Abstract
Improving the capacity of detecting positive severe acute respiratory syndrome coronavirus 2 is critical for identifying the infection of coronavirus disease 2019 (COVID-19) precisely and thereby curbing the pandemic. Cross-disciplinary approaches may improve the efficiency of COVID-19 diagnosis by compensating to some extent the limitations encountered by traditional test methods during the COVID-19 pandemic. Combining computed tomography (CT), serum-specific antibody detection, and nanopore sequencing with nucleic acid testing for individual testing may improve the accuracy of identifying COVID-19 patients. At community or even regional/national levels, the combination of pooled screening and spatial epidemiological strategies may enable the detection of early transmission of epidemics in a cost-effective way, which is also less affected by restricted access to diagnostic tests and kit supplies. This would significantly advance our capacity of curbing epidemics as soon as possible, and better prepare us for entering a new era of high-impact and high-frequency epidemics.
Keywords: SARS-CoV-2, COVID-19, Nucleic acid testing, Sampling, CT, Spatial epidemiology
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has rapidly spread to more than 200 countries worldwide and been identified by the World Health Organization (WHO) as a pandemic (Wiersinga et al. 2020). Although SARS-CoV-2 is similar to other human coronaviruses, such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), COVID-19 has exhibited higher infectivity and lower mortality (Sah et al. 2020). The basic reproduction number (R0) of the COVID-19 infection has varied from 2.2 to 3.9 by location (Sanche et al. 2020), which is higher than that of SARS-CoV (1.7–1.9) and MERS-CoV (< 1) (Yan et al. 2020b). At present, it is believed that SARS-CoV-2 has been mainly spread by COVID-19 patients with symptoms (Qu and Cong 2020), although asymptomatic infections have also accounted for an uncertain portion of the total infections due to their similar viral loads of SARS-CoV-2 as that in symptomatic patients (Nishiura et al. 2020). For example, it was recently estimated that symptoms in about 31–60% of all COVID-19 infections might be covert or mild (Nishiura et al. 2020), where they may have not been ill enough to seek medical care, and probably slipped past traditional screening methods, such as temperature check (Qiu 2020). Although the roles played by asymptomatic infections in virus transmission remain elusive (Qiu 2020), it is vital to identify and isolate both symptomatic and asymptomatic infections as early and accurately as possible, which is a major way to prevent the spread of infectious diseases including COVID-19 (Qiu 2020).
This review offers an overview of the current methods for SARS-CoV-2 nucleic acid testing (NAT), discusses how sampling time and sample types could influence SARS-CoV-2 NAT, identifies challenges for improving SARS-CoV-2 NAT, introduces other test methods that may assist with SARS-CoV-2 NAT for overcoming the false negative issue, and provides an outlook on the potential use of spatial technologies in furthering strengthening traditional test methods at individual and community levels during the COVID-19 pandemic.
Current methods for SARS-CoV-2 NAT
The currently available methods for SARS-CoV-2 NAT can be generally grouped into three categories: reverse transcriptase polymerase chain reaction (RT-PCR), reverse transcription loop-mediated isothermal amplification (RT-LAMP), and metagenome sequencing. RT-PCR is the most widely used and also considered as a “gold standard” (Baron et al. 2020), which consists of viral extraction, reverse transcription, PCR amplification, and result interpretation, requiring well-equipped laboratory-based facilities and skilled technicians. Numerous laboratory-based and commercial assays have been developed based on RT-PCR, each with adequate sensitivity and specificity claimed for evaluation on different sample types. The sensitivity of these assays has varied from 66.7 to 100% (Chan et al. 2020), mainly across different SARS-CoV-2 genome targets, testing procedures, and references for sensitivity validation. Although RT-PCR has been approved with the capability for sensitive detection, the implementation of RT-PCR still suffers from a time-consuming detection process (usually more than 2 h or longer, depending on the procedures of sample processing, shipment, and handling in a clinical diagnostic laboratory) and strict requirements on the facilities and technicians’ skills (Dao Thi et al. 2020). Moreover, the false negative rates associated with RT-PCR have been high due to the low viral load during both initial and recovery stages of patients (Pokhrel et al. 2020).
RT-LAMP assays for SARS-CoV-2 have emerged as an alternative NAT method (Ali et al. 2020; Kashir and Yaqinuddin 2020). Based on the isothermal amplification employing reverse transcriptase and DNA polymerase with strong activities of strand replacement, RT-LAMP, especially the RNA extraction-free colorimetric RT-LAMP assay, possesses advantages including rapid diagnostic time (usually less than 60 min) and not requiring special equipment, such as thermal cyclers (reaction at a constant temperature, e.g., 65 °C or 63 °C), allowing point-of-care diagnostic testing (Dao Thi et al. 2020; Kashir and Yaqinuddin 2020). RT-LAMP assays can be assisted by artificial intelligence (e.g., using smart diagnostic devices for automated image acquisition and data processing) (Rohaim et al. 2020), enhanced by coupling the detection of amplified targets with CRISPR-Cas12 module (Ali et al. 2020), and optimized by two-stage isothermal methods (El-Tholoth et al. 2020). The sensitivity was reported from 75 to 97.5% by validation using clinical samples or RNA extracts (Ali et al. 2020). However, several studies reported that RT-LAMP assays were less analytically sensitive than RT-PCR. For example, the limit of detection (LOD) of RT-LAMP assays is about ten-fold higher in copies per reaction than that of RT-PCR (Baek et al. 2020).
