Abstract
Objective
To assess the role of versican (VCAN) in uveal melanoma (UVM) from its expression, prognostic value and biological function.
Methods
The general profile of VCAN mRNA and protein expression levels were obtained using bioinformatic approaches. Then, UALCAN database was adopted to examine the association of VCAN mRNA expression and clinical factors in UVM. The prognostic value of VCAN was assessed by UALCAN, GEPIA and TISIDB databases. Besides, Cox regression analysis was performed to predict the independent prognostic factors for UVM. Further, functional enrichment analysis was conducted to reveal the biological functions of VCAN involved in UVM through DAVID, Cytoscape and GSEA analyses.
Results
VCAN showed a relative low expression level in normal eye but was highly expressed in UVM cell lines. Tumor histology and stage in UVM were significantly related to VCAN mRNA expression (all P <0.05). Besides, high VCAN mRNA expression led to unfavorable prognosis of UVM patients, especially in female patients and those aged <60 years (all P <0.05). Cox regression analysis indicated that VCAN mRNA expression was an independent prognostic factor for overall survival in UVM. Enrichment analysis suggested that VCAN was mainly involved in cytokine–cytokine receptor interaction, chemokine signaling pathway and T cell receptor signaling pathway (all P <0.05). Meanwhile, hyaluronic acid was revealed to be a potential drug for the UVM treatment.
Conclusion
VCAN served as an independent prognostic factor for UVM. Further analysis found that VCAN was positively correlated with metastasis-related pathway, which might imply the metastasis risk of UVM. Our study initially revealed the vital role of VCAN in the process of UVM and provided a therapeutic target for UVM treatment.
Keywords: bioinformatic approach, versican, uveal melanoma, prognosis, pathway
Introduction
Uveal melanoma (UVM) is the most common malignancy originating from the uveal tract.1 It often occurs in adults and has a high metastasis rate, especially metastasizes to liver and lung.2,3 Approximately half of the patients are at risk of developing metastasis, which lead to an unfavorable prognosis and high mortality in UVM.3,4 UVM can be histologically classified into spindle, epithelioid and the mixed type, and epithelioid patients had the shortest survival time.5 Some methods including excision, radiotherapy and chemotherapy are available for the UVM treatment, but the 5-year overall survival (OS) remains low and metastatic UVM still lacks effective therapy.6 Therefore, it is of urgency to identify reliable biomarkers and drugs for improving the clinical prognosis of UVM.
As a chondroitin sulfate proteoglycan (CSPG), versican (VCAN) is a component of the extracellular matrix.7 VCAN exerts a regulatory role in cell adhesion, proliferation, migration, and angiogenesis.8 Besides, VCAN accelerates distant metastasis via reduction of cell-matrix adhesion.9 Studies have reported that abnormal VCAN expression was observed in colorectal and breast cancers.10,11 And VCAN expression was up-regulated in ovarian cancer compared with the normal tissue.12 It has also been demonstrated that high VCAN expression was associated with poor prognosis in gastric cancer.13 Nevertheless, the role of VCAN expression in UVM has never been reported.
Immune cells are closely associated with the progression of various cancers. Immune cells account for a large proportion of normal cells in the tumor tissue microenvironment, and tumor‐infiltrating immune cells are considered as powerful biomarkers for cancer treatment.14 Lymphocytes (T cells) also exist in the microenvironment of tumors including CD8 + cytotoxic T cells (CD8 + T cell) and CD4 + helper T cells (CD4 + T cell), which are tumor-promoting/-suppressive.5 In addition, macrophages are monocytes differentiated by the blood circulation extravasation and are involved in tumor growth as well as metastasis.15 Crosstalk in the tumor microenvironment may facilitate the tumor cell inflammatory response, and hence promoting the differentiation of neutrophils and macrophages.1,16 B cells and Dendritic cells serve as T cell activation and antigen presentation, which might provide anticancer immunity.17 There were several researches reporting the correlation between VCAN and immune infiltration. Previous study has demonstrated a positive correlation between VCAN high expression and high immune infiltration in gastric cancer patients,13 and VCAN may affect the tumor microenvironment of gastric cancer by regulating the immune infiltration.18 In addition, the proteolysis of VCAN was strongly correlated with CD8 T cell infiltration in colorectal cancer, and tumors displaying active VCAN proteolysis and VCAN low expression were associated with robust (10-fold) CD8 T cell infiltration.19 Thus, we also put the emphasis on the correlation of VCAN with immune cell infiltration level in UVM progression.
In our study, the gene and protein expression levels of VCAN were obtained in human protein atlas (HPA) and gene expression omnibus (GEO) databases. Then, we assessed the clinical factors influencing VCAN mRNA expression in UALCAN database. The prognostic significance of VCAN was investigated and validated through three different databases. Cox regression was performed for exploration of independent prognostic factor. Next, significant pathways associated with VCAN in UVM were evaluated by KEGG and GSEA enrichment analyses.
Materials and Methods
HPA Database Analysis
HPA database (https://www.proteinatlas.org/) was utilized to obtain the general profile of VCAN expressions in human normal organs by searching VCAN in “TISSUE” tab, and VCAN protein localization in tumor cells were derived in “CELL” column. The database aims to map all human proteins in cells, tissues and organs by integrating various omics technologies.
GEO Database Analysis
GEO database (https://www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository for users to query and download experiments and curated gene expression profiles. The database was used to retrieve VCAN mRNA expression profiles in UVM and normal cell lines of GSE27831 and GSE62075 datasets. Due to insufficient sample size, we combined GSE27831 and GSE62075 to enlarge samples for expression analysis, which contained 25 normal and 36 UVM samples. The combat method in R-sva package was used to remove the batch effects between 2 datasets.
UALCAN Database Analysis
UALCAN database (http://ualcan.path.uab.edu/) is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. The database was employed to study the association of VCAN gene expression with clinical pathological parameters in UVM. We entered the gene symbol “VCAN” and chose the “Uveal Melanoma” for exploration. Then, the VCAN mRNA expression profile based on individual cancer stages, patient’s gender, weight, age and histology in UVM were shown by clicking the “Expression”. Besides, to investigate the prognostic significance of VCAN mRNA level in UVM patients, the effect of VCAN mRNA expression on UVM patient survival, the effect of VCAN mRNA expression level and body weight on UVM patient survival, and the effect of VCAN mRNA expression level and gender on UVM patient survival were presented in “Survival”.
