acquisitions42,43. Furthermore, we tested our model in a range of different scenarios and on different independent test datasets. Importantly, the model maintained high sensitivity on external imaging, suboptimal imaging and imaging from fetal surveys and fetal echocardiograms, datasets with community-level CHD prevalence and those with high CHD prevalence. When a test dataset approximately 10% of the size of the training dataset has arisen as an informal rule of thumb for adequate testing in the data science community, we tested on over 350% of the number of studies in the training set and over 4,000% the number of images. While UCSF is a referral center for fetal CHD, it also provides community care for obstetrics, and so the fetal screening ultrasounds in OB-4000 represent a cross-section of the community with an expected incidence of CHD that mirrors that of the population at large.
Our approach to both model design and testing ensured interpretability at several levels, which can help with clinical adoption. Choosing to use an ensemble of classifiers (first a view detector and then per-view diagnostic classifiers and finally a classifier for composite diagnosis) allowed us to incorporate clinical view recommendations into our model and to demonstrate that model performance per view and per CHD lesion were consistent with clinical knowledge about which views were most likely to aid in detection of specific lesions.
Analysis of confusion matrices, ROC curves and incorrectly classified images helped determine that model error mirrored uncertainties in clinical practice. Saliency mapping and Grad-CAM for both view and diagnostic classifications demonstrated that model predictions relied on cardiac structures. The prominence of the aorta, the right heart and the stomach as distinguishing features among the five target views is both new and makes sense. A comparison of the different testing scenarios and visualization of per-image misclassifications suggests that both the quality of images and the number of available images per study contribute to the best overall performance.
As mentioned above, we incorporated two similar study types, fetal echocardiograms and fetal surveys, in a multimodal approach to model training that harnessed more specialized imaging in service of improving performance on screening imaging. By feeding only target views into the diagnostic classifier step, we took a more data-efficient approach to the diagnostic classifier compared to using the entire ultrasound. We also took a new approach to addressing variation in image quality that relied on human experts to agree only on labeling diagnostic-quality images for training (in testing, the model analyzed all images). This approach economized on human labor, consolidating inter-expert agreement on diagnostic-quality images, while providing fewer constraints to the model training, as some aspects that make an image low in quality to a human eye may not matter as much to a computer ‘eye’ (image contrast is a good example of this). We found that prediction probability was an indirect representation of the model’s quality assessment and that using cutoffs for high-prediction-probability images improved model performance.
While it is the most common birth defect, CHD is still relatively rare. Moreover, unlike modalities such as photographs21,23, electrocardiograms40 or chest X-rays, each ultrasound study contains thousands of image frames. Therefore, designing a model that could work on a large number of non-independent images from a dataset with relatively few individuals was an important challenge to overcome. In sum, the strengths above allowed us to find diagnostic signals for rare diseases and allowed computational efficiency both in training and in subsequent predictions on new data, which is key to translating this work toward real-world and resource-poor settings where it is needed44.
While 4,108 fetal surveys is a substantial test set, especially when considering the size of each ultrasound exam, hundreds of millions of fetal surveys are performed annually at many thousands of medical centers and clinics worldwide. Furthermore, while the OB-4000 test set represented community fetal screening ultrasounds, they were still drawn from a center with experience in fetal screening. Therefore, expanded testing of the model prospectively on consecutive cases and in multiple centers, including rural community and/or non-expert centers and including more multiple gestations, will be important moving forward. It will also be important to test the model on imaging that includes a range of non-cardiac malformations. Several small improvements in model algorithms, as well as more training data from more centers, may further boost performance and may allow for diagnosis of specific lesion types. Similarly, more training data for image segmentation, including segmenting additional CHD lesions, will improve segmentation model performance and allow those results to be integrated into the composite diagnostic classifier. Further clinical validation of segmentation-derived fetal biometrics will be needed, particularly when metrics on particular CHD lesions have not yet been described elsewhere.
We look forward to testing and refining ensemble learning models in larger populations in an effort to democratize the expertise of fetal cardiology experts to providers and patients worldwide and to applying similar techniques to other diagnostic challenges in medical imaging.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-021-01342-5.
Methods
The methods below describe datasets, data processing, algorithms and performance measures46.
Datasets.
All datasets were obtained retrospectively and de-identified, with waived consent in compliance with the Institutional Review Board at the UCSF and the Institutional Review Board at Boston Children’s Hospital.
Inclusion, exclusion and definitions of normal heart and CHD.
Fetal echocardiograms (dedicated ultrasounds of the fetal heart, performed by fetal cardiologists and sonographers with specific expertise in fetal cardiology)19 and fetal surveys (second-trimester obstetric anatomy scans performed by sonographers, radiologists, obstetricians and/or maternal–fetal-medicine physicians)11 performed between 2000 and 2019 were used. Images came from GE (67%), Siemens (27%), Philips (5%) and Hitachi (<1%) ultrasound machines. Inclusion criteria were fetuses of 18–24 weeks of gestational age. Fetuses with clinically significant non-cardiac malformations (for example, congenital diaphragmatic hernia, congenital airway malformation, congenital cystic adenomatoid malformation, meningomyelocele) were excluded. Gold-standard definitions of normal heart versus CHD were made as follows. CHD pathology was determined by review of the clinical report as well as visual verification of the CHD lesion for each ultrasound by clinician experts (A.J.M.-G., J.C.L. and Y.Z., with over 60 years of combined experience in fetal cardiology). Additionally, for studies performed in or after 2012, we were able to validate the presence, absence and type of cardiac findings in the ultrasound studies with electronic health-record codes for CHD in the resulting neonates (ICD-9 codes 745*, 746* and 747* and ICD-10 code Q2* and ICD procedure codes 02*, 35*, 36*, 37* 38*, 88* and 89*). Studies for which clinician experts did not agree on the lesion and for which no postnatal diagnosis was present were not included. Normal fetal hearts were defined as negative for structural heart disease, fetal arrhythmia, maternal diabetes, maternal lupus, maternal Sjögren syndrome or the presence or history of abnormal nuchal translucency measurement, non-cardiac congenital malformations or CHD as defined above. Abnormal fetal studies had any of the following lesions: TOF, pulmonary atresia with ventricular septal defect (VSD) or double-outlet right ventricle with VSD committed to aorta (TOF); critical aortic stenosis and HLHS, including critical aortic stenosis with dilated left ventricle (HLHS); isolated aortic stenosis; atrioventricular septal defect; d-transposition of the great arteries; l-transposition of the great arteries; single ventricle, including heterotaxy with left or right atrial isomerism; double-outlet right ventricle with uncommitted, doubly committed or subpulmonary VSD (double-outlet right ventricle); aortic coarctation; total anomalous pulmonary venous return; truncus arteriosus; Ebstein’s anomaly (tricuspid dysplasia); tricuspid atresia; and pulmonary atresia with intact interventricular septum. Isolated VSDs were not included as they only rarely require perinatal intervention.
