Abstract
The ability of Gram-negative bacteria to resist killing by antimicrobial agents and to avoid detection by host immune systems often entails modifications of the lipopolysaccharide (LPS). The PmrA/PmrB two-component system is the major regulator of these modifications. In this review, we describe the signals that activate the PmrA/PmrB system and the PmrA-regulated gene products that mediate chemical decoration of the LPS in the enteric pathogen Salmonella enterica. We then discuss the variety of feedback mechanisms that modulate the activity and thus the output of the PmrA/PmrB system, dictating when, where and to what extent bacteria modify their LPS. Finally, we explore how the evolution of orthologous PmrA/PmrB regulatory circuits among closely related bacteria has led to qualitative and quantitative differences in gene expression outputs, promoting survival in distinct ecological niches.
INTRODUCTION
Gram-negative bacteria are characteristically surrounded by an outer membrane, which consists of phospholipids in the inner leaflet and the glycolipid lipopolysaccharide (LPS) in the outer leaflet (Nikaido, 2003). The asymmetric nature of the outer membrane makes it an effective permeability barrier that protects bacteria from noxious compounds (Nikaido, 2003). The ability of bacteria to modify their LPS in response to signals experienced in host and non-host environments is critical for resistance to bactericidal agents (Shai, 1999; Vaara et al., 1979) and for evasion of host immune defenses (Raetz et al., 2007; Takeda et al., 2003). Many of these modifications are mediated by the PmrA/PmrB regulatory system, which is the major regulator of gene products that chemically alter the LPS in a wide variety of bacterial species, such as Salmonella enterica (Gunn, 2008), Escherichia coli (Hagiwara et al., 2004; Winfield and Groisman, 2004), Klebsiella pneumoniae (Mitrophanov et al., 2008), Yersinia pestis (Winfield et al., 2005), Citrobacter rodentium (Viau et al., 2011) and Pseudomonas aeruginosa (McPhee et al., 2006). The expression of these PmrA-dependent genes promotes resistance to antibiotics such as polymyxin B (Gunn, 2008; Raetz et al., 2007) and to toxic metals like Fe3+ (Chamnongpol et al., 2002).
The pmrA locus (for polymyxin resistance A) was first identified using spontaneous or chemical mutagenesis to isolate S. enterica serovar Typhimurium mutants that displayed enhanced resistance to polymyxin B (Makela et al., 1978; Roland et al., 1993). These mutants also exhibited increased resistance to several other cationic agents, including protamine, lysine polymers, the bulky amine Tris and antimicrobial peptides derived from human neutrophils (Roland et al., 1993; Shafer et al., 1984a; Shafer et al., 1984b; Vaara, 1981, 1992). LPS isolated from polymyxin B-resistant mutants bound less polymyxin B than LPS from the wild-type strain, suggesting that PmrA controls resistance to cationic antimicrobial peptides by altering one or more properties of the LPS (Vaara et al., 1981; Vaara et al., 1979). Genetic mapping and DNA sequence analysis later revealed that PmrA is encoded by the pmrCAB operon (Roland et al., 1993). PmrA is expressed from both a PmrA-activated promoter upstream of the pmrC gene (Wosten and Groisman, 1999) and a constitutive promoter located within the pmrC coding region (Gunn and Miller, 1996; Soncini and Groisman, 1996).
The PmrA response regulator and PmrB sensor constitute a two-component regulatory system. Such systems are key mediators of signal transduction that allow bacteria to alter gene expression programs in response to physical and/or chemical cues in the environment. The physiological functions regulated by two-component systems are diverse and include antibiotic resistance, virulence, chemotaxis, cell cycle progression and quorum sensing (Gao et al., 2007; Mascher et al., 2006). Classical two-component systems are usually encoded in operons and typically consist of both an integral membrane sensor kinase that responds to an input signal and a response regulator that determines the output (Mascher et al., 2006). Activation of the sensor kinase results in autophosphorylation from ATP at a conserved histidine residue followed by transfer of the phosphoryl group to a conserved aspartate residue on its cognate response regulator. The response regulator is often a transcription factor whose affinity for target promoters is modulated by phosphorylation, allowing the organism to alter its gene expression profile in response to environmental changes (Gao et al., 2007). In many cases, sensor kinases are bifunctional and exhibit phosphatase activity towards their cognate phosphorylated response regulators when inducing signals are absent (Figure 1). As such, the signaling output is proportional to the levels of phosphorylated response regulator. Because the majority of response regulators are active only when phosphorylated (Hoch, 2000), the ability of a response regulator to activate or repress its target genes is controlled not only by the presence of specific signals detected by its cognate sensor kinase but also by any factor that modulates the response regulator’s phosphorylation state. Bacteria control the various steps leading up to response regulator phosphorylation, allowing their transcriptional responses to be tightly regulated upon encountering environmental changes.
Figure 1. Model for Activation of the PmrA/PmrB Two-Component System by Various Signals and Selected Targets of PmrA Control.
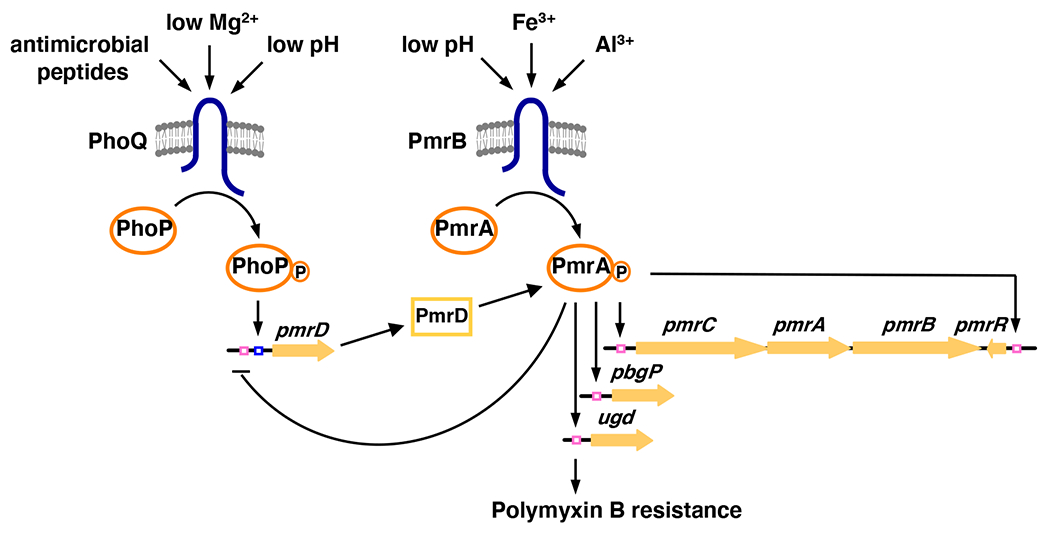
In S. typhimurium, transcription of PmrA-activated genes is promoted during growth in low Mg2+ via the PhoP/PhoQ system, the PmrD protein and the PmrA/PmrB system, and in the presence of Fe3+ via the PmrA/PmrB system and independently of PhoP/PhoQ and PmrD. Apart from the direct transcriptional control of LPS-modification loci (i.e., pbgP, ugd and pmrC), the phosphorylated PmrA protein also controls its own levels by positively autoregulating transcription of the pmrCAB operon, by repressing transcription of the pmrD gene and by promoting the expression of pmrR, which specifies a membrane peptide that downregulates the activity of the PmrA/PmrB system. PmrA-controlled modifications of the LPS confer resistance to the antibiotic polymyxin B.
In this review, we describe how the PmrA/PmrB two-component system responds to particular environmental cues and promotes synthesis of gene products required for modification of the LPS. We then highlight the striking wealth of feedback mechanisms operating on the PmrA/PmrB system, each with a distinct role in controlling PmrA-dependent gene expression over time, thereby dictating when, where and to what extent bacteria modify their LPS. The Salmonella-centric tone of this review reflects that most of the work on PmrA/PmrB has been carried out with this enteric pathogen. However, many of the findings about the Salmonella PmrA/PmrB system apply to PmrA/PmrB homologs in other gram-negative species, and we discuss the latter where appropriate. In particular, we focus on three general principles of regulatory circuit evolution that are emerging from investigations of the PmrA/PmrB system in enteric bacteria. First, the integration of a horizontally acquired gene product into an ancestral regulatory circuit can impact the evolutionary trajectories of pre-existing proteins within that circuit. Second, subtle differences in ancestral genes can affect the ability of horizontally acquired genes to confer new properties. Third, variation among orthologous regulatory proteins can lead to quantitative differences in their biochemical activities and thus, qualitatively and quantitatively distinct gene expression profiles within and between closely related species.
THE PMRA/PMRB REGULATORY SYSTEM RESPONDS TO MULTIPLE ENVIRONMENTAL SIGNALS
PmrB Directly Detects the Presence of Fe3+, Al3+ and Low pH
In S. enterica, activation of the PmrA/PmrB system occurs when bacteria experience environments containing high Fe3+ (i.e. 100 μM) (Wosten et al., 2000), high Al3+ (i.e. 100 μM) (Wosten et al., 2000) or mild acid pH (i.e., pH 5.8) (Perez and Groisman, 2007; Soncini and Groisman, 1996), all of which are detected by the PmrB sensor (Figure 1). In response to these inducing signals, PmrB autophosphorylates and then transfers the phosphoryl group to its cognate response regulator PmrA. Phosphorylated PmrA (PmrA-P) is the active form of the protein that binds to DNA (Shin and Groisman, 2005), promoting expression of PmrA-activated genes (Shin et al., 2006) and repression of PmrA-repressed genes (Kato et al., 2003). In the absence of inducing signals, the PmrB protein acts primarily as a PmrA-P phosphatase (Kato and Groisman, 2004).
The PmrA/PmrB two-component system represents the first example of a signal transduction cascade that responds to extracytoplasmic Fe3+ (Wosten et al., 2000). The Fe3+ signal binds directly to the periplasmic region of the Salmonella PmrB in a manner dependent on the glutamate residues in the twice-repeated ExxE motif and the serine residue at position 37 (Wosten et al., 2000). This iron-binding motif, which is also present in yeast and fungal iron transporters (Ramanan and Wang, 2000; Stearman et al., 1996) and in the mammalian ferritin light chain (Trikha et al., 1995), is specific for Fe3+ (as opposed to Fe2+) (Wosten et al., 2000), thereby distinguishing the PmrB sensor from other iron-sensing proteins that are typically cytosolic and respond to Fe2+ (Fleischhacker and Kiley, 2011). Interestingly, the BqsR/BqsS two-component system in the opportunistic pathogen P. aeruginosa was recently demonstrated to respond specifically to extracellular Fe2+, suggesting that bacteria have evolved mechanisms to distinguish between the two forms of iron and to elicit the appropriate transcriptional response (Kreamer et al., 2012). The BqsS periplasmic domain contains a single ExxE motif that is postulated to directly sense Fe2+ (Kreamer et al., 2012). If so, other amino acid residues surrounding the conserved glutamates might dictate the distinct specificity of sensor proteins for Fe3+ versus Fe2+. The ExxE motif also mediates PmrB’s ability to recognize and respond to Al3+ even though the level of activation is 50% of that achieved with the same concentration of Fe3+ (Wosten et al., 2000). PmrB is responsible for detecting Fe3+ and Al3+ in other enteric bacteria (Hyytiainen et al., 2003; Mitrophanov et al., 2008; Viau et al., 2011; Winfield and Groisman, 2004; Winfield et al., 2005), consistent with the conservation of the ExxE motif in PmrB orthologs from these species (Wosten et al., 2000). However, PmrB orthologs differ in their ability to respond to additional signals. Zn2+ activates the PmrA/PmrB system in E. coli but not in S. enterica (Lee et al., 2005; Wosten et al., 2000).
PmrA-dependent genes are activated when S. enterica experience mild acid pH (i.e. 5.8) in a manner that requires the PmrB sensor (Perez and Groisman, 2007; Soncini and Groisman, 1996). Substitution of the single histidine residue or of any one of four glutamate residues located in PmrB’s periplasmic domain, which is 31 amino acids long, lowered the mild acid-promoted transcription of PmrA-activated genes, suggesting that PmrB directly senses extracytoplasmic acid pH via changes in the protonation state of these conserved amino acids (Perez and Groisman, 2007). Consistent with this proposed mechanism, the pKaof free histidine is ~6. Although the pKaof a free glutamic acid residue is ~4, which is much lower than the mild acid pH condition that activates PmrB, protein folding might change the pKa of these residues (Tanford and Roxby, 1972) and lead to their protonation at pH ~ 5.8 (Perez and Groisman, 2007). The histidine and glutamate residues are conserved in the PmrB periplasmic domain of other enteric species, so it is likely that mild acid pH activates transcription of PmrA-regulated genes in these organisms (Perez and Groisman, 2007). The periplasmic histidine and/or glutamic acid residues in the Helicobacter pylori ArsS sensor kinase (Muller et al., 2009) and the E. coli CadC protein (Haneburger et al., 2012) are crucial for their ability to detect extracytoplasmic pH changes. Collectively, these findings point towards a common pH sensing mechanism whereby the protonation state of these amino acids dictates the pH sensing ability of integral membrane proteins.
Addition of the catecholamine adrenaline results in a ~2-fold reduction in the levels of several PmrA-dependent transcripts in S. typhimurium grown in LB media (Karavolos et al., 2008). It is unclear how adrenaline represses PmrA-dependent gene transcription: the PmrB sensor might directly detect adrenaline (Karavolos et al., 2008), or alternatively, adrenaline might indirectly reduce Fe3+ levels in the growth media because cathecolamines are capable of binding iron (Paris et al., 2005; Siraki et al., 2000). In support of the latter notion, adrenaline also stimulates the expression of iron acquisition systems (Karavolos et al., 2008) that scavenge iron from various sources when this metal is limiting (Faraldo-Gomez and Sansom, 2003).
