Highlights
-
•
Traditional and modern methods of diagnosing bacterial wilt disease of potato.
-
•
Evaluate the susceptibility of some Egyptian potato cultivars against to R. solanacearum.
-
•
Differentiate potato cultivars using molecular genetics methods such as SDS-PAGE profile by protein content.
Keywords: Potato, Ralstonia solanacearum, IFAS, Tag-Man (PCR), Protein, SDS-PAGE
Abstract
Bacterial wilt caused by Ralstonia solanacearum (Smith), is one of the chief severe diseases of potato in warm temperate regions, tropics and subtropics of the world. The study was conducted to isolate and identify bacterial pathogens and select the most resistant cultivars and avoid the decrease in the total value of Egyptian potato exports to the European Union (EU) due to the quarantine restrictions imposed by the EU on potato tubers exported from Egypt affected by bacterial wilt. The results of traditional identification through morphological and serological studies showed that the five isolates were isolated and identified as Ralstonia solanacearum. Furthermore, the results illustrated that RS5 isolate showed the lowest percentage of disease incidence reduction on the three tested potatoes cultivar Bellini, Spunta and Mondial recorded 9.64%, 15.41% and 34.12%, respectively. While, RS8 isolate exhibited the highest effective one the percentage of disease reduction on all tested potato cultivars. This isolate reduced disease incidence 60.60%, 63.21% and 71.66%, compering to the healthy control treatment. The result of molecular identification represent that the probe used in Taq-man (PCR) was of the type (B2) capable to detect only biovar 2 of R. solanacearum bacterial wilt. Furthermore, primer and probe are specific for detection of the race 3 biovar 2 strain. Positive results were obtained in all assays used including IFAS, protein content and SDS-PAGE with all five isolates. So the isolate (RS5) was the most virulence one, followed by RS1, RS3, RS2 and RS8, registered that the tested isolates were R. solanacearum race 3, biovar 2. Also, studies focused on the form of genetic distances and similarities based on pathogenic and plant growth parameters. The results illustrate that the highest genetic similarity (0.998) was found between Bellini and Spunta cultivars as the closest but the lowest value (0.946) was found between Mondial and Bellini as most distant. These results were similarity with genetic distances and SDS-PAGE profile of the three tested potato cultivars.
1. Introduction:
Potato (Solanum tuberosum L.) is take up a high class among essential vegetable crops in the world and consistently ranks fourth among the most important staple food crops after wheat, corn and rice. (Birch et al., 2012). Potato is a steady nutrition safety crop with great achievable for difficulty alleviation and combating malnutrition in the developing world (Devaux et al., 2020). Currently, the total value of Egyptian potato exports lurch due to quarantine constraint on the potato bacterial wilt by the EU (Makled and Elkodosy, 2018). The bacterial wilt disease was first recorded in Egypt from potato tubers that had symptoms of rot (Sabet, 1961).
Bacterial wilt of potato plants and brown rot on potato tubers induced by Ralstonia solanacearum Smith, this bacterium is one of the furthermost threatening diseases of potato production in Asia, Africa, and Central and South America (Charkowski et al., 2020). These bacteria are considered soil borne, so it's usually attack plants through the roots and colonizes xylem vessels (García et al., 2019). As the vascular bundles in the diseased plants are filled with bacteria impede that transport of water and nutrients, and thus the symptoms appear, which are yellowing of the leaves, redness of the vascular bundles, necrosis, and ultimately the total wilt of the infected plants, then followed by physiological modifications in the diseased plants, such as increasing the respiratory rate and reducing transpiration and photosynthesis (Karim and Hossain, 2018).
The bacterium that causes wilt in the potato plant is classified into five races according to the host range, and into five biovars according to the capability of races to oxidize three hexose sugar alcohols and three disaccharides (Hayward, 1964). The race (3) and biovar (2) of R. solanacearum is responsible for attacking the potato crop, as it was discovered in the Andean region of South America (Prior and Fegan, 2005). R3bv2 was introduced to northern Europe around 1972, which increased the incidence and severity of disease and caused significant damage (Chávez et al., 2012). Consequently, governments passed quarantine legislation requiring the destruction of the affected crops and the rest of the affected fields (Janse, 1996). Serological methods can deficiency particularity because of cross-reactions of polyclonal antibodies with other bacteria and may have defined susceptibility (Puaprasert et al., 2018). Immunofluorescent antibody stain and Tag-Man (PCR) were used by Janse et al. (2004). A fluorogenic (TaqMan) PCR analysis was developed to detect R. solanacearum strains. Two fluorogenic probes were utilized in a multiplex response; one broad-range probe (RS) revealed all biovars of R. solanacearum, and a second further specific probe (B2) was applied to only detect biovar 2A (Chen et al., 2010).
Bacterial wilt attacks more than two hundred plant species, including more than fifty plant families (Aslam et al., 2017, Uwamahoro et al., 2020). The pathogen of bacterial wilt is the most destructive bacterial pathogen due to its infestation, uncommonly wide host range, perseverance, and wide topographical distribution (Wei et al., 2018). The disease is common to occur in the wet equatorial areas, sub-equatorial and some temperate areas of the world which has been estimated to affect about 3.75 million acres in relatively 80 countries throughout the world with international damage estimates actually over $950 million per year (Charkowski et al., 2020).
Bacterial wilt is the most significant one of bacterial disease on potato, and announced many years ago in Egypt (Farag et al., 2017). As the disease incidence and severity varies between potato cultivars, due to the presence of genetic differences between ones. Studies have shown that Spunta and Bellini cultivars are among the most susceptible to the disease, unlike Mondial cultivar (Karim and Hossain, 2018). So the studies illustrate that the highest genetic similarity was found between Bellini and Spunta as the closest but the lowest value was found between Mondial and Bellini as most distant (ELHetawy, 2018). Genetic distance and Similarity coefficient were estimated between the three cultivars of potato by d determining the protein contents (Amelio and Tagarelli, 2018). So, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) is the most one techniques to differ the protein. This technique is most one used to evaluate the proteins in complex extracts, and resulting from the discontinuous SDS-PAGE system first mention by Laemmli (1970). This system relies on two types of gels - a solvent gel (aka running) in which the proteins are crucial based on their molecular weights (MWs) and a stacking gel in which proteins are centered prior to entering the resolving gel (Pavlova et al., 2018). Genetic multiplicity is the source for the existence of plants in nature and for improved yields. By studying the genetic differences between potato varieties, it helps the breeder to obtain information on diversity in plant genetic resources, thus increases the chance to improve and produce new varieties with desirable characteristics (Priya et al., 2018). Therefore, the research aimed to isolate the causal pathogen of bacterial wilt and differentiate between three varieties of potatoes that are widely cultivated in Egypt in terms of their susceptibility infection and resistance to benefit from these results in the breeding programs.
2. Materials and Methods:
2.1. Isolation of R. Solanacearum from different potato fields in Egypt:
Brown rot samples of potato tuber cultivars (Spunta, Bellini, Lady rosetta and Mondial) collected from different potato districts at different governorates i.e. Beni-Suef (Al Maymun village), which located between the latitude: 29.0667° N, longitude: 31.0833° E, Menoufia (Telia1, 2 and El-Basha village), which located between the latitude: 30.5972° N, longitude: 30.9876° E, Beheira (El-Bostan and Om Sabir village), which located between the latitude: 30.8481° N, longitude: 30.3436° E, and El- Sharqia (El-salhia city), which located between the latitude: 30.7327° N, longitude:31.7195° E, were washed in running spout water, surface disinfected by submersion in ethyl alcohol 95% and flamed. For each sample cores of 5–10 mm in diameter and 5 mm in length, containing main vascular and cortical tissues were macerated in 10 ml sterilized water. Then serial-diluted and streaked on triphenyl tetrazolium chloride medium (Kelman’TZC, 1954). Petri-dishes were incubated at 28 ± 2 °C for 24–48 h. This medium was proposed by Kelman (1954), and modified by Sujeet et al., 2017.
2.2. Morphological and physiological properties of isolated Ralstonia solanacearum isolates:
Previously isolated bacteria from potato brown rot tubers were identified based on the morphological, physiological and cultural studies, as described by Kelman (1954). Cultural and physiological properties, as well as morphological characterizations were studied through grown on three types of media SMSA, King's B and TZC petri-dishes at 28 ± 2 °C for 48 h (Mutimawurugo et al., 2019). The colony's colour, shape, size, surface, margin, elevation, sporulation, gram reaction and motility were esteemed. As stated by Timila and Manandhar (2016) the isolates of R. solanacearum were characterizes into biovars on the basis of oxidation of disaccharides and hexose alcohols including (Lactose, Maltose, Cellobiose, Mannitol, Sorbitol and Dulcitol.
The biochemical properties of the isolated bacterial isolates were submited to various biochemical tests including Gram's stain, oxidase test (Pradhanang et al., 2000), gelatin and starch hydrolysis (Fahy and Persley, 1983), and arginine dihydrolase activity (Zhou et al., 2012).
