Abstract
Despite the fact that AML is the most common acute leukemia in adults, patient outcomes are poor necessitating the development of novel therapies. We identified that inhibition of Thioredoxin Reductase (TrxR) is a promising strategy for AML and report a highly potent and specific inhibitor of TrxR, S-250. Both pharmacologic and genetic inhibition of TrxR impairs the growth of human AML in mouse models. We found that TrxR inhibition leads to a rapid and marked impairment of metabolism in leukemic cells subsequently leading to cell death. TrxR was found to be a major and direct regulator of metabolism in AML cells through impacts on both glycolysis and the TCA cycle. Studies revealed that TrxR directly regulates GAPDH leading to a disruption of glycolysis and an increase in flux through the pentose phosphate pathway (PPP). The combined inhibition of TrxR and the PPP led to enhanced leukemia growth inhibition. Overall, TrxR abrogation, particularly with S-250, was identified as a promising strategy to disrupt AML metabolism.
Keywords: securinine, thioredoxin reductase, AML, cancer cell metabolism, mitochondrial metabolism
Introduction:
Acute Myeloid Leukemia (AML) is characterized by aberrant proliferation of immature myeloid cells for which the defining feature is a block in myeloid differentiation 1. It is estimated that AML will affect approximately 20,000 new patients in 2017 with only 24% surviving 5 years 2. The standard treatment used for most AML patients has not changed in over four decades. This regimen includes cytarabine and an anthracycline which is unfortunately known for high toxicity and poor efficacy particularly in adults 3.
Due to specific metabolic dependencies in cancer cells as compared to normal cells, metabolic inhibitors have been successful in several cancers and recent efforts have seen promise as a safer therapeutic approach in AML. AML cells in particular have been found to be sensitive to impairment of multiple branches of metabolism including oxidative phosphorylation and glycolysis4. Isocitrate dehydrogenase (IDH) inhibitors for IDH mutant AML patients for example has been a successful strategy to target metabolism in AML 5,6.In addition, the recent success of the combination of venetoclax and azacytidine in some AML patients has been reported to be due to disruption of metabolism in the AML cells 7. While targeting metabolism in AML has shown promise, the ideal strategies to accomplish this goal are not yet known.
Here we report a promising strategy to target AML metabolism through inhibition of thioredoxin reductase using a novel small molecule inhibitor based upon the natural product securinine. We previously reported that securinine, a plant-derived alkaloid from Securinega suffruticosa, is a promising AML differentiation agent though the underlying mechanism of action was unclear 8,9. Though the parent securinine compound showed high promise in AML cell and animal models, it exhibits low potency (~20μM) and unacceptable toxicity for further clinical development 8. The major apparent toxicity of securinine is related to its role as a γ-aminobutyric acid (GABA) receptor antagonist which leads to seizures 10. We have previously demonstrated that the anti-AML properties of securinine are independent of GABA receptor activity 8. We performed lead optimization chemistry on securinine that led to derivatives with nanomolar efficacy on AML cells that also do not induce seizures10.
Although a potential role for securinine in cancer has been explored for over a decade, the direct mechanism by which it functions to target cancer cells is still unclear. Several proteins have been explored as potential mediators of securinine’s anti-cancer activity 11. These include PDIA6, NF-κB12, topoisomerase I 13, p73 14 and thioredoxin reductase. Thioredoxin Reductasereductase (TrxR), an oxidoreductase that is essential for redox homeostasis in mammalian cells 15. There are three isoenzymes found in cells that include cytosolic TrxR1, mitochondrial TrxR2, and TrxR3 16. TrxR functions as a homodimer with the primary catalytic site in the N-terminus and a selenocysteine containing second active site in the C-terminus of the protein 17,18. Thioredoxin reductase is highly expressed in several cancers and its role in maintaining redox homeostasis is thought to offer survival advantage to cancer cells 19. Therefore, TrxR has been explored as an anti-cancer target although efforts to identify specific inhibitors have not been very successful 20.
We show here that our optimized securinine analog S-250 is a potent and highly specific inhibitor of TrxR. This degree of potency and selectivity is in high contrast to previously identified TrxR inhibitors 20. Importantly, our studies also revealed that TrxR is an important modulator of metabolism in AML and TrxR inhibition leads to the rapid and direct impairment of metabolism leading to cell death.
Results:
New securinine analogs effectively inhibit various AML cell lines and patient samples
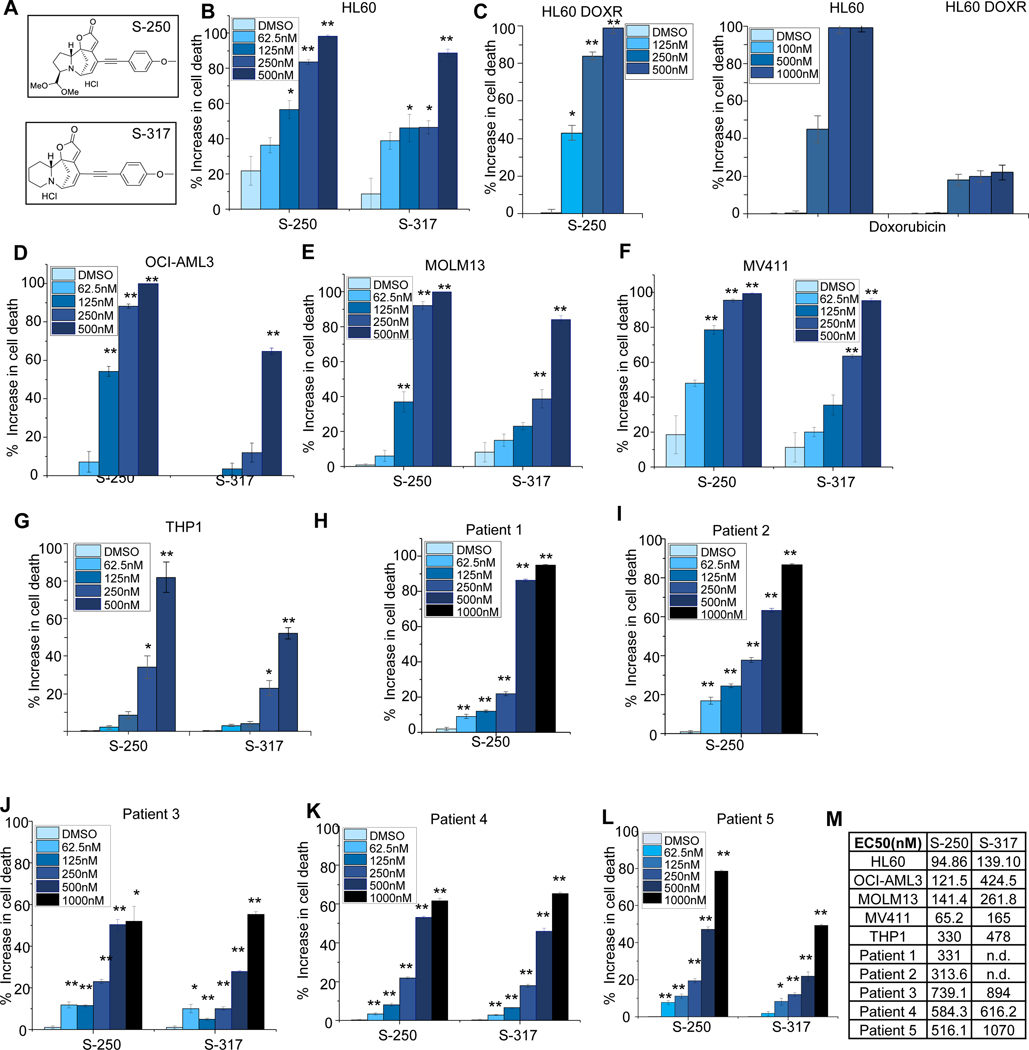
Screening for securinine derivatives with increased potency against AML cells and improved drug-like properties revealed the securinine analog S-317 and the closely related nor-securinine derivative S-250 (Figure 1A and 10). While the parental compound securinine exhibits an EC50 of ~20μM on AML cells 8, S-250 and S-317 show about ~100 fold in increase in efficacy (<150nM and <300nM respectively) when tested against a panel of AML cell lines, HL60, OCI-AML3, MOLM13, MV411 and THP-1 (Figure 1B-F and 1M). In the case of HL-60 cells, both parental and HL-60 Doxorubicin resistant cells (HL-60 DOX) were utilized. While HL-60 DOX cells showed a significant reduction in sensitivity to Doxorubicin treatment, S-250 treatment lead to similar effects on parental and HL-60 DOX cells (Figure 1C).
Figure 1: S-250 potently decreases the viability of AML cell lines and patient samples.
