Summary
The process of pyroptosis is mediated by inflammasomes and a downstream effector known as gasdermin D (GSDMD). Upon cleavage by inflammasome-associated caspases, the N-terminal domain of GSDMD forms membrane pores that promote cytolysis. Numerous proteins promote GSDMD cleavage, but none are known to be required for pore formation after GSDMD cleavage. Herein, we report a forward genetic screen that identified the Ragulator-Rag complex as being necessary for GSDMD pore formation and pyroptosis in macrophages. Mechanistic analysis revealed that Ragulator-Rag is not required for GSDMD cleavage upon inflammasome activation, but rather promotes GSDMD oligomerization in the plasma membrane. Defects in GSDMD oligomerization and pore formation can be rescued by mitochondrial poisons that stimulate reactive oxygen species (ROS) production, and ROS modulation impacts the ability of inflammasome pathways to promote pore formation downstream of GSDMD cleavage. These findings reveal an unexpected link between key regulators of immunity (inflammasome-GSDMD) and metabolism (Ragulator-Rag).
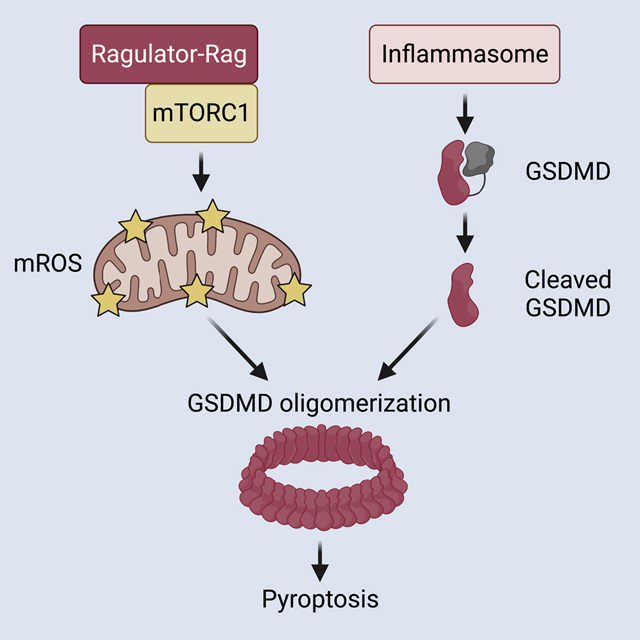
Graphical Abstract
The Ragulator-Rag complex is required for the oligomerization of gasdermin D at the plasma membrane and for pore formation and pyroptosis in macrophages.
Introduction
The link between cell death and inflammation has long-been recognized, with certain death processes inducing immuno-suppressive responses (e.g. apoptosis) and others inducing inflammatory responses (e.g. pyroptosis). Central to the inflammatory process of pyroptosis is the protein gasdermin D (GSDMD) (Kayagaki et al., 2015; Shi et al., 2015), which forms pores in the plasma membrane of cells that can result in lysis and the release of intracellular inflammatory mediators (Kovacs and Miao, 2017; Lieberman et al., 2019). Among these inflammatory mediators are interleukin (IL)-1 family cytokines (Dinarello, 2018), which are cytosolic proteins that can be released from living (Evavold et al., 2018; Heilig et al., 2018) or pyroptotic (Kayagaki et al., 2015; Shi et al., 2015) cells after GSDMD pore formation. In contrast to their pyroptotic counterparts, apoptotic cells typically maintain plasma membrane integrity and therefore do not release inflammatory mediators. Distinctions in plasma membrane integrity therefore (in part) explain inflammatory or non-inflammatory consequences of the different processes of cell death.
Two known mechanisms explain GSDMD pore formation and inflammation. The first mechanism involves the actions of inflammasomes, which are supramolecular organizing centers that function as the subcellular sites of caspase-1 activation (Chan and Schroder, 2020). Caspase-1 is a dormant enzyme in resting cells. Upon infection or select disruptions of cellular homeostasis, inflammasomes are assembled in the cytosol that recruit and activate caspase-1. This enzyme cleaves GSDMD into two fragments. The N terminal fragment oligomerizes and inserts into the plasma membrane where it forms pores of 10–20 nm in inner diameter (Aglietti et al., 2016; Ding et al., 2016; Liu et al., 2016; Ruan et al., 2018; Sborgi et al., 2016). The second means by which pyroptosis is induced is by the actions of murine caspase-11 (or caspase-4 and -5 in humans). The catalytic activity of these caspases is not stimulated by recruitment into inflammasomes. Rather, catalytic activity is stimulated upon binding to bacterial lipopolysaccharides (LPS) (Hagar et al., 2013; Kayagaki et al., 2013; Shi et al., 2014). Upon LPS binding, active caspase-11 cleaves GSDMD in a manner similar to caspase-1 (Wang et al., 2020), leading to pore formation and pyroptosis (Aglietti et al., 2016; Kayagaki et al., 2015; Liu et al., 2016; Shi et al., 2015). Caspase-8 also cleaves GSDMD to induce pyroptosis in certain contexts (Orning et al., 2018; Rogers et al., 2017; Wang et al., 2017).
Despite the importance of GSDMD in pyroptosis, mechanisms regulating its activity are largely focused on upstream factors that influence its cleavage. For example, genetic deficiencies in components of inflammasomes (e.g. NLRP3 or ASC) prevent caspase-1 activation, resulting in inefficient GSDMD cleavage and defects in pyroptosis. Moreover, certain stimuli and cell types may differentially impact the magnitude and duration of caspase activation, with a direct effect on the rate of GSDMD cleavage and pore formation (Boucher et al., 2018; Chen et al., 2014). TCA cycle intermediates modify a reactive cysteine within GSDMD resulting in reduced cleavage and pore formation (Humphries et al., 2020). While the above-mentioned mechanisms of regulation are diverse, they all operate upstream of GSDMD cleavage. Whether factors exist that promote pore formation at the plasma membrane, after GSDMD cleavage has occurred, is less clear.
In this study, we sought to fill this gap in our knowledge by performing a genome-wide genetic screen to identify factors which are required for pore forming activity at the plasma membrane downstream of GSDMD cleavage. We identified components of the Ragulator-Rag complex, a mediator of mTOR activities, as necessary for GSDMD pore forming activity in macrophages. Subsequent analysis revealed that the Ragulator-Rag complex controls mTORC1-dependent events that operate downstream of GSDMD cleavage to promote GSDMD oligomerization and pore formation, by a process mediated by mitochondrial reactive oxygen species (ROS). These studies therefore provide a link between key metabolic and cell death regulators to control pyroptosis.
Results
An experimental platform that bypasses natural regulatory mechanisms to specifically stimulate GSDMD pore forming activity
We created a genetic screening system to identify genes that act after GSDMD cleavage to promote pore formation and pyroptosis. This goal was accomplished by taking advantage of the fact that the expression of the N-terminal caspase-cleavage product of GSDMD is sufficient to induce pore formation in the plasma membrane (Kayagaki et al., 2015; Shi et al., 2015). Thus, expression of the N-terminal GSDMD fragment (NT-GSDMD) bypasses upstream regulatory events that naturally lead to pyroptosis.
Using immortalized bone marrow-derived macrophages (iBMDMs) from Cas9-expressing mice, we stably expressed the Tet3G transactivator, which enabled the doxycycline (Dox)-inducible expression of transgenes encoding NT-GSDMD and full length GSDMD (FL-GSDMD) both of which contained a C-terminal BFP tag (Figure 1A and 1B). We utilized a variant of the NT-GSDMD that contains an I105N mutation, as this mutant is more readily detected within cells than its wild type (WT) counterpart (Aglietti et al., 2016).
Figure 1. Engineered macrophages model pyroptosis through expression of the GSDMD N-terminal domain.
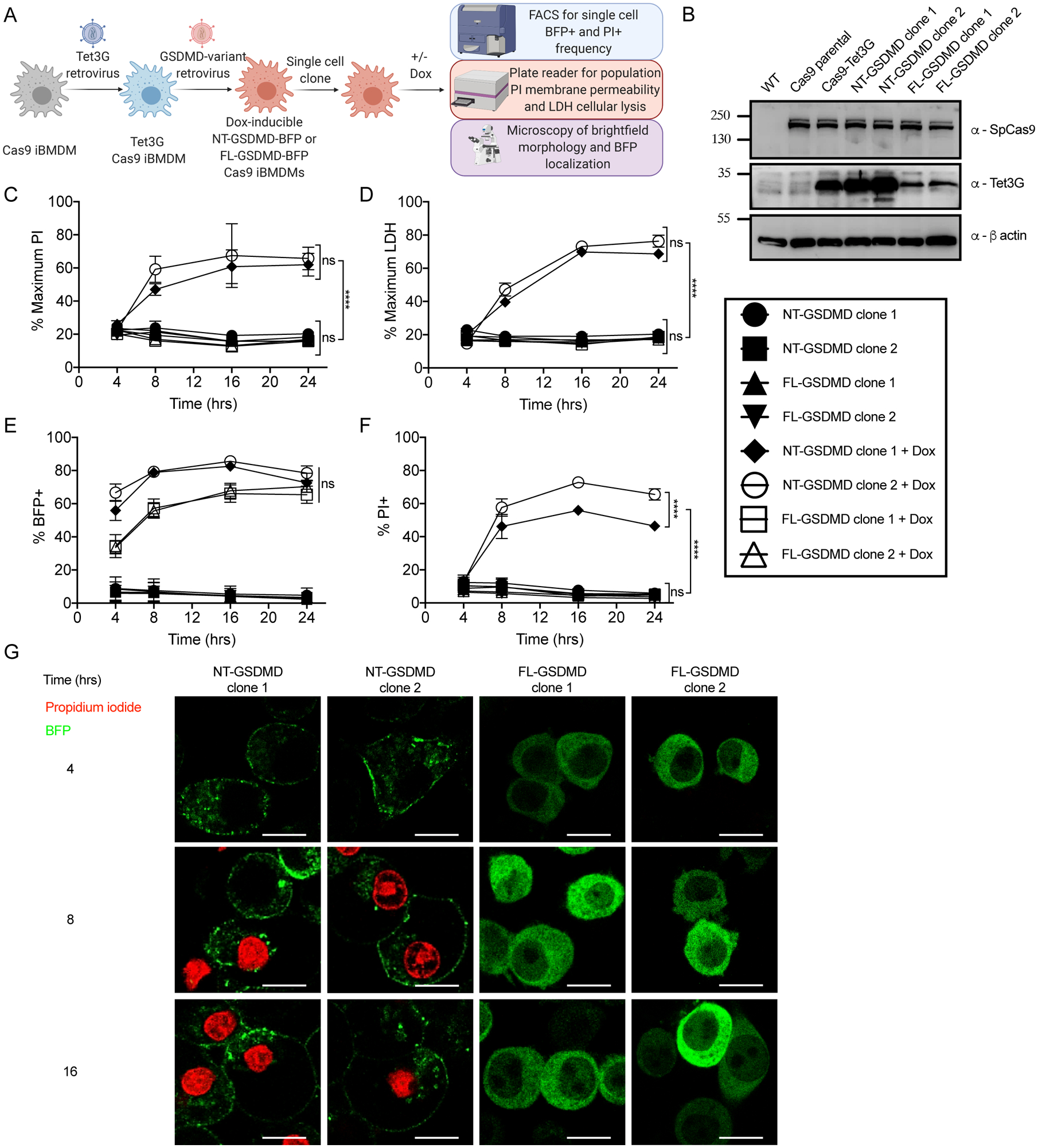
(A) Retroviral transduction workflow to generate Tet3G transactivator expressing Dox-inducible fluorescently tagged variants of GSDMD in iBMDMs from the Cas9 knock-in mouse and downstream characterization.
(B) Western blot of stable Cas9 expression in parental and progeny iBMDMs and stable expression of Tet3G transactivator in progeny iBMDM clones with β-actin loading control.
(C) Kinetic analysis of PI uptake by plate reader to measure bulk membrane permeability in populations of uninduced or Dox-induced (0.5 μg/ml) cells expressing NT-GSDMD-BFP or FL-GSDMD-BFP.
(D) Time course end point analysis of LDH release into cell free supernatants to measure cell lysis in populations of uninduced or Dox-induced (0.5 μg/ml) cells expressing NT-GSDMD-BFP or FL-GSDMD-BFP.
(E, F) Time course end point analysis by flow cytometry of the frequency of BFP+ (E) or PI+ cells (F) using uninduced or Dox-induced (0.5 μg/ml) cells expressing NT-GSDMD-BFP or FL-GSDMD-BFP.
(G) Time course live cell imaging of Dox-induced cells expressing NT-GSDMD-BFP or FL-GSDMD-BFP noting localization of BFP signal and PI uptake. Scale bar indicates 10 μm.
All quantification represents the mean and SEM of three independent experiments. Two-way ANOVA was used for analysis. See also Figure S1.
We characterized the ability of NT-GSDMD and FL-GSDMD to induce plasma membrane pore formation and lysis after Dox treatment. Pore formation was assessed by treatment with propidium iodide (PI), a membrane impermeable dye that can pass through GSDMD pores and enter cells. PI fluoresces upon binding to intracellular nucleic acids. Cytolysis was assessed by the release of the cytosolic enzyme lactate dehydrogenase (LDH) into the extracellular space. LDH is only released from cells post-lysis (Evavold et al., 2018).
Dox-mediated induction of NT-GSDMD, but not FL-GSDMD, resulted in pore formation and cell lysis, as revealed by population level PI fluorescence and LDH release (Figure 1C and 1D). Induction of both GSDMD transgenes corresponded with BFP induction, as evidenced by an increase in frequency of BFP positive cells over time in a Dox-dependent manner (Figure 1E). Flow cytometry demonstrated that the frequency of PI positive cells increased over time in a Dox-dependent manner for cells expressing NT-GSDMD (Figure 1F). Confocal microscopy demonstrated that NT-GSDMD is localized primarily to the plasma membrane, whereas FL-GSDMD was distributed throughout the cell (Figure 1G). Live cell microscopy demonstrated that PI staining occurred in a time-dependent manner in cells expressing the NT-GSDMD, but not in cells expressing FL-GSDMD (Figure 1G).
Finally, we sought to determine if NT-GSDMD-mediated pyroptosis induced by our Dox system was impacted by the endogenous inflammasome machinery within the cell. We found that Dox-induced pore forming activity of NT-GSDMD remained intact in LPS primed or unprimed cells that were treated with the NLRP3-specific inhibitor MCC950 (Coll et al., 2019; Tapia-Abellan et al., 2019) (Figures S1A and S1B). In parallel stimulations, MCC950 prevented PI uptake and LDH release that was induced by the treatment of LPS-primed cells with the NLRP3 agonist Nigericin (Figures S1C and S1D). These collective results establish an experimental system to induce GSDMD pore forming activity directly, independent of inflammasomes, thus enabling the identification of factors that specifically mediate these events.
A forward genetic screen identifies the Ragulator-Rag complex as necessary for GSDMD pore formation
To identify genes that regulate GSDMD pore formation, a genome-wide CRISPR-Cas9 screen was performed. iBMDMs encoding NT-GSDMD were verified to produce similar amounts of Cas9 protein, as compared to their parental line (Figure 1B). We lentivirally transduced iBMDMs encoding NT-GSDMD with the Brie genome-wide single guide RNA (sgRNA) library (Doench et al., 2016). Cells were treated with Dox to induce NT-GSDMD expression for 16 hours and then stained with PI to label cells containing GSDMD pores. The top 15% of BFP-positive cells, PI-negative cells were isolated by FACS. These cells expressed the NT-GSDMD transgene (as identified by the BFP tag) but have not formed pores. These cells were therefore predicted to contain mutations in genes that promote GSDMD pore formation.
After PCR amplification of sgRNA sequences from genomic DNA, and subsequent next generation sequencing, the log-normalized sgRNA abundance from the input sample was subtracted from the log-normalized sgRNA abundance of BFP-positive survivors. Using hypergeometric analysis, average p-values were generated and plotted against average log-fold change (LFC) at the gene level (Figure 2A). Gene ontology analysis of the rank list of genes from most positive LFC to most negative LFC identified functional annotations that were enriched in our dataset. The most significant function was attributed to positive regulation of mTOR signaling (Figure 2B). Many of the genes with the highest positive LFC and most significant p-values were part of the Ragulator-Rag complex (Figures 2A and 2C). Highlighted potential positive regulators of GSDMD pore formation are indicated in red on the volcano plot (Figure 2A) and on a schematized cryo-EM structure of this protein complex (Figure 2C) (Shen et al., 2019). Specifically, we identified the GTPases RagA and RagC and their functionally-related GTPase activating protein (GAP) FLCN, as well as the major components of the Ragulator complex—Lamtor-1, -2, -3, -4. These proteins are best known for their metabolic activities (Liu and Sabatini, 2020), but a role in GSDMD activities is unprecedented. We also noted that several of the genes with the highest negative LFC and most significant p-values correspond to members of the autophagy pathway, including E2-like conjugating enzymes Atg10 and Atg3 and cysteine protease Atg4b. Highlighted potential negative regulators of GSDMD pores are indicated in green on the volcano plot (Figure 2A). Recent work has begun to uncover that the Ragulator-Rag complex can serve as an organelle quality control mechanism for lysosomes and mitochondria that likely operates through induction of autophagy (Condon et al., 2021; Jia et al., 2018). Based on the concordance of biological processes between our potential positive and negative regulators of GSDMD pore formation, we focused our subsequent studies on the functions of Ragulator-Rag in pyroptosis.
