Abstract
Background
Obesity, one of the most common chronic health conditions worldwide, is a multifactorial disease caused by complex genetic and environmental interactions. Several association studies have revealed a considerable number of candidate loci for obesity; however, the genotype–phenotype correlations remain unclear. To date, no comprehensive systematic review has been conducted to investigate the genetic risk factors for obesity among Arabs.
Objectives
This study aimed to systematically review the genetic polymorphisms that are significantly associated with obesity in Arabs.
Methods
We searched four literature databases (PubMed, Science Direct, Scopus, and Google Scholar) from inception until May 2020 to obtain all reported genetic data related to obesity in Arab populations. Quality assessment and data extraction were performed individually by three investigators.
Results
In total, 59 studies comprising a total of 15,488 cases and 9,760 controls were included in the systematic review. A total of 76 variants located within or near 49 genes were reported to be significantly associated with obesity. Among the 76 variants, two were described as unique to Arabs, as they have not been previously reported in other populations, and 19 were reported to be distinctively associated with obesity in Arabs but not in non-Arab populations.
Conclusions
There appears to be a unique genetic and clinical susceptibility profile of obesity in Arab patients.
Subject terms: Obesity, Genetics, Diseases
Introduction
Obesity is a chronic health condition in which excessive body fat has built up to a level that may cause negative health consequences. People are considered obese when their body mass index (BMI, kg/m2) exceeds 30 kg/m2 [1]. Our knowledge of human obesity has progressed beyond the simple generalization that obesity is fully explained by inappropriate eating. It is now believed that obesity is a complex and multifactorial disease caused by several interactions that involve multiple factors, including genetic, metabolic, environmental, behavioral, sociodemographic, and economic [2]. Excessive fat deposition in obesity is widely considered the result of disequilibrium between energy expenditure (i.e., lack of physical activity) and energy intake (i.e., diet), resulting in excess adiposity and accumulation of lipids, primarily triglycerides, in skeletal muscle, liver, and other organs [3, 4].
The prevalence of obesity worldwide approximately tripled between 1975 and 2018 according to the World Health Organization (WHO) [1]. In 2016, over 650 million adults were obese, and 41 million children were either overweight or obese [1]. It is expected that by 2025, 17% of the world’s adults or almost 1 billion of the population will be obese, with an estimated 177 million adults predicted to become morbidly obese [5]. In the Arab world, the prevalence of obesity has drastically increased during the last three decades [6]. According to the WHO, the average prevalence of obesity in the Arab world increased from ~6.5% in 1975 to 20% in 2016 [6]. The prevalence of obesity varies significantly across the 22 Arab countries, ranging from as low as 7.8% in Comoros to as high as 37.9% in Kuwait [6]. The highest number of obesity-attributable deaths in 2016 among the 22 Arab countries was reported in Bahrain (25.69%) [6].
Arabs are a major panethnic group comprising 22 countries [7]. The ethnic composition of the Arab world has historically been altered, yielding a high degree of genetic heterogeneity [8]. It is noteworthy to mention that consanguinity rates are high in most Arab countries, with first-cousin marriage rates reaching 30% [9], this makes the architecture of the Arab genome unique with regard to their susceptibility to different diseases, including both Mendelian and complex diseases [10–20]. Given that certain ethnic groups and specific populations residing in particular geographic areas in the Arab world are more prone to obesity than others, genetic factors are believed to play a key role in predisposing certain Arab populations to obesity [21]. While genes play a fundamental role in predisposing a person to obesity, the environment can influence these genes both positively and negatively. In fact the dramatic increase in the numbers of obese people in the Arab world is believed to be largely driven by the rapid environmental changes in the Arab world that have occurred in the last two decades, which have brought significant prosperity, lifestyle changes, and urbanization to the Arab world [22]. These factors have created an “obesogenic environment” in Arab countries, and thus, genetic and environmental factors are believed to have a balance that variably intertwines in the development of obesity [22].
With the rising prevalence of obesity in the Arab world, there has been a growing interest in understanding the genetic architecture that renders Arabs susceptible to obesity. Over the past decade, researchers have started to sequence Arab genomes through national projects in hopes of defining disease-associated genetic polymorphisms for gene disorders in Arabs and to establish meaningful genotype–phenotype correlations. Starting with Saudi Arabia [23], followed by Qatar [24], and currently the United Arab Emirates [25], these countries are establishing their 1000 Genomes Projects. These projects are crucial for mapping potential obesity-associated genetic variants among Arabs to help characterize Arab patients suffering from obesity, which may ultimately improve premarital genetic counseling, health outcomes and quality of life in obese Arab patients.
Obesity etiology is known to be multifactorial, involving a complex interplay between genes and environment [26]. While a genetic basis for obesity does exist, defining the exact genetic contribution has proven to be a difficult task [26]. The genetic heritability of obesity is estimated to account for 30–50% of the age- and gender-adjusted phenotypic variances [26]. Approximately 31–90% of interindividual body weight variability is attributed to genetic factors [27, 28]. As many as 200 genetic variants in various genome-wide association studies (GWASs) have been associated with obesity in many different populations, predominantly Europeans. Nevertheless, only about 3% of the heritability can be explained by variants currently known to be associate with BMI [29].
According to the genetic criteria, obesity is classified into: (i) monogenic, when a mutated gene is responsible for the phenotype; (ii) syndromic, when a set of specific symptoms are present, and a small group of genes is involved; and (iii) polygenic, also referred to as “common” obesity, which accounts for 95% of obese cases, with many genes adding up to provide a further risk to the individual [30].
Given the complex nature of obesity and the fact that it does not follow a typical Mendelian transmission pattern, it is believed that several susceptibility genes with low or moderate effects play a role in predisposition to the disease [31, 32]. There is firm evidence that genes influencing energy homeostasis and thermogenesis, adipogenesis, leptin-insulin signaling transduction, and hormonal signaling peptides play a role in the development of obesity [33–37]. Several potential obesity candidate genes and variants have been documented, including LEP and LEPR, which have been mainly implicated in monogenic obesity, as well as FTO, APOE, PPARG, and PPARA which have been mainly implicated in polygenic/common obesity [31, 32]. However, the genotype–phenotype correlations remain unclear. In addition, there has been relatively little attention devoted to comprehensively investigating genetic polymorphisms associated with obesity in Arab populations. Therefore, in this study, we aimed to systematically review the current evidence on genetic variants associated with monogenic and polygenic obesity in the Arab world.
Methods
To ensure the rigor of the current systematic review, it was designed and implemented based on the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [38] (Table S1). The described search method and selection strategy were used to identify studies investigating the effect of obesity-associated genetic variants, including single nucleotide polymorphisms (SNPs), variable number tandem repeats (VNTRs), deletions, insertions, and copy-number variants, on obesity risk in Arab patients residing in any of the 22 Arab countries.
Search strategy
We used four literature databases (PubMed, Scopus, Science Direct, and Google Scholar) to retrieve all studies related to obesity genetics in Arab populations up to May 2020. In addition, cross-referencing from the bibliographies of all retrieved articles (citation tracking) was performed. We utilized several Boolean operators and search strings for the different electronic database searches (Table S2).
To gain a better understanding of the ethnic distribution of the captured variants and whether they are distinctive to Arab populations (i.e., if they circulate only among Arabs but not in other ethnic groups), we individually searched the captured variants in the following databases: Leiden Open Variation Database (LOVD) (http://www.lovd.nl/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), dbSNP (https://www.ncbi.nlm.nih.gov/snp/), Human Genome Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/ac/index.php), Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), PubMed, and Google Scholar.
Study selection
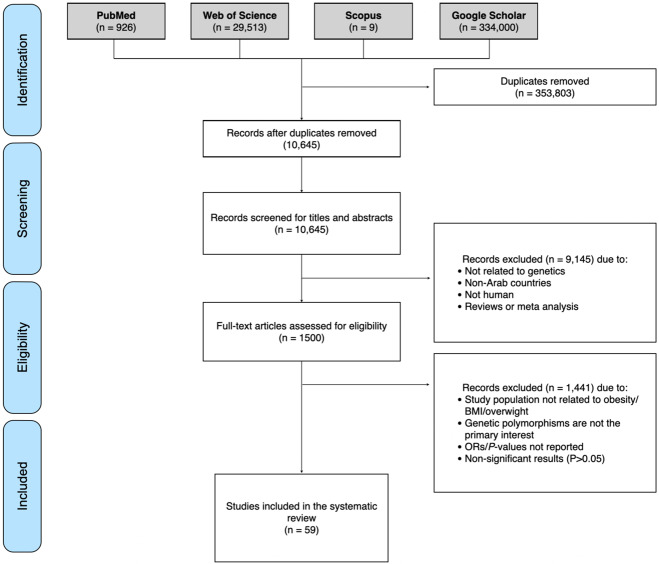
The inclusion and exclusion criteria were developed using a PECO(T) [participants, exposure, comparator, outcome(s), and type of study] structure. However, as we are looking for observational studies, we only considered participants, exposure and the outcome of interest. For inclusion, studies had to meet all the following criteria: (1) Population: Arabs residing in Arab countries. (2) Exposure: genetic polymorphisms significantly associated with obesity (P < 0.05). (3) Outcome: obesity (BMI ≥ 30 kg/m2). We included all studies reporting obesity indices or anthropometric measures, such as BMI, fat mass, waist circumference (WC), hip circumference (HC), and waist-hip ratio (WHC). (4) Type of study: no study type limits were applied. Articles published in peer-reviewed journals were included. We excluded duplicate publications, studies on animal subjects, studies conducted on non-Arabs, case series, review articles, and articles not in the English language. The PRISMA flow chart for study selection is shown in Fig. 1.
Fig. 1. PRISMA flow diagram of the included studies.
