Abstract
Two-component systems are a dominant form of bacterial signal transduction. The prototypical two-component system consists of a sensor that responds to a specific input(s) by modifying the output of a cognate regulator. Because the output of a two-component system is the amount of phosphorylated regulator, feedback mechanisms may alter the amount of regulator, and/or modify the ability of a sensor or other proteins to alter the phosphorylation state of the regulator. Two-component systems may display intrinsic feedback whereby the amount of phosphorylated regulator changes under constant inducing conditions and without the participation of additional proteins. Feedback control allows a two-component system to achieve particular steady state levels, to reach a given steady state with distinct dynamics, to express coregulated genes in a given order, and to activate a regulator to different extents, depending on the signal acting on the sensor.
Keywords: expression dynamics, phosphorylation, signal access, transcription surge
INTRODUCTION
The two-component system is a family of signal transduction proteins present in all domains of life (97). In bacteria, two-component systems constitute a dominant form of gene control in response to changes in an organism’s surroundings. This protein family has been implicated in the adaptation to a variety of stress conditions (23, 25, 30, 37, 44, 81), in pathogenic (30, 51, 84) and symbiotic (65, 79) interactions with eukaryotic hosts, and in essential cellular pathways (23, 78). The prototypical two-component system responds to a change in an environmental condition by modifying gene expression; alternatively or in addition, the output may be modification of the biochemical activities of target proteins.
Gene control systems often use feedback mechanisms to adjust their outputs (61). That is to say, they make the output part of the input. Negative feedback is observed when a deviation in the output results in changes in the direction opposite to the original deviation. Positive feedback, by contrast, occurs when a deviation in the output gives rise to changes in the same direction as the original deviation. Negative feedback generally serves to stabilize the state of a system, whereas positive feedback moves the system away from the original state.
Feedback mechanisms play critical roles in the response to stress conditions (30, 37) and in developmental pathways (13). Moreover, they are essential for pathogens to cause disease (89); for symbiotic organisms to reside within their eukaryotic hosts (100); and for the synthesis of (87), resistance to (19), and persistence in the presence of (5) antibacterial agents. In this article, I review the mechanisms and physiological significance of feedback regulation of bacterial two-component systems. I begin with an introduction to the architectural classes of two-component systems. Then, I describe the mechanisms by which these systems adjust their output, and the significance of feedback-promoted adjustments. Finally, I discuss how the investigation of feedback control of two-component systems has uncovered novel principles about organismal responses to a change in conditions. Much of my perspective on these feedback control systems derives from my own work on the PhoP/PhoQ system (30), which I reference thoughout this review.
VARIATION IN THE ARCHITECTURE OF TWO-COMPONENT SIGNAL TRANSDUCTION
Classical two-component systems consist of a sensor protein that responds to a physical or chemical signal by modifying the phosphorylated state of a cognate regulatory protein (Figure 1a). Regulators are often DNA-binding transcriptional repressors and/or activators, and the consequence of an inducing condition for a two-component system is typically a change in the transcriptional profile of an organism. Phosphorylation usually increases the affinity of a regulator for its DNA target (28, 113, 114). Certain regulators lack DNA-binding domains and exert their regulatory effects by establishing direct interactions with protein (34) or RNA (91) targets.
Figure 1.
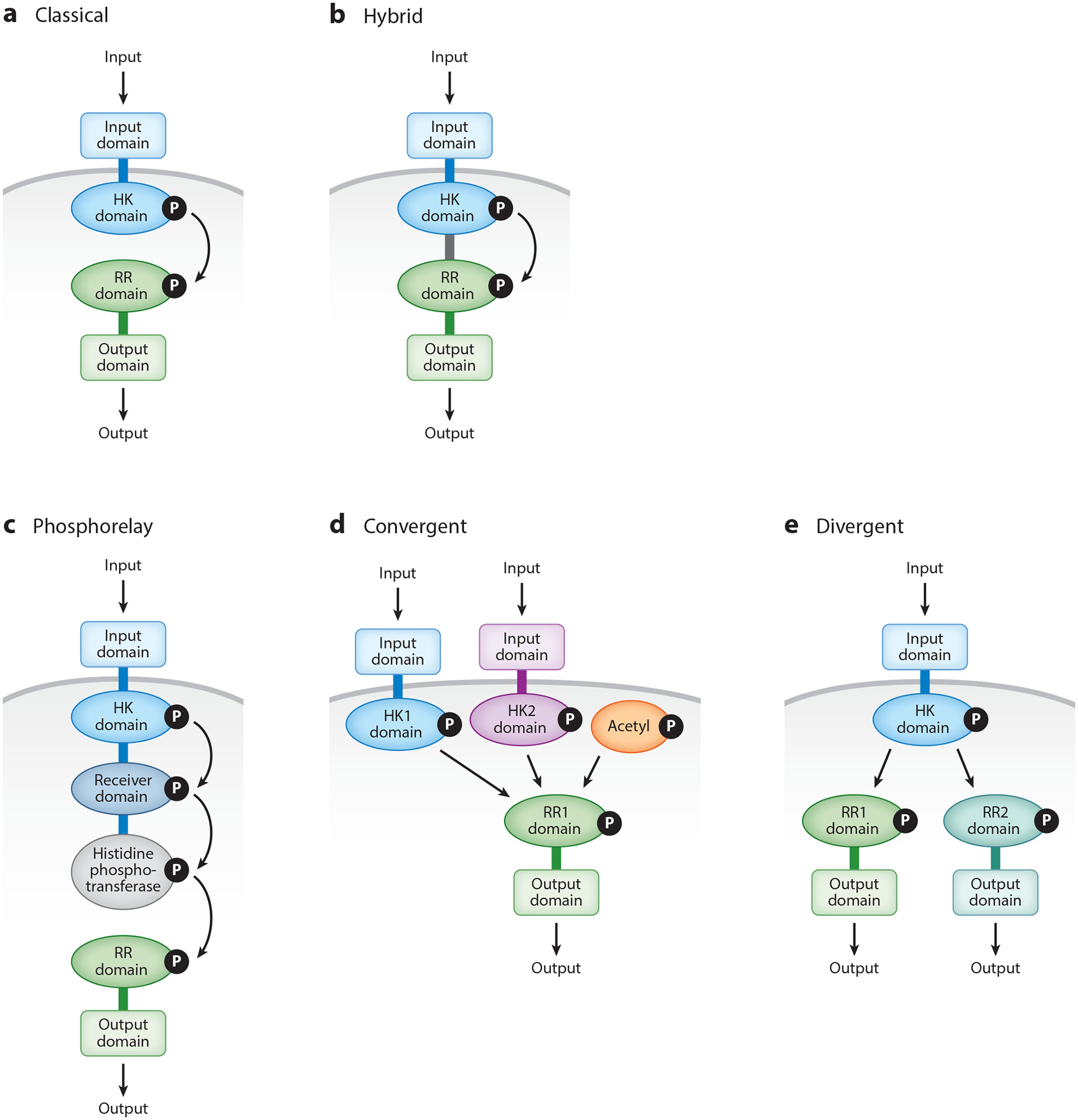
Architectural classes of two-component signal transduction systems, and of signaling pathways using two-component system proteins. (a) In classical systems, the sensor harboring an input domain and a histidine kinase (HK) domain responds to an input by autophosphorylating at a conserved histidine residue. The phosphorylated sensor serves as a phosphoryl donor to the regulator harboring a response regulator (RR) domain and an output domain. The phosphorylated regulator generates an output. (b) A hybrid system harbors all the domains described for the classical system in panel a, in a single polypeptide. (c) In a phosphorelay, there are four consecutive phosphotransfer events, starting when a sensor responds to an input by autophosphorylating from ATP at a conserved histidine residue. The phosphoryl group is then transferred to a conserved aspartate in the RR domain and from there to a histidine residue in the histidine-containing phosphotransferase domain, and ultimately to an RR domain, which upon phosphorylation modifies the output domain to generate an output. The sequential phosphotransfer may occur between different domains of a single polypeptide, as depicted here, or between separate proteins harboring these domains. (d) In a convergent signaling pathway, distinct signals activate different sensors that modify the phosphorylation state of a single regulator to generate an output. The phosphorylation state of a regulator may also be modified by metabolites such as acetyl phosphate. (e) In a divergent signaling pathway, a sensor responds to a signal by modifying the phosphorylation state of two regulators, thereby generating two different outputs.
Sensors use ATP to autophosphorylate at a conserved histidine residue. In addition to using ATP, the senor MprB from Mycobacterium tuberculosis can autophosphorylate with the high-energy molecule polyphosphate (). The phosphorylated sensor serves as a phosphodonor to its partner regulator, which is phosphorylated at a conserved aspartic acid residue. The vast majority of sensors also display phosphatase activity toward their respective phosphorylated regulators. The recognition surfaces that confer specificity between sensor and regulator as well as the biochemical properties of these proteins are largely conserved across systems and species (49). By contrast, the signals acting on sensors, and the genes regulated by the regulators, typically differ among two-component systems (57). Differences in input signals and regulated genes/proteins have been reported even for homologous systems of closely related bacterial species (19, 76).
