Abstract
Purpose
Molecular imaging has provided unparalleled opportunities to monitor disease processes, although tools for evaluating infection remain limited. Coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is mediated by lung injury that we sought to model. Activated macrophages/phagocytes have an important role in lung injury, which is responsible for subsequent respiratory failure and death. We performed pulmonary PET/CT with 124I-iodo-DPA-713, a low-molecular-weight pyrazolopyrimidine ligand selectively trapped by activated macrophages cells, to evaluate the local immune response in a hamster model of SARS-CoV-2 infection.
Procedures
Pulmonary 124I-iodo-DPA-713 PET/CT was performed in SARS-CoV-2-infected golden Syrian hamsters. CT images were quantified using a custom-built lung segmentation tool. Studies with DPA-713-IRDye680LT and a fluorescent analog of DPA-713 as well as histopathology and flow cytometry were performed on post-mortem tissues.
Results
Infected hamsters were imaged at the peak of inflammatory lung disease (7 days post-infection). Quantitative CT analysis was successful for all scans and demonstrated worse pulmonary disease in male versus female animals (P < 0.01). Increased 124I-iodo-DPA-713 PET activity co-localized with the pneumonic lesions. Additionally, higher pulmonary 124I-iodo-DPA-713 PET activity was noted in male versus female hamsters (P = 0.02). DPA-713-IRDye680LT also localized to the pneumonic lesions. Flow cytometry demonstrated a higher percentage of myeloid and CD11b + cells (macrophages, phagocytes) in male versus female lung tissues (P = 0.02).
Conclusion
124I-Iodo-DPA-713 accumulates within pneumonic lesions in a hamster model of SARS-CoV-2 infection. As a novel molecular imaging tool, 124I-Iodo-DPA-713 PET could serve as a noninvasive, clinically translatable approach to monitor SARS-CoV-2-associated pulmonary inflammation and expedite the development of novel therapeutics for COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11307-021-01638-5.
Key words: SARS-CoV-2, COVID-19, PET/CT, Macrophage, Immune response, Molecular imaging, Sex difference
Introduction
Compared with conventional technologies currently utilized to investigate the pathogenesis of infectious diseases, imaging can evaluate disease processes deep within the body noninvasively and relatively rapidly. Tomographic imaging can provide whole-body, three-dimensional assessments, enabling a complete view of the disease process, which is less affected by sampling bias (e.g., with biopsy or resected tissues). Molecular imaging can also provide detailed spatiotemporal visualization of molecular events noninvasively and longitudinally in the same individual [1]. While continued advances in molecular imaging have provided unparalleled opportunities to monitor disease progression in oncology and neurology, their application for the evaluation of infections remains limited.
Coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has affected over 191 million people worldwide and led to over 4 million deaths. Severe COVID-19 is worse for males than females [2] and can lead to dysregulated inflammation through the activation of pro-inflammatory pathways and recruitment of immune cells leading to lung disease [3]. While the incidence of COVID-19 is similar between the sexes, adult males are almost 3 times more likely to be admitted into ICUs and twice as likely to die as females [4, 5]. Therefore, understanding the inflammatory and immunological responses to SARS-CoV-2 infection is fundamental and could assist in the development of novel therapeutic interventions. The golden Syrian hamster (Mesocricetus auratus) inoculated with SARS-CoV-2 develops pulmonary disease, including bilateral and peripherally distributed regions of lung consolidation which can be assessed with computed tomography (CT) [6, 7] with infiltration by myeloid cells (neutrophils, monocytes, and macrophages) at infection sites [8]. Adult male golden Syrian hamsters also develop worse morbidity and pulmonary disease than females [9]. Here, we evaluated 124I-iodo-DPA-713 PET/CT as a noninvasive molecular imaging tool to detect pulmonary inflammation in SARS-CoV-2-infected hamsters. Radioiodinated DPA-713, which targets the translocator protein (TSPO), a mitochondrial protein upregulated in activated macrophages/phagocytic cells, has been utilized to image pulmonary infections and inflammatory diseases [10]. Recently, 124I-iodo-DPA-713 PET has also been translated to the clinic [11].
Materials and Methods
Ethics
All animal protocols were approved by the Johns Hopkins University Biosafety, Radiation Safety, and Animal Care and Use Committees.
Viral Propagation
VeroE6-TMPRSS2 cells (National Institute of Infectious Diseases, Japan) were cultured at 37 °C with 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, and 100 units/ml of penicillin–streptomycin. The SARS-CoV-2/USA-WA1/2020, NR-52281 isolate was obtained from BEI Resources (National Institutes of Allergy and Infectious Diseases). Infectious stocks were grown by infecting VeroE6-TMPRSS2 cells and collecting supernatant upon observation of cytopathic effect; debris were removed by centrifugation and passage through a 0.22-μm filter (Sigma-Aldrich). The supernatant was then aliquoted and stored at − 80 °C. Infectious virus titers were determined by tenfold serially diluting and plating the supernatant onto 96-well plates with monolayers of Vero E6-TMPRSS2 cells in replicates of six for 5 days at 37 °C. Incubation was followed by fixation with 4% formaldehyde, staining with naphthol blue-black solution, and scoring of cytopathic effects.