Figure 2. Analysis of survivor cells sgRNA enrichment and validation of screen hits.
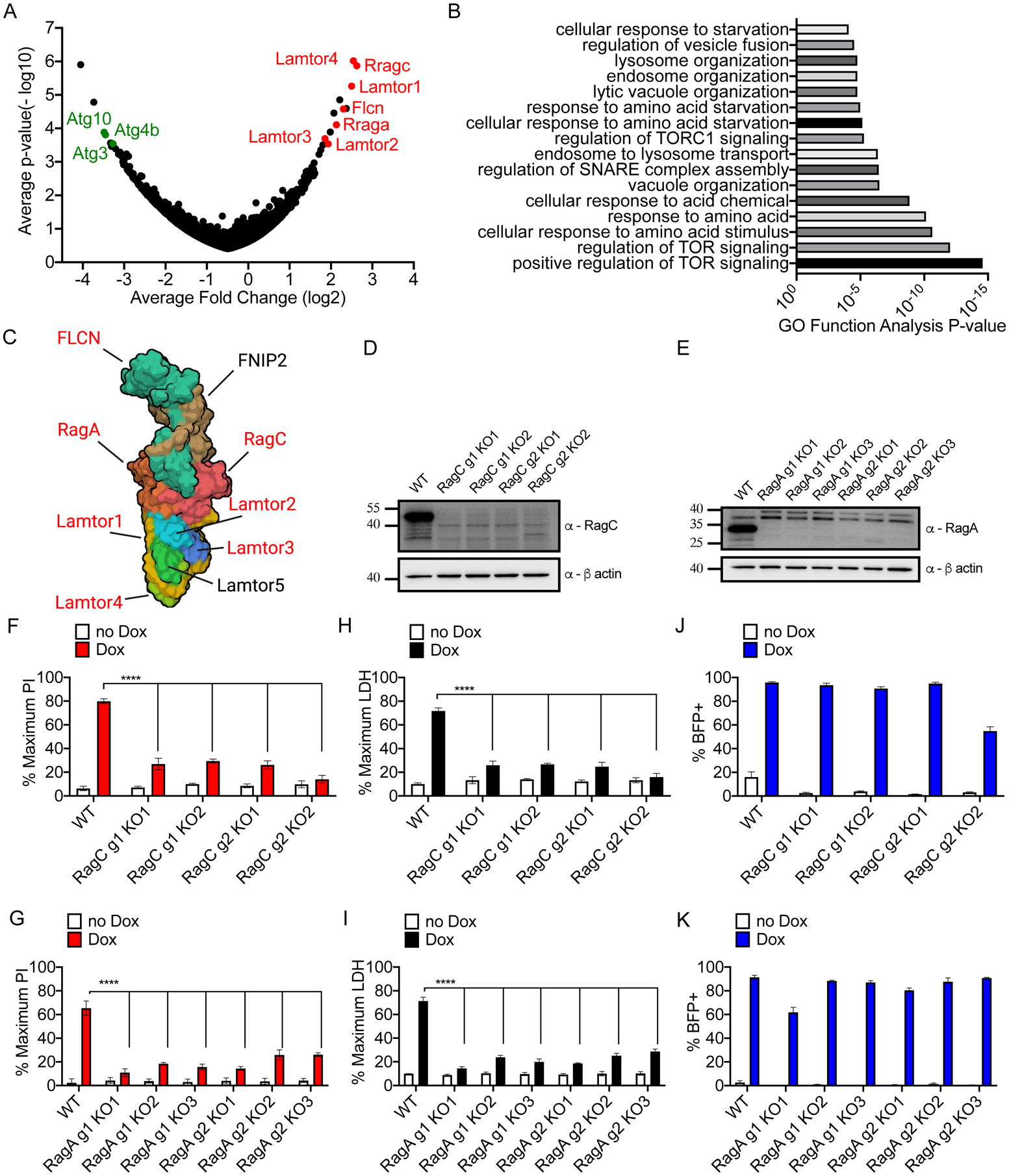
(A) Hypergeometric analysis of log-normalized guide abundance of survivor cells subtracted by the log-normalized abundance of input cells plotted as gene level average p-value versus gene level average LFC.
(B) Gene ontology functional annotation enrichment analysis for ranked hit list using GOrilla web analysis tool.
(C) Cryo-EM structure of the Ragulator-Rag complex cartoon schematic with top hits identified from CRISPR screen highlighted in red.
(D, E) Western blot of RagC (D) or RagA (E) protein ablation comparing empty vector transduced or cells expressing sgRNA guides targeting RagC and RagA.
(F, G) PI uptake analysis of cells of the genotypes indicated, left uninduced or Dox-induced (2 μg/ml) for 16 hours.
(H, I) LDH release from cells of the genotypes indicated, left uninduced or Dox-induced (2 μg/ml) for 16 hours.
(J, K) Frequency of BFP+ cells by flow cytometry of cells of the genotypes indicated, left uninduced or Dox-induced (2 μg/ml) for 16 hours.
All quantification represents the mean and SEM of three independent experiments. Two-way ANOVA was used for analysis. See also Figure S2.
To validate our top screen hits, we targeted specific components of the Ragulator-Rag complex for genetic ablation. Using independent sgRNAs and single cell clones from each sgRNA, we ablated protein production of RagA and RagC, as compared to empty vector transduced cells (Figure 2D and 2E). As expected (Rogala et al., 2019), ablation of RagA and RagC reduced activity of mTOR, based on reduced phosphorylation of S6 Kinase (S6K) (Figure S2A). Phospho-S6K abundance was not altered by Dox-induction (Figure S2A). After induction of NT-GSDMD, RagA- or RagC-deficient clones displayed defects in PI staining (Figures 2F and 2G) and LDH release (Figures 2H and 2I). The percentage of BFP-positive cells was comparable between Rag-deficient cells and cells containing an empty vector control (Figures 2J and 2K). Live cell imaging verified these results, as empty vector control cells displayed high frequencies of PI uptake and disintegration of morphology after NT-GSDMD expression (Video S1). In contrast, a low frequency of PI uptake and maintenance of cellular morphology was observed in two different lines of RagA-deficient iBMDMs (Video S2–S3). These data validate the importance of Ragulator-Rag in GSDMD-mediated pore formation and pyroptosis.
RagA regulates NT-GSDMD pore activity in primary murine cells and a human cell line
To explore the role of RagA in inflammasome signaling in primary immune cells, we employed the CHimeric IMmune Editing (CHIME) protocol to create control untargeted and RagA targeted bone marrow chimeras to ablate genes of interest in hematopoietic cells (LaFleur et al., 2019). We depleted the RagA protein in primary BMDMs derived from these mice, as compared to control untargeted BMDMs (Figure 3A). To activate inflammasome signaling in unprimed cells, we treated BMDMs with the synthetic toxin system consisting of anthrax protective antigen (PA) and the fusion protein of anthrax lethal factor with bacterial flagellin (LFn-Fla) that is collectively termed FlaTox (von Moltke et al., 2012; Zhao et al., 2011). Delivery of flagellin to the cytosol activates the NAIP-NLRC4 inflammasome, caspase-1, and subsequent GSDMD cleavage (Kofoed and Vance, 2011; Zhao et al., 2011). WT and RagA-deficient BMDMs displayed similar GSDMD cleavage after FlaTox administration, suggesting that RagA does not affect inflammasome activities (Figure 3A). In contrast, RagA depletion resulted in diminished pore formation, as observed by the frequency of PI positive cells (Figure 3B), or population level PI fluorescence (Figure S3A). Similarly, bulk populations of primary splenocytes from WT bone marrow chimeras treated with FlaTox displayed CD11b+ phagocyte depletion, as compared to LFn-Fla treated splenocytes from the same mice (Figure 3C). In contrast, splenocytes from RagA deficient bone marrow chimeras demonstrated resistance to CD11b+ depletion by FlaTox (Figure 3C). These data in primary BMDMs and splenic phagocytes support the importance of RagA in GSDMD-mediated pyroptosis.
Figure 3. The role of Ragulator-Rag and mTOR activity in NT-GSDMD pore formation.
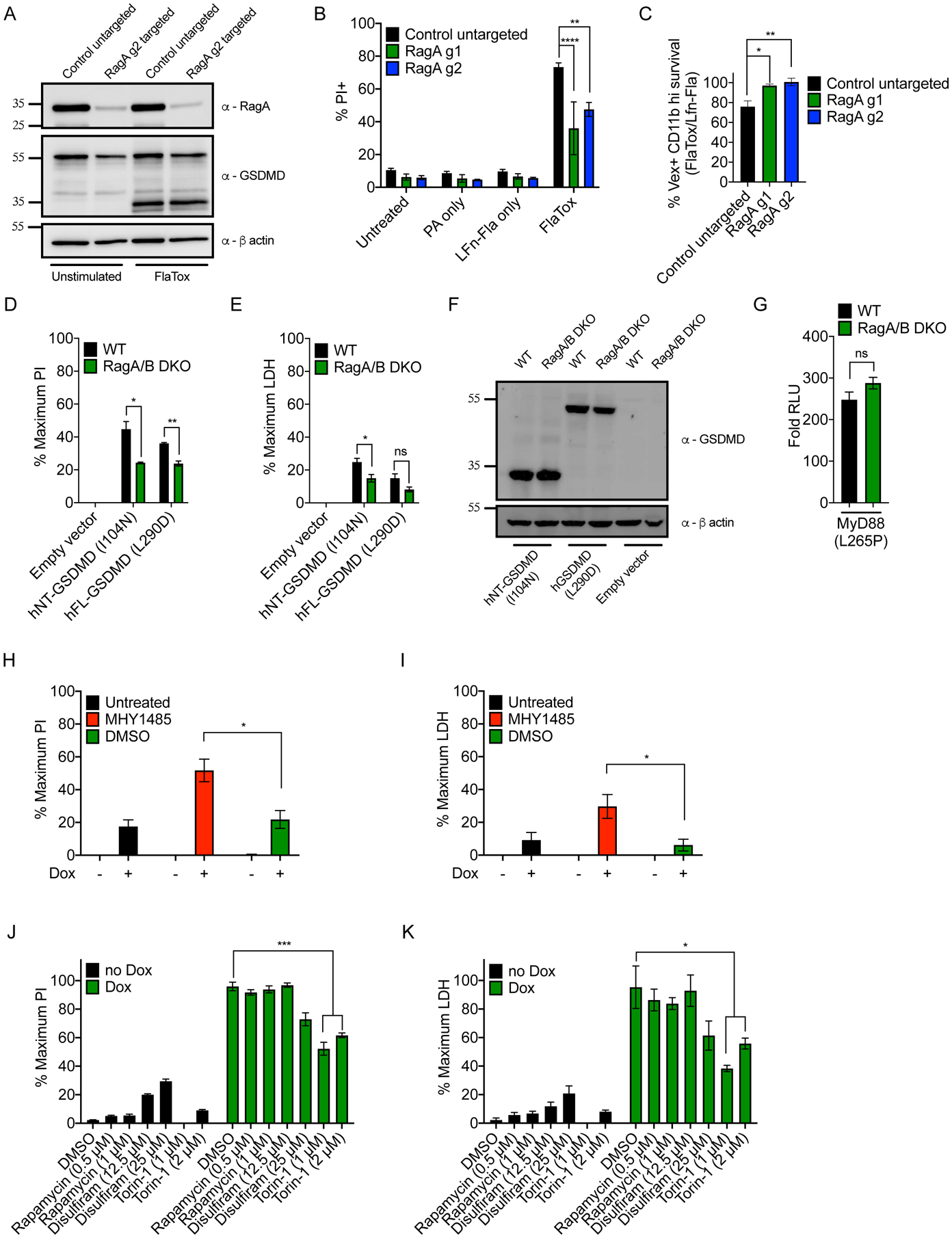
(A) Western blot of RagA and GSDMD cleavage in WT control untargeted and RagA targeted primary BMDMs after 1 hour of FlaTox treatment with β-actin loading control.
(B) Frequency of PI+ cells by flow cytometry of WT and RagA targeted primary BMDMs after 1 hour of FlaTox treatment.
(C) Ratio of depletion of Vex+, CD11b high splenocytes after 1 hour FlaTox treatment over LFn-Fla alone treatment.
(D, E) WT and RagA/B DKO 293T cells were transfected with plasmids encoding hGSDMD variants and normalized PI fluorescence (D) and cell-free supernatant LDH activity (E) were analyzed 20 hours after transfection.
(F) Western blot of hGSDMD variants in transfected WT and RagA/B DKO 293T cells.
(G) MyD88 L265P-driven NF-κB reporter activity in WT and RagA/B DKO 293T cells by dual-luciferase system 24 hours after transfection. Response of MyD88 L265P transfected cells over empty vector transfected cells is shown.
(H, I) Dox-inducible NT-GSDMD-BFP macrophages were treated with MHY1485 (10 μM) and Dox (0.5 μg/ml). Normalized PI fluorescence (H) and LDH activity in supernatants (I) were measured 6 hours later.
(J, K) Dox-inducible NT-GSDMD-BFP macrophages were treated with different compounds and Dox (0.5 μg/ml). DMSO dose corresponds to DMSO content present in 2 μM Torin-1 samples. Normalized PI fluorescence (J) and extracellular LDH activity (K) were measured 14 hours later. Blots (A, F) are representative of two independent experiments. Means and SEM of three (B, C, D, E, G, J, K) or four (H, I) independent experiments are shown. Unpaired two-tailed t-test was used for pairwise comparison (D, E, G, H, I). One-way ANOVA (C) and two-way ANOVA (B, J, K) were used for analysis. See also Figure S3.
To extend our analysis of Ragulator-Rag control of GSDMD pore formation and pyroptosis, we examined human GSDMD alleles in human cells. We analyzed the ability of GSDMD to form pores in WT and RagA/B double KO (DKO) 293T cells (Rogala et al., 2019). In addition to the caspase cleavage product of human GSDMD (hNT-GSDMD), we examined a destabilized full-length variant GSDMD L290D, which can form pores as a full-length protein (Liu et al., 2019). We found that GSDMD-mediated PI uptake and LDH release was diminished in RagA/B DKO cells, as compared to WT cells (Figure 3D and 3E). No genotype-specific differences in GSDMD expression were observed (Figure 3F). In addition, RagA/B DKO cells were not generally defective for immune signaling events, as these cells were not deficient in MyD88 L265P-induced NF-κB reporter activation (Figure 3G).
mTOR activity is required for NT-GSDMD pore formation
Ragulator-Rag is best-known for its functions in mTOR regulation. To determine the role of mTOR in GSDMD pore formation, we examined chemical agonists or inhibitors of mTOR activity. To exclude complications in data interpretation associated with known mTOR functions upstream of inflammasome activation (Moon et al., 2015), we used Dox-inducible cells for this analysis. We found that the mTOR activator MHY1485 promoted GSDMD pore formation, as treatments with this chemical at the time of Dox treatment led to an increase in PI uptake and LDH release, as compared to untreated controls (Figures 3H and 3I). We note that because we were examining a boost in pore formation (not an inhibition) by MHY1485, we used a time point where uninhibited cells contained evidence of <20% pores. The mTOR inhibitor Torin-1 had the opposite effect from MHY1485, in that Torin-1 suppressed NT-GSDMD-induced PI uptake and LDH release (Figures 3J and 3K). Although incomplete, the inhibitory effect of Torin-1 on GSDMD pore formation was comparable to that observed with the GSDMD inhibitor disulfiram (Hu et al., 2020). In contrast to the inhibition of GSDMD pore activity with Torin-1 treatment, the mTOR inhibitor rapamycin, which is not as effective as Torin-1 (Thoreen et al., 2009), was unable to prevent GSDMD induced pore forming activities (Figures 3J and 3K). Rapamycin and Torin-1 suppressed phosphorylation of the mTOR target S6K (Figure S3B). No effects of these inhibitors on the expression of GSDMD were observed (Figure S3B). The lack of phenotype with rapamycin on GSDMD pore formation, while unexplained, may be related to observations that this drug does not affect all branches of mTORC1 signaling equally (Howell and Manning, 2011).
To better define the role of mTOR in GSDMD activities, we focused on the two protein complexes that harbor this enzyme, mTORC1 and mTORC2. mTORC1 is controlled by several upstream pathways, including the Ragulator-Rag complex in response to amino acid availability (Sancak et al., 2010). To test whether mTORC1 stimulation by amino acids affects NT-GSDMD activities, we controlled GSDMD cleavage through co-expression of a small molecule dimerizable caspase-1 and hFL-GSDMD in 293T cells. After amino acid starvation, dimerizer was added to cells that were either stimulated with amino acids or not. Amino acid treatments led to an increase in mTORC1 activity, as observed by increased S6K phosphorylation (Figure S3C). While addition of dimerizer induced comparable reduction in FL-GSDMD band intensity, indicating similar GSDMD cleavage (Figure S3C), pyroptosis was most increased in amino acid stimulation conditions (Figure S3D). Thus, stimulation of mTORC1 activities promote GSDMD pore forming activity.