GEPIA and TISIDB Database Analyses
GEPIA (http://gepia.cancer-pku.cn/) and TISIDB (http://cis.hku.hk/TISIDB/) databases were adopted to validate the prognostic value of VCAN mRNA expression level in patients with UVM. GEPIA database provides customizable functions such as tumor/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis. The survival curves were plotted by using “Median” of the VCAN mRNA expression in UVM patients as the group cutoff in GEPIA dataset.
TISIDB database is a web portal for tumor and immune system interaction, which integrates multiple heterogeneous data types. First, we entered the “clinical” tab and chose “Uveal Melanoma” to generate the survival curve. In addition, we chose “Lymphocyte” tab to obtain the Spearman correlations between VCAN mRNA expression and tumor-infiltrating lymphocytes in UVM for identifying the relationship between neutrophil and VCAN gene expression. The details on drugs targeting VCAN were derived from “Drug” tab.
cBioPortal Database Analysis
VCAN gene expression data along with the clinical information of UVM patients including age, clinical stage, gender, histology was downloaded from cBioPortal database (https://www.cbioportal.org/) by searching UVM and choosing “Uveal Melanoma” (TCGA, Firehose Legacy) with 80 samples. Patients without survival information were excluded. Therefore, totally 69 patients with complete clinical data were enrolled in the Kaplan–Meier plotter analysis and the construction of nomogram. Moreover, VCAN co-expressed genes in UVM were retrieved from cBioPortal database. A total of 19,470 co-expressed genes were shown and finally 512 genes which met the threshold of absolute value of Spearman’s r >0.5 and q-value <0.05 were selected for functional enrichment analysis.
TIMER Database Analysis
TIMER database (https://cistrome.shinyapps.io/timer/) is a web server for comprehensive analysis of immune infiltrates across diverse cancer types. Cox univariate and multivariate analyses were performed to identify the clinical risk factors for OS in UVM. In addition, to understand the significance of immune cell infiltration, the database was applied for figuring out the correlation between immune infiltrates and prognosis of UVM by clicking “Survival” and inputting UVM in “Cancer Types”, followed by plotting “KM curve”. After that, multivariate analysis of VCAN, clinical pathological characteristics and immune infiltration cells was performed in the same module to explore whether the immune infiltration cells had an impact on the independent prognostic value of VCAN in UVM. Female is the reference level for gender, stage 1 for stage as well.
DAVID Database Analysis
DAVID (https://david.ncifcrf.gov/tools.jsp) is a free online bioinformatics resource database for annotation, visualization, and integrated discovery, providing functional annotation and visualization of large-scale lists of genes. The selected 512 genes that co-expressed with VCAN were loaded into DAVID database for analyses of Gene Ontology (GO) annotation and KEGG pathways.
GeneMANIA Database Analysis
Apart from the significant co-expressed genes of VCAN in UVM, we also generated a protein/gene interaction network for VCAN with production of 20 proteins/genes via GeneMANIA plugin of Cytoscape. GeneMANIA searches many large, publicly available biological datasets to find related genes. These datasets include protein–protein, protein-DNA and genetic interactions, pathways, reactions, gene and protein expression data, protein domains and phenotypic screening profiles.
Besides, the associated biological process of VCAN and its 20 related genes was analyzed and visualized by BiNGO plug-in of Cytoscape. Benjamini & Hochberg false discovery rate (FDR) was selected as the multiple testing method. Organism referred to Homo sapiens. The significant level was set as 0.001.
Gene Set Enrichment Analysis (GSEA)
In order to reveal the underlying mechanism of VCAN involved in UVM, we further performed the GSEA to explore VCAN-related positive and negative KEGG pathways. The analysis criteria for GSEA were set as follows: number of permutations (1000), permutations type (gene_set), enrichment statistic (weighted), metric for ranking genes (Pearson), genes list ordering mode (descending). The terms with FDR <0.25 and P <0.05 were considered as significant pathways.
Statistical Analysis
Statistical analyses were performed by SPSS23.0 (SPSS, Inc., Chicago, IL, USA) software. Comparisons of VCAN expression between two different groups were conducted using the Mann–Whitney test in GraphPad Prism version 8. Survival curves were plotted using Kaplan–Meier and the Log rank test was used to compare the differences between groups. P <0.05 was considered as statistically significant.
Results
The Expression Analysis of VCAN
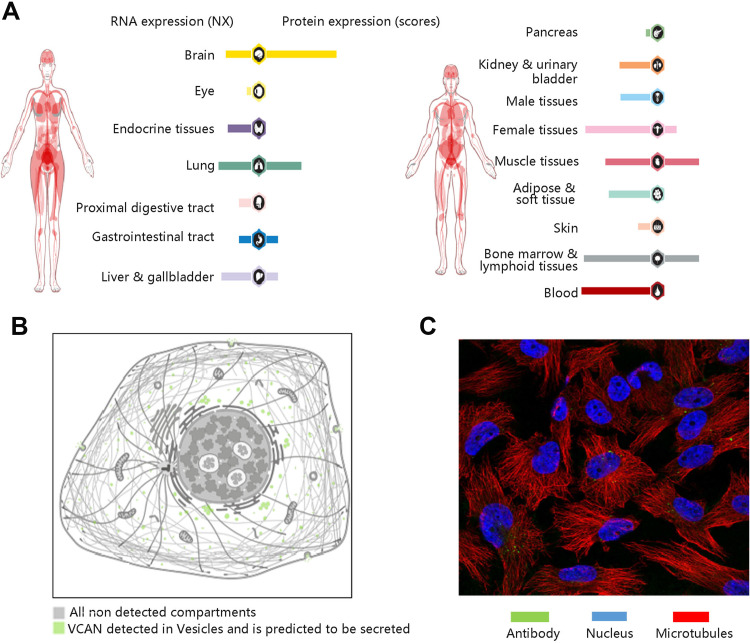
HPA database was used to visualize the expression levels of VCAN in human normal tissues and its protein localization in tumor cells. Among the human normal tissues, VCAN expression is lowly expressed in normal eye (Figure 1A). Figure 1B shows that VCAN protein was expressed in vesicles, and is predicted to be secreted, but no VCAN protein expression was observed in nucleus in human tumor cells. The immunofluorescence staining result in U-251 MG cell line was presented in Figure 1C.
