Abstract
Within their natural habitat, plants are subjected to abiotic stresses that include heat stress. In the current study, the effect of 4 h, 24 h, and 48 h of heat stress on Tetraena propinqua ssp. migahidii seedling’s protein profile and proteomic analyses were investigated. Total soluble protein SDS-PAGE (Sodium dodecyl sulfate–polyacrylamide gel electrophoresis) profile showed 18-protein bands, the newly synthesized protein band (with molecular weights 86.5, 30.2 and 31.4 KD) at 24 h of heat stress and 48 of normal conditions. Proteomic analysis showed that 81 and 930 targets are involved in gene and protein expression respectively. At 4 h, 57 genes and 110 proteins in C4 reached 56 genes and 173 proteins in T4. At 24 h, 63 genes and 180 proteins in C24 decreased to 54 genes and 151 protein in T24. After 48 h, 56 genes and 136 proteins in C48 increased to 64 genes and 180 proteins in T48. The genes and proteins involved in transcription, translation, photosynthesis, transport, and other unknown metabolic processes, were differentially expressed under treatments of heat stress. These findings provide insights into the molecular mechanisms related to heat stress, in addition to its influence on the physiological traits of T. propinqua seedlings. Heat stress-mediated differential regulation genes indicate a role in the development and stress response of T. propinqua. The candidate dual-specificity genes and proteins identified in this study paves way for more molecular analysis of up-and-down-regulation.
Keywords: Heat stress, Tetraena propinqua, Protein SDS-PAGE profile, Proteomics
1. Introduction
The natural environment for plants involves a series of abiotic and biotic stresses. The plant's response to these stresses are combinations of changes in various processes (Cramer et al., 2011). Among the common abiotic stresses, high temperature is considered a major factor that substantially affects plant growth and developmental processes (Alafari and Abd-Elgawad, 2021, Rodríguez Graña and del Soengas Fernández, 2015).
Heat stress creates changes in physiological processes, such as increased lipid peroxidation, which decreases the thermal stability of the cell membrane (Narayanan et al., 2016) and excess generation of reactive oxygen species (ROS), which produce oxidative stress (Hasanuzzaman et al., 2012). The reactive oxygen species (ROS)-scavenging enzymes and heat shock proteins (HSPs) are major functional proteins that are induced by heat stress and are well-known target genes of heat stress responsive transcription factors (TFs) (Kotak et al., 2007, Qu et al., 2013).
Heat stress also induces the production of compatible solutes which are able to organize proteins and cellular structures, maintain cell turgor by osmotic adjustment, and alter the antioxidant system to return the cellular redox balance and homeostasis (Hasanuzzaman et al., 2013). At the gene expression levels, heat stress is associated with multiple processes involving upregulation and downregulation of genes controlling various proteins particularly heat shock proteins (HSPs), transcription factors (TFs), and other stress-related genes (Guo et al., 2016, Liu et al., 2012). These genes are in control of the expression of osmoprotectants, detoxifying enzymes, transporters, and regulatory proteins involved in essential processes like photosynthesis (Krasensky and Jonak, 2012), changes in protein domains, gene and protein structures (Cao et al., 2016). The transcriptional regulatory network and post-translational regulation of the transcription factors involved in the heat stress response (Ohama et al., 2017).
Proteomics is defined as the large-scale study of proteins. A proteome is a set of proteins generated in an organism or biological system. The proteome varies from cell to cell and alters over time. Additionally, protein activity is also modified by various factors in addition to the expression level of the relevant gene (Anderson and Anderson, 1998, Blackstock and Weir, 1999). Proteomics discovers heat stress-responsive proteins (HRPs) and explains pathways that are critical for heat tolerance (Wang et al., 2017). Various proteins linked to signal transduction, photosynthesis, antioxidant defense, transcriptional and post-transcriptional regulation, protein synthesis and turnover, carbohydrate, and energy metabolism play vital roles in preserve leaves against heat stress (Hasanuzzaman et al., 2013, Hu et al., 2015, Wahid et al., 2007). Proteomic responses to heat stress were investigated in Arabidopsis leaves (Higashi et al., 2015), maize (Li et al., 2017), spinach (Zhao et al., 2018), perennial ryegrass (Lolium perenne) (Wang et al., 2017), Rhazya stricta (Obaid et al., 2016), grape (Jiang et al., 2017). Proteomics can be done by optimizing the sample preparation as well as by the LC-MS (Liquid chromatography-mass spectrometry) method for specific proteins (Mitchell, 2010). Several programs for protein identification take the output of peptide sequences from mass spectrometry and get back information about matching identical proteins from famous databases such as UniProt (Consortium, 2007) to predict proteins in the sample.
Tetraena. propinqua (Decne.) is a species of Tetraena. It is a perennial and low shrub with green, opposite, 2-foliolate, terete, glabrous, fleshy leaves, white-pubescent, pentamerous flowers, and brown, glabrescent, oblong to obovate, schizocarpic fruits. It grows in the various habitats of sandy and gravel desert, sandy depressions, sandy shores, and low coastal dunes. It is widely distributed in Sinai of Egypt, Arabian Peninsula of Saudi Arabia and Yemen, Iraq, S Iran, Afghanistan, and Pakistan (Ghazanfar and Osborne, 2015). It belongs to the subfamily Zygophylloideae; the largest of the family Zygophyllaceae (Beier et al., 2003), which includes about 180 species of shrubs, subshrubs and herbs mainly distribute in arid and semi-arid areas mostly in tropics and subtropics saline regions. Alzahrani (Alzahrani, 2019) reported that Tetraena propinqua ssp. propinqua, and Tetraena propinqua ssp. migahidii are a new subspecies combination nova from Saudi Arabia in this subfamily.
In the previous study, the effect of heat stress on three populations of Tetraena propinqua ssp. Migahidii caused protein degradation and conjointly induced expression of new synthesized HSPs throughout heat acclimatization in total soluble protein and superoxide dismutase may be related to heat injury and the improved thermotolerance in early hours of germination (Alafari and Abd-Elgawad, 2021). In the present study, we examine the influence of heat stress on protein expression using proteomic methods in T. propinqua seedlings exposed to different durations of high-temperature stress, to identify proteins and genes closely related to heat stress, which enables T. propinqua to adapt to heat tolerance in Saudi Arabia.
2. Materials and methods
2.1. Samples preparation
Seeds of T. propinqua ssp. migahidii were collected from plants growing in Al Narjas (Riyadh 13362) of central Saudi Arabia. Seeds of three plants were used for three germination experiments. Fifty (50) seeds were germinated by placing in a Petri dish, moistened with 5 mL of distilled water, and incubated at 25 °C (C; control) and 40 °C (T; heat stress) for 4, 24, and 48 h (Alafari and Abd-Elgawad, 2021). The samples were collected for extraction.
2.2. Extraction of protein
Protein was extracted by phosphate buffer (Alafari and Abd-Elgawad, 2021). Supernatants were used for total soluble proteins SDS-PAGE profile and proteomics analysis.
2.3. SDS PAGE profile of total soluble proteins
Protein extract samples were mixed with loading buffer and 10 µl samples were loaded onto a gel in a Bio-Rad apparatus according to Laemmli (Alafari and Abd-Elgawad, 2021, Laemmli, 1957). The gel was removed from the apparatus gently and placed in a plastic tank, and covered with a fixing solution of 30% ethanol, 10% acetic acid overnight, followed by rinsing briefly in 5% acetic acid, then treated with a sensitizer working solution of 25 µl composed of 12.5 mL Sensitizer and 12.5 mL water and stained using “Pierce Silver Stain Kit” (Catalog number: 24612), using silver stain enhancer (0.125 mL of enhancer and 12.5 mL stain). Developer working solution (0.125 mL of enhancer and 12.5 mL developer). The 10% SDS gel was washed with ultrapure water for 5 min twice. Then, the fixing solution was added to the gel for 15 min at room temperature. This step was repeated twice followed by gel washing with the 10% ethanol for 5 min. The gel was washed again with ultrapure water for 5 min twice, incubated in Sensitizer working solution for exactly 1 min, and then washed twice with ultrapure water for 1 min. Silver stain enhancer was added for 5 min followed by gel wash with ultrapure water for 20 s. Developer working solution was added until protein bands appear in 2–3 min. When the desired band intensity was reached, the developer working solution was replaced with a stopping solution. The gel was washed briefly and then replaced by incubation in acetic acid for 10 min. The protein ladder used as a molecular weights marker (MW) ranged between 10 and 250 KD “Precision Plus Protein™ Unstained Protein Standards” (Catalog number: 1610396) according to manufacturer instructions. The stained gel was photographed while wet.