2.3. Molecular characteristics:
Serological characteristics such as Immunofluorescence antibody stains (IFAS) and Quantitative, Real-time, Fluorogenic PCR (Taq-Man) were carried out as stated by Puaprasert et al., 2018, García et al., 2019 .This procedure was conducted at Potato Brown Rot Project (PBRC), Agric. Res. Center, Giza- Egypt. Taq-Man probe and primers (thermo scientific cat No. 0721), Bangalore, India, were used as shown in Table 1. Primers and probe were prepared by using Primer Express version 1.0 software by adapting previous R. solanacearum PCR protocols. Taq-Man probe and primers for biovar 2A-specified assess were illustrate from the biovar 2A-specified DNA sequence. The system use an oligonucleotide identified probe, prepared to link to amplified target DNA between a pair of target-specified PCR primers. The probe is marked at each end with two dyes: a reporter and a quencher. While, the probe is flawless, any fluorescences released by the reporter dye was absorbed by the quencher. When the two dyes be spatially separated (e.g. if the probe was degraded), then fluorescence was released. This generic precept is named Fluorescence Resonance Energy Transfer (FRET). Throughout a Taq-Man assess, if target DNA is ready, the primers bind to it and DNA amplification occurs, as in PCR. This releases the reporter dye from the quencher, and rises in fluorescence is identified, detection the sample is positive for the target organism.
Table 1.
list of primers and TaqMan probe used to detect pathogenic Ralstonia solanacearum (biovar 2/race 3).
| Primers | |
|---|---|
| B2-1-FS‘ | TGG CGC ACT GCA CTC AAC − 3‘ |
| B2- 11- R5‘ | AAT CAC ATG CAA TTC GCC TAC G − 3‘ |
| Probe | |
| B2-P5‘ - [VIC] | TTC AAG CCG AAC ACC TGC TGC AAG - [TAMRA]-3 ‘. |
2.4. Susceptibility of Spunta, Bellini and Mondial three potato cultivars against bacterial wilt, caused by Ralstonia solanacearum.
Inoculum preparation: The inoculums of previously identified five pathogenic isolates of R. solanacearum (RS1, RS2, RS3, RS5, and RS8) were used. The inoculum was prepared by incubating the bacteria for a day in nutrient broth on a shaker running at 120 rpm. Then the bacteria concentration was adjusted to 108 cells ml -1 by spectrophotometer measures. All suspensions were directed to the cell count technique which gives a reliable count for only life cells (Andreas et al., 2009).
Potato tubers of Spunta, Bellini and Mondial cultivars were obtained from Potato Brown Rot Project (PBRC), Agric. Res. Center, Giza- Egypt, to the assess their susceptibility. The inoculum of these isolates was adjusted to 108 and inoculated four weeks old healthy seedlings of potato plant cultivars grown in 25 cm diameter plastic pots containing 10 kg pest-free sandy-clay soil (1/1, v/v). Soil used in the experiment was previously bacteriologically checked to ensure R. solanacearum free. Inoculation was done using the previously prepared bacterial suspension 108cfu/ml by the stem puncture technique described by Kumari and Ranjan (2019), Artificial infection of soil was done by soaking 250 ml of bacterial suspension per potting soil (25 ml / kg soil). Similarly, control treatments were carried out using sterile water instead of bacterial inoculum. The inoculated plants in pots were coated with polyethylene bags for three days, at 28 ± 2 °C under green-house conditions, then bags were detached and the pots were irrigated daily. Four pots as replicates were used for each source of tested isolate and arranged in a randomized complete block design. Inoculated potato seedlings were observed for disease assessment plant growth parameters such as number of tubers and weight of tubers / plant, as well as percent reduction of disease incidence and disease severity were calculated as follows:
Disease assessment: The percentage of disease incidence (DI) was recorded 40 days after inoculation with the pathogen.
The percentage of disease severity (DS) was calculated from disease rating for each plant periodically according to the scale proposed by Messiha et al. (2007) describing the wilt symptoms in the plant foliage as follows:
0 = no symptoms 1 = up to 25% 2 = 26–50%
3 = 51–75% 4 = 76–100% 5 = dead plants
DS was calculated by the following formula:
T = Total number of wilted plants with each category
R = Disease ratting scale R (R = 0, 1, 2, 3, 4 and 5)
N = Total number of tested plants
Percent reduction of disease incidence and severity were calculated by the following formula:
C = control T = treatment
2.5. Assessment of potato cultivars by evaluate protein contents using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE):
This procedure was conducted at the Biotechnology Lab at Cairo University Research Park, Faculty of Agriculture, (CURP). SDS-PAGE is an electrophoresis method that allows protein separation by weight according to (Laemmli, 1970). To evaluate the three cultivars of potato which used in this experiment, the test was carried out in several steps.
Newly grown leaves were taken from the control treatment in this experiment for the three potato cultivars and extracted by using 1.0 ml of 0.05 M Tris –Hcl buffer. Twenty microliters of β mercapto ethanol added to partitionate the proteins to small sub-units. The mixture was centrifuged at 12000 rpm for five minutes.
Fifteen microliters of each protein extracts of each potato cultivars genotype were resolved in 12% poly acrylamide –SDS resolving gel, to separate the protein sub-units according to their molecular weights.
Sample loading, electrophoresis and gel staining: For each one of slot gels, 15 μl of each leaves extract sample was loaded, electrophoresis and gel staining according to (Kim and Cho, 2019). The gels were photographed and the protein bands were scored. The electrophoretic products compared with protein bands of a ladder, from the Bio- Rad manual for the Gel Slab Dryers model 483.
2.6. Genetic distances and similarities based on pathogenic and plant growth parameters:
The genetic similarity coefficient among potato cultivars based on differences in pathogenic and plant growth parameters was calculated according to the Pearson coefficient using the IBM SPSS statistics program (George and Mallery, 2019). The genetic distance was estimated, and the phylogenetic dendrogram was generated by the clustering analysis method (Amelio and Tagarelli, 2018) using STATISTICA 8 program (Weiβ, 2007).
2.7. Genetic distances and similarities based on SDS-PAGE profile:
Patterns of the SDS-PAGE bands were recorded as present (1) or absent (0), and each was treated as independent. Genetic diversity was determined by comparing the banding patterns of all genotypes at the standard level of protein banding patterns. Genetic similarity among genotypes was calculated according to Dice coefficient measurement (Dice, 1945) using the same program as previously mentioned in Genetic distances and similarities based on pathogenic and plant growth parameters.
2.8. Statistical analysis:
The results of the previous experiments were statistically analyzed according to the procedures reported by Sneedecor and Cochran (1980) and Statistix 9 computer program was then used to analyze statistically the obtained data and mean separation as stated by Duncan's multiple range test (p < 0.5).
3. Results:
3.1. Isolation of R. Solanacearum from different potato fields in Egypt:
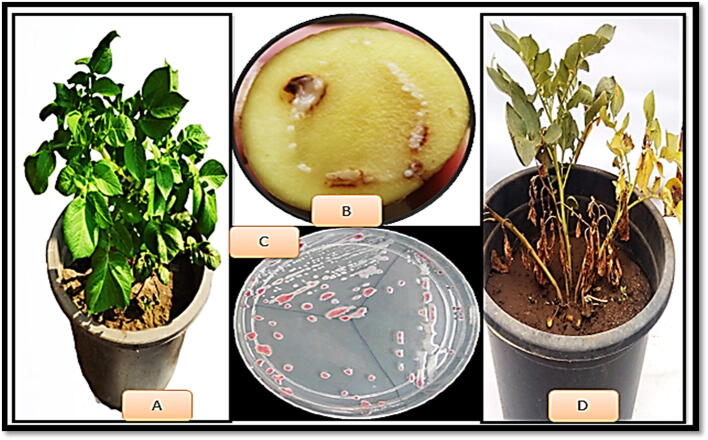
Eighty samples were obtained from different naturally diseased potato plants screening symptoms of bacterial wilt collected from Spunta, Bellini, Lady-rosetta and Mondial cultivars at different localities in four governorates (Beni-suef, Menoufia, Beheira and El- sharqia). Data in table 2 show the number of isolates obtained from different localities. Only nine isolates among the collected samples were positive against Ralstonia solanacearum. Four virulence isolates from Spunta cultivars, including three isolates achieved of Al- Maymun village, Beni-suef governorate and one isolate acquired from Telia (1), Menoufia governorate. Four isolates from Bellini cultivars, two from Al Maymun village, Beni-suef governorate and two from Om Sabir village, Beheira governorate and one isolate from Lady-rosetta obtained from Telia (2), Menoufia governorate. On the other hand, 71 isolates were negative. Fig. 1 show the different between healthy and artificially typical infected wilted potato plants (Spunta cv.) as well as the bacterial ooze and the character of virulence colonies isolated from infected naturally plants. As the vascular bundles in the diseased plants are filled with bacteria that impede the transport of water and nutrients, and thus the symptoms appear, which are yellowing of the leaves, redness of the vascular bundles, necrosis, and ultimately the total wilt of the infected plants.