Animal Studies
Golden Syrian hamsters (male and female, 6–8 weeks old; Envigo, Indianapolis, IN) with surgically implanted central venous catheters were utilized. The animals were housed individually with ad libitum access to food, water, and cage enrichment. After 1 week of acclimatization in the animal biosafety level-3 (ABSL-3) facility, the animals were anesthetized with ketamine and xylazine for intranasal infection with 1.5 × 105 50% tissue culture infectious dose (TCID50) of SARS-CoV-2, delivered in 100 µL of DMEM.
Imaging
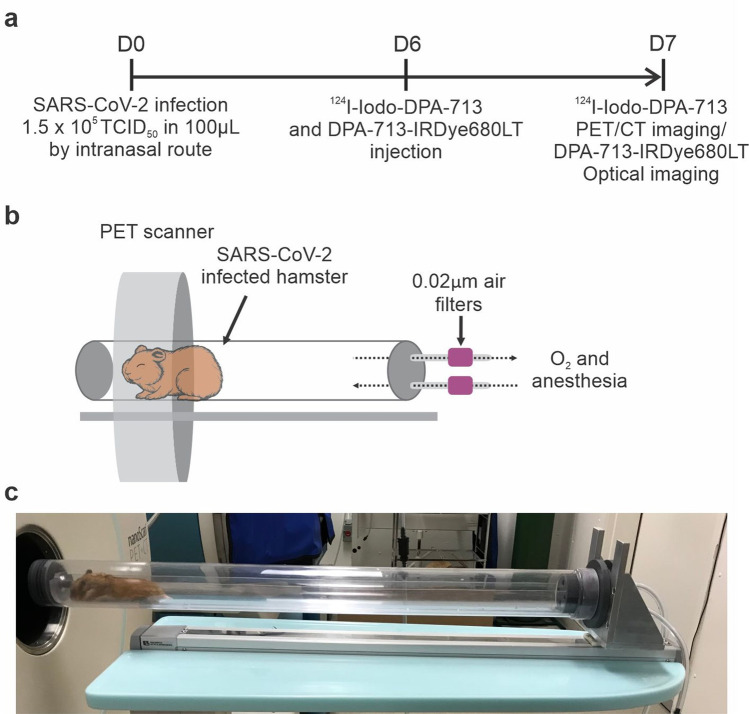
124I-iodo-DPA-713 was synthesized in-house as described previously yielding > 90% radiochemical purity [12]. SARS-CoV-2-infected hamsters (n = 3 male, n = 4 female) underwent PET/CT at 7 days post-infection, and 24 h after intravenous administration of 124I-iodo-DPA-713 (10.22 ± 0.14 MBq/hamster) via the surgically implanted central venous catheter (Fig. 1a). A 20-min PET acquisition and subsequent CT were performed using the nanoScan PET/CT (Mediso, Arlington, VA). An additional set of animals (n = 15 male, n = 11 female) underwent CT scans only [9, 13]. Given that SARS-CoV-2 is designated as a BSL-3 pathogen, live SARS-CoV-2-infected animals were imaged inside transparent and sealed biocontainment cells developed in-house, compliant with BSL-3 containment and capable of delivering air-anesthetic mixture to sustain live animals during imaging [13–15] (Fig. 1b, c). DPA-713-IRDye680LT, a fluorescent analog of DPA-713, was intravenously administered 24 h prior euthanasia. Lungs were collected, formalin-fixed, and imaged with the Peral® Impulse Imager (LI-COR, Lincoln, NE).
Fig. 1.
Experimental scheme. a Golden Syrian hamsters were intranasally challenged with SARS-CoV-2 and imaging performed as outlined. b In-house developed, transparent, and sealed biocontainment cells with 0.02-µm filters attached to the inlet and outlet for O2 and anesthesia and compliant with BSL-3 containment. c SARS-CoV-2-infected hamster inside a biocontainment bed being imaged at Ci3R imaging facility (Johns Hopkins University).
Lung Segmentation Tool and Image Analysis
A multi-atlas lung segmentation (MALS) algorithm was used to create the whole lung volumes of interest (VOI) [16, 17]. A reference library was generated from a selection of study images that included SARS-CoV-2-infected hamsters in various stages of lung disease. A bounding box for the lung VOI was generated using a combination of rigid and affine transformations followed by a high-dimensional deformable registration technique inside this bounding box to efficiently refine the linear mapping accuracy [18]. The propagated labels were merged using a weighted voting-based label fusion technique [19]. A local search algorithm was also used to improve robustness against registration errors [20]. Pulmonary lesions were defined using a global Hounsfield units (HU) threshold ≥ 0 [21]. The data are represented as CT score [(pulmonary lesions volume/whole lung volume) × 100]. The investigators analyzing the CT were blinded to the group assignments.
VivoQuant™ 2020 (Invicro, Boston, MA, USA) was used for visualization and quantification. Scatter and attenuation corrections were applied to the PET data and multiple VOIs were manually drawn per animal using the CT as a reference.
