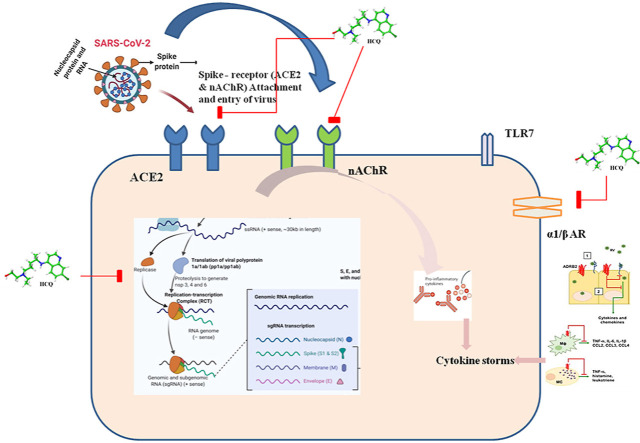
Abstract
COVID-19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has posed a global health emergency. Repurposing of existing drugs can be a rapid and effective strategy to fight the infection. Clinical trials have reported reduction or elimination of viral load when patients were treated with the anti-malarial drug Hydroxychloroquine (HCQ). To understand the molecular mechanism of action for effective repurposing of this drug we have carried out in silico docking and dynamics studies on complexes between HCQ and target proteins, which were identified through both literature survey and structural similarity searches in databases of small molecule – protein complexes. The proteins identified as binding HCQ are: Angiotensin Converting Enzyme 2 (ACE2), α7 nicotinic AcetylCholine Receptor (α7 nAChR), α1D-adrenergic receptor (α1D-AR), Histamine N- Methyl Transferase (HNMT) and DNA gyrase/Topoisomerase III β (Top3β). The majority of these proteins are novel and have not been used before, in docking studies. Our docking and simulation results support action of HCQ both at the entry and post-entry stages of SARS-CoV2 infection. The mechanism of action at the entry stage is through blocking the virus-binding sites on the two receptors, ACE2 & α7 nAChR, by binding directly at those sites. Our computational studies also show that the action of HCQ at the post-entry stage is to prevent both viral replication and generation of ‘cytokine storm’ by inhibiting host Top3β enzyme and α1D-AR, respectively. Binding of HCQ to HNMT is not a desired binding, and therefore this should be reduced during repurposing of HCQ.
Keywords: Coronavirus, SARS-COV-2, Hydroxychloroquine, ACE2, Nicotinic acetylcholine receptor, Spike, Adrenergic receptor, Cytokine storm, Topoisomerase III beta, Replication
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) has spread relatively quickly resulting in a world-wide pandemic [1]. This new SARS-COV-2 can infect the lower respiratory tract and cause pneumonia in humans. The most common symptoms reported include headache, fever, diarrhea, haemoptysis, runny nose, phlegm-producing cough, and lymphopenia affecting heart, kidney, liver, gastrointestinal system and the central nervous system [1]. One important adverse effect is the inflammatory response of ‘cytokine storm’ that can lead to multiple organ disorders [2].
The entry of the virus into the host cell by endocytosis is mediated by the viral spike protein (S), which is cleaved by the host protease furin into S1 and S2 subunits. The receptor binding domain (RBD) in the S1 subunit mediates viral attachment to host receptor, Angiotensin converting enzyme 2 (ACE2) expressed on the surfaces of a variety of cells (e.g. renal, cardiac, pulmonary and gastro-intestinal system) [3]. The S2 subunit mediates the post-fusion steps in the endocytosis [4]. After the virus entry and an incubation period of 5.2 days, the symptoms of SARS-COV-2 infection begin to appear [1].
The clinical management of COVID-19 is presently through symptomatic therapy as no specific drugs are yet available to cure the disease. The anti-malarial drug Hydroxychloroquine (HCQ), which has been shown to be effective against Human Immunodeficiency virus (HIV), SARS-COV-1, influenza virus, hepatitis C virus etc. [5], is found to have a significant success rate in the control of SARS-COV-2 infection also [6]. The benefits of treatment with HCQ are in the reduction of respiratory symptoms, pulmonary inflammation and nasopharyngeal clearance. In combination with azithromycin, HCQ was shown to reduce viral load and mortality [7]. Some other studies, however, showed HCQ to be ineffective for COVID 19 patients [8], thus leading to a controversy about the efficacy of HCQ [8,9]. A recent survey of reports on HCQ treatment of covid-19 patients reveals the efficacy of HCQ to be dependent on the stage of the disease when the drug is administered, being more effective at the early disease stage [6,10,11]. Experimental studies suggest that the anti-viral and anti-inflammatory properties of HCQ are based on multiple mechanisms [[11], [12], [13]].
HCQ interferes with the glycosylation of cellular receptors ACE2 and sialic acid receptor required for efficient entry of the virus into host cell. By increasing the endosomal pH, HCQ interferes with the fusion of viral and cellular endosomal membranes. HCQ also adversely affects virion assembly and budding, and reduces cytokine storm [5,6,13]. However, these mechanisms of action are not understood at the molecular level.
Therefore, in our study reported here, we have investigated the molecular mechanisms of HCQ by finding target molecules and then studying the interactions of targets with the drug using in-silico techniques of molecular docking and molecular dynamics simulation. The target-finding is based on molecular similarities only, without there being any bias toward molecules of relevance to SARS-COV-2. One of the targets found was nicotinic acetylcholine receptor, which was recently reported to be involved in the pathophysiology of SARS-COV-2 [14,15]. Our results show that HCQ binds directly to α7 nicotinic acetylcholine (α7 nAChR) and ACE2 receptors [3] in a way that would interfere with the binding of the viral spike protein to these receptors. Our calculations also show that HCQ binds directly to α1D-adrenergic receptor (α1D-AR), which may affect the anti-inflammatory response, such as the cytokine storm [2,5]. Our results also show that HCQ can interfere with virus replication by inhibiting the activity of the topoisomerase III beta (Top3β) that is required for SARS-COV-2 replication. Our results thus show the potential of HCQ to affect the entry and replication of SARS-CoV-2, and the anti-inflammatory response from the host. By giving atomic-level structural details of the drug-target binding, our results also provide template for chemical modifications that would make modified HCQ more specific to COVID-19.
2. Materials and Methods
2.1. Drug target prediction
Several software tools are available to predict proteins that can bind a given drug molecule. Some of these tools used in the present study on HCQ are: SwissTargetPrediction (http://www.swisstargetprediction.ch/), [16], Targetmine, Promiscous [17], Similarity Ensemble Approach (SEA) [18], and SuperTarget [19]. Information from the drug databases DrugBank, ChEMBL [20] was also used to compile target molecules. DrugBank (https://www.drugbank.ca/), an online free web server was used for determining chemical, biological, and structural details of the drugs [21]. The SMILE (Simplified Molecular Input Line Entry System) representation of drugs obtained from the DrugBank (DrugBank ID for HCQ is DB01611) is given as input to target-finding software.
2.2. Protein structure modelling
The amino acid sequence of the protein whose three-dimensional structures is to be modeled was retrieved from UniProt database [22] and used to query against Protein Data Bank (PDB) in BLASTp (Basic Logic Alignment Sequence Tool) [23]. From the BLASTp output the structure with higher sequence identity was selected as template for homology modeling, which was carried out using the tool – Robetta homology modeler [24]. The stereochemical quality of the models was evaluated using a variety of tools: calculation of target-template (structure used for homology modelling) RMSD using PyMOL [25], Ramachandran plot analysis via VADAR (Volume, Area, Dihedral Angle Reporter) [26], ProSA (Protein Structure Analysis) [27], ERRAT and Verify3D [28]. The best model was subjected to Molecular Dynamics simulation using GROMACS [29] and the quality of the output energy minimized structure was further confirmed using the same tools as above.
2.3. Docking - protein preparation
Three dimensional structures of the following target proteins were retrieved from RCSB Protein Data Bank (rcsb.org): native ACE2 (PDB ID: 1R42), α7 nAChR (3SQ9), DNA gyrase (4KFG), HNMT (2AOT), TOP3β (5GVC). The protein models were prepared for docking by using the protein preparation wizard (Schrödinger Suite 2019–2 Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2019; Impact, Schrödinger, LLC, New York, NY, 2019; Prime, Schrödinger, LLC, New York, NY, 2019) [30], and the procedure involved addition of the hydrogen atoms covalently bound to appropriate atoms, optimization of hydrogen bonds, removal of atomic clashes, assignment of partial charges to the hetero groups and then energy-optimization at neutral pH. The same procedure was followed for all the proteins selected as targets of HCQ, in the present study.
2.4. Binding site analysis and grid generation
Receptor grid was generated using Receptor grid generation option in the Glide application (Glide, Schrödinger, LLC, and New York-2) of Maestro (Schrödinger, LLC, New York, NY, 2019–2). The receptor grid for the protein targets was generated so as to enclose the binding (active) site residues that were identified by the SiteMap tool (SiteMap, Schrödinger, LLC, New York, NY, 2019) [31]. The drug binding sites were ranked based on the site score and BSA (binding surface area). Cubic grid boxes of length 20 Å were generated at each site and molecular docking at each of these sites was carried out. In the case of target proteins HNMT and DNA gyrase, the cubic grid box was positioned with its centre coinciding with the centroid of the ligand molecule in the crystal structure of the ligand-protein complex. Partial atomic charge was 0.25 units.