Table 2.
Number of positive Ralstonia solanacearum samples collected from different cultivars at different localities in four Egyptation governorates.
| Governorate | No. of sample | Cultivar |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Spunta |
Bellini |
Lady rosetta |
Mondial |
||||||
| (+) | (-) | (+) | (-) | (+) | (-) | (+) | (-) | ||
| Beni-suef | 15 | ||||||||
| Al Maymun | 3 | 1 | 2 | 3 | – | – | * | 6 | |
| Menoufia | 30 | ||||||||
| Telia (1) | 1 | * | * | 4 | * | 6 | * | * | |
| Telia (2) | * | * | * | 6 | 1 | * | – | – | |
| El-Basha | * | 8 | * | * | * | * | * | 4 | |
| Beheira | 25 | ||||||||
| El-Bostan | * | 7 | * | 9 | * | 5 | – | – | |
| Om Sabir | * | * | 2 | * | * | 2 | – | – | |
| El-Sharkia | 10 | ||||||||
| El-salhia city | * | 4 | – | – | – | – | * | 6 | |
| Total | 80 | 4 | 20 | 4 | 22 | 1 | 13 | 0 | 16 |
(*) Samples don't collected from this site.
(-) Negative reaction during isolation.
(+) Number of positive reaction during isolation
Fig. 1.
Typical symptoms of bacterial wilt disease on potato plants (Spunta cv.), caused by R. solanacearum. (A) Healthy plant, (B) bacterial ooze and colony growth (C) virulence colonies of bacteria and (D) wilted plant.
3.2. Detection of pathogenic isolates:
3.2.1. Morphological and physiological properties of the isolated Ralstonia solanacearum.
Nine isolates from different habitats were subjected to complete identification up to species level. All identified isolates were Gram negative, short rods, motility and oxidase positive reaction. These isolates were non-sporulation, negative reaction for Arginine dihydrolase, starch hydrolysis and gelatin liquefaction. Fluorescent pigments could be detected with UV on King's B medium, but production of brown pigments on glucose nutrient agar medium was detected. No variation could be detected among the isolates in these characteristics except for the motility test in which the phenotype conversion mutants (PC- type) variant isolates were more motile, and for the oxidation/fermentation (O/F) test in which the virulent isolates were aerobic while the PC-type variant ones were microaerophilic. As for the utilization of carbon sources is concerned all the pathogenic isolates mentioned were utilized glucose, lactose, maltose and cellbiose. On the other hand, none of the examined isolates was able to utilize sorbitol, mannitol and dulcitol.
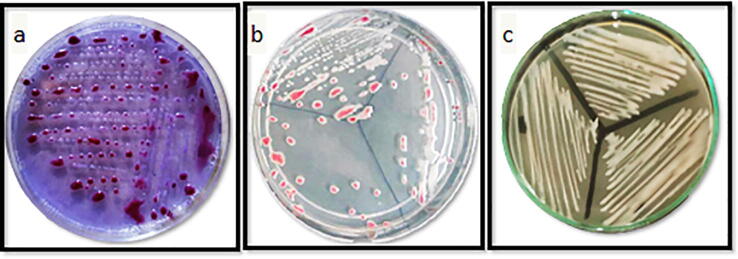
Cultivation on SMSA, King's B and TZC media: To differentiate between virulence (RS5) and avirulence (RS7) isolate of R. solanacearum, the isolates were cultivated on SMSA, King's B and TZC media. Data in table 3 and Fig. 2 explained the characters of RS5. Virulent colony isolate developed on the SMSA medium were slimy, white, irregular and fluidal with pink-red coloration in the center, whereas the PC-type variant ones was less fluidal or a fluidal, uniformly round, butyrous, dry and ultimately deep pink to red as shown in Fig. 3. Triphenyl tetrazolium chloride medium (TTC or TZC), containing 2, 3, 5 triphenyl tetrazolium chloride was used to differentiate between the virulent (wild-type) and avirulent (PC-type variant) isolates of the bacterium. Differentiation between the two forms were easily noticed on media containing 2,3,5 triphenyltetrazolium chloride. Colonies of the avirulent isolate (RS7) was uniformly round, butyrous and deep red in color due to the formation of formozan on tetrazolium-containing medium, contrary to the virulent ones. But the virulent one was highly fluidal, slimy, irregular, mucous creamy-white with pink centers. The virulence colonies on King's B medium had the same characteristics on modified SMSA and TZC media. But the coloration of virulence and non– virulence colonies on King's B medium were brownish and white, respectively.
Table 3.
Cultural characterization of virulent (v) and avirulent (av) Ralstonia solanacearum isolates on the used three different media.
| Colony character | Medium |
|||||
|---|---|---|---|---|---|---|
| SMSA |
TZC |
K. B |
||||
| v | av | v | av | V | av | |
| Margin | Entire | Entire | Entire | Entire | Entire | Entire |
| surface | smooth surface | smooth surface | smooth surface | smooth surface | smooth surface | smooth surface |
| Form | Irregular | Circular | Irregular | Circular | Irregular | Circular |
| Elevation | Convex | Convex | Convex | Convex | Convex | Convex |
| colour | White with pink centers | Red | White with pink centers | Red | Brownish | white |
Fig. 2.
Colony characters of virulent Ralstonia solanacearum isolate (RS5) on different media (a) modified SMSA, (b) TZC and (c) King’s B medium.
Fig. 3.
Colony characters of avirulent Ralstonia solanacearum isolate (RS7) on different media (a) modified SMSA, (b) TZC and (c) King’s B medium.
3.3. Molecular characteristics:
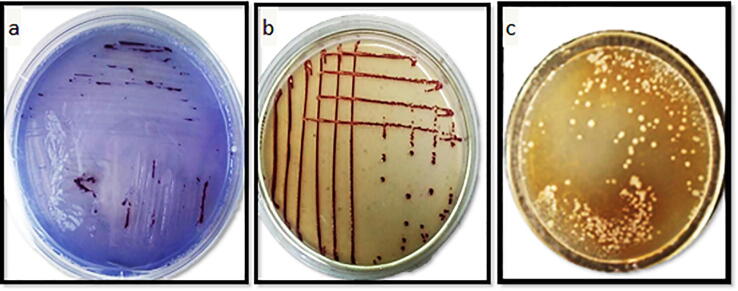
Identification methods for the pathogenic isolates were carried out through using Immunofluorescence antibody stains (IFAS) and Quantitative, Real-time, Fluorogenic PCR (Taq-Man). The most five R. solanacearum pathogenic isolates showed fluorescent short rod shape cells and evenly stained as bright green fluorescent under immunofluorescence microscope (Fig. 4). Also, the result represent that the probe used in Taq-man was of the type (B2) capable to detect only biovar 2 of R. solanacearum bacterial wilt. Furthermore, primer and probe are specific for detection of the race 3 biovar 2 strain. Positive results were obtained in both assays with all five isolates. The curve approached from number (1) on horizontal axis which indicates to cycle number was the most virulence isolate compared to negative control. . So the isolate (RS5) was the most virulence one, followed by RS1, RS3, RS2 and RS8, representative that the tested isolates were R. solanacearum race 3, biovar 2.
Fig. 4.
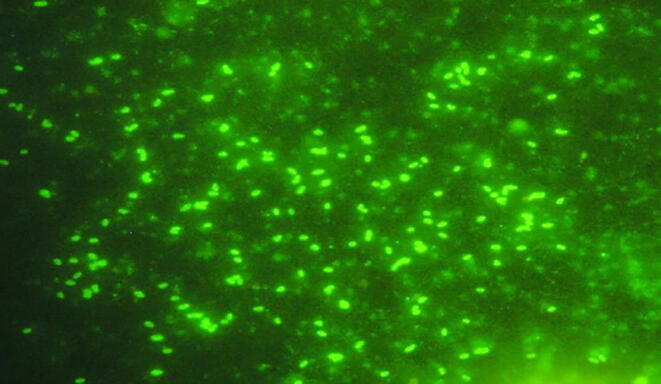
Positive reaction of race (3), biovar (2) of R. solanacearum isolate (RS5) in the serological immunofluorescent antibody staining (IFAS) test.