When an organism experiences inducing conditions for a given two-component system, the levels of phosphorylated regulator increase. This increase may come about from several nonmutually exclusive effects exerted by the signal on the sensor. For example, an inducing signal may promote sensor autophosphorylation, thereby elevating the concentration of phosphorylated sensor, which constitutes the usual substrate for regulator phosphorylation. Alternatively or in addition, the signal may inhibit the sensor’s phosphatase activity toward the phosphorylated regulator. A regulator may phosphorylate not only from a sensor that autophosphorylates continuously but also from a small-molecular-weight phosphodonor, such as acetyl phosphate (58a), the levels of which are modulated by the metabolic status of the bacterial cell (47, 85). Furthermore, a signal may alter the synthesis and/or activity of a protein(s) that modifies the ability of a sensor to act as an autokinase or phosphatase (39), or of a phosphorylated regulator to be dephosphorylated by its cognate sensor (43).
Hybrid two-component systems constitute a protein family that harbors the sensor and regulator domains of a classical two-component system fused in a single polypeptide (79) (Figure 1b). That is to say, a hybrid two-component system comprises a signal-sensing domain at the amino terminus followed by the sensor kinase domain, which, in turn, is followed by a regulator domain containing the conserved aspartate site of phosphorylation, and then by a DNA-binding domain at the carboxy terminus. Therefore, a hybrid two-component system is typically an integral membrane protein with a periplasmic signal-sensing domain and a DNA-binding domain.
The best characterized hybrid two-component systems are found in the mammalian symbiotic bacterium Bacteroides thetaiotaomicron, where they control the expression of genes involved in the uptake and utilization of carbohydrates. B. thetaiotaomicron harbors over 30 hybrid two-component systems (79a and 110), each mediating growth on a different carbohydrate as the sole carbon source. In addition to the hybrid two-component systems, B. thetaiotaomicron harbors ~40 classical two-component systems, which are predicted to mediate the response to stress conditions. Thus, hybrid and classical two-component systems may be found in the same organism.
The phosphorelay is a version of the two-component system involving four sequential phosphoryl transfer events (Figure 1c). In the phosphorelay, a sensor autophosphorylates at a conserved histidine residue and serves as phosphoryl donor to a protein (or a domain of the same protein) that becomes phosphorylated at an aspartate residue; the latter phosphorylated protein or domain, in turn, donates the phosphoryl group to a third protein or domain, which is phosphorylated at a histidine residue and ultimately donates the phosphoryl group to a regulatory protein or domain whose activity changes when it is phosphorylated at a conserved aspartate residue. The existence of multiple phosphorylated intermediates provides ample opportunities to abort the phosphotransfer process in response to additional inputs (70). In a phosphorelay, there are four sequential steps of phosphotransfer that take place between different proteins and/or within individual domains of a given protein.
In the vast majority of two-component systems, the sensor only affects the phosphorylated state of its cognate regulator (Figure 1a) (49, 101). However, certain regulators do phosphorylate from both cognate and noncognate sensors (Figure 1d), enabling regulator output to reflect the different inputs operating on cognate and noncognate sensors (14). Thus, different signals acting on distinct sensors or altering the levels of particular metabolites can converge on a given regulator (Figure 1d). In other cases, a given sensor may serve as phosphodonor for multiple regulators (Figure 1e), thereby expanding the spectrum of genes, proteins, and activities affected by the signal acting on a given sensor (59).
For two-component systems that regulate gene expression, the enzymatic activity of a reporter gene product or the mRNA levels of the gene of interest are typically used as surrogates for the output of a two-component system. These approaches are used because measuring mRNA levels or determining enzymatic activities is less laborious than determining the amount of phosphorylated regulator, and/or because of interest in a particular gene or protein controlled by the regulator. However, ascribing properties of a downstream output to a two-component system can be confounding because such properties may only apply to a particular target of the system. As stated above, the phosphorylated form of the regulator is the actual output of a two-component system. The in vivo levels of phosphorylated regulator can be measured using cells grown with radiolabeled phosphate or using polyacrylamide gels containing 1,3-bis[bis(pyridin-2-ylmethyl) amino]propan-2-olato dizinc(II) complex, which allow the separation of the phosphorylated and unphosphorylated forms of the regulator (7, 105).
Below, I discuss how two-component system proteins control their output by altering the amount of active regulator. This control is exerted at the level of transcription of sensor and regulator genes, by the sensor and regulator proteins modifying their biochemical activities, or conducted by the products of the genes they control (Figure 2a).
Figure 2.
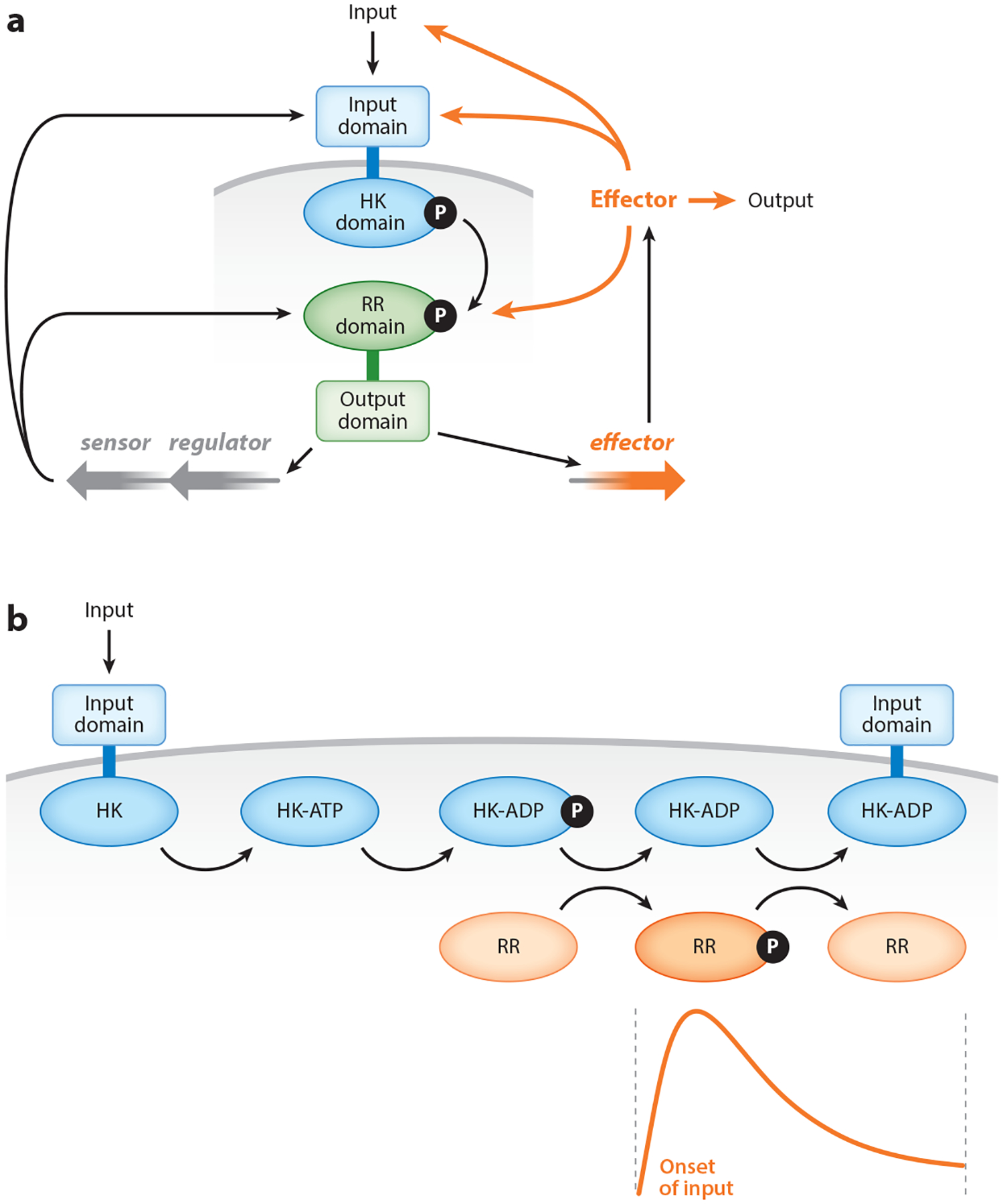
Feedback control of two-component systems. (a) Extrinsic control of two-component systems by a regulator governing transcription of the genes specifying the sensor and regulator, and by an Effector encoded by a gene under transcriptional control of the regulator. The Effector may affect input access to the sensor (42, 68, 80), bind to the sensor modifying its enzymatic activities (36, 40, 52, 82, 86), or modify the phosphorylation state of the regulator in a direct or indirect fashion (104). Effectors may act at other levels and are not necessarily proteins. (b) Intrinsic feedback by a sensor (from left to right) that responds to an input by binding to ATP, autophosphorylating at a histidine residue, and leaving ADP in the nucleotide-binding pocket. The regulator (RR) phosphorylates from the phosphorylated sensor. The ADP-bound sensor is in the phosphatase mode that promotes dephosphorylation of the phosphorylated regulator. The inset graph depicts the changes in the levels of phosphorylated regulator when an organism is switched from noninducing to inducing conditions for the sensor. Abbreviation: HK, histidine kinase.