2.4. Protein sample preparation for proteomics
2.4.1. Protein extraction from gel and denaturation
Sixty μl of lysis buffer containing 8 M Urea, 500 mM Tris pH 8.5, and protease inhibitor (1:100) was added to the samples. Then samples were shaken vigorously and centrifuged at 10,000 rpm for 30 min at 4 °C, followed by BCA (Bicinchoninic acid) protein quantification using “Pierce Detergent Compatible Bradford Assay Kit” (Catalog number: 23246) according to manufacturer instructions.
2.4.2. Protein digestion
Each protein sample was reduced using 2 µl of 200 mM DTT (Dithiothreitol) with shaking using a vortex, and spin down and incubated for 45 min at room temperature, and then alkylated using 2 µl of 1 M IAA (Iodoacetamide) and incubated at room temperature for 45 in the dark followed by 102 μl of 100 mM Tris pH 8.5. Trypsin was added to digest the samples at 6 μl trypsin containing 1ug procaine enzyme and incubated overnight at 37 °C with shaking at 900 rpm then the sample was acidified to pH 2–3 by adding 6ul of 100% formic acid, followed by spin down for 30 min at room temperature.
2.4.3. The phosphorylated peptides
By using the stage tip “Pierce™ C18 Spin Tips” (Catalog number: 84850) the peptides were activated by adding 15 ul methanol on the tip, the initialization was washed by using 15 ul from “solution B” (0.2% FA (Formic acid) + 80% ACN (Acetonitrile)). The re-equilibration was washed twice using 15 ul from “solution A” (0.2% FA) and the samples were added to a new Eppendorf tube, and then washed twice with 15 µl “solution A”, respectively. Then, the phosphorylated peptides were eluted with elution buffer “solution B“ in a collection tube 3 times followed by speed-vac followed by re-constituting the samples in 22 µl of “solution B”. In this stage, the samples were centrifuged between each step at 3000 RPM. Inject the samples on Mass spectrometry.
2.4.4. The peptides quantification
Peptides were quantified by measuring the concentration by using the bicinchoninic acid assay (BCA assay), incubated at 95 °C for 5 min, followed by adding 1000 µl from prepared BCA followed by incubating at 60 °C for 30 min, and then cooled down at room temperature for 20 min. The samples were measured at A 562.
2.5. Chromatography
The LC (Liquid chromatography) system using a Nano-LC system consisted of Eksigent nano-LC 400 autosamplers attached with an Ekspert nano-LC425 pump. Chromatographic separation of samples was performed on a trapping cartridge CHROMXP C18CL 5 um (10x0.5 mm) pumped at a flow rate of 10 ul/min for 3 min using mobile phase A. Chrom-XP C18CL column (3 um, 120 A, 150 × 0.3 mm) at a flow rate of 5 ul/min. The injection volume of all samples was 5 µl. Trap and elute needle wash was done two cycles using 10% isopropanol in an injection mechanism with an analysis time of 55 min. Mobile phases were MilliQ containing 0.1% FA (A) and acetonitrile containing 0.1% FA (B). The following gradient profile of mobile phase was used: 0.0 min, 97% A, 3% B; 38 min, 70% A, 30% B; 43 min, 60% A, 40% B; 45 min, 20% A, 80% B; 48 min, 20% A, 80% B; 49 min, 97% A, 3% B; 57 min, 97% A, 3% B.
2.5.1. Mass spectrometry
The LC-QTOF system analysis was carried out using a Sciex TripleTOFTM 5600+ in a positive acquisition mode with TOF MS survey scan followed by product ion scan for the most abundant 40 ions (High-resolution mode). The source conditions were as follows: cycle time is 1.5 sec; TOF mass range, 400 – 1250 m/z; MS2 range (product ion), 170 – 1500 m/z; ion selection threshold, 150 cps; Total run time, 55 min and MS calibration, Sciex tuning solution (P/N 4457953).
2.6. The search parameters
The measured parameters were as follows: Trypsin digestion, identification algorithms X! Tandem. Max missed cleavages 2. Precursor m/z tolerance 20.0 ppm. Fragment m/z tolerance 10.0 ppm. Precursor charge 2–5 isotopes 0–1. modification: 1- fixed modification; carbamidomethylation of C (Mass; 57.02). 2- variable modification; acetylation of K (Mass; 42.01), acetylation of protein N-term (Mass; 42.01), deamidation of N (Mass; 0.98), deamidation of Q (Mass; 0.98), oxidation of M (Mass; 15.99).
2.7. Statistical analysis
The LC-QTOF system analysis was carried out using a Sciex TripleTOFTM 5600 + in a positive acquisition mode with a TOF MS survey scan. All MS parameters were controlled by Analyst TF 1.7.1(Sciex software) for data acquisition. Raw MS files from the TripleTOFTM 5600 + were converted into MGF files then analyzed by Peptide shaker (v1.16.43). Database used is Uniprot Zygophyllum organism (swiss-prot containing 930 proteins).
Each experiment was performed in three replicates and statistical analysis was made using Excel software. The distinct proteins and genes were scored as (1) for the presence or (0) for absence for all samples. The cluster analysis and distance tree were performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm. In addition, the principal component analysis (PCA) scatter diagram was constructed based on the Dice coefficients genetic similarity matrix by using PAST, ver. 3.22 software (Hammer et al., 2001). The PCA applied to assign the variables to genes and proteins based on their tolerance to heat stress. The multivariate analysis was performed by constructing a Heatmap matrix using the module of R software.
3. Results
3.1. SDS-PAGE profile of total soluble proteins
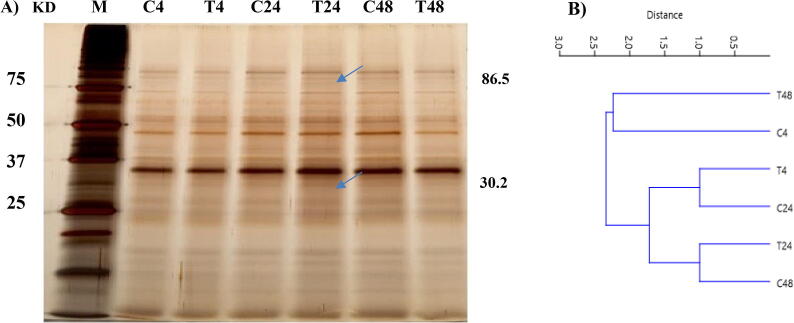
The protein pattern of T. propinqua seedlings in SDS-PAGE showed remarkable differences (Fig. 1). The relative expression of 18 protein bands (with molecular weights of about 102.3, 93.8, 71.4, 66, 55, 53.2, 47.1, 41.3, 40.2, 38.4, 35.1, 32, 26.8, 25, 10.9, 10.5, 10, 9.4 KD) appeared in the control and heat treatments. The relative intensity of these bands was often downregulated after 4 h and 48 h at high temperature, while upregulated after 24 h of high temperature. The expression of protein band (with molecular weights of about 86.5 and 30.2 KD) newly appeared at 24 h of high temperature. On the other hand, the protein band of 31.4 KD appeared at 48 h under normal conditions then disappeared under a high temperature of 48 h.
Fig. 1.
Changes in the protein profile of T. propinqua seedlings under heat stress among control (C, 25 °C) and treated (T, 40 °C) for 4, 24 and 48 h, respectively. (A) SDS-PAGE, (B) Cluster analysis of SDS-PAGE.
3.2. Heat stress-responsive proteome in T. Propinqua
Eighty-one (81) genes were identified in control and treated T. propinqua seedlings depended on the appearance and disappearance of genes they code for (Table 1, Fig. 4, Fig. 5). At 4 h of germination, Fifty-seven (57) genes were scored in control (C4) then decreased to 56 under heat stress (T4). After 24 h, the identified genes were increased in control (C24) to 63 genes, but it was decreased to 54 genes under heat stress (T24). After 48 h, the identified genes reached 56 genes in control (C48) then it was increased to 64 genes under heat stress (T48).
