Abstract
Background
Three tools are currently available to predict the risk of contralateral breast cancer (CBC). We aimed to compare the performance of the Manchester formula, CBCrisk, and PredictCBC in patients with invasive breast cancer (BC).
Methods
We analyzed data of 132,756 patients (4682 CBC) from 20 international studies with a median follow-up of 8.8 years. Prediction performance included discrimination, quantified as a time-dependent Area-Under-the-Curve (AUC) at 5 and 10 years after diagnosis of primary BC, and calibration, quantified as the expected-observed (E/O) ratio at 5 and 10 years and the calibration slope.
Results
The AUC at 10 years was: 0.58 (95% confidence intervals [CI] 0.57–0.59) for CBCrisk; 0.60 (95% CI 0.59–0.61) for the Manchester formula; 0.63 (95% CI 0.59–0.66) and 0.59 (95% CI 0.56–0.62) for PredictCBC-1A (for settings where BRCA1/2 mutation status is available) and PredictCBC-1B (for the general population), respectively. The E/O at 10 years: 0.82 (95% CI 0.51–1.32) for CBCrisk; 1.53 (95% CI 0.63–3.73) for the Manchester formula; 1.28 (95% CI 0.63–2.58) for PredictCBC-1A and 1.35 (95% CI 0.65–2.77) for PredictCBC-1B. The calibration slope was 1.26 (95% CI 1.01–1.50) for CBCrisk; 0.90 (95% CI 0.79–1.02) for PredictCBC-1A; 0.81 (95% CI 0.63–0.99) for PredictCBC-1B, and 0.39 (95% CI 0.34–0.43) for the Manchester formula.
Conclusions
Current CBC risk prediction tools provide only moderate discrimination and the Manchester formula was poorly calibrated. Better predictors and re-calibration are needed to improve CBC prediction and to identify low- and high-CBC risk patients for clinical decision-making.
Keywords: Contralateral breast cancer, Risk prediction, Validation, Clinical decision-making
Introduction
A rising number of women with breast cancer (BC) are at risk to develop a new primary tumor in the contralateral breast (CBC) with consequently another cancer treatment and potentially less favorable prognosis [1]. Although CBC incidence is low (~ 0.4% per year) in the general BC population, contralateral preventive mastectomy (CPM) is increasing, also among women with low-CBC risk [2–5].
Three tools are tools currently available to predict the risk of CBC, although probably none are widely used: (1) the Manchester formula; (2) CBCrisk, and (3) PredictCBC [6–8]. The Manchester group in the United Kingdom (UK) proposed a set of guidelines for counseling women about CPM [8]. Based on a systematic review of the literature, they devised a formula to estimate lifetime CBC risk based on age at first primary BC, family history of BC, estrogen-receptor (ER) status, diagnosis of ductal carcinoma in situ (DCIS), and oophorectomy.
The second tool, CBCrisk, was developed using data on 1921 CBC cases and 5763 matched controls with primary BC [7]. The model uses data on age at first BC diagnosis, age at first birth, first degree family history of BC, high-risk pre-neoplasia, breast density (obtained using the BI-RADS system), ER status, first BC type (pure invasive, pure DCIS, a mix of the two, unknown), and adjuvant endocrine therapy. External validation was performed using two independent studies in the United States (US) of 5185 and 6035 patients with 111 and 117 CBC events [7, 9]. A web-based application provides individualized prediction of CBC risk [10].
Third, PredictCBC was developed, cross-validated and evaluated using data from 132,756 patients with first BC and 4672 CBC events, as part of an international collaboration [5]. PredictCBC predicts CBC risk as a function of family history (first degree) of primary BC, and information of primary BC diagnosis: age, nodal status, size, grade, morphology, ER status, human epidermal growth factor receptor 2 (HER2) status, administration of adjuvant or neoadjuvant chemotherapy, adjuvant endocrine therapy, adjuvant trastuzumab therapy, and radiotherapy. Two versions were developed: PredictCBC version 1A includes presence or absence of a mutation in the BRCA1 or BRCA2 genes, an important determinant of CBC [5, 11, 12], while PredictCBC version 1B was developed for untested patients.
External validation in different studies is relevant to assess the prediction performance of prediction models [13]. Our aim was to perform a head-to-head comparison between CBCrisk, PredictCBC and the Manchester formula. We hereto used several large population- and hospital-based studies used to develop and cross-validate the PredictCBC models.
Material and methods
External validation of CBCrisk and the Manchester formula was performed in 20 studies: four with individual patient data from the Netherlands [the Amsterdam Breast Cancer Study (ABCS), the Breast Cancer Outcome Study of Mutation carriers (BOSOM), the Erasmus MC Breast Cancer Registry (EMC), the Netherlands Cancer Registry (NCR)]; and 16 other studies of the Breast Cancer Association Consortium (BCAC). The latter is an international consortium of 102 studies comprising 182,898 patients (data version: January 2017) with a primary BC diagnosed between 1939 and 2016 [14]. Of these, 16 non-familial BC BCAC studies including invasive non-metastatic European-descent female patients with first primary invasive BC diagnosed from 1990 onwards, and with at least 10 CBC events, were included in the analyses [14]. Details about studies and patient selection, and data imputation were described previously [5].
The outcome was in situ or invasive metachronous CBC. Follow-up started 3 months after invasive first primary BC diagnosis, to exclude synchronous CBCs, and ended at date of CBC, distant metastasis (but not at loco-regional relapse), CPM or last date of follow-up (due to death, being lost to follow-up, or end of study), whichever occurred first. In the BCAC, 27,155 patients were recruited more than 3 months after diagnosis of the first primary BC (prevalent cases); for these patients, follow-up started at date of recruitment (left truncation). Distant metastasis and death due to any cause were competing events.
The Manchester formula provides an estimate of a woman’s individual lifetime CBC risk. To assess the prediction performance, we translated the lifetime CBC risk to 5- and 10-year CBC risks (see Supplementary Material). The predictors included in the CBC risk estimation in the Manchester formula, CBCrisk and PredictCBC models are provided in Table 1. Predictors that were sporadically missing were multiply imputed as described elsewhere [5].
Table 1.