Study design and training and test sets.
Overview: source data from the UCSF were split into the training set (a mix of fetal echocardiograms and fetal screening ultrasounds) and the FETAL-125 test set of fetal echocardiograms (first test set). A second test set, OB-125, was obtained from UCSF fetal screening ultrasounds corresponding to those echocardiograms from FETAL-125. Additional normal fetal screening ultrasounds were added to OB-125 to create a third test set, OB-4000. Next, fetal echocardiograms were obtained from Boston Children’s Hospital to make the fourth test set, BCH-400. A fifth test set comprised ten sets of twins (TWINS-10). Training and test sets did not overlap by image, patient or study. Among the test datasets, FETAL-125 and OB-125 test sets represent fetal echocardiograms and fetal screening ultrasounds from the same patients by design, and OB-125 is part of OB-4000. This information is summarized in Supplementary Table 1; details on these datasets follow below.
UCSF source data for the training set for all models and for FETAL-125 (first test set).
Our source data began with all UCSF fetal echocardiograms fitting the inclusion and exclusion criteria above for CHD (n = 437 studies). We then added a random sample of normal UCSF fetal echocardiograms (n = 875 studies), such that CHD studies comprised approximately 30% of individuals. This was carried out to maintain relatively balanced classes of CHD versus normal-heart studies. We used the axial video and still-image clips from the fetal echocardiograms. We also included all images from 139 randomly chosen normal fetal screening ultrasound studies to include a range of non-target images found in screening. Together, these data made up the UCSF source data for training and initial testing (first test set).
From this source dataset, we used the 139 fetal screening ultrasounds, 787 normal fetal echocardiograms and 400 CHD echocardiograms for training (n = 1,326 studies in total; 69,841 images from normal studies and 102,974 images from abnormal studies; Table 1). The remaining 88 normal echocardiograms and 37 CHD echocardiograms made up the first test set, FETAL-125 (n = 125; 11,445 normal images and 8,377 abnormal images).
OB-125 test set (second test set).
We obtained the corresponding fetal screening ultrasounds from the same patients in FETAL-125 to create the OB-125 test set. All image frames from each study were used (220,990 normal images and 108,415 abnormal images).
OB-4000 test set (third test set).
To create a UCSF testing set of fetal screening ultrasounds with a CHD prevalence similar to that of the standard population, we started with OB-125 and added an additional 3,983 normal fetal surveys, such that the CHD lesions in OB-125 comprised 0.9% of an overall dataset totaling 4,108 surveys. The result was OB-4000 (4,473,852 images; 4,365,437 normal and 108,415 abnormal).
BCH-400 test set (fourth test set).
As an external testing set, we received 423 fetal echocardiograms (4,389 images from 32 studies of normal hearts and 40,123 images from 391 studies of abnormal hearts) from Boston Children’s Hospital.
TWINS-10 test set (fifth test set).
Separately, we obtained a test set of ten twin ultrasounds between 18–24 weeks of gestational age (5,754 echocardiogram images). Eight sets of twins had normal hearts; one set of twins had one normal and one TOF heart; and one set of twins had one normal and one HLHS heart.
Use of imaging from the training set to train different models in the ensemble.
Images from the overall training dataset above were used to train (1) a view classifier, (2) normal versus abnormal diagnostic classifiers for each target view and (3) a segmentation model. For all trainings, roughly equal proportions of data classes were used. For the view classifier, 53,532 images from the 926 normal hearts were used. For the per-view diagnostic classifiers, 46,498 of the above images from 916 normal hearts were combined with an additional 54,291 images from 400 abnormal hearts (for a total of 1,316 studies and 100,789 images) (ten of the studies used to train the view classifier only had non-target views and so were not used to train the diagnostic classifiers). For segmentation of cardiac chambers, 1,248 A4C images from 186 studies (122 normal, 25 HLHS, 39 TOF) were used. For segmentation of the heart and thorax, 952 A4C images from 223 studies (157 normal, 25 HLHS, 41 TOF) were used.
Image labeling by clinicians.
Every image frame of the training set, FETAL-125, OB-125 and BCH-400 were view labeled by clinician experts (for the training data, images for which clinician experts did not agree on the view were excluded from training. For test sets, a portion of the dataset was independently scored by both labelers to ensure agreement (Extended Data Fig. 5). Because OB-4000 was too large for this approach, experts instead only verified that the top five predicted views for each ultrasound study did in fact contain views of interest (and were normal hearts) before that study underwent diagnostic classification. For view labeling, as per clinical guidelines, 3VT, 3VV, A4C and abdomen views were from axial sections of the fetal thorax, while the LVOT view included both axial and sagittal LVOT images. For segmentation labeling, clinicians manually traced cardiothoracic structures (thorax, heart, spine and each of the four cardiac chambers) on A4C images.
Data processing.
DICOM-formatted images were de-identified and converted to grayscale as previously described27. Axial sweeps of the thorax were split into constituent frames at a resolution of 300 by 400 pixels. For view classification tasks, images were labeled as 3VT, 3VV, LVOT, A4C and ABDO. A sixth category, called non-target, comprised any fetal image that was not one of the five cardiac views of interest. For disease-classification tasks, studies were labeled to correspond to normal hearts or CHD lesions as mentioned above.
For input into classification networks, each image was cropped to 240 × 240 pixels centered on the ultrasound window and downsampled to 80 × 80 pixels and scaled with respect to grayscale value (rescale intensity). For input into segmentation networks, images were cropped to 272 × 272 pixels centered on the ultrasound window and scaled with respect to grayscale value. All preprocessing steps made use of open-source Python libraries OpenCV (https://opencv.org/), scikit-image (https://scikit-image.org/) and NumPy (https://numpy.org). For training fetal structural and functional measurements, OpenCV was used to label the thorax, heart, right atrium, right ventricle, left atrium, left ventricle and spine from A4C images.
Model architecture and training parameters.
