Abstract
To date, the locus with the most robust human genetic association to COVID-19 severity is 3p21.31. Here, we integrate genome-scale CRISPR loss-of-function screens and eQTLs in diverse cell types and tissues to pinpoint genes underlying COVID-19 risk. Our findings identify SLC6A20 and CXCR6 as putative causal genes that modulate COVID-19 risk and highlight the usefulness of this integrative approach to bridge the divide between correlational and causal studies of human biology.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-021-02454-4.
Keywords: COVID-19, SARS-CoV-2, GWAS, eQTL, CRISPR
Background
COVID-19, which is caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection, results in diverse individual disease courses from asymptotic carriers to severe disease with respiratory failure [1, 2]. A growing body of evidence suggests also an important role of genetic factors in COVID-19 susceptibility and severity [3–5]. Most notably, the chromosome 3p21.31 locus was one of the first genome-wide significant loci found to be associated with COVID-19 [3]. To date, 3p21.31 encompasses the strongest and the most replicated signal for more severe outcomes of SARS-CoV-2 infection. However, the functional mechanisms of this association are unclear. The locus includes multiple protein-coding genes, for example, LIMD1, SACM1L, SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1, many of which have a potentially relevant role in the pathophysiology of COVID-19. As genetic variants have been shown to often exert their effect on complex traits or disease via cis-regulation of gene expression [6, 7], expression quantitative trait locus (eQTL) mapping could serve as a means to pinpoint candidate genes for traits or diseases of interest.
We previously performed a genome-scale CRISPR loss-of-function screen to identify genes required for SARS-CoV-2 viral infection in human lung epithelial-like cells expressing ACE2 (A549ACE2) [8]. Top-ranked genes from this CRISPR screen have an established link in SARS-CoV-2 infection in a cell line and could be informative for prioritizing host genes for COVID-19. In this work, we present an integrative approach using the results of the CRISPR screen and eQTLs in various cell types and tissues from the eQTL Catalogue [9] and Genotype Tissue Expression (GTEx) v8 [7] data release to pinpoint genes underlying COVID-19 risk in the 3p21.31 locus. Our results suggest that genes enriched in the in vitro CRISPR screen contribute to COVID-19 risk in humans, and highlight SLC6A20 and CXCR6 as the putative causal genes in the 3p21.31 COVID-19 risk locus.
Results and discussion
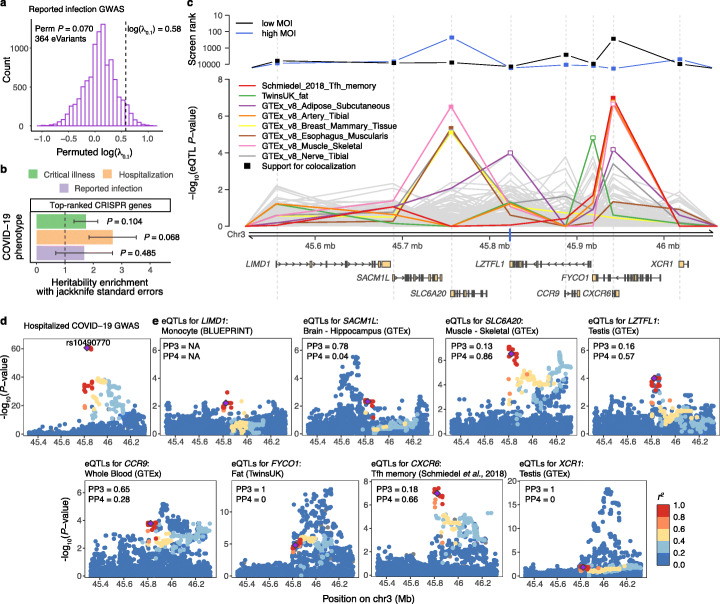
To set the stage for identifying the causal genes for COVID-19 in the 3p21.31 locus, we first evaluated whether the top-ranked genes from the CRISPR screen in human lung cells contribute to COVID-19 risk in humans. We hypothesized that genetic regulatory variants for genes pinpointed by the CRISPR screen would show an increased signal for the genetic association for COVID-19 in genome-wide association studies (GWAS) in the human population. To this end, we analyzed if cis-eQTLs from lung tissue [7] for these genes are enriched for overall association signal in data from the COVID-19 Host Genetics Initiative (HGI) [5] for the three main COVID-19-related phenotypes: critical illness (A2), hospitalization (B2), reported infection (C2) as compared to population controls without known SARS-CoV-2 infection. Indeed, we observed a trend towards significance in inflation of the GWAS association signal for variants that are cis-eQTLs for the top-ranked genes from the CRISPR screen in the SARS-CoV-2 reported infection GWAS, compared to all lung cis-eQTLs (λ0.1 = 1.78, permutation P value = 0.070, Fig. 1a). We did not see a significant signal for the other two COVID-19 phenotypes in the present data (Additional file 1: Figure S1). Additionally, by using stratified linkage disequilibrium (LD) score regression [10], we noticed a suggestive signal for enriched heritability in the top-ranked genes for hospitalization conditional on the 96 annotations in the baseline model and the set of all protein-coding genes (P = 0.068, Fig. 1b). Thus, our results suggest the link between genes enriched in the in vitro CRISPR screen and the genetic component of COVID-19 risk in humans, but future COVID-19 GWAS with larger sample size are needed to refine this suggested association.