Tuberous sclerosis complex is an endogenous negative regulator of mTORC1 activation (Yang et al., 2021). We found that overexpression of TSC-1, a component of tuberous sclerosis complex, decreased NT-GSDMD-induced PI uptake and LDH release (Figures 4A and 4B). NT-GSDMD protein abundance was unaffected by TSC-1 overexpression (Figure 4C). In addition, TSC-1 overexpression did not affect NF-κB reporter activation by MyD88 L265P (Figure 4D). Thus, several strategies of mTOR modulation impact GSDMD pore forming activity in a manner consistent with a role of mTORC1 in promoting pyroptosis.
Figure 4. mTORC1 acts downstream of GSDMD cleavage to promote pore formation and pyroptosis.
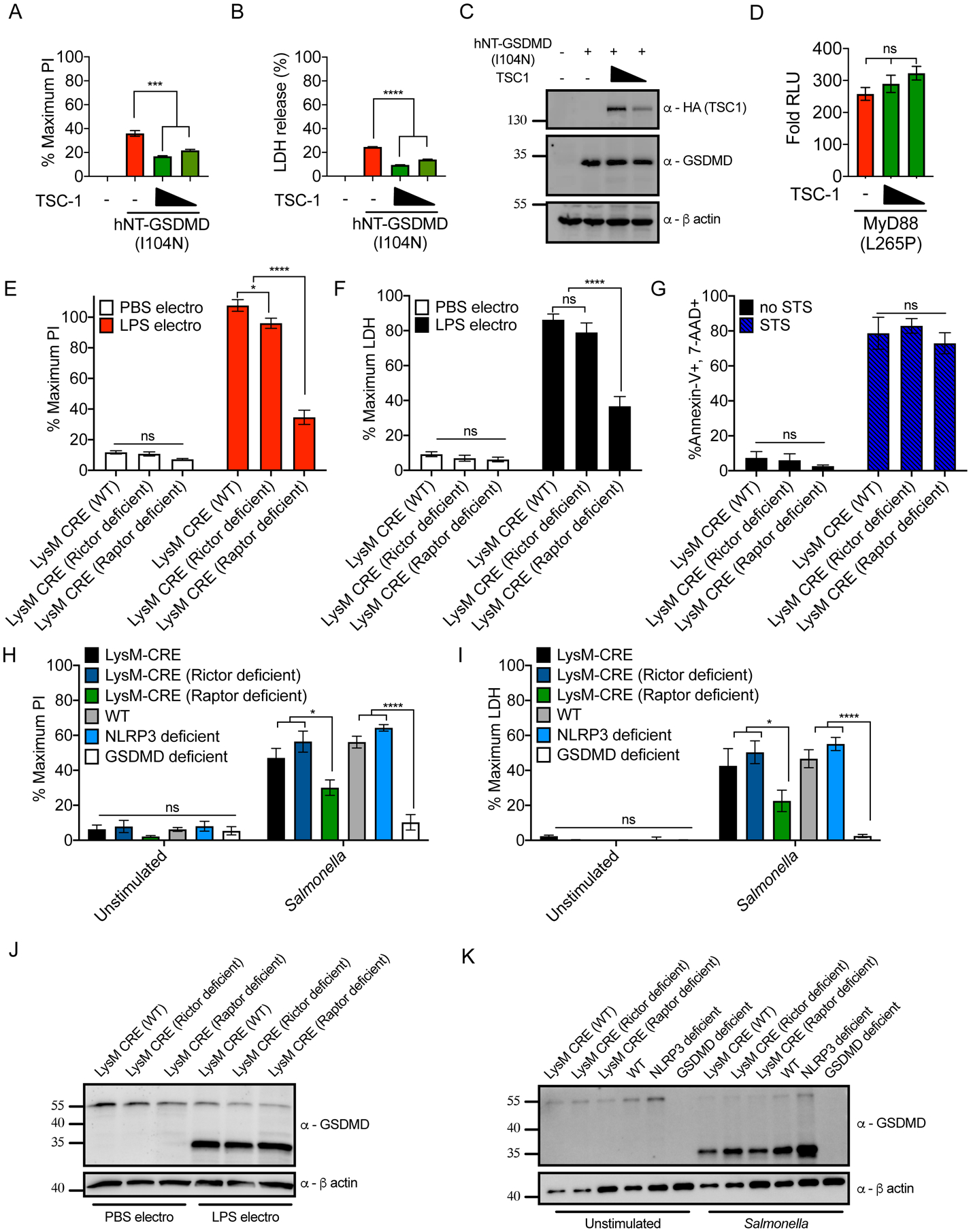
(A–C) HEK293 cells were co-transfected with plasmids encoding hNT-GSDMD (I104N) and TSC-1. Normalized PI fluorescence (A), extracellular LDH activity (B) and protein abundance (C) were measured 20 hours post-transfection.
(D) MyD88 L265P-driven NF-κB dual-luciferase system response in HEK293 cells transfected with TSC-1 24 hours after transfection. Response of MyD88 L265P transfected cells over empty vector transfected cells is shown.
(E, F) PI uptake (E) and LDH release assessment (F) of LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs, primed with recombinant IFNβ for 3 hours then electroporated with PBS or LPS.
(G) Frequency of Annexin-V and 7-AAD positive cells by flow cytometry from LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs after 8 hours of 1 μM Staurosporine (STS) treatment.
(H, I) PI uptake (H) and LDH release (I) from LysM-CRE WT, Rictor-deficient, Raptor-deficient, C57/BL6J WT, NLRP3-deficient, and GSDMD-deficient iBMDMs, unstimulated or infected with S. Typhimurium at an MOI of 10 for 1 hour.
(J) Western blot of the proteins indicated in LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs, after these cells were primed with IFNβ and then electroporated with PBS or LPS.
(K) Western blot of the proteins indicated in LysM-CRE WT, Rictor-deficient, Raptor-deficient, C57/BL6J WT, NLRP3-deficient, and GSDMD-deficient iBMDMs, after these cells were unstimulated or infected with S. Typhimurium at an MOI of 10 for 1 hour.
All quantifications represent mean and SEM of 3 independent experiments. Two-way ANOVA was used for analysis. See also Figure S4.
mTORC1, but not mTORC2, is required for GSDMD pore forming activity and pyroptosis
To determine the relative roles of mTORC1 and mTORC2 in pyroptosis, we examined the function of the mTORC1 component Raptor and the mTORC2 component Rictor in iBMDMs. iBMDMs were generated from mice encoding CRE recombinase whose expression is driven by the macrophage-specific LysM promoter (hereafter referred to as LysM-CRE iBMDMs). Similarly, iBMDMs were derived from mice dually encoding LysM-CRE and floxed alleles of Raptor or Rictor (hereafter referred to as Raptor-deficient or Rictor-deficient iBMDMs). We verified protein ablation in Raptor- and Rictor-deficient iBMDMs (Figure S4A). These cells were subjected to stimulations that activate pathways leading to cleavage and pore formation by endogenous GSDMD.
We first focused on GSDMD pore forming activity that is stimulated upon detection of cytosolic LPS by caspase-11. LysM-CRE WT iBMDMs were primed with type I interferon (IFNβ) to induce caspase-11 expression and then electroporated with PBS or LPS to activate caspase-11. Under these conditions, we observed LPS-induced pyroptosis of LysM-CRE WT and Rictor-deficient iBMDMs, as evidenced by PI incorporation and LDH release (Figures 4E and 4F). In contrast, Raptor-deficient cells were defective for LPS-induced pore formation and lysis (Figures 4E and 4F).
To complement these studies, we replaced the IFNβ priming step with a priming step using LPS. As pro-IL-1β is upregulated by TLR4 signaling, we expanded our analysis to include IL-1β release after pyroptosis. LPS electroporation of LPS-primed LysM-CRE WT iBMDMs stimulated PI staining, LDH, and IL-1β release, all of which were reduced in Raptor-deficient cells (Figures S4B–S4D). In contrast, Rictor-deficient cells displayed no defects in PI staining or LDH release upon LPS electroporation (Figures S4B and S4C). Interestingly, whereas Raptor-deficient cells released less IL-1β than WT counterpart cells, Rictor-deficient cells released a greater amount of IL-1β than WT counterparts (Figure S4D). This increased secretion of IL-1β is likely attributed to increased pro-IL-1β expression after LPS priming in Rictor-deficient cells (Figure S4E). Thus, mTORC1 and mTORC2 have disparate functions in the pathways to pyroptosis. Live cell imaging confirmed these findings, as most LysM-CRE WT iBMDMs (Video S4) and Rictor-deficient iBMDMs (Video S5) experienced PI uptake and disintegration of morphology after LPS electroporation. In contrast, fewer Raptor-deficient cells converted to PI positivity after LPS electroporation (Video S6). In contrast to the requirement of Raptor for pyroptosis, WT, Raptor- and Rictor-deficient cells similarly committed to death upon treatment with the apoptosis-inducer staurosporine (STS) (Figure 4G).
To complement the above studies, we performed infections with Salmonella enterica serovar Typhimurium. S. Typhimurium infections activate inflammasome pathways mediated by caspase-11, NLRP3 and NAIP-NLRC4, all of which converge on GSDMD to induce pyroptosis (Broz et al., 2010; Franchi et al., 2006; Man et al., 2014). Within one hour of infection, PI staining and LDH release occurred in LysM-CRE WT and in C57BL/6J WT iBMDMs (Figures 4H and 4I). NLRP3 deficiency did not protect iBMDMs from pyroptosis during infection, whereas GSDMD-deficient cells were defective for these responses (Figures 4H and 4I). Notably, Raptor-deficient cells displayed lower PI staining and LDH release upon S. Typhimurium infection, as compared to Rictor-deficient or LysM-CRE WT cells (Figures 4H and 4I).
To determine the stage of the pyroptotic process Raptor is necessary to execute, we examined early and late events. An early event is the upregulation of caspase-11 induced by extracellular LPS or IFNβ (Rathinam et al., 2012). We found that the abundance of caspase-11 marginally differed when comparing LPS- or IFNβ-treated LysM-CRE WT iBMDMs to Raptor- and Rictor-deficient cells (Figures S4F and S4G). However, within unstimulated cells, caspase-11 abundance differed between these genotypes (Figure S4F and S4G). The significance of this phenotype is unclear. Thus, we sought to examine the question of cell priming in a functional manner. We reasoned that if there were functional differences associated with Raptor or Rictor deficiency, in terms of priming events, then cleavage of GSDMD should be impacted. To address this possibility, we monitored GSDMD cleavage after the stimulations described above. Raptor-deficient cells were not defective for GSDMD cleavage when IFNβ-primed cells were electroporated with LPS or when cells were infected with S. Typhimurium (Figure 4J and 4K). The Ragulator-Rag complex and its downstream effector mTORC1 therefore operate downstream of GSDMD cleavage to promote pore formation and pyroptosis.
RagA or RagC are not required for NT-GSDMD plasma membrane localization, but are required for oligomerization and pore formation
To determine the mechanisms by which Ragulator-Rag-mTORC1 controls GSDMD pore formation, we examined the two primary GSDMD activities that occur post-cleavage—membrane localization and oligomerization (Ding et al., 2016; Liu et al., 2016).
We first investigated the trafficking of NT-GSDMD to the plasma membrane. WT cells displayed NT-GSDMD at the plasma membrane and co-stained with PI (Figure 5A). RagA- and RagC-deficient cells also displayed NT-GSDMD at the cell surface, yet stained negative for PI (Figure 5A). Similarly, biochemical fractionations revealed no defects in the localization of NT-GSDMD to membrane fractions in RagA- or RagC-deficient macrophages (Figure 5B). Thus, RagA and RagC do not control the localization or membrane binding properties of NT-GSDMD.
Figure 5. RagA or RagC are not required for NT-GSDMD plasma membrane localization, but are required for oligomerization and pore formation.
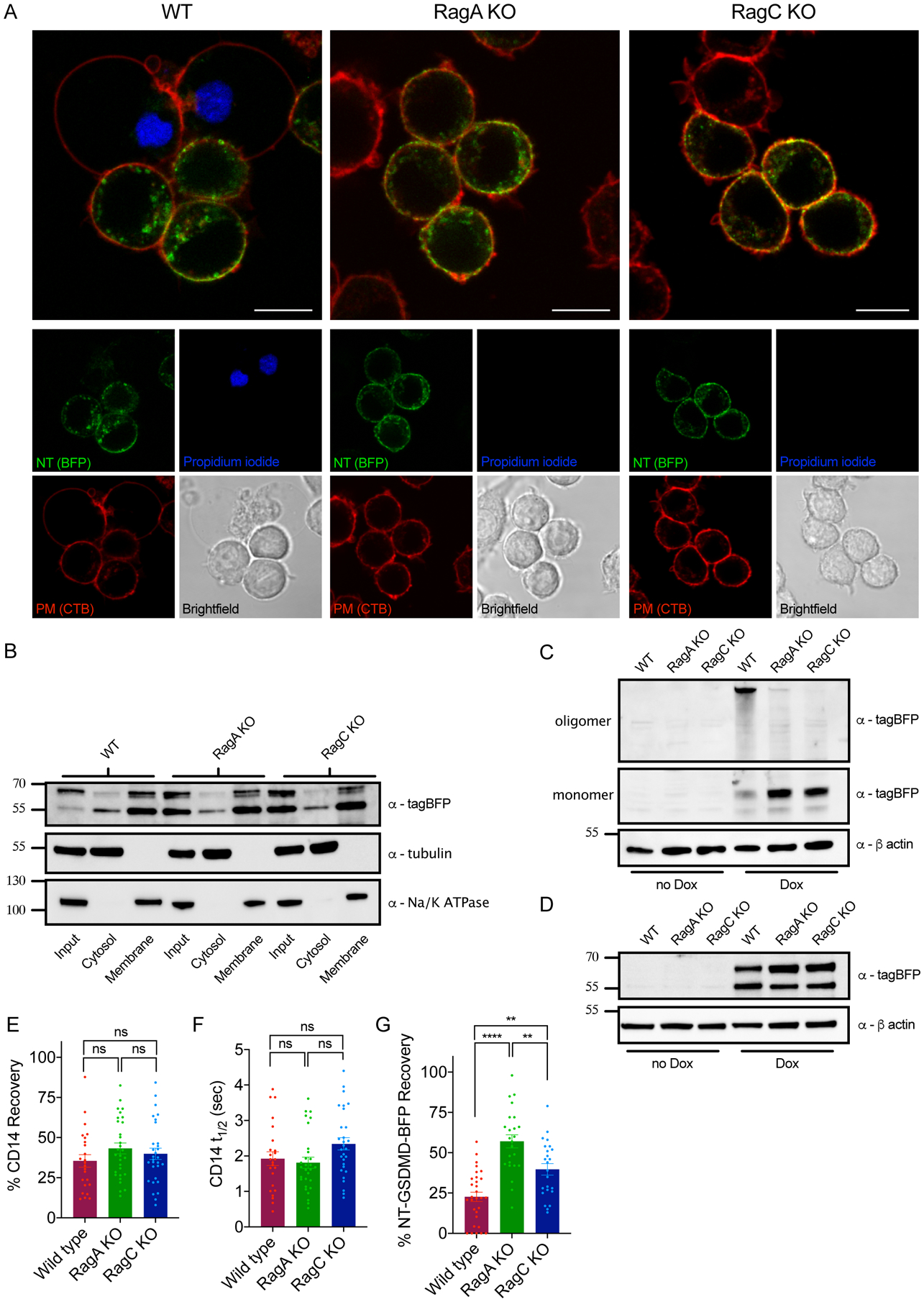
(A) Confocal microscopy of NT-GSDMD-BFP (green) at 8 hours post Dox-induction (0.5 μg/ml) in WT, RagA KO, and RagC KO cells. Plasma membrane was labeled with CTB-AF647 (red) and membrane permeability was assessed by PI (blue). Scale bar indicates 10 μm.
(B) Western blot of NT-GSDMD-BFP from post-nuclear input, cytosolic, and bulk membrane fractions of WT, RagA KO, and RagC KO cells, after Dox-induction (2 μg/ml) for 8 hours. Tubulin represents a cytosol marker and Na+/K+ ATPase represents a membrane marker.
(C, D) Western blot of NT-GSDMD-BFP from combined cell lysate and supernatant under non-reducing conditions (C) or reducing conditions (D) from WT, RagA KO, and RagC KO cells after Dox-induction for 16 hours.
(E) FRAP proportion of fluorescence recovery of CD14 labeled with PE-conjugated anti-CD14 antibody in WT, RagA KO, and RagC KO macrophages.
(F) FRAP half-life of fluorescence recovery of CD14 labled with PE-conjugated anti-CD14 antibody in WT, RagA KO, and RagC KO macrophages.
(G) FRAP proportion of fluorescence recovery of BFP signal from NT-GSDMD-BFP in WT, RagA KO, and RagC KO macrophages after Dox-induction (2 μg/ml) for 8 hours.
Each data point (E, F, G) represents parameters from a fitted FRAP curve from individual cells (n ≥ 24). One-way ANOVA was used for analysis.
To examine GSDMD oligomerization, which correlates with pore formation, we used a protocol that distinguishes monomeric and oligomeric GSDMD. Specifically, NT-GSDMD oligomers (i.e. pores) are resistant to SDS destabilization unless a reducing agent is present (Liu et al., 2016). Using this principle, we subjected WT, RagA-deficient, and RagC-deficient cells to SDS lysis after Dox-induction of NT-GSDMD. When no reducing agent was added to the lysate, WT cells had mostly oligomerized NT-GSDMD (Figure 5C). In contrast, GSDMD oligomers were nearly absent from RagA- and RagC-deficient cells, yet NT-GSDMD monomeric species were abundant (Figure 5C). Treatment of the same lysates with the reducing agent TCEP dissociated the oligomerized NT-GSDMD in WT cells to the monomer phase (Figure 5D). These data demonstrate that RagA and RagC promote NT-GSDMD oligomerization.