Table 1.
List of expressed genes by heat stress in T. propinqua seedlings depending on the number of identified proteins/genes under normal conditions and heat stress treatments using MALDI-Triple-TOF.
| No. | Gene ID | Gene Description | NO. of Proteins/Gene |
|||||
|---|---|---|---|---|---|---|---|---|
| C4 | T4 | C24 | T24 | C48 | T48 | |||
| rpoA | DNA-directed RNA polymerase subunit alpha | 2 | 3 | 3 | 2 | 3 | 2 | |
| rpoC1 | DNA-directed RNA polymerase subunit beta' | 6 | 6 | 3 | 2 | 7 | 4 | |
| rpoC2 | DNA-directed RNA polymerase subunit beta'' | 5 | 9 | 8 | 6 | 9 | 9 | |
| rpoB | DNA-directed RNA polymerase subunit beta | 3 | 5 | 5 | 3 | 5 | 6 | |
| rpb2 | DNA-directed RNA polymerase subunit B | 1 | 0 | 0 | 0 | 1 | 1 | |
| WRKY21 | WRKY transcription factor 21 | 1 | 1 | 1 | 1 | 1 | 1 | |
| matK | Maturase K | 6 | 6 | 7 | 8 | 10 | 12 | |
| matR | Maturase | 2 | 3 | 3 | 2 | 3 | 2 | |
| EMB2765 | Embryo defective 2765 | 1 | 1 | 1 | 1 | 0 | 1 | |
| PHYC | Phytochrome C | 1 | 2 | 1 | 0 | 0 | 1 | |
| rpl14 | 50S ribosomal protein L14, chloroplastic | 1 | 2 | 3 | 4 | 3 | 4 | |
| rpl23 | 50S ribosomal protein L23, chloroplastic | 2 | 3 | 2 | 3 | 1 | 2 | |
| rpl22 | 50S ribosomal protein L22, chloroplastic | 3 | 2 | 2 | 1 | 2 | 2 | |
| rpl20 | 50S ribosomal protein L20, chloroplastic | 1 | 0 | 0 | 0 | 0 | 0 | |
| rpl2 | 50S ribosomal protein L2, chloroplastic | 2 | 2 | 2 | 2 | 2 | 0 | |
| rpl16 | 50S ribosomal protein L16, chloroplastic | 0 | 1 | 1 | 0 | 0 | 2 | |
| rpl36 | 50S ribosomal protein L36, chloroplastic | 0 | 0 | 1 | 0 | 0 | 0 | |
| rpl32 | 50S ribosomal protein L32, chloroplastic | 1 | 1 | 2 | 1 | 1 | 1 | |
| rpl33 | 50S ribosomal protein L33, chloroplastic | 0 | 1 | 0 | 0 | 0 | 1 | |
| rps18 | 30S ribosomal protein S18, chloroplastic | 0 | 2 | 4 | 0 | 1 | 1 | |
| rps8 | 30S ribosomal protein S8, chloroplastic | 1 | 2 | 2 | 0 | 2 | 0 | |
| rps4 | 30S ribosomal protein S4, chloroplastic | 1 | 3 | 4 | 1 | 4 | 3 | |
| rps19 | 30S ribosomal protein S19, chloroplastic | 1 | 0 | 0 | 1 | 3 | 0 | |
| rps7 | 30S ribosomal protein S7, chloroplastic | 1 | 0 | 2 | 1 | 0 | 0 | |
| rps11 | 30S ribosomal protein S11, chloroplastic | 0 | 1 | 1 | 2 | 2 | 1 | |
| rps14 | 30S ribosomal protein S14, chloroplastic | 0 | 0 | 1 | 0 | 0 | 0 | |
| rps2 | 30S ribosomal protein S2, chloroplastic | 1 | 3 | 1 | 2 | 0 | 1 | |
| rps15 | 30S ribosomal protein S15, chloroplastic | 1 | 0 | 1 | 1 | 2 | 2 | |
| rps3 | Ribosomal protein S3 | 5 | 5 | 7 | 5 | 5 | 5 | |
| petA | Cytochrome f | 2 | 2 | 3 | 1 | 2 | 4 | |
| petD | Cytochrome b6-f complex subunit 4 | 1 | 0 | 0 | 1 | 1 | 0 | |
| psbE | Cytochrome b559 subunit alpha | 0 | 0 | 1 | 0 | 0 | 0 | |
| ycf3 | Photosystem I assembly protein Ycf3 | 2 | 2 | 2 | 1 | 3 | 1 | |
| ycf4 | Photosystem I assembly protein Ycf4 | 1 | 2 | 1 | 1 | 1 | 4 | |
| psbL | Photosystem II reaction center protein L | 1 | 0 | 0 | 0 | 1 | 1 | |
| psbI | Photosystem II reaction center protein I | 0 | 0 | 0 | 0 | 1 | 0 | |
| psbK | Photosystem II reaction center protein K | 0 | 2 | 2 | 0 | 0 | 2 | |
| psaA | Photosystem I P700 chlorophyll a apoprotein A1 | 1 | 1 | 1 | 6 | 2 | 1 | |
| psaB | Photosystem I P700 chlorophyll a apoprotein A2 | 1 | 1 | 0 | 0 | 1 | 0 | |
| psaC | Photosystem I iron-sulfur center | 0 | 0 | 0 | 0 | 0 | 1 | |
| psbB | Photosystem II CP47 reaction center protein | 2 | 2 | 2 | 2 | 3 | 2 | |
| psbH | Photosystem II reaction center protein H | 1 | 0 | 1 | 0 | 1 | 1 | |
| psbD | Photosystem II D2 protein | 0 | 0 | 0 | 0 | 1 | 0 | |
