Abstract
Background:
We evaluated the feasibility of mitochondrial DNA (mtDNA) copy number measurement in dried blood spots (DBS), its comparability with measurement in whole blood samples, and stability of mtDNA copy number from DBS over time.
Methods:
Women in this pilot study were participants in the Sister Study, a large prospective cohort. Sister Study participants provided a whole blood sample and DBS at enrollment. A second DBS sample was collected 5–10 years later from a subcohort of women with and without an incident breast cancer diagnosis between collections. Among 54 women (27 with breast cancer, 27 without) we measured mtDNA copy number from whole blood at enrollment and from DBS at both time points.
Results:
The average age at enrollment was 58.7 years (range:50–69). Values of mtDNA copy number measured in whole blood samples and DBS from enrollment were moderately correlated (Spearman R=0.45;p=0.005). Stability of mtDNA copy number in DBS from the two time points was moderate overall (ICC=0.50) and similar between women with (ICC=0.50) and without (ICC=0.51) a breast cancer diagnosis between the two collections.
Conclusions:
Our results suggest that measurement of mtDNA copy number in DBS is feasible and may be a valid alternative to measurement in whole blood samples.
Keywords: mitochondrial DNA copy number, humans, dried blood spots
1. Introduction
Mitochondria are membrane-bound organelles that perform several essential functions in eukaryotic cells, including production of energy via oxidative phosphorylation. Hundreds to thousands of mitochondria are present in all cell types except red blood cells, and each mitochondrion contains multiple copies of the mitochondrial genome (mitochondrial DNA [mtDNA]). The high number of copies is considered a compensatory mechanism for the greater susceptibility of mtDNA to genetic damage due to limited repair capacity, exposure to reactive oxidative species (ROS) as oxidative phosphorylation byproducts, and the absence of introns and histone caps.[1] Depletion of mtDNA copy number is an indicator of mitochondrial dysfunction, and has been proposed as a marker of biologic aging given its reported associations with lifestyle factors and the development of neurodegenerative diseases, cardiovascular disease, diabetes, and some cancers.[2–5]
In most studies to date, mtDNA copy number has been measured in buffy coat or whole blood samples. However, dried blood spots, obtained by spotting a small amount of blood on filter paper, may present an attractive alternative medium for measurement in large-scale prospective studies, as they may be easier to collect and store than whole blood samples. In this study, we compared mtDNA copy number values obtained from dried blood spots to those obtained from whole blood samples in 54 women enrolled in the Sister Study cohort. We also examined the correlation between mtDNA copy number values from two dried blood spot samples collected 5–10 years apart.
2. Materials and methods
2.1. Parent study cohort
Women included in these analyses were participants in the Sister Study, a prospective cohort of 50,884 women that was designed to evaluate genetic and environmental risk factors for breast cancer. Details of the design, recruitment, and methods have been described elsewhere.[6] Briefly, between 2003 and 2009, women aged 35 to 74 years from the U.S. and Puerto Rico were recruited through a national multi-media campaign and a network of recruitment volunteers. Eligible women had a full or half-sister who had been diagnosed with breast cancer, but were themselves free of breast cancer at enrollment. The study was approved by the Institutional Review Board of the National Institute of Environmental Health Sciences, the National Institute of Health, and the Copernicus Group, and all participants provided written informed consent.
At enrollment, Sister Study participants completed extensive questionnaires for assessment of demographic and lifestyle information, medical history, and other characteristics. Biological samples, including blood samples, were collected during an enrollment home visit. In 2014, approximately 5–10 years after enrollment, a subset of Sister Study participants was invited to participate in a second home visit for biological sample collection; all women who had been diagnosed with breast cancer since enrollment were invited to participate (N=1,909), as was a randomly selected 4% sample of the entire cohort (N=1,828). A total of 1,215 and 1,207 women with and without an incident breast cancer diagnosis, respectively, completed the second home visit. During each home visit, fasting blood samples were collected by a trained examiner following standardized protocols. Whole blood was collected in BD Vacutainer ® EDTA and ACD tubes and shipped at ambient temperature to the Sister Study biorepository. The EDTA whole blood was then spotted in 60 μL spots on Whatman FTA Classic Cards ® (chemically impregnated to lyse cells and stabilize DNA). FTA cards were stored at an ambient humidity-controlled temperature. [6] As a part of a case-cohort study nested within the Sister Study,[7] DNA from enrollment whole blood samples was extracted for a subset of the full cohort, including women with and without an incident breast cancer diagnosis since enrollment.