Predictors included in current contralateral breast cancer risk prediction tools
| List of predictors | CBCriskb | Manchester formulac | PredictCBC version 1Ad | PredictCBC version 1Bd |
|---|---|---|---|---|
| Age at diagnosis | ✔ | ✔ | ✔ | ✔ |
| Age at first birth | ✔ | |||
| First-degree family history | ✔ | ✔ | ✔ | ✔ |
| BRCA1/2 germline mutation | ✔ | ✔ | ||
| First breast cancer behavior typea | ✔ | ✔ | ||
| Lymph node status | ✔ | ✔ | ||
| Breast density | ✔ | |||
| Tumor size | ✔ | ✔ | ||
| Morphology | ✔ | ✔ | ||
| Tumor grade | ✔ | ✔ | ||
| High-risk pre-neoplasia | ✔ | |||
| ER status | ✔ | ✔ | ✔ | ✔ |
| HER2 status | ✔ | ✔ | ||
| Chemotherapy | ✔ | ✔ | ||
| Endocrine therapy | ✔ | ✔ | ✔ | |
| Radiation to the breast | ✔ | ✔ | ||
| Trastuzumab | ✔ | ✔ | ||
| Oophorectomy under 40 years | ✔ |
Statistical analysis
Discrimination, the ability of the model to differentiate between patients who experienced CBC and those who did not, was calculated by time-dependent Area-Underthe-Curve (AUCs) based on Inverse Censoring Probability Weighting at 5 and 10 years [15, 16]. Values of AUCs close to 1 indicate good discrimination while values close to 0.5 indicate poor discrimination (a coin flip). Calibration is the agreement between observed and predicted risk and is commonly characterized by calibration-in-the-large and slope statistic. Calibration-in-the-large characterizes the overall difference between the observed and predicted risks. It was calculated using the expected/observed (E/O) ratio. An E/O less than 1 indicates that the model systematically underestimates CBC risk, while an E/O above 1 indicates that the model systematically overestimates CBC risk. The expected number of cases was calculated by summing the individual predicted probabilities at 5 and 10 years, based on the patient-specific covariate values [17]. The observed number of cases was estimated by the non-parametric CBC cumulative incidence at 5 and 10 years. The calibration slope was estimated using a Fine and Gray regression model using the linear predictor of the prediction tools. The linear predictor was vs constructed as the sum of the factors included in each model weighted by the corresponding regression coefficients (or parameters), and then computed in the validation dataset exactly as reported for the development set. The calibration slope is determined as the regression coefficient for this linear predictor when fitted as a single covariate in a regression model of disease outcome in the validation dataset. A well-calibrated model should have a calibration slope of 1; slopes < 1 indicate that coefficients were too optimistic for the validation setting [18]. Calibration results were graphically displayed.
Analyses were stratified by geographic groups of studies, since stratification by individual studies would provide too few events in some strata [5, 13, 19]. To allow for heterogeneity across multiple studies, random-effect meta-analyses were performed. We calculated 95% confidence intervals (CI) and 95% prediction intervals (PI), which indicate the likely range for prediction accuracy of the model in a new dataset, for discrimination and calibration measures. A sensitivity analysis was performed to check the consistency of CBCrisk performance measures when metachronous CBC was defined as an event after 6 instead of 3 months since the first BC diagnosis. More details are provided in the Supplementary Material. All analyses were implemented using SAS (SAS Institute Inc., NC, USA) and R software [20].
Results
We included 132,756 patients from 20 studies who experienced 4862 CBC events during a median follow-up of 8.8 years. The main patient and clinical characteristics across studies and geographic areas are shown in Table 2.
Table 2.
Description of main patient and clinical factors used for evaluation of the models and formula
| Studya/geographic area | Europe —otherb | Europe—Scandinavia | Europe—United Kingdom | Netherlands— BOSOM | Netherlands— EMC | Netherlands— NCR | United States and Australia |
|---|---|---|---|---|---|---|---|
| N | 15,183 | 12,928 | 11,921 | 3760 | 3390 | 83,138 | 2436 |
| Age at first diagnosis, years (%) | |||||||
| < 30 | 152 (1.0) | 46 (0.4) | 156 (1.3) | 108 (2.9) | 46 (1.4) | 388 (0.5) | 41 (1.7) |
| 30–39 | 1252 (8.2) | 489 (3.8) | 1811 (15.2) | 842 (22.4) | 374 (11.0) | 4241 (5.1) | 494 (20.3) |