2.5. Ligand library preparation
Information about the ligand molecule, 4-Hydroxychloroquine was downloaded in the Spatial Data File (.SDF) file format from the PubChem Compound Database (PubChem CID: 3652) (National Center for Biotechnology Information; https://pubchem.ncbi.nlm.nih.gov/). The 3D structure prepared with the help of Marwin sketch (Marwin 15.12.17, ChemAxon (https://www.chemaxon.com) tool was then put through LigPrep (LigPrep, Schrödinger, LLC, New York, NY, 2019) before docking. The preparation of molecules via LigPrep involves preserving the definite chiralities to generate minimum of five low-energy stereoisomers per ligand, using default conditions at pH 7.0 ± 2.0. LigPrep corrects the protonation, and ionization states of the compounds, and assigns proper bond orders. Afterwards, the tautomeric and ionization states were created for each ligand.
2.6. Molecular docking
The docking of ligand 4-Hydroxychloquine to target proteins was done using two software tools Glide program of Schrodinger [32] and AutoDock Vina [33]. The ligand library output file and the receptor grid file were used as input files. The ligand was treated as flexible while the receptor was treated as rigid object in the docking process. The ligands were docked using XP –Extra precision mode in Glide docking and the final selection of ligand poses was based on the docking score, Glide score and interaction studies. Best poses were independently verified by using the second software tool AutoDock Vina. The protein and ligand preprocessings via H-bond optimization, charge addition were done before the docking, followed by grid box generation similar to site in Glide grid box. Lamarckian Genetic Algorithm (GA) in combination of grid based energy evaluation method was used for docking [33]. Other parameters were set at default values and the final result obtained was analyzed manually using PyMOL [25] and LigPlot+ [34].
2.7. Molecular dynamics simulation
The molecular dynamics (MD) simulation was performed for protein and protein-ligand complexes using GROMACS 2018.1 [29]. The ligand parameters were analyzed using ATB online server [35] in the framework of the GROMOS force-field 43a1. The ligand-protein complex was placed at the centre of a cubic box of dimensions 10*10*10 Å, which was then solvated by SPC/E water molecules. The complex was charge-neutralized by addition of counter ions, and energy minimized by the steepest gradient method to remove steric clashes. The energy - minimized system was further analyzed with NVT and then NPT ensemble for 100 ps (ps) simulations at 300 K temperature. The final production MD runs were performed for 50 ns (2fs per step) with periodic boundary conditions keeping the temperature of the system at 300 K. Each trajectory produced was analyzed using 2D plots of energy, root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg) and the number of hydrogen bonds (HB) calculated using option xmgrace in the software package. The stability of the protein-HCQ complexes is reflected in the binding free energies calculated by the Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method [36].
2.8. Protein-protein docking
The Protein-Protein dockings (mainly of receptor-binding domain of the viral spike protein with receptor proteins) were done using the tools - HADDOCK and ClusPro. The three-dimensional structures of necessary proteins and the identity of predicted interface residues were used as input in the docking calculations. The docked structures were ranked on the basis of interaction-energy values (ClusPro) and HADDOCK score (HADDOCK), which gives the cumulative score of various interaction energies [37,38]. All the docked complexes were analyzed also using PDBsum and PDBePISA servers for the exploration of macromolecular interface [39,40].
2.9. Other tools used
Molecular graphics software PyMOL and COOT were utilized for the visualization and superposition of molecular structures [25,41]. Protein BLAST (https://www.ncbi.nlm.nih.gov/) and Clustal O (clustal omega) were used for pairwise sequence alignment studies.
3. Results and discussion
3.1. HCQ targets
The protein targets identified by different software tools as binding to HCQ are given in Table 1 .
Table 1.
Predictions by different software of target proteins to which HCQ can bind with high affinity.(The software tool is given in bold and the proteins identified by that tool are listed immediately below).
| SWISS TARGET PREDICTION | TARGETMINE |
|---|---|
| Muscarine acetylcholine receptor M2 | Toll like receptor 7 |
|
Alpha-1D-Adrenergic receptor |
Toll like receptor 9 |
| SEA SERVER |
Cytochrome P450 |
| Muscarine acetylcholine receptor M2 | DRUGBANK/PUBCHEM |
| Alpha-1D-Adrenergic receptor | DNA |
| Histamine N methyltransferase | Toll like receptor7 |
| DNA gyrase subunit B | Toll like receptor 9 |
|
C–C Chemokine receptor type 4 |
Cytochrome P450 |
| PROMISCOUS |
Angiotensin converting enzyme 2 |
| Caspase3 | CHEMBL |
| Caspase 8 | Coagulation factor X |
|
Interleukin-6 |
Carbonic anhydrase |
| SUPERTARGET |
Neuronal acetylcholine receptor alpha7 (Nicotinic acetylcholine receptor) |
| Caspase3 | Acetylcholinesterase |
| Caspase 8 | |
| Interleukin-6 |
Interestingly, the receptor ACE2 was not identified by any of the target finder software. In addition to those listed in Table 1, the following proteins were identified as possible binders of HCQ by searching through the literature: MAPK [42], ACE2, Spike protein, NSP3 protease, furin, TMPRSS2 [13], and polymerase protein [43]. All these proteins identified through literature survey and target finding software were used as targets in our molecular docking calculations.
3.2. Molecular docking studies
Grid generation and preparation of the target and ligand molecules as described in Methods preceded the molecular docking calculations. The results of docking HCQ on each of the identified protein targets are given in Table 2 a. Table 2b gives for comparison docking scores obtained when known inhibitors/substrates are docked into corresponding receptor proteins. Only those targets that gave a docking score of −8 kcal/mol or better (Table 2a), were pursued further, and these targets are: ACE2, α7 nAChR, α1D-AR, DNA gyrase and HNMT (Table 2a). Among the two types of acetylcholine receptors, nAChR was relevant to SARS-COV-2 infection and hence it was taken-up for our studies [14,15]. Many other targets that have given good docking scores with HCQ are found to be directly relevant to SARS-COV-2 infection [5,13]. For example, the docking score with ACE2, which is a known viral receptor, is the third highest docking score in Table 2a. Binding of HCQ to ACE2 has the potential to affect viral spike protein binding in two ways: 1) by affecting the glycosylation of ACE2 and 2) by blocking the spike protein binding site on ACE2 [44]. Interestingly, an equally high docking score is obtained for the target α7 nAChR, which has only recently been shown to be involved in the pathophysiology of SARS-COV-2 [14,15]. The high score for the adrenergic receptor, which is part of the cholinergic pathway, is significant because recently α1D-AR antagonists are found to reduce mortality in COVID-19 patients, and HCQ binding can affect catecholamine signaling pathways during inflammatory cytokine production that is linked to many deaths [45]. Another novel target with a high docking score is the topoisomerase protein homologous to DNA gyrase, which is shown to be involved in SARS-COV-2 replication [46]. The possible inhibition of topoisomerase activity may be one of the mechanisms of post-entry action of HCQ observed in in vitro studies [6].
Table 2.
Docking scores for molecular docking of (a) HCQ (b) HCQ/inhibitors/substrates to different protein targets using Glide docking software (controls for comparison in docking).
| Rank | PROTEIN TARGETS | DOCKING SCORE (kcal/mol) |
|---|---|---|
| 1 | Histamine N methyltransferase | −15.08 |
| 2 | α 7 Nicotinic acetylcholine receptor | −11.9 |
| 3 | Angiotensin converting enzyme2 | −10.059 |
| 4 | Muscarine Acetylcholine receptor | −9.804 |
| 5 | DNA gyrase | −9.33 |
| 6 | Beta 2 -Adrenergic receptor | −9.16 |
| 7 | MAPK P38 | −7.564 |
| 8 | Cytochrome P450 D26 | −7.369 |
| 9 | Chemokine receptor 4 | −7.08 |
| 10 | Caspase 8 | −7.033 |
| 11 | NSP3 protease | −7 |
| 12 | Toll like receptor 7 | −6.366 |
| 13 | Toll like receptor 9 | −5.772 |
| 14 | Nsp12 RdRp (RNA dependent RNA polymerase) | −5.6 |
| 15 | Furin | −5.558 |
| 16 | Caspase3 | −2.617 |
| (a) | ||
| PROTEIN TARGETS |
LIGAND |
DOCKING SCORE (kcal/mol) |
| α 7 Nicotinic acetylcholine receptor | HCQ | −11.9 |
| Nicotine | −5.172 | |
| Acetylcholine | −7.668 | |
| Lobeline | −7.80 | |
| Angiotensin converting enzyme2 | HCQ | −10.059 |
| Quinapril | −7.5 | |
| α1D- Adrenergic receptor | HCQ | −8.8 |
| Prazosin | −9.1 | |
| DNA Topoisomerase IIIβ | HCQ | −9.23 |
| Doxorubicin | −9.4 | |
| (b) | ||
A detailed characterization of the environment around the HCQ molecule bound to each of these targets will enable efficient repurposing of HCQ for treating COVID-19 patients. Earlier studies have sought to repurpose drugs targeted towards host molecules ACE2, quinone reductase and viral molecules nsp12 RNA polymerase, nsp3 protease, nucleocapsid, spike, capping machinery nsp14/nsp16 etc. [[47], [48], [49], [50]] and these are all different, except ACE2, from the molecules we have found as targets for docking in the present study. Further, the target proteins identified in this study are shown experimentally to play critical roles in the SARS-COV-2 infection process [15,44,45,51] and therefore, are appropriate targets for inhibition by HCQ [44,52,53].