Figure 1.
General profile of VCAN gene and protein expressions. (A) VCAN gene and protein expression in human normal organs. (B) The localization of VCAN protein expression in tumor cells. (C) Immunofluorescence staining in U-251 MG cell line.
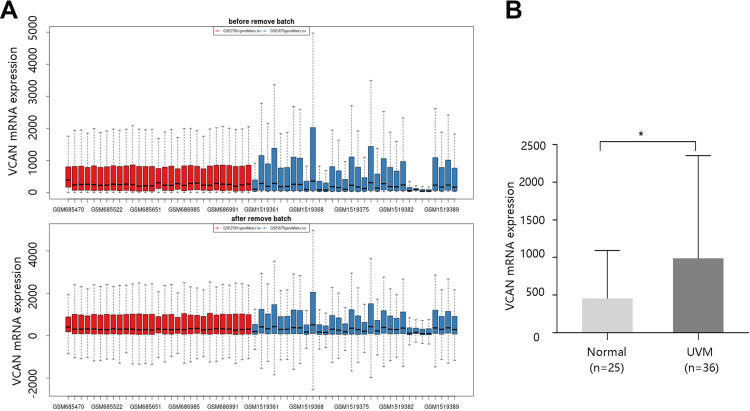
We subsequently obtained the VCAN mRNA expression in UVM and normal cell lines through GEO database. Figure 2A exhibits the sample distribution before and after removing batch effect from GSE27831 and GSE62075 datasets. As shown in Figure 2B, VCAN mRNA expression was significantly higher in UVM than that in normal cell lines (P <0.05).
Figure 2.
High VCAN mRNA expression in UVM. (A) Samples distribution before and after removing batch effect obtained from GEO database. Red: GSE27831 dataset; Blue: GSE62075 dataset. (B) VCAN mRNA expression in normal and uveal melanoma (UVM) cell lines. *P <0.05.
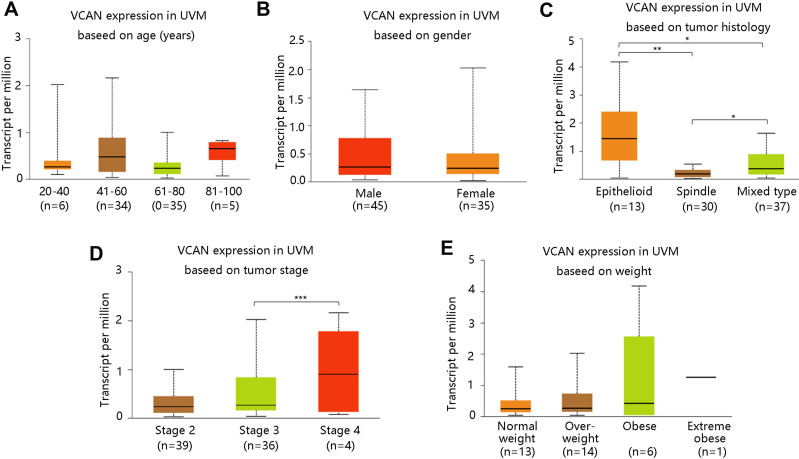
Then, we assessed the relationship between VCAN mRNA expression and clinical pathological factors including age, gender, tumor histology, tumor stage and weight. Patients were classified as normal weight/overweight/obese/extreme obese based on the body-mass-index. The age, gender, and weight of the patients were not significantly correlated with VCAN mRNA expression in UVM (all P >0.05) (Figure 3A–B and E). However, patients with epithelioid tumors had highest VCAN mRNA expression, followed by the mixed type, and spindle tumors. There were statistical differences among three groups (all P <0.05) (Figure 3C). Patients in stage 4 were found to have notably higher VCAN mRNA expression compared with those in stage 3 (P <0.001) (Figure 3D).
Figure 3.
Association of VCAN mRNA expression with clinicopathologic characteristics in UVM. (A) Age. (B) Gender. (C) Tumor histology. (D) Tumor stage. (E) Weight. Normal weight: 18.5≤BMI<25; Overweight: 25≤BMI<30; Obese: 30≤BMI<40; Extreme obese: BMI>40. BMI: body-mass-index. *P <0.05, **P <0.01, ***P <0.001.
Prognostic Value of VCAN mRNA Expression
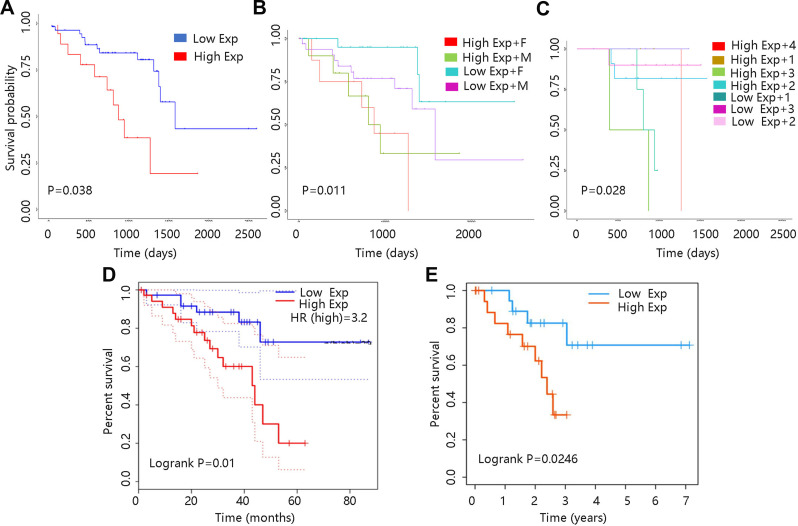
The relationship between VCAN mRNA expression and OS in UVM was investigated via UALCAN database. As shown in Figure 4A, VCAN mRNA expression at a higher level predicted poor prognosis in UVM. Besides, patient’s gender and weight in combination with VCAN mRNA expression also made a difference to OS (all P <0.05). In low expression + male group, the OS seemed to be shorter than that in low expression + female group. And patients with low expression + obese had worse prognosis compared to those with low expression + normal weight (Figure 4B–C). These results indicated that VCAN mRNA expression played an important role in prognosis of UVM, and gender as well as weight might have an impact on the clinical outcome of UVM.