Metagenome sequencing, including next generation sequencing (NGS) and nanopore sequencing, have been used to obtain the full genome of SARS-CoV-2 to facilitate better understanding of its evolution, transmission, and pathogenicity (Garces-Ayala et al. 2020; Laamarti et al. 2020). However, NGS techniques are costly, time-consuming, low-throughput, and require large amounts of viral templates (e.g., viral load ≥ 105 copies/mL) (Xiao et al. 2020b), and hence are usually not adopted as the initial NAT method(s) for virus detection. Although resource-intensive, NGS techniques are essential for monitoring emerging SARS-CoV-2 variants and updating the genome resources of SARS-CoV-2, which is critical for optimization of primers and probes to improve the capability of RT-PCR and RT-LAMP assays. The SWIFT SARS-CoV-2 panel (a commercial panel) for NGS enables high-throughput performance and preparation of viral sequence libraries from samples with low viral titers, which provides a potential solution to optimize NGS for regular SARS-CoV-2 diagnosis. A nanopore sequencing assay with the efficacy of targeting amplification and long-read and real-time sequencing, called nanopore targeted sequencing (NTS), has recently been developed to detect SARS-CoV-2 within 6–10 h (Wang et al. 2020). By parallel testing with RT-PCR assays, NTS was found to have increased sensitivity and specificity for SARS-CoV-2 detection (Wang et al. 2020).
Sampling time and sample types influencing SARS-CoV-2 NAT
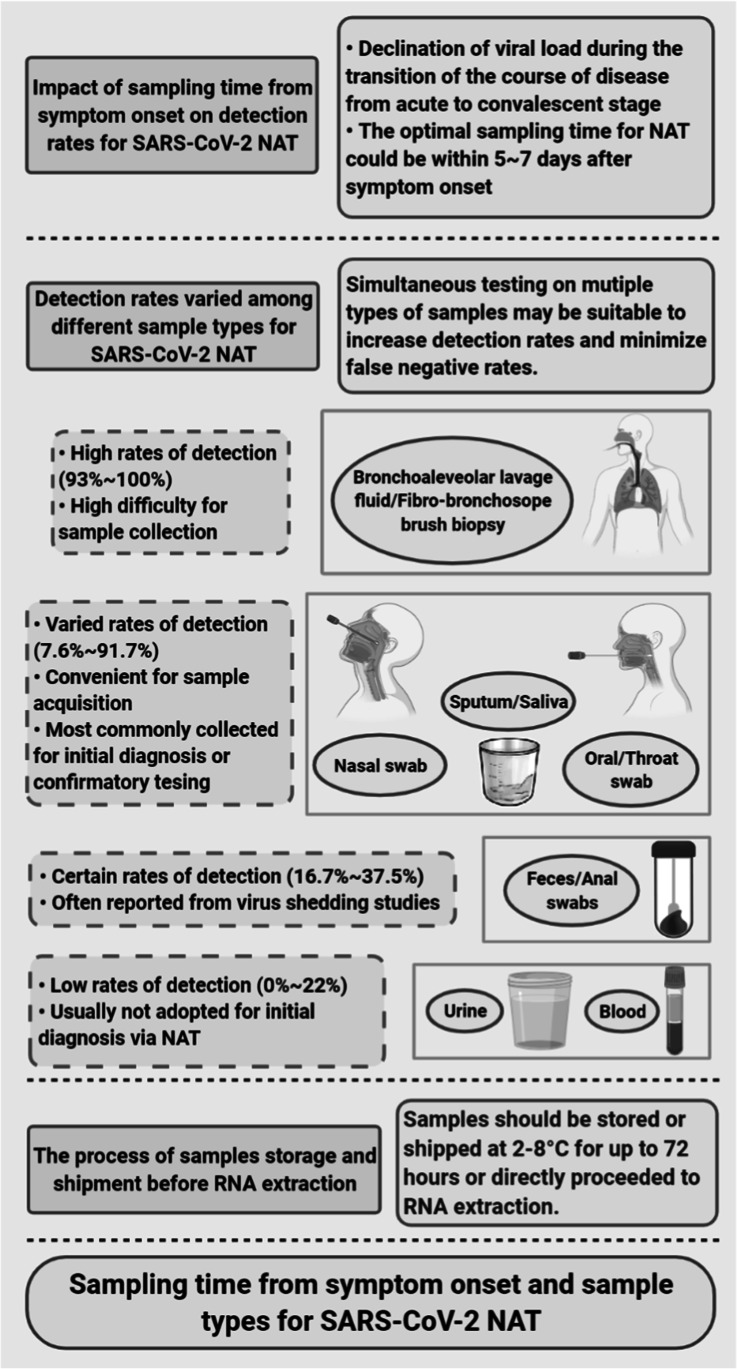
Sampling time from the onset of symptoms is one of the important factors affecting the detection rate of SARS-CoV-2 (Fig. 1). Several studies have described high detection rates (> 90%) using real-time quantitative RT-PCR for samples collected within 5–7 days from the onset of symptoms (Xiao et al. 2020a). The detection rates of samples diminished as the days went by after the onset of symptoms, even becoming zero after 6 weeks (Xiao et al. 2020a). Another similar study described increased RT-PCR cycle threshold (Ct) values (i.e., declines of viral load) for up to 20 days after the onset of COVID-19 symptoms (Singanayagam et al. 2020). Therefore, the optimal sampling time for NAT seems to be within 5–7 days after the onset of symptoms, owing to a higher viral load during this acute stage of the disease (Wang 2020).