A total of 1500 full-text articles were assessed for eligibility, of which 59 studies met our inclusion criteria and were included in the systematic review.
Three scientists (SY, AI, and R.A.-J) worked independently in identifying, screening, and performing the quality control of the extracted data from the four literature databases. All citations were exported to Endnote X9, and duplicate citations were removed. Articles were screened in two stages: [1] the first stage involved performing the initial screening of the titles and abstracts, and assessing relevance for the scope of the systematic review according to our inclusion/exclusion criteria, [2] the second stage involved retrieval of the full text of each potentially relevant study and screening for content to decide on its inclusion. Articles for which full-text articles could not be retrieved or only abstracts were available were reviewed for content and were included if they met the inclusion criteria. Points of discrepancies were resolved through discussion between SY, AI, and R.A.-J, and additional points of debate were further resolved with HZ until a consensus was reached.
Data extraction
The following information was extracted and recorded: genes associated with obesity, chromosomal location, gene function, significantly associated variant (P < 0.05), SNP ID, country of origin, clinical phenotype reported in the study, cases (n), controls (n), gender and age (if mentioned), the number of individuals screened for gene variants vs number of obese individuals carrying variants, associated allele/genotype, methods (including baseline characteristics and genotyping techniques) and statistical data [crude odds ratios (cOR) and adjusted (aOR), 95% confidence interval (CI), P value]. Finally, the ethnic distribution of each variant was recorded. A thorough search was conducted in public databases (ClinVar and dbSNP) for any missing information.
Quality assessment
Quality assessment was performed using the assessment tools of the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH): [1] the NIH Quality Assessment tool for Observational Cohort and Cross-Sectional Studies; and [2] the NIH Quality Assessment tool for Case-Control Studies [39]. The NIH tools categorizes studies as either good, fair, or poor. Table S4 summarizes the quality assessments of the 59 studies that were included in this systematic review.
Results
A total of 1500 full-text articles were assessed for eligibility, among which 59 studies met our inclusion criteria and were included in the systematic review (Fig. 1). Most of the studies were case-control studies (n = 49). The remaining were cross-sectional (n = 9) and cohort GWA analysis (n = 1). The studies included in the systematic review comprised a total of 15,488 cases and 9760 controls. The studies captured Arabs from 12 Arab countries, and most of the studies were conducted in Saudi Arabia (n = 15) and Egypt (n = 15), followed by Tunisia (n = 14). Fewer studies were conducted in the United Arab Emirates (n = 4), Kuwait (n = 2), Iraq (n = 2), Morocco (n = 2), Bahrain (n = 1), Qatar (n = 1), Lebanon (n = 1), Jordan (n = 1), and Algeria (n = 1) (Fig. 2). No genetic data were captured from the remaining ten Arab countries (Yemen, Oman, The Comoros Islands, Djibouti, Mauritania. Somalia, Sudan, Libya, Syria or Palestine). The number of captured variants comprised 76 located within or near 49 genes (Table 1, Table S3, and Fig. 2). Data are summarized in Table 1 and Table S3. Among the 49 genes included in this systematic review, FTO was the most frequently studied gene in Arab patients with obesity (reported in 14 different articles) (Table 1, Table S3). In this systematic review, variants in the FTO gene were captured from six Arab countries (Saudi Arabia, Tunisia, Egypt, Iraq, Kuwait, and United Arab Emirates) (Table 1, Table S3). Other commonly studied genes were LEP (n = 7), ADIPOQ (n = 4), and LEPR (n = 3). All remaining genes were reported only once or twice (Table 1).
Fig. 2. Classification of the included studies according to country.
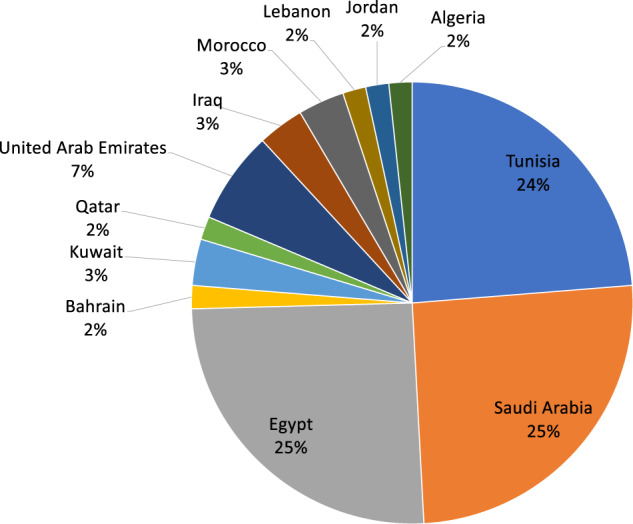
Most of the studies were conducted in Saudi Arabia (25%) and Egypt (25%), followed by Tunisia (24%). Fewer studies were conducted in the United Arab Emirates (7%), Kuwait (3%), Iraq (3%), Morocco (3%), Bahrain (2%), Qatar (2%), Lebanon (2%), Jordan (2%), and Algeria (2%). No genetic data were captured from the remaining 10 Arab countries (Yemen, Oman, The Comoros Islands, Djibouti, Mauritania. Somalia, Sudan, Libya, Syria, or Palestine). The number of captured variants was 76 located within or near 49 genes.
Table 1.
Genetic polymorphisms associated with obesity retrieved from the 59 included studies.
| Gene | Variant | SNP ID | Country | Cases (n) | Controls (n) | Clinical phenotype | OR | 95% CI | P value | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| ABCA1 | G > A | rs2230806 | Egypt | 128 | 128 | Obesity | 2.75 | 1.19–6.36 | 0.042 | [69] |
| ACE | 287-bp Alu ins/del in intron 16 | – | Tunisia | 259 | 302 | Obesity | NR | NR | NR | [70] |
| ACE | 287-bp Alu ins/del in intron 16 | – | Egypt | 70 | 72 | Obesity | 3.45 | 1.26–9.48 | 0.005 | [71] |
| ADIPOQ | +2019delA | – | Tunisia | 160 | 169 | Obesity | NR | NR | NR | [72] |
| ADIPOQ | c.11374 C > G | – | Tunisia | 160 | 169 | Obesity | NR | NR | NR | [72] |
| ADIPOQ | c.164 C > T | – | Jordan | 389 | – | Obesity | NR | NR | 0.000 | [73] |
| ADIPOQ | G > T | rs1501299 | Egypt | 100 | 97 | Obesity | 4.23 | 2.8–6.4 | 0.000 | [74] |
| ADIPOQ | G > T | rs1501299 | Jordan | 389 | – | Obesity | NR | NR | 0.000 | [73] |
| ADIPOQ | G > T | rs1501299 | Tunisia | 160 | 169 | Obesity | 0.64 | 0.409–0.917 | 0.039 | [72] |
| ADIPOQ | G > T | rs1501299 | Tunisia | 400 | 721 | Obesity | 0.603 | 0.467–0.779 | 0.001 | [75] |
| ADIPOQ | −11391 G/A | rs17300539 | Tunisia | 160 | 169 | Obesity | 1.68 | 1.040–3.110 | 0.044 | [72] |
| ADIPOQ | c.639 C > T | rs3821799 | Tunisia | 160 | 169 | Obesity | 1.85 | 1.110–3.100 | 0.018 | [72] |
| ADIPOQ | c.4552 C > T | rs822393 | Tunisia | 400 | 721 | Obesity | 1.52 | 1.230–1.994 | 0.001 | [75] |
| ADRB2 | Arg16Gly | rs1042713 | Saudi Arabia | 214 | 115 | Obesity | 3.38 | 1.456–7.822 | 0.0001 | [76] |
| ADRB3 | Trp64Arg | rs4994 | Saudi Arabia | 214 | 115 | Obesity | 7 | 2–24 | 0.0004 | [77] |
| AGT | M235T | rs699 | Tunisia | 259 | 302 | Obesity | 0.29 | 0.19–0.44 | 0.001 | [70] |
| APLN | – | rs3115757 | Egypt | 112 | 39 | Obesity | 12.09 | 1.39–104.85 | 0.024 | [78] |
| APOA2 | c.−492T > C | – | Egypt | 196 | 107 | Obesity | NR | NR | 0.001 | [79] |
| APOA2 | c.−265T > C | rs5082 | Egypt | 240 | 260 | Obesity | NR | NR | 0.001 | [80] |
| APOA5 | c.56 C > G | rs3135506 | Morocco | 164 | 295 | Obesity | 3.16 | 1.52–6.62 | 0.002 | [81] |
| APOA5 | c.−1131T > C | rs662799 | Morocco | 164 | 295 | Obesity | 4.13 | 1.86–9.03 | 0.0001 | [81] |
| ARAP1 | G > T | rs1552224* | Saudi Arabia | 1800 | 423 | Obesity | 1.5 | 1.0–2.3 | 0.04 | [82] |
| ASAH1 | C > T | rs17126232 | United Arab Emirates | 80 | – | Obesity | NR | NR | NR | [83] |
| BDNF | T > A | rs10767664 | Saudi Arabia | 204 | NR | Obesity | 1.923 | 1.32–2.81 | 0.00072 | [84] |
| CD36 | G > A | rs1761667 | Algeria | 83 | 82 | Obesity | 1.63 | 1.04–2.55 | 0.041 | [85] |
| ENPP1 | K121Q | rs1044498 | Morocco | 503 | 412 | Obesity | 1.91 | 1.27–2.85 | 0.002 | [86] |
| EXT2 | – | rs3740878* | Egypt | 37 | 37 | Obesity | 6 | 2.0–17.6 | 0.001 | [87] |