Classification models.
Classification models were based on the ResNet architecture28 with the following modifications. For view classification, the batch size was 32 samples, and training was over 175 epochs using the Adam optimizer and an adaptive learning rate (0.0005 for epochs 1–99; 0.0001 for epochs 100–149 and 0.00005 at 150+ epochs). Dropout of 50% was applied before the final fully connected layer. Data were augmented at run time by randomly applying rotations of up to 10°, Gaussian blur, width and height shifts of up to 20% of the total length, zooms of up to 50%, rescaling image intensity between the second and 98th percentiles and vertical and/or horizontal flips. For diagnostic classification, transfer learning was applied to the previously described view classification model as follows: the first 18 layers were frozen. Additional training used the above settings, except that epochs ranged from 12 to 60, learning rate was constant for each model, no adaptive learning was used, and the learning rate ranged from 0.00001 to 0.0001. The loss function was categorical cross-entropy (view classifier) or binary cross-entropy (diagnostic classifiers). The classification network architecture is shown in Extended Data Fig. 1a. During model optimization, training data were subdivided (by study) into training and validation sets using threefold cross-validation. Training and validation datasets in which view labels were randomized were used as a negative control, resulting in an F score commensurate with random chance among classes.
Segmentation model.
A4C images with clinician-labeled cardiothoracic structures (thorax, heart, spine and each of the four cardiac chambers) were used as training inputs to a U-Net36 neural network architecture with modifications as in Extended Data Fig. 1b. Two different models were trained to detect (1) the heart, spine and thorax and (2) the four cardiac chambers. Batch size was two, models were trained for 300–500 epochs, and an Adam optimizer was used with adaptive learning rates of 0.0001 to 0.00001. For data augmentation, width (shift) was set at 20%, zoom was 15%, random rotations of up to 25° were permitted, and horizontal and/or vertical flips were used. The loss function was categorical cross-entropy.
Framework and training and prediction times.
All models were implemented in Python using Keras (https://keras.io/, GitHub,2015 ) with TensorFlow (https://www.tensorflow.org/) backend. Trainings were performed on Amazon’s EC2 platform with a GPU instance ‘p2.xlarge’ and lasted about 1.95–5 h for segmentation models and between 6 min and 4.6 h for classification models. Prediction times per image averaged 3 ms for classification and 50 ms for segmentation on a standard laptop (2.6-GHz Intel core, 16 GB of RAM).
Probabilities and calculations.
Use of prediction probabilities in classification.
For each classification decision on a given image, the model calculates a probability of the image belonging to each of the possible output classes; as a default, the image is automatically assigned to the class with the highest probability. In certain testing scenarios, a threshold of acceptable prediction probability was applied to view classifications as follows. For OB-4000 ‘high-confidence’ views, diagnostic classification was performed only on images with view-prediction probabilities greater than the first quartile of correctly predicted views from the FETAL-125 test set. For OB-125 ‘low-quality’ views, model-predicted views that human labelers did not choose as diagnostic quality were used (Results and Supplementary Table 2). A probability threshold for diagnostic classifications was also used in the rule-based composite diagnostic classifier, described below.
Composite diagnostic classification.
A rule-based classifier (‘composite dx classifier,’ Fig. 1b) was developed to unite per-view, per-image predictions into a single composite decision of normal heart versus CHD. The rule-based composite diagnostic classifier sums prediction probabilities for CHD and for normal heart, across images within a view and then across views, as follows (Extended Data Fig. 1c).
Only views with AUC > 0.85 on validation data were used. For each of the cardiac views of interest, a variable number of images each held a probability PCHD of CHD for each image; the probability of normal heart for each image was also recorded, where PNL = 1 − PCHD. A high-pass threshold was determined from validation data for three of the four views (3VT, 3VV, LVOT; A4C was excluded from this due to high performance during cross-validation). The thresholds were an averaged Youden’s J statistic, calculated across the threefold cross-validation for each of these three per-view models. PCHD values below the high-pass threshold were reset to 0 to avoid overscoring CHD.
The resulting PNL and PCHD values for each view were then summed separately, to maintain a distinction between a view being present and normal versus being missing from a study, and each sum was normalized by the total sum of all predictions to account for different numbers of images in each view class to obtain the view-specific prediction values.
Note that ‘view’ is either 3VT, 3VV, LVOT or A4C. These prediction values for each view were in turn summed for a composite classification. Evaluating true positives, false positives, true negatives and false negatives with different values for the offset number allowed construction of an ROC curve (Fig. 3e).
Quantification of cardiothoracic ratio, chamber fractional area change and cardiac axis.
The CTR was measured as the ratio of the heart circumference to the thorax circumference. FAC for each of the four cardiac chambers was calculated as (maximum pixel area − minimum pixel area)(maximum pixel area)−1. CA was calculated as the angle between a line centered on the spine and thorax and a line centered on either the left chambers or the right chambers, whichever side had the greatest area (the line centered on the cardiac chambers was chosen as a computational method of finding a line parallel to the intraventricular septum, used clinically and in ground-truth labeling). Various checks were implemented to prevent calculation of clinical values from images with poor segmentation results. Concordance of predicted quantitative measurements were compared to ground-truth measures (labeled images and clinical measurements when available) using the Mann–Whitney U test. Measurements among normal, TOF and HLHS groups were compared using the Kruskal–Wallis test.
Model evaluation.
Overall accuracy, per-class accuracy, average accuracy, confusion matrices, F scores, receiver operator characteristics, C statistics and saliency maps (guided backpropagation) were calculated as previously described27,47. Grad-CAM was also used as previously described30. For performance analysis of segmentation models, Jaccard similarities were calculated in the standard fashion as the intersection of predicted and labeled structures divided by their union.
Comparison to human experts.
Clinicians with expertise in fetal cardiology (fetal cardiology and maternal–fetal medicine attendings, experienced fetal cardiology sonographers, fetal cardiology fellows, n = 7), were shown up to one image per view for the studies in the OB-125 test set and asked whether that study was normal or not. For segmentation, clinical labelers segmented a subset of images multiple times, and intra-labeler Jaccard similarities were calculated as a benchmark. Use of clinicians for validation was deemed exempt research by the UCSF CHR.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Due to the sensitive nature of patient data, we are not able to make these data publicly available at this time. Source data are provided with this paper.