Histopathology and Immunofluorescence
Formalin-fixed and paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E) or anti-Iba-1 antibody (Wako; 019–19,741 Richmond, VA; at a dilution of 1:2000). Morphometric analyses were performed on affected lung tissues using Image J software (NIH, USA) [22]. A minimum of three fields of view were obtained from each animal (n = 6 animals; 3 male and 3 female hamsters). Heat-induced epitope retrieval was conducted by heating slides to 95 °C for 20 min in sodium citrate–based ER1 buffer (Leica Biosystems, Richmond, IL) before immunostaining. Immunostaining was performed using the Bond RX automated system (Leica Biosystems, Richmond, IL). Positive immunostaining was visualized using diaminobenzidine tetrahydrochloride (DAB) and slides were counterstained with hematoxylin.
Flow Cytometry
Lungs were harvested and homogenized, the cells filtered and washed with PBS followed by RBC lysis [23]. Cell viability was determined with trypan blue dye staining. A single-cell suspension was stained with Zombie Aqua Fixable Viability Kit (Biolegend), resuspended in FACS buffer (1% bovine serum albumin in saline), and incubated in block buffer prior to surface staining with anti-CD11b antibody (Novus Biologicals, #NB110-89474PECY7, polyclonal) for macrophages, monocytes, and granulocytes. Cells were fixed using intracellular fixation buffer and data acquired and analyzed using FACSDiva software and FlowJo (v10). The gating strategy is reported in Figure S1. Myeloid and lymphoid cells were identified based on their scatter characteristics.
Virus Titration and Anti-spike Receptor-Binding Domain IgG ELISA
All procedures handling live virus were performed in a biosafety level-3 (BSL-3) facility. The TCID50 of the virus in the supernatant and lung tissues was calculated using the Reed-Muench method [9]. Hamster antibody titers from plasma were performed using ELISA protocols adapted from human COVID-19 antibody assays described previously [9, 24]. Briefly, ELISA plates (96-well plates, Immunol 4HBX, Thermo Fisher Scientific) were coated with anti-spike receptor-binding domain (S-RBD). An HRP-conjugated secondary IgG (Abcam, MA, USA) was used with titer cutoff of three times the absorbance of first dilution of mock (uninfected) animal samples.
Statistical Analysis
The data are represented on a linear scale as median and interquartile range (IQ). Statistical comparisons were performed using a two-tailed Mann–Whitney U test. P values of < 0.05 were considered statistically significant. All statistical analyses were performed using Prism 8 (GraphPad Software, San Diego, CA, USA).
Results
Lung Pathology in SARS-CoV-2
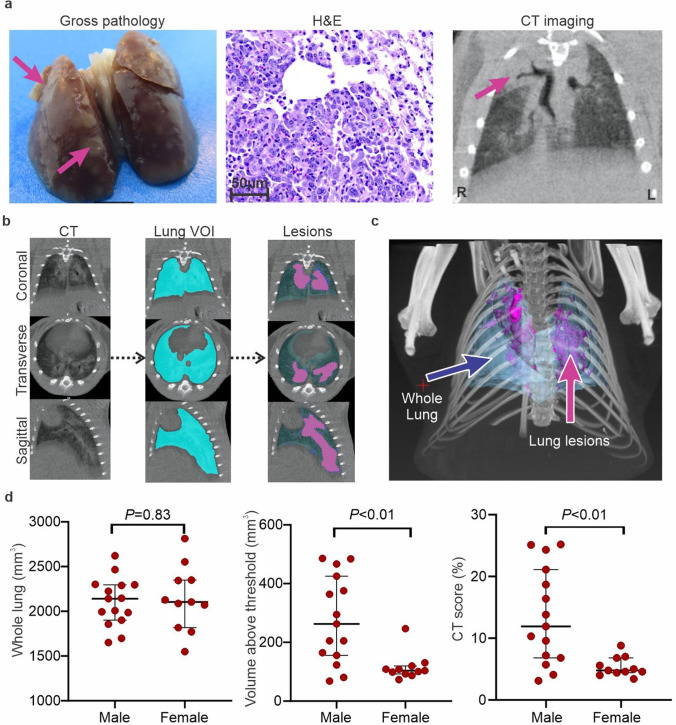
Seven days post-infection, pulmonary CT demonstrated extensive peripheral, bilateral, and multi-lobular ground-glass opacities (GGO), and mixed GGO with consolidation (Figure S2). Lungs were examined after necropsy showing focal areas of spotted brown coloration distributed around all lobules. Histopathology findings were consistent with previous reports [7, 25], showing multiple areas of consolidation characterized by abundant infiltration of inflammatory cells of predominantly macrophages, intermixed with neutrophils and lymphocytes, as well as type II pneumocyte hyperplasia and multinucleated cells (Fig. 2a). To reduce bias in the visual assessment of lung disease, a custom-built lung segmentation tool was utilized to quantify pathology of the whole lung on chest CT (Fig. 2b and Movie S1). Our segmentation tool allowed a semi-automated quantification of the lung volume and determined regions of increased tissue density (HU > 0). Quantitative CT analysis was performed for all scans with an average total lung volume of 2.1 ± 0.3 mL. While the whole lung volume was similar between sexes (P = 0.83), the CT score (disease severity) was higher in males compared to females (P < 0.01) (Fig. 2d). Additionally, no difference were noted in the CT score (disease severity) in males compared to females in uninfected hamsters (Figure S3).