| FTO | G > T | rs17817449 | Egypt | 37 | 37 | Obesity | 2.4 | 1.0–5.5 | 0.036 | [87] |
| FTO | G > T | rs17817449 | Saudi Arabia | 106 | 106 | Obesity | 1.75 | 1.02–3.03 | 0.043 | [88] |
| FTO | G > T | rs3751812 | United Arab Emirates | 318 | 392 | Obesity | 1.47 | 1.09–1.99 | 0.011 | [89] |
| FTO | G > T | rs3751812 | Saudi Arabia | 204 | NR | obesity | 1.523 | 1.08–2.15 | 0.016 | [84] |
| FTO | C > A | rs11642841 | Saudi Arabia | 1800 | 423 | Obesity | 1.5 | 1.1–2.1 | 0.01 | [82] |
| FTO | T > C | rs1421085 | United Arab Emirates | 915 | – | Obesity | NR | NR | 0.0041 | [90] |
| FTO | T > C | rs1421085 | Saudi Arabia | 136 | 104 | Obesity | 2.5 | 1.13–5.55 | 0.023 | [91] |
| FTO | T > A | rs9939609 | Egypt | 110 | 122 | Obesity | 1.89 | 1.29–2.78 | 0.001 | [32] |
| FTO | T > A | rs9939609 | Saudi Arabia | 20 | 166 | Obesity | NR | NR | [92] | |
| FTO | T > A | rs9939609 | Kuwait | 674 | 214 | Obesity | 1.47 | 1.01–2.12 | 0.041 | [93] |
| FTO | T > A | rs9939609 | United Arab Emirates | 315 | 98 | Obesity | NR | NR | 0.027 | [90] |
| FTO | T > A | rs9939609 | Iraq | 120 | 50 | Obesity | NR | NR | NR | [94] |
| FTO | T > A | rs9939609 | Iraq | 120 | 60 | Obesity | NR | NR | 0.031 | [95] |
| FTO | T > A | rs9939609 | Saudi Arabia | 136 | 104 | Obesity | 2.97 | 1.30–6.79 | 0.009 | [91] |
| FTO | T > A | rs9939609 | Tunisia | 494 | 334 | Obesity | 1.7 | 1.29–2.25 | 0.001 | [96] |
| GC | GT | rs7041* | Bahrain | 406 | – | Obesity | NR | NR | 0.012 | [97] |
| GCKR | T > C | rs780094 | Egypt | 37 | 37 | Obesity | 3.6 | 1.6–7.9 | 0.001 | [87] |
| GNB3 | c.825 C > T | rs5443 | Saudi Arabia | 106 | 106 | Obesity | 6.68 | 3.26–13.69 | 0.0001 | [88] |
| GNPDA2 | A > G | rs10938397 | Qatar | 614 | 190 | Obesity | 2.4 | 1.27–4.56 | 0.0266 | [98] |
| IGF2BP2 | G > T | rs4402960 | Tunisia | 256 | 90 | Obesity and T2D | 2.06 | 1.40–3.03 | 0.0001 | [99] |
| IL-6 | G > T | rs1554606* | Saudi Arabia | 204 | – | Obesity | 1.55 | 1.08–2.22 | 0.024 | [100] |
| IL-6 | c.174 G/C | rs1800795* | Egypt | 85 | 64 | Obesity | NR | NR | 0.000 | [101] |
| LEP | c.104 T > G** | – | Egypt | 80 | – | Obesity | NR | NR | NR | [36] |
| LEP | c.34delC** | – | Egypt | 80 | – | Obesity | NR | NR | NR | [36] |
| LEP | c.313 C > T | rs104894023 | Egypt | 80 | – | Obesity | NR | NR | NR | [36] |
| LEP | 3’ UTR A/C | rs11761556* | Tunisia | 160 | 169 | Obesity | 2.63 | 1.13–6.11 | 0.025 | [102] |
| LEP | G/A | rs1349419 | Tunisia | 27 | 6 | Obesity | NR | NR | 0.039 | [65] |
| LEP | G > A | rs2167270 | Tunisia | 27 | 6 | Obesity | NR | NR | 0.039 | [65] |
| LEP | c.2548 G > A | rs7799039* | Tunisia | 160 | 169 | Obesity | 1.87 | 1.106–2.78 | 0.028 | [102] |
| LEP | c.2548 G > A | rs7799039* | Tunisia | 400 | 721 | Obesity | 2.36 | 1.75–3.18 | 0.001 | [75] |
| LEP | c.2548 G > A | rs7799039* | Tunisia | 160 | 169 | Obesity and MetS | 3.405 | 1.78–6.48 | 0.001 | [103] |
| LEP | c.2548 G > A | rs7799039* | Tunisia | 229 | 251 | Obesity | NR | NR | 0.001 | [64] |
| LEP | Tetranucleotide repeat (TTTC)n | Egypt | 120 | 83 | Obesity and MS | 14.6 | 5.5–38.6 | 0.000 | [66] | |
| LEPR | p.Lys109Arg | rs1137100 | Tunisia | 160 | 169 | Obesity | 0.399 | 0.179–0.892 | 0.025 | [102] |
| LEPR | p.Gln223Arg | rs1137101 | Tunisia | 160 | 169 | Obesity | 1.41 | 1.035–1.85 | 0.045 | [102] |
| LEPR | p.Gln223Arg | rs1137101 | Egypt | 110 | 122 | Obesity | 2.21 | 1.30–3.74 | 0.004 | [32] |
| LEPR | c.3057 G > A | rs62589000 | Tunisia | 393 | 317 | Obesity | 2.73 | 1.03–7.21 | 0.042 | [31] |
| LOC284260, RIT2 | G/A | rs7239883 | United Arab Emirates | 915 | – | Obesity | NR | NR | 0.0075 | [104] |
| LPL | C > T | rs285* | Egypt | 120 | 83 | Obesity | 6.6 | 1.9–22.9 | 0.000 | [105] |
| MC4R | T > C | rs17782313 | United Arab Emirates | 318 | 392 | Obesity | 1.35 | 1.00–1.81 | 0.048 | [89] |
| MC4R | T > C | rs17782313 | Saudi Arabia | 136 | 104 | Obesity | 1.72 | 1.02–2.89 | 0.038 | [91] |
| MC4R | C > A | rs571312 | United Arab Emirates | 318 | 392 | Obesity | 1.39 | 1.03–1.87 | 0.03 | [89] |
| MMP-1 | −519 A/G | rs1144393* | Tunisia | 168 | 202 | Obesity | 1.61 | 1.07–2.44 | 0.02 | [106] |
| MMP-12 | c.82 A > G | rs2276109* | Tunisia | 168 | 202 | Obesity | 2.39 | 1.36–4.2 | 0.002 | [106] |
| MMP-3 | Lys45Glu (A > G) | rs679620 | Tunisia | 168 | 202 | Obesity | 4.93 | 1.84–13.32 | 0.002 | [106] |
| MMP-7 | −181 A/G | rs11568818* | Tunisia | 168 | 202 | Obesity | 3.68 | 1.40–6.71 | 0.01 | [106] |
| MTHFD1 | c.1958G > A | rs2236225* | Egypt | 37 | 37 | Obesity | 4.6 | 0.9–23.6 | 0.050 | [87] |
| MTHFR | c.677 C > T | rs1801133 | Egypt | 51 | 30 | Obesity | 13.5 | 2.82–64.67 | 0.001 | [107] |
| NOS3 | c.894 G > T | rs1799983 | Tunisia | 183 | 211 | Obesity | 2.62 | 0.99–6.92 | 0.04 | [108] |
| NOS3 | 4a/b* | Tunisia | 183 | 211 | Obesity | 1.72 | 1.16–2.56 | 0.004 | [108] | |
| PPARG | C > G | rs1801282 | Egypt | 37 | 37 | Obesity | 3.4 | 1.6–7.2 | 0.001 | [87] |
| PPARG | C > G | rs1801282 | Tunisia | 262 | 83 | Obesity | 3.4 | 1.6–7.2 | 0.001 | [109] |
| PRDM16 | – | rs2651899* | Saudi Arabia | 89 | 84 | Obesity | 44.6 | 11.5984–172.0157 | 0.0001 | [110] |
| PROX1 | – | rs1704198 | United Arab Emirates | 80 | – | Obesity | NR | NR | NR | [83] |
| PTGS1 | – | rs5788* | Egypt | 37 | 37 | Obesity | 3.3 | 1.4–7.9 | 0.006 | [87] |
| RETN | −420 C > G | rs1862513* | Egypt | 145 | 155 | Obesity | 3.06 | 1.49–6.26 | 0.014 | [111] |
| RETN | −420 C > G | rs1862513* | Tunisia | 400 | 721 | Obesity | 1.48 | 1.09–2 | 0.011 | [75] |
| RETN | −420 C > G | rs1862513* | Tunisia | 160 | 169 | Obesity and MetS | 2.22 | 1.39–3.56 | 0.001 | [112] |
| RETN | c.+299 G > A | rs3745367 | Egypt | 145 | 155 | Obesity | 3.53 | 1.65–7.52 | 0.005 | [111] |
| STAT4 | G > T | rs7574865* | Egypt | 37 | 37 | Obesity and diabetes | 4 | 1.7–9.5 | 0.001 | [87] |
| TCF7L2 | C > T | rs7903146 | Saudi Arabia | 1800 | 423 | Obesity | 1.4 | 1.0–1.8 | 0.03 | [82] |
| TCF7L2 | C > T | rs7903146 | Lebanon | 308 | – | Obesity | NR | NR | 0.001 | [113] |
| TCN2 | c.67 A > G | rs9606756* | Kuwait | 1260 | 1272 | T1D | NR | NR | 0.0132 | [114] |
| TFAP2B | A > G | rs987237 | Qatar | 614 | 190 | Obesity | 0.79 | 0.54–1.15 | 0.0014 | [98] |
| TMEM18 | A > C | rs12463617 | United Arab Emirates | 318 | 392 | Obesity | 0.7 | 0.51–0.95 | 0.023 | [89] |
| TP53 | C > G | rs1042522* | Saudi Arabia | 136 | 122 | Obesity | 2.169 | 1.086–4.334 | 0.02716 | [115] |
| UCP1 | A > G | rs1800592 | Saudi Arabia | 337 | 155 | Moderate and extreme obesity | 1.52 | 1.10–2.08 | 0.009 | [116] |
| UCP1 | – | rs3811791 | Saudi Arabia | 337 | 155 | Moderate obesity | 2.89 | 1.33–6.25 | 0. 007 | [116] |
| UCP2 | 45-bp ins/del | – | Saudi Arabia | 86 | 65 | Severe obesity | 0.18 | 0.07–0.44 | 0.0004 | [117] |
| UCP2 | 45-bp ins/del | – | Saudi Arabia | 84 | 29 | Obesity and T2D | 0.18 | 0.07–0.44 | 0.0004 | [118] |
| USP37 | – | rs492400 | United Arab Emirates | 915 | – | Obesity | NR | NR | 0.0096 | [104] |
| VDR | Bsm-I | rs1544410 | Saudi Arabia | 402 | 489 | Obesity | NR | NR | 0.028 | [119] |
| VDR | Bsm-I | rs1544410 | Saudi Arabia | 200 | 100 | Obesity | NR | NR | 0.042 | [120] |
| VDR | TaqI | rs731236 | Saudi Arabia | 402 | 489 | Obesity | NR | NR | 0.009 | [119] |
| VDR | TaqI | rs731236 | Saudi Arabia | 200 | 100 | Obesity | NR | NR | 0.021 | [120] |
| VDR | TaqI | rs731236 | Egypt | 50 | 60 | Obesity | 2.33 | 1.34–4.1 | 0.003 | [121] |
CI confidence interval, OR odds ratio, NR not reported
*Unique genotype–phenotype correlation.