Low Mg2+ Indirectly Activates the PmrA/PmrB System via the PmrD Protein
Growth of S. typhimurium in low Mg2+ (i.e. 10 μM) also promotes PmrA-dependent gene expression (Garcia Vescovi et al., 1996) (Figure 1). In this case, activation of the PmrA/PmrB system occurs indirectly via the low Mg2+-responsive PhoP/PhoQ two-component system through a process that requires the PhoP-activated gene pmrD (Kox et al., 2000; Soncini and Groisman, 1996). The pmrD locus was first identified in a genetic screen as a gene that conferred polymyxin B resistance in a PmrA-dependent manner when expressed from a medium copy number plasmid (Roland et al., 1994). PmrD is a basic, 85 amino acid protein that post-translationally regulates PmrA activity by targeting the N-terminal domain of PmrA-P (Kato and Groisman, 2004), which harbors the putative phosphorylation site required for PmrD-mediated activation of PmrA in vivo (Kox et al., 2000) and for phosphotransfer from phosphorylated PmrB protein in vitro (Kato and Groisman, 2004). Consequently, PmrD stabilizes the phosphorylated form of PmrA, protecting it from dephosphorylation by PmrB (Kato and Groisman, 2004; Kox et al., 2000). Such protection promotes the accumulation of active PmrA (i.e., PmrA-P) and the expression of PmrA-dependent genes when bacteria experience low Mg2+. These genes are expressed in the absence of Fe3+, a condition known to stimulate PmrB’s phosphatase activity (Kato and Groisman, 2004). Therefore, PmrD defines a class of proteins – termed connectors – that promote activation of two-component systems in the absence of their cognate signal(s), creating regulatory links between two otherwise independent signal transduction pathways (Mitrophanov and Groisman, 2008b).
Certain cationic antimicrobial peptides – polymyxin B, C18G, LL-37 and protegrin – are believed to directly activate the PhoP/PhoQ system (Bader et al., 2005; Shprung et al., 2012), and thus promote transcription of PmrA-dependent genes (Richards et al., 2012). In contrast to the low Mg2+ signal, these cationic antimicrobial peptides only activate a subset of genes in the PhoP and PmrA regulons, and the extent of this activation differs among the various peptides (Bader et al., 2005; Richards et al., 2012). However, the PhoP/PhoQ and PmrA/PmrB systems do not necessarily have to be activated antimicrobial peptides even though they control resistance to antimicrobial peptides and it remains unclear whether such activation occurs under physiological conditions (Groisman and Mouslim, 2006).
Small molecules such as acetyl phosphate can serve as phosphodonors to PmrA, giving rise to target gene expression in a manner independent of the cognate PmrB sensor (Perez and Groisman, 2007), as previously demonstrated for other two-component systems (Lukat et al., 1992; Wolfe, 2005). In support of this notion, the residual PmrA-dependent gene expression exhibited by a pmrB mutant grown in mild acid conditions was abrogated in an isogenic strain defective for the production of acetyl phosphate (Perez and Groisman, 2007). Nevertheless, it is unlikely that acetyl phosphate contributes to PmrA phosphorylation under normal conditions when PmrB is present because a strain that lacks the ability to synthesize acetyl phosphate but harbors a functional pmrB gene produces wild-type levels of PmrA-dependent mRNAs (Perez and Groisman, 2007).
In sum, multiple stimuli promote activation of the PmrA/PmrB system: the PmrB protein directly senses extracytoplasmic Fe3+ and Al3+ or mild acid pH, whereas low Mg2+ activation occurs via the PhoP/PhoQ system and PmrD protein (Figure 1). These different inputs activate the PmrA/PmrB system to varying extents (Kox et al., 2000; Perez and Groisman, 2007; Wosten et al., 2000), leading to the production of appropriate amounts of PmrA-regulated gene products that contribute to S. enterica’s survival in various environments.
S. enterica Encounters Multiple Environments that Contain PmrA-Activating Signals
S. enterica experience a number of diverse environments throughout their lifecycle that induce the PmrA/PmrB system. Acquisition of S. enterica usually occurs orally from the ingestion of contaminated food or water (Pang et al., 1995). Bacteria need to survive the acidic pH of the stomach as well as killing by cationic antimicrobial peptides and bile in the small intestine as they transit through the gastrointestinal tract (Winfield and Groisman, 2003). The stomach and small intestine are considered high iron environments because mice fed a standard rodent breeding diet contained 29–733 μM and 10–100 μM of iron in the stomach and duodenum, respectively (Simpson and Peters, 1990), and healthy humans are believed to have similar iron concentrations (Goddard et al., 1997). Indeed, PmrA-activated promoter expression has been detected in the murine intestinal lumen by resolvase-in vivo expression technology (RIVET), a method used to analyze spatial-temporal patterns of gene expression in vivo (Merighi et al., 2005).
In the small intestine, S. enterica invade the intestinal epithelium and enter the deeper submucosal tissues, where they are engulfed by resident macrophages and dendritic cells (Mastroeni et al., 2009). S. enterica can replicate within macrophages in which they must survive the acidic, nutrient-limiting conditions and the bactericidal environment created by the host production of cationic antimicrobial peptides (Mastroeni et al., 2009). The phagosomal environment that S. enterica experience within the macrophage is believed to contain low levels of Mg2+, a condition that activates the PhoP/PhoQ system and thus the PmrA/PmrB system (Groisman, 1998; Mouslim and Groisman, 2003). Accordingly, S. typhimurium grown in murine macrophages express PmrA-activated genes (Merighi et al., 2005) and display PmrA-dependent lipid A modifications (Gibbons et al., 2005). Escape of bacteria from these phagocytic cells allows further systemic dissemination to the bloodstream (Mastroeni et al., 2009), where bacteria have to avoid killing by complement and other host antimicrobial mechanisms.
Some S. enterica serovars, such as Typhi and Paratyphi, can further colonize the human gallbladder, which is the site of bile production, resulting in a chronic carrier state (Gunn, 2000). In this case, the expression of PmrA-dependent genes may not be beneficial to these serovars because polymyxin-resistant mutants of S. typhimurium display heightened sensitivity to the bile detergents sodium deoxycholate and sodium cholate (Makela et al., 1978) and because constitutive expression of PmrA-dependent LPS-modifying genes increases susceptibility of E. coli to deoxycholate (Froelich et al., 2006). Furthermore, an S. paratyphi B pmrA mutant exhibited enhanced biofilm formation compared to wild-type bacteria on cholesterol-coated surfaces (Chen et al. submitted), an assay that mimics the ability of bacteria to attach to cholesterol-coated gallstones (Crawford et al., 2010; Gonzalez-Escobedo et al., 2010; Schioler et al., 1983). This suggests that the expression of a PmrA-regulated gene product(s) interferes with biofilm formation.
S. enterica that disseminate back into the environment can survive in soil and water, environments that likely contain low Mg2+ and/or high Fe3+ levels (Winfield and Groisman, 2003). A functional PmrA/PmrB system is required for growth in soil, where the concentrations of free Fe3+ can be as high as 350 μmol cm−3 (Ratering and Schnell, 2000), because a pmrA mutant lacking PmrA-regulated LPS decorations is hypersusceptible to killing by Fe3+ (Chamnongpol et al., 2002). PmrA-controlled membrane remodeling may be particularly important in soil because it renders S. enterica resistant to polymyxin B, an antibiotic produced by the soil bacterium Paenibacillus polymyxa (Girardin et al., 2002; Storm et al., 1977).
PMRA-REGULATED PROMOTERS
When bacteria experience the presence of inducing signals for the PmrA/PmrB system, the PmrB sensor phosphorylates PmrA, generating the active form of PmrA that binds target promoters and promotes gene transcription in vivo (Shin and Groisman, 2005). Phosphorylation of PmrA likely promotes its dimerization and thus increases its affinity for PmrA-dependent promoters (Wosten and Groisman, 1999), as is the case for several regulators from the OmpR/PhoB family (Liu and Hulett, 1997). Conditions that promote PmrA dimerization bypass the requirement for PmrA phosphorylation in DNA binding: for example, in vitro experiments demonstrated that unphosphorylated PmrA dimerizes and specifically binds to DNA fragments containing PmrA-dependent promoters, albeit with lower affinity than PmrA-P (Wosten and Groisman, 1999).
PmrA activates transcription by binding to a conserved DNA motif within the promoters of target genes (Aguirre et al., 2000; Marchal et al., 2004; Wosten and Groisman, 1999). Such binding generally increases the affinity of RNA polymerase for PmrA-regulated promoters, as previously demonstrated for other transcription factors belonging to the OmpR family (Browning and Busby, 2004; Stock et al., 2000). The sequence to which the PmrA protein binds consists of a hexanucleotide repeat CTTAAG separated by 5 nucleotides – termed the PmrA box – that is typically located upstream from the transcription start sites of PmrA-regulated genes (Aguirre et al., 2000; Marchal et al., 2004; Wosten and Groisman, 1999). The PmrA box is located upstream of the −35 element (i.e., the sequence recognized by the RNA polymerase σ70 subunit) in the promoters of PmrA-dependent genes whose transcription start sites have been experimentally defined to date (Aguirre et al., 2000; Delgado et al., 2006; Pescaretti et al., 2011; Wosten and Groisman, 1999). This location is consistent with PmrA activating transcription via a Class I mechanism, whereby the transcription factor binds upstream of the −35 element and contacts the C-terminal domain of the α subunit of RNA polymerase (α-CTD), thereby recruiting the polymerase to the promoter (Browning and Busby, 2004). Binding sites for PmrA-dependent genes similar to those defined in S. typhimurium have been identified in the enteric species Escherichia, Klebsiella, Yersinia, Citrobacter, Serratia and Erwinia, all of which also encode pmrAB genes (Aguirre et al., 2000; Hyytiainen et al., 2003; Marchal et al., 2004; Mitrophanov et al., 2008; Winfield and Groisman, 2004; Winfield et al., 2005; Wosten and Groisman, 1999), and PmrA likely promotes transcription through similar mechanisms in these species.
Several PmrA-dependent genes are adjacent to each other in the S. typhimurium genome. For example, the pbgPE and pmrG genes are divergently transcribed from each other (Wosten and Groisman, 1999). The single PmrA-binding site in the pbgPE-pmrG intergenic region promotes expression from both the pbgPE and pmrG promoters (Wosten and Groisman, 1999). This site is closer to the pbgPE promoter than to the pmrG promoter. However, it is unclear whether one promoter is preferentially activated over the other (Wosten and Groisman, 1999). Still, other PmrA-dependent genes are convergently transcribed from opposing promoters located on opposite strands of the DNA, as exemplified by the pbgPE and pmrD (Kox et al., 2000) as well as the pmrCAB and pmrR gene pairs (Kato et al., 2012). Because no intrinsic terminator has been identified in the intergenic regions of these genes, convergent transcription of these genes might give rise to transcripts with sense-antisense interactions in the overlapping region that impact their stability or to collisions between RNA polymerases transcribing from opposite directions, thus affecting the levels of these transcripts. Such antisense regulation and/or transcription interference mechanisms potentially constitute an additional layer of control for the expression of these PmrA-dependent gene pairs, as previously shown for other convergently transcribed genes in bacteria (Georg and Hess, 2011).
PmrA also functions as a repressor. The binding of PmrA-P to the PmrA box located 59 bp upstream from the pmrD transcription start site reduces the levels of pmrD transcript (Kato et al., 2003). This binding does not prevent PhoP from associating with the pmrD promoter, in agreement with the finding that PmrA-mediated repression does not eliminate PhoP-dependent pmrD transcription (Kato et al., 2003). Likewise, transcription of the E. coli yrbL gene is repressed by PmrA and activated by PhoP (Zwir et al., 2005). The S. typhimurium and E. coli yrbL promoters contain PmrA and PhoP boxes with an arrangement similar to that of the S. typhimurium pmrD promoter (Zwir et al., 2005). However, the mechanism by which PmrA represses pmrD and yrbL transcription is currently unknown.
In the following sections, we discuss the target genes that harbor a PmrA box in their promoter regions and which have been grouped into three categories: 1) loci specifying LPS-modifying enzymes (Table 1), 2) genes that have no known function in LPS modification (Table 2), and 3) genes encoding products that modify the level and activity of the PmrA/PmrB system.
Table 1.
PmrA-regulated genes specifying products involved in LPS modifications in Salmonella enterica serovar Typhimurium.
| Gene | Function | PmrA-binding site | Reference |
|---|---|---|---|
| cptA/yijP | Phosphoethanolamine addition to the core region | CTTCAgattctTTTAA | (Tamayo et al., 2005a; Tamayo et al., 2005b) |
| pbgPE/arn/pmrHFIJKL operon | L-4-aminoarabinose modification of the lipid A | CTTAAtgttaaTTTAA | (Breazeale et al., 2002; Gunn et al., 1998; Marchal et al., 2004; Soncini and Groisman, 1996; Wosten and Groisman, 1999) |
| pmrC/yjdB | Phosphoethanolamine modification of the lipid A | CTTAAggttcaCTTAA | (Lee et al., 2004; Marchal et al., 2004; Wosten and Groisman, 1999) |
| pmrG | Dephosphorylation of the Hep (II) phosphate in the core region | ATTAAattaacATTAA | (Marchal et al., 2004; Nishino et al., 2006; Wosten and Groisman, 1999) |
| ugd | UDP-glucose dehydrogenase that converts UDP-D-glucose into UDP-D-glucuronic acid, L-4-aminoarabinose modification of the lipid A | CTTAAtattaCTTAA | (Gunn et al., 1998; Marchal et al., 2004; Soncini and Groisman, 1996; Wosten and Groisman, 1999) |
| wzzfepE | Very long O-antigen chains | GCATATAtttgcTTTAT | (Pescaretti et al., 2011) |
| wzzST | Long O-antigen chains | CATAATAattacTAATT | (Delgado et al., 2006) |
Table 2.