Code availability
ResNet and U-Net are publicly available and can be used with the settings described in the Methods and in Extended Data Fig. 1. The model weights that support this work are copyright of the Regents of the University of California and are available upon request. Additional code will be available upon publication at https://github.com/ArnaoutLabUCSF/cardioML.
Extended Data
Extended Data Fig. 1 |. Neural network architectures and schematic of rules-based classifier.
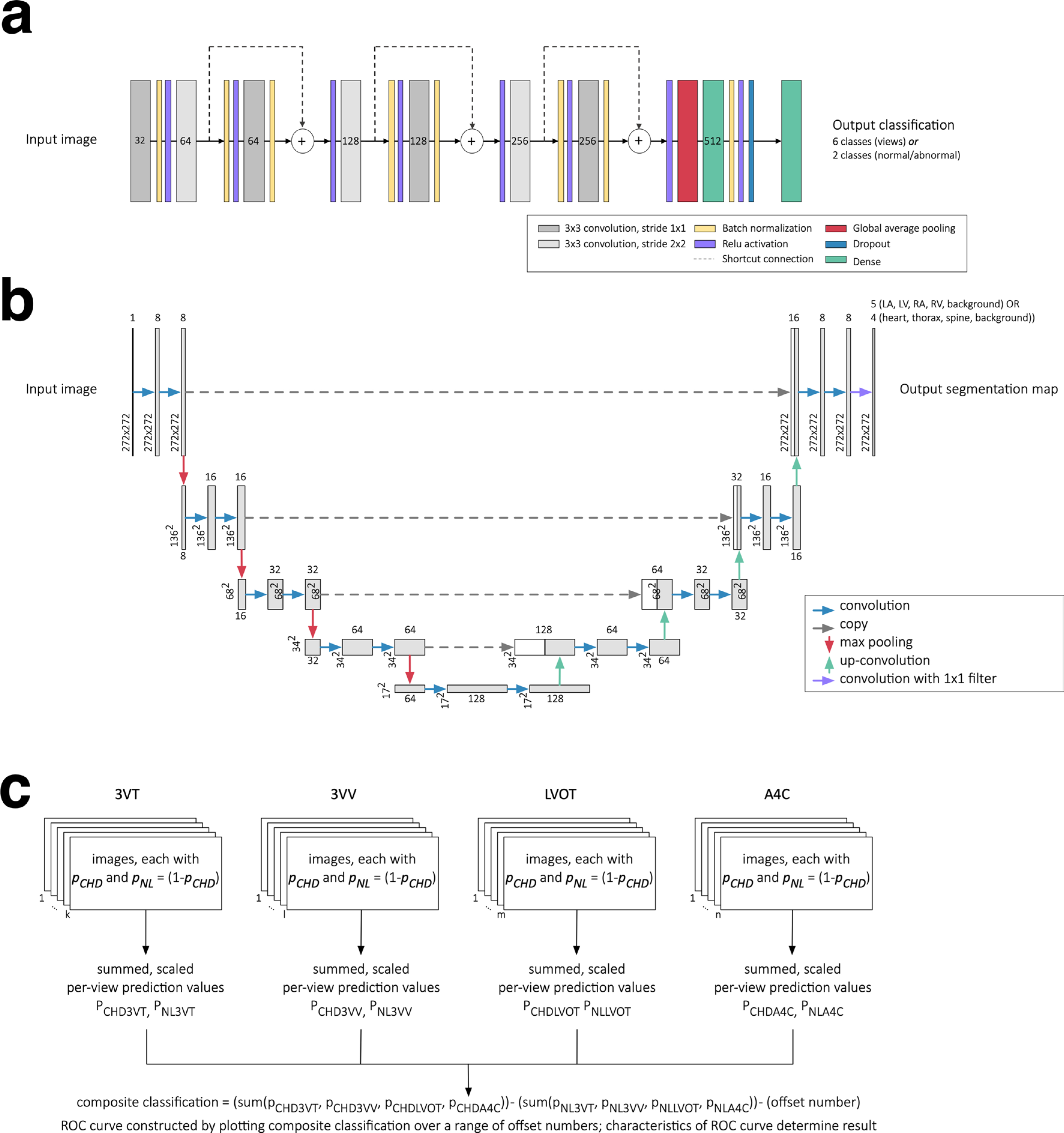
a, Neural network architecture used for classification, based on ResNet (He et. al. 2015). Numbers indicate the number of filters in each layer, while the legend indicates the type of layer. For convolutional layers (grey), the size and stride of the convolutional filters is indicated in the legend. b, Neural network architecture used for segmentation, based on UNet (Ronneberger et. al. 2015). Numbers indicate the pixel dimensions at each layer. c, A schematic for the rules-based classifier (‘Composite dx classifier,’ Figure 1b) used to unite per-view, per-image predictions from neural network classifiers into a composite (per-heart) prediction of normal vs. CHD. Only views with AUC > 0.85 on validation data were used. For each view, there are various numbers of images k,l,m,n, each with a per-image prediction probability pCHD and pNL. For each view, per-image pCHD and pNL were summed and scaled (see Methods) into a pair of overall prediction values for each view (for example PCHD3VT and PNL3VT). These are in turn summed for a composite classification. Evaluating true positive, false positive, true negative, and false negative with different offset numbers allowed construction of an ROC curve for each test dataset (Figure 3e). 3VT, 3-vessel trachea. 3VV, 3-vessel view. LVOT, left ventricular outflow tract. A4C, axial 4-chamber.
Extended Data Fig. 2 |. Bland-Altman plots comparing cardiac measurements from labeled vs. predicted structures.