**Unique variant.
According to the number of variants reported per gene, most variants were captured in LEP, with eight obesity-associated variants reported among Arabs (rs11761556, rs7799039, c.104 T > G, c.34delC, rs104894023, rs1349419, rs2167270, and tetranucleotide repeat (TTTC) n) from 7 studies that were conducted in two Arab countries (Tunisia and Egypt) (Table 1, Table S3). The remaining variants were primarily captured in ADIPOQ (7 variants), FTO (5 variants), and LEPR (3 variants). The remaining genes had one or two variants (Table 1, Table S3). Among all 76 variants captured in this study, FTO rs9939609 was the most commonly studied variant, reported in eight studies conducted in six different countries (Egypt, Saudi Arabia, Tunisia, Iraq, Kuwait, and United Arab Emirates) in association with obesity (Table 1, Table S3).
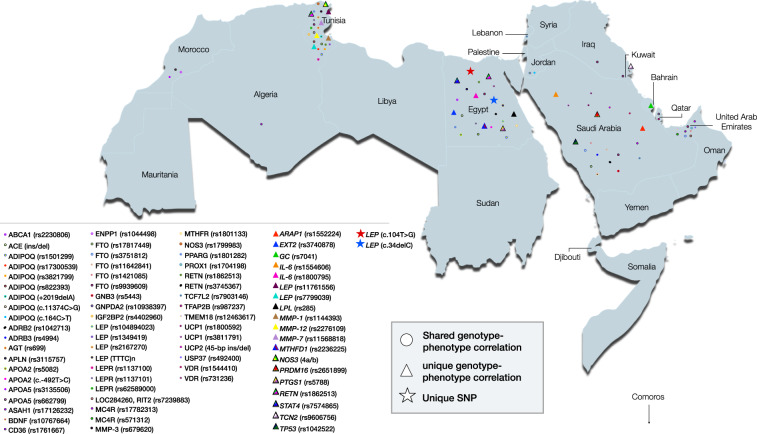
Among the 76 variants in 49 different genes, two variants were unique to Arabs (LEP: c.104 T > G, c.34delC) (i.e., reported in Arabs and not reported in any other ethnic groups) (Fig. 3). The remaining 74 variants were classified into two different categories based on their genotype–phenotype correlation. The first category included variants with unique genotype–phenotype correlations in Arabs; i.e., they are distinctively associated with obesity among Arabs but have been reported in association with clinical conditions other than obesity among different ethnicities (19 variants) (Fig. 3). The second category included variants that were reported in association with obesity in Arabs as well as other non-Arab ethnic populations and were thus considered common variants, i.e., shared with other ethnicities (55 variants) (Fig. 3). Data on the ethnic distribution of the captured variants is summarized in Table S3.
Fig. 3. Geographic distribution of obesity-associated genetic polymorphisms in the Arab world.
A total of 76 variants were reported to be significantly associated with obesity. Among the 76 variants, 55 exhibited shared genotype–phenotype correlation, 19 showed unique genotype–phenotype correlation, and two were described as unique to Arabs, as they were not previously reported in other ethnicities.
Statistical data from the 59 individual studies included in this systematic review are shown in Table S3. Overall, 57 studies reported cORs, which ranged from 0.18 to 44.60, with a median of 2.39 (interquartile range 1.62–3.57), and 49 studies reported aORs that ranged from 0.07 to 13.76, with a median of 1.65 (interquartile range 1.29–2.18) (Table S3). Among the 57 studies which reported cORs, PRDM16: rs2651899 conferred the strongest association with obesity (cOR = 44.60, 95% CI: 11.60–172.02, P = 0.0001). Among the 49 studies, which adjusted for confounders, MTHFR: rs1801133 conferred the strongest adjusted association (aOR = 13.5, 95% CI: 2.82–64.67, P = 0.001).
Discussion
To our knowledge, this is the first systemic review to comprehensively summarize all genetic polymorphisms associated with the risk of developing obesity in Arab patients. In this systematic review, we captured 76 genetic variants from 59 eligible studies, comprising 15,488 cases and 9760 controls. We identified 76 variants in 49 genes (Table 1, Table S3). Among the 49 genes that have been reported, the FTO gene, reported in 14 different articles, was the most frequently studied gene among Arab patients with obesity. The FTO gene is one of the most common genes that has been studied around the world in association with obesity [40]. FTO is highly expressed in the hypothalamus and functions as a regulator of appetite and energy expenditure [41–44] (Table S3). In this study, five captured variants were located in the FTO gene, all of which are intronic variants (rs11642841, rs1421085, rs17817449, rs3751812, rs9939609), reported in six Arab countries (Saudi Arabia, Tunisia, Egypt, Iraq, Kuwait, and United Arab Emirates) (Table 1, Table S3). The variant FTO: rs9939609 was the most commonly studied variant among all 76 variants captured in this study (Table 1), reported eight times in six different countries (Egypt, Saudi Arabia, Tunisia, Iraq, Kuwait, and United Arab Emirates) in association with obesity (Table 1, Table S3). FTO; rs9939609 has been previously reported in several other Non-Arab ethnicities in association with energy intake, eating habits, and susceptibility to obesity [45–51].
According to the number of variants reported per gene, most were captured in LEP, with eight obesity-associated variants reported among Arabs (rs11761556, rs7799039, c.104 T > G, c.34delC, rs104894023, rs1349419, rs2167270, and tetranucleotide repeat (TTTC) n) from seven studies conducted in two Arab countries (Tunisia and Egypt) (Table 1). The remaining variants were primarily captured in ADIPOQ (7 variants), FTO (5 variants), and LEPR (3 variants). The remaining genes had one or two variants each (Table 1).
Among the 76 variants captured in this systematic review, two were unique to Arabs, as they were not previously reported in any other ethnic groups and were identified in the original study as novel, both of which are located in LEP (c.104 T > G, c.34delC) (Fig. 3).
Variants unique to Arab countries
Two variants were found to be unique to Arabs, both of which are located in LEP (c.104 T > G and c.34delC) [36] (Table 1). These two variants were identified as novel homozygous variants and were not previously reported in any database. The missense variant c.104 T > G was identified in a 7‐month‐old boy who presented with excessive weight gain. This variant resulted in an amino acid substitution from isoleucine to serine at position 35 (p. Ile35Ser) and was predicted to likely be pathogenic [36] (Table 1, Table S3). The variant c.34 delC is a deletion of a cytosine nucleotide at position 34, causing a frameshift in the protein and changing the leucine at position 12 of the protein [36] (Table 1, Table S3). Functional analysis revealed that these two variants are likely to be disease‐causing [36]. The LEP gene has globally been reported to contribute to the pathogenesis of obesity [52]. LEP regulates the leptin-melanocortin signaling pathway, which regulates appetite and energy homoeostasis (Table S3).
Variants with unique genotype–phenotype correlations in the Arab world
Out of the 76 captured variants that have been reported to be significantly associated with obesity in Arab world countries, 19 variants were found to exhibit genotype-phenotype correlations that are unique to Arabs, given the fact that they were reported to be distinctively associated with obesity among Arab, but not non-Arab populations. Notably, these variants have been associated with other clinical conditions (i.e., different from obesity), in non-Arab populations.
The variants with unique genotype–phenotype correlations in Arabs were primarily located in LEP and IL-6, with two variants in each of the two genes. The two LEP variants rs7799039 and rs11761556, which have been associated with obesity in Tunisia (Table 1), were reported to be associated with breast cancer and systemic lupus erythematosus in Chinese and Mexican populations (Table 1). The two IL6 variants, rs1800795 and rs1554606, reported in Egypt and Saudi Arabia, respectively (Table 1, Table S3, Fig. 3), are considered unique to the Arab region according to genotype–phenotype correlation, as they were only reported in other ethnic populations with inflammatory conditions, such as osteoporosis in Taiwan [53] (Table 1, Table S3). The IL6 gene encodes interleukin-6 (IL-6), an immune modulator and proinflammatory cytokine involved in regulation of the acute phase response [54]. IL6 levels increase with increasing body fat content, and it plays an important role in glucose disposal, lipolysis, and fat oxidation. Therefore, impaired IL6 represents a risk factor for increased body weight (Table S3).
