Abstract
Eosinophils accumulate adjacent to epithelial cells in the mucosa of patients with eosinophilic esophagitis (EoE), yet the bidirectional communication between these cells is not well understood. Herein, we investigated the crosstalk between human eosinophils and esophageal epithelial cells. We report that blood-derived eosinophils have prolonged survival when cocultured with epithelial cells; 96±1%, and 30±6% viability was observed after 7 and 14 days of coculture, respectively, compared with 1±0% and 0±0% of monoculture. In the presence of IL-13 and epithelial cells, eosinophils had greater survival (68±1%) at 14 days compared with cocultures lacking IL-13. Prolonged eosinophil viability did not require cellular contact and was observed when eosinophils were cultured in conditioned media from esophageal epithelial cells; neutralizing GM-CSF attenuated eosinophil survival. The majority of eosinophil transcripts (58%) were dysregulated in cocultured eosinophils compared with freshly isolated cells. Analysis of epithelial cell transcripts indicated that exposure to eosinophils induced differential expression of a subset of genes that were part of the EoE esophageal transcriptome. Collectively, these results uncover a network of crosstalk between eosinophils and esophageal epithelial cells involving epithelial mediated eosinophil survival and reciprocal changes in cellular transcripts, events likely to occur in EoE.
Introduction
An array of immune cells, including eosinophils, mast cells, and lymphocytes, accumulate in the esophageal epithelium in a variety of diseases, including Barrett’s esophagitis, gastroesophageal reflux disease, esophageal cancer and eosinophilic esophagitis (EoE). Recent attention has focused on the emerging disease EoE, a food antigen-driven chronic inflammatory disease associated with overproduction of IL-13 by pathogenic effector type 2 T cells (Th2 cells). IL-13 overexpression is sufficient to induce EoE-like features in mice, and blockade of IL-13 signaling in mice and humans attenuates EoE1–4. IL-13 triggers a variety of transcriptional responses in epithelial cells, including induction of CCL26, which encodes the eosinophil chemokine eotaxin-3, as well as a set of genes that overlap with the esophageal transcriptome seen in patients with EoE5, 6. Genetic studies have identified a key role for the esophageal epithelium in disease pathoetiology, as the gene products of the top two susceptibility loci encode for esophageal epithelial gene products, thymic stromal lymphopoietin (TSLP) and calpain-147–9. The multifaceted relationship between inflammation and epithelial alterations is central to disease pathology and presents opportunities for therapeutic intervention.
A growing body of evidence illustrates a positive role of long-lived, “tissue-resident” eosinophils in developmental and homeostatic responses10–16. These tissue-resident functions require prolonged survival of otherwise short-lived eosinophils, which is mediated by one of three known eosinophilopoietins—IL-3, IL-5 or GM-CSF. Eosinophilopoietins may be produced in either an autocrine or paracrine fashion that prevents intrinsic apoptosis17–21. In the small intestine, where eosinophils are constitutively present, epithelial-derived GM-CSF is required for eosinophil survival22, and in turn eosinophil-derived factors are required to downregulate activation of Th17 cells, providing evidence of an anti-inflammatory and/or immunomodulatory role of gastrointestinal eosinophils11. We have previously reported transcriptional changes to esophageal eosinophils with respect to bone marrow–derived eosinophils in a murine model of EoE2; these changes are consistent with an activation phenotype in tissue and included chemokine and cytokine receptors, adhesion and migration factors and secreted profibrogenic factors. However, currently there is limited information about how the unique environment of the esophagus impacts human eosinophils and conversely how eosinophils modify the surrounding tissue.
Herein, we hypothesized that the environment of the esophageal epithelium in patients with active EoE may induce specific changes to eosinophil activation and that this in turn promotes the epithelial cell gene expression associated with barrier dysfunction and fibrosis. We investigated human peripheral blood eosinophils upon coculture with esophageal epithelial cells and furthermore examined the role of IL-13. We observed changes to eosinophil viability and activation during coculture and evaluated reciprocal changes to gene expression in both eosinophils and epithelial cells. Our data demonstrate bidirectional crosstalk that alters survival and activation of eosinophils and also promotes EoE-like changes in epithelial gene expression.
Results
Esophageal epithelial cells extend eosinophil viability
Purified human blood-derived eosinophils (Siglec-8+, CCR3+, and CD44+) were cocultured with esophageal epithelial cells. Following coculture, eosinophil viability was 94 ± 1%, 96 ± 1% and 30 ± 6% after 4, 7 and 14 days, respectively. In contrast, within 4 days of culture in media alone, over 95% of the eosinophils stained positively for both the viability exclusion dye and Annexin V (Figure 1A). As a positive control, IL-5 (10 ng/mL) supplemented in the media preserved eosinophil viability at day 4 (96 ± 1%), 7 (94 ± 2%), and 14 (52 ± 11%) (Figure 1A). We evaluated the epithelial-to-eosinophil cell ratio that would support eosinophil survival. We noted an increased eosinophil survival at a ratio of 1 epithelial cell to 8000 eosinophils, which plateaued at 1:800 (Figure 1B). These results collectively demonstrate that esophageal epithelial cells promote human eosinophil survival and that this is an efficient process that requires only a small number of epithelial cells relative to eosinophils.
Figure 1. Eosinophil survival and surface marker expression in coculture.
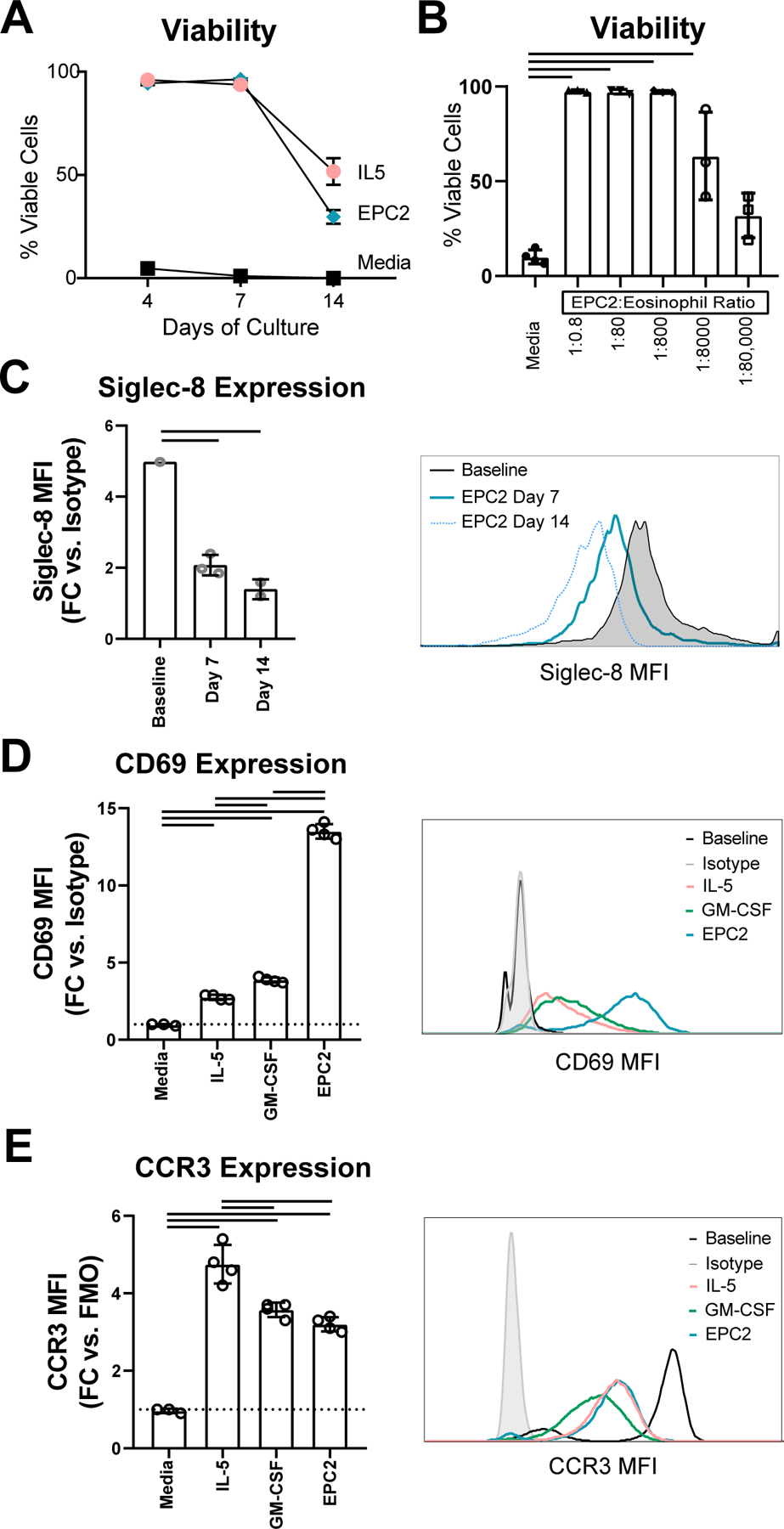
(A) The percentage of AnnexinV-/LiveDead- eosinophils was assessed by flow cytometry following monoculture or coculture (B) Eosinophil viability was assessed following 96 h of culture with media alone or coculture with varying quantities of EPC2 cells. (C) Surface expression of Siglec-8 was assessed at baseline and following 7 or 14 days of EPC2 coculture. (D) CD69 and (E) CCR3 expression were quantified on eosinophils at baseline or following 96 h of culture with IL-5 or GM-CSF or EPC2 coculture. Fold change (FC) in mean fluorescence intensity (MFI) was relative to control samples as measured by isotype or fluorescence minus one (FMO). Data shown are representative of at least 3 biological replicates, each with 3–4 technical replicates per condition and timepoint. Data shown are mean ± SD, and individual symbols represent technical replicates; bars signify p < 0.05.