2.2. Pilot sample selection
For the current pilot study, we included only women who had completed both home visits, had DNA extracted from enrollment whole blood samples, and were age 50 years or older and either naturally postmenopausal or postmenopausal with both ovaries removed at enrollment. We first selected all breast cancer cases who were diagnosed with stage I-III disease at least 1 year after enrollment and at least one year before the second home visit, and who had received an anthracycline as part of their first adjuvant chemotherapy regimen (N=27). These women were then matched on age at enrollment in 2003–2009 (within 8 months) and time between enrollment and the second home visit in 2014 (within 8 months) to an equal number of eligible women who remained cancer-free as of the second home visit.
2.3. DNA extraction
DNA from whole blood samples was extracted using an automated system (Autopure LS, Gentra Systems) in the NIEHS Molecular Genetics Core Facility or using DNAQuik at BioServe Biotechnologies LTD (Beltsville, MD). Extracted DNA was aliquoted into screw top cryo-tubes and stored in liquid nitrogen vapor phase. DNA yield and A260/280 was measured and tracked. DNA was isolated from the dried blood spots using the QIAmp DNA Blood Mini kit and isolated DNA concentrations were measured using the Nanodrop in the Fry Laboratory at the University of North Carolina at Chapel Hill.
2.4. MtDNA copy number analysis
Using predesigned TaqMan Copy Number Assays (Applied Biosystems) for human mitochondriallyencoded 12S rRNA and nuclear-encoded Ribonuclease P RNA component H1 (RNase P H1) RNA sequences, we measured the copy number of target and references sequences. RNase P is an endogenous reference gene present in two copies in the human genome. The TaqMan Genotyping Master Mix was added for the DNA polymerase and dNTPs to make the PCR reaction possible. The copy number of mtDNA was determined by calculating the delta CT by subtracting the average CT of the RNase P from the average CT of the 12S rRNA.[8] Each sample was run in triplicate. A no template control and five reference DNA samples were also run in triplicate on each 96-well plate. Women with and without breast cancer, and the two time points for the same woman, were blinded and analyzed together on the same plate. DBS samples that had low DNA concentrations following extraction using the Qiagen kit were cleaned and concentrated using Zymo DNA Clean and Concentrator-25 kit. Following this step, all DNA samples were analyzed by real-time polymerase chain reaction (RTPCR).
2.5. Statistical analysis
We calculated the Spearman correlation coefficient to assess the comparability between mtDNA copy number values at enrollment measured from the whole blood sample and from the dried blood spot. To assess stability of the dried blood spot measure over time between the two home visits, we calculated both the Pearson correlation coefficient and the intra-class correlation coefficient (ICC). The ICC is calculated as the between-person variance divide by the total variance (sum of within- and between-person variances). We considered an ICC of >0.90 to indicate excellent stability, 0.75 to 0.90 to indicate good stability, 0.50 to 0.74 to indicate moderate stability, and <0.50 to indicate poor stability.[9] Analyses were performed using SAS software, version 9.4 (SAS Institute, Cary, NC).
3. Results
A total of 54 women had mtDNA copy number measured in the whole blood sample from enrollment and in dried blood spots from both home visits. Thirteen dried blood spot samples, collected from 12 women, required an additional step for clean-up. After this additional step, all samples had measurable mtDNA copy number in whole blood and DBS at enrollment, and DBS at the second visit.