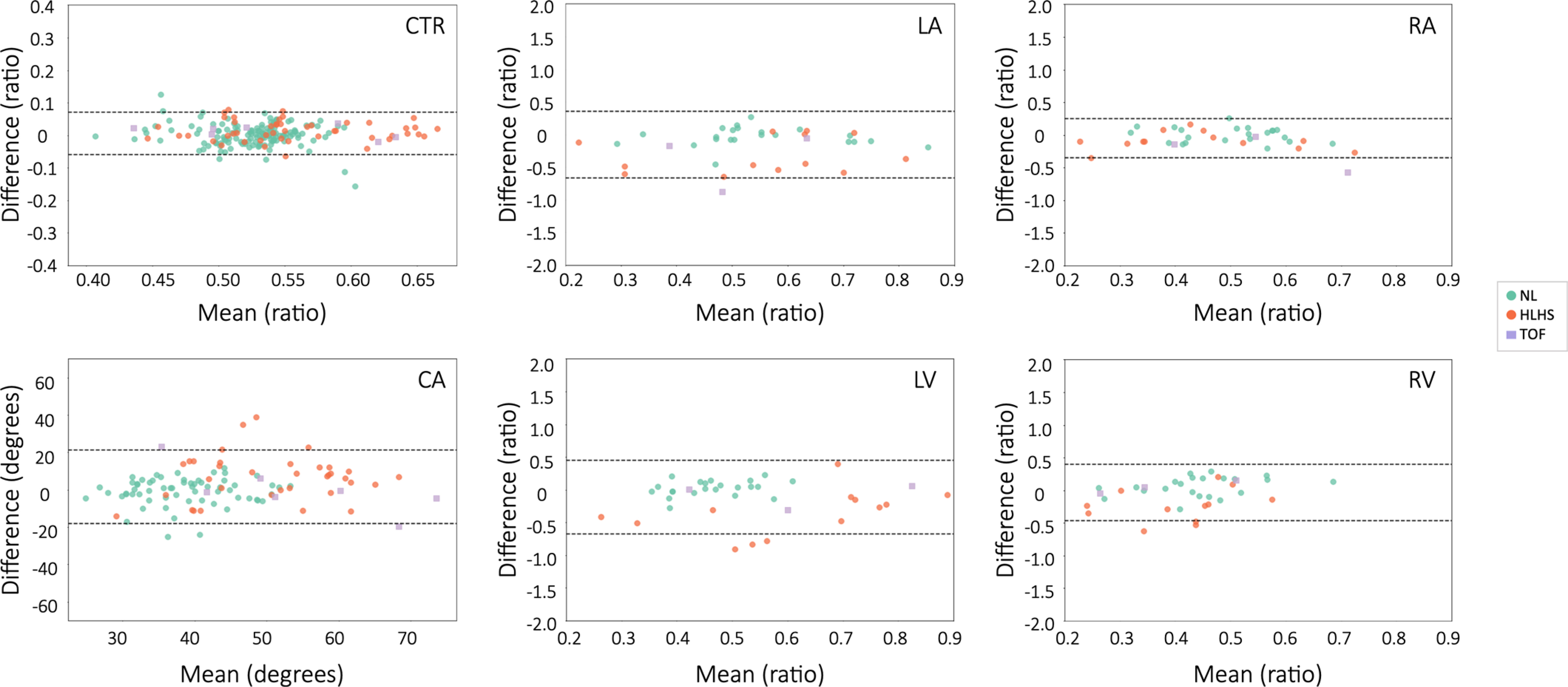
CTR, cardiothoracic ratio; CA, cardiac axis; LV, left ventricle; RV, right ventricle; LA, left atrium, RA, right atrium. Legend indicates measures for normal hearts (NL), hypoplastic left heart syndrome (HLHS), and tetralogy of Fallot (TOF).
Extended Data Fig. 3 |. Model confidence on sub-optimal images.
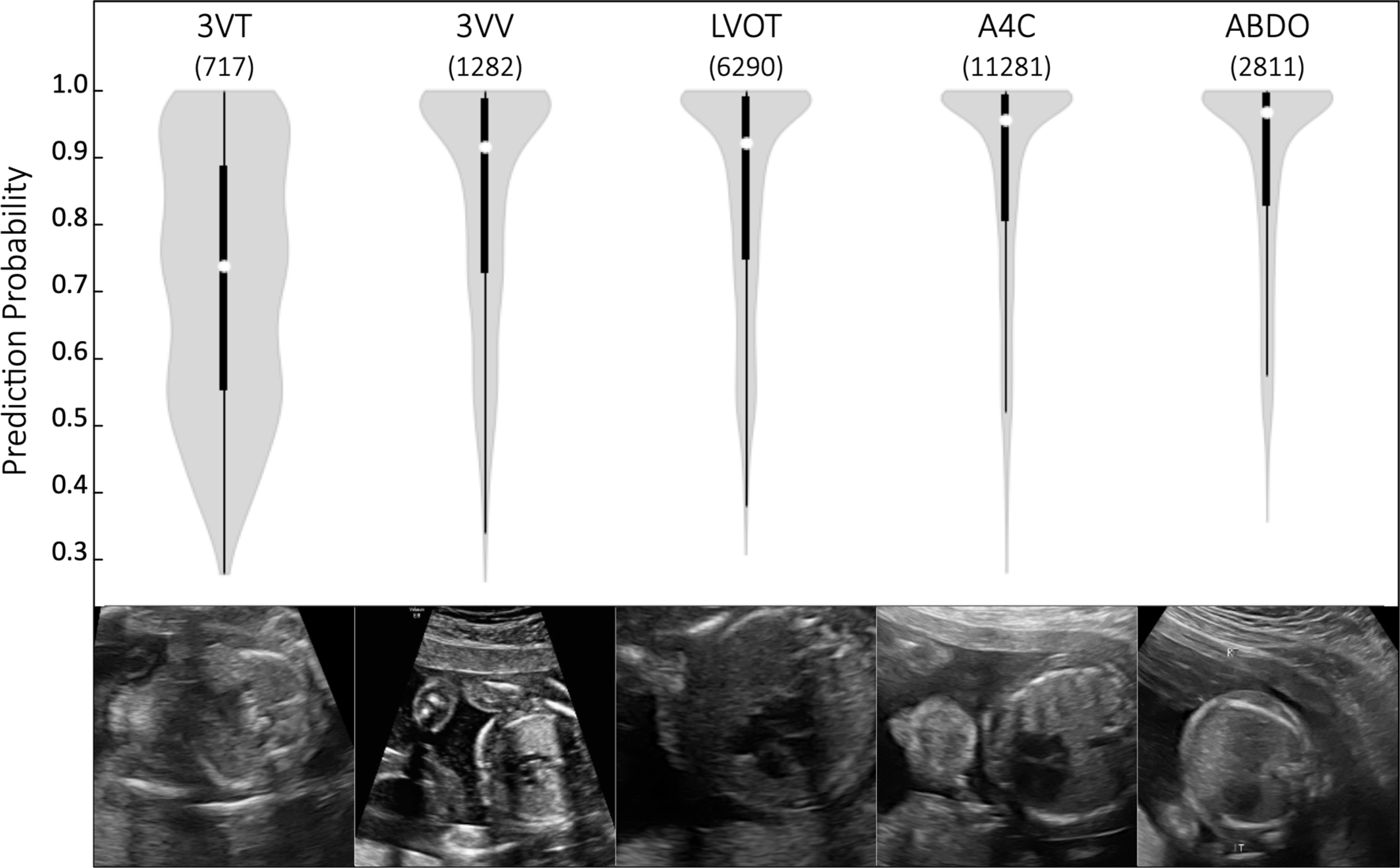
Examples of sub-optimal quality images (target views found by the model but deemed low-quality by human experts) are shown for each view, along with violin plots showing prediction probabilities assigned to the sub-optimal target images (White dots signify mean, thick black line signifies 1st to 3rd quartiles). Numbers in parentheses on top of violin plots indicate the number of independent images represented in each plot. For 3VT images, minimum, Q1, median, Q3, and maximum prediction probabilities are 0.27, 0.55, 0.74, 0.89, and 1.0, respectively. For 3VV images, minimum, Q1, median, Q3, and maximum prediction probabilities are 0.27, 0.73, 0.91, 0.99 and 1.0, respectively. For LVOT images, minimum, Q1, median, Q3, and maximum prediction probabilities are 0.31, 0.75, 0.92, 0.99, and 1.0, respectively. For A4C images, minimum, Q1, median, Q3, and maximum prediction probabilities are 0.28, 0.80, 0.95, 0.99, and 1.0, respectively. For ABDO images, minimum, Q1, median, Q3, and maximum prediction probabilities are 0.36, 0.83, 0.97, 1.0, and 1.0, respectively. Scale bars indicate 5mm. 3VT, 3-vessel trachea. 3VV, 3-vessel view. LVOT, left ventricular outflow tract. A4C, axial 4-chamber; ABDO, abdomen.
