ABSTRACT
Because both dearth and overabundance of histones result in cellular defects, histone synthesis and demand are typically tightly coupled. In Drosophila embryos, histones H2B, H2A and H2Av accumulate on lipid droplets (LDs), which are cytoplasmic fat storage organelles. Without LD binding, maternally provided H2B, H2A and H2Av are absent; however, how LDs ensure histone storage is unclear. Using quantitative imaging, we uncover when during oogenesis these histones accumulate, and which step of accumulation is LD dependent. LDs originate in nurse cells (NCs) and are transported to the oocyte. Although H2Av accumulates on LDs in NCs, the majority of the final H2Av pool is synthesized in oocytes. LDs promote intercellular transport of the histone anchor Jabba and thus its presence in the ooplasm. Ooplasmic Jabba then prevents H2Av degradation, safeguarding the H2Av stockpile. Our findings provide insight into the mechanism for establishing histone stores during Drosophila oogenesis and shed light on the function of LDs as protein-sequestration sites.
KEY WORDS: Histones, Protein sequestration, Lipid droplets, Drosophila oogenesis, Protein turnover, Proteasome
Summary: During Drosophila oogenesis, lipid droplets promote the intercellular transport of the histone anchor Jabba to the oocyte, where it prevents histone degradation.
INTRODUCTION
Histones perform crucial functions in eukaryotic cells, from promoting genome stability (Herrero and Moreno, 2011) to regulating gene expression (Celona et al., 2011). These roles are sensitive to histone levels: both too few and too many histones can result in severe defects (Gunjan and Verreault, 2003; Han et al., 1987; Meeks-Wagner and Hartwell, 1986; Singh et al., 2009). Therefore, histone abundance is typically closely coupled to histone demand, e.g. synthesis of canonical histones occurs almost exclusively during DNA replication, ensured via multiple regulatory mechanisms that control transcription and mRNA turnover (Marzluff et al., 2008).
However, these strategies are unavailable during developmental stages that are transcriptionally silent, such as early embryonic. Histone messages cannot be synthesized, and their destruction at the end of one S phase would prevent their translation in the next one. Many animal eggs therefore stockpile excess histones (O'Farrell et al., 2004). For Drosophila, it has been estimated that newly laid eggs have a 1000-fold excess of histones over DNA (Cermelli et al., 2006). But as excess histones are toxic, embryos have to prevent the dangers of histone overabundance.
Two strategies are known to solve this problem. In Xenopus, excess histones form complexes with cytosolic chaperones; histone release is developmentally controlled by post-translational modifications (Onikubo et al., 2015). In Drosophila, certain histones (H2B, H2A and H2Av) are sequestrated on lipid droplets (LDs) (Cermelli et al., 2006): the cellular organelles for fat storage (Walther and Farese, 2012). LD-bound histones can be transferred to nuclei (Johnson et al., 2018) and support early development (Li et al., 2012). Although it is unclear how widespread these strategies are, histones on LDs have been detected in embryos and oocytes of house flies and mice (Cermelli et al., 2006; Kan et al., 2012), and in somatic cells (Kan et al., 2012; Nagai et al., 2005; see also Welte, 2015).
LDs play crucial roles in lipid metabolism and energy homeostasis (Walther and Farese, 2012; Yu and Li, 2017; Zechner et al., 2012). They also perform regulatory functions in the life cycles of many proteins (Filipe and McLauchlan, 2015; Ohsaki et al., 2006, 2008; Welte and Gould, 2017), e.g. sequestering proteins involved in transcriptional regulation (Aramburu et al., 2006; Gallardo-Montejano et al., 2016; Mejhert et al., 2020; Romanauska and Köhler, 2018; Zhang et al., 2017). However, the function of such sequestration is molecularly understood in only a few cases.
In Drosophila embryos, newly translated H2Av transiently binds to LDs; LD binding retains it in the cytoplasm and slows its nuclear import (Johnson et al., 2018). This ‘buffering’ role of LDs prevents H2Av overaccumulation on chromatin (Li et al., 2014). As newly laid embryos from mothers lacking Jabba, the histone anchor on LDs, have reduced levels of H2B, H2A and H2Av (Li et al., 2012), LDs somehow promote the build-up of maternally provided histones. In contrast to short-term buffering, the molecular basis of this long-term ‘storage’ is unknown. These two roles of histone sequestration likely require distinct mechanisms as buffering changes the intracellular distribution of histones, not their levels. Additionally, LD buffering in early embryos has no detectable effects on H2A and H2B nuclear accumulation (Li et al., 2014), but LD sequestration is crucial to maintain H2A and H2B stores, and thus applies to a broader set of histones.
During Drosophila oogenesis, mature eggs are produced by egg chambers (ECs), which consist of follicle cells, the germline-derived oocyte and its 15 sister cells [nurse cells (NCs)]. The oocyte initiates meiosis, is arrested in metaphase (Ables, 2015; Von Stetina and Orr-Weaver, 2011) and is transcriptionally silent (Navarro-Costa et al., 2016). The NCs manufacture many components needed for oocyte growth. As NCs and oocyte remain connected through cytoplasmic bridges, NC contents can be transferred to the oocyte, initially slowly and later rapidly by NC contraction during stages 10B (S10B) and S11, which squeezes the remaining NC cytoplasm into the oocyte (Hudson and Cooley, 2014); the latter transfer is called ‘dumping’. This structure of ECs suggests three possibilities for how lack of Jabba might lead to reduced H2Av in newly laid eggs. First, Jabba may be necessary for histone biosynthesis, by promoting transcription or translation. Second, if histones are made in NCs, Jabba may facilitate their transport to the oocyte by attaching them to LDs in the NCs. Finally, Jabba may prevent histone turnover. LD binding can indeed prevent degradation of proteins (Masuda et al., 2006; Takahashi et al., 2016; Xu et al., 2006).
Using quantitative imaging, we determined the developmental time course of H2Av accumulation in oocytes and find that the bulk of oocyte H2Av does not depend on Jabba-mediated transport from NCs. We detect no evidence for Jabba-dependent H2Av biosynthesis but show that, without Jabba, H2Av is turned over in a proteasome-dependent manner. Unlike transport of H2Av, oocyte accumulation of Jabba depends on its LD-binding ability. We propose that LDs retain Jabba in the NC cytoplasm, thus allowing its transport into the oocyte, where it then protects H2Av from degradation. Taken together, our results argue that, during oogenesis, LD sequestration is necessary for moving Jabba into the oocyte, where Jabba then promotes histone stability.
RESULTS
Histones localize to NC LDs in a Jabba-dependent manner
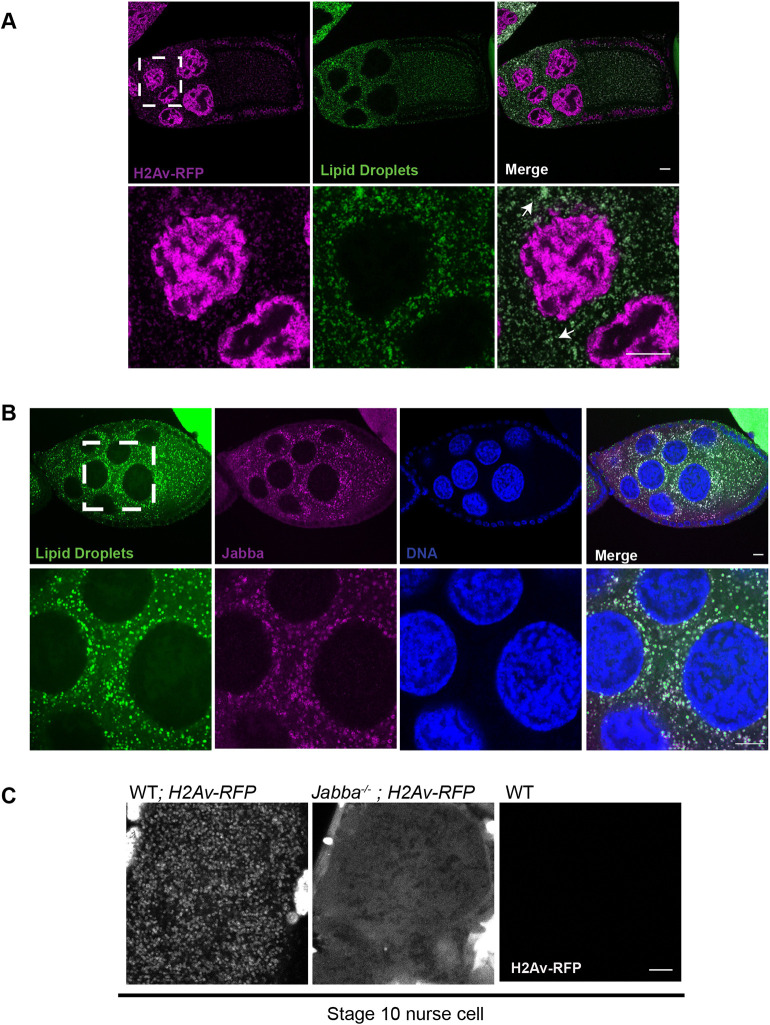
Jabba might promote histone storage by boosting synthesis, mediating histone transport to oocytes or preventing histone degradation. To start distinguishing between these models, we asked when histones associate with LDs, using females expressing H2Av-RFP and co-labeling samples with BODIPY (Fig. 1A). In agreement with earlier studies (summarized by Welte, 2015), we found that LDs are sparse until S8. LDs are present in massive amounts in NCs from S9 onwards until dumping (not shown). We observed H2Av-RFP puncta in the cytoplasm of NCs of the same stages (Fig. 1C), and they colocalized with LDs (Fig. 1A). This result is consistent with earlier observations (Cermelli et al., 2006).
Fig. 1.
Histones localize to NC LDs in a Jabba-dependent manner. (A) H2Av (magenta) is abundant in nuclei and cytoplasmic puncta on LDs (green). Arrows indicate H2Av-Jabba colocalization. (B) Jabba (magenta) localizes to LDs. ECs are stained with Nile Red (green) and DAPI (blue). (C) Jabba anchors H2Av to LDs in ovaries. NCs of S10 wild-type and Jabba−/− ECs expressing H2Av-RFP. H2Av (white) is present in distinct puncta in the wild type (left) and diffusely through the cytoplasm in Jabba−/− (center); wild-type NCs not expressing fluorescently tagged H2Av (right) show no signal. Dashed boxes indicate the area shown at higher magnification in the lower panels. Scale bars: 10 μm.
In the embryo, H2Av-LD association is mediated by Jabba, likely via direct physical interactions (Kolkhof et al., 2017; Li et al., 2012). Immunostaining revealed that Jabba was already present on LDs in NCs (Fig. 1B). By live imaging, H2Av-RFP was present in distinct particles (Fig. 1C). In NCs from Jabba−/− ECs, no accumulation was detectable; rather, H2Av-RFP signal was diffuse through the cytoplasm (Fig. 1C). Comparison with a strain not expressing H2Av-RFP (Fig. 1C) demonstrates that this is not background fluorescence. Thus, as in embryos, Jabba is necessary to recruit H2Av to LDs. However, unlike in embryos, where cytoplasmic signal is negligible in Jabba−/− (Johnson et al., 2018; Li et al., 2012), cytoplasmic H2Av was abundant, suggesting that, without Jabba, H2Av synthesis is not completely abolished.