The average age among participants was 58.7 years (SD=4.5) at enrollment, and the average time between the two visits was 7.6 years (SD=1.3) (Table 1). Most women were non-Hispanic white (89%), had at least some college education (85%), and were not current smokers (94%). The median mtDNA copy number among participants was 277.6, 671.8, and 973.8, as measured from the enrollment visit whole blood sample, the enrollment visit dried blood spot, and the second visit dried blood spot, respectively. Participant age was not strongly associated with mtDNA copy number from the whole blood sample (Spearman R=0.09, p=0.534) or from the dried blood spot at enrollment (Spearman R=−0.27, p=0.047) or the second visit (Spearman R=−0.13, p=0.357). There was no significant difference in values of mtDNA copy number from DBS at the second visit between women with and without a breast cancer diagnosis (t-test p-value=0.918).
Table 1.
Descriptive characteristics of 54 women from the Sister Study cohort
| Age at enrollment (years), mean (SD) | 58.7 | (4.5) |
| Time between enrollment and second visit (years), mean (SD) | 7.6 | (1.3) |
| Race/ethnicity, N(%) | ||
| Non-Hispanic White | 48 | 89% |
| Other | 6 | 11% |
| Education, N(%) | ||
| High school or less | 8 | 15% |
| Some college | 20 | 37% |
| Bachelor’s degree or higher | 26 | 48% |
| BMI at enrollment (kg/m 2 ), mean (SD) | 28.1 | (5.8) |
| Smoking status at enrollment, N(%) | ||
| Never | 25 | 46% |
| Former | 26 | 48% |
| Current | 3 | 6% |