Fig. 2.
Lung pathology in SARS-CoV-2-infected hamsters. a, Gross pathology, histopathology (H&E, 40 × , scales bars of 50 µm), and CT findings at 7 days post-infection. Ground glass opacities (GGO) and consolidations were detected by CT scan (pink arrows indicates affected area). b Automated lung segmentation of CT images from a SARS-CoV-2-infected hamster is shown. c Maximum intensity projection of SARS-CoV-2-infected hamster demonstrating the lung (blue) and pneumonic areas (pink). d Whole lung, lung volume above threshold, and CT score for male (n = 15) and female (n = 11) hamsters. While no differences are noted in the whole lung volumes, the lung disease (CT score) was substantially worse in male compared to female animals. Data represented as median ± interquartile range.
124I-Iodo-DPA-713 PET/CT
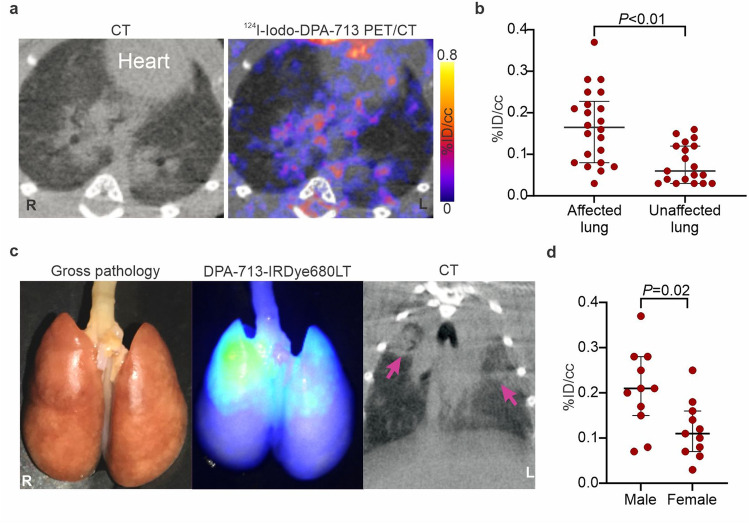
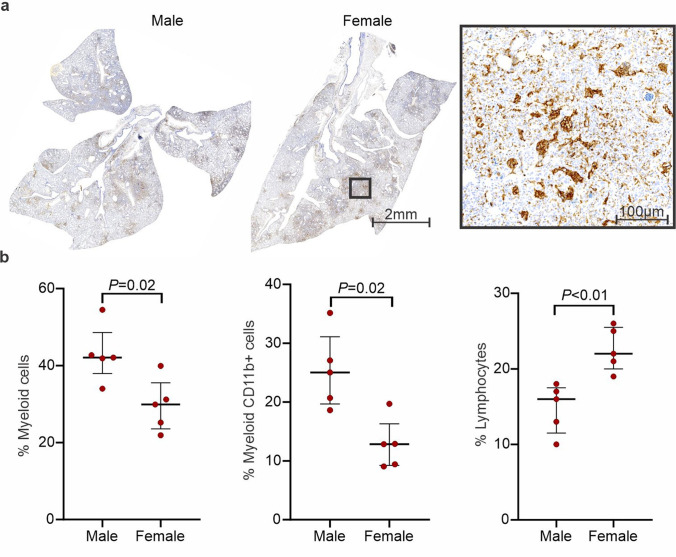
Whole-body 124I-iodo-DPA-713 PET/CT of a representative SARS-CoV-2 hamster is shown (Figure S4a-b). As previously reported in other mammalian species [11, 20, 26], 124I-iodo-DPA-713 followed urinary and hepatobiliary excretion, with the signal localizing to the liver, gastrointestinal tract, and kidneys. Specific uptake in macrophage-rich brown fat and metabolized iodine in the thyroid was also noted, with minimal PET activity in the brain parenchyma. In the lungs, 124I-iodo-DPA-713 PET signal co-localized with the pneumonic lesions noted on the CT (Fig. 3a) and quantification of the signal demonstrated that 124I-iodo-DPA-713 uptake was higher in the pneumonic regions, compared to the unaffected areas of the lung (P < 0.01) (Fig. 3b). Additionally, post-mortem analysis demonstrated that DPA-713-IRDye680LT fluorescence also co-localized with the pneumonic regions in the lung (Fig. 3c). 124I-iodo-DPA-713 PET activity was significantly higher in males compared to females (P < 0.05) (Fig. 3d). However, no differences were noted in the 124I-iodo-DPA-713 PET activity in males compared to females in unaffected areas of the lung (Figure S4c). Post-mortem analysis was consistent with imaging findings demonstrating worse lung disease in male versus female hamsters (Figure S5a). Immunostaining demonstrated a rich cluster of Iba-1 + macrophages within the consolidated areas in the lung parenchyma in both sexes at the peak of lung disease (7 days post-infection) (Fig. 4a). To characterize the immune response, multicolor flow-cytometric analyses were performed. The percentage of lung myeloid and specifically CD11b + cells (macrophages, phagocytes) was higher in male versus female animals (P < 0.05) (Fig. 4b). Conversely, the percentage of pulmonary lymphocytes was lower in males versus females (P < 0.01).