LDs make at most a modest contribution to H2Av accumulation in oocytes
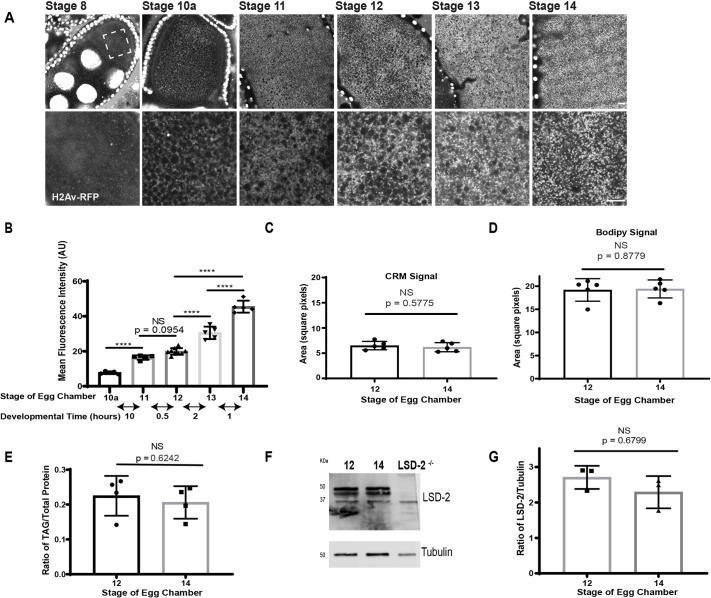
Our data show that LDs and H2Av are produced in NCs, but do not address what fraction of LDs/H2Av in oocytes comes from NCs. To address the contribution of LD-mediated H2Av transport, we determined the time course for oocyte H2Av accumulation. We quantitated the H2Av-RFP ooplasmic concentration at various stages by measuring mean fluorescence intensity (Fig. 2A,B, Fig. S1). Ooplasmic H2Av-RFP in S8 ECs was not quantified because signal was barely above background. From S10A (before dumping, mid-oogenesis) to S14 (late oogenesis), H2Av concentration increased steadily, even though total oocyte volume increases substantially between S10 and S12 (Jia et al., 2016). After S12, oocyte volume is fairly stable (King and Koch, 1963); nevertheless, H2Av-RFP concentration increased further, about 2.5-fold by S14 (Fig. 2A,B). Similar accumulation patterns were observed using H2Av-GFP or H2B-mEos3.2 (Fig. S2A,B). Thus, the concentrations of H2Av and H2B rise significantly in the ooplasm late in oogenesis.
Fig. 2.
Ooplasmic H2Av levels increase even when LDs remain constant. (A) ECs of flies expressing H2Av-RFP (white) at two magnifications: lower magnification above and higher magnification below. Dashed box indicates the area shown at higher magnification in the lower panel. Scale bars: 10 μm. (B) Quantitation of A showing the mean fluorescence intensity (AU). Each data point represents the value from a single EC. Double-headed arrows indicate the approximate time (in hours) taken to traverse from the middle of one stage to the middle of the next (Jia et al., 2016). S10a versus S11, P<0.0001; S11 versus S12, P=0.0954; S12 versus S13, P<0.0001; S13 versus S14, P<0.0001; S14 versus S12, P<0.0001 (one-way ANOVA followed by Tukey's test); n=5-9. (C,D) Quantification of the area covered by LDs in the ooplasm of S12 and S14 ECs. Images were acquired by confocal reflection microscopy (CRM) (C) or after BODIPY staining (D). n=5. (E) The ratio of triglyceride levels relative to total protein in S12 and S14 ECs (n=4). (F) Western analysis of LSD-2 levels in S12 and S14 ECs. (G) Quantitation of F expressed as the H2Av:tubulin ratio (n=3). Data are mean±s.d. In C-E,G, an unpaired, two-tailed Student's t-test was used.
Although dumping is essentially complete by S12, some LDs might still be funneled from NCs to the oocyte, bringing in more H2Av. We quantified ooplasmic LD concentrations from S12 to S14. Using two methods, we measured the area covered by LDs in a single focal plane of the oocyte (Fig. 2C,D). We also determined total triglyceride (TAG) levels relative to total protein content (Fig. 2E). All measurements indicate that the net amount of LDs is unchanged from S12 onwards. The abundance of the LD-bound protein LSD-2 (Fig. 2F,G) did not change between S12 and S14. These observations indicate that the majority of oocyte LDs is generated prior to dumping.
The increased H2Av concentrations from S12 to S14 and unchanged LD concentration indicate that H2Av is newly synthesized in the ooplasm and loaded onto pre-existing LDs. Our quantitation suggests that 56% of the H2Av pool in mature oocytes originates from synthesis in the oocyte after dumping is complete. This is most likely an underestimate as some H2Av may already have been synthesized in the ooplasm in S10 and S11.
To test the contribution of LD-mediated transport to the remaining 44% of the H2Av oocyte pool, we compared H2Av-RFP of wild-type and Jabba−/− oocytes immediately after dumping (S12). The Jabba−/− ooplasm displayed considerable H2Av-RFP signal, about 55% of wild type (Fig. 3A,B). Thus, the contribution of Jabba-dependent transport to the final H2Av pool at S14 is no more than 20% (=45% of 44%) and possibly much less (see Discussion). As LDs lacking Jabba have no detectable H2Av accumulation by imaging (in NCs, Fig. 1C) or biochemically (in embryos, Li et al., 2012), these observations argue that LD-mediated transport of H2Av from NCs to oocytes plays a minor role.
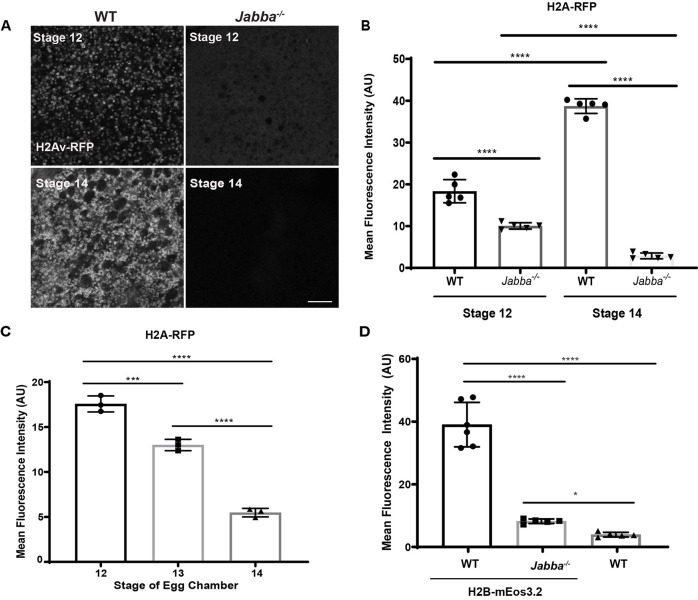
Fig. 3.
Jabba maintains the maternal supply of H2Av and H2B. (A) Ooplasmic H2Av-RFP (white) in S12 and S14 wild-type and Jabba−/− ECs. Scale bar: 10 μm. (B) Quantitation of A showing mean fluorescence intensity (AU). Wild type S12 versus Jabba−/− S12, P<0.0001; wild type S12 versus wild type S14, P<0.0001; wild type S14 versus Jabba−/− S14, P<0.0001; Jabba−/− S12 versus Jabba−/−S14, P<0.0001 (n=5). (C) Mean fluorescence intensity (AU) of H2Av-RFP in S12 to S14 Jabba−/− ECs. S12 versus S13, P=0.0005; S12 versus S14, P<0.0001; S13 versus S14, P<0.0001 (n=3). (D) Quantitation of the mean fluorescence intensity (AU) of H2B-mEos3.2 in ooplasms of S14 wild-type, Jabba−/− and wild type; H2B-mEos3.2 ECs. Wild-type ECs lacking fluorescently tagged H2B were analyzed to determine background signal. Wild type; H2B-mEos3.2 versus wild type, P<0.0001; wild type; H2B-mEos3.2 versus Jabba−/−; H2B-mEos3.2, P<0.0001; Jabba−/−; H2B-mEos3.2 versus wild type, P=0.0149; n=5 or 6 (one-way ANOVA followed by Tukey's test). Data are mean±s.d.
Jabba prevents the loss of H2Av as oocytes mature
Previous analysis found that newly laid Jabba−/− eggs have reduced total H2Av (Li et al., 2012). We find that Jabba−/− S12 oocytes display a modest reduction in H2Av and that this reduction represents a small fraction of the total H2Av in mature oocytes. This disparity could arise from technical differences in how we detect H2Av (western versus imaging) or because H2Av levels in the two genotypes diverge between S12 and egg laying. Western analysis of S12 oocytes is challenging as it would require removing both follicle cells and NC nuclei. Those cells contain high histone levels (see Discussion), so that even mild contamination would likely overwhelm the oocyte histone signal. We therefore assessed the developmental time course of ooplasmic H2Av levels by quantitative imaging.
H2Av-RFP signal in wild-type oocytes increases from S12 to S14 (Fig. 3A, left). S12 Jabba−/− oocytes showed substantial signal, although it was broadly diffuse instead of punctate. Thus, as in NCs, Jabba is required to recruit H2Av to LDs. By S14, Jabba−/− had lower H2Av levels (Fig. 3A, right). Quantitation revealed that levels had dropped to 28% of the Jabba−/− S12 levels and to 7% of the wild-type S14 levels (Fig. 3B), and that the drop was gradual, with S13 at intermediate levels (Fig. 3C). Thus, in Jabba−/−, H2Av stores in the oocyte are not stably maintained, and this loss counteracts the rise observed in the wild type. We observed a similar pattern using an H2B construct (Fig. 3D). We conclude that, in Jabba−/−, the maternal histone pool is already reduced during oogenesis.
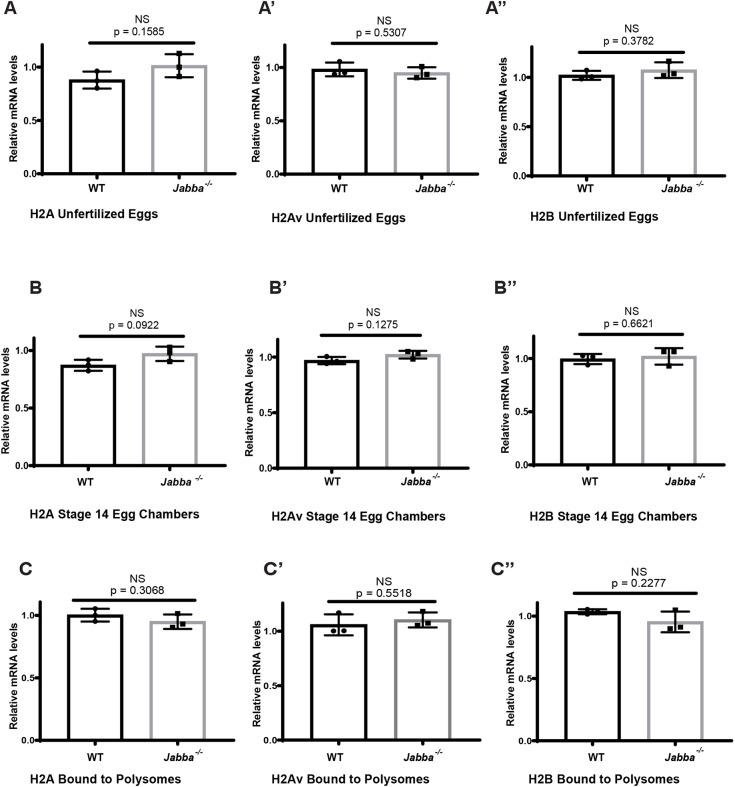
The divergence in H2Av levels between wild type and Jabba−/− could be due to decreased H2Av synthesis, enhanced H2Av degradation or a combination of both. However, there is no evidence that lack of Jabba alters the transcription or stability of histone messages: levels of H2A, H2B and H2Av messages were indistinguishable between wild type and Jabba−/− in S14 ECs (Fig. 4B-B″), or in unfertilized embryos (Fig. 4A-A″; Li et al., 2012). In addition, immunostaining revealed no appreciable presence of Jabba in nuclei, as expected if it were directly involved in transcription regulation (Fig. 1B).