Fig. 1.
Key factors for high detection rate of SARS-CoV-2 by nucleic acid testing (NAT). Timely sampling in acute phases of SARS-CoV-2, simultaneous testing on multiple types of samples, and proper storage and shipping of the samples to RNA extraction in the laboratories are the keys for high detection rates of SARS-CoV-2
The detection rates (mostly based on RT-PCR) for initial diagnosis of SARS-CoV-2 varied among different (collected) sample types. Samples from bronchoalveolar lavage fluid and fibro-bronchoscope brush biopsy, highly demanding of sampling facilities, were reported to have detection rates that were as high as 93–100% (Liu et al. 2020). Convenience samples, such as nasopharyngeal swabs, oral pharyngeal swabs, throat swabs, sputum, and saliva, were the most commonly used for initial diagnosis or confirmatory testing for SARS-CoV-2, with detection rates of 7.6–91.7% (Wolfel et al. 2020). Among these, nasopharyngeal swabs were the most abundantly used and represented the recommended convenience sample for screening owing to moderate detection rates (Bwire et al. 2020). Feces/stool samples or anal swabs were adopted in several studies related to virus shedding, with detection rates of 16.7–37.5% (To et al. 2020a). These were usually not the first choice for initial diagnosis or confirmatory testing. Similarly, low detection rates of 0–22% were observed among urine and blood samples (Wolfel et al. 2020).
Both sampling time and sample types should be considered for increasing detection rates. Timely sampling in acute phases of the disease is the key for high detection rates regardless of sample types. For example, a detection rate of 91.7% was observed among saliva samples that were collected on the second day from the onset of symptoms (To et al. 2020b). Also, simultaneous testing on multiple types of samples may be suitable to increase detection rates and decrease false negative rates (Woloshin et al. 2020). Further, samples should be directly proceeded, or properly stored and shipped at 2–8 °C within 72 h, to RNA extraction in the laboratories, as improper operations during the process of sample storage and shipment may lead to the degradation of RNA, possibly resulting in false negative outcomes via NAT.
Challenges for improving SARS-CoV-2 NAT
The primary concern for improving the sensitivity of NAT is to minimize the false negative rate for SARS-CoV-2 detection. Although more COVID-19 cases could be identified by repeated and/or simultaneous testing on multiple types of samples, two major intrinsic factors for false negative detection remained. One factor is the genome variants or deletion of genomic fragments of SARS-CoV-2. The current RT-PCR and RT-LAMP assays target specific genomic/protein-encoding regions, such as ORF1ab, RdRP (RNA-dependent RNA polymerase), S (surface glycoprotein), E (envelope protein), M (membrane glycoprotein), and N (nucleocapsid phosphoprotein). A recent analysis of 10,022 SARS-CoV-2 genomes from the four national and global databases identified 5775 distinct genome variants, belonging to five major clades (i.e., D614G, L84S, L3606F, D448del, and G392D) (Koyama et al. 2020). These variants were described to be associated with different levels of influences on pathogenicity, but rarely with increased disease severity. Nevertheless, it should be noted that the sensitivity of NAT may suffer from these variants, especially in the case of mutations within the regions that are targeted by the primers and probes used for RT-PCR or RT-LAMP assays. Also, the genome variation and the reported deletion of a certain length of the virus genome may make the existing primers or probes lose the target, consequently increasing the chance of false negative detection (Su et al. 2020).