Fig. 1.
Genetic regulatory effects of top-ranked CRISPR genes and prioritization of genes in the 3p21.31 locus associated with COVID-19 GWAS. a Histogram of the permuted log(λ0.1) to test the significance of the inflation in reported SARS-CoV-2 infection GWAS for variants that are cis-eQTLs for top-ranked CRISPR genes in Lung (n = 364). Vertical dashed line denotes the observed log(λ0.1) value. b Heritability enrichment in top-ranked genes from the CRISPR screen for the three main COVID-19 phenotypes. Vertical dashed line denotes no enrichment. c Prioritization of genes in the 3p21.31 locus by integrating CRISPR screens and cis-eQTLs. The top panel shows the ranking of the genes in the locus according to the second-best guide RNA score in the low MOI (black) and high MOI (blue) pooled CRISPR screens. The middle panel shows the cis-eQTL P values for the lead GWAS variant in the 3p21.31 locus (rs10490770, denoted as a blue tick on the x-axis) in different cell types and tissues from the eQTL Catalogue and GTEx, 112 eQTL data sets in total. Highlighted are eight cell types/tissues, where the cis-eQTL P value for the lead GWAS variant is < 10−4 for at least one gene in the region. Filled square denotes support for colocalization between the GWAS and cis-eQTL signal (posterior probability for one shared causal variant (PP4) > 0.5). The bottom panel depicts the transcripts of the eight protein-coding genes in the locus. Ranks and cis-eQTL P values are aligned to match the start of the gene which is shown as a gray dashed line across the panels. d, e Regional association plots of the hospitalized COVID-19 GWAS (d) and cis-eQTLs for the eight genes (e) from the associated locus in the cell type/tissue where the lead GWAS variant has the lowest cis-eQTL P value. Purple diamond denotes the lead GWAS variant, and the data points are colored based on the (weighted average) LD between the lead GWAS variant and other variants in the region in the respective study population. PP3 and PP4—posterior probability for two different variants or one shared causal variant in coloc, respectively
To elucidate which genes in the chromosome 3 locus might mediate the genetic association, we first observed that the Solute Carrier Family 6 Member 20 (SLC6A20) gene and the C-X-C Motif Chemokine Receptor 6 (CXCR6) gene have a relatively high rank in our CRISPR screen (Fig. 1c). Next, we analyzed if the COVID-19 hospitalization GWAS lead variant rs10490770, where the alternative allele increases risk, affects expression of any of these genes in the locus. Using data from the eQTL Catalogue [9] and GTEx v8 [7], we analyzed all cis-eQTL associations for this variant and performed colocalization analysis [11, 12] to assess if the cis-eQTL and GWAS signal share a genetic cause. CXCR6 and SLC6A20 stood out, with the eQTL data indicating that the COVID-19 severity variant affects the expression of these two genes (Fig. 1c, Additional file 1: Figure S2). The cis-eQTL associated with lower expression of CXCR6 is active in memory T follicular helper (Tfh) CD4+ T cells [13] and colocalizes with the GWAS (PP4 = 0.66, Fig. 1d–e). The colocalization signal was very strong for the cis-eQTL associated with higher expression of SLC6A20 in four tissues from GTEx - breast mammary tissue, esophagus muscularis, skeletal muscle, and tibial nerve (0.70 < PP4 < 0.90, Additional file 1: Figure S3, Additional file 2: Table S1). Other genes in the region, such as FYCO1 and XCR1, also have regulatory variants affecting their expression, but two distinct variants likely drive the GWAS and eQTL signals (Fig. 1d–e). Also, we applied joint likelihood mapping (JLIM) [14] and corroborated the evidence for shared genetic effects of cis-eQTLs for CXCR6 and SLC6A20 and COVID-19 hospitalization (Additional file 1: Figure S4, Additional file 2: Table S2). Of note, colocalization of cis-eQTLs for LZTFL1 in the testis and hospitalization GWAS with coloc was not replicated using JLIM (PJLIM > 0.05/8). Additionally, we did not observe evidence for colocalization signal with splicing QTLs (sQTLs) nor transcript usage QTLs (tuQTLs) in cis (Additional file 2: Table S3). To confirm the robustness of the observed colocalization signal, we showed that the COVID-19 hospitalization signal was observed in the reported infection GWAS (Additional file 1: Figure S5). Given that both the CRISPR screen and eQTL data support the causal role of SLC6A20 and CXCR6, it is possible that the COVID-19 GWAS association in the 3p21.31 locus is driven by pleiotropic effects of the same variant on multiple genes in different cell types.
