Abstract
Cyclocarya paliurus (Batal.) Iljinskaja, a monotypic species in Cyclocarya of Juglandaceae, is regarded as one of important medical plants in China. In order to reveal the alterations in chloroplast (cp) genome with nuclear genome duplication, we presented the complete cp genomes of C. paliurus, and firstly analyzed on the basis of ploidy type (tetraploid and diploid C. paliurus). The total length of the cp genome of tetraploid and diploid C. paliurus is 160,938 and 161,105 bp, respectively. Both type genome consist of a large single-copy (LSC) region (90,221 and 90,391 bp), a small single-copy (SSC) region (18,593 and 18,590 bp), and an pair of invert repeats (IRs) regions (26,062 and 26,062 bp). Tetraploid and diploid plastid genome contain 132 and 137 genes, 87 and 88 protein-coding genes, 37 and 39 tRNA genes, and both eight rRNA genes, respectively. Closely phylogenetic relationship by analyzing 23 cp genomes suggests that tetraploid C. paliurus probably originated from diploid C. paliurus.
Keywords: Cyclocarya paliurus, sequencing, chloroplast genome, phylogeny
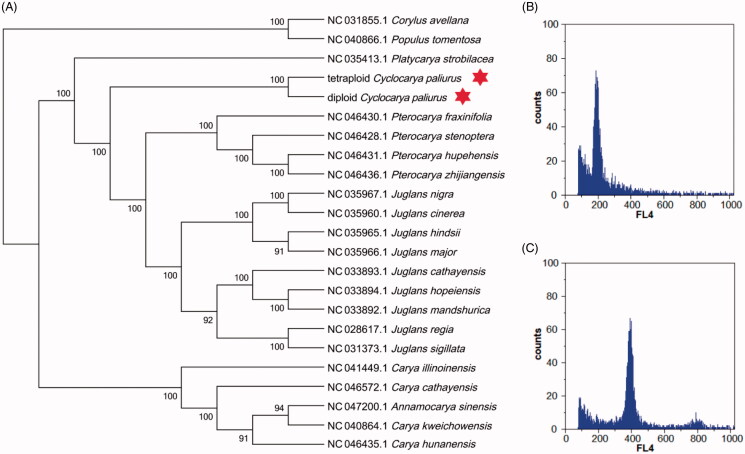
Cyclocarya paliurus (Batal.) Iljinskaja is generally known as an important medicinal plant, and its leaves are used to cure fever, detoxify, and alleviate pain (Xie and Li 2001). It is distributed in subtropical mountainous areas of China (Fang and Fu 2007). Additionally, two ploidy types of C. paliurus were identified: (i) diploid C. paliurus (Figure 1(B)) and (ii) tetraploid C. paliurus (Figure 1(C)). To understand genetic diversity of the two types related to cell ploidy differences, for the first time, we reported the complete chloroplast (cp) genomes of diploid and tetraploid C. paliurus based on Illumina pair-end sequencing data. Fresh leaves were collected from tetraploid and diploid plants growing in germplasm bank of C. paliurus, which locate in Baima experimental, Nanjing, Jiangsu province, China (N 31°35′, E 119°09′), immediately frozen in liquid nitrogen and stored at −80 °C. The specimen was deposited at Southern Tree Seed Testing Center (https://linxue.njfu.edu.cn/zzjg/yjzx/20210322/i206237.html, Xiangxiang Fu and xxfu@njfu.edu.cn) under the voucher number 201910tetra-Cp and 201910dip-Cp, respectively. The total genomic DNA was extracted from fresh leaves tissues, with 500 bp randomly interrupted sequence by the Covaris ultrasonic breaker for library construction. Approximately, 2.0 GB of raw data were generated using the Illumina Hiseq X Ten platform. The Illumina raw reads were filtered using Trimmomatic (Bolger et al. 2014). The complete plastid genome of Cyclocarya paliurus (GeneBank accession: NC_034315.1) as reference and plastid genome of C. paliurus was assembled by get_organelle_from_reads.py runscript of getorganelle (https://github.com/Kinggerm/GetOrganelle), which can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. 2015). The cp genome annotation was assembled and the result was drawn based on online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html).