| RCA1 | Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplastic | 1 | 1 | 1 | 1 | 1 | 1 | |
| RCA2 | Ribulose bisphosphate carboxylase/oxygenase activase 2, chloroplastic | 1 | 1 | 0 | 1 | 1 | 1 | |
| rbcL | Ribulose bisphosphate carboxylase large chain | 6 | 36 | 33 | 29 | 4 | 40 | |
| atpE | ATP synthase epsilon chain, chloroplastic | 2 | 3 | 2 | 2 | 1 | 2 | |
| atp1 | ATP synthase subunit alpha | 1 | 1 | 2 | 3 | 1 | 2 | |
| atpB | ATP synthase subunit beta, chloroplastic | 1 | 3 | 3 | 2 | 2 | 2 | |
| atpA | ATP synthase subunit alpha, chloroplastic | 2 | 3 | 3 | 2 | 3 | 2 | |
| atpI | ATP synthase subunit a, chloroplastic | 1 | 1 | 2 | 2 | 1 | 0 | |
| atpF | ATP synthase subunit b, chloroplastic | 1 | 1 | 1 | 0 | 1 | 1 | |
| ndhE | NAD(P)H-quinone oxidoreductase subunit 4L, chloroplastic | 1 | 2 | 2 | 1 | 1 | 2 | |
| ndhD | NAD(P)H-quinone oxidoreductase chain 4, chloroplastic | 0 | 0 | 0 | 1 | 0 | 1 | |
| ndhB | NADH-quinone oxidoreductase subunit N | 0 | 1 | 1 | 0 | 1 | 1 | |
| ndhF | NAD(P)H-quinone oxidoreductase subunit 5, chloroplastic | 1 | 3 | 3 | 3 | 2 | 2 | |
| ndhA | NAD(P)H-quinone oxidoreductase subunit 1, chloroplastic | 1 | 3 | 3 | 4 | 2 | 3 | |
| ndhJ | NAD(P)H-quinone oxidoreductase subunit J, chloroplastic | 1 | 1 | 1 | 0 | 1 | 1 | |
| ndhH | NAD(P)H-quinone oxidoreductase subunit H, chloroplastic | 2 | 3 | 2 | 3 | 2 | 2 | |
| ndhI | NAD(P)H-quinone oxidoreductase subunit I, chloroplastic | 0 | 0 | 0 | 1 | 0 | 1 | |
| ndhK | NAD(P)H-quinone oxidoreductase subunit K, chloroplastic | 1 | 1 | 0 | 0 | 1 | 1 | |
| ycf1 | Protein TIC 214 | 3 | 3 | 4 | 2 | 3 | 3 | |
| ycf2 | Hypothetical chloroplast RF21 | 3 | 2 | 2 | 2 | 2 | 2 | |
| cemA | envelope membrane protein, chloroplastic | 0 | 2 | 2 | 3 | 0 | 1 | |
| AKT1 | Potassium channel | 1 | 1 | 1 | 1 | 0 | 1 | |
| NHX | Vacuolar Na+/H + antiporter | 1 | 1 | 1 | 1 | 1 | 1 | |
| DGAT1a | O-acyltransferase | 0 | 0 | 1 | 0 | 0 | 0 | |
| DGAT1b | O-acyltransferase | 0 | 1 | 0 | 0 | 0 | 0 | |
| CAAT1 | Cinnamyl alcohol acyltransferase 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| CAAT2 | Cinnamyl alcohol acyltransferase 2 | 1 | 0 | 1 | 1 | 0 | 1 | |
| accD | Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, chloroplastic | 1 | 2 | 2 | 1 | 3 | 2 | |
| ccsA | Cytochrome c biogenesis protein CcsA | 2 | 2 | 1 | 0 | 3 | 1 | |
| ccmB | Cytochrome c biogenesis | 0 | 0 | 1 | 1 | 0 | 1 | |
| VP1-1 | Vacuolar H + -pyrophosphatase | 0 | 0 | 1 | 0 | 0 | 1 | |
| VP1-2 | Vacuolar H + -pyrophosphatase | 0 | 0 | 1 | 0 | 0 | 0 | |
| clpP | ATP-dependent Clp protease proteolytic subunit | 1 | 1 | 1 | 1 | 0 | 1 | |
| Xdh | Xanthine dehydrogenase | 1 | 1 | 1 | 1 | 1 | 1 | |
| APS1 | Allylphenol synthase 1 | 0 | 0 | 0 | 1 | 0 | 1 | |
| APS2 | Allylphenol synthase 2 | 0 | 0 | 1 | 1 | 1 | 1 | |
| PPS1 | Propenylphenol synthase 1 | 1 | 1 | 0 | 1 | 1 | 1 | |
| psbA | PsbA | 0 | 0 | 2 | 2 | 1 | 0 | |
| Unknown gene | 11 | 16 | 17 | 16 | 11 | 14 | ||
| Total | 81 gene | 110 | 173 | 180 | 151 | 136 | 180 | |
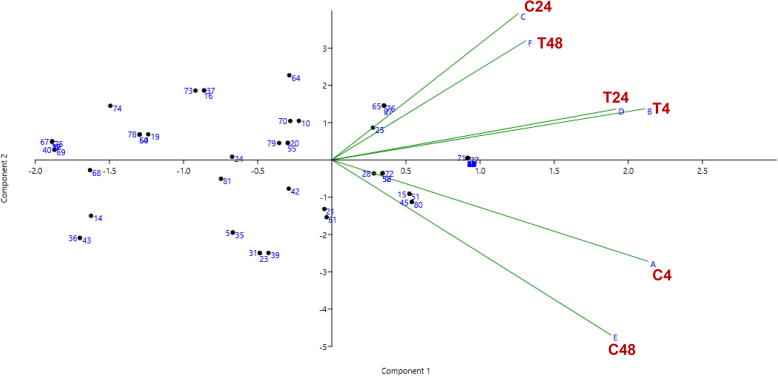
Fig. 4.
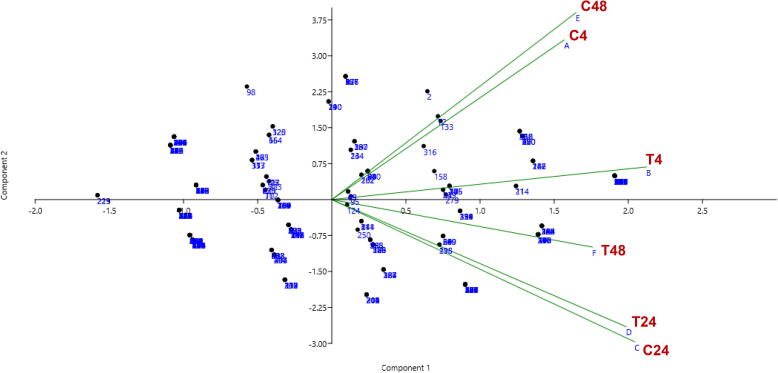
The PCA-Biplot of the 81 genes was differentially expressed in T. propinqua under heat stress. Control (C, 25 °C) and treated (T, 40 °C) for 4, 24 and 48 h, respectively.
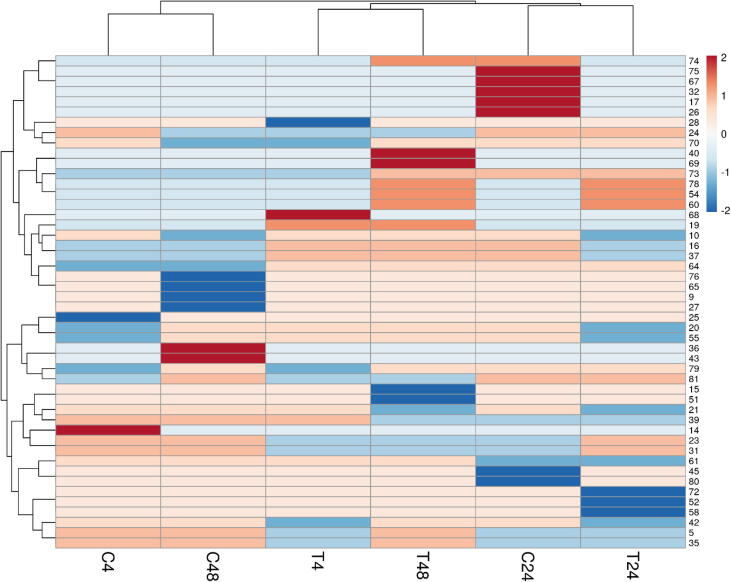
Fig. 5.
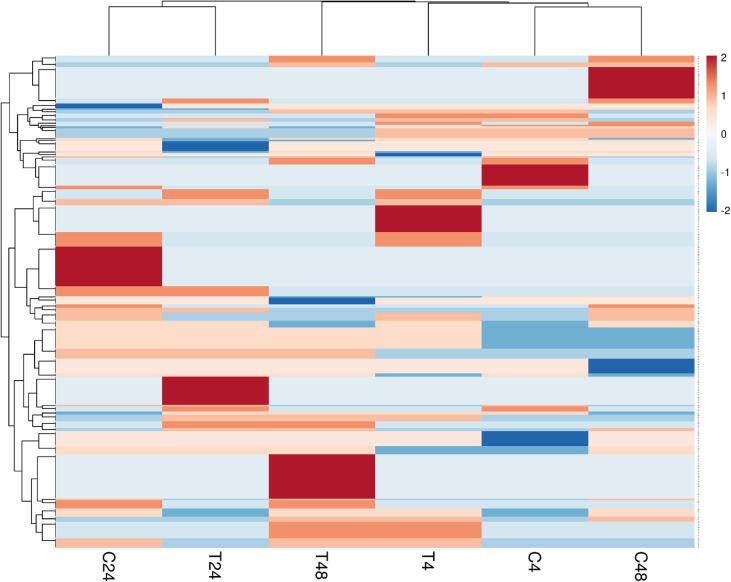
The Heatmap of the 81 genes was differentially expressed in T. propinqua seedlings under heat stress. Control (C, 25 °C) and treated (T, 40 °C) for 4, 24 and 48 h, respectively.