Other variants that were found to be associated with obesity only in Arab countries include MTFHD1 rs2236225, EXT: rs3740878, PRDM16 rs2651899, and TP53 rs1042522 (Table 1, Table S3, and Fig. 3). These variants have been significantly associated with congenital heart defects in Iran, type 2 diabetes mellitus in China, migraine in China, and cervical cancer in China [55–58] (Table S3). This is not surprising because obesity is known to progress or cause other diseases. Therefore, it is not unexpected to identify common genes and mechanisms between obesity and other associated diseases.
Notably, PRDM16: rs2651899 variant that conferred the strongest association with obesity (cOR = 44.60, 95% CI: 11.60–172.02, P = 0.0001) (Table S3), has not been associated with obesity in any non-Arab ethnic population. It has been primarily associated with a higher risk of migraines, as revealed by a large GWAS of over 5000 patients [59].
Finally, we investigated the significant and unique genes and variants associated with obesity in Arab countries and further classified them as common or unique in terms of the manifested phenotype among other ethnic groups. Nevertheless, further studies need to be performed to highlight the significance of these genes in contributing to the direct pathogenesis of obesity. This review may help build a platform for designing a gene panel to test the susceptibility of Arab patients to obesity and will also be useful globally for molecular diagnostic purposes.
Variants shared with other ethnic groups in terms of genotype–phenotype correlation
Out of the 76 captured variants that have been reported to be significantly associated with obesity in Arab countries worldwide, 55 variants located in 35 different genes were found to exhibit genotype-phenotype correlations that are shared between Arab and non-Arab populations, given the fact that they were reported in association with obesity in both Arabs and non-Arabs.
Most of the variants in this category were located in ADIPOQ, the second most common gene according to the number of obesity-associated variants among Arabs. ADIPOQ had seven variants, all of which were found to be shared with other ethnic populations according to genotype–phenotype correlation (Table S3). These seven variants were reported in Tunisia, Egypt, and Jordan (Table 1, Fig. 3). Among them, is the intron variant rs1501299, captured in obese patients from Egypt, Jordan, and Tunisia (Table 1). This variant was previously reported to be among the top-ten obesity risk-associated SNPs in a randomly recruited cohort of Spanish children and adolescents aged 5–17-years-old [60]. ADIPOQ gene is a critical gene that contributes to obesity and has been investigated worldwide, having been reported in other ethnic groups, such as Japanese and Portuguese populations in association with obesity [61, 62], as well as being reportedly associated with both type 2 diabetes mellitus and obesity in South India [63] (Table S3).
The remaining variants in this category were primarily located in FTO and LEP, with five and four variants in each gene, respectively. FTO is the third most common gene according to the number of obesity-associated identified variants in Arabs (Table 1, Table S3). FTO variants (rs11642841, rs1421085, rs17817449, rs3751812, rs9939609), which have been linked to obesity in six Arab countries (Table 1, Fig. 3), and according to the clinical phenotype, they are considered as “common variants”; as they were found to be associated with obesity in other non-Arab countries, including China, Croatia, and Brazil (Table S3). LEP variants (c.313 C > T, rs1349419, rs2167270, (TTTC) n polymorphism) have been associated with obesity in other non-Arab ethnic countries, including Pakistan, Mexico, Malaysia, and India [36, 64–66] (Table S3).
Strengths and limitations
The strengths of this study lie in the fact that it is the first systematic study to comprehensively summarize all reported obesity-associated genetic polymorphisms in the Arab world. We conducted a comprehensive search using stringent predetermined inclusion and exclusion criteria and thorough analyses of each article included. Despite precluding a meta-analysis, key findings were reported narratively. Nevertheless, our study has some limitations. First, it is noteworthy to mention that obesity is a complex multifactorial disease that can progress or cause other diseases, such as type 2 diabetes mellitus hypertension, dyslipidemia, cardiovascular disease, and nonalcoholic fatty liver disease [67, 68]. Therefore, most studies in the field of obesity include other metabolic syndromes, and most of the time, the patient group had obesity as well as other metabolic syndromes. This makes investigating the genetic polymorphisms that are only linked to obesity more challenging; thus, many of the significantly reported genes will also be associated with other metabolic syndromes. Moreover, because of recently available genome-wide association studies, over 227 genetic variants have been associated with polygenic obesity. However, the available studies are often heterogeneous with regard to their design and operational quality, which adds to the complexity of evidence and conclusion synthesis. Furthermore, the included studies also varied vastly in sample size and variables measured, as well as the degree to which they controlled for confounders. In addition, despite thoroughly searching the existing literature on obesity-associated genetic polymorphisms in the Arab world, we found relatively few genetic association studies on obesity risk in the Arab world.
Conclusions
The epidemic of obesity has been largely attributed to changes in lifestyle habits established over the past three decades. These changes are mainly attributed to dietary and behavioral trends such as decline in physical activity. However, the obesogenic environment is not sufficient to determine the presence of obesity, a combination with genetic predisposition is required. This systematic review was designed to comprehensively assess all genetic variations significantly associated with obesity risk in Arab countries. Our findings indicate that Arabs have distinct disease susceptibility genotypes that are responsible for obesity. Although some of the obesity-associated variants mentioned in this study have been reported in other ethnic groups, the complex gene-environment interactions allow for enrichment of these genotypes, thus predisposing individuals of these ethnic groups to obesity. Our study creates a paradigm for future well-controlled epidemiological studies that will allow dissection of the genetic architecture that renders Arabs susceptible to obesity and thus may in the future serve as a platform to design a gene panel for early, accurate, and presymptomatic diagnosis of obesity. Despite our comprehensive search strategy, the dearth of genetic association studies related to obesity in the Arab world suggests a need for additional well-designed genetic association studies to serve as the basis for understanding the genetic architecture that renders Arab populations susceptible to obesity.
Supplementary information
Acknowledgements
Open access funding provided by the Qatar National Library.
Funding
No funding was provided for this study.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Salma Younes, Amal Ibrahim, Rana Al-Jurf
Supplementary information
The online version contains supplementary material available at 10.1038/s41366-021-00867-6.
References
- 1.WHO. Obesity and overweight. 2019. Available from: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
- 2.Hruby A, Hu FB. The epidemiology of obesity: a big picture. Pharmacoeconomics. 2015;33:673. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lara-Castro C, Garvey WT. Intracellular lipid accumulation in liver and muscle and the insulin resistance syndrome. Endocrinol Metabol Clin North Am. 2008;37:841. doi: 10.1016/j.ecl.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galgani J, Ravussin E. Energy metabolism, fuel selection and body weight regulation. Int J Obes. 2008;32 (Suppl 7):S109.. doi: 10.1038/ijo.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Obesity Federation. Prevalence of obesity. 2019. Available from: https://www.worldobesity.org/about/about-obesity/prevalence-of-obesity.
- 6.Ritchie H. Obesity [Published online at OurWorldInData.org]. 2017. Available from: https://ourworldindata.org/obesity [Online Resource].
- 7.Hajjej A, Almawi WY, Arnaiz-Villena A, Hattab L, Hmida S. The genetic heterogeneity of Arab populations as inferred from HLA genes. PLoS ONE. 2018;13:e0192269-e. doi: 10.1371/journal.pone.0192269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim Z. Linguistics in an Age of Globalization: Perspectives on Arabic Language and Teaching. American Univ in Cairo Press; 2008.
