Abstract
Background
Germline ATM mutations are suggested to contribute to predisposition to prostate cancer (PrCa). Previous studies have had inadequate power to estimate variant effect sizes.
Objective
To precisely estimate the contribution of germline ATM mutations to PrCa risk.
Design, setting, and participants
We analysed next-generation sequencing data from 13 PRACTICAL study groups comprising 5560 cases and 3353 controls of European ancestry.
Outcome measurements and statistical analysis
Variant Call Format files were harmonised, annotated for rare ATM variants, and classified as tier 1 (likely pathogenic) or tier 2 (potentially deleterious). Associations with overall PrCa risk and clinical subtypes were estimated.
Results and limitations
PrCa risk was higher in carriers of a tier 1 germline ATM variant, with an overall odds ratio (OR) of 4.4 (95% confidence interval [CI]: 2.0–9.5). There was also evidence that PrCa cases with younger age at diagnosis (<65 yr) had elevated tier 1 variant frequencies (pdifference = 0.04). Tier 2 variants were also associated with PrCa risk, with an OR of 1.4 (95% CI: 1.1–1.7).
Conclusions
Carriers of pathogenic ATM variants have an elevated risk of developing PrCa and are at an increased risk for earlier-onset disease presentation. These results provide information for counselling of men and their families.
Patient summary
In this study, we estimated that men who inherit a likely pathogenic mutation in the ATM gene had an approximately a fourfold risk of developing prostate cancer. In addition, they are likely to develop the disease earlier.
Keywords: Prostate cancer, ATM gene mutations, Genetic predisposition, Targeted screening and therapy
Introduction
In 2018, prostate cancer (PrCa) was the second most common cancer diagnosed in men worldwide, with over 1.2 million new cases [1]. The disease has high heritability [2]; family history is a well-known risk factor and genome-wide association studies (GWASs) have identified nearly 200 common germline risk variants [3], [4], [5]. Several genes have also been proposed to harbour rare moderate penetrance variants that may contribute to an elevated risk of PrCa. There is convincing evidence that rare loss-of-function variants in BRCA2 contribute to the development of PrCa [6] and additionally to an aggressive phenotype [7]. Rare variants in several other genes, primarily ATM, BRCA1, CHEK2, NBN, PALB2, and the mismatch repair genes, have also been proposed to increase PrCa risk [8], [9], [10], [11], [12], [13], [14], [15], [16], [17].
Most prior germline sequencing studies reporting ATM mutation data have been relatively small (up to a few hundred PrCa cases), lacked control cohorts, or were conducted in non-European ancestral populations for whom the frequency of pathogenic ATM mutations may differ from those of Europeans [9], [11], [12], [13], [14], [16], [18], [19]. Precise estimates of risk for ATM mutation carriers have not been established. In a few small clinical studies, ATM has also been shown to be linked with a more aggressive subtype of PrCa. In The Cancer Genome Atlas, about 7% of PrCa primary tumour samples had either somatic or germline alterations in ATM [18], and it has also been shown that germline ATM variant carriers have reduced survival [13].
In this study, we collected, harmonised, and analysed available ATM sequencing data from 14 study groups within The PRACTICAL Consortium and report on the overall association of rare germline ATM variants with PrCa from over 8000 European samples.
Participants and methods
Study groups
We collected individual-level data for 10 404 participants from 14 PRACTICAL Consortium study groups across North America (five study groups), Europe (six study groups), Asia (one study group), and Australia (two study groups). We excluded 459 samples of non-European or unknown ethnicity and an additional 1032 samples due to interstudy duplicates, relatedness, or unavailability of phenotype data, leaving 8913 participants from 13 study groups available for analysis (Supplementary Table 1).
Full details of each study group, including recruitment criteria, data collection, and sequencing methods, are given in Supplementary Tables 2 and 3, and the Supplementary material.
Quality control and processing of sequence data
Variant Call Format (VCF) files of the ATM gene region (chr11:108093211–108239829 GRCh37/hg19) were standardised to allow consistent variant- and sample-level quality control and variant annotation. VCF files aligned to GRCh38 were converted to GRCh37 using LiftoverVcf. BCFtools [20] norm was utilised to split multiallelic sites to multiple rows, left align variants, and normalise to the reference. Variants with low coverage (depth <10), with low quality (GQ < 20 or equivalent), situated within repeat regions, with an allelic ratio <30% or >70%, or which were monomorphic were excluded. Variant annotation was performed on multisample VCF files using variant effect predictors (VEP; ClinVar classification, ExAC MAF, CADD scores, Impact, and REVEL scores).
Variant categorisation
Only rare variants (defined as ExAC non-Finnish European MAF < 0.01) were included in downstream analyses. Rare variants were categorised into two classes. Tier 1 variants were defined as variants with either a pathogenic or likely pathogenic ClinVar https://www.ncbi.nlm.nih.gov/clinvar/ [21] classification or a “high” VEP [22] impact score; these included transcript ablation, splice acceptor/donor, stop gained/lost, frameshift, and some missense variants. Tier 2 included variants that had a “moderate” VEP impact score (in-frame insertion/deletion [indels], missense, and protein-altering variants) and were also predicted to be potentially deleterious by at least one of the following two algorithms: combined annotation dependent depletion (CADD [23]; Phred-scaled score >20) or rare exome variant ensemble learner (REVEL [24]; score >0.60).
Statistical analysis
We calculated the prevalence of variants in PrCa cases and controls. Owing to the rarity of individual variants (Table 1 and Supplementary Table 4), mutation status was defined as a binary variable, indicating the presence of at least one variant in the ATM gene. Analyses were conducted for tier 1 and 2 variants independently and for both combined.