Extended Data Fig. 4 |. Misclassifications from per-view diagnostic classifiers.
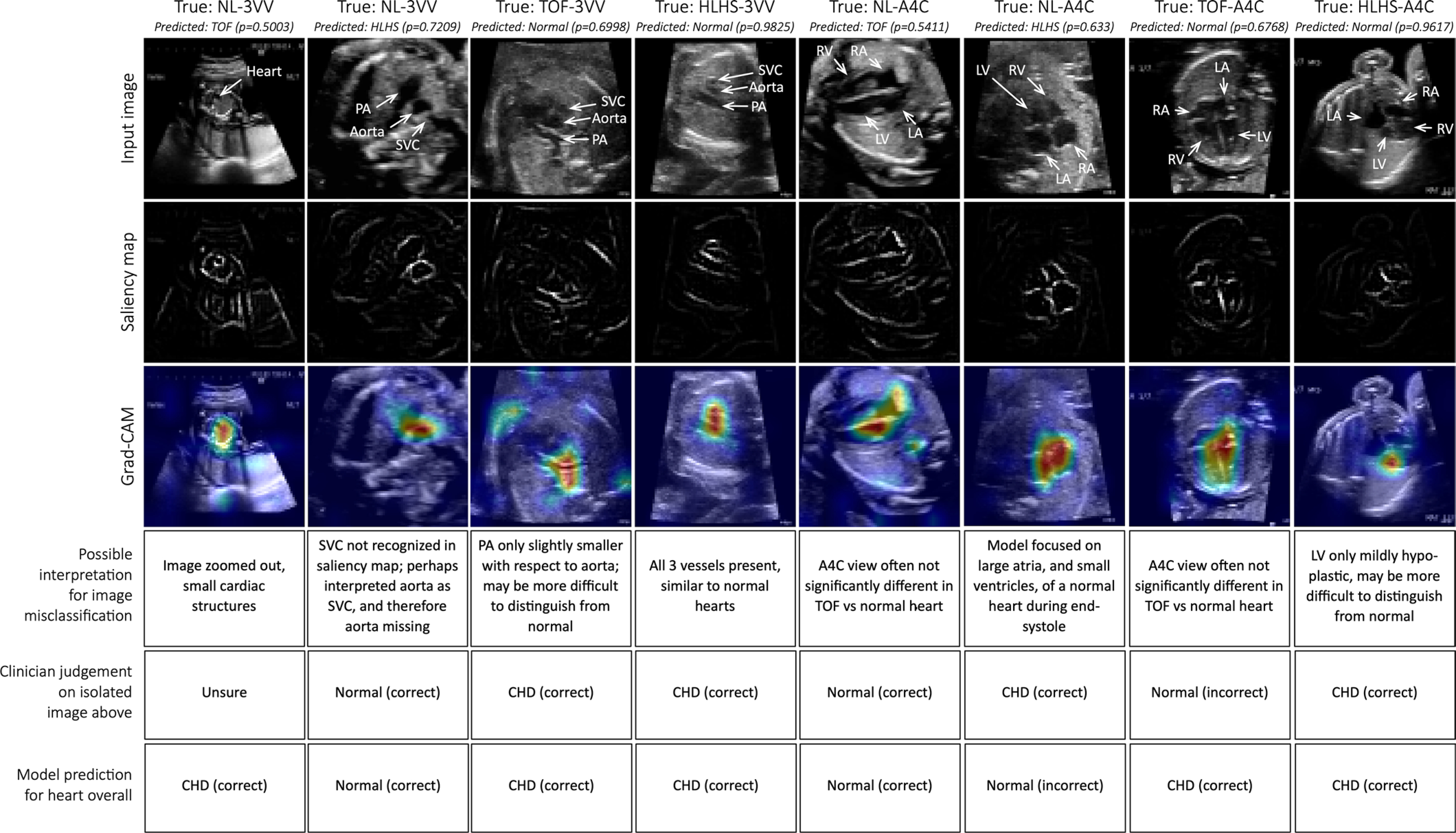
Top row: Example images misclassified by the diagnostic classifiers, with probabilities for the predicted class. Relevant cardiac structures are labeled. Second row: corresponding saliency map. Third row: Grad-CAM. Fourth row: possible interpretation of model’s misclassifications. Importantly, this is only to provide some context for readers who are unfamiliar with fetal cardiac anatomy; formally, it is not possible to know the true reason behind model misclassification. Fifth row: Clinician’s classification (normal vs. CHD) on the isolated example image. Sixth row: Model’s composite prediction of normal vs. CHD using all available images for the given study. For several of these examples, the composite diagnosis per study is correct, even when a particular image-level classification was incorrect. Scale bars indicate 5 mm. 3VV, 3-vessel view. A4C, axial 4-chamber. SVC, superior vena cava. PA, pulmonary artery. RA, right atrium. RV, right ventricle. LA, left atrium. LV, left ventricle.
