Abstract
This commentary provides background, historical context, and a critical assessment of the concept that microbial dysbiosis drives the pathogenesis of periodontal diseases. It is long known that periodontal pathogenesis is dependent on tooth-borne microbial biofilms (dental plaque) that trigger host inflammation resulting in periodontal destruction and tooth loss in some patients. Ecological principles governing plaque biofilm development, along with localized host responses, are both rooted in evolution. Interpretation of available evidence suggests that, in most patients, alveolar bone loss results from interactions of a highly diverse commensal microbiota with the host, and not from “overgrowth” of a few “pathobionts” that results in a “dysbiosis.” Most previously described dysbiotic chronic diseases, for example, inflammatory bowel diseases and dermatitis, are characterized by decreased microbial diversity (likely due to frank overgrowth of one or a few microbial taxa). Most common forms of periodontitis do not appear to conform to this general principle, and the associated microbiome in fact almost always shows increased bacterial diversity compared with periodontal health. This diversity is driven by interactions of genetic and environmental factors working in concert within specific windows of time. Periodontal pathogenesis is likely the result of “personalized pathology,” insofar as each patient likely has a variable constellation of microbes and host risk factors influencing specific tissue sites where disease activity occurs, and during a limited window of time (a tissue-destructive “burst”). The concept of cooperative virulence of higher abundance commensals in periodontal pathogenesis, which does not conform to the model of dysbiosis observed for other diseases, is discussed.
Keywords: calculus, genomics, host-parasite interactions, microbiology, pathogenesis of periodontal diseases
1 ∣. INTRODUCTION
Our understanding of dental biofilms (dental plaque), a complex microbial ecosystem that drives the pathogenesis of periodontitis, has dramatically evolved over the past century. Human periodontitis results from a polymicrobial insult, including as yet unidentified and never cultivated bacterial species,5 and possibly viruses, fungi, and parasites. A recent model to explain periodontal pathogenesis involves microbial dysbiosis initiated by keystone pathogens,1 which describes a state of resident microbiota imbalances that are incompatible with health. However, the number and identities of member bacterial species of a dysbiotic periodontal microbiome often differ between individuals within study groups, and between groups with similar disease types in different studies.2-4 Also, microbial community structure shifts due to host or environmental pressures do not always accompany tissue destruction.5-7 Thus, the term dysbiosis may not apply to common cases of periodontal disease.
Detailed and accurate knowledge of the pathogenesis of periodontitis has important clinical implications with respect to diagnosis and treatment of this complex and consequential disease. The goal of this broad view (“from 35,000 feet”) commentary is to provide historical context and a critical assessment of the recent concept that microbial dysbiosis explains the etiology of periodontitis.
2 ∣. TRADITIONAL IDEAS OF INFECTION AND ORAL DISEASES
Research methods devised by pioneer scientists such as Pasteur and Koch in the mid-1800s showed that several diseases result from infection by a single, virulent pathogen (e.g., tuberculosis, cholera, rabies, Streptococcus pneumoniae infection, etc.).8 From this work, a “pathogen” was defined as a biological agent causing disease or illness to its host, disrupting normal physiology or tissue integrity. Oral microbiologists have since searched to identify one or a few pathogenic species that cause the most common oral diseases, dental caries, and periodontitis.
Starting in the 1950s, research focused on how dental plaque was causally linked to gingivitis and periodontitis.9 By 1970, Socransky summarized the evidence on the role of plaque bacteria in the initiation, progression, and experimental transmission of periodontal disease.10 The extent of gingival inflammation and bone loss was already known to be related to the amount of dental plaque, and removing plaque from teeth slowed, and often prevented, the progression of disease. Other studies found dental plaque-induced periodontal bone loss in animals, and pure cultures of (often) anaerobic bacteria isolated from human periodontitis plaque colonized animals resulting in a “syndrome” similar to human periodontitis. The consensus view was that all bacteria in plaque contribute to tissue destruction supporting the “non-specific plaque hypothesis.”