TRANSCRIPTIONAL AUTOREGULATION OF TWO-COMPONENT SYSTEMS AND ITS CONSEQUENCES
Transcriptional Control of Two-Component Systems
Two-component system proteins are often encoded in operons specifying cognate sensor-regulator pairs. This gene arrangement makes sense physiologically, given that, in most cases, sensors exclusively govern the phosphorylated state of their cognate regulators. Yet, bona fide two-component system proteins are sometimes independently transcribed (12, 24, 54) and may be encoded in distant regions of a genome.
Two-component system genes must be transcribed at some basal level so that an organism has enough sensor to detect a specific signal(s), and enough regulator to initiate a response. In addition to the constitutive promoter providing basal levels of expression, many two-component systems harbor a second, autoregulated, promoter. The latter promoter is responsible for generating the amounts of active regulator necessary to carry out the genetic program that will help the organism prosper in the environment denoted by the signal.
The location of the constitutive and autoregulated promoters often varies across systems and/or species. For example, the two-component system operon phoPQ is transcribed from both a constitutive promoter and a positively regulated promoter in Salmonella enterica serovar Typhimurium (96) and Escherichia coli (60). These two species differ in the relative position of the two promoters: the constitutive promoter is proximal to the phoP coding region in S. enterica serovar Typhimurium but distal in E. coli, and the converse is true for the autoregulated promoter. Autoregulation of the phoPQ operon, which has been experimentally demonstrated in S. enterica serovar Typhimurium (96), E. coli (60), and Yersinia pestis (72) species, is likely conserved in other members of the family Enterobacteriaceae, because all investigated species carrying phoP and phoQ genes harbor a binding site for the PhoP protein upstream of the phoP gene or of a gene upstream of phoP and cotranscribed with phoP (72). For example, the autoregulated phoP promoter is located upstream of a gene preceding phoP and phoQ in both Y. pestis (72) and Pseudomonas aeruginosa (53).
For certain two-component system operons, a single promoter is used to provide basal and autoregulated expression. This is the case of the genes specifying the ComD/ComE two-component system governing competence in the gram-positive bacterium Streptococcus pneumoniae (56). Transcription of the comD and comE genes under noninducing conditions requires both the sensor ComD and the ability of ComE to become phosphorylated. Therefore, the regulator ComE is phosphorylated from the sensor ComD even under noninducing conditions, and phosphorylated ComE is the form that promotes gene transcription.
The RcsC/RcsD/RcsB system of enteric bacteria is a phosphorelay that controls several properties, including motility and the properties of the bacterial cell surface (54). Curiously, the rcsC, rcsD, and rcsB genes are transcribed in distinct fashions. Although rcsD is followed by rcsB in what appears to be an operon, a separate promoter located within the rcsD coding region drives rcsB transcription under conditions distinct from those operating on the rcsD promoter (75). rcsB overexpression from a heterologous promoter (75) or conditions that activate the Rcs system (74) repress transcription from the rcsD promoter. RcsB represses the rcsD promoter directly by binding to the predicted −35 region (74). By contrast, rcsB overexpression has little effect on the rcsB promoter and no effect on the promoter of the rcsC gene (75), which is convergently transcribed toward rcsB (54).
That the rcsB, rcsC, and rcsD genes are independently transcribed may allow gene activation by RcsB in atypical ways. For example, unphosphorylated RcsB generated in the absence of RcsC and RcsD may regulate gene expression by forming heterodimers with other DNA-binding proteins of the FixJ/NarL family (67). Moreover, in addition to the demonstrated phosphotransfer from RcsD to RcsB (98) and anticipated phosphotransfer from RcsC to RcsD, it has recently been reported that RcsB can phosphorylate directly from RcsC (73). Therefore, the production of RcsC and RcsD, as well as the signals acting on these proteins, likely modifies transcription of RcsB-dependent targets based on whether they require the phosphorylated form of RcsB or not.
The gene arrangement and transcriptional autoregulation of the rcs system described above for S. enterica serovar Typhimurium appear to be conserved in other enteric bacteria (75). However, autoregulation of two-component systems may differ even among closely related bacterial species. For example, the osmolarity-responding OmpR/EnvZ two-component system is autoregulated in S. enterica serovar Typhimurium (6, 18) but not in E. coli, from which it diverged ~100 Mya (22, 66).
Transcriptional Autoregulation Controls Both Steady State Levels and Kinetics of Expression
Transcriptional autoregulation of a two-component system provides opportunities for fine-tuning expression among genes controlled by a given regulator. For instance, when the positively autoregulated phoP promoter from S. enterica serovar Typhimurium was inactivated, transcription of some PhoP-activated genes was reduced and of others completely eliminated (62). This finding indicates that the basal levels of regulator PhoP and sensor PhoQ originating from the constitutively produced mRNA generate PhoP-P amounts that activate only a limited subset of PhoP-dependent promoters. In agreement with this notion, transcription of PhoP-dependent genes was restored to wild-type levels upon replacement of the PhoP-activated phoP promoter by a strong constitutive promoter (62).
Autoregulation makes it possible for an organism to time expression of a group of target genes controlled by a given regulator. For instance, when an organism faces conditions inducing a positively autoregulated two-component system after experiencing noninducing conditions for such a system, the number of phosphorylated regulator molecules will be low. That low number will increase as the regulator promotes its own transcription, thereby creating additional substrates to phosphorylate from the sensor. In the case of the PhoP/PhoQ system, early on, PhoP binds to only those promoters with high-affinity binding sites and/or those that are not subjected to strong silencing by nucleoid-associated proteins, such as the histone-like nucleoid structuring protein H-NS (113, 114). As the concentration of PhoP-P increases, promoters with lower-affinity binding sites and/or subjected to H-NS-mediated silencing will be occupied, and transcription of the corresponding genes will take place.
Positive feedback can create an expression hierarchy that reflects the number of active regulator molecules and the properties of its regulated promoters within a given regulon. This hierarchy may reflect physiological responses. For instance, in the phosphate limitation–activated PhoB/PhoR system of E. coli, the autoregulated PhoB protein promotes expression first of genes directly involved in phosphate uptake, and then of those that carry out phosphate scavenging (28). By contrast, the expression hierarchy in the Salmonella PhoP regulon reflects gene ancestry (i.e., whether the regulated gene is horizontally acquired or ancestral to the Enterobacteriaceae family), as opposed to the function of the corresponding gene products (114).
In the Bacillus subtilis phosphorelay, lower levels of the phosphorylated regulator Spo0A are required for expression of genes involved in bacterial cannibalism than are required for expression of genes involved in sporulation (27). Moreover, a combination of experimental and computational studies established that the gradual accumulation of Spo0A-P levels is critical for the ordered expression of Spo0A-dependent genes and sporulation efficiency (102). In other words, the fate of B. subtilis depends not only on achieving specific Spo0A-P levels but also on the timing with which such levels are achieved.
The BvgA/BvgS phosphorelay system governs virulence in the mammalian pathogens Bordetella pertussis and Bordetella bronchiseptica (10). Positive feedback on BvgA dictates the steady state levels of expression of BvgA-regulated genes, thereby increasing the sensitivity of the system to subtle changes in the intensity of the signal controlling the sensor BvgS (106). In addition, positive feedback is required for the precise temporal expression of four distinct phenotypic phases exhibited by Bordetella spp. upon a shift from noninducing (i.e., Bvg−) to inducing (i.e., Bvg+) conditions. These phenotypic phases reflect the action of the regulator BvgA at the individual promoters. BvgA-regulated promoters differ in the affinity of the BvgA-binding sites for phosphorylated BvgA (BvgA-P), as well as the number and location of such sites, which, in turn, determine their roles in activation or repression (106). The requirement for BvgA/BvgS autoregulation was bypassed if these proteins were expressed constitutively at high levels. Curiously, positive autoregulation of the BvgA/BvgS system is not essential for the correct temporal expression when bacteria experience a shift in the opposite direction: from inducing (Bvg+) to repressing (Bvg−) conditions (106).