Esophageal epithelial cells activate eosinophils
In order to understand phenotypic changes to eosinophils caused by exposure to esophageal epithelial cells, we quantified changes in surface marker expression in eosinophils following coculture. Focusing on the inhibitory receptor Siglec-8, we observed that surface expression of Siglec-8 decreased by day 7 of culture (mean fold change (MFC) 4.9 vs. 2.1 p < 0.01; Figure 1C). The activation marker CD69 was upregulated in coculture (MFC 13.5 vs. 1.0, p < 0.01; Figure 1D). For comparison, eosinophils cultured for 4 days in IL-5 or GM-CSF exhibited only slight increases in CD69 expression (MFC 2.8 and 3.9; Figure 1D). Expression of a key chemokine receptor, CCR3, was comparable on eosinophils cocultured with EPC2 cells with respect to GM-CSF alone and was slightly elevated on eosinophils cultured with IL-5 (Figure 1E).
Eosinophil-epithelial contact is not required to maintain eosinophil survival
Epithelial cells produce both secreted and surface-bound molecules that may act on eosinophils; we therefore set out to distinguish between contact-dependent and -independent effects of esophageal epithelial cell coculture on eosinophils. First, we compared the effects of standard coculture, in which cell:cell contact occured, with the effects of transwell cocultures, in which eosinophils and epithelial cells were separated by cell-impermeable porous inserts. We observed comparable viability in contact and transwell cocultures (93.3% vs. 96.7%; p > 0.05; Figure 2A). In contrast, induction of CD69 on cocultured eosinophils was only observed when cells were seeded in contact with EPC2 cells and not when cultured in transwell (MFC 1.6 vs. 3.8; p < 0.01; Figure 2B), supporting epithelial cell contact–dependent activation of eosinophils.
Figure 2. Contact-independent effect of coculture on eosinophil viability.
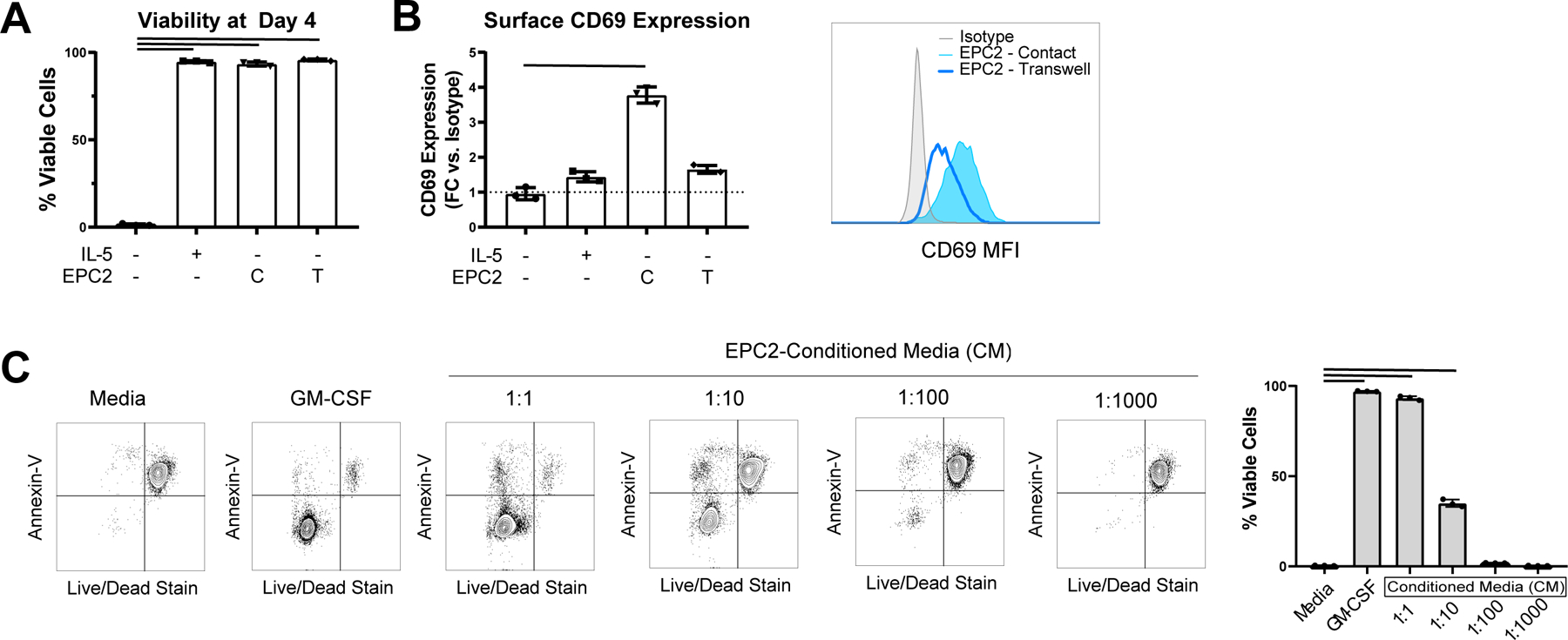
(A) Eosinophil survival and (B) CD69 MFI were assessed on eosinophils cultured for 96 h with media or media with IL-5 or cocultured in the bottom well in contact with EPC2s (“C”) or in the transwell insert with EPC2s in the bottom well (“T”). (C) Eosinophil viability was measured after culture with media alone, media with GM-CSF, or dilutions of EPC2-conditioned media (CM); results were anayzed by ANOVA with Dunnett’s post-test, and bars indicate p < 0.05. Data are representative of 3 biological replicates, each with 3–4 technical replicates per condition. In A-C, data shown are mean ± SD, and individual symbols represent technical replicates.
Next, we generated conditioned media derived from confluent esophageal epithelial cells. The conditioned media promoted eosinophil survival in a dose-dependent fashion (Figure 2C). However, no changes in surface expression of CD69 were observed in eosinophils cultured with the conditioned media (data not shown). Collectively, these results demonstrate that contact-independent factors account for eosinophil survival in coculture and furthermore that this effect is mediated by soluble mediators constitutively produced by confluent esophageal epithelial cells, yet the soluble factors are not sufficient to fully activate the eosinophils.
Secreted factors in coculture
Having observed that soluble factors are critical to the crosstalk between eosinophils and esophageal epithelial cells, we used a 65-analyte multiplex array of inflammatory mediators to identify soluble factors in supernatants after 7 days. As a control and to maintain viability, eosinophils in monoculture were treated with IL-5. Monocyte chemotactic protein 1 (MCP1; 33 ± 5 pg/mL), fibroblast growth factor 2 (FGF2; 119 ± 7 pg/mL), IL-16 (4 ± 1 pg/mL) and IL-23 (19 ± 7 pg/mL) were identified in supernatants of eosinophils but not epithelial cell monocultures (Figure 3A–B). Seven factors were identified in supernatants of esophageal epithelial cells but not eosinophil monocultures including fractalkine (40 ± 6 pg/mL), IL-15 (15 ± 1 pg/mL), IL-8 (>8 ng/mL), and tumor necrosis factor alpha (TNF-α; 193.6 ± 0.4 pg/mL) (Figure 3A–B). These factors were also detectable in coculture.
Figure 3. Soluble factors observed in eosinophil and epithelial cell coculture.
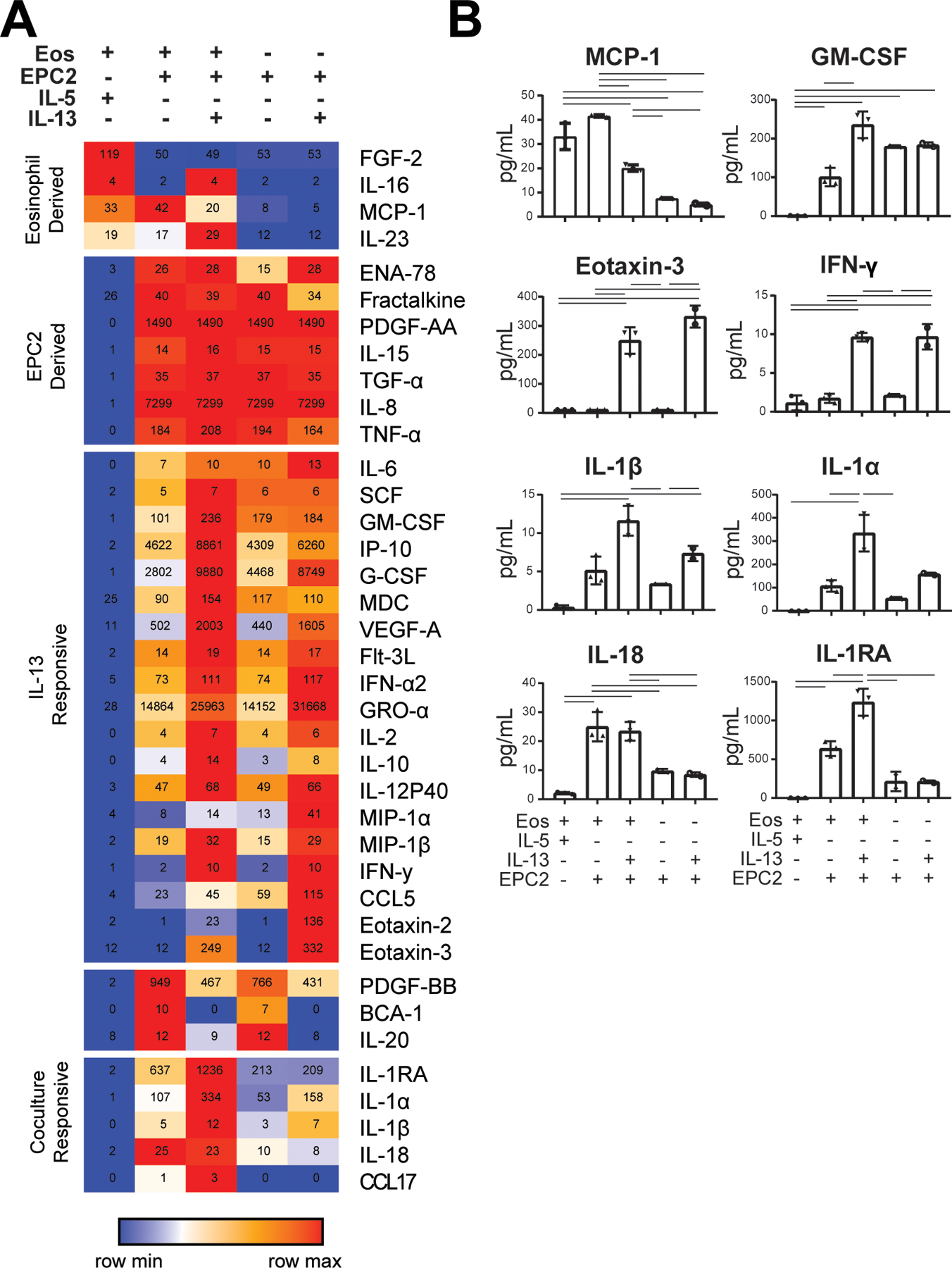
(A) A 65-analyte multiplex array was used to identify inflammatory mediators in culture supernatants from 7 days of IL-5-treated eosinophil monocultures, cocultures, cocultures with IL-13, EPC2 monocultures with EPC2 monocultures with IL-13. Values within the heatmap represent the mean concentration (pg/mL) for each treatment group. (B) Individual analytes are plotted. Significant differences were assessed with one-way ANOVA and Tukey post-test -bars indicate p < 0.01. Samples from three biological replicates were analyzed, with 2–3 technical replicates of each culture condition for each biological replicate. Representative data shown are mean ± SD, and individual symbols represent technical replicates.