Socransky also cited evidence that mixed infections with fusobacteria and spirochetes, as well as monocultures of specific strains could induce periodontitis in animals.10 He suggested these bacteria release cytotoxic products that injure the gingival tissues, or induce inflammation resulting in periodontal destruction. Löe et al. developed the human experimental gingivitis model and showed that removal of tooth biofilm is followed by recolonizing bacteria, with Gram-positive cocci (Firmicutes), Gram-positive rods (Actinomyces), and Gram-negative cocci (Veillonella) dominating. Within 48 to 96 hours, transitional species appear (Fusobacteria, Corynebacteria, and other species), coinciding with the initiation of gingivitis.11 Recent microbiome studies of experimental gingivitis found enrichment of TM7, Tannerella, Cardiobacterium, Campylobacter, Porphyromonas, Leptotrichia, and Selenomonas spps.,12 whereas others reported Actinomyces, Rothia, Veillonella, and Spirochetes associated with the climax community.13 Consistent associations between specific bacterial species and clinical disease parameters reported in the 1970s led to the proposed “specific plaque hypothesis” by Loesche.14 Indeed, many papers to this day often cite specific plaque bacterial species as “the” cause of periodontitis and associated systemic diseases.
3 ∣. ECOLOGIC MODELS OF DENTAL PLAQUE BIOFILM DEVELOPMENT
Stable microbial colonization of host habitats, including tooth surfaces, takes place by selective adaptation over the course of evolution, in a bidirectional relationship with the host defense system,15,16 following principles of natural selection. Ecological succession orchestrates changes in species membership within a community over time. In the case of the natural history of a forest, the time for succession scale can be decades (e.g., after a wildfire), or even millions of years (after a mass extinction). For a microbial habitat, the time scale can be considerably shorter, starting with a small number of pioneer species, followed by community succession with increasing complexity until the community becomes stable and self-perpetuating. The “engines” of succession are the impact of established organisms upon their own environments, together with environmental or host defense-mediated perturbations. When ≥2 species use the same source of nutrients, some will be compromised, and others excluded. Ultimately, a steady state results where multiple species with mutualistic relationships coexist over long periods of time. Variations in environmental conditions may allow one or another species in a mixed population to “bloom,” and new species sometimes enter the competition, and others die off. Processes such as synergism (one species provides nutrients to another) and antagonism (one species produces products toxic to another) also contribute to the resulting equilibrium.
A variety of mechanisms have been proposed to explain microbial succession in dental plaque. In the early 1960s, Loesche proposed that nutritional networks between bacteria control plaque development, with some bacteria producing substances used as nutrients by other bacteria.17 Later, Gibbons and co-workers discovered that the attachment of bacteria to teeth was essential for bacterial colonization and plaque development.18
4 ∣. PERIODONTITIS AS THE RESULT OF POLYMICROBIAL ACTIVITY
Socransky and coworkers proposed the red complex, a group of Gram negative anaerobic species (Porphyromonas gingivalis, Tanerella forsythia, and Treponema denticola) that together cause periodontal disease.19 There is continued interest to identify “specific pathogens” or “pathobionts,” and “keystones,” low abundant but particularly subversive species, that initiate the process.1,20 One hope is for a vaccine or drug therapy specific for one or only a few bacteria to prevent periodontitis.