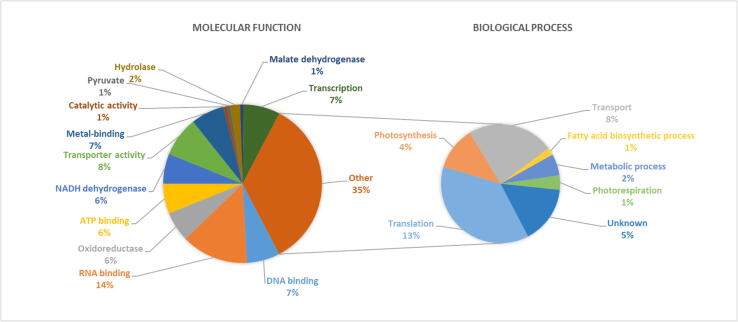
The genes of proteins were classified by gene ontology (GO) annotation and then classified into three functional groups: molecular function, biological process, and cellular component. The results of the GO analyses for the various treatments are shown in (Table 1, Fig. 2, Fig. 3). Ten genes are involved in transcription, encoding rpoA, rpoC1, rpoC2, rpoB, rpb2, WRKY21, matK, matR, MB2765, and PHYC, among them rpb2 and PHYC disappeared after 4 and 24 h of heat treatment, but EMB2765 and PHYC appeared after 48 h under 40 °C heat stress. 19 genes for rpl14, rpl23, rpl22, rpl20, rpl2, rpl16, rpl36, rpl32, rpl33, rps18, rps8, rps4, rps19, rps7, rps11, rps14, rps2, rps15 and rps3 are involved in translation (Fig. 2). Among them, some genes for rpl and rps disappeared such as rpl20, rpl2, rpl36, rps8, rps7, rps14, rps15, however, other genes were observed like rpl33, rpl16, rps11, rps2 under different exposure time of heat stress treatments. However, some genes were observed after 4 h and 48 h and disappeared after 24 h for example the rpl16 and rps18, but rps19 disappeared after 4 h and 48 h and appeared after 24 h under 40 °C of heat stress in T. propinqua seedlings (Table 1).
Fig. 2.
Histogram illustrating the functional classification of identified and unidentified genes from T. propinqua categorized by their major function based on information from Uni-Prot databases and gene ontology.
Fig. 3.
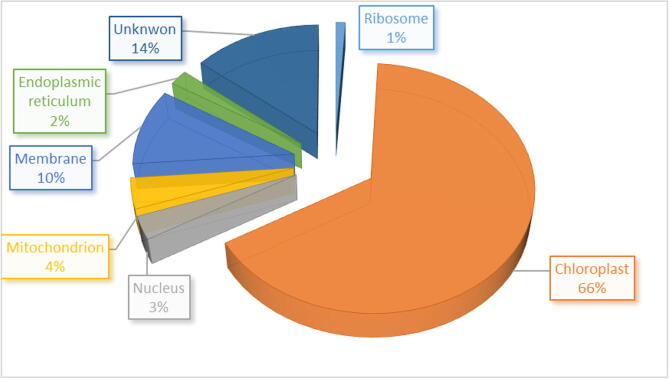
Diagram illustrating the percentage of the major functional genes in cellular components.
In T. propinqua seedlings, 15 genes are involved in the transport process (Table 1, Fig. 2), among them, ycf1 is involved in protein transport, but cemA is involved in hydrogen ion transport, AKT1 involved in the regulation of ion transmembrane transport, and NHX involved in the regulation of pH. DGAT1a and DGAT1b involved triglyceride biosynthetic process. CAAT1 and CAAT2 are involved in acyltransferase. accD involved in the fatty acid biosynthetic process and malonyl-CoA biosynthetic process. ccsA and ccmB are involved in cytochrome complex assembly. VP1-1, VP1-2, and clpP are involved in hydrolase. Xdh is involved in the oxidation–reduction process. After 4 h of heat stress, cemA, DGAT1b observed, but CAAT2 disappeared. At 24 h of heat stress, DGAT1a, ccsA, VP1-2, VP1-1 disappeared. At 48 h of heat stress, cemA, AKT1, CAAT1, ccmB, VP1-1, clpP, and CAAT2 appeared under 40 °C with compared control. APS1, APS2, PPS1, and psbA genes were unknown functions. Among them, APS1 appeared at 24 and 48 h, and PPS1 was observed at 24 h, however, psbA disappeared after 48 h under 40 °C of heat stress compared to control. The cellular components of proteins were in the chloroplast, nucleus, mitochondrion, membrane, endoplasmic reticulum, ribosome, and unknown components (Fig. 3).
On the other hand, 17 genes for petA, petD, psbE, ycf3, ycf4, psbL, psbI, psbK, psaA, psaB, psaC, psbB, psbH, psbD, RCA1, RCA2, and rbcL are involved in photosynthesis and photorespiration (Table 1; Fig. 2). The proteins encoded by some genes disappeared after 4 h under 40 °C of heat stress like psbE, psaC, psbH, but the expression of other genes appeared such as ycf4. After 24 h of heat stress, the expression of the psbE, ycf4, psaC, and psbD genes disappeared, but psbH and RCA2 were observed compared to control. At 48 h of heat stress, ycf3, psbK, psaB, and psbH genes disappeared, but the protein encoded by ycf4 and psaA appeared in T. propinqua seedlings. In T. propinqua seedlings, the ycf2, atpE, atp1, atpB, atpA, atpI, atpF, ndhE, ndhD, ndhB, ndhF, ndhA, ndhJ, ndhH, ndhI, and ndhK are involved in the metabolic process such as ATP synthesis, coupled proton transport, photosynthesis and light reaction (Fig. 2). Among them, the proteins encoded by atpF and ndhJ genes disappeared after 24 h of heat stress; however, the proteins encoded by ndhD and ndhI genes appeared after 24 h, 48 h under 40 °C of heat stress compared to the control. On the other hand, the protein encoded by the ndhB gene was observed at 4 h then disappeared after 24 h 40 °C of heat stress compared to control (Table 1, Fig. 2). A PCA-biplot of the 81 genes differentially expressed in T. propinqua seedlings under heat stress is shown in Fig. 4 and Heat Map (heatmap) in Fig. 5. The first PCA has clarified about 47.1% of all-out genes, while the second PCA was settled 15.6% of all-out genes.
In total, 930 protein species were identified at germinating stage. After 4 h of germination, 110 proteins were identified in C4 and 173 proteins were scored in T4. However, after 24 h, 180 proteins were identified in C24 decreased to 151 proteins in T24. After 48 h, 136 proteins were identified in C48 increased to 180 proteins in T48. All proteins of T. propinqua seedlings were differentially expressed in control and heat exposed seedlings based on abundance (NSAF; protein expression) (Table 1, Fig. 6, Fig. 7). The first PCA-biplot has clarified about 33.5% of all-out proteins, while the second PCA was settled 16% of all-out proteins was shown in Fig. 6 and Heat Map (heatmap) in Fig. 7.
Fig. 6.
PCA-Biplot for the 930 protein species were identified in T. propinqua seedlings under heat stress. Control (C, 25 °C) and treated (T, 40 °C) for 4, 24 and 48 h, respectively.
Fig. 7.
Heatmap for the 930 protein species was identified in T. propinqua seedlings under heat stress. Control (C, 25 °C) and treated (T, 40 °C) for 4, 24 and 48 h, respectively.
4. Discussion
Eighteen (18) protein expressed in SDS-PAGE of T. propinqua seedlings under normal conditions and heat stress. The relative intensity of these bands was often downregulated after 4 h and 48 h at high temperature, while upregulated after 24 h of high temperature. The proteins of molecular weights 86.5 and 30.2 KD were newly synthesized at 24 h of heat stress, and 31.4 KD at 48 h under normal conditions. The response to heat stress, at the molecular level, is associated with multiple processes involving heat shock proteins (HSPs) and other stress-related genes (Qin et al., 2008). The protein with molecular weight 86.5 KD in T. propinqua seedlings might be the Hsp90 family of molecular weights (82–90 KD) is for heat stress tolerance. These results agreed with various reports, as in Arabidopsis (Yamada et al., 2007), it was recorded that AtHsp90s are fundamental for tolerance to biotic and abiotic stresses, and in Pennisetum glaucum (Reddy et al., 2011) was reported PgHsp90 played a protecting role in counter stress-induced protein injury. The proteins with a molecular weight of 30.2 and 31.4 KD in T. propinqua seedlings might be the small heat shock proteins (sHsps) with molecular weights (15–42 KD). This study agreed with the report the sHsps may bind to partly folded or denatured proteins, that prevents irreversible unfolding or incorrect protein aggregation, or binds to unfolded proteins and permits additional refolding by Hsp70/Hsp100 complexes (Al-Whaibi, 2011).