3.2.1. Angiotensin Converting Enzyme 2 (ACE2) - HCQ
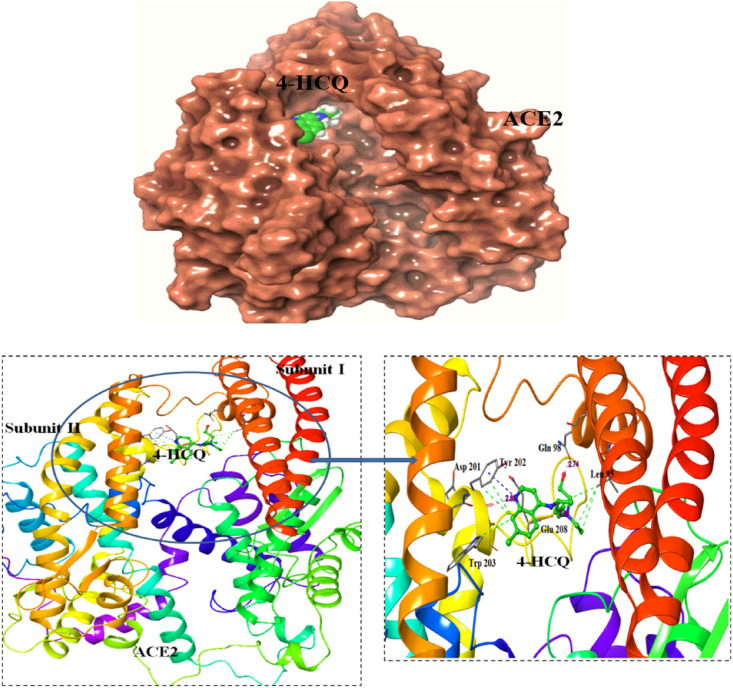
Docking of HCQ to ACE2 was directed at each site identified by the sitemap analysis. In the best pose with a docking score of −10.059 kcal/mol (Table 2a), HCQ is located close to the active site cavity, as shown in Fig. 1 . This binding site for HCQ is different from the allosteric site binding to ACE2 reported in an earlier docking study [47,48]. The difference could be because of use of different software or due to the use of ACE2-spike crystal complex for HCQ docking in their studies. Additionally, the site of chloroquine binding to ACE2 found in recent calculations [50] is also found in our calculations on HCQ, but with a lower ranking.
Fig. 1.
Docking of HCQ to ACE2 : HCQ binds at the interface of the two domains of ACE2. Surface view of the protein with the ligand docked poses (top). The interacting residues are labelled and the hydrogen bonding interactions are shown (bottom fig).
The binding of HCQ observed here is stronger than that of Quinapril, a known ACE2 inhibitor, for which the docking score, under identical parameter values, was −7.5 kcal/mol. The HCQ interaction at this site was further confirmed with docking studies via AutoDock Vina [33] and the docking score obtained was −6.2 kcal/mol (Table 4 ). The HCQ-binding site is near the active site located at the interface of subdomains I and II of ACE2, with HCQ forming hydrogen bonds to residues from subdomain I (Gln 98) and II (Tyr 202, Glu 208) (Fig. 1), as given in Table 3. Binding of HCQ in the cleft may interfere with the hinge movement of the two subdomains required for catalytic activity of ACE2 [54]. The amino acid residues Gln 98, Tyr 196, Gln 102 and Glu 208 are predicted, by both Glide and Vina, to be involved in binding HCQ (Table 4). The effect of HCQ binding to ACE2, at the newly discovered site, on the interaction between ACE2 and viral spike protein is investigated, and the results are reported below.
Table 4.
HCQ docking studies with the protein targets using AutoDock Vina: score & interacting residues details.
| PROTEIN TARGETS | AFFINITY (kcal/mol) | HYDROGEN BONDS | HYDROPHOBIC INTERACTING RESIDUES |
|---|---|---|---|
| ACE2 | −6.2 | Glu 208 | Gln 102, Tyr 196, Gln 98, Leu 95, Pro 565, Asp 206, Ala 396 |
| α7 nAChR | −6.2 | Ser 77, Asp 87 | Trp 145, Ile 80, Val 85, Tyr 116 |
| α1D-AR | −6.4 | Asp 80, Cys 150 | Lys 289, Phe 288, Asp 80, Trp 76, Ala 77, Phe 292, Ser 283 |
| TOP 3β | −6.1 | Asp 185, Phe 214 | Arg 181, Gly 189, Gln 182, Ala 512, Gln 218, Arg 524, His 517 |
Table 3.
HCQ interactions with different proteins in best docked pose.
| PROTEIN TARGETS | HYDROGEN BONDS | HYDROPHOBIC INTERACTION | SALT BRIDGE |
|---|---|---|---|
| ACE2 | Gln 98 (2.74), Tyr 202 (2.85), Glu 208 (2.69) | Leu 95, Trp 566, Gln 98,Tyr 196, Trp 203, Asp 201 | Tyr 202 |
| α 7 nAChR | A:Trp 145 (2.75), B:Gln 55 (2.79), B: Asp 160 (3.08) | A:Tyr 191, B:Trp 53, B:Leu 116, A:Tyr 184, B: Leu 116, B:Lys 139 | A:Trp 145 |
| α1D-AR | Ile 152 (2.94), Glu 154 (3.43) | Trp 76, Met 60, Phe 292, Phe 288, Tyr 296 | Asp 80 |
| TOP 3β | Ala 384 (3.08), Asp 511 (2.93), Arg 181 (2.94) | Ala 384, Ala 512, Val 178 | Asp 386, Asp 119, Glu 121 |
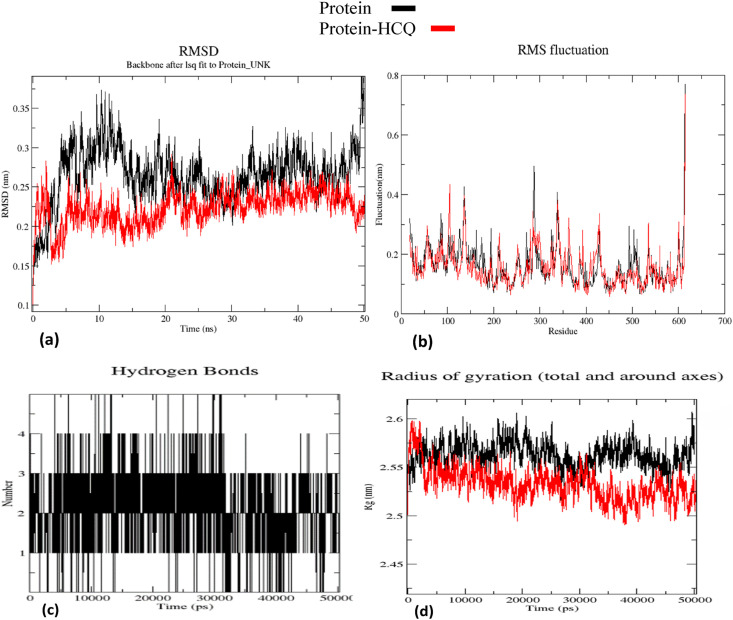
The stability of HCQ-ACE2 complex was investigated by running MD simulation for 50 ns using the software GROMACS [29], and the results are shown as plots of RMSD, RMSF, Rg and number of ligand-protein hydrogen bonds as a function of time (Fig. 2 ). It may be seen that the RMSD of the protein-HCQ complex changes significantly initially, before 10 ns, and has become stable after 12 ns, converging to an average value of 0.25 nm until 50ns of the run. In contrast, for the apo protein this plot shows continuous variation and shows no convergence even after 50 ns (Fig. 2a). The RMSF plot shows lesser fluctuations for the 596 amino acids of ACE2 with an average value of 0.1 nm, when HCQ is bound to the protein (Fig. 2b). The number of hydrogen bonds between HCQ and the protein hovers between three and five throughout the simulation showing stability of the hydrogen bonding interactions (Fig. 2c). The Rg plot shows that the HCQ-bound protein is more compact and stable without fluctuations unlike the unliganded protein (Fig. 2d). The MM-PBSA [36] binding energy calculations were performed using 2 ns MD trajectories, and the ACE2-HCQ complex shows a binding energy of −1090.881 +/- 57.615 kJ/mol. The binding energy, and the polar and apolar solvation energy plots are given as supplementary material (Supplementary material: Table 1 & Fig. 1a and b).
Fig. 2.
ACE2- HCQ complex MD simulation output: (a) RMSD plot (b) RMSF plot (c) Hydrogen bond plot (d) Radius of gyration plot.
The hydrogen bonds from HCQ with Gln 102, Tyr 196 and Glu 398 were stably maintained throughout the MD simulation of the ACE2 HCQ docked complex. The amino acid residues Gln 102 and Tyr 196 are displaced from their native positions by 1.5–6 Å in the crystal structure of ACE2-spike protein complex [55](Fig. 2). Interactions of these residues with HCQ may not allow such movements to enable interaction with the viral spike protein. Additional residues, mainly from S1 subdomain, have been displaced by as much as 3–5 Å in the HCQ-ACE2 docked complex when compared with native ACE2 [54]. Since these residues are also involved in the binding of spike protein to ACE2 [55], our results suggest that HCQ binding to ACE2 can adversely affect the affinity of spike protein toward ACE2. The Rg plot shows (Fig. 2d) that ACE2 becomes conformationally more stable and compact on binding HCQ.