The idea that periodontitis is the result of true polymicrobial activity is supported by much evidence. Molecular microbiome studies show both cultivable and never-cultivated bacteria amongst the diverse group of microbes associated with disease.3,4,21,22 While reported findings are in general agreement at the phylum level, high abundance bacteria associated with periodontitis vary at the species level.2-4,21 Metatranscriptomic analysis reveals that certain metabolic signatures are consistent with disease progression, suggesting the whole community, and not just a handful of oral pathogens, drive disease progression.23 Also, not every tooth site with tissue destruction harbor suspected pathogens, and longitudinal studies report no major differences between the cultivable subgingival microbiota of periodontal sites with recent increased pocket depth compared with stable sites.21 To further complicate matters, recent microbiome studies from multiple groups have established that the most abundant species, such as Filifactor alocis, P. gingivalis, and T. denticola, which increase in disease, still represent <4% of the total microbial species involved.2,21
Hajishengallis and Lamont proposed a model whereby periodontitis is initiated by a dysbiotic and synergistic microbiota, as opposed to a conventional infectious disease caused by a single or a few select species, such as the "red complex."20 They argued that diverse bacteria (or specific combinations of genes within the community) fulfill distinct roles that converge to form a stable, disease-provoking microbiota. The microbes within the biofilm work together to resist the host responses and drive inflammation, with concomitant increased proteolytic activity and cytokine induction that results in tissue destruction.
Yet, many papers continue to promote one species or another as the cause of disease or the major driver of dysbiosis (often red complex species or Fusobacterium nucleatum). Some investigators, and dental practitioners, still monitor only one or a few species as indicators of disease activity. Indeed, treatment in the form of antibiotics, is sometimes prescribed based on the use of these species as biomarkers, even in the absence of strong evidence of efficacy.
5 ∣. ORAL BIOFILMS AND PERIODONTITIS FROM AN EVOLUTIONARY PERSPECTIVE
Almost all bacteria found in dental biofilms are commensals, or members of the health-associated microbiota, that have evolved with the human host over millions of years. Thus, none of these organisms should a priori be considered true pathogenic species. High-throughput sequencing studies of human dental calculus (tartar, or calcified dental plaque) from ancient remains, which serve as a long-term biorepository for microorganisms,24 show that the microbiome in ancient calculus broadly resembles that of contemporary humans.24
The transition from human hunter-gatherer to farming practice substantially shifted the oral microbial community, where more Gram-negative species become common.25 The composition of the oral microbiota remained unexpectedly constant between Neolithic and medieval times, after which (the now ubiquitous) cariogenic bacteria became dominant, apparently as a result of the increased exposure to a refined-sugar containing diet.26
Modern gut microbial ecosystems are markedly less diverse than those from prehistoric populations.27 This reduced diversity may result from the fiber rich diet of ancient humans compared with modern humans, and that ancient people were not exposed to commercial antimicrobials, which diminish microbial diversity. Similar observations have been noted for the oral microbiome.28 While we now consider periodontal bone loss as a pathogenic process, it is possible that periodontal inflammation evolved as a “normal” response to contain the oral microbiota, and protect the host from assault by diverse indigenous microbes. Since the average life-span of ancient humans was considerably shorter than that of present humans, alveolar bone loss likely did not reduce the survival of most prehistoric humans. Since periodontitis often manifests in older individuals, it probably did not impact individual evolutionary fitness. Interestingly, Manji et al. have recently argued that progression and regression of both caries and periodontitis may be considered as “natural,” rather than pathological events, even if left unchecked over long periods of time, and they can result in tissue destruction.29 The likelihood of progression or regression of tissue damage can be influenced by other determinants, but these processes could nevertheless occur in the absence of such influences.29
6 ∣. PERIODONTITIS AS A CHRONIC DYSBIOTIC DISEASE
The term dysbiosis was first mentioned in a 1959 paper addressing gastrointestinal disturbance in infants.30 By the 1980s, dysbiosis was implicated in the pathogenesis of a number of gut diseases, for example., inflammatory bowel disease, ulcerative colitis, and Crohn’s disease.31 In fact, the dysbiosis described for these conditions appears as enrichment of the gastrointestinal microbiota (including in the stomach, small intestine, and bowel) by unusual species such as Proteus, coincident with a sharp decrease in intestinal absorption. Disease signs and symptoms were mimicked in mice infected with Proteus.32
While early studies were limited by the use of culture techniques, more recent studies using molecular methods that assess cultivable and “non-cultivable” gut microbes corroborate earlier impressions. Thus, in most cases of chronic GI tract diseases, microbial dysbiosis is accompanied by reductions in microbiome diversity, likely due to overgrowth of select species.33,34 Other diseases such as dermatitis are also characterized by decreased microbial diversity.35