In addition to human genetic evidence and support from experimental data, both SLC6A20 and CXCR6 have a plausible biological function that could affect COVID-19. Notably, SLC6A20 functionally interacts with ACE2 [15], the receptor of the SARS-CoV-2 Spike protein that is the key host gene for viral entry [16]. In contrast, CXCR6 regulates the localization of resident memory T (TRM) cells in the lung and maintains a pool of airway TRM cells, critical for cellular immunity against respiratory pathogens [17]. It is also an alternate coreceptor for HIV [18, 19], raising a hypothesis of a similar function in lung cells for SARS-CoV-2. The fact that we observed cis-eQTLs for SLC6A20 and CXCR6 in different tissues and cell types other than lung highlights the complexity and possibility of extrapulmonary spread of SARS-CoV-2 [20], as well as potential pleiotropic effects of these genes in multiple cell types and physiological processes leading to COVID-19. However, further research is needed to establish whether all these cis-eQTLs have a causal role in COVID-19 risk. Importantly, GWAS can only show associations for common variants in the human population, and thus, the functional role of these genes in lung cells, captured by the screen, may not be captured by GWAS and cis-eQTL data.
Notably, in addition to the well-replicated signal for COVID-19 severity in the 3p21.31 locus, other independent signals more strongly associated with susceptibility to SARS-CoV-2 infection in the same region have been identified as sample sizes for COVID-19 GWAS have grown [21]. The two candidate genes based on our integrative analysis, SLC6A20 and CXCR6, have been prioritized for reported infection and more severe outcomes of SARS-CoV-2 infection, respectively [5]. Corroborating evidence from entirely independent approaches pinpointing SLC6A20 and CXCR6 presented here, together with additional evidence from other studies, warrants further research into exploring the virological or mechanistic mode of action of these two genes in understanding their role in COVID-19 susceptibility and severity.
Conclusions
In this work, our genome-wide analysis suggested that genes required for SARS-CoV-2 infection in vitro also contribute to COVID-19 susceptibility in humans. By integrating the results of CRISPR screen and cis-eQTLs, we have identified SLC6A20 and CXCR6 as potential protein-coding genes in the 3p21.31 locus through which noncoding variants associated with COVID-19 risk in human patients may function. This integrative approach should prove useful for other human diseases and pathogens to bridge the divide between correlational and causal studies of human biology.
Methods
Genome-wide CRISPR screen and guide RNA analysis
Details regarding the SARS-CoV-2 CRISPR screen in A549 human lung epithelial cells that over-express ACE2 (A549ACE2) have been described before [8]. Briefly, human GeCKOv2 A and B libraries (Addgene, 1000000048) were used for the genome-wide CRISPR-Cas9 screen [22]. After transduction and selection for the GeCKOv2 library (maintaining ~ 1000-fold library representation), we infected 125 million A549ACE2 cells with SARS-CoV-2 virus (NIAID BEI isolate USA-WA1/2020) at two multiplicity of infections (MOIs): 0.01 (low MOI) and 0.3 (high MOI), which differ by the amount of virus used to infect the cells. Surviving cells were collected on day 6 post-infection, when genomic DNA was isolated, guide RNAs were recovered by PCR, and guide representation was determined by next-generation sequencing (Illumina).