3.2.2. ACE2 - spike & HCQ-ACE2 – spike complexes
The crystal structure of the spike-ACE2 complex is now available (PDB ID: 6lzgg), thus revealing amino acid residues at the binding interface [55]. With this input, ACE2-spike dockings were conducted via HADDOCK and ClusPro [37,38]. The best ACE2–spike complex pose in our docking studies was having HADDOCK and weighted ClusPro scores of −132.4 +/- 8.4 and −1432.5 respectively (Table 5 & Fig. 3 a). In order to examine if binding of HCQ to ACE2 inhibits spike binding, the coordinates of ACE2-HCQ complex after simulation were extracted for docking with the spike protein. For this docking the corresponding HADDOCK and ClusPro scores were −74.7 +/- 8.7 and −1372.5 respectively (Table 5). It is interesting that the HADDOCK score is substantially reduced suggesting that HCQ-binding to ACE2 interfered with spike-ACE2 binding.
Table 5.
Protein-protein docking studies of protein targets with spike protein.
| PROTEINS | HADDOCK |
ClusPro |
||
|---|---|---|---|---|
| HADDOCK score | Buried Surface Area | |||
| ACE2 | SPIKE | −132.4 +/- 8.4 | 2009.8 | −1432.5 |
| ACE2-HCQ | SPIKE | −74.7 +/- 8.7 | 1888.4 | −1377.4 |
| α7 nAChR | SPIKE | −87.2 +/- 2.3 | 2112.9 | −1432. |
| α7 nAChR - HCQ | SPIKE | −64.8 +/- 6.0 | 1998.7 | −759 |
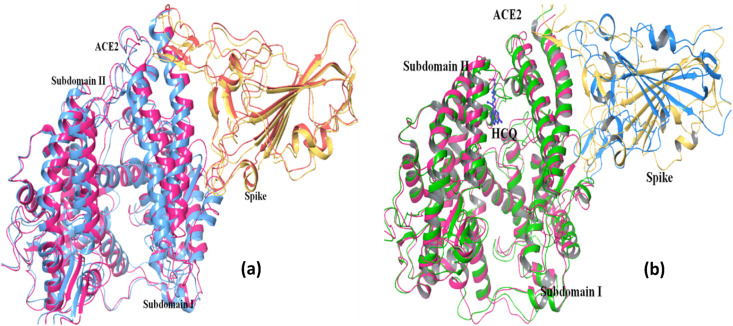
Fig. 3.
ACE2 ~ Spike docking studies: Superimposition of ace2 (magenta)- spike (yellow) docked complex with (a) crystal ACE2 (blue)- spike (orange) complex structure (6LZG) and b) with HCQ-ACE2 (green) ~ Spike (blue) docked complex. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The superposition of ACE2-spike docked complex on the recently determined crystal structure of ACE2-spike complex is shown in Fig. 3a. As may be seen, the structural agreement is excellent providing confidence in the correctness of the parameters used during protein-protein docking calculations. The same parameters were used during docking the spike protein to the HCQ-ACE2 complex, and this HCQ-ACE2-spike docked complex is shown structurally superposed on ACE2-spike docked complex in Fig. 3b. It may be seen that in the HCQ-bound ternary complex, the orientation of the spike protein relative to S1 subdomain of ACE2 is significantly different. Interestingly after HCQ binding there are some conformational changes in the helices and the loop regions of the ACE2 that interact with the spike protein, as discussed above. These conformational changes might be responsible for the loss of inter-protein interactions and the reduction in the HADDOCK score. The hydrogen bonding and salt-bridge interactions at the protein – protein interface was analyzed using the PDBePISA server [40]. Details of these chemical interactions are given in Table 6, Table 7 .
Table 6.
Interactions between ACE2 and Spike proteins: (a) hydrogen bonds, (b) salt bridges.
| ACE2 | DISTANCE (Ao) | SPIKE |
|---|---|---|
| Lys 353 [NZ] | 2.65 | Asp 320 [OD1] |
| Lys 68 [NZ] | 2.87 | Tyr 449 [OH] |
| Gln 24 [NE2] | 3.01 | Ala 475 [O] |
| Asp 30 [OD2] | 2.71 | Lys 417 [NZ] |
| Asp 38 [OD2] | 3.07 | Tyr 453 [OH] |
| Gln 24 [OE1] | 3.86 | Ser 477 [OG] |
| Tyr 83 [OH] | 2.92 | Asn 487 [ND2] |
| Glu 35 [OE2] | 2.99 | Gln 493 [NE2] |
| Tyr 41 [OH] | 2.68 | Gly 502 [N] |
| Asp 355 [OD1] | 2.67 | Val 503 [N] |
| Asp 38 [OD1] | 2.63 | Tyr 505 [OH] |
| Gln 42 [OE1] |
3.50 |
Tyr 449 [OH] |
| ACE2 |
DISTANCE (Ao) |
SPIKE |
| Lys 353 [NZ] | 2.65 | Asp 405 [OD1] |
| Lys 353 [NZ] | 3.48 | Asp 405 [OD2] |
| Asp 38 [OD1] | 3.64 | Asp 403 [NH2] |
| Asp 30 [OD1] | 3.32 | Lys 417 [NZ] |
| Asp 30 [OD2] | 2.71 | Lys 417 [NZ] |
Table 7.
Interactions between HCQ-bound ACE2 and Spike proteins: (a) hydrogen bonds, (b) salt bridges.
| ACE2-HCQ | DISTANCE (Ao) | SPIKE |
|---|---|---|
| Asn 322 [ND2] | 2.76 | Ser 477 [OG] |
| Gln 325 [N] | 3.16 | Ala 475 [O] |
| Lys 353 [NZ] | 2.91 | Tyr 453 [OH] |
| Asp 30 [OD1] | 2.76 | Gln 498 [NE2] |
| Asp 30 [OD2] | 2.62 | Tyr 499 [OH] |
| Glu 35 [OE2] | 2.88 | Gly 502 [N] |
| Glu 35 [OE2] | 2.91 | Asn 501 [ND2] |
| Asp 38 [OD1] | 2.69 | Arg 403 [NH1] |
| Asp 38 [OD2] | 2.71 | Arg 403 [NH2] |
| Lys 353 [O] | 2.9 | Tyr 453 [NZ] |
| His 34 [NE2] |
2.88 |
Tyr 505 [O] |
| ACE2-HCQ |
DISTANCE (Ao) |
SPIKE |
| Asp 38 [OD1] | 3.55 | Arg 403 [NH2] |
| Asp 38 [OD1] | 2.69 | Arg 403 [NH1] |
| Asp 38 [OD2] | 2.71 | Arg 403 [NH2] |
| Asp 38 [OD2] | 3.45 | Arg 403 [NH1] |
In the ACE2-spike docked complex there are 12 hydrogen bonding and 5 salt-bridge interactions between ACE2 and Spike proteins (Tables 6a & 6b). In the case of HCQ-bound ACE2-spike complex, there are 11 hydrogen bonding and four salt-bridge interactions (Tables 7a & 7b). Comparison of Table 6, Table 7a shows that there is a difference in the two residues hydrogen bonding at the inter-protein interface. For example, Lys 353 from ACE2 forms a hydrogen bond with Asp 320 from the spike protein in the ACE2-spike binary complex, whereas in the ternary HCQ-ACE2-spike complex, Lys 353 hydrogen bonds with Tyr 453 from the spike protein. These differences are due to subtle conformational changes induced by HCQ binding to ACE2. While the position of ACE2 is similar, the orientation of the spike protein in the complex with HCQ-ACE2 is significantly different from that in the complex with native ACE2 (Fig. 3b). This feature predicts that the binding of spike protein to sialic acid co-receptors [56] might be substantially affected by the presence of HCQ. This result implies that HCQ interferes with the binding of the virus to the cell receptor through its spike protein. This result is also consistent with the suggestions made based on in-vitro experimental studies [5,6,57].
