Abstract
The ongoing Covid-19 pandemic has spurred research in the biology of the nidovirus severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Much focus has been on the viral RNA synthesis machinery due to its fundamental role in viral propagation. The central and essential enzyme of the RNA synthesis process, the RNA-dependent RNA polymerase (RdRp), functions in conjunction with a coterie of viral-encoded enzymes that mediate crucial nucleic acid transactions. Some of these enzymes share common features with other RNA viruses, while others play roles unique to nidoviruses or CoVs. The RdRps are proven targets for viral pathogens, and many of the other nucleic acid processing enzymes are promising targets. The purpose of this review is to summarize recent advances in our understanding of the mechanisms of RNA synthesis in CoVs. By reflecting on these studies, we hope to emphasize the remaining gaps in our knowledge. The recent onslaught of structural information related to SARS-CoV-2 RNA synthesis, in combination with previous structural, genetic and biochemical studies, have vastly improved our understanding of how CoVs replicate and process their genomic RNA. Structural biology not only provides a blueprint for understanding the function of the enzymes and cofactors in molecular detail, but also provides a basis for drug design and optimization. The concerted efforts of researchers around the world, in combination with the renewed urgency toward understanding this deadly family of viruses, may eventually yield new and improved antivirals that provide relief to the current global devastation.
Keywords: Backtracking, Coronavirus, NiRAN domain, Proofreading, Remdesivir, Replication, RNA capping, RNA synthesis, RNA-dependent RNA polymerase, SARS-CoV-2
1. Introduction
The ongoing Covid-19 pandemic has claimed millions of lives and disrupted countless more as of April 2021, spurring interest in understanding the lifecycle of the causative agent: the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Prior to the Covid-19 pandemic, CoVs were responsible for numerous pathogenic events in both avian and mammalian species, the most notable being the 2003 SARS-CoV and 2012 MERS-CoV outbreaks [1]. Such events exemplified the threat of zoonotic outbreaks from circulating viruses in reservoir species like bats. However, these threats were largely unheeded until the Covid-19 pandemic unfolded and brought the world to a standstill. An improved understanding of the virus life cycle will be crucial to combating this current (and future) pandemics. Pivotal to viral propagation is the replication of the viral genome and transcriptiona of viral genes necessary to form new infectious viral particles. Here, we provide an overview on CoV replication and transcription, with a focus on recent results elucidating the enzyme mechanisms involved.
In addition to the family Coronaviridae, the order Nidovirales comprises the families Arteriviridae, Mesoniviridae, and Roniviridae [2]. Prior to the SARS-CoV outbreak in 2003, early studies focused on the Arteriviridae family, resulting in the characterization of the genome organization of nidoviruses as well as identifying the function and essentiality of many viral genes [3]. These early efforts set the platform for the current torrent of studies addressing major gaps in our understanding of the causative agent of the Covid-19 pandemic.
An essential element of the viral lifecycle involves the replication and transcription of the viral RNA genome. The central enzyme of the replication/transcription process is the RNA-dependent RNA polymerase (RdRp) [4]. At approximately 30 kb in length, the CoV family encodes among the largest genomes of RNA viruses [5]. To successfully replicate and transcribe its genome, CoVs evolved an arsenal of nonstructural proteins (nsps) to allow for enhanced processivity and proofreading during replication/transcription, protection against the host innate immune system, and translation. Many of these nsps function in concert with the RdRp in a so-called replication/transcription complex (RTC). Several of these processes mediated by the nsps are unique to the Nidoviral order or even to the CoV family. Understanding the underlying mechanisms involved in replication/transcription can facilitate the identification of novel pharmacological treatments to stymie future outbreaks caused by zoonotic transmission of circulating CoV viruses.
Pioneering studies characterizing CoV replication/transcription were driven by the 2003 SARS-CoV outbreak. These studies led to the identification of the factors responsible for replication/transcription along with functional roles for many of the factors. A focus of recent studies has been to unravel the structural basis by which these factors interact with their substrates and with each other, in the hope of detailing druggable pockets of essential enzymatic complexes. Herein, we will summarize what is known about the CoV replication/transcription machinery as revealed by biochemical and structural investigations, with the added aim of illustrating how furthering our understanding of the basic biology can aid the drug development process.
While the mechanisms of replication/transcription appear to be universal across the CoV family, most biochemical studies have used recombinant proteins of the pathogenic CoV family members SARS-CoV-1, MERS-CoV, or SARS-CoV-2. As such, our discussions will be drawn primarily from studies utilizing a subset of CoV family members, although extrapolations across the CoV family can be made where evidence exists for the model CoVs murine hepatitis virus (MHV) along with the less virulent strains human CoV-229E (HCoV-229E) and HCoV-NL63.
2. Coronavirus genome organization
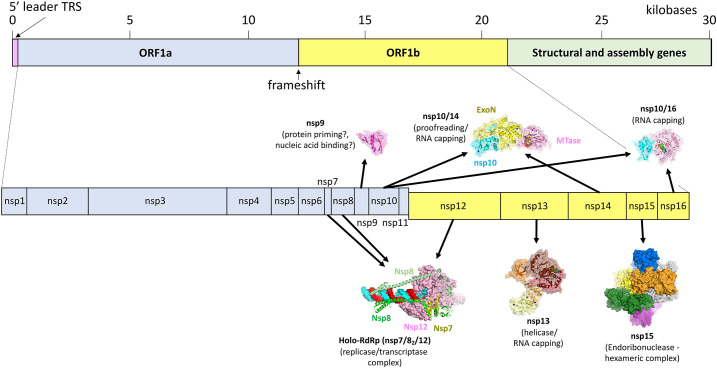
Inherent to our understanding of CoV replication/transcription is the organization of the CoV genome. CoV genomes are polycistronic, with the 5′ proximal two-thirds containing two overlapping ORFs (Orfs1a and 1b; Fig. 1 ) [11]. Orf1ab of SARS-CoV-2 encodes a polypeptide containing 16 nsps with diverse functions including host membrane reorganization, proteases, host immune system suppression, RNA capping, and replication/transcription of the viral genome. CoVs regulate the relative stoichiometries of the nsps through the presence of a -1-frameshift element (FSE) in between ORFs1a/1b, which enables the production of the ORF1a encoded nsps (nsps 1–11) at ~ 1.4–2.2 increased fold over the ORF1b nsps (nsps 12–16) [12]. Proximal to the genome 3′-end are genes encoding the structural proteins that package the viral RNA and form the viral particles.
Fig. 1.
Schematic depicting general organization of the Coronavirus genome. The top panel shows the layout of the two open reading frames (Orf1a and b) and the 3′ structural genes. The bottom panel illustrates that Orf1a and b are divided into 16 genes numbered nsp1–16. Surface rendered models are shown for nsp9 (PDB 6W9Q[6]), holo-RdRp (6XEZ [7]), nsp10/14 (5C8S [8]), nsp13 (6XEZ [7]), nsp15 (6VWW [9]) and nsp10–16 (6W4H [10]).
In this review, we focus on nsps with functions directly related to nucleic acid transactions directed toward replication of the viral genome and for correct processing of genomic and subgenomic transcripts. At least nine of the nsps encoded in ORF1ab play known or suspected roles in replication/transcription (Fig. 1), orchestrating replication of the entire positive-strand RNA genome via a negative-strand intermediate (continuous replication) as well as mediating the unique process of discontinuous subgenomic transcription (Fig. 2 ).
Fig. 2.
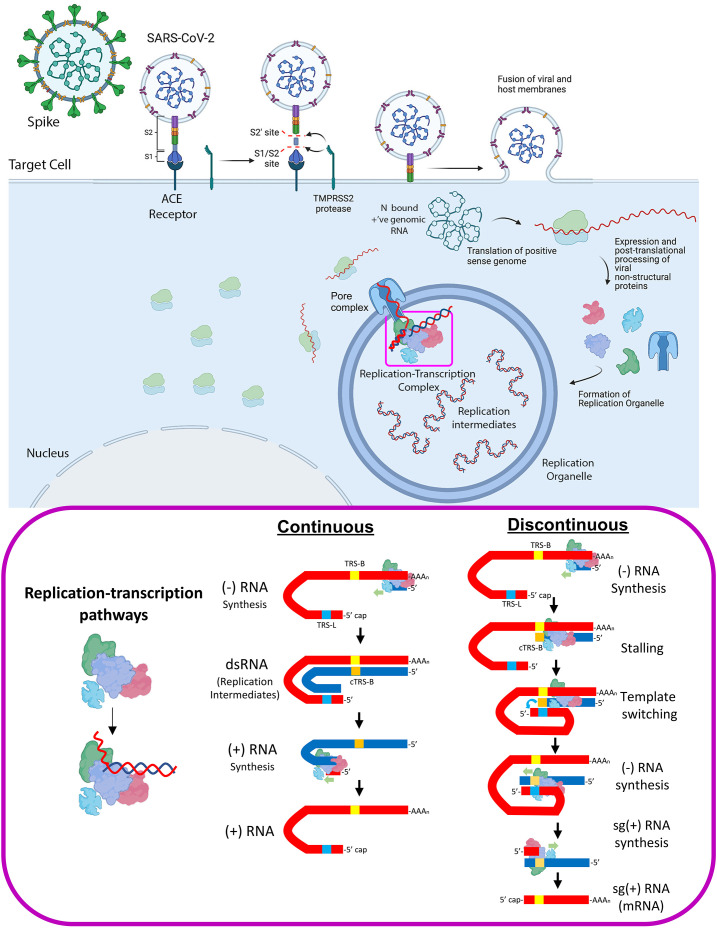
Cellular processes involved in SARS-CoV-2 infection. The interaction of the viral Spike protein with the host ACE receptor mediates cell entry. The packaged viral genome is translated to produce the viral nonstructural proteins that mediate the production of genomic RNAs in a specialized, viral induced double membraned compartment known as the Replication Organelle (RO). Top panel was created with BioRender.com. CoV RNA synthesis may be characterized as continuous (replication of full-length genomic RNA) or discontinuous (transcription or more appropriately described as subgenomic RNA synthesis) (bottom panel).