In addition to the impact of genome variants or deletion of genomic fragments of SARS-CoV-2, the other factor for false negative detection for SARS-CoV-2 is the LOD of NAT. The viral loads in patients during the latent or early convalescent periods of SARS-CoV-2 infection may be lower than the LOD of the selected NAT, which may result in false negative results. To date, a vast range of LODs have been reported among various brands of assays and laboratory settings and in different countries. For example, the LODs of RT-PCR were reported between 5 and 7740 copies/mL (Norz et al. 2020; Sahajpal et al. 2020) or between 3.8 and 5.2 copies/reaction (Corman et al. 2020), while the LODs of RT-LAMP assays ranged from 2 to 304 copies/reaction (Lamb et al. 2020; Yan et al. 2020a). Such large variations in LODs may be due to multiple aspects during the NAT process that affect the stability of testing performance, which includes viral extraction, volume of RNA templates required for amplification, efficiency of reverse transcription (and amplification), conditions and settings of the reaction, and the effect of inhibitors within the reaction. However, despite these limitations, some less sensitive NAT is easy to implement on site, and hence still strongly encouraged to assist as an important diagnostic tool with the control of the pandemic.
Other test methods assisting with SARS-CoV-2 NAT
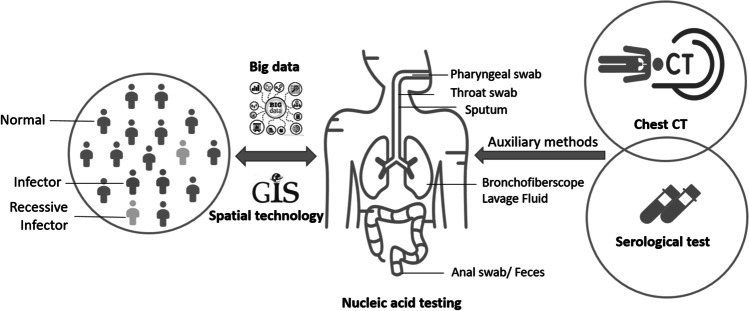
The detection rate of SARS-CoV-2 by NAT is usually difficult to improve during the epidemics. Given the persistence of a certain rate of false negative detection and thus missed diagnoses for any single method of NAT, other test methods should be combined with NAT to improve the rate of diagnosis of SARS-COV-2 infection (Fig. 2). Computed tomography (CT) scanning can be considered an auxiliary diagnostic tool by assessing the nature and extent of lesions and discovering subtle lung parenchymal changes in the early stage of COVID-19 (Ai et al. 2020). Several studies have indicated that combining CT with RT-PCR could increase the sensitivity of the detection, compared to using RT-PCR alone (Table 1). Moreover, CT can help evaluate the severity of COVID-19 in patients (De Smet et al. 2021). However, CT scanning is less cost-effective and not capable of detecting asymptomatic patients. For those with positive CT results (e.g., ground-glass opacity, consolidation, reticulation and/or thickened interlobular septa, nodules) but negative RT-PCR results, repeated RT-PCR tests on a different type of samples or using a different test assay are recommended.
Fig. 2.
Cross-disciplinary methods to assist with nucleic acid testing (NAT) for SARS-CoV-2. Combining SARS-CoV-2 NAT with traditional auxiliary methods, such as computed tomography (CT) and serological tests, and spatial technologies could improve the diagnostic accuracy of COVID-19 patients
Table 1.
Sensitivity of RT-PCR and CT test methods for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
| First author (year) | Number of patients | Sensitivity of RT-PCR and CT | Difference of sensitivity | |
|---|---|---|---|---|
| RT-PCR | CT scans | |||
| (Ai et al. 2020) | 1014 | 59% | 88% | 29% |
| (Bernheim et al. 2020) | 121 | 74% | 83% | 9% |
| (Falaschi et al. 2020) | 773 | 60% | 63% | 3% |
| (Fang et al. 2020) | 51 | 71% | 98% | 27% |
| (Feng et al. 2020) | 38 | 24% | 97% | 73% |
| (Gietema et al. 2020) | 193 | 43% | 57% | 14% |
| (He et al. 2020) | 34 | 79% | 77% | 2% |
| (Long et al. 2020) | 36 | 83% | 97% | 13% |
| (Mei et al. 2020) | 905 | 46% | 75% | 29% |
| (Yamamoto et al. 2020) | 36 | 14% | 44% | 30% |
CT computed tomography, RT-PCR real-time quantitative reverse transcriptase PCR
Antibody tests, a widely used rapid test method, have also been commonly used for diagnosis for SARS-CoV-2, with detection rates ranging from 55 to 100% (Mahase 2020). Antibodies, such as immunoglobulin G (IgG), M (IgM), and A (IgA) generated by the immune system in response to SARS-CoV-2 infection, are used as potential markers. Current evidence has suggested that IgM may be a good indicator of early SARS-CoV-2 infection, and a higher level of IgG has often been observed in late stages (or during the post-recovery period) of SARS-CoV-2 infection (Lv et al. 2020). The most common antibody tests are based on colloidal gold-based immunochromatographic assay (GICA) and enzyme-linked immunosorbent type assays (ELISA); other types of antibody tests include lateral flow type assays (LFA) and chemiluminescent immunoassays (CLIA). The optimal sampling time for antibody tests was found to be 7 days after the onset of symptoms, considering the time required for antibody production (Mallapaty 2020). Despite the low sensitivity of antibody tests (e.g., 30.1% in the first week since the onset of symptoms (Deeks et al. 2020)), which may have limited its application as the primary diagnostic tool for COVID-19, they have played an important role in complementing NAT to improve the detection rate, especially when NAT results are negative for those presenting COVID-19 symptoms (Petherick 2020). However, developing accurate antibody tests remains a challenge due to various levels of antibody response and cross-reaction of antibody assays.