Figure 4.
Survival curves of UVM patients with different VCAN mRNA expression levels. (A) UALCAN database. (B) Based on VCAN expression and gender. F: female; M: male (C) Based on VCAN expression and weight. 1: Normal weight; 2: Overweight; 3: Obese; 4: Extreme obese. The classification was based on their body-mass-index. (D): Gepia database. (E): TISIDB database.
Moreover, Gepia and TISIDB databases were employed to verify the effect of VCAN mRNA expression on the UVM prognosis. UVM patients in high VCAN mRNA expression group had shorter survival time compared with those in low VCAN mRNA expression group, revealing that high VCAN mRNA expression contributed to worse prognosis in UVM (all P <0.05) (Figure 4D–E).
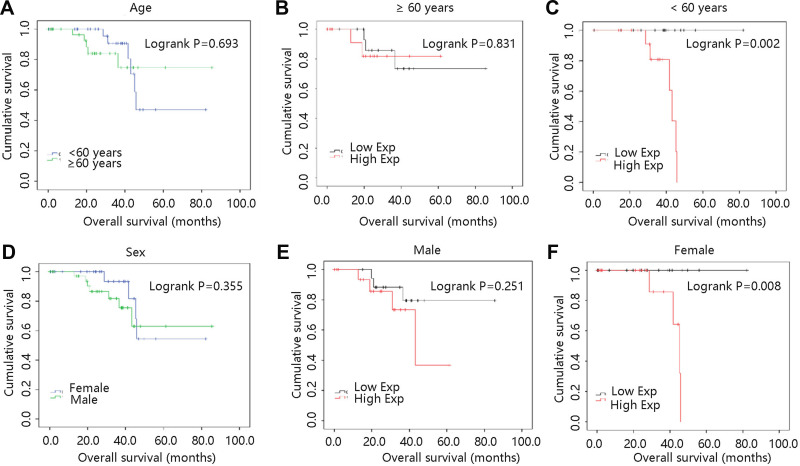
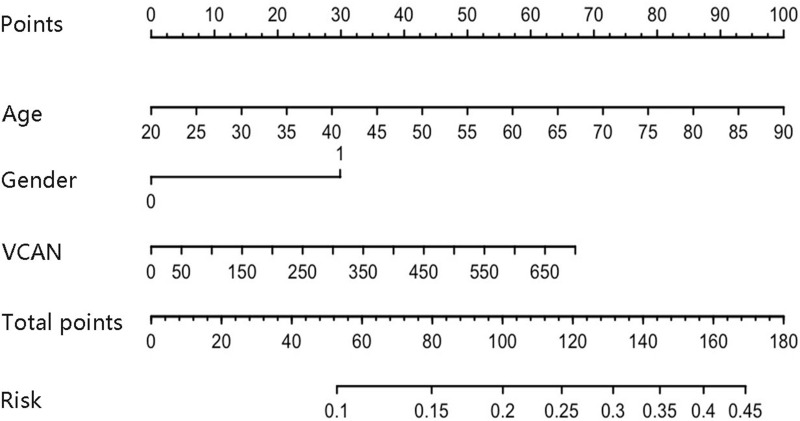
Age and gender were essential in clinical prognosis, and hence we assessed the two factors in survival analysis. As shown in Figure 5A, age had no significant relation with survival time (P >0.05). And OS was not different between two VCAN mRNA expression groups in UVM patients aged ≥60 years (P >0.05) (Figure 5B). However, high VCAN mRNA expression significantly shortened the OS time in patients aged <60 years (P <0.01) (Figure 5C). In terms of gender, male and female had similar OS time in UVM (P >0.05) (Figure 5D). And high VCAN mRNA expression was not related to UVM prognosis in male patients (P >0.05) (Figure 5E), while high mRNA expression of VCAN was remarkably associated with worse prognosis in female patients (P <0.01) (Figure 5F). In order to further clarify the survival result, age, gender and VCAN were enrolled for the nomogram construction. As shown in Figure 6, age was the biggest risk contributing factor in UVM patients, followed by VCAN mRNA expression. There was almost no risk when age was less than 60 years, and possible risk was largely originated from VCAN expression. However, the risk in patients with age greater than 60 years was largely from age factor, because of limited contribution of gene expression for risk.
Figure 5.
Survival analyses on basis of clinical pathological factors in UVM patients. (A) Age. (B) ≥ 60 years. (C) < 60 years. (D) Sex. (E) Male. (F) Female.
Figure 6.
Construction of nomogram prediction model for UVM.
Cox regression analysis was performed to explore the clinical factors which may influence the UVM prognosis. Both univariate and multivariate analyses indicated that VCAN mRNA expression was notably associated with OS of UVM patients (HR =4.352, P =0.031; HR =4.566, P =0.045, respectively). Besides, age, stage 2 and stage 3 were also significantly related to OS of UVM patients (all P <0.05), but gender had no significant relationship with OS in UVM (P >0.05) (Table 1). The results revealed that VCAN mRNA expression served as an independent prognostic factor for OS in UVM patients.
Table 1.