Not all chronic infectious diseases show reduced microbial diversity. Bacterial vaginosis shows an increase in vaginal mucosal bacterial diversity, which in health is dominated by Lactobacillus species maintaining a low vaginal pH.36 Vaginal dysbiosis-related changes can vary among individuals, but typically involve enrichment with anaerobic bacterial pathobionts that outcompete lactobacilli,36 and thus increase the local diversity of the microbiome. Similarly, there is greater bacterial diversity in the lower airways of the asthmatic patient compared with health.37 However, in these cases, pathobionts mostly replace commensal species, whereas in the periodontium health-associated species are not generally lost, but represent a smaller fraction of the total community as the plaque biomass increases.2,21 Importantly, compared with the oral cavity and GI tract, the healthy vaginal and lower respiratory mucosa are sparsely colonized and have low bacterial diversity.38 It is not surprising that disease-associated dysbiotic shifts manifest as an increase in diversity since expansion of a smaller number of bacterial species in these mucosal sites represent dysbiotic shifts, compared with the periodontium. Although proinflammatory activities of some of these species was confirmed experimentally, whether or not an altered microbiome contributes to pathogenesis or is only a marker of inflammation remains unresolved.39,40 Note that diversity of the upper respiratory tract mucosa (where diversity in health resembles the oral cavity) is significantly reduced in patients with pneumonia compared with healthy controls,39 demonstrating another site where high microbial diversity in health changes to reduced diversity in disease. In contrast, the microbiota associated with periodontal inflammation is almost always increased in diversity.2-4,21
Most studies that describe dysbiosis in periodontal diseases often have methodologic and analytical limitations. First and foremost, as there is no formal accepted definition of active periodontitis, and disease activity occurs intermittently in most patients,41 it is likely that sampling is often performed when the disease is not active. Thus, correlating a microbiota to disease activity is problematic. Also, most studies are cross-sectional, and therefore results from them cannot define causality. It is interesting to note that recent longitudinal studies that combine metagenomics and metatranscriptomics analyses to compare health with disease, or progressing and not progressing sites, report that a large number of bacterial taxa are increased in patients with periodontitis when compared with those with periodontal health.3,42 These findings support the concept of synergistic virulence of metabolically active commensals and not expansion of specific pathobionts, which is the hallmark of most dysbiotic diseases.
Most cross-sectional oral microbiome studies have analyzed samples between groups, not within-groups, and used pooled samples, either from a tooth, from multiple teeth, from saliva, or from a mucosal swab. Pooling of samples may dilute the proportion of taxa in a sample. As few studies assess quantities or proportions of taxa at specific sites, it is not possible to assess variation in quantities of species between tooth sites.43 Studies are also lacking that localize specific species at microscopic scale within a site, and then correlate findings to disease activity at that site.
While microbiome studies typically provide relative abundance data for each species as a percentage of their community members, few microbiome studies validate their high-throughput sequencing data with quantitative polymerase chain reaction or cultivation methods to confirm higher biomass in species that exist in higher relative abundance. Traditional plaque sampling methods (using curets or paper points) do not allow for standardization of the amount or the location (more superficial biofilm versus deeper biofilm layers) of the sample, and therefore biomass estimates may not be comparable between sites. However, an increase in relative abundance of a species in a disease site may be a result of a decrease in the biomass of other species in the community and not expansion of the species.21 Similarly, another microbiome study debunked the idea that Gram-negative biomass increases in disease since an increase in their relative abundance could be explained by a decrease in the proportions of health-associated species of streptococci.2
Thus, it appears that periodontal pathogenesis is much more complicated than what could be addressed by any single study. Indeed, it is likely that pathogenesis is the result of “personalized pathology,” insofar as each patient likely has a variable constellation of microbes and host risk factors influencing specific tissue sites where disease activity occurs, and during a limited window of time (a tissue-destructive “burst”).41 The traditional concept of dysbiosis, as described for other diseases, does not seem to adequately describe the situation for most common forms of periodontitis (chronic or adult periodontitis) at the individual subject or site level. Studies of longitudinal, integrated multiomics data including site-specific host and microbiota profiles are needed to further clarify this issue.