To determine if cytokine secretion by eosinophils in monoculture may occur specifically in response to IL-5 compared to other pro-survival factors, we compared culture supernatants from monocultures treated with IL-5 or GM-CSF for 7 days. Of the detectable analytes, we observed that GM-CSF-treated monocultures secreted less IL-27 (45 vs. 5 pg/mL; p < 0.01) and more macrophage-derived cytokine (MDC) (0 vs. 5 pg/mL; p < 0.01), MCP-1 (21 vs. 65 pg/mL; p < 0.01), and IL-8 (2 vs. 9 pg/mL; p < 0.01) compared with IL-5-treated monocultures.
In order to understand unique components of eosinophil and epithelial cell crosstalk, we examined factors that were upregulated in coculture. In order to approximate the environment of the inflamed esophagus, we included cocultures that were supplemented with IL-13. Nineteen factors were modulated by addition of IL-13, including GM-CSF, CCL26, and IFN-γ. Among these cytokines, GM-CSF was produced by high-density esophageal epithelial cell monocultures (179 ± 3 pg/mL), detected in cocultures (100 ± 24 pg/mL) and increased by the addition of IL-13 in cocultures (235 ± 34 pg/mL; p < 0.05). Other factors that were similarly upregulated in the presence of IL-13 in both esophageal epithelial cell monocultures and cocultures included CCL26 (not detected (ND) vs. 249 ± 46 pg/mL) and IFN-γ (1.7 ± 1 vs. 10 ± 1 pg/mL; p < 0.05). Importantly, multiple factors were robustly detected only in the eosinophil–epithelial cell coculture, suggesting that their secretion is responsive to intercellular crosstalk. Specifically, several IL-1 family molecules were detected in coculture and were increased with IL-13, including IL-1RA (637 ± 96 vs. 1236 ± 175 pg/mL; p < 0.05), IL-1B (5 ± 2 vs. 12 ± 2 pg/mL; p < 0.05), and IL-1α (107 ± 25 vs. 335 ± 79 pg/mL; p < 0.05). IL-18 was the only IL-1 family cytokine whose expression in coculture was not increased in the presence of IL-13 (25 ± 5 vs. 23 ± 3 pg/mL; p = 0.67).
In order to evaluate the impact of soluble factors in coculture, we performed functional assays to ascertain the role of epithelial-derived factors on eosinophil migration and survival. Multiple eosinophil chemoattractants were detected by multiplex, including CCL11 (eotaxin) and CCL26 (eotaxin-3); thus, we performed migration assays to determine whether eosinophils migrate in response to conditioned media generated from IL-13–treated esophageal epithelial cells. A greater fraction of IL-5-treated eosinophils migrated in response to IL-13–treated conditioned media compared with untreated conditioned media (12% vs. 20%, p < 0.01; Figure 4A). Because factors that impact eosinophil survival and activation were upregulated in IL-13–treated cocultures, we compared eosinophil survival in monoculture and coculture with and without IL-13 (Figure 4B). After 14 days, we did not observe any viable eosinophils in monoculture with or without IL-13; however, in coculture, IL-13 supplementation led to a significant increase in viability versus coculture alone (68 ± 1 vs. 30 ± 6%; p < 0.01). Collectively, these results identify specific changes to eosinophil function when they are cocultured with IL-13–treated epithelial cells.
Figure 4. Functional impact of soluble factors in coculture on eosinophil migration and viability.
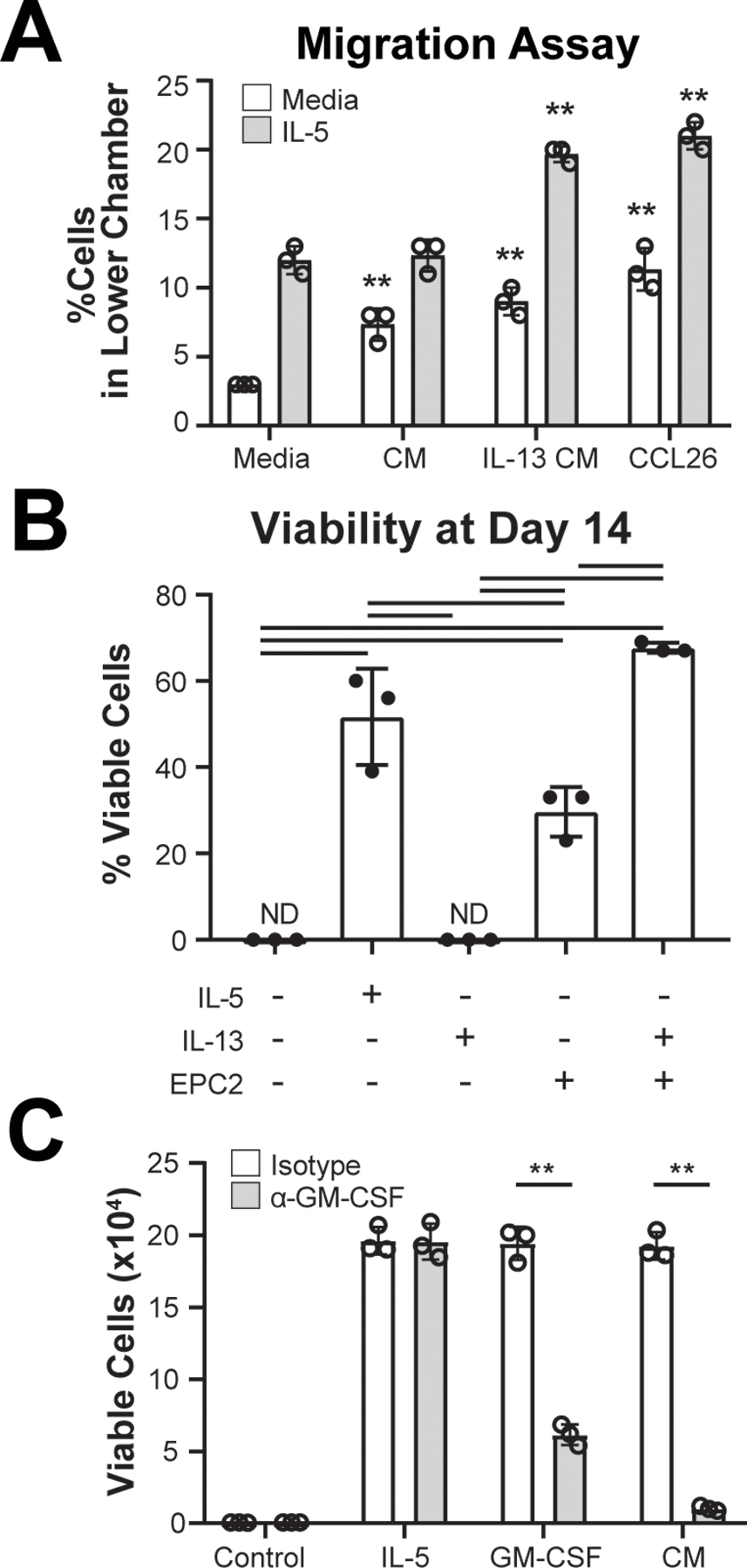
(A) Eosinophils cultured for 24 h with media alone or media with IL-5 were seeded in porous inserts and allowed to migrate for 4 h in response to media, EPC2-conditioned media (CM), IL-13–treated EPC2–conditioned media or CCL26. (B) Viability was assessed after 14 days of culture with media alone or media with IL-5 or IL-13 or EPC2 coculture with or without IL-13. (C) Eosinophil viability was measured by Annexin-V staining and viability dye after 96 h of culture with either IL-5, GM-CSF, or EPC2-conditioned media (CM) with neutralizing antibody against GM-CSF or IgG control (Isotype). Data are representative of experiments that were repeated 2–3 times with 3 technical replicates per condition. Data shown are mean ± SEM, and individual symbols represent technical replicates. Significant differences were assessed with one-way ANOVA and Tukey post-test. Bars indicate p < 0.01, ** p < 0.001. ND, not detected.
Identification of GM-CSF as key esophageal epithelial cytokine driving eosinophil survival
GM-CSF has a well-characterized role in promoting eosinophil survival and was produced by epithelial cells in monoculture and in coculture; whereas, IL-5, another established pro-survival factor, was not detected (Table S1). To test the hypothesis that GM-CSF was necessary for the pro-survival effect on eosinophils in coculture, we tested the effect of GM-CSF blockade on eosinophil viability. We found that a neutralizing antibody against GM-CSF, but not IgG control, was sufficient to attenuate eosinophil survival in the presence of EPC2-conditioned media (1.9 ± 0.1 × 105 vs. 1.0 ± 0.2 × 104 live cells; p < 0.001; Figure 4C).