Cox Regression Analysis of VCAN and Other Clinicopathological Factors in UVM Patients
| Covariates | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI_l | 95% CI_u | P-value | HR | 95% CI_l | 95% CI_u | P-value | |
| Age | 1.046 | 1.008 | 1.085 | 0.019 | 1.045 | 1.006 | 1.085 | 0.023 |
| Male | 1.542 | 0.651 | 3.652 | 0.325 | 1.492 | 0.575 | 3.870 | 0.410 |
| Stage 2 | 0.014 | 0.001 | 0.148 | <0.001 | 0.021 | 0.002 | 0.223 | 0.001 |
| Stage 3 | 0.017 | 0.002 | 0.171 | 0.001 | 0.015 | 0.001 | 0.171 | 0.001 |
| Stage 4 | NA | NA | NA | NA | NA | NA | NA | NA |
| VCAN | 3.115 | 1.693 | 5.734 | <0.001 | 3.485 | 1.740 | 6.979 | < 0.001 |
Due to the vital role of immune infiltration cells in the tumor progression, the association of six immune cells with 5-year cumulative survival in UVM was first explored. As shown in Figure S1, only CD8 + T cell and neutrophil were correlated with OS of patients (all P<0.05). We further predicted the independent prognostic value of VCAN in UVM after taking immune factors into consideration. The multivariate analysis results revealed that VCAN still served as an independent prognostic factor for UVM (P <0.05) (Table 2). Besides, neutrophil was also significantly related to the survival probability of UVM patients in multivariate analysis (P <0.05) (Table 2), and hence we explored the relationship between neutrophil and VCAN in UVM through TISIDB database. Interestingly, VCAN had a positive correlation with neutrophil with a rho =0.462 (P <0.001) (Figure S2).
Table 2.
Multivariate Analysis of VCAN, Clinicopathological Factors and Immune Infiltration Cells
| Covariates | HR | 95% CI_l | 95% CI_u | P-value |
|---|---|---|---|---|
| Age | 1.055 | 0.998 | 1.116 | 0.059 |
| Male | 1.757 | 0.576 | 5.361 | 0.322 |
| Stage2 | 0.019 | 0.001 | 0.232 | 0.002 |
| Stage3 | 0.014 | 0.001 | 0.198 | 0.002 |
| Stage4 | NA | NA | NA | NA |
| B cell | 0.013 | 0.000 | 11.393 | 0.209 |
| CD8 + T cell | 0.016 | 0.000 | 5.109 | 0.159 |
| CD4 + T cell | 0.000 | 0.000 | 0.833 | 0.045 |
| Macrophage | 20.390 | 0.063 | 6617.825 | 0.307 |
| Neutrophil | 0.000 | 0.000 | 0.000 | 0.000 |
| Dendritic | 0.146 | 0.007 | 3.176 | 0.221 |
| VCAN | 2.505 | 1.052 | 5.965 | 0.038 |
Enrichment Analysis of VCAN
To clarify the biological functions of VCAN in UVM, the genes that co-expressed with VCAN in UVM were selected for analyses of GO annotation containing biological process (BP), cellular component (CC) and molecular function (MF) and KEGG pathways according to the selection criteria via DAVID database. The BP that VCAN and its co-expressed genes were mainly involved in inflammatory response, cell adhesion, and immune response. In terms of CC, genes that co-expressed with VCAN were mainly enriched in extracellular matrix, extracellular space, and extracellular region. MF were mainly enriched in extracellular matrix structural constituent, collagen binding, and integrin binding. As for KEGG pathway, genes co-expressed with VCAN were mainly participated in ECM-receptor interaction, focal adhesion, cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, and pathways in cancer (Table 3). The results suggested that VCAN may play a vital role in tumor cell adhesion and inflammation.
Table 3.
Functional Enrichment Analyses of VCAN and Its Co-Expressed Genes in UVM
| Category | ID | Term | Count | P-value |
|---|---|---|---|---|
| GO BP | GO: 0006954 | Inflammatory response | 47 | 3.09E-18 |
| GO: 0007155 | Cell adhesion | 44 | 5.18E-13 | |
| GO: 0006955 | Immune response | 40 | 9.32E-12 | |
| GO: 0001525 | Angiogenesis | 25 | 5.60E-09 | |
| GO: 0031295 | T cell costimulation | 14 | 1.17E-07 | |
| GO: 0045766 | Positive regulation of angiogenesis | 14 | 1.07E-05 | |
| GO: 0007267 | Cell-cell signaling | 21 | 1.40E-05 | |
| GO: 0043066 | Negative regulation of apoptotic process | 29 | 3.11E-05 | |
| GO CC | GO: 0031012 | Extracellular matrix | 45 | 5.18E-21 |
| GO: 0005615 | Extracellular space | 86 | 1.91E-14 | |
| GO: 0005576 | Extracellular region | 89 | 1.88E-11 | |
| GO: 0005578 | Proteinaceous extracellular matrix | 31 | 2.17E-11 | |
| GO: 0005925 | Focal adhesion | 33 | 1.33E-08 | |
| GO: 0009986 | Cell surface | 35 | 2.55E-06 | |
| GO: 0005840 | Ribosome | 16 | 3.19E-05 | |
| GO MF | GO: 0005201 | Extracellular matrix structural constituent | 15 | 1.47E-09 |
| GO: 0005518 | Collagen binding | 12 | 3.53E-07 | |
| GO: 0005178 | Integrin binding | 14 | 3.31E-06 | |
| GO: 0050840 | Extracellular matrix binding | 7 | 4.42E-05 | |
| GO: 0005102 | Receptor binding | 24 | 5.42E-05 | |
| GO: 0004872 | Receptor activity | 18 | 5.52E-05 | |
| GO: 0008009 | Chemokine activity | 6 | 1.62E-03 | |
| KEGG pathway | hsa: 04512 | ECM-receptor interaction | 18 | 3.40E-09 |
| hsa: 04510 | Focal adhesion | 23 | 1.50E-06 | |
| hsa: 04060 | Cytokine-cytokine receptor interaction | 24 | 6.79E-06 | |
| hsa: 04514 | Cell adhesion molecules | 17 | 2.17E-05 | |
| hsa: 04151 | PI3K-Akt signaling pathway | 28 | 3.64E-05 | |
| hsa: 04064 | NF-kappa B signaling pathway | 12 | 1.53E-04 | |
| hsa: 05200 | Pathways in cancer | 26 | 1.64E-03 | |
| hsa: 04660 | T cell receptor signaling pathway | 11 | 1.96E-03 | |
| hsa: 04620 | Toll-like receptor signaling pathway | 11 | 3.02E-03 | |
| hsa: 04062 | Chemokine signaling pathway | 15 | 4.12E-03 | |
| hsa: 05205 | Proteoglycans in cancer | 15 | 7.76E-03 | |
| hsa: 04014 | Ras signaling pathway | 14 | 4.28E-02 |
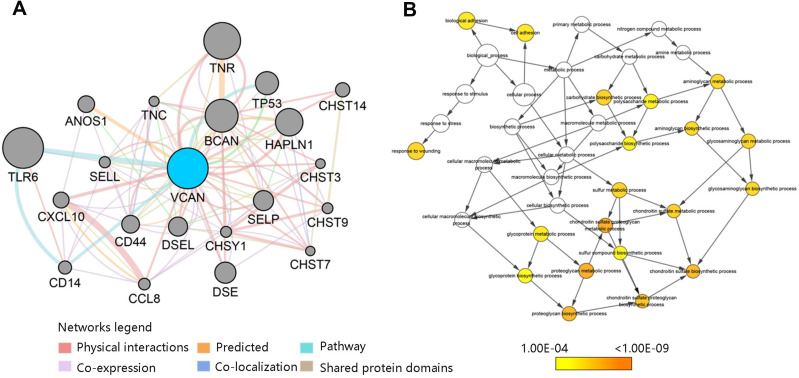
To elucidate the biological process of the genes most associated with VCAN, the protein/gene interaction work for VCAN were generated by GeneMANIA plugin of Cytoscape. As shown in Figure 7A, a total of 20 most relevant genes were derived. Then, BiNGO plug-in of Cytoscape was used to analyze and visualize the associated biological process of VCAN and its 20 related genes. We found that the biological process included proteoglycan metabolic process, chondroitin sulfate biosynthetic process and cell adhesion (Figure 7B). Combined with the results of GO annotation, it could be concluded that VCAN participated in cell adhesion in UVM, indicating that VCAN may be involved in the UVM occurrence and metastasis.