Extended Data Fig. 5 |. Inter-observer agreement on a subset of labeled data.
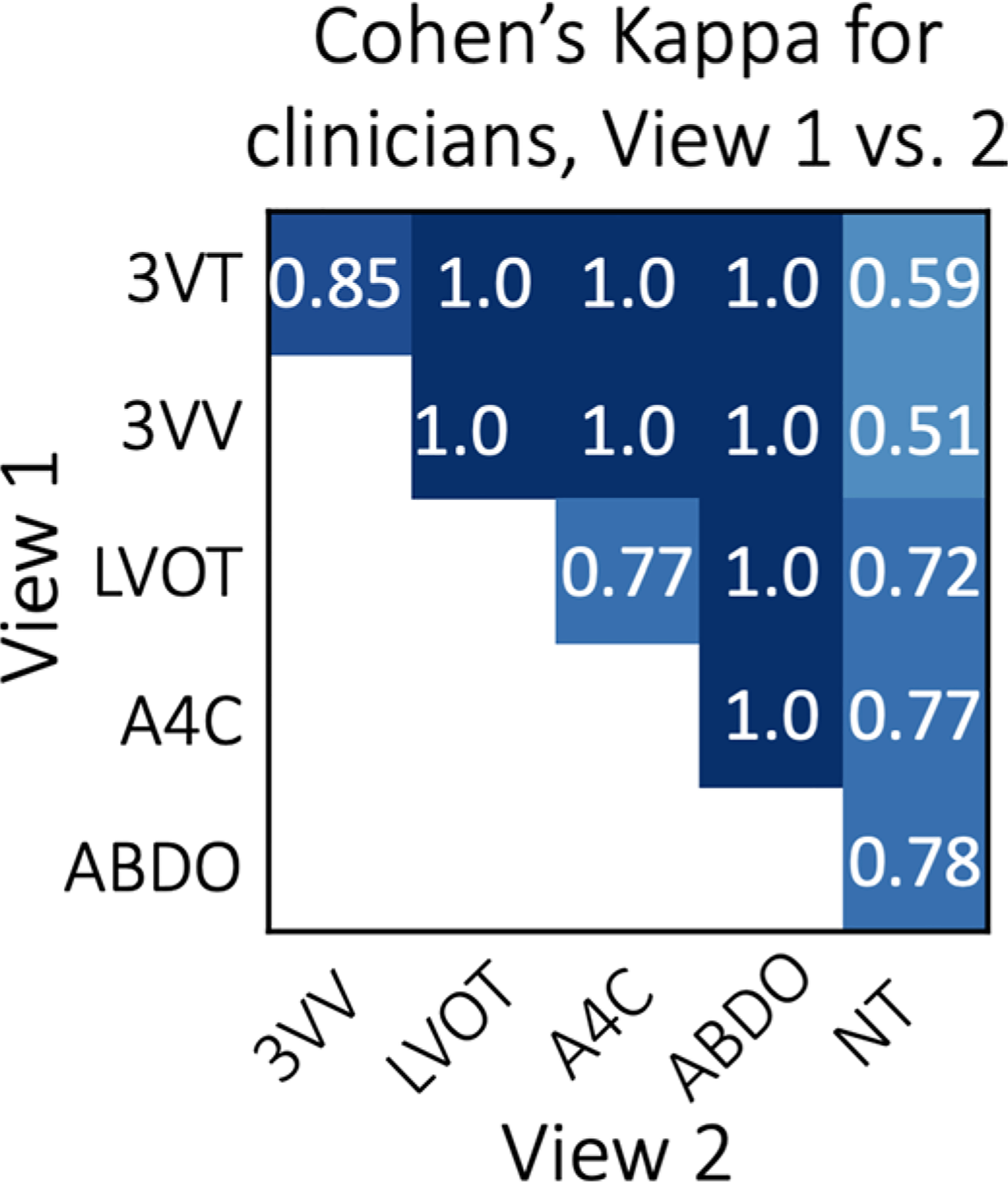
Inter-observer agreement on a sample of FETAL-125 is shown as Cohen’s Kappa statistic across different views, where poor agreement is 0–0.20; fair agreement is 0.21–0.40; moderate agreement is 0.41–0.60; good agreement is 0.61–0.80 and excellent agreement is 0.81–1.0. Of note, images where clinicians did not agree were not included in model training (see Methods). Most agreement is good or excellent, with moderate agreement on including 3VT and 3VV views as diagnostic-quality vs. non-target. 3VT, 3-vessel trachea. 3VV, 3-vessel view. LVOT, left ventricular outflow tract. A4C, axial 4-chamber, ABDO, abdomen, NT, non-target.
Supplementary Material
Acknowledgements
We thank A. Butte and D. Srivastava for critical reading of the manuscript and M. Brook, M. Kohli, W. Tworetzky and K. Jenkins for facilitating data access. We thank all clinicians who served as human participants, including C. Springston, K. Kosiv, C. Tai and D. Abel; others wished to remain anonymous. The American Heart Association Precision Medicine Platform (https://precision.heart.org/) was used for data analysis. This project was also supported by the UCSF Academic Research Systems and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI grant UL1 TR991872. R.A., Y.Z., J.C.L., E.C. and A.J.M.-G. were supported by the National Institutes of Health (R01HL150394) and the American Heart Association (17IGMV33870001) and the Department of Defense (W81XWH-19-1-0294), all to R.A.
Footnotes
Competing interests
Some methods used in this work have been filed in a provisional patent application.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-021-01342-5.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-021-01342-5.