3.2.3. Nicotinic acetylcholine receptor (nAChR) - HCQ
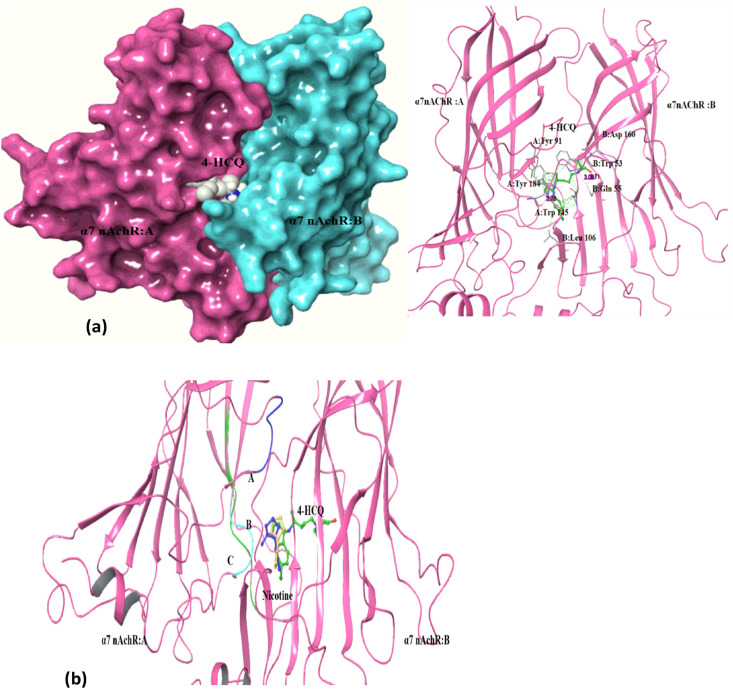
nAChRs form pentameric ligand-gated ion channels that mediate fast chemical neurotransmission at the neuromuscular junction. Ligands such as nicotine, acetylcholine, antagonist bungarotoxin and snake toxins bind to the orthosteric and allosteric sites in the extra-cellular or transmembrane domain of this receptor and trigger the activation/blocking of ion channels and anti-inflammatory signaling [[58], [59], [60]]. Based on the observation that the RBD domain of the viral spike protein (SARS-COV-2) contains amino acid sequence motif similar to the known nAChR antagonists, such as α-bungarotoxin, nAChR receptor is suggested as a secondary receptor for SARS-COV-2 entry. This hypothesis suggests not only nAChR mediated virus entry but also dysregulation of the nicotinic cholinergic system (NCS) leading to a cytokine storm along with failure of the immune response to return to homeostasis [14,15,58,61]. The high docking score of HCQ to α7 nAChR (Table 2a) at the toxin binding site (PDB: 3sq9) suggests that HCQ may impact nAChR-mediated entry and pathophysiology of SARS-CoV2 [59,60]. The docking score for the best pose for HCQ was −11.5 kcal/mol with a binding prime energy of −17292.2, and this site is at the interface region between two α7 subunits (Table 2a, Fig. 4 a and b). Docking with AutoDock Vina also yielded a high score of −6.2 kcal/mol at this binding site (Table 4). The ligands nicotine and acetylcholine were also docked separately at the same site and under identical parameter values, for purposes of comparison. The best docking score for nicotine, the agonist, is much lower with a value of −5.17 kcal/mol (Table 2b). HCQ interacts with residue Trp 145 in the B loop and with residues Tyr 184 and Tyr 191 in the C loop through hydrogen bonds and hydrophobic interactions (Table 3). AutoDock Vina also predicted involvement of residue Trp 145 in binding HCQ (Table 4).
Fig. 4.
Docking of α7 nAChR with HCQ: (a) Docked pose: α7 nAChR bound HCQ towards interface of two subunit at the orthosteric site (left – surface view, right – ribbon representation of protein); (b) Superimposed image view: nicotine (blue-docked ligand, yellow-crystal structure ligand) interacting with α7 nAChR protein in the same cleft as HCQ (within A, B, and C loop). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
HCQ-binding site in the docked complex is separated by 3.32 Å from the position of nicotine in the nicotine- α7 nAChR docked complex (Fig. 4b), and by 4.11 Å from nicotine in the crystal structure of nicotine-AChBP complex. This short separation suggests that HCQ-binding can prevent simultaneous nicotine binding. Absence of agonist nicotine binding may lead to dysfunctioning of the cholinergic anti-inflammatory pathways [59].
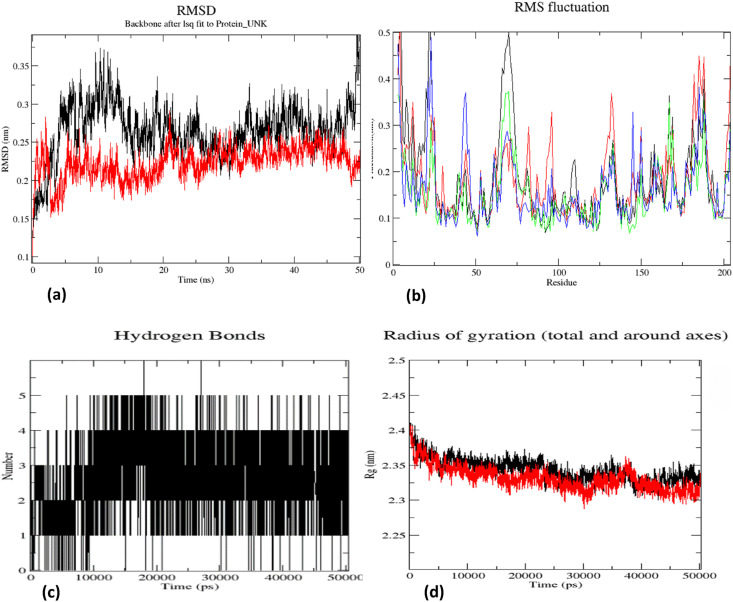
The stability of HCQ– α7 nAChR complex was examined in a 50 ns MD simulation using GROMACS [29]. Fig. 5 shows the results in the form of plots of variation of protein-RMSD, protein-RMSF and the number of ligand-protein hydrogen bonds as a function of time. As the HCQ and other ligands are targeted to substrate binding site, the focus of simulation analysis was towards the principal interacting regions (A, B and C loops) of α7 subunits [60,62]. The RMSD values for the protein-ligand complex were stable and lower (average value 0.25 nm) compared to those of unliganded protein at all time points (Fig. 5a). The RMSF plot of α7 nAChR shows an average difference of ~0.02 nm between homomeric chains A & B (red and green for unbound and HCQ-bound chain A; black and blue for unbound and HCQ-bound chain B in Fig. 5b) of the receptor. Further, the RMSF plot shows lesser fluctuation of 0.15 nm for protein-HCQ complex in the region involved in binding HCQ (A,B C loop regions) (Fig. 5b). This shows the contribution of each chain towards interactions with HCQ [62]. The protein-HCQ complex becomes more compact as may be seen from the lower Rg value, especially after converging beyond 40 ns of dynamics (Fig. 5d). The hydrogen bond plot shows that hydrogen bonds between the ligand and the protein are stable through-out the simulation period (Fig. 5c). HCQ interactions with Trp 145 from B-loop and Tyr 191 from C-loop are maintained during MD simulation with an additional bond with Tyr 91 of loop A. Binding of HCQ to loops A, B and C might block the binding site on nAChR that facilitates virus entry into the cell. Experimental validation is, however, needed of this prediction. The binding energy calculated using MMPBSA [36] shows the free energy value of −2921.473 +/- 57.100 kJ/mol. The binding energy along with the polar and apolar solvation energy graphs are given as supplementary material (Supplementary material: Table 1 & Fig. 2a and b).
Fig. 5.
α7 nAChR – HCQ complex MD simulation output : (a) RMSD plot (b) RMSF plot (red and green for unbound and HCQ-bound chain A; black and blue for unbound and HCQ-bound chain B) (c) Hydrogen bond plot (d) Radius of gyration plot (Protein Protein – HCQ). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.4. α7 nAChR – spike & HCQ- α7 nAChR - spike
The MD simulations carried out by A. Sofia F. Oliveira et al. support the interaction of the SARS-CoV-2 spike protein with nicotinic acetylcholine receptors, particularly α7 nAChRs which are present in human bronchial epithelial and endothelial cells [15,61]. Guided by the crystal structure of toxin-α7 nAChR complex (PDBID:4hqp), we have explored the interaction of spike protein with α7 nAChR, since a region in the RBD domain of the viral spike protein has amino acid sequence matching that of the toxin. As already stated, we have used for this purpose the software tools HADDOCK and ClusPro [37,38]. While the blind docking gave a HADDOCK score of −35, a better score of −87.2 +/- 2.3 was obtained after specifying toxin-interacting residues during docking (Table 4). Analysis of the interactions at the interface using PDBePISA shows that the spike protein binds at the same site as the toxins [40] through hydrogen bonding and salt-bridge interactions given in Table 8 .
Table 8.
Interactions from docking: Hydrogen bonds (a) and salt bridges (b) between α7 nAChR –Spike interactions.
| α7 nAChR | DISTANCE (Ao) | SPIKE |
|---|---|---|
| A:Arg 182 [NH1] | 3.22 | B:Tyr 380 [O] |
| A:Arg 182 [NH1] | 2.79 | B:Gly 381 [O] |
| A:Lys 139 [NZ] | 2.74 | B:Thr 385 [O] |
| A:Lys 139 [NZ] | 2.74 | B:Lys 386 [O] |
| A:Ser 124 [OG] | 2.74 | B:Asn 388 [O] |
| A:Trp 145 [NE1] | 3.82 | B:Thr 393 [OG1] |
| A:Arg 182 [NH2] | 2.70 | B:Thr 430 [OG1] |
| A:Trp 145 [NE1] | 3.02 | B:Ala 520 [O] |
| A:Ser 124 [OG] | 3.60 | B:Gly 526 [O] |
| A:Tyr 91 [O] | 3.02 | B:Cys 391 [N] |
| A:Asp 126 [OD1] | 2.64 | B:Lys 386 [NZ] |
| A:Glu 185 [OE1] | 2.65 | B:Arg 355[NH1] |
| A:Tyr 191 [OH] |
3.33 |
B:His 519 [NE2] |
| α7 nAChR |
DISTANCE (Ao) |
SPIKE |
| A:Asp 182 [NH1] | 2.64 | B:Lys 386 [NZ] |
| A:Glu 185[OE1] | 3.60 | B:Arg 35[NH2] |
| A:Glu 185[OE1] | 2.65 | B:Arg 35[NH1] |
| A:Glu 185[OE2] | 3.36 | B:Arg 35[NH1] |
Similarly, the stable HCQ- α7 nAChR complex structure obtained after 50 ns MD-simulation was used for docking with the viral spike protein under conditions identical to those used for docking the spike protein with α7 nAChR alone. The HADDOCK score obtained was −64.8 +/- 6, which is worse than the score of −87.2 +/- 2.3 obtained for α7 nAChR-spike docked complex (Table 5). The actual residues involved in inter-protein hydrogen bonding are shown in Table 9 . It may be seen that some of the residues at the interaction interface in the binary complex are changed in the ternary complex, HCQ-α7 nAChR-spike. HCQ interaction with the A, B and C loops can block/reduce the nAChR interface residues from interacting with the SARS-COV-2 spike protein for virus entry. Contribution of residues Trp 145 of B loop and Tyr 191 of C loop towards spike binding has been lost in the ternary complex (after HCQ inhibition). Hydrogen bonding and salt bridge interactions from residue Glu 185 with α7 nAChR were also lost after HCQ interaction at substrate binding site. These results are similar to earlier findings that the α7 nAChRs - SARS-CoV-2 S1 interaction is significantly disturbed by the binding of AchR-agonists nicotine, and acetylcholine [14].