We addressed whether membrane fluidity might underlie the defects in NT-GSDMD oligomerization in RagA- and RagC-deficient cells. Toward this end, we performed fluorescence recovery after photobleaching (FRAP) studies of the GPI-anchored membrane protein CD14 within the plasma membranes of WT, RagA- and RagC-deficient macrophages. We noted no significant changes in either the fraction of recovery of fluorescence signal or half-life of recovery for the genotypes examined (Figure 5E and 5F). These results suggest that bulk membrane fluidity and diffusion of proteins within the plasma membrane are unaffected by RagA and RagC deficiency. We also performed FRAP studies on the plasma membrane localized BFP-tagged NT-GSDMD. Intriguingly, the fraction of BFP recovery was lower in WT cells than in RagA- and RagC-deficient cells (Figure 5G). This observation may represent in situ measurements of oligomerization of NT-GSDMD in the plasma membrane, as an oligomer would be expected to be more immobile than a monomer. These results support the conclusion that RagA and RagC regulate GSDMD pore formation because they control the necessary step of oligomerization in the plasma membrane.
Mitochondrial dysfunction and ROS mediate GSDMD oligomerization
To further define the link between Ragulator-Rag and GSDMD oligomerization, we considered another phenomenon co-incident with pore formation by GSDMD — the disruption of mitochondrial activities (de Vasconcelos et al., 2019; Kayagaki et al., 2021), which are also linked to Ragulator-Rag functions (Condon et al., 2021).
We quantified mitochondria abundance and membrane potential using MitoTracker and the potentiometric dye TMRM, along with PI positivity after Dox-induced NT-GSDMD expression. MitoTracker and TMRM staining was diminished in WT cells upon NT-GSDMD expression but was maintained upon NT-GSDMD expression in RagA-deficient cells (Figures S5A–S5G). In addition, mitochondrial ROS abundance increased in WT cells, but not RagA-deficient cells, upon NT-GSDMD expression (Figure S5H). Similar findings were made when we measured total cellular ROS in WT, RagA- and RagC-deficient cells using the reagent CellROX (Figure 6A). While diminished mitochondrial activities and increased ROS production may be expected to occur after pore formation, we were intrigued by the basal differences in ROS production observed. ROS levels were higher in resting WT cells than RagA- and RagC-deficient cells (Figure 6A), an observation that cannot be explained by NT-GSDMD pore activities. Thus, in cells with high ROS (WT cells), GSDMD pores are formed more efficiently than in cells with low ROS (Rag-deficient cells). We therefore determined if the correlation between ROS and GSDMD pore formation might be causal.
Figure 6. Mitochondrial dysfunction and ROS mediate GSDMD oligomerization.
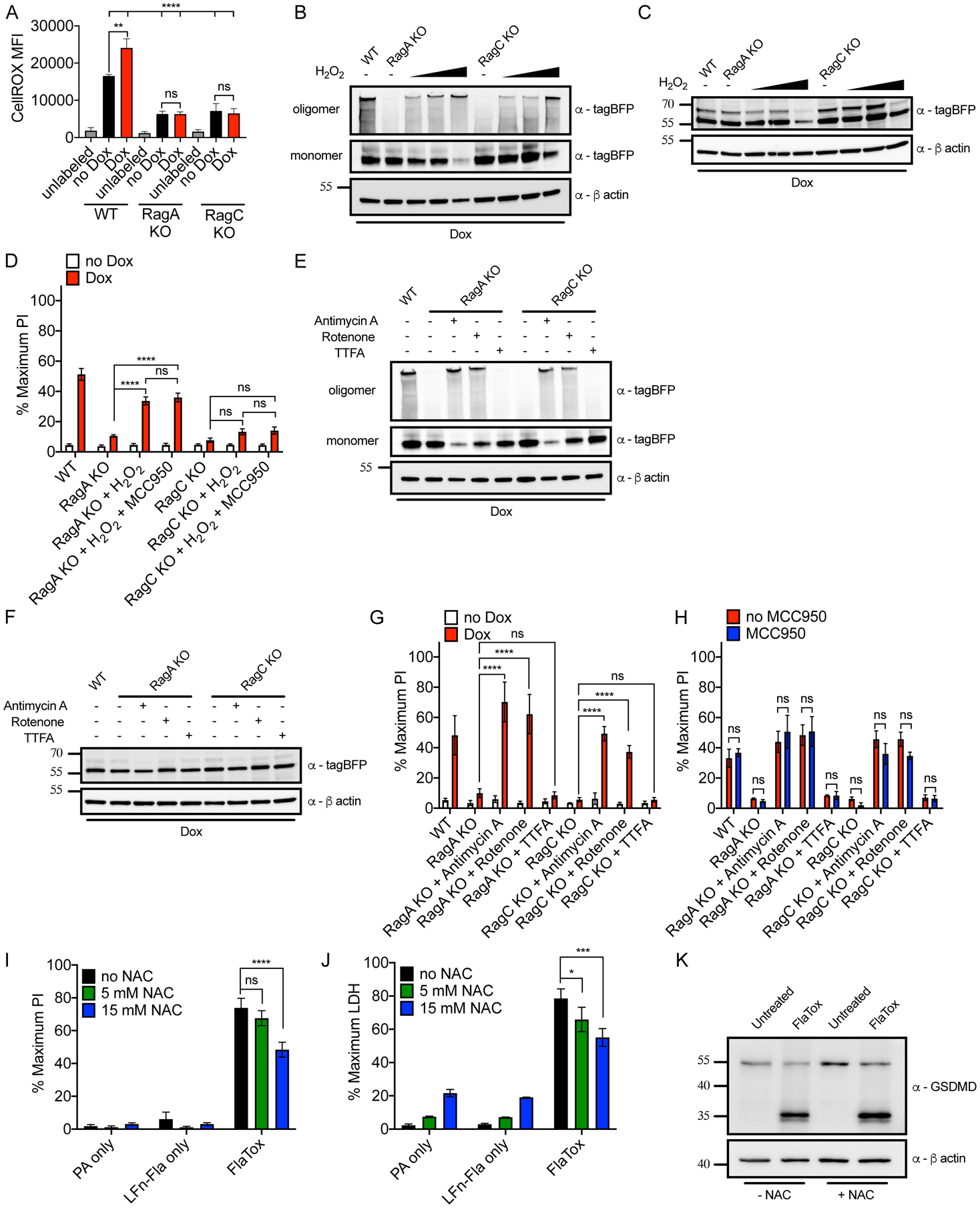
(A) Intracellular ROS levels in live cells of the genotypes indicated by CellRox Deep Red fluorescence after Dox induction (0.5 μg/ml) for 8 hours.
(B, C) Western blot of the proteins indicated from combined cell lysates and supernatants under non-reducing (B) or reducing (C) conditions from from WT, RagA KO, and RagC KO cells after Dox induction for 8 hours and treated with increasing concentrations of H2O2 for the last 4 hours.
(D) PI uptake by WT, RagA KO, and RagC KO cells after Dox induction for 8 hours and treated with 625 μM of H2O2 for the last 4 hours. MCC950 (10 μM) was added to cells at the start of Dox induction.
(E, F) Western blot of NT-GSDMD-BFP from combined cell lysates and supernatants under non-reducing (E) or reducing (F) conditions from WT, RagA KO, and RagC KO cells after Dox induction for 8 hours and treated with Antimycin A (10 μg/ml), Rotenone (10 μM) or TTFA (100 μM) for the last 4 hours.
(G) PI uptake by WT, RagA KO, and RagC KO cells after Dox induction for 8 hours and treated with Antimycin A (10 μg/ml), Rotenone (10 μM) or TTFA (100 μM) for the last 4 hours.
(H) PI uptake by MCC950-treated cells of the genotypes indicated after Dox induction for 8 hours and treated with Antimycin A (10 μg/ml), Rotenone (10 μM) or TTFA (100 μM) for the last 4 hours.
(I, J) PI uptake (I) and LDH release into cell-free supernatant (J) from WT iBMDMs treated with FlaTox and indicated concentrations of NAC for 2 hours.
(K) Western blot of the proteins indicated from combined cell lysates and supernatants from WT iBMDMs treated with FlaTox and 15 mM NAC for 2 hours.
Means and SEM of three (A, D, G, H, J) or four (I) independent experiments are shown. Two-way ANOVA was used for analysis.
See also Figures S5 and S6.
To test whether ROS might affect the pore forming ability of NT-GSDMD, we treated RagA- and RagC-deficient cells with increasing concentrations of H2O2. Upon addition of H2O2, RagA- and RagC-deficient cells displayed rescue of their oligomerization defect, as seen by increased NT-GSDMD oligomers (Figure 6B). Addition of reducing agent to these lysates reduced these oligomers back to the monomer phase, with the exception of a fraction of oligomers resisting reduction for the highest H2O2 dose (Figure 6C). The rescue of GSDMD oligomerization by H2O2 correlated with pore formation, as H2O2 treatment led to PI staining in RagA-deficient cells (Figure 6D). We excluded the possibility that H2O2-induced PI staining could be explained by ROS stimulating endogenous NLRP3 activities (Zhou et al., 2011), since NLRP3 inhibition by MCC950 did not impact PI staining (Figure 6D). Despite the observed restoration of NT-GSDMD pore forming activities by H2O2, the extent of pore formation rescue was incomplete, in particular in the case of RagC-deficient cells (Figure 6D). We considered the possibility that an endogenous source of ROS (from mitochondria) may better impact NT-GSDMD pore forming activity. Inhibition of the electron transport chain (ETC) can promote ROS production (Zhou et al., 2011). The complex I inhibitor Rotenone and complex III inhibitor Antimycin A alter the ETC in a manner that drives ROS production, whereas the complex II inhibitor TTFA does not induce ROS production (Zhou et al., 2011). Consistent with a role for ROS in GSDMD oligomerization, treatment with the mitochondrial ROS fluxing agents Rotenone and Antimycin A stimulated oligomerization of NT-GSDMD in RagA- and RagC-deficient cells (Figure 6E). In contrast, TTFA, which does not flux mitochondrial ROS, did not rescue NT-GSDMD oligomerization in RagA- and RagC-deficient cells. Addition of reducing agent to these lysates reduced oligomers into monomers, and treatment with these mitochondrial poisons did not alter NT-GSDMD expression (Figure 6F). Importantly, we observed a complete rescue of NT-GSDMD pore forming activities in RagA- and RagC-deficient cells, but only with mitochondrial disrupting agents that also rescued oligomerization (Figure 6G). All PI staining observed was NT-GSDMD-dependent, as under no conditions were PI staining observed in the absence of Dox treatments (Figure 6G). Finally, MCC950 did not impact PI staining in Rotentone or Antimycin A treated RagA- or RagC-deficient cells expressing NT-GSDMD (Figure 6H). These data provide causal links between Ragulator-Rag, mitochondrial ROS, and NT-GSDMD oligomerization and pore formation. To investigate whether endogenous ROS promotes pyroptosis in natural inflammasome signaling, we again used the FlaTox system. We chose FlaTox (von Moltke et al., 2012) to avoid the roles of ROS in NLRP3 inflammasome activities (Bauernfeind et al., 2011; Dostert et al., 2008; Shimada et al., 2012; Zhou et al., 2011). Treatment of iBMDMs with increasing doses of the antioxidant NAC resulted in a reduction in pore formation and cytolysis (Figure 6I and 6J), while cleavage of GSDMD remained unchanged (Figure 6K). Mitochondrial health and ROS production therefore promote GSDMD oligomerization and pore formation.
The protein Ninjurin-1 (NINJ1) controls pyroptosis at a late stage (Kayagaki et al., 2021). NINJ1 is reportedly required to induce plasma membrane rupture downstream of GSDMD pore formation. To compare the phenotypes of NINJ1 deficient cells to our results, we generated NINJ1 KOs using Cas9 expressing iBMDMs after electroporation of two independent sgRNAs against NINJ1 (referred to as g1 and g2). After subjecting LPS-primed WT, NINJ1 g1, and NINJ1 g2 iBMDMs to the NLRP3 agonist Nigericin, we observed PI staining that was similar between these genotypes (Figure S6A), but reduction of LDH release in NINJ1-deficient cells (Figure S6B). Similarly, LPS electroporation of LPS-primed NINJ1-deficient cells displayed a marginal reduction in PI staining, but a substantial reduction in LDH release, as compared to WT cells (Figure S6C and S6D). These data position the actions of Ragulator-Rag downstream of GSDMD cleavage and plasma membrane localization, but upstream of NT-GSDMD oligomerization and NINJ1-induced plasma membrane rupture.
Discussion
In this study, a genetic screen was designed to specifically interrogate a single stage in the pyroptosis pathway. This approach was distinct from other genetic screens, where natural upstream stimuli were used (Kayagaki et al., 2019; Kayagaki et al., 2015; Napier et al., 2016; Shi et al., 2015). By focusing on a single (and poorly characterized) stage, we bypassed the possibility of identifying previously defined genes that are necessary for pyroptosis, such as regulators of cell priming or inflammasome assembly. Moreover, our screen employed an internal control (the BFP tag on the GSDMD transgene) to ensure the continued expression of the pore-forming molecule throughout our assays. This quality-control strategy likely explains why we did not identify regulators of transcription and translation in our dataset. Thus, we were able to focus on potential regulators of a process that was considered to be poorly regulated, namely the steps between cleavage of GSDMD and plasma membrane pore formation. Current models of pyroptosis invoke the process of membrane repair as a negative regulatory mechanism to limit GSDMD activities after this pore forming factor has been cleaved (Ruhl et al., 2018). It was unexpected that we could identify factors that promote GSDMD pore forming activities post-cleavage, yet our forward genetic screen and subsequent analysis revealed Ragulator-Rag-mTORC1 as factors that act at this stage.
In the metabolic pathways, Ragulator-Rag operates on lysosomes to sense amino acid availability and induce mTOR-dependent cellular responses that depend on mTORC1 (Liu and Sabatini, 2020). Despite its lysosomal localization, mTORC1 has roles in controlling mitochondrial activities (Chen et al., 2008; Cunningham et al., 2007; Schieke et al., 2006). One of these activities appears to be the control of ROS production, which we found is critical for GSDMD oligomerization and pore formation, but not plasma membrane localization of GSDMD. This finding is noteworthy, as biophysical studies have been unable to determine if GSDMD pores assemble from monomers within the plasma membrane, or if monomers pre-assemble and then insert an oligomer into the plasma membrane. Our studies suggest the former, as membrane-associated NT-GSDMD monomers can convert into pore forming oligomers when exposed to ROS inducing agents. However, more work is needed to define these events.
Our results suggest that GSDMD pore forming activity is dependent on the actions of the mTORC1 component Raptor and the upstream Ragulator-Rag complex. However, this link between GSDMD and Ragulator-Rag-mTORC1 does not require direct interactions between these proteins. Indeed, the ability of ROS to restore GSDMD oligomerization and pore forming activity in Rag-deficient cells suggests that the principal function of Ragulator-Rag in pyroptosis may be in ROS regulation. If this model is correct, one would expect that other stimuli that promote ROS production would promote GSDMD oligomerization and may even bypass the need for Ragulator-Rag in this process. Additional activators of the mTOR pathway exist, which operate independent of Ragulator-Rag, such as glucose, growth factors, cytokine receptors, and pattern recognition receptors (Liu and Sabatini, 2020). We propose that these types of stimuli may be sufficient to activate mTORC1 and generate ROS that promotes GSDMD oligomerization. Moreover, some inflammasome triggers and microbial invasion itself may serve as a source of mTOR activation or ROS production to prime GSDMD for subsequent oligomerization (West et al., 2011). The role of additional endogenous or microbial activators of these pathways should be investigated in the context of control of GSDMD pore formation in future studies.
A link between metabolism and cell death has been investigated in prior studies, including work demonstrating that NLRP3 and GSDMD can be post-translationally modified by TCA cycle intermediates (Hooftman et al., 2020; Humphries et al., 2020). Moreover, metabolic dysfunction and mitochondrial damage have been investigated in the initiation of death pathway signaling, such as formation of the apoptosome and NLRP3 inflammasomes (Andersen and Kornbluth, 2013; Prochnicki and Latz, 2017). A notable outcome of our study is that our data suggest a metabolic checkpoint exists after activation of the inflammasome and after cleavage of GSDMD. The discovery that upstream regulators of the mTOR pathway define a checkpoint for pore formation and pyroptosis suggests that we can no longer conclude that biochemical assessment of GSDMD cleavage is equivalent to pore formation.
In summary, this study broadens the regulated stages in the pyroptosis pathway to include activities downstream of GSDMD cleavage. These regulatory events are likely specific to activities within cells, as GSDMD has the intrinsic capacity to insert into liposomal membranes upon cleavage by caspase-1 or caspase-11 (Aglietti et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). Based on this idea, it stands to reason that additional cellular factors may exist that control pore formation, pyroptosis and other innate immune signaling pathways. Our study provides a mandate for “precision screening strategies” to be performed in the innate immune system, where assays for single stages in a pathway may allow for the complexity of cellular activities to be condensed, thereby revealing unexpected regulation of innate immune signal transduction.