2.1. Continuous and discontinuous RNA synthesis
Although replication and transcription are performed by the same central enzyme, the viral RdRp, there are some fundamental differences to consider. The (+)-strand RNA genome serves as a template for: (i) translation of the nsp's, (ii) continuous replication to generate full-length (−)-strand copies of the genome, and (iii) discontinuous transcription (sg-transcription) to generate (−)-strand templates for subsequent synthesis of mRNAs encoding for the viral structural proteins (Fig. 2).
Sg-transcription includes a discontinuous step that involves a template-switching step unique to nidoviruses [13], producing sg-RNAs that are 5′- and 3′-coterminal with the virus genome (Fig. 2; for further reading see Ref. [14]). The current understanding of the template switching process is incomplete. The process is controlled by transcription-regulating sequences (TRSs); one TRS is located near the 3′-end of the positive-strand RNA genome 5′-leader sequence (TRS-L), and others precede each of the viral structural genes (TRS-B). Each TRS contains an identical core sequence of 6–7 nucleotides [15], [16], [17], [18].
The process begins when the RdRp initiates transcription from the 3′-poly(A) tail of the positive-strand RNA genome. Transcription proceeds until the RdRp transcribes a TRS-B, where the RdRp stalls due to an unknown mechanism [15]. At this point, two scenarios are possible, the RdRp can continue elongating until the next TRS-B is encountered or the RdRp can switch transcription templates resulting in transcription of the positive-strand RNA genome 5′-leader (Fig. 2). The 5′- and 3′-ends of the positive-strand RNA genome are known to interact [19]. Specifically, the TRS-L is thought to be held in close proximity to the TRS-B sequences, probably assisted by unknown RNA-protein complexes. Template switching requires base pairing between the TRS-L [on the (+)-strand RNA genome] and a complementary (−)-strand sequence resulting from transcription of the TRS-B (cTRS-B) [16], [17], [18], [20]. The 3′-end of the cTRS-B sequence hybridized to TRS-L then serves as a primer for continued extension of the (−)-strand RNA, but with the 5′-leader as the new template. These nested RNAs then serve as the templates for (+)-strand sg-mRNAs synthesis.
Erroneous template switching has been observed with the aid of direct RNA sequencing approaches, pointing to the potential for dysregulated sg-transcription that leads to gene recombination [21], [22]. Although most notably leading to the formation of nonviable viral particles [22], rare recombination events may be involved in CoV evolution as postulated in the acquisition of a furin cleavage site in the SARS-CoV-2 spike (S) protein [23], [24]. Template switching at non-TRS sites may also promote the overexpression of Orf1a encoded genes as evident from the abundance of transcripts containing in frame nsp1-nsp2(NTD)-N open reading frames. With up to 30% of all sg-transcripts represented by noncanonical fusions, their presence may skew the production of viral proteins nsp1 and N, which serve essential roles in infectivity [21]. As subsequent sections will focus on the canonical TRS-dependent pathway, we will not discuss TRS-independent template switching further. However, these intramolecular fusions may have an under-studied importance during infection, with potential repercussions for zoonosis.
2.2. Subcellular compartmentalization of replication and transcription
Continuous and discontinuous replication of the viral genome would be futile if the newly produced viral RNAs were exposed to host RNA decay mechanisms. A major feature of CoV infection is the formation of a network of double membrane lipid vesicles (DMVs) that exist as interconnected appendages to host organelles or as separate compartments in the cytosol [25], [26]. As the site of RNA synthesis, it is postulated that DMVs exist to both protect the newly synthesized viral RNAs as well as to provide a platform to coordinate the localization of the RTC, supporting the DMVs role as a replication organelle (RO; Fig. 2).
A combination of fluorescence microscopy, negative stain electron microscopy, and cryo-electron tomography (cryo-ET) studies revealed the presence of double-stranded viral RNA in the RO, illustrating the role of the RO as a repository of replication/transcription substrates, intermediates, and products [25], [27], [28]. In particular, cryo-ET datasets collected on MHV and SARS-CoV-2 infected cells show the double-stranded nature of the RNA as well as identified branched structures that may represent intermediates of sg-transcription [28]. These studies also identified a proteinaceous pore in the RO membrane that co-localizes with RTC components [27], [29], [30], [31].
Critical for the localization of the RTC in the RO are three components embedded in the DMV membrane, nsps 3, 4, and 6 [32], [33]. Their presumed role in RO biogenesis was confirmed upon observing that ROs formed when nsps 3, 4, and 6 were ectopically expressed in mammalian cell culture [34]. It is unknown whether these subunits commandeer host factors and pathways to establish the RO. CRISPR based screens indicate a stringent requirement of the cholesterol biosynthetic pathway for SARS-CoV-2 infectivity, which may indicate the utilization of cholesterol stores for RO biogenesis [35], [36]. In addition, the host secretory and autophagy pathways that mediate ER membrane remodeling may be subverted during infection to trigger the production of the ROs, as indicated by the colocalization of the autophagy marker LC3 with viral replicase proteins during infection by other DMV forming viruses [37], [38]. Further experiments will hopefully define the repertoire of viral and host factors needed to form the RO, which will improve our understanding of the CoV infection process.
The application of cryo-ET revealed that nsp3 is assembled as a hexameric ring with a solvent accessible pore at its center that enables transport into and out of the RO [27], [39]. Additionally, nsps 2, 4, and 6 appear to anchor RTC components and host factors to the DMV membrane surface as evident from proximity based labeling studies [29]. Tethered at the ROs pore, the RTC may be able to direct newly synthesized viral RNAs to the cytosol for translation. By storing replication intermediates and products in the DMV lumen, CoVs mitigate decay by the host nonsense mediate decay (NMD) pathway prior to completion of the viral post-transcriptional processing events, such as capping and proofreading [40]. It is unclear how the double-stranded RNAs observed in the DMVs are separated so that the newly synthesized (+)-stranded genomes can be exported to the cytosol for packaging into new viral particles; the fate of the (−)-strand intermediates is also unknown.
3. The RNA-dependent RNA polymerase
3.1. The holo-RdRp, the central enzyme for replication/transcription
Integral to both replication and transcription is nsp12, encoding for the viral RdRp, which catalyzes the synthesis of new RNAs. We call this the core RdRp or nsp12. Nsp12 functions in combination with the viral cofactors nsp7 and two copies of nsp8 in the holo-RdRp (nsp7/82/12) [4], [41]. Initial biochemical characterization of nsp12 revealed that it inefficiently catalyzed the elongation of a primer/template RNA scaffold in the absence of the cofactors nsp7 and nsp8 [41]. The presence of both cofactors dramatically increased the elongation activity which instigated comparisons between the cofactors and the eukaryotic PCNA sliding clamp [41], [42], [43]. In addition to its role in processivity, nsp8 is known to mediate interactions between other viral nsps and the holo-RdRp, acting as a “hub” of the RTC interactome [32], [44]. Interactions have been identified between nsp8 and the viral helicase, exonuclease, capping complex, as well as with a plethora of host proteins. Pivotal to nsp8 function is the binding interface with nsp7/12 and RNA in which loss of function point mutations lead to pronounced replication defects for both model CoVs and SARS-CoV in plaque assays [41].
While structural and biochemical evidence has cemented the role of nsp7/8 as nsp12 cofactors [41], [45], early studies following the 2003 SARS-CoV outbreak postulated that nsp7/8 may act as a primase [46], [47], [48]. Nsp8 was suggested to function as an RdRp with de novo initiating capability but very low processivity. This activity was proposed to prime RNA synthesis for subsequent extension by the holo-RdRp, which has weak de novo initiation activity of its own [41], [46]. This observation gained traction from crystal structures of nsp7/8, which form oligomeric assemblies in crystal packing environments [48], [49]. The nsp7/8 assemblies contain positively charged grooves or channels, suggesting RNA interacting determinants. Subsequent studies have not always confirmed template-directed primase activity for nsp7/8 [41], [50]; instead Tvarogova et al. [50] observed a 3′-terminal adenylyltransferase activity (TATase) for nsp8 that was suggested to polyadenylate the 3′-end of positive-strand RNAs. Due to: (i) the contradictory functional observations, (ii) the nsp7/8 sequences lacking characteristic sequence motifs of enzyme families known to catalyze RNA polymerase or adenylyltransferase reactions, and (iii) the structures of the nsp7/8 oligomeric assemblies lacking putative sites with characteristics of enzyme active sites of these enzyme families [48], [49], it must be considered that the nsp7/8 oligomeric assemblies observed in crystal structures may be crystallization artifacts. Thus, the primase model is difficult to reconcile with the conformations of nsp7/8 observed in recent single particle cryo-EM datasets, and biochemical data supporting this model remains inconclusive [7], [42], [45], [50], [51]. While in vivo genetic evidence points to the presence of nsp8 at the 3′ UTR, a point used to justify the primase model, the role of nsp8 in priming remains to be verified [52], [53].
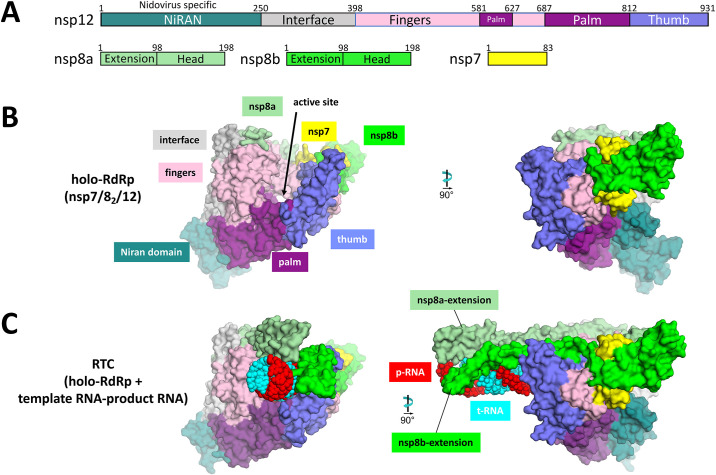
Our understanding of the structural basis for the enhanced processivity of the holo-RdRp complex expanded with the advent of cryo-EM. Seminal structural work was conducted by Kirchdoerfer and Ward, who obtained the cryo-EM structure of the SARS-CoV holo-RdRp complex in the absence of an RNA substrate [45]. The structure revealed that, similar to other viral RdRps, the nsp12 polymerase domain (amino acids 398–932) resembles a cupped right hand comprising a fingers subdomain (amino acids 398–581, 628–687), a palm subdomain (amino acids 582–627, 688–815) and a thumb subdomain (amino acids 816–919) (Fig. 3 ) [55], [56]. The structure also showed the disposition of the nsp7/8 cofactors; nsp7 associated with the nsp8 C-terminal domain (CTD) as a heterodimer, interacting with the nsp12 thumb subdomain and index fingers proximal to the NTP-entry tunnel, and a second nsp8-CTD associated with the fingers subdomain. Notably, the nsp8 N-terminal extensions were disordered and not observed in the absence of an RNA substrate (Fig. 3).