We aligned trimmed sequencing reads to the GeCKOv2 reference using bowtie v1.1.2 [-a --best --strata -v 1 –norc] allowing 1 nucleotide mismatch to determine the read counts per guide. Alignment rates were ~ 80% for all samples. We normalized read counts between biological samples and computed a fold-change by comparison of SARS-CoV-2-infected samples to the uninfected control. To identify genes where loss-of-function mutations reduce SARS-CoV-2 infection resulting in enrichment within the pool, the genes were ranked based on three methods: robust-rank aggregation (RRA) [23], RIGER (weighted-sum), and second-best rank method. For example, according to the second-best rank method, each gene in the human GeCKOv2 A+B library is targeted by 6 different guides. Genes are ranked based on the guide RNA with second highest fold-change (sbScore), as described before [24–26].
For enrichment analyses, we used a set of top-ranked (top500) genes in low MOI condition by each of three ranking methods (RRA, RIGER, sbScore), see Figure S1C in Daniloski et al. [8], resulting in n = 890 unique protein-coding genes across the methods.
COVID-19 phenotypes and GWAS
We used summary statistics of the three main COVID-19 GWAS generated by the COVID-19 HGI [27] based on round 5, worldwide meta-analyses without 23andMe, released January 18, 2021 [5]: (1) A2 (critical illness)—very severe respiratory confirmed COVID vs. population (ncases = 5582, ncontrols = 709,010), (2) B2 (hospitalization)—hospitalized covid vs. population (ncases = 12,888, ncontrols = 1,295,966), (3) C2 (reported infection)—COVID vs. population (ncases = 36,590, ncontrols = 1,668,938). As the lead GWAS variant in the 3p21.31 locus to highlight on figures, we used rs10490770 (3:45823240:T:C), which had the lowest P value in the hospitalization COVID-19 worldwide meta-analysis with 23andMe as reported by the COVID-19 HGI [5].
Inflation of COVID-19 GWAS signal for variants that are cis-eQTLs
To estimate the importance of the genes enriched in the CRISPR screen in modulating the COVID-19 risk in humans, we tested if we observe inflation of signal in the aforementioned COVID-19 GWAS for variants that are cis-eQTLs in GTEx v8 lung [7] for the top-ranked genes from the CRISPR screen. We measured inflation using the lambda inflation statistic relative to the chi-square quantile function of 0.1, λ0.1, i.e., estimating the inflation among 10% of the most significant tests. We calculated λ0.1 for two sets of cis-eQTLs: (1) all lead cis-eQTLs in the lung (9557 protein-coding genes with significant cis-eQTLs in the lung at false discovery rate (FDR) < 0.05, serving as a background set), (2) all lead cis-eQTLs in the lung for the top-ranked genes from the CRISPR screen (439 out of 890 top-ranked protein-coding genes have cis-eQTLs in the lung with FDR < 0.05).
To test the significance of λ0.1, we used a permutation-based test. We selected n number of lead cis-eQTLs from the background set k = 10,000 times, where n is the number of lead cis-eQTLs for the top-ranked genes from the CRISPR screen tested in a given COVID-19 GWAS, and calculated λ0.1 on the permuted data. To calculate two-sided permutation P values, we applied log-transformation on the permuted λ0.1 values to get a symmetric null distribution. We then calculated permutation P value as the proportion of permuted log(λ0.1) as extreme as or more extreme than the observed log(λ0.1).