(A) Chemical structures of securinine derivatives, S-250 and S-317. (B-G) HL60, HL-60DOXR, OCI-AML3, THP-1 MOLM13 and MV411 cells were treated with the indicated doses of S-250, S-317 or Doxorubicin. After 72h of treatment, cells were stained with prestoblue and the percent increase in cell death as compared to vehicle is shown. n=4. A representative image of at least 3 independent experiments is shown. (H-L) Primary patient AML cells were treated with the indicated doses of S-250 or S-317 as described in Figure 1B-E. Increase in cell death in comparison to DMSO treatment are shown. (M) Effective concentration (EC50) of S-250 and S-317 on the indicated cells. n.d.- not determined. Unless otherwise indicated, data are presented as mean ± SE; *p<0.05; **p<0.01.
While securinine itself exhibits significant AML differentiation activity 8, these securinine derivatives exhibited their inhibitory properties primarily through cytotoxic effects (Figure 1) and only induced modest levels of monocytic differentiation as evidenced by the induced expression of CD11b and CD14 (Figure S1). In addition to activity on AML cell lines, these analogs also potently impact the survival of primary patient AML samples (Figure 1H-M). Despite the fact that AML is highly heterogenous, S-250 shows high activity across cell lines and patient samples containing various genetic abnormalities (Table S1). As the nor-securinine analog (S-250) showed higher potency as compared to the securinine derivative, S-317, it was primarily used in subsequent studies.
S-250 targets thioredoxin reductase
The direct target through which the parental compound securinine exhibits its anti-cancer properties is not well established. Several candidate proteins have been reported as targets of securinine including protein disulfide isomerase 21, thioredoxin reductase 15 and others such as Hsp70, Hsp90 and Fabp5 11. Further, the mechanism of action of analogs such as S-250 that exhibit a dramatically lower EC50 than securinine on AML cells is completely unknown.
To identify the target of S-250, we performed Drug Affinity Responsive Target Screening (DARTS) 22. DARTS utilizes the principle that target proteins are stabilized by ligand interactions rendering them resistant to digestion by proteases such as pronase 22. This approach identified several proteins as potential targets for S-250 (Table S2). In order to ascertain any functional, structural, genetic or physical interaction patterns among the candidate proteins, STRING, a protein interaction database was used to identify any protein networks involved 23. STRING analysis revealed that proteins belonging to the class of oxidoreductase enzymes were clustered together and were the most represented (Figure S2A) and therefore we focused on this cluster in our secondary analyses.
Interestingly, although several thioredoxin-domain containing proteins were present in the oxidoreductase enzyme cluster, two primary members of this class, thioredoxin reductase 1 and 2 were unaffected by pronase treatment and therefore it was not possible to assess their interaction with S-250 through DARTS analysis. We therefore included these two candidates to our STRING analysis and secondary screening (Figure S2A).
To validate direct S-250 targets, we performed secondary screening on a large panel of enzymes including several oxidoreductases in vitro. S-250 was found to inhibit thioredoxin reductase 1 (TrxR1) at nanomolar concentrations in a dose and time-dependent manner (Figure S2B-C). Interestingly none of the other enzymes such as malate dehydrogenases (MDH), lactate dehydrogenase (LDH) or GAPDH were inhibited in vitro at doses up to the highest tested (10μM) (Table S3). This panel also included the thioredoxin domain containing protein PDI, the structurally related glutathione reductase (GR) and the other human selenocysteine containing enzyme glutathione peroxidase (GPX) (Table S3). In contrast, known inhibitors of TrxR1 including a widely used inhibitor, Auranofin, inhibits similar enzymes such as glutathione peroxidase 24. Additionally, to further assess its specificity and potential safety, S-250 was tested using the SafetyScreen 44 panel to assess for interactions with a panel of proteins that commonly lead to adverse reactions during drug development. Supporting the specificity of its interaction with TrxR1, S-250 did not impact the activity of the vast majority of these proteins on this panel at 10μM S-250 (Table S4). Next we assessed the effects of S-250 on binding to the GABAA Receptor as the parental compound securinine interacts with the GABAA Receptor leading to seizures in animals. Utilizing an in vitro radioactive binding assay, S-250 unlike securinine, did not interact with the GABAA Receptor at the highest dose tested (100μM) (Table S5).
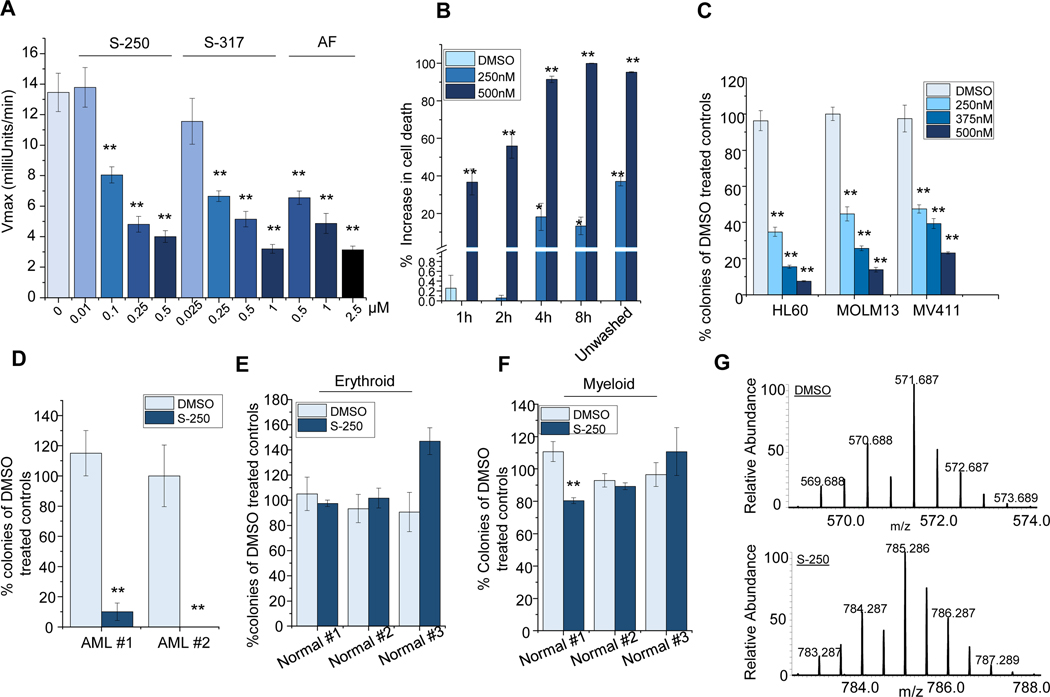
In addition to testing the inhibition of TrxR1 using purified enzyme, we investigated whether S-250 treatment inhibited TrxR1 in intact cells. S-250 was found to rapidly and potently inhibit TrxR1 at nanomolar concentrations in HL60 cells in a dose-responsive fashion as measured by DTNB reduction (Figure 2A). In order to ensure that the DTNB reduction observed was due to changes in TrxR1 activity, the cell lysates were treated with a commonly used TrxR inhibitor, Sodium Aurothiomalate. There was no further reduction in TrxR activity when combined with S-250 (Figure S2D) validating that S-250 significantly inhibits TrxR in AML cells. The activity of S-250 was also directly compared to auranofin, one of the most commonly utilized TrxR inhibitors. S-250 demonstrated greater than 5-fold higher potency in inhibiting TrxR activity in AML cells (Figure 2A). To explore the mode of activity of S-250 in cells, washout experiments were performed on AML cells. Treatments with S-250 as short as 4 hours led to nearly the same activity as continuous treatment for 72 hours suggesting irreversible inhibition of TrxR1 in cultured cells (Figure 2B).
Figure 2: S-250 irreversibly targets Thioredoxin reductase.
(A) HL60 cells were treated with varying concentrations of S-250, S-317 or Auranofin for 4h. Thioredoxin reductase activity in the cell lysates was measured using DTNB reduction assay. Average Vmax of TrxR from at least 3 independent experiments is shown. (B) HL60 cells were treated with DMSO or 250nM or 500nM S-250 for 1–8h. Cells were washed and viability was measured after 72h using the prestoblue assay. Unwashed control represents cells treated with S-250 continually for 72h. n=4. (C) Colony formation assay: HL60, MOLM13 and MV411 cells were treated with the indicated doses of S-250 for 4h, washed and plated in 0.3% soft agar. Colony counts after 10 days were normalized to DMSO treated controls. n=4. (D-F) AML patient samples (D) or normal human bone marrow samples (E-F) were treated with 500nM S-250 for 4h, washed and plated in methylcellulose. Colonies formed after 10 days were counted and plotted as percentage of the DMSO treated controls. n=4. (G) LC-MS analysis of the S-250 modified TrxR active-site peptide (SGGDILQSGCysSecG). Top- A doubly charged peptide was observed in the unmodified sample (DMSO) that has an average m/z ratio of 571.687 Da. The [M+H] for this peptide is 1142.374 Da. Bottom- A doubly charged peptide was observed in the S-250 modified sample that has an average m/z ratio of 785.286 Da. The [M+H] for this peptide is 1569.572 Da. Unless otherwise indicated, data are presented as mean ± SE; *p<0.05; **p<0.01.