Fig. 3.
(A) Schematic illustrating the structural domains of the holo-RdRp subunits nsp7, 8 and 12. (B) View of the apo holo-RdRp showing the “cupped right hand” structure of nsp12 (left). A 90° rotation reveals the NiRAN domain and nsp7/8 heterodimer interface with nsp12 (right). (C) The nsp8 N-terminal helices are ordered in the presence of an upstream RNA duplex substrate. Figures in panels A–C were generated using PDB 6XEZ[7]. The original apo holo-RdRp structure was from SARS-CoV-1 (6NUR) [45]. Published RNA bound Holo-RdRp structures include 6YYT [42] and 7BV2 [54].
3.2. The RdRp active site
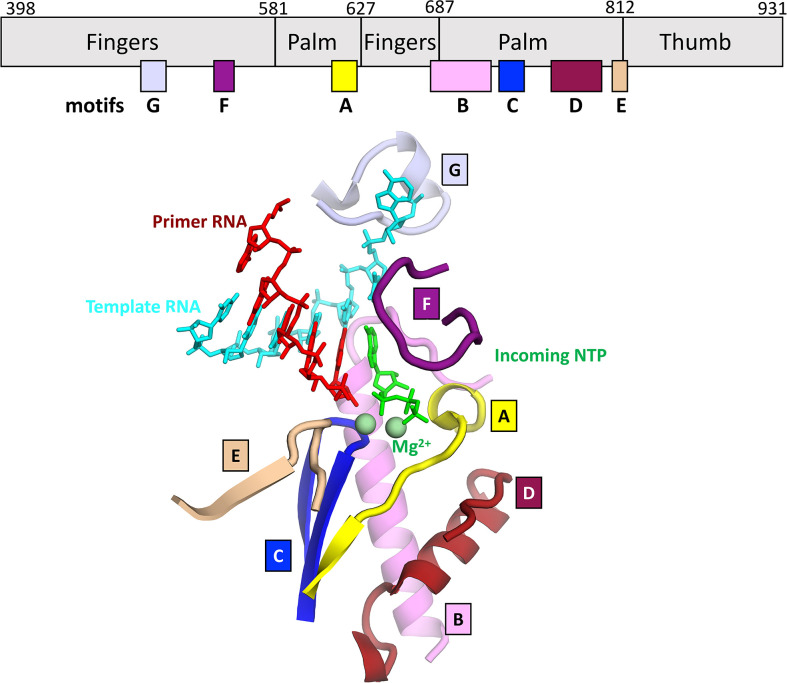
The active site features the conserved structural motifs (A–G) that are common to all viral RdRps and mediate nucleotide binding and catalysis [57], [58], [59] (Fig. 4 ). Nucleotide binding occurs via direct interactions with residues in motifs A, B, C, and F that are further stabilized by motifs D and E. The key motif “SDD,” which coordinates two Mg2 +-ions both required for catalysis [61], is positioned in motif C [41], [61]. Mutating either Asp residue (760 or 761) results in complete loss of the replicase activity and viral infectivity [41]. The incoming NTP diffuses through the NTP-entry tunnel while maintaining nonspecific interactions with motifs E and F. Molecular Dynamics simulations performed with the HCV RdRp suggest that the NTP enters “phosphate first,” reaching the mouth of the active site, at which point the base flips into the active site cleft [62]. Following localization to the active site cleft, ribonucleotides are selected via 3′-OH interactions with motif A residue D623 and 2′-OH interactions with motif B residues T680 and N691, rendering the RdRp specific for RNA synthesis. A conformational change of the motif F loop may orchestrate the correct positioning of the + 1-template base with the incoming NTP through template-base interactions with V557 and NTP interactions with K545, K551 and R553. The incoming NTP coordinates the two metal ions with the catalytic Asps through its α- and β-phosphates. The Mg2 + coordination enhances the nucleophilicity of the nascent RNA 3′-OH which facilitates the attack on the NTP α-phosphate. Residue R553 may act as a general acid that facilitates the release of the pyrophosphate through protonation of the α-phosphate, as described for the poliovirus RdRp [63]. Structural evidence, based on a preinsertion HCV RdRp structure (PDB 4WTA), indicates that HCV-R158 (homologous to R553) interacts with the incoming NTP α-phosphate; potentially facilitating its protonation [60]. Capturing a snapshot of the CoV RdRp in a similar pre-insertion state is of immediate interest for the design of nucleotide analog inhibitors whose use will be discussed in Section 5.
Fig. 4.
Active site of the RdRp, comprising seven conserved sequence motifs denoted as motifs A–G (top). Motifs A–F mediate nucleotide recognition, active site orientation of the NTP, and catalysis. Motif G regulates RNA translocation [46]. The structure shown here is a model derived from SARS-CoV-2 nsp12 with t-RNA and p-RNA (PDB 6YYT[42]), but the 3′-nucleotide of the p-RNA, the incoming NTP, and the two metal ions were modeled with the help of PDB 4WTJ[60].
3.3. The replication/transcription complex
At the onset of the COVID-19 pandemic, several structures were determined in quick succession that established the structural basis of RNA recognition by the holo-RdRp [42], [51], [54]. Here, we call the holo-RdRp-RNA complex the replication/transcription complex (RTC). In the presence of a product-RNA strand (p-RNA) which forms an RNA-RNA duplex with the template-RNA strand (t-RNA), N-terminal helical extensions of each nsp8 monomer reach back and interact with the upstream duplex RNA like “sliding poles” (Fig. 3) [42]. These contacts increase the stability of the RTC and explain the deleterious effects of previously observed point mutations such as nsp8 K58A [41]. Deciphering the network of protein–RNA interactions in the RTC provides new perspectives into how functional motifs are modulated by RNA binding, which is important for structure-based drug design.
3.4. The nsp12-NiRAN domain
Nidoviral RdRps uniquely possess an N-terminal nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain that catalyzes an essential nucleotidylation activity [53], [64]. The NiRAN domain is a genetic marker unique to nidoviruses [64]. Biochemical experiments with the equine arteritis virus (EAV) RdRp nsp9 (homolog of SARS-CoV-2 nsp12) discovered the titular nucleotidylation activity and prompted further analysis to gauge how widespread the activity is in nidoviruses. Comparative sequence analysis characterized three putative NiRAN domain sequence motifs (AN, BN, CN) likely mediating nucleotide binding and nucleotidylation activity. Using both EAV and SARS-CoV, loss of function mutations crippled viral replication in cell culture infectivity assays. Such mutant constructs lacked the in vitro nucleotidylation activity, thereby demonstrating its importance for viral replication. In particular, the EAV NiRAN domain utilizes UTP as its preferred substrate with a lower specificity toward GTP [64]. These early observations sparked intense discussions focused on the NiRANs potential role as an RNA ligase that could utilize ATP or as a Guanylyltransferase (GTase) that could act in the RNA capping pathway. We will address the potential NiRAN domain role in capping in a later section. A third hypothesis, accounting for the preference for UTP, proposed that the nucleotidylation activity plays a role in protein dependent replication/transcription priming at the 3′ UTR poly(A) tail [53].
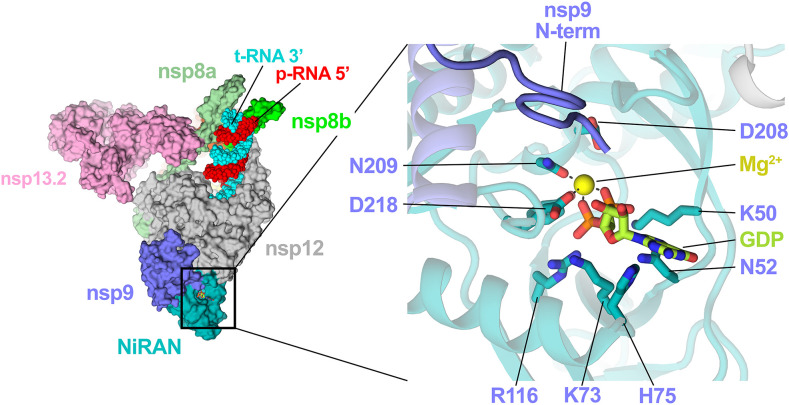
Recent structural work utilizing the SARS-CoV-2 RdRp characterized the NiRAN nucleotide binding pocket in detail [7] and revealed a potential binding interface for nsp9 [65] (Fig. 5 ). The latter study revealed the nsp9 N-terminus snaking into the NiRAN active site and identified nsp9 as an inhibitor of NiRAN function [65]. However, the finding that nsp9 inhibited NiRAN function was confounded by the presence of a two amino acid N-terminal scar from the nsp9 purification tag [65]. Ziebuhr and colleagues subsequently demonstrated that the NiRAN domain enzymatic activity efficiently transfers UMP to the native nsp9 N-terminus through a phosphoramidate bond in vitro [53], an activity termed UMPylation. GMPylation of the nsp9 N-terminus was observed somewhat less efficiently while AMPylation and CMPylation were not observed.
Fig. 5.
Nsp9 acts as a substrate for the nsp12 NiRAN. The nsp9 N-terminus snakes into the NiRAN active site proximal to the α/β phosphates of the bound nucleotide (GDP in this case). Conserved NiRAN catalytic residues are depicted as sticks (right inseam). This figure was generated using PDB 7CYQ[65].