| 40 + | 13,779 (90.8) | 12,393 (95.9) | 9954 (83.5) | 2810 (74.7) | 2970 (87.6) | 78,509 (94.4) | 1901 (78.0) |
| Age at first birth = unknown (%) | 15,183 (100.0) | 12,928 (100.0) | 11,921 (100.0) | 3760 (100.0) | 3390 (100.0) | 83,138 (100.0) | 2436 (100.0) |
| Family history (%) | |||||||
| Yes | 2123 (14.0) | 818 (6.3) | 1371 (11.5) | 737 (19.6) | 591 (17.4) | 0 (0.0) | 319 (13.1) |
| No | 8057 (53.1) | 3158 (24.4) | 8210 (68.9) | 1177 (31.3) | 2482 (73.2) | 0 (0.0) | 1498 (61.5) |
| Unknown | 5003 (33.0) | 8952 (69.2) | 2340 (19.6) | 1846 (49.1) | 317 (9.4) | 83,138 (100.0) | 619 (25.4) |
| First BC type = Pure invasive (%) | 15,183 (100.0) | 12,928 (100.0) | 11,921 (100.0) | 3760 (100.0) | 3390 (100.0) | 83,138 (100.0) | 2436 (100.0) |
| Breast density = unknown (%) | 15,183 (100.0) | 12,928 (100.0) | 11,921 (100.0) | 3760 (100.0) | 3390 (100.0) | 83,138 (100.0) | 2436 (100.0) |
| ER status (%) | |||||||
| Negative | 3387 (22.3) | 1746 (13.5) | 1718 (14.4) | 896 (23.8) | 842 (24.8) | 14,591 (17.6) | 445 (18.3) |
| Positive | 10,071 (66.3) | 9401 (72.7) | 7175 (60.2) | 2024 (53.8) | 2427 (71.6) | 64,790 (77.9) | 1572 (64.5) |
| Unknown | 1725 (11.4) | 1781 (13.8) | 3028 (25.4) | 840 (22.3) | 121 (3.6) | 3757 (4.5) | 419 (17.2) |
| High-risk pre-neoplasia = unknow n (%) | 15,183 (100.0) | 12,928 (100.0) | 11,921 (100.0) | 3760 (100.0) | 3390 (100.0) | 83,138 (100.0) | 2436 (100.0) |
| Anti-estrogen therapy (%) | |||||||
| Yes | 7868 (51.8) | 6434 (49.8) | 8712 (73.1) | 809 (21.5) | 1559 (46.0) | 40,214 (48.4) | 363 (14.9) |
| No | 4570 (30.1) | 1947 (15.1) | 2046 (17.2) | 2739 (72.8) | 1821 (53.7) | 42,924 (51.6) | 8 (0.3) |
| Unknown | 2745 (18.1) | 4547 (35.2) | 1163 (9.8) | 212 (5.6) | 10 (0.3) | 0 (0.0) | 2065 (84.8) |
| CBC cumulative incidence (%) | |||||||
| 3-year (95% CI) | 1.0 (0.8–1.2) | 0.7 (0.5–0.9) | 0.5 (0.3–0.7) | 1.7 (1.3–2.1) | 1.7 (1.2–2.1) | 1.3 (1.2–1.4) | 1.8 (0.8–2.8) |
| 5-year (95% CI) | 1.6 (1.4–1.9) | 1.0 (0.8–1.3) | 1.0 (0.8–1.3) | 3.0 (2.5–3.6) | 2.6 (2.1–3.2) | 2.4 (2.3–2.5) | 2.8 (1.7–3.8) |
| 10-year (95% CI) | 3.5 (3.1–3.9) | 2.1 (1.7–2.4) | 1.3 (1.0–1.5) | 5.5 (4.7–6.2) | 5.7 (4.9–6.6) | 4.6 (4.5–4.8) | 4.1 (3.0–5.3) |
More details about the main patient and clinical characteristics by study are available in the supplementary information of [5]
BOSOM Breast Cancer Outcome Study of Mutation carriers, EMC Erasmus Medical Center, NCR Netherlands Cancer Registry, BC breast cancer, ER estrogen receptor, CBC contralateral breast cancer, CI confidence interval
The studies denoted with Europe and United States and Australia are part of the Breast Cancer Association Consortium
Europe—other geographic area included studies from Belgium (1), Germany (2), Netherlands (2) and Poland (2)
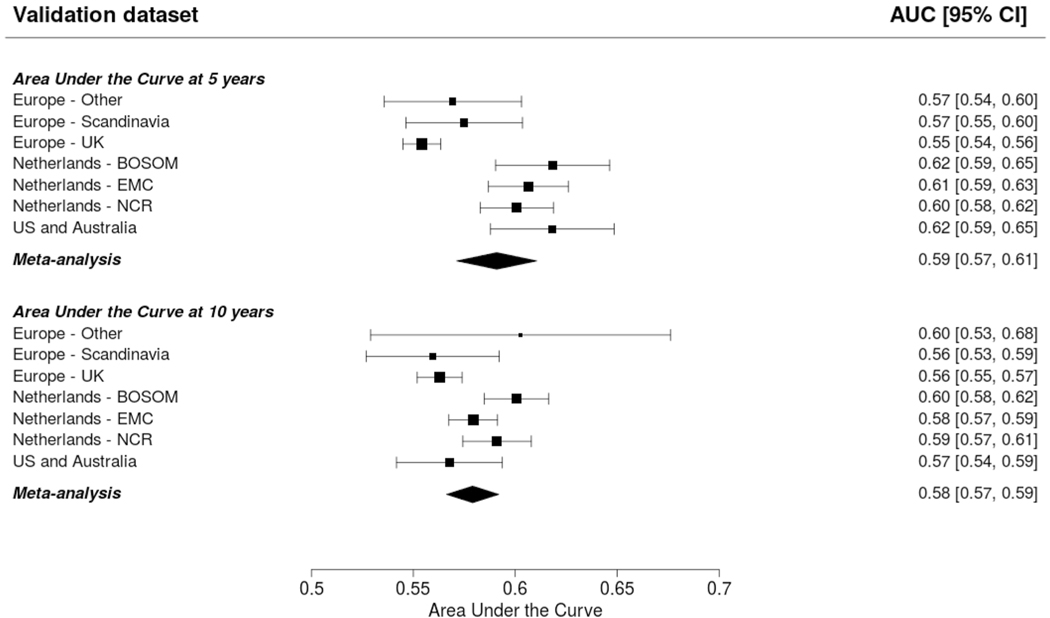
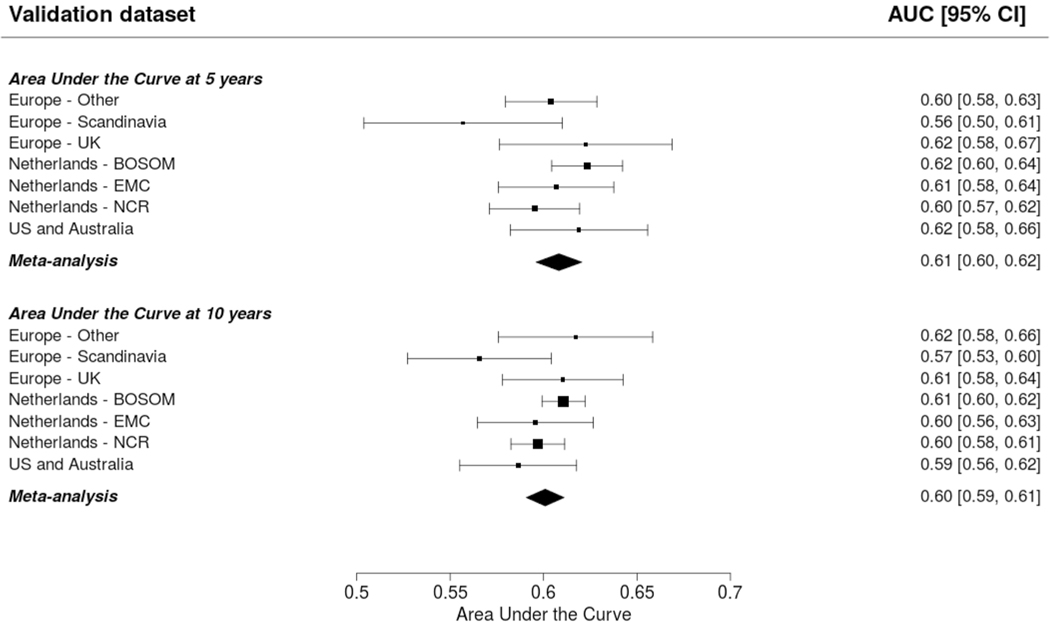
The AUCs at 5 and 10 years was around 0.6: 0.59 (95% CI 0.57–0.61; 95% PI 0.54–0.64) and 0.58 (95% CI 0.57–0.59; 95% PI 0.55–0.61) for CBCrisk (Fig. 1); 0.61 (95% CI 0.60–0.62; 95% PI 0.59–0.63) and 0.60 (95% CI 0.59–0.61; 95% PI 0.58–0.62) for the Manchester formula (Fig. 2). The E/O ratio at 5 and 10 years was close to 1 for all models: 0.86 (95% CI 0.50–1.46; 95% PI 0.20–3.75) and 0.82 (95% CI 0.51–1.32; 95% PI 0.21–3.14) for CBCrisk (Table 3); 1.54 (95% CI 0.61–3.92; 95% PI 0.11–20.72, Table 4), and 1.53 (95% CI 0.63–3.73; 95% PI 0.13–18.52) for the Manchester formula (Table 4); 1.26 (95% CI 0.57–2.77; 95% PI 0.14–11.34), and 1.28 (95% CI 0.63–2.58; 95% PI 0.18–9.18) for PredictCBC-1A (Table 5); 1.33 (95% CI 0.59–2.99, 95% PI 0.14–12.76), 1.35 (95% CI 0.65–2.77; 95% PI 0.19–10.24) for PredictCBC-1B (Table 5) [5]. The calibration slope was close to 1 for CBCrisk (1.26, 95% CI 1.01–1.50 and 95% PI 1.01–1.50, Tables 3, 4, 5), and PredictCBC-1A and 1B 0.90 (95% CI 0.79–1.02; 95% PI 0.73–1.08), and 0.81 (95% CI 0.63–0.99; 95% PI 0.50–1.12) (Table 5), while prognostic effects were far too large for the Manchester formula (slope: 0.39, 95% CI 0.34–0.43, 95% PI 0.34–0.43, Tables 4, 5). Calibration plots of CBCrisk at 5 and 10 years are shown in Supplementary Fig. 1 and Supplementary Fig. 2. As reported previously [5], the AUCs at 5 and 10 years for PredictCBC-1A were 0.63 (95% CI 0.58–0.67, 95% PI 0.52–0.74), and 0.63 (95% CI 0.59–0.66, 95% PI 0.53–0.72), respectively; for PredictCBC-1B 0.59 (CI 0.54–0.63, 95% PI 0.46–0.71, Table 5), and 0.59 (95% CI 0.56–0.62, 95% PI 0.52–0.66, Table 5), respectively.