PmrA-regulated genes specifying products not known to be involved in LPS modifications in Salmonella enterica serovar Typhimurium.
| Gene | Function | PmrA-binding site | Reference |
|---|---|---|---|
| aroQ | Periplasmic chorismate mutase | CTTAAtgttatCTTAAT | (Marchal et al., 2004) |
| deoA | Thymidine phosphorylase | Not detected | (Tamayo et al., 2005b) |
| dgoA | 2-oxo-3-deoxygalactonate 6-phosphate aldolase, catalyzes the final reaction in the degradation of D-galactonate | Not detected | (Tamayo et al., 2002) |
| mig-13/ybjG | Undecaprenyl pyrophosphate phosphatase | CTTTAAggttaaTTTAA | (Marchal et al., 2004) |
| pmrAB | Two-component regulatory system responding to Fe3+ | Follows pmrC in the pmrCAB operon | (Gunn et al., 1998; Roland et al., 1994; Wosten and Groisman, 1999; Wosten et al., 2000) |
| pmrD | Connects the PhoP/PhoQ and PmrA/PmrB systems | ATTAATgttagGTTAAT | (Kox et al., 2000) |
| pmrR | Negative regulator of the PmrA/PmrB system | CTTAAGgttcgCTTAAT | (Kato et al., 2012) |
| sseJ | SPI-2 secreted effector | CTTAAgaaataTTTAAT | (Marchal et al., 2004) |
| STM1253 | Putative inner membrane protein, cytochrome b homolog | TTTAAggttctGTTAAG | (Tamayo et al., 2005b) |
| udp | Uridine phosphorylase | Not detected | (Tamayo et al., 2005b) |
| yibD | Putative glycosyltransferase | CTTAAtagtttCTTAAT | (Marchal et al., 2004; Tamayo et al., 2002) |
| yrbL | Unknown function | ACATTAAgaaaacCTTAAA | (Zwir et al., 2005) |
PMRA CONTROLS LPS MODIFICATIONS IN S. ENTERICA AND OTHER ENTERIC BACTERIA
The LPS consists of three structurally distinct regions: 1) the innermost lipid A, which is a glucosamine-based phospholipid that anchors LPS to the outermost layer of the bacterial outer membrane, 2) the central core region, a phosphorylated nonrepeating oligosaccharide, and 3) the distal O-antigen, an oligosaccharide polymer that consists of a variable number of repeat units and differs greatly among bacteria (Raetz et al., 2007; Raetz and Whitfield, 2002) (Figure 2). Gene products whose expression is governed by the PmrA/PmrB regulatory system mediate chemical alterations in each of these three LPS regions as described in the following sections.
Figure 2. Schematic Representation of the LPS Structure and the PmrA-Regulated LPS Modifications in S. typhimurium.

The glucosamine-based lipid A serves as a hydrophobic anchor for the LPS. In S. typhimurium, the predominant species is hexa-acylated and phosphorylated at the 1 and 4′ positions. The 6′-position of lipid A is linked by two Kdo residues to the core region, which is followed by the outermost O-antigen. The lipid A phosphates can be modified with phosphoethanolamine (pEtN) by PmrC or with L-4-aminoarabinose (L-Ara4N) by the Ugd and PbgP proteins. LpxT adds a second phosphate group to the 1-position, resulting in a 1-disphosphate species and PmrR inhibits LpxT activity. The Hep(I) phosphate group in the inner core can be modified with pEtN by the CptA protein and the core Hep(II) phosphate can be dephosphorylated by PmrG. The formation of long or very long O-antigen is controlled by the WzzST and WzzFepE proteins, respectively.
Modifications of the Lipid A
In S. typhimurium, the 1- and 4′-phosphate groups of lipid A can be covalently modified with phosphoethanolamine (pEtN) or with L-4-aminoarabinose (L-Ara4N) (Figure 2). The pmrC gene product (also referred to as eptA (Doerrler et al., 2004)), which is the first gene of the pmrCAB operon, catalyzes the addition of pEtN to lipid A (Lee et al., 2004; Zhou et al., 2001). Incorporation of L-Ara4N into lipid A is mediated by the PmrA-activated pbgPE seven-gene operon (Groisman et al., 1997) (also referred to as pmrHFIJKLM (Gunn et al., 1998) or arn (Breazeale et al., 2003; Raetz et al., 2007)) as well as by the ugd gene (also referred to as pmrE) (Gunn et al., 2000). This pathway starts with the oxidation of UDP-glucose to UDP-glucuronic acid by Ugd (Breazeale et al., 2002). The C-terminal domain of ArnA (PbgP3) then catalyzes the oxidative decarboxylation of UDP-glucuronic acid (Breazeale et al., 2002; Williams et al., 2005). The resulting UDP-4-ketopentose is transaminated by ArnB (PbgP1) to produce UDP-L-Ara4N (Breazeale et al., 2003), which is formylated by the N-terminal domain of ArnA (Breazeale et al., 2005; Williams et al., 2005). ArnC (PbgP2) subsequently transfers this derivative to undecaprenyl phosphate (Breazeale et al., 2005). This product is later deformylated by ArnD (PbgP4), rendering the pathway irreversible (Breazeale et al., 2005). ArnE (PbgE2) and ArnF (PbgE3) are responsible for transporting undecaprenyl phosphate-α-L-Ara4N across the membrane (Yan et al., 2007). Finally, ArnT (PbgE1) transfers the L-Ara4N moiety to the 4′-phosphate of the core-lipid A (Trent et al., 2001b; Trent et al., 2001c).
Transfer of the L-Ara4N and pEtN groups occurs on newly synthesized core-lipid A molecules (Doerrler et al., 2004; Raetz et al., 2007), consistent with findings that the catalytic domains of the enzymes mediating these transfers are on the periplasmic face of the inner membrane (Lee et al., 2004; Trent et al., 2001c). PmrC-mediated incorporation of pEtN takes place predominantly at the 1-position of lipid A (Lee et al., 2004), whereas L-Ara4N is preferentially added to the lipid A 4′-phosphate group, at least under the investigated conditions (Breazeale et al., 2002; Gunn et al., 1998; Trent et al., 2001c; Zhou et al., 2001). However, the 1- and 4′ lipid A positions can also bear these decorations because minor lipid A species with two L-Ara4N or two pEtN substituents have been detected (Zhou et al., 2001). Bacteria defective in the ability to incorporate L-Ara4N decorations accumulate higher levels of pEtN-modified lipid A than the wild-type strain and vice versa (Herrera et al., 2010; Zhou et al., 2001). These results imply that when bacteria lack the ability to perform a particular type of lipid A modification, a different type of modification is enhanced.
Together, the covalent modifications of lipid A with L-Ara4N and pEtN neutralize the net negative charge on the lipid A and decrease binding between cationic antimicrobial agents and the bacterial surface (Gunn et al., 1998; Lee et al., 2004; Zhou et al., 2001). As expected, the enhanced survival of polymyxin-resistant pmrA mutants in the presence of polymyxin B is derived from the increased amounts of L-Ara4N and pEtN substitutions in their lipid A compared to wild-type S. typhimurium (Helander et al., 1994; Vaara et al., 1981). However, the L-Ara4N and pEtN moieties differ in their abilities to neutralize the negative charges on lipid A: pEtN decreases the net charge of lipid A from −1.5 to −1, while L-Ara4N brings it down to 0 (Nikaido, 2003). Consequently, a mutant deficient in the L-Ara4N modification is > 500-fold more sensitive to polymyxin B and binds 4-fold more Fe3+, whereas bacteria defective in generating pEtN-decorated lipid A are only 3-5-fold more susceptible to and bind 1.5-fold more Fe3+ than the wild-type strain (Lee et al., 2004) (Kato et al, 2012). The L-Ara4N species also promotes swarming motility in S. typhimurium, a phenomenon that has been linked with elevated resistance to a variety of antibiotics, including polymyxin B (Kim et al., 2003).
Several lines of evidence suggest that the PmrA-controlled lipid A modifications contribute to S. typhimurium pathogenesis. First, inactivation of the pmrA and/or pmrB genes attenuates virulence in Balb/c mice (Gunn et al., 2000). Interestingly, a pmrF mutant that lacks the L-Ara4N modification is more attenuated for virulence than the pmrA mutant (Gunn et al., 2000), perhaps because the pbgP operon is expressed in vivo by a PmrA-independent mechanism or because particular byproducts of L-Ara4N metabolism are detrimental to S. typhimurium. By contrast, the pmrC gene does not appear to be a major contributor towards virulence in mice (Tamayo et al., 2005a). Second, an S. typhimurium mutant that constitutively synthesizes PmrA-regulated LPS modifications displays heightened resistance toward antimicrobial peptides from human polymorphonuclear leukocytes (Shafer et al., 1984a). Third, S. enteriditis pmrB, ugd and pmrF mutants were defective in infection of chicken macrophages (Zhao et al., 2002).
Analyses of lipid A isolated from bacteria grown in non-inducing conditions for the PmrA/PmrB system demonstrated that ~ 30% of lipid A is diphosphorylated at the 1-position of lipid A (1-PP) (Zhou et al., 2001), increasing the overall negative charge on the bacterial surface and compromising bacterial resistance to polymyxin B (Herrera et al., 2010). The PmrA/PmrB system indirectly inhibits the activity of LpxT (Herrera et al., 2010), the inner membrane enzyme responsible for generating 1-PP (Touze et al., 2008b). This inhibition is accomplished by a PmrA-activated membrane peptide – termed PmrR – that interacts with and hinders LpxT’s activity, thereby lowering the net negative charge on the LPS (Kato et al., 2012). Thus, apart from the transcriptional regulation of LPS-modifying enzymes, the PmrA/PmrB system also modulates the activities of these enzymes at the post-translational level.
The fatty acyl chains of the lipid A moiety can also be modified. These modifications are not regulated by the PmrA/PmrB system, unlike the covalent alterations that neutralize the negative charges of the lipid A phosphates. Some of the changes to the lipid A acylation pattern are controlled by the PhoP/PhoQ system: PagP catalyzes the addition of palmitate (Gibbons et al., 2005; Guo et al., 1998), and PagL removes the R-3-hydroxymyristoyl chain at position 3 of the lipid A (Trent et al., 2001a). Other changes, such as hydroxylation of the 3′ secondary acyl chain by LpxO (Gibbons et al., 2000) and cleavage of the intact 3′-acyloxyacyl moiety by LpxR (Reynolds et al., 2006), are independent of the PhoP/PhoQ system.
The ability of LPS-modifying enzymes to alter the LPS and thus change outer membrane properties can be affected by pre-existing lipid A decorations. For instance, addition of L-Ara4N to lipid A requires myristoylated LPS (Tran et al., 2005). The presence of L-Ara4N occludes PagL-mediated deacylation of lipid A (Kawasaki et al., 2005), and the incorporation of pEtN to the Kdo residue of LPS is postulated to interfere with other LPS modifications (Moon and Gottesman, 2009). In addition, the 1-position of lipid A is predominantly modified with pEtN and to a lesser extent with L-Ara4N in the absence of 1-PP (Herrera et al., 2010) (Kato et al., 2012), reflecting competition among the various LPS-modifying enzymes for a given lipid A substrate. Collectively, these results indicate that the relative proportions of each lipid A species are further controlled by complex regulatory mechanisms occurring at the level of activity of the various LPS-modifying proteins.
Modifications of the LPS Core
PmrA-regulated remodeling of the LPS also entails modifications to the inner core that are mediated by the CptA and PmrG enzymes (Figure 2). The cptA gene product is required for pEtN addition to the Hep(I) phosphate group in the core region (Tamayo et al., 2005a). A pmrC cptA double mutant was 3-5-fold less effective in competing against wild-type S. typhimurium when mice were orally inoculated with both strains even though the single pmrC and cptA mutants had no defect in competition experiments against the wild-type strain (Tamayo et al., 2005a). However, it is unclear how pEtN addition to the Hep(I) phosphate group contributes to mouse virulence because this modification is required neither for Fe3+ resistance (Nishino et al., 2006) nor for survival in the presence of polymyxin B (Tamayo et al., 2005a).
The periplasmic PmrG protein is a phosphatase that acts on the core Hep(II) phosphate (Nishino et al., 2006). Resistance to Fe3+ toxicity requires dephosphorylation of the Hep(II) phosphate as well as modification of the two lipid A phosphates, which collectively reduce Fe3+ association with the bacterial cell (Nishino et al., 2006). Yet, the PmrG-mediated core modification does not contribute to polymyxin B resistance, unlike the L-Ara4N and pEtN lipid A modifications (Nishino et al., 2006). Together, these findings indicate that there is only a partial overlap between the sites targeted by Fe3+ and by polymyxin B.