The analysis by Slanina et al. [53] indicated that the nsp9 N-terminal tripeptide motif NNE was required for nsp9 to act as a substrate; Alanine or Serine substitutions at residues 1 and 3 mitigated the activity while position 2 mutants fully ablated the in vitro UMPylation [53]. These mutations hampered HCoV-229E viral replication in cell culture, highlighting that the native nsp9 N-terminus is essential during the viral lifecycle [53]. With the preference for UTP shared between EAV and HCoV-229E NiRAN domains, the authors favor the hypothesis that nsp9 UMPylation may be involved in a protein-dependent priming mechanism analogous to the role of VPg in picornaviral replication [66]. In vitro results by Yan et al. [65] demonstrated that the NiRAN GTase activity guanylates the 5′-end of RNA substrates [65]. Importantly, this reaction required the presence of nsp13 which would serve to remove the 5′-γ-phosphate prior to the guanylation, suggesting relevance to the RNA capping mechanism.
4. Concerted enzymatic functions vital to replication/transcription
The holo-RdRp is the central enzymatic complex of the replication/transcription process. Other nsps involved in nucleic acid transactions (Fig. 1) mediate essential processes of their own which likely occur in coordination with the holo-RdRp. Protein–protein interaction studies suggest that constituents of the holo-RdRp may physically interact with other viral RTC subunits. Nsp8 has been shown to interact with nsp9, nsp10, nsp13, nsp14, nsp15 and nsp16 in pull-down assays based on a reticulocyte lysate in vitro transcription/translation system in which nsp12 was also characterized to interact with the above RTC subunits [32], [44]. A potential caveat is that insufficient data are present to rule out that these interactions occur indirectly through nucleic acid binding. Subsequent work expanded on the functional significance of some of these interactions; for example, nsp12 can enhance the viral nsp13 helicase unwinding activity [67], suggesting a physical interaction. Still to be discerned in detail is how the exonuclease, nsp14, functions in conjunction with the RdRp during proofreading [68]. Also, of great interest would be to elucidate how the separate nsps mediating individual enzymatic steps of RNA capping function in concert with each other to regulate this process [69], [70]. Given the essentiality of both proofreading and capping for viral fitness, elucidating the underlying mechanisms will be informative in exploring the druggability of both pathways as well as revealing the mechanisms particular to CoVs. Below, we describe what is known about the molecular basis underlying these pathways.
4.1. Proofreading and nsp14 ExoN activity
A combination of comparative sequence and mutational analyses of nsp14 led to the prediction that it contained an exonuclease (ExoN) domain at its N-terminus, linked to the C terminal MTase domain [71]. The MTase domain has been implicated in the capping process that will be discussed later. The ExoN function of nsp14 is unique to nidoviruses having a genome size > 20 kb, being notably absent in the Arterivirdae family with genome sizes of ~ 13–16 kb [5]. This suggested a link between genome size and the requirement for a proofreading machinery [5]. Initial genetic evidence indicated that loss of function mutations in the ExoN active site were deleterious for viral replication but yielded viable progeny [72], [73]. Bouvet et al. [74] reconstituted SARS-CoV nsp14 biochemical activity, highlighting a 3′–5′ ExoN activity on a forked dsRNA substrate that is enhanced in the presence of its cofactor nsp10. In other studies, it was observed that loss of ExoN function led to mutator phenotypes and/or enhanced susceptibility to nucleotide analogues such as 5-Fluorouracil and Ribavirin [68], [75], [76], [77]. Both analogues are incorporated into RNA chains by the RdRp and act as chain terminators, indicating that the nsp14 ExoN activity can excise misincorporated nucleotides from nascent RNA strands. A recent report found that the set of nsp14 ExoN inactivating mutations that were deleterious but nonlethal for Murine Hepatitis Virus (MHV) and SARS-CoV [76] led to nonviable viral progeny in SARS-CoV-2 and MERS-CoV [72]. Interestingly this suggests that some β-CoVs utilize the ExoN in an essential aspect of the viral lifecycle other than proofreading, or that the requirement for proofreading is more stringent in SARS-CoV-2 and MERS-CoV and as such these latter viruses are more sensitive to ExoN inhibition.
The contrasting effects of loss of function mutations in β-CoV family members coincide with subtle differences in the reconstituted biochemical activity. While bulkier adducts are excised inefficiently by the SARS-CoV complex as is evident through the protection conferred by the presence of puromycin at the RNA 3′ end [74], in studies utilizing reconstituted SARS-CoV-2 nsp10–14, the excision of bulky moieties like biotin was observed in an apparent endoribonucleolytic cleavage [78]. The authors concluded that these discrepancies may be a result of differences in the handling and storage of the purified enzymes although the difference may amount to intrinsic enzymatic differences. Characterizing the respective enzymatic activities across the spectra of the CoV family will help unravel the role of the exonuclease in the viral lifecycle. Remaining key questions focus on how the signal from the mis-incorporated product is propagated through the RTC to the exonuclease complex. There are two schools of thought on this process; one mandates that the nascent primer strand must be transferred directly from the active site of the RdRp to the active site of the exonuclease whereas the second mechanism argues that nascent strand could be extruded through a periphery channel on the RdRp to the mouth of the exonuclease active site. Both postulations are precedented given similar mechanisms demonstrated for other DNA and RNA polymerases. The former mechanism is best described for phage DNA polymerases such as T4, T7 and phi29 that are known to transfer the primer strand approximately 40 Å or more to the ExoN domain encoded at the polymerase N-terminus [79], [80], [81]. The basis of the latter mechanism is detailed in Section 4.3.
Both postulations are based on the reasoning that the 3′-p-RNA is sterically inaccessible to the nsp14 active site when engaged with the RdRp active site. As such, either the RdRp must dissociate when stalled or the p-RNA needs to be repositioned to allow for nsp14 engagement. Single molecule experiments that track the behavior of the RdRp during elongation will enhance our understanding in relation to whether the RdRp dissociates following nucleotide misincorporation or if it remains bound in an arrested state.
4.2. Nsp13: An essential CoV helicase
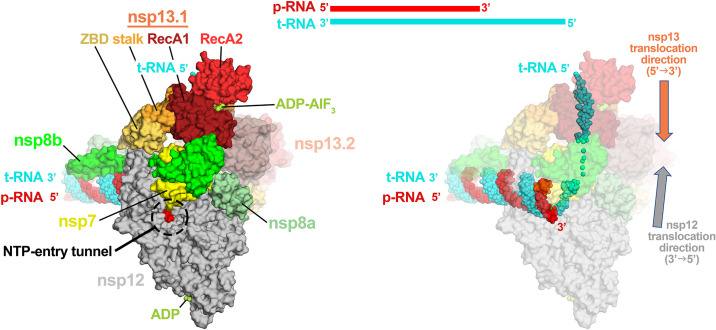
Inherent to the mechanism of proofreading may be role of the viral helicase, nsp13. Nsp13 is a superfamily 1B (SF1B) helicase that mediates nucleic acid unwinding with a 5′ → 3′ polarity [82], [83], [84], [85]. Nsp13 can unwind both dsDNA and dsRNA substrates that have a 5′ single stranded overhang in an NTP dependent manner [67], [86], [87]. Besides its role as a helicase, nsp13 harbors RNA 5′-triphosphatase activity that is thought to play a role in viral RNA capping [69], [88], [89]. The helicase is essential for replication in the nidovirus EAV [90], [91], [92] and in the β-CoV MHV [93], and is postulated to be essential in all nidoviruses [4], [83], [89], [92]. As such, it is a promising pan-CoV drug target. With early work showing a twofold enhanced unwinding rate in the presence of the RdRp [67], there was evidence that the ternary RdRp-nsp13 complex, if characterizable, would be functionally significant. Recent work determined that nsp13 could form a stable complex with the RTC in which cryo-EM structures revealed that nsp13 forms interactions with nsp7, nsp8, nsp12, and the t-strand RNA downstream of the RdRp active site [7], [65]. Surprisingly, two copies of nsp13 were bound to the RTC, with only one of the nsp13 protomers interacting with the t-strand RNA (Fig. 6 ).
Fig. 6.
Surface rendered model of the nsp13-RTC [7]. Nsp13 binds to the 5′ t-RNA strand downstream of the RdRp active site. Nsp13 structural domains are highlighted to indicate the domains involved in holo-RdRp and RNA binding (left). Nsp13 translocates on the t-RNA (cyan) in the 5′-3′ direction, whereas the RdRp translocates in the 3′-5′ direction on the t-RNA.
These structural results posed a conundrum since the RdRp and nsp13 have opposing translocation polarities (Fig. 6). The RdRp translocates on the t-strand RNA in the 3′ → 5′ direction whereas nsp13 bound to the t-strand RNA translocates in the 5′ → 3′ direction, leading to a translocation conflict. To reconcile this conundrum, Chen et al. [7] proposed that, under certain circumstances, the translocation activity of nsp13 may push the RdRp backwards on the RNA template. The backwards motion of cellular DNA-dependent RNA polymerases (DdRps), a process known as “backtracking,” has been well described. In DdRps, the 3′-end of the nascent RNA transcript forms a 9 or 10 base pair RNA-DNA hybrid with the t-strand DNA [94]. During backtracking, the RNA maintains base pairing register with the t-strand DNA, resulting in the RNA transcript reverse-threading through the complex. This activity disengages the RNA 3′-OH from the DdRp active site, generating a 3′-single stranded RNA fragment that extrudes out the DdRp NTP-entry channel [94], [95], [96], [97].