Fig. 1.
Prediction performance of the CBCrisk model (Chowdhury et al. [7]). The upper and lower panel show the discrimination assessed by a time-dependent Area-Under-the-Curve at 5 and 10 years, respectively. The black squares indicate the estimated accuracy of a model built on all remaining studies or geographic areas. The black horizontal lines indicate the corresponding 95% confidence intervals of the estimated accuracy (interval whiskers). The black diamonds indicate the mean with the corresponding 95% confidence interval of the predictive accuracy
Fig. 2.
Prediction performance of the Manchester formula (Basu et al. [8]) The upper and lower panel show the discrimination assessed by a time-dependent Area-Under-the-Curve at 5 and 10 years, respectively. The black squares for each dataset indicate the estimated accuracy of a model built on all remaining studies or geographic areas. The black horizontal lines indicate the corresponding 95% confidence intervals of the estimated accuracy (interval whiskers). The black diamonds indicate the mean with the corresponding 95% confidence interval of the predictive accuracy
Table 3.
Calibration performance of the CBC risk model
| Validation dataset | E/O ratio at 5 years (95% CI) | E/O ratio at 10 years (95% CI) | Calibration slope (95% CI) |
|---|---|---|---|
| Europe—Other | 0.87 (076 to 0.98) | 0.75 (0.68 to 0.81) | 1.11 ( 0.40 to 1.83) |
| Europe—Scandinavia | 1.59 (1.28 to 1.91) | 1.23 (1.08 to 1.38) | 0.86 ( 0.16 to 1.57) |
| Europe—UK | 1.35 (1.38 to 2.17) | 1.82 (1.53 to 2.11) | 0.85 (− 0.03 to 1.73) |
| Netherlands—BOSOM | 0.45 (0.37 to 0.53) | 0.50 (0.43 to 0.57) | 1.34 ( 0.76 to 1.93) |
| Netherlands—EMC | 0.48 (0.38 to 0.57) | 0.43 (0.37 to 0.50) | 1.19 ( 0.65 to 1.73) |
| Netherlands—NCR | 0.57 (0.54 to 0.59) | 0.54 (0.52 to 0.56) | 1.40 ( 1.11 to 1.68) |
| US and Australia | 0.43 (0.33 to 0.54) | 0.56 (0.45 to 0.67) | 1.13 ( 0.25 to 2.00) |
| Meta-analysis | 0.86 (0.50 to 1.46) | 0.82 (0.51 to 1.32) | 1.26 ( 1.01 to 1.50) |
| 95% PI | 0.20 to 3.75 | 0.21 to 3.14 | 1.01 to 1.50 |
Chowdhury et al. [7]
E/O expected-observed, CI confidence interval, UK United Kingdom, BOSOM Breast Cancer Outcome Study of Mutation carriers, EMC Erasmus Medical Center, NCR Netherlands Cancer Registry, PI prediction interval
Table 4.
Calibration performance of the Manchester formula
| Validation dataset | E/O ratio at 5 years (95% CI) | E/O ratio at 10 years (95% CI) | Calibration slope (95% CI) |
|---|---|---|---|
| Europe—Other | 1.64 (1.44 to 1.85) | 1.46 (1.34 to 1.58) | 0.40 (0.29 to 0.50) |
| Europe—Scandinavia | 2.61 (2.09 to 3.12) | 2.11 (1.85 to 2.37) | 0.35 (0.13 to 0.57) |
| Europe—UK | 3.34 (2.60 to 4.08) | 3.49 (2.93 to 4.05) | 0.42 (0.23 to 0.61) |
| Netherlands—BOSOM | 0.81 (0.66 to 0.96) | 0.92 (0.79 to 1.05) | 0.45 (0.33 to 0.56) |
| Netherlands—EMC | 0.94 (0.75 to 1.14) | 0.87 (0.75 to 1.00) | 0.35 (0.21 to 0.49) |
| Netherlands—NCR | 1.00 (0.95 to 1.04) | 1.01 (0.98 to 1.05) | 0.37 (0.33 to 0.42) |
| US and Australia | 0.77 (0.58 to 0.96) | 1.02 (0.82 to 1.23) | 0.51 (0.33 to 0.68) |
| Meta-analysis | 1.54 (0.61 to 3.92) | 1.53 (0.63 to 3.73) | 0.39 (0.34 to 0.43) |
| 95% PI | 0.11 to 20.72 | 0.13 to 18.52 | 0.34 to 0.43 |
Basu et al. [8]
E/O expected-observed, CI confidence interval, UK United Kingdom, BOSOM Breast Cancer Outcome Study of Mutation carriers, EMC Erasmus Medical Center, NCR Netherlands Cancer Registry, PI prediction interval
Table 5.