Limitations of the Study
Our screen identified regulators of GSDMD that operate (post-cleavage) to promote oligomerization and pore formation that leads to pyroptosis. We focused our mechanistic analysis on the Rag GTPases and downstream mTORC1 effectors, but survival enrichment suggests that FLCN and Lamtor1–4 also serve a similar role in promoting mitochondrial ROS and GSDMD pore formation to enforce pyroptosis. In other contexts, RagA and RagB have redundant roles in mTOR activation (Liu and Sabatini, 2020). Thus, other Rag-Ragulator complex members as well as other prominent identified screen hits related to ROS biology, autophagy or organelle homeostasis should be investigated further. Moreover, our results suggest that the genetic association between the Ragulator-Rag complex and GSDMD may act through downstream effectors of the mTORC1 metabolic pathway, namely mitochondrial quality control and ROS metabolite production. Mechanistic insights into how mitochondrial ROS may affect GSDMD oligomerization potential await further characterization.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan C. Kagan (jonathan.kagan@childrens.harvard.edu).
Materials availability
All materials generated in this study are available from the Lead Contact.
Data availability
De-multiplexed FASTQ files and corresponding raw read counts for uninduced input and sorted BFP+ samples have been deposited at the Sequence Read Archive (SRA) and are publicly available as of the date of publication. PRJNA739880 is the BioProject number associated with data from this manuscript.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Immortalized macrophages, HEK293, HEK293T cell lines were cultured in complete DMEM with 10% FBS (Gibco) and Penicillin Streptomycin (Gibco) with added supplements of L-glutamine (Gibco) and sodium pyruvate (Gibco) at 37°C and 5% CO2.
METHOD DETAILS
Construct preparation
For inducible expression upon retroviral transduction, all constructs were inserted into pRETROX Tre3G plasmid (Clontech). Gene encoding mouse full-length GSDMD was ordered from Synobiological and tagBFP-encoding plasmid was from Evrogen. I105N point mutation was introduced by site-directed mutagenesis. tagBFP-tagged GSDMD variants and Dmrb-caspase-1 construct were prepared by overlap PCR using Phusion HF polymerase (Thermo Fisher Scientific). All mouse GSDMD gene variants were restriction digested by BamHI and EcoRI restriction nucleases and inserted into pRETROX Tre3G at BamHI/EcoRI sites.
Dimerizable caspase-1 construct Dmrb-caspase-1 (DMRB-DMRB-casp1ΔCARD) was digested by NotI and XbaI restriction nucleases and inserted into pcDNA3. I104N and L290D point mutations were introduced by site directed mutagenesis in ORFs encoding human NT and full-length GSDMD in pcDNA3.1, respectively.
For CRISPR/Cas9 targeting parental empty vectors pXPR-054-Blast or pXPR-053-Vex were digested with BsmbI restriction enzyme and the cut vector was purified by gel extraction. SgRNA-corresponding DNA oligos were ordered from IDT and designed based on the targeting sequence of guides found in the Brie library. Forward oligo sequences had the sequence CACCG added 5’ to the sgRNA targeting sequence. Reverse complement oligo sequences had the sequence AAAC added to the 5’ end and C added to the 3’ end. Equal molar quantities of forward and reverse oligos were allowed to anneal to each other and were phosphorylated for efficient ligation to the purified and digested parental vectors.
Primary BMDM generation and immortalization
Bone marrow derived macrophages (BMDMs) were differentiated from whole bone marrow derived from flushed tibia and femurs from C57BL/6J wild type, NLRP3-deficient, gasdermin D-deficient, LysM CRE-expressing, LysM CRE × flox/flox Raptor, LyM CRE × flox/flox Rictor, and Cas9 knock in mice. BMDMs were also differentiated from CHIME reconstituted chimeras of WT control untargeted mice, RagA g1 targeted mice, and RagA g2 targeted mice. In brief, leg bones were surgically removed and cleaned of surrounding tissue. The bones were washed with sterile PBS and left in complete DMEM ((Gibco) with 10% FBS (Gibco) and Penicillin Streptomycin (Gibco) with added supplements of L-glutamine (Gibco) and sodium pyruvate (Gibco) during extraction). The ends of tibia and femur were cut, and the bones were flushed with sterile PBS in a 10 ml syringe per bone and bone marrow from each individual mouse was pooled and pelleted at 400 × g for 5 minutes. ~10E6 bone marrow cells were used per non-TC coated 10 cm plate for differentiation. Differentiation was carried out in complete DMEM supplemented with 30% L929 supernatants containing M-CSF. J2 retrovirus secreting cells were passaged in complete DMEM and split every 2–3 days to prevent acidification and confluency. For immortalization viral supernatant production, flasks of J2 secreting cells were allowed to reach confluency and left to accumulate virus in their supernatants for 24 hours after reaching 100% confluency. Viral supernatants were spun at 400 × g to remove cell debris and passed through a 0.45 μm syringe filter to ensure no cellular cross-contamination. Two rounds of immortalization viral supernatants were placed on differentiating macrophage cultures and supplemented with 30% L929 supernatants. Transduced cells were allowed to recover in fresh complete DMEM with 30% L929 supernatants then split 1:2 or 1:3 as immortalized macrophages began to form clusters / colonies in non-TC coated 10 cm plates. Cells were progressively starved of M-CSF by reducing the L929 supernatant supplementation concentration over approximately 1.5–2 months until cells were able to grow without M-CSF and be split 1:10 every 2–3 days. Immortalized bone marrow derived macrophages (iBMDMs) were cultured in complete DMEM without L929 supernatant supplementation. Cells were washed in PBS pH 7.4 containing 2.5 mM EDTA to detach cells for passage. Cells were passaged 1:10 every 2–3 days to prevent overcrowding and media acidification.
Retroviral/Lentiviral transduction
For generating stable expression of Tet3G in Cas9 KI iBMDMs, pantrophic retrovirus particles were produced by transfecting plasmids pCMV-VSV-G (1.5 μg) and pRETROX Tet3G (2.5 μg) (Clontech) in Platinum-GP (Cell Biolabs) packaging cell line with Lipofectamine 2000 (10 μL). 3 days after transduction media was swapped for complete DMEM with 1.5 mg/ml G-418 for selection of productively transduced cells. For generating doxycycline inducible GSDMD variants in Cas9 KI Tet3G stable cells, amphotrophic retrovirus particles were produced by transfecting the respective pRETROX Tre3G plasmid (4 μg) into Gryphon Ampho (Alelle Biotechnology) packaging cell line with Lipofectamine 2000 (10 μL). Media was swapped for complete DMEM with 10 μg/ml puromycin and 1.5 mg/ml G-418 3 days after transduction. These cells were further single cell cloned by limiting serial dilution to yield clonal populations for purposes of magnitude and synchronization of doxycycline induction responses.
For generating CRISPR/Cas9 targeted cell lines, lentivirus particles were produced by transfecting plasmids psPAX2, pMD2.G, and the transfer plasmid pXPR-054-Blast containing the sgRNA sequence of interest. We targeted RagA with two independent guide RNA sequences derived from the Brie library (g1: GGTTCCCCAAGAATCGGACG and g2: GATCAGCTGATAGACGATGC). We targeted RagC with two independent guide RNA sequences derived from the Brie library (g1: CTTCCAGGACGACTACATGG and g2: TTTCTGTACCACCTTACTGA). Plasmids were transfected into HEK 293T cells in 10 cm dishes at a confluency of 50–70% with Lipofectamine 2000 at a DNA to Lipofectamine ratio of 1:3. Media was changed 8–12 hours post DNA transfection and viral supernatants were collected 24 hours post media change. Viral supernatants were spun at 400 × g to remove cellular debris then passed through a 0.45 μm PVDF filter via syringe. Viral supernatants were placed directly on growing Cas9 expressing iBMDMs and spinfected at 1250 × g for 45 minutes at 30°C. These cell lines were selected with 10 μg/ml of Blasticidin to select for cells expressing sgRNA stably after transduction.
CHimeric IMmune Editing
For generating CRISPR/Cas9 edited bone marrow chimeras for ablation of genes of interest in primary immune cells, we adapted a protocol known as CHimeric IMmune Editing (CHIME) as previously described (LaFleur et al., 2019). We targeted RagA with two independent guide RNA sequences derived from the Brie Library (g1: GGTTCCCCAAGAATCGGACG and g2: GATCAGCTGATAGACGATGC). These correspond to the same guide RNA sequences used in the above pXPR-054-Blast vector for generation of RagA-deficient iBMDMs. We also utilized a control untargeted guide RNA sequence (GCGAGGTATTCGGCTCCGCG). These guide RNAs were cloned into a lentiviral expression vector pXPR-053, which expresses an sgRNA and the fluorophore Vex (a violet laser excited variant of GFP). Lentivirus was prepared by PEI-based transfection of HEK-293x cells with Optimem containing pXPR-053 plasmids with described gRNAs and the packaging plasmids psPAX2 and pMD2.G. Lentivirus was isolated 72 hours post transfection, centrifuged at 2000 rpm for 5 minutes, and passed through a 0.45 μm filter to remove cells. The lentivirus was then concentrated by ultracentrifugation (20000 rpm, 2 hours, 4°C) using an SW28 rotor. Viral pellets were resuspended overnight in Ham’s F12 media and frozen at −80°C. Viral titer was assessed on 293x cells. The mice used in these experiments were generated by crossing loxP-Stop-loxP-Cas9 mice (Jackson #026175) (Platt et al., 2014) to Zp3-Cre (Jackson #003651) to delete the Stop cassette in the germline and create a constitutive Cas9-expressing mouse strain. This resulting strain loxP-Cas9 was backcrossed greater than 10 generations to Jackson C57BL6/J mice (Jackson #000664). LSK bone marrow cells were isolated from femurs, tibias, and spines from Cas9-expressing mice. Briefly, bones were mechanically dissociated and LSK were enriched using a CD117 isolation kit (Miltenyi Biotec). Enriched bone marrow was stained with cKit (CD117), Sca-1, and lineage markers (Ter-119, CD3e, CD11b, Gr-1, B220, and CD5) and sorted on a BD AriaII for Lineage– Sca-1+ cKit+. LSK were plated overnight in PVA-containing media (500 mL Ham’s F12 media with 10 mM HEPES, 1% ITSX, 1% Penicillin-Streptomycin, 1 mg/mL 87–90% hydrolyzed polyvinyl alcohol, 10 ng/mL murine recombinant stem cell factor, and 100 ng/mL murine recombinant thrombopoietin). Non-TC coated plates were coated with retronectin overnight at 4°C. The next day, LSK were then spin transduced (1800 rpm for 90 minutes at 27°C) with lentiviral constructs on the retronectin-coated plates (MOI of 30). The transduced LSK cells were then cultured for one week in PVA-containing media splitting the cells every other day (Wilkinson et al., 2019). LSK were then sorted for Vex positivity and 50K cells per mouse were intravenously transferred into irradiated (2 × 600 rad 3 hours apart) CD45.2 WT mice. These mice were administered Sulfamethoxazole-Trimethoprim in their drinking water for 2 weeks. After 8 weeks, Vex+ cells from the spleens and bone marrow of recipient mice were used for in vitro differentiation and ex vivo inflammasome assays.
For ex vivo splenocyte stimulations, spleens were surgically removed from deceased mice from WT control untargeted, RagA g1 targeted, and RagA g2 targeted bone marrow chimeras. Spleens were kept on ice in complete DMEM after removal. Whole spleens were homogenized using the plunger of a 10 ml syringe through a 70 μm cell strainer atop a 50 ml falcon tube. The cell strainer was washed with complete DMEM to collect residual single cells. Cells were pelleted at 400 × g for 5 minutes then subjected to ACK lysis buffer for 5 minutes. Cells were again pelleted and resuspended in complete DMEM and passed through a 40 μm cell strainer atop a 50 ml conical tube. Bulk splenocyte cells were counted, and 2E5 cells were dispensed into sterile FACS tubes for FlaTox stimulations. After stimulation, cells were stained with anti-CD11b antibody (Thermo, 1:200) and TruStain FcX (Biolegend) according to the manufacturer’s protocol. These stained splenocytes were then analyzed on a Fortessa FACS machine. Cells were analyzed for frequency of Vex+ (productive transduction of sgRNA construct) and CD11b high (splenic phagocyte). The ratio of Vex+, CD11b high splenocytes were compared from FlaTox-treated cells and single LFn-Fla component treated conditions to yield a ratio of depletion of splenic phagocytes after FlaTox treatment.
Brie cellular library and screening assay
Brie sgRNA only lentiviral library was obtained from the Broad Institute Genome Perturbation Platform. The Brie library contains expression constructs for sgRNA with coverage of 4 guides per gene in the murine genome and non-targeting controls. Our viral library coupled sgRNA expression to the expression of a blasticidin resistance gene for selection of productive transduction. Viral tittering prior to bulk library creation optimized transduction conditions. Increasing amounts of Brie library virus stock (0–500 μL) were added to 3E6 cells in prewarmed, complete DMEM with 5 μg/ml polybrene (in a constant final volume of 2 ml) in a 12-well plate. Spinfection transduction was performed by centrifugation at 1250 × g for 45 minutes at 30°C. After spinfection, 2 ml of complete DMEM was added and cells were incubated until the next day. Each condition was re-plated at lower density (1E5 cells/well of 6-well plate) in two wells to allow for ample room to grow during expansion and selection. In half of the wells, cells were selected with 4 μg/ml blasticidin for 4 days. At the end of selection and expansion, cell viability was measured to determine infection efficiency through a ratio of the number of blasticidin-resistant cells and the number of non-selected cells for each condition. The target efficiency is 30–50% transduction efficiency by this assay to yield primarily 0.5–1 virus per productively transduced cell to ensure our downstream enrichment analysis could correlate guide abundance to a single perturbation (Doench et al., 2016).
Downstream screening assays should have 400 cells per single guide perturbation representation for statistical robustness during enrichment analysis (Doench et al., 2016). For our library creation, we utilized 9450 μl of stock virus on 189E6 engineered macrophages. After spinfection as described above, 2 ml of fresh complete DMEM was added to each well and cells were incubated for one day. 24 hours after spinfection, cells were detached with cold sterile PBS with 2.5 mM EDTA and replated into 175 cm2 TC filter flasks (12E6 starting cells / flask). After one more day, selection was initiated using 4 μg/ml of blasticidin in complete DMEM that also contained 10 μg/ml puromycin and 1.5 mg/ml G418 to ensure our synthetic pathway was still maintained and productively transduced cells expressing sgRNA would also be selected and expanded. After 6 additional days, surviving cells were pooled and plated into 10 cm TC-coated dishes. ~150E6 cells were induced with doxycycline to express the toxic NT-GSDMD molecule for 16 hours. Cells were detached with cold PBS with 2.5 mM EDTA and sequentially washed with MACS buffer (1–2% FBS, 2.5 mM EDTA in pH 7.4 sterile PBS) to get rid of cellular debris and focus sorting analysis on intact and surviving cells. Cells were stained with PI solution (3.3 μg/ml) and sorted on an Aria cell sorter (BD). Cells were assayed for BFP fluorescence in the PacBlue channel and PI florescence in the PE-A channel. The top 15% BFP+, PI− cells were sorted as cells that may be defective for genes that promote GSDMD pore formation as they expressed the NT-GSDMD pore forming domain to high levels, but showed defective functional pore formation. Cells were sorted into PBS EDTA with 30% FBS to ensure efficient capture of cells/nuclei for downstream genomic DNA extraction. ~200E6 cells were collected that were left uninduced to serve as an input control to account for potential skewing of guide abundance in our population during the expansion and screening assay (for example for guides targeting known tumor suppressors that may outgrow in the population). Genomic DNA was isolated using Nucleospin Blood XL (Clontech) according to Broad Genome Perturbation Platform protocols. The gDNA was also subjected to PCR inhibitor clean up columns from Zymo Research (D6030) then diluted to the desired concentration and number of wells and sent on dry ice to the Broad Institute for PCR amplification and next generation Illumina Sequencing. After PCR amplification of sgRNAs from genomic DNA, and subsequent next generation sequencing, the log-normalized sgRNA abundance from the input sample was subtracted from the log-normalized sgRNA abundance of BFP-positive survivors.
Doxycycline induction in engineered cell lines
Cell lines of different genotypes were counted using a Luna 2 cell counter and plated at 1E5 cells / well in a black 96 well plate with optically clear bottoms in 200 μl of complete DMEM. After cells adhered (>6 hours resting in an incubator), media was replaced with warm complete DMEM or complete DMEM with doxycycline (0.5 μg/ml for initial characterization and 2 μg/ml for analysis of CRISPR/Cas9 edited clones). Cells were stained with diluted PI solution (final concentration 1:300) and lysis buffer or additional media were added to respective wells 30 minutes prior to a time point. The plate was spun at 400 × g for 5 minutes to ensure all cells were at the bottom of the well prior to adding the plate to the plate reader. Population PI staining was assayed with a Tecan plate reader or Biotek Synergy Mx. The program settings for PI inclusion assay were bottom reading of fluorescence with an excitation wavelength of 530 nm and emission wavelength of 617 nm. Supernatants from the spun down plate were taken for CyQuant LDH colormetric enzyme assay (Thermo) per the manufacturer’s instructions. Cells were lifted directly with MACS buffer (1–2% FBS, 2.5 mM EDTA in sterile pH 7.4 PBS) and assayed for frequency of PI+ cells and frequency of BFP+ cells on a Fortessa FACS machine (BD). Associated flow cytometry data was analyzed using FlowJo 10.0. For live cell microscopy, diluted PI (final concentration 1:300) was added to the wells right after doxycycline addition. The plate was allowed to equilibrate with PI for 30 minutes prior to imaging to allow for setting of exposure, gain, and focus settings based on lysed wells. The plate was placed in the Biotek Cytation with 5% CO2 and 37°C incubation with bright field and red channel images taken every 30 minutes for 8 hours.