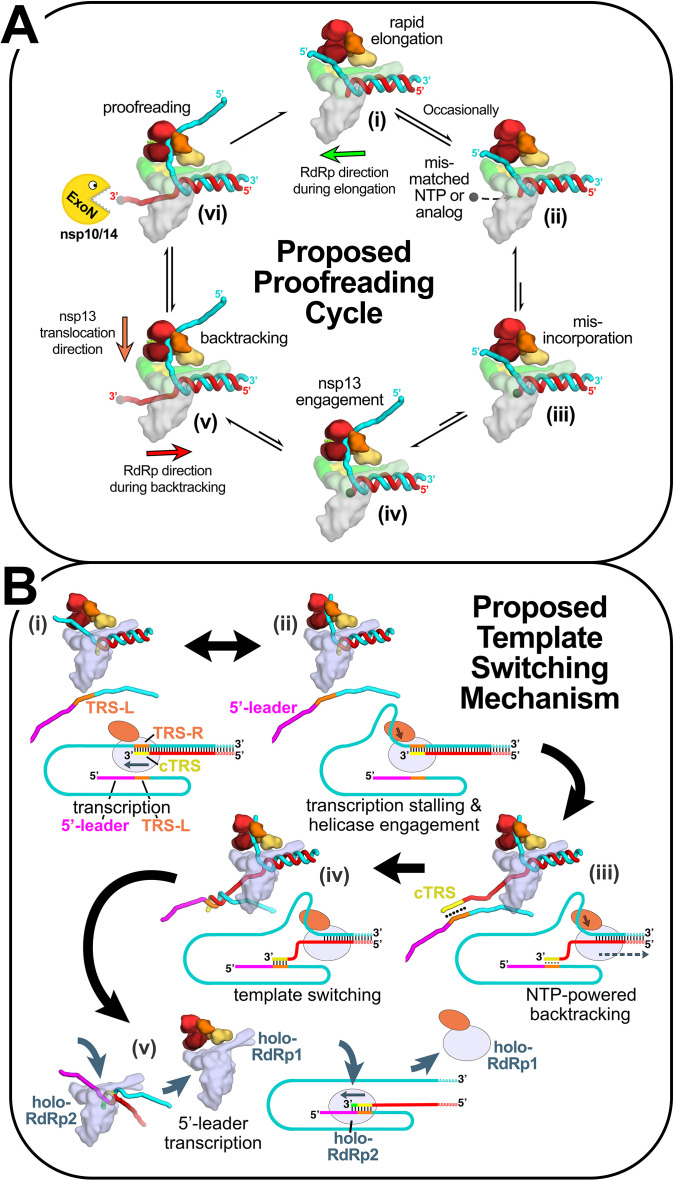
Although the cellular DdRps and viral RdRps are not evolutionarily related, their active sites have architectural similarities [7]. Like the DdRp NTP-entry tunnel, the viral RdRp NTP-entry tunnel is positioned to accommodate the 3′-single-stranded p-RNA fragment that would arise during backtracking [37]. Indeed, single molecule observations of elongating F6, poliovirus, rhinovirus C, and SARS-CoV-2 RdRps have characterized an arrested complex consistent with backtracking [98], [99], [100]. Additional new structural evidence supports the role of the SARS-CoV-2 RdRp NTP-entry tunnel in accommodating the backtracked RNA [101]. Given the functional and structural evidence suggesting that backtracking is relevant for the SARS-CoV-2 RdRp and that nsp13 may facilitate backtracking, two potential roles for backtracking in the viral life cycle were proposed (Fig. 7 ), one in “proofreading” as discussed in Section 4.1, and the other in template-switching during sg-transcription [7].
Fig. 7.
Structural basis for the proposed role of nsp13-mediated backtracking during proofreading and template switching during sg-transcription [7], [101]. Structural models are shown as cartoons (holo-RdRp, light blue or gray; nsp13.1 helicase, orange shades; RNA strands, colored tubes). The nsp13.2 helicase is not shown for clarity (all of the models are compatible with the presence of nsp13.2). With each structural diagram is a schematic cartoon illustrating the arrangement of RNA strands. Additional proteins involved in these processes are omitted. The product RNA (p-RNA) being elongated by the RdRp is shown in red. (A) Structural basis for the proposed role of nsp13-mediated backtracking during proofreading: (i) In most cases, the RdRp elongates the p-RNA rapidly and processively [99] (in this orientation, the RdRp moves from right to left; green arrow). The single-stranded 5′-t-RNA segment (cyan) is not engaged with nsp13; (ii), (iii) Infrequently, a mis-matched NTP or a nucleotide analog (represented by the black dot) binds in the RdRp active site (ii) and is incorporated at the p-RNA 3′-end (iii); (iv) The misincorporation causes the RdRp to pause or stall, allowing nsp13 to engage with the single-stranded 5′-t-RNA segment; (v) nsp13 translocation in the 5′ → 3′ direction (orange arrow) and the hindered RdRp elongation activity (due to the mismatch) results in backtracking (backwards motion of the RdRp, red arrow) with extrusion of the p-RNA 3′-end out the RdRp NTP-entry tunnel [101]; (vi) The p-RNA 3′-end is exposed to the nsp10/14 ExoN proofreading activity, resulting in removal of the mis-incorporation and return to the elongating state. (B) Proposed role of nsp13-mediated backtracking in template-switching associated with sg-transcription [see Refs. [4], [14], [83], [102]: (i) (−)-strand RNA synthesis proceeds from the genomic 3′-poly(A)-tail until a Transcription-Regulating Sequence [TRS-R, orange; [15]] is transcribed (cTRS, yellow); (ii) The TRS causes transcription complex stalling; (iii) Helicase function acting on the (+)-strand RNA (cyan) causes backtracking of the transcription complex [101], freeing the pRNA 3′-end; (iv) The p-RNA 3′-end cTRS (yellow) hybridizes with the complementary TRS-L (orange) following the genomic 5′-leader sequence [magenta; [15], [16], [18]; (v) Processive helicase function backtracks the RdRp complex and unwinds the p-RNA from the genomic 3′-end. A second holo-RdRp (holo-RdRp2) can load into the p-RNA 3′-end and continue transcription using the 5′-leader as template.
This figure is adapted from Fig. 6c of Chen, J., B. Malone, E. Llewellyn, M. Grasso, P.M.M. Shelton, P.D.B. Olinares, K. Maruthi, E.T. Eng, H. Vatandaslar, B.T. Chait, T.M. Kapoor, S.A. Darst, and E.A. Campbell, Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell, 2020 182, 6, 1560–1573.
4.3. Template switching and helicase function
In the model for template switching (described in Section 2.1), it must be explained how the cTRS-B, which has just been transcribed by the RdRp and is therefore enclosed in the stalled RdRp active site, can become available for base pairing to TRS-L. One model suggests that template switching involves a termination event, meaning the RdRp releases from the RNA, freeing the cTRS-B to base pair with TRS-L. An alternative model builds on the backtracking premise as shown in Fig. 7B [7], [14], [16], [102]. Stalling of the RdRp after transcription of the TRS-B could allow nsp13 to engage with the downstream t-strand RNA, and the nsp13 translocation activity could mediate backtracking, which would expel the 3′-end of the p-RNA strand containing the cTRS-B sequence out through the NTP-entry tunnel. The exposed cTRS-B could then base pair with the nearby TRS-L, and a second RdRp could then load onto the cTRS-B 3′-end to complete transcription of the 5′-leader.
Complicating the situation further, the host helicase DDX1 has been shown to be crucial for sg-transcription. In studies on MHV, DDX1 was present in pull-downs of the Nucleocapsid (N) protein, pointing to its potential role during infection. In particular, DDX1 binds to phosphorylated Ser197 N protein (pS197-N) [103]. Disruption of the DDX1-pS197-N interaction led to a pronounced loss in longer sg-RNA transcripts as revealed by pharmacological perturbations and DDX1 knockdowns [103]. The N protein-DDX1 interaction was also described in IBV [104] as well as more recently in SARS-CoV-2 [105], suggesting a conserved role across CoVs. With DDX1 possessing both 5′-3′ and 3′-5′ polarities [106], its role in template switching is not antagonistic to the described hypothesis of nsp13. Mechanistically it may act to destabilize the TRS-L:cTRS-B interaction and thereby prevent the association of a second Holo-RdRp. With DDX1 additionally shown to interact with nsp14, the exonuclease may be recruited to excise the backtracked cTRS-B restoring the RdRp to a catalytically competent state [107]. Future experiments will hopefully unravel the function of both host and viral helicases in template switching.
4.4. RNA capping
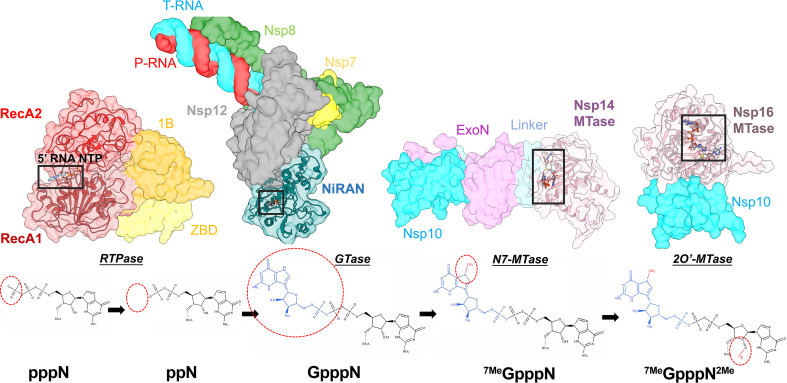
Alongside the proofreading pathways, targeting the CoV RNA capping pathway is promising in drug discovery. RNA capping is required for both translation initiation as well as to dampen the host immune response to uncapped RNA. The host cell RNA capping machinery is restricted to the nucleus, while viral replication, transcription, and capping must occur in the ROs and/or in the cytoplasm. Therefore, CoVs have evolved their own RNA capping machinery that is distinct from the host. The niche assembly of factors required to produce the Guanine cap have been structurally defined in isolation (Fig. 1), with a remaining goal being to decipher the potential for higher order complexes that could coordinate and regulate the individual capping enzymes. With evidence suggesting an interactome of the individual factors [44], [47], their activities may be modulated through the formation of a ternary complex analogous to a viral RNA capping assembly line [108]. The RNA capping complex is postulated to minimally comprise the holo-RdRp alongside nsp13, nsp10/14, and the nsp10/16 complex. These factors perform a concerted set of enzymatic activities that begins with the synthesis of RNA by the RdRp. This is followed by hydrolysis of the 5′ triphosphate by an RNA triphosphatase (RTPase), addition of the GMP cap in a 5′-5′ orientation by a guanyltransferase (GTase), methylation of the cap at position N7 by a N7 methyltransferase (N-7 MTase), and finally formation of 2′ hydroxy methyl cap via a 2′-O-methyltransferase (2′-O-MTase) (see Fig. 8 ). Below, we describe the roles of the viral nsps in this process. (See Fig. 8.)