Eosinophil gene expression in coculture
In order to comprehensively assess the impact of epithelial coculture, RNA sequencing was performed on the eosinophils. Cultured eosinophils (7 days) were compared to baseline eosinophils. We examined eosinophils cultured with esophageal epithelial cells with and without IL-13; eosinophils cultured in IL-5 served as a positive pro-survival control. We observed high correlation values between eosinophils in coculture with and without IL-13, suggesting similar transcriptional responses to these two culture conditions (Figure S1A). Using ANOVA to identify genes differentially expressed by any condition (FDR < 0.05), we found that 58% of expressed genes in eosinophils exhibited differential expression after 7 days of culture with either IL-5, esophageal epithelial cells without IL-13, or esophageal epithelial cells with IL-13 (Figure 5A). We performed K means clustering to obtain functional groups of differentially expressed genes with similar expression patterns and analyzed those groups for functional enrichment via Gene Ontology (GO). In freshly isolated eosinophils, we observed preferential expression of genes involved in defense response, response to mechanical stimulus and response to cytokine; these genes included IL5RA and CASP1 (Figure 5B–C). Coculture led to upregulation of genes involved in inflammation, secretion, leukocyte migration and response to IL-1 (e.g., ICAM1, CCL2, NFKB1, CD83 and TLR1) and downregulation of genes involved in differentiation and development (e.g., CASP3, ANXA6, APAF1 and ARHGAP3). We examined gene expression in eosinophils cultured with IL-5 compared to GM-CSF by RT-PCR; whereas, we did not observe a difference in expression of CD83, CASP3, or ANXA6 (data not shown), we observed a 1.9-fold decrease in expression of IL5RA by eosinophils cultured with GM-CSF compared with IL-5 (p < 0.01).
Figure 5. Tissue-specific responses of eosinophil gene expression to epithelial coculture.
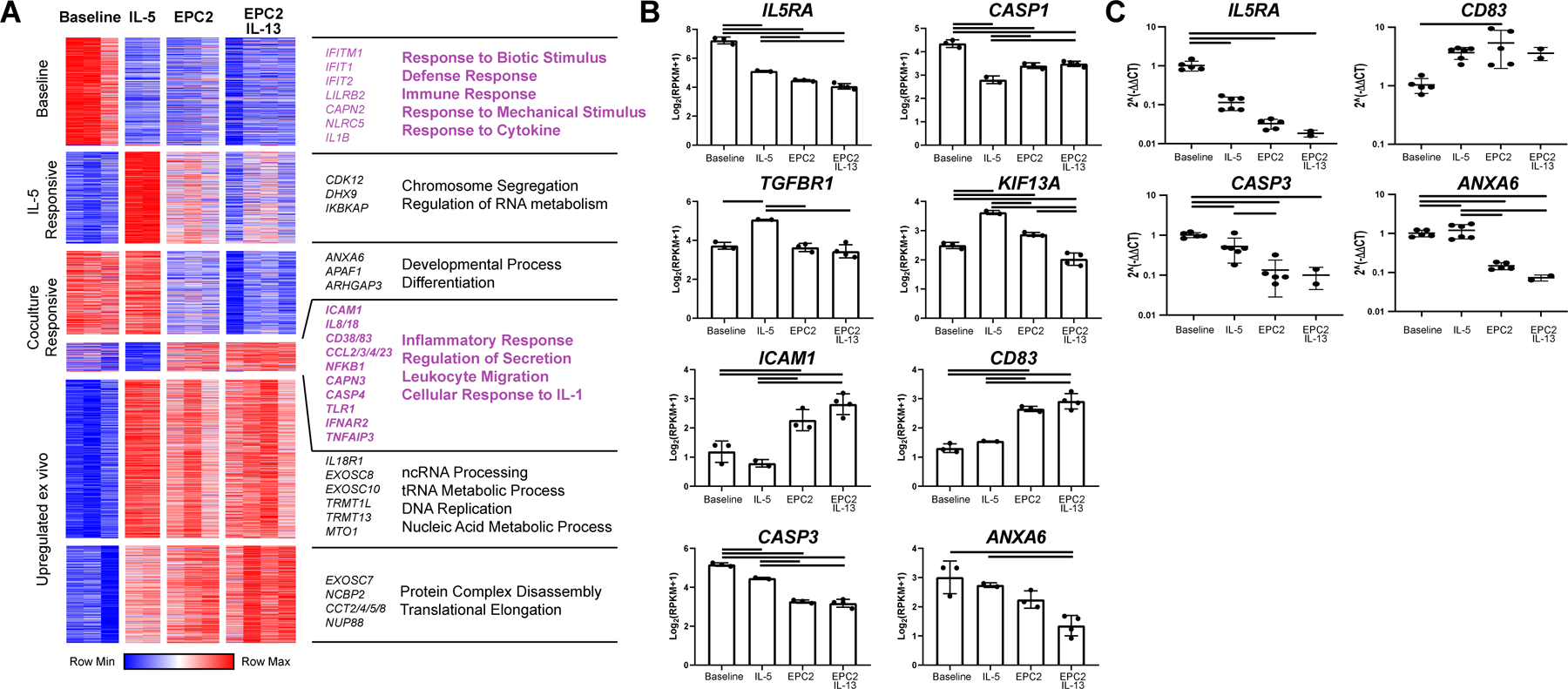
RNA extracted from freshly isolated eosinophils or non-adherent, CD45-enriched cells after 7 days of culture in media alone (baseline) or with IL-5 (IL-5) or cocultured with EPC2s (EPC2) or EPC2s with IL-13 (EPC2 IL-13) was sequenced. In the dataset, 6591 genes were expressed. (A) Of these, 3652 genes identified as significant by ANOVA (FDR < 0.05) and were clustered via K means (K = 6). Clusters were entered into GOrilla to obtain enrichments using total expressed genes as background. Selected processes and representative genes are annotated. (B) Representative individual genes are plotted. Data represent a single biological replicate with 2–4 technical replicates per condition. (C) Selected genes were quantified by RT-PCR using RNA from 2–5 technical replicates and 3 biological replicates. Significant differences were assessed with one-way ANOVA and Tukey post-test. Bars indicate p < 0.01. Data shown in B-C are mean ± SD, and individual symbols represent technical replicates.
To comprehensively define unique and shared gene expression changes in monoculture and coculture, we used DEseq to identify differentially expressed genes in each culture condition, individually, compared to baseline (Figure S1B). There were 199 genes commonly dysregulated by Day 7 versus freshly isolated eosinophils regardless of the culture conditions (i.e., coculture alone, coculture with IL-13, IL-5 alone), suggesting that these genes were characteristic of post-mitotically differentiated eosinophils; these genes were enriched for pathways involved in immune response (e.g., IL5RA, IL1RN, IL1A), leukocyte migration (e.g., CCL2, CXCL1, CXCR1), and response to lipid (e.g., CD68, CD180, FPS, FOSB). By comparing the gene expression of eosinophils cocultured with esophageal epithelial cells with or without IL-13, we identified 66 genes that were preferentially upregulated (e.g., SPP1, IL1RL1, and IFI44) and 63 genes that were downregulated by presence of IL-13 in coculture (e.g., CCL2, IFI6, KIT; Figure 6A, C).
Figure 6. Tissue-specific responses of eosinophil gene expression to epithelial coculture.
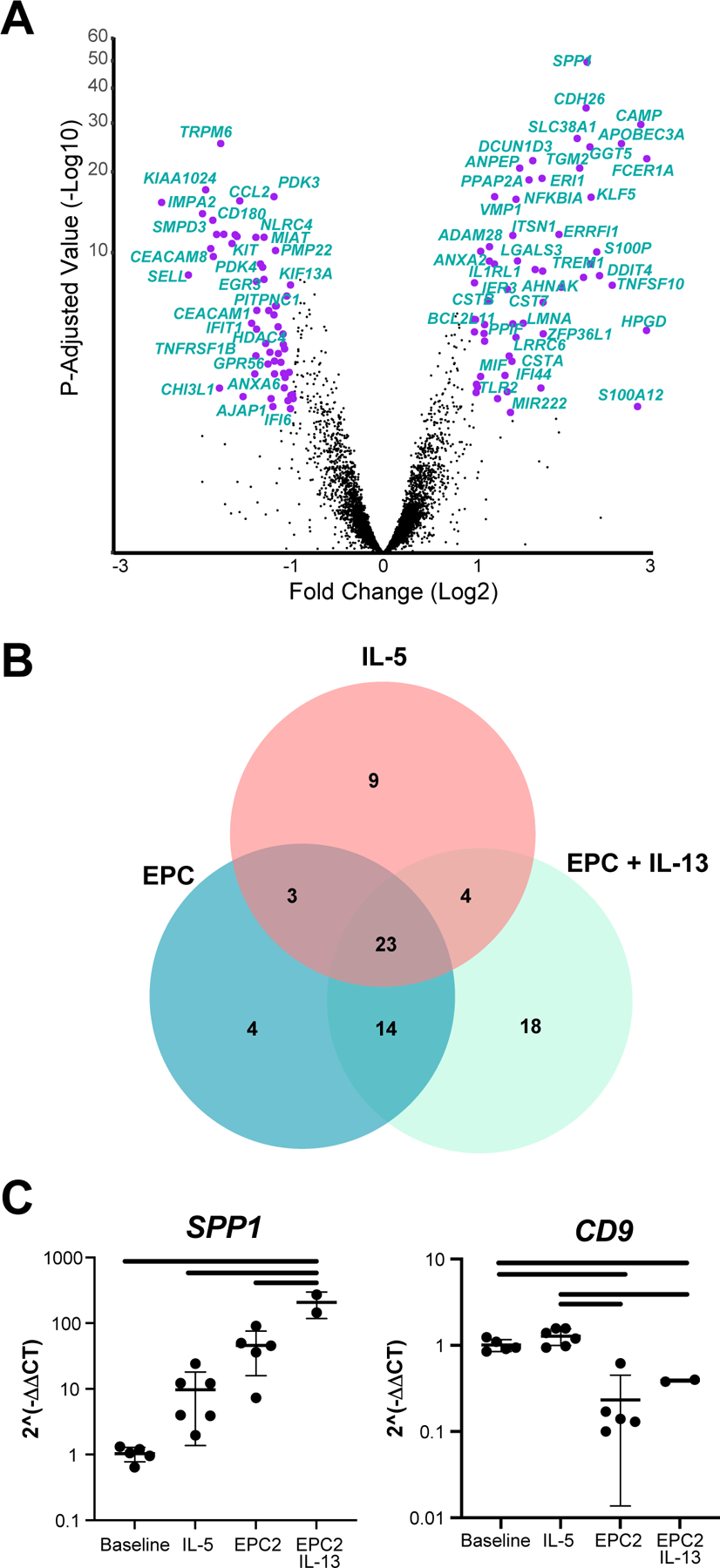
(A) DEseq was used to compare gene expression in EPC2 cocultured eosinophils with and without IL-13. Genes with a fold change > 2 and an adjusted p value < 0.05 are annotated in purple, and select genes are labeled. (B) Genes that were identified as upregulated in esophageal eosinophils in a murine model of EoE were compared with the genes that were differentially expressed in our three culture conditions with respect to baseline state when isolated from human blood. Data represent a single biological replicate with 2–4 technical replicates per condition. (C) Selected genes were quantified by RT-PCR using RNA from 2–5 technical replicates and 3 biological replicates. Significant differences were assessed with one-way ANOVA and Tukey post-test. Bars indicate p < 0.01.