Partitioned heritability analysis
We performed stratified LD score regression (LDSC) [10] to calculate the single nucleotide polymorphism (SNP) heritability of COVID-19 phenotypes in the top-ranked genes from the CRISPR screen using GWAS summary statistics from the European-only meta-analysis (round 5, excluding 23andMe): A2—critical illness (ncases = 4606, ncontrols = 702,801), B2—hospitalization (ncases = 9373, ncontrols = 1,197,256), and C2—reported infection (ncases = 29,071, ncontrols = 1,559,712). We generated genome-wide custom annotation files and LD scores using the top-ranked genes identified in the CRISPR screen (n = 890 protein-coding genes across the three ranking methods) and used all protein-coding genes as background (--gene-coord-file in make_annot.py from LDSC package). We added 100 kb windows on either side of the transcribed region of each gene. Next, we jointly modeled the annotation corresponding to the top-ranked genes and the 96 annotations in the “baseline model” (baselineLD_v2.2.tgz, https://alkesgroup.broadinstitute.org/LDSCORE/GRCh38/). We have used regression weights (weights.tgz, https://alkesgroup.broadinstitute.org/LDSCORE/GRCh38/) from HapMap3 SNPs, excluding the HLA region and restricted to SNPs with minor allele frequency (MAF) > 5% (plink_files.tgz, https://alkesgroup.broadinstitute.org/LDSCORE/GRCh38/).
Prioritization of genes in the 3p21.31 locus based on CRISPR screen and eQTL data
We focused on eight genes, LIMD1, SACM1L, SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1, in the 3p21.31 locus that has been shown to robustly associate with COVID-19 severity [3–5]. Firstly, we ranked the genes based on the sbScore in the low MOI and high MOI CRISPR screen. Secondly, we gathered summary statistics for 112 cis-eQTL data sets from the eQTL Catalogue [9] (63 data sets, mostly immune cell types with and without treatment) and GTEx v8 [7] (49 tissues). We ranked the genes based on the P value in eQTL studies for the lead GWAS variant in the 3p21.31 locus. Since observing low eQTL P value for the lead GWAS variant does not necessarily translate into shared genetic causality, we further performed colocalization analysis.
Colocalization analysis of COVID-19 GWAS and cis-eQTLs in the 3p21.31 locus with coloc
To assess evidence for shared causal variant of a cis-eQTL and a COVID-19 GWAS (hospitalized and reported infection GWAS, round 5, worldwide meta-analysis without 23andMe), we used the Bayesian statistical test for colocalization, coloc [11], assuming one causal variant per trait. We only included cis-eQTLs for genes for colocalization test, if there was a cis-eQTL with a nominal P value < 10−4 within 100 kb of the lead GWAS variant in the 3p21.31 locus. Coloc was run on a 1 Mb region centered on the lead GWAS variant (+/− 500 kb from the variant) with prior probabilities set to p1 = 10−4, p2 = 10−4, p3 = 5 × 10−6. We defined suggestive support for colocalization between the COVID-19 GWAS and cis-eQTL signal if the posterior probability for one shared causal variant (PP4) > 0.5.
Allelic heterogeneity of gene expression in cis is widespread [7], and it violates the assumption of one causal variant per trait. Thus, we also used the development version of coloc [12] (https://github.com/chr1swallace/coloc/tree/condmask) in a wider 2 Mb region centered at the lead hospitalized COVID-19 GWAS variant. The enhanced version of coloc allows conditioning and masking to overcome one single causal variant assumption. For eQTL data sets from the eQTL Catalogue, we used method = mask with LD data from the 1000 Genomes Project [28] EUR population that matches the genetic ancestry of the study population in the majority of the studies in the eQTL Catalogue. For eQTL data sets from the GTEx Project, we used method = cond with LD calculated from the individuals that had gene expression data in the given tissue in GTEx. We used the mode = iterative and allowed for a maximum of three variants to condition/mask. We set the r2 threshold to 0.01 to call two signals independent when masking, and we set the P value threshold to 10−4 to call the secondary signal significant. We used method = single for the hospitalized COVID-19 GWAS. As a result, the maximum PP4 (posterior probability for shared causal variants) estimates with conditioning/masking were similar to PP4 estimates from the standard coloc run (Additional file 2: Table S1), suggesting no additional colocalization events with secondary independent cis-eQTLs.
Additionally, standard coloc with the options mentioned above was used to test for colocalization between COVID-19 hospitalization and cis-sQTLs from GTEx v8 and cis-tuQTLs from the eQTL Catalogue (including both transcript usage and txrevise event usage). Note that cis-tuQTLs were mapped also for the GTEx tissues by the eQTL Catalogue.