3.4. Susceptibility of Spunta, Bellini and Mundial cultivars to bacterial wilt, caused by Ralstonia solanacearum.
Results in Table 4 demonstrate that significant differences between the treatments. The isolate RS5 showed the lowest percentage of disease incidence reduction on the three potatoes cultivar Bellini, Spunta and Mondial recorded 9.64%, 15.41% and 34.12%, respectively. Followed by the isolate RS1that recorded 18.14%, 23.78% and 45.32%, respectively, while RS3 isolate registered 29.71%, 40.43% and 52.79, respectively. RS2 isolate calculated 45.25%, 56.36% and 60.77%. Finally, RS8 isolate exhibited the highest effective one the percentage of disease reduction on all tested potato cultivars. This isolate recorded 60.60%, 63.21% and 71.66%, compering to the healthy control treatment. Results presented that potato cultivars reacted differently and varied significantly in their degree of susceptibility. Bellini cv. was the highest in susceptibility, so decreased the average of percentage disease incidence and severity reduction which showed 32.66% and 38.88%, respectively. Followed by Spunta cv. calculated 39.81% and 45.62%. While Mondial cv. was the moderately susceptible this registered 52.94% and 60.16%. Furthermore, the quality of potatoes cultivars reduced with increasing the disease incidence and severity. So the production of potatoes cultivars decreased and thus associated with the number and weight of tubers decreased. The data revealed that the number of tubers in the three cultivars of potato was close, while differences were observed in the weights of the tubers due to their size. Mondial cultivar was the largest average of weight and recorded that 86.50 g, followed by Spunta cultivar was calculated 71.84 g, and finally Bellini cultivar was the lowest one and recorded 64.37 g.
Table 4.
Pathogenic and plant growth parameter of the three potato cultivars inoculated with the five virulent Ralstonia solanacearum measured as percentage reduction of disease incidence and disease severity, number of tubers and weight of tubers.
| Treatment | Pathogenic parameter |
Plant growth parameter |
||
|---|---|---|---|---|
| DI redution (%) | DS reduction (%) | No. of tubers | Weight of tubers (g/plant) | |
| Spunta + RS1 | 23.68 ± 1.1 | 31.80 ± 1.5 | 3.25 ± 0.03 | 62.43 ± 1.2 |
| Spunta + RS2 | 56.36 ± 2.3 | 60.46 ± 1.6 | 4.25 ± 0.02 | 89.85 ± 1.3 |
| Spunta + RS3 | 40.43 ± 1.2 | 45.11 ± 1.4 | 3.00 ± 0.04 | 63.12 ± 1.9 |
| Spunta + RS5 | 15.41 ± 0.9 | 20.39 ± 0.7 | 3.00 ± 0.03 | 55.12 ± 1.5 |
| Spunta + RS8 | 63.21 ± 2.5 | 70.68 ± 2.2 | 4.25 ± 0.05 | 88.70 ± 2.1 |
| Average | 39.81 ± 1.6 | 45.62 ± 0.8 | 3.55 ± 0.04 | 71.84 ± 2.6 |
| Spunta | 100 ± 2.5 | 100 ± 3.5 | 4.50 ± 0.03 | 128.18 ± 4.2 |
| Bellini + RS1 | 18.14 ± 0.8 | 23.58 ± 1.1 | 3.25 ± 0.04 | 59.23 ± 2.1 |
| Bellini + RS2 | 45.25 ± 1.4 | 51.10 ± 2.1 | 4.00 ± 0.02 | 73.03 ± 2.6 |
| Bellini + RS3 | 29.71 ± 1.5 | 35.02 ± 2.3 | 3.75 ± 0.03 | 64.50 ± 2.9 |
| Bellini + RS5 | 9.64 ± 0.7 | 17.25 ± 0.9 | 2.75 ± 0.01 | 43.97 ± 2.4 |
| Bellini + RS8 | 60.60 ± 2.2 | 67.86 ± 3.2 | 4.50 ± 0.04 | 81.16 ± 2.8 |
| Average | 32.66 ± 1.2 | 38.88 ± 2.8 | 3.65 ± 0.03 | 64.37 ± 2.8 |
| Bellini | 100 ± 1.9 | 100 ± 2.5 | 4.75 ± 0.03 | 127.53 ± 3.9 |
| Mondial + RS1 | 45.32 ± 1.8 | 50.90 ± 1.6 | 3.00 ± 0.02 | 71.08 ± 2.8 |
| Mondial + RS2 | 60.77 ± 1.6 | 70.24 ± 2.1 | 4.25 ± 0.03 | 112.31 ± 4.6 |
| Mondial + RS3 | 52.79 ± 1.7 | 61.02 ± 2.3 | 3.50 ± 0.01 | 81.65 ± 3.1 |
| Mondial + RS5 | 34.17 ± 1.2 | 40.49 ± 2.2 | 2.25 ± 0.02 | 47.39 ± 1.5 |
| Mondial + RS8 | 71.66 ± 2.8 | 78.14 ± 3.1 | 4.25 ± 0.03 | 120.08 ± 4.2 |
| Average | 52.94 ± 2.4 | 60.16 ± 2.9 | 3.45 ± 0.04 | 86.50 ± 3.5 |
| Mondial | 100 ± 3.6 | 100 ± 2.92.8 | 5.00 ± 0.03 | 145.26 ± 4.9 |
| LSD 0.05 for Cultivar 2.319 Isolates 2.082 DI for cultivar 1.059 DI for isolates 1.293 No. of tuber for cultivar 1.924 No. of tuber for isolates 2.035 weight for cultivar 12.684 weight for isolates 9.572 |
Interaction cultivar*isolates 5.036 DI for cultivar*isolates 1.037 No. of tuber for cultivar*isolates 1.380 weight for cultivar*isolates 15.729 |
|||
3.5. Genetic distances and similarities based on pathogenic and plant growth parameters:
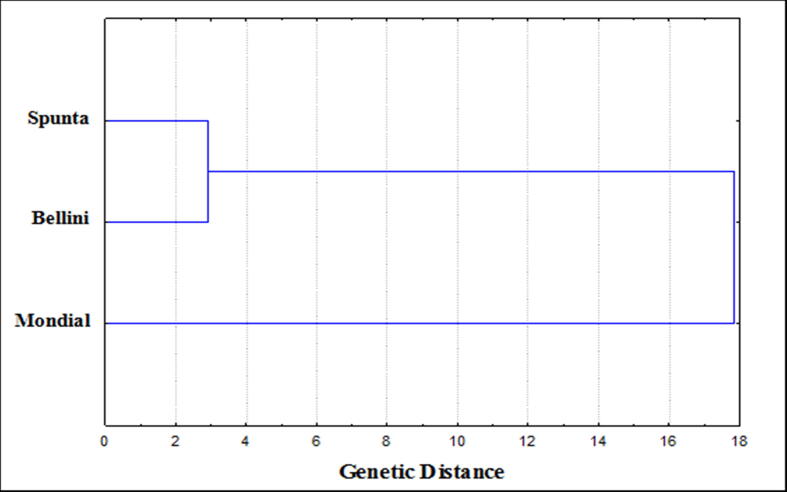
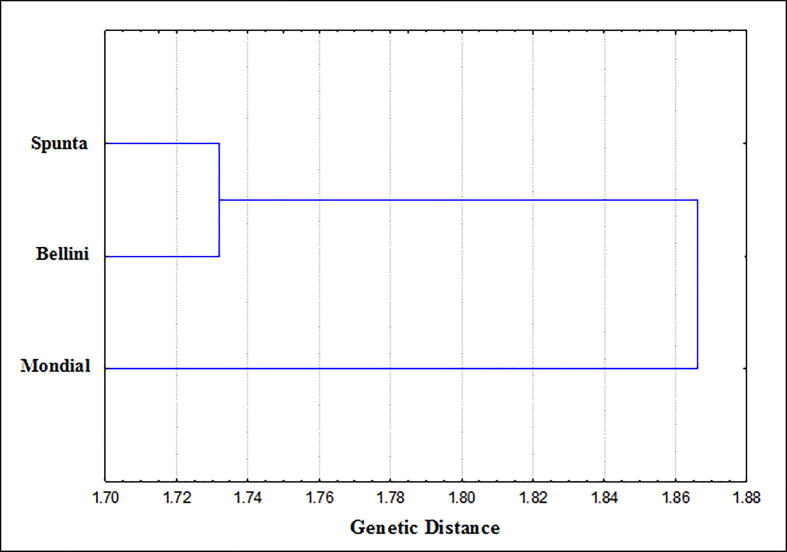
The genetic distance and genetic similarity among potato cultivars based on pathogenic and plant growth parameters were indicated in Table 5. The results illustrate that the highest genetic similarity (0.998) was found between Bellini and Spunta as the closest but the lowest value (0.946) was found between Mondial and Bellini as most distant. The same results show that the highest genetic distances value (19.0) was found between Bellini and Mondial as the most distant genetically, while the lowest value (2.9) found between Bellini and Spunta cultivars.
Table 5.