Modulating the Length of the O-Antigen
The length of the O-antigen polymer is controlled by the PmrA-activated wzzST (Delgado et al., 2006) (also referred to as cld (Morona et al., 1995)) and wzzfepE genes (Pescaretti et al., 2011), which encode products responsible for long and very long O-antigen, respectively (Morona et al., 1995; Murray et al., 2003) (Figure 2). Transcription of wzzST (but not wzzfepE) is also promoted by the RcsC/YojN/RcsB regulatory system in a manner independent of the PmrA/PmrB system (Delgado et al., 2006; Pescaretti et al., 2011). Consistent with findings that the RcsC/YojN/RcsB modulates the swarming behavior of bacteria grown on an agar surface (Takeda et al., 2001) and that the S. typhimurium O-antigen is required for swarm colony expansion (Toguchi et al., 2000), inactivation of the wzzST gene prevented the enhanced swarming behavior of an rcsB mutant (Delgado et al., 2006). The length of the O-antigen must be tightly regulated due to its conflicting impact on S. typhimurium’s ability to resist antimicrobial agents and to gain access to host cells. On the one hand, expression of wzzST and wzzfepE leads to heightened resistance to complement (Delgado et al., 2006; Pescaretti et al., 2011) and to polymyxin B (Holzer et al., 2009; Pescaretti et al., 2011). On the other hand, the incorporation of long and very long O-antigen into the LPS reduces the ability of S. typhimurium to invade epithelial cells (Holzer et al., 2009), a process critical for S. typhimurium to successfully infect its host.
Partially Overlapping Functions of the Various PmrA-Dependent LPS Modifications
The findings described above reveal a role for the different PmrA-regulated LPS decorations in promoting S. typhimurium’s survival when it experiences distinct environmental stresses. In some cases, inactivation of a single target gene is sufficient to recapitulate the phenotype of a pmrA mutant. For example, resistance to complement is due entirely to PmrA-dependent transcription of wzzST because the pmrA mutant was as susceptible to serum as one unable to synthesize long O-antigen (Delgado et al., 2006). In other cases, the inactivation of several PmrA-target gene products is required to reproduce the phenotype displayed by a pmrA mutant. For instance, resistance to polymyxin B requires modification of the lipid A with both L-Ara4N and pEtN (Lee et al., 2004). Mutants defective in the L-Ara4N or the pEtN modification are ~1000- and 3-fold more susceptible to polymyxin B than the wild-type strain, significantly less than the ~10,000-fold susceptibility displayed by the pmrA mutant (Lee et al., 2004). However, this susceptibility is amplified in a double mutant lacking both modifications such that it is as sensitive to killing by the antibiotic as the pmrA mutant (Lee et al., 2004). Likewise, several LPS modifications govern resistance towards Fe3+ toxicity: apart from the L-Ara4N and pEtN modifications to the lipid A, this resistance also requires dephosphorylation of the Hep(II) phosphate in the core region (Nishino et al., 2006). Together, these results suggest that the inability of bacteria to synthesize a particular LPS modification may be compensated for by the enhancement of a different modification and that the role of a given modification in mediating a particular phenotype might only be uncovered in the absence of other contributing modifications.
PmrA-Dependent LPS Modifications in Other Bacterial Species
Many Gram-negative bacteria, like S. typhimurium, modify their LPS to withstand environmental stresses. The PmrA/PmrB system governs these modifications in a wide variety of enteric bacteria, albeit with several differences from those in S. typhimurium. First, the genes mediating chemical decorations of the LPS in S. typhimurium are not necessarily present in the genomes of other bacteria. Decoration of the lipid A with L-Ara4N has been shown to protect K. pneumoniae (Cheng et al., 2010; Helander et al., 1996), Y. pestis (Rebeil et al., 2004), E. coli (Nummila et al., 1995) and E. carotovora (Hyytiainen et al., 2003) from killing by polymyxin B. The genes specifying the enzymes responsible for L-Ara4N synthesis are absent from the C. rodentium genome. Instead, this bacterium encodes homologs of the S. typhimurium pmrC and cptA genes, which are required for maintaining outer membrane integrity and resistance to lipophilic antibiotics (Viau et al., 2011). Similarly, the pmrC gene is absent from the Yersinia genome (Marceau et al., 2004) whereas the pmrG gene is absent from the Klebsiella genome (our own observations). Second, the amount of each modification varies among enteric bacteria: in E. coli, the 1- and 4′-phosphates of lipid A are predominantly decorated with pEtN, whereas L-Ara4N-modified lipid A is prevalent in S. typhimurium (Herrera et al., 2010; Zhou et al., 2001). Third, modification of the lipid A in some bacterial species, such as Helicobacter pylori (Stead et al., 2005) and Burkholderia cenocepacia (Hamad et al., 2012), appears constitutive, unlike other bacteria in which the majority of LPS modifications are synthesized in response to environmental cues that activate PmrB. Together, these findings suggest that variation in the nature and levels of LPS modifications among bacterial species contributes to their survival in distinct environmental niches and raises questions as to the molecular mechanisms underlying this variation.
Additional Regulators Independently Control the Expression of PmrA-Dependent Genes
Other two-component systems promote the transcription of several PmrA-target genes independently of the PmrA protein. That multiple regulatory systems control the expression of these genes indicates that they are required in a variety of environments in which these systems are active. For instance, the RcsC/YojN/RcsB system, which regulates colanic acid capsule synthesis and swarming behavior (Majdalani and Gottesman, 2005), activates ugd transcription independently of the PhoP/PhoQ and PmrA/PmrB systems (Mouslim and Groisman, 2003). This activation reflects that the end product (i.e., UDP-D-glucuronic acid) of the Ugd-catalyzed reaction is used as a precursor not only for L-Ara4N biosynthesis (Gunn et al., 1998) but also for extracellular polysaccharide production (Stevenson et al., 1996). Similarly, the RcsC/YojN/RcsB system activates transcription of wzzST to promote swarming behavior (Delgado et al., 2006). In addition, as discussed earlier, PhoP directly activates the expression of the pmrD and yrbL genes, which are also repressed by PmrA (Kato et al., 2003; Zwir et al., 2005).
The response regulator PreA can activate the expression of a subset of genes in the PmrA regulon in a PmrA-independent fashion (Merighi et al., 2006; Merighi et al., 2009). PreA and its cognate sensor PreB form a two-component system that is orthologous to the QseB/QseC quorum-sensing system in E. coli. Microarray analyses demonstrated that these genes were ~2-4-fold upregulated in a preA preB double mutant overexpressing the PreA response regulator but not in the isogenic strain harboring the empty vector (Merighi et al., 2009), consistent with findings that PreA is required for this activation (Merighi et al., 2006). This activation occurs only in the preB sensor mutant but not the wild-type strain, possibly because PreB’s phosphatase activity limits cross-phosphorylation of its cognate PreA regulator by other sources (Merighi et al., 2006). These findings are reminiscent of cross-talk between noncognate sensor kinase and response regulator pairs, typically seen only after introducing various genetic perturbations to the wild-type organisms (Laub and Goulian, 2007). Because the cognate signal(s) that induces the S. typhimurium PreA/PreB system remains undefined (Merighi et al., 2009), it is not known whether PreA’s control of PmrA-dependent genes is physiologically relevant in vivo.
An additional layer of regulation further controls the expression of PmrA-activated loci in several Yersinia species. Y. enterocolitica and Y. pestis modify the lipid A with L-Ara4N and display resistance to antimicrobial peptides only when grown at room temperature (21-26°C) but not at temperatures encountered within the mammalian hosts (37°C) (Rebeil et al., 2004; Reines et al., 2012). This is because RovA is more highly expressed at room temperature, and RovA appears to antagonize repression of LPS-modifying loci and/or at the pmrAB operon by the histone-like nucleoid structuring protein (H-NS) (Reines et al., 2012). However, the fact that Y. enterocolitica mutants defective in L-Ara4N biosynthesis are attenuated for mouse colonization implies that these lipid A modifications are synthesized even at 37°C (Reines et al., 2012) and raises questions as to the regulatory mechanism(s) controlling the expression of these LPS-modifying enzymes at this temperature. The ability to remodel the LPS in a temperature-dependent manner is not conserved within all members of the Yersinia genus (Marceau et al., 2004; Rebeil et al., 2004), raising the possibility that such variation in gene expression contributes to distinct transmission and pathogenesis among Yersinia species.
OTHER PMRA TARGET GENES
The PmrA/PmrB regulatory system controls the expression of additional loci that are not known to be involved in LPS modifications (Table 2) (Hagiwara et al., 2004; Marchal et al., 2004; Ogasawara et al., 2012; Tamayo et al., 2002). Some of these genes regulate membrane homeostasis as described below. Other genes appear to modulate various aspects of cellular metabolism. However, it is largely unclear how these genes relate to the known functions of the PmrA regulon.
PmrA-Dependent Regulation of Membrane Homeostasis
PmrA regulates the synthesis of undecaprenyl phosphate (C55-P), a universal lipid carrier of glycan biosynthetic intermediates that are required for the synthesis of peptidoglycan, lipid A, O-antigen, and other bacterial carbohydrate polymers (Hartley and Imperiali, 2011; Tatar et al., 2007). This regulation is brought about by transcriptional control of ybjG (Marchal et al., 2004) (also referred to as Mig-13 (Valdivia and Falkow, 1997)) and by inhibition of LpxT’s activity via the PmrA-activated pmrR gene product (Herrera et al., 2010) (Kato et al., 2012). Both YbjG and LpxT recycle C55-P from preformed undecaprenyl pyrophosphate (C55-PP) (El Ghachi et al., 2005; Hartley and Imperiali, 2011; Touze et al., 2008a). Whereas LpxT specifically transfers the distal phosphate group from C55-PP to the 1-position of lipid A to generate 1-PP lipid A (El Ghachi et al., 2005; Herrera et al., 2010), it is unclear whether YbjG phosphorylates other bacterial components within the periplasm. The levels of C55-P appear to be tightly regulated in bacteria since this lipid carrier is made in very small amounts, limiting its availability for the various glycan biosynthetic pathways (Barreteau et al., 2009). Inhibition of LpxT’s activity (Kato et al., 2012) likely decreases its capacity to regenerate C55-P, raising the possibility that PmrA compensates for this decrease by upregulating the expression of other C55-PP phosphatases (i.e., YbjG).
PmrA transcriptionally controls the expression of genes modulating membrane composition in other enteric bacteria. In E. coli, PmrA activates expression of dgkA, which specifies diacylglycerol kinase, an enzyme that breaks down diacylglycerol for re-entry into the phospholipid biosynthesis pathway (Badola and Sanders, 1997; Wahl et al., 2011). The S. typhimurium dgkA promoter also harbors a PmrA box, to which PmrA might bind and promote transcription of this gene (Wahl et al., 2011). Interestingly, diacylglycerol is a byproduct of the PmrC-mediated pEtN lipid A decoration (Lee et al., 2004; Reynolds et al., 2005), leading to the proposal that PmrA-dependent upregulation of dgkA recycles the diacylglycerol generated from PmrA-activated LPS modifications (Wahl et al., 2011). In Yersinia spp., the pmrA and pmrB genes are cotranscribed with dacB, which specifies a D-alanyl-D-alanine carboxypeptidase involved in peptidoglycan biosynthesis (Reines et al., 2012). These results reveal a wider role beyond control of LPS modifications for the PmrA/PmrB regulatory system in governing cell envelope homeostasis and remodeling.
MULTIPLE REGULATORY MECHANISMS DICTATE THE LEVELS AND ACTIVITY OF THE PMRA/PMRB SYSTEM OVER TIME
The regulatory networks controlling gene expression are remarkably dynamic in that they constantly respond to specific signals as well as adapt their transcriptional outputs to physiological changes that have already occurred within the cell (Brandman and Meyer, 2008; Yosef and Regev, 2011). These dynamic changes can be generated by interactions between distinct regulatory circuits or among different layers of a regulatory circuit whereby target genes modulate the levels or activities of regulatory proteins through feedback loops (Thomas and D’Ari, 1990). Although such multicomponent feedback loops are typically a characteristic of eukaryotic gene regulatory circuits (Shen-Orr et al., 2002), it is increasingly clear that multiple regulatory loops can alter the quantitative properties of transcriptional circuits in bacteria (Goulian, 2010; Perez and Groisman, 2009; Tu et al., 2010). In this section, we describe how the levels and activity of the PmrA/PmrB two-component system are tightly regulated by a variety of transcriptional and post-transcriptional mechanisms, which together dictate when, where and to what extent S. typhimurium modifies its LPS.
The PmrA/PmrB System Positively Autoregulates Its Activity
Positive autoregulation has been shown to frequently modulate the sensitivity of two-component systems to input stimulus (Williams and Cotter, 2007), and to temporally control the activation and inactivation of two-component systems (Fujita and Losick, 2005; Hoffer et al., 2001; Shin et al., 2006) (Goulian, 2010; Mitrophanov and Groisman, 2008a). Similarly, phosphorylated PmrA autogenously controls its own expression and that of its cognate sensor PmrB. The pmrA and pmrB genes are transcribed from two promoters: a PmrA-activated promoter located upstream from the pmrC gene in the pmrCAB operon and a constitutive promoter located within the pmrC open reading frame (Gunn and Miller, 1996; Lee et al., 2004; Soncini and Groisman, 1996). Transcription from the latter provides the basal levels of the PmrA and PmrB proteins required for detecting and responding to PmrA-inducing signals (Lee et al., 2004). Positive autoregulation controls the output of the PmrA/PmrB system at early and late activation times (Figure 3). On the one hand, positive feedback dictates the steady-state expression levels of PmrA-activated genes (Mitrophanov and Groisman, 2008a; Wosten and Groisman, 1999). On the other hand, this autoregulation likely governs the transient increase in PmrA activity within 20 min of bacteria experiencing PmrB-inducing conditions. This increase leads to a surge in PmrA-activated mRNAs, which peak and then decrease to reach new steady-state levels by 60 min (Shin et al., 2006) in a manner reflecting changes in the amount of PmrA-P protein and correlating with PmrA-P binding to PmrA-dependent promoters (Shin et al., 2006).