Table 9.
Interactions between HCQ-bound α7 nAChR and Spike protein from docking via PDBePISA.
| α7 nAChR - HCQ | DISTANCE (Ao) | SPIKE |
|---|---|---|
| A:Gln 46 [NE2] | 2.80 | B:Asp 360 [OD1] |
| A:Thr 195 [OG1] | 2.70 | B:Asp 389 [O] |
| A:Lys 139 [NZ] | 2.78 | B:Leu 390 [O] |
| A:Arg 182 [NH2] | 2.81 | B:Thr 430 [OG1] |
| A:Ala 93 [N] | 3.81 | B:Pro 521 [O] |
| A:Ser 124 [OG] | 2.76 | B:Pro 521 [O] |
| A:Lys 139 [NZ] | 2.86 | B:Cys 525 [O] |
| A:Asn 92 [ND2] | 3.63 | B:Cys 525 [SG] |
| A:Lys 139 [NZ] | 2.75 | B:Gly 526 [O] |
| A:Asp 126 [O] | 3.17 | B:Thr 333 [N] |
| A:Ser 128 [O] | 2.81 | B:Thr 333 [OG1] |
| A:Phe 183 [O] | 3.55 | B:Val 382 [N] |
| A:Asp 126 [OD2] | 2.70 | B:Gly 526 [N] |
3.2.5. α1D- adrenergic receptors - HCQ
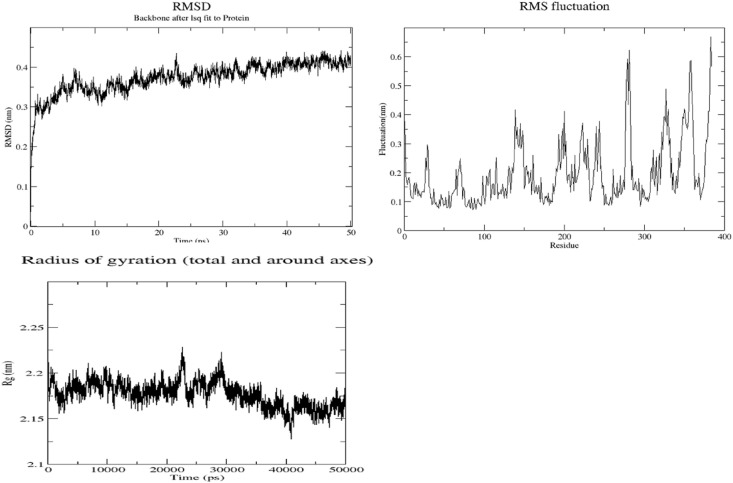
Treatment of COVID-19 patients with α1-adrenergic receptor antagonist reduces both the risk of adverse outcome in the lower respiratory track, and the cytokine storm [45]. As may be seen from Table 1, α1D-AR is also predicted as a high-affinity target for HCQ binding. Since crystal structure of this receptor was not available in the PDB, its 3D structure was modeled using Robetta homology-modelling tool [24]. The human beta-2-adrenergic receptor (β2-AR) with a sequence similarity of 33.9% over a coverage of 91% (36–280 amino acids) served as the best template (PDBID:6mxt) (Supplementary Material Fig. 6) [63] for the modelling. The molecular models were validated using software Verify3D, ERRAT and ProSa, and also through analysis of Ramachandran plot (VADAR) (Table 10 ) [[26], [27], [28]]. It may be seen that the model built by Robetta is of very good quality with a quality factor of 98.14%, while a generally accepted range is >50% for a high-quality model [22] (Protein model deposited in the Data base PMDB under the Id: PM0083525) [64]. This modeled structure was further subjected to 50 ns MD simulation using GROMACS. During this simulation, the RMSD and Rg values converged after 40 ns showing stability of the structure (Fig. 6 ). The molecular model obtained by averaging the structures during the MD trajectory was validated using the tools mentioned above. This MD-simulated model was assigned a quality factor of 95.75% by ERRAT, and further details are given in Table 10, and also as supplementary material (Fig. 3, Fig. 4, Fig. 5). This molecular model was used to study interactions with HCQ.
Table 10.
Validation of α 1D – adrenergic receptor models.
| PROTEIN MODELS |
RAMACHANDRAN PLOT |
VERIFY 3D |
ERRAT (Quality factor) |
PROSA |
RMSD |
|
|---|---|---|---|---|---|---|
| Favoured region | Outlier | |||||
| Robetta model | 94.5% | 1.05% | 82.21% of residue have average 3D-1D score > = 0.2 (Pass) | 98.138% | −4.04 | 0.509 |
| Robetta model after simulation | 84.23% | 3.66% | 84.23% of residue have average 3D-1D score > = 0.2 (Pass) | 95.75% | −4.95 | 2.113 |
Fig. 6.
α1D-Adrenergic receptor simulation output plots: RMSD, RMSF & Rg plots.
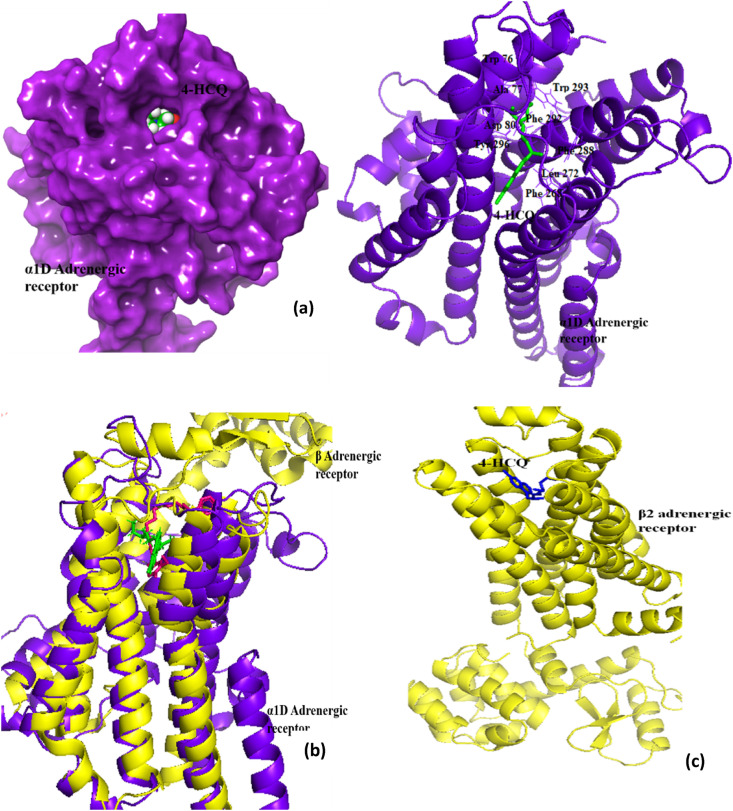
Docking scores of HCQ binding to α1D-AR and β2-AR are: −8.8 kcal/mol and −9.16 kcal/mol (Table 2b), respectively. These values are comparable to the score of −9.1 kcal/mol for prazosin, which is a known α1D-AR inhibitor. The docking studies using AutoDock Vina also gave a good docking score of −6.4 kcal/mol, thus confirming HCQ interaction at this site (Table 4). Fig. 7 a shows that many amino acid residues from the receptor (Asp 80, Trp 76, Ile 152, Phe 288, Phe 292) are predicted as interacting residues, by both docking software tools. In the case of α1D-AR, HCQ targets the membrane spanning domain (TM1-7 helix) which contains both the agonist binding site and the G protein interacting region (Fig. 7a). This site is very close to AR antagonist binding site for β2-AR(~3.04Ao between HCQ and β AR antagonist), which has a structure similar to that of α1D-AR (Fig. 7b & c) [63]. When docked explicitly to β2-AR, the HCQ binding almost overlaps the antagonist binding site. By interfering with the substrate binding site, HCQ may be blocking the signaling pathway that leads to adverse conditions such as hyper inflammation and imbalance in pulmonary regulation [45]. This may be a mechanism of action of HCQ toward anti inflammation, in addition to its other known modes of action to block the cytokine release [12,13].
Fig. 7.