Figure 7.
VCAN-related genes analysis (A) Interaction analysis of VCAN with relevant genes. (B) Biological process of VCAN and its related genes.
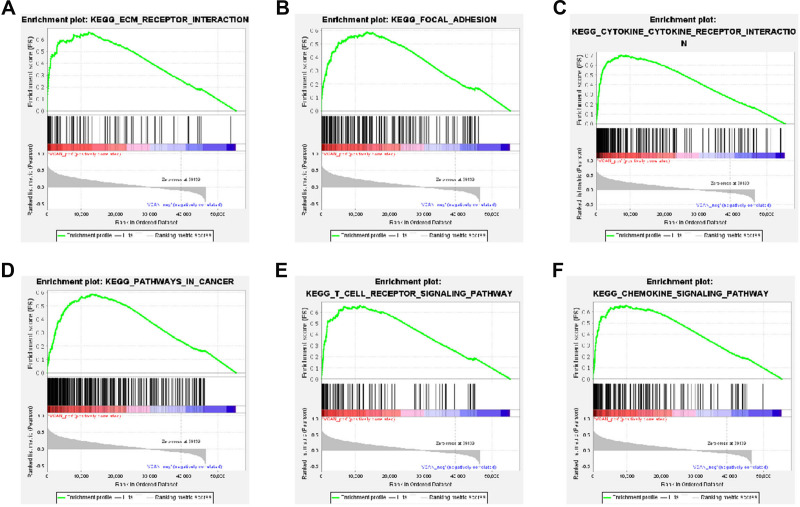
Finally, GSEA was performed to explore the biological pathogenesis of VCAN in UVM. The results displayed that 123 positive KEGG pathways were screened out in accordance with the criterion of FDR < 0.25 and P <0.05 but there was no negative pathway. GSEA analysis shared 6 positive significant pathways with DAVID database analysis, including ECM-receptor interaction, focal adhesion, cytokine-cytokine receptor interaction, pathways in cancer, T cell receptor signaling pathway, and chemokine signaling pathway (Figure 8A–F and Table 4). These findings revealed that VCAN may be essential in the progression of UVM.
Figure 8.
Enrichment plots of VCAN in UVM by GSEA. (A) ECM-receptor interaction. (B) Focal adhesion. (C) Cytokine-cytokine receptor interaction. (D) Pathways in cancer. (E) T cell receptor signaling pathway. (F) Chemokine signaling pathway.
Table 4.
GSEA Analysis of VCAN in UVM
| KEGG Name | ES | NES | Nominal p-value | FDR q-value |
|---|---|---|---|---|
| ECM-receptor interaction | 0.6633031 | 1.765032 | <0.001 | <0.001 |
| Focal adhesion | 0.5886187 | 1.6274207 | <0.001 | <0.001 |
| Cytokine-cytokine receptor interaction | 0.7012275 | 1.9448571 | <0.001 | <0.001 |
| Pathways in cancer | 0.5882056 | 1.6359161 | <0.001 | <0.001 |
| T cell receptor signaling pathway | 0.6602689 | 1.7828739 | <0.001 | <0.001 |
| Chemokine signaling pathway | 0.6563821 | 1.8080744 | <0.001 | <0.001 |
Abbreviations: ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate.
Potential Drug of UVM
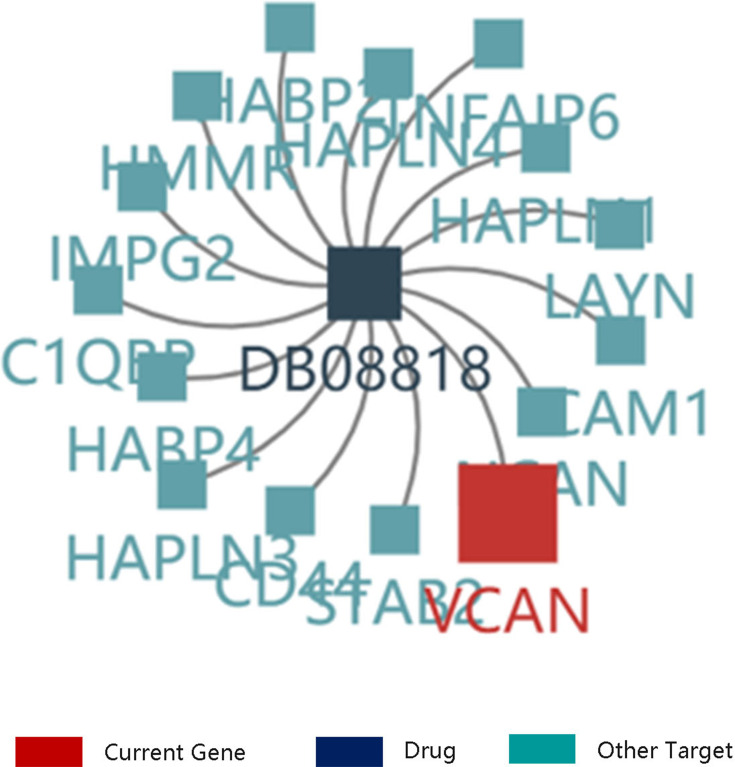
VCAN was proved to be critical in UVM growth and metastasis. Therefore, to explore the potent drug for the treatment of UVM, we searched the details on the drug targeting VCAN through TISIDB database. The result disclosed that VCAN was considered as a target of hyaluronic acid. The other targets of hyaluronic acid included C1QBP, CD44, HABP2 and HABP4 (Figure 9). Therefore, hyaluronic acid may be a potential drug for the UVM treatment.