7 ∣. PERIODONTAL “DYSBIOSIS” REVISITED
One idea to explain microbial shifts is that inflammation in certain individuals triggers microbiome composition changes by altering the growth environment.20 A positive feedback cycle amplifies the inflammatory response to promote further inflammation and additional changes to microbiome composition leading to release of potent host proteases and the loss of tooth connective tissue attachment and alveolar bone resorption. This model assumes that certain bacteria trigger a cascade of events that enriches putative periodontal pathogens. This model may involve specific “keystone” pathogens (e.g., P. gingivalis).20 But are these truly pathogens? If so, how many pathogenic taxa influence disease?
The idea that the local inflammatory or hypoxic microenvironment alters the composition of the microbiome is not supported by most human studies. In patients with periodontitis, neither increasing probing depth, which reduces local oxygen tension,2,4 nor presence of bleeding, a marker of inflammation,21 have been associated with differences in microbial community relative abundance or structure. Thus, while periodontitis may result from a whole-mouth dysbiosis rather than site-specific changes, this cannot be reconciled with the fact that periodontitis is sometimes a localized disease.
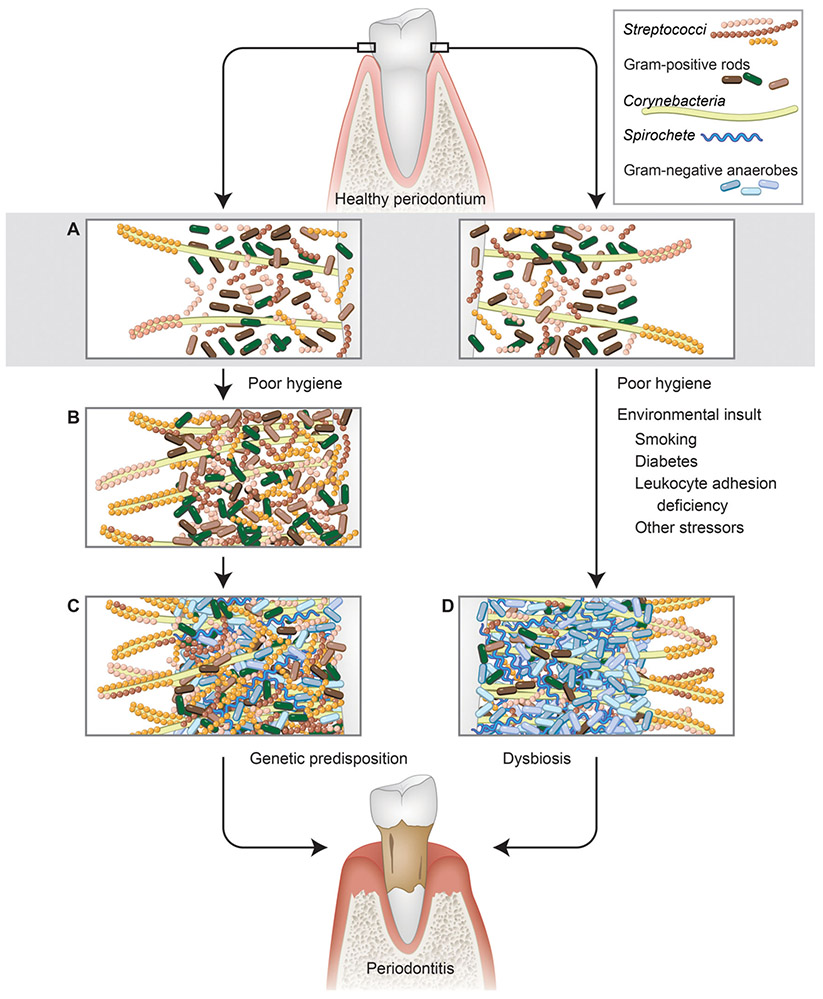
Microbiome analyses of patients with systemic diseases report that the host background and systemic host status influences the microbiome composition more than disease severity.4,6,44 For example, dental plaque in patients with rheumatoid arthritis with a healthy periodontium is enriched with Gram-negative anaerobes that are compatible with periodontal homeostasis. This is an example where the host systemic background drives microbiome changes that are not consequential to periodontal condition and thus, at least based on cross-sectional data, cannot be defined as dysbiosis. Along the same lines, diabetes triggers changes in subgingival community structure, notably characterized by a reduction in diversity (Fig. 1), which in certain individuals does not cause clinical evidence of disease.4,44,45
FIGURE 1.