Fig. 3.
Pulmonary 124I-iodo-DPA-713 PET/CT of SARS-CoV-2-infected hamsters. a Transverse lung sections of a representative hamster imaged with 124I-iodo-DPA-713 showing higher uptake within the GGO and pneumonic areas. b 124I-iodo-DPA-713 PET activity is higher in pneumonic regions compared to unaffected lung. c DPA-713-IRDye680LT fluorescence co-localizes with the diseased lung. d 124I-iodo-DPA-713 PET signal is higher in male versus female hamsters. A VOI was created for each lung lesion (GGO and consolidations) in males (n = 11), females (n = 11) and unnaffected lung (n = 19). Data represented as median ± interquartile range.
Fig. 4.
Iba-1 staining and flow cytometry. a Immunohistochemistry demonstrates clusters of Iba-positive cells within pneumonic tissues. b Flow cytometry demonstrates a higher percentage of myeloid cells and CD11b + cells (macrophages, phagocytes) in males (n = 5), than females (n = 5). Data represented as median ± interquartile range.
To explore other biological differences during SARS-CoV-2 infection, antibody responses in plasma and viral titers in lungs were evaluated at the time of imaging (7 days post-infection). Anti-S-RBD IgG titers in plasma were noted between male and female hamsters and viral titers in the lungs were undetectable (Figure S5b-c). Both findings are consistent with previous reports at this time point after infection [9].
Discussion
There has been increasing interest in developing molecular imaging probes to assist in the diagnosis and management of infections [1, 27]. In COVID-19, findings from human autopsies, plasma, and bronchoalveolar lavage samples have demonstrated dysregulation of the myeloid compartment, peripheral lymphopenia, and neutrophil activation [3, 8, 28, 29]. SARS-CoV-2-infected golden Syrian hamsters develop mild-to-moderate clinical manifestations and recreate radiological and pathological features similar to those noted in human disease [9], with areas of consolidation and ground-glass opacities (GGO), and infiltration of neutrophil, monocytes, and macrophages in the lung [8]. Emerging literature also suggests that both the numbers and pro-inflammatory responses of lung monocytes and macrophages increase with disease severity [30]. Sex is a crucial variable that influences the innate and adaptive host immune responses to viruses [31] and there is substantial data demonstrating more severe disease in male versus female patients with COVID-19 [32–39] as well as hamster models [9].
Chest CT has been utilized in patients with SARS-CoV-2 for the diagnosis and monitoring of lung disease [40, 41]. Additionally, efforts are being made to develop artificial intelligence-enabled analysis of chest CT for rapid diagnosis of patients with COVID-19 [42, 43]. Prior studies with preclinical animal models of SARS-CoV-2 have relied on semiquantitative or non-automated methods to score disease severity on CT [7, 44]. We developed and utilized a semi-automated lung segmentation tool for rapid, reliable, and unbiased analysis of chest CT in SARS-CoV-2-infected hamsters. Our total lung volume data (without positive pressure) is consistent with prior published data about lung volumes in hamsters [21, 45] and thus validates this technique. Moreover, similar to what has been reported in patients with COVID-19 [9, 34], male hamsters had worse pulmonary disease compared to females.
Radioiodinated DPA-713 has been validated in several animal models as a selective marker for activated macrophages/phagocytes, but not lymphoid cells [10, 12, 20, 26, 46, 47]. As a small molecular imaging probe, iodo-DPA-713 has an excellent pharmacokinetic profile with low background in pulmonary tissues [11, 20]. In this study, the in vivo biodistribution of 124I-iodo-DPA-713 in hamsters was comparable to prior studies with other mammalian species, with slightly higher hepatobiliary elimination [11, 20, 26]. 124I-Iodo-DPA-713 activity colocalized with the pulmonary infection sites and analysis disaggregated by sex demonstrated that male hamsters had higher PET activity compared to females. Post-mortem analysis using optical imaging and DPA-713-IRDye680LT confirmed these findings. Additionally, multicolor flow cytometry demonstrated that the lungs of male hamsters had a higher percentage of CD11b + myeloid cells (macrophages, phagocytes) versus females, but higher lymphocyte infiltration was detected in females, which is consistent with human studies [39, 48]. Given the limited availability of reagents, a more detailed flow cytometric analysis could not be performed. Finally, while the long physical half-life of I-124 allows for an ideal imaging time-point after tracer injection (typically > 4 h), other PET isotopes such as F-18, which provide higher positron yield, should be considered in the design of new tracers for these applications.