Fig. 4.
H2A, H2Av and H2B biosynthesis is unaffected in Jabba−/−. (A-C″) There is no significant change in relative mRNA levels of H2A, H2Av and H2B in wild-type and Jabba−/− unfertilized eggs (A-A″), in S14 ECs (B-B″) or bound to polysomes (C-C″) (unpaired, two-tailed Student's t-test; n=3). Data are mean±s.d.
To determine whether the reduced histone pool in Jabba−/− arises due to impaired translation, we examined the histone mRNAs bound to polysomes. There was no significant change in H2A, H2Av and H2B mRNA levels being actively translated in wild-type and Jabba−/− S14 ECs (Fig. 4C-C″). Therefore, we have no evidence that histone synthesis is appreciably altered in Jabba−/−.
H2Av is degraded in the absence of Jabba
Our analysis indicates that Jabba dependence of histone levels in mature oocytes is not due to major roles of Jabba in histone synthesis or transport, suggesting that Jabba counteracts histone turnover. If so, inhibiting histone degradation should reverse the effects of a lack of Jabba. As excess histones are often turned over by proteasome-mediated degradation (Imschenetzky et al., 1997; Lin et al., 1991; Morin et al., 2012; Singh et al., 2009), this pathway is an obvious candidate.
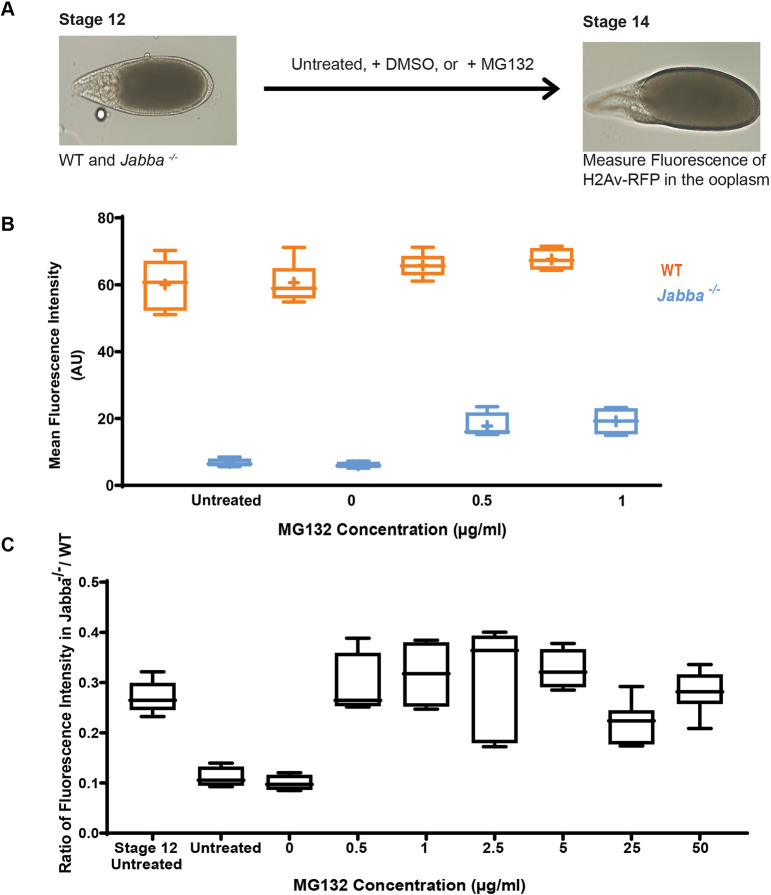
We employed in vitro egg chamber maturation (IVEM) (Spracklen and Tootle, 2013) and inhibited the proteasome pharmacologically. S12 ECs isolated from H2Av-RFP-expressing females were incubated in IVEM media to allow for progression to S14 (Fig. 5A). We measured mean ooplasmic H2Av-RFP fluorescence either in newly dissected ECs or after 6 h, when full maturation had occurred. The disparity of H2Av-RFP signal between S14 wild-type and Jabba−/− ECs observed in vivo was also present under in vitro conditions (Fig. 5B, condition labeled ‘untreated’), even when the solvent dimethyl sulfoxide was added to the culture media (Fig. 5B, condition labeled ‘0 µg/ml’). These observations set the stage to test the effects of the proteasome inhibitor MG132.
Fig. 5.
H2Av is degraded in the absence of Jabba. (A) Schematic depiction of IVEM. (B) The mean fluorescence intensity (AU) in S14 wild-type (orange) or Jabba−/− (blue) ECs after IVEM. Length of box plot: the 25th and 75th percentile; line indicates median; cross indicates mean (n=4-8). (C) The ratio of the average mean fluorescence intensity (AU) of Jabba−/−:wild type for MG132 concentrations. Stage 12 untreated indicates S12 ECs analyzed immediately after dissection; untreated indicates cultured in IVEM media.
Full proteasome inhibition arrests oogenesis (Velentzas et al., 2011). It is therefore not surprising that, at high concentrations (50 µg/ml), MG132 interfered with maturation and ECs never reached S14. Consistent with arrest, H2Av-RFP levels were comparable with those in S12 ECs (untreated) for both wild type and Jabba−/− (Fig. S3).
We titered MG132 concentrations to a point where development to S14 was supported (0.5 and 1 µg/ml). Presumably, under these conditions, proteasome function is only partially abolished. H2Av-RFP levels in wild-type ECs were unaltered. In contrast, H2Av-RFP intensity was increased in Jabba−/− ECs. This difference between genotypes is particularly obvious when we compute the ratio of H2Av-RFP signal in mutant ooplasm to that in wild-type ooplasm (Fig. 5C). Over a range of inhibitor concentrations, this ratio remains close to the S12 ratio and higher than in S14, whether the inhibitor still allowed morphological maturation (0.5 and 1 µg/ml) or arrested it (2.5, 5, and 25 µg/ml). We conclude that, without Jabba, H2Av is prone to turnover, consistent with the model that Jabba protects H2Av from degradation.
The data from Fig. S3 allow a quantitative estimate of the contribution of the proteasome to H2Av turnover. At 2.5 µg/ml (Fig. S4A), H2Av levels in the wild type rise by 1839 AU from S12 to S14. A similar rise in Jabba−/− ECs would lead to 3481 AU. With no drug, levels in Jabba−/− instead fall to 606 AU. Thus, if the drug had no effect on turnover, H2Av levels would be close to 606 AU (0% effect). If MG132 treatment completely inhibited turnover, we would expect the same rise as in the wild type (100% drug effect). The observed value after MG132 treatment represents 42% of maximal inhibition. Fig. S4B shows this analysis for all concentrations. The effect is 0% without drug, steadily increased to 42% at 2.5 µg/ml and is then decreased, likely because MG132 starts interfering with development. We conclude that even partial proteasome inhibition prevents almost half of H2Av turnover normally seen in Jabba−/−.
As an independent approach, we used a mutant form of proteasome subunit β2: DTS7. DTS7 acts as an antimorph to inhibit proteasome activity, especially at elevated temperatures (Smyth and Belote, 1999). When expressed in the Jabba−/− background, H2Av levels were increased at 29°C relative to 25°C (Fig. S5). This supports our conclusions that, without Jabba, H2Av is turned over by the proteasome.
H2Av levels in S14 oocytes scale with Jabba protein levels
How might Jabba prevent histone degradation? It might trigger a molecular switch that regulates the degradation machinery or it might physically protect histones from degradation. We tested whether protection by Jabba scales with the amount of Jabba present and whether it depends on the ability of Jabba to physically bind histones.
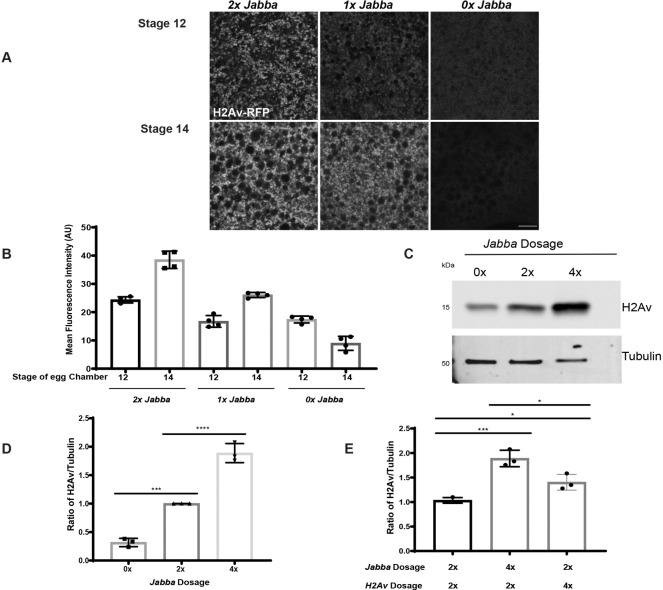
If Jabba directly protects histones, then H2Av levels in S14 should depend on how much Jabba protein is present; if Jabba acts as a regulatory switch, no such dependency is expected. Jabba levels were previously shown to scale with Jabba dose in early embryos (Johnson et al., 2018), and we find the same dependency in oocytes (Fig. 6). Quantitative imaging revealed that oocytes from mothers with one copy of Jabba have H2Av-RFP levels about half-way between those lacking Jabba and those with two gene copies (Fig. 6A,B). Remarkably, H2Av levels are elevated beyond wild-type levels by increasing Jabba expression. S14 ECs from mothers with two additional copies of Jabba (4x Jabba) accumulated more Jabba (Fig. S6C,D) and also twice as much H2Av as the wild type (Fig. 6C,D). Thus, Jabba limits histone accumulation. In fact, an increased Jabba dose is more effective at increasing total H2Av levels than an increased H2Av dose (Fig. 6E); the fact that H2Av dose has any effect on H2Av levels may be due to extra H2Av displacing H2A from a common binding site on Jabba (Kolkhof et al., 2017). These results suggest that Jabba directly protects H2Av from degradation.
Fig. 6.
H2Av supply scales with Jabba dose. (A) Ooplasmic H2Av-RFP (white) in ECs expressing zero, one or two copies of Jabba during S12 and S14. Scale bar: 10 μm. (B) Quantitation of A showing mean fluorescence intensity (AU). n=4. (C) Western analysis of H2Av levels in S14 ECs expressing zero, two or four copies of Jabba. (D) Quantitation of C expressed as the H2Av:tubulin ratio (n=3). 0x Jabba versus 2x Jabba, P=0.0002; 2x Jabba versus 4x Jabba, P<0.0001. (E) Quantitation of anti-H2Av western analysis for indicated genotypes. Total H2Av is expressed as the ratio of H2Av:tubulin. 2x Jabba;2x H2Av versus 4x Jabba;2x H2Av, P=0.0007; 2x Jabba;2x H2Av versus 2x Jabba;4x H2Av, P=0.0388; 4x Jabba;2x H2Av versus 2x Jabba;4x H2Av, P=0.012; n=3 (one-way ANOVA followed by Tukey's test). Data are mean±s.d.