Colocalization analysis of COVID-19 GWAS and cis-eQTLs in the 3p21.31 locus with JLIM
To corroborate the observed colocalization signal found with coloc, we used JLIM (version 2.5) [14] with default options (https://github.com/cotsapaslab/jlim/). We treated hospitalized COVID-19 GWAS (round 5, European-only meta-analysis without 23andMe) as the primary trait and cis-eQTLs from GTEx v8 (mapped in European-American subjects) and the eQTL Catalogue (note that some studies are not from the European population) as the secondary trait. Reference genotype panel was created by including all the European subpopulations from the 1000 Genomes Project. By default, the analysis window was set to 200 kb, centered at the lead GWAS variant in the 3p21.31 locus.
Regional association plots for the 3p21.31 locus
Data points in the locuszoom-like regional association plots are colored by the LD between the lead GWAS variant in the 3p21.31 locus (rs10490770) and other variants in the region. For plotting data for COVID-19 GWAS and cis-eQTLs from the eQTL Catalogue, we used genotype data from the 1000 Genomes Project, and calculated weighted average r2 value based on the counts of global ancestral populations in the analysis (if multiple global ancestry populations were studied). Note that for European populations, we only included non-Finnish populations in the 1000 Genomes Project. For plotting data for cis-eQTLs from the GTEx project, we used genotype data available via dbGaP, accession phs000424.v8, and calculated r2 using the set of individuals that had expression data from the given tissue.
Supplementary Information
Additional file 1:. Supplementary figures S1-S5.
Additional file 2:. Supplementary tables S1-S3.
Acknowledgements
We thank the COVID-19 Host Genetics Initiative for making the results from meta-analyses public. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal and dbGaP accession number phs000424.v8.
Peer review information
Andrew Cosgrove was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3.
Authors’ contributions
S.K., Z.D., N.E.S., and T.L. designed the study. Z.D., T.X.J., B.R.t., and N.E.S. designed and performed the genome-scale CRISPR screen. S.K. and S.B. performed the eQTL and GWAS integration analyses. T.L. and N.E.S. supervised the work. S.K. and T.L. wrote the manuscript. Z.D. and N.E.S. contributed to the editing of the manuscript. The authors read and approved the final manuscript.
Authors’ information
Twitter handles: @silvakasela (Silva Kasela); @ZDaniloski (Zharko Daniloski); @s_bollepalli (Sailalitha Bollepalli); @virusninja (Benjamin R. tenOever); @nevillesanjana (Neville E. Sanjana); @tuuliel_lab (Tuuli Lappalainen).
Funding
S.K. and T.L. are supported by NIH/NHLBI (R01HL142028). Z.D. is supported by an American Heart Association postdoctoral fellowship (grant no. 20POST35220040). S.B. and T.L. are supported by NIH/NIMH (R01MH106842). Postdoctoral fellowship support for T.X.J. is provided by the NIH (grant no. R01AI123155). B.R.t. is supported by the Marc Haas Foundation, the National Institutes of Health, and DARPA’s PREPARE Program (HR0011-20-2-0040). N.E.S. is supported by NYU and NYGC startup funds, NIH/NHGRI (DP2HG010099), NIH/NCI (R01CA218668), DARPA (D18AP00053), the Sidney Kimmel Foundation, and the Brain and Behavior Foundation. T.L. is supported by NIH/NHGRI (UM1HG008901).
Availability of data and materials
The data sets analyzed during the current study are available from the following repositories: CRISPR screen data can be accessed from GEO repository under an accession number GSE158298 [29]), summary statistics of the COVID-19 GWAS (round 5) by the COVID-19 Host Genetics Initiative are available at https://www.covid19hg.org/results/r5/ [30], eQTL summary statistics can be downloaded from the eQTL Catalogue [31] and the GTEx Portal [32], and the variant calls from 1000 Genomes Project on the GRCh38 reference assembly from the EBI FTP site [33]. Analysis code has been deposited to the GitHub repository at https://github.com/LappalainenLab/covid-crispr-eqtl [34] under an Apache 2.0 License and at the repository Zenodo [35].
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
N.E.S. is an advisor to Vertex and Qiagen. T.L. advises and has equity in Variant Bio, and is a member of the scientific advisory board of Goldfinch Bio. The remaining authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Silva Kasela, Email: skasela@nygenome.org.
Tuuli Lappalainen, Email: tlappalainen@nygenome.org.