Fig. 8.
The CoV RNA capping pathway. Surface rendered models of nsp13 (PDB 6XEZ[7]), nsp12-NiRAN (6XEZ [7]), nsp10–14 (5C8S [8]) and nsp10–16 (6WKS [109]) (top panel—left to right). The active site domains are transparent to expose cartoon models and substrate binding sites. The reaction scheme depicts the chemical structures of the reactants & products in which the modification is highlighted with a red dotted circle (bottom panel).
The RTPase activity is mediated by nsp13, in which the binding pocket for the viral RNA 5′-triphosphate is coincident with the ATPase active site [83], [89]. Conserved residues important for ATPase activity have been shown to also be important for RTPase activity [88]. Given the shared binding site for the NTPase & RTPase activities, the authors further showed that the presence of ATP competitively inhibited the RTPase activity. Pharmacological inhibitors that bind in this pocket are likely to affect viral replication by perturbing both the NTPase & RTPase activities highlighting it as a possible druggable site.
Next, a GMP is transferred to the 5′-diphosphate of the RNA by a GTase, generating GpppN-RNA. The enzyme responsible for this crucial step of viral RNA capping has yet to be identified, but a leading candidate is the nsp12 NiRAN domain. The NiRAN domain has demonstrated GTase activity when modifying a model RNA substrate [65]. This reaction was dependent on the presence of nsp13, consistent with the expected requirement for the nsp13 RTPase activity. Slanina et al. [53], however, found that UTP was the preferred NiRAN substrate (at least for nsp9) and argued this was inconsistent with the NiRAN domain acting as the RNA capping GTase [65]. Given the accepted pleiotropic activities of the viral nsps, it would not be surprising if the NiRAN enzymatic activity played multiple roles. To further develop our understanding of the NiRAN, future studies will hopefully expand on the substrate preferences (nucleic acid vs protein) of the targeted nucleotidylation activity. These studies are likely to take advantage of a reconstituted biochemical system composed of all the capping complex components which would allow for monitoring of the production of the Guanine cap through highly sensitive readouts like MS or autoradiographs. Ultimately, in vivo studies will be required to fully establish the identity of the GTase that caps the viral RNAs in infected cells.
Following the putative GTase activity, the guanine base is methylated at N7 by the nsp14 N7-MTase in an S-adenosyl methionine (SAM) dependent manner, producing the “cap-0 structure” 7MeGpppN (Fig. 8). Nsp14 was determined to specifically methylate GpppN rather than 7MeGpppN, indicating it lacked discernable 2′O-MTase activity [110]. In contrast to the nsp14 ExoN activity, the methylation activity is not significantly enhanced by the presence of the cofactor nsp10 [110]. This is unsurprising as nsp10 is bound directly to the N-terminal ExoN domain, which is connected through a flexible “hinge” to the MTase domain [8], [111]. Structurally, the MTase active site tightly couples the association of the SAM donor methyl group and the GpppN acceptor in an electrostatic environment amenable for methylation [8], [111].
The substrate and donor pockets of the nsp14 MTase are accessible to natural inhibitors such as S-adenosyl-homocysteine and are plausible therapeutic targets [112]. The pleotropic activities of nsp14 are functionally entwined through the “hinge,” which may mediate intermolecular interactions with other viral RTC components and host proteins. With previous discussion points delving into the interactions between nsp14, the holo-RdRp and nsp13; the capping substrates may be channeled to the respective RTC subunits for increased efficiency [113]. Linked to this is the observation that nsp14 interacts with inosine monophosphate dehydrogenase (IMPDH) which mediates the rate limiting step in Guanine biosynthesis [114]. This interaction may allow CoVs to prioritize Guanine usage by streamlining the conversion of purine precursors in the vicinity of the RTC.
In the final step of capping, nsp10/16 methylates the ribosyl 2′-hydroxyl of the Guanine cap linked nucleotide to form the “cap-1 structure” 7MeGpppN2me (Fig. 8). Similar to the nsp14 N7-MTase, the 2′O-MTase catalyzes a SAM dependent methylation that is indispensable to the viral lifecycle. This is in-part due to the increased stability that cap modifications confer to cytosolic mRNAs by mitigating detection by the innate immune effectors such as IFIT [115], [116]. Recent advancements in our structural understanding of nsp10/16 have been spurred by high resolution crystal structures of nsp10/16 bound to its cofactor SAM with a cap-0 RNA and bound to an inhibitor Sinefungin [10], [109], [117], [118], [119]. A tentative path for the RNA is found in a positively charged “canyon” that is nestled before the 5′ Guanine cap and SAM binding pockets [10], [109], [119]. A rearrangement of two gating loops in the cap pocket favors the interaction with the cap and repositions the 2′-OH of the next nucleotide for an inline attack on the SAM methyl donor group [109]. The reported bias for methylation of adenosyl 2′-OH may stem from an electrostatic clash with the guanine N2 amine and the SAM sulfur [109]. Structural studies also aided our understanding of the mode of inhibition of the natural compound, Sinefungin, which is bound in the SAM pocket [10], [119]. By providing the basis for its recognition, optimizing the pharmacokinetic properties of Sinefungin may benefit from bioisosteric replacements [120].
5. Antivirals targeting the RdRp
5.1. Remdesivir
Targeting the holo-RdRp has gained significant traction in drug development due to the early signs of clinical success with Remdesivir (Rdv) [121]. Rdv was initially trialed as a treatment against Ebola, where it showed moderate preclinical success but lacked efficacy in a phase III clinical trial [122].
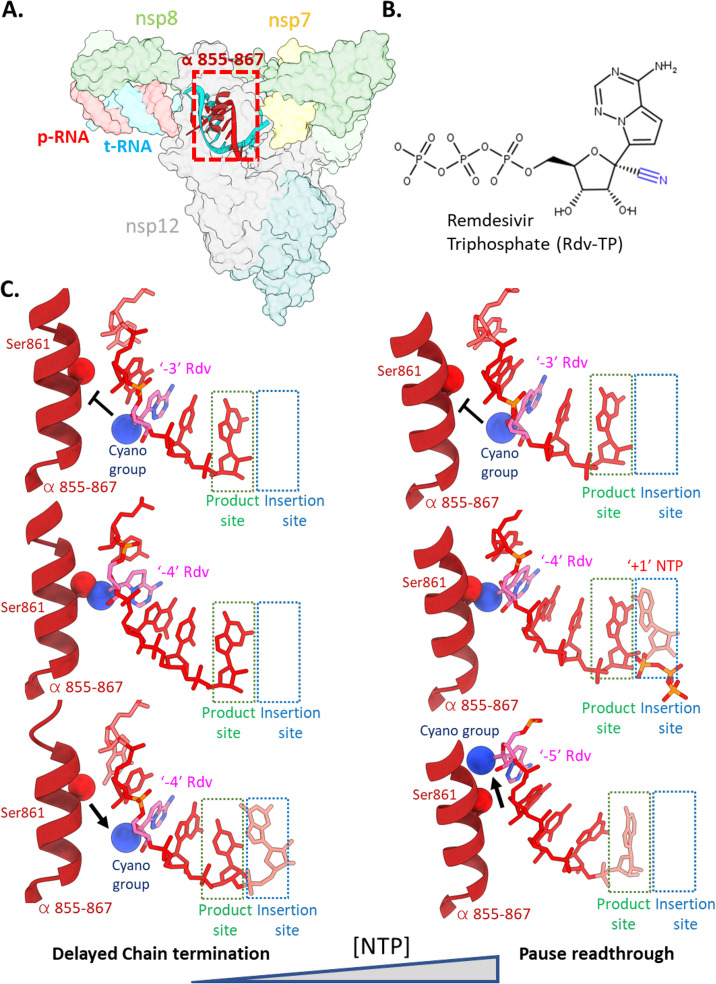
The hunt for effective therapeutics at the onset of the Covid-19 pandemic led to a potential role for Rdv, which was later established to improve clinical outcomes and shorten hospitalization stays [121]. Rdv is a nucleotide analog prodrug that is metabolized to a triphosphate form resembling ATP (Rdv-TP). Early insights into the mechanism of action of Rdv confirmed the RdRp as the target and identified resistance mutations in the active site following selection [123]. The authors also demonstrated that the nsp14 ExoN activity, an obstacle in the search for effective nucleotide analogues, reduced the effectiveness of Rdv but to a smaller degree than other nucleotide analogues such as 5-fluorouracil and Ribavirin [76], [111], [123]. A key advantage for Rdv over other nucleotide analogues is its enhanced selectivity over ATP by SARS/MERS-CoVs RdRps [124], [125]. Following incorporation of Rdv-TP, the RdRp elongates until stalling when the Rdv 1′-ribose cyano group clashes with the sidechain of nsp12 S861, hindering further translocation [124], [125], [126], [127], [128] (Fig. 9 ). This observation led to initial descriptions of the mechanism of inhibition as delayed chain termination. However, under the saturating nucleotide concentrations occurring in the cellular milieu, the translocation block is surmounted [99], [126], [127], [128], suggesting that Rdv is incorporated throughout the nascently synthesized CoV genome [124], [127].
Fig. 9.
(A) The holo-RdRp is the target of Remdesivir (Rdv). (B) Chemical structure of Rdv-triphosphate (Rdv-TP) with the 1-cyano group highlighted in blue. (C) (Left) At low nucleotide concentrations, Rdv inhibits RNA synthesis via the delayed chain termination mechanism [124], [125], [126]. Following incorporation of Rdv at the 3′-end of the RNA p-strand and translocation, two more cycles of NTP addition and translocation can occur rapidly (top panel). A third NTP following the Rdv can be incorporated by the RdRp, but further translocation is hindered by steric clash of the Rdv 1-cyano group with the side chain of nsp12 Ser861 (PDB 7B3B[127]), forcing the RdRp into a pretranslocated stalled state (7B3C [127]). (Right) At high nucleotide concentrations, the presence of a nucleotide substrate in the “insertion” site prohibits the reversion from post to pre-translocated states. The p-RNA can thus be extended past the pause induced by Rdv at the p-RNA “− 3” position [99], [126].