α 1D-AR and HCQ docking studies : (a) 3D interaction showing HCQ-bound to the membrane spanning domain of the receptor protein at the orthosteric site (left side– surface view, right side – ribbon representation of protein); (b) Superimposition of α1D-AR and β2-AR (template with 38% similarity), ligand in stick representation, green: HCQ and pink: AR antagonist (c) 3D interaction showing HCQ bound to the membrane spanning domain of the β2-AR protein at the substrate binding site. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.6. DNA gyrase - HCQ
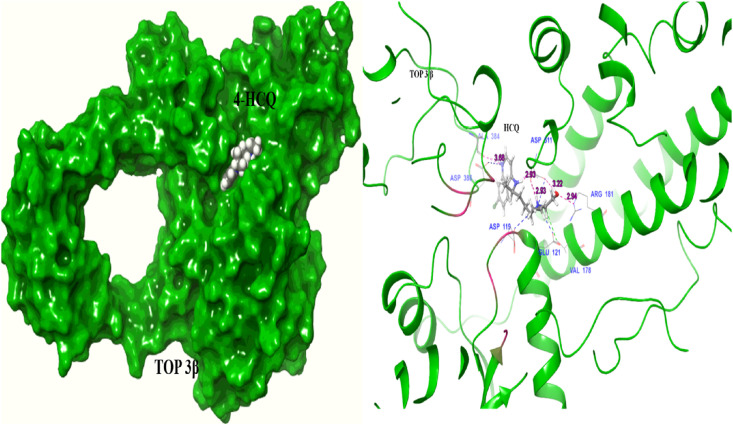
As may be seen from Table 1, DNA gyrase, a bacterial type IIA topoisomerase is identified as a potential binder of HCQ, and the docking score of −9.23 kcal/mol is quite high (Table 2a). HCQ, therefore, can act as an inhibitor of prokaryotic DNA gyrase as suggested in previous studies [53]. The human protein topoisomerase III-beta (TOP3β), which has both DNA and RNA topoisomerase activities, is required for efficient replication of positive-sense RNA viruses, including SARS-COV-2. It is shown that TOP3β directly acts on the viral genome to facilitate viral replication [46]. In vitro studies show that the topoisomerase inhibition therapy provides protection from SARS-COV-2 induced inflammation [51]. Thus, TOP3β may be an important target for developing antiviral drugs to treat COVID-19. Therefore, we explored binding of HCQ to TOP3β using two docking software tools (Glide and AutoDock Vina), and both gave good docking scores of −9.2 kcal/mol and −6.1 kcal/mol respectively, for the same site (Table 2b, Table 4). The docking score is comparable to the value of −9.4 kcal/mol obtained for doxorubicin, a known topoisomerase inhibitor. The interactions between HCQ and TOP3β are shown in Fig. 8 . It is perhaps functionally significant that the interactions involve amino acid residues Asp 119, Asp 386 and Asp 384 (Table 3) which are located near the catalytic pocket, and which also undergo conformational changes during DNA/RNA binding [65]. These findings suggest that HCQ can interfere with virus replication by inhibiting the essential topoisomerase activity.
Fig. 8.
TOP3β interaction with HCQ towards the site near to catalytic pocket (left – surface view, right – ribbon representation of protein) (Asp119, Asp 386) and important DNA/RNA binding regulatory regions (pink color marked region: catalytic residues) (Hydrogen bond –pink color line, salt bridge-blue, hydrophobic interaction –green). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.7. Histamine N-Methyltransferase (HNMT) - HCQ
HNMT is an important protein that inactivates histamine by methylation, and regulates the airway response to histamine [66]. Recent studies on SARS-COV-2 show histamine as an important inflammatory mediator initiating abnormal immune response leading to cytokine storm and multi organ failure [67]. Further, respiratory virus infection characterized by broncho-constriction response, breathing difficulty etc. is found to be associated with a decreased expression of HNMT. An increase in the expression of HNMT can reduce the infectious conditions [68]. These observations suggest that HNMT protein can help in regulating histamine mediated abnormal immune response and pulmonary infection in SARS-COV-2. Thus, inhibition of HNMT by HCQ suggested by our docking study (Table 2a) is an undesired effect, and HCQ should be modified to eliminate its binding to HNMT.
4. Conclusion
We have computationally identified proteins that can bind, with high affinity, the antimalarial drug HCQ, and then, from among these, have focused on docking HCQ to those targets relevant to the treatment of COVID-19. Many targets we have identified are novel and have not been studied before in the context of COVID-19. For the receptor protein ACE2, the binding site we have obtained is different from earlier finding. By directly binding to ACE2 receptor, the drug HCQ causes movement of key residues that are involved in the binding of the viral spike protein to the ACE2 receptor. The affinity of spike protein toward ACE2 receptor, we suggest, would be diminished because of this HCQ-induced movement. This effect is therefore likely to decrease the entry of the virus into target cells, thereby reducing the infection. Our in-silico studies also show that HCQ binds at the same site as nicotine to α7 nAChR, which is thought to be the second entry point for the SARS-CoV2 virus. This binding also, therefore, has the effect of blocking viral entry into the cell. This binding may also affect nAChR-mediated pathophysiology of the virus. Apart from interfering with the entry, our studies suggest that HCQ may be interfering in virus replication by inhibiting TOP3β activity. Our studies also indicate that HCQ, through direct binding to the α1D-AR, will reduce cytokine storm and other inflammatory conditions by interfering with catecholamine–cytokines pathways. The chemical environment of the binding sites of HCQ on each of these target proteins identified here would enable repurposing of HCQ for more effective treatment of COVID-19. Apart from increasing affinity toward desired targets the affinity towards HNMT needs to be eradicated by proper chemical modification of HCQ.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to Director, National Institute of Advanced Studies, Bangalore, India for support. MVH is thankful to Indian National Science Academy, New Delhi, for the award of Senior Scientist Fellowship post-retirement. MVH is thankful to Open Source Pharma Foundation for a Junior Research Fellowship award to Ms. Navya Baburaj.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100714.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guo G., Ye L., Pan K., Chen Y., Xing D., Yan K., Chen Z., Ding N., Li W., Huang H., Zhang L., Li X., Xue X. New insights of emerging SARS-CoV-2: epidemiology, etiology, clinical features, clinical treatment, and prevention. Front. Cell Dev. Biol. 2020;8:1–22. doi: 10.3389/fcell.2020.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Yang C., feng Xu X., Xu W., wen Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arshad S., Kilgore P., Chaudhry Z.S., Jacobsen G., Wang D.D., Huitsing K., Brar I., Alangaden G.J., Ramesh M.S., McKinnon J.E., O'Neill W., Zervos M., Nauriyal V., Hamed A.A., Nadeem O., Swiderek J., Godfrey A., Jennings J., Gardner-Gray J., Ackerman A.M., Lezotte J., Ruhala J., Fadel R., Vahia A., Gudipati S., Parraga T., Shallal A., Maki G., Tariq Z., Suleyman G., Yared N., Herc E., Williams J., Lanfranco O.A., Bhargava P., Reyes K. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. Int J Infect Dis. 2020;97:396–403. doi: 10.1016/j.ijid.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fragkou P.C., Belhadi D., Peiffer-Smadja N., Moschopoulos C.D., Lescure F.X., Janocha H., Karofylakis E., Yazdanpanah Y., Mentré F., Skevaki C., Laouénan C., Tsiodras S. Review of trials currently testing treatment and prevention of COVID-19. Clin Microbiol Infect. 2020;26:988–998. doi: 10.1016/j.cmi.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh H., Chauhan P., Kakkar A.K. Hydroxychloroquine for the treatment and prophylaxis of COVID-19: the journey so far and the road ahead. Eur J Pharmacol. 2021;890:173717. doi: 10.1016/j.ejphar.2020.173717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prodromos C., Rumschlag T. Hydroxychloroquine is effective, and consistently so when provided early, for COVID-19: a systematic review. New Microbes New Infect. 2020;38 doi: 10.1016/j.nmni.2020.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 13.Quiros Roldan E., Biasiotto G., Magro P., Zanella I. The possible mechanisms of action of 4-aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): a role for iron homeostasis? Pharmacol Res. 2020;158 doi: 10.1016/j.phrs.2020.104904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandris N., Lagoumintzis G., Chasapis C.T., Leonidas D.D., Papadopoulos G.E., Tzartos S.J., Tsatsakis A., Eliopoulos E., Poulas K., Farsalinos K. Nicotinic cholinergic system and COVID-19: in silico evaluation of nicotinic acetylcholine receptor agonists as potential therapeutic interventions. Toxicol. Reports. 2021;8:73–83. doi: 10.1016/j.toxrep.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagoumintzis G., Chasapis C.T., Alexandris N., Kouretas D., Tzartos S., Eliopoulos E., Farsalinos K., Poulas K. Nicotinic cholinergic system and COVID-19: in silico identification of interactions between α7 nicotinic acetylcholine receptor and the cryptic epitopes of SARS-Co-V and SARS-CoV-2 Spike glycoproteins. Food Chem Toxicol. 2021;149:112009. doi: 10.1016/j.fct.2021.112009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357. doi: 10.1093/nar/gkz382. –W364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Eichborn J., Murgueitio M.S., Dunkel M., Koerner S., Bourne P.E., Preissner R. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keiser M.J., Roth B.L., Armbruster B.N., Ernsberger P., Irwin J.J., Shoichet B.K. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 19.Hecker N., Ahmed J., Von Eichborn J., Dunkel M., Macha K., Eckert A., Gilson M.K., Bourne P.E., Preissner R. SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res. 2012;40:D1113. doi: 10.1093/nar/gkr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., MacIejewski A., Gale N., Wilson A., Chin L., Cummings R., Le Di, Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium T.U. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190. doi: 10.1093/NAR/GKM895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gm B., Aa S., R A., Sf A., Dj L., Tl M. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7 doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano W. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 26.Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R.F., Sykes B.D., Wishart D.S. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/NAR/GKG565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407. doi: 10.1093/NAR/GKM290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 30.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 31.Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 32.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 33.Trott O., Olson A. Autodock vina: improving the speed and accuracy of docking. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334.AutoDock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/CI200227U. [DOI] [PubMed] [Google Scholar]
- 35.Malde A.K., Zuo L., Breeze M., Stroet M., Poger D., Nair P.C., Oostenbrink C., Mark A.E. An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theor Comput. 2011;7:4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- 36.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez C., Boelens R., Bonvin A.M.J.J., HADDOCK A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 38.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 40.G. Battle, PDBePISA : Identifying and interpreting the likely biological assemblies of a protein structure What is PDBePISA ? Where does the data come from ? PDBePISA for analysing the NGF structure 1bet Starting the PDBePISA service, (n.d.) 1–12.