Conclusion
Our findings demonstrate that 124I-iodo-DPA-713 accumulates within lung lesions in a hamster model of SARS-CoV-2 infection. Subtle differences between the host response of males and females can be quantified by using an unbiased CT lung segmentation tool as well as by 124I-iodo-DPA-713 PET. This study provides proof of concept for the application of specific noninvasive biomarkers to image SARS-CoV-2-associated pulmonary inflammation. Novel molecular imaging agents could provide new insights into the pathophysiology of COVID-19, expedite the development and clinical translation of novel therapeutics against COVID-19, and also serve as clinically translatable tools for patient care.
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1. Gating strategy for multicolor flow cytometric analyses. Representative gating strategy used to illustrate the myeloid versus lymphoid population based on their forward (FSC) and side (SSC) scatter characteristics. Single cells were sub gated for CD11b+ myeloid cells (macrophages, phagocytes). (PNG 172 KB)
Figure S2. CT features in SARS-CoV-2-infected hamsters. Hamster developed multiple, bilateral, and peripheral ground-glass opacities (GGO) consolidations and air-bronchograms. Some males developed pneumomediastinum. (JPG 353 KB)
Figure S3. CT imaging in uninfected hamsters. a, Whole lung, lung volume above threshold and CT score for healthy male (n=3) and female (n=4) hamsters. b, Whole lung, lung volume above threshold and CT score for healthy controls (n=7) and infected male (n=15) and female hamsters (n=11). Data represented as median ± interquartile range. (JPG 316 KB)
Figure S4. 124I-iodo-DPA-713 organ biodistribution. a, 124I-iodo-DPA-713 signal is noted within the hepatobiliary tract, thyroid, and brown fat, with very low activity in the brain (n = 7 animals). b, maximum intensity projection (MIP). c, No differences are noted in the 124I-iodo-DPA-713 PET activity in unaffected areas of the lungs in infected male versus female hamsters. Data represented as median ± interquartile range. (JPG 208 KB)
Figure S5. Histopathology, virus titers and antibody responses in SRAS-CoV-2 infected males and females. a, Histopathology (H&E) of a representative male and female SARS-CoV-2-infected lungs at 2X magnification are shown, scale bars of 2mm. Percentage of affected area show higher lung consolidation in males compare to females. Pink arrows show areas of consolidation. b, S-RBD-specific IgG antibodies were determined. c, Infectious virus titers in the homogenates of lungs were determined by TCID50 at day seven post-infection. Histopathology data is represented as median ± interquartile range. ELISA and TCID50 are represented as mean ± SD. (JPG 322 KB)
Video S1. Multi-atlas lung segmentation (MALS) tool in a SARS-CoV-2-infected hamster. (GIF 42625 KB)
Acknowledgements
We thank the National Institute of Infectious Diseases, Japan, for providing VeroE6TMPRSS2 cells and FastGrants/Emergent Ventures for the financial support. We also acknowledge the Centers for Disease Control and Prevention, BEI Resources, NIAID, NIH for SARS-related coronavirus 2, isolate USA-WA1/2020, NR-5228.
Author contribution
C. A. R. B. and S. K. J. conceptualized and designed the study. C. A. R. B., F. M., A. A. O., K. F., P. D. J., S. D., and M. B. performed animal experiments. C. A. F. synthesized 124I-iodo-DPA-713. C. S., K. M., and J. L. M. performed H&E review and immunohistochemistry. W. R. B. provided reagents and F. J. M., A. K., and M. P. performed multicolor flow cytometry. S. D. and S. L. K. provided expertise in sex difference analyses and performed ELISAs. R. Z. and A. P. performed viral quantification. A. P., J. L. M., J. V., and S.L.K provided expertise in the hamster model and virology. A. G. developed the MALS tool. C. A. R. B., F. M., K. F., and M. B. analyzed the imaging data. C. A. R. B. and S. K. J. wrote the initial draft and all co-authors participated in editing the final manuscript. S. K. J. provided funding and supervised the project.
Funding
This study was supported by the National Institutes of Health (R01-AI153349, R01-AI145435-A1, and R21-AI149760 to S. K. J.; T32-OD011089 to J. L. M.), the Center for Infection and Inflammation Imaging Research (Johns Hopkins University, S. K. J.). These studies were also supported through the NIH/NCI COVID-19 Serology Center of Excellence U54CA260492 (S. L. K.), the NIH/ORWH/NIA Specialized Center of Research Excellence in Sex Differences U54AG062333 (S. L. K.), and the generosity of the collective community of donors to the Johns Hopkins University School of Medicine for COVID research.
Declarations
Conflict of Interest
Ali Ghayoor works at Invicro, Boston, MA, USA. A. A. O. receives consulting fees from Cubresa Inc. S. K. J. is a Senior Editor for Molecular Imaging and Biology.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Camilo A. Ruiz-Bedoya and Filipa Mota are co-first authors. Sabra L. Klein and Sanjay K. Jain contributed equally.