Table 1.
Numbers contributed, by study
| Study | Total participants | Cancers | Noncancers | ||||
|---|---|---|---|---|---|---|---|
| Noncarriers (N) | Carriers (N) | Prevalence (%) | Noncarriers (N) | Carriers (N) | Prevalence (%) | ||
| Tier 1 | |||||||
| CAPS | 267 | 168 | 5 | 2.89 | 94 | 0 | 0.00 |
| CeRePP | 347 | 295 | 2 | 0.67 | 49 | 1 | 2.00 |
| FHCRC | 370 | 259 | 5 | 1.89 | 105 | 1 | 0.94 |
| Finland | 291 | 212 | 3 | 1.40 | 76 | 0 | 0.00 |
| Germany | 318 | 188 | 3 | 1.57 | 127 | 0 | 0.00 |
| ICR | 3350 | 1990 | 22 | 1.09 | 1336 | 2 | 0.15 |
| JHU | 186 | 98 | 1 | 1.01 | 87 | 0 | 0.00 |
| MAYO | 971 | 386 | 5 | 1.28 | 577 | 3 | 0.52 |
| MCCS | 1313 | 1258 | 16 | 1.26 | 39 | 0 | 0.00 |
| MD_Anderson | 449 | – | – | – | 448 | 1 | 0.22 |
| Porto | 479 | 476 | 3 | 0.63 | – | – | – |
| TASPRAC | 26 | 18 | 0 | 0.00 | 8 | 0 | 0.00 |
| UTAH | 546 | 147 | 0 | 0.00 | 399 | 0 | 0.00 |
| All | 8913 | 5495 | 65 | 1.17 | 3345 | 8 | 0.24 |
| Tier 2 | |||||||
| CAPS | 267 | 166 | 7 | 4.05 | 86 | 8 | 8.51 |
| CeRePP | 347 | 272 | 25 | 8.42 | 48 | 2 | 4.00 |
| FHCRC | 370 | 243 | 21 | 7.95 | 96 | 10 | 9.43 |
| Finland | 291 | 206 | 9 | 4.19 | 76 | 0 | 0.00 |
| Germany | 318 | 174 | 17 | 8.90 | 117 | 10 | 7.87 |
| ICR | 3350 | 1896 | 116 | 5.77 | 1278 | 60 | 4.48 |
| JHU | 186 | 85 | 14 | 14.14 | 83 | 4 | 4.60 |
| MAYO | 971 | 368 | 23 | 5.88 | 556 | 24 | 4.14 |
| MCCS | 1313 | 1199 | 75 | 5.89 | 38 | 1 | 2.56 |
| MD_Anderson | 449 | – | – | – | 438 | 11 | 2.45 |
| Porto | 479 | 407 | 72 | 15.03 | – | – | – |
| TASPRAC | 26 | 14 | 4 | 22.22 | 7 | 1 | 12.50 |
| UTAH | 546 | 143 | 4 | 2.72 | 395 | 4 | 1.00 |
| All | 8913 | 5173 | 387 | 6.96 | 3218 | 135 | 4.03 |
| Tier 1 + 2 | |||||||
| CAPS | 267 | 161 | 12 | 6.94 | 86 | 8 | 8.51 |
| CeRePP | 347 | 270 | 27 | 9.09 | 47 | 3 | 6.00 |
| FHCRC | 370 | 238 | 26 | 9.85 | 95 | 11 | 10.38 |
| Finland | 291 | 203 | 12 | 5.58 | 76 | 0 | 0.00 |
| Germany | 318 | 171 | 20 | 10.47 | 117 | 10 | 7.87 |
| ICR | 3350 | 1875 | 137 | 6.81 | 1276 | 62 | 4.63 |
| JHU | 186 | 84 | 15 | 15.15 | 83 | 4 | 4.60 |
| MAYO | 971 | 363 | 28 | 7.16 | 553 | 27 | 4.66 |
| MCCS | 1313 | 1185 | 89 | 6.99 | 38 | 1 | 2.56 |
| MD_Anderson | 449 | – | – | – | 437 | 12 | 2.67 |
| Porto | 479 | 405 | 74 | 15.45 | – | – | – |
| TASPRAC | 26 | 14 | 4 | 22.22 | 7 | 1 | 12.50 |
| UTAH | 546 | 143 | 4 | 2.72 | 395 | 4 | 1.00 |
| All | 8913 | 5112 | 448 | 8.06 | 3210 | 143 | 4.26 |
Odds ratios (ORs) were estimated for the association between mutation status and PrCa diagnosis, and also after stratifying cases by first-degree family history of PrCa, metastatic PrCa, Gleason score (≥8, 7, and ≤6), PrCa aggressiveness (aggressive, intermediate, and nonaggressive), age at diagnosis (<65 and ≥65 yr), and death from PrCa (death from PrCa and non-PrCa death/alive). Cases were defined as “aggressive” if they had at least one of stage T4, N1, Gleason score ≥8, metastatic PrCa, or death from PrCa; as “nonaggressive” if they had stage T1–T2 and Gleason score ≤6 disease plus, if deceased, death was not due to PrCa; and finally as “intermediate” aggressive if they failed to fulfil either other criteria (ie, had stage T3 and/or Gleason score 7 disease).