H2Av binding is necessary for Jabba's protective effect
Using a split luciferase protein-protein interaction assay in Drosophila Kc167 cells, we showed that Jabba-H2Av interactions require a short, positively charged stretch of amino acids in Jabba (Kolkhof et al., 2017). We therefore asked whether the same mutation abolishes H2Av interactions in vivo to then test how that affects H2Av accumulation in oogenesis.
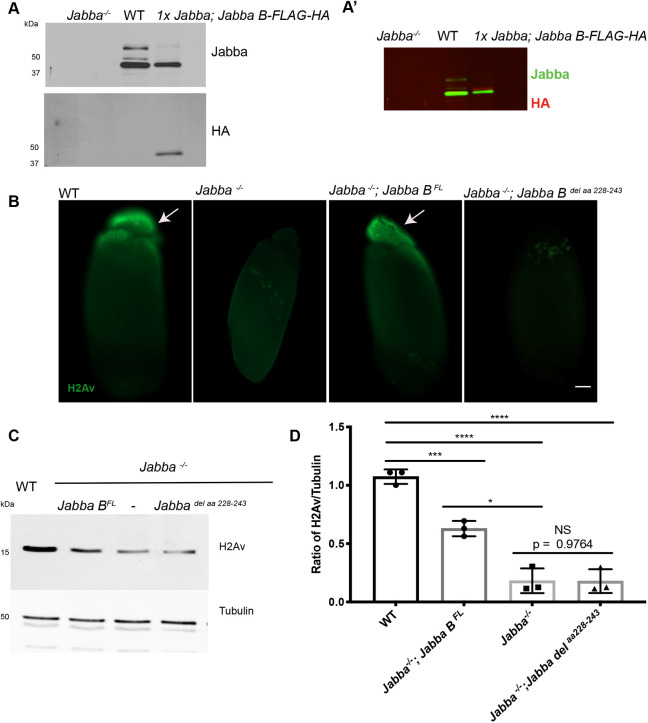
We first developed a simplified platform to analyze the ability of Jabba constructs to localize to LDs and recruit histones. The endogenous Jabba locus gives rise to eight messages predicted to generate seven distinct proteins (Li et al., 2012) with a common N-terminal region of 316 amino acids and varying C-terminal tails. Publicly available RNA-seq data from FlyBase (Thurmond et al., 2019) of late ECs and early embryos indicate that the predominant isoforms are Jabba B, Jabba G and Jabba H, with Jabba B most abundant. Consistent with this notion, Western analysis of these stages reveals three major Jabba bands, and the most abundant one co-migrates with ectopically expressed HA-tagged Jabba B (Fig. 7A).
Fig. 7.
Jabba protects H2Av from degradation. (A) Embryo lysates from Jabba−/−, wild type and 1xJabba; Jabba B-FLAG-HA mothers were probed with anti-Jabba (top) and anti-HA (bottom). In the wild type, three major Jabba bands are visible. (A′) Overlay of the anti-Jabba (green) and anti-HA signal (red). This demonstrates that Jabba B-FLAG-HA migrates immediately above the lowest of the three endogenous Jabba bands, consistent with the addition of 1.9 kDa due to the tag. Based on the apparent molecular weight of Jabba B, the molecular weights of the top bands are consistent with them representing Jabba H and Jabba G (the molecular weights of which are predicted to be 7.1 and 13.3 kDa larger, respectively). (B) Anti-H2Av immunostaining of centrifuged embryos. H2Av is in the LD layers (arrows) in wild-type and Jabba−/−; Jabba BFL embryos. Histones are absent from LD layers in Jabba−/− and Jabba−/−; Jabba Bdel aa228-243 embryos. Scale bar: 50 μm. (C) H2Av western analysis in S14 ECs of wild-type, Jabba−/−, Jabba−/−; Jabba BFL and Jabba−/−; Jabba Bdel aa228-243 flies. (D) Quantitation of C expressed as the H2Av:tubulin ratio. Wild type versus Jabba−/−; Jabba BFL, P=0.0002; wild type versus Jabba−/−, P<0.0001; wild type versus Jabba−/−;Jabba Bdel aa228-243, P<0.0001; Jabba−/−; Jabba BFL versus Jabba−/−, P=0.0124; Jabba−/− ;Jabba BFL versus Jabba−/−;Jabba Bdel aa228-243, P=0.0118; Jabba−/− versus Jabba−/−;Jabba Bdel aa228-243, P>0.9999; n=3 (one-way ANOVA followed by Tukey's test). Data are mean±s.d.
Using the Gal4/UAS system, we next expressed an mCherry-tagged full length Jabba B (Jabba BFL) in an otherwise Jabba−/− background and analyzed its ability to bind to LDs using in vivo embryo centrifugation. In this assay, cellular components separate with respect to their densities within an intact embryo and LDs are enriched at the tip of the embryo (Tran and Welte, 2010). Jabba BFL prominently accumulated in this LD layer (Fig. S8), demonstrating LD association. Immunostaining such embryos also revealed that the construct rescued H2Av recruitment to LDs (Fig. 7B). Because tagged Jabba BFL and endogenous Jabba have very different molecular weights and thus might behave differently during western transfer, we compared their expression levels using immunostaining. Jabba BFL levels were comparable with those with a single copy of Jabba (1x Jabba, Fig. S7A,B). Finally, we measured H2Av levels in S14 ECs by western analysis and found that Jabba BFL was able to maintain H2Av to a similar level as 1x Jabba (Fig. 7C,D). These observations suggest that Jabba B can protect histones from degradation in the absence of other Jabba isoforms, providing a platform to introduce mutations into Jabba and test their effect on histone metabolism.
We generated a similar transgene, but with the positive stretch deleted: Jabba Bdel aa 228-243. The mutant protein was expressed at similar levels to Jabba B (Fig. S7A,B) and was localized to LDs (Fig. S8). However, it failed to restore H2Av recruitment to LDs (Fig. 7B), suggesting that amino acids 228-243 are necessary for histone binding in vivo. Additionally, H2Av protection in oogenesis was not rescued (Fig. 7C,D). Thus, it is unlikely that Jabba acts as a switch for maintaining histones; we propose that Jabba physically protects H2Av from degradation.
LD binding allows Jabba to be transported into the oocyte
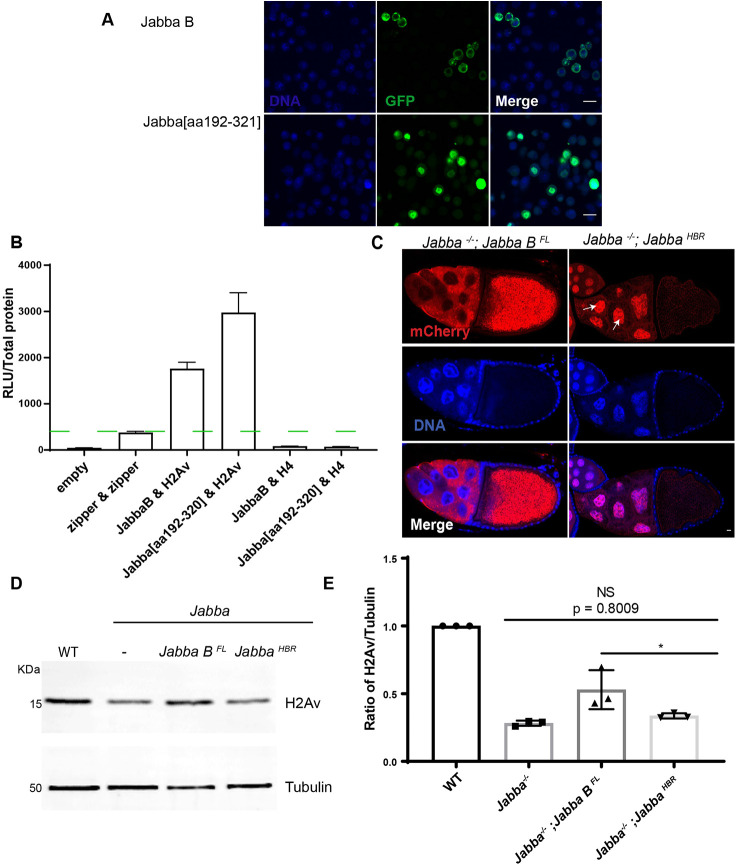
If the primary role of Jabba is to protect histones from degradation, it is unclear why Jabba is present specifically on LDs. In principle, Jabba that is free in the ooplasm or not bound to LDs should be sufficient to protect the histone pool. We therefore designed a Jabba truncation that lacks the motif for LD recruitment, but retains amino acids 228-243. When analyzed in Kc167 cells, this fragment [Jabba(aa192-321)] is not localized to LDs but broadly throughout the cytoplasm and the nucleus (Fig. 8A; Fig. S9). Using a luciferase complementation assay, we determined that it interacts with H2Av (Fig. 8B, Fig. S10). We will refer to this fragment in the following as JabbaHBR, for histone-binding region.
Fig. 8.
LD binding allows Jabba to be transported into the oocyte. (A) Jabba[aa192-321] localization in Drosophila Kc167 cells. Jabba B (green, top) is in the cytoplasm. Jabba[aa192-321] (green, bottom) is in nuclei (blue). Scale bars: 10 μm. (B) Split luciferase complementation assay showing relative light units (RLU) per μg total protein for the indicated proteins. Green dashed line indicates threshold for positive interactions (n=3). (C) DAPI staining (blue) of ECs expressing mCherry-tagged Jabba BFL and JabbaHBR in a Jabba−/− background. mCherry signal (red) is enriched in NC cytoplasm and ooplasm in Jabba−/−;Jabba BFL. In Jabba−/−; JabbaHBR ECs, mCherry signal is in NC nuclei (arrows). (D) Western analysis of H2Av levels in S14 ECs of wild-type, Jabba−/−, Jabba−/−; Jabba BFL and Jabba−/−; JabbaHBR flies. (E) Quantitation of D expressed as the H2Av:tubulin ratio. Jabba−/− versus Jabba−/−; JabbaHBR, P=0.8009; Jabba−/−; Jabba BFL versus Jabba−/−; JabbaHBR, P=0.0486; n=3 (one-way ANOVA followed by Tukey's test). Data are mean±s.d.
Jabba BFL expressed during oogenesis mimicked the distribution of endogenous Jabba: Jabba BFL was detected in the NC cytoplasm and the ooplasm. In contrast, JabbaHBR was highly enriched in NC nuclei, with only a small amount present in the oocyte nucleus or diffusely in the NC cytoplasm (Fig. 8C).
It is unknown why JabbaHBR accumulates in nuclei, but we speculate that it might reflect its histone-binding ability. Our observations suggest that Jabba-LD association prevents Jabba from becoming trapped in NC nuclei, which would abrogate its transport into the oocyte.
JabbaHBR can bind to H2Av but is not present in the ooplasm. The amount of H2Av detected in S14 JabbaHBR oocytes was comparable with that for Jabba−/− and lower than that for Jabba BFL (Fig. 8D,E), consistent with the notion that Jabba has to be in the ooplasm to physically protect H2Av.