In order to establish the relevance of these gene expression data in esophageal eosinophils, we compared genes that were differentially expressed in our model system with genes previously reported to be differentially expressed by esophageal eosinophils in a murine model of EoE driven by IL-13 overexpression2. A set of genes (n = 75) that was preferentially expressed by esophageal eosinophils in the EoE model was also differentially expressed in at least one of the culture conditions (monoculture with IL-5 and coculture with or without IL-13; Figure 6B). A set of 23 experimental EoE genes were differentially expressed by eosinophils in all 3 culture conditions, including IL1A, ITGA4, and CD68. Importantly, 14 of the experimental EoE genes were specifically dysregulated by coculture, including caspase 3 (CASP3) and CD9 (Figure 6C). A further 18 genes, such as nucleic acid binding protein 1 (NABP1), the antimicrobial cathelicidin antimicrobial peptide (CAMP), and the complement C1q binding protein (C1QBP), were exclusively dysregulated in IL-13–supplemented coculture.
Eosinophil-dependent modulation of epithelial gene expression
In order to determine whether eosinophils induced changes in esophageal epithelial gene expression, RNA sequencing was performed on esophageal epithelial cells following 7 days of monoculture or eosinophil coculture with or without IL-13. By examining genes with an FDR p < 0.05 by ANOVA, we identified 2,828 genes whose expression varied by culture condition (Figure 7A). K means was used to obtain clusters of genes whose expression was dictated by culture condition. This resulted in one small cluster containing genes that were upregulated in monoculture with IL-13 and coculture with or without IL-13 as well as two large clusters that were upregulated and downregulated, respectively, by the presence of IL-13 in both monoculture and coculture (Figure 7A). There was no difference in IL-13 detected in the media of cocultures or EPC2 monocultures (Supplemental Table 1); therefore, IL-13 is unlikely to account for the pattern of gene expression observed in cluster 1. We identified a cluster of 262 genes that were upregulated specifically with eosinophil coculture, including the pro-inflammatory genes NEK7 and IL18, as well as a component of the IL-13 receptor, IL13RA1 (Figure 7C and S2). Additionally, the gene expression of a key transcription factor that acts downstream of TGF-β signaling, SMAD4, was induced by eosinophil exposure (Figure S2).
Figure 7. Eosinophil-dependent changes to epithelial gene expression.
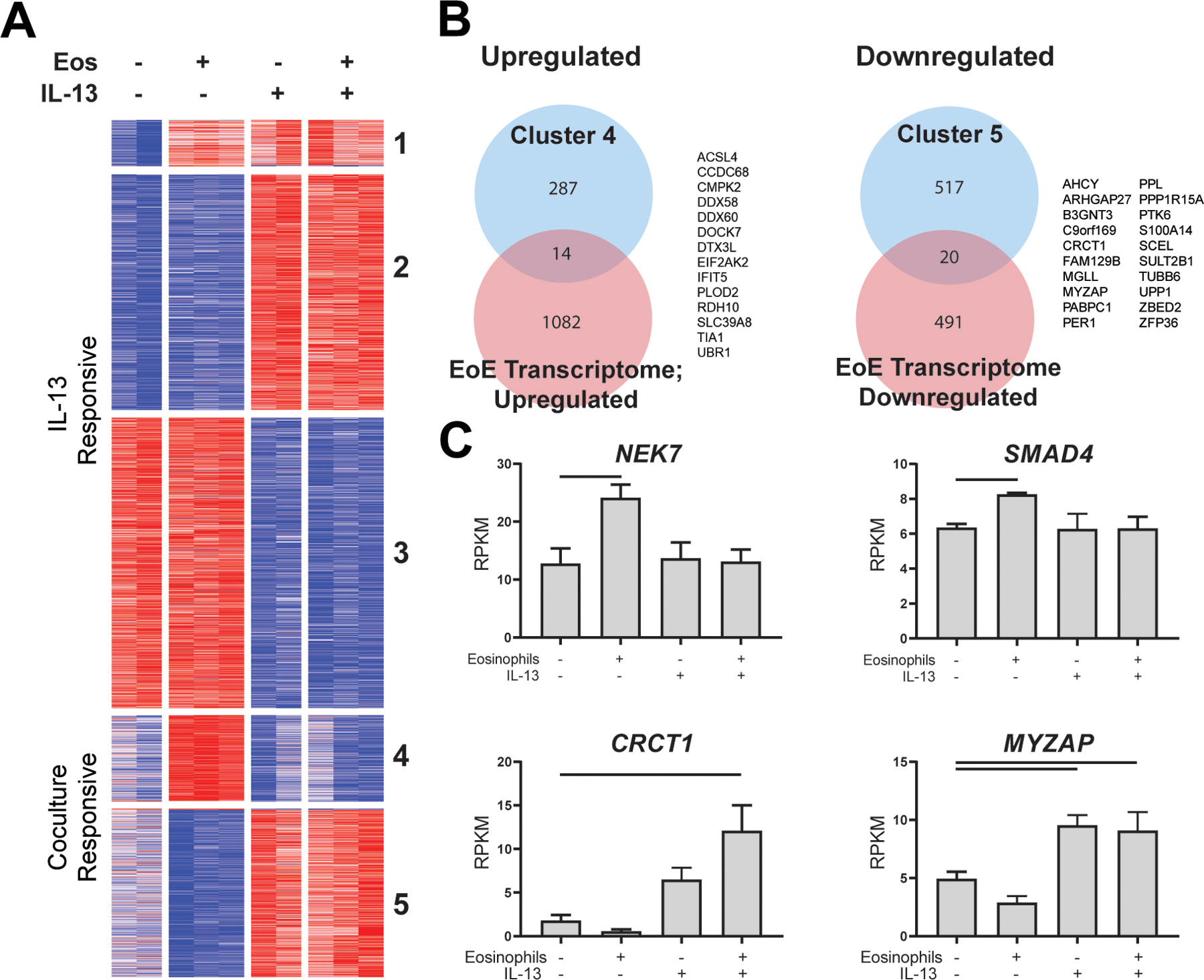
RNA from adherent epithelial cells following 7 days of culture with or without eosinophils (Eos) and/or IL-13 was sequenced, and 7457 genes were detected in the dataset. (A) Of these, 2828 genes were identified as significant by ANOVA (FDR < 0.05) and were clustered via K means (K = 5). (B) Clusters of significant genes that were upregulated (cluster 4) or downregulated (cluster 5) in the presence of eosinophils were compared with differentially expressed genes in EoE biopsy tissue (EoE Transcriptome). (C) Representative genes that were significantly upregulated or downregulated with coculture. Data represent a single biological replicate with 2–3 technical replicates per condition. Data were analyzed by ANOVA with Tukey post-test; bars represent p < 0.05. Data shown in C are mean ± SD.
The transcriptional changes specifically attributable to eosinophil coculture were modest compared with the changes attributable to IL-13 (Figure 7A). These results were reinforced by pairwise differential expression analysis. Only 27 genes were differentially expressed (|FC| > 2, adjusted p < 0.05) in cocultured vs. monocultured esophageal epithelial cells, whereas 536 genes were differentially expressed by IL-13–treated vs. untreated esophageal epithelial cell monocultures. To obtain a more sensitive readout of functional changes to esophageal epithelial cell gene expression in coculture, we performed preranked gene set enrichment analysis (GSEA) and tested for enrichment of GO terms (Table 1). Gene sets that were positively enriched were involved in maintenance of the extracellular matrix and basement membrane, as well as transmembrane receptor protein tyrosine kinase activity. Negatively enriched pathways included GDP binding and ubiquitin-like enzyme activity.
Table 1.
Significantly enriched gene sets in cocultured epithelial cells
| Gene Ontology (GO) Process Name | Gene Set Size | Enrichment Score | Normalized Enrichment Score | Nominal p-value | FDR q-value | Rank at Maximum |
|---|---|---|---|---|---|---|
| Extracellular Matrix Structural Constituent | 52 | 0.764 | 2.868 | 0.000 | 0.000 | 692 |
| Basement Membrane | 38 | 0.659 | 2.338 | 0.000 | 0.013 | 692 |
| Collagen Containing Extracellular Matrix | 137 | 0.513 | 2.278 | 0.000 | 0.027 | 818 |
| Transmembrane Receptor Protein Tyrosine Kinase Activity | 24 | 0.693 | 2.258 | 0.000 | 0.025 | 1349 |
| Collagen Trimer | 15 | 0.807 | 2.237 | 0.003 | 0.027 | 542 |
| Extracellular Matrix | 160 | 0.472 | 2.226 | 0.000 | 0.027 | 1438 |
| Peptide Cross Linking | 15 | 0.747 | 2.171 | 0.000 | 0.046 | 983 |
| GDP Binding | 50 | −0.712 | −2.260 | 0.003 | 1.000 | 1327 |
| Ubiquitin Like Protein Conjugating Enzyme Activity | 33 | −0.694 | −2.041 | 0.004 | 1.000 | 849 |
GO Processes with a |normalized enrichment score| > 2 and an FDR-adjusted significance < 0.05 are included in this table.
Overlap of eosinophil-regulated epithelial genes with EoE transcriptome
We compared the K means clusters of coculture-responsive genes with genes previously reported as differentially expressed in esophageal tissue from patients with EoE compared with controls23. There were 14 epithelial genes induced by eosinophil exposure that were also upregulated in the EoE transcriptome, including the interferon-responsive gene IFIT5, collagen-modifying enzyme PLOD2 and cell-fate factors ASCL4 and TIA1 (Figure 7B–C, S2). Conversely, the 20 EoE-associated genes that were downregulated in esophageal epithelial cells by eosinophil exposure included the epithelial signal transducer PTK6, cytoskeletal component TUBB6, transcription factor ZNF36, serum response factor MYZAP and epithelial metabolic regulator CRCT1 (Figure 7C).