To account for possible heterogeneity between study groups in recruitment and sequencing procedures, we generated study-specific ORs and obtained a pooled estimate using a two-stage model [25]. First, we meta-analysed study-specific estimates using the fixed-effect Mantel–Haenszel method with continuity correction [26]. This method has been shown to perform better than inverse variance methods when events are rare [27]. Our analyses were restricted to those of European ancestry and assumed a common effect across study groups. If no appreciable between-study heterogeneity was detected using the I2 statistic [28], we analysed the data in a pooled data set using Firth logistic regression [29], controlling for study. Prior to each stratified analysis, we removed studies that did not vary in outcome or variant status (ie, contained only cases or only controls, or individuals with no variants). The number of studies excluded varied, depending on the particular analysis (Supplementary Table 5). Owing to the rarity of mutation carriers, we mainly conducted univariate analyses controlling for study, but we also investigated the effect of controlling for age at diagnosis/interview on our results.
We also calculated hazard ratios (HRs) for the association between mutation status and risk of death from PrCa in cases. HRs were estimated from Cox proportional hazard regression models, with time since diagnosis as the underlying timescale. Cases became at risk at their age at PrCa diagnosis and came under observation at their age at consent or first interview. The time to event was calculated from age at diagnosis to death from PrCa. Cases that did not die from PrCa were censored at the age of death from other causes or age of last follow-up, whichever was earliest. For these analyses, we included studies with available information on follow-up, and restricted the analysis to those studies that had more than five deaths due to PrCa in variant positive and negative strata. Models were adjusted for age at diagnosis and study.
All analyses were performed with Stata 16.0 (StataCorp LLC, College Station, TX, USA).
Results
The analyses included 8913 individuals of European ancestry; among them, 65 (1.2%) of 5560 PrCa cases carried a tier 1 ATM variant, compared with eight (0.24%) of 3353 controls. For tier 2 variants, 387 (7.0%) cases were carriers compared with 135 (4.0%) controls. For tiers 1 and 2 combined, 448 (8.1%) cases and 143 (4.3%) controls were variant carriers (Table 1). The prevalence of tier 1 variants in individual studies ranged from 0.6% to 2.9% in cases and from 0% to 2.0% for controls. For tier 2 variants, prevalence ranged from 2.7% to 22.2% for cases and from 0% to 12.5% for controls. For tiers 1 and 2 combined, the interstudy range was 2.7–22.2% for cancer cases and 0–12.5% for controls (Table 1).
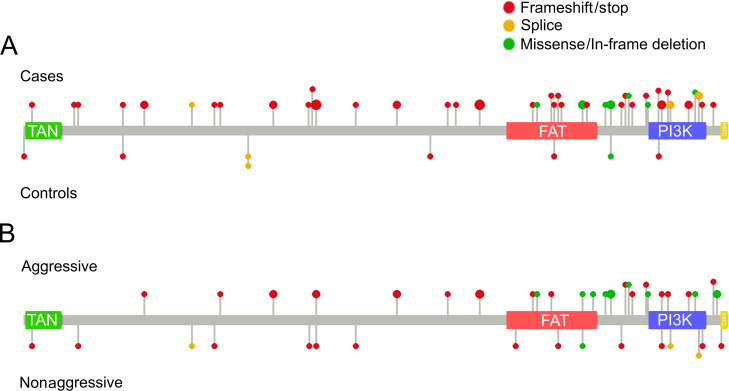
No sample had more than one tier 1 ATM variant. Four cases and no controls had tier 2 variants in addition to a tier 1 variant. Twenty-three cases and two controls had more than one tier 2 variant. Tier 1 variants consisted of frameshift indels (n = 19), stop-gain mutations (n = 16), splice site variants (n = 6), missense variants (n = 8), one in-frame deletion, and one start lost variant (Fig. 1 and Supplementary Table 4). All missense variants were listed as pathogenic or likely pathogenic in ClinVar, and five of these (observed in seven cases) were in 3′ end functional domains. Fourteen variants were observed in more than one sample, of which 12 were identified in multiple studies. Three stop-gain mutations, V1268*, E1978*, and W2769*, were each observed in four samples.
Fig. 1.
Tier 1 mutations identified within the ATM gene. Position of tier 1 ATM mutations in (A) case and control samples and (B) aggressive and nonaggressive cancer cases. Lollipop size is relative to the number of samples.
Of 5560 cases, information on family history of PrCa was available for 4663 (83.9%), metastatic disease for 4432 (79.7%), Gleason score for 4788 (86.1%), and aggressiveness for 5093 (91.6%) cases (Table 2 and Supplementary Table 5). Of the 5560 cancer cases, 1203 were listed to have lethal PrCa, 603 died of non–PrCa-related causes, and 69 deaths had an unknown relationship between PrCa and cause of death (Supplementary Table 6). Two study groups (MCCS and ICR) contributed to the time-to-event analysis for risk of PrCa death. Information on age at diagnosis/interview was available for 7474 of 8913 participants. (Supplementary Table 7). The median age at diagnosis was 60 yr (interquartile range [IQR]: 56–67). For controls, the median age at interview was 60 yr (IQR: 55–73). The interstudy range for age at diagnosis was 54–67 yr, whilst for age at interview this was 46–75 yr.
Table 2.