To further investigate the ability of limited exposures of S-250 to impair AML growth, HL60, MOLM13 and MV411 cells were assessed for colony formation in semi-solid media after 4 hours of treatment. S-250 led to a dramatic reduction in the ability of AML cells to grow after this short treatment exposure (Figure 2C). Similarly, short S-250 exposures also inhibited colony formation of primary AML cells derived from patients (Figure 2D). As most AML therapeutics lead to significant hematopoietic cell toxicity, normal bone marrow cells were also treated and found to be unaffected by similar exposures to S-250 (Figure 2E-F). In addition to bone marrow, the liver is also a major site of drug toxicity, S-250 treatment did not have significant effects on a liver cell line, HepG2, at dose levels that lead to significant effects on leukemic cells (Figure S2E).
Finally, to confirm irreversible binding of S-250 to TrxR1, we performed mass spectrometric analyses utilizing S-250 and purified TrxR1 protein. This analysis revealed a covalent interaction between S-250 and the C-terminus active site of TrxR1 suggesting S-250 may act as an irreversible inhibitor. LC/MS analysis revealed an interaction at the active site peptide SGASILQAGCUG (Figure 2G), with a mass addition of 409.165 or 427.198 Da, corresponding to addition of S-250 possibly through Michael Addition of selenocysteine (U) at the 15th position of S-250, with a molecular weight increase of 409 or 427 (S-250+H2O) (Figure S2F). This mass addition was specific to S-250 treated sample as expected.
Securinine-250 significantly decreases AML burden in mice
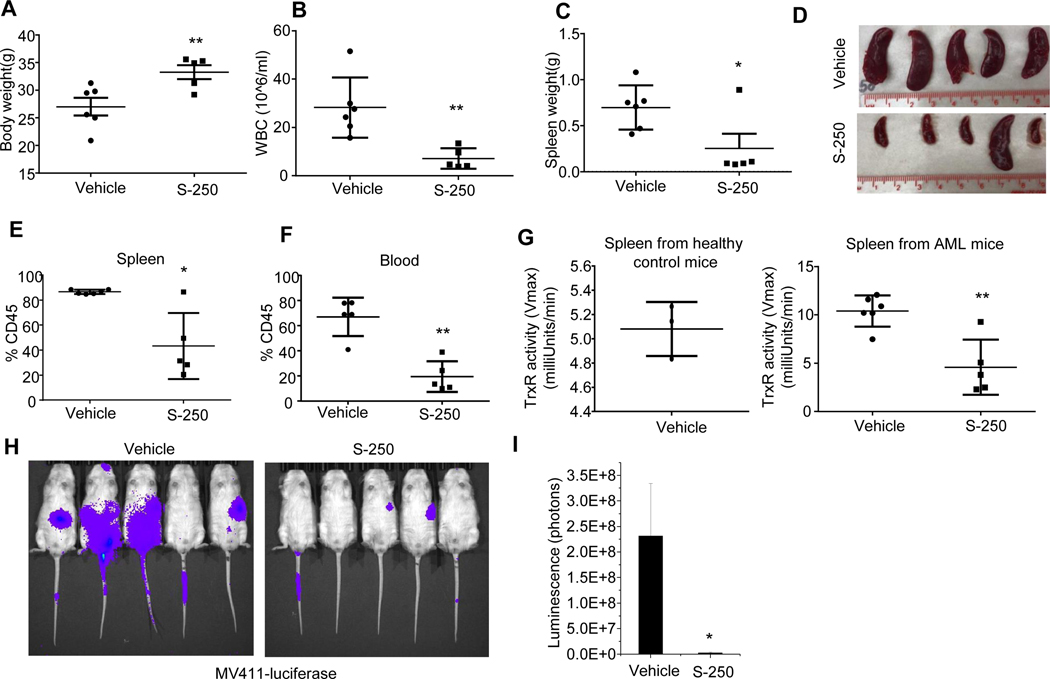
To investigate the effect of securinine derivatives in vivo, we used a highly aggressive circulating model of an AML patient derived xenograft using NOD-SCID IL2Rγ (NSG) mice. These human AML cells were shown to have an EC50 of ~500nM when tested with S-250 treatment in culture (Patient #5, Figure 1L-M). AML cells were injected intravenously into NSG mice in order to establish a circulating AML model and were treated with either vehicle or S-250. While the vehicle treated mice exhibited significant weight loss and an elevated WBC count concurrent with disease progression, mice treated with S-250 exhibited reduced weight loss and a lower peripheral blood WBC count (Figure 3A-B and Figure S3). S-250 treated mice showed reduced spleen sizes and weights (Figure 3C-D) and markedly decreased leukemic cell burden in the blood and spleen (Figure 3E-F and S3A-B).
Figure 3: S-250 decreases leukemic burden in circulating AML mice models.
(A) Body weights of NSG mice at the end of the 5 week study period that were injected with patient-derived leukemic cells and treated with either vehicle or S-250. (B) White blood cell (WBC) count in the mice in (A). (C-D) Weight and image of the spleens in the mice in (A). (E-F) Percent human CD45 positive cells in the spleen (E) or blood (F). (G) (Left) C57B/6 mice were injected with vehicle and splenic cells were isolated. TrxR activity in the normal spleen cells was measured by the DTNB reduction assay. n=3. (Right) TrxR activity in splenic cells from vehicle or S-250 treated mice as measured by DTNB reduction assay. Data are presented as mean ± Standard Deviation. n=5–6 mice per group. (H) Bioluminescent imaging on day 39 of NSG mice that were injected with MV411-luciferase cells and treated with either vehicle or S-250. (I) Quantification of images in (H). n=5 mice. Data are presented as mean ± SE. *p<0.05; **p<0.01.
We next investigated if S-250 treatment in mice led to inhibition of TrxR in AML cells in vivo. As the AML model leads to the virtual replacement of normal cells in the spleen with leukemic cells, we assessed TrxR activity in lysates from isolated spleen cells. Initially, we compared the TrxR activity of cells isolated from the spleen in healthy mice (C57BL/6) as compared to spleen cells isolated from mice with a high leukemic burden representing predominantly leukemic cells. The spleen cells from healthy controls exhibited reduced levels of TrxR activity (Figure 3G). Importantly, S-250 treated mice exhibited significantly reduced TrxR activity in splenic derived AML cells as compared to similar cells from vehicle-treated mice (Figure 3G) further supporting TrxR as a major target of S-250 in vivo.
To further explore the in vivo activity of S-250 in another highly aggressive model of human AML, MV411-luciferase cells were injected intravenously into NSG mice. S-250 caused a 50-fold reduction in tumor burden in this model as measured by bioluminescent imaging (Figure 3H-I). We confirmed that the normal hematopoietic progenitor cells were only minimally affected at the dose levels used in mice to cause AML cell death (Table S6).
TrxR1 expression levels inversely correlate with the efficacy of S-250.