Alternative models of plaque biofilm development in health and disease. A) Early biofilm dominated by initial colonizers such as streptococci and Gram-positive rods such as Actinomyces. B) Mature plaque, with ecological succession that allows for colonization of diverse organisms, including multiple streptococcal and Gram-positive rod species, and later introduction of filamentous forms such as Corynebacteria that serve as scaffolds for streptococci and other species. This stage of development, which results from poor hygiene that allows biofilm development, is associated with gingivitis. C) Continued succession allowing for colonization by diverse Gram-negative anaerobic species and spirochetes. This stage of development is associated with periodontitis. D) Aberrant development resulting in dysbiotic plaque biofilm with reduced diversity, as influenced by various environmental perturbations (smoking, diabetes, stress, etc.). Both outcomes require a permissible host genetic background. 1) Streptococci. 2) Gram-positive rods. 3) Corynebacteria. 4) Spirochetes. 5) Gram-negative anaerobe. (Illustration by Heather McDonald, BioSerendipity, LLC, Elkridge, MD, USA.)
The subgingival plaque from smokers with periodontitis is less diverse than that of periodontally healthy smokers or non-smokers, suggesting a true dysbiosis (Fig. 1).4 However, subgingival Gram-negative anaerobe enrichment caused by smoking did not lead to periodontal changes in a different cohort studied by the same investigative team.5 Similar findings have been described for e-cigarette use.7 Although disease-associated changes in microbial gene expression are observed, these changes were attributed to species that do not differ in abundance between health and disease.23 This suggests that it is the bacterial community metabolic activity rather than species composition that drives disease. Prospective evidence linking subgingival microbiome changes to disease development is required to prove dysbiosis as a cause of disease.
An increase in bacterial diversity in periodontitis in systemically healthy hosts may not reflect true dysbiosis, but the succession of microbiota over time to an ecologically stable state, without overgrowth of pathobionts. High-throughput sequencing studies reproduced earlier culture findings showing a diverse subgingival microbiome, and also expanded the spectrum of disease-associated phyla (e.g., Synergistes) or species (e.g., Filifactor alocis). Shifts in community membership and structure from health to periodontitis suggests ecological succession to potentially more diverse and stable communities.2,21 High-throughput studies show an increase in the biomass of health-associated core species and periodontitis-associated species.2,21 Thus, total microbial biomass and not community membership emerges again as a significant disease driver, in harmony with the non-specific plaque hypothesis of periodontal pathogenesis. These findings discourage the use of systemic antibiotics to treat most cases of periodontitis in systemically healthy patients.46
8 ∣. PERIODONTAL DYSBIOSIS AND HOST GENETICS
Host-genetic determinants that increase periodontal disease susceptibility may influence the subgingival microbiome to drive dysbiosis. As an example, the subgingival microbiome of patients with leukocyte adhesion deficiency shows significantly reduced diversity, with fewer, unique and more invasive species associated with severe periodontal destruction. Such a microbiome is not found in common forms of periodontitis in healthy hosts, and is thus more consistent with dysbiosis.47 Similarly, a reduction in diversity has been reported in patients with aggressive periodontitis with tissue-invasive bacteria thought to overwhelm other members of the microbiota.23
9 ∣. SUMMARY
We posit that periodontitis in systemically healthy adults is not a result of a dysbiosis but an increase in the biomass of a highly diverse microbiota that has evolved over time in each individual. In genetically susceptible or systemically compromised individuals, environmental perturbations sometimes drives a reduction in diversity which results in a more severe disease phenotype that represents true dysbiosis. In all cases the commensal microbiome, through cooperative virulence, influences periodontal disease phenotypes. To better define the role for bacteria in disease pathogenesis, more accurate models for periodontal pathogenesis are required, and new methods to accurately identify periodontal disease activity are essential to pinpoint the character of the microbiota present at the site at the time of disease activity. Microbial spatial patterning present within subgingival plaque may influence site-specific disease activity.48 Further development of imaging methods to characterize the biogeography of plaque biofilm development in situ during human disease might localize different taxa of microbes within plaque biofilm to compare healthy and disease sites, and sites that break down over time. Finally, detailed study of a larger number of species, including heretofore uncultivated microbial species, in periodontal pathogenesis would help to illuminate their role in disease pathogenesis.