Numbers contributed to each subtype analysis, by study
| Noncancers | Cancers |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | Family history |
Metastatic |
Gleason |
Aggressive |
Age at diagnosis |
Death | ||||||||||||||
| FH+ | FH– | U | M1 | M0 | U | >7 | 7 | <7 | U | Agg. | Int. | Non. | U | <65 | ≥65 | U | PrCa | |||
| Tier 1 status | ||||||||||||||||||||
| +ve | 8 | 65 | 26 | 27 | 12 | 8 | 43 | 14 | 18 | 14 | 21 | 12 | 35 | 11 | 15 | 4 | 50 | 14 | 1 | 21 |
| –ve | 3345 | 5495 | 2053 | 2557 | 885 | 496 | 3885 | 1114 | 1283 | 1350 | 2102 | 760 | 2202 | 1178 | 1652 | 463 | 3660 | 1714 | 121 | 1182 |
| +ve Prev (%) | (0.24) | (1.17) | (1.25) | (1.04) | (1.59) | (1.09) | (1.38) | (1.03) | (0.99) | (1.56) | (0.93) | (0.90) | (1.35) | (0.81) | (1.75) | |||||
| Total | 3353 | 5560 | 2079 | 2584 | 897 | 504 | 3928 | 1128 | 1301 | 1364 | 2123 | 772 | 2237 | 1189 | 1667 | 467 | 3710 | 1728 | 122 | 1203 |
| Tier 2 status | ||||||||||||||||||||
| +ve | 135 | 387 | 153 | 169 | 65 | 35 | 272 | 80 | 81 | 106 | 153 | 47 | 143 | 99 | 120 | 25 | 265 | 117 | 5 | 69 |
| –ve | 3218 | 5173 | 1926 | 2415 | 832 | 469 | 3656 | 1048 | 1220 | 1258 | 1970 | 725 | 2094 | 1090 | 1547 | 442 | 3445 | 1611 | 117 | 1134 |
| +ve Prev (%) | (4.03) | (6.96) | (7.36) | (6.54) | (6.94) | (6.92) | (6.23) | (7.77) | (7.21) | (6.39) | (8.33) | (7.20) | (7.14) | (6.77) | (5.74) | |||||
| Total | 3353 | 5560 | 2079 | 2584 | 897 | 504 | 3928 | 1128 | 1301 | 1364 | 2123 | 772 | 2237 | 1189 | 1667 | 467 | 3710 | 1728 | 122 | 1203 |
| Tier 1 + 2 status | ||||||||||||||||||||
| +ve | 143 | 448 | 178 | 193 | 77 | 43 | 312 | 93 | 99 | 120 | 171 | 58 | 176 | 109 | 134 | 29 | 312 | 130 | 6 | 88 |
| –ve | 3210 | 5112 | 1901 | 2391 | 820 | 461 | 3616 | 1035 | 1202 | 1244 | 1952 | 714 | 2061 | 1080 | 1533 | 438 | 3398 | 1598 | 116 | 1115 |
| +ve Prev (%) | (4.26) | (8.06) | (8.56) | (7.47) | (8.53) | (7.94) | (7.61) | (8.80) | (8.05) | (7.87) | (9.17) | (8.04) | (8.41) | (7.52) | (7.32) | |||||
| Total | 3353 | 5560 | 2079 | 2584 | 897 | 504 | 3928 | 1128 | 1301 | 1364 | 2123 | 772 | 2237 | 1189 | 1667 | 467 | 3710 | 1728 | 122 | 1203 |
Agg. = aggressive; FH = family history; Int. = intermediate aggressive; Non. = nonaggressive; PrCa = prostate cancer; Prev = prevalence; U = unknown.
The likelihood of carrying a tier 1 ATM variant was greater in PrCa cases than in controls (OR = 4.4, 95% confidence interval [CI]: 2.0–9.5, p = 2.3 × 10–4; Table 3). Comparing subtypes, ORs were higher in all clinically significant disease subgroups (positive family history, metastatic disease, Gleason score ≥8, and aggressive disease); however, we could not conclude that any differences within the stratified subtype analyses were significant, except for age at diagnosis (p = 0.037). Cases diagnosed before age 65 yr were more likely to carry a tier 1 variant (OR = 4.9, 95% CI: 2.2–11.1, p = 1.3 × 10–4) than those diagnosed after age 65 yr (OR = 3.8, 95% CI: 1.4–10.4, p = 0.010). Finally, we found no appreciable heterogeneity in the various associations when looking at individual study estimates (Supplementary Fig. 1A–C and 2A–C). There were also no appreciable differences in results when tier 1 results were adjusted for age at diagnosis/interview (Supplementary Table 10) or if Finnish populations were excluded (Supplementary Table 13).
Table 3.
Primary results, tier 1
| Studies (N) | Cancers (N) | Ref. groupa (N) | OR | 95% CI | p value | |
|---|---|---|---|---|---|---|
| Tier 1 | ||||||
| Overall | 9 | 4916 | 2497 | 4.4 | (2.0, 9.5) | 2.3 × 10–4 |
| Subtypes | ||||||
| Family history | ||||||
| FH + cancers vs noncancers | 7 | 1708 | 2283 | 5.6 | (2.3, 13.9) | 2.0 × 10–4 |
| FH– cancers vs noncancers | 6 | 2289 | 2207 | 3.3 | (1.4, 7.9) | 0.008 |
| FH + vs FH– cancers | 8 | 1955 | 2543 | 1.3 | (0.8, 2.3) | 0.305 |
| Metastatic | ||||||
| M1 cancers vs noncancers | 4 | 378 | 2074 | 6.4 | (2.0, 20.6) | 0.002 |
| M0 cancers vs noncancers | 8 | 3491 | 2410 | 3.8 | (1.7, 8.5) | 0.001 |
| M1 vs M0 cancers | 9 | 504 | 3910 | 1.8 | (0.8, 4.0) | 0.146 |
| Gleason | ||||||
| Gleason ≥8 vs noncancers | 6 | 1153 | 2207 | 5.5 | (2.2, 13.8) | 2.3 × 10–4 |
| Gleason 7 vs noncancers | 7 | 1064 | 2316 | 3.9 | (1.5, 10.4) | 0.006 |
| Gleason ≤6 vs noncancers | 5 | 1743 | 2113 | 3.1 | (1.2, 8.1) | 0.018 |
| Gleason ≥8 vs Gleason ≤6 | 7 | 1220 | 1952 | 1.3 | (0.7, 2.5) | 0.431 |
| Aggressive | ||||||
| Agg. vs noncancers | 8 | 2108 | 2410 | 5.4 | (2.4, 12.5) | 7.4 × 10–5 |
| Non-Agg. vs noncancers | 5 | 1412 | 2113 | 3.2 | (1.1, 9.2) | 0.028 |
| Agg. vs non-Agg. Cancers | 9 | 2184 | 1613 | 1.6 | (0.9, 3.0) | 0.135 |
| Age at diagnosis | ||||||
| <65 cancers vs noncancers | 8 | 3095 | 2410 | 4.9 | (2.2, 11.1) | 1.3 × 10–4 |
| ≥65 cancers vs noncancers | 8 | 1652 | 2421 | 3.8 | (1.4, 10.4) | 0.010 |
| <65 cancers vs ≥65 cancers | 10 | 3623 | 1650 | 2.0 | (1.0, 3.7) | 0.037 |
Agg. = aggressive; CI = confidence interval; FH = family history; OR = odds ratio; Ref. = reference.