The results of this study were recorded that 81 genes and 930 proteins were differentially expressed in T. propinqua seedlings. Some genes were downregulated under heat stress such as; rpb2, rpl20, rps19, rps7, rps15, petD, psbL, psbH, CAAT2 gene at 4 h, PHYC, rpl16, rpl36, rps18, rps8, rps14, psbE, psbK, psbH, atpF, ndhB, ndhJ, DGAT1a, ccsA, VP1-1, VP1-2 gene at 24 h and rpl2, rps8, rps19, petD, psbI, psaB, psbD, atpI, psbA gene at 48 h. Other genes were upregulated under heat stress as; rpl16, rpl33, rps18, rps11, psbK, ndhB, cemA, DGAT1b gene at 4 h, rps19, petD, RCA2, ndhD, ndhI, APS1, PPS1 gene at 24 h and EMB2765, PHYC, rpl16, rpl33, rps2, psbK, psaC, ndhD, ndhI, cemA, AKT1, CAAT1, CAAT2, ccmB, VP1-1, clpP, APS1 gene at 48 h. These results showed that the tolerance to heat stress during germination and seedling establishment might be supported by downregulation or upregulation of some genes with different biological processes, molecular functions, and cellular components. These findings agreed with the report of Benincasa hispida Cogn. var. recorded various genes induced after 4 days of heat stress (Wang et al., 2019). However, in Populus tomentosa Carr under heat stress upregulated and downregulated of various transcription factors, most downregulated genes were related to the light-harvesting complexes and photosynthetic electron transport system and the upregulated expression of some transcription factors at 12 h of heat stress (Ren et al., 2019).
The results of this study revealed that 10 transcriptional genes were differentially expressed in T. propinqua seedlings. Some genes were downregulated under heat stress for 4 h and 24 h such as rpb2 and PHYC, but others were upregulated under heat stress at 48 h like EMB2765 and PHYC. The regulation of transcriptional genes (rpb2, EMB2765, PHYC) might be to tolerate heat stress in T. propinqua seedlings by transcription regulation of RNA, heat shock protein (HSP), heat shock transcription factors (HSF), and chaperones associated with increasing heat tolerance, these results were confirmed in SDS-PAGE by induction of proteins with molecular weight 86.5, 31.4 and 30.2 KD. Transcriptional profile of some genes downregulated under drought stress such as proteins in roots of Arachis duranensis was identified as Cht2, MLP-34, heat shock proteins (HS70 and HS17.3), DOT-1, and MatK related to drought stress (Carmo et al., 2019). The changes in the transcription regulation of RNA processing-related proteins suggest that the variation and complexity of these proteins control the plants’ ability to overcome drought stress (Wang et al., 2016).
The proteome analysis indicated that Ribosomal proteins (RP) translational genes were differentially expressed in T. propinqua under different exposure times of heat stress including 6 rpl and 10 rps genes. The downregulated genes were identified as rpl2, rpl20, rpl36, rps7, rps8, rps14, rps15, and the upregulated genes were coded as rpl33, rps2, rps11. On the other hand, some genes were upregulated at 4 h and 48 h and downregulated at 24 h for example rpl16, rps18, but rps19 downregulated at 4, 48 h and upregulated at 24 h under heat stress. The regulation of translation genes of (rpl2, rpl16, rpl20, rpl33, rpl36, rps2, rps7, rps8, rps11, rps14, rps15, rps18, rps19) might be to acclimate the heat stress in T. propinqua seedlings by the transcription regulation and ribosome synthesis, heat shock protein (HSP), heat shock transcription factors (HSF), and chaperones associated with increasing heat tolerance. The RP gene family has global roles in forming and stabilizing the ribosomal complex and protein synthesis and has been reported to regulate transcription and synthesis of the ribosome and are differentially regulated by abiotic and biotic stresses that affect the growth of plants (Fromont-Racine et al., 2003). The current findings agree with some reports that Arabidopsis has 247 RP genes including 98 RPS and 143 RPL genes (Wang et al., 2013), whereas rice has 57 RPS and 123 RPL genes copies, the rpl gene upregulated under abiotic and biotic stress (Moin et al., 2016).
Some genes were downregulated in T. propinqua at 4 h like psbL, psbH but others upregulated such as psbK under heat stress. Genes of psbE, psbK, psbH were downregulated, but RCA2 upregulated at 24 h of heat stress. Other genes were downregulated at 48 h of heat stress as psbI, psaB and psbD, but psbK and psaC were upregulated. Gene of rbcl was recorded the highest number of proteins in T. propinqua seedlings. In T. propinqua seedlings, the protein expression of rbcl gene was increased from 6 to 36 proteins at 4 h of heat stress compared to control. However, 33 proteins were decreased to 29 proteins at 24 h under normal conditions and heat stress, respectively. After 48 h, the protein expression was increased from 4 to 40 proteins under heat stress compared with control. The regulation of photosynthetic genes (psbL, psbH, psbK, psbE, psbK, psbH, RCA2, psbI, psaB, psbD, psbK, psaC) and (1 4 8) proteins of rbcl gene might be to tolerate heat stress in T. propinqua seedlings by electron transport, Calvin cycle, photorespiration, photosynthesis, carbon fixation, reductive pentose-phosphate cycle, the oxidation–reduction process. This is in agreement with the basic aim of regulated proteins and metabolites linked with photosynthetic pathways are chloroplast, thylakoid membrane, and nucleus (Nouri et al., 2015). The differentially expressed 25 photosynthesis-related proteins in two soybean varieties affecting RuBisCO regulation, electron transport, Calvin cycle, and carbon fixation during drought and heat stress (Das et al., 2016). In the drought-tolerant cultivar of wheat, two proteins were down-regulated and three proteins of Rubisco-related proteins were not expressed (Cheng et al., 2015). In both wheat and barley, the expression of Rubisco large subunit under water deficit condition decreased (Chmielewska et al., 2016, Michaletti et al., 2018). In the thylakoid membrane of rice, Rubisco plays a role in the defense and regulation of photosynthesis under moderate heat stress (Shayan et al., 2020).
In T. propinqua seedlings, four differentially expressed genes are involved in the metabolic process. Genes of atpF, ndhB, and ndhJ were downregulated at 24 h of heat stress; but ndhB was upregulated at 4 h, however, ndhD and ndhI were upregulated at 24, 48 h of heat stress compared with the control. The regulation ADP and NAD genes (atpF, ndhB, ndhD, ndhJ) could be to acclimate heat stress in T. propinqua seedlings by increasing ADP and NADP + rates under heat stress to protect photosynthetic activity by decreasing ROS production. The results agreed with (Chang et al., 2015) that recorded the highest rates of ADP and NADP + produced in Arabidopsis thaliana under stress conditions, to protect photosynthetic activity by decreasing ROS production. NDH activity in chloroplasts increased in Nicotiana tabacum after exposure to heat stress at 50 °C in the light (Ju and Yu, 2001).
In our study, 15 genes were differentially expressed in T. propinqua seedlings involved in transport processes. Some genes are upregulated such as cemA at 4 h and 48 h of stress, AKT1, CAAT1, ccmB, and clpP were upregulated at 48 h of heat stress. However, other genes were downregulated like DGAT1a, ccsA, and VP1-2 at 24 h of heat stress. The DGAT1b was upregulated at 4 h of stress, but CAAT2 was downregulated at 4 h of stress then upregulated at 48 h of stress. On the other hand, VP1-1 was downregulated at 24 h of heat stress then upregulated at 48 h of heat stress with compared control. The regulation of transported genes (AKT1, ccsA, cemA, ccmB, clpP, CAAT1, CAAT2, DGAT1a, DGAT1b, VP1-1, VP1-2) were recorded in down or upregulation for tolerance to heat stress in T. propinqua seedlings by the regulation of the antioxidant system, stress defense through proton transporter, hydrolases, and acetyltransferase. The findings agreed with that reported, The AKT1 Arabidopsis is pivotal under low-potassium conditions for seedling establishment and post-germination growth (Pyo et al., 2010). CcmF messenger reacted through three-day-long cold exposure in maize embryos (Naydenov et al., 2010). The upregulation of DGAT1 was under freezing stress in Boechera stricta lines might provide a common mechanism to allow freezing stress tolerance (Arisz et al., 2018). The structural/functional homolog of ABI3 in maize indicates that VP1 and ABI3 play roles in the stabilization and activation of regulatory complexes involved in the transcription of target genes (Hill et al., 1996).