Computer simulations of the BvgA/BvgS system support the notion that the four phenotypic Bvg classes behave in distinct manners. For example, class 1 and class 3 genes exhibit minimal dependence on positive feedback for their correct response to inducing signals, a property ascribed to the high affinity of the corresponding promoters for BvgA-P. Class 1 and class 3 genes are BvgA-repressed and -activated genes, respectively. By contrast, class 2 and class 4 genes demand positive feedback to generate the levels of BvgA-P necessary to bind to the low-affinity binding sites in their promoters (77). The single class 2 gene described to date harbors a high-affinity binding site that promotes activation and a low-affinity binding site that stimulates repression (77), thereby providing Bordetella spp. with a defined window of opportunity in which to express this particular gene. Class 4 genes correspond to BvgA-activated genes harboring only low-affinity binding sites, and their expression is delayed until enough BvgA-P is accumulated (77).
The need to promote transcription from multiple targets is not the sole reason for transcriptional autoregulation of two-component systems. For example, the VanR/VanS two-component system encoded in the transposon Tn1516 is autoregulated even though the regulator VanR appears to regulate a single transcriptional unit specifying proteins mediating resistance to glycopeptide antibiotics (4).
Positive feedback has been associated with switch-like responses and bimodality (i.e., the presence of two phenotypically distinct populations in an genetically identical culture). However, no such behavior has been reported for the well-characterized E. coli PhoB/PhoR (28), Salmonella PhoP/PhoQ (89) and Bordetella BvgA/BvgS (106) systems.
As discussed above, many two-component systems positively regulate their own transcription. How, then, does a bacterium prevent the accumulation of potentially toxic amounts of sensor and regulator proteins once the organism successfully adapts to the stress condition that activated the two-component system? As discussed below, the output of a system (i.e., the amount of phosphorylated regulator) may decrease under constant inducing conditions by mechanisms that are intrinsic to the sensor and regulator proteins, and/or mediated by the products under control of the regulator.
Role of Transcriptional Autoregulation Under Different Stimulus Intensity
Under which conditions does autoregulation of a two-component system exert its strongest effects? A combination of modeling and experimental approaches led to the proposal that positive autoregulation of the E. coli phoPQ system has no effect over a large range of input intensities. However, in the presence of a strong stimulus, there was an effect on the steady state behavior of the PhoP/PhoQ system (63). By contrast, investigation of the Salmonella PhoP/PhoQ autoregulation revealed that positive feedback does have a significant impact on the system’s steady state behavior under both low and high levels of stimulus (62). Moreover, the impact of positive feedback is dependent on the strength of the constitutive promoter providing basal transcription of the phoPQ operon (62). When the constitutive promoter is weak, positive feedback influences strongly the output of the system (62). And when the constitutive promoter is strong, the dependence of the output on the positive feedback is minimal for both low and high stimulus levels (62), presumably because the PhoP and PhoQ proteins are already produced at high levels.
The constitutive promoters providing basal expression for two-component system genes are typically weak (62). This property is critical to avoid spurious activation of a two-component system in the absence of the specific inducing signal acting on the sensor, which can take place by two nonmutually exclusive mechanisms. First, regulators typically function as homodimers and regulator phosphorylation often favors regulator dimerization (8, 15). Therefore, conditions promoting regulator dimerization independently of phosphorylation will activate a regulator. In agreement with this notion, expression of the Salmonella PhoP protein at 11 times its normal levels resulted in transcription of PhoP-activated genes in a mutant lacking the sensor PhoQ, and with a PhoP variant where the aspartate site of phosphorylation was substituted by a nonphosphorylatable amino acid (50). By contrast, when the PhoP protein was produced at up to four times the physiological levels, transcription of PhoP-activated genes required both a PhoQ-inducing signal and the conserved aspartate site of phosphorylation in PhoP (88).
Second, elevated levels of sensor and/or regulator can result in nonphysiological cross talk across two-component systems because of competition between cognate and noncognate proteins, thereby altering the output of a system. It appears that during evolution, cross talk between two-component system proteins has been avoided in two ways: by keeping basal levels of expression of two-component system proteins low, and by utilizing feedback to increase the levels of cognate sensor and regulator proteins only when the signal operating on the sensor is present.
Transcriptional Autoregulation May Alter the Ratio of Sensor to Regulator
The number of sensor and regulator molecules for a given two-component system is not constant under inducing and noninducing conditions, even when the corresponding genes are part of an operon. In the vast majority of examined cases, the number of regulator molecules is larger than the number of sensor molecules. This may reflect that a regulator, often having multiple DNA, RNA, and/or protein targets, operates in a stoichiometric fashion, whereas the sensor functions catalytically as an intermediate in the phosphorylation of the regulator.
The use of different promoters to transcribe a two-component system operon can result in different ratios of regulator to sensor when an organism experiences noninducing versus inducing conditions. For example, the ratio of regulator PhoP to sensor PhoQ was 6.8:1 under noninducing conditions but increased to 18.3:1 under inducing conditions (112). Keeping a physiological ratio of regulator to sensor is critical for the normal operation of the PhoP/PhoQ system, as increased expression of the phoQ gene without a concomitant increase in the expression of phoP abolished transcription of PhoP-activated genes in S. enterica serovar Typhimurium (93). In E. coli, the PhoP:PhoQ ratio at steady state–inducing conditions is 50:1 (63), and it is 35:1 for the OmpR:EnvZ system (17a).
Unlike other two-component system proteins where the regulator is present at higher levels than the sensor, the ratio of regulator LiaR to sensor LiaS is 1:4 in B. subtilis (86). LiaS exhibits both autokinase and phosphotransferase activity toward LiaR, and phosphatase activity toward LiaR-P. Yet, an increase in LiaR levels results in increased transcription of LiaR-activated genes owing to LiaR phosphorylation from acetyl phosphate even when LiaS is present (86). By contrast, the OmpR/EnvZ system from E. coli is reported to be robust to changes in the amounts of OmpR and EnvZ (9).
Transcriptional Autoregulation Is Necessary for a Surge in Phosphorylated Regulator of Classical Two-Component Systems
When an organism experiences an inducing signal for an autoregulated classical two-component system there is a surge in the levels of phosphorylated response regulator, which peaks and then decreases to levels that are 25–30% of the maximum (89) (Figure 2b). The surge in phosphorylated regulator results in corresponding surges in promoter occupancy by the regulator, and in mRNA levels of the target genes (89). The surge requires the ability of a two-component system to positively regulate its own expression as well as the phosphatase activity of the sensor toward its cognate phosphorylated regulator.
The physiological role of the surge was examined in the Salmonella PhoP/PhoQ system by comparing two isogenic strains that differed only in the nature of the promoters driving transcription of the phoPQ operon from its normal chromosomal location. One strain harbored the constitutive and the positively autoregulated phoPQ promoters of wild-type S. enterica serovar Typhimurium. The other strain harbored two constitutive promoters: the original constitutive promoter from the wild-type strain and a promoter with a consensus −35 sequence replacing the PhoP binding site required for positive regulation by PhoP. The latter strain produced PhoP protein amounts constitutively at levels that were equivalent to those of PhoP protein made by the wild-type strain after induction of the PhoP/PhoQ system by low Mg2+ (89).
Both strains reached the same steady state levels of promoter occupancy by the PhoP-P protein as well as PhoP-activated mRNAs within 60 minutes (89). However, they differed in that the strain with the constitutive promoters reached steady state levels monotonically whereas the one with the autoregulated promoter did so following a surge (89). The organism expressing PhoP constitutively was highly attenuated for virulence—as attenuated as mutants lacking the phoP or phoQ genes (89). This striking result established that the surge in PhoP-P is required to jump-start the virulence circuit of S. enterica serovar Typhimurium. Moreover, it indicated that, as discussed above for the developmental pathway controlled by Spo0A in B. subtilis, it is important for an organism not only what particular steady state is achieved upon activation of a given regulatory system, but also how a system reaches that steady state.
A surge has also been observed for the regulator PmrA when Salmonella experienced Fe3+ (89), a signal that activates PmrA’s cognate sensor PmrB (108). These results indicate that the surge behavior is not exclusive to a particular system or inducing signal. The only two properties that the PhoP/PhoQ and PmrA/PmrB systems have in common are that both systems positively regulate their own transcription (93, 109) and the sensors PhoQ and PmrB are bifunctional: they autophosphorylate from ATP and exhibit phosphatase activity toward their respective phosphorylated regulators (112). These results suggest that other autoregulated two-component systems with bifunctional sensors may also display a surge behavior upon experiencing their specific inducing signals. This appears to be the case of the VanR/VanS system from vancomycin-resistant Streptomyces coelicolor (38) and the CovR/CovS system from S. pneumoniae (3), as indicated by the surge in mRNA levels for the genes regulated by the VanR and CovR proteins, respectively.
How does the surge in phosphorylated regulator benefit an organism experiencing an inducing condition for its cognate sensor? It may help an organism to establish a new phenotypic state by producing large amounts of the transcripts encoding proteins that mount a response to the new conditions. The levels of phosphorylated regulator achieved after the surge may allow the organism to maintain the newly established phenotypic state (89).