Figure 9.
The drug targeting VCAN. DB08818: hyaluronic acid.
Discussion
VCAN was a component of extracellular matrix belonging to CSPG family, which were essential in the tumor microenvironment.20 It has also been exhibited that VCAN was implicated in tumor progression and abnormal VCAN expression were observed in various cancers.8 High mRNA level of VCAN has been demonstrated to be correlated with the poor prognosis of bladder cancer and the patients with luminal infiltration exhibited a high VCAN gene expression.21 Whereas, Haferlach et al showed that VCAN mRNA expression was lower in leukemia group than that in normal group.22 Therefore, the authors speculated that the VCAN mRNA expression may play an essential role in tumor development. Besides, we found that VCAN mRNA expression had a significant relationship with tumor histology and epithelioid patients presented the highest VCAN mRNA expression compared with those with spindle and mixed type (P <0.05), which may be a cause for that epithelioid patient had shortest survival time.5 In addition, Folberg et al conducted a study on tumor cell plasticity in UVM, and they have observed the down-regulation of several metastasis-associated genes including VCAN in the UVM cells that could form vasculogenic mimicry (VM) patterns in 3D-structures. However, after the formation of such VM-patterns, the UVM cells indeed transformed into a less invasive phenotype with a spindle-like morphology, which are in accordance with the findings of this manuscript.23 And VCAN mRNA expression was proved to predict poor prognosis in UVM, which were validated by Gepia and TISIDB databases. Due to the possible differences in time unit and analytic approaches of Gepia and TISIDB databases, the survival curves obtained for VCAN expression from two databases were slightly different. The results also showed that gender and weight might be important factors in UVM prognosis, which should be studied in the future. To analyze whether the important clinical factors including age and sex had an impact on OS in UVM, Kaplan–Meier plotter analysis was performed and the results showed that age was not related to OS, but among the patients aged <60 years, high VCAN mRNA expression showed a significant lower survival time (P <0.01). As for gender, high VCAN mRNA expression predicted poor prognosis among female patients (P <0.01). Previous studies have proved that the ovarian hormones estrogen and progesterone could indeed promote the up-regulation of VCAN expression in mouse models in a tissue-specific manner,24,25 which may result in the association of a worse survival rate with the higher VCAN expression in females but not males. Moreover, the normal average body fat percentage is usually higher in females, which may also lead to the survival difference since VCAN-mRNA is relatively up-regulated in the adipose tissue. Cox regression analyses containing clinicopathological factors and VCAN expression showed that VCAN mRNA expression was an independent factor for OS in UVM. Meanwhile, we found that CD8 + T cell with a higher immune infiltration level led to poor prognosis in UVM patients, which was consistent with the finding of Bronkhorst et al that CD8 + T cell was significantly related to UVM unfavorable prognosis.26 It has also been demonstrated that CD8 infiltration would shorten OS in gastric carcinoma.27 On the contrary, high infiltration level of neutrophil resulted in longer survival time of UVM patients. However, the prognostic significance of neutrophil in UVM has never been reported. After integrating VCAN, clinicopathological factors and immune infiltration cells into multivariate analysis, VCAN remained an independent prognostic factor for UVM, which further supported the importance of VCAN on patient’s survival.
Moreover, we performed the functional enrichment analysis of genes that co-expressed in UVM. The DAVID database analysis showed that VCAN and its co-expressed genes participated in various biological process including inflammatory response and cell adhesion. They were mainly enriched in extracellular matrix, extracellular space, and extracellular matrix structural constituent. The VCAN and its co-expressed genes were involved in the pathways such as ECM-receptor interaction and focal adhesion. The result of GeneMANIA analysis on the VCAN-related genes presented that the biological process that they were participated in were chondroitin sulfate biosynthetic process and cell adhesion. In addition, GSEA analysis disclosed that VCAN mainly participated in the positive pathways such as cytokine-cytokine receptor interaction, chemokine signaling pathway, and T cell receptor signaling pathway. Cytokines are important extracellular components in the tumor microenvironment, which was related to tumorigenesis, progression and metastasis.28,29 Besides, chemokine receptors are regarded as molecules associated with tumor metastasis.30 It has also been evidenced that many T cell sub sets were observed in some tumors and they were participated in tumor metastasis.31 Therefore, VCAN may play an important role in UVM progression and metastasis by regulating positive pathways including cytokine-cytokine receptor interaction, chemokine signaling pathway, and T cell receptor signaling pathway. For exploring the usage of VCAN in clinical therapy, TISIDB database has demonstrated that VCAN was a binding target of hyaluronic acid. With a similar structure to hyaluronic acid-binding proteins in the amino-terminal domain, VCAN protein with a B-loop domain, was considered to perform a binding function towards hyaluronic acid in human.32
Our study initially investigated the significance of VCAN in UVM, providing a potent therapeutic target for the UVM treatment. Nevertheless, the role of VCAN mRNA expression in combination with gender or weight should be investigated in the future. And the underlying mechanism predicted from our study requires further study to be validated by in vitro and in vivo experiments.
Conclusions
VCAN mRNA high expression was observed in UVM and led to poor prognosis, which might be involved in the positive regulation of ECM-receptor interaction, focal adhesion, cytokine-cytokine receptor interaction, pathways in cancer, T cell receptor signaling pathway, and chemokine signaling pathway. Therefore, VCAN may be related to the UVM occurrence and progression, and it may be a reliable marker for UVM clinical prognosis. Besides, hyaluronic acid may be an effective drug in the clinical UVM treatment.