Peer review informationNature Medicine thanks Zachi Attia, Declan O’Regan and Shaine Morris for their contribution to the peer review of this work. Editor recognition statement: Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
References
- 1.Donofrio MT et al. Diagnosis and treatment of fetal cardiac disease: a scientific statement from the American Heart Association. Circulation 129, 2183–2242 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Holland BJ, Myers JA & Woods CR Jr. Prenatal diagnosis of critical congenital heart disease reduces risk of death from cardiovascular compromise prior to planned neonatal cardiac surgery: a meta-analysis. Ultrasound Obstet. Gynecol. 45, 631–638 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Wright LK et al. Relation of prenatal diagnosis with one-year survival rate for infants with congenital heart disease. Am. J. Cardiol. 113, 1041–1044 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Bensemlali M et al. Neonatal management and outcomes of prenatally diagnosed CHDs. Cardiol. Young 27, 344–353 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Li YF et al. Efficacy of prenatal diagnosis of major congenital heart disease on perinatal management and perioperative mortality: a meta-analysis. World J. Pediatr. 12, 298–307 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Oster ME et al. A population-based study of the association of prenatal diagnosis with survival rate for infants with congenital heart defects. Am. J. Cardiol. 113, 1036–1040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freud LR et al. Fetal aortic valvuloplasty for evolving hypoplastic left heart syndrome: postnatal outcomes of the first 100 patients. Circulation 130, 638–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sizarov A & Boudjemline Y Valve interventions in utero: understanding the timing, indications, and approaches. Can. J. Cardiol. 33, 1150–1158 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Committee on Practice, B.-O., the American Institute of Ultrasound in, M. Practice bulletin no. 175: ultrasound in pregnancy. Obstet. Gynecol. 128, e241–e256 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Tuncalp et al. WHO recommendations on antenatal care for a positive pregnancy experience—going beyond survival. BJOG 124, 860–862 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Carvalho JS et al. ISUOG Practice Guidelines (updated): sonographic screening examination of the fetal heart. Ultrasound Obstet. Gynecol. 41, 348–359 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Bak GS et al. Detection of fetal cardiac anomalies: is increasing the number of cardiac views cost-effective? Ultrasound Obstet. Gynecol. 55, 758–767 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Friedberg MK et al. Prenatal detection of congenital heart disease. J. Pediatr. 155, 26–31 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Sekar P et al. Diagnosis of congenital heart disease in an era of universal prenatal ultrasound screening in southwest Ohio. Cardiol. Young 25, 35–41 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Sklansky M & DeVore GR Fetal cardiac screening: what are we (and our guidelines) doing wrong? J. Ultrasound Med. 35, 679–681 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sun HY, Proudfoot JA & McCandless RT Prenatal detection of critical cardiac outflow tract anomalies remains suboptimal despite revised obstetrical imaging guidelines. Congenit. Heart Dis. 13, 748–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corcoran S et al. Prenatal detection of major congenital heart disease—optimising resources to improve outcomes. Eur. J. Obstet. Gynecol. Reprod. Biol. 203, 260–263 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Letourneau KM et al. Advancing prenatal detection of congenital heart disease: a novel screening protocol improves early diagnosis of complex congenital heart disease. J. Ultrasound Med. 37, 1073–1079 (2018). [DOI] [PubMed] [Google Scholar]
- 19.AIUM practice parameter for the performance of fetal echocardiography. J. Ultrasound Med. 39, E5–E16 (2020). [DOI] [PubMed] [Google Scholar]
- 20.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chilamkurthy S et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 392, 2388–2396 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Gulshan V et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Baumgartner CF et al. SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans. Med. Imaging 36, 2204–2215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnaout R Toward a clearer picture of health. Nat. Med. 25, 12 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Ouyang D et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madani A, Arnaout R, Mofrad M & Arnaout R Fast and accurate view classification of echocardiograms using deep learning. npj Digital Med. 1, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He K, Zhang X, Ren S & Sun J Identity mappings in deep residual networks. Preprint at https://arxiv.org/abs/1603.05027 (2016). [Google Scholar]
- 29.Lee W et al. AIUM practice guideline for the performance of fetal echocardiography. J. Ultrasound Med. 32, 1067–1082 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Selvaraju RR et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Preprint at https://arxiv.org/abs/1610.02391 (2016).
- 31.Liu H et al. Fetal echocardiography for congenital heart disease diagnosis: a meta-analysis, power analysis and missing data analysis. Eur. J. Prev. Cardiol. 22, 1531–1547 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Pinheiro DO et al. Accuracy of prenatal diagnosis of congenital cardiac malformations. Rev. Bras. Ginecol. Obstet. 41, 11–16 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Chu C et al. Prenatal diagnosis of congenital heart diseases by fetal echocardiography in second trimester: a Chinese multicenter study. Acta Obstet. Gynecol. Scand. 96, 454–463 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zech JR et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 15, e1002683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miceli F A review of the diagnostic accuracy of fetal cardiac anomalies. Australas. J. Ultrasound Med. 18, 3–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronneberger O, Fischer P & Brox T U-Net: convolutional networks for biomedical image segmentation. Preprint at https://arxiv.org/abs/1505.04597 (2015).
- 37.Zhao Y et al. Fetal cardiac axis in tetralogy of Fallot: associations with prenatal findings, genetic anomalies and postnatal outcome. Ultrasound Obstet. Gynecol. 50, 58–62 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Goldinfeld M et al. Evaluation of fetal cardiac contractility by two-dimensional ultrasonography. Prenat. Diagn. 24, 799–803 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Best KE & Rankin J Long-term survival of individuals born with congenital heart disease: a systematic review and meta-analysis. J. Am. Heart Assoc. 5, e002846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attia ZI et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 394, 861–867 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Peahl AF, Smith RD & Moniz MH Prenatal care redesign: creating flexible maternity care models through virtual care. Am. J. Obstet. Gynecol. 10.1016/j.ajog.2020.05.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo L, Markush D & Romero R Prenatal diagnosis of tetralogy of Fallot with pulmonary atresia using: Fetal Intelligent Navigation Echocardiography (FINE). J. Matern. Fetal Neonatal Med. 32, 3699–3702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen L et al. Three-dimensional fast acquisition with sonographically based volume computer-aided analysis for imaging of the fetal heart at 18 to 22 weeks’ gestation. J. Ultrasound Med. 29, 751–757 (2010). [DOI] [PubMed] [Google Scholar]
- 44.World Health Organization. WHO Guideline: Recommendations on Digital Interventions for Health System Strengthening (2019). [PubMed]
- 45.Yagel S, Cohen SM & Achiron R Examination of the fetal heart by five short-axis views: a proposed screening method for comprehensive cardiac evaluation. Ultrasound Obstet. Gynecol. 17, 367–369 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Norgeot B et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat. Med. 26, 1320–1324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Springenberg JT, Dosovitskiy A, Brox T & Riedmiller M Striving for simplicity: the all convolutional net. Preprint at https://arxiv.org/abs/1412.6806 (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the sensitive nature of patient data, we are not able to make these data publicly available at this time. Source data are provided with this paper.