FEEDBACK INTRINSIC TO SENSOR AND REGULATOR PROTEINS
The interactions that cognate sensor and regulator proteins establish with one another, and/or with small nucleotides modifying their biochemical activities, can alter the output of a two-component system. This form of control is intrinsic to the proteins that constitute a two-component system because it does not require the participation of the genes specifying sensor and regulator, or gene products regulated by the system.
A bifunctional sensor can display intrinsic feedback, modifying the levels of phosphorylated regulator. For example, dynamic modifications in the levels of phosphorylated regulator result from the opposing biochemical activities of the sensor, which changes from being in an autokinase and phosphotransferase mode to being in a phosphatase mode (112) (Figure 2b). These modifications are triggered by the nucleotide occupying the nucleotide-binding pocket in the catalytic subdomain of the sensor. Whereas a sensor typically binds ATP and ADP with similar affinities, ATP promotes sensor autophosphorylation and phosphotransfer, which increases the levels of phosphorylated regulator. By contrast, ADP favors the unphosphorylated state of the regulator in two ways: first, by stimulating the sensor’s phosphatase activity toward the phosphorylated regulator, and second, by inhibiting the autokinase reaction when ADP prevents ATP binding to the nucleotide-binding pocket (112). The ADP is generated endogenously from ATP in vitro. ADP is likely to be derived from ATP in vivo as well, because the physiological ATP:ADP ratio is 10:1 (11, 16), which would favor binding of ATP over ADP if these nucleotides were to diffuse into the nucleotide-binding pocket of the sensor. Because the biochemical activities that promote and hinder the phosphorylated state of the regulator reside within the same protein (i.e., the sensor), two-component systems with bifunctional sensors may be more easily tuned during evolution. Such tuning may alter the intrinsic biochemical properties of the sensor, including binding of allosteric regulators such as ADP and/or interaction with protein factors.
An emerging theme in two-component signal transduction is the allosteric regulation of sensor and regulator proteins. For example, the crystal structure of the complex between the sensor kinase ThkA and the regulator TrrA from Thermotoga maritima uncovered interactions suggesting that the regulator affects the sensing and/or kinase activities of the sensor (111). In the case of the cell fate–determining phosphorylation network of Caulobacter crescentus, the regulator DivK allosterically switches the sensor PleC from a phosphatase into an autokinase mode and stimulates the autokinase activity of the sensor DivJ (69). These DivK activities have two important consequences. First, DivK, which lacks an output domain, stimulates phosphorylation of the regulator PleD indirectly, by stimulating the autokinase activity of the sensor PleC. And second, phosphorylated DivJ serves as a phosphodonor for DivK, which favors DivK polar localization.
Certain interactions among sensors or between sensor and regulator have phenotypic consequences independent of phosphorylation (79). For instance, in the DesK/DesR two-component system from B. subtilis, the sensor DesK allosterically activates its cognate regulator DesR in a process that is independent of phosphorylation (99). Interestingly, this activation is regulated by temperature (99), which constitutes the physiological signal sensed by DesK (1). The allosteric activation of the sensor facilitates regulator phosphorylation. Moreover, it provides a plausible explanation why regulator phosphorylation is more efficient when sensors are used as phosphodonors instead of small–molecular weight phosphoryl donors (99).
FEEDBACK CONTROL BY GENE PRODUCTS REGULATED BY TWO-COMPONENT SYSTEMS
When an organism experiences an inducing signal for a two-component system mediating a response to a particular stress, a bacterium expresses proteins designed to help the organism cope with such a stress. These proteins alleviate the stress, enabling the organism to prosper in a stressful environment without destroying the stress. Feedback mechanisms enable organisms to reduce expression of the stress response proteins once they reach a particular level. In other cases, the opposite takes place: Production of stress response proteins further enhances activation of a two-component system. Genes controlled by a given system may target different steps in a signal transduction cascade—from signal perception to direct interaction with sensor or regulator proteins.
Modifying Signal Access to the Sensor
The PmrA/PmrB two-component system controls the chemical modification of the lipopolysaccharide (LPS) in the outer membranes of S. enterica serovar Typhimurium and E. coli (19). The sensor PmrB is activated by a mildly acidic pH (71) and/or the presence of Fe3+ in the periplasm (108). The regulator PmrA governs transcription of genes specifying products involved in the chemical modification of the LPS in the outer membrane (109), rendering the organism resistant to killing by Fe3+ (64). The PmrA-dependent modification of phosphate residues in the LPS hinders Fe3+ binding to the bacterial cell surface, thereby decreasing Fe3+ availability for activation of PmrB (42) (Figure 3a). This regulatory mechanism enables the bacterium to adjust the levels of PmrA-P dynamically, based on the degree of LPS modification. In other words, the PmrB protein responds to the downstream effects of the PmrA-regulated changes taking place on the bacterial cell surface. In addition to the negative feedback exerted by the PmrA-regulated LPS-modifying gene products, the sensor PmrB exhibits intrinsic negative feedback (112), and the regulator PmrA positively regulates its own transcription (109).
Figure 3.
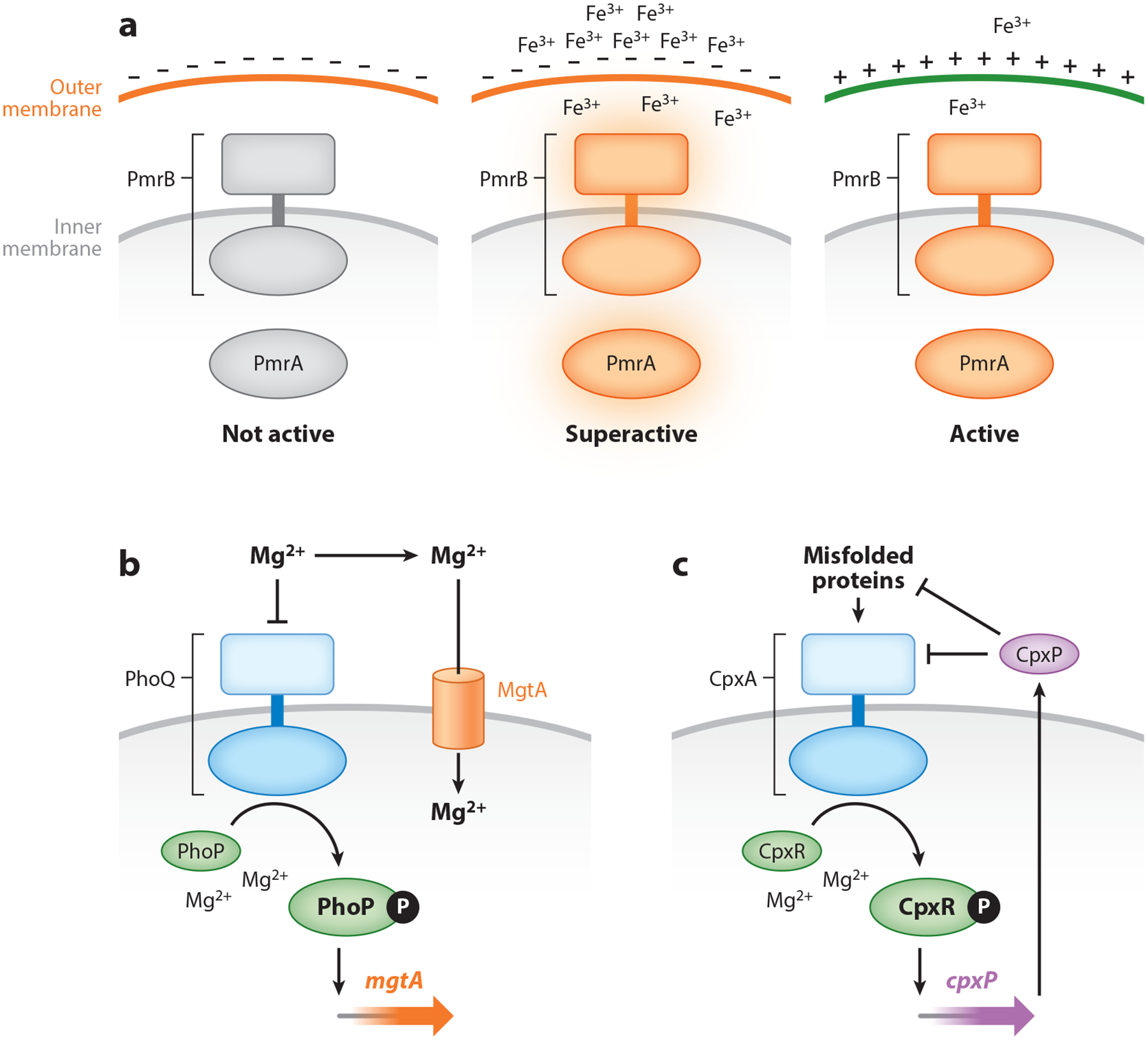
Feedback mechanisms altering sensor access of an inducing signal, a repressing signal, and a protein with regulatory properties. (a) Under noninducing conditions for the sensor PmrB, the regulator PmrA is not active and the outermost part of the outer membrane is negatively charged owing to the phosphate residues in the lipopolysaccharide. The positively charged Fe3+ binds to the negatively charged outer membrane, which increases Fe3+ levels in the periplasm, where it serves as an inducing ligand for PmrB and results in a superactive PmrA. The covalent modification of the negative charges in the lipopolysaccharide by PmrA-activated gene products reduces the negative charge of the outer membrane, which decreases Fe3+ binding, further diminishing access to PmrB and activation of PmrA. (b) The sensor PhoQ is activated when periplasmic Mg2+ levels are low, resulting in phosphorylation of the regulator PhoP and transcription of PhoP-activated genes. The PhoP-activated mgtA gene specifies a Mg2+ transporter that imports Mg2+ from the periplasm into the cytoplasm, thereby removing an inhibitory signal for PhoQ. Thus, MgtA increases the levels of phosphorylated PhoP. (c) Misfolded proteins in the periplasm activate the sensor CpxA, favoring phosphorylation of the regulator CpxR and transcription of the CpxR-activated genes, including cpxP. The CpxP protein binds to misfolded proteins in the periplasm; as the levels of these proteins decrease, CpxP binds to the CpxA, decreasing its activation.