Controls for case/control analyses. Lower-risk subcategory for case-only analyses.
Analysis of tier 2 ATM variants also revealed a positive association with PrCa risk, albeit smaller than that observed for tier 1 (OR = 1.4, 95% CI: 1.1–1.7, p = 0.008; Table 4). In subgroup analyses, ORs were elevated for cases with a first-degree family history of PrCa (OR = 1.6, 95% CI: 1.2–2.1) and cases with metastatic disease (OR = 1.5, 95% CI: 1.0–2.3); however, we again could not conclude that there were appreciable differences within a subgroup. When we combined tier 1 and 2 variants, a positive association between PrCa risk and carrier status was observed once more (OR = 1.5, 95% CI: 1.2–1.9, p = 9.3 × 10–5; Table 5). Subtype analyses revealed trends broadly similar to those seen for tier 1 variants alone, with none of these differences being statistically significant. We also investigated the effect of classifying tier 1 variants as protein truncating variants (PTVs) or non-PTVs, and found that the association with PTVs for overall PrCa risk was stronger (OR = 5.7, 95% CI: 2.1–15.3; Supplementary Table 9).
Table 4.
Primary results, tier 2
| Studies (N) | Cancers (N) | Ref. groupa (N) | OR | 95% CI | p value | |
|---|---|---|---|---|---|---|
| Tier 2 | ||||||
| Overall | 11 | 5081 | 2904 | 1.4 | (1.1, 1.7) | 0.008 |
| Subtypes | ||||||
| Family history | ||||||
| FH + cancers vs noncancers | 9 | 1832 | 2690 | 1.6 | (1.2, 2.1) | 0.002 |
| FH– cancers vs noncancers | 9 | 2352 | 2690 | 1.2 | (0.9, 1.6) | 0.206 |
| FH + vs FH– cancers | 10 | 2079 | 2584 | 1.2 | (0.9, 1.5) | 0.242 |
| Metastatic | ||||||
| M1 cancers vs noncancers | 8 | 497 | 2410 | 1.5 | (1.0, 2.3) | 0.065 |
| M0 cancers vs noncancers | 9 | 3509 | 2809 | 1.3 | (1.0, 1.7) | 0.047 |
| M1 vs M0 cancers | 9 | 504 | 3910 | 1.2 | (0.8, 1.7) | 0.422 |
| Gleason | ||||||
| Gleason ≥8 vs noncancers | 11 | 1234 | 2904 | 1.3 | (0.9, 1.8) | 0.135 |
| Gleason 7 vs noncancers | 10 | 1148 | 2896 | 1.5 | (1.1, 2.1) | 0.008 |
| Gleason ≤6 vs noncancers | 11 | 1937 | 2904 | 1.3 | (1.0, 1.8) | 0.052 |
| Gleason ≥8 vs Gleason ≤6 | 11 | 1296 | 2114 | 0.9 | (0.7, 1.2) | 0.384 |
| Aggressive | ||||||
| Agg. vs noncancers | 11 | 2161 | 2904 | 1.3 | (1.0, 1.8) | 0.053 |
| Non Agg. vs noncancers | 9 | 1487 | 2820 | 1.4 | (1.0, 1.9) | 0.058 |
| Agg. vs non-Agg. Cancers | 11 | 2231 | 1667 | 0.9 | (0.7, 1.2) | 0.674 |
| Age at diagnosis | ||||||
| <65 cancers vs noncancers | 11 | 3272 | 2904 | 1.3 | (1.1, 1.7) | 0.018 |
| ≥65 cancers vs noncancers | 12 | 1809 | 2904 | 1.4 | (1.0, 1.9) | 0.054 |
| <65 cancers vs ≥65 cancers | 12 | 3710 | 1728 | 0.9 | (0.7, 1.2) | 0.370 |
Agg. = aggressive; CI = confidence interval; FH = family history; OR = odds ratio; Ref. = reference.
Controls for case/control analyses. Lower-risk subcategory for case-only analyses.
Table 5.