Summary of prediction performance of CBCrisk, Manchester formula, and PredictCBC version 1A and version 1B with the corresponding 95% prediction intervals (PI)
| Characteristics | CBCriska | Manchester formulab | PredictCBC version 1Ac,d | PredictCBC version 1Bc,d |
|---|---|---|---|---|
| Discrimination | ||||
| AUC at 5 years (95% PI) | 0.59 (0.54 to 0.64) | 0.61 (0.59 to 0.63) | 0.63 (0.52 to 0.74) | 0.59 (0.46 to 0.71) |
| AUC at 10 years (95% PI) | 0.58 (0.55 to 0.61) | 0.60 (0.58 to 0.62) | 0.63 (0.53 to 0.72) | 0.59 (0.52 to 0.66) |
| Calibration | ||||
| E/O ratio at 5 years (95% PI) | 0.86 (0.20 to 3.75) | 1.54 (0.11 to 20.72) | 1.26 (0.14 to 11.34) | 1.33 (0.14 to 12.76) |
| E/O ratio at 10 years (95% PI) | 0.82 (0.21 to 3.14) | 1.53 (0.13 to 18.52) | 1.28 (0.18 to 9.18) | 1.35 (0.19 to 10.24) |
| Slope (95% PI) | 1.26 (1.01 to 1.50) | 0.39 (0.34 to 0.43) | 0.90 (0.73 to 1.08) | 0.81 (0.50 to 1.12) |
Sensitivity analysis showed that the performance measures of CBCrisk did not change when metachronous CBC was defined after 6 months since first BC diagnosis (see Supplementary Materials, Supplementary Tables 1, 2 and Supplementary Fig. 3).
Discussion
Accurate CBC risk predictions are essential in clinical decision-making around CPM or tailored surveillance among patients with first primary BC. In particular, overestimation of risk can lead to recommending CPM among BC patients with low risks. Underestimation can lead to suboptimal surveillance or hesitance about recommending CPM for patients with substantial risk. Using individual patient data from multiple studies with long follow-up, we externally evaluated the prediction performance accuracy of CBCrisk, a tool developed and validated to provide individualized CBC risk prediction, and the Manchester formula, a heuristically derived calculation of CBC lifetime risk [6–9]. In addition, the availability of different European-descendent studies allowed heterogeneity in the performance by geographic area to be assessed.
CBCrisk under-predicted the risk of CBC and had moderate discrimination ability with considerable heterogeneity between studies. The Manchester formula was empirically derived from a systematic review, and its discrimination accuracy was higher than CBCrisk. This may be explained by the inclusion of BRCA1/2 mutation carrier information, an important determinant of CBC risk [21]. With the same large individual patient data sets, PredictCBC models had been developed and validated [5]. In particular, PredictCBC version 1A includes information of BRCA1/2 mutation carriers and extensive information about the primary BC including treatments. The discrimination of all three prediction models was moderate, with AUC values around 0.6.
CBCrisk was previously externally validated using two independent clinical studies from Johns Hopkins University (JH) and MD Anderson Cancer Center (MDA) in the US [9]. Discrimination ability was 0.61 and 0.65 at 3 years, and 0.62 and 0.61 at 5 years for JH and MDA, respectively. The risk of CBC was overestimated in JH with E/O ratios of 2.02 and 1.56 at 3 and 5 years, while underestimated in MDA with E/O ratios of 0.61 and 0.62, respectively.
The considerable heterogeneity in all CBC risk calculators, especially in the CBCrisk and the Manchester formula, reflects the different CBC incidences in every study [13]. Another potential source of heterogeneity is the carrier frequency of germline mutations associated with CBC that may vary among studies, especially in the CBC calculators not including information of BRCA1/2 mutation as CBCrisk and the PredictCBC-1B [22]. In addition, heterogeneity may be due to the different proportions of the use of (neo)adjuvant systemic therapies explained by the different distribution of tumor subtypes among studies [4]. Besides, inter-observer variation in pathological examination of BC among studies may lead to different adjuvant systemic therapy advice and, consequently, prediction of CBC risk [23]. Variation in prediction performance and limited generalizability of CBC risk calculators can also be partially explained by differences in how predictors are measured among studies [24, 25]. For example, lack of family history knowledge may lead to uncertainty in risk prediction and varies according to demographics of the patients [26]. In particular, if in some studies BC patients misreported information about family history, the CBC risk would be over(under)estimated causing inappropriate decision-making regarding CPM or tailored surveillance. Some limitations of our study must be recognized. First, our dataset, while large, had missing data for three covariates that were used in the CBCrisk model: breast density, age at first birth, and high-risk pre-neoplasia. The authors of CBCrisk estimated the relative risks for patients with the unknown characteristics, but the use of the missing indicator variable is suboptimal compared to having the prognostic information available. It may lead to over or under-estimation of absolute CBC risk [27]. For this reason, we suggest that it is preferable to use multiple imputation of missing data, as is done in the PredictCBC models [28, 29]. In addition, investigation of the potential source of model misspecification due to possible different definitions or measurement error was not possible [30–32].
In conclusion, current statistical risk prediction models and heuristic formulas provided moderate CBC individualized prediction performance. Careful re-calibration is required before considering these models for clinical decision-making. A more direct comparison between the current CBC risk prediction models using a large external dataset with complete information on all factors included in all CBC prediction models would be ideal, but is currently unavailable. There is an ongoing debate about improvements of clinical prediction performance using machine learning approaches compared to standard regression approaches for risk prediction [33, 34]. However, irrespective of the methodology, better predictors are needed to predict CBC more accurately. Deeper biological insights and potential inclusion of other genetic markers such as CHEK2 c.1100del mutation status and polygenic risk scores based on common genetic variants may improve CBC risk prediction, although rare mutations are unlikely to contribute substantially to CBC risk in the general population [35, 36]. Life-style factors such as body mass index, alcohol consumption, and smoking also may help to better stratify high- and low-CBC risk patients even though these factors are difficult to measure accurately. Moreover, breast density may be important. More detailed information about adjuvant systemic therapies may better identify patients with low- and high-CBC risk since chemotherapy and especially endocrine therapy reduce CBC risk [4]. After extension and further external validation of prediction models for CBC risk, investigation of their potential clinical utility is an important future step.
Supplementary Material
Acknowledgements
We thank all individuals who took part in these studies and all researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. ABCFS thank Maggie Angelakos, Judi Maskiell, Gillian Dite. ABCS and BOSOM thanks all the collaborating hospitals and pathology departments and many individual that made this study possible, specifically, we wish to acknowledge: Annegien Broeks, Sten Cornelissen, Frans Hogervorst, Laura van ‘t Veer, Floor van Leeuwen, Emiel Rutgers. EMC thanks J.C. Blom-Leenheer, P.J. Bos,C.M.G. Crepin and M. van Vliet for data management. CGPS thanks staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen. HEBCS thanks Taru A. Muranen, Kristiina Aittomäki, Karl von Smitten, Irja Erkkilä. KARMA thanks the Swedish Medical Research Counsel. LMBC thanks Gilian Peuteman, Thomas Van Brussel, EvyVanderheyden and Kathleen Corthouts. MARIE thanks Petra Seibold, Dieter Flesch-Janys, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. ORIGO thanks E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The authors thank the registration team of the Netherlands Comprehensive Cancer Organisation (IKNL) for the collection of data for the Netherlands Cancer Registry as well as IKNL staff for scientific advice. PBCS thanks Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. The ethical approval for the POSH study is MREC /00/6/69, UKCRN ID: 1137. We thank the SEARCH team.