Genetic distance and similarity coefficient (Pearson similarity) calculated based on differences in pathogenic and plant growth parameters between the three potato cultivars.
| Cultivar | Spunta | Bellini | Mondial |
|---|---|---|---|
| Genetic distance | |||
| Spunta | 0.0 | ||
| Bellini | 2.9 | 0.0 | |
| Mondial | 16.7 | 19.0 | 0.0 |
| Similarity coefficient | |||
| Spunta | 1.000 | ||
| Bellini | 0.998 | 1.000 | |
| Mondial | 0.958 | 0.946 | 1.000 |
The phylogenetic tree based on the genetic distance (Fig. 5) showed two clusters according to the data scored from pathogenic and plant growth parameters, clustering analysis subdivided the investigated cultivars into two groups. Each of Spunta and Bellini cultivars formed cluster I, and Mondial formed an independent cluster (cluster II).
Fig. 5.
Phylogenetic tree (Linkage dendrogram) of studied potato cultivars based on their pathogenic and plant growth parameters.
3.6. Genetic distances and similarities based on SDS-PAGE profile:
SDS-PAGE profiles of total dissolved proteins contents obtained from the resistant and susceptible plants of three potato cultivars, as well as the control is presented in Fig. 6. SDS-PAGE analysis illustrated that nine protein bands with various molecular weights (MWs) reached from 275 to 6.5 kDa. The results in Table 6 explain that the genetic distance and similarity coefficient (Dice similarity) calculated based on differences in protein banding patterns between the three potato cultivars. The highest genetic distances value (2.00) was found between Mondial and Bellini as the most distant genetically, while the lowest value (1.73) found between Bellini and Spunta cultivars, but the highest similarity value (0.842) was found between Bellini and Spunta as the closest but the lowest value (0.778) was found between Mondial and Bellini as most distant.
Fig. 6.
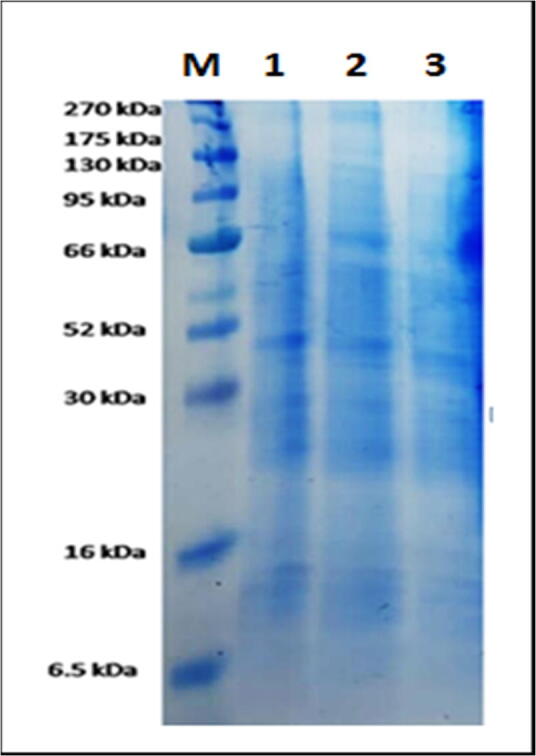
Differentiate between potato cultivars by protein SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis. (M) Marker, (1) Spunta cultivar, (2) Bellini cultivar and (3) Mondial cultivar.
Table 6.
Genetic distance and similarity coefficient (Dice similarity) calculated based on differences in protein banding patterns between the three potato cultivars.
| Cultivar | Spunta | Bellini | Mondial |
|---|---|---|---|
| Genetic distance | |||
| Spunta | 0.00 | ||
| Bellini | 1.73 | 0.00 | |
| Mondial | 1.73 | 2.00 | 0.00 |
| Similarity coefficient | |||
| Spunta | 1.000 | ||
| Bellini | 0.842 | 1.000 | |
| Mondial | 0.800 | 0.778 | 1.000 |
The phylogenetic tree based on the genetic distance between three cultivars of potato by banding patterns of protein SDS-PAGE is shown in Fig. 7. the dendrogram separated all cultivars into two clusters. First cluster divided into two sub-clusters formed a separate sub-cluster with Spunta, and the second sub-cluster included Belini. While the second cluster included one cultivar Mondial. These results were consistent with the pathological studies, as they showed that the potato cultivars Spunta and Bellini were the closest to each other genetically. Therefore, their sensitivity to disease was the closest while it was high, as was shown by the pathological studies.
Fig. 7.
Phylogenetic tree (Linkage dendrogram) of studied potato cultivars based on their protein banding patterns.
4. Discussion:
Ralstonia solanacearum the causal pathogen of potato bacterial wilt, has been recorded earlier in Egypt many years ago (Farag et al., 2017) based on symptomology only. R. solanacearum is measured one of the greatest serious problems of potato production (Ramesh et al., 2009). In Egypt disease caused economic losses that affect potato yield and threatens potato exportation to European Union (Farag et al., 1999).
Under those circumstances, R. solanacearum is listed as a regulatory pathogen in A2 group (the group of quarantine pest which can be present in the location but cannot be widely distributed there and has to be officially controlled) quarantine organism (Database, 2017) and is listed by Asia and Pacific Plant Protection Commission (APPPC) and International Association of Professional Security Consultants (IAPSC). The occurrence of different races and strains of the pathogen with varying virulence under different environmental conditions presents a serious danger to European and Mediterranean potato and tomato production. Therefore, the absence of the bacterium is an important consideration for countries exporting seed potatoes (CABI, 2017). Moreover, these bacteria attacks more than two hundred plant species, including more than fifty plant families (Aslam et al., 2017, Uwamahoro et al., 2020). The pathogen of bacterial wilt is the most destructive bacterial pathogen due to its infestation, uncommonly wide host range, perseverance, and wide topographical distribution (Wei et al., 2018). Therefore, we are confronted to study this disease. The disease was limited to some governorates of Egypt where potato cultivation is abundant.
In this study eighty samples were accessed from different naturally infected potato plants exhibition symptoms of bacterial wilt obtained from different cultivars (Spunta, Bellini, Lady-rosetta and Mondial) at different localities in four governorates (Beni-suef, Menoufia, Beheira and El-Sharqia). The results showed that the highest number isolates were obtained from Spunta and Bellini cultivars, due to their highly susceptible to bacterial wilt pathogen. The highest percentage of positive samples were obtained from in Beni-suef governorate. Isolates obtained during isolation were detected as Ralstonia solanacearum by using morphological and physiological properties (Kubota et al., 2008).
Using the traditional identification methods based on morphological, physiological and biochemical characteristics of bacteria. The results indicate that all tested isolates were non-spore, short rods, motile bacteria and Gram negative. The isolates exhibited positive reaction for oxidase activity and negative reaction for Arginine dihydrolase, starch hydrolysis and gelatin liquefaction. As for the utilization of carbon sources is concerned all pathogenic isolates were capale to utilize glucose, lactose, maltose and cellbiose. On the other hand, none of the observed isolates were capable to utilize sorbitol, mannitol and dulcitol. Such result was identical with (Pradhanang et al., 2000). R. solanacearum divided into five biovars based on ability of races to oxidize three hexose sugar alcohols (dulcitol, mannitol, and sorbitol), and three other of disaccharides (cellobiose, lactose, and maltose), according to Mutimawurugo et al., 2019. Accordingly, biovar (1) can't analyze these sugars, while biovar (2) can analyze only disaccharides, biovar (3) can analyze all of them and biovar (4) analyze only hexose alcohols, while biovar (5) can analyze all sugars except dulcitol and sorbitol as stated by Sagar et al. (2014).
Growth of pathogenic R. solanacearum on three differentiated media as used to between virulence and avirulence. The characters of virulent isolate (RS5) colony developed on SMSA medium were slimy, white, irregular and fluid with pink-red coloration in the center. Whilst using TZC medium, to differentiate between virulent and avirulent isolates. Colonies of the avirulent isolate (RS7) were consistently round, butyrous and deep red in color due to formozan formation on tetrazolium-containing medium, contrary to the virulent ones. But the virulent ones were highly fluidal, slimy, irregular, mucoid creamy-white with pink centers. The virulence colonies on King's B medium had the same characteristics on modified SMSA and TZC media. But the coloration of virulence and non– virulence colonies on King's B medium were brownish and white, respectively. The methods newly advanced for the revealing of the pathogen in different habitats have qualitatively improved the pathological studies. The obtained results were consistent with Sujeet et al. (2017) results showed that single colonies of these bacteria appear after their growth for 36 to 48 h at a temperature of 28 °C and the virulence colonies were characterized by white or cream colored, irregularly shaped, highly fluidal and opaque. Infrequently, these types of non-virulence colonies appear and were characterized by round, smaller and butyrous or dry. A Kelman’s selective nutrient tetrazolium chloride (TZC) medium (Kelman, 1954) capable to discriminate between the two colony categories on this medium. Strains of R. solanacearum have been classified as mentioned before into five biovars (Santiago et al., 2017), and five races (Bin et al., 2010).
The causal pathogen of potato brown rot R. solanacearum is classified nowadays into five races affecting hosts in different plant families (Ravelomanantsoa et al., 2018). Race 3 biovar 2, the so- called potato race, is the dominant strain in Egypt (Serag et al., 2020) which is characterized by low virulence to tobacco and lower optimum temperature (Buddenhagen and Kelman, 1964).