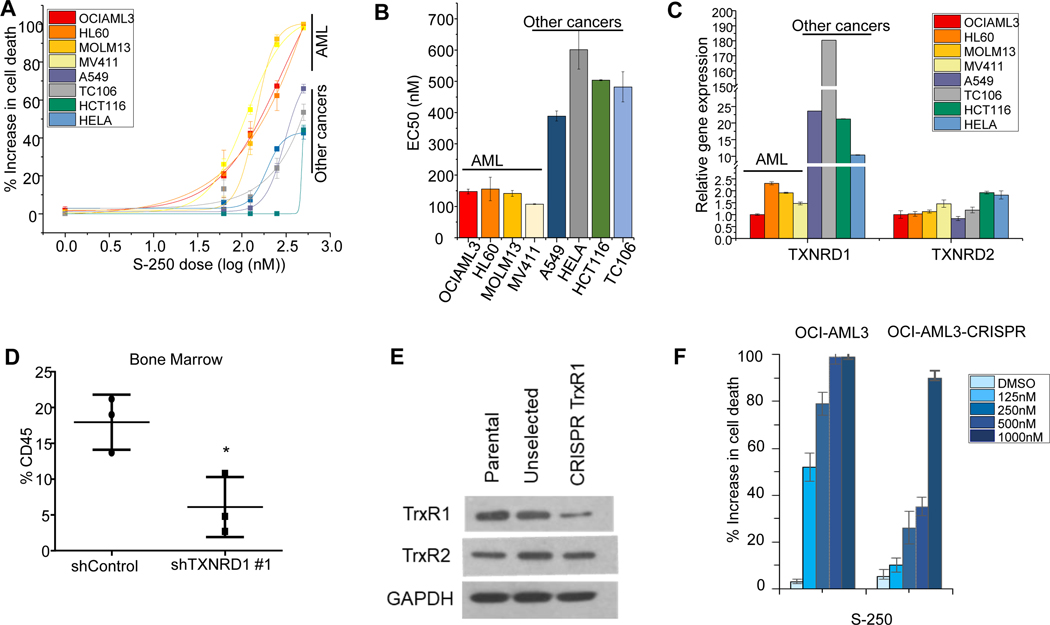
We hypothesized that if TrxR inhibition was the primary mechanism by which S-250 imparted its anti-cancer properties, then changes in TrxR expression would reflect responses to S-250 treatment. We screened a panel of solid tumor and AML cell lines for sensitivity to S-250 and found that solid tumors are more resistant to S-250 with at least a two-fold increase in EC50 (Figure 4A-B). Interestingly, when we analyzed TrxR expression in these cell lines, we observed that solid cancer lines express significantly higher levels of TXNRD1 as compared to AML cell lines likely leading to the requirement of higher concentrations of S-250 to impact their growth and viability (Figure 4C). This finding further supports AML as an excellent target for TrxR inhibition. TXNRD2 is a paralog of TXNRD1 and is primarily expressed in mitochondria. Unlike TXNRD1, TXNRD2 expression was unchanged across solid cancer and AML cell lines (Figure 4C) suggesting that TrxR1 is playing a more predominant role in accounting for the differences in sensitivity to S-250. These results are consistent with RNA expression studies performed on the NCI-60 cell line panel 25, wherein TXNRD1 expression across several solid tumor cell lines is significantly higher than hematopoietic malignancy derived cell lines while TXNRD2 mRNA levels are unchanged (Figure S4A-B).
Figure 4: TrxR1 levels determine sensitivity to S-250.
(A) Response of various cell lines to increasing doses of S-250 are plotted as logarithmic dose response curves. Indicated cells were treated with S-250 for 72h and cell viability was measured. n=4. (B) EC50 of dose curves in (A). (C) Expression levels of TXNRD1 and TXNRD2 in the cell lines in (A). n=4. (D) HL60 cells stably expressing either Control shRNA or TXNRD1 shRNA were injected into NSG mice. Leukemic burden at the end of the experiment was assessed by staining for human CD45 in the bone marrow. (D) Expression levels measured by western blot of TrxR1 and TrxR2 in OCI-AML3 control cells, OCI-AML3 cells transduced with TrxR1 CRISPR (pre-selection) and OCI-AML3 cells stably expressing a CRISPR construct targeting TrxR1 (post-selection). (E) OCI-AML3 parental cells or TrxR1 CRISPR expressing cells were treated with the indicated doses of S-250 and the percent increase in cell death relative to DMSO control was assessed. n=3. *p<0.05; **p<0.01.
We next assessed whether knockdown of TXNRD1 in HL60 cell line impacted the sensitivity to S-250 treatment. While knockdown of TXNRD1 leading to a reduced expression of TrxR1 did not demonstrate an increased sensitivity of S-250 treated AML cells in vitro, TXNRD1 knockdown significantly slowed the progression of circulating leukemic burden in NSG mice (Figure 4D and Figure S4C). TXNRD1 knockdown did not lead to a decrased in TXNRD2 expression (Figure S4D) implying that the effects are specific to TXNRD1 downregulation. In order to better understand the importance of TrxR1 in AML cell growth, we knocked out its expression using CRISPR in OCI-AML3 cells. In contrast to partial knockdown of TXNRD1, disruption of the utilizing CRISPRled to a significant reduction in sensitivity to S-250 (Figure 4E-F). Similar to knockdown of TXNRD1, CRISPR-mediated disruption of TXNRD1 did not lead to any significant change in TXNRD2 expression (Figure 4E). This results further supports TrxR1 as an important target of S-250.
Thioredoxin reductase inhibition markedly alters metabolism in AML cells
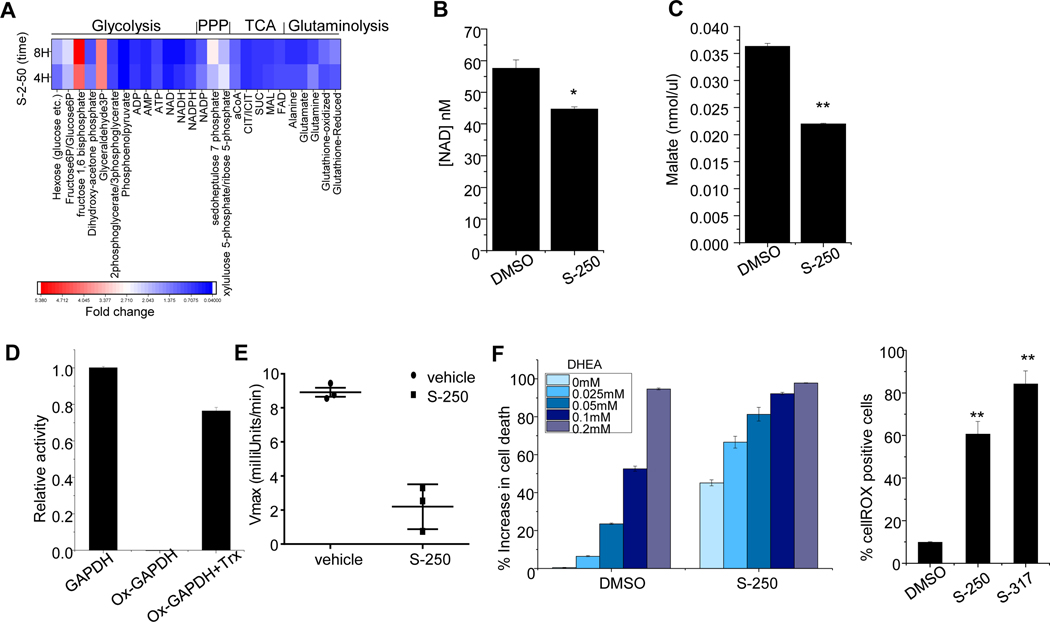
Previous studies primarily in plants demonstrate TrxR is a regulator of cell metabolism 26,27,28. Therefore, we performed global metabolomic studies on HL-60 cells treated with S-250 or vehicle for 4h or 8h. We observed an accumulation of glycolytic metabolites leading up to glyceraldehyde 3-phosphate and reduction of glycolytic metabolites downstream of 2,3-phosphoglycerate indicating a block at the level of GAPDH (Figure 5A). This accumulation of metabolites upstream of GAPDH was accompanied by an increase in Pentose Phosphate Pathway (PPP) metabolites, likely due to shunting from glycolysis. Furthermore, we identified that while the glycolytic block was compensated at the level of acetyl coA, several TCA cycle intermediates were rapidly decreased upon S-250 treatment (Figure 5A). Notably, NADH levels were decreased by ~50% in 4h and ~80% in 8h while malate levels went down by ~25% in 4h and ~40% in 8h (Figure 5A). We confirmed the results from the LC/MS metabolomics screen by demonstrating that the levels of malate and NAD decrease after 4h of S-250 treatment in lysates of S-250 treated HL60 cells using colorimetric assays (Figure 5B-C). We further validated that TrxR knockdown itself can lead to changes in TCA metabolites by confirming a decrease in malate levels in lysates from HL60 cells in which TrxR1 was downregulated as compared to vector control cells (Figure S5A). We further confirmed by an independent analysis in both HL-60 and OCI-AML3 cells using U-13C6 glucose metabolic flux that metabolites accumulated before GAPDH, decreased downstream of GAPDH followed by compensation at the level of pyruvate (Figure S5B-D and Figure S6). The results revealed significant accumulation of PPP metabolites and depletion of TCA metabolites (Figure S5E-K and Figure S6). To further assess metabolic effects of S-250 in AML cells, we also performed metabolic flux studies on HL-60 and OCI-AML3 cells using L-13C5 glutamine. S-250 led to impairment of the ability of glutamine to feed the U-13C6 glucose (Figure S7)
Figure 5: S-250 treatment induces metabolic changes in AML cells.