- 9.Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:17. doi: 10.1186/1742-4755-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Younes N, Younes S, Alsharabasi OA, El Zowalaty ME, Mustafa I, Jahromi M, et al. Immunogenetics of celiac disease: a focus on Arab Countries. Curr Mol Med. 2020;20:275–85. doi: 10.2174/1566524019666191024104930. [DOI] [PubMed] [Google Scholar]
- 11.Abuhendi N, Qush A, Naji F, Abunada H, Al Buainain R, Shi Z, et al. Genetic polymorphisms associated with type 2 diabetes in the Arab world: a systematic review and meta-analysis. Diabetes Res Clin Pract. 2019;151:198–208. doi: 10.1016/j.diabres.2019.03.037. [DOI] [PubMed] [Google Scholar]
- 12.Zayed H. Genetic epidemiology of type 1 diabetes in the 22 Arab Countries. Curr Diab Rep. 2016;16:37. doi: 10.1007/s11892-016-0736-4. [DOI] [PubMed] [Google Scholar]
- 13.Sidenna M, Bux R, Fadl T, Ozbek U, Zayed H. Association of genetic variants with colorectal cancer in the extended MENA region: a systematic review. Curr Mol Med. 2020;20:286–98. doi: 10.2174/1566524019666191014170136. [DOI] [PubMed] [Google Scholar]
- 14.Younes N, Zayed H. Genetic epidemiology of ovarian cancer in the 22 Arab countries: a systematic review. Gene. 2019;684:154–64. doi: 10.1016/j.gene.2018.10.044. [DOI] [PubMed] [Google Scholar]
- 15.Sidenna M, Fadl T, Zayed H. Genetic epidemiology of hearing loss in the 22 Arab Countries: a systematic review. Otol Neurotol. 2020;41:e152–62. doi: 10.1097/MAO.0000000000002489. [DOI] [PubMed] [Google Scholar]
- 16.Khan AM, Al-Sulaiti AM, Younes S, Yassin M, Zayed H. The spectrum of beta-thalassemia mutations in the 22 Arab countries: a systematic review. Expert Rev Hematol. 2021;14:109–22. doi: 10.1080/17474086.2021.1860003. [DOI] [PubMed] [Google Scholar]
- 17.Al-Sadeq D, Abunada T, Dalloul R, Fahad S, Taleb S, Aljassim K, et al. Spectrum of mutations of cystic fibrosis in the 22 Arab countries: a systematic review. Respirology. 2019;24:127–36. doi: 10.1111/resp.13437. [DOI] [PubMed] [Google Scholar]
- 18.Doss CG, Alasmar DR, Bux RI, Sneha P, Bakhsh FD, Al-Azwani I, et al. Genetic epidemiology of glucose-6-phosphate dehydrogenase deficiency in the Arab World. Sci Rep. 2016;6:37284. doi: 10.1038/srep37284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zayed H. Propionic acidemia in the Arab World. Gene. 2015;564:119–24. doi: 10.1016/j.gene.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Alhababi D, Zayed H. Spectrum of mutations of familial hypercholesterolemia in the 22 Arab countries. Atherosclerosis. 2018;279:62–72. doi: 10.1016/j.atherosclerosis.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Alzaman N, Ali A. Obesity and diabetes mellitus in the Arab world. J Taibah Univ Med Sci. 2016;11:301–9. [Google Scholar]
- 22.Badran M, Laher I. Obesity in arabic-speaking countries. J Obes. 2011;2011:686430. doi: 10.1155/2011/686430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zayed H. The Arab genome: health and wealth. Gene. 2016;592:239–43. doi: 10.1016/j.gene.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Zayed H. The Qatar genome project: translation of whole-genome sequencing into clinical practice. Int J Clin Pract. 2016;70:832–4. doi: 10.1111/ijcp.12871. [DOI] [PubMed] [Google Scholar]
- 25.Al-Ali M, Osman W, Tay GK, AlSafar HS. A 1000 Arab genome project to study the Emirati population. J Hum Genet. 2018;63:533–6. doi: 10.1038/s10038-017-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouchard C. Genetic determinants of regional fat distribution. Hum Reprod (Oxford, England) 1997;12:1–5. doi: 10.1093/humrep/12.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 27.Goni L, García-Granero M, Milagro FI, Cuervo M, Martínez JA. Phenotype and genotype predictors of BMI variability among European adults. Nutr Diabetes. 2018;8:27. doi: 10.1038/s41387-018-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min J, Chiu DT, Wang Y. Variation in the heritability of body mass index based on diverse twin studies: a systematic review. Obes Rev. 2013;14:871–82. doi: 10.1111/obr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moustafa JSE-S, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol. 2013;9:402. doi: 10.1038/nrendo.2013.57. [DOI] [PubMed] [Google Scholar]
- 30.Pigeyre M, Meyre D. Monogenic obesity. In: Freemark MS, editors. Pediatric Obesity Etiology, Pathogenesis and Treatment. 2nd ed. New York, USA: Humana Press; 2018. p. 135–52. 10.1007/978-3319-68192-4.
- 31.Ben Ali S, Sediri Y, Kallel A, Ftouhi B, Haj-Taib S, Omar S, et al. The G3057A LEPR polymorphism is associated with obesity in Tunisian women. Nutr Metab Cardiovasc Dis. 2011;21:591–6. doi: 10.1016/j.numecd.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Ali EMM, Diab T, Elsaid A, Abd El Daim HA, Elshazli RM, Settin A. Fat mass and obesity-associated (FTO) and leptin receptor (LEPR) gene polymorphisms in Egyptian obese subjects. Arch Physiol Biochem. 2021;127:28–36. doi: 10.1080/13813455.2019.1573841. [DOI] [PubMed] [Google Scholar]
- 33.Bordoni L, Marchegiani F, Piangerelli M, Napolioni V, Gabbianelli R. Obesity-related genetic polymorphisms and adiposity indices in a young Italian population. IUBMB life. 2017;69:98–105. doi: 10.1002/iub.1596. [DOI] [PubMed] [Google Scholar]
- 34.Yang MM, Wang J, Fan JJ, Ng TK, Sun DJ, Guo X, et al. Variations in the Obesity Gene “LEPR” Contribute to Risk of Type 2 Diabetes Mellitus: Evidence from a Meta-Analysis. J Diabetes Res. 2016;2016:5412084. [DOI] [PMC free article] [PubMed]
- 35.Shabana HasnainS. The p. N103K mutation of leptin (LEP) gene and severe early onset obesity in Pakistan. Biol Res. 2016;49:23. doi: 10.1186/s40659-016-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ElSaeed G, Mousa N, El-Mougy F, Hafez M, Khodeera S, Alhelbawy M, et al. Monogenic leptin deficiency in early childhood obesity. Pediatr Obes. 2020;15:e12574. doi: 10.1111/ijpo.12574. [DOI] [PubMed] [Google Scholar]
- 37.Loktionov A. Common gene polymorphisms and nutrition: emerging links with pathogenesis of multifactorial chronic diseases (review) J Nutr Biochem. 2003;14:426–51. doi: 10.1016/S0955-2863(03)00032-9. [DOI] [PubMed] [Google Scholar]
- 38.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.NIH National Heart, Lung and Blood Institute. Study quality assessment tools [cited 2017, March 30]. Available from: https://www.nhlbi.nih.gov/health-pro/guidelines/in-develop/cardiovascular-risk-reduction/tools.
- 40.Hubacek JA, Bohuslavova R, Kuthanova L, Kubinova R, Peasey A, Pikhart H, et al. The FTO gene and obesity in a large Eastern European population sample: the HAPIEE study. Obesity (Silver Spring. Md) 2008;16:2764–6. doi: 10.1038/oby.2008.421. [DOI] [PubMed] [Google Scholar]
- 41.Woods SC, D’Alessio DA. Central control of body weight and appetite. J Clin Endocrinol Metab. 2008;93:S37–50. doi: 10.1210/jc.2008-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (New York, NY) 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature genetics. 2007;39:724–6. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 44.Peters T, Ausmeier K, Rüther U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mammalian genome: official journal of the International Mammalian Genome. Society. 1999;10:983–6. doi: 10.1007/s003359901144. [DOI] [PubMed] [Google Scholar]
- 45.Prakash J, Mittal B, Srivastava A, Awasthi S, Srivastava N. Association of FTO rs9939609 SNP with obesity and obesity- associated phenotypes in a North Indian Population. Oman Med J. 2016;31:99–106. doi: 10.5001/omj.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ursu RI, Badiu C, Cucu N, Ursu GF, Craciunescu I, Severin E. The study of the rs9939609 FTO gene polymorphism in association with obesity and the management of obesity in a Romanian cohort. J Med Life. 2015;8:232–8. [PMC free article] [PubMed] [Google Scholar]
- 47.Rees SD, Islam M, Hydrie MZ, Chaudhary B, Bellary S, Hashmi S, et al. An FTO variant is associated with Type 2 diabetes in South Asian populations after accounting for body mass index and waist circumference. Diabet Med. 2011;28:673–80. doi: 10.1111/j.1464-5491.2011.03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, et al. Meta-analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity (Silver Spring. Md) 2010;18:1619–24. doi: 10.1038/oby.2009.469. [DOI] [PubMed] [Google Scholar]
- 49.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring. Md) 2008;16:1961–5. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 50.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. The. N Engl J Med. 2008;359:2558–66. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 51.Gulati S, Misra A, Tiwari R, Sharma M, Pandey RM, Upadhyay AD. The influence of polymorphisms of fat mass and obesity (FTO, rs9939609) and vitamin D receptor (VDR, BsmI, TaqI, ApaI, FokI) genes on weight loss by diet and exercise interventions in non-diabetic overweight/obese Asian Indians in North India. Eur J Clin Nutr. 2020;74:604–12. doi: 10.1038/s41430-020-0560-4. [DOI] [PubMed] [Google Scholar]
- 52.Mazen I, El-Gammal M, Abdel-Hamid M, Amr K. A novel homozygous missense mutation of the leptin gene (N103K) in an obese Egyptian patient. Mol Genet Metab. 2009;97:305–8. doi: 10.1016/j.ymgme.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Lin C-C, Li T-C, Liu C-S, Yang C-W, Lin C-H, Hsiao J-H, et al. Associations of TNF-α and IL-6 polymorphisms with osteoporosis through joint effects and interactions with LEPR gene in Taiwan: Taichung Community Health Study for Elders (TCHS-E) Mol Biol Rep. 2016;43:1179–91. doi: 10.1007/s11033-016-4037-4. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295–a. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khatami M, Ratki FM, Tajfar S, Akrami F. Relationship of the MTHFD1 (rs2236225), eNOS (rs1799983), CBS (rs2850144) and ACE (rs4343) gene polymorphisms in a population of Iranian pediatric patients with congenital heart defects. Kaohsiung J Med Sci. 2017;33:442–8. doi: 10.1016/j.kjms.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 56.Ren Q, Xiao J, Han X, Yang W, Ji L. Impact of variants of the EXT2 gene on Type 2 diabetes and its related traits in the Chinese han population. Endocr Res. 2015;40:79–82. doi: 10.3109/07435800.2014.952015. [DOI] [PubMed] [Google Scholar]