| mtDNA copy number from whole blood at enrollment, median (IQR) | 277.6 | (108.9, 455.1) |
| mtDNA copy number from dried blood spot at enrollment, median (IQR) | 671.8 | (406.4, 944.5) |
| mtDNA copy number from dried blood spot at second visit, median (IQR) | 973.8 | (627.4, 1502.7) |
Values of mtDNA copy number measured in the whole blood sample and the dried blood spot from enrollment were moderately correlated (Spearman R=0.45, p=0.005; Figure 1), with dried blood spot values generally exceeding whole blood values. The analysis comparing mtDNA copy number measured in blood spots from the two time points indicated moderate stability (ICC=0.50; Pearson R=0.66, p<0.0001; Figure 2). The ICC was similar between women with (ICC=0.50) and without (ICC=0.51) an incident breast cancer diagnosis between the two visits. When samples that required an additional step for clean-up (N=13) were excluded, the correlation between the dried blood spot and whole blood mtDNA copy number values was similar (Spearman R=0.47, p=0.001), as was the ICC for the two dried blood spot measures over time (ICC=0.53).
Figure 1.
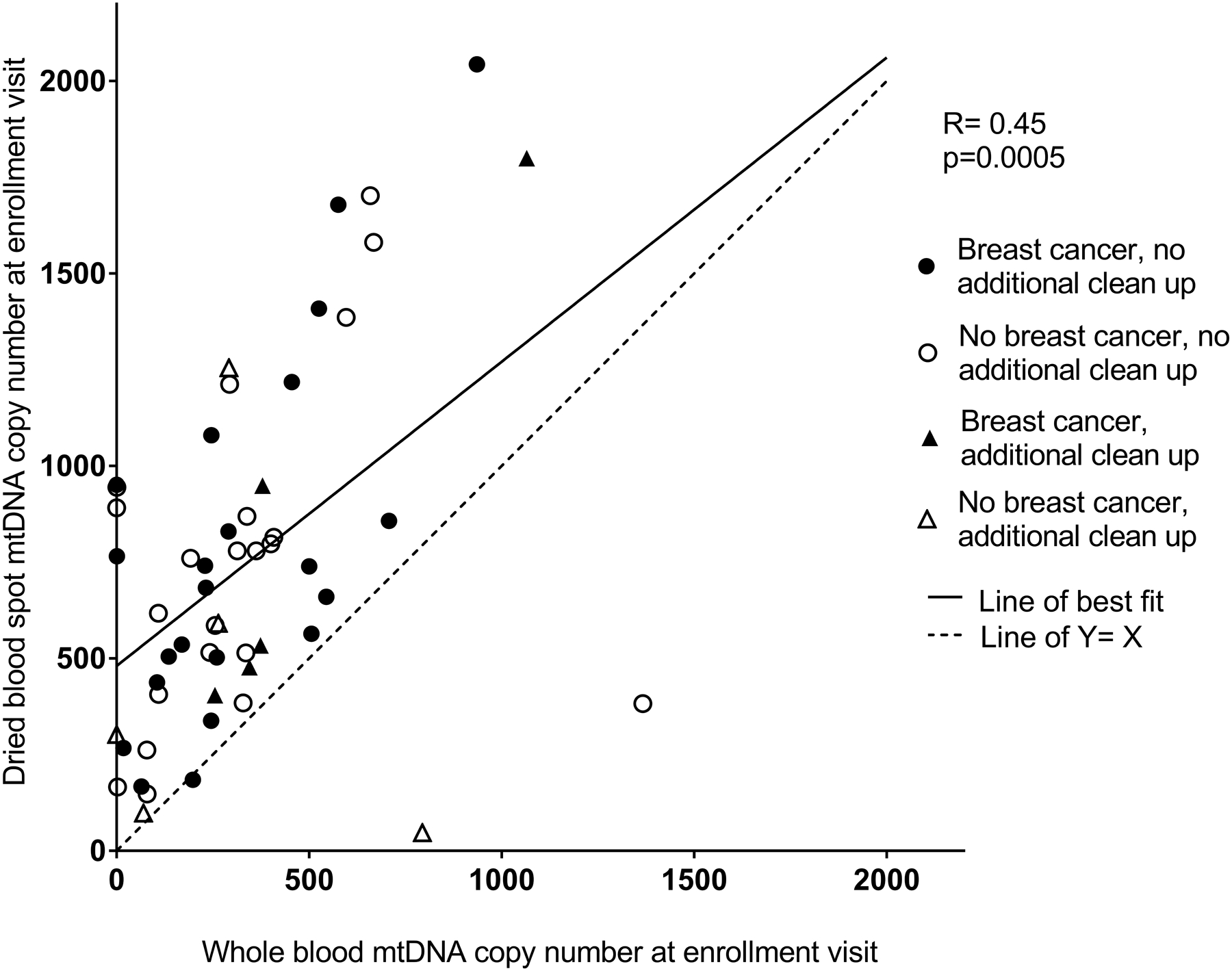
Comparison of mtDNA copy number from the whole blood sample from enrollment versus the dried blood spot sample from enrollment. Spearman correlation: R=0.45, p=0.0005. The coordinates of the two points below the line of Y=X are (794, 47) and (1367, 383) and are from women without a breast cancer diagnosis. Samples which required an additional clean-up step are indicated with triangles.
Figure 2.