References
- 1.Ordonez AA, Sellmyer MA, Gowrishankar G, et al (2019) Molecular imaging of bacterial infections: overcoming the barriers to clinical translation. Sci Transl Med 11 [DOI] [PMC free article] [PubMed]
- 2.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL (2020) Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol 1–6 [DOI] [PMC free article] [PubMed]
- 3.Schulte-Schrepping J, Reusch N, Paclik D, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419-+. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GlobalHealth5050 The sex, gender and COVID-19 Project. https://globalhealth5050.org/the-sex-gender-and-covid-19-project/ (accessed Dec 28, 2020)
- 5.Peckham H, de Gruijter NM, Raine C, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awulachew E, Diriba K, Anja A, Getu E, Belayneh F. Computed tomography (CT) imaging features of patients with COVID-19: systematic review and meta-analysis. Radiol Res Pract. 2020;2020:1023506. doi: 10.1155/2020/1023506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai M, Iwatsuki-Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. P Natl Acad Sci USA. 2020;117:16587–16595. doi: 10.1073/pnas.2018975117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carsana L, Sonzogni A, Nasr A, et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhakal S, Ruiz-Bedoya CA, Zhou R, et al (2021) Sex differences in lung imaging and SARS-CoV-2 antibody responses in a COVID-19 golden Syrian hamster model. mBio:e0097421 [DOI] [PMC free article] [PubMed]
- 10.Sanchez-Bautista J, Foss CA, Ordonez AA, Klunk MH, Jain SK. Imaging pulmonary foreign body reaction using [I-125]iodo-DPA-713 SPECT/CT in mice. Mol Imaging Biol. 2019;21:228–231. doi: 10.1007/s11307-018-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foss CA, Plyku D, Ordonez AA, et al. Biodistribution and radiation dosimetry of (124)I-DPA-713, a PET radiotracer for macrophage-associated inflammation. J Nucl Med. 2018;59:1751–1756. doi: 10.2967/jnumed.117.207431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ordonez AA, Pokkali S, DeMarco VP, et al. Radioiodinated DPA-713 imaging correlates with bactericidal activity of tuberculosis treatments in mice. Antimicrob Agents Chemother. 2015;59:642–649. doi: 10.1128/AAC.04180-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordonez AA, Wintaco LM, Mota F, et al (2021) Imaging Enterobacterales infections in patients using pathogen-specific positron emission tomography. Sci Transl Med [DOI] [PMC free article] [PubMed]
- 14.Ordonez AA, Wang H, Magombedze G, et al. Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat Med. 2020;26:529–534. doi: 10.1038/s41591-020-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker EW, Guglieri-Lopez B, Ordonez AA, et al (2018) Noninvasive (11)C-rifampin positron emission tomography reveals drug biodistribution in tuberculous meningitis. Sci Transl Med 10 [DOI] [PMC free article] [PubMed]
- 16.Hesterman J, Ghayoor A, Novicki A, et al. Multi-atlas approaches for image segmentation across modality, species and application area. Future. 2019;6:7. [Google Scholar]
- 17.Wang X, Ghayoor A, Novicki A, Holmes S, Seibyl J, Hesterman J. Application of multi-atlas segmentation tool to hippocampus, ventricle and whole brain segmentation. Alzheimers Dement. 2017;13:P79–P80. [Google Scholar]
- 18.Jean Philippe T (1996) Non-rigid matching using demons [abstract]. 245P
- 19.Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell. 2012;35:611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foss CA, Harper JS, Wang H, Pomper MG, Jain SK. Noninvasive molecular imaging of tuberculosis-associated inflammation with radioiodinated DPA-713. J Infect Dis. 2013;208:2067–2074. doi: 10.1093/infdis/jit331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boudewijns R, Thibaut HJ, Kaptein SJ, et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skerry C, Harper J, Klunk M, Bishai WR, Jain SK. Adjunctive TNF inhibition with standard treatment enhances bacterial clearance in a murine model of necrotic TB granulomas. PLoS ONE. 2012;7:e39680. doi: 10.1371/journal.pone.0039680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerbeck CD, Hooper JW. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol. 2011;85:9929–9944. doi: 10.1128/JVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein SL, Pekosz A, Park HS, et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest. 2020;130:6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber AD, Osterrieder N, Bertzbach LD, et al. Standardization of reporting criteria for lung pathology in SARS-CoV-2–infected hamsters: what matters? Am J Respir Cell Mol Biol. 2020;63:856–859. doi: 10.1165/rcmb.2020-0280LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker EW, Pokkali S, Zhang Z, et al. Microglia activation in a pediatric rabbit model of tuberculous meningitis. Dis Model Mech. 2016;9:1497–1506. doi: 10.1242/dmm.027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mota F, Ordonez AA, Firth G, Ruiz-Bedoya CA, Ma MT, Jain SK. Radiotracer development for bacterial imaging. J Med Chem. 2020;63:1964–1977. doi: 10.1021/acs.jmedchem.9b01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 29.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Alessio FR, Heller NM (2020) COVID-19 and myeloid cells: complex interplay correlates with lung severity. J Clin Investig 130 [DOI] [PMC free article] [PubMed]