The PhoP/PhoQ system governs Mg2+ homeostasis in most enteric species (72). The sensor PhoQ responds to low Mg2+ in the periplasm by activating the regulator PhoP, which promotes transcription of different genes, including two specifying Mg2+ transport proteins (31). In S. enterica serovar Typhimurium and E. coli, the PhoP protein governs transcription initiation of the Mg2+ transporter mgtA gene (60, 94). By importing Mg2+ into the cytoplasm (92), MgtA removes a repressing signal away from PhoQ’s Mg2+-sensing domain in the periplasm, thereby increasing PhoP-P levels (68) (Figure 3b).
Transcription elongation into the mgtA coding region is controlled by the leader region of the mgtA transcript, which operates as a Mg2+-responding riboswitch that allows transcription elongation into the mgtA coding region when cytosolic Mg2+ levels drop below a certain threshold (20). The MgtA-dependent activation of PhoP specifically takes place when the inducing signal acting on PhoQ is low Mg2+. By contrast, MgtA had no effect on PhoP-dependent gene transcription when PhoQ was activated by a mildly acidic pH or a cationic antimicrobial peptide (68), because MgtA does not, it appears, transport H+ or antimicrobial peptides. The MgtA-dependent positive feedback on PhoP-P enables timing expression of a subset of PhoP-dependent genes to follow the cytosolic signal advancing mgtA expression (68) and activity (Figure 3b).
The hybrid two-component system BT3334 governs the uptake and breakdown of chondroitin in B. thetaiotaomicron. A regulatory scheme allows B. thetaiotaomicron to respond to the rate at which a chondroitin is utilized as opposed to monitoring its presence/absence. This scheme entails BT3334, which is activated in the periplasm by a disaccharide generated during chondroitin breakdown, rather than by chondroitin itself (80), and a periplasmic glycosyl hydrolase that splits the disaccharide into two monosaccharides. Even though BT3334 and the glycosyl hydrolase exhibit similar affinities for the disaccharide, the disaccharide binds preferentially to BT3334 upon exposure to chondroitin, because BT3334 is constitutively produced whereas the glycosyl hydrolase requires transcriptional activation by BT3334. As the level of the glycosyl hydrolase increases, the amount of disaccharide decreases, thereby reducing the concentration of the inducing ligand for BT3334, which turns down expression of BT3334-activated genes (80).
Unlike the autoregulated PhoP/PhoQ and PmrA/PmrB systems, the BT3334 hybrid two-component system is constitutively expressed. Yet, BT3334 displays a surge behavior that requires a BT3334-regulated gene product (i.e., the glycosyl hydrolase). This is in contrast to the surge in PhoP/PhoQ and PmrA/PmrB, which is intrinsic to these proteins (80).
The abundance of hybrid two-component systems devoted to polysaccharide utilization within and across Bacteroides species (110) raises the possibility of the response to metabolic intermediates governing the activity of other hybrid two-component systems operating in a manner analogous to that of BT3334.
Modifying the Activity of the Sensor
The CpxR/CpxA two-component system of E. coli is activated by a variety of membrane stresses, including the presence of misfolded proteins (81). The regulator CpxR responds to these stresses by promoting expression of proteins that facilitate protein folding and/or degradation in the periplasm. The positively autoregulated CpxR/CpxA system is negatively regulated by the CpxR-activated periplasmic protein CpxP (82), which exerts its effect by binding to the sensor CpxA (36) (Figure 3c). When CpxP is titrated by misfolded proteins, it is no longer available to silence CpxA, allowing expression of CpxR-activated genes (21). As the bacterium recovers from the stress, the number of misfolded proteins decreases, liberating CpxP to exert its repressing action on CpxA.
Two PhoP-activated gene products exert negative feedback on the sensor PhoQ: MgrB and SlyB. The 47–amino acid inner membrane peptide MgrB binds to PhoQ, thereby decreasing transcription of PhoP-activated genes in E. coli (52). MgrB appears to target the periplasmic domain of PhoQ, because MgrB no longer exerted its repressive effect in a strain with a PhoQ variant harboring the PhoQ periplasmic domain from P. aeruginosa instead of its own (52). The MgrB-mediated action is likely to take place in other members of the family Enterobacteriaceae, because highly conserved mgrB homologs are found in several species and also because the Yersinia mgrB gene rescued an E. coli mgrB mutant (52).
Second, inactivation of the Salmonella slyB gene derepressed transcription of PhoP-dependent genes by a factor of 1.5 to 2, including that of the mgrB (also designated yobG) gene (72). SlyB is a lipoprotein predicted to localize to the outer membrane. Overexpression of the slyB gene inhibited Salmonella growth in low but not in high Mg2+, presumably because the PhoP/PhoQ system failed to fully express the regulated target(s) necessary for growth in low Mg2+ (72). Despite the high degree of amino acid identity between the SlyB proteins from S. enterica serovar Typhimurium and E. coli, inactivation of the slyB gene did not alter PhoP-dependent gene transcription in the latter species (52). These results suggest that certain feedback mechanisms are species specific even when mediated by highly conserved genes.
The SaeR/SaeS two-component system controls virulence gene expression in the gram-positive pathogen Staphylococcus aureus. It is encoded in an operon that also includes two additional genes specifying the SaeP and SaeQ proteins, which form a complex with the sensor SaeS. SaeP and SaeQ are required for SaeS’s phosphatase activity toward SaeR-P, suggesting they help S. aureus decrease the output of the system (40).
In B. subtilis, the LiaR/LiaS two-component system is activated by particular antibiotics that perturb cell wall synthesis (58). In addition to the positive transcriptional autoregulation exerted by LiaR, two products encoded in the operon of liaR and liaS (i.e., LiaH and LiaF) (41) exert negative control on the activity of the system (86). LiaF exerts its effects via LiaS, because it had no effect on LiaR-dependent gene transcription when LiaR was activated by acetyl phosphate (86).
Several two-component systems depend on auxiliary factors for sensing particular signals (17). These auxiliary factors typically associate with the sensor, often operating as cosensors, and are under transcriptional control of the regulator. For example, the DcuR/DcuS system from E. coli governs the uptake and breakdown of C4-dicarboxylates such as succinate and fumarate. DcuR controls transcription of the dctA and dcuB genes, which specify a C4-dicarboxylate transporter and a fumarate/succinate antiporter, respectively. Whereas the sensor DcuS can respond to the presence of C4-dicarboxylates, the transporters DctA and DcuB enable tuning of the DcuR/DcuS system to the flux of C4-dicarboxylates (107). A flux-sensing mechanism also controls resistance to antimicrobial peptides in several gram-positive species. The two-component system BceR/BceS associates with a transporter specified by the BceR-activated bceA and bceB genes and confers resistance to bacitracin. The behavior of the system reflects the concentration of the antibiotic outside the bacterium as well as dynamic changes in the levels of transporter proteins (26).
Modifying the Activity and/or Levels of the Regulator
The regulator PmrA is normally activated by a mildly acidic pH (71) or Fe3+ (108) sensed by its cognate sensor PmrB. In addition, transcription of PmrA-dependent genes is induced when S. enterica serovar Typhimurium experiences the signals activating the noncognate sensor PhoQ (43, 48, 95). The PhoP-activated gene product PmrD is responsible for the latter activation: PmrD binds to PmrA-P, preventing dephosphorylation by PmrB (46, 48, 95). PmrA, in turn, represses pmrD transcription by binding to the pmrD promoter (45).