Spatial technologies to further strengthen traditional test methods
Although traditional test methods could harmonize for better testing than using each method alone, all of them may suffer from limited testing capacity and materials (e.g., reagents) during public health emergencies, especially at the early stages of the epidemic outbreak. Advanced spatial technologies may help us take fuller advantage of limited testing resources to monitor the infection status of a larger population in a cost-effective manner (Yang et al. 2020). Spatial epidemiological and big data approaches, such as spatial clustering analysis in a Geographic Information Systems (GIS) environment, have been used to detect local clustering of cases in previous infectious disease and epidemic research; spatiotemporal clustering analysis may further reveal space–time clusters of cases (Cuadros et al. 2017). They have played a critical role in curbing COVID-19 epidemic, and may play an even bigger role if data-sharing infrastructures and confidentiality protection mechanisms were set up more properly ahead. For example, combining one’s movement trajectory and that of individual infected cases, from mobile service providers and/or smartphone-based apps, could determine one’s contact history precisely and hence estimate the level of risk for infection (Yang et al. 2021b). Such estimation may to some extent improve the sensitivity and specificity of traditional tests for SARS-CoV-2. That is to say, one with recent exposure to COVID-19 cases, if tested positive for SARS-CoV-2, is more likely to be true positive. One without recent exposure to COVID-19 cases, if tested negative for SARS-CoV-2, is more likely to be true negative (Yang et al. 2021a). The integration of spatial epidemiological methods with RT-PCR detection may be realized without violating the confidentiality requirements in an intelligent syndromic surveillance system (Jia and Yang 2020a, b). Such spatial approaches would also overcome recall bias to a large extent, which is a key limitation of the traditional epidemiological survey (Budd et al. 2020).
Spatial technologies may also help select sites of greatest need for interventions, such as individual NAT for SARS-CoV-2. For example, if the potential environmental determinants of COVID-19 incidence are identified (e.g., air pollutants), remote sensing could simultaneously acquire those environmental data at large scales over a short period (e.g., acquiring air pollutant concentration over the whole of China within 2 h), to detect the regions with specific environments (Jia et al. 2019; Jiang et al. 2020). Such large-scale data acquisition methods are extremely useful when healthcare resources (e.g., NAT reagents) are limited during emergency events, especially when the epidemic has become a pandemic at the global scale. Moreover, agent-based modeling may further integrate environmental data with individual characteristics over time, to simulate a dynamic process of host-agent-environment interaction in a lifecourse epidemiological triad (Jia 2019, 2020; Jia et al. 2020). This would enable the precise assignment of priority for SARS-CoV-2 tests at the individual level.
More cost-effectively than most, if not all, approaches, spatial epidemiological approaches may leverage individuals’ movement and contact history and environmental information from multiple sources (e.g., population/syndromic/environmental surveillance systems, social networks, web searches) to improve early warning of epidemics and identify covert COVID-19 cases, who may otherwise escape laboratory and clinical tests due to lack of any symptoms (Hu and Ci 2020). For example, spatial technologies can assist in building global databases of SARS-CoV-2 on the basis of the data from wastewater surveillance, which has been used by myriad universities and cities worldwide during COVID-19 to monitor the presence of infection within a given region, and to track and infer the spreading of infection across regions. Tests of historical wastewater samples have shown that SARS-CoV-2 had existed at different places of the world in 2019, for example, in Barcelona of Spain on 12 March, in Florianópolis of Brazil on 27 November, and in Milan and Turin of Italy on 18 December. Such databases of SARS-CoV-2 could supplement other COVID-19 surveillance systems to inform timely public health actions, such as screening for infections at targeted sites at early stages.
Concluding remarks
Cross-disciplinary approaches may improve the efficiency of COVID-19 diagnosis. To improve the sensitivity of NAT at the individual level, we need to select appropriate diagnostic site(s) and use a standardized sampling process at different stages of infection, and follow standardized operation procedures in laboratories. Combining CT, serum-specific antibody detection, and nanopore sequencing with NAT may improve the accuracy of identifying COVID-19 patients. At community or even regional/national levels, the combination of pooled screening and spatial epidemiological strategies might enable the detection of early transmission of epidemics in a cost-effective way, which is also less affected by restricted access to diagnostic tests and kit supplies. This would significantly advance our capacity of curbing epidemics as soon as possible, and better prepare us for entering a new era of high-impact and high-frequency epidemics.
Author contribution
PJ, SY, and PZ conceived and designed research. SY, XP, and DY conducted research. SY, XP, PZ, and PJ wrote the manuscript. All authors read and approved the manuscript.