Figure 3. Schematic of PmrA-Dependent Gene Expression over Time and the Various Feedback Mechanisms Operating at Each Stage.
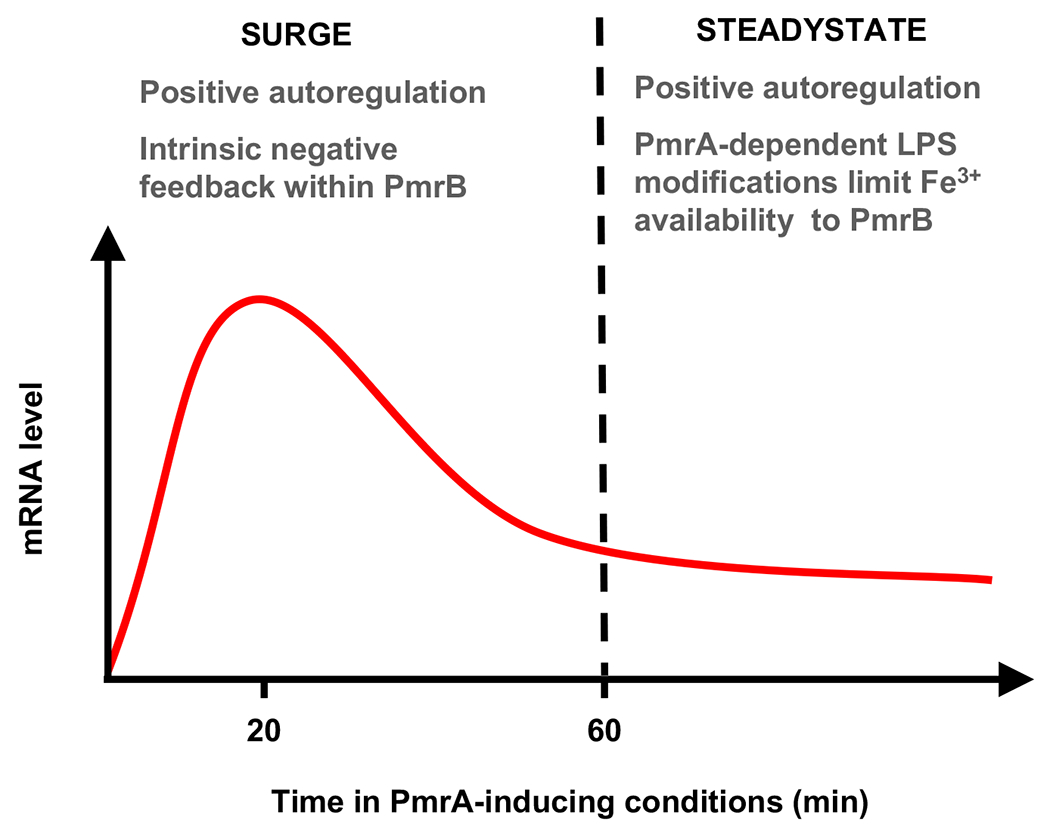
The levels of PmrA-dependent mRNAs peak at 20-30 minutes upon activation (surge), and then decrease to reach a steady state by ~60 min when bacteria are shifted from repressing to inducing conditions for the PmrA/PmrB system. The feedback mechanisms operating at each stage are indicated on the figure.
Intrinsic Negative Feedback within the PmrB Sensor
The biochemical activities of a sensor protein can control a two-component system’s output via a negative feedback mechanism intrinsic to the protein itself (Yeo et al., 2012). PmrB is a bifunctional enzyme that displays opposing kinase and phosphatase activities, like many known sensors (Gao and Stock, 2009; Stock et al., 2000). The change experienced by PmrB from being predominantly in the kinase state to the phosphatase state depends on the trapping of ADP, endogenously generated from ATP during the kinase reaction, within the enzyme’s nucleotide-binding pocket by a flexible loop (referred to as the ATP lid) (Yeo et al., 2012). This lid is critical for regulating PmrB’s phosphatase activity because a lid mutant with lower affinity for ADP was defective in PmrA-P desphosphorylation compared to the wild-type PmrB protein (Yeo et al., 2012). The kinase-to-phosphatase shift in PmrB likely controls the decline in PmrA-dependent mRNAs after the initial activation surge (Figure 3), as was demonstrated for the PhoQ sensor (Shin et al., 2006; Yeo et al., 2012). Thus, both positive feedback on the pmrCAB promoter and intrinsic negative feedback within the PmrB sensor give rise to the PmrA/PmrB activation surge even as bacteria experience constant inducing conditions (Shin et al., 2006; Yeo et al., 2012). These temporal changes in gene expression may represent an adaptation phase during which bacteria rapidly respond to the environmental stimuli triggering PmrA activation and then transition to a different phenotypic state optimized for survival in the new milieu.
PmrA-Dependent Lipid A Modifications Reduce the Activity of the PmrA/PmrB System
Physiological changes in the cell surface brought about by a regulatory system can subsequently mediate feedback on that system’s activity in a time-dependent manner, ensuring that the amount of active transcription factor, which impacts the level and duration of a transcriptional response, is optimal for the needs of the cell. Such a feedback mechanism is exerted by the PmrA-dependent gene products controlling chemical decorations to the lipid A moiety on the activity of the PmrA/PmrB system (Kato et al., 2012). This downregulation is accomplished by the PmrA-activated PmrR membrane peptide, which inhibits LpxT’s ability to make the 1-PP modification, and by the PmrA-mediated incorporation of L-Ara4N and pEtN (Kato et al., 2012). The actions of these PmrA-dependent gene products collectively reduce the net negative charge on the bacterial surface and decrease the amount of Fe3+ bound to the LPS (Kato et al., 2012). As a result, the activity of the PmrA/PmrB system is dampened because less Fe3+ makes it to the periplasm to be directly detected by the PmrB sensor (Wosten et al., 2000). The negative feedback exerted by PmrA-regulated lipid A modifications on the PmrA/PmrB system is time-dependent: a ugd mutant defective for L-Ara4N biosynthesis binds more Fe3+ and displays enhanced expression of PmrA-activated transcripts than the wild-type strain when grown in Fe3+-containing media for 120 min but displays similar levels at 20 min (Kato et al., 2012). This feedback is manifested only after a significant portion of the cell surface has been modified (Figure 3), distinguishing it from intrinsic negative feedback within the PmrB sensor which takes place almost immediately after bacteria encounter PmrB-activating conditions (Yeo et al., 2012). Such a negative feedback regulatory mechanism might enable Salmonella to monitor its LPS modification status and adjust the activity of the regulatory system governing expression of the loci that bring about these LPS modifications.
PmrD Impacts the Duration and Levels of PmrA-Dependent Gene Expression
By modulating the activity of sensor kinases or response regulators, connector proteins not only promote the activation of two-component regulatory systems in response to noncognate signals but also endow these regulatory circuits with specific kinetic properties. The PmrD connector binds to PmrA-P and inhibits its dephosphorylation by PmrB when bacteria experience PmrD-inducing conditions (i.e., low Mg2+) (Kato and Groisman, 2004). In doing so, PmrD expands the spectrum of environments where PmrA-regulated gene products are expressed as well as dictates the levels and timing of their expression. An S. typhimurium strain deleted for the pmrD gene produces lower amounts of PmrA-activated transcript compared to the wild-type when bacteria are incubated with low Mg2+ and high Fe3+ (Kox et al., 2000). This result indicates that PmrD amplifies the levels of PmrA-dependent mRNAs even when bacteria encounter Fe3+, a signal that directly activates the PmrA/PmrB system (Wosten et al., 2000).
Distinct circuit properties brought about by the PmrD protein were further revealed through a comparison of gene transcription levels promoted by the indirect connector-mediated pathway, in which the PhoP-activated PmrD promotes the accumulation of PmrA-P to high enough levels to stimulate target gene expression, versus those of a direct regulation circuit, whereby the PhoP protein binds to the target promoter and activates transcription. First, target gene expression via the PhoP-dependent PmrD connector promotes higher output levels than does direct activation by the PhoP protein (Kato et al., 2007). Second, there is a longer time delay before genes are transcribed via the PmrD-mediated pathway than through direct gene activation by the PhoP protein (Kato et al., 2007), likely because pmrD must be transcribed and translated and because the PmrD protein has to find PmrA-P and bind to it. Third, the PmrD-mediated pathway promotes long deactivation delays when bacteria are shifted from inducing to non-inducing conditions, whereby target mRNAs persist for longer periods of time compared to the pathway where a transcription factor directly controls a given target gene (Kato et al., 2007). This is probably because any remaining PmrD will bind to PmrA-P, promoting the expression of PmrA-dependent genes (Kato et al., 2007). Collectively, these findings demonstrate that PmrD confers particular dynamic features upon the PmrA/PmrB regulatory circuit and impacts the intensity and timing of PmrA-P’s output.
PmrA Represses pmrD Expression
Activation of the PmrA/PmrB system by cognate signals directly detected by the PmrB sensor and in a manner independent of the PmrD protein results in reduced levels of pmrD transcript (Kato et al., 2003). This reduction is accomplished through binding of PmrA-P to the PmrA box upstream from the pmrD transcription start site (Kato et al., 2003). The presence of PmrA does not exclude PhoP’s ability to associate to the pmrD promoter and to activate pmrD transcription (Kato et al., 2003). Therefore, the PmrA-mediated repression of pmrD might provide a means to maintain optimal cellular quantities of PmrA-P protein and to avoid the potential detrimental effect of PmrD overproduction in S. typhimurium (Kato et al., 2003).
In sum, the PmrA/PmrB two-component system in S. typhimurium constitutes a singular example whereby multiple overlapping feedback mechanisms coordinate the activity of a regulatory system. These feedback loops have different temporal characteristics that appear to shape the signaling and transcriptional responses of the PmrA/PmrB system over time.
EVOLUTION OF THE PMRA/PMRB SYSTEM IN ENTERIC BACTERIA
Resistance to polymyxin B is conserved in a number of enteric bacterial species that also specify orthologs of the PmrA/PmrB two-component system, including Y. pestis (Rebeil et al., 2004; Winfield et al., 2005), K. pneumoniae (Helander et al., 1996; Mitrophanov et al., 2008), E. coli (Winfield and Groisman, 2004), C. rodentium (Viau et al., 2011) and E. carotovora (Hyytiainen et al., 2003). However, the orthologous PmrA/PmrB transcriptional regulatory circuits have undergone extensive modifications, resulting in qualitative and quantitative differences in gene expression outputs and thus, in phenotypic variation among closely related bacteria. In this section, we explore how the PmrA/PmrB regulatory circuit has diverged in enteric bacteria.
PmrA/PmrB Regulatory Architectures Differ among Related Enteric Bacteria
Y. pestis promotes transcription of the PmrA-activated pbgP and ugd genes during growth in low Mg2+ despite the fact that it lacks a pmrD gene (Winfield et al., 2005) (Figure 4A). This is because the promoters of these polymyxin resistance genes harbor PhoP-binding sites such that PhoP directly activates transcription, rendering bacteria resistant to killing by the antibiotic in PhoP-inducing environments (Winfield et al., 2005). Such direct, PhoP-activated expression of polymyxin B resistance genes distinguishes the Y. pestis regulatory circuit from that of S. typhimurium, where transcription of the pbgP and ugd genes in low Mg2+ occurs through an indirect pathway involving the PhoP-dependent horizontally acquired gene product PmrD (Kox et al., 2000). The K. pneumoniae regulatory circuit appears to be an intermediate between those of Y. pestis and S. typhimurium: it consists of direct PhoP-dependent activation of pbgP, as in Y. pestis, as well as the indirect PmrD-mediated pathway, as in S. typhimurium (Mitrophanov et al., 2008) (Figure 4B). However, PmrA does not repress pmrD expression in K. pneumoniae experiencing low Mg2+, unlike in S. typhimurium (Mitrophanov et al., 2008). Such restructuring of the interactions between orthologous regulatory proteins and their target genes in closely related organisms results in dissimilar gene expression outputs for the same target genes activated by the same signal (Mitrophanov et al., 2010; Mitrophanov et al., 2008). Thus, subtle changes in the levels and/or kinetics with which target gene products are produced may endow bacteria with the ability to occupy particular niches, leading to phenotypic diversity among related species.
Figure 4. Model of the Interactions between the PhoP/PhoQ and PmrA/PmrB Systems in Y. pestis, K pneumoniae and E. coli.

(A) In Y. pestis, the PmrA-activated pbgP gene is expressed in low Mg2+ directly via the PhoP/PhoQ regulatory system or in the presence of Fe3+ directly via the PmrA/PmrB regulatory system. The pmrD gene is absent from Y. pestis.
(B) In K. pneumoniae, the PmrA-activated pbgP gene is expressed in low Mg2+ directly via the PhoP/PhoQ regulatory system or indirectly, via the PhoP/PhoQ system, the PmrD protein and the PmrA/PmrB system. Expression of pbgP is also directly activated by the PmrA/PmrB system when bacteria encounter Fe3+. The PmrA protein does not regulate pmrD transcription.
(C) In E. coli, transcription of the PmrA-activated gene pbgP is promoted in the presence of Fe3+ via the PmrA/PmrB system. The PmrD protein is produced in low Mg2+ in a PhoP-dependent manner but fails to activate the PmrA/PmrB system.