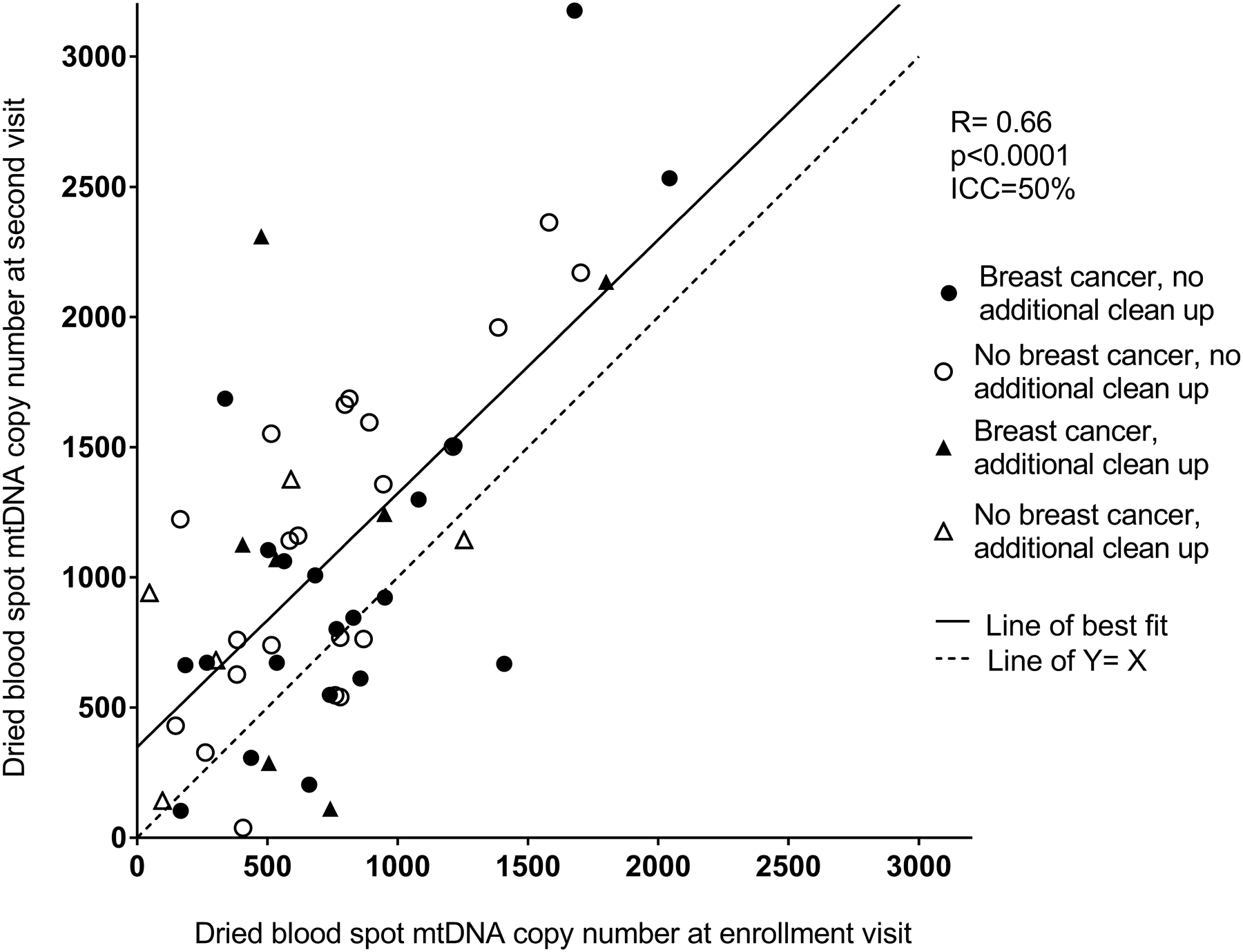
Comparison of mtDNA copy number from the dried blood spot sample from enrollment versus the dried blood spot sample at the second visit. Pearson correlation: R=0.66, p<0.0001; Intra-class correlation coefficient=50%. Samples which required an additional clean-up step are indicated with triangles.
4. Discussion
In this study, we examined the feasibility of mtDNA copy number measurement in dried blood spots, its comparability with whole blood samples, and stability of mtDNA copy number from dried blood spots over time. We found that mtDNA copy number values were higher, on average, when measured in the dried blood spots than in the whole blood samples, and that measurements from the two sample types were moderately correlated. In two samples collected 5–10 years apart, we observed moderate stability of mtDNA copy number from dried blood spots. Our results thus suggest that dried blood spots may be a feasible option for measurement of mtDNA copy number in future studies.
Growing recognition of the many advantages of dried blood spots has led to their increasing use in large research studies. In some settings, dried blood spot samples can be obtained through a simple finger prick, a less invasive and less costly method than traditional venipuncture for whole blood collection, and one that does not require medically trained personnel.[10] After collection, dried blood spots are much easier to transport and store than whole blood samples, as they do not require freezing and can be stored for long periods at room temperature. This makes them ideal for archiving in epidemiologic cohorts and other large-scale studies.