Discussion
Herein, we have examined changes that occur in epithelial cells and eosinophils as a result of their interaction. We observed a striking pro-survival effect on eosinophils in coculture that does not require direct cell:cell contact and demonstrated a role of epithelial-derived GM-CSF in eosinophil survival. We identified increased production of several IL-1 family members in coculture and observed that the transcriptome of cocultured eosinophils was enriched for genes involved in IL-1 signaling as well migration and secretory pathways. This profile was not observed in eosinophils cultured with IL-5, a pro-survival cytokine, underscoring a unique response to epithelial signals. We propose that the unique activation state of cocultured eosinophils mimicks a tissue-resident phenotype. Finally, we demonstrated that epithelial cells respond to eosinophil coculture by upregulating genes involved in the interferon response and moreover that genes involved in EoE pathogenesis were dysregulated in epithelial cells in response to coculture with eosinophils. Collectively, these findings establish evidence of crosstalk between esophageal epithelial cells and eosinophils and resultant functional changes in both cell types.
Recently, we have reported heterogeneity of type 2 gene expression in active EoE, observing that inflammatory gene expression is not lost in patients with fibrostenotic characteristics24. This supports a role of eosinophils in the epithelial remodeling process. Data presented in this report substantiate multi-faceted crosstalk between cell types that results in changes to epithelial function that might accompany remodeling events, including desmosome assembly and extracellular matrix (ECM) remodeling. ECM and basement membrane remodeling are functions involved in EoE pathogenesis23, 25. We also observed upregulation of SMAD4, which encodes a TGF-β–responsive transcription factor, specifically in coculture. This may be important as TGF-β signaling and mutations in the TGF-β receptor (TGFB1R) and genetic variants in the region of the TGF-β signaling activator (LRRC32) have been implicated in EoE7, 26, 27. It is interesting to speculate that eosinophil-derived factors, including TGF-β, may be responsible for tissue pathology at later stages of disease by promoting epithelial-to-mesenchymal transition, wherein epithelial cells adopt a fibroblast phenotype and remodel the ECM. Whereas, the current study was primarily focused on changes in eosinophil function and gene expression during coculture, future studies will explore changes in epithelial cell differentiation, barrier establishment and maintenance, especially the role of TGF-β in coculture.
Previous studies have reported changes to human eosinophil gene expression in response to different eosinophilopoietins, including GM-CSF, IL-5 and IL-3. These studies have examined the transcriptome of freshly isolated eosinophils2, 28 or examined survival and activation of cultured murine eosinophils; however, to our knowledge this is the first study to describe global changes in human eosinophil gene expression following prolonged culture with esophageal epithelial cells. Given the need to understand eosinophil responses to unique environments in situ, several groups have attempted to obtain single cell RNA sequencing (scRNAseq) data on eosinophils from allergic tissues without success. This result has been attributed to the high RNAse content in eosinophils, which overcomes the RNAse inhibitor concentration in microdroplets generated during the scRNAseq workflow29. Recently, one group reported distinct inflammatory and regulatory gene expression profiles by eosinophils sorted from murine lungs and arthritic joints using scRNAseq; these results corroborate our finding of a dynamic transcriptional response of eosinophils to unique environments and suggest that continued efforts to analyze eosinophil gene expression at the single cell level may produce valuable findings30. Until technological advances are able to reliably address the high RNAse content of eosinophils, bulk sequencing of cultured and stimulated eosinophils remains the best way to gain insight into diverse and heterogeneous functions of these cells. The findings described herein underscore the dynamic capacity of human eosinophils, despite their post-mitotic state.
Several studies have examined the activation state of eosinophils in the circulation and in esophageal tissue of EoE patients using surface staining to quantify expression of activation markers. CD69 is upregulated on human eosinophils in response to a variety of signals including IL-1331, has been identified on circulating eosinophils in EoE, and its strong upregulation in the esophageal tissue of EoE patients has been observed histologically, though the increase was attributed to T-cells.32 Herein, we show that CD69 is upregulated in cocultured eosinophils in a cell-contact-dependent manner. The increased surface expression of CCR3 that was observed in cocultured eosinophils is reminiscent of increased CCR3 expression on eosinophils in blood and biopsies from EoE patients33–35. Several studies have illustrated the necessity of CCR3 ligation in eosinophil migration in response to a variety of ligands during allergic inflammation, including several ligands that were identified in the multiplex experiment including CCL11, CCL26, and RANTES36–39. Finally, changes to Siglec-8 expression have been associated with eosinophil maturation, and internalization of Siglec-8 resulting in decreased surface expression is associated with a pre-apoptotic phenotype, which may account for the decreased surface expression of Siglec-8 over time in culture40. Taken together, the observed changes in eosinophil expression of CD69, CCR3, and Siglec-8 echo trends that have been observed in EoE and other allergic conditions.
Whereas in this report we have presented compelling evidence that GM-CSF is necessary and sufficient for a pro-survival effect of epithelial cells on cocultured eosinophils, it is probable that there is an additional role of T-cell derived IL-5 in allergic tissue. The respective contributions of these two cytokines is of particular interest, especially given the association of genetic polymorphisms in IL5 with EoE susceptibility27, as well as ongoing trials of IL-5 signaling antagonists in the treatment of EoE and other EGID41–44. Moreover, it is possible that the relative importance of IL-5 and GM-CSF signaling changes over time in situ, as we have observed downregulation of the IL-5 receptor subunit IL5RA following 7 days of coculture. Given the potential for both redundant and non-redundant roles of IL-5 and GM-CSF on eosinophil survival and activation in EoE, further studies should compare the role of their individual and synergistic signaling and incorporate each cytokine into culture experiments.
Increasingly, a role for long-lived, tissue-resident eosinophils in tissue immunity and structural integrity has been established in allergy, cancer and gut homeostasis; the critical role of eosinophils in mucosal homeostasis was recently reviewed by Shah et al. (2020)16. Widespread differences in eosinophil gene and protein expression have been observed in response to differential cytokine or growth factor exposure or in specific tissue environments2, 40, 45–48. Specialized changes to gene expression and granule secretion, combined with prolonged survival in non-inflammatory diseases, such as cancer, suggest highly specialized and diverse eosinophil functions and populations of eosinophils in tissue49. The Local Immunity And/Or Remodeling/Repair (LIAR) hypothesis advances a critical role of eosinophils in maintaining healthy tissue function under homeostatic conditions and returning to homeostasis following inflammatory insult14. This function of long-lived tissue eosinophils is consistent with established roles of other granulocytes in tissue homeostasis and represents an opportunity to develop therapeutic strategies that harness pro-repair functions of eosinophils in the treatment of hypereosinophilic disorders.
It is interesting to note that the cytokines detected specifically in coculture supernatants belong to a family of proteins (e.g., IL-1β, IL-18, IL-33) whose secretion is primarily regulated posttranslationally through proteolytic cleavage of precursor proteins. It is possible that the effect of eosinophil coculture on epithelial activity and function is largely exerted at the posttranslational level. This may explain why the transcriptional effects induced by the coculture were modest compared with the effects of IL-13. Further investigation will be needed to evaluate changes to enzymatic and proteolytic activity, as well as cytoskeletal reorganization and cellular morphology, in epithelial cells following coculture with eosinophils. Moreover, it is possible that the effects of coculture may be observed in epithelial cells during differentiation from basal to suprabasal phenotype or while establishing a robust epithelial barrier. These effects may be measured using primary esophageal epithelial cells either in air-liquid interface cultures or in more natural conditions, such as esophageal organoids, and will be the subject of future investigation.
In this study, IL-13 was used to approximate the cytokine profile that is observed in esophageal tissue during active EoE disease, particularly as IL-13 has a robust effect on epithelial cell gene expression. Whereas eosinophils may secrete IL-13, the multiplex data does not support a role for eosinophis as the key cellular source of this cytokine, and IL13 was not detected in eosinophils by RNAseq. Indeed, single cell sequencing of esophageal biopsies has identified T-helper type 2 (Th2) cells as a primary sources of IL-1350.
We propose that the environment of the EoE epithelium is the primary driver of disease pathoetiology. Genetic polymorphisms associated with risk of EoE are primarily expressed by epithelial cells as opposed to myeloid cells. Eosinophil-centric and epithelial-centric disease mechanisms are not mutually exclusive; therefore, ongoing studies will examine the roles of both mechanisms. Future work will leverage primary cells from patients and controls to define a role of disease-dependent phenotypes such as increased activation of circulating eosinophils or impaired epithelial differentiation on intercellular crosstalk during coculture.
In summary, we have established evidence of a pro-survival effect of esophageal epithelial cells on human eosinophils that was accompanied by a unique eosinophil activation state, as indicated by changes to surface expression of Siglec-8, CD69 and CCR3. By multiplex we identified the eosinophilopoietin GM-CSF, several eosinophil chemoattractants, and IL-1 family cytokines. These factors likely exert functional changes on eosinophils, as we observed that GM-CSF neutralization was sufficient to attenuate eosinophil survival in coculture and furthermore that eosinophils migrated in response to esophageal epithelial cell–conditioned media. We also observed a transcriptional response of esophageal epithelial cells in eosinophil coculture that was enriched for extracellular matrix remodeling and collagen production and that included genes that we have shown to be differentially expressed in biopsies of patients with EoE. Collectively, these results identify reciprocal crosstalk between eosinophils and esophageal epithelial cells that likely has consequences in the inflammatory and remodeling stages of EoE pathogenesis. Given the involvement of eosinophils in homeostatic and remodeling processes in mucosal tissue, a greater understanding of this crosstalk will have significant implications in an array of eosinophilic gastrointestional disorders.