References
- 1.Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, et al. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed]
- 4.Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Somnez T, Coker D, et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. Preprint at medRxiv https://doi.org/10.1101/2020.09.04.20188318. 2020.
- 5.The COVID-19 Host Genetics Initiative, Ganna A. Mapping the human genetic architecture of COVID-19 by worldwide meta-analysis. Preprint at medRxiv 10.1101/2021.03.10.21252820. 2021.
- 6.Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniloski Z, Jordan TX, Wessels H-H, Hoagland DA, Kasela S, Legut M, et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2021;184:92-105.e16. [DOI] [PMC free article] [PubMed]
- 9.Kerimov N, Hayhurst JD, Manning JR, Walter P, Kolberg L, Peikova K, et al. eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs. Preprint at bioRxiv. 10.1101/2020.01.29.924266. 2020.
- 10.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16:e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of genetic polymorphisms on human immune cell gene expression. cell. 2018;175:1701-1715.e16. [DOI] [PMC free article] [PubMed]
- 14.Chun S, Casparino A, Patsopoulos NA, Croteau-Chonka DC, Raby BA, De Jager PL, et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuille-dit-Bille RN, Camargo SM, Emmenegger L, Sasse T, Kummer E, Jando J, et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47:693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-280.e8. [DOI] [PMC free article] [PubMed]
- 17.Wein AN, McMaster SR, Takamura S, Dunbar PR, Cartwright EK, Hayward SL, et al. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J Exp Med. 2019;216:2748–2762. doi: 10.1084/jem.20181308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao F, Alkhatib G, Peden KW, Sharma G, Berger EA, Farber JM. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaak H, Boers PHM, Gruters RA, Schuitemaker H, van der Ende ME, Osterhaus ADME. CCR5, GPR15, and CXCR6 Are Major Coreceptors of Human Immunodeficiency Virus Type 2 Variants Isolated from Individuals with and without Plasma Viremia. J Virol. 2005;79:1686. doi: 10.1128/JVI.79.3.1686-1700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts GHL, Partha R, Rhead B, Knight SC, Park DS, Coignet MV, et al. Novel COVID-19 phenotype definitions reveal phenotypically distinct patterns of genetic association and protective effects. Preprint at medRxiv https://doi.org/10.1101/2021.01.24.21250324. 2021.
- 22.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinforma Oxf Engl. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.COVID-19 Host Genetics Initiative. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet EJHG. 2020;28:715–8. [DOI] [PMC free article] [PubMed]
- 28.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed]
- 29.Daniloski Z, Jordan TX, tenOever BR, Sanjana NE. Identification of required host factors for SARS-CoV-2 infection in human cells. GSE158298. Gene Expression Omnibus. 2020 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158298. [DOI] [PMC free article] [PubMed]
- 30.COVID19-hg GWAS meta-analyses round 5. 2021 https://www.covid19hg.org/results/r5/.
- 31.eQTL Catalogue. 2021 https://www.ebi.ac.uk/eqtl/.
- 32.GTEx Portal. 2021 https://gtexportal.org/home/.
- 33.Variant calls from 1000 Genomes Project on the GRCh38 reference assembly. 2021 http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000_genomes_project/release/20190312_biallelic_SNV_and_INDEL/.
- 34.Kasela S, Bollepalli S. covid-crispr-eqtl. Github. 2021 https://github.com/LappalainenLab/covid-crispr-eqtl.
- 35.Kasela S, Bollepalli S. covid-crispr-eqtl. Zenodo. 2021 10.5281/zenodo.5144601.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1:. Supplementary figures S1-S5.
Additional file 2:. Supplementary tables S1-S3.
Data Availability Statement
The data sets analyzed during the current study are available from the following repositories: CRISPR screen data can be accessed from GEO repository under an accession number GSE158298 [29]), summary statistics of the COVID-19 GWAS (round 5) by the COVID-19 Host Genetics Initiative are available at https://www.covid19hg.org/results/r5/ [30], eQTL summary statistics can be downloaded from the eQTL Catalogue [31] and the GTEx Portal [32], and the variant calls from 1000 Genomes Project on the GRCh38 reference assembly from the EBI FTP site [33]. Analysis code has been deposited to the GitHub repository at https://github.com/LappalainenLab/covid-crispr-eqtl [34] under an Apache 2.0 License and at the repository Zenodo [35].