Figure 1.
(A) Phylogenetic analysis of 21 species from Juglandaceae and 2 taxa (Populus tomentosa, Corylus avellana) as outgroups based on plastid genome sequences by RAxML, bootstrap support value near the branch. (B) Analysis of cell ploidy of diploid C. paliurus. (C) Analysis of cell ploidy of tetraploid C. paliurus; FL4: fluorescence intensity.
The complete plastid genome sequence of tetraploid C. paliurus (GenBank accession: MW118603.1) and diploid C. paliurus (GenBank accession: MW531677.2) are 160,938 and 161,105 bp in length, with a large single-copy (LSC) region of 90,221 and 90,391 bp, a small single-copy (SSC) region of 18,593 and 18,590 bp, and a pair of inverted repeats (IRs) regions of both 26,062 bp, respectively. Tetraploid and diploid types complete cp genomes contain 132 and 137 genes, 87 and 88 protein-coding genes, 37 and 39 tRNA genes, and both eight rRNA genes. The complete genome GC content in both ploidy was 36.0 and 36.1%, respectively. Comparing analysis of cp genomes from two types of C. paliurus, five single nucleotide polymorphisms (SNPs) were identified, one SNP was synonymous in accD and four SNPs were non-synonymous in atpA, ycf4, rpoA, and rps3, respectively. In addition, we found a slight difference in junction position between two cp genomes: the ndhF gene varied from 106 bp from the IRb/SSC junction in diploid C. paliurus, while in tetraploidy, it varied from 111 bp from the junction. To reveal the phylogenetic position of C. paliurus with other members of Juglandaceae, we performed a phylogenetic analysis based on 21 complete cp genomes of species in Juglandaceae, and took 2 taxa (Populus tomentosa, Corylus avellana) as outgroups. All data were downloaded from NCBI GenBank. The sequences were aligned by MAFFT version 7.307 (Katoh and Standley 2013). The maximum likelihood (ML) tree was constructed with RAxML version 8.2.12 (Stamatakis 2014) and performed using MEGA version 6.0 (Tamura et al. 2013) with the GTRCAT mode. The phylogenetic tree revealed that tetraploid and diploid C. paliurus formed a sister clade (Red pentagram in Figure 1), and both types were most closely related to P. strobilacea with 100% bootstrap support (Figure 1). The results provide strong support that tetraploidy could derive from diploidy with slight alteration in cp genome. Prospectively, valuable genetic information based on this result could support the development of molecular markers from cp genome; moreover, further investigation should be considered to find the connection between two ploidy types.
This work revealed the effects of polyploidy of nuclear genome on cp genome. Thus, variation on the molecular level could expound the differences of morphological traits and medicinal values between two ploidy types of C. paliurus.
Acknowledgments
The author thanks anonymous reviewers who provided helpful suggestions and critical comments on this manuscript.
Funding Statement
This work was supported by the Jiangsu Provincial Key Research and Development Program [Grant No. BE2019388], the National Natural Science Foundation of China [Project numbers: 31470637], and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW118603.1 (tetraploid C. paliurus) and MW531677.2 (diploid C. paliurus). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA718351 and PRJNA719912, SRS8600730 and SRS8641412, and SAMN18529737 and SAMN18623874, respectively.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SZ, Fu XX.. 2007. Progress and prospects on silviculture and utilization of Cyclocarya paliurus resources. J Nanjing For Univ. 31(1):95–100. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE.. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie MY, Li L.. 2001. Review in studies on chemical constituents and bioactivities of Cyclocarya paliurus. Chin Trad Herbal Drugs. 32(4):365–366. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW118603.1 (tetraploid C. paliurus) and MW531677.2 (diploid C. paliurus). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA718351 and PRJNA719912, SRS8600730 and SRS8641412, and SAMN18529737 and SAMN18623874, respectively.