DISCUSSION
Histone stores accumulate from mid- to late oogenesis
Early animal embryogenesis often exhibits rapid cell cycles dominated by DNA replication, mitosis and little to no transcription. Drosophila is a dramatic case where the first 13 nuclear divisions occur every 8-20 min (Foe and Alberts, 1983). This speed poses a challenge for histone biology: demand increases exponentially, yet major regulatory mechanisms that control histone expression are unavailable. To meet this demand, many embryos inherit maternally synthesized histones. Here, we examined the origin of H2Av, H2B and H2A stockpiles in Drosophila.
Expression of histone messages is increased during S10 (Ambrosio and Schedl, 1985; Ruddell and Jacobs-Lorena, 1985), but it was unclear when the complementary maternally deposited histone proteins accumulate. A methodological challenge is that ECs have polyploid nuclei. Each of the NC nuclei are estimated to contain about 500 times more DNA than diploid nuclei (King, 1970); the 900 follicle cells (Fadiga and Nystul, 2019) contain DNA levels about eight times higher than a diploid nucleus (Mulligan and Rasch, 1985). Thus, the histones needed to package NC and follicle cell chromatin are at least tenfold more abundant than the histone stockpile of newly laid embryos, which are estimated to be the equivalent of 1000 diploid nuclei (Cermelli et al., 2006). Detecting accumulation of the oocyte histone stockpile in addition to other histones in the EC is therefore challenging.
We followed H2Av accumulation using an imaging approach that specifically quantifies histone signal in the NC and oocyte cytoplasm. H2Av already accumulates in the cytoplasm of S9 NCs and is associated with LDs. An intriguing possibility is that this LD-associated H2Av pool in the early-stage NCs might support NC endoreplication. This H2Av-LD association likely brings some H2Av into the oocytes, but our quantitation indicates that transfer from NCs contributes at most one fifth of the final H2Av pool in mature oocytes. These data are consistent with the possibility that the majority of the ooplasmic H2Av is synthesized in the oocyte; reduced H2Av levels in Jabba−/− S12 oocytes may represent early loss by degradation rather than defective transfer from NCs.
After dumping, total H2Av levels continue to rise from S12 through S14. We propose that two mechanisms contribute to this rise. First, the translational efficiency of H2Av mRNA is upregulated from S12 to S14 (Eichhorn et al., 2016). Second, Jabba increases from S12 to S14 (Fig. S6A,B). As more Jabba protein becomes available, it can presumably recruit more H2Av to LDs and protect it from degradation (Fig. 9A′).
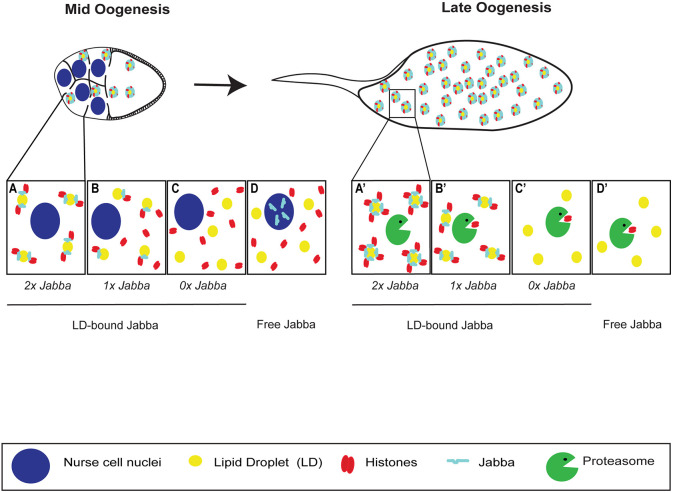
Fig. 9.
LD association promotes intercellular transport of Jabba to oocytes where it protects histone H2Av from degradation. LD-histone interaction is dependent upon Jabba dose and Jabba localization. (A) In wild-type NCs, Jabba (cyan) is LD bound and recruits H2Av (red). LDs (yellow) and LD-bound Jabba are transferred to the oocyte. (A′) In the ooplasm, Jabba levels further increase and Jabba protects H2Av from degradation. (B) Decreasing Jabba dose (1x Jabba) reduces LD-binding capacity and likely increases the free histone pool in the NC cytoplasm. (B′) In oocytes, Jabba levels are limiting, leading to partial H2Av degradation. (C,C′) In 0x Jabba, H2Av is no longer localized to LDs (C). H2Av is unable to evade the degradation machinery and H2Av stores are not maintained in late oogenesis (C′). (D,D′) When Jabba is not LD bound (free Jabba), it relocalizes to NC nuclei (D) and is not in the oocyte; thus, H2Av degradation is not prevented (D′).
H2A and H2B levels in embryos are Jabba dependent (Li et al., 2012), and the pattern of H2B accumulation during oogenesis (Fig. S2B) resembles that of H2Av (Fig. 2A,B). We currently lack good tools to determine H2A accumulation, but predict it will follow the same pattern, as canonical histones are typically similarly regulated (Marzluff et al., 2008). We propose that increasing H2Av, H2B and H2A accumulation during late oogenesis establishes an LD-bound histone depot for the early embryo.
It will be interesting to determine whether H3 and H4 accumulation follows a similar pattern. As canonical histones, their transcriptional and translational regulation is likely similar to that of H2A and H2B. However, they are not LD associated (Cermelli et al., 2006) nor are their embryonic levels Jabba dependent (Li et al., 2012), so there must be mechanistic differences.
Degradation of excess histones
Newly laid Jabba−/− eggs have lower H2A, H2B and H2Av levels than wild type. We find that this divergence is established during late oogenesis; H2Av and H2B levels rise in the wild type but drop in Jabba−/− (Fig. 3). Experiments to inhibit proteasome activity reveal that histone degradation plays a major role in bringing about this difference (Fig. 5, Fig. 9C-C′). As high MG132 concentrations arrest development, we infer that under conditions that allow EC development to S14, degradation is only partially inhibited. Yet even with partial inhibition, almost half of the H2Av normally lost in Jabba−/− is retained. We conclude that Jabba prevents histone turnover and that a major contributor to turnover is a proteasome-dependent pathway. The remaining turnover not prevented by drug treatment may be due to a proteasome-independent mechanism (e.g. Schulze et al., 2020).
In the wild type, proteasome inhibition did not increase the H2Av pool beyond levels in untreated ECs. This observation was surprising as 4x Jabba females can accumulate more H2Av than wild type, which presumably indicates that even the wild type produces excess H2Av that is degraded if not protected by Jabba. We reason that the direct effects on proteasome turnover are balanced out by indirect effects that compromise histone or Jabba synthesis. Indeed, high concentrations of the inhibitor abolish any rise in H2Av (Fig. S3).
Turnover of excess histones by the proteasome is well established in yeast (Gunjan and Verreault, 2003). It is one of numerous mechanisms that prevent accumulation of free histones and the resulting cytotoxicity (Bannister and Kouzarides, 2011; Kaygun and Marzluff, 2005). We propose that H2Av turnover in late oogenesis is due to this general protective machinery; Jabba allows oocytes to deploy this safety feature while also accumulating H2Av needed to provision the embryo. Accordingly, Jabba determines the H2Av pool that is protected and any excess is recognized as a potential hazard. Because the mechanism that targets excess H2Av for proteasomal degradation is not known, we cannot yet test what type of damage this mechanism guards against.
Our data suggest that Jabba prevents degradation by physically protecting H2Av. First, H2Av levels scale with Jabba dose, and we can achieve H2Av levels beyond normal by simply doubling Jabba levels (Fig. 6C,D). Second, a version of Jabba that is mislocalized to the NC nuclei and not present in oocytes is unable to support high H2Av levels (Figs 8D,E and 9D,D′). Finally, a Jabba mutant unable to bind to H2Av resulted in H2Av levels indistinguishable from those in Jabba−/− (Fig. 7D,E).
To further unravel how Jabba prevents degradation, it will be necessary to identify the machinery that targets excess H2Av to the proteasome. Previous work has identified E3 ligases promoting H3 and H4 turnover (Singh et al., 2012), but those for other histones remain uncharacterized. Jabba may physically protect H2Av by shielding a ubiquitylation site. This remains to be tested. An alternate hypothesis is that histone degradation in oocytes occurs independently of ubiquitylation. Intriguingly, during development of the mammalian male germline, histone turnover occurs via ubiquitin-independent proteasome-dependent degradation (Huang et al., 2016; Khor et al., 2006; Qian et al., 2013).
In embryos, H2Av exchanges between LDs. If similar exchange occurs in oocytes, it is unclear how transient interactions with Jabba are sufficient to prevent H2Av degradation. We speculate that either the transit time is negligible relative to the time H2Av spends interacting with Jabba or that cytosolic H2Av is accompanied by a chaperone.
LD binding promotes availability of Jabba and, consequently, of histones
Our analysis provides a first answer to why some histones are stored on LDs, while others are apparently stored in the cytoplasm. Jabba, the protein necessary to stabilize the H2A, H2B and H2Av ooplasmic pool, becomes trapped in NC nuclei if it is not LD bound. Thus, it is absent from the oocyte and cannot perform its protective function. We propose that LD binding ensures proper intercellular transport of Jabba, safeguarding its ability to function in the ooplasm (Fig. 9).
H3 and H4 ooplasmic stores are presumably bound to a partner that prevents their degradation, perhaps the histone chaperone NASP (Campos et al., 2010; Finn et al., 2012). As these histones are apparently cytoplasmic, it is unclear how they and their binding partners avoid mislocalization to NC nuclei.
It is also unclear why histones are stored on LDs rather than another cytoplasmic structure. A priori, any cytoplasmic organelle that is transferred to oocytes could suppress the NC nuclear import of the histone anchor and promote transport into the oocyte. The large surface area of LDs may provide a readily available and, at these developmental stages, metabolically inert platform for recruitment. Other organelles may not have enough storage capacity or might be functionally impaired by histones on their surface. Alternatively, LD association may be an evolutionary accident and many other organelles might in principle be able to store histones. Our identification of a Jabba region sufficient for histone binding will make it possible to address this question in the future.
But why is JabbaHBR mislocalized to NC nuclei? We suspect that the histone-binding ability of JabbaHBR leads to its mislocalization, dragged along by histone nuclear import or retained in nuclei via chromatin binding. As histone binding is important to protect against degradation, mislocalization cannot be avoided unless JabbaHBR is anchored outside the nucleus. We have not yet been able to test this idea, as JabbaHBR that is unable to interact with histones still accumulates in nuclei, in cultured cells and in NCs (M.B., P.K. and R.A.S., unpublished observations). We hypothesize that a cryptic nuclear localization signal in JabbaHBR promotes nuclear transport even without histone binding.
LDs and protein sequestration
Our analysis may shed light on how LDs regulate other proteins. LDs can transiently accumulate proteins from other cellular compartments (Welte, 2007). This has been particularly documented for proteins involved in nuclei acid binding and/or transcriptional regulation: MLX and Perilipin 5 can either be present on LDs or move into the nucleus to regulate transcription (Gallardo-Montejano et al., 2016; Mejhert et al., 2020). The bacterial transcriptional regulator MLDSR is sequestered to LDs under stress conditions (Zhang et al., 2017). As for Jabba and H2Av, LD association of such ‘refugee proteins’ (Welte, 2007) may prevent their premature turnover or may promote their delivery to the correct intra- or intercellular location (Beller et al., 2006; Camus et al., 2013; McLauchlan et al., 2002). A role for LDs in protein delivery to distant cellular compartments might be important in neurons where recent discoveries suggest important, but largely uncharacterized, roles for LDs (Pennetta and Welte, 2018; Wat et al., 2020).