Certain regulators are active in the unphosphorylated form. For example, the regulator SreR from the plant-associated bacterium Xanthomonas campestris binds to some of its targets when unphosphorylated. The unphosphorylated form of SreR is promoted by the SreS protein, which acts as a phosphate sink by competing with SreR for the phosphoryl group of the sensor SreK (104).
As discussed above, RcsB can regulate gene transcription in both its phosphorylated and its unphosphorylated forms. RcsB phosphorylation is required when RcsB operates as a homodimer or RcsB-RcsA heterodimer, but not when it functions as a heterodimeric protein with BglJ, MatA, or GadE (67). This raises the possibility of conditions that promote expression of the latter proteins preventing RcsB-P-dependent gene control by titrating RcsB away in heterodimeric complexes. Heterodimer formation is not exclusive to the regulator RcsB from enteric bacteria, as the orphan regulators BldM and WhiI from Streptomyces coelicolor form a heterodimer that controls transcription of a different set of genes than the BldM homodimer (2).
Certain two-component systems are feedback regulated by small regulatory RNAs (see for References 29 and 55 for reviews). For example, the regulator OmpR of E. coli promotes transcription of the sRNAs OmrA and OmrB, which, when overproduced, decrease the levels of the ompR transcript, thereby creating a negative feedback loop (33). Likewise, the regulator CpxR protein is a direct transcriptional activator of the gene rprA, which specifies an sRNA that, upon overexpression, downregulates cpxR mRNA levels (103).
There are two consequences of feedback control by gene products controlled by a given regulator. First, it helps maintain and/or establish particular levels of activation of a given two-component regulatory system. Of course, the protein products exerting feedback by direct interaction with a sensor and/or regulator proteins operate faster on the output of a two-component system than sRNAs controlling the stability and/or translation of the corresponding transcripts. And second, the gene products mediating feedback are often subjected to regulation by signals other than those acting on the sensor and/or regulator of a two-component system. This raises the possibility of integration of additional inputs into the output of a two-component system.
FINAL CONSIDERATIONS
The information developed over the last 25 years on feedback control of two-component systems has enabled the development of regulatory circuits with defined properties. For example, the incorporation of positive feedback to the constitutively expressed copper-responding CusR/CusS system from E. coli had three consequences (83). First, it increased the sensitivity to copper, enabling activation at copper concentrations 10 times lower than those activating the original system present in wild-type organisms. Second, it amplified the output of the system twofold. And third, it extended the duration of the response to high copper concentrations.
The positive feedback that two-component systems exert on their own expression can give rise to what has been referred to as a learning behavior. That is to say, an organism that autoregulates a two-component system responds faster and to a greater degree when exposed for a second time to the signal activating the sensor. In the case of the phosphate limitation–inducible system PhoB/PhoR of E. coli, a faster response was observed in organisms that had experienced low phosphate up to 2 h before (35). The learning behavior reflects that PhoB/PhoR positively activates transcription of the phoBphoR operon (32, 90), and that the initial activation created a pool of PhoB protein that could be readily phosphorylated when the organism experienced the PhoR inducing signal a second time.
Finally, feedback mechanisms operating on two-component signal transduction pathways provide organisms with the ability to continuously adjust their behavior, even in the presence of a constant stress signal. Therefore, they enable bacteria to incorporate information about the status of the cell into the gene control triggered by an extracellular stress.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Bassam MM, Bibb MJ, Bush MJ, Chandra G, Buttner MJ. 2014. Response regulator heterodimer formation controls a key stage in Streptomyces development. PLOS Genet. 10:e1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alloing G, Martin B, Granadel C, Claverys JP. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol 29:75–83 [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol 179:97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban NQ. 2011. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev 21:768–75 [DOI] [PubMed] [Google Scholar]
- 6.Bang IS, Audia JP, Park YK, Foster JW. 2002. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol 44:1235–50 [DOI] [PubMed] [Google Scholar]
- 7.Barbieri CM, Stock AM. 2008. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem 376:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbieri CM, Wu T, Stock AM. 2013. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J. Mol. Biol 425:1612–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batchelor E, Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. PNAS 100:691–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier D, Gross R. 2008. The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv. Exp. Med. Biol 631:149–60 [DOI] [PubMed] [Google Scholar]
- 11.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol 5:593–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bijlsma JJ, Groisman EA. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol 57:85–96 [DOI] [PubMed] [Google Scholar]
- 13.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, et al. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904 [DOI] [PubMed] [Google Scholar]
- 14.Botella E, Devine SK, Hubner S, Salzberg LI, Gale RT, et al. 2014. PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol 94:1242–59 [DOI] [PubMed] [Google Scholar]
- 15.Boudes M, Sanchez D, Graille M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S. 2014. Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae. Nucleic Acids Res. 42:5302–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckstein MH, He J, Rubin H. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol 190:718–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buelow DR, Raivio TL. 2010. Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol. Microbiol 75:547–66 [DOI] [PubMed] [Google Scholar]
- 17a.Cai S, Inouye M. 2002. EnvZ-OmpR interactions and osmoregulation in Escherichia coli. J. Biol. Chem 277:24155–61 [DOI] [PubMed] [Google Scholar]
- 18.Cameron AD, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLOS Genet. 8:e1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol 67:83–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cromie MJ, Shi Y, Latifi T, Groisman EA. 2006. An RNA sensor for intracellular Mg2+. Cell 125:71–84 [DOI] [PubMed] [Google Scholar]
- 21.DiGiuseppe PA, Silhavy TJ. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol 185:2432–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doolittle RF, Feng DF, Tsang S, Cho G, Little E. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470–77 [DOI] [PubMed] [Google Scholar]
- 23.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol 70:1307–22 [DOI] [PubMed] [Google Scholar]
- 24.Feng X, Oropeza R, Kenney LJ. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol 48:1131–43 [DOI] [PubMed] [Google Scholar]
- 25.Freeman ZN, Dorus S, Waterfield NR. 2013. The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLOS Pathog. 9:e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fritz G, Dintner S, Treichel NS, Radeck J, Gerland U, et al. 2015. A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6:e00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol 187:1357–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao R, Stock AM. 2015. Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. mBio 6:e00686–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopel Y, Gorke B. 2012. Rewiring two-component signal transduction with small RNAs. Curr. Opin. Microbiol 15:132–39 [DOI] [PubMed] [Google Scholar]
- 30.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol 183:1835–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groisman EA, Hollands K, Kriner MA, Lee EJ, Park SY, Pontes MH. 2013. Bacterial Mg2+ homeostasis, transport, and virulence. Annu. Rev. Genet 47:625–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan C-D, Wanner B, Inouye H. 1983. Analysis of regulation of phoB using a phoB-cat fusion. J. Bacteriol 156:710–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillier M, Gottesman S. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 36:6781–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengge R 2008. The two-component network and the general stress sigma factor RpoS (σS) in Escherichia coli. Adv. Exp. Med. Biol 631:40–53 [DOI] [PubMed] [Google Scholar]
- 35.Hoffer SM, Westerhoff HV, Hellingwerf KJ, Postma PW, Tommassen J. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J. Bacteriol 183:4914–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornschemeyer P, Liss V, Heermann R, Jung K, Hunke S. 2016. Interaction analysis of a two-component system using nanodiscs. PLOS ONE 11:e0149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutchings MI, Hong HJ, Buttner MJ. 2006. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol 59:923–35 [DOI] [PubMed] [Google Scholar]
- 39.Ishii E, Eguchi Y, Utsumi R. 2013. Mechanism of activation of PhoQ/PhoP two-component signal transduction by SafA, an auxiliary protein of PhoQ histidine kinase in Escherichia coli. Biosci. Biotechnol. Biochem 77:814–19 [DOI] [PubMed] [Google Scholar]
- 40.Jeong DW, Cho H, Jones MB, Shatzkes K, Sun F, et al. 2012. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol 86:331–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol 188:5153–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato A, Chen HD, Latifi T, Groisman EA. 2012. Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol. Cell 47:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato A, Groisman EA. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 18:2302–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato A, Groisman EA. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol 631:7–21 [DOI] [PubMed] [Google Scholar]
- 45.Kato A, Latifi T, Groisman EA. 2003. Closing the loop: The PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. PNAS 100:4706–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato A, Mitrophanov AY, Groisman EA. 2007. A connector of two-component regulatory systems promotes signal amplification and persistence of expression. PNAS 104:12063–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosono S, Tamura M, Suzuki S, Kawamura Y, Yoshida A, et al. 2015. Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLOS ONE 10:e0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kox LFF, Wosten MMSM, Groisman EA. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet 41:121–45 [DOI] [PubMed] [Google Scholar]