Compound treatment
1E5 cells per well were seeded into 96 well black plates. Inhibitors/activators of mTOR pathway were added to cells at the same time as doxycycline (0.5 μg/ml). For inhibitors 14 hours endpoint PI and LDH measurement was selected. For activator MHY1486 early timepoint of 6 hours was selected to determine PI uptake and LDH release. Response was normalized to untreated (0%) and uninhibited, Dox-treated samples (100%).
Neon electroporation
Electroporation was conducted using the Neon transfection machine, pipette, tips, and associated commercial buffers (ThermoFisher Scientific). For experiments involving electroporation of LPS into cells, 1.2E6 cells were suspended in 120 μl of R buffer from the Neon transfection kit. Either 1.2 μl of PBS or with 1.2 μl of 1 mg/ml LPS (effective dose of 1 μg of LPS per 1E6 cells in a 100 μl Neon pipette tip) were added to the cell suspension prior to electroporation. Cells were drawn into the Neon transfection pipette and electroporated with the parameters of Voltage 1400, pulse width 10 ms, and pulse number 2. For protein isolation, 1E6 cells were dispensed into 500 μl of L-glutamine and sodium pyruvate supplemented, serum-free OptiMEM to allow for analysis of whole lysate and supernatant combined through direct lysis with 5X SDS Laemmli buffer and TCEP reducing agent. For PI incorporation, LDH release, and supernatant collection for ELISA analysis of cytokines, the 100 μl containing 1E6 electroporated cells were dispensed into 900 μl of complete DMEM lacking antibiotics and 100 μl of this cell solution was dispensed into 6 wells for each condition in a black 96 well plate with optically clear plastic bottom. Of these wells, 3 wells served as internal lysis controls and 3 wells served as experimental conditions. Diluted PI solution (final concentration 1:300) was added to every well 30 minutes prior to the end time point. Lysis buffer or complete DMEM with no antibiotics were added to corresponding wells for PI and LDH normalization. Population PI staining was measured with a Tecan plate reader. For live cell microscopy, diluted PI (final concentration 1:300) was added to the wells right after plating. The plate was allowed to equilibrate with PI for 30 minutes prior to imaging to allow for setting of exposure, gain, and focus settings based on lysed wells. The plate was placed in the Biotek Cytation with 5% CO2 and 37°C incubation with bright field and red channel images taken every 10 minutes for 1 hour.
To generate NINJ1 knock out macrophages via electroporation, 1E6 Cas9-expressing iBMDMs were electroporated with 2 μl of sgRNAs (sgRNA1: TGCCAACAAGAAGAGCGCTG; sgRNA2: GACCACAAGGGGCACGAAGA; pre-designed and ordered from IDT, diluted to 100 μM in nuclease-free H2O) using the Neon transfection system (electroporation conditions as described above for LPS electroporations, Voltage 1400, pulse width 10 ms, and pulse number 2). After 3 days, gDNA was extracted from 1E5 cells using 50 μl of QuickExtract DNA extraction solution (Lucigen) according to manufacturer’s protocol. The genomic region around exon 2 of NINJ1 was amplified by PCR (Fwd: TGTGTGCAGAGGTTGGTGTT; Rev: AGTCACAGCTTGGGCCATAC), and the resulting PCR product was sequenced by Sanger sequencing using the Fwd PCR primer. Sequencing traces from WT, guide 1 targeted, and guide 2 targeted cells were uploaded into ICE software tool by Synthego (https://ice.synthego.com/#/) to evaluate knock out efficiency, which was determined to be >70% for both sgRNAs used.
Characterization of apoptosis
The PacBlue Annexin-V and 7-AAD apoptosis detection kit from Biolegend was used to characterize the apoptotic ability of LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs in response to 1 μM of Staurosporine (STS) for 8 hours. The manufacturer’s staining protocol was followed. Cells were analyzed by a Fortessa FACS machine (BD). Double-positive (Annexin-V+, 7-AAD+) cells were quantified by quartile gating comparing unstimulated control cells and STS stimulated experimental conditions using FlowJo.
Bacterial infection
Salmonella enterica serovar Typhimurium (SL1344) was streaked on Luria Broth (LB) agar plates with no antibiotic selection to yield single colonies. Single colonies were picked with a sterile tip and used to inoculate a liquid culture of normal LB in 2.5 ml of a 14 ml bacterial culture tube and left to shake at 37°C for 12 hours. Cultures were back-diluted 1:20 into 2.5 ml of high salt LB (LB + 0.3M NaCl) to encourage an invasive phenotype for 3 hours shaking at 37°C. Liquid culture was analyzed undiluted and 1:10 diluted in high salt LB compared to a blank of high salt LB alone to measure an accurate OD600. It was assumed that an OD600 corresponded to roughly 1E9 bacteria / ml for purposes of MOI calculations. 1 ml of liquid culture was spun at 10,000 × g in a microcentrifuge tube for 5 minutes at 4°C to pellet bacteria. This pellet was sequentially washed in sterile PBS pH 7.4 for 3 washes and spins. Calculated amounts of bacteria to yield an MOI of 10 were added to either L-glutamine / sodium pyruvate supplemented antibiotic free Opti-MEM for protein isolation experiments or complete DMEM lacking antibiotics for plate reader experiments in a black 96 well plate with optically clear bottoms. Bacteria were spun down onto cells at 400 × g for 5 minutes to synchronize bacterial uptake by macrophage lines. 5X SDS laemmli buffer with TCEP reducing agent was added directly to protein isolation wells to yield a cellular lysate and low serum supernatant protein sample for western blot analysis. 30 minutes prior to end point, dilute PI solution (final concentration 1:300) was added to all wells in the 96 well plate. Lysis buffer or complete DMEM with no antibiotics were added to respective wells for PI and LDH normalization. At the end point time point of 1 hour, the plate was spun at 400 × g for 5 minutes to ensure all cells were at the bottom of the wells and PI fluorescence was read with a Tecan plate reader. Samples of supernatant were taken for LDH colorimetric enzyme assay.
Confocal microscopy
To follow localization of tagBFP-tagged gasdermin variants cells were seeded into μ-slide (Ibidi). Cells were non-treated or treated with doxycycline (0.5 μg/ml) for the defined time. 30 minutes prior the end of incubation period PI (3.33 μg/ml) was added. Cholera toxin subunit B-Alexa Fluor 647 (2 μg/ml) was added as well in the experiment to follow plasma membrane localization. Cells were observed under a Leica TCS SP5 laser scanning microscope mounted on a Leica DMI 6000 CS inverted microscope (Leica Microsystems, Germany) with an HCX plan apo 63X (NA 1.4) oil immersion objective used for imaging. A 405 nm laser line of 20 mW diode laser was used for tagBFP excitation, a 543 nm laser was used to follow propidium iodide, and a 633 nm laser was used for Cholera toxin subunit B-Alexa Fluor 647 excitation. To observe mitochondria, cells were followed at different time points after addition of doxycycline (0.5 μg/ml), Mitotracker Deep Red FM (30 nM) and PI (3.33 μg/ml).
Detection of cellular ROS
A day before stimulation, 3E5 cells were seeded per well of 24-well plate. For detection of intracellular ROS, CellRox Deep Red reagent (final concentration of 5 μM) was added to cells 7.5 hours after doxycycline stimulation. After 30 mins incubation in CO2-incubator, cells were washed with PBS and detached with PBS/EDTA. Samples were analyzed by flow cytometry using Cytek Aurora and FlowJo. Mean fluorescence intensity of morphologically intact population was analyzed.
Mitochondrial ROS and mitochondrial potential
For detection of mitochondrial ROS, cells were detached after 8 h-stimulation with doxycycline and incubated with MitoSOX (5 μM) for 20 minutes at 37°C in PBS/2% FBS. After washing step, samples were followed by flow cytometry using Cytek Aurora and analyzed using FlowJo. Fraction of MitoSOX-positive cells from morphologically intact population was analyzed.
Detection of change in mitochondrial potential was followed with TMRM (tetramethylrhodamine, methyl ester, perchlorate). Cells in 96 well black microtiter plates were stimulated with doxycycline (0.5 μg/ml) for 7 hours after which TMRM (final concentration 100 nM) was added for one hour. Medium was replaced with PBS/2% FBS and TMRM fluorescence was measured with Synergy Mx (Biotek) with 535 nm excitation and 600 nm emission settings. TMRM fluorescence was normalized to untreated control as 100%. To follow TMRM fluorescence using flow cytometry, cells were detached after 7.5 hour stimulation with doxycycline and incubated with TMRM (100 nM) for 30 minutes at 37°C in PBS/2% FBS. After washing step, samples were followed by flow cytometry using Cytek Aurora and analyzed using FlowJo. Mean fluorescence intensity of morphologically intact population was analyzed. MFI of samples 8 hours after doxycycline addition was normalized to non-treated (100%) and unlabeled (0%) control.
Activation assays in HEK293T and HEK293 cells
To follow human GSDMD variants’ constitutive pore-forming activity and Myd88 L265P stimulation of NF-κB activation, control HEK293T (WT) and RagA/B-DKO HEK293T cells (a kind gift from D.M. Sabatini (Rogala et al., 2019)) were seeded (25–55 E4/well) into black or white 96 well plate (Corning). The next day, cells were transfected with plasmids containing human GSDMD variants using Lipofectamine 2000. 20 hours later cells were analyzed for PI uptake and LDH release using Synergy Mx microtiter plate reader (Biotek). To determine % of PI uptake and LDH release, results were normalized to control plasmid transfected cells (0%) and lysed cells (100%). To follow Myd88 L265P activity, cells were transfected with pcDNA3 Myd88 L265P or empty vector, pELAM1-F-luciferase, phRL-TK Renilla luciferase (R-luc) plasmid mixture with Lipofectamine 2000. The total DNA in transfection mixture was kept constant in all conditions. Cells were lysed 24 hours later in Passive lysis buffer (Promega). Luciferase activity was followed upon coelentrazine and luciferin addition on Orion microtiter plate reader. Relative luminescence units were determined as ratio between F-luciferase and R-luciferase activity and normalized to control response (empty vector (pcDNA3) transfected cells).
4E4 HEK293 cells were seeded per well of 96 well black or white microtiter plate. The next day, cells were transfected with plasmid encoding Nt I104N hGSDMD (30 ng) and different amounts of plasmid encoding TSC-1 (170 ng, 100 ng, 0 ng). 20 hours later cells were analyzed for PI uptake and LDH release using Synergy Mx microtiter plate reader (Biotek). To follow Myd88 L265P activity, cells were transfected with Myd88 L265P or empty vector and pELAM1-F-luciferase/phRL-TK Renilla luciferase plasmid mixture and different amounts of plasmid encoding TSC-1 (140 ng, 100 ng, 0 ng). The total DNA in transfection mixture was kept constant in all conditions. Fold RLUs were calculated by normalizing Myd88 L265P response to empty vector control.
Starvation experiment
Dialyzed FBS was prepared by dialysis against PBS using 3500 MWCO dialysis tubes. 10X amino acid (AA) solution was prepared by mixing essential and nonessential AA solutions and L-glutamine and adjusting pH to 7.2.
3.5E4 HEK293T cells were seeded per well of black microtiter well plate. The next day cells were transfected with 70 ng of Dmrb-caspase-1 plasmid, 10 ng of full-length human GSDMD plasmid and 20 ng of pcDNA3 and 0.5 μL of Lipofectamine 2000. For western blot analysis experiment was performed in 24-well plate format. The number of cells and amount of transfection mixture was adjusted accordingly. The next day, cells were washed with DMEM without amino acids and glucose. After wash, medium containing dialized FBS and glucose was added, and cells were incubated for 50 minutes. 10X amino acid solution or DMEM without AA was added to final concentration of 1X. Homodimerizing molecule A20187 was added at the same time (final concentration 500 nM). LDH release was measured 4 hours later. To determine % of PI uptake and LDH release, results were normalized to control plasmid transfected cells (0%) and lysed cells (100%).
Subcellular fractionation
WT, RagA KO, and RagC KO iBMDMs (10E6 cells) were seeded in 15 cm tissue culture plates and incubated overnight at 37°C and 5% CO2. On the next day, transgene expression was induced by culturing cells in complete DMEM containing 2 μg/ml of doxycycline for 8 hours. Cells were lifted with PBS with 2.5 mM EDTA and washed once with 1 ml of hypotonic buffer (10 mM Tris pH 7.4, 1.5 mM MgCl2, 10 mM KCl, complete mini protease inhibitors). Cells were centrifuged (400 × g for 3 min at 4°C) and pellet was resuspended in 750 μl hypotonic buffer followed by incubation on ice for 30 min. Swelled cells were then homogenized by passing them 50 times through a 26-gauge needle. Nuclei and cell debris were pelleted by centrifugation at 2,500 × g for 5 min at 4°C. A small fraction of the post-nuclear supernatant was removed and used as an input control for western blotting. Supernatants were transferred to a new set of tubes and centrifuged again at 17,000 × g for 30 min at 4°C to pellet cellular membranes. The supernatant (cytosolic fraction) was removed, and membrane pellet was washed three times with 1 ml of hypotonic buffer. Samples for immunoblotting were prepared by adding an appropriate amount of 5X SDS loading buffer + 10X TCEP to cytosolic fraction and resuspending membrane pellet in an equal amount of 1X SDS loading buffer + TCEP. These protein samples were heated to 65°C for 10 min. Presence of NT-GSDMD-BFP fusion protein in each fraction was determined by western blot using a rabbit anti-tagBFP primary antibody. Tubulin (mouse primary antibody from DSHB, 1:100 dilution) and Na/K-ATPase (rabbit primary antibody from CST, diluted 1:1000) served as control markers for the cytosol and membrane fraction, respectively.
GSDMD oligomerization assay
WT, RagA KO, and RagC KO iBMDMs (1E6 cells per condition in 2 ml of complete DMEM) were seeded in 12-well plates and incubated at 37°C and 5% CO2 for 1 hour to allow cells to attach. Media was exchanged for 1 ml of Opti-MEM containing 2 μg/ml of doxycycline to induce the expression of NT-GSDMD-BFP for 8 hours at 37°C and 5% CO2. As indicated, 50 μl of Opti-MEM containing mitochondrial poisons (10 μM rotenone, 10 μg/ml antimycine A, 100 μM TTFA final diluted concentration) or H2O2 (625 μM, 1.25 mM, or 3.125 mM final diluted concentrations) were added to cell culture media after 4 hours to induce ROS. 250 μl of 5X SDS loading buffer was added directly to each well to capture proteins present in both the cell lysates and cell culture supernatants. Samples were split equally into two tubes and 75 μl of TCEP or dH2O was added to generate non-reducing and reducing western blot samples. After heating to 65°C for 10 min, proteins were separated by SDS-PAGE on a 4–12% acrylamide gradient gel (ThermoFisher Scientific) and NT-GSDMD-BFP transgene was detected by western blot. The assembled GSDMD pore is resistant to SDS under non-reducing conditions, thus oligomerization of NT-GSDMD-BFP was indicated by a gel shift towards higher molecular weight.
FRAP of CD14 and NT-GSDMD-BFP
Fluorescence recovery after photobleaching (FRAP) experiments were performed on a Zeiss 880 laser scanning confocal microscope using the in-built FRAP module within the microscope control software Zen Black. Empty vector WT Dox-inducible NT-GSDMD-BFP or RagA KO and RagC KO cells were plated on the 35 mm μ-Dish (Ibidi; Munich, Germany) and stained with PE conjugated anti-CD14 antibody (1:200) for 20 minutes at 37°C. Staining media was aspirated, and cells were washed twice with warm complete DMEM. After 8 hours of doxycycline induction of NT-GSDMD-BFP, endogenous BFP was also subjected to FRAP analysis in these genotypes. Cells were scanned using a 63X oil immersion lens with the 405 nm laser. Regions of interest (ROI) were created at cell membranes and fluorescence bleached by rapid scanning of increased laser power (5–10%) to a bleach depth of 40–60%. Time-lapse images were acquired over a 3-minute time course post-bleaching at 2-second intervals. Images were processed in Zen and FRAP data were fit to a single exponential model using GraphPad Prism.
Data analysis was performed using previously published methods (Haggie and Verkman, 2008). Fluorescence intensities of ROI in the bleaching area (ROIb = bleached area) were recorded for each time point. The final data was normalized to pre-bleached intensities of the ROIs data and fitted to a single exponential recovery curve. Percent fluorescence recovery (mobile fraction) was calculated from the plateau (Vmax) of the fitted curves normalized to the total bleached fluorescence.