Primary results, tier 1 + tier 2
| Studies (N) | Cancers (N) | Ref. groupa (N) | OR | 95% CI | p value | |
|---|---|---|---|---|---|---|
| Tier 1 + 2 | ||||||
| Overall | 11 | 5081 | 2904 | 1.5 | (1.2, 1.9) | 9.3 × 10–5 |
| Subtypes | ||||||
| Family history | ||||||
| FH + cancers vs noncancers | 9 | 1832 | 2690 | 1.8 | (1.4, 2.4) | 1.4 × 10–5 |
| FH– cancers vs noncancers | 9 | 2352 | 2690 | 1.3 | (1.0, 1.8) | 0.039 |
| FH + vs FH– cancers | 10 | 2079 | 2584 | 1.2 | (1.0, 1.5) | 0.120 |
| Metastatic | ||||||
| M1 cancers vs noncancers | 8 | 497 | 2410 | 1.8 | (1.2, 2.7) | 0.005 |
| M0 cancers vs noncancers | 9 | 3509 | 2809 | 1.5 | (1.1, 1.8) | 0.002 |
| M1 vs M0 cancers | 9 | 504 | 3910 | 1.3 | (0.9, 1.8) | 0.197 |
| Gleason | ||||||
| Gleason ≥8 vs noncancers | 11 | 1234 | 2904 | 1.6 | (1.1, 2.2) | 0.005 |
| Gleason 7 vs noncancers | 10 | 1148 | 2896 | 1.7 | (1.2, 2.2) | 0.001 |
| Gleason ≤6 vs noncancers | 11 | 1937 | 2904 | 1.5 | (1.1, 2.0) | 0.009 |
| Gleason ≥8 vs Gleason ≤6 | 11 | 1296 | 2114 | 1.0 | (0.7, 1.2) | 0.715 |
| Aggressive | ||||||
| Agg. vs noncancers | 11 | 2161 | 2904 | 1.6 | (1.2, 2.1) | 7.8 × 10–4 |
| Non-Agg. vs noncancers | 9 | 1487 | 2820 | 1.5 | (1.1, 2.1) | 0.013 |
| Agg. vs non-Agg. Cancers | 11 | 2231 | 1667 | 1.0 | (0.8, 1.3) | 0.850 |
| Age at diagnosis | ||||||
| <65 cancers vs noncancers | 11 | 3272 | 2904 | 1.6 | (1.2, 2.0) | 2.0 × 10–4 |
| ≥65 cancers vs noncancers | 11 | 1809 | 2904 | 1.5 | (1.1, 2.1) | 0.008 |
| <65 cancers vs ≥65 cancers | 12 | 3710 | 1728 | 1.0 | (0.8, 1.3) | 0.995 |
Agg. = aggressive; CI = confidence interval; FH = family history; OR = odds ratio; Ref. = reference.
Controls for case/control analyses. Lower-risk subcategory for case-only analyses.
Lastly, we investigated the relationship between ATM variants and PrCa-specific death in more detail. Compared with a prevalence of 1.1% in overall PrCa, tier 1 variants were slightly enriched in lethal PrCa cases at 1.7% (95% CI: 1.1–2.7). The prevalence in lethal PrCa cases by categories of age at death was 2.3% (95% CI: 1.2–3.9), 2.0% (95% CI: 0.7–4.2), and 0.4% (95% CI: 0.0–2.0) for those who died at ages <65, 65–74, and ≥75 yr. For tier 2 variants, the prevalence for the age categories was 6.3% (95% CI: 4.4–8.6), 4.9% (95% CI: 2.8–8.0), and 5.7% (95% CI: 3.3–9.1), respectively. For tier 1 and 2 variants combined, the overall prevalence was 7.3% (95% CI: 5.9–8.9), and prevalence by the categories of age at death was 8.4% (95% CI: 6.2–11.0), 6.6% (95% CI: 4.1–10.0), and 6.0% (95% CI: 3.6–9.5), respectively (Supplementary Table 8). In the time-to-event case-only analysis, which was restricted to the MCCS and ICR cohorts, the risk of dying from PrCa for tier 1 variant carriers compared with that for noncarriers was as follows: HR: 1.3 (95% CI: 0.8–2.3, p = 0.3). For tier 2, the HR was 1.0 (95% CI: 0.7–1.3, p = 0.8), and for tiers 1 and 2 combined, HR was 1.0 (95% CI: 0.8–1.3, p = 0.9; Supplementary Table 11).
Discussion
Although GWASs have identified many common, low penetrance PrCa susceptibility loci, no genes have consistently been demonstrated to either have a large effect on risk or contribute to aggressive disease presentation aside from BRCA2 and HOXB13. In the large germline sequencing study presented here, our aim was to determine the contribution of rare ATM variants to PrCa predisposition and risk of aggressive disease. We focused primarily on tier 1 variants, which included all rare predicted loss-of-function variants and any nontruncating variants listed as pathogenic or likely pathogenic in ClinVar, and demonstrated their substantial contribution to PrCa susceptibility. We further demonstrated that tier 2 ATM variants (rare nontruncating variants predicted to be deleterious by CADD or REVEL) also showed a lower magnitude of association with PrCa risk, suggesting that a subset of rare tier 2 variants of uncertain significance also contributes to PrCa predisposition.
To date, BRCA2 is the only gene in which tier 1 variants have consistently been linked to aggressive PrCa. We also attempted to establish whether ATM carrier status is associated with family history, age of onset, or other clinical features of disease, as previously suggested in smaller studies [13]. We observed a higher tier 1 mutation prevalence in cases diagnosed at <65 yr of age, and this finding would support that screening should be started at an earlier age for men carrying a mutation. However, we could not conclude that ATM mutations, either tier 1 or tier 2, predispose specifically to, or are sufficient to distinguish, more aggressive phenotypes.