MATERIALS AND METHODS
Fly stocks
Oregon R was used as the wild-type strain. Alleles JabbaDL and Jabbazl01 were used to analyze ovaries and embryos lacking Jabba (Li et al., 2012). To analyze embryos and ovaries with varying Jabba levels, these additional genotypes were used. Df(2R)Exel7158/CyO [Bloomington Drosophila Stock Center (BDSC: 7895; FLYB: FBab0038053)] carries a large deletion that encompasses Jabba and is used to reduce Jabba dosage; for simplicity, it is referred to as JabbaDF in the figures. Jabbalow was derived from Jabbad03001 (a PBac insertion, P[XP]d03001, obtained from The Exelixis Collection at Harvard Medical School) by imprecise P-element excision. To generate 4x Jabba, the genomic transgene gJabba (previously described by Johnson et al., 2018) was introduced into an otherwise wild-type background. H2Av dose was increased using the genomic transgene gH2Av (Johnson et al., 2018), which expresses H2Av under endogenous control at normal levels. The following fluorescently tagged histone stocks were used to obtain H2Av-RFP, H2Av-GFP and H2B-mEos3.2 ovaries with varying Jabba dose: H2Av-RFP Bloomington Drosophila Stock Center (BDSC: 23650; FLYB: FBst0023650), H2Av-GFP Bloomington Drosophila Stock Center (BDSC: 24163; FLYB: FBst0024163) and H2B-mEos3.2, a gift from Michael Eisen, The University of California, Berkeley, CA, USA (Mir et al., 2018). The LSD-2 null allele was previously described (Welte et al., 2005). Jabba B FLAG-HA encodes Jabba B with a C-terminal FLAG tag under UAST control and was generated by the Drosophila Protein Interaction Map consortium (Guruharsha et al., 2012) and obtained from the Bangalore Fly Facility. The generation of the following mCherry-tagged Jabba lines is described below: Jabba BFL, Jabba Bdel aa 228-243 and JabbaHBR. Expression of all Jabba constructs was driven using one copy of P{mat4-GAL-VP16} V2H [Bloomington Drosophila Stock Center (BDSC: 7062; FLYB: FBst0007062)] and either a single copy of the mCherry-tagged transgenes or two copies of Jabba B FLAG-HA. To inhibit the proteasome, we expressed a dominant-negative form of the proteasome subunit β2, called DTS7, using UAS-DTS7 (BDSC: 6785; FLYB: FBst0006785) or w[*]; DTS7/Sb (Smyth and Belote, 1999) (a kind gift from John Belote, Syracuse University, NY, USA).
Transgene generation/molecular biology
The JabbaHBR construct (amino acids 192-321) was generated using the oligonucleotides JabbaB192_321forward (5′-CAGGGTTTAAGCAATTTCGTAGT) and Vector_Start_re (5′-P-CATGGTGGCGGCCGCGGAGC), standard in vitro mutagenesis and the JabbaB full-length construct as a template. GFP or luciferase-tagged versions of the proteins were generated using the Gateway recombination cloning procedure (Invitrogen) and custom-made vectors.
To generate mCherry-tagged Jabba BFL, Jabba Bdel aa 228-243 and JabbaHBR transgenes, the desired sequences were amplified from cDNA samples and donor plasmids (Kolkhof et al., 2017), and then cloned into the pENTR Gateway plasmid system (Invitrogen). Entry plasmids were recombined using the Gateway recombination cloning system (Invitrogen) into pPTAPmChW attB (described by Hudson and Cooley, 2010) destination plasmids. Transgenic lines were created by BestGene using PhiC31 integrase-mediated transgenesis. All insertions were incorporated onto the third chromosome at site 68A4.
Western analysis
ECs and embryos were heat-fixed in 1×Triton Salt Solution (embryos were devitellinized), sorted by stage and boiled in Laemmli buffer (Bio-Rad) for 15 min. Proteins were separated using 4-15% SDS-PAGE gels (Bio-Rad) and transferred to PVDF membranes (Immobilon-FL, EMD Millipore). Transfers were performed in N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (10 mM CAPS and 10% methanol) at 50 V for 40 min (Jabba) or Towbin buffer (10% 10×Tris-Glycine solution and 20% methanol) at 80 V for 30 min (HA and H2Av). Immunodetection was carried out using the following primary antibodies: rabbit anti-Jabba (1:5000) (previously described by Johnson et al., 2018), rabbit anti-H2Av (1:2500) (Active Motif), rat anti-HA (1:1000) (Clone 3F10; Roche Diagnostics) and mouse anti-α-Tubulin (1:10,000) (Cell Signaling); and the following secondary antibodies: IRDye 800CW goat anti-rabbit IgG, IRDye 680RD goat anti-mouse IgG and IRDye 680RD goat anti-rat IgG (1:10,000) (Li-COR). The same membranes were probed with the indicated primary antibody and mouse anti-α-tubulin as a loading control. Images were acquired using Li-COR Odyssey CLx and fluorescence was quantified using Image StudioLite Software.
Temperature shift assays
We expressed DTS7 either endogenously or using the Gal4/UAS system. Drosophila females were kept at 25°C with dry yeast paste for 48 h. Then flies were further incubated at 25°C or moved to 29°C for 24 h after the initial incubation. Ovaries were dissected and then used for western analysis.
Immunostaining and staining
Embryos were collected on apple juice agar plates and de-chorionated with 50% bleach. In vivo centrifugation was then performed (Tran and Welte, 2010) and embryos were subsequently fixed with 4% formaldehyde in 1×phosphate-buffered saline (PBS) for 20 min. Embryos were devitellenized using heptane/methanol and subsequently washed in 1×PBS/0.1% Triton X-100.
Ovaries were dissected from females maintained on yeast at 25°C overnight. Samples were then fixed with 4% formaldehyde in 1×phosphate-buffered saline (PBS) for 15 min and subsequently washed in 1×PBS/0.1%Triton X-100.
Embryo and ovary samples were sorted, counted (to ensure equal numbers per tube) and blocked overnight at 4°C in 10% BSA, 0.1% Triton X-100 and 1×PBS solution. Incubation with primary antibodies (Jabba, H2Av) was performed at 1:1000 overnight. Samples were washed and probed with secondary antibodies at a final concentration of 1:1000 (goat anti-rabbit Alexa 488, goat anti-rabbit Alexa 594 and goat anti-mouse Alexa 488). For DNA staining, embryos/ECs were stained using 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Lipid droplets were stained using either Nile Red (Sigma-Aldrich) or BODIPY 558/568 C12 [4,4-Difluoro-5-(2-Thienyl)-4-Bora-3a,4a-Diaza-s-Indacene-3-Dodecanoic Acid] (Thermo Fisher Scientific).
For Fig. S1 ECs were isolated from the ovaries and incubated for 20 min in BODIPY. ECs were then mounted using the technique adapted from Prasad et al. (2007).
Microscopy/imaging
Centrifuged embryos were imaged with a Nikon Eclipse E600 epifluorescence microscope, using a 20× objective. For live imaging, ovaries were dissected from flies maintained on yeast overnight at 25°C. ECs were placed on a coverslip and overlaid with Oil 10 S, VOLTALEF (VWR Chemicals). Imaging of both live and fixed samples was performed using a Leica SP5 confocal microscope. All images were assembled using Abode Illustrator.
RNA extraction and qPCR
S14 ECs were dissected from females maintained on yeast at 25°C for 48 h. Unfertilized eggs were collected on apple juice plates and de-chorionated in 50% bleach. Trizol Reagent (Invitrogen) was used according to the manufacturer's instructions to extract total RNA. Recovered RNA was measured to ensure that equal concentrations of RNA samples were digested with DNAse (Thermo Fisher Scientific). cDNA was then synthesized using Maxima H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific). To estimate the H2B, H2A and H2Av mRNA levels, Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) was used and qPCR was performed on a StepOnePlus Real time PCR System (Applied Biosystems). Primers for qPCR were as follows: H2A Fw, 5′-CGCAACGACGCGGCGTTAAA-3′; Rv, 5′-GCCTTCTTCTCGGTCTTCTTG-3′; H2Av Fw, 5′-GTGTACTCCGCTGCCATATT-3′; Rv, 5′-CGAATGGCGAGCTGTAAGT-3′; H2B Fw, 5′-CCATCCTGACACCGGAATTT-3′; Rv, 5′-TCGAGCGCTTGTTGTAGTG-3′. H2B, H2A and H2Av mRNA levels were normalized to the reference gene 60S Ribosomal ProteinL24 (RPL24): Fw, 5′-TTTCTACGCCAGCAAGATGAAAA-3′; Rv, 5′-ATTGCGCTTCATCAGGTAGG-3′.
Triglyceride (TAG) assays
Triglyceride amounts were measured using approaches adapted from Palanker et al. (2009). 100 ECs were homogenized in 100 μl PBS, 0.5% Tween 20 and immediately incubated at 70°C for 5 min. 20 μl of the homogenate was incubated with either 20 μl PBS or 20 μl triglyceride reagent (Sigma-Aldrich) for 30 min at 37°C. The samples were then centrifuged using an Eppendorf Microcentrifuge (Model 5415D) at maximum speed for 3 min. 30 μl of each sample was transferred to a 96-well plate and incubated with 100 μl of free glycerol reagent (Sigma-Aldrich) for 5 min at 37°C. Samples were assayed using a SpectraMax M2e spectrophotometer at 540 nm. TAG levels were determined by subtracting the amount of free glycerol in the PBS-treated sample from the total glycerol present in the sample treated with triglyceride reagent. TAG amounts were normalized to the total protein amounts in each sample using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The data were analyzed using an unpaired, two-tailed Student's t-test.
Polysome analysis
Ovaries from flies (maintained on yeast paste overnight at 25°C) were dissected in PBS. S14 ECs were treated with 10 μg/ml puromycin dihydrochloride from Streptomyces alboniger (Millipore Sigma) in PBS on ice. ECs were then lysed [lysis buffer: 20 mM Tris-HCl, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 1% Triton X-100, 0.1 mg/ml cycloheximide (Sigma-Aldrich), 1 mg/ml heparin, 50 units/ml RNAsin (Applied Biosystems)] and equal amounts of lysate were cleared by centrifugation at 9391 g for 5 min at 4°C. Sucrose sedimentation profiles were performed as described previously (Connolly et al., 2008; Maki et al., 2002), onto a 10-50% sucrose gradient [containing 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 100 mM KCl, 2 mM DTT, 100 μg/ml cycloheximide and 10-50% sucrose]. Samples were spun using a Beckman Coulter Optima L-90K Ultracentrifuge in an SW41 rotor (221,831 g) for 2.5 h at 4°C. Gradients were analyzed using a Biocomp Piston Gradient Fractionator with a BioRad Econo UV Monitor with a Full Scale of 1.0. Data was recorded using DataQ DI-158-UP data acquisition software. Samples were collected from fractions containing the 80S peak and polysome peaks. RNA was then extracted using Trizol Reagent.