- 50.Lejona S, Castelli ME, Cabeza ML, Kenney LJ, Garcia Vescovi E, Soncini FC. 2004. PhoP can activate its target genes in a PhoQ-independent manner. J. Bacteriol 186:2476–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin YH, Gao R, Binns AN, Lynn DG. 2008. Capturing the VirA/VirG TCS of Agrobacterium tumefaciens. Adv. Exp. Med. Biol 631:161–77 [DOI] [PubMed] [Google Scholar]
- 52.Lippa AM, Goulian M. 2009. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLOS Genet. 5:e1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol 34:305–16 [DOI] [PubMed] [Google Scholar]
- 54.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol 59:379–405 [DOI] [PubMed] [Google Scholar]
- 55.Mandin P, Guillier M. 2013. Expanding control in bacteria: interplay between small RNAs and transcriptional regulators to control gene expression. Curr. Opin. Microbiol 16:125–32 [DOI] [PubMed] [Google Scholar]
- 56.Martin B, Granadel C, Campo N, Henard V, Prudhomme M, Claverys JP. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol. Microbiol 75:1513–28 [DOI] [PubMed] [Google Scholar]
- 57.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev 70:910–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol 50:1591–604 [DOI] [PubMed] [Google Scholar]
- 58a.McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem 269:31657–72 [PubMed] [Google Scholar]
- 59.Mika F, Hengge R. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev. 19:2770–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, et al. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol 185:3696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitrophanov AY, Groisman EA. 2008. Positive feedback in cellular control systems. BioEssays 30:542–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitrophanov AY, Hadley TJ, Groisman EA. 2010. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J. Mol. Biol 401:671–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyashiro T, Goulian M. 2008. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. PNAS 105:17457–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishino K, Hsu FF, Turk J, Cromie MJ, Wosten MM, Groisman EA. 2006. Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol. Microbiol 61:645–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Norsworthy AN, Visick KL. 2015. Signaling between two interacting sensor kinases promotes biofilms and colonization by a bacterial symbiont. Mol. Microbiol 96:233–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ochman H, Wilson AC. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol 26:74–86 [DOI] [PubMed] [Google Scholar]
- 67.Pannen D, Fabisch M, Gausling L, Schnetz K. 2016. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem 291:2357–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park SY, Groisman EA. 2014. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol. Microbiol 91:135–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, et al. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perego M, Brannigan JA. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22:1541–47 [DOI] [PubMed] [Google Scholar]
- 71.Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol 63:283–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez JC, Shin D, Zwir I, Latifi T, Hadley TJ, Groisman EA. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLOS Genet. 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pescaretti M, Farizano JV, Morero R, Delgado MA. 2013. A novel insight on signal transduction mechanism of RcsCDB system in Salmonella enterica serovar Typhimurium. PLOS ONE 8:e72527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pescaretti M, Lopez FE, Morero RD, Delgado MA. 2010. Transcriptional autoregulation of the RcsCDB phosphorelay system in Salmonella enterica serovar Typhimurium. Microbiology 156:3513–21 [DOI] [PubMed] [Google Scholar]
- 75.Pescaretti M, Morero R, Delgado MA. 2009. Identification of a new promoter for the response regulator rcsB expression in Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett 300:165–73 [DOI] [PubMed] [Google Scholar]
- 76.Pontes MH, Smith KL, De Vooght L, Van Den Abbeele J, Dale C. 2011. Attenuation of the sensing capabilities of PhoQ in transition to obligate insect-bacterial association. PLOS Genet. 7:e1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prajapat MK, Saini S. 2013. Role of feedback and network architecture in controlling virulence gene expression in Bordetella. Mol. Biosyst 9:2635–44 [DOI] [PubMed] [Google Scholar]
- 78.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93 [DOI] [PubMed] [Google Scholar]
- 79.Raghavan V, Groisman EA. 2010. Orphan and hybrid two-component system proteins in health and disease. Curr. Opin. Microbiol 13:226–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raghavan V, Lowe EC, Townsend GE 2nd, Bolam DN, Groisman EA. 2014. Tuning transcription of nutrient utilization genes to catabolic rate promotes growth in a gut bacterium. Mol. Microbiol 93:1010–25 [DOI] [PubMed] [Google Scholar]
- 81.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 1843:1529–41 [DOI] [PubMed] [Google Scholar]
- 82.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol 181:5263–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravikumar S, Pham VD, Lee SH, Yoo IK, Hong SH. 2012. Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop. J. Ind. Microbiol. Biotechnol 39:861–68 [DOI] [PubMed] [Google Scholar]
- 84.Ryndak M, Wang S, Smith I. 2008. PhoP, a key player in Mycobacterium tuberculosis virulence. Trends Microbiol. 16:528–34 [DOI] [PubMed] [Google Scholar]
- 85.Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, et al. 2015. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol 98:847–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schrecke K, Jordan S, Mascher T. 2013. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol. Microbiol 87:769–88 [DOI] [PubMed] [Google Scholar]
- 87.Sherwood EJ, Bibb MJ. 2013. The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. PNAS 110:E2500–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shin D, Groisman EA. 2005. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem 280:4089–94 [DOI] [PubMed] [Google Scholar]
- 89.Shin D, Lee EJ, Huang H, Groisman EA. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607–9 [DOI] [PubMed] [Google Scholar]
- 90.Shinagawa H, Makino K, Nakata A. 1983. Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J. Mol. Biol 168:477–88 [DOI] [PubMed] [Google Scholar]
- 91.Shu CJ, Zhulin IB. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci 27:3–5 [DOI] [PubMed] [Google Scholar]
- 92.Snavely MD, Florer JB, Miller CG, Maguire ME. 1989. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtA systems. J. Bacteriol 171:4761–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Soncini FC, García Véscovi E, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol 177:4364–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soncini FC, García Véscovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol 178:5092–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soncini FC, Groisman EA. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol 178:6796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella Typhimurium phoPQ operon. J. Bacteriol 177:4364–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem 69:183–215 [DOI] [PubMed] [Google Scholar]
- 97a.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodreigue S, Basu J, Kundu M. 2007. Polysphosphate kinase is involved in stress-induced mprAB-sigE-rel signaling in mycobacteria. Mol. Microbiol 65:261–76 [DOI] [PubMed] [Google Scholar]
- 98.Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol 40:440–50 [DOI] [PubMed] [Google Scholar]
- 99.Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, et al. 2014. Allosteric activation of bacterial response regulators: The role of the cognate histidine kinase beyond phosphorylation. mBio 5:e02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. 2010. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell 37:567–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148:69–78 [DOI] [PubMed] [Google Scholar]
- 102.Vishnoi M, Narula J, Devi SN, Dao HA, Igoshin OA, Fujita M. 2013. Triggering sporulation in Bacillus subtilis with artificial two-component systems reveals the importance of proper Spo0A activation dynamics. Mol. Microbiol 90:181–94 [DOI] [PubMed] [Google Scholar]
- 103.Vogt SL, Evans AD, Guest RL, Raivio TL. 2014. The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J. Bacteriol 196:4229–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang FF, Deng CY, Cai Z, Wang T, Wang L, et al. 2014. A three-component signalling system fine-tunes expression kinetics of HPPK responsible for folate synthesis by positive feedback loop during stress response of Xanthomonas campestris. Environ. Microbiol 16:2126–44 [DOI] [PubMed] [Google Scholar]
- 105.Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. 2012. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol. Microbiol 86:645–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williams CL, Cotter PA. 2007. Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J. Bacteriol 189:1974–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Witan J, Monzel C, Scheu PD, Unden G. 2012. The sensor kinase DcuS of Escherichia coli: two stimulus input sites and a merged signal pathway in the DctA/DcuS sensor unit. Biol. Chem 393:1291–97 [DOI] [PubMed] [Google Scholar]
- 108.Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–25 [DOI] [PubMed] [Google Scholar]
- 109.Wösten MMSM, Groisman EA. 1999. Molecular characterization of the PmrA regulon. J. Biol. Chem 274:27185–90 [DOI] [PubMed] [Google Scholar]
- 109a.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–76 [DOI] [PubMed] [Google Scholar]
- 110.Xu J, Chiang HC, Bjursell MK, Gordon JI. 2004. Message from a human gut symbiont: Sensitivity is a prerequisite for sharing. Trends Microbiol. 12:21–28 [DOI] [PubMed] [Google Scholar]
- 111.Yamada S, Akiyama S, Sugimoto H, Kumita H, Ito K, et al. 2006. The signaling pathway in histidine kinase and the response regulator complex revealed by X-ray crystallography and solution scattering. J. Mol. Biol 362:123–39 [DOI] [PubMed] [Google Scholar]
- 112.Yeo WS, Zwir I, Huang HV, Shin D, Kato A, Groisman EA. 2012. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol. Cell 45:409–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. 2012. The promoter architectural landscape of the Salmonella PhoP regulon. Mol. Microbiol 84:463–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zwir I, Yeo WS, Shin D, Latifi T, Huang H, Groisman EA. 2014. Bacterial nucleoid-associated protein uncouples transcription levels from transcription timing. mBio 5:e01485–14 [DOI] [PMC free article] [PubMed] [Google Scholar]