Although mtDNA copy number values from dried blood spots have not been previously compared, to our knowledge, with those from whole blood samples or other standard biological samples, prior studies have reported high validity of other biomarkers from dried blood spots.[11–15] For example, a validation study based in the Melbourne Collaborative Cohort Study (N=62) found good agreement between measurements of 25-hydroxyvitamin D from dried blood spots and plasma.[12] Likewise, another study of 150 adults reported high validity of several biomarkers of cardiovascular disease risk, including total cholesterol, high-density lipoprotein cholesterol, and C-reactive protein, measured in dried blood spots as compared to those measured in concurrently collected serum samples.[13] These and other studies have concluded that use of dried blood spots may be an acceptable method of sample collection in studies of certain critical biomarkers of human health and disease.
It is unclear why the mtDNA copy number was higher, on average, in the dried blood spot samples, than in the whole blood samples collected at the same time point. There are several hypotheses that we have considered for this difference. Among these considerations is the potential for disproportionate abundance of cells that contain mtDNA that could have been impacted by differences in analytical processing between the dried blood spots and whole blood samples. However, as the analytical processing was similar between the dried blood spots and the whole blood samples, sample processing seems an unlikely reason for this difference. Nevertheless, the moderate correlation that we observed between values in the two sample types suggests that dried blood spots may be a valid alternative to conventional samples for measuring mtDNA copy number. Given our small sample size, further examination of mtDNA copy number values from dried blood spots compared to those from other blood sample types is warranted in larger studies.
To our knowledge, our study is the first to examine the stability of mtDNA copy number measured in two dried blood spots collected several years apart. However, others have assessed stability of mtDNA copy number over time using whole blood or buffy coat samples, with mixed results. Among 96 healthy adults enrolled in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort, the ICC for mtDNA copy number measured in blood samples collected 15 years apart was 0.6, indicating moderate stability over the study period.[16] In contrast, large temporal variation in mtDNA copy number was observed in buffy coat samples collected 6 years apart from 91 women in the UK-based Breakthrough Generations Study (ICC=0.38).[17] Our finding of an ICC of approximately 0.5 for mtDNA copy number in dried blood spots over a 5–10 year period, regardless of breast cancer status during this interval, is within the range of that reported in studies using whole blood or buffy coat samples. Thus the stability of mtDNA copy number obtained from dried blood spots may be comparable to that obtained from more conventional samples.
This study has some limitations. The small sample size limited our ability to examine associations between mtDNA copy number and participant characteristics. Half of our sample had an incident breast cancer diagnosis between enrollment and the second visit, though the ICC for stability of mtDNA copy number over time did not appear to differ according to breast cancer status. Our sample was also comprised entirely of postmenopausal women, most of whom were non-Hispanic White. Therefore our findings may not be generalizable to more diverse populations.
In summary, dried blood spot samples have several favorable properties for use in large cohort studies, including ease of collection and storage. Our results suggest that measurement of mtDNA copy number in dried blood spot samples is feasible and may be a valid alternative to measurement in whole blood samples. Replication in a larger sample is warranted to strengthen these conclusions.
Acknowledgements
This research was supported in part by the UNC Center for Women’s Health Research and the Intramural Research Program of the National Institutes of Health, the National Institute of Environmental Health Sciences (Z01-ES044005). C.A. was supported by the UNC Lineberger Cancer Control Education Program (T32 CA057726).