(A) HL60 cells were treated with 500nM S-250 for 4h or 8h. Targeted LC/MS based metabolomics analyses was performed and fold changes in intermediates of glycolysis, TCA cycle and pentose phosphate pathways are shown. The experiment was conducted in triplicates and the average is shown. (B-C) HL60 cells were treated with DMSO or 500nM S-250 for 4h. NAD levels (B) or malate levels (C) in the cell lysates were quantitated. n=3. (D) In vitro restoration of oxidized GAPDH activity using reduced thioredoxin. Oxidized GAPDH was incubated with 8.3 μM thioredoxin and activity was measured. GAPDH activity relative to non-oxidized controls are plotted as average of triplicate measurements. (E) GAPDH activity in splenic cells from vehicle or S-250 treated mice described in Figure 3. Data are presented as mean ± Standard Deviation. n=3 mice per group. (F) HL60 cells were treated with DMSO or 250nM S-250 together with indicated doses of DHEA. Cell viability was assessed after 72h. n=4. *p<0.05; **p<0.01.
In plants, TrxR is known to be a major regulator of specific metabolic enzymes by controlling the oxidation/reduction status of thioredoxin which then directly regulates the redox status and activity level of the target metabolic enzyme. We investigated whether or not human TrxR could also serve as a direct regulator of metabolism through thioredoxin. Due to the metabolomics studies suggesting inhibition specifically at the level of GAPDH, these studies were focused on GAPDH. First we confirmed as previously reported that GAPDH enzymatic activity is lost upon oxidation. Next we assessed if reduced thioredoxin, the primary substrate of thioredoxin reductase, could restore GAPDH activity. Reduced thioredoxin was able to restore ~75% of the activity of oxidized GAPDH demonstrating that it can directly modulate the activity of GAPDH (Figure 5D). Next we assessed whether TrxR inhibition could regulate GAPDH activity in vivo. Utilizing spleen lysates from leukemic mice treated with S-250, we found that GAPDH activity (as well as TrxR activity) were diminished (Figure 3G and Figure 5E).
Interestingly, a combination of inhibitors of glycolysis, PPP and thioredoxin reductase system has previously been suggested to increase cancer cell death 29. Since the S-250 mediated inhibition of TrxR causes a block in GAPDH activity and a resultant increase in PPP activity as evidenced by an increase in its metabolites (Figure 5A), we hypothesized that combined inhibition of TrxR and the PPP pathway may lead to additive cell killing of AML cells. We co-treated HL-60 cells with S-250 and DHEA, an inhibitor of Glucose 6-phosphate dehydrogenase (G6PDH) which is a rate-limiting enzyme in PPP. S-250 combined with DHEA increased leukemic cell killing efficiency (Figure 5F). Therefore, S-250 treatment results in marked changes in AML metabolism by affecting glycolysis, TCA cycle and PPP.
TrxR inhibition affects mitochondrial metabolism independently of mitochondrial damage and ROS production
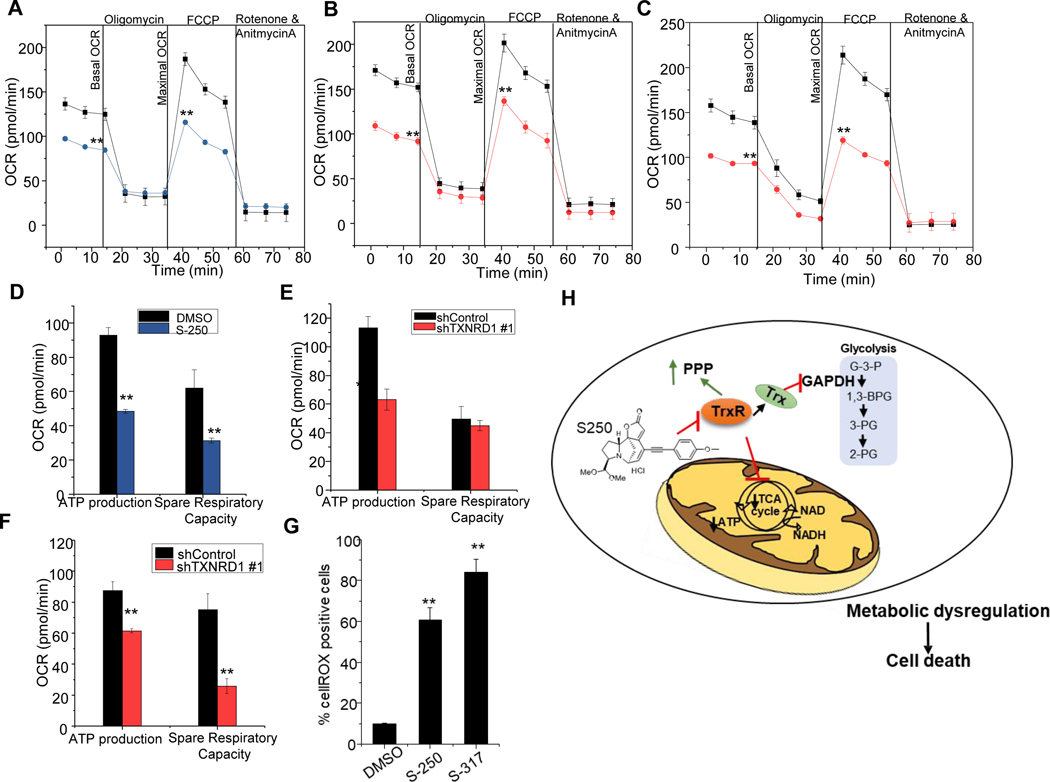
LC/MS metabolomic profiling suggest that TrxR inhibition affects metabolism at the level of GAPDH as well as at one or multiple points in the TCA cycle. To further investigate the effect of TrxR inhibition on mitochondrial metabolism, we explored the impact on oxidative phosphorylation using seahorse assay. When HL60 cells were challenged with mitochondrial stress, S-250 treatment compromised ATP production, basal and maximal OCR and spare respiratory capacity (Figure 6A and 6D). Importantly, S-250 leads to rapid (detectable within 3h) and marked changes in metabolism in AML cells as measured by the seahorse platform and LC/MS metabolomics studies that precedes the cell death process. The changes observed in mitochondrial respiration were not a result of mitochondrial damage or function as confirmed by lack of changes in mitogreen and mitoorange that measure mitochondrial mass and potential respectively (Figure S8A-D).
Figure 6: S-250 treatment impairs mitochondrial metabolism.
(A) OCR changes during a mitochondrial stress test with sequential addition of oligomycin, FCCP and rotenone/antimycin in HL60 cells treated with DMSO or 0.5μM S-250 for 3h. n=8. (B-C) HL60 (B) or OCI-AML3 (C) cells stably expressing shControl (black lines) or shTXNRD1 (red lines) were assessed by mitostress test. Changes in OCR during sequential addition of complex inhibitors are shown. n=8. (D-F) ATP production and spare respiratory capacity of 0.5μM S-250 (3h) treated HL60 cells (D) or of HL60 (E) and OCI-AML3 (F) cells expressing shTXNRD1. n=8. Unless otherwise indicated, data are presented as mean ± SE. (G) HL60 cells were treated with 375nM S-250 or 750nM S-317 overnight and stained with cellROX. Percent cellROX positive cells were quantitated by flow cytometry. n=3. Unless otherwise indicated, data are presented as mean ± SE; *p<0.05; **p<0.01. (H) Schematic of inhibition of thioredoxin reductase by S-250 leading to metabolic dysregulation and cell death in AML cells.
To confirm the metabolic effects were due to TrxR inhibition, HL60 and OCI-AML3 cells with TXNRD1 knockdown were also tested. In both cases knockdown of TXNRD1 led to similar effects as S-250 treatment and exhibited a reduction in basal and maximal OCR as well as spare respiratory capacity (Figure 6B-C and 6E-F). Importantly, this genetic study reveals that inhibition of TXNRD1, without concurrent inhibition of TXNRD2, leads to impairment of mitochondrial metabolism.
TrxR1 has been reported to be a regulator of redox homeostasis in cells 30. It has been widely claimed that inhibition of TrxR leads to cancer cell death due to reactive oxygen species (ROS) induction 31. Therefore, we tested if treatment with securinine derivatives increased the production of ROS. If ROS production was a direct mediator of cell death in response to TrxR inhibition then it would be anticipated to occur rapidly as AML cells exhibit metabolic dysfunction, metabolite depletion and undergo the initiation of irreversible cell death within hours of S-250 or S-317 treatment. Surprisingly, we did not observe any increase in ROS production with shorter treatments (up to 3h) of securinine analogs when AML cells were stained with cellROX, a fluorescent probe sensitive to oxidation by ROS (Figure S8F). In contrast, when HL60 cells were treated with S-250 or S-317 for 16h an increase in percentage of cellROX positive cells was observed (Figure 6G). Due to the kinetics of ROS induction, ROS production is presumably a by-product of the cell death process and not a major contributor to the mechanism of TrxR inhibition-induced cell death. To further confirm that ROS is not required for TrxR inhibition-induced AML cell growth inhibition/cell death, we pre-treated cells with Tert-ButylHydroquinone (TBHQ) followed by S-250. TBHQ acts as a scavenger of free radicals and is commonly used to mitigate the ROS dependent effects of compounds 32. TBHQ did not impact the cytotoxic effects of S-250 further showing that ROS induction is not the direct cause of AML cell death in response to S-250 (Figure S8F) and supporting our model (Figure 6H) that TrxR inhibition leads to AML cell death through perturbation of AML cell metabolism.