Purification of LFn-Fla from insect cells
We adapted the protocol detailed from (Rauch et al., 2017; Rauch et al., 2016) to produce LFn-Fla in insect cells using the Bac-to-Bac baculoviral expression system (ThermoFisher Scientific). In brief, Sf9 cells were initially seeded at a density of 1E6 cells/ml in 750 ml of Hyclone SFX-insect media (GE Life Sciences) and grown overnight at 27°C at 110 rpm. 10 ml of P2 or P3 virus for expression of LFn-Fla was added with an additional 250 ml of media and grown at 27°C at 110 rpm for 72 hours. The insect cell pellets were collected by first spinning in large buckets at 1000 × g for 20–30 minutes. The cells were resuspended in PBS and transfered into a 50 ml conical tube (rated for high-speed centrifugations). All following steps were performed at 4°C and / or on ice. The 50 ml conical tube of cells was centrifuged at 1000 × g for 10 minutes. We resuspended the insect cell pellet in 50 ml of lysis buffer (25 mM HEPES-NaOH pH 7.4, 150 mM NaCl, 10 mM Imidazole) supplemented with complete protease inhibitor tablets and split into two 50 ml falcon tubes. We lysed these 25 ml fractions via sonication. An additional 25 ml of lysis buffer + 1 mM PMSF + 2 mM Benzamidine were added to each tube. The tubes were centrifuged at 20,000 × g for 45 minutes at 4°C to clarify the lysate. We equilibrated 1 ml of Ni-NTA agarose beads in lysis buffer (corresponds to 2 ml of a 50% ethanol slurry). Beads were added to the clarified lysate and incubated at 4°C for 1 hour while rotating. We washed the beads with a total of 50 ml of lysis buffer and combined into one tube. After a subsequent spin, we resuspended the beads in 25 ml of lysis buffer and poured into a gravity flow column (BioRad polyprep column). We washed the beads on the column with one column volume (~25 ml) of wash buffer (pH 7.4 25 mM HEPES-NaOH, 400 mM NaCl, 25 mM Imidazole).
Elution was performed with 5X the bead volume (~5 ml) of elution buffer (pH 7.4 25 mM HEPES-NaOH, 150 mM NaCl, 250 mM Imidazole). Buffer was exchanged into ion exchange chromatorgraphy (IEX) buffer A (25 mM Tris-HCl, pH 7.5) with PD-10 Desalting columns (GE Healthcare). IEX was performed on BioRad FPLC system (using HiTrap Q HP 1 ml column and loaded with a 10 ml loop) with a gradient from 0–100% of IEX buffer B (25 mM Tris-HCl pH 7.5, 1 M NaCl). The protein of interest was eluted as one single peak. Presence of protein of interest in peak fractions was confirmed by SDS-PAGE followed by InstantBlue staining (Expedeon). Protein concentration in pooled peak fractions was determined by measuring absorbance at 280 nm on a nanodrop device. 10% glycerol was added to pure protein and snap-frozen aliquots were stored at −80°C.
FlaTox stimulation in unprimed phagocytes
LFn-Fla was purified from insect cultures as described above. Aliquots of LFn-Fla were snap-frozen at a concentration of 0.5 mg/ml. Protective antigen (PA) was bought from a commercial vendor (List Biological Laboratories) and resuspended in sterile PBS pH 7.4 containing 10% glycerol to a concentration of 1 mg/ml and snap-frozen. For all stimulations, LFn-Fla and PA were diluted to a 2X working concentration in complete DMEM. To create 1X PA only treatments an equal volume of complete DMEM and 2X PA were combined. To create 1X LFn-Fla only treatments an equal volume of complete DMEM and 2X LFn-Fla were combined. To create 1X FlaTox treatments an equal volume of 2X PA and 2X LFn-Fla were combined. For iBMDM and BMDM stimulations, a final concentration of 250 ng/ml of PA and 100 ng/ml of LFn-Fla were used. For splenocyte stimulation, a final concentration of 250 ng/ml of PA and 500 ng/ml of LFn-Fla were used. For NAC experiments with iBMDM, NAC (Sigma) was reconstituted in sterile PBS pH 7.4 and pH was adjusted to pH 7.2–7.4 with NaOH. The final concentration of NAC stocks after pH adjustment was 200 mM. NAC stock solution was aliquoted into sterile tubes and kept at −20°C. For PI plate reader experiments, 1E5 cells were treated with 0 mM, 5 mM, or 15 mM NAC with respective stimulations in complete DMEM for 2 hours. For protein isolations, 1E6 cells were treated with 0 mM or 15 mM NAC with respective stimulations in supplemented Opti-MEM for 2 hours and directly lysed (supernatant and cells) with corresponding amounts of 5X SDS + 10X TCEP. Samples were heated for 10 minutes at 65°C, needled, then run on SDS-PAGE to investigate GSDMD cleavage by western blot.
Antibodies and reagents
LPS from E. coli, serotype 0111:B4 was supplied by Enzo (ALX-581-012-L002). Nigericin was purchased from Invivogen (tlrl-nig-5) or Sigma (N7143). Propidium iodide (PI) was purchased from Sigma (P4864-10ML). LDH activity was assayed using legacy Pierce™ LDH Cytotoxicity Assay Kit from ThermoFisher Scientific (88954) and replaced by CyQuant LDH Cytotoxicity Assay Kit from ThermoFisher Scientific according to the manufacturer’s protocol (C7026). Mouse IL-1β release was quantitatively measured from cell-free culture supernatants using the Invitrogen IL-1β Mouse Uncoated ELISA Kit (88-7013-88) according to the manufacturer’s protocol. For FACS analysis, the APC-conjugated antibody against CD11b from ThermoFisher Scientific (formerly eBioscience, 17-0112-82, 1:200) was used. For bone marrow FACS sorting, the APC-conjugated antibody against CD117 from Biolegend (135108, 1:100), the BV421-conjugated antibody against Sca-1 from Biolegend (108128, 1:100), the PE-conjugated antibody against Ter-119 from Biolegend (116208, 1:100), the PE-conjugated antibody against CD3ε from Biolegend (100308, 1:100), the PE-conjugated antibody against CD11b from Biolegend (101208, 1:100), the PE-conjugated antibody against Gr-1 from Biolegend (108408, 1:100), the PE-conjugated antibody against B220 from Biolegend (103208, 1:100), and the PE-conjugated antibody against CD5 from Biolegend (100608, 1:100) were used. For FRAP imaging analysis, the PE-conjugated antibody against CD14 from ThermoFisher Scientific (formerly eBioscience, 12-0141-82, 1:200) was used. For western blots, primary antibodies were used to detect proteins of interest including antibodies to tagBFP from Evrogen (AB233, 1:1000), Tet3G transactivator from Takara (631131, 1:1000), spCas9 from Cell Signaling Technologies (65832S, 1:1000) and from Abcam (ab191468, 1:1000), RagA from Cell Signaling Technologies (4357S, 1:1000), RagC from Cell Signaling Technologies (5466S, 1:1000), Raptor from Cell Signaling Technologies (2280S, 1:1000), Rictor from Cell Signaling Technologies (2114S, 1:1000), GSDMD from Abcam (ab209845, 1:1000), caspase-11 from Biolegend (647202, 1:500), human cleaved GSDMD from Abcam (ab210070, 1:1000), human GSDMD from Abcam (ab215203, 1:1000), phospho-p70 S6 kinase (Thr389) (1:1000) and p70 S6 kinase (49D7) (1:2000), β tubulin from DHSB (clone E7, 1:100), pro-IL-1β from GeneTex (GTX74034, 1:1000), Na/K ATPase from Cell Signaling Technologies (3010S, 1:1000), and β actin from Sigma (A5441, 1:5000) and Cell Signaling Technologies (3700S and 4967, 1:5000). Secondary antibodies to respective species with conjugated HRP were from Jackson ImmunoResearch or Abcam and used at 1:5000 (anti-mouse HRP 115-035-003, anti-rat HRP 112-035-003, anti-rabbit 111-035-003, anti-rabbit ab6721). Chemoluminescent signal substrate for HRP was SuperSignal West Pico (34577) and Femto Chemiluminescent Substrate (34095) from ThermoFisher Scientific and ECL from Amersham, GE Healthcare Life Sciences (RPN2232).
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of screening results
Log-normalized sgRNA abundance was provided by the Broad GPP from input and BFP+/PI− survivor cells populations. Population bias during expansion and assay were corrected for by subtraction of input log-normalized sgRNA abundance from log-normalized sgRNA abundance from the BFP+/PI− survivor cell population. We used the Broad GPP web portal for further analysis: https://portals.broadinstitute.org/gpp/public/. A strict Chip file (CP1139 from Broad GPP) was used to map perturbations to sites in the genome, and the normalized data was subjected to hypergeometric analysis without replacement. In this method, the rank of sgRNAs is used to calculate gene p-values using the probability mass function of a hypergeometric distribution. P-values are generated by calculating the average −log10(p-value) in both directions and picking the more significant one. The top n% of sgRNAs per gene can be used to calculate the average p-value with this method. The average log-fold change per gene is also reported and this can be used to assess the magnitude of effect. Data displayed on the volcano plot requires a minimum of 3 out of 4 guides being present.
For gene ontology functional analysis of our screen results, the complete rank list based on most positive log-fold change (LFC) to most negative LFC was input into the GOrilla web tool: http://cbl-gorilla.cs.technion.ac.il/.
Microscopy image processing
For acquisition and image processing, Leica LAS AF software was used where threshold, brightness and contrast were adjusted. All images from the same experiment were processed in the same way using the same parameters. Mitotracker Deep Red FM fluorescence was calculated with particle analysis tool in ImageJ where also initial thresholding to reduce non-mitochondrial stain was performed. Cells from 3–5 246×246 μm2 frames per experimental point were counted with CellCounter plugin in ImageJ. All images from the same experiment were processed in the same way using the same parameters. Composite figures were assembled in Microsoft Powerpoint.
Statistical analysis
Statistical significance for experiments with multiple groups and multiple independent variables were tested with two-way ANOVA with Tukey, Dunnett, or Sidak multiple comparison test corrections. One-way ANOVA with Dunnett multiple comparison test correction was used for analysis of multiple groups with a single independent variable. Unpaired two-tailed t-test was used for comparison of two groups. Adjusted p values were calculated with Prism 8.0 from GraphPad and the designation of * corresponds to p ≤ 0.05, ** corresponds to p ≤ 0.01, *** corresponds to p ≤ 0.001, and **** corresponds to a p ≤ 0.0001 in the figures. Data presented is representative of at least 3 independent experiments for western blots unless otherwise designated in figure legends. Data presented as quantified bar graphs or time course analysis is the combined means of 3 independent experiments unless otherwise designated in figure legends. Data with error bars are represented as mean ± SEM.
Supplementary Material
Supplemental Video Legends
Supplemental Video 1. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in WT macrophages.
Related to Figure 2.
Time-lapse live cell imaging of empty vector transduced iBMDMs during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 2. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in RagA guide 1 macrophages.
Related to Figure 2.
Time-lapse live cell imaging of RagA guide 1 transduced iBMDMs (g1, KO3) during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 3. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in RagA guide 2 macrophages.
Related to Figure 2.
Time-lapse live cell imaging of RagA guide 2 transduced iBMDMs (g2, KO3) during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 4. Cellular morphology and membrane permeability of primed WT macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE WT iBMDMs primed with IFNβ for 3 hours then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Video 5. Cellular morphology and membrane permeability of primed Rictor-deficient macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE Rictor deficient iBMDMs primed with IFNβ for 3 hours and then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Video 6. Cellular morphology and membrane permeability of primed Raptor-deficient macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE Raptor deficient iBMDMs primed with IFNβ for 3 hours and then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Figure Legends
Figure S1. Inhibition of NLRP3 inflammasome does not affect pore formation and pyroptosis induced by NT-GSDMD Related to Figure 1
(A, B) PI uptake by plate reader to measure bulk membrane permeability (A) and LDH release into cell free supernatants to measure bulk cellular lysis (B) after Dox induction (0.5 μg/ml) of NT-GSDMD-BFP in iBMDMs with or without MCC950 pretreatment (5 μM for 30 minutes) for 13 hours. Cells were primed for 5 hours with LPS (100 ng/ml) prior to Dox addition.
(C, D) PI uptake (C) and LDH release into cell free supernatants (D) in LPS primed (100 ng/ml for 5 hours) and Nigericin stimulated (5 μM for 1 hour) WT iBMDMs with or without MCC950 pretreatment (5 μM for 30 minutes).
All quantifications represent the mean and SEM of two independent experiments. Unpaired two-tailed t-test was used for analysis.
Figure S2. mTORC1 activity is decreased in RagA-deficient and RagC-deficient cells, Related to Figure 2
(A) Western blot of phosphorylated S6K (Thr389) and total S6K with β actin loading control in WT, RagA KO, and RagC KO iBMDM lysates.
Figure S3. Amino acid stimulation of mTORC1 promotes NT-GSDMD mediated pyroptosis, Related to Figure 3
(A) PI uptake analysis of WT and RagA targeted primary BMDMs after 2 hours of FlaTox treatment.
(B) Western blot of the proteins indicated after Dox induction (0.5 μg/ml) and compound treatment for 7 hours. Note that two forms of S6K are detected, p85 and p70.
(C) Western blot of transfected 293T cells expressing a small molecule dimerizable caspase-1 (Dmrb-caspase-1) and hFL-GSDMD. Cells were starved of amino acids for 50 min then treated with amino acids and dimerizer compound. Phosphorylated S6K, total S6K, and hFL-GSDMD were assessed with corresponding β actin loading controls 4 hours after amino acid supplementation and addition of the dimerizer compound A20187.
(D) LDH release into cell-free supernatants from cells transfected and treated as in (C).
Mean and SEM of three (A, D) independent experiments are shown. Two-way ANOVA with Tukey multiple comparison test (A) and unpaired two-tailed t-test (D) were used for analysis.
Figure S4. Analysis of LysM CRE, Rictor-deficient, and Raptor-deficient iBMDMs Related to Figure 4
(A) Western blot of the proteins indicated from lysates of unstimulated iBMDMs of LysM CRE, Rictor deficient, and Raptor deficient cells.
(B, C) PI uptake analysis (B) and LDH release into cell-free supernatants (C) from LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells plated for 3 hours before analysis.
(D) IL-1β ELISA measurements of cell-free culture supernatants from LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells plated for 3 hours before analysis.
(E) Western blot analysis of the proteins indicated from lysates of unstimulated or LPS (1 μg/ml for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
(F) Western blot analysis of the proteins indicated from lysates of unstimulated or LPS (1 μg/ml for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
(G) Western blot analysis of the proteins indicated from lysates of unstimulated or IFNβ (1000U for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
Mean and SEM of three independent experiments are shown. Two-way ANOVA was used for analysis.
Figure S5. RagA-deficient cells are resistant to mitochondrial damage Related to Figure 6
(A) Live cell confocal microscopy of mitochondria labeled with Mitotracker Deep Red stain (white), nuclei of porous cells with PI (red) and NT-GSDMD expression (green) in WT and RagA KO cells. Images were taken at defined times after Dox addition. Scale bar indicates 10 μm.
(B–E) To quantify cells enriched in Mitotracker Deep red stain (B), PI+ cells (C), Mitotracker Deep Red highly fluorescent and BFP+ cells (D) as well as BFP+ and PI+ cells (E) 3–5 246×246 μm2 frames were analyzed per experimental point. Mitotracker fluorescence was normalized to uninduced controls.
(F) TMRM fluorescence analysis by plate reader of WT and RagA KO iBMDMs after 8 hours of Dox induction (0.5 μg/ml). Values were normalized to uninduced controls.
(G) TMRM MFI in WT and RagA KO iBMDMs after 8 hours of Dox induction. MFI normalized to uninduced controls.
(H) Frequency of MitoSOX high after 8 hours of Dox induction in WT and RagA KO cells. Mean and SEM of MitoSOX-enriched fraction of intact (live) cells is presented.
Mean and the SEM of three (B, C, D, E, G, H) or five (F) independent experiments are shown. Unpaired two-tailed t-test (B, C, D, E, F, G) or two-way ANOVA (H) were used for analysis.
Figure S6. NINJ1 deficiency affects lysis but not GSDMD pore formation Related to Figure 6
(A, B) PI uptake analysis by plate reader (A) and LDH release into cell-free supernatants (B) was assessed from Cas9-WT, NINJ1 g1, and NINJ1 g2 iBMDMs primed with LPS (1 μg/ml) for 3 hours then treated with Nigericin (10 μM) for 2 hours.
(C, D) PI uptake analysis by plate reader (C) and LDH release into cell-free supernatants (D) was assessed from Cas9-WT, NINJ1 g1, and NINJ1 g2 iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells were plated for 2 hours before analysis.
Mean and SEM of 3 independent experiments are shown. Two-way ANOVA was used for analysis.
Highlights.