- 31.Klein SL. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. BioEssays. 2012;34:1050–1059. doi: 10.1002/bies.201200099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunders MJ, Altfeld M (2020) Implications of sex differences in immunity for SARS-CoV-2 pathogenesis and design of therapeutic interventions. Immunity [DOI] [PMC free article] [PubMed]
- 33.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 34.Moradi B, Ghanaati H, Kazemi MA, et al. Implications of sex difference in CT scan findings and outcome of patients with COVID-19 pneumonia. Radiology: Cardiothoracic Imaging. 2020;2:e200248. doi: 10.1148/ryct.2020200248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dangis A, De Brucker N, Heremans A, et al. Impact of gender on extent of lung injury in COVID-19. Clin Radiol. 2020;75:554–556. doi: 10.1016/j.crad.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang Y, Xu C, Jiang F, et al. Clinical characteristics and changes of chest CT features in 307 patients with common COVID-19 pneumonia infected SARS-CoV-2: a multicenter study in Jiangsu, China. Int J Infect Dis. 2020;96:157–162. doi: 10.1016/j.ijid.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spagnolo PA, Manson JE, Joffe H (2020) Sex and gender differences in health: what the COVID-19 pandemic can teach us. American College of Physicians [DOI] [PMC free article] [PubMed]
- 38.Chen T, Wu D, Chen H, et al (2020) Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. bmj 368 [DOI] [PMC free article] [PubMed]
- 39.Jin S, An H, Zhou T, et al. Sex-and age-specific clinical and immunological features of coronavirus disease 2019. PLoS Pathog. 2021;17:e1009420. doi: 10.1371/journal.ppat.1009420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020;296:E55–E64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei X, Lee H-C, Diao K-y, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gieraerts C, Dangis A, Janssen L, et al. Prognostic value and reproducibility of AI-assisted analysis of lung involvement in COVID-19 on low-dose submillisievert chest CT: sample size implications for clinical trials. Radiology: Cardiothoracic Imaging. 2020;2:e200441. doi: 10.1148/ryct.2020200441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaptein SJ, Jacobs S, Langendries L, et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2− infected hamsters, whereas hydroxychloroquine lacks activity. Proc Natl Acad Sci. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldstein RH. Response of the aging hamster lung to elastase injury. Am Rev Respir Dis. 1982;125:295–298. doi: 10.1164/arrd.1982.125.3.295. [DOI] [PubMed] [Google Scholar]
- 46.Foss CA, Bedja D, Mease RC, et al. Molecular imaging of inflammation in the ApoE -/- mouse model of atherosclerosis with IodoDPA. Biochem Biophys Res Commun. 2015;461:70–75. doi: 10.1016/j.bbrc.2015.03.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foss CA, Liu L, Mease RC, Wang H, Pasricha P, Pomper MG. Imaging macrophage accumulation in a murine model of chronic pancreatitis with (125)I-Iodo-DPA-713 SPECT/CT. J Nucl Med. 2017;58:1685–1690. doi: 10.2967/jnumed.117.189571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi T, Wong P, Ellingson MK, et al (2020) Sex differences in immune responses to SARS-CoV-2 that underlie disease outcomes. Nature [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating strategy for multicolor flow cytometric analyses. Representative gating strategy used to illustrate the myeloid versus lymphoid population based on their forward (FSC) and side (SSC) scatter characteristics. Single cells were sub gated for CD11b+ myeloid cells (macrophages, phagocytes). (PNG 172 KB)
Figure S2. CT features in SARS-CoV-2-infected hamsters. Hamster developed multiple, bilateral, and peripheral ground-glass opacities (GGO) consolidations and air-bronchograms. Some males developed pneumomediastinum. (JPG 353 KB)
Figure S3. CT imaging in uninfected hamsters. a, Whole lung, lung volume above threshold and CT score for healthy male (n=3) and female (n=4) hamsters. b, Whole lung, lung volume above threshold and CT score for healthy controls (n=7) and infected male (n=15) and female hamsters (n=11). Data represented as median ± interquartile range. (JPG 316 KB)
Figure S4. 124I-iodo-DPA-713 organ biodistribution. a, 124I-iodo-DPA-713 signal is noted within the hepatobiliary tract, thyroid, and brown fat, with very low activity in the brain (n = 7 animals). b, maximum intensity projection (MIP). c, No differences are noted in the 124I-iodo-DPA-713 PET activity in unaffected areas of the lungs in infected male versus female hamsters. Data represented as median ± interquartile range. (JPG 208 KB)
Figure S5. Histopathology, virus titers and antibody responses in SRAS-CoV-2 infected males and females. a, Histopathology (H&E) of a representative male and female SARS-CoV-2-infected lungs at 2X magnification are shown, scale bars of 2mm. Percentage of affected area show higher lung consolidation in males compare to females. Pink arrows show areas of consolidation. b, S-RBD-specific IgG antibodies were determined. c, Infectious virus titers in the homogenates of lungs were determined by TCID50 at day seven post-infection. Histopathology data is represented as median ± interquartile range. ELISA and TCID50 are represented as mean ± SD. (JPG 322 KB)
Video S1. Multi-atlas lung segmentation (MALS) tool in a SARS-CoV-2-infected hamster. (GIF 42625 KB)