Our analysis included only samples of European ancestry, as data from other ethnicities were available only in limited numbers. Whilst differences in mutation prevalence were observed between study groups, overall, ATM carrier frequencies appear to be higher in Europeans than in other ancestral populations. A recent PrCa sequencing study in Japanese men [12] reported a significant association between rare pathogenic ATM variants and PrCa risk despite much lower frequencies in both cases and controls (0.5% vs 0.2%; OR: ∼3), whilst a significant association was also reported for African ancestry cases and controls (0.48% vs 0.3%; OR: ∼2) [30].
The importance of identifying DNA repair gene mutation carriers is becoming increasingly evident in the era of precision medicine and targeted therapies [31]. Currently, germline testing of PrCa patients is recommended only for metastatic disease or a family history suggestive of hereditary PrCa, with BRCA2 considered a priority gene to screen for in all settings. ATM sequencing is considered for informing clinical trial eligibility or active surveillance decisions, and for screening of healthy men in high-risk PrCa families [32]. BRCA1/2 mutation carriers with breast, ovarian, and other cancer types have been shown to benefit from PARP inhibitor treatment in clinical trial studies [33], [34], whilst initial studies in metastatic castrate-resistant PrCa patients, which included ATM mutation carriers (some with missense variants), have demonstrated evidence that a significant proportion of these patients also responded favourably to this treatment [19], [35]. A recent report has also found that for men with localised PrCa, rates of germline pathogenic ATM mutations were significantly enriched in cases with Gleason score >7 tumours compared with the rates in cases with Gleason score 6 tumours [36]. In conjunction with our findings that men with PrCa are at substantially higher risk of harbouring germline ATM mutations and that these men are possibly also at increased risk of younger-onset and more aggressive clinical presentation, this suggests that larger clinical trials may be warranted to further clarify which patients would benefit from targeted screening and personalised treatments, in addition to whether ATM mutation carriers with localised or locally advanced PrCa may also benefit from earlier treatment with PARP inhibitors, prior to progression to incurable metastatic castrate-resistant disease spread. Opposing evidence has also been reported with respect to the effectiveness and toxicity of radiotherapy in individuals with ATM sequence variants. In PrCa patients, ATM sequence variants have previously been implicated in a greater likelihood of adverse responses to radiotherapy [37], [38], [39], although an enhanced response to radiotherapy with no apparent increase in toxicity has also been reported for patients with pathogenic ATM mutations [40]. In the context of breast cancer, there is also conflicting evidence around the role of pathogenic ATM variants on the effectiveness of radiation therapy [41]. The implications regarding the use of radiotherapy in PrCa patients who are carriers of germline ATM mutations would therefore warrant further investigation.
To our knowledge, this analysis represents the largest ATM sequencing study to estimate PrCa risk in men of European ancestry to date. Despite our large sample size, ATM carrier numbers remained relatively modest, especially within individual study groups. As the frequency of each single variant was low, we could not make direct conclusions in relation to the specific effect of individual variants on overall PrCa risk or clinical subtypes of the disease. It is also important to note that each study group recruited men according to different criteria, some enriching for aggressive or younger age at diagnosis disease, in addition to using different sequencing technologies and analysis pipelines. For this reason, whilst analyses showed that, in aggregate, ATM tier 1 variants, and to a lesser extent tier 2 variants, have a relatively large and significant association with PrCa predisposition, our overall estimates of association could potentially be inflated. In an attempt to discern and control for heterogeneity that may have been introduced by these differences, we performed a two-stage analysis, but due to small sample sizes within individual studies these analyses may still have been underpowered. We were also limited in our ability to detect differences in mutation prevalence between samples or study sites.
Conclusions
Our study provides strong support for ATM as a moderate penetrance PrCa risk gene for men of European ancestry. Men who had developed any form of PrCa had an approximately fourfold risk of carrying a protein truncating or likely pathogenic nontruncating germline ATM variant. There was evidence that men who carry a tier 1 mutation had a higher risk for early-onset disease. This result provides more robust OR estimates for use in genetic counselling of male carriers, and targeted screening studies will be needed to determine whether genetic-based PrCa screening identifies a higher number of younger patients. These results also provide further information for the selection of relevant therapeutic options for PrCa patients and the management of high-risk disease [32].
Author contributions: Zsofia Kote-Jarai had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kote-Jarai, Eeles, The PRACTICAL Consortium.
Acquisition of data: Wakerell, Saunders, Muir, Neal, Giles, MacInnis, Thibodeau, McDonnell, Cannon-Albright, Teixeira, Paulo, Cardoso, Huff, Li, Yu, Scheet, Permuth, Stanford, Dai, Ostrander, Cussenot, Cancel-Tassin, Hoegel, Herkommer, Schleutker, Tammela, Rathinakannan, Sipeky, Wiklund, Grönberg, Aly, Isaacs, Dickinson, FitzGerald, Chua, Nguyen-Dumont.
Analysis and interpretation of data: Kote-Jarai, Karlsson, Brook.
Drafting of the manuscript: Kote-Jarai, Eeles, Karlsson, Brook, Saunders, Southey, Schaid.
Critical revision of the manuscript for important intellectual content: Kote-Jarai, Eeles, Karlsson, Brook, Saunders, Southey, Schaid.
Statistical analysis: Brook, Dadaev.
Obtaining funding: Kote-Jarai, Eeles, Southey, Schaid.
Administrative, technical, or material support: None.
Supervision: Kote-Jarai, Eeles.
Other: None.
Financial disclosures: Zsofia Kote-Jarai certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This study was supported by The PRACTICAL Consortium. We acknowledge support from Cancer Research UK (C5047/A17528), from National Cancer Institute at the National Institutes of Health (R01 CA196931) the Bob Champion Trust, and the Global Challenges Research Fund (GCRF).