ACKNOWLEDGMENTS
Anna Dongari-Bagtzoglou received funding by grant RO1 DE013986 from the National Institutes of Health. The authors report no conflicts of interest related to this commentary.
REFERENCES
- 1.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganesan SM, Joshi V, Fellows M, et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11:2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paropkari AD, Leblebicioglu B, Christian LM, Kumar PS. Smoking, pregnancy and the subgingival microbiome. Sci Rep. 2016;6:30388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Oliva I, Paropkari AD, Saraswat S, et al. Dysbiotic subgingival microbial communities in periodontally healthy patients with rheumatoid arthritis. Arthritis Rheumatol. 2018;70:1008–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganesan SM, Dabdoub SM, Nagaraja HN, et al. Adverse effects of electronic cigarettes on the disease-naive oral microbiome. Sci Adv. 2020;6:eaaz0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossel PP. Pasteur, Koch and American bacteriology. Hist Philos Life Sci. 2000;22:81–100. [PubMed] [Google Scholar]
- 9.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000. 2013;62:95–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Socransky SS. Relationship of bacteria to the etiology of periodontal disease. J Dent Res. 1970;49:203–222. [DOI] [PubMed] [Google Scholar]
- 11.Löe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177–187. [DOI] [PubMed] [Google Scholar]
- 12.Al-Kamel A, Baraniya D, Al-Hajj WA, et al. Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J Oral Microbiol. 2019;11:1608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schincaglia GP, Hong BY, Rosania A, et al. Clinical, immune, and microbiome traits of gingivitis and peri-implant mucositis. J Dent Res. 2017;96:47–55. [DOI] [PubMed] [Google Scholar]
- 14.Loesche WJ. Clinical and microbiological aspects of chemotherapeutic agents used according to the specific plaque hypothesis. J Dent Res. 1979;58:2404–2412. [DOI] [PubMed] [Google Scholar]
- 15.Alexander M Microbial Ecology. New York: John Wiley; 1971. [Google Scholar]
- 16.Barton LL, Northrup DE. Microbial Ecology. Hoboken, NJ: Wiley-Blackwell; 2011. [Google Scholar]
- 17.Loesche WJ. Importance of nutrition in gingival crevice microbial ecology. Periodontics. 1968;6:245–249. [PubMed] [Google Scholar]
- 18.Gibbons RJ, Houte Jv. Bacterial adherence in oral microbial ecology. Ann Rev Microbiol. 1975;29:19–44. [DOI] [PubMed] [Google Scholar]
- 19.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. [DOI] [PubMed] [Google Scholar]
- 20.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartold PM, Van Dyke TE. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J Clin Periodontol. 2019;46:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. mBio. 2014;5:e01012–01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warinner C, Rodrigues JF, Vyas R, et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genetics. 2014;46:336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adler CJ, Dobney K, Weyrich LS, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and industrial revolutions. Nat Genetics. 2013;45:450–455.455e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanzer JM. Dental caries is a transmissible infectious disease: the keyes and fitzgerald revolution. J Dent Res. 1995;74:1536–1542. [DOI] [PubMed] [Google Scholar]
- 27.Moeller AH. The shrinking human gut microbiome. Curr Opin Microbiol. 2017;38:30–35. [DOI] [PubMed] [Google Scholar]