Analyses of several Citrobacter genomes identified a pmrD homolog which specifies a gene product that has 70% identity to the S. enterica PmrD protein (Chen et al., 2011), and it is plausible that these Citrobacter strains exhibit PmrA-dependent gene expression when grown in PhoP-inducing conditions. However, pmrD is absent from the C. rodentium genome, resulting in PmrA-dependent but PhoP-independent expression of the LPS-modifying genes pmrC and cptA (Viau et al., 2011). C. rodentium also lacks homologs to the pbgPE operon that governs L-Ara4N biosynthesis (Viau et al., 2011), unlike other strains of Citrobacter. Given that C. rodentium is suggested to be a recently evolved pathogen undergoing large-scale genomic rearrangements and functional gene loss (Petty et al., 2011), the absence of the pmrD and pbgPE genes might be a derived state that occurred after the ancestral organism to the Klebsiella, Citrobacter, Salmonella and Escherichia lineages acquired pmrD. These observations raise the possibility of the production of PmrA-dependent transcripts in low Mg2+ environments and modification of the LPS with L-Ara4N being detrimental to C. rodentium’s lifestyle as an enteric pathogen that causes colonic hyperplasia in mice.
Allelic Differences between the Ancestral PmrB or PmrA Proteins Dictate the Functionality of the Horizontally Acquired PmrD Gene Product
E. coli induces expression of PmrA-dependent genes upon encountering the presence of Fe3+, like S. typhimurium (Winfield and Groisman, 2004). However, E. coli does not transcribe PmrA-activated genes and is sensitive to polymyxin B when grown in low Mg2+ even though it harbors the horizontally acquired PhoP-dependent pmrD gene (Winfield and Groisman, 2004) (Figure 4C). Moreover, the PmrA-P protein does not repress transcription of the pmrD gene in E. coli, unlike what happens in S. typhimurium (Winfield and Groisman, 2004). The disparate expression outputs between E. coli and S. typhimurium arise from differences between both the orthologous PmrD proteins (Winfield and Groisman, 2004) and the PmrB sensors (Chen et al., 2011).
The E. coli and S. typhimurium PmrD proteins are highly divergent and display only 55.3% amino acid identity (Winfield and Groisman, 2004), which is significantly lower than the 90% median identity between homologous proteins in these two species (McClelland et al., 2001). Indeed, an engineered E. coli strain expressing the S. typhimurium pmrD gene expressed PmrA-activated genes when grown in low Mg2+, unlike the isogenic strain harboring E. coli’s own pmrD gene (Winfield and Groisman, 2004). Analysis of the nucleotide sequences corresponding to the pmrD gene from a large collection of E. coli and S. enterica natural isolates suggested that the pmrD gene was evolving in a non-neutral fashion (Winfield and Groisman, 2004). However, the E. coli PmrD was as effective as the S. typhimurium ortholog in protecting PmrA-P from PmrB-mediated dephosphorylation in vitro (Chen et al., 2011) and promoted transcription of PmrA-activated genes in S. typhimurium grown in low Mg2+ (Chen et al., 2011). Together, these results established that the E. coli PmrD protein is functional and that its ability to connect the PhoP/PhoQ and PmrA/PmrB systems is context-dependent: it functions in S. typhimurium but, paradoxically, not in E. coli. Moreover, they indicated that E. coli and S. typhimurium must differ at an additional genetic locus, resulting in lower levels of PmrA-P – the target of PmrD – in E. coli than in S. typhimurium.
The ancestral PmrB proteins from E. coli and S. typhimurium display a high level of amino acid identity (~83%). However, even subtle differences between these PmrB orthologs are sufficient to bring about quantitative differences in their biochemical activities and in resistance of these bacteria to killing by polymyxin B in low Mg2+ (Chen et al., 2011). The E. coli PmrB dephosphorylates PmrA-P at a 10-fold higher rate than does the S. typhimurium ortholog (Chen et al., 2011). This higher phosphatase activity renders the horizontally acquired PmrD ineffective in promoting the accumulation of high enough levels of PmrA-P to activate the ancestral PmrA/PmrB system when E. coli experience PmrD-inducing conditions (Chen et al., 2011). Consistent with this notion, an E. coli strain expressing the S. typhimurium pmrB gene produced PmrA-activated mRNAs and displayed polymyxin B resistance during growth in low Mg2+ (Chen et al., 2011). Therefore, when a horizontally acquired gene product acts on an ancestral pathway, its ability to confer a new function can be affected by quantitative differences in the biochemical activities of orthologous ancestral proteins.
The ability to express PmrA-activated genes via the PmrD protein varies even within species, with some E. coli isolates displaying an S. typhimurium-like phenotype and vice versa (Winfield and Groisman, 2004). For example, most S. enterica isolates activate the PmrA/PmrB system and are resistant to polymyxin B in the presence of low Mg2+ (Winfield and Groisman, 2004). However, strains belonging to the serovar Paratyphi B, which causes paratyphoid fever in humans (Beltran et al., 1991; Prager et al., 2003), fail to do so (Chen et al., submitted). This is because a single amino acid substitution in the S. paratyphi B PmrA diminished its affinity for target promoters, abolishing PmrA-activated gene expression in low Mg2+ (Chen et al., submitted). Furthermore, S. paratyphi B transcribes PmrA-activated genes only when both PmrD- and PmrA-inducing signals are present (Chen et al., submitted), whereas S. typhimurium can do so in the presence of the low Mg2+ and/or the high Fe3+ signal (Kox et al., 2000; Wosten et al., 2000). These results present a singular example whereby a natural allele of an ancestral transcription factor is rendered dependent on the presence of a horizontally transferred gene product.
Allelic variation between orthologous regulatory proteins not only alters the steady-state levels of target genes but also results in distinct expression kinetics between closely related bacteria. Quantitative differences between the phosphatase activities of the S. typhimurium and E. coli PmrB sensors lead to disparate temporal activation profiles of PmrA and hence to expression of PmrA-activated genes upon induction of the PmrA/PmrB system by Fe3+ (Chen et al., 2011). Likewise, the affinities of response regulators for their promoter sequences control gene expression dynamics because the S. paratyphi B PmrA promotes gene expression with slower kinetics than does the S. typhimurium PmrA (Chen et al., submitted). Hence, the distinct expression dynamics in closely related organisms are not limited to differences in the genetic architectures of their signaling systems (Alon, 2007; Perez and Groisman, 2009) but rather, can also arise from subtle amino acid differences between orthologous regulatory proteins.
Proposed Evolutionary Trajectories of the PmrA/PmrB Regulatory System in Enteric Bacteria
When did the genetic changes leading to variation in the PmrA/PmrB regulatory circuits occur in the evolutionary history of enteric species? The pmrA and pmrB genes are widely distributed among enteric bacteria (Gunn, 2008; Hyytiainen et al., 2003; Mitrophanov et al., 2008; Viau et al., 2011; Winfield and Groisman, 2004; Winfield et al.,2005). By contrast, the horizontally transferred pmrD gene appears limited to enteric bacteria of the Klebsiella, Citrobacter, Salmonella and Escherichia lineages, suggesting that pmrD was acquired by the ancestral strain giving rise to these lineages (Chen et al., 2011; Mitrophanov et al., 2008). This ancestral organism likely transcribed PmrA-activated genes in low Mg2+ via the PmrD protein because this ability is retained in Klebsiella and most Salmonella strains (Kox et al., 2000; Mitrophanov et al., 2008; Winfield and Groisman, 2004). We propose that the ability to make low Mg2+-dependent, PmrA-controlled modifications of the LPS was retained by S. typhimurium and the vast majority of S. enterica strains (Winfield and Groisman, 2004) but lost in E. coli and S. paratyphi B, perhaps contributing towards their survival in distinct host and non-host environments. In the case of E. coli, selection for a PmrB protein with heightened phosphatase activity might prevent overaccumulation of active (i.e., phosphorylated) PmrA in low Mg2+, which might be detrimental to E. coli’s lifestyle because a mutant that constitutively expresses PmrA-dependent genes is hypersusceptible to the bile detergent deoxycholate (Froelich et al., 2006). In S. paratyphi B, selection for a pmrA allele that activates transcription only when both low Mg2+ and high Fe3+ signals are present (but not when each signal is sensed individually) increases biofilm formation (Chen et al., submitted) and might enhance the ability to cause chronic infection of the gallbladder, a property of the serovar Paratyphi B (Gonzalez-Escobedo et al., 2010; Gunn, 2000; Ristori et al., 1982a; Ristori et al., 1982b; Vogelsang and Boe, 1948). These results suggest that the integration of horizontally acquired gene products into ancestral genetic networks can impact the evolutionary trajectories of pre-existing regulatory proteins.
The P. aeruginosa PmrA/PmrB System Controls Resistance to Polymyxin B
The opportunistic nosocomial pathogen P. aeruginosa modifies its LPS with L-Ara4N and displays resistance to polymyxin B when grown in Mg2+-limiting conditions (McPhee et al., 2003; Moskowitz et al., 2004). The PmrA/PmrB system appears to respond to low Mg2+ because the transcription of gene products responsible for this L-Ara4N modification is 2-fold lower in a P. aeruginosa pmrB mutant compared to the wild-type strain (McPhee et al., 2003). However, it is presently unclear whether PmrB directly detects the low Mg2+ signal or whether this signal indirectly activates the PmrA/PmrB system, perhaps via the PhoP/PhoQ system and a PmrD-like protein. The P. aeruginosa PmrA/PmrB system is not activated when bacteria are grown in the presence of Fe3+ or in low pH, consistent with the finding that the P. aeruginosa PmrB periplasmic sensing domain exhibits low identity with the S. typhimurium ortholog (McPhee et al., 2003). The presence of PmrA- and PhoP-binding sites within the promoter of the operon encoding gene products mediating the L-ARa4N decoration suggests that both PmrA and PhoP directly promote transcription of these genes (McPhee et al., 2006), like in Y. pestis (Winfield et al., 2005) and K. pneumoniae (Mitrophanov et al., 2008).
Increased modification of the lipid A with L-Ara4N has been implicated in the emergence of polymyxin B-resistant P. aeruginosa clinical isolates (Ernst et al., 1999). This heightened antibiotic resistance is attributed, in part, to amino acid substitutions in their PmrA and/or PmrB proteins that likely result in hyperactivation of the PmrA/PmrB system and elevated expression of LPS-modifying genes (Abraham and Kwon, 2009; Moskowitz et al., 2011). Likewise, mutations in the pmrA and/or pmrB genes were shown to augment the ability of the opportunistic pathogen Acinetobacter baumannii to resist killing by polymyxins (Arroyo et al., 2012). Together, these findings emphasize the need to further understand the mechanisms through which the PmrA/PmrB system controls resistance to polymyxins if these antibiotics are to be used as a therapeutic option against multidrug-resistant Gram-negative bacteria (Yahav et al., 2012).
The PmrA/PmrB System Regulates Virulence Properties of Legionella pneumophila
The PmrA/PmrB regulatory system is conserved among strains of Legionella pneumophila (Al-Khodor et al., 2009), the causative agent of Legionnaires’ disease. The L. pneumophila PmrB is predicted to detect Fe3+ because its periplasmic domain harbors the ExxE motif (Al-Khodor et al., 2009), which is required for Fe3+ to activate the PmrA/PmrB system (Wosten et al., 2000). PmrA recognizes DNA sequences similar to the PmrA box in S. typhimurium (Marchal et al., 2004; Wosten and Groisman, 1999; Zusman et al., 2007) and directly binds to the promoters of several genes encoding substrates of the icm/dot Type IV secretion system, the major virulence system in L. pneumophila (Zusman et al., 2007). Genome-wide microarray analyses further suggested that PmrA is a global regulator of virulence and metabolic genes in this intracellular pathogen, including those encoding the Type IV secretion system and flagellar biosynthesis, metabolic and stress response genes (Al-Khodor et al., 2009). The PmrA regulon appears to have undergone extensive transcriptional rewiring in L. pneumophila because PmrA does not regulate expression of LPS-modifying genes in this species, which distinguishes it from other bacteria encoding PmrA homologs (Al-Khodor et al., 2009). The PmrA/PmrB system contributes to L. pneumophila’s proliferation within human macrophages and its amoeba hosts (Al-Khodor et al., 2009; Zusman et al., 2007), likely by dictating the expression of gene products responsible for generating a niche suitable for replication.
A Putative PmrA Homolog in Francisella Controls Polymyxin B Resistance
Bioinformatic analyses of the sequenced genomes of the gram-negative Francisella spp., which includes the facultative intracellular pathogen F. tularensis that causes tularemia in humans and F. novicida that causes lethal infection in mice, found a dearth of two-component systems in this species (Larsson et al., 2005; Mohapatra et al., 2007). However, an orphan response regulator with 44% amino acid identity to the S. typhimurium PmrA protein was identified (Mohapatra et al., 2007). (Note that the putative F. tularensis PmrA homolog also displays 42% and 32% identity to the S. typhimurium QseB and PhoP proteins, respectively.) The Francisella pmrA gene is not encoded in an operon with its cognate sensor kinase (Mohapatra et al., 2007), unlike the vast majority of genes specifying two-component systems (Mascher et al., 2006). Instead, in vitro biochemical assays demonstrated that phosphorylation of the F. novicida PmrA at its conserved aspartate residue appears to be largely mediated by the KdpD sensor, which is located in an operon with the response regulator KdpE in this bacterium (Bell et al., 2010). Consistent with the finding that PmrA regulates a large number of genes within the Francisella pathogenicity island, an F. novicida pmrA mutant is defective for intramacrophage survival and mouse virulence (Mohapatra et al., 2007). This mutant is also 32-fold more susceptible than the wild-type strain to killing by polymyxin B (Mohapatra et al., 2007). However, PmrA does not activate the expression of LPS-modification genes, and no differences were detected in lipid A isolated from wild-type or pmrA mutant bacteria, suggesting that PmrA-mediated resistance toward cationic antimicrobial peptides in F. novicida is independent of modifications to its lipid A (Mohapatra et al., 2007).