- 57.Liu GC, Zhou YF, Su XC, Zhang J. Interaction between TP53 and XRCC1 increases susceptibility to cervical cancer development: a case control study. BMC Cancer. 2019;19:24. [DOI] [PMC free article] [PubMed]
- 58.Fu X, Yang J, Wu X, Lin Q, Zeng Y, Xia Q, et al. Association between PRDM16, MEF2D, TRPM8, LRP1 gene polymorphisms and migraine susceptibility in the She ethnic population in China. Clin Invest Med. 2019;42:21–30. doi: 10.25011/cim.v42i1.32389. [DOI] [PubMed] [Google Scholar]
- 59.Chasman DI, Schürks M, Anttila V, de Vries B, Schminke U, Launer LJ, et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nature genetics. 2011;43:695–8. doi: 10.1038/ng.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cortés-Martín A, Colmenarejo G, Selma MV, Espín JC. Genetic polymorphisms, mediterranean diet and microbiota-associated urolithin metabotypes can predict obesity in childhood-adolescence. Sci Rep. 2020;10:7850. doi: 10.1038/s41598-020-64833-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nascimento H, Vieira E, Coimbra S, Catarino C, Costa E, Bronze-da-Rocha E.et al. Adipokine Gene Single-Nucleotide Polymorphisms in Portuguese Obese Adolescents: Associations with Plasma Concentrations of Adiponectin, Resistin, IL-6, IL-1β, and TNF-α. Child Obes. 2016;12:300–13.. [DOI] [PubMed]
- 62.Kotani K, Saiga K, Kurozawa Y, Sakane N, Tsuzaki K, Hamada T. Adiponectin I164T gene polymorphism and the obesity-related effects on the Japanese female population. Clin Chim Acta. 2007;384:182–3. doi: 10.1016/j.cca.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 63.Ramya K, Ayyappa KA, Ghosh S, Mohan V, Radha V. Genetic association of ADIPOQ gene variants with type 2 diabetes, obesity and serum adiponectin levels in south Indian population. Gene. 2013;532:253–62. doi: 10.1016/j.gene.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 64.Ben Ali S, Kallel A, Ftouhi B, Sediri Y, Feki M, Slimane H, et al. Association of G-2548A LEP polymorphism with plasma leptin levels in Tunisian obese patients. Clin Biochem. 2009;42:584–8. [DOI] [PubMed]
- 65.Fourati M, Mnif M, Kharrat N, Charfi N, Kammoun M, Fendri N. Association between Leptin gene polymorphisms and plasma leptin level in three consanguineous families with obesity. Gene. 2013;527:75–81. doi: 10.1016/j.gene.2013.05.064. [DOI] [PubMed] [Google Scholar]
- 66.Elbaz R, Dawood N, Mostafa H, Zaki S, Wafa A, Settin A. Leptin gene tetranucleotide repeat polymorphism in obese individuals in Egypt. Int J Health Sci (Qassim). 2015;9:63–71. [PMC free article] [PubMed]
- 67.Yao MH, He J, Ma RL, Ding YS, Guo H, Yan YZ, et al. Association between Polymorphisms and Haplotype in the ABCA1 Gene and Overweight/Obesity Patients in the Uyghur Population of China. Int J Environ Res Public Health. 2016;13:220. doi: 10.3390/ijerph13020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diels S, Vanden Berghe W, Van, Hul W. Insights into the multifactorial causation of obesity by integrated genetic and epigenetic analysis. Obes Rev. 2020;21:e13019. doi: 10.1111/obr.13019. [DOI] [PubMed] [Google Scholar]
- 69.Fawzy MS, Alhadramy O, Hussein MH, Ismail HM, Ismail NM, Biomy NM, et al. Functional and Structural Impact of ATP-Binding Cassette Transporter A1 R219K and I883M Gene Polymorphisms in Obese Children and Adolescents. Mol Diagn Ther. 2015;19:221–34. [DOI] [PubMed]
- 70.Khamlaoui W, Mehri S, Hammami S, Elosua R, Hammami M. Association of angiotensin-converting enzyme insertion/deletion (ACE I/D) and angiotensinogen (AGT M235T) polymorphisms with the risk of obesity in a Tunisian population. J Renin Angiotensin Aldosterone Syst. 2020;21:1470320320907820. doi: 10.1177/1470320320907820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Kabbany ZA, Hamza RT, Shinkar DM, Kamal TM, Abdelmageed RI, Said MS, et al. Screening of Egyptian obese children and adolescents for insertion/deletion (I/D) polymorphism in angiotensin-converting enzyme gene. Int J Pediatr Adolesc Med. 2019;6:21–4. doi: 10.1016/j.ijpam.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boumaiza I, Omezzine A, Rejeb J, Rebhi L, Rejeb NB, Nabli N, et al. Association between eight adiponectin polymorphisms, obesity, and metabolic syndrome parameters in Tunisian volunteers. Metab Syndr Relat Disord. 2011;9:419–26. doi: 10.1089/met.2011.0035. [DOI] [PubMed] [Google Scholar]
- 73.Alomari A, Khabour OF, Obaid AAJG The association between adiponectin and ADIPOQ gene polymorphisms with obesity among young Jordanian women. Genetika. 2016;48:363–72.
- 74.Elghazy AM, Elsaeid AM, Refaat M, Youssef MM. Biochemical studies of adiponectin gene polymorphism in patients with obesity in Egyptians. Arch Physiol Biochem. 2019:1–8. 10.1080/13813455.2019.1662451. [DOI] [PubMed]
- 75.Zayani N, Omezzine A, Boumaiza I, Achour O, Rebhi L, Rejeb J, et al. Association of ADIPOQ, leptin, LEPR, and resistin polymorphisms with obesity parameters in Hammam Sousse Sahloul Heart Study. J Clin Lab Anal. 2017;31. [DOI] [PMC free article] [PubMed]
- 76.Daghestani MH, Warsy A, Daghestani MH, Al-Odaib AN, Eldali A, Al-Eisa NA, et al. Arginine 16 Glycine Polymorphism in β2-Adrenergic Receptor Gene is Associated with Obesity, Hyperlipidemia, Hyperleptinemia, and Insulin Resistance in Saudis. Int J Endocrinol. 2012;2012:945608. [DOI] [PMC free article] [PubMed]
- 77.Daghestani M, Daghestani M, Daghistani M, Eldali A, Hassan ZK, Elamin MH, et al. ADRB3 polymorphism rs4994 (Trp64Arg) associates significantly with bodyweight elevation and dyslipidaemias in Saudis but not rs1801253 (Arg389Gly) polymorphism in ARDB1. Lipids Health Dis. 2018;17:58. [DOI] [PMC free article] [PubMed]
- 78.Aboouf MA, Hamdy NM, Amin AI, Mesallamy El. Genotype screening of APLN rs3115757 variant in Egyptian women population reveals an association with obesity and insulin resistance. Diabetes Res Clin Pract. 2015;109:40–7. [DOI] [PubMed]
- 79.Zaki ME, Amr KS, Abdel-Hamid M. APOA2 polymorphism in relation to obesity and lipid metabolism. Cholesterol. 2013;2013:289481. doi: 10.1155/2013/289481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaki ME, Amr KS, Hamid MA. Evaluating the association of APOA2 polymorphism with insulin resistance in adolescents. Meta Gene. 2014;2:366–73. doi: 10.1016/j.mgene.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.El Yaagoubi FL, Charoute H, Bakhchane A, Ajjemami M, Benrahma H, Errouagui A, et al. Association analysis of APOA5 rs662799 and rs3135506 polymorphisms with obesity Moroccan patients. Pathol Biol. 2015;63:243–7. doi: 10.1016/j.patbio.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Al-Daghri NM, Alkharfy KM, Al-Attas OS, Krishnaswamy S, Mohammed AK, Albagha OM, et al. Association between type 2 diabetes mellitus-related SNP variants and obesity traits in a Saudi population. Mol Biol Rep. 2014;41:1731–40. doi: 10.1007/s11033-014-3022-z. [DOI] [PubMed] [Google Scholar]
- 83.Hachim MY, Aljaibeji H, Hamoudi RA, Hachim IY, Elemam NM, Mohammed AK, et al. An Integrative Phenotype-Genotype Approach Using Phenotypic Characteristics from the UAE National Diabetes Study Identifies HSD17B12 as a Candidate Gene for Obesity and Type 2 Diabetes. Genes. 2020;11. [DOI] [PMC free article] [PubMed]
- 84.Alharbi KK, Richardson TG, Khan IA, Syed R, Mohammed AK, Boustred CR, et al. Influence of adiposity-related genetic markers in a population of saudi arabians where other variables influencing obesity may be reduced. Dis Markers. 2014;2014:758232. [DOI] [PMC free article] [PubMed]
- 85.Daoudi H, Plesník J, Sayed A, Šerý O, Rouabah A, Rouabah L, et al. Oral Fat Sensing and CD36 Gene Polymorphism in Algerian Lean and Obese Teenagers. Nutrients. 2015;7:9096–104. [DOI] [PMC free article] [PubMed]
- 86.El Achhab Y, Meyre D, Bouatia-Naji N, Berraho M, Deweirder M, Vatin V, et al. Association of the ENPP1 K121Q polymorphism with type 2 diabetes and obesity in the Moroccan population. Diabetes Metab. 2009;35:37–42. doi: 10.1016/j.diabet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 87.Attia HRM, Kamel SA, Ibrahim MH, Farouk HA, Rahman AHA, Sayed GH, et al. Open-array analysis of genetic variants in Egyptian patients with type 2 diabetes and obesity. Egypt J Med Hum Genet. 2017;18:341–8. doi: 10.1016/j.ejmhg.2017.03.002. [DOI] [Google Scholar]
- 88.Moselhy SS, Alhetari YA, Iyer A, Huwait EA, Al-Ghamdi MA, Al-Ghamdi S, et al. Analysis of SNPs of MC4R, GNB3 and FTO gene polymorphism in obese Saudi subjects. African Health Sci. 2017;17:1059–69. doi: 10.4314/ahs.v17i4.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El Hajj Chehadeh S, Osman W, Nazar S, Jerman L, Alghafri A, Sajwani A, et al. Implication of genetic variants in overweight and obesity susceptibility among the young Arab population of the United Arab Emirates. Gene. 2020;739:144509. doi: 10.1016/j.gene.2020.144509. [DOI] [PubMed] [Google Scholar]
- 90.Khan SM, El Hajj Chehadeh S, Abdulrahman M, Osman W, Al Safar H. Establishing a genetic link between FTO and VDR gene polymorphisms and obesity in the Emirati population. BMC Medical Genetics. 2018;19:11. doi: 10.1186/s12881-018-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cyrus C, Ismail MH, Chathoth S, Vatte C, Hasen M, Al Ali AJGt, et al. Analysis of the impact of common polymorphisms of the FTO and MC4R Genes with the risk of severe obesity in Saudi Arabian population. Genet Test Mol Biomarkers. 2018;22:170–7. doi: 10.1089/gtmb.2017.0218. [DOI] [PubMed] [Google Scholar]
- 92.El Nashar DE, Alananbeh KM, Al, Hassan N. Genetic, dietary, and non-dietary risk factors of obesity among preparatory-year female students at Taibah University, Saudi Arabia. J Taibah Univ Sci. 2017;11:408–21. doi: 10.1016/j.jtusci.2016.06.003. [DOI] [Google Scholar]
- 93.Al-Serri A, Al-Bustan SA, Kamkar M, Thomas D, Alsmadi O, Al-Temaimi R, et al. Association of FTO rs9939609 with Obesity in the Kuwaiti population: a public health concern? Med Princ Pract. 2018;27:145–51. doi: 10.1159/000486767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jumaa MN, Yaseen NY, Shehab AF, Karim RM. Sagban LHJIJoMS. Analysis of Single Nucleotide Polymorphism rs9939609 in FTO Gene of Obese Males in Iraqi Population. Iraqi J Med Sci. 2016;14:252–8. [Google Scholar]
- 95.Al-Tu’ma FJ, Obed KHJIMJ. Association between Fat Mass and Obesity Associated (FTO) gene polymorphism (rs9939609) and lipid profile in type 2 diabetic obese Iraqi male. Iraq Med J. 2018;2:15–9.