The Ragulator-Rag-mTORC1 pathway is required for pyroptosis induced by gasdermin D
Ragulator-Rag promotes gasdermin D oligomerization, but not membrane localization
Ragulator-Rag promotes reactive oxygen species (ROS) production in macrophages
ROS promotes gasdermin D oligomerization, pore formation and pyroptosis
Acknowledgements
This work was supported by NIH grants AI133524, AI093589, AI116550 and P30DK34854 to J.C.K, AI138369 to C.L.E., and DK125630 and DK113106 to J.R.T. J.C.K. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. C.L.E. is supported by the Harvard Herchel Smith and Landry Cancer Biology Fellowships. P.D. is supported by the Boehringer Ingelheim Fonds. I.H.B. received a Fulbright visiting scholar grant. The work was supported by Slovenian Research Agency (grant ARRS J3-1746 to I.H.B. and core funding P4-0176). I.H.B. is a recipient of the ICGEB grant (CRP SVN18-01). We thank members of the Kagan lab, Prof. Roman Jerala and members of Department of Synthetic Biology and Immunology for helpful discussions. We thank Mojca Benčina and Nataša Kopitar Jerala for methodological advice. Biorender generated schematics in Figure 1A, 2C, and the graphic abstract. We thank Ivan Zanoni for bone marrow and David Sabatini for critical cell lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
J.C.K. holds equity and consults for IFM Therapeutics and Corner Therapeutics. None of these relationships influenced the work performed in this study. J.G.D. consults for Microsoft Research, Agios, Phenomic AI, Maze Therapeutics, BioNTech, and Pfizer; JGD consults for and has equity in Tango Therapeutics. J.G.D.’s interests were reviewed and are managed by the Broad Institute in accordance with its conflict of interest policies.
References
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, and Dueber EC (2016). GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 113, 7858–7863. 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, and Kornbluth S (2013). The tangled circuitry of metabolism and apoptosis. Mol. Cell 49, 399–410. 10.1016/j.molcel.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avbelj M, Wolz OO, Fekonja O, Bencina M, Repic M, Mavri J, Kruger J, Scharfe C, Delmiro Garcia M, Panter G, et al. (2014). Activation of lymphoma-associated MyD88 mutations via allostery-induced TIR-domain oligomerization. Blood 124, 3896–3904. 10.1182/blood-2014-05-573188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, and Hornung V (2011). Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol 187, 613–617. 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher D, Monteleone M, Coll RC, Chen KW, Ross CM, Teo JL, Gomez GA, Holley CL, Bierschenk D, Stacey KJ, et al. (2018). Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med 215, 827–840. 10.1084/jem.20172222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, and Monack DM (2010). Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med 207, 1745–1755. 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, and Schroder K (2020). Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med 217. 10.1084/jem.20190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, and Zheng P (2008). TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med 205, 2397–2408. 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KW, Gross CJ, Sotomayor FV, Stacey KJ, Tschopp J, Sweet MJ, and Schroder K (2014). The neutrophil NLRC4 inflammasome selectively promotes IL-1beta maturation without pyroptosis during acute Salmonella challenge. Cell Rep. 8, 570–582. 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- Coll RC, Hill JR, Day CJ, Zamoshnikova A, Boucher D, Massey NL, Chitty JL, Fraser JA, Jennings MP, Robertson AAB, and Schroder K (2019). MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol 15, 556–559. 10.1038/s41589-019-0277-7. [DOI] [PubMed] [Google Scholar]
- Condon KJ, Orozco JM, Adelmann CH, Spinelli JB, van der Helm PW, Roberts JM, Kunchok T, and Sabatini DM (2021). Genome-wide CRISPR screens reveal multitiered mechanisms through which mTORC1 senses mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 118. 10.1073/pnas.2022120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, and Puigserver P (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740. 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- de Vasconcelos NM, Van Opdenbosch N, Van Gorp H, Parthoens E, and Lamkanfi M (2019). Single-cell analysis of pyroptosis dynamics reveals conserved GSDMD-mediated subcellular events that precede plasma membrane rupture. Cell Death Differ. 26, 146–161. 10.1038/s41418-018-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA (2018). Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev 281, 8–27. 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, and Shao F (2016). Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116. 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, and Tschopp J (2008). Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320, 674–677. 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, and Kagan JC (2018). The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity 48, 35–44 e36. 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. (2006). Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol 7, 576–582. 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, and Miao EA (2013). Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253. 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggie PM, and Verkman AS (2008). Monomeric CFTR in plasma membranes in live cells revealed by single molecule fluorescence imaging. J. Biol. Chem 283, 23510–23513. 10.1074/jbc.C800100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R, Dick MS, Sborgi L, Meunier E, Hiller S, and Broz P (2018). The Gasdermin-D pore acts as a conduit for IL-1beta secretion in mice. Eur. J. Immunol 48, 584–592. 10.1002/eji.201747404. [DOI] [PubMed] [Google Scholar]
- Hooftman A, Angiari S, Hester S, Corcoran SE, Runtsch MC, Ling C, Ruzek MC, Slivka PF, McGettrick AF, Banahan K, et al. (2020). The Immunomodulatory Metabolite Itaconate Modifies NLRP3 and Inhibits Inflammasome Activation. Cell Metab. 32, 468–478 e467. 10.1016/j.cmet.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, and Manning BD (2011). mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol. Metab 22, 94–102. 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, and Xiong Y (2008). WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 22, 866–871. 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JJ, Liu X, Xia S, Zhang Z, Zhang Y, Zhao J, Ruan J, Luo X, Lou X, Bai Y, et al. (2020). FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol 21, 736–745. 10.1038/s41590-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries F, Shmuel-Galia L, Ketelut-Carneiro N, Li S, Wang B, Nemmara VV, Wilson R, Jiang Z, Khalighinejad F, Muneeruddin K, et al. (2020). Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637. 10.1126/science.abb9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, Peters R, Mudd MH, Allers L, Salemi M, et al. (2018). Galectins Control mTOR in Response to Endomembrane Damage. Mol. Cell 70, 120–135 e128. 10.1016/j.molcel.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Kornfeld OS, Lee BL, Stowe IB, O’Rourke K, Li Q, Sandoval W, Yan D, Kang J, Xu M, et al. (2021). NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136. 10.1038/s41586-021-03218-7. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Lee BL, Stowe IB, Kornfeld OS, O’Rourke K, Mirrashidi KM, Haley B, Watanabe C, Roose-Girma M, Modrusan Z, et al. (2019). IRF2 transcriptionally induces GSDMD expression for pyroptosis. Sci. Signal 12. 10.1126/scisignal.aax4917. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, et al. (2013). Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249. 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Kofoed EM, and Vance RE (2011). Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595. 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs SB, and Miao EA (2017). Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 27, 673–684. 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MW, Nguyen TH, Coxe MA, Yates KB, Trombley JD, Weiss SA, Brown FD, Gillis JE, Coxe DJ, Doench JG, et al. (2019). A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun 10, 1668. 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yin HL, and Yuan J (2008). Flightless-I regulates proinflammatory caspases by selectively modulating intracellular localization and caspase activity. J. Cell Biol 181, 321–333. 10.1083/jcb.200711082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J, Wu H, and Kagan JC (2019). Gasdermin D activity in inflammation and host defense. Sci. Immunol 4. 10.1126/sciimmunol.aav1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol 21, 183–203. 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J (2016). Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158. 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang C, Yang J, Zhou B, Yang R, Ramachandran R, Abbott DW, and Xiao TS (2019). Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization. Immunity 51, 43–49 e44. 10.1016/j.immuni.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, and Bryant CE (2014). Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA 111, 7403–7408. 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JS, Hisata S, Park MA, DeNicola GM, Ryter SW, Nakahira K, and Choi AMK (2015). mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep. 12, 102–115. 10.1016/j.celrep.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Napier BA, Brubaker SW, Sweeney TE, Monette P, Rothmeier GH, Gertsvolf NA, Puschnik A, Carette JE, Khatri P, and Monack DM (2016). Complement pathway amplifies caspase-11-dependent cell death and endotoxin-induced sepsis severity. J. Exp. Med 213, 2365–2382. 10.1084/jem.20160027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, et al. (2018). Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069. 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnicki T, and Latz E (2017). Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab. 26, 71–93. 10.1016/j.cmet.2017.06.018. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, and Fitzgerald KA (2012). TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150, 606–619. 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Deets KA, Ji DX, von Moltke J, Tenthorey JL, Lee AY, Philip NH, Ayres JS, Brodsky IE, Gronert K, and Vance RE (2017). NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and -8. Immunity 46, 649–659. 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Tenthorey JL, Nichols RD, Al Moussawi K, Kang JJ, Kang C, Kazmierczak BI, and Vance RE (2016). NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J. Exp. Med 213, 657–665. 10.1084/jem.20151809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala KB, Gu X, Kedir JF, Abu-Remaileh M, Bianchi LF, Bottino AMS, Dueholm R, Niehaus A, Overwijn D, Fils AP, et al. (2019). Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475. 10.1126/science.aay0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, and Alnemri ES (2017). Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun 8, 14128. 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, and Wu H (2018). Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67. 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S, Shkarina K, Demarco B, Heilig R, Santos JC, and Broz P (2018). ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 362, 956–960. 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L, Ruhl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Muller DJ, Broz P, and Hiller S (2016). GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35, 1766–1778. 10.15252/embj.201694696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP Jr., Aponte AM, Shen RF, Balaban RS, and Finkel T (2006). The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem 281, 27643–27652. 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Shen K, Rogala KB, Chou HT, Huang RK, Yu Z, and Sabatini DM (2019). Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex. Cell 179, 1319–1329 e1318. 10.1016/j.cell.2019.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, and Shao F (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192. 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, et al. (2012). Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414. 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Abellan A, Angosto-Bazarra D, Martinez-Banaclocha H, de Torre-Minguela C, Ceron-Carrasco JP, Perez-Sanchez H, Arostegui JI, and Pelegrin P (2019). MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol 15, 560–564. 10.1038/s41589-019-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, and Gray NS (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem 284, 8023–8032. 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, and Vance RE (2012). Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490, 107–111. 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, Li Z, Wang Y, Zhao Q, Shao F, and Ding J (2020). Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis. Cell 180, 941–955 e920. 10.1016/j.cell.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, and Shao F (2017). Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547, 99–103. 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, and Ghosh S (2011). TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480. 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson AC, Ishida R, Kikuchi M, Sudo K, Morita M, Crisostomo RV, Yamamoto R, Loh KM, Nakamura Y, Watanabe M, et al. (2019). Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121. 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yu Z, Chen X, Li J, Li N, Cheng J, Gao N, Yuan HX, Ye D, Guan KL, and Xu Y (2021). Structural insights into TSC complex assembly and GAP activity on Rheb. Nat. Commun 12, 339. 10.1038/s41467-020-20522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, and Shao F (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600. 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, and Tschopp J (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225. 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video Legends
Supplemental Video 1. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in WT macrophages.
Related to Figure 2.
Time-lapse live cell imaging of empty vector transduced iBMDMs during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 2. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in RagA guide 1 macrophages.
Related to Figure 2.
Time-lapse live cell imaging of RagA guide 1 transduced iBMDMs (g1, KO3) during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 3. Cellular morphology and membrane permeability after Dox-induction of NT-GSDMD in RagA guide 2 macrophages.
Related to Figure 2.
Time-lapse live cell imaging of RagA guide 2 transduced iBMDMs (g2, KO3) during 8 hours of Dox (2 μg/ml) induction of NT-GSDMD. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 30 minutes at 37°C in 5% CO2.
Supplemental Video 4. Cellular morphology and membrane permeability of primed WT macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE WT iBMDMs primed with IFNβ for 3 hours then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Video 5. Cellular morphology and membrane permeability of primed Rictor-deficient macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE Rictor deficient iBMDMs primed with IFNβ for 3 hours and then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Video 6. Cellular morphology and membrane permeability of primed Raptor-deficient macrophages after LPS electroporation.
Related to Figure 4.
Time-lapse live cell imaging of LysM CRE Raptor deficient iBMDMs primed with IFNβ for 3 hours and then electroporated with PBS or 1 μg of LPS in 100 μl of R buffer containing 1E6 cells for 1 hour following equilibration with PI containing media. Left panel is brightfield. Middle panel is PI fluorescence. Right panel is a merge of these two channels. Images were captured every 10 minutes at 37°C in 5% CO2.
Supplemental Figure Legends
Figure S1. Inhibition of NLRP3 inflammasome does not affect pore formation and pyroptosis induced by NT-GSDMD Related to Figure 1
(A, B) PI uptake by plate reader to measure bulk membrane permeability (A) and LDH release into cell free supernatants to measure bulk cellular lysis (B) after Dox induction (0.5 μg/ml) of NT-GSDMD-BFP in iBMDMs with or without MCC950 pretreatment (5 μM for 30 minutes) for 13 hours. Cells were primed for 5 hours with LPS (100 ng/ml) prior to Dox addition.
(C, D) PI uptake (C) and LDH release into cell free supernatants (D) in LPS primed (100 ng/ml for 5 hours) and Nigericin stimulated (5 μM for 1 hour) WT iBMDMs with or without MCC950 pretreatment (5 μM for 30 minutes).
All quantifications represent the mean and SEM of two independent experiments. Unpaired two-tailed t-test was used for analysis.
Figure S2. mTORC1 activity is decreased in RagA-deficient and RagC-deficient cells, Related to Figure 2
(A) Western blot of phosphorylated S6K (Thr389) and total S6K with β actin loading control in WT, RagA KO, and RagC KO iBMDM lysates.
Figure S3. Amino acid stimulation of mTORC1 promotes NT-GSDMD mediated pyroptosis, Related to Figure 3
(A) PI uptake analysis of WT and RagA targeted primary BMDMs after 2 hours of FlaTox treatment.
(B) Western blot of the proteins indicated after Dox induction (0.5 μg/ml) and compound treatment for 7 hours. Note that two forms of S6K are detected, p85 and p70.
(C) Western blot of transfected 293T cells expressing a small molecule dimerizable caspase-1 (Dmrb-caspase-1) and hFL-GSDMD. Cells were starved of amino acids for 50 min then treated with amino acids and dimerizer compound. Phosphorylated S6K, total S6K, and hFL-GSDMD were assessed with corresponding β actin loading controls 4 hours after amino acid supplementation and addition of the dimerizer compound A20187.
(D) LDH release into cell-free supernatants from cells transfected and treated as in (C).
Mean and SEM of three (A, D) independent experiments are shown. Two-way ANOVA with Tukey multiple comparison test (A) and unpaired two-tailed t-test (D) were used for analysis.
Figure S4. Analysis of LysM CRE, Rictor-deficient, and Raptor-deficient iBMDMs Related to Figure 4
(A) Western blot of the proteins indicated from lysates of unstimulated iBMDMs of LysM CRE, Rictor deficient, and Raptor deficient cells.
(B, C) PI uptake analysis (B) and LDH release into cell-free supernatants (C) from LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells plated for 3 hours before analysis.
(D) IL-1β ELISA measurements of cell-free culture supernatants from LysM-CRE WT, Rictor-deficient, and Raptor-deficient iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells plated for 3 hours before analysis.
(E) Western blot analysis of the proteins indicated from lysates of unstimulated or LPS (1 μg/ml for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
(F) Western blot analysis of the proteins indicated from lysates of unstimulated or LPS (1 μg/ml for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
(G) Western blot analysis of the proteins indicated from lysates of unstimulated or IFNβ (1000U for 3 hours) stimulated iBMDMs from LysM CRE, Rictor deficient, and Raptor deficient genotypes.
Mean and SEM of three independent experiments are shown. Two-way ANOVA was used for analysis.
Figure S5. RagA-deficient cells are resistant to mitochondrial damage Related to Figure 6
(A) Live cell confocal microscopy of mitochondria labeled with Mitotracker Deep Red stain (white), nuclei of porous cells with PI (red) and NT-GSDMD expression (green) in WT and RagA KO cells. Images were taken at defined times after Dox addition. Scale bar indicates 10 μm.
(B–E) To quantify cells enriched in Mitotracker Deep red stain (B), PI+ cells (C), Mitotracker Deep Red highly fluorescent and BFP+ cells (D) as well as BFP+ and PI+ cells (E) 3–5 246×246 μm2 frames were analyzed per experimental point. Mitotracker fluorescence was normalized to uninduced controls.
(F) TMRM fluorescence analysis by plate reader of WT and RagA KO iBMDMs after 8 hours of Dox induction (0.5 μg/ml). Values were normalized to uninduced controls.
(G) TMRM MFI in WT and RagA KO iBMDMs after 8 hours of Dox induction. MFI normalized to uninduced controls.
(H) Frequency of MitoSOX high after 8 hours of Dox induction in WT and RagA KO cells. Mean and SEM of MitoSOX-enriched fraction of intact (live) cells is presented.
Mean and the SEM of three (B, C, D, E, G, H) or five (F) independent experiments are shown. Unpaired two-tailed t-test (B, C, D, E, F, G) or two-way ANOVA (H) were used for analysis.
Figure S6. NINJ1 deficiency affects lysis but not GSDMD pore formation Related to Figure 6
(A, B) PI uptake analysis by plate reader (A) and LDH release into cell-free supernatants (B) was assessed from Cas9-WT, NINJ1 g1, and NINJ1 g2 iBMDMs primed with LPS (1 μg/ml) for 3 hours then treated with Nigericin (10 μM) for 2 hours.
(C, D) PI uptake analysis by plate reader (C) and LDH release into cell-free supernatants (D) was assessed from Cas9-WT, NINJ1 g1, and NINJ1 g2 iBMDMs primed with LPS (1 μg/ml) for 3 hours then electroporated with PBS or 1 μg of LPS. Electroporated cells were plated for 2 hours before analysis.
Mean and SEM of 3 independent experiments are shown. Two-way ANOVA was used for analysis.
Data Availability Statement
De-multiplexed FASTQ files and corresponding raw read counts for uninduced input and sorted BFP+ samples have been deposited at the Sequence Read Archive (SRA) and are publicly available as of the date of publication. PRJNA739880 is the BioProject number associated with data from this manuscript.