Funding
The authors thank the International Institute of Spatial Lifecourse Epidemiology (ISLE) for research support.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shujuan Yang and Xiongfeng Pan contributed equally to this work.
Contributor Information
Peibin Zeng, Email: zengpeibin@live.cn.
Peng Jia, Email: jiapengff@hotmail.com.
References
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Ziyong S, Liming X. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;22(1):11–19. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, Pain A, Alofi FS, Alsomali A, Hashem AM, Khogeer A, Almontashiri NAM, Abedalthagafi M, Hassan N, Mahfouz MM. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek YH, Um J, Antigua KJC, Park JH, Kim Y, Oh S, Kim YI, Choi WS, Kim SG, Jeong JH, Chin BS, Nicolas HDG, Ahn JY, Shin KS, Choi YK, Park JS, Song MS. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron RC, Risch L, Weber M, Thiel S, Grossmann K, Wohlwend N, Lung T, Hillmann D, Ritzler M, Bigler S, Egli K, Ferrara F, Bodmer T, Imperiali M, Heer S, Renz H, Flatz L, Kohler P, Vernazza P, Kahlert CR, Paprotny M, Risch M. Frequency of serological non-responders and false-negative RT-PCR results in SARS-CoV-2 testing: a population-based study. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0978. [DOI] [PubMed] [Google Scholar]
- Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3):200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd J, Miller BS, Manning EM, Lampos V, Zhuang M, Edelstein M, Rees G, Emery VC, Stevens MM, Keegan N, Short MJ, Pillay D, Manley E, Cox IJ, Heymann D, Johnson AM, McKendry RA. Digital technologies in the public-health response to COVID-19. Nat Med. 2020;26(8):1183–1192. doi: 10.1038/s41591-020-1011-4. [DOI] [PubMed] [Google Scholar]
- Bwire GM, Majigo MV, Njiro BJ, Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: a systematic review and meta-analysis. J Med Virol. 2020 doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WM, Ip JD, Chu AW, Yip CC, Lo LS, Chan KH, Ng AC, Poon RW, To WK, Tsang OT, Leung WS, Kwan MY, Chua GT, Chung TW, Hung IF, Kok KH, Cheng VC, Chan JF, Yuen KY, To KK. Identification of nsp1 gene as the target of SARS-CoV-2 real-time RT-PCR using nanopore whole-genome sequencing. J Med Virol. 2020 doi: 10.1002/jmv.26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros DF, Li J, Branscum AJ, Akullian A, Jia P, Mziray EN, Tanser F. Mapping the spatial variability of HIV infection in Sub-Saharan Africa: effective information for localized HIV prevention and control. Sci Rep. 2017;7:9093. doi: 10.1038/s41598-017-09464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, Freistaedter A, Papagiannidis D, Galmozzi C, Stanifer ML, Boulant S, Klein S, Chlanda P, Khalid D, Barreto Miranda I, Schnitzler P, Krausslich HG, Knop M, Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12(556):eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet K, De Smet D, Ryckaert T, Laridon E, Heremans B, Vandenbulcke R, Demedts I, Bouckaert B, Gryspeerdt S, Martens GA. Diagnostic performance of chest CT for SARS-CoV-2 Infection in individuals with or without COVID-19 symptoms. Radiology. 2021;298(1):E30–e37. doi: 10.1148/radiol.2020202708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Dittrich S, Emperador D, Hooft L, Leeflang MM, Van den Bruel A, Cochrane C-DTAG. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tholoth M, Bau HH, Song J. A single and two-stage, closed-tube, molecular test for the 2019 Novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11860137.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi Z, Danna PSC, Arioli R, Pasche A, Zagaria D, Percivale I, Tricca S, Barini M, Aquilini F, Andreoni S, Carriero A. Chest CT accuracy in diagnosing COVID-19 during the peak of the Italian epidemic: a retrospective correlation with RT-PCR testing and analysis of discordant cases. Eur J Radiol. 2020;130:109192. doi: 10.1016/j.ejrad.2020.109192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yuan LF, Zheng XX, Liu WS, Osric YRC, Li LL. Novel coronavirus pneumonia diagnosis by CT and nucleic acid detection. Guangdong Med. 2020;22(3):1–4. [Google Scholar]