Funding This work is supported by the Alpe d’HuZes/Dutch Cancer Society (KWF Kankerbestrijding) project 6253. BCAC is funded by Cancer Research UK [C1287/A16563, C1287/A10118], the European Union’s Horizon 2020 Research and Innovation Programme (Grant Nos. 634935 and 633784 for BRIDGES and B-CAST respectively), and by the European Community’s Seventh Framework Programme under grant agreement number 223175 (Grant No. HEALTHF2-2009-223175) (COGS). The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. The Australian Breast Cancer Family Study (ABCFS) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. M.C.S. is a NHMRC Senior Research Fellow. The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. BOSOM was supported by the Dutch Cancer Society grant numbers DCS-NKI 2001-2423, DCS-NKI 2007-3839, and DCSNKI 2009-4363; the Cancer Genomics Initiative; and notary office Spier & Hazenberg for the coding procedure. The EMC was supported by grants from Alpe d’HuZes/Dutch Cancer Society NKI2013-6253 and from Pink Ribbon 2012.WO39.C143. The HEBCS was financially supported by the Helsinki University Hospital Research Fund, the Finnish Cancer Society, and the Sigrid Juselius Foundation. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation. The KARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. LMBC is supported by the ‘Stichting tegen Kanker’. The MARIE study was supported by the Deutsche Krebshilfe e.V.[70-2892-BR I, 106332, 108253, 108419, 110826, 110828], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MEC was support by NIH grants CA63464, CA54281, CA098758, CA132839 and CA164973. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Genotyping for PLCO was supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The POSH study is funded by Cancer Research UK (Grants C1275/A11699, C1275/C22524, C1275/A19187, C1275/A15956 and Breast Cancer Campaign 2010PR62, 2013PR044. PROCAS is funded from NIHR grant PGfAR 0707-10031. SEARCH is funded by Cancer Research UK [C490/A10124, C490/A16561] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. SKKDKFZS is supported by the DKFZ. The SZBCS (Szczecin Breast Cancer Study) was supported by Grant PBZ_KBN_122/P05/2004 and The National Centre for Research and Development (NCBR) within the framework of the international ERANET TRANSAN JTC 2012 application no. Cancer 12-054 (Contract No. ERA-NET-TRANSCAN / 07/2014).
Footnotes
Compliance with ethical standards
Conflict of interest Author DG, MH, EW, MAA, DA, JCB, CB, SEB, MKB, JCC, KC, PD, AMD, DFE, JF, HF, MGC, LH, CAH, PH, UH, JLH, AJ, AJ2, AJ3, RK, LBK, IK, DL, LLN, AL, JL, MM, LM, HN, HSAO, SP, PDPP, MS, SS, VTHBMS, MCS, WJT, RAEMT, AJvdB, CHMvD, FEvL, CvO, LvV, QW, CW, PJW, MJH declares that he has no conflict of interest. Author DMM declares that she receives a lecture fee from Pierre Fabre and personal fees for consultancy from Astra Zeneca. Author PAF reports grants from Novartis, grants from Biontech, personal fees from Novartis, personal fees from Roche, personal fees from Pfizer, personal fees from Celgene, personal fees from Daiichi-Sankyo, personal fees from TEVA, personal fees from Astra Zeneca, personal fees from Merck Sharp & Dohme, personal fees from Myelo Therapeutics, personal fees from Macrogenics, personal fees from Eisai, personal fees from Puma, grants from Cepheid.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of international, national, and institutional research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10549-020-05611-8) contains supplementary material, which is available to authorized users.
References
- 1.Langballe R, Frederiksen K, Jensen MB, Andersson M, CroninFenton D, Ejlertsen B, Mellemkjaer L (2018) Mortality after contralateral breast cancer in Denmark. Breast Cancer Res Treat 171(2):489–499. 10.1007/s10549-018-4846-3 [DOI] [PubMed] [Google Scholar]
- 2.Xiong Z, Yang L, Deng G, Huang X, Li X, Xie X, Wang J, Shuang Z, Wang X (2018) Patterns of occurrence and outcomes of contralateral breast cancer: analysis of SEER data. J Clin Med. 10.3390/jcm7060133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SM, Freedman RA, Sagara Y, Aydogan F, Barry WT, Golshan M (2017) Growing use of contralateral prophylactic mastectomy despite no improvement in long-term survival for invasive breast cancer. Ann Surg 265(3):581–589. 10.1097/SLA.0000000000001698 [DOI] [PubMed] [Google Scholar]
- 4.Kramer I, Schaapveld M, Oldenburg HSA, Sonke GS, McCool D, van Leeuwen FE, Van de Vijver KK, Russell NS, Linn SC, Siesling S, der Houven M-V, van Oordt CW, Schmidt MK (2019) The influence of adjuvant systemic regimens on contralateral breast cancer risk and receptor subtype. J Natl Cancer Inst. 10.1093/jnci/djz010 [DOI] [PubMed] [Google Scholar]
- 5.Giardiello D, Steyerberg EW, Hauptmann M, Adank MA, Akdeniz D, Blomqvist C, Bojesen SE, Bolla MK, Brinkhuis M, Chang-Claude J, Czene K, Devilee P, Dunning AM, Easton DF, Eccles DM, Fasching PA, Figueroa J, Flyger H, Garcia-Closas M, Haeberle L, Haiman CA, Hall P, Hamann U, Hopper JL, Jager A, Jakubowska A, Jung A, Keeman R, Kramer I, Lambrechts D, Le Marchand L, Lindblom A, Lubinski J, Manoochehri M, Mariani L, Nevanlinna H, Oldenburg HSA, Pelders S, Pharoah PDP, Shah M, Siesling S, Smit V, Southey MC, Tapper WJ, Tollenaar R, van den Broek AJ, van Deurzen CHM, van Leeuwen FE, van Ongeval C, Van’t Veer LJ, Wang Q, Wendt C, Westenend PJ, Hooning MJ, Schmidt MK (2019) Prediction and clinical utility of a contralateral breast cancer risk model. Breast Cancer Res 21(1):144. 10.1186/s13058-019-1221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell M (2018) Estimating Contralateral Breast Cancer Risk. Curr Breast Cancer Rep 10(2):91–97 [Google Scholar]