Furthermore, the results revealed that fluorescent short rod shape cells of pathogenic bacteria stained evenly as bright green fluorescent under the immunofluorescence antibody stain (IFAS) to confirm the characterization of R. solanacearum in potato tissue and is considered as another achievement technique (Janse, 1988). According to Janse et al. (2004), using an immunofluorescence antibody stain to R. solanacearum biovar 2 was shown to be detectable as few as104 CFU per mL in potato tissue. The sensitivity of serological methods for the detection of R. solanacearum in potato or in soil can be increased by applying an enrichment procedure, for example by inoculating extracts in a selective broth (Shahbaz et al., 2015, Sujeet et al., 2017).
The hierarchical classification is sub-divided into four phylotypes (genetic groups) according to analysis of the 16S-to-23S internal transcribed spacer region, and each of them is further subdivided into smaller groups called sequevars. Every phylotype reverse the topographical origin of varieties. The fraction of phylotype II embraces race 3 biovar 2 (Williamson et al., 2002). The strain obtained results indicated that R. solanacearum in Egypt belong to phylotype II sequevar I, which is also included the American strains (Pardo et al., 2019).
The result represent that the probe used in Taq-man was of the type (B2) capable to detect only biovar 2 of R. solanacearum bacterial wilt. The isolate (RS5) was the most virulente one, followed by RS1, RS3, RS2 and RS8, representative that the tested isolates were R. solanacearum race 3, biovar 2. Tag-Man is a molecular revelation method to join polymerase chain reaction (PCR) with fluorescent detection of the amplicon (Puaprasert et al., 2018, Birhane et al., 2020). The RS primers and probe detected all biovars and races of virulent R.solanacearum whereas the B2 primer and probe are specific for identification of the phylotype II sequevar I, equivalent to Race 3 biovar 2 strain. These results agree with the finding of (Malko et al., 2019).
This study planned to clear up the sensibility of potato varieties to R. solanacearum was applied to screen variety Egyptian potato varieties for their sensibility to the pathogen as foundation to recover the administration of potato bacterial wilt. No resistance against R. solanacearum is obtainable in the three tested potato cultivars in the present research, however the level of susceptibility diverse significantly amongst them. As all known potato cultivars in Egypt are susceptible; breeding programs should to research of resistance in the wild solanum types that display tolerance or resistance the disease to accomplish positive results (Poudel and Neupane, 2018).
The results of the study aimed that there are losses in potato production in infected plants compared to healthy ones, due to the severity of the disease and spreading the bacterial pathogen into plant tissues. R. solanacearum is a soil-dwelling bacterium that invades plants through roots and colonizes xylem vessels (Karim and Hossain, 2018). This study was compatible with some other studies, the potato production and yield losses due to bacterial wilt as high as 100 per cent have been reported in parts of tropical Africa (Shivani, 2016). In Kenya, the potato industry is threatened by bacterial wilt (BW) because soils in most production areas are infested with the wilt causing bacterium and over 50% yield losses have been reported (IPDN, 2014). The farmers reported experiencing yield losses ranging from 5% to 80% due to bacterial wilt. According to some recent studies, the disease is found in all the potato growing areas of Kenya and the country is affecting 77% of potato farms which had been introduced with tuber seeds imported from Europe (Kaguongo et al., 2010). Potato yield losses in Uganda estimated about 30% (IPDN, 2014)), with more severe losses being 100%. Kabeil et al. (2008) reported that potatoes were one of the largest exported crops in Egypt. Yet, the total value of Egyptian potato exports fell from a peak value of US$ 102.12 million in 1995 to $US 7.7 million in 2000 mainly due to this organism related quarantine restrictions imposed by the European Union (EU) which used to account for about 70–90% of Egyptian potato exports and it represented a drop from approximately 419,000 metric tons to 48,500 tons.
The results illustrated that through the phylogenetic tree based on the genetic distance of both the recorded data from the pathogen and plant growth parameters, as well as from the results of the protein analysis (SDS-PAGE) of the three potato cultivars. Data showed two groups from clustering analysis divided the studied items into two groups. The Spunta and Bellini varieties each formed the first group, and Mondial formed an independent group (Group II). Therefore, their sensitivity to disease was the closest while it was high, as was shown by the pathological studies. This is due to the similarity in terms of upbringing and also some genetic characteristic, so slight significant differences in disease severity between three cultivars were detected and calculated, these due to the common between the varieties in some characteristics. Bellini cultivar was crossed by (Felsina × Mondial), medium early for maturity and Netherlands for origin according to Spanoghe et al. (2015).While, spunta cultivar was crossed by (Bea × USDA 96–56), intermediate early for maturity and Netherlands for origin, as mentioned by Khidr et al. (2017). Mondial cultivar was crossed by (Spunta × SVP VE 66–295), intermediate late for maturity and Netherlands for origin, as stated by Eriksson et al. (2016).
Meanwhile, the potato varieties with the least susceptibility to the disease would be studied when implanting potatoes in fields with a history of bacterial wilt. Therefore, we must go to build up programs to discover resistant and / or tolerant potato varieties (Shimira et al., 2020). SDS-PAGE is an electrophoresis method that allows protein by mass to be separated (Mouzo et al., 2018).This recent method of differentiation between plants. Constructed on the results of this research, we can conclude that the intra-cultivar variances in protein polymorphism may be detected through the electrophoresis patterns applied to content protein through SDS-PAGE in potato varieties. For efficient breeding program information, genetic diversity within crop varieties is fundamental. It is especially appropriate for describing individual approached and cultivars and as a general guide in the chosen of the parents for hybridization. Genetic fingerprinting has been adopted traditionally through the use of isozymes, total seed proteins, tuber storage proteins and further newly through various types of molecular markers (Zhang et al., 2017, Sogbohossou et al., 2018). The diversity within and between identified species is the basis of all crop amelioration programs. Whether all the individuals in the species would have been analogous, thereafter possibly there could not have been any scope for amelioration in plant for different traits. Since the inception of plant breeding programs, the natural variability and divergence between crops has been widely defined and used in the advancement of crop cultivars. However, as time progressed, the natural variability was reduced due to unbalanced breeding practices centering on the progression of only a few traits, the continued use of minor selected genotypes as heterogeneous parents of the amelioration program and the addition of a few salient lines to Many countries through this broaden the genetic similarities between current crop cultivars (Bhandari et al., 2017).
5. Conclusion
In this study, bacterial pathogens was isolated and defined, as well as select the most resistant cultivars, thus avoiding a decrease in the total value of Egyptian potato exports. Molecular genetics methods such as SDS-PAGE profile was used to differentiate between varieties by protein content and determine the genetic diversity between them, which is useful in breeding programs to produce disease-resistant varieties.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgment
All thanks to God and then to my professors and everyone who supported this research. I would like to thank the head of the plant diseases laboratory, Faculty of Agriculture, Zagazig University and the director of Potato Brown Rot Project (PBRC), Agric. Res. Center, Giza- Egypt, as well as the laboratory Biotechnology Lab at Cairo University Research Park, Faculty of Agriculture.
Author Contributions
Ahmed M. Khairy, Mohamed R.A. Tohamy, Mohamed A. Zayed and Mohamed A.S. Ali.
Data curation, AM Khairy, MRA Tohamy, MAS Ali; Investigation, MRA Tohamy, MA Zayed, MAS Ali ; Methodology, AM Khairy, MRA Tohamy; Resources, AM Khairy, MRA Tohamy, MAS Ali; Software, MA Zayed; Supervision, MRA Tohamy, MA Zayed; Validation, AM Khairy, MRA Tohamy, MAS Ali; Writing – original draft, AM Khairy, MRA Tohamy, MAS Ali.
Footnotes
Peer review under responsibility of King Saud University.
References
- Amelio A., Tagarelli A. Data mining: clustering. Encyclopedia of Bioinformatics and Computational Biology. 2018:437–448. [Google Scholar]
- Andreas O., Florian D., Wolfgang F., Axel S. Growth inhibition of Staphylococcus aureus induced by low-frequency electric and electromagnetic Fields. Bem. 2009;30:270–279. doi: 10.1002/bem.20479. [DOI] [PubMed] [Google Scholar]
- Aslam M.N., Mukhtar T., Hussain M.A., Raheel M. Assessment of resistance to bacterial wilt incited by Ralstonia solanacearum in tomato germplasm. Journal of Plant Disease Protection. 2017;124:585–590. doi: 10.1007/s41348-017-0100-1. [DOI] [Google Scholar]
- Bhandari H.R., Bhanu A.N., Srivastava K. Assessment of genetic diversity in crop plants - an overview. Adv. Plants Agric. Res. 2017;7(3):279–286. doi: 10.15406/apar.2017.07.00255. [DOI] [Google Scholar]
- Bin L.I., Ting S.U., Rongrong Y.U., Zhongyun T., Zhiyi W., Algam S.A., Guanlin X., Yanli W., Guochang S. Inhibitory activity of Paenibacillus macerans and Paenibacillus polymyxa against Ralstonia solanacearum. African Journal of Microbiology Research. 2010;4(19):2048–2054. ISSN 1996–0808 ©2010 Academic Journals. [Google Scholar]
- Birch P.R., Bryan G., Fenton B., Gilroy E.M., Hein I., Jones J.T., Toth I.K. Crops that feed the world 8: Potato: are the trends of increased global production sustainable. Food Security. 2012;4(4):477–508. doi: 10.1007/s12571-012-0220-1. [DOI] [Google Scholar]
- Birhane E., Hailemariam M., Gebresamuel G. Source of mycorrhizal inoculum influences growth of Faidherbia albida seedlings. Journal of Forestry Research. 2020;31(1):313–323. [Google Scholar]
- Buddenhagen, I., Kelman, A., 1964. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annual Review of Phytopathology, 2(1),203-230. DOI.org/10.1146/annurev.py.02.090164.001223.