In this study, 4 genes were identified in T. propinqua seedlings with unknown function. Among them, the APS1 was upregulated at 24 h and 48 h, and PPS1 was upregulated at 24 h, however, psbA was downregulated at 48 h of heat stress compared with the control. These genes of (APS1, PPS1, psbA) might be related to heat stress in T. propinqua seedlings by miRNA induction, antioxidant system regulating, and stress defense gene expression. In Arabidopsis, miRNAs target superoxide dismutases, laccases, and ATP sulfurylases (APS) genes (Jones-Rhoades and Bartel, 2004), one particular miRNA (miR395) increased under sulfate deprivation, indicating that miRNAs may be induced by environmental conditions and not only by developmental processes (Jones-Rhoades and Bartel, 2004). In tobacco, the Maize psbA gene was upregulated under drought stress tolerance out of antioxidant system regulating, photosynthetic capability, and stress defense gene expression (Huo et al., 2016).
In this study, one actin protein of an unknown gene was upregulated after 4 h of heat stress in T. propinqua seedlings to adapt to the heat stress by the regulation of developmental, abiotic, and biotic stress responses. This opinion agrees with different reports that actin is considered a fragment of 70-kDa heat shock to adapt to heat stress (Flaherty et al., 1991). Experiments with heat-sensing mutant Arabidopsis thaliana lines resulted in the identification of an actin-related protein 6 (ARP6) gene which plays a role in heat shock response (Sajid et al., 2018). In tobacco BY-2 cultured cells, heat shock stimulates accumulation of the HSP70 binding protein were reduced at (35 °C, 45 °C), but enhanced at 50 °C in the responses to heat stress (Malerba et al., 2010).
5. Conclusions
Short exposure to heat stress alters the proteome profile in T. propinqua seedlings. In SDS-PAGE, 18 protein bands differentially expressed under heat stress, the bands with 86.5, 31.4 and 30.2 KD were differntially expressed to adapt heat stress. Differential expression of 81 heat stress-responsive genes and 930 proteins suggests that various signaling transduction pathways and molecular processes are affected due to heat stress. The highest expression of genes and proteins were 64 and 180 under heat stress at 48 h to adapt seedlings to a long time of heat stress. The Differential expression of rpb2, EMB2765, PHYC transcriptional genes, and rpl2, rpl16, rpl20, rpl33, rpl36, rps2, rps7, rps8, rps11, rps14, rps15, rps18, rps19 translational genes were associated with increased heat tolerance. The photosynthetic genes of psbL, psbH, psbK, psbE, psbK, psbH, RCA2, psbI, psaB, psbD, psbK, psaC, and (1 4 8) proteins of rbcl gene are involved in the oxidation–reduction process, and atpF, ndhB, ndhD, ndhJ metabolic genes were differentially expressed by increasing ADP and NADP + rates under heat stress to protect photosynthetic activity by decreasing ROS production. In addition, AKT1, ccsA, cemA, ccmB, clpP, CAAT1, CAAT2, DGAT1a, DGAT1b, VP1-1, VP1-2 transported genes were differentially expressed by stress defense through proton transporter, hydrolases, and APS1, PPS1, psbA genes were identified with unknown function, such as the regulation of the antioxidant system, and stress defense gene expression. Actin proteins may be involved in many plant processes and majorly found in the regulation of developmental, abiotic, and biotic stress responses. These results are helpful to understand the plant’s adaptation to heat stress.
Funding
This research was funded by Deanship of Scientific Research at Princess Nourah bint Abdulrahman University. (Grant No 39-S-268).
Declaration of Competing Interest
The author declare that there is no conflict of interest.
Acknowledgments
This research was funded by Deanship of Scientific Research at Princess Nourah bint Abdulrahman University. (Grant No 39-S-268).
Footnotes
Peer review under responsibility of King Saud University.
References
- Alafari H., Abd-Elgawad M. Heat-Induced Protein and Superoxide Dismutase Changes in Wild Tetraena propinqua ssp. Migahidii Seedlings. Pak. J. Biol. Sci. 2021;24:310–318. doi: 10.3923/pjbs.2021.310.318. [DOI] [PubMed] [Google Scholar]
- Al-Whaibi M.H. Plant heat-shock proteins: a mini review. Journal of King Saud University-Science. 2011;23:139–150. [Google Scholar]
- Alzahrani D.A. Systematic studies on the Zygophyllaceae of Saudi Arabia: Two new subspecies combination in Tetraena Maxim. Saudi journal of biological sciences. 2019;26:57–65. doi: 10.1016/j.sjbs.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N.L., Anderson N.G. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- Arisz S.A., Heo J.-Y., Koevoets I.T., Zhao T., van Egmond P., Meyer A.J., Zeng W., Niu X., Wang B., Mitchell-Olds T., Schranz M.E., Testerink C. DIACYLGLYCEROL ACYLTRANSFERASE1 Contributes to Freezing Tolerance. Plant Physiol. 2018;177:1410–1424. doi: 10.1104/pp.18.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier B.-A., Chase M.W., Thulin M. Phylogenetic relationships and taxonomy of subfamily Zygophylloideae (Zygophyllaceae) based on molecular and morphological data. Plant Syst. Evol. 2003;240:11–39. [Google Scholar]
- Blackstock W.P., Weir M.P. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–127. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- Cao J., Jiang M., Li P., Chu Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genomics. 2016;17:175. doi: 10.1186/s12864-016-2526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo L.S.T., Martins A.C.Q., Martins C.C.C., Passos M.A.S., Silva L.P., Araujo A.C.G., Brasileiro A.C.M., Miller R.N.G., Guimarães P.M., Mehta A. Comparative proteomics and gene expression analysis in Arachis duranensis reveal stress response proteins associated to drought tolerance. J. Proteomics. 2019;192:299–310. doi: 10.1016/j.jprot.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Chang L., Guo A., Jin X., Yang Q., Wang D., Sun Y., Huang Q., Wang L., Peng C., Wang X. The beta subunit of glyceraldehyde 3-phosphate dehydrogenase is an important factor for maintaining photosynthesis and plant development under salt stress—Based on an integrative analysis of the structural, physiological and proteomic changes in chloroplasts in Thellungiella halophila. Plant Sci. 2015;236:223–238. doi: 10.1016/j.plantsci.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Dong K., Ge P., Bian Y., Dong L., Deng X., Li X., Yan Y. Identification of Leaf Proteins Differentially Accumulated between Wheat Cultivars Distinct in Their Levels of Drought Tolerance. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0125302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewska K., Rodziewicz P., Swarcewicz B., Sawikowska A., Krajewski P., Marczak Lukasz, Ciesiolka D., Kuczyńska A., Mikolajczak K., Ogrodowicz P. Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Frontiers in plant science. 2016;7:1108. doi: 10.3389/fpls.2016.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium U. The universal protein resource (UniProt) Nucleic Acids Res. 2007;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer G.R., Urano K., Delrot S., Pezzotti M., Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Eldakak M., Paudel B., Kim D.-W., Hemmati H., Basu C., Rohila J.S. Leaf Proteome Analysis Reveals Prospective Drought and Heat Stress Response Mechanisms in Soybean. Biomed Res. Int. 2016;2016 doi: 10.1155/2016/6021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K.M., McKay D.B., Kabsch W., Holmes K.C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. PNAS. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Ghazanfar S.A., Osborne J. Typification of Zygophyllum propinquum Decne. and Z. coccineum L. (Zygophyllaceae) and a key to Tetraena in SW Asia. Kew Bull. 2015;70:38. [Google Scholar]
- Guo M., Liu J.-H., Ma X., Luo D.-X., Gong Z.-H., Lu M.-H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D.A., Ryan P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electronica. 2001;4:9. [Google Scholar]