- 28.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manji F, Dahlen G, Fejerskov O. Caries and periodontitis: contesting the conventional wisdom on their aetiology. Caries Res. 2018;52:548–564. [DOI] [PubMed] [Google Scholar]
- 30.Haenel H, Schmidt EF, Feldheim G. Fecal dysbiosis in infancy. Z Kinderheilkd. 1959;82:595–603. [PubMed] [Google Scholar]
- 31.Godoy-Vitorino F Human microbial ecology and the rising new medicine. Ann Transl Med. 2019;7:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanareykina SK, Misautova AA, Zlatkina AR, Levina EN. Proteus dysbioses in patients with ulcerative colitis. Die Nahrung. 1987;31:557–561. [DOI] [PubMed] [Google Scholar]
- 33.Magro DO, Santos A, Guadagnini D, et al. Remission in Crohn’s disease is accompanied by alterations in the gut microbiota and mucins production. Sci Rep. 2019;9:13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colquhoun C, Duncan M, Grant G. Inflammatory bowel diseases: host-microbial-environmental interactions in dysbiosis. Diseases. 2020;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilera AC, Dagher IA, Kloepfer KM. Role of the microbiome in allergic disease development. Curr Allergy Asthma Rep. 2020;20:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgdorff H, Gautam R, Armstrong SD, et al. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol. 2016;9:621–633. [DOI] [PubMed] [Google Scholar]
- 37.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. Plos One. 2010;5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez R, Banerjee S, Bhattacharya SK, Banerjee S. Lung inflammation and disease: a perspective on microbial homeostasis and metabolism. Iubmb Life. 2019;71:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. Plos Pathogens. 2015;11:e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Teijlingen NH, Helgers LC, Zijlstra-Willems EM, et al. Vaginal dysbiosis associated-bacteria megasphaera elsdenii and prevotella timonensis induce immune activation via dendritic cells. J Reprod Immunol. 2020;138:103085. [DOI] [PubMed] [Google Scholar]
- 41.Socransky SS, Haffajee AD, Goodson JM, Lindhe J. New concepts of destructive periodontal disease. J Clin Periodontol. 1984;11:21–32. [DOI] [PubMed] [Google Scholar]
- 42.Duran-Pinedo AE, Chen T, Teles R, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proctor DM, Shelef KM, Gonzalez A, et al. Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontol 2000. 2020;82:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabharwal A, Ganley K, Miecznikowski JC, Haase EM, Barnes V, Scannapieco FA. The salivary microbiome of diabetic and non-diabetic adults with periodontal disease. J Periodontol. 2019;90:26–34. [DOI] [PubMed] [Google Scholar]
- 45.Farina R, Severi M, Carrieri A, et al. Whole metagenomic shotgun sequencing of the subgingival microbiome of diabetics and non-diabetics with different periodontal conditions. Arch Oral Bioly. 2019;104:13–23. [DOI] [PubMed] [Google Scholar]
- 46.Smiley CJ, Tracy SL, Abt E, et al. Evidence-based clinical practice guideline on the nonsurgical treatment of chronic periodontitis by means of scaling and root planing with or without adjuncts. J Am Dent Assoc. 2015;146:525–535. [DOI] [PubMed] [Google Scholar]
- 47.Moutsopoulos NM, Chalmers NI, Barb JJ, et al. Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. Plos Pathogens. 2015;11:e1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mark Welch JL, Ramirez-Puebla ST, Borisy GG. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe. 2020;28:160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]