Methods
Human peripheral blood eosinophil isolation
Blood was collected from 13 control donors (IRB 2008-0090; Supplemental Table 2), and 150–250 mL of blood was collected in heparin-coated tubes at each draw. Red blood cells were removed with a 4% dextran gradient, and the remaining buffy coat was diluted with EDTA in PBS and layered over a percoll gradient. After 30 minutes of centrifugation at 1300 rpm, buffy coat, serum, and percoll layers were removed, and the remaining granule/erythrocyte pellet was resuspended and washed with 2% FBS (Atlanta Biologicals #S11150). Remaining erythrocytes were lysed with hypotonic buffer, and eosinophils were purified from the granulocyte pellet by negative immunomagnetic purification (Miltenyi #130-092-010) per the manufacturer’s instructions. Cytospins of the granulocyte and eosinophil fraction were stained with HEMA 3 to confirm eosinophil purity and recovery. Eosinophil purity and viability were regularly >95% and >98%, respectively. Eosinophils were resuspended and cocultured in a 1:1 mixture of eosinophil media (RPMI-1640, Invitrogen #SH30027.01 + 10% FBS) and keratinocyte serum-free media (KSFM; Invitrogen cat no. 17005042) supplemented with bovine pituitary extract (BPE, 12.5 mg/L) and epithelial growth factor (EGF, 1 ng/mL).
EPC2 validation and culture conditions
An immortalized human esophageal epithelial cell line (EPC2, a kind gift of Dr. Anil Rustgi) was subjected to short tandem repeat (STR) profiling (Genetica Inc.). The STR profile exhibited a 100% match to the reference profile, and the cells were mycoplasma negative. Banked cells from the same batch as those authenticated were used in these studies, and cultures were discarded after 2–3 months of passages. EPC2s were cultured in a humidified incubator at 37°C with 5% CO2 with keratinocyte serum-free media (KSFM) supplemented with BPE and EGF. Two days prior to eosinophil isolation, EPC2s were seeded at high density (5×105 cells/well of a 24-well plate). Media was changed the following day to remove nonadherent cells. To generate conditioned media, EPC2s were seeded at high density and media was replaced at 24 h. At 72 h, conditioned media was recovered and frozen after cells and debris were removed by centrifugation.
For coculture, a 1:1 mixture of eosinophil culture media and KSFM was used. Eosinophils were added at a concentration of 1×106 cells per mL (4×105 cells/well), and half of the media was replaced every 48 h for the duration of the experiment. To preserve survival in eosinophil monocultures, media was supplamented with either IL-5 (10ng/mL; PreproTech cat no. 200-05) or GM-CSF (50 ng/mL; PreproTech cat no. 300-03). In neutralization studies, polyclonal anti-GM-CSF (Abcam cat no. ab9741) and immunoglobulin control (Abcam cat no. ab37415) were used at 100 ng/mL.
Flow cytometry staining
Nonadherent cells were collected, pelleted and washed with PBS prior to viability staining (Invitrogen cat no. L34957, 1:1000) for 10 minutes; cells were washed with 2% FBS in PBS for antibody staining. Cells were incubated with Fc Block (BD Biosciences cat no. 564219, 1:50) for 15 minutes prior to the addition of labeled antibodies. Cells were stained for Siglec-8 (PE-Cy7, Biolegend cat no. 347112, 1:50), CCR3 (Alexa647, Biolegend cat no. 310710, 2.5:50), CD69 (BV421, Biolegend cat no. 310930, x:50), and CD44 (PerCP-Cy5.5, Biolegend cat no. 103032, 0.5:50) for 30 minutes. Stained cells were washed with PBS and stained with Annexin V (1:100) in Annexin-V Binding Buffer (BD cat no. 556454) for 10 minutes. Cells were washed and stored in fixation buffer (eBioscience cat no. 8222-49) for 1–3 days. All staining procedures were carried out at 4°C. Prior to analysis, samples were resuspended in 2% FBS and analyzed on an LSRII or Fortessa cytometer in the Cincinnati Children’s Hospital Medical Center (CCHMC) Research Flow Cytometry Core (RFCC). Single-color compensation was calculated, and data were analyzed in FlowJo (Version 10.6.1).
Multiplex array
Culture supernatants were centrifuged to remove cells and debris. Supernatant aliquots were analyzed for a 65-plex Human Cytokine/Chemokine Array (Eve Technologies; Supplemental Table S1). Concentration values above or below the reported limit of detection were manually adjusted to those limits, and analytes for which no sample was measured above the limit of detection were excluded. Results were confirmed with two independent biological replicates and representative data are shown.
Migration assay
Following overnight culture with- and without IL-5, 2×104 eosinophils were resuspended in coculture media and seeded into well inserts with 5.0-μm pore size and increasing concentrations of EPC2-conditioned media or 5 ng/mL CCL26 (Preprotech cat no. 300-48) in coculture media were added to lower wells. Cells were incubated at 37°C for 4 h, at which time all cells in lower wells were collected and fixed. Using count beads (Invitrogen cat no. C36950), total cell counts were enumerated by flow cytometry. Percent of migrated cells in response to chemokine were normalized to the number of cells recovered from the lower well when no chemokine was present.
RNA isolation, sequencing, and analysis
After 7 days of culture, nonadherent cells were removed and subjected to positive immunomagnetic separation using CD45 beads (Miltenyi cat no. 130-045-801) per manufacturer’s instructions. Purity was >98%. CD45+ nonadherent cells (eosinophils) and adherent cells (EPC2s) were lysed with Trizol, and lysates were frozen prior to RNA extraction with chloroform. For eosinophil samples, the chloroform extraction step was repeated in order to attenuate RNA degradation51, 52. Aqueous phases mixed 1:1 with ethanol were added to Zymogen Quick RNA Microprep kit (Zymo Research R1051) and RNA was purified per manufacturer’s instructions and eluted with water. cDNA libraries were prepared and quality control measures provided by the CCHMC Gene Expression Core, and libraries of acceptable quality were submitted for sequencing at the DNA Sequencing and Genotyping Core. Resulting FASTQ files were trimmed, aligned and normalized using BioWardrobe53, 54.
Normalized data were analyzed in R-Studio (R Version 3.6.1). By plotting gene count at escalating thresholds in each sample, we set a threshold of Log2(RPKM+1)>2 in 2 or more samples in eosinophils and Log2(RPKM+1)>2 in 2 or more samples in EPC2s, resulting in 6,591 and 7,457 genes in each data set, respectively. Pearson’s correlation coefficients were calculated using normalized counts of expressed genes. To obtain a list of genes whose expression varied across more than two treatment groups, analysis of variance (ANOVA) was performed on normalized values of all expressed genes, and p-values were corrected for a false discovery rate (q = 0.05). For pairwise comparisons, DEseq2 was used to calculate fold change and FDR-adjusted p values on the basis of total gene counts.
Gene ontology (GO) analysis was performed using the two-list setting in GOrilla (http://cbl-gorilla.cs.technion.ac.il/), and the full set of expressed genes as background. Gene set enrichment analysis (GSEA; Broad Institute, version 4.0.3) preranked analysis was used to compare gene expression in EPC2s cultured with and without eosinophils. Heatmaps were prepared and K means clustering were performed using Morpheus (https://software.broadinstitute.org/morpheus/). Gene expression data in this study were compared to genes identified in a murine model of EoE (GSE81135) and in a database of biopsies from EoE and control individuals (GSE58640)2, 23.
For the quantitative PCR analysis of gene expression, RNA was reverse transcribed with the ProtoScript II Reverse Transcriptase kit (New England Biotech, cat no. M0368) and gene expression was determined using a QuantStudio™ 7 Flex Real-Time PCR System from Applied Biosystems (Life Technologies) with PowerUp™ SYBR™ Green Master Mix (Thermo Fisher, cat no. A25742). Expression of ANXA6 (fwd 5’-TTACGATGCCAAGCAGTTGA-3’; rev 5’-GATTTCAGCATTGGTCCGAG-3’), CASP3 (fwd 5’- AAAATGGATTATCCTGAGATGGG-3’, rev 5’- CGACATCTGTACCAGACCGAG-3’), CD9 (fwd 5’-GACCAAGAGCATCTTCGAGC-3’; rev 5’-CGGCTCCGATCAGAATATAGAC-3’), CD83 (fwd 5’-CTCCGAAGATGTGGACTTGC-3’; rev 5’-GGGGTGTCTCCATCCTCTCT-3’), IL5RA (fwd 5’-AGCTGGGCTTCTGCTGAACT-3’; rev 5’-TTCTGTAGTGTTTGTGGTGCAAG-3’), NEK7 (fwd 5’-ACTAGCAGATGCTGGCGACC-3’; rev 5’-TCTTTCAGGAATTAGCCTCTTTTG-3’), SMAD4 (fwd 5’-AGCCTCCCATTTCCAATCAT-3’; rev 5’-CAATAGGGCAGCTTGAAGGA-3’), SPP1 (fwd 5’- CTGGAAGTTCTGAGGAAAAGCA-3’, rev 5’- AGTCAATGGAGTCCTGGCTG-3’), and TIA1 (fwd 5’-AACCGCTTAAACGATTTGGG-3’; rev 5’-CTCTGGAAAGGTTACCGACGTA-3’) were normalized to GAPDH (fwd 5’- TGGAAATCCCATCACCATCT-3’; rev 5’- GTCTTCTGGGTGGCAGTGAT-3’). Gene expression changes were calculated using the 2^(-delta delta CT) method using baseline expression in eosinophils and unstimulated monoculture of EPC2 cells for reference values.
Flow cytometry data and individual genes or multiplex analytes were plotted in GraphPad Prism (Version 8.0.1). Significance was determined by student’s t test or one-way ANOVA, as appropriate. P values were calculated using Graphpad Prism, and p < 0.05 was considered significant.
Supplementary Material
Acknowledgements
This study was supported by the NIH R01 A1045898. All flow cytometric data were acquired using equipment maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, and this work was supported in part by the Digestive Health Center (NIDDK P30 DK078392). The authors would like to thank Shawna Hottinger for editorial support.
Footnotes
Conflict of Interest: M.E.R. is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, Allakos, Celgene, Astra Zeneca, Arena Pharmaceuticals, Glaxo Smith Kline, Guidepoint and Suvretta Capital Management and has an equity interest in the first five listed and royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust) and UpToDate. M.E.R. is an inventor of patents owned by Cincinnati Children’s Hospital.
Disclosures
M.E.R. is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, Allakos, Celgene, Astra Zeneca, Arena Pharmaceuticals, Glaxo Smith Kline, Guidepoint and Suvretta Capital Management and has an equity interest in the first five listed and royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust) and UpToDate. M.E.R. is an inventor of patents owned by Cincinnati Children’s Hospital.