Discussion:
Our studies identified TrxR as a major regulator of metabolism in AML cells and a promising AML therapeutic target. An established function of TrxR is to control the redox status of thioredoxin 33. Thioredoxin whose redox status is primarily regulated by TrxR, directly modulates the oxidation/reduction status of its target proteins. Interestingly, it has been previously reported TrxR functions as a “master regulator” of metabolism and operates by directly regulating the oxidation/reduction status of key metabolic enzymes involved in glycolysis and the TCA cycle through thioredoxin 34. The redox status of these particular enzymes is a major regulator of their activity. In humans, a similar role of TrxR has not been described.
Here we report that TrxR is also a major regulator of mammalian cell metabolism through thioredoxin and inhibition of TrxR leads to effects on glycolysis, the TCA cycle, and the PPP. We found that TrxR can directly impact GAPDH activity through modulating the redox status of GAPDH, decreasing its activity and channeling substrates to the PPP. While further studies will be aimed at uncovering other direct targets of TrxR, metabolomics studies suggest TrxR may be impacting a step of the TCA cycle at the level of citrate synthase whose redox status is known to be regulated by TrxR in plants 34,35. Subsequently, the decreased TCA metabolite levels may impact spare respiratory capacity as they are linked to the succinate dehydrogenase/complex II of the electron transport chain 36.
We have identified that metabolic perturbation is the primary mechanism through which TrxR inhibition impairs AML growth. ROS production has previously been reported to be the primary mechanism of action through which TrxR inhibition leads to cancer cell death. However, metabolic changes were observed significantly earlier (2–4h) than changes in oxidative stress (16h) when leukemic cells were treated with securinine derivatives. Therefore, it is unlikely that ROS itself is a primary driver of the observed cell death. In fact, blocking ROS with TBHQ, had no effect on the cell death induced by S-250. Of note, genetic abrogation of TXNRD1 caused similar downregulation of glycolytic and TCA metabolites, and mitochondrial respiration that preceded ROS production.
Targeting thioredoxin reductase as a therapeutic strategy for cancer has been an area of interest 37. However, a major challenge in developing successful TrxR inhibitors has been their lack of specificity19,20. Most reported thioredoxin reductase inhibitors target the selenocysteine residue in TrxR causing them to cross-react with several related thiol-containing enzymes reducing their clinical applicability 19. We identified that the securinine derivative, S-250, is a highly specific and potent inhibitor of TrxR. S-250 does not inhibit related oxidoreductases such as glutathione peroxidase and glutathione reductase. In particular, glutathione peroxidase, like TrxR is a unique enzyme in that it contains a highly reactive selenocysteine residue in its active site. Therefore, agents such as auranofin which is considered a relatively specific TrxR also leads to inhibition of glutathione peroxidase 24.
Our studies revealed that AML is a highly desirable indication for TrxR inhibition strategies. TrxR is highly expressed in several cancers such as pancreatic, thyroid, breast and colorectal cancers 38. Interestingly, TrxR1 expression was found to be significantly lower in leukemic cells as opposed to solid tumor cells. Further the sensitivity of leukemia cells to S-250 was markedly enhanced as compared to solid tumors possibly as less drug is required to overcome TrxR1 enzyme activity. A major challenge faced by current AML chemotherapy is high toxicity particularly on normal bone marrow. S-250 has a favorable toxicity profile as it exhibits minimal effects on the growth of normal hematopoietic progenitor cells under similar treatment conditions that lead to marked anti-leukemic effects in vitro and in mice. Besides TrxR expression, the reduced spare respiratory capacity of AML cells as compared to normal blood cells also likely improves the therapeutic window in AML 39. Additionally, AML cells are highly susceptible to metabolic and mitochondrial modulators 39,40.
Overall, we have identified a potent and highly specific TrxR inhibitor that exhibits potential for AML therapy and identified a novel mechanism through which TrxR regulates AML metabolism leading to cell death.
Materials and Methods:
Cell lines and reagents:
Lenti-X 293T cells were purchased from Clontech. Lenti-X 293T and AML cell lines (HL60, OCIAML3, THP-1, MOLM13, MV411) were maintained in RPMI media supplemented with 10% cosmic serum (GE Healthcare) and 100U/ml Penicillin with 100μg/ml Streptomycin (GE Healthcare). MV411-luciferase cells were generated by stably transfecting MV411 cells using the plasmid pLenti-CMV V5-luciferase (Addgene). Patient samples were obtained from Hematopoietic biorepository and cellular therapy core at Case Western Reserve University. All work on human patient samples were approved by Institutional Review Board at University Hospitals Cleveland Medical Center. All patient samples were derived from bone marrow, had >60% leukemic blasts, and underwent ficoll purification to isolate mononuclear cells prior to testing. Primary leukemic cells were cultured in IMDM media (GE Healthcare) with 20% serum supplemented with 20ng/ml Granulocyte Stem cell factor (Gold Biotechnology). Transient transfections were performed using Turbofect (ThemoFisher Scientific) following manufacturer’s protocols. Stable transfections were obtained by using shRNA constructs - Control shRNA (SHC002) and TXNRD1 shRNA (TRCN0000046533 (#1) and TRCN0000046535 (#2)) from Sigma as well as the CRISPR construct pLV[CRISPR]-hCas9:T2A:Puro-U6>hTXNRD with the following guide sequence TATGTCGCTTTGGAGTGCGC from VectorBuilder.
Cell viability assay:
Cell viability and proliferation in cell lines were estimated using a resazurin based assay (Prestoblue, Thermo scientific). Cell viability in patient samples were measured using propidium iodide cell exclusion test using flow cytometric analysis. Cell viability measurements were used to calculate cell death and percent increase in cell death from DMSO treated controls was plotted for treatment groups.
Antibodies and flow cytometry:
Cells were labelled with indicated antibodies following manufacturer’s instructions and analyzed by flow cytometry. Antibodies to CD11b-FITC (catalog # 562793), CD14-PE (catalog #562691), CD45-APC-H7 (catalog #560178), CD33-PE (catalog #555450) were purchased from BD Biosciences. TrxR1 (catalog #28321), TrxR2 (catalog #365714), and GAPDH antibodies (catalog #47724) were from Santa Cruz Biotech.
Mice experiments:
NOD-SCID IL2Rgamma (NSG) mice were injected with 3*10^6 MV411 cells labelled with luciferase. Mice were randomly split into two groups and were given intraperitoneal injections of vehicle or S-250 at 12.5mg/kg of mice twice daily for three days a week. Circulating AML progression was followed weekly by bioluminescence imaging using Spectrum Imager. For experiments with patient derived xenografts, 2*10^6 leukemic cells were injected into NSG mice and treated with vehicle or S-250 as described above. AML progression was assessed weekly by monitoring body weight of mice and white blood cell (WBC) counts intermittently. For assessment of the effect of downregulation of TrxR1, female NSG mice were irradiated (200 Gy) and injected intravenously with 3*10^6 HL60 cells stably transfected with shC002 or shTXNRD1 #1 were injected into NSG mice and leukemic burden was assessed at the end of the experiment. Paralysis or loss of >15% body weight was determined as end of experiment and mice were sacrificed. Blood, spleen and bone marrow were isolated as indicated, cells were separated by tissue homogenization and analyzed by flow cytometry or lysed for enzymatic assays.
DTNB reduction assay:
DTNB reduction assay was adapted from Liu et al, 201141. Briefly, 85nM rat Thioredoxin Reductase (Cayman Chemical) was pre-reduced with 0.2mM NADPH for 10 min at room temperature. Reduced TrxR was incubated with DMSO or varying concentrations of drug for 2h in TE buffer (50mM Tris, 1mM EDTA, pH 7.4). DTNB reduction upon addition of 0.2mM NADPH and 2mM DTNB was followed by measuring absorbance at 412nM in kinetic mode for 7 minutes using SpectraMax M3 microplate reader. Maximum velocity was calculated from linear phase of the reaction. For measuring TrxR activity in cell lysates, cells were incubated with the drug at indicated doses for 4h. Cells were lysed in TE buffer and TrxR activity was quantitated using DTNB reduction. To measure the percentage of DTNB reduction caused by TrxR activity, lysates were treated with sodium authiomalate (Cayman Chemical) as per manufacturer’s protocol. TrxR activity in mice spleens was measured by isolating splenic cells followed by lysis and DTNB reduction assay.