- 7.Chowdhury M, Euhus D, Onega T, Biswas S, Choudhary PK (2017) A model for individualized risk prediction of contralateral breast cancer. Breast Cancer Res Treat 161(1):153–160. 10.1007/s10549-016-4039-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu NN, Ross GL, Evans DG, Barr L (2015) The Manchester guidelines for contralateral risk-reducing mastectomy. World J Surg Oncol 13:237. 10.1186/s12957-015-0638-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury M, Euhus D, Arun B, Umbricht C, Biswas S, Choudhary P (2018) Validation of a personalized risk prediction model for contralateral breast cancer. Breast Cancer Res Treat. 10.1007/s10549-018-4763-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury M, Euhus D, Onega T, Choudhary P (2017) CBCRisk: contralateral breast cancer (CBC) risk predictor. https://cbc-predictor-utd.shinyapps.io/CBCRisk/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Broek AJ, Van’t Veer LJ, Hooning MJ, Cornelissen S, Broeks A, Rutgers EJ, Smit VT, Cornelisse CJ, van Beek M, Janssen-Heijnen ML, Seynaeve C, Westenend PJ, Jobsen JJ, Siesling S, Tollenaar RA, van Leeuwen FE, Schmidt MK (2016) Impact of age at primary breast cancer on contralateral breast cancer risk in BRCA1/2 mutation carriers. J Clin Oncol 34(5):409–418. 10.1200/JCO.2015.62.3942 [DOI] [PubMed] [Google Scholar]
- 12.Malone KE, Begg CB, Haile RW, Borg A, Concannon P, Tellhed L, Xue S, Teraoka S, Bernstein L, Capanu M, Reiner AS, Riedel ER, Thomas DC, Mellemkjaer L, Lynch CF, Boice JD Jr, Anton-Culver H, Bernstein JL (2010) Population-based study of the risk of second primary contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2. J Clin Oncol 28(14):2404–2410. 10.1200/JCO.2009.24.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Austin PC, van Klaveren D, Vergouwe Y, Nieboer D, Lee DS, Steyerberg EW (2016) Geographic and temporal validity of prediction models: different approaches were useful to examine model performance. J Clin Epidemiol 79:76–85. 10.1016/j.jclinepi.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A, Bolla MK, Wang Q, Tyrer J, Dicks E, Lee A, Wang Z, Allen J, Keeman R, Eilber U, French JD, Qing Chen X, Fachal L, McCue K, McCart Reed AE, Ghoussaini M, Carroll JS, Jiang X, Finucane H, Adams M, Adank MA, Ahsan H, Aittomaki K, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Arun B, Auer PL, Bacot F, Barrdahl M, Baynes C, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Borresen-Dale AL, Brand JS, Brauch H, Brennan P, Brenner H, Brinton L, Broberg P, Brock IW, Broeks A, Brooks-Wilson A, Brucker SY, Bruning T, Burwinkel B, Butterbach K, Cai Q, Cai H, Caldes T, Canzian F, Carracedo A, Carter BD, Castelao JE, Chan TL, David Cheng TY, Seng Chia K, Choi JY, Christiansen H, Clarke CL, Collaborators N, Collee M, Conroy DM, CordinaDuverger E, Cornelissen S, Cox DG, Cox A, Cross SS, Cunningham JM, Czene K, Daly MB, Devilee P, Doheny KF, Dork T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Elvira M, Engel C, Eriksson M, Fasching PA, Figueroa J, Flesch-Janys D, Fletcher O, Flyger H, Fritschi L, Gaborieau V, Gabrielson M, Gago-Dominguez M, Gao YT, Gapstur SM, Garcia-Saenz JA, Gaudet MM, Georgoulias V, Giles GG, Glendon G, Goldberg MS, Goldgar DE, GonzalezNeira A, Grenaker Alnaes GI, Grip M, Gronwald J, Grundy A, Guenel P, Haeberle L, Hahnen E, Haiman CA, Hakansson N, Hamann U, Hamel N, Hankinson S, Harrington P, Hart SN, Hartikainen JM, Hartman M, Hein A, Heyworth J, Hicks B, Hillemanns P, Ho DN, Hollestelle A, Hooning MJ, Hoover RN, Hopper JL, Hou MF, Hsiung CN, Huang G, Humphreys K, Ishiguro J, Ito H, Iwasaki M, Iwata H, Jakubowska A, Janni W, John EM, Johnson N, Jones K, Jones M, Jukkola-Vuorinen A, Kaaks R, Kabisch M, Kaczmarek K, Kang D, Kasuga Y, Kerin MJ, Khan S, Khusnutdinova E, Kiiski JI, Kim SW, Knight JA, Kosma VM, Kristensen VN, Kruger U, Kwong A, Lambrechts D, Le Marchand L, Lee E, Lee MH, Lee JW, Neng Lee C, Lejbkowicz F, Li J, Lilyquist J, Lindblom A, Lissowska J, Lo WY, Loibl S, Long J, Lophatananon A, Lubinski J, Luccarini C, Lux MP, Ma ESK, MacInnis RJ, Maishman T, Makalic E, Malone KE, Kostovska IM, Mannermaa A, Manoukian S, Manson JE, Margolin S, Mariapun S, Martinez ME, Matsuo K, Mavroudis D, McKay J, McLean C, Meijers-Heijboer H, Meindl A, Menendez P, Menon U, Meyer J, Miao H, Miller N, Taib NAM, Muir K, Mulligan AM, Mulot C, Neuhausen SL, Nevanlinna H, Neven P, Nielsen SF, Noh DY, Nordestgaard BG, Norman A, Olopade OI, Olson JE, Olsson H, Olswold C, Orr N, Pankratz VS, Park SK, Park-Simon TW, Lloyd R, Perez JIA, Peterlongo P, Peto J, Phillips KA, Pinchev M, Plaseska-Karanfilska D, Prentice R, Presneau N, Prokofyeva D, Pugh E, Pylkas K, Rack B, Radice P, Rahman N, Rennert G, Rennert HS, Rhenius V, Romero A, Romm J, Ruddy KJ, Rudiger T, Rudolph A, Ruebner M, Rutgers EJT, Saloustros E, Sandler DP, Sangrajrang S, Sawyer EJ, Schmidt DF, Schmutzler RK, Schneeweiss A, Schoemaker MJ, Schumacher F, Schurmann P, Scott RJ, Scott C, Seal S, Seynaeve C, Shah M, Sharma P, Shen CY, Sheng G, Sherman ME, Shrubsole MJ, Shu XO, Smeets A, Sohn C, Southey MC, Spinelli JJ, Stegmaier C, Stewart-Brown S, Stone J, Stram DO, Surowy H, Swerdlow A, Tamimi R, Taylor JA, Tengstrom M, Teo SH, Beth Terry M, Tessier DC, Thanasitthichai S, Thone K, Tollenaar R, Tomlinson I, Tong L, Torres D, Truong T, Tseng CC, Tsugane S, Ulmer HU, Ursin G, Untch M, Vachon C, van Asperen CJ, Van Den Berg D, van den Ouweland AMW, van der Kolk L, van der Luijt RB, Vincent D, Vollenweider J, Waisfisz Q, Wang-Gohrke S, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett W, Winqvist R, Wolk A, Wu AH, Xia L, Yamaji T, Yang XR, Har Yip C, Yoo KY, Yu JC, Zheng W, Zheng Y, Zhu B, Ziogas A, Ziv E, Investigators A, ConFab AI, Lakhani SR, Antoniou AC, Droit A, Andrulis IL, Amos CI, Couch FJ, Pharoah PDP, Chang-Claude J, Hall P, Hunter DJ, Milne RL, Garcia-Closas M, Schmidt MK, Chanock SJ, Dunning AM, Edwards SL, Bader GD, Chenevix-Trench G, Simard J, Kraft P, Easton DF (2017) Association analysis identifies 65 new breast cancer risk loci. Nature 551(7678):92–94. 10.1038/nature24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanche P, Dartigues JF, Jacqmin-Gadda H (2013) Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 32(30):5381–5397. 10.1002/sim.5958 [DOI] [PubMed] [Google Scholar]