- Garces-Ayala F, Araiza-Rodriguez A, Mendieta-Condado E, Rodriguez-Maldonado AP, Wong-Arambula C, Landa-Flores M, Del Mazo-Lopez JC, Gonzalez-Villa M, Escobar-Escamilla N, Fragoso-Fonseca DE, Esteban-Valencia MDC, Lloret-Sanchez L, Arellano-Suarez DS, Nunez-Garcia TE, Contreras-Gonzalez NB, Cruz-Ortiz N, Ruiz-Lopez A, Fierro-Valdez MA, Regalado-Santiago D, Martinez-Velazquez N, Mederos-Michel M, Vazquez-Perez J, Martinez-Orozco JA, Becerril-Vargas E, Salas J, Hernandez-Rivas L, Lopez-Martinez I, Alomia-Zegarra JL, Lopez-Gatell H, Barrera-Badillo G, Ramirez-Gonzalez JE. Full genome sequence of the first SARS-CoV-2 detected in Mexico. Arch Virol. 2020;165(9):2095–2098. doi: 10.1007/s00705-020-04695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietema HA, Zelis N, Nobel JM, Lambriks LJG, van Alphen LB, Oude Lashof AML, Wildberger JE, Nelissen IC, Stassen PM. CT in relation to RT-PCR in diagnosing COVID-19 in The Netherlands: a prospective study. PLoS ONE. 2020;15(7):e0235844. doi: 10.1371/journal.pone.0235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JL, Luo L, Luo ZD, Lyu JX, Ng MY, Shen XP, Wen Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan. China Respir Med. 2020;168:105980. doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZB, Ci C. Screening and management of asymptomatic infection of corona virus disease 2019 (COVID-19) Zhonghua Yu Fang Yi Xue Za Zhi [Chin J Prev Med] 2020;54:E025–E025. doi: 10.3760/cma.j.cn112150-20200229-00220. [DOI] [PubMed] [Google Scholar]
- Jia P. Spatial lifecourse epidemiology. Lancet Planet. Health. 2019;3(2):e57–e59. doi: 10.1016/S2542-5196(18)30245-6. [DOI] [PubMed] [Google Scholar]
- Jia P. Understanding the epidemic course in order to improve epidemic forecasting. Geohealth. 2020;4(10):e2020GH000303. doi: 10.1029/2020GH000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Dong W, Yang S, Zhan Z, Tu LS. Spatial lifecourse epidemiology and infectious disease research. Trends Parasitol. 2020 doi: 10.1016/j.pt.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Stein A, James P, Brownson RC, Wu T, Xiao Q, Wang L, Sabel CE, Wang Y. Earth observation: investigating noncommunicable diseases from space. Annu Rev Public Health. 2019 doi: 10.1146/annurev-publhealth-040218-043807. [DOI] [PubMed] [Google Scholar]
- Jia P, Yang S (2020a) China needs a national intelligent syndromic surveillance system. Nat Med 26(7):990. 10.1038/s41591-020-0921-5 [DOI] [PubMed]
- Jia P, Yang S (2020b) Early warning of epidemics: towards a national intelligent syndromic surveillance system (NISSS) in China. BMJ Glob Health 5(10):e002925. 10.1136/bmjgh-2020-002925 [DOI] [PMC free article] [PubMed]
- Jiang Y, Wu XJ, Guan YJ. Effect of ambient air pollutants and meteorological variables on COVID-19 incidence. Infect Control Hosp Epidemiol. 2020;41(9):1011–1015. doi: 10.1017/ice.2020.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashir J, Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98(7):495–504. doi: 10.2471/blt.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laamarti M, Chemao-Elfihri MW, Kartti S, Laamarti R, Allam L, Ouadghiri M, Smyej I, Rahoui J, Benrahma H, Diawara I, Alouane T, Essabbar A, Siah S, Karra M, El Hafidi N, El Jaoudi R, Sbabou L, Nejjari C, Amzazi S, Mentag R, Belyamani L, Ibrahimi A. Genome sequences of six SARS-CoV-2 strains isolated in Morocco, obtained using Oxford nanopore MinION technology. Microbiol Resour Announc. 2020;9(32):e00767–20. doi: 10.1128/MRA.00767-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus/Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE. 2020;15(6):e0234682. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Zeng B, Li Z, Li X, Li H. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H, Wu NC, Tsang OT, Yuan M, Perera R, Leung WS, So RTY, Chan JMC, Yip GK, Chik TSH, Wang Y, Choi CYC, Lin Y, Ng WW, Zhao J, Poon LLM, Peiris JSM, Wilson IA, Mok CKP. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31(9):107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: Antibody test that claims to be 99% accurate is certified by EU. BMJ (Clin Res Ed) 2020;369:m1742. doi: 10.1136/bmj.m1742. [DOI] [PubMed] [Google Scholar]
- Mallapaty S. Antibody tests suggest that coronavirus infections vastly exceed official counts. Nature. 2020 doi: 10.1038/d41586-020-01095-0. [DOI] [PubMed] [Google Scholar]
- Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, Xie Z, Ma Y, Robson PM, Chung M, Bernheim A, Mani V, Calcagno C, Li K, Li S, Shan H, Lv J, Zhao T, Xia J, Long Q, Steinberger S, Jacobi A, Deyer T, Luksza M, Liu F, Little BP, Fayad ZA, Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H, Kobayashi T, Suzuki A, Jung SM, Hayashi K, Kinoshita R, Yang Y, Yuan B, Akhmetzhanov AR, Linton NM, Miyama T. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norz D, Fischer N, Schultze A, Kluge S, Mayer-Runge U, Aepfelbacher M, Pfefferle S, Lutgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J Clin Virol. 2020;128:104390. doi: 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick A. Developing antibody tests for SARS-CoV-2. Lancet (London, England) 2020;395(10230):1101–1102. doi: 10.1016/s0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel P, Hu C, Mao H. Detecting the coronavirus (COVID-19) ACS Sensors. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J (2020) Covert coronavirus infections could be seeding new outbreaks. Nature. 10.1038/d41586-020-00822-x [DOI] [PubMed]