- CABI., 2017. Invasive Species Compandium (Datasheets, maps, images, abstracts and full text on invasive species of the world: Ralstonia solanacearum).
- Charkowski A., Sharma K., Parker M.L., Secor G.A., Elphinstone J. Bacterial diseases of potato. The potato crop. 2020:351–388. [Google Scholar]
- Chávez P., Yarlequé C., Loayza H., Mares V., Hancco P., Priou S., Quiroz R. Detection of bacterial wilt infection caused by Ralstonia solanacearum in potato (Solanum tuberosum L.) through multifractal analysis applied to remotely sensed data. Precision agriculture. 2012;13(2):236–255. doi: 10.1007/s11119-011-9242-5. [DOI] [Google Scholar]
- Chen Y., Zhang W.Z., Liu X., Ma Z.H., Li B., Allen C., Guo J.H. A real-time PCR assay for the quantitative detection of Ralstonia solanacearum in the horticultural soil and plant tissues. J. Microbiol. Biotechnol. 2010;20(1):193–201. doi: 10.4014/jmb.0906.06019. [DOI] [PubMed] [Google Scholar]
- Database O.G. Ralstonia solanacearum (RALSSO) (Associated EPPO Standards. EPPO A1 and A2 Lists of pests recommended for regulation as quarantine pests. 2017;2(25):1–17. [Google Scholar]
- Devaux A., Goffart J.P., Petsakos A., Kromann P., Gatto M., Okello J., Hareau G. The Potato Crop. Springer; Cham: 2020. Global food security, contributions from sustainable potato agri-food systems; pp. 3–35. [Google Scholar]
- Dice, R.L., 1945. Measures of the Amount of Ecologic Association between Species. Ecology. 26 (3), 297–302. DOI:org/10.2307/1932409
- ELHetawy D. In vitro, induction of salt tolerant potato (solanum tuberosum l.) Plants with gamma irradiation and characterization of genetic variations through sds-page and issr-pcr analysis. Annals of Agricultural Science, Moshtohor. 2018;56(4th ICBAA):167–176. doi: 10.21608/ASSJM.2018.65134. [DOI] [Google Scholar]
- Eriksson D., Carlson-Nilsson U., Ortíz R., Andreasson E. Overview and breeding strategies of table potato production in Sweden and the Fennoscandian region. Potato Research. 2016;59(3):279–294. doi: 10.1007/s11540-016-9328-6. [DOI] [Google Scholar]
- Fahy, P.C., Persley, G.J., 1983. Plant bacterial disease. A Diagnostic Guide. Academic Press, New York, 393p.
- Farag N.S., Stead D.F., Janse J.D. Ralstonia (Pseudomonas) solanacearum race 3, biovar 2, detected in surface (irrigation) water in Egypt. J. Phytopathol. 1999;147:485–487. doi: 10.1111/j.1439-0434.1999.tb03854.x. [DOI] [Google Scholar]
- Farag S.M., Elhalag K.M., Hagag M.H., Khairy A.S.M., Ibrahim H.M., Saker M.T., Messiha N.A. Potato bacterial wilt suppression and plant health improvement after application of different antioxidants. Journal of Phytopathology. 2017;165(7–8):522–537. doi: 10.1111/jph.12589. [DOI] [Google Scholar]
- García R.O., Kerns J.P., Thiessen L. Ralstonia solanacearum species complex: a quick diagnostic guide. Plant Health Progress. 2019;20(1):7–13. doi: 10.1094/PHP-04-18-0015-DG. [DOI] [Google Scholar]
- George, D., Mallery, P., 2019. IBM SPSS Statistics 26 step by step: A simple guide and reference. Routledge.
- Hayward A.C. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 1964;27:265–277. doi: 10.1111/j.1365-2672.1964.tb04912.x. [DOI] [Google Scholar]
- IPDN., 2014. Bacterial Wilt Disease Ralstonia solanacearum: Standard Operating Procedure for Use in Diagnostic Laboratories. International Plant Diagnostic Network EA-SOP-RS1,1–24.
- Janse J.D. A detection method for Pseudomonas solanacearum in symptomless potato tubers and some data on its sensitivity and specificity. Bull. OEPP. 1988;18:343–351. doi: 10.1111/j.1365-2338.1988.tb00385.x. [DOI] [Google Scholar]
- Janse J.D. Potato brown rot in Western Europe–history, present occurrence and some remarks on possible origin, epidemiology and control strategies. EPPO Bulletin. 1996;26(3–4):679–695. doi: 10.1111/j.1365-2338.1996.tb01512.x. [DOI] [Google Scholar]
- Janse J.D., van den Beld H.E., Elphinstone J., Simpkins S., Tjou-Tam-Sin N.A.A., van Vaerenbergh J. Introduction to Europe of Ralstonia solanacearum biovar 2, race 3 in Pelargonium zonale cuttings. J. Plant Pathol. 2004;86:147–155. doi: 10.4454/jpp.v86i2.950. [DOI] [Google Scholar]
- Kabeil S.S., Lashin S.M., El-Masry M.H., El-Saadani M.A., Abd-Elgawad M.M., Aboul-Einean A.M. Potato brown rot disease in Egypt: current status and prospects. Am. Eurasian J. Agric. Environ. Sci. 2008;4:44–54. [Google Scholar]
- Kaguongo, W.P., Ng’ang’a, N.M., Muthoka, N., Muthami, F., Maingi, G., 2010. Seed potato subsector master plan for Kenya. GTZ-PSDA, USAID, CIP and Government of Kenya. Ministry of Agriculture. Kenya
- Karim Z., Hossain M.S. Management of bacterial wilt (Ralstonia solanacearum) of potato: focus on natural bioactive compounds. Journal of Biodiversity Conservation and Bioresource Management. 2018;4(1):73–92. doi: 10.3329/jbcbm.v4i1.37879. [DOI] [Google Scholar]
- Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appreance on a tetrazolium medium. Phytopathology. 1954;44:693–695. doi: 10.4236/fns.2012.37125. [DOI] [Google Scholar]
- Khidr, Y., Arafa, M., EL-demery, S., EL-Sanhoty, R., 2017. Molecular and morphological evaluation of potato genotypes cultivated in sandy soil. Egyptian Journal of Genetics and Cytology, 46(1), 1-17. DOI: 10.21608/EJGC.2018.9487.