References
- 1.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol 2010; 185(1): 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben Baruch-Morgenstern N, Mingler MK, Stucke E, Besse JA, Wen T, Reichman H et al. Paired Ig-like Receptor B Inhibits IL-13-Driven Eosinophil Accumulation and Activation in the Esophagus. J Immunol 2016; 197(3): 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol 2015; 135(2): 500–507. [DOI] [PubMed] [Google Scholar]
- 4.Hirano I, Collins MH, Assouline-Dayan Y, Evans L, Gupta S, Schoepfer AM et al. RPC4046, a Monoclonal Antibody Against IL13, Reduces Histologic and Endoscopic Activity in Patients With Eosinophilic Esophagitis. Gastroenterology 2019; 156(3): 592–603 e510. [DOI] [PubMed] [Google Scholar]
- 5.Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS One 2015; 10(6): e0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol 2017; 140(3): 738–749 e733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet 2014; 46(8): 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight 2016; 1(4): e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. J Allergy Clin Immunol 2017; 139(6): 1762–1771 e1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masterson JC, McNamee EN, Fillon SA, Hosford L, Harris R, Fernando SD et al. Eosinophil-mediated signalling attenuates inflammatory responses in experimental colitis. Gut 2015; 64(8): 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara R, Lee EJ, Jang MS, Jeun EJ, Hong CP, Kim JH et al. Small intestinal eosinophils regulate Th17 cells by producing IL-1 receptor antagonist. J Exp Med 2016; 213(4): 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weller PF, Spencer LA. Functions of tissue-resident eosinophils. Nat Rev Immunol 2017; 17(12): 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozenberg P, Reichman H, Zab-Bar I, Itan M, Pasmanik-Chor M, Bouffi C et al. CD300f:IL-5 crosstalk inhibits adipose tissue eosinophil homing and subsequent IL-4 production. Sci Rep 2017; 7(1): 5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy 2010; 40(4): 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobsen EA, Lesuer WE, Willetts L, Zellner KR, Mazzolini K, Antonios N et al. Eosinophil activities modulate the immune/inflammatory character of allergic respiratory responses in mice. Allergy 2014; 69(3): 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah K, Ignacio A, McCoy KD, Harris NL. The emerging roles of eosinophils in mucosal homeostasis. Mucosal Immunol 2020; 13(4): 574–583. [DOI] [PubMed] [Google Scholar]
- 17.Simon HU, Yousefi S, Dibbert B, Levi-Schaffer F, Blaser K. Anti-apoptotic signals of granulocyte-macrophage colony-stimulating factor are transduced via Jak2 tyrosine kinase in eosinophils. Eur J Immunol 1997; 27(12): 3536–3539. [DOI] [PubMed] [Google Scholar]
- 18.Nutku-Bilir E, Hudson SA, Bochner BS. Interleukin-5 priming of human eosinophils alters siglec-8 mediated apoptosis pathways. Am J Respir Cell Mol Biol 2008; 38(1): 121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson MW, Gunderson KA, Kelly EA, Denlinger LC, Jarjour NN, Mosher DF. Anti-IL-5 attenuates activation and surface density of beta(2) -integrins on circulating eosinophils after segmental antigen challenge. Clin Exp Allergy 2013; 43(3): 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J Immunol 2014; 193(8): 4043–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han ST, Mosher DF. IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am J Respir Cell Mol Biol 2014; 50(3): 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egea L, Hirata Y, Kagnoff MF. GM-CSF: a role in immune and inflammatory reactions in the intestine. Expert Rev Gastroenterol Hepatol 2010; 4(6): 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol 2014; 7(3): 718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn JLM, Shoda T, Caldwell JM, Wen T, Aceves SS, Collins MH et al. Esophageal type 2 cytokine expression heterogeneity in eosinophilic esophagitis in a multisite cohort. J Allergy Clin Immunol 2020; 145(6): 1629–1640 e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu S, Kouzaki H, Ogawa T, Takezawa K, Tojima I, Shimizu T. Eosinophil-epithelial cell interactions stimulate the production of MUC5AC mucin and profibrotic cytokines involved in airway tissue remodeling. Am J Rhinol Allergy 2014; 28(2): 103–109. [DOI] [PubMed] [Google Scholar]
- 26.Kottyan LC, Rothenberg ME. Genetics of eosinophilic esophagitis. Mucosal Immunol 2017; 10(3): 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namjou B, Marsolo K, Caroll RJ, Denny JC, Ritchie MD, Verma SS et al. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to Eosinophilic Esophagitis. Front Genet 2014; 5: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnig C, Alsaleh G, Jung N, Dembele D, Paul N, Poirot A et al. Circulating Human Eosinophils Share a Similar Transcriptional Profile in Asthma and Other Hypereosinophilic Disorders. PLoS One 2015; 10(11): e0141740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 2020; 38(6): 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreev D, Liu M, Kachler K, Llerins Perez M, Kirchner P, Kolle J et al. Regulatory eosinophils induce the resolution of experimental arthritis and appear in remission state of human rheumatoid arthritis. Ann Rheum Dis 2020. [DOI] [PubMed] [Google Scholar]
- 31.Luttmann W, Knoechel B, Foerster M, Matthys H, Virchow JC Jr., Kroegel C. Activation of human eosinophils by IL-13. Induction of CD69 surface antigen, its relationship to messenger RNA expression, and promotion of cellular viability. J Immunol 1996; 157(4): 1678–1683. [PubMed] [Google Scholar]
- 32.Le-Carlson M, Seki S, Abarbanel D, Quiros A, Cox K, Nadeau KC. Markers of antigen presentation and activation on eosinophils and T cells in the esophageal tissue of patients with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2013; 56(3): 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullock JZ, Villanueva JM, Blanchard C, Filipovich AH, Putnam PE, Collins MH et al. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2007; 45(1): 22–31. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya B, Carlsten J, Sabo E, Kethu S, Meitner P, Tavares R et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol 2007; 38(12): 1744–1753. [DOI] [PubMed] [Google Scholar]
- 35.Johnsson M, Bove M, Bergquist H, Olsson M, Fornwall S, Hassel K et al. Distinctive blood eosinophilic phenotypes and cytokine patterns in eosinophilic esophagitis, inflammatory bowel disease and airway allergy. J Innate Immun 2011; 3(6): 594–604. [DOI] [PubMed] [Google Scholar]
- 36.Penido C, Castro-Faria-Neto HC, Vieira-de-Abreu A, Figueiredo RT, Pelled A, Martins MA et al. LPS induces eosinophil migration via CCR3 signaling through a mechanism independent of RANTES and Eotaxin. Am J Respir Cell Mol Biol 2001; 25(6): 707–716. [DOI] [PubMed] [Google Scholar]
- 37.Fulkerson PC, Fischetti CA, McBride ML, Hassman LM, Hogan SP, Rothenberg ME. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci U S A 2006; 103(44): 16418–16423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trautmann A, Schmid-Grendelmeier P, Kruger K, Crameri R, Akdis M, Akkaya A et al. T cells and eosinophils cooperate in the induction of bronchial epithelial cell apoptosis in asthma. J Allergy Clin Immunol 2002; 109(2): 329–337. [DOI] [PubMed] [Google Scholar]
- 39.Pope SM, Zimmermann N, Stringer KF, Karow ML, Rothenberg ME. The eotaxin chemokines and CCR3 are fundamental regulators of allergen-induced pulmonary eosinophilia. J Immunol 2005; 175(8): 5341–5350. [DOI] [PubMed] [Google Scholar]
- 40.Johansson MW, Kelly EA, Nguyen CL, Jarjour NN, Bochner BS. Characterization of Siglec-8 Expression on Lavage Cells after Segmental Lung Allergen Challenge. Int Arch Allergy Immunol 2018; 177(1): 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fulkerson PC, Rothenberg ME. Eosinophil Development, Disease Involvement, and Therapeutic Suppression. Adv Immunol 2018; 138: 1–34. [DOI] [PubMed] [Google Scholar]
- 42.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut 2010; 59(1): 21–30. [DOI] [PubMed] [Google Scholar]
- 43.Assa’ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology 2011; 141(5): 1593–1604. [DOI] [PubMed] [Google Scholar]
- 44.Otani IM, Anilkumar AA, Newbury RO, Bhagat M, Beppu LY, Dohil R et al. Anti-IL-5 therapy reduces mast cell and IL-9 cell numbers in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol 2013; 131(6): 1576–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson RK, Brickner H, Panwar B, Ramirez-Suastegui C, Herrera-de la Mata S, Liu N et al. Human Eosinophils Express a Distinct Gene Expression Program in Response to IL-3 Compared with Common beta-Chain Cytokines IL-5 and GM-CSF. J Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol 2008; 121(6): 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto K, Appiah-Pippim J, Schleimer RP, Bickel CA, Beck LA, Bochner BS. CD44 and CD69 represent different types of cell-surface activation markers for human eosinophils. Am J Respir Cell Mol Biol 1998; 18(6): 860–866. [DOI] [PubMed] [Google Scholar]
- 48.Barthel SR, Jarjour NN, Mosher DF, Johansson MW. Dissection of the hyperadhesive phenotype of airway eosinophils in asthma. Am J Respir Cell Mol Biol 2006; 35(3): 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichman H, Itan M, Rozenberg P, Yarmolovski T, Brazowski E, Varol C et al. Activated Eosinophils Exert Antitumorigenic Activities in Colorectal Cancer. Cancer Immunol Res 2019; 7(3): 388–400. [DOI] [PubMed] [Google Scholar]
- 50.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest 2019; 130: 2014–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodavanti UP, Jaskot RH, Bonner J, Badgett A, Dreher KL. Eosinophilic lung inflammation in particulate-induced lung injury: technical consideration in isolating RNA for gene expression studies. Exp Lung Res 1996; 22(5): 541–554. [DOI] [PubMed] [Google Scholar]
- 52.Toni LS, Garcia AM, Jeffrey DA, Jiang X, Stauffer BL, Miyamoto SD et al. Optimization of phenol-chloroform RNA extraction. MethodsX 2018; 5: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouffi C, Kartashov AV, Schollaert KL, Chen X, Bacon WC, Weirauch MT et al. Transcription Factor Repertoire of Homeostatic Eosinophilopoiesis. J Immunol 2015; 195(6): 2683–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallabh S, Kartashov AV, Barski A. Analysis of ChIP-Seq and RNA-Seq Data with BioWardrobe. Methods Mol Biol 2018; 1783: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