Funding Statement
This study is supported by Grants from Natural Science Foundation of Zhejiang Province (No. LQ18H120002).
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Oliva M, Rullan AJ, Piulats JM. Uveal melanoma as a target for immune-therapy. Ann Trans Med. 2016;4(9):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaliki S, Shields C, Shields JJ. Uveal melanoma: estimating prognosis. Indian J Ophthalmol. 2015;63(2):93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souto EB, Zielinska A, Luis M, et al. Uveal melanoma: physiopathology and new in situ-specific therapies. Cancer Chemother Pharmacol. 2019;84(1):15–32. [DOI] [PubMed] [Google Scholar]
- 4.Martel A, Oberic A, Moulin A, et al. Eyelids metastases from uveal melanoma: clinical and histopathologic features of two cases and literature review. Eye. 2018;33(5):767–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronkhorst I, Jager MJ. Inflammation in uveal melanoma. Eye. 2013;27(2):217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bol KF, Mensink HW, Aarntzen EH, et al. Long overall survival after dendritic cell vaccination in metastatic uveal melanoma patients. Am J Ophthalmol. 2014;158(5):939–947.e5. [DOI] [PubMed] [Google Scholar]
- 7.Wight TN. Provisional matrix: a role for versican and hyaluronan. Matrix Biol. 2016;60:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricciardelli C, Sakko AJ, Ween MP, et al. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009;28(1–2):233. [DOI] [PubMed] [Google Scholar]
- 9.Theocharis AD. Versican in health and disease. Connect Tissue Res. 2008;49(3):230–234. [DOI] [PubMed] [Google Scholar]
- 10.Chida S, Okayama H, Noda M, et al. Stromal VCAN expression as a potential prognostic biomarker for disease recurrence in stage II-III colon cancer. Carcinogenesis. 2016;37(9):878–887. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zou X, Qian W,et al. Enhanced PAPSS2/VCAN sulfation axis is essential for Snail-mediated breast cancer cell migration and metastasis. Cell Death Differ. 2018;26(Pt 15):565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh S, Albitar L, LeBaron R, et al. Up-regulation of stromal versican expression in advanced stage serous ovarian cancer. Gynecol Oncol. 2010;119(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X, Liu JJ, Liu X, et al. Bioinformatics analysis of the prognosis and biological significance of VCAN in gastric cancer. Immun Inflamm Dis. 2021;9(2):547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Lu W, Dong ZJ. Tumor-infiltrating macrophages are involved in suppressing growth and metastasis of human prostate cancer cells by inf-β gene therapy in nude mice. Clin Cancer Res. 2002;8(9):2942. [PubMed] [Google Scholar]
- 16.Barnes TA, Amir EJ. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117(4):451–460. doi: 10.1038/bjc.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath WR, Kato Y, Steiner TM, et al. Antigen presentation by dendritic cells for B cell activation. Curr Opin Immunol. 2019;58:44–52. [DOI] [PubMed] [Google Scholar]
- 18.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. doi: 10.1186/s13045-018-0643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hope C, Emmerich PB, Papadas A, et al. Versican-derived matrikines regulate batf3-dendritic cell differentiation and promote T cell infiltration in colorectal cancer. J Immunol. 2017;199(5):1933–1941. doi: 10.4049/jimmunol.1700529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng W, Wang G, Wang Y, et al. The roles of versican V1 and V2 isoforms in cell prolifer1ation and apoptosis. Cell. 2005;16(3):1330–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Wu J, Chen X, et al. Upregulation of versican associated with tumor progression, metastasis, and poor prognosis in bladder carcinoma. BioMed Res Int. 2021;2021(4):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haferlach T, Kohlmann A, Wieczorek Let al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the international microarray innovations in leukemia study group. J Clin Oncol. 2010;28(15):2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folberg R, Arbieva Z, Moses J, et al. Tumor cell plasticity in uveal melanoma: microenvironment directed dampening of the invasive and metastatic genotype and phenotype accompanies the generation of vasculogenic mimicry patterns. Am J Pathol. 2006;169(4):1376–1389. doi: 10.2353/ajpath.2006.060223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salgado RM, Capelo LP, Favaro RR, Glazier JD, Aplin JD, Zorn TM. Hormone-regulated expression and distribution of versican in mouse uterine tissues. Reprod Biol Endocrinol. 2009;7(1):60. doi: 10.1186/1477-7827-7-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salgado RM, Covarrubias AC, Favaro RR, Serrano-Nascimento C, Nunes MT, Zorn TMT. Estradiol induces transcriptional and posttranscriptional modifications in versican expression in the mouse uterus. J Mol Histol. 2013;44(2):221–229. doi: 10.1007/s10735-012-9476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bronkhorst I, Ly LV, Jordanova ES, et al. Detection of M2-macrophages in uveal melanoma and relation with survival. Invest Ophthalmol Vis Sci. 2011;52(2). 643650. [DOI] [PubMed] [Google Scholar]
- 27.Thompson E, Zahurak M, Murphy A, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2017;66(5):794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–1492. [DOI] [PubMed] [Google Scholar]
- 29.Wu T, Dai YJ. Tumor microenvironment and therapeutic response. Cancer Lett. 2016;387:S0304383516300155. [DOI] [PubMed] [Google Scholar]
- 30.Koizumi K, Hojo S, Akashi T, et al. Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci. 2007;98(11):1652–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bronkhorst I, Vu TK, Jordanova ES, et al. Different subsets of tumor-infiltrating lymphocytes correlate with macrophage influx and monosomy 3 in uveal melanoma. Investig Ophthalmol Vis Sci. 2012;53(9):5370–5378. [DOI] [PubMed] [Google Scholar]
- 32.Bignami A, Lane WS, Andrews D, Dahl D. Structural similarity of hyaluronate binding proteins in brain and cartilage. Brain Res Bull. 1989;22(1):67–70. doi: 10.1016/0361-9230(89)90129-9 [DOI] [PubMed] [Google Scholar]