The incorporation of Rdv in the (−)-sense t-strand has implications for a second proposed mechanism of antiviral activity—template dependent inhibition. The premise of this mechanism is that the presence of Rdv in the RNA t-strand hinders incorporation of the opposing UTP [126]. Supporting this mode of inhibition is the finding that a much higher concentration of UTP is required to alleviate this block, indicating that this mechanism of inhibition may be more relevant for RdRp inhibition in vivo [126]. The in vitro characterized resistance mutation V557L lowered the UTP concentrations needed to surmount this barrier fivefold, reflecting that this mechanism may exert a selective pressure leading to escape mutations [123], [126]. Incorporation of the next nucleotide was also rate-limiting, demonstrating that Rdv in the − 1 position of the t-strand RNA hinders binding and/or catalysis of the active site NTP [126]. Single molecule magnetic tweezer experiments demonstrated that Rdv-TP incorporation did not lead to delayed chain termination at saturating NTP concentrations but instead led to an increased frequency of pauses, consistent with a rate-limiting translocation step [99]. As such, a combinatorial set of biochemical, single molecule and structural results point to the occurrence of several major rate-limiting steps in the presence of Rdv that perturb the normal replication/transcription cycle.
5.2. Favipiravir
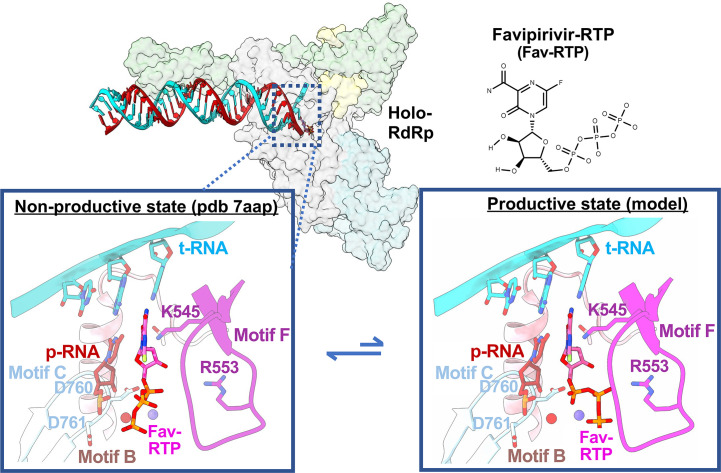
Another promising inhibitor of the RdRp is the nucleotide analog Favipiravir (Fvp). Fvp, a purine base analog licensed in Japan for use in the treatment against influenza virus, has been the focus of several clinical trials that are testing its efficacy for the treatment of COVID-19. In contrast to Rdv, Fvp exhibits relatively poor selectivity compared to ATP, rendering it an unlikely competitor [124]. Gel based primer elongation assays indicate that Fvp is incorporated more efficiently opposite a cytosine, whereas it induces chain termination events if used as an ATP analog [131]. Interestingly, magnetic tweezer experiments demonstrated that Fvp triggered backtracking of the poliovirus RdRp when used as an ATP analog [98]. Should Fvp induce a similar backtrack in CoVs, the ExoN may excise the incorporated nucleotides as discussed in Section 4.1. The in vivo relevant mechanism of inhibition likely involves its role as a Guanosine analog, through which it hampers viral replication fidelity via lethal mutagenesis [131]. Two recent studies have provided structural snapshots of RdRp bound to Fvp [129], [132]. In these structures, Fvp binds in a catalytically inactive state (Fig. 10 ), which is consistent with its relatively poor selectivity [129].
Fig. 10.
Favipiravir (Fvp) targets the holo-RdRp active site. Fvp bound in the active site readily adopts a conformation incapable of undergoing catalysis (left; PDB 7AAP[129]). By contrast, the correct conformation of the incoming nucleotide features a 120° rotation around the ribose O5′-α-phosphate bond, setting up an in-line nucleophilic attack by the p-RNA 3′-OH (right—modeled using PDB 6SZV[130]).
6. Conclusion
The urgency of mitigating the COVID-19 pandemic has led to a surge of basic research to understand the complex SARS-CoV-2 viral life cycle. Structural biology is playing a pivotal role in many areas, including contributing to a basic understanding of the viral replication/transcription program as well as providing insights for antiviral development. Future work promises to provide increasingly informative views of the RdRp and other critical enzymes with known and novel inhibitors. Most excitingly, developments in cryo-electron microscopy (single-particle analysis as well as tomography) are enabling structural studies with the potential for revealing how the SARS-CoV-2 nsps coordinate with each other to accomplish their functions, both in vitro and in vivo.
Footnotes
Here, we use the term “transcription” to conform with the standard terminology in the field.
References
- 1.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Krupovic M., Mushegian A., Kropinski A.M., Siddell S.G., Varsani A., Adams M.J., Davison A.J., Dutilh B.E., Harrach B., Harrison R.L., Junglen S., King A.M.Q., Knowles N.J., Lefkowitz E.J., Nibert M.L., Rubino L., Sabanadzovic S., Sanfaçon H., Simmonds P., Walker P.J., Zerbini F.M., Kuhn J.H., International Committee on Taxonomy of Viruses Executive Committee The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020;5(5):668–674. doi: 10.1038/s41564-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94(Pt 10):2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 4.Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23(7) doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182(6):1560–1573. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. 2015;112(30):9436. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J.F. High-resolution structures of the SARS-CoV-2 2′-<em > O </em >−methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020;13(651) doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 13.Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81(1):20. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annual Review of Virology. 2015;2(1):265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso S., Izeta A., Sola I., Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 2002;76(3):1293. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20(24):7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sola I., Mateos-Gomez P.A., Almazan F., Zuñiga S., Enjuanes L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011;8(2):237–248. doi: 10.4161/rna.8.2.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zúñiga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78(2):980. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huston N.C., Wan H., Strine M.S., de Cesaris Araujo R., Tavares C.B.W., Pyle A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021;81(3):584–598.e5. doi: 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013;87(1):177–186. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. (e10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nomburg J., Meyerson M., DeCaprio J.A. Pervasive generation of non-canonical subgenomic RNAs by SARS-CoV-2. Genome Med. 2020;12(1):108. doi: 10.1186/s13073-020-00802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(1):68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28(12):1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grünewald K., Koster A.J., Snijder E.J., Bárcena M. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369(6509):1395. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11(1):5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.V'Kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., Bruggmann R., Stoffel M.H., Heller M., Dijkman R., Thiel V. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife. 2019;8 doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex Association of the Putative Mouse Hepatitis Virus RNA-dependent RNA polymerase. J. Virol. 2003;77(19):10515. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbert I., Snijder E.J., Dimitrova M., Guillemot J.-C., Lécine P., Canard B. The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133(2):136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P.J., te Lintelo E.G., Rottier P.J.M., de Haan C.A.M. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82(24):12392. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with Coronaviral RNA replication. MBio. 2017;8(6) doi: 10.1128/mBio.01658-17. p. e01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann H.H., Schneider W.M., Sánchez-Rivera F.J., Luna J.M., Ashbrook A.W., Soto-Feliciano Y.M., Leal A.A., Le Pen J., Ricardo-Lax I., Michailidis E., Hao Y., Stenzel A.F., Peace A., Allis C.D., Lowe S.W., MacDonald M.R., Poirier J.T., Rice C.M. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2020;29:1–14. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., Stenzel A.F., Lowe S.W., MacDonald M.R., Rice C.M., Poirier J.T. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184(1):120–132. doi: 10.1016/j.cell.2020.12.006. (e14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monastyrska I., Ulasli M., Rottier P.J.M., Guan J.-L., Reggiori F., de Haan C.A.M. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9(2):164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A.M., Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting Short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7(6):500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koning R.I., Koster A.J., Sharp T.H. Advances in cryo-electron tomography for biology and medicine. Annals of Anatomy - Anatomischer Anzeiger. 2018;217:82–96. doi: 10.1016/j.aanat.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Hyde J.L., Diamond M.S. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology. 2015;479–480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. 2014;111(37):E3900. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 43.Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 44.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.te Velthuis A.J.W., van den Worm S.H.E., Snijder E.J. The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2011;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211(2):107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal Adenylyltransferase activity. J. Virol. 2019;93(12) doi: 10.1128/JVI.00291-19. p. e00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Züst R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ < em > cis </em >-acting RNA pseudoknot, Replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008;82(3):1214. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slanina H., Madhugiri R., Bylapudi G., Schultheiß K., Karl N., Gulyaeva A., Gorbalenya A.E., Linne U., Ziebuhr J. Coronavirus replication–transcription complex: vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. 2021;118(6) doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.-C., Tian G., Jiang H.-W., Tao S.-C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonald S.M. RNA synthetic mechanisms employed by diverse families of RNA viruses. WIREs RNA. 2013;4(4):351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peersen O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017;234:4–20. doi: 10.1016/j.virusres.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorbalenya A.E., Pringle F.M., Zeddam J.-L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H.J., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324(1):47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruenn J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31(7):1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., Wetmore D.R., McGrath M.E., Ray A.S., Sofia M.J., Swaminathan S., Edwards T.E. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347(6223):771. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 61.Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274(25):17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 62.Ben Ouirane K., Boulard Y., Bressanelli S. The hepatitis C virus RNA-dependent RNA polymerase directs incoming nucleotides to its active site through magnesium-dependent dynamics within its F motif. J. Biol. Chem. 2019;294(19):7573–7587. doi: 10.1074/jbc.RA118.005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16(2):212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184(1):184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paul A.V., Wimmer E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015;206:12–26. doi: 10.1016/j.virusres.