In vitro egg maturation
In vitro egg maturation (IVEM) experiments were adapted from Spracklen and Tootle (2013). Drosophila females were fed yeast overnight and maintained at 25°C. Ovaries were dissected into freshly prepared IVEM media [1× Schneider's Drosophila media (Gibco, Thermo Fisher Scientific), heat-inactivated 10% fetal bovine serum (Invitrogen) and 1× penicillin/streptomycin glutamine (100×, Gibco, Thermo Fisher Scientific)]. Individual ovarioles were isolated so that morphological stages of EC development were easily distinguishable. ECs of interest were then transferred into a multi-well plate with 500 μl fresh media with the addition of MG132 (EMD Millipore) or DMSO. Plates were placed in the dark for in vitro development. After 6 h, ECs were mounted in Oil 10 S, VOLTALEF (VWR Chemicals) for live imaging.
Tissue culture
Drosophila Kc167 cells were cultured in Schneider's Drosophila medium with L-glutamine (PAN Biotech) including 10% heat-inactivated fetal calf serum (PAN Biotech) and penicillin/streptomycin according to general culturing procedures.
For the images shown in Fig. 8A, cells transiently transfected with expression plasmids encoding GFP-tagged versions of JabbaB or JabbaB [amino acids 192-321] were transferred to cover slips, fixed with 4% paraformaldehyde, counterstained with Hoechst33342 for DNA and embedded with Mowiol. Images were recorded using a Zeiss LSM710 confocal microscope.
For Fig. S9, Drosophila S2R+ cells transiently expressing GFP-tagged Jabba B or Jabba (aa 192-321) were plated in CellCarrier Ultra (PerkinElmer) 96-well plates and incubated with 400 µM oleic acid including 0.5 µg/ml of the fluorescently labeled C12 BODIPY558/568 fatty acid (Invitrogen/Molecular Probes). The cells were imaged live with an Operetta CLS confocal high-content-screening microscopy system (PerkinElmer) using a 40× objective in confocal mode.
Luciferase complementation assay
Protein-protein interactions were detected using a split-luciferase complementation assay as described previously (Kolkhof et al., 2017). In brief, putative interactors were tagged at the N or the C terminus with either the N-or C-terminal half of the Gaussia princeps luciferase enzyme [termed ‘hGluc(1)’ or ‘hGluc(2)’ fragments, respectively]. Cells were seeded in transparent tissue-culture 96-well plates (Sarstedt, Thermo Fisher Scientific) and allowed to adhere overnight. The next day, plasmids encoding putative interactors were transfected in triplicate using the Effectene transfection reagent (Qiagen) and BSA-bound oleic acid was added to a final concentration of 400 µM. After 4 days, the medium was removed from the cells, and the cells were – after one PBS-washing step – frozen at −80°C to facilitate homogenization. Lysis and luciferase activity detection was performed using the Gaussia-Juice Luciferase Assay (PJK Biotec) according to the manufacturer's description in white opaque 96-well plates (OptiPlate, PerkinElmer) and using a Synergy Mx microplate reader (BioTek). Part of the lysate was used to measure protein content using standard BCA protein assay (Pierce).
As controls, each assay included untransfected cells (baseline based on the background luciferase activity) and cells transfected with plasmids encoding the yeast GCN4 leucine zipper protein fused to hGluc(1) as well as hGluc(2), which is known to dimerize. The threshold for positive interactions was the average of two ‘zipper-zipper’ controls. Proteins are indicated in the graphs (Fig. 8B and Fig. S10) as hGluc(1) and hGluc(2).
Quantitation and image analysis
Image J was used to quantitate all images. Statistics were performed using an unpaired two-tailed Student's t-test or ANOVA followed by Tukey's test [****P<0.0001, ***P<0.001, **P<0.01, *P<0.05; NS, not significant (P≥0.05)] (Prism8, GraphPad). All graphs were generated using Prism 8. Data are mean±s.d.
To determine mean fluorescence intensity (AU) of histones, the fluorescence intensity of RFP, GFP or mEos3.2 (in a constant ROI) was measured. Because fluorescent intensity in NCs was quite variable from EC to EC, we chose not to quantify the signal in NCs and concentrated on oocytes. Oocytes were imaged immediately below the follicle nuclei, where LDs were most abundant. To determine the area covered by lipid droplets, ECs were imaged using confocal reflection microscopy (CRM) (excitation 633 nm/emission 623-643 nm) (Gáspár and Szabad, 2009). The boundaries of the area covered by lipid droplets was identified via thresholding and the area was measured. This analysis was repeated for ECs stained with BODIPY.
For IVEM experiments in Fig. 5, the average mean fluorescence intensity (AU) of wild type; H2Av-RFP was calculated. To obtain each data point for the ratio of Jabba−/−:wild type, individual Jabba−/−; H2Av-RFP mean fluorescence intensity (AU) was divided by the previously calculated average mean fluorescence intensity (AU) of wild type; H2Av-RFP. This was repeated for each treatment condition.
To determine the expression levels of Jabba B and Jabba Bdel aa 228-243, embryos were immunostained using anti-Jabba antibody and then imaged using confocal microscopy. For quantitation of anti-Jabba signal (Fig. S7), the embryo boundary was determined using thresholding and the image calculator tool in Image J. The fluorescence intensity of the embryo was then measured by subtracting the background signal in a region outside the embryo (using the original image) from the anti-Jabba signal of the embryo. Summed fluorescence intensity was then divided by the area of the embryo (Fig. S7). To quantify Jabba levels of Jabba B and Jabba Bdel aa 228-243, the fluorescence intensity/area of Jabba B and Jabba Bdel aa 228-243 was compared with the fluorescence intensity/area of embryos expressing varying levels of endogenous Jabba. (The expression levels of these embryos were determined by western analysis in Fig. S7.) Using this comparison, embryos of similar Jabba expression patterns were identified. This method of quantitation was employed instead of Western analysis to avoid errors due to potential differential transfer of endogenous and mCherry-tagged Jabba.
Supplementary Material
Acknowledgements
We thank Michael Eisen, John Belote, the Bloomington Stock Center, the Exelixis Drosophila Collection at Harvard Medical School and the Bangalore Fly Facility for fly stocks. We are grateful to Sina Ghaemmaghami for discussion and advice, and to Xin Bi, Daniel Bergstralh and Jeffrey Hayes for comments on the manuscript. We also thank Tina Tootle and Roxanne Keltz for technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.A.S., M.A.W.; Methodology: R.A.S., L.C., P.K., M.B.; Formal analysis: R.A.S., P.K., M.B.; Investigation: R.A.S., J.M.T., L.C., P.K., R.P.W.; Writing - original draft: R.A.S., M.A.W.; Writing - review & editing: R.A.S., J.M.T., M.B., M.A.W.; Visualization: R.A.S., R.P.W., M.B.; Supervision: M.A.W.; Project administration: M.A.W.; Funding acquisition: M.A.W.
Funding
This work was supported by the National Institutes of Health [RO1 GM102155 to M.A.W.]. This work was funded in part by the Deutsche Forschungsgemeinschaft (INST 208/760-1FUGG to M.B.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199381
References
- Ables, E. T. (2015). Drosophila oocytes as a model for understanding meiosis: an educational primer to accompany “corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila”. Genetics 199, 17-23. 10.1534/genetics.114.167940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio, L. and Schedl, P. (1985). Two discrete modes of histone gene expression during oogenesis in Drosophila melanogaster. Dev. Biol. 111, 220-231. 10.1016/0012-1606(85)90447-6 [DOI] [PubMed] [Google Scholar]
- Aramburu, J., Drews-Elger, K., Estrada-Gelonch, A., Minguillón, J., Morancho, B., Santiago, V. and López-Rodríguez, C. (2006). Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem. Pharmacol. 72, 1597-1604. 10.1016/j.bcp.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Bannister, A. J. and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381-395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller, M., Riedel, D., Jänsch, L., Dieterich, G., Wehland, J., Jäckle, H. and Kühnlein, R. P. (2006). Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5, 1082-1094. 10.1074/mcp.M600011-MCP200 [DOI] [PubMed] [Google Scholar]
- Campos, E. I., Fillingham, J., Li, G., Zheng, H., Voigt, P., Kuo, W.-H. W., Seepany, H., Gao, Z., Day, L. A., Greenblatt, J. F.et al. (2010). The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343-1351. 10.1038/nsmb.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus, G., Herker, E., Modi, A. A., Haas, J. T., Ramage, H. R., Farese, R. V., Jr. and Ott, M. (2013). Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J. Biol. Chem. 288, 9915-9923. 10.1074/jbc.M112.434910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celona, B., Weiner, A., Di Felice, F., Mancuso, F. M., Cesarini, E., Rossi, R. L., Gregory, L., Baban, D., Rossetti, G., Grianti, P.et al. (2011). Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 9, e1001086. 10.1371/journal.pbio.1001086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli, S., Guo, Y., Gross, S. P. and Welte, M. A. (2006). The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783-1795. 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- Connolly, K., Rife, J. P. and Culver, G. (2008). Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 70, 1062-1075. 10.1111/j.1365-2958.2008.06485.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn, S. W., Subtelny, A. O., Kronja, I., Kwasnieski, J. C., Orr-Weaver, T. L. and Bartel, D. P. (2016). mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5, e16955. 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadiga, J. and Nystul, T. G. (2019). The follicle epithelium in the Drosophila ovary is maintained by a small number of stem cells. eLife 8, e49050. 10.7554/eLife.49050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe, A. and McLauchlan, J. (2015). Hepatitis C virus and lipid droplets: finding a niche. Trends Mol. Med. 21, 34-42. 10.1016/j.molmed.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Finn, R. M., Ellard, K., Eirín-López, J. M. and Ausió, J. (2012). Vertebrate nucleoplasmin and NASP: egg histone storage proteins with multiple chaperone activities. FASEB J. 26, 4788-4804. 10.1096/fj.12-216663 [DOI] [PubMed] [Google Scholar]
- Foe, V. E. and Alberts, B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31-70. 10.1242/jcs.61.1.31 [DOI] [PubMed] [Google Scholar]
- Gallardo-Montejano, V. I., Saxena, G., Kusminski, C. M., Yang, C., McAfee, J. L., Hahner, L., Hoch, K., Dubinsky, W., Narkar, V. A. and Bickel, P. E. (2016). Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat. Commun. 7, 12723. 10.1038/ncomms12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáspár, I. and Szabad, J. (2009). In vivo analysis of MT-based vesicle transport by confocal reflection microscopy. Cell Motil. Cytoskeleton 66, 68-79. 10.1002/cm.20334 [DOI] [PubMed] [Google Scholar]
- Gunjan, A. and Verreault, A. (2003). A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537-549. 10.1016/S0092-8674(03)00896-1 [DOI] [PubMed] [Google Scholar]
- Guruharsha, K. G., Obar, R. A., Mintseris, J., Aishwarya, K., Krishnan, R. T., Vijayraghavan, K. and Artavanis-Tsakonas, S. (2012). Drosophila Protein interaction Map (DPiM): a paradigm for metazoan protein complex interactions. Fly 6, 246-253. 10.4161/fly.22108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., Chang, M., Kim, U. J. and Grunstein, M. (1987). Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48, 589-597. 10.1016/0092-8674(87)90237-6 [DOI] [PubMed] [Google Scholar]