GAPDH activity assay:
GAPDH activity in tissue lysates was measured using glyceraldehyde 3-phosphate (G3P) as substrate in the presence of NAD. Absorbance measurements at 340nM were recorded in kinetic mode. For in vitro restoration assays, 300nM GAPDH was oxidized in the presence of hydrogen peroxide for 20 minutes and excess hydrogen peroxide was cleared by the addition of catalase. Following this, the mixture was incubated with 8.3 μM thioredoxin for 2h and assayed in the presence of G3P and NAD.
ROS measurements:
Reactive oxygen species (ROS) was measured in HL60 cells after S-250 treatment at varying time points as indicated. Cells were stained with cellROX reagent (ThemoFisher Scientific) as per manufacturer’s protocol. Mitochondrial mass and potential were measured in HL60 cells after S-250 treatment for 3h using mitoTracker green (ThemoFisher Scientific) and mitoTracker orange (ThemoFisher Scientific) respectively following manufacturer’s instructions. Auranofin (Tocris Biosciences) was used at indicated concentrations as a control for known inhibitor for TrxR.
RNA isolation and quantitative PCR:
RNA isolation was carried out using RNeasy mini kit (Qiagen). RNA was reverse transcribed using High Capacity RNA-to-cDNA kit (ThermoFisher Scientific). Expression analysis was performed using EvaGreen dye (Biotium) in a BioRad CFX96 Real-Time system with following primers from Sigma: ACTB (F: 5’-AGAGCTACGAGCTGCCTGAC-3’ and R: 5’-AGCACTGTGTTGGCGTACAG-3’), TXNRD1 (F: 5’-AGACAGTTAAGCATGATTGG −3’ and R: 5’-AATTGCCCATAAGCATTCTC −3’) and TXNRD2 (F: 5’-ACTTTAACATCAAAGCCAGC −3’ and R: 5’-GTAGCAATGATGATGTGATCG −3’).
Seahorse Assays:
Oxygen consumption rate (OCR) and Extracellular Acidification Rate (ECAR) were measured using a Seahorse XF96 analyzer (Agilent Technologies). HL60 or OCI-AML3 cells were treated with S-250 or S-317 as indicated and 1*10^5 cells were seeded on Cell-Tak (Corning) coated wells in manufacturer recommended assay medium. Basal and stressed measurements of OCR and ECAR were obtained after treating cells with 1μM Oligomycin and 0.125μM Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP). ATP production, spare respiratory capacity and oxygen consumption over time was measured by sequential addition of oligomycin (1μM), FCCP (0.125μM) and rotenone/antimycin A (0.5μM).
Enzymatic assays:
Enzyme assays were performed following manufacturer’s protocols. GAPDH activity assay kit was purchased from Biovision. Glutathione peroxidase activity was measured using the Gpx assay kit (Cayman Chemical). Enzymatic activity of PDIA1 and PDIA6 (Novus Biologicals) were measured using the PDI assay kit (Enzo Lifesciences). Enzymes MDH1, MDH2, LDHA, and GR were from Novus. The CEREP safety panel (Safetyscreen 44) was performed by Eurofins Panlabs Discovery Service, Taiwan to identify any off-target effects of S-250 at 10μM.
DARTS screen:
Protocol for performing DARTS analysis was adapted from Lomenick et al, 2012 42. Briefly, HL60 cells were lysed in MPER lysis buffer and equal amount of protein containing lysates were incubated with DMSO or 50μM S-250 for 1h at room temperature. Lysates were digested with 1.25mg/ml Pronase for 30 minutes. Small fragments generated during pronase digestion were filtered using 10KDa spin columns. Samples were trypsin digested and peptides were identified using Multidimensional protein identification technology (MudPIT). Proteins enriched in S-250 digested sample compared to that of DMSO were included in further analysis. Functional, structural or genetic interactions between enriched proteins were assessed using STRING database 23.
Metabolomic studies:
HL60 cells (10*10^6) were treated in triplicates with DMSO or 0.5μM S-250 for 4h or 8h and were harvested and washed with 150mM ammonium acetate and flash frozen. Targeted metabolomics of glycolytic and tricarboxylic acid cycle (TCA) intermediates in these samples were performed by the metabolomics core at the University of Michigan using LC/MS/MS. Differential changes in the metabolite levels were identified by comparing S-250 treatments to DMSO treated samples. Fold changes in the levels of metabolites were plotted as heat maps. NAD levels in cells were quantitated using NAD/NADH cell-based assay kit from Cayman Chemicals using manufacturer’s protocols. Malate levels were measured using malate colorimetric assay kit from BioVision following the manufacturer’s protocols.
Metabolic flux analysis:
HL60 or OCI-AML3 cells were starved in DMEM no glucose containing dialyzed FBS overnight. After 16h, cells were treated with DMSO or 500 nM 250 for 3 hours. Following this, cells were incubated with 12mM 12C glucose (12C control) or 12mM [U-13C6] Glucose for 3h or glutamine or 2mM L-13C5 glutamine (Cambridge Isotope Laboratories). Cells were then spun down and snap frozen. Metobolomic flux analysis was performed using agilent 6490 Triple Quadruple mass spectrometry as described earlier 43,44.
Mass spectrometric analysis:
Pre-reduced rat TrxR1 (10μg) was incubated with 75μM S-250 for 2h at room temperature. The samples were digested in-solution with trypsin and the digests were analyzed by LC/MS/MS using the Fusion Lumos instrument. The data were analyzed using Mascot to search the human UniProtKB database and Proteome Discoverer and Batch Tag (http://prospector2.ucsf.edu/prospector/cgibin/msform.cgi?form=batchtagweb) to search the sequence of TrxR1.
Clonogenic assays:
Colony formation assays were performed as described previously 8. HL60, MOLM13 and MV411 cells were treated with varying doses of S-250 for 4 hours. After 4 hours, cells were washed to remove the drug and plated in media containing 20% serum and 0.3% soft agar (Noble agar, Sigma). Colonies were allowed to form for 10 days and were counted under a light microscope. For normal bone marrow samples, cells were treated with 500nM S-250 for 4h, washed and plated on methylcellulose (Methocult Classic, Stem cell technologies) containing 0.25mM hemin (Sigma). For AML patient samples, cells were likewise treated for 4h with 500nM S-250 and seeded on methylcellulose supplemented with 20% serum and 20ng/ml G-CSF (GoldBio). Colonies were counted after incubating for 10 days.
Western blot analysis:
Cells were washed with PBS, centrifuged and lysed with a triton containing lysis buffer. Protein lysates (50μg/lane) were resolved on appropriate SDS-PAGE gel and transferred to PVDF membrane (Millipore) using BioRad transfer apparatus. The membranes were blocked, incubated with the indicated primary antibodies at horseradish peroxidase–conjugated secondary antibodies. Immunoreactive protein bands were detected by enhanced chemiluminescence (Pierce) using XAR-5 film.
Bioinformatics and Statistical analysis:
Unpaired student t-tests were used for comparison of two groups. One-way ANOVA was used when more than two groups were analyzed for statistical significance. Gene expression data for TXNRD1 and TXNRD2 were downloaded from cBioportal (Gao et al., 2012).
Supplementary Material
Acknowledgements:
We thank the Proteomics facility at the Cleveland Clinic Lerner Research Institute for analysis of the mass spectrometry data. This research was supported by the Hematopoietic Biorepository and Cellular Therapy, Athymic Animal and Preclinical Therapeutics, Proteomics and Cytometry and Imaging Microscopy Shared Resources of the Case Comprehensive Cancer Center (P30CA043703). The research was also supported by the NIH grant R43CA22870 (D.N.W. and Y.H.) and VA medical center grant I01BX004995. Targeted metabolomics performed at the University of Michigan metabolomics core was supported by the NIH grant U24DK097153. This research was supported by American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N.P.), NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.). W81XWH-18-1-0035 and W81XWH-18-1-0084 to SMK .
Footnotes
Conflicts of Interest: DNW is an inventor on patents related to S-250 and his employerCase Western Reserve University is the owner of patents related to S-250. SMK is a stakeholder in NeoZenome Therapeutics Inc. No other authors have any conflicts of interest with this study.