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adedeji A.O., Marchand B., te Velthuis A.J.W., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLOS ONE. 2012;7(5):e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferron F., Decroly E., Selisko B., Canard B. The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res. 2012;96(1):21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94(23) doi: 10.1128/JVI.01246-20. p. e01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of < em > S </em >−Adenosyl-<span class ="sc">l </span >−methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90(16):7248. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. 2012;109(24):9372. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81(22):12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith E.C., Sexton N.R., Denison M.R. Thinking outside the triangle: replication Fidelity of the largest RNA viruses. Annual Review of Virology. 2014;1(1):111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- 78.Baddock H., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., Swift L., Cruz-Migoni A., Morris G., Schofield C., Gileadi O., McHugh P. Characterisation of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. bioRxiv. 2020 doi: 10.1101/2020.08.13.248211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darmawan H., Harrison M., Reha-Krantz L.J. DNA polymerase 3′→5′ exonuclease activity: different roles of the beta hairpin structure in family-B DNA polymerases. DNA Repair. 2015;29:36–46. doi: 10.1016/j.dnarep.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 80.Wuite G.J.L., Smith S.B., Young M., Keller D., Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404(6773):103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 81.Kamtekar S., Berman A.J., Wang J., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16(4):609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 82.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235(1–2):16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6(7):1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278(41):39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee N.-R., Kwon H.-M., Park K., Oh S., Jeong Y.-J., Kim D.-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38(21):7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mickolajczyk K.J., Shelton P.M.M., Grasso M., Cao X., Warrington S.E., Aher A., Liu S., Kapoor T.M. Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase. Biophys. J. 2020;120:1–11. doi: 10.1016/j.bpj.2020.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside Triphosphatase, and RNA 5′-Triphosphatase activities. J. Virol. 2004;78(14):7833. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78(11):5619. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74(11):5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seybert A., van Dinten L.C., Snijder E.J., Ziebuhr J. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between Arterivirus and coronavirus helicases. J. Virol. 2000;74(20):9586. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of Nidovirus helicases. J. Virol. 2005;79(2):696. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang R., Li Y., Cowley T.J., Steinbrenner A.D., Phillips J.M., Yount B.L., Baric R.S., Weiss S.R. The nsp1, nsp13, and M proteins contribute to the Hepatotropism of murine coronavirus JHM.WU. J. Virol. 2015;89(7):3598. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nudler E., Mustaev A., Lukhtanov E., Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89(1):33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 95.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149(7):1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheung A.C., Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 98.Dulin D., Arnold J.J., van Laar T., Oh H.-S., Lee C., Perkins A.L., Harki D.A., Depken M., Cameron C.E., Dekker N.H. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 2017;21(4):1063–1076. doi: 10.1016/j.celrep.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seifert M., Bera S.C., van Nies P., Kirchdoerfer R.N., Shannon A., Le T.-T.-N., Grove T.L., Papini F.S., Arnold J.J., Almo S.C., Canard B., Depken M., Cameron C.E., Dulin D. Inhibition of SARS-CoV-2 polymerase by nucleotide analogs: a single molecule perspective. bioRxiv. 2020 doi: 10.1101/2020.08.06.240325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seifert M., van Nies P., Papini F.S., Arnold J.J., Poranen M.M., Cameron C.E., Depken M., Dulin D. Temperature controlled high-throughput magnetic tweezers show striking difference in activation energies of replicating viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2020;48(10):5591–5602. doi: 10.1093/nar/gkaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., Olinares P.D.B., Cao X., Hernandez C., Eng E.T., Chait B.T., Shaw D.E., Landick R., Darst S.A., Campbell E.A. Structural basis for backtracking by the SARS-CoV-2 replication–transcription complex. Proc. Natl. Acad. Sci. 2021;118(19) doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Enjuanes L., Almazán F., Sola I., Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 103.Wu C.-H., Chen P.-J., Yeh S.-H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16(4):462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Emmott E., Munday D., Bickerton E., Britton P., Rodgers M.A., Whitehouse A., Zhou E.-M., Hiscox J.A. The cellular Interactome of the coronavirus infectious bronchitis virus Nucleocapsid protein and functional implications for virus biology. J. Virol. 2013;87(17):9486. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fairman-Williams M.E., Guenther U.-P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu L., Khadijah S., Fang S., Wang L., Tay F.P.L., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84(17):8571. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sutton G., Grimes J.M., Stuart D.I., Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat. Struct. Mol. Biol. 2007;14(5):449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 109.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez-Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11(1):3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M., Canard B., Vasseur J.-J., Decroly E., Debart F. Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase. Eur. J. Med. Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sweetlove L.J., Fernie A.R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 2018;9(1):2136. doi: 10.1038/s41467-018-04543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., Kaake R.M., Weckstein A.R., Owens T.W., Gupta M., Pourmal S., Titus E.W., Cakir M., Soucheray M., McGregor M., Cakir Z., Jang G., O’Meara M.J., Tummino T.A., Zhang Z., Foussard H., Rojc A., Zhou Y., Kuchenov D., Hüttenhain R., Xu J., Eckhardt M., Swaney D.L., Fabius J.M., Ummadi M., Tutuncuoglu B., Rathore U., Modak M., Haas P., Haas K.M., Naing Z.Z.C., Pulido E.H., Shi Y., Barrio-Hernandez I., Memon D., Petsalaki E., Dunham A., Marrero M.C., Burke D., Koh C., Vallet T., Silvas J.A., Azumaya C.M., Billesbølle C., Brilot A.F., Campbell M.G., Diallo A., Dickinson M.S., Diwanji D., Herrera N., Hoppe N., Kratochvil H.T., Liu Y., Merz G.E., Moritz M., Nguyen H.C., Nowotny C., Puchades C., Rizo A.N., Schulze-Gahmen U., Smith A.M., Sun M., Young I.D., Zhao J., Asarnow D., Biel J., Bowen A., Braxton J.R., Chen J., Chio C.M., Chio U.S., Deshpande I., Doan L., Faust B., Flores S., Jin M., Kim K., Lam V.L., Li F., Li J., Li Y.-L., Li Y., Liu X., Lo M., Lopez K.E., Melo A.A., Moss F.R., Nguyen P., Paulino J., Pawar K.I., Peters J.K., Pospiech T.H., Safari M., Sangwan S., Schaefer K., Thomas P.V., Thwin A.C., Trenker R., Tse E., Tsui T.K.M., Wang F., Whitis N., Yu Z., Zhang K., Zhang Y., Zhou F., Saltzberg D., Hodder A.J., Shun-Shion A.S., Williams D.M., White K.M., Rosales R., Kehrer T., Miorin L., Moreno E., Patel A.H., Rihn S., Khalid M.M., Vallejo-Gracia A., Fozouni P., Simoneau C.R., Roth T.L., Wu D., Karim M.A., Ghoussaini M., Dunham I., Berardi F., Weigang S., Chazal M., Park J., Logue J., McGrath M., Weston S., Haupt R., Hastie C.J., Elliott M., Brown F., Burness K.A., Reid E., Dorward M., Johnson C., Wilkinson S.G., Geyer A., Giesel D.M., Baillie C., Raggett S., Leech H., Toth R., Goodman N., Keough K.C., Lind A.L., Klesh R.J., Hemphill K.R., Carlson-Stevermer J., Oki J., Holden K., Maures T., Pollard K.S., Sali A., Agard D.A., Cheng Y., Fraser J.S., Frost A., Jura N., Kortemme T., Manglik A., Southworth D.R., Stroud R.M., Alessi D.R., Davies P., Frieman M.B., Ideker T., Abate C., Jouvenet N., Kochs G., Shoichet B., Ott M., Palmarini M., Shokat K.M., García-Sastre A., Rassen J.A., Grosse R., Rosenberg O.S., Verba K.A., Basler C.F., Vignuzzi M., Peden A.A., Beltrao P., Krogan N.J. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370(6521) doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.-Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2'-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5):e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11(1):3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lima L.M., Barreiro E.J. Bioisosterism: a useful strategy for molecular modification and drug design. Curr. Med. Chem. 2005;12(1):23–49. doi: 10.2174/0929867053363540. [DOI] [PubMed] [Google Scholar]
- 121.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-d., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 — final report. New England Journal of Medicine. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mulangu S., Dodd L.E., Davey R.T., Jr., Mbaya O.T., Proschan M., Mukadi D., Manzo M.L., Nzolo D., Oloma A.T., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Lane H.C., Muyembe-Tamfum J.J., Sivahera B., Camara M., Kojan R., Walker R., Dighero-Kemp B., Cao H., Mukumbayi P., Mbala-Kingebeni P., Ahuka S., Albert S., Bonnett T., Crozier I., Duvenhage M., Proffitt C., Teitelbaum M., Moench T., Aboulhab J., Barrett K., Cahill K., Cone K., Eckes R., Hensley L., Herpin B., Higgs E., Ledgerwood J., Pierson J., Smolskis M., Sow Y., Tierney J., Sivapalasingam S., Holman W., Gettinger N., Vallée D., Nordwall J. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral Remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2) doi: 10.1128/mBio.00221-18. p. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Götte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295(47):16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12(1):279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS CoV-2 replication. Mol. Cell. 2021;81(7):1548–1552. doi: 10.1016/j.molcel.2021.01.035. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Naydenova K., Muir K.W., Wu L.-F., Zhang Z., Coscia F., Peet M.J., Castro-Hartmann P., Qian P., Sader K., Dent K., Kimanius D., Sutherland J.D., Löwe J., Barford D., Russo C.J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. 2021;118(7) doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wandzik J.M., Kouba T., Karuppasamy M., Pflug A., Drncova P., Provaznik J., Azevedo N., Cusack S. A structure-based model for the complete transcription cycle of influenza polymerase. Cell. 2020;181(4):877–893. doi: 10.1016/j.cell.2020.03.061. (e21) [DOI] [PubMed] [Google Scholar]
- 131.Shannon A., Selisko B., Le N.-T.-T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B., Peersen O., Canard B. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11(1):4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peng Q., Peng R., Yuan B., Wang M., Zhao J., Fu L., Qi J., Shi Y. Structural basis of SARS-CoV-2 polymerase inhibition by Favipiravir. The Innovation. 2021;2(1):100080. doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]