- Kim Y.I., Cho J.Y. Gel-based proteomics in disease research: Is it still valuable. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2019;1867(1):9–16. doi: 10.1016/j.bbapap.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Kubota R., Vine B.G., Alvarez A.M., Jenkins D.M. Detection of Ralstonia solanacearum by loop-mediated isothermal amplification. Phytopathology. 2008;98(9):1045–1051. doi: 10.1094/PHYTO-98-9-1045. [DOI] [PubMed] [Google Scholar]
- Kumari R., Ranjan R.K. Characterization of Ralstonia solanacearum causing bacterial wilt of potato. Journal of Pharmacognosy and Phytochemistry. 2019;8(5):1762–1767. [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Makled S.M., Elkodosy E.F. Analytical study of Egyptian potato exports, the most important markets of the European Union. Arab Universities Journal of Agricultural Sciences. 2018;26(1):61–73. doi: 10.21608/ajs.2018.13866. [DOI] [Google Scholar]
- Malko A., Frantsuzov P., Nikitin M., Statsyuk N., Dzhavakhiya V., Golikov A. Potato pathogens in Russia’s regions: an instrumental survey with the use of real-time PCR/RT-PCR in matrix format. Pathogens. 2019;8(1):18–32. doi: 10.3390/pathogens8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiha N.A., van Bruggen A.H., van Diepeningen A.D., de Vos O.J., Termorshuizen A.J., Tjou-Tam-Sin N.N.A., Janse J.D. Potato brown rot incidence and severity under different management and amendment regimes in different soil types. European Journal of Plant Pathology. 2007;119(4):367–381. doi: 10.1007/s10658-007-9167-z. [DOI] [Google Scholar]
- Mouzo D., Bernal J., López-Pedrouso M., Franco D., Zapata C. Advances in the biology of seed and vegetative storage proteins based on two-dimensional electrophoresis coupled to mass spectrometry. Molecules. 2018;23(10):2462–2490. doi: 10.3390/molecules23102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutimawurugo M.C., Wagara I.N., Muhinyuza J.B., Ogweno J.O. Virulence and characterization of isolates of potato bacterial wilt caused by Ralstonia solanacearum (Smith) in Rwanda. African Journal of Agricultural Research. 2019;14(6):311–320. doi: 10.5897/AJAR2018.13686. [DOI] [Google Scholar]
- Pardo J.M., López-Alvarez D., Ceballos G., Alvarez E., Cuellar W.J. Detection of Ralstonia solanacearum phylotype II, race 2 causing Moko disease and validation of genetic resistance observed in the hybrid plantain FHIA-21. Tropical Plant Pathology. 2019;44(4):371–379. doi: 10.1007/s40858-019-00282-3. [DOI] [Google Scholar]
- Pavlova A.S., Dyudeeva E.S., Kupryushkin M.S., Amirkhanov N.V., Pyshnyi D.V., Pyshnaya I.A. SDS-PAGE procedure: Application for characterization of new entirely uncharged nucleic acids analogs. Electrophoresis. 2018;39(4):670–674. doi: 10.1002/elps.201700415. [DOI] [PubMed] [Google Scholar]
- Poudel N.S., Neupane S. Bacterial Diseases of Plants in Nepal: A Review. Asian Journal of Agricultural and Horticultural Research. 2018;2(1):1–10. doi: 10.9734/AJAHR/2018/42455. [DOI] [Google Scholar]
- Pradhanang P.M., Elphinstone J.G., Fox R.T.V. Sensitive detection of Ralstonia solanacearum in soil: a comparison of different detection techniques. Plant Pathology. 2000;49(4):414–422. doi: 10.1046/j.1365-3059.2000.00481.x. [DOI] [Google Scholar]
- Prior P., Fegan M. Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta. Hortic. 2005;695:127–136. doi: 10.17660/ActaHortic.2005.695.14. [DOI] [Google Scholar]
- Priya M., Siddique K.H.M., Dhankhar O.P., Prasad P.V., Rao B.H., Nair R.M., Nayyar H. Molecular breeding approaches involving physiological and reproductive traits for heat tolerance in food crops. Indian Journal of Plant Physiology. 2018;23(4):697–720. doi: 10.1007/s40502-018-0427-z. [DOI] [Google Scholar]
- Puaprasert K., Chu C., Saralamba N., Day N.P., Nosten F., White N.J., Imwong M. Real time PCR detection of common CYP2D6 genetic variants and its application in a Karen population study. Malaria Journal. 2018;17(1):427. doi: 10.1186/s12936-018-2579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R., Joshi A.A., Ghanekar M.P. Pseudomonads: major antagonistic endophytic bacteria to suppress bacterial wilt pathogen, Ralstonia solanacearum in the eggplant (Solanum melongena L.) World Journal of Microbiology and Biotechnology. 2009;25(1):47–55. doi: 10.1007/s11274-008-9859-3. [DOI] [Google Scholar]
- Ravelomanantsoa S., Vernière C., Rieux A., Costet L., Chiroleu F., Arribat S., Guérin F. Molecular epidemiology of bacterial wilt in the Madagascar highlands caused by Andean (Phylotype IIB-1) and African (Phylotype III) brown rot strains of the Ralstonia solanacearum species complex. Frontiers in Plant Science. 2018;8:2258–2275. doi: 10.3389/fpls.2017.02258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet K.A. The occurrence of bacterial wilt of potatoes caused by Pseudomonas solanacearum (EF Smith) EF. Sm. in Egypt. Min. of Agric., Extension Dept. Tech. Bull. 1961;112:116–119. [Google Scholar]
- Sagar V., Jeevalatha A., Mian S., Chakrabarti S.K., Gurjar M.S., Arora R.K., Singh B.P. Potato bacterial wilt in India caused by strains of phylotype I, II and IV of Ralstonia solanacearum. European Journal of Plant Pathology. 2014;138(1):51–65. doi: 10.1007/s10658-013-0299-z. [DOI] [Google Scholar]
- Santiago T.R., Lopes C.A., Caetano-Anollés G., Mizubuti E.S.G. Phylotype and sequevar variability of Ralstonia solanacearum in Brazil, an ancient centre of diversity of the pathogen. Plant Pathology. 2017;66(3):383–392. doi: 10.1111/ppa.12586. [DOI] [Google Scholar]
- Serag A.M., Salim T.M., Farid M.A., Elsisi A.A. Molecular Characteristics of Ten Ralstonia solanacearum Strains of Brown Rot Disease in Potato from three Governorates in Egypt. Journal of Agricultural Chemistry and Biotechnology. 2020;11(1):29–37. doi: 10.21608/jacb.2020.73048. [DOI] [Google Scholar]
- Shahbaz M.U., Mukhtar T., Ul-Haque M.I., Begum N. Biochemical and serological characterization of Ralstonia Solanacearum associated with chilli seeds from Pakistan. Int. J. Agric. Biol. 2015;17:31–40. [Google Scholar]
- Shimira, F., Afloukou, F., Maniriho, F., 2020. A review on challenges and prospects of potato (Solanum tuberosum) production systems in Rwanda. Journal of Horticulture and Post-harvest Research, 3(Special Issue-Abiotic and Biotic Stresses), 97-112. DOI: 10.22077/jhpr.2020.2854.1099.
- Shivani, R., 2016. Bacterial wilt disease of potato caused by Ralstonia Solanacearum: Plant disease.
- Sneedecor G.W., Cochran W.G. Statistical methods. Oxford and J. PJ. Publishing Com. 1980 [Google Scholar]
- Sogbohossou E.D., Achigan-Dako E.G., Maundu P., Solberg S., Deguenon E.M., Mumm R.H., Schranz M.E. A roadmap for breeding orphan leafy vegetable species: a case study of Gynandropsis gynandra (Cleomaceae) Horticulture research. 2018;5(1):1–15. doi: 10.1038/s41438-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanoghe M., Marique T., Rivière J., Lanterbecq D., Gadenne M. Investigation and development of potato parentage analysis methods using multiplexed SSR fingerprinting. Potato Research. 2015;58(1):43–65. doi: 10.1007/s11540-014-9271-3. [DOI] [Google Scholar]
- Sujeet K., Kedarnath N.H., Ramanjini P.H.G., Rohini I.B., Rangaswamy K.T., Achari R. Isolation and characterization of Ralstonia solanacearum causing bacterial wilt of solanaceae crops. International Journal of Current Microbiology and Applied Sciences. 2017;6(5):1173–1190. doi: 10.31018/jans.v10i3.1747. [DOI] [Google Scholar]
- Timila R.D., Manandhar S. Biovar Differentiation and Variation in Virulence of Ralstonia solanacearum Isolates Infecting Solanaceous Vegetables. Journal of Nepal Agricultural Research Council. 2016;2:22–26. doi: 10.3126/jnarc.v2i0.16117. [DOI] [Google Scholar]
- Uwamahoro F., Berlin A., Bucagu C., Bylund H., Yuen J. Ralstonia solanacearum causing potato bacterial wilt: host range and cultivars’ susceptibility in Rwanda. Plant Pathology. 2020;69(3):559–568. doi: 10.1111/ppa.13140. [DOI] [Google Scholar]
- Wei Y., Moreno C.C., Gongora T.J., Wang K., Sang Y., Duran R.L., Macho A.P. The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the solanaceae family. Journal of Plant Biotechnology. 2018;16(7):1–14. doi: 10.1111/pbi.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß, Christian., 2007. AStA Advances in Statistical Analysis. tatSoft, Inc., Tulsa, OK.: STATISTICA, Version 8. 91(3),339-341. DOI 10.1007/s10182-007-0038-x.
- Williamson L., Nakaho K., Hudelson B., Allen C. Ralstonia solanacearum race 3, biovar 2 strains isolated from geranium are pathogenic on potato. Plant Dis. 2002;86:987–991. doi: 10.1094/PDIS.2002.86.9.987. [DOI] [PubMed] [Google Scholar]
- Zhang H., Mittal N., Leamy L.J., Barazani O., Song B.H. Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evolutionary Applications. 2017;10(1):5–24. doi: 10.1111/eva.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T., Chen, D., Li ,C., Sun, Q., Li, L., Liu, F., ... Shen, B., 2012. Isolation and characterization of Pseudomonas brassicacearum J12 as an antagonist against Ralstonia solanacearum and identification of its antimicrobial components. Microbiological research, 167(7), 388-394. https://doi.org/10.1016/j.micres.2012.01.003. [DOI] [PubMed]