References:
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med 2015; 373: 1136–52. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2018. Am Cancer Soc 2018. doi: 10.1182/blood-2015-12-687814. [DOI] [Google Scholar]
- 3.De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J 2016; 6: e441–e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapuis N, Poulain L, Birsen R, Tamburini J, Bouscary D. Rationale for Targeting Deregulated Metabolic Pathways as a Therapeutic Strategy in Acute Myeloid Leukemia. Front Oncol 2019. doi: 10.3389/fonc.2019.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi A, Maria M, Cruz A, Jyotsana N, Sharma A, Yun H et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood 2013. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 6.Levis M Targeting IDH: The next big thing in AML. Blood 2013. doi: 10.1182/blood-2013-09-522441. [DOI] [PubMed] [Google Scholar]
- 7.Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 2018. doi: 10.1038/s41591-018-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta K, Chakrabarti A, Rana S, Ramdeo R, Roth BL, Agarwal MKML et al. Securinine, a myeloid differentiation agent with therapeutic potential for AML. PLoS One 2011; 6: e21203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong NZ, Gu ZL, Chou WH, Kwok CY. Securinine induced apoptosis in human leukemia HL-60 cells. Zhongguo Yao Li Xue Bao 1999; 20: 267–70. [PubMed] [Google Scholar]
- 10.Beutler JA, Karbon EW, Brubaker AN, Malik R, Curtis DR, Enna SJ. Securinine alkaloids: A new class of GABA receptor antagonist. Brain Res 1985; 330: 135–40. [DOI] [PubMed] [Google Scholar]
- 11.Shipman M, Lubick K, Fouchard D, Guram R, Grieco P, Jutila M et al. Proteomic and Systems Biology Analysis of Monocytes Exposed to Securinine, a GABAA Receptor Antagonist and Immune Adjuvant. PLoS One 2012; 7: e41278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonoudakis D, Rane A, Angeli S, Lithgow GJGJ, Andersen JKJK, Chinta SJSJSJ. Anti-Inflammatory and Neuroprotective Role of Natural Product Securinine in Activated Glial Cells: Implications for Parkinson’s Disease. Mediators Inflamm 2017; 2017: 8302636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou W, Wang Z-YY, Peng C-KK, Lin J, Liu X, Chang Y-QQ et al. Novel securinine derivatives as topoisomerase I based antitumor agents. Eur J Med Chem 2016; 122: 149–163. [DOI] [PubMed] [Google Scholar]
- 14.Rana S, Gupta K, Gomez J, Matsuyama S, Chakrabarti A, Agarwal ML et al. Securinine induces p73-dependent apoptosis preferentially in p53-deficient colon cancer cells. FASEB J 2010; 24: 2126–34. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Yao J, Peng S, Li X, Fang J. Securinine disturbs redox homeostasis and elicits oxidative stress-mediated apoptosis via targeting thioredoxin reductase. Biochim Biophys Acta - Mol Basis Dis 2017; 1863: 129–138. [DOI] [PubMed] [Google Scholar]
- 16.Arnér ESJ. Focus on mammalian thioredoxin reductases - Important selenoproteins with versatile functions. Biochim Biophys Acta - Gen Subj 2009; 1790: 495–526. [DOI] [PubMed] [Google Scholar]
- 17.Fritz-Wolf K, Kehr S, Stumpf M, Rahlfs S, Becker K. Crystal structure of the human thioredoxin reductase-thioredoxin complex. Nat Commun 2011; 2: 383. [DOI] [PubMed] [Google Scholar]
- 18.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A 2001; 98: 9533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Li X, Han X, Liu R, Fang J. Targeting the Thioredoxin System for Cancer Therapy. Trends Pharmacol. Sci. 2017; 38: 794–808. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Zhang J, Peng S, Liu R, Li X, Hou Y et al. Thioredoxin reductase inhibitors: a patent review. Expert Opin. Ther. Pat. 2017; 27: 547–556. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan A, Stockwell BR. Structural elucidation of a small molecule inhibitor of protein disulfide isomerase. ACS Med Chem Lett 2015; 6: 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S et al. Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci 2009; 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radenkovic F, Holland O, Vanderlelie JJ, Perkins A V. Selective inhibition of endogenous antioxidants with Auranofin causes mitochondrial oxidative stress which can be countered by selenium supplementation. Biochem Pharmacol 2017; 146: 42–52. [DOI] [PubMed] [Google Scholar]
- 25.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J et al. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res 2012; 72: 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng X, Giménez-Cassina A, Petrus P, Conrad M, Rydén M, Arnér ESJJ. Thioredoxin reductase 1 suppresses adipocyte differentiation and insulin responsiveness. Sci Rep 2016; 6: 28080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cassidy PB, Honeggar M, Poerschke RL, White K, Florell SR,Andtbacka RHII et al. The role of thioredoxin reductase 1 in melanoma metabolism and metastasis. Pigment Cell Melanoma Res 2015; 28: 685–95. [DOI] [PubMed] [Google Scholar]
- 28.Stafford WC, Peng X, Olofsson MH, Zhang X, Luci DK, Lu L et al. Irreversible inhibition of cytosolic thioredoxin reductase 1 as a mechanistic basis for anticancer therapy. Sci Transl Med 2018; 10: eaaf7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Fath MA, Scarbrough PM, Watson WH, Spitz DR. Combined inhibition of glycolysis, the pentose cycle, and thioredoxin metabolism selectively increases cytotoxicity and oxidative stress in human breast and prostate cancer. Redox Biol 2015. doi: 10.1016/j.redox.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustacich D, Powis G. Thioredoxin reductase. Biochem J 2000; 346 Pt 1: 1–8. [PMC free article] [PubMed] [Google Scholar]
- 31.Urig S, Becker K. On the potential of thioredoxin reductase inhibitors for cancer therapy. Semin Cancer Biol 2006; 16: 452–465. [DOI] [PubMed] [Google Scholar]
- 32.Yu R, Tan TH, Kong AN. Butylated hydroxyanisole and its metabolite tert-butylhydroquinone differentially regulate mitogen-activated protein kinases. The role of oxidative stress in the activation of mitogen-activated protein kinases by phenolic antioxidants. J Biol Chem 1997; 272: 28962–70. [DOI] [PubMed] [Google Scholar]
- 33.Arnér ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 34.Daloso DM, Müller K, Obata T, Florian A, Tohge T, Bottcher A et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc Natl Acad Sci 2015. doi: 10.1073/pnas.1424840112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidtmann E, König AC, Orwat A, Leister D, Hartl M, Finkemeier I. Redox regulation of arabidopsis mitochondrial citrate synthase. Mol Plant 2014. doi: 10.1093/mp/sst144. [DOI] [PubMed] [Google Scholar]
- 36.Pfleger J, He M, Abdellatif M. Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell Death Dis 2015; 6: e1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Zhang B, Li X, Han X, Liu R, Fang J. Small molecule inhibitors of mammalian thioredoxin reductase as potential anticancer agents: An update. Med Res Rev 2018. doi: 10.1002/med.21507. [DOI] [PubMed] [Google Scholar]
- 38.Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. The thioredoxin-thioredoxin reductase system: Over-expression in human cancer. Anticancer Res 2003; 23: 2425–33. [PubMed] [Google Scholar]
- 39.Sriskanthadevan S, Jeyaraju D V., Chung TE, Prabha S, Xu W, Skrtic M et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 2015; 125: 2120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Škrtić M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z et al. Inhibition of Mitochondrial Translation as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 2011; 20: 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Pietsch KE, Sturla SJ. Susceptibility of the antioxidant selenoenyzmes thioredoxin reductase and glutathione peroxidase to alkylation-mediated inhibition by anticancer acylfulvenes. Chem Res Toxicol 2011; 24: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomenick B, Jung G, Wohlschlegel JA, Huang J. 3.4.2_Target identification using drug affinity responsive target stability (DARTS). Curr Protoc Chem Biol 2011; 3: 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vantaku V, Dong J, Ambati CR, Perera D, Donepudi SR, Amara CS et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in Bladder Cancer. Clin Cancer Res 2019. doi: 10.1158/1078-0432.CCR-18-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vantaku V, Putluri V, Bader DA, Maity S, Ma J, Arnold JM, Rajapakshe K, Donepudi SR, von Rundstedt FC, Devarakonda V, Dubrulle J, Karanam B, McGuire SE, Stossi F, Jain AK, Coarfa C, Cao Q, Sikora AG, Villanueva H, Kavuri SM, Lotan Y, Sreekumar A PN. Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene 2019; PMID: 3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Jianjiong, Aksoy Bülent Arman, Dogrusoz Ugur, Dresdner Gideon, Gross Benjamin, Sumer S. Onur, Sun Yichao, Jacobsen Anders, Rileen Sinha EL, Cerami Ethan, Sander Chris and NS. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013; 6: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.