- 16.Blanche P, Kattan MW, Gerds TA (2018) The c-index is not proper for the evaluation of $t$-year predicted risks. Biostatistics. 10.1093/biostatistics/kxy006 [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer RM, Park Y, Kreimer AR, Lacey JV Jr, Pee D, Greenlee RT, Buys SS, Hollenbeck A, Rosner B, Gail MH, Hartge P (2013) Risk prediction for breast, endometrial, and ovarian cancer in white women aged 50 y or older: derivation and validation from population-based cohort studies. PLoS Med 10(7):e1001492. 10.1371/journal.pmed.1001492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Calster B, Nieboer D, Vergouwe Y, De Cock B, Pencina MJ, Steyerberg EW (2016) A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol 74:167–176. 10.1016/j.jclinepi.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 19.Collins GS, Ogundimu EO, Altman DG (2016) Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med 35(2):214–226. 10.1002/sim.6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.RDC Team (2017) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- 21.Akdeniz D, Schmidt MK, Seynaeve CM, McCool D, Giardiello D, van den Broek AJ, Hauptmann M, Steyerberg EW, Hooning MJ (2018) Risk factors for metachronous contralateral breast cancer: a systematic review and meta-analysis. Breast 44:1–14. 10.1016/j.breast.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 22.Armstrong N, Ryder S, Forbes C, Ross J, Quek RG (2019) A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol 11:543–561. 10.2147/CLEP.S206949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bueno-de-Mesquita JM, Nuyten DS, Wesseling J, van Tinteren H, Linn SC, van de Vijver MJ (2010) The impact of inter-observer variation in pathological assessment of node-negative breast cancer on clinical risk assessment and patient selection for adjuvant systemic treatment. Ann Oncol 21(1):40–47. 10.1093/annonc/mdp273 [DOI] [PubMed] [Google Scholar]
- 24.Whittle R, Peat G, Belcher J, Collins GS, Riley RD (2018) Measurement error and timing of predictor values for multivariable risk prediction models are poorly reported. J Clin Epidemiol 102:38–49. 10.1016/j.jclinepi.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 25.Luijken K, Groenwold RHH, Van Calster B, Steyerberg EW, van Smeden M (2019) Impact of predictor measurement heterogeneity across settings on the performance of prediction models: a measurement error perspective. Stat Med 38(18):3444–3459. 10.1002/sim.8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflieger LT, Mason CC, Facelli JC (2017) Uncertainty quantification in breast cancer risk prediction models using self-reported family health history. J Clin Transl Sci 1(1):53–59. 10.1017/cts.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groenwold RH, White IR, Donders AR, Carpenter JR, Altman DG, Moons KG (2012) Missing covariate data in clinical research: when and when not to use the missing-indicator method for analysis. CMAJ 184(11):1265–1269. 10.1503/cmaj.110977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen KJ, Donders AR, Harrell FE Jr, Vergouwe Y, Chen Q, Grobbee DE, Moons KG (2010) Missing covariate data in medical research: to impute is better than to ignore. J Clin Epidemiol 63(7):721–727. 10.1016/j.jclinepi.2009.12.008 [DOI] [PubMed] [Google Scholar]
- 29.Janssen KJ, Vergouwe Y, Donders AR, Harrell FE Jr, Chen Q, Grobbee DE, Moons KG (2009) Dealing with missing predictor values when applying clinical prediction models. Clin Chem 55(5):994–1001. 10.1373/clinchem.2008.115345 [DOI] [PubMed] [Google Scholar]
- 30.Royston P, Altman DG (2013) External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol 13:33. 10.1186/1471-2288-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Houwelingen HC (2000) Validation, calibration, revision and combination of prognostic survival models. Stat Med 19(24):3401–3415 [DOI] [PubMed] [Google Scholar]
- 32.Pajouheshnia R, van Smeden M, Peelen LM, Groenwold RHH (2019) How variation in predictor measurement affects the discriminative ability and transportability of a prediction model. J Clin Epidemiol 105:136–141. 10.1016/j.jclinepi.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 33.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B (2019) A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 110:12–22. 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 34.Ming C, Viassolo V, Probst-Hensch N, Chappuis PO, Dinov ID, Katapodi MC (2019) Machine learning techniques for personalized breast cancer risk prediction: comparison with the BCRAT and BOADICEA models. Breast Cancer Res 21(1):75. 10.1186/s13058-019-1158-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torkamani A, Wineinger NE, Topol EJ (2018) The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19(9):581–590. 10.1038/s41576-018-0018-x [DOI] [PubMed] [Google Scholar]
- 36.Mellemkjaer L, Dahl C, Olsen JH, Bertelsen L, Guldberg P, Christensen J, Borresen-Dale AL, Stovall M, Langholz B, Bernstein L, Lynch CF, Malone KE, Haile RW, Andersson M, Thomas DC, Concannon P, Capanu M, Boice JD Jr, Group WSC, Bernstein JL (2008) Risk for contralateral breast cancer among carriers of the CHEK2*1100delC mutation in the WECARE Study. Br J Cancer 98(4):728–733. 10.1038/sj.bjc.6604228 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.