- 96.Ben Halima M, Kallel A, Baara A, Ben Wafi S, Sanhagi H, Slimane H, et al. The rs9939609 polymorphism in the fat mass and obesity associated (FTO) gene is associated with obesity in Tunisian population. Biomarkers. 2018;23:787–92. doi: 10.1080/1354750X.2018.1499129. [DOI] [PubMed] [Google Scholar]
- 97.Almesri N, Das NS, Ali ME, Gumaa K, Giha HA. Independent associations of polymorphisms in vitamin D binding protein (GC) and vitamin D receptor (VDR) genes with obesity and plasma 25OHD3 levels demonstrate sex dimorphism. Appl Physiol Nutr Metab. 2015;41:345–53. [DOI] [PubMed]
- 98.Tomei S, Mamtani R, Al Ali R, Elkum N, Abdulmalik M, Ismail A, et al. Obesity susceptibility loci in Qataris, a highly consanguineous Arabian population. J Transl Med. 2015;13:119. [DOI] [PMC free article] [PubMed]
- 99.Lasram K, Ben Halim N, Benrahma H, Mediene-Benchekor S, Arfa I, Hsouna S, et al. Contribution of CDKAL1 rs7756992 and IGF2BP2 rs4402960 polymorphisms in type 2 diabetes, diabetic complications, obesity risk and hypertension in the Tunisian population. J Diabetes. 2015;7:102–13. doi: 10.1111/1753-0407.12147. [DOI] [PubMed] [Google Scholar]
- 100.Alharbi KK, Syed R, Khan IA. Association of interleukin-6 polymorphisms with obesity and metabolic alterations in young Saudi population. Mol Biol Rep. 2014;41:1519–23. [DOI] [PubMed]
- 101.Ibrahim OM, Gabre AA, Sallam SF, El-Alameey IR, Sabry RN, Galal EM, et al. Influence of Interleukin-6 (174G/C) Gene Polymorphism on Obesity in Egyptian Children. Open Access Maced J Med Sci. 2017;5:831–5. doi: 10.3889/oamjms.2017.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nesrine Z, Haithem H, Imen B, Fadoua N, Asma O, Fadhel NM, et al. Leptin and Leptin receptor polymorphisms, plasma Leptin levels and obesity in Tunisian volunteers. Int J Exp Pathol. 2018;99:121–30. doi: 10.1111/iep.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boumaiza I, Omezzine A, Rejeb J, Rebhi L, Ouedrani A, Ben Rejeb N, et al. Relationship between leptin G2548A and leptin receptor Q223R gene polymorphisms and obesity and metabolic syndrome risk in Tunisian volunteers. Genet Test Mol Biomarkers. 2012;16:726–33. doi: 10.1089/gtmb.2011.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Osman W, Tay GK, Alsafar H. Multiple genetic variations confer risks for obesity and type 2 diabetes mellitus in arab descendants from UAE. Int J Obes. 2018;42:1345. [DOI] [PubMed]
- 105.Hendawy A, HasanM ER, El-Kannishy G, Elshaer S, Settin A. Physiological Study of Lipoprotein Lipase Gene Pvu II Polymorphism in Cases of Obesity in Egypt. Int J Zool Res. 2012;8:98–105.
- 106.Boumiza S, Bchir S, Ben Nasr H, Abbassi A, Jacob MP, Norel X, et al. Role of MMP-1 (-519A/G, -1607 1G/2G), MMP-3 (Lys45Glu), MMP-7 (-181A/G), and MMP-12 (-82A/G) Variants and Plasma MMP Levels on Obesity-Related Phenotypes and Microvascular Reactivity in a Tunisian Population. Dis Markers. 2017;2017:6198526. [DOI] [PMC free article] [PubMed]
- 107.Borai IH, Soliman AF, Ahmed HM, Ahmed GF, Kassim SK. Association of MTHFR C677T and ABCA1 G656A polymorphisms with obesity among Egyptian children. Gene Rep. 2018;11:143–9.
- 108.Nasr HB, Dimassi S, M'Hadhbi R, Debbabi H, Kortas M, Tabka Z, et al. Functional G894T (rs1799983) polymorphism and intron-4 VNTR variant of nitric oxide synthase (NOS3) gene are susceptibility biomarkers of obesity among Tunisians. Obes Res Clin Pract. 2016;10:465–75. doi: 10.1016/j.orcp.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 109.Berhouma R, Kouidhi S, Ammar M, Abid H, Ennafaa H, Benammar-Elgaaied A. Correlation of peroxisome proliferator-activated receptor (PPAR-γ) mRNA expression with Pro12Ala polymorphism in obesity. Biochem Genet. 2013;51:256–63. [DOI] [PubMed]
- 110.AlAmrani A, AbdelKarim M, AlZoghaibi M. PRDM16 gene polymorphism is associated with obesity and blood lipids profiles in Saudi population. J Clin Med. 2018;7:141. [DOI] [PMC free article] [PubMed]
- 111.Amal S, Pasha HF, Rashad NM. Association of resistin gene polymorphisms with insulin resistance in Egyptian obese patients. Gene. 2013;515:233–8. [DOI] [PubMed]
- 112.Boumaiza I, Omezzine A, Rejeb J, Rebhi L, Ben Rejeb N, Nabli N, et al. Association between four resistin polymorphisms, obesity, and metabolic syndrome parameters in Tunisian volunteers. Genet Test Mol Biomarkers. 2012;16:1356–62. doi: 10.1089/gtmb.2012.0156. [DOI] [PubMed] [Google Scholar]
- 113.Nasreddine L, Akika R, Mailhac A, Tamim H, Zgheib NK. The Interaction between Genetic Polymorphisms in FTO and TCF7L2 Genes and Dietary Intake with Regard to Body Mass and Composition: An Exploratory Study. J Pers Med. 2019;9:11. [DOI] [PMC free article] [PubMed]
- 114.Hebbar P, Alkayal F, Nizam R, Melhem M, Elkum N, John SE, et al. The TCN2 variant of rs9606756 [Ile23Val] acts as risk loci for obesity‐related traits and mediates by interacting with Apo‐A1. Obesity. 2017;25:1098–108. doi: 10.1002/oby.21826. [DOI] [PubMed] [Google Scholar]
- 115.Sabir JSM, Omri AE, Shaik NA, Banaganapalli B, Hajrah NH, Zrelli H, et al. The genetic association study of TP53 polymorphisms in Saudi obese patients. Saudi J Biol Sci. 2019;26:1338. doi: 10.1016/j.sjbs.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chathoth S, Ismail MH, Vatte C, Cyrus C, Al Ali Z, Ahmed KA, et al. Association of Uncoupling Protein 1 (UCP1) gene polymorphism with obesity: a case-control study. BMC Med Genet. 2018;19:203. doi: 10.1186/s12881-018-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaabi YA. The deletion polymorphism in exon 8 of uncoupling protein 2 is associated with severe obesity in a Saudi Arabian case-control study. Indian J Endocrinol Metab. 2019;22:200–3. doi: 10.4103/ijem.IJEM_655_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiffri E. Association of the UCP2 45-bp insertion/deletion polymorphism with diabetes type 2 and obesity in Saudi population. Egypt J Med Hum Genet. 2012;13:257–62. doi: 10.1016/j.ejmhg.2012.04.002. [DOI] [Google Scholar]
- 119.Al-Daghri NM, Guerini FR, Al-Attas OS, Alokail MS, Alkharfy KM, Draz HM, et al. Vitamin D receptor gene polymorphisms are associated with obesity and inflammosome activity. PLoS ONE. 2014;9:e102141. doi: 10.1371/journal.pone.0102141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Al-Hazmi AS, Al-Mehmadi MM, Al-Bogami SM, Shami AA, Al-Askary AA, Alomery AM, et al. Vitamin D receptor gene polymorphisms as a risk factor for obesity in Saudi men. Electronic Physician. 2017;9:5427–33. doi: 10.19082/5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mohamed S, El-Askary A. Vitamin D receptor gene polymorphism among Egyptian obese children. Asian J Clin Nutr. 2017;9:24–9.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.