- Qu Y-M, Cong H-Y. Positive result of Sars-Cov-2 in sputum from a cured patient with COVID-19. Travel Med Infect Dis. 2020;34:101619. doi: 10.1016/j.tmaid.2020.101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim MA, Clayton E, Sahin I, Vilela J, Khalifa ME, Al-Natour MQ, Bayoumi M, Poirier AC, Branavan M, Tharmakulasingam M, Chaudhry NS, Sodi R, Brown A, Burkhart P, Hacking W, Botham J, Boyce J, Wilkinson H, Williams C, Whittingham-Dowd J, Shaw E, Hodges M, Butler L, Bates MD, La Ragione R, Balachandran W, Fernando A, Munir M. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020;12(9):972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R, Rodriguez-Morales AJ, Jha R, Chu DKW, Gu H, Peiris M, Bastola A, Lal BK, Ojha HC, Rabaan AA, Zambrano LI, Costello A, Morita K, Pandey BD, Poon LLM. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol Resour Announc. 2020;9(11):e00169–20. doi: 10.1128/mra.00169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahajpal NS, Mondal AK, Njau A, Ananth S, Jones K, Ahluwalia PK, Ahluwalia M, Jilani Y, Chaubey A, Hegde M, Kota V, Rojiani A, Kolhe R. Proposal of RT-PCR-based mass population screening for severe acute respiratory syndrome coronavirus 2 (Coronavirus Disease 2019) J Mol Diagn. 2020 doi: 10.1016/j.jmoldx.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, Zhuang Y, Kalimuddin S, Low JGH, Tan CW, Chia WN, Mak TM, Octavia S, Chavatte JM, Lee RTC, Pada S, Tan SY, Sun L, Yan GZ, Maurer-Stroh S, Mendenhall IH, Leo YS, Lye DC, Wang LF, Smith GJD. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11(4):e01610–20. doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, Leung WS, Chik TS, Choi CY, Kandamby DH, Lung DC, Tam AR, Poon RW, Fung AY, Hung IF, Cheng VC, Chan JF, Yuen KY. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z, Gu J, Xiang B, Liu J, Jiang W, Shen G, Zhao W, Men D, Deng Z, Yu L, Wei W, Li Y, Liu T. Nanopore Targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small. 2020;16(32):e2002169. doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J Virol Methods. 2020;283:113919. doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a Review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woloshin S, Patel N, Kesselheim AS. False negative tests for SARS-CoV-2 infection - challenges and implications. N Engl J Med. 2020;383(6):e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- Xiao AT, Tong YX, Gao C, Zhu L, Zhang YJ, Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J Clin Virol. 2020;127:104346. doi: 10.1016/j.jcv.2020.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Liu X, Ji J, Li M, Li J, Yang L, Sun W, Ren P, Yang G, Zhao J, Liang T, Ren H, Chen T, Zhong H, Song W, Wang Y, Deng Z, Zhao Y, Ou Z, Wang D, Cai J, Cheng X, Feng T, Wu H, Gong Y, Yang H, Wang J, Xu X, Zhu S, Chen F, Zhang Y, Chen W, Li Y, Li J. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Medicine. 2020;12(1):57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Saito S, Hayakawa K, Hashimoto M, Takasaki J, Ohmagari N. When should clinicians repeat SARS-CoV-2 RT-PCR?: repeat PCR testing targeting patients with pulmonary CT findings suggestive of COVID-19. Jpn J Infect Dis. 2020 doi: 10.7883/yoken.JJID.2020.531. [DOI] [PubMed] [Google Scholar]
- Yan C, Cui J, Huang L, Du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, Yao H, Li N, Zhao H, Feng Y, Liu S, Zhang Q, Liu D, Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Chang L, Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev Med Virol. 2020;30(3):e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xu T, Jia P, Xia H, Guo L, Zhang L, Ye K (2020) Transportation, germs, culture: a dynamic graph model of COVID-19 outbreak. Quantitative biology (Beijing, China): 1–7. 10.1007/s40484-020-0215-4 [DOI] [PMC free article] [PubMed]
- Yang S, Pan X, Zeng P, Jia P. Spatial technologies to strengthen traditional testing for SARS-CoV-2. Trends Microbiol. 2021 doi: 10.1016/j.tim.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yu C, Jia P. Spatiobehavioral characteristics - defining the epidemiology of new contagious diseases at the earliest moment possible. Trends Parasitol. 2021;37(3):179–181. doi: 10.1016/j.pt.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]