Acknowledgements
A full list of acknowledgements for each study, and The PRACTICAL Consortium, can be found in the Supplementary material. There are restrictions to the availability of datasets and code because the data are not public.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.euo.2020.12.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Mucci L.A., Hjelmborg J.B., Harris J.R. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matejcic M., Saunders E.J., Dadaev T. Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat Commun. 2018;9:4616. doi: 10.1038/s41467-018-06863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dadaev T., Saunders E.J., Newcombe P.J. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat Commun. 2018;9:2256. doi: 10.1038/s41467-018-04109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumacher F.R., Al Olama A.A., Berndt S.I. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kote-Jarai Z., Leongamornlert D., Saunders E. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro E., Goh C., Olmos D. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cybulski C., Wokolorczyk D., Kluzniak W. An inherited NBN mutation is associated with poor prognosis prostate cancer. Br J Cancer. 2013;108:461–468. doi: 10.1038/bjc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leongamornlert D., Saunders E., Dadaev T. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110:1663–1672. doi: 10.1038/bjc.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leongamornlert D.A., Saunders E.J., Wakerell S. Germline DNA repair gene mutations in young-onset prostate cancer cases in the UK: evidence for a more extensive genetic panel. Eur Urol. 2019;76:329–337. doi: 10.1016/j.eururo.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mijuskovic M., Saunders E.J., Leongamornlert D.A. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br J Cancer. 2018;119:96–104. doi: 10.1038/s41416-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Momozawa Y., Iwasaki Y., Hirata M. Germline pathogenic variants in 7,636 Japanese patients with prostate cancer and 12,366 controls. J Natl Cancer Inst. 2020;112:369–376. doi: 10.1093/jnci/djz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Na R., Zheng S.L., Han M. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71:740–747. doi: 10.1016/j.eururo.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard C.C., Mateo J., Walsh M.F. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusak B., Kluzniak W., Wokolorczykv D. Inherited NBN mutations and prostate cancer risk and survival. Cancer Res Treat. 2019;51:1180–1187. doi: 10.4143/crt.2018.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang K.L., Mashl R.J., Wu Y. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–370. doi: 10.1016/j.cell.2018.03.039. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darst B.F., Dadaev T., Saunders E., et al. Germline sequencing DNA repair genes in 5,545 men with aggressive and non-aggressive prostate cancer. J Natl Cancer Inst. In press. https://doi.org/10.1093/jnci/djaa132. [DOI] [PMC free article] [PubMed]
- 18.Lu C., Xie M., Wendl M.C. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J., Carreira S., Sandhu S. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landrum M.J., Lee J.M., Benson M. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W., Gil L., Hunt S.E. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis N.M., Rothstein J.H., Pejaver V. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stukel T.A., Demidenko E., Dykes J., Karagas M.R. Two-stage methods for the analysis of pooled data. Stat Med. 2001;20:2115–2130. doi: 10.1002/sim.852. [DOI] [PubMed] [Google Scholar]
- 26.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 27.Bradburn M.J., Deeks J.J., Berlin J.A., Russell Localio A. Much ado about nothing: a comparison of the performance of meta-analytical methods with rare events. Stat Med. 2007;26:53–77. doi: 10.1002/sim.2528. [DOI] [PubMed] [Google Scholar]
- 28.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 29.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 30.Matejcic M., Patel Y., Lilyquist J. Pathogenic variants in cancer predisposition genes and prostate cancer risk in men of African ancestry. JCO Precis Oncol. 2020;4:32–43. doi: 10.1200/PO.19.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglehart JD, Silver DP. Synthetic lethality—a new direction in cancer-drug development. N Engl J Med. 2009;361:189–191. doi: 10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- 32.Giri V.N., Knudsen K.E., Kelly W.K. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol. 2020;38:2798–2811. doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caulfield SE, Davis CC, Byers KF. Olaparib: a novel therapy for metastatic breast cancer in patients with a BRCA1/2 mutation. J Adv Pract Oncol. 2019;10:167–174. [PMC free article] [PubMed] [Google Scholar]
- 34.McLachlan J., George A., Banerjee S. The current status of PARP inhibitors in ovarian cancer. Tumori. 2016;102:433–440. doi: 10.5301/tj.5000558. [DOI] [PubMed] [Google Scholar]
- 35.de Bono J., Mateo J., Fizazi K. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y., Yu H., Li S. Rare germline pathogenic mutations of DNA repair genes are most strongly associated with grade group 5 prostate cancer. Eur Urol Oncol. 2020;3:224–230. doi: 10.1016/j.euo.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Andreassen CN, Rosenstein BS, Kerns SL. Individual patient data meta-analysis shows a significant association between the ATM rs1801516 SNP and toxicity after radiotherapy in 5456 breast and prostate cancer patients. Radiother Oncol. 2016;121:431–439. doi: 10.1016/j.radonc.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cesaretti J.A., Stock R.G., Lehrer S. ATM sequence variants are predictive of adverse radiotherapy response among patients treated for prostate cancer. Int J Radiat Oncol Biol Phys. 2005;61:196–202. doi: 10.1016/j.ijrobp.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Hall E.J., Schiff P.B., Hanks G.E. A preliminary report: frequency of A-T heterozygotes among prostate cancer patients with severe late responses to radiation therapy. Cancer J Sci Am. 1998;4:385–389. [PubMed] [Google Scholar]
- 40.Lu C., Pitter K.L., Casey D.L. Pathogenic mutations in ATM predict for enhanced local control in prostate cancers treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2018;102:e126–7. [Google Scholar]
- 41.Jerzak K.J., Mancuso T., Eisen A. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer: a narrative review. Curr Oncol. 2018;25:e176–80. doi: 10.3747/co.25.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.