- 41.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Kono M., Tatsumi K., Imai A.M., Saito K., Kuriyama T., Shirasawa H. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antivir Res. 2008;77:150–152. doi: 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen S.N., Yielding K.L. Inhibition of DNA and RNA polymerase reactions by chloroquine. Proc Natl Acad Sci USA. 1965;54:521–527. doi: 10.1073/pnas.54.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L., Chen H., Dong S., Huang W., Chen L., Wei Y., Shi L., Li J., Zhu F., Zhu Z., Wang Y., Lv X., Yu X., Li H., Wei W., Zhang K., Zhu L., Qu C., Hong J., Hu C., Dong J., Qi R., Lu D., Wang H., Peng S., Hao G. The effects of chloroquine and hydroxychloroquine on ACE2-related coronavirus pathology and the cardiovascular system: an evidence-based review. Function. 2020;1 doi: 10.1093/function/zqaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose L., Graham L., Koenecke A., Powell M., Xiong R., Shen Z., Mench B., Kinzler K.W., Bettegowda C., Vogelstein B., Athey S., Vogelstein J.T., Konig M.F., Wagner T.H. The association between alpha-1 adrenergic receptor antagonists and in-hospital mortality from COVID-19. Front Med. 2021;8:637647. doi: 10.3389/fmed.2021.637647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasanth K.R., Hirano M., Fagg W.S., McAnarney E.T., Shan C., Xie X., Hage A., Pietzsch C.A., Bukreyev A., Rajsbaum R., Shi P.Y., Bedford M.T., Bradrick S.S., Menachery V., Garcia-Blanco M.A. Topoisomerase III-ß is required for efficient replication of positive-sense RNA viruses. BioRxiv. 2020 doi: 10.1101/2020.03.24.005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skariyachan S., Gopal D., Chakrabarti S., Kempanna P., Uttarkar A., Muddebihalkar A.G., Niranjan V. Structural and molecular basis of the interaction mechanism of selected drugs towards multiple targets of SARS-CoV-2 by molecular docking and dynamic simulation studies- deciphering the scope of repurposed drugs. Comput Biol Med. 2020;126:104054. doi: 10.1016/j.compbiomed.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celı̇k I., Onay-Besı̇kcı̇ A., Ayhan-Kilcigı̇l G. Approach to the mechanism of action of hydroxychloroquine on SARS-CoV-2: a molecular docking study. J Biomol Struct Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1792993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentile D., Fuochi V., Rescifina A., Furneri P.M. New anti sars-cov-2 targets for quinoline derivatives chloroquine and hydroxychloroquine†. Int J Mol Sci. 2020;21:1–16. doi: 10.3390/ijms21165856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baildya N., Ghosh N.N., Chattopadhyay A.P. Inhibitory capacity of chloroquine against SARS-COV-2 by effective binding with angiotensin converting enzyme-2 receptor: an insight from molecular docking and MD-simulation studies. J Mol Struct. 2021;1230:129891. doi: 10.1016/j.molstruc.2021.129891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho J.S.Y., Mok B.W.-Y., Campisi L., Jordan T., Yildiz S., Parameswaran S., Wayman J.A., Gaudreault N.N., Meekins D.A., Indran S.V., Morozov I., Trujillo J.D., Fstkchyan Y.S., Rathnasinghe R., Zhu Z., Zheng S., Zhao N., White K., Ray-Jones H., Malysheva V., Thiecke M.J., Lau S.-Y., Liu H., Zhang A.J., Lee A.C.-Y., Liu W.-C., Jangra S., Escalera A., Aydillo T., Melo B.S., Guccione E., Sebra R., Shum E., Bakker J., Kaufman D.A., Moreira A.L., Carossino M., Balasuriya U.B.R., Byun M., Albrecht R.A., Schotsaert M., Garcia-Sastre A., Chanda S.K., Miraldi E.R., Jeyasekharan A.D., TenOever B.R., Spivakov M., Weirauch M.T., Heinz S., Chen H., Benner C., Richt J.A., Marazzi I. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell. 2021;184:2618–2632. doi: 10.1016/j.cell.2021.03.051. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballestero J.A., Plazas P.V., Kracun S., Gómez-Casati M.E., Taranda J., Rothlin C.V., Katz E., Millar N.S., Elgoyhen A.B. Effects of quinine, quinidine, and chloroquine on α9α10 nicotinic cholinergic receptors. Mol Pharmacol. 2005;68:822–829. doi: 10.1124/mol.105.014431. [DOI] [PubMed] [Google Scholar]
- 53.Mitton-Fry M.J., Brickner S.J., Hamel J.C., Brennan L., Casavant J.M., Chen M., Chen T., Ding X., Driscoll J., Hardink J., Hoang T., Hua E., Huband M.D., Maloney M., Marfat A., McCurdy S.P., McLeod D., Plotkin M., Reilly U., Robinson S., Schafer J., Shepard R.M., Smith J.F., Stone G.G., Subramanyam C., Yoon K., Yuan W., Zaniewski R.P., Zook C. Novel quinoline derivatives as inhibitors of bacterial DNA gyrase and topoisomerase IV. Bioorg Med Chem Lett. 2013;23:2955–2961. doi: 10.1016/j.bmcl.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 54.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matrosovich M., Herrler G., Klenk H.D. Sialic acid receptors of viruses. Top Curr Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:6–9. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinbach J.H. Mechanism of action of the nicotinic acetylcholine receptor. Ciba Found Symp. 1990;152:53–61. doi: 10.1002/9780470513965.ch4. discussion 61. [DOI] [PubMed] [Google Scholar]
- 59.Nirthanan S., Gwee M.C.E. Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty Years on. J Pharmacol Sci. 2004;94:1–17. doi: 10.1254/jphs.94.1. [DOI] [PubMed] [Google Scholar]
- 60.Delbart F., Brams M., Gruss F., Noppen S., Peigneur S., Boland S., Chaltin P., Brandao-Neto J., Von Delft F., Touw W.G., Joosten R.P., Liekens S., Tytgat J., Ulens C. An allosteric binding site of the α7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein. J Biol Chem. 2018;293:2534–2545. doi: 10.1074/jbc.M117.815316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveira A.S.F., Ibarra A.A., Bermudez I., Casalino L., Gaieb Z., Shoemark D., Gallagher T., Sessions R., Amaro R., Mulholland A. Simulations support the interaction of the SARS-CoV-2 spike protein with nicotinic acetylcholine receptors. BioRxiv Prepr. Serv. Biol. 2020:1–14. doi: 10.1101/2020.07.16.206680. [DOI] [Google Scholar]
- 62.Sine S.M., Engel A.G. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 63.Masureel M., Zou Y., Picard L.P., van der Westhuizen E., Mahoney J.P., Rodrigues J.P.G.L.M., Mildorf T.J., Dror R.O., Shaw D.E., Bouvier M., Pardon E., Steyaert J., Sunahara R.K., Weis W.I., Zhang C., Kobilka B.K. Structural insights into binding specificity, efficacy and bias of a β 2 AR partial agonist. Nat Chem Biol. 2018;14:1059–1066. doi: 10.1038/s41589-018-0145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castrignanò T., De Meo P.D.O., Cozzetto D., Talamo I.G., Tramontano A. The PMDB protein model database. Nucleic Acids Res. 2006;34:306–309. doi: 10.1093/nar/gkj105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goto-Ito S., Yamagata A., Takahashi T.S., Sato Y., Fukai S. Structural basis of the interaction between Topoisomerase IIIβ and the TDRD3 auxiliary factor. Sci Rep. 2017;7:1–10. doi: 10.1038/srep42123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., Thomae B., Eckloff B., Wieben E., Weinshilboum R. Human histamine N-methyltransferase pharmacogenetics: gene resequencing, promoter characterization, and functional studies of a common 5′-flanking region single nucleotide polymorphism (SNP) Biochem Pharmacol. 2002;64:699–710. doi: 10.1016/S0006-2952(02)01223-6. [DOI] [PubMed] [Google Scholar]
- 67.Eldanasory O.A., Eljaaly K., Memish Z.A., Al-Tawfiq J.A. Histamine release theory and roles of antihistamine in the treatment of cytokines storm of COVID-19, Travel Med. Inf Disp. 2020;37 doi: 10.1016/j.tmaid.2020.101874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakazawa H., Sekizawa K., Morikawa M., Yamauchi K., Satoh M., Maeyama K., Watanabe T., Sasaki H. Viral respiratory infection causes airway hyperresponsiveness and decreases histamine N-methyltransferase activity in Guinea pigs. Am J Respir Crit Care Med. 1994;149:1180–1185. doi: 10.1164/ajrccm.149.5.8173757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.