Footnotes
Declarations of interest: None
References
- [1].Taylor RW and Turnbull DM, Mitochondrial DNA mutations in human disease, Nat Rev Genet 6 (2005), 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wu S, Li X, Meng S, Fung T, Chan AT, Liang G, Giovannucci E, De Vivo I, Lee JH and Nan H, Fruit and vegetable consumption, cigarette smoke, and leukocyte mitochondrial DNA copy number, Am J Clin Nutr 109 (2019), 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chang YK, Kim DE, Cho SH and Kim JH, Association between Leukocyte Mitochondrial DNA Copy Number and Regular Exercise in Postmenopausal Women, Korean J Fam Med 37 (2016), 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clay Montier LL, Deng JJ and Bai Y, Number matters: control of mammalian mitochondrial DNA copy number, J Genet Genomics 36 (2009), 125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Meyer JN, Leuthner TC and Luz AL, Mitochondrial fusion, fission, and mitochondrial toxicity, Toxicology 391 (2017), 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D’Aloisio AA, Suarez LM, Kleeberger CA, Shore DL, DeRoo LA, Taylor JA, Weinberg CR and Sister Study Research T, The Sister Study Cohort: Baseline Methods and Participant Characteristics, Environ Health Perspect 125 (2017), 127003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kresovich JK, Xu Z, O’Brien KM, Weinberg CR, Sandler DP and Taylor JA, Methylation-based biological age and breast cancer risk, J Natl Cancer Inst (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mondal R, Ghosh SK, Choudhury JH, Seram A, Sinha K, Hussain M, Laskar RS, Rabha B, Dey P, Ganguli S, Nathchoudhury M, Talukdar FR, Chaudhuri B and Dhar B, Mitochondrial DNA copy number and risk of oral cancer: a report from Northeast India, PLoS One 8 (2013), e57771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koo TK and Li MY, A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research, J Chiropr Med 15 (2016), 155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Crimmins E, Kim JK, McCreath H, Faul J, Weir D and Seeman T, Validation of blood-based assays using dried blood spots for use in large population studies, Biodemography Soc Biol 60 (2014), 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ladror D, Pitt B and Funk W, Quantification of cotinine in dried blood spots as a biomarker of exposure to tobacco smoke, Biomarkers 23 (2018), 44–50. [DOI] [PubMed] [Google Scholar]
- [12].Heath AK, Williamson EJ, Ebeling PR, Kvaskoff D, Eyles DW and English DR, Measurements of 25-hydroxyvitamin D concentrations in archived dried blood spots are reliable and accurately reflect those in plasma, J Clin Endocrinol Metab 99 (2014), 3319–24. [DOI] [PubMed] [Google Scholar]
- [13].Samuelsson LB, Hall MH, McLean S, Porter JH, Berkman L, Marino M, Sembajwe G, McDade TW and Buxton OM, Validation of Biomarkers of CVD Risk from Dried Blood Spots in Community-Based Research: Methodologies and Study-Specific Serum Equivalencies, Biodemography Soc Biol 61 (2015), 285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Funk WE, Waidyanatha S, Chaing SH and Rappaport SM, Hemoglobin adducts of benzene oxide in neonatal and adult dried blood spots, Cancer Epidemiol Biomarkers Prev 17 (2008), 1896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McDade TW, Woodruff TK, Huang YY, Funk WE, Prewitt M, Kondapalli L and Gracia CR, Quantification of anti-Mullerian hormone (AMH) in dried blood spots: validation of a minimally invasive method for assessing ovarian reserve, Hum Reprod 27 (2012), 2503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Campa D, Barrdahl M, Santoro A, Severi G, Baglietto L, Omichessan H, Tumino R, Bueno-de-Mesquita HBA, Peeters PH, Weiderpass E, Chirlaque MD, Rodriguez-Barranco M, Agudo A, Gunter M, Dossus L, Krogh V, Matullo G, Trichopoulou A, Travis RC, Canzian F and Kaaks R, Mitochondrial DNA copy number variation, leukocyte telomere length, and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study, Breast Cancer Res 20 (2018), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lemnrau A, Brook MN, Fletcher O, Coulson P, Tomczyk K, Jones M, Ashworth A, Swerdlow A, Orr N and Garcia-Closas M, Mitochondrial DNA Copy Number in Peripheral Blood Cells and Risk of Developing Breast Cancer, Cancer Res 75 (2015), 2844–50. [DOI] [PubMed] [Google Scholar]