- Herrero, A. B. and Moreno, S. (2011). Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 30, 2008-2018. 10.1038/emboj.2011.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Haratake, K., Miyahara, H. and Chiba, T. (2016). Proteasome activators, PA28γ and PA200, play indispensable roles in male fertility. Sci. Rep. 6, 23171. 10.1038/srep23171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. M. and Cooley, L. (2010). Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. J. Cell Biol. 188, 29-37. 10.1083/jcb.200909017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. M. and Cooley, L. (2014). Methods for studying oogenesis. Methods 68, 207-217. 10.1016/j.ymeth.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imschenetzky, M., Díaz, F., Montecino, M., Sierra, F. and Puchi, M. (1997). Identification of a cysteine protease responsible for degradation of sperm histones during male pronucleus remodeling in sea urchins. J. Cell. Biochem. 67, 304-315. 10.1002/(SICI)1097-4644(19971201)67:3<304::AID-JCB3>3.0.CO;2-# [PubMed] [Google Scholar]
- Jia, D., Xu, Q., Xie, Q., Mio, W. and Deng, W.-M. (2016). Automatic stage identification of Drosophila egg chamber based on DAPI images. Sci. Rep. 6, 18850. 10.1038/srep18850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M. R., Stephenson, R. A., Ghaemmaghami, S. and Welte, M. A. (2018). Developmentally regulated H2Av buffering via dynamic sequestration to lipid droplets in Drosophila embryos. eLife 7, e36021. 10.7554/eLife.36021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, R., Jin, M., Subramanian, V., Causey, C. P., Thompson, P. R. and Coonrod, S. A. (2012). Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev. Biol. 12, 19. 10.1186/1471-213X-12-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun, H. and Marzluff, W. F. (2005). Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12, 794-800. 10.1038/nsmb972 [DOI] [PubMed] [Google Scholar]
- Khor, B., Bredemeyer, A. L., Huang, C.-Y., Turnbull, I. R., Evans, R., Maggi, L. B., White, J. M., Walker, L. M., Carnes, K. and Hess, R. A. (2006). Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 26, 2999-3007. 10.1128/MCB.26.8.2999-3007.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R. C. (1970). Ovarian Development in Drosophila melanogaster. Academic Press. [Google Scholar]
- King, R. and Koch, E. A. (1963). Studies on the ovarian follicle cells of Drosophila. J. Cell Sci. 3, 297-320. 10.1242/jcs.s3-104.67.297 [DOI] [Google Scholar]
- Kolkhof, P., Werthebach, M., van de Venn, A., Poschmann, G., Chen, L., Welte, M., Stühler, K. and Beller, M. (2017). A luciferase-fragment complementation assay to detect lipid droplet-associated protein-protein interactions. Mol. Cell. Proteomics 16, 329-345. 10.1074/mcp.M116.061499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Thiel, K., Thul, P. J., Beller, M., Kühnlein, R. P. and Welte, M. A. (2012). Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104-2113. 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Johnson, M. R., Ke, Z., Chen, L. and Welte, M. A. (2014). Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr. Biol. 24, 1485-1491. 10.1016/j.cub.2014.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., Cook, R. G. and Allis, C. D. (1991). Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymena macronuclear development. Genes Dev. 5, 1601-1610. 10.1101/gad.5.9.1601 [DOI] [PubMed] [Google Scholar]
- Maki, J. A., Schnobrich, D. J. and Culver, G. M. (2002). The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell 10, 129-138. 10.1016/S1097-2765(02)00562-2 [DOI] [PubMed] [Google Scholar]
- Marzluff, W. F., Wagner, E. J. and Duronio, R. J. (2008). Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843-854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, Y., Itabe, H., Odaki, M., Hama, K., Fujimoto, Y., Mori, M., Sasabe, N., Aoki, J., Arai, H. and Takano, T. (2006). ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J. Lipid Res. 47, 87-98. 10.1194/jlr.M500170-JLR200 [DOI] [PubMed] [Google Scholar]
- McLauchlan, J., Lemberg, M. K., Hope, G. and Martoglio, B. (2002). Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21, 3980-3988. 10.1093/emboj/cdf414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner, D. and Hartwell, L. H. (1986). Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43-52. 10.1016/0092-8674(86)90483-6 [DOI] [PubMed] [Google Scholar]
- Mejhert, N., Kuruvilla, L., Gabriel, K. R., Elliott, S. D., Guie, M.-A., Wang, H., Lai, Z. W., Lane, E. A., Christiano, R., Danial, N. N.et al. (2020). Partitioning of MLX-family transcription factors to lipid droplets regulates metabolic gene expression. Mol. Cell 77, 1251-1264. 10.1016/j.molcel.2020.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir, M., Stadler, M. R., Ortiz, S. A., Hannon, C. E., Harrison, M. M., Darzacq, X. and Eisen, M. B. (2018). Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7, e40497. 10.7554/eLife.40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, V., Sanchez-Rubio, A., Aze, A., Iribarren, C., Fayet, C., Desdevises, Y., Garcia-Huidobro, J., Imschenetzky, M., Puchi, M. and Genevière, A.-M. (2012). The protease degrading sperm histones post-fertilization in sea urchin eggs is a nuclear cathepsin L that is further required for embryo development. PLoS ONE 7, e46850. 10.1371/journal.pone.0046850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, P. K. and Rasch, E. M. (1985). Determination of DNA content in the nurse and follicle cells from wild type and mutant Drosophila melanogaster by DNA-Feulgen cytophotometry. Histochemistry 82, 233-247. 10.1007/BF00501400 [DOI] [PubMed] [Google Scholar]
- Nagai, A., Sato, T., Akimoto, N., Ito, A. and Sumida, M. (2005). Isolation and identification of histone H3 protein enriched in microvesicles secreted from cultured sebocytes. Endocrinology 146, 2593-2601. 10.1210/en.2004-1478 [DOI] [PubMed] [Google Scholar]
- Navarro-Costa, P., McCarthy, A., Prudêncio, P., Greer, C., Guilgur, L. G., Becker, J. D., Secombe, J., Rangan, P. and Martinho, R. G. (2016). Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nat. Commun. 7, 12331. 10.1038/ncomms12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell, P. H., Stumpff, J. and Su, T. T. (2004). Embryonic cleavage cycles: how is a mouse like a fly? Curr. Biol. 14, R35-R45. 10.1016/j.cub.2003.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki, Y., Cheng, J., Fujita, A., Tokumoto, T. and Fujimoto, T. (2006). Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol. Biol. Cell 17, 2674-2683. 10.1091/mbc.e05-07-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki, Y., Cheng, J., Suzuki, M., Fujita, A. and Fujimoto, T. (2008). Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J. Cell Sci. 121, 2415-2422. 10.1242/jcs.025452 [DOI] [PubMed] [Google Scholar]
- Onikubo, T., Nicklay, J. J., Xing, L., Warren, C., Anson, B., Wang, W.-L., Burgos, E. S., Ruff, S. E., Shabanowitz, J., Cheng, R. H.et al. (2015). Developmentally regulated post-translational modification of nucleoplasmin controls histone sequestration and deposition. Cell Rep 10, 1735-1748. 10.1016/j.celrep.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker, L., Tennessen, J. M., Lam, G. and Thummel, C. S. (2009). Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metab. 9, 228-239. 10.1016/j.cmet.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetta, G. and Welte, M. A. (2018). Emerging links between lipid droplets and motor neuron diseases. Dev. Cell 45, 427-432. 10.1016/j.devcel.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, M., Jang, A. C.-C., Starz-Gaiano, M., Melani, M. and Montell, D. J. (2007). A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat. Protoc. 2, 2467-2473. 10.1038/nprot.2007.363 [DOI] [PubMed] [Google Scholar]
- Qian, M.-X., Pang, Y., Liu, C. H., Haratake, K., Du, B.-Y., Ji, D.-Y., Wang, G.-F., Zhu, Q.-Q., Song, W., Yu, Y.et al. (2013). Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 153, 1012-1024. 10.1016/j.cell.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska, A. and Köhler, A. (2018). The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell 174, 700-715.e718. 10.1016/j.cell.2018.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddell, A. and Jacobs-Lorena, M. (1985). Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 82, 3316-3319. 10.1073/pnas.82.10.3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, R. J., Krueger, E. W., Weller, S. G., Johnson, K. M., Casey, C. A., Schott, M. B. and McNiven, M. A. (2020). Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl. Acad. Sci. USA 117, 32443-32452. 10.1073/pnas.2011442117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. K., Kabbaj, M.-H. M., Paik, J. and Gunjan, A. (2009). Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11, 925-933. 10.1038/ncb1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. K., Gonzalez, M., Kabbaj, M.-H. M. and Gunjan, A. (2012). Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS ONE 7, e36295. 10.1371/journal.pone.0036295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, K. A. and Belote, J. M. (1999). The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome β-type subunit. Genetics 151, 211-220. 10.1093/genetics/151.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen, A. J. and Tootle, T. L. (2013). The utility of stage-specific mid-to-late Drosophila follicle isolation. J. Vis. Exp., 50493. 10.3791/50493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Shinoda, A., Kamada, H., Shimizu, M., Inoue, J. and Sato, R. (2016). Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 6, 20975. 10.1038/srep20975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond, J., Goodman, J. L., Strelets, V. B., Attrill, H., Gramates, L. S., Marygold, S. J., Matthews, B. B., Millburn, G., Antonazzo, G., Trovisco, V.et al. (2019). FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759-D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, S. L. and Welte, M. A. (2010). In-vivo centrifugation of Drosophila embryos. J. Vis. Exp. 10.3791/2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velentzas, P. D., Velentzas, A. D., Mpakou, V. E., Papassideri, I. S., Stravopodis, D. J. and Margaritis, L. H. (2011). Proteasome inhibition induces developmentally deregulated programs of apoptotic and autophagic cell death during Drosophila melanogaster oogenesis. Cell Biol. Int. 35, 15-27. 10.1042/CBI20100191 [DOI] [PubMed] [Google Scholar]
- Von Stetina, J. R. and Orr-Weaver, T. L. (2011). Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 3, a005553. 10.1101/cshperspect.a005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T. C. and Farese, R. V.Jr. (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687-714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat, L. W., Chao, C., Bartlett, R., Buchanan, J. L., Millington, J. W., Chih, H. J., Chowdhury, Z. S., Biswas, P., Huang, V., Shin, L. J.et al. (2020). A role for triglyceride lipase brummer in the regulation of sex differences in Drosophila fat storage and breakdown. PLoS Biol. 18, e3000595. 10.1371/journal.pbio.3000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A. (2007). Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17, 363-369. 10.1016/j.tcb.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Welte, M. A. (2015). As the fat flies: The dynamic lipid droplets of Drosophila embryos. Biochim. Biophys. Acta 1851, 1156-1185. 10.1016/j.bbalip.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A. and Gould, A. P. (2017). Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862(10 Pt B), 1260-1272. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A., Cermelli, S., Griner, J., Viera, A., Guo, Y., Kim, D.-H., Gindhart, J. G. and Gross, S. P. (2005). Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr. Biol. 15, 1266-1275. 10.1016/j.cub.2005.06.062 [DOI] [PubMed] [Google Scholar]
- Xu, G., Sztalryd, C. and Londos, C. (2006). Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1761, 83-90. 10.1016/j.bbalip.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Yu, J. and Li, P. (2017). The size matters: regulation of lipid storage by lipid droplet dynamics. Sci. China Life Sci. 60, 46-56. 10.1007/s11427-016-0322-x [DOI] [PubMed] [Google Scholar]
- Zechner, R., Zimmermann, R., Eichmann, T. O., Kohlwein, S. D., Haemmerle, G., Lass, A. and Madeo, F. (2012). FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279-291. 10.1016/j.cmet.2011.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Yang, L., Ding, Y., Wang, Y., Lan, L., Ma, Q., Chi, X., Wei, P., Zhao, Y., Steinbüchel, A.et al. (2017). Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 8, 15979. 10.1038/ncomms15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.