CONCLUSIONS AND PERSPECTIVES
The extent of LPS decorations in bacteria must be finely tuned. On the one hand, these decorations are critical for resistance against bactericidal agents (Shai, 1999; Vaara et al., 1979) and for evasion of the host immune system (Takeda et al., 2003). On the other hand, excessive decorations render bacteria hypersusceptible to the bile detergent deoxycholate (Froelich et al., 2006). The PmrA/PmrB two-component system is the major regulator of these LPS modifications, and we have focused here on the PmrA-dependent gene products that mediate alterations to the LPS. However, it was recently demonstrated that a pEtN transferase in C. jejuni – EptC – modifies not only the lipid A with pEtN but also the flagellar rod protein FlgG, establishing a link between membrane biogenesis and motility (Cullen et al., 2012; Cullen and Trent, 2010); these findings raise the possibility that the LPS-modifying enzymes in other bacteria might similarly possess additional functional and/or regulatory roles. Furthermore, the PmrA regulon extends beyond LPS-modifying genes (Hagiwara et al., 2004; Marchal et al., 2004; Ogasawara et al., 2012; Tamayo et al., 2005b; Tamayo et al., 2002). Thus, understanding the functions of these other PmrA-dependent genes and how they promote the survival of bacteria in environments containing PmrA-activating signals may provide insights into a broader regulatory role for the PmrA/PmrB system.
That the gene expression output of the PmrA/PmrB system has to be tightly regulated is underscored by the presence of multiple feedback mechanisms operating at the transcriptional, translational and post-translational levels. Each feedback module affects the output of the system on a broad range of timescales, from rapid responses once bacteria experience PmrA-inducing conditions (Shin et al., 2006; Yeo et al., 2012) to changes that occur only after bacteria have undergone several cell divisions (Kato et al., 2012). We anticipate that the integration of computational analyses and experimental studies will offer a more comprehensive understanding of how these various feedback loops act in concert to modulate the amplitude and duration of PmrA-dependent responses and how such elaborate feedback loops enable bacteria to coordinate the levels of cell surface modifications with the amount of PmrA-inducing signals in their surroundings.
Finally, the evolution of orthologous PmrA/PmrB regulatory circuits has occurred through changes in the ancestral genome or by the effect that horizontally acquired genes have on the properties of ancestral circuits, giving rise to phenotypic variation within and across bacterial species. Some of these changes arise from subtle amino acid differences among orthologous proteins (Chen et al., 2011) or from modifications of cis regulatory sequences (Winfield et al., 2005), both of which cannot be readily predicted from sequence conservation and computational comparisons of related genomes. The qualitative and quantitative differences in PmrA-dependent gene expression reflect the need to tightly control the activity of the PmrA/PmrB system to levels optimal for the survival of individual organisms in their particular ecological niches. This raises questions as to the selective pressures that drove the emergence of these disparate regulatory phenotypes and that determined the distinct spectrum of PmrA-dependent targets in different bacteria.
REFERENCES
- Abraham N, and Kwon DH (2009). A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol Lett 298, 249–254. [DOI] [PubMed] [Google Scholar]
- Aguirre A, Lejona S, Vescovi EG, and Soncini FC (2000). Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar typhimurium. J Bacteriol 182, 3874–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodor S, Kalachikov S, Morozova I, Price CT, and Abu Kwaik Y (2009). The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect Immun 77, 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U (2007). Network motifs: theory and experimental approaches. Nat Rev Genet 8, 450–461. [DOI] [PubMed] [Google Scholar]
- Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, and Hancock RE (2012). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55, 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, and Miller SI (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. [DOI] [PubMed] [Google Scholar]
- Badola P, and Sanders CR 2nd (1997). Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J Biol Chem 272, 24176–24182. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, and Blanot D (2009). Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J Chromatogr B Analyt Technol Biomed Life Sci 877, 213–220. [DOI] [PubMed] [Google Scholar]
- Bell BL, Mohapatra NP, and Gunn JS (2010). Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect Immun 78, 2189–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P, Plock SA, Smith NH, Whittam TS, Old DC, and Selander RK (1991). Reference collection of strains of the Salmonella typhimurium complex from natural populations. J Gen Microbiol 137, 601–606. [DOI] [PubMed] [Google Scholar]
- Brandman O, and Meyer T (2008). Feedback loops shape cellular signals in space and time. Science 322, 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breazeale SD, Ribeiro AA, McClerren AL, and Raetz CR (2005). A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J Biol Chem 280, 14154–14167. [DOI] [PubMed] [Google Scholar]
- Breazeale SD, Ribeiro AA, and Raetz CR (2002). Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose. J Biol Chem 277, 2886–2896. [DOI] [PubMed] [Google Scholar]
- Breazeale SD, Ribeiro AA, and Raetz CR (2003). Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-L-arabinose. J Biol Chem 278, 24731–24739. [DOI] [PubMed] [Google Scholar]
- Browning DF, and Busby SJ (2004). The regulation of bacterial transcription initiation. Nat Rev Microbiol 2, 57–65. [DOI] [PubMed] [Google Scholar]
- Chamnongpol S, Dodson W, Cromie MJ, Harris ZL, and Groisman EA (2002). Fe(III)-mediated cellular toxicity. Mol Microbiol 45, 711–719. [DOI] [PubMed] [Google Scholar]
- Chen HD, Jewett MW, and Groisman EA (2011). Ancestral genes can control the ability of horizontally acquired loci to confer new traits. PLoS Genet 7, e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Chen YF, and Peng HL (2010). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Reeve KE, and Gunn JS (2010). Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol 192, 2981–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Madsen JA, Ivanov PL, Brodbelt JS, and Trent MS (2012). Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J Biol Chem 287, 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, and Trent MS (2010). A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci U S A 107, 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MA, Mouslim C, and Groisman EA (2006). The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol Microbiol 60, 39–50. [DOI] [PubMed] [Google Scholar]
- Doerrler WT, Gibbons HS, and Raetz CR (2004). MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem 279, 45102–45109. [DOI] [PubMed] [Google Scholar]
- El Ghachi M, Derbise A, Bouhss A, and Mengin-Lecreulx D (2005). Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J Biol Chem 280, 18689–18695. [DOI] [PubMed] [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, and Miller SI (1999). Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286, 1561–1565. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gomez JD, and Sansom MS (2003). Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol 4, 105–116. [DOI] [PubMed] [Google Scholar]
- Fleischhacker AS, and Kiley PJ (2011). Iron-containing transcription factors and their roles as sensors. Curr Opin Chem Biol 15, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich JM, Tran K, and Wall D (2006). A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol 188, 1180–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, and Losick R (2005). Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19, 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Mack TR, and Stock AM (2007). Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, and Stock AM (2009). Biological insights from structures of two-component proteins. Annu Rev Microbiol 63, 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Vescovi E, Soncini FC, and Groisman EA (1996). Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174. [DOI] [PubMed] [Google Scholar]
- Georg J, and Hess WR (2011). cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75, 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons HS, Kalb SR, Cotter RJ, and Raetz CR (2005). Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol Microbiol 55, 425–440. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Lin S, Cotter RJ, and Raetz CR (2000). Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem 275, 32940–32949. [DOI] [PubMed] [Google Scholar]
- Girardin H, Albagnac C, Dargaignaratz C, Nguyen-The C, and Carlin F (2002). Antimicrobial activity of foodborne Paenibacillus and Bacillus spp. against Clostridium botulinum. J Food Prot 65, 806–813. [DOI] [PubMed] [Google Scholar]
- Goddard WP, Coupland K, Smith JA, and Long RG (1997). Iron uptake by isolated human enterocyte suspensions in vitro is dependent on body iron stores and inhibited by other metal cations. J Nutr 127, 177–183. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G, Marshall JM, and Gunn JS (2010). Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M (2010). Two-component signaling circuit structure and properties. Curr Opin Microbiol 13, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA (1998). The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays 20, 96–101. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Kayser J, and Soncini FC (1997). Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol 179, 7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, and Mouslim C (2006). Sensing by bacterial regulatory systems in host and non-host environments. Nat Rev Microbiol 4, 705–709. [DOI] [PubMed] [Google Scholar]
- Gunn JS (2000). Mechanisms of bacterial resistance and response to bile. Microbes Infect 2, 907–913. [DOI] [PubMed] [Google Scholar]
- Gunn JS (2008). The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16, 284–290. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, and Miller SI (1998). PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27, 1171–1182. [DOI] [PubMed] [Google Scholar]
- Gunn JS, and Miller SI (1996). PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol 178, 6857–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, and Miller SI (2000). Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun 68, 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, and Miller SI (1998). Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Yamashino T, and Mizuno T (2004). A genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem 68, 1758–1767. [DOI] [PubMed] [Google Scholar]
- Hamad MA, Di Lorenzo F, Molinaro A, and Valvano MA (2012). Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia. Mol Microbiol. [DOI] [PubMed] [Google Scholar]
- Haneburger I, Eichinger A, Skerra A, and Jung K (2012). New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli. J Biol Chem 286, 10681–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley MD, and Imperiali B (2011). At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch Biochem Biophys 517, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander IM, Kato Y, Kilpelainen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, and Yokochi T (1996). Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem 237, 272–278. [DOI] [PubMed] [Google Scholar]
- Helander IM, Kilpelainen I, and Vaara M (1994). Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol 11, 481–487. [DOI] [PubMed] [Google Scholar]
- Herrera CM, Hankins JV, and Trent MS (2010). Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol 76, 1444–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA (2000). Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3, 165–170. [DOI] [PubMed] [Google Scholar]
- Hoffer SM, Westerhoff HV, Hellingwerf KJ, Postma PW, and Tommassen J (2001). Autoamplification of a two-component regulatory system results in “learning” behavior. J Bacteriol 183, 4914–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer SU, Schlumberger MC, Jackel D, and Hensel M (2009). Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica. Infect Immun 77, 5458–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytiainen H, Sjoblom S, Palomaki T, Tuikkala A, and Tapio Palva E (2003). The PmrA-PmrB two-component system responding to acidic pH and iron controls virulence in the plant pathogen Erwinia carotovora ssp. carotovora. Mol Microbiol 50, 795–807. [DOI] [PubMed] [Google Scholar]
- Karavolos MH, Spencer H, Bulmer DM, Thompson A, Winzer K, Williams P, Hinton JC, and Khan CM (2008). Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics 9, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Chen HD, Latifi T, and Groisman Eduardo A. (2012). Reciprocal Control between a Bacterium’s Regulatory System and the Modification Status of Its Lipopolysaccharide. Molecular Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, and Groisman EA (2004). Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev 18, 2302–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Latifi T, and Groisman EA (2003). Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci U S A 100, 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Mitrophanov AY, and Groisman EA (2007). A connector of two-component regulatory systems promotes signal amplification and persistence of expression. Proc Natl Acad Sci U S A 104, 12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Ernst RK, and Miller SI (2005). Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J Bacteriol 187, 2448–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Killam T, Sood V, and Surette MG (2003). Swarm-cell differentiation in Salmonella enterica serovar typhimurium results in elevated resistance to multiple antibiotics. J Bacteriol 185, 3111–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox LF, Wosten MM, and Groisman EA (2000). A small protein that mediates the activation of a two-component system by another two-component system. EMBO J 19, 1861–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreamer NN, Wilks JC, Marlow JJ, Coleman ML, and Newman DK (2012).BqsR/BqsS constitute a two-component system that senses extracellular Fe(II) in Pseudomonas aeruginosa. J Bacteriol 194, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, et al. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet 37, 153–159. [DOI] [PubMed] [Google Scholar]
- Laub MT, and Goulian M (2007). Specificity in two-component signal transduction pathways. Annu Rev Genet 41, 121–145. [DOI] [PubMed] [Google Scholar]
- Lee H, Hsu FF, Turk J, and Groisman EA (2004). The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 186, 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LJ, Barrett JA, and Poole RK (2005). Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J Bacteriol 187, 1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, and Hulett FM (1997). Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol 179, 6302–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, and Stock JB (1992). Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A 89, 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, and Gottesman S (2005). The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59, 379–405. [DOI] [PubMed] [Google Scholar]
- Makela P, Sarvas M, Calcagno S, and Lounatmaa K (1978). Isolation and genetic characterization of polymyxin-resistant mutants of Salmonella. FEMS Microbiol Lett 3, 323–326. [Google Scholar]
- Marceau M, Sebbane F, Ewann F, Collyn F, Lindner B, Campos MA, Bengoechea JA, and Simonet M (2004). The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP-PhoQ two-component system but not by PmrA-PmrB, and is not required for virulence. Microbiology 150, 3947–3957. [DOI] [PubMed] [Google Scholar]
- Marchal K, De Keersmaecker S, Monsieurs P, van Boxel N, Lemmens K, Thijs G, Vanderleyden J, and De Moor B (2004). In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol 5, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, and Unden G (2006). Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70, 910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni P, Grant A, Restif O, and Maskell D (2009). A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol 7, 73–80. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. [DOI] [PubMed] [Google Scholar]
- McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, and Hancock RE (2006). Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol 188, 3995–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, and Hancock RE (2003). Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50, 205–217. [DOI] [PubMed] [Google Scholar]