- Hasanuzzaman M., Hossain M.A., da Silva J.A.T., Fujita M. Plant Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In: Venkateswarlu B., Shanker A.K., Shanker C., Maheswari M., editors. Crop Stress and Its Management: Perspectives and Strategies. Springer; Netherlands, Dordrecht: 2012. pp. 261–315. [DOI] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M.M., Roychowdhury R., Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y., Okazaki Y., Myouga F., Shinozaki K., Saito K. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 2015;5:10533. doi: 10.1038/srep10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Nantel A., Rock C.D., Quatrano R.S. A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 1996;271:3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- Hu X., Wu L., Zhao F., Zhang D., Li N., Zhu G., Li C., Wang W. Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y., Wang M., Wei Y., Xia Z. Overexpression of the maize psbA gene enhances drought tolerance through regulating antioxidant system, photosynthetic capability, and stress defense gene expression in tobacco. Front. Plant Sci. 2016;6:1223. doi: 10.3389/fpls.2015.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Liu X., Liu C., Liu G., Li S., Wang L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017;173:1502–1518. doi: 10.1104/pp.16.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Ju Y.Z., Yu Y.J. The role of chloroplast Ndh complex in resisting heat stress in tobacco strain. Science. Access. 2001;3 [Google Scholar]
- Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K.-D. Complexity of the heat stress response in plants. Current Opinion in Plant Biology, Physiology and Metabolism. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K., 1957. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. nature 227, 680–685 (1970). CAS PubMed Article. [DOI] [PubMed]
- Li P., Cao W., Fang H., Xu S., Yin S., Zhang Y., Lin D., Wang J., Chen Y., Xu C., Yang Z. Transcriptomic Profiling of the Maize (Zea mays L.) Leaf Response to Abiotic Stresses at the Seedling Stage. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.-T., Wang J.-F., Cramer G., Dai Z.-W., Duan W., Xu H.-G., Wu B.-H., Fan P.-G., Wang L.-J., Li S.-H. Transcriptomic analysis of grape (Vitis viniferaL.) leaves during and after recovery from heat stress. BMC Plant Biol. 2012;12:174. doi: 10.1186/1471-2229-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba M., Crosti P., Cerana R. Effect of heat stress on actin cytoskeleton and endoplasmic reticulum of tobacco BY-2 cultured cells and its inhibition by Co2+ Protoplasma. 2010;239:23–30. doi: 10.1007/s00709-009-0078-z. [DOI] [PubMed] [Google Scholar]
- Michaletti A., Naghavi M.R., Toorchi M., Zolla L., Rinalducci S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018;8:5710. doi: 10.1038/s41598-018-24012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Proteomics retrenches. Nat. Biotechnol. 2010;28:665–670. doi: 10.1038/nbt0710-665. [DOI] [PubMed] [Google Scholar]
- Moin M., Bakshi A., Saha A., Dutta M., Madhav S.M., Kirti P.B. Rice Ribosomal Protein Large Subunit Genes and Their Spatio-temporal and Stress Regulation. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S., Tamura P.J., Roth M.R., Prasad P.V.V., Welti R. Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant, Cell Environ. 2016;39:787–803. doi: 10.1111/pce.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov N.G., Khanam S., Siniauskaya M., Nakamura C. Profiling of mitochondrial transcriptome in germinating wheat embryos and seedlings subjected to cold, salinity and osmotic stresses. Genes & Genetic Systems. 2010;85:31–42. doi: 10.1266/ggs.85.31. [DOI] [PubMed] [Google Scholar]
- Nouri M.-Z., Moumeni A., Komatsu S. Abiotic Stresses: Insight into Gene Regulation and Protein Expression in Photosynthetic Pathways of Plants. Int. J. Mol. Sci. 2015;16:20392–20416. doi: 10.3390/ijms160920392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaid A.Y., Sabir J.S.M., Atef A., Liu X., Edris S., El-Domyati F.M., Mutwakil M.Z., Gadalla N.O., Hajrah N.H., Al-Kordy M.A., Hall N., Bahieldin A., Jansen R.K. Analysis of transcriptional response to heat stress in Rhazya stricta. BMC Plant Biol. 2016;16:252. doi: 10.1186/s12870-016-0938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Pyo Y.J., Gierth M., Schroeder J.I., Cho M.H. High-Affinity K+ Transport in Arabidopsis: AtHAK5 and AKT1 Are Vital for Seedling Establishment and Postgermination Growth under Low-Potassium Conditions. Plant Physiol. 2010;153:863–875. doi: 10.1104/pp.110.154369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D., Wu H., Peng H., Yao Y., Ni Z., Li Z., Zhou C., Sun Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics. 2008;9:432. doi: 10.1186/1471-2164-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu A.-L., Ding Y.-F., Jiang Q., Zhu C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013;432:203–207. doi: 10.1016/j.bbrc.2013.01.104. [DOI] [PubMed] [Google Scholar]
- Reddy P.S., Thirulogachandar V., Vaishnavi C.S., Aakrati A., Sopory S.K., Reddy M.K. Molecular characterization and expression of a gene encoding cytosolic Hsp90 from Pennisetum glaucum and its role in abiotic stress adaptation. Gene. 2011;474:29–38. doi: 10.1016/j.gene.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Ren S., Ma K., Lu Z., Chen G., Cui J., Tong P., Wang L., Teng N., Jin B. Transcriptomic and Metabolomic Analysis of the Heat-Stress Response of Populus tomentosa Carr. Forests. 2019;10:383. doi: 10.3390/f10050383. [DOI] [Google Scholar]
- Rodríguez Graña, V.M., Soengas Fernández, M. del P., Alonso-Villaverde Iglesias, V., Sotelo Pérez, T., Cartea González, M.E., Velasco Pazos, P., 2015. Effect of temperature stress on the early vegetative development of Brassica oleracea L. http://dx.doi.org/10.13039/501100003329 [DOI] [PMC free article] [PubMed]
- Sajid M., Rashid B., Ali Q., Husnain T. Mechanisms of heat sensing and responses in plants. It is not all about Ca2+ ions. Biologia plant. 2018;62:409–420. doi: 10.1007/s10535-018-0795-2. [DOI] [Google Scholar]
- Shayan S., Norouzi M., Moghaddam Vahed M., Mohammadi S.A., Toorchi M. Leaf proteome pattern of two bread wheat varieties under water deficit stress conditions. Curr. Plant Biol. 2020;100146 doi: 10.1016/j.cpb.2020.100146. [DOI] [Google Scholar]
- Wahid A., Gelani S., Ashraf M., Foolad M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- Wang J., Lan P., Gao H., Zheng L., Li W., Schmidt W. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics. 2013;14:783. doi: 10.1186/1471-2164-14-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu Y., Tian J., Huang K., Shi T., Dai X., Zhang W. Transcriptional profiling and identification of heat-responsive genes in perennial ryegrass by RNA-sequencing. Front. Plant Sci. 2017;8:1032. doi: 10.3389/fpls.2017.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Jiang B., Liu W., Lin Y., Liang Z., He X., Peng Q. Transcriptome Analyses Provide Novel Insights into Heat Stress Responses in Chieh-Qua (Benincasa hispida Cogn. var. Chieh-Qua How) Int. J. Mol. Sci. 2019;20:883. doi: 10.3390/ijms20040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cai X., Xu C., Wang Q., Dai S. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016;17:1706. doi: 10.3390/ijms17101706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Fukao Y., Hayashi M., Fukazawa M., Suzuki I., Nishimura M. Cytosolic HSP90 Regulates the Heat Shock Response That Is Responsible for Heat Acclimation in Arabidopsis thaliana. J. Biol. Chem. 2007;282:37794–37804. doi: 10.1074/jbc.M707168200. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Chen W., Bian J., Xie H., Li Y., Xu C., Ma J., Guo S., Chen J., Cai X., Wang X., Wang Q., She Y., Chen S., Zhou Z., Dai S. Proteomics and Phosphoproteomics of Heat Stress-Responsive Mechanisms in Spinach. Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]