- Merighi M, Carroll-Portillo A, Septer AN, Bhatiya A, and Gunn JS (2006). Role of Salmonella enterica serovar Typhimurium two-component system PreA/PreB in modulating PmrA-regulated gene transcription. J Bacteriol 188, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Ellermeier CD, Slauch JM, and Gunn JS (2005). Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J Bacteriol 187, 7407–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Septer AN, Carroll-Portillo A, Bhatiya A, Porwollik S, McClelland M, and Gunn JS (2009). Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, and Groisman EA (2008a). Positive feedback in cellular control systems. Bioessays 30, 542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, and Groisman EA (2008b). Signal integration in bacterial two-component regulatory systems. Genes Dev 22, 2601–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Hadley TJ, and Groisman EA (2010). Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J Mol Biol 401, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Jewett MW, Hadley TJ, and Groisman EA (2008). Evolution and dynamics of regulatory architectures controlling polymyxin B resistance in enteric bacteria. PLoS Genet 4, e1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, Carlson RW, and Gunn JS (2007). Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun 75, 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, and Gottesman S (2009). A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74, 1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, van den Bosch L, and Manning PA (1995). Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol 177, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, et al. (2011). PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK, and Miller SI (2004). PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186, 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouslim C, and Groisman EA (2003). Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol 47, 335–344. [DOI] [PubMed] [Google Scholar]
- Muller S, Gotz M, and Beier D (2009). Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One 4, e6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray GL, Attridge SR, and Morona R (2003). Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47, 1395–1406. [DOI] [PubMed] [Google Scholar]
- Nikaido H (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Hsu FF, Turk J, Cromie MJ, Wosten MM, and Groisman EA (2006). Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol Microbiol 61, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nummila K, Kilpelainen I, Zahringer U, Vaara M, and Helander IM (1995). Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol 16, 271–278. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Shinohara S, Yamamoto K, and Ishihama A (2012). Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiology 158, 1482–1492. [DOI] [PubMed] [Google Scholar]
- Pang T, Bhutta ZA, Finlay BB, and Altwegg M (1995). Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol 3, 253–255. [DOI] [PubMed] [Google Scholar]
- Paris I, Martinez-Alvarado P, Perez-Pastene C, Vieira MN, Olea-Azar C, Raisman-Vozari R, Cardenas S, Graumann R, Caviedes P, and Segura-Aguilar J (2005). Monoamine transporter inhibitors and norepinephrine reduce dopamine-dependent iron toxicity in cells derived from the substantia nigra. J Neurochem 92, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Perez JC, and Groisman EA (2007). Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol 63, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, and Groisman EA (2009). Evolution of transcriptional regulatory circuits in bacteria. Cell 138, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescaretti MM, Lopez FE, Morero RD, and Delgado MA (2011). The PmrA/PmrB regulatory system controls the expression of wzzfepE gene involved in the O-antigen synthesis of Salmonella enterica serovar Typhimurium. Microbiology. [DOI] [PubMed] [Google Scholar]
- Petty NK, Feltwell T, Pickard D, Clare S, Toribio AL, Fookes M, Roberts K, Monson R, Nair S, Kingsley RA, et al. (2011). Citrobacter rodentium is an unstable pathogen showing evidence of significant genomic flux. PLoS Pathog 7, e1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager R, Rabsch W, Streckel W, Voigt W, Tietze E, and Tschape H (2003). Molecular properties of Salmonella enterica serotype paratyphi B distinguish between its systemic and its enteric pathovars. J Clin Microbiol 41, 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Reynolds CM, Trent MS, and Bishop RE (2007). Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76, 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, and Whitfield C (2002). Lipopolysaccharide endotoxins. Annu Rev Biochem 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N, and Wang Y (2000). A high-affinity iron permease essential for Candida albicans virulence. Science 288, 1062–1064. [DOI] [PubMed] [Google Scholar]
- Ratering S, and Schnell S (2000). Localization of iron-reducing activity in paddy soil by profile studies. Biogeochemistry 48, 341–365. [Google Scholar]
- Rebeil R, Ernst RK, Gowen BB, Miller SI, and Hinnebusch BJ (2004). Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol 52, 1363–1373. [DOI] [PubMed] [Google Scholar]
- Reines M, Llobet E, Llompart CM, Moranta D, Perez-Gutierrez C, and Bengoechea JA (2012). Molecular basis of Yersinia enterocolitica temperature-dependent resistance to antimicrobial peptides. J Bacteriol 194, 3173–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CM, Kalb SR, Cotter RJ, and Raetz CR (2005). A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem 280, 21202–21211. [DOI] [PubMed] [Google Scholar]
- Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CR, and Trent MS (2006). An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem 281, 21974–21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SM, Strandberg KL, Conroy M, and Gunn JS (2012). Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front Cell Infect Microbiol 2, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristori C, Rodriguez H, Vicent P, Ferreccio C, Garcia J, Lobos H, and D’Ottone K (1982a). Persistence of the Salmonella typhi-paratyphi carrier state after gallbladder removal. Bull Pan Am Health Organ 16, 361–366. [PubMed] [Google Scholar]
- Ristori C, Rodriguez H, Vicent P, Lobos H, D’Ottone K, Garcia J, Pinto ME, Nercelles P, and Cisneros L (1982b). Investigation of the Salmonella typhi-paratyphi carrier state in cases of surgical intervention for gallbladder disease. Bull Pan Am Health Organ 16, 161–171. [PubMed] [Google Scholar]
- Roland KL, Esther CR, and Spitznagel JK (1994). Isolation and characterization of a gene, pmrD, from Salmonella typhimurium that confers resistance to polymyxin when expressed in multiple copies. J Bacteriol 176, 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland KL, Martin LE, Esther CR, and Spitznagel JK (1993). Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol 175, 4154–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schioler H, Christiansen ED, Hoybye G, Rasmussen SN, and Greibe J (1983). Biliary calculi in chronic Salmonella carriers and healthy controls: a controlled study. Scand J Infect Dis 15, 17–19. [DOI] [PubMed] [Google Scholar]
- Shafer WM, Casey SG, and Spitznagel JK (1984a). Lipid A and resistance of Salmonella typhimurium to antimicrobial granule proteins of human neutrophil granulocytes. Infect Immun 43, 834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer WM, Martin LE, and Spitznagel JK (1984b). Cationic antimicrobial proteins isolated from human neutrophil granulocytes in the presence of diisopropyl fluorophosphate. Infect Immun 45, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai Y (1999). Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta 1462, 55–70. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, and Alon U (2002). Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31, 64–68. [DOI] [PubMed] [Google Scholar]
- Shin D, and Groisman EA (2005). Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem 280, 4089–4094. [DOI] [PubMed] [Google Scholar]
- Shin D, Lee EJ, Huang H, and Groisman EA (2006). A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314, 1607–1609. [DOI] [PubMed] [Google Scholar]
- Shprung T, Peleg A, Rosenfeld Y, Trieu-Cuot P, and Shai Y (2012). Effect of PhoP-PhoQ activation by broad repertoire of antimicrobial peptides on bacterial resistance. J Biol Chem 287, 4544–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RJ, and Peters TJ (1990). Forms of soluble iron in mouse stomach and duodenal lumen: significance for mucosal uptake. Br J Nutr 63, 79–89. [DOI] [PubMed] [Google Scholar]
- Siraki AG, Smythies J, and O’Brien PJ (2000). Superoxide radical scavenging and attenuation of hypoxia-reoxygenation injury by neurotransmitter ferric complexes in isolated rat hepatocytes. Neurosci Lett 296, 37–40. [DOI] [PubMed] [Google Scholar]
- Soncini FC, and Groisman EA (1996). Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol 178, 6796–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead C, Tran A, Ferguson D Jr., McGrath S, Cotter R, and Trent S (2005). A novel 3-deoxy-D-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J Bacteriol 187, 3374–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, and Dancis A (1996). A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557. [DOI] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, and Reeves PR (1996). Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol 178, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, and Goudreau PN (2000). Two-component signal transduction. Annu Rev Biochem 69, 183–215. [DOI] [PubMed] [Google Scholar]
- Storm DR, Rosenthal KS, and Swanson PE (1977). Polymyxin and related peptide antibiotics. Annu Rev Biochem 46, 723–763. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, and Akira S (2003). Toll-like receptors. Annu Rev Immunol 21, 335–376. [DOI] [PubMed] [Google Scholar]
- Takeda S, Fujisawa Y, Matsubara M, Aiba H, and Mizuno T (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC --> YojN --> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol 40, 440–450. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, and Gunn JS (2005a). Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J Bacteriol 187, 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Prouty AM, and Gunn JS (2005b). Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA-regulated genes. FEMS Immunol Med Microbiol 43, 249–258. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Ryan SS, McCoy AJ, and Gunn JS (2002). Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect Immun 70, 6770–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C, and Roxby R (1972). Interpretation of protein titration curves. Application to lysozyme. Biochemistry 11, 2192–2198. [DOI] [PubMed] [Google Scholar]
- Tatar LD, Marolda CL, Polischuk AN, van Leeuwen D, and Valvano MA (2007). An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology 153, 2518–2529. [DOI] [PubMed] [Google Scholar]
- Thomas R, and D’Ari R (1990). Biological Feedback (CRC, Boca Raton, FL: ). [Google Scholar]
- Toguchi A, Siano M, Burkart M, and Harshey RM (2000). Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J Bacteriol 182, 6308–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touze T, Blanot D, and Mengin-Lecreulx D (2008a). Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J Biol Chem 283, 16573–16583. [DOI] [PubMed] [Google Scholar]
- Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, and Trent MS (2008b). Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol 67, 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Lester ME, Stead CM, Raetz CR, Maskell DJ, McGrath SC, Cotter RJ, and Trent MS (2005). Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J Biol Chem 280, 28186–28194. [DOI] [PubMed] [Google Scholar]
- Trent MS, Pabich W, Raetz CR, and Miller SI (2001a). A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem 276, 9083–9092. [DOI] [PubMed] [Google Scholar]
- Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, and Raetz CR (2001b). Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J Biol Chem 276, 43132–43144. [DOI] [PubMed] [Google Scholar]
- Trent MS, Ribeiro AA, Lin S, Cotter RJ, and Raetz CR (2001c). An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem 276, 43122–43131. [DOI] [PubMed] [Google Scholar]
- Trikha J, Theil EC, and Allewell NM (1995). High resolution crystal structures of amphibian red-cell L ferritin: potential roles for structural plasticity and solvation in function. J Mol Biol 248, 949–967. [DOI] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, and Bassler BL (2010). Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M (1981). Increased outer membrane resistance to ethylenediaminetetraacetate and cations in novel lipid A mutants. J Bacteriol 148, 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M (1992). Agents that increase the permeability of the outer membrane. Microbiol Rev 56, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Vaara T, Jensen M, Helander I, Nurminen M, Rietschel ET, and Makela PH (1981). Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett 129, 145–149. [DOI] [PubMed] [Google Scholar]
- Vaara M, Vaara T, and Sarvas M (1979). Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J Bacteriol 139, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, and Falkow S (1997). Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Viau C, Le Sage V, Ting DK, Gross J, and Le Moual H (2011). Absence of PmrAB-mediated phosphoethanolamine modifications of Citrobacter rodentium lipopolysaccharide affects outer membrane integrity. J Bacteriol 193, 2168–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelsang TM, and Boe J (1948). Temporary and chronic carriers of Salmonella typhi and Salmonella paratyphi B. J Hyg (Lond) 46, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A, My L, Dumoulin R, Sturgis JN, and Bouveret E (2011). Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol Microbiol 80, 1260–1275. [DOI] [PubMed] [Google Scholar]
- Williams CL, and Cotter PA (2007). Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J Bacteriol 189, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Breazeale SD, Raetz CR, and Naismith JH (2005). Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. J Biol Chem 280, 23000–23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, and Groisman EA (2003). Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69, 3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, and Groisman EA (2004). Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A 101, 17162–17167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, Latifi T, and Groisman EA (2005). Transcriptional regulation of the 4-amino-4-deoxy-L-arabinose biosynthetic genes in Yersinia pestis. J Biol Chem 280, 14765–14772. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ (2005). The acetate switch. Microbiol Mol Biol Rev 69, 12–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM, and Groisman EA (1999). Molecular characterization of the PmrA regulon. J Biol Chem 274, 27185–27190. [DOI] [PubMed] [Google Scholar]
- Wosten MM, Kox LF, Chamnongpol S, Soncini FC, and Groisman EA (2000). A signal transduction system that responds to extracellular iron. Cell 103, 113–125. [DOI] [PubMed] [Google Scholar]
- Yahav D, Farbman L, Leibovici L, and Paul M (2012). Colistin: new lessons on an old antibiotic. Clin Microbiol Infect 18, 18–29. [DOI] [PubMed] [Google Scholar]
- Yan A, Guan Z, and Raetz CR (2007). An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem 282, 36077–36089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo WS, Zwir I, Huang HV, Shin D, Kato A, and Groisman EA (2012). Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol Cell 45, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, and Regev A (2011). Impulse control: temporal dynamics in gene transcription. Cell 144, 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jansen R, Gaastra W, Arkesteijn G, van der Zeijst BA, and van Putten JP (2002). Identification of genes affecting Salmonella enterica serovar enteritidis infection of chicken macrophages. Infect Immun 70, 5319–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, and Raetz CR (2001). Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J Biol Chem 276, 43111–43121. [DOI] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, and Segal G (2007). The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63, 1508–1523. [DOI] [PubMed] [Google Scholar]
- Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, and Groisman EA (